Abstract
The herpesvirus entry mediator C (HveC), previously known as poliovirus receptor-related protein 1 (PRR1), and the herpesvirus Ig-like receptor (HIgR) are the bona fide receptors employed by herpes simplex virus-1 and -2 (HSV-1 and -2) for entry into the human cell lines most frequently used in HSV studies. They share an identical ectodomain made of one V and two C2 domains and differ in transmembrane and cytoplasmic regions. Expression of their mRNA in the human nervous system suggests possible usage of these receptors in humans in the path of neuron infection by HSV. Glycoprotein D (gD) is the virion component that mediates HSV-1 entry into cells by interaction with cellular receptors. We report on the identification of the V domain of HIgR/PRR1 as a major functional region in HSV-1 entry by several approaches. First, the epitope recognized by mAb R1.302 to HIgR/PRR1, capable of inhibiting infection, was mapped to the V domain. Second, a soluble form of HIgR/PRR1 consisting of the single V domain competed with cell-bound full-length receptor and blocked virion infectivity. Third, the V domain was sufficient to mediate HSV entry, as an engineered form of PRR1 in which the two C2 domains were deleted and the V domain was retained and fused to its transmembrane and cytoplasmic regions was still able to confer susceptibility, although at reduced efficiency relative to full-length receptor. Consistently, transfer of the V domain of HIgR/PRR1 to a functionally inactive structural homologue generated a chimeric receptor with virus-entry activity. Finally, the single V domain was sufficient for in vitro physical interaction with gD. The in vitro binding was specific as it was competed both by antibodies to the receptor and by a mAb to gD with potent neutralizing activity for HSV-1 infectivity.
The receptors that mediate herpes simplex virus (HSV) entry into cells have remained elusive for a long time for several reasons. Cell lines lacking receptors are very rare, hampering a genetic approach to the search of the receptors. The virus appears to be able to use alternative receptors (1). Cellular proteins that are able to act as mediators of virus entry when transfected in cells that do not express any other suitable receptor have such a narrow distribution that their actual usage is limited to very specialized cell types. This appears to be the case for herpesvirus entry mediator A (HveA), previously designated HVEM (for herpesvirus entry mediator), which appears to be expressed and functional only in T lymphocytes (2). Recently, the bona fide receptors that mediate HSV-1 entry into human cells were identified as a cluster of molecules belonging to the IgG superfamily (3–5). They have a common structure defined by six conserved cysteines in the ectodomain, which form three domains, one V-like and two C2-like. There are three members known to date: the herpesvirus entry mediator C (HveC) (3), previously known as PRR1, for poliovirus receptor-related protein 1 (6), and HIgR, for herpesvirus Ig-like receptor (5), both of which enable entry of all HSV-1 and -2 strains tested, and HveB (or PRR2) (7), which enables entry of a subset of HSV strains, namely HSV-2 and some HSV-1 gD mutants, but not wild-type HSV-1 strains (4). HIgR and PRR1(HveC) share an identical ectodomain, differ in the transmembrane and cytoplasmic regions, and appear to be splice-variant isoforms (5). Evidence that they can be considered as the bona fide receptors that mediate HSV-1 entry into the most frequently used human cell lines rested on the expression of HIgR/PRR1 proteins in cell lines like HEp-2, HeLa, human fibroblasts, etc., as detected by reactivity to mAb R1.302 to PRR1, and on the ability of the same antibody to block HSV-1 infection in these cells (5). The high level of mRNA expression in samples from nervous system suggests possible usage in humans in the path of neuron infection by HSV (5). The finding that two isoforms—HIgR and PRR1(HveC)—sharing the ectodomain can both mediate HSV entry mapped the functional region of the receptors to their ectodomain (5).
At least four virion glycoproteins, gB, gD, and the heterodimer gH/gL, participate in HSV-1 entry into cells (8–11). Work of the past decade has pointed to gD as the virion component that interacts with cellular receptor(s). The initial observation that expression of gD rendered cells resistant to infection led to the proposal that gD sequesters a putative receptor able to bind the glycoprotein (12). The notion subsequently was strengthened by the findings that incubation of gD-expressing cells with antibodies to gD released the block (1, 13), that viral unrestricted mutants able to overcome the gD-mediated block carry mutations in gD (1, 13, 14), that antiidiotypic antibodies mimicking gD could bind the surface of commonly used cell lines and blocked virus infectivity and cell-to-cell spread of virus (15), that cells susceptible to HSV infection were able to bind gD in a saturable manner (16), and that soluble forms of gD inhibited virion infectivity (16, 17). No such evidence for involvement of cellular receptors exists for the other glycoproteins, gB and gH/gL.
HSV-1 penetration into cells occurs through a pH-independent fusion event (18, 19). The molecular mechanisms underlying this process remain, in part, obscure, and a model is still lacking that shows how the interaction of gD with its cellular receptor triggers the fusion of the virion envelope with the plasma membrane and recruits the other virion glycoproteins. The aim of the present work was to identify regions of HIgR/PRR1 that are functional in HSV-1 entry and interact with HSV-1 gD. We demonstrate that (i) a major functional region of HIgR/PRR1 with HSV-1 entry activity resides in the V domain of the molecules; (ii) when anchored to cell membrane either through its own transmembrane and cytoplasmic domains, or through transplantation to a structurally related, functionally inactive molecule, the V domain was sufficient to mediate HSV-1 entry into cells, although the collapsed receptor displayed a reduced efficiency compared with full-length molecules; and (iii) the single V domain was sufficient for binding to gD in vitro in a specific fashion, providing evidence for a direct physical interaction between the major functional region of HIgR/PRR1 in virus entry and its viral ligand, gD.
MATERIALS AND METHODS
Cells and Viruses.
Cells were grown in DMEM supplemented with 5% fetal calf serum. HIgR/cl 11, PRR1/cl 5, and V-TM(PRR1)/Q were obtained by lipofectamine transfection of J1.1–2 cells (5) with pCF18 (HIgR), pLX1.12 (PRR1), or pCDTMR1V.3 and neomycin G418 selection. The HSV-1 recombinant R8102, which carries the lacZ gene under the control of the α27 promoter inserted between UL3 and UL4 genes, was a gift of B. Roizman and will be described elsewhere. Pelleted extracellular virions were used in all experiments. Infectivity of R8102 was assayed as described (2).
Antibodies.
mAb R1.302 to PRR1 and mAb 30 to gD were described (1, 20). mAb HD1 (21) to gD was from Goodwin Institute (Plantation, FL).
Construction, Production, and Purification of Soluble Forms of HIgR and PVR Receptors sVCC(HIgR)-Fc, sV(HIgR)-Fc, and sVCC(PVRα)-Fc.
The entire extracellular region of HIgR (amino acids 1–334) was amplified by PCR with primers CFLPRR15 (CCGG AGAT ATCA TGGC TCGG ATGG GGCT TG) and CFLPRR13 (CCGA TCGG CCGA TGTG ATAT TGAC CTCC AC). The V domain (amino acids 1–144) was amplified with CFLPRR15 and CFLR1V (GTTG CGGC CGCC ATCA CCGT GAGA TTGA GCTG GC. The extracellular region of PVR (amino acids 1–330) was amplified with primers SBPVR5 (TTGA TCTG CAGA TGGC CCGA GCCA TGGC CGCC) and SBPVR3 (ATTT CTTT GCGG CCGC TTTG ACCT GGAC GGTC AGTT C). The PCR products were cloned in the Cos Fc Link (CFL) (SmithKline Beecham) vector (22) and transfected in COS 1 with Fugene 6 (Boehringer Mannheim). The proteins were purified on Affigel protein A (22). Purification was monitored by the sandwich ELISA in 96 wells coated with antibody against human Fc (Sigma) and biotinylated R1.302 antibody. The CTLA4-Fc was provided by R. Sweet (SmithKline Beecham).
Construction of V-TM(PRR1) and V(HIgR)-PVRα Transmembrane Receptors.
The V domain of HIgR was amplified with PRR1V5 (TAAT AAGC TTAT GGCT CGGA TGGG GCTT GCGG GC) and PRR1V3(GGTG TAGG GGAA TTCC ATCA CCGT GAGA TTG). The transmembrane and intracytoplasmic region was amplified by using primers PRR1IC5 (CAAT CTCA CGGT GATG GAAT TCCC CTAC ACC) and PRR1IC3 (ATTA GGAT CCCT ACAC GTAC CACT CCTT CTTG G). Both PCR products were mixed in a second PCR to get the final cDNA with primers PRR1V5 and PRR1IC3 (23), cloned in the BamHI-HindIII sites of pcDNA3. For the chimeric receptor V(HIgR)-PVRα, the V domain was amplified with primers PRR1V5 and R1VRV3 (GTGT TCTG GGGC TTGG CCAT CACC GTGA GATT G). The two C domains and transmembrane and intracytoplasmic regions of PVRα were amplified with primers R1VRCC5 (CAAT CTCA CGGT GATG GCCA AGCC CCAG AACA C) and R1VRCC3 (GTTA GGAT CCTC ACCT TGTG CCCT CTGT CTG). The 1,253-bp cDNA fragment was cloned in BamHI/HindIII sites of the pcDNA3.
Sandwich ELISA for the Soluble Forms of HIgR/PRR1, sVCC(HIgR)-Fc, and sV(HIgR)-Fc.
sVCC(HIgR)-Fc and sV(HIgR)-Fc were bound to microwells by means of anti-hIgG-Fc (Sigma) and reacted with biotinylated mAb R1.302, followed by streptavidin-peroxidase and One Step ABTS (Pierce).
Competition by sVCC(HIgR)-Fc and sV(HIgR)-Fc on HIgR-Mediated HSV-1 Infectivity.
Aliquots of R8102 were reacted with sVCC(HIgR)-Fc, sV(HIgR)-Fc, or CTLA4-Fc for 1 h at 37°C and absorbed to cells for 2 h at 4°C. Virus was removed. Cells were overlaid with medium containing the sVCC(HIgR)-Fc and sV(HIgR)-Fc at the same concentrations used in the inoculum and incubated for 16 h at 37°C. β-Galactosidase (β-gal) was assayed as described (2).
Competition of HSV-1 Infectivity by Soluble gD.
HIgR/cl 11 cells in 96-well plates were preincubated for 2 h at 4°C with gD-1(Δ290–299t) (24). Viral inoculum in 7.5 μl was added for 90 min at 4°C, removed, and cells were overlaid with DMEM containing glycoprotein. Infection was monitored as above.
In Vitro Binding of gD to Soluble Forms of HIgR/PRR1 by ELISA.
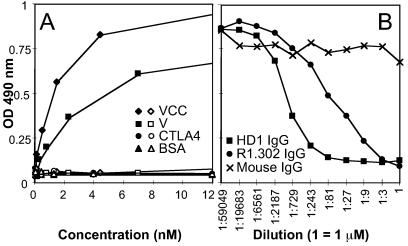
gD was purified to homogeneity from HSV-1-infected BHK cells by affinity chromatography to mAb 30 (1) immobilized to Affigel. Microwell plates were coated with 16 nM gD and reacted with sVCC(HIgR)-Fc or sV(HIgR)-Fc, followed by anti-human peroxidase (1:6,000) and o-phenylenediamine (Sigma). For competition ELISA, microwells were coated with gD. Ten nanomolar sV(HIgR)-Fc [representing the saturating amount of sV(HIgR)-Fc for the gD-coated microwell (see Fig. 6A)] were mixed with increasing concentrations of purified IgG of mAbs HD1, R1.302, or mouse IgG, and binding to gD was detected as described above. In Fig. 6B, dilution 1 corresponds to 1 μM purified IgG.
Figure 6.
(A) In vitro binding of HSV gD to soluble forms of HIgR receptor sV(HIgR)-Fc (V, ■, □) or sVCC(HIgR)-Fc (VCC, ⧫, ◊) or to CTLA4-Fc (CTLA4, •, ○) or BSA (▴, ▵). gD (solid symbols) or fetuin (open symbols) were immobilized to microwells and then allowed to react with increasing concentrations of the indicated proteins. Binding was detected with anti-human IgG-peroxidase (1:6,000). (B) Competition of the in vitro binding of HSV gD to sV(HIgR)-Fc by mAb R1.302 to PRR1 (R1.302, •), mAb to gD HD1 (HD1, ■), or purified mouse IgGs (IgG, ×). A fixed amount of sV(HIgR)-Fc (10 nM), giving saturable binding to gD in A, was mixed with increasing amounts of IgGs from the indicated antibodies and then allowed to react with gD, preimmobilized to microwells. Binding was revealed by incubation with anti-human IgG-peroxidase as above. Dilution 1 corresponds to 1 μM purified IgG for each antibody.
RESULTS
Construction and Purification of Soluble Forms of HIgR/PRR1 and of Human Poliovirus Receptor-α (hPVRα).
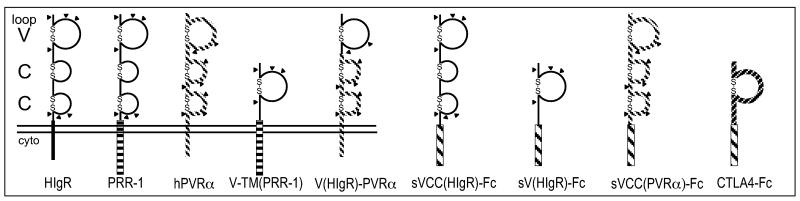
Two soluble forms of HIgR/PRR1 and one of hPVRα were constructed. In sVCC(HIgR)-Fc, the three domains (one V and two C2) that constitute the ectodomain of HIgR and of PRR1 were fused to the Fc domain of human IgG1. In sV(HIgR)-Fc, the single V domain of HIgR/PRR1 was fused to IgG Fc. In sVCC(PVR)-Fc, the ectodomain of hPVRα was fused to the IgG1 Fc. Schematic representation is shown in Fig. 1. COS1 cells were transfected with the plasmid DNAs, and the soluble receptors were purified from the culture medium by affinity chromatography to protein A. The purity of the protein preparations was checked by silver staining of proteins separated by SDS/PAGE and by immunoblotting with a peroxidase-labeled antibody directed to the human Fc portion of IgG (data not shown). Under reducing conditions, the apparent Mr of purified sVCC(HIgR)-Fc and sV(HIgR)-Fc was 70,000 and 45,000, respectively, whereas the predicted Mr values were 59,600 and 38,300, possibly reflecting glycosylation of some of the predicted glycosylation sites present in the ectodomain of HIgR/PRR1. The V domain, as defined here and below, comprises the N-terminal 114 residues, after cleavage of the predicted signal sequence.
Figure 1.
Schematic representation of constructs employed in this study. HIgR, PRR1, and hPVRα represent the full-length molecules encoded by cDNAs. V-TM(PRR1) is an internally deleted version of PRR1, where the two C domains were deleted. V(HIgR)-PVRα is a chimeric protein formed of the V domain of HIgR fused to the CC domains and transmembrane and cytoplasmic region of hPVRα. sVCC(HIgR)-Fc, sV(HIgR)-Fc, sVCC(PVRα)-Fc, and CTLA4-Fc represent soluble forms of receptors constructed by fusion of the VCC or V domains of HIgR, or the V domain of CTLA4, with the Fc portion of human IgG1. Construction is described in Materials and Methods.
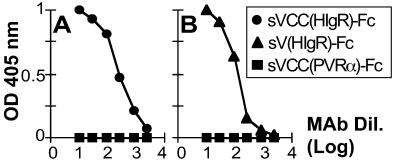
The HIgR/PRR1 Epitope Recognized by mAb R1.302 Resides in the V Domain.
mAb R1.302 is capable of blocking HSV-1 infectivity in hamster cell lines that express either HIgR or PRR1 from transfected plasmids as well as in human cell lines such as HEp-2, HeLa, human fibroblasts, U937, TF-1, etc., which express either one or both isoforms (5). As HIgR and PRR1 share the ectodomain, the epitope recognized by mAb R1.302 must reside in the ectodomain of the molecules. Fig. 2 shows that the epitope recognized by mAb R1.302 resides in the V domain of the HIgR/PRR1. In a sandwich ELISA that measured the binding of mAb R1.302 to sVCC(HIgR)-Fc or sV(HIgR)-Fc, the antibody was capable of binding both molecules. The binding was highly specific as mAb R1.302 failed to bind soluble forms of hPVRα (Fig. 2) and of PRR2(HveB) (not shown), two structurally related receptors belonging to the same Ig cluster (6, 7, 22), and sCTLA4-Fc (data not shown), a chimeric protein carrying the V domain of the T cell costimulatory protein CTLA4 (25) fused to IgG1 Fc. These results suggest that a major functional region of HIgR/PRR1 involved in HSV-1 entry into cells resides in the V domain.
Figure 2.
(A) Binding of mAb R1.302 to the soluble form of HIgR—sVCC(HIgR)-Fc—containing the entire ectodomain (VCC) fused to Fc. (B) Mapping of the epitope recognized by the mAb R1.302 to the V domain of HIgR. sVCC(HIgR)-Fc (•), sV(HIgR)-Fc (▴), or sVCC(PVRα)-Fc (■), immobilized to microwells through anti-human IgGs, were reacted with serial dilutions of biotinylated mAb R1.302. Abscissa represents the mAb dilutions (mAb Dil.) expressed as log10.
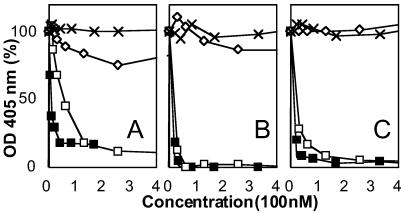
A Soluble Form of HIgR/PRR1 Consisting of the Single V Domain Inhibits HSV-1 Infectivity.
It has been reported that a soluble form of HveC(PRR1) blocks HSV-1 infectivity (3). The following experiments show that the soluble truncated form containing the single V domain, sV(HIgR)-Fc, competed successfully with the full-length, cell-bound HIgR, resulting in inhibition of HSV-1 infectivity. Replicate aliquots of the HSV-1 recombinant virus R8102, which carries a lacZ reporter gene fused to the immediate early α-27 promoter, were preincubated with increasing amounts of sV(HIgR)-Fc for 1 h at 37°C and absorbed to the HIgR/cl 11 cells. Infection was quantified after 16 h as β-gal activity. Positive control consisted of the soluble full-length receptor sVCC(HIgR)-Fc. Negative controls consisted of sVCC(PVR)-Fc and sCTLA4-Fc. As shown in Fig. 3A, the sV(HIgR)-Fc inhibited HSV-1 infectivity in a dose-dependent manner, with an inhibition curve similar, although not exactly overlapping, that obtained with sVCC(HIgR)-Fc. The inhibition was specific since sVCC(PVR)-Fc or sCTLA4-Fc had no significant inhibitory effect. The results in Fig. 3 B and C show that in HEp-2 and HeLa cells, the sV(HIgR)-Fc also competed with the resident cellular receptors and blocked R8102 infectivity. The reason for the slight difference between the blocking effect of sVCC(HIgR)-Fc and of sV(HIgR)-Fc in HIgR- or PRR1-transformed cells and not in HeLa or HEp-2 cells is not clear at the moment. Overall, the results are consistent with those above showing that the V domain contains the epitope recognized by the mAb R1.302, capable of inhibiting virus infectivity, and allow to draw two conclusions. A major functional region in HSV-1 entry is located in the V domain of HIgR/PRR1. The interaction between HSV-1 and the functional region encoded in the V domain is crucial for virus infectivity.
Figure 3.
HSV-1 infectivity is competitively blocked by soluble forms of receptors, sVCC(HIgR)-Fc and sV(HIgR)-Fc, and not by sVCC(PVRα)-Fc or CTLA4-Fc. Replicate aliquots of R8102 were preincubated with the indicated amounts of sVCC(HIgR)-Fc (■), sV(HIgR)-Fc (□), sVCC(PVRα)-Fc (×), and CTLA4-Fc (⋄) for 1 h at 37°C and allowed to absorb to the HIgR-expressing cells HIgR/cl 11 (A), HeLa (B), or HEp-2 (C) for 2 h at 4°C. Infection was quantified at 16 h after infection as β-gal activity. Note that sVCC(PVRα)-Fc, closely related to HIgR, and CTLA4-Fc did not affect R8102 infectivity. Each point represents the average of triplicate assays. One hundred percent indicates the optical density measured in untreated, virus-infected cultures.
The Single V Domain of PRR1 Fused to Its Transmembrane Domain or Transferred to the CC-Transmembrane–Cytoplasmic Domains of hPVRα Is Sufficient for HSV-1 Infectivity.
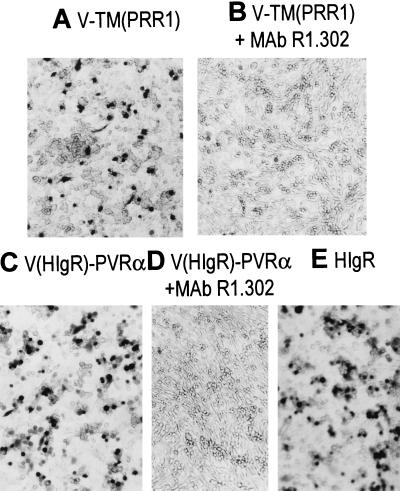
To ascertain whether the V domain of HIgR/PRR1 was sufficient to mediate HSV-1 entry into cells, a construct was generated in which the two C2 domains of PRR1 were deleted and the single V domain was fused directly to its transmembrane–cytoplasmic domains, designated V-TM(PRR1). The J1.1-2 cells, resistant to HSV infection because of lack of suitable receptors (5), were transfected with the plasmid DNA, subjected to neomycin G418 selection for 2 weeks, and assayed for susceptibility to R8102. Fig. 4A shows that V-TM(PRR1)/Q cells acquired susceptibility to R8102 infection, as detected by in situ 5-bromo-4-chloro-3-indolyl β-d-galactoside staining. The susceptibility correlated specifically with the V domain, as infectivity was abolished by mAb R1.302, which is capable of inhibiting infection and whose reactive epitope maps to the V domain (Fig. 4B). The number of cells acquiring susceptibility increased when cells were infected at higher moltiplicity of infection (100 plaque-forming units per cell) (data not shown). The number of cells acquiring susceptibility was much lower in cultures expressing V-TM(PRR1) than in cultures expressing the full-length HIgR or PRR1. This may be due to either a lower extent of expression of V-TM(PRR1) relative to the full-length molecules or a lower efficiency of the truncated molecule in conferring susceptibility to infection. To discriminate between these two possibilities the extent of expression was compared in V-TM(PRR1)- and HIgR-transfected cells by immunofluorescence with mAb R1.302. The number of fluorescent cells was found to be practically the same (data not shown), suggesting that the deleted version of PRR1 lacking the two C domains is less effective than the full-length counterpart in conferring susceptibility to HSV-1 infection. Parenthetically, the reactivity of mAb R1.302 to cells expressing the deleted and the full-length versions of HIgR/PRR1 confirms that mAb R1.302 is directed to an epitope present in the V domain. Altogether, the results demonstrate that the V domain of HIgR/PRR1 was sufficient to mediate HSV-1 entry into cells, although at reduced efficiency relative to the full-length receptor, and that susceptibility conferred by V-TM(PRR1) correlated specifically with the presence of the V domain.
Figure 4.
The V domain of HIgR is sufficient to mediate HSV-1 entry into J1.1–2 cells. J1.1–2 cells were transfected with a deleted form of PRR1 consisting of the V domain fused to its transmembrane and cytoplasmic regions, designated as V-TM(PRR1) (A and B), or with a chimeric receptor consisting of the V domain of HIgR/PRR1 fused to the CC-transmembrane–cytoplasmic regions of hPVRα, designated as V(HIgR)-PVRα (C and D), and for comparison with HIgR (E). (B and D) Blocking effect of mAb R1.302 on R8102 infectivity.
To confirm this and to investigate the reasons for the lower efficiency of V-TM(PRR1), a second construct was generated in which the V domain of HIgR/PRR1 was transferred to CC-transmembrane–cytoplasmic regions of hPVRα. This receptor was chosen as acceptor of the HIgR/PRR1 V domain because it has an overlapping structure to that of HIgR (6) but fails to mediate entry of any HSV-1 and -2 tested (3). Therefore, it represents the available receptor that is functionally inactive but structurally closer to HIgR/PRR1. Fig. 4C shows that V(HIgR)-PVRα transfected into the resistant J1.1–2 cells conferred susceptibility to HSV-1 infection and had an efficiency comparable to that of full-length HIgR (compare Fig. 4 C and E). Infectivity was abolished by exposure of cells expressing V(HIgR)-PVRα to mAb R1.302 (Fig. 4D), demonstrating that the susceptibility acquired by V(HIgR)-PVRα was a result of transfer of the V domain of HIgR/PRR1. The results confirm that the V domain of HIgR was sufficient to confer susceptibility and, in addition, suggest that the CC backbone of this cluster of molecules augments the virus entry activity located in the V domain and/or participates in virus entry activity with other mechanisms, as noted in the Discussion.
Soluble gD Competes with HSV-1 Infectivity in Cells in Which HIgR Is the Only Available Receptor.
Soluble forms of gD inhibit HSV-1 infectivity in Vero cells (16). As the receptors employed by HSV in these cells have not yet been identified, the cellular protein binding gD required for the gD inhibitory effect is unknown. In HveA-transformed cells, gD blocked HSV-1 infectivity (26), consistent with the ability of gD to bind HveA (27). Cells expressing HIgR or PRR1 acquire the ability to bind gD (5, 27). In this series of experiments we verified whether soluble gD can compete with HSV-1 infectivity in cells in which HIgR was the only available receptor. HIgR/cl 11 cells were preincubated with increasing amounts of a soluble recombinant form of gD [gD-1(Δ290–299t)] (24), or with fetuin, as a control, unrelated glycoprotein, and subsequently infected with R8102. Results in Fig. 5 show that gD inhibited R8102 infectivity in a dose-dependent manner, while fetuin had no significant effect. The experiment demonstrates that in cells where HIgR was the only available receptor, the binding of HIgR with gD was crucial for HSV-1 entry.
Figure 5.
R8102 infectivity in HIgR-expressing cells is competitively blocked by a soluble form of gD. HIgR/cl 11 cells were preincubated with the indicated amounts of gD-1(Δ290–299t) (■) or fetuin (□), expressed as nanomolar concentrations (nM), and then infected with R8102. Infection was quantified in triplicates as β-gal assay at 16 h. Each point represents the average of a triplicate assay.
The Single V Domain Is Sufficient for Physical Interaction with gD.
The above data demonstrate that a major region of HIgR/PRR1 functional in HSV-1 entry resides in the V domain and that this domain is sufficient to mediate HSV-1 infectivity. gD binds to a soluble form of HveC(PRR1) containing the entire ectodomain (27). Here we investigated whether the single V domain of HIgR/PRR1 was sufficient for the physical interaction with gD. For this assay, gD was immobilized to microwells and then reacted with the soluble receptor consisting of the single V-domain, sV(HIgR)-Fc, or with the full-length sVCC(HIgR)-Fc as a positive control. The results in Fig. 6A demonstrate that sV(HIgR)-Fc bound gD in a dose-dependent manner, with a curve essentially similar to that obtained with sVCC(HIgR)-Fc. There was an ≈30% reduction in the level of saturable binding with sV(HIgR)-Fc relative to sVCC(HIgR)-Fc, suggesting a somewhat higher efficiency in the binding to gD for the full-length molecule. The control, unrelated molecules CTLA4-Fc or BSA did not bind gD. Specificity of the binding next was measured in a competitive ELISA. A fixed amount of sV(HIgR)-Fc, giving a saturable binding to immobilized gD (see Fig. 6A), was preincubated with increasing concentrations of IgG from mAb R1.302 to HIgR/PRR1, or from mAb HD1, a mAb to HSV gD with potent neutralizing activity on HSV infectivity (21) and with the ability to compete with the binding of PRR1(HveC) to virions in vitro (27), or with purified mouse IgGs, as control. As can be seen from Fig. 6B, both the mAb to HIgR and the neutralizing mAb HD1 to gD competed with the binding of gD to the soluble V domain of HIgR/PRR1, demonstrating that the in vitro binding of the V domain to gD was highly specific. The results of these two assays indicate that the V domain of HIgR/PRR1 was sufficient for specific binding to gD. The results confirm and extend the finding that the gD region recognized by HIgR/PRR1 contains the antigenic site Ia (27, 28).
DISCUSSION
To understand how the interaction of HSV-1 with HIgR or PRR1 leads to virus entry into the cell, it is necessary to obtain a detailed picture of the domains of HIgR/PRR1 that are functional in HSV-1 entry and of the physical interaction between the functional domains and gD, the virion component engaged in virus entry. For many viruses and cognate receptor systems, genetic analyses coupled to demonstration of physical interaction between the two partner molecules provided a basis for unraveling this interaction. This paper shows that a major region of HIgR/PRR1 with HSV-1 entry activity resides in the V domain. We show that (i) the V domain is a major determinant of HIgR/PRR1 in mediating HSV-1 entry, (ii) the V domain anchored to cell membrane is sufficient to mediate HSV-1 entry into cells, and (iii) the single V domain is sufficient for the in vitro physical interaction with gD in a specific manner. It is convenient to discuss each issue separately.
The V Domain Is a Major Determinant of HIgR/PRR1 in Mediating HSV-1 Entry.
Evidence for this conclusion rests cumulatively on two series of experiments. First, the mAb R1.302 to HIgR/PRR1, capable of inhibiting infection, reacted with an epitope mapped to the V domain of HIgR/PRR1. Second, a soluble form of the HIgR/PRR1 containing the single V domain competed in a dose-dependent manner with full-length, cell-bound receptor and blocked virus infectivity. Thus, HIgR adds a new member to the list of Ig-like viral receptors whose major functional domains reside in the more external regions. The list includes PVR, the poliovirus receptor, ICAM, the rhinovirus receptor, and CD4, the HIV receptor (29–36). That the N-terminal domains carry functional regions is consistent with the view that, even in the folded molecules, these regions are located more externally and therefore are more likely to interact with the appropriate regions of the virions.
The V Domain Anchored to Cell Membrane Is Sufficient to Mediate HSV-1 Entry into Cells.
This conclusion rests on two experiments. First, the receptor-deficient J1.1–2 cells resistant to HSV-1 infection were rendered susceptible when transfected with an engineered form of PRR1 in which the V domain and transmembrane and cytoplasmic regions were retained and fused together but the two C2 domains were deleted. Second, the V domain of HIgR/PRR1 was transplanted to CC-transmembrane–cytoplasmic portion of hPVRα, a structural homologue of HIgR and PRR1 not functional in HSV-1 and -2 entry, generating a chimeric receptor that acquired HSV-1 entry activity. As infectivity was abolished by neutralizing mAb R1.302 in both cases, the receptor activity resided in the V domain. We conclude that the V domain is sufficient to act as a receptor as long as it is anchored to the membrane by suitable transmembrane/cytoplasmic domains.
The Single V Domain Is Sufficient for the in Vitro Physical Interaction of HIgR/PRR1 with gD.
gD is a key component of the viral machinery essential for virus entry into susceptible cells, as outlined in the Introduction. Furthermore, gD can interact physically with full-length ectodomain of HveC(PRR1) (27). It was of interest to determine whether the V domain, herein shown to encode a major region functional in HSV-1 entry, was sufficient for in vitro binding to gD, or whether other portions of the ectodomain would mediate this binding. Here we show that a soluble form of HIgR/PRR1 consisting of the single V domain interacted physically with gD in an in vitro binding assay. The binding was specific as it was competed by mAbs to each partner with the ability to neutralize HSV infectivity. Thus, mAb R1.302 to HIgR/PRR1 competed with the ability of gD to bind to the receptor. In a similar fashion, the mAb HD1 to gD with potent neutralizing activity on virion infectivity competed with the binding of gD to its receptor. It can be noted that mAb HD1 recognizes antigenic site Ia of gD, according to the classification of Cohen and Eisenberg (28). The results further indicate that the gD region that interacts with the V domain of HIgR/PRR1 contains the antigenic site Ia, in agreement with the finding that mAbs to this site block binding of PRR1(HveC) to virions (27), and that the gD region interacting with HIgR/PRR1 does not exactly overlap with that recognized by HveA, which recognizes antigenic sites Ib and VII, but not Ia (26).
Current data provide several lines of evidence that the V domain of HIgR/PRR1 contains the major functional region involved in and sufficient for HSV-1 entry into cells and for physical interaction with gD. Notwithstanding this, we noticed a reduction in the efficiency of the V domain relative to the full-length molecule to interact with virions and with gD, particularly evident in the lower ability of V-TM(PRR1) to mediate susceptibility to HSV-1 infection. Several explanations may account for these reductions, which remain to be investigated further. Thus, for example, the CC domains may provide a spacer that enhances accessibility of virions to the V domain, and/or the CC domains may form a scaffolding for a conformation of the V domain suitable to its interaction with virions and gD, as proposed for PVR (34). Alternatively, homo- or heterooligomer formation or association with additional cell surface proteins may be altered as a consequence of the removal of the CC domains. Further yet, the CC domains may have secondary sites that bind gD or may be able to bind the additional glycoproteins, gB and gH/gL, that participate in HSV-1 entry process and thus recruit them to the fusion complex. Consistent with current findings, in the case of poliovirus or HIV interaction with engineered forms of PVR or CD4, respectively, the N-terminal domain was sufficient to render cells susceptible to infection although the virus yields were reduced (32–34). In addition, in the case of poliovirus and rhinovirus receptors, the functional domains need to have a certain degree of flexibility, which may be provided by the CC and interdomain portions, as their cognate binding sites are located in a canyon embedded in the capsid.
Resolution of the interaction at the molecular level of HSV-1 with its bona fide receptors has two major implications. On one hand, it is instrumental to define the interaction between gD and HIgR/PRR1 in terms of minimal size of the functional domain, structural requirements, key residues, etc. In the present study the major functional region of HIgR/PRR1 with HSV-1 entry activity able to physically interact with the viral gD was narrowed to a region of about 114 aa residues. Ultimately, these studies could lead to a model that would allow critical tests of the fusion event between HSV-1 envelope and the plasma membranes and the mode of recruitment of the other viral glycoproteins involved in the process. On another hand, analyses of the interaction of gD with the functional regions of its receptor may lead to practical applications. Infections with HSV-1 are highly prevalent among humans. Pathologic manifestations vary from mucocutaneous lesions of the mouth, face, eyes, or genitals, to involvement of central nervous system, resulting sometimes in encephalitis (see ref. 37). The identification that a major functional domain of HIgR/PRR1 is encoded in the V domain coupled with the demonstration that this domain is sufficient for physical binding to gD and to compete with virion infectivity may provide a basis for the future development of a novel class of anti-HSV agents designed specifically to block HSV-1 infection.
Acknowledgments
We thank colleagues Sara Di Gaeta and José Adelaide for generous help and are indebted to Elisabetta Romagnoli for assistance with cell cultures and to Angela Algeri for invaluable suggestions. We thank G. Cohen and R. Eisenberg, Philadelphia, for recombinant gD; B. Roizman, Chicago, for gifts of R8102; and R. Sweet, SmithKline Beecham, for CTLA4-Fc. The work done at the University of Bologna was supported by grants from Target Project in Biotechnology, AIDS Project from Istituto Superiore di Sanità, BIOMED2 BMH4 CT95 1016 Grant from UE, Ministero dell’Università e della Ricerca Scientifica e Tecnologica (40%), University of Bologna (60%), and pluriannual plan.
ABBREVIATIONS
- gD
glycoprotein D
- HSV-1
herpes simplex virus-1
- HIgR
herpesvirus receptor/herpes virus Ig-like receptor
- hPVRα
human poliovirus receptor-α
- PRR1
poliovirus receptor-related protein 1
- HveC
herpesvirus entry mediator C
- β-gal
β-galactosidase
References
- 1.Brandimarti R, Huang T, Roizman B, Campadelli Fiume G. Proc Natl Acad Sci USA. 1994;91:5406–5410. doi: 10.1073/pnas.91.12.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montgomery R I, Warner M S, Lum B J, Spear P G. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 3.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 4.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 5.Cocchi F, Menotti L, Mirandola P, Campadelli-Fiume G. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez M, Eberlé F, Mattei M G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 7.Eberlé F, Dubreuil P, Mattei M G, Devilard E, Lopez M. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 8.Cai W H, Gu B, Person S. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis Poynter N, Minson T. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roop C, Hutchinson L, Johnson D C. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ligas M W, Johnson D C. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campadelli-Fiume G, Qi S, Avitabile E, Foà-Tomasi L, Brandimarti R, Roizman B. J Virol. 1990;64:6070–6079. doi: 10.1128/jvi.64.12.6070-6079.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean H J, Terhune S S, Shieh M T, Susmarski N, Spear P G. Virology. 1994;199:67–80. doi: 10.1006/viro.1994.1098. [DOI] [PubMed] [Google Scholar]
- 15.Huang T, Campadelli Fiume G. Proc Natl Acad Sci USA. 1996;93:1836–1840. doi: 10.1073/pnas.93.5.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson D C, Burke R L, Gregory T. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicola A V, Peng C, Lou H, Cohen G H, Eisenberg R J. J Virol. 1997;71:2940–2946. doi: 10.1128/jvi.71.4.2940-2946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittels M, Spear P G. Virus Res. 1991;18:271–290. doi: 10.1016/0168-1702(91)90024-p. [DOI] [PubMed] [Google Scholar]
- 19.Spear P G. Semin Virol. 1993;4:167–180. [Google Scholar]
- 20.Lopez M, Jordier F, Bardin F, Coulombel L, Chabannon C, Dubreuil P. In: Leukocyte Typing VI, White Cells Differentiation Antigens. Kishomoto T, Kikutani H, von dem Borne A, Mason D Y, Miyasaka M, Moretta A, Okumura K, Shaw S, Springer T A, Sugumura K, et al., editors. New York: Garland; 1997. pp. 1081–1083. [Google Scholar]
- 21.Pereira L, Klassen T, Baringer J R. Infect Immun. 1980;29:724–732. doi: 10.1128/iai.29.2.724-732.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez M, Aoubala M, Jordier F, Isardon D, Gomez S, Dubreuil P. Blood. 1998;92:4602–4611. [PubMed] [Google Scholar]
- 23.Maroc N, Rottapel R, Rosnet O, Marchetto S, Lavezzi C, Mannoni P, Birnbaum D, Dubreuil P. Oncogene. 1993;8:909–918. [PubMed] [Google Scholar]
- 24.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linsley P S, Brady W, Urnes M, Grosmaire L S, Damle N K, Ledbetter J A. J Exp Med. 1991;174:561–569. doi: 10.1084/jem.174.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muggeridge M I, Isola V J, Byrn R A, Tucker T J, Minson A C, Glorioso J C, Cohen G H, Eisenberg R J. J Virol. 1988;62:3274–3280. doi: 10.1128/jvi.62.9.3274-3280.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landau N R, Warton M, Littman D R. Nature (London) 1988;334:159–162. doi: 10.1038/334159a0. [DOI] [PubMed] [Google Scholar]
- 30.Mendelsohn C L, Wimmer E, Racaniello V R. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 31.Diamond D C, Finberg R, Chaudhuri S, Sleckman B P, Burakoff S J. Proc Natl Acad Sci USA. 1990;87:5001–5005. doi: 10.1073/pnas.87.13.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jasin M, Page K A, Littman D R. J Virol. 1991;65:440–444. doi: 10.1128/jvi.65.1.440-444.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koike S, Ise I, Nomoto A. Proc Natl Acad Sci USA. 1991;88:4104–4108. doi: 10.1073/pnas.88.10.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selinka H C, Zibert A, Wimmer E. Proc Natl Acad Sci USA. 1991;88:3598–3602. doi: 10.1073/pnas.88.9.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernhardt G, Harber J, Zibert A, deCrombrugghe M, Wimmer E. Virology. 1994;203:344–356. doi: 10.1006/viro.1994.1493. [DOI] [PubMed] [Google Scholar]
- 36.Arita M, Koike S, Aoki J, Horie H, Nomoto A. J Virol. 1998;72:3578–3586. doi: 10.1128/jvi.72.5.3578-3586.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitley R J. In: Virology. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. New York: Raven; 1996. pp. 2297–2342. [Google Scholar]