Abstract
Abscisic acid (ABA) is a ubiquitous hormone that regulates plant growth, development, and responses to environmental stresses. Its action is mediated by the PYR/PYL/RCAR family of START proteins, but it remains unclear how these receptors bind ABA and in turn, how hormone binding leads to inhibition of the downstream type 2C protein phosphatase (PP2C) effectors. Here we report crystal structures of apo and ABA-bound receptors as well as a ternary PYL2-ABA-PP2C complex. The apo receptors contain an open ligand-binding pocket flanked by a gate that closes in response to ABA via conformational changes in two highly conserved β-loops that serve as a gate and latch. Moreover, ABA-induced closure of the gate creates a surface that enables the receptor to dock into and competitively inhibit the PP2C active site. A conserved tryptophan in the PP2C inserts directly between the gate and latch, which functions to further lock the receptor in a closed conformation. Together, our results identify a conserved gate-latch-lock mechanism underlying ABA signaling.
Abscisic acid (ABA) is a vital hormone used by plants in their responses to environmental stresses such as drought, cold, and salinity. Genetic studies have established a canonical signaling pathway mediated by type 2C protein phosphatases (PP2Cs) and subfamily 2 of SNF1-related kinases (SnRK2 kinases), which act downstream of ABA sensing1-5. However, identification of bona fide ABA receptors has been a daunting task full of controversy and frustration6. A new class of START protein receptors called the PYR/PYL/RCARs has recently been described7-9. ABA binding to members of this protein family enables them to bind to and inhibit PP2Cs such as HAB1, leading to the activation of the SnRK2 kinases that phosphorylate downstream effectors such as the basic leucine-zipper transcription factors called ABFs/AREBs, thus switching on stress response programs1,10-13. The mechanisms of ABA binding and PP2C inhibition by the PYR/PYL/RCARs are currently unknown. In addition, the binding of ABA to RCAR1 (PYL9) and PYL5 is stimulated >10 fold by the presence of a PP2C and many of the PYR/PYL/RCAR proteins show constitutive (i.e. unliganded) binding to PP2Cs7,8. These observations have led to an important but currently unresolved question: does ABA bind to a PYR/PYL/RCAR-PP2C co-receptor, or does ABA bind first to the PYR/PYL/RCAR receptors followed by secondary PP2C interactions that stabilize ligand binding? Many of the most critical questions regarding how ABA receptors function will require structural elucidation of ligand-bound and ligand-free receptors, and a receptor-PP2C complex. Here we report the crystal structures of apo- and ABA-bound forms of PYL2, and the ABA-PYL2-HAB1 complex at resolutions of 1.95Å, 1.85Å, and 1.95 Å, respectively, as well as the apo-PYL1 structure at a resolution of 2.40Å. Comparison of the apo- and ABA-bound structures shows that ABA binding induces closure of two highly conserved β-loops that function as a gate and latch to sense ligand entry. In the apo form of the receptor, the gate and latch reside in an open conformation, which permits access of ABA to the ligand-binding pocket. ABA binding induces the closure of the gate and formation of a gate-latch interface, which in turn binds to the PP2C active site and converts the ABA-bound PYL2 into a competitive PP2C inhibitor. The interactions between PYL2 and HAB1 further induce conformational changes in both proteins that lock the gate into the closed position, which is primarily mediated by a conserved tryptophan of HAB1. Importantly, this residue also makes a water-mediated interaction with ABA, thus allowing HAB1 to sense the presence of ABA in the PYL2 ligand-binding pocket. The critical residues in ligand binding and the PYL2/HAB1 interface are highly conserved, suggesting that the gate-latch-lock mechanism of ligand sensing and signal transduction is likely utilized by the entire PYR/PYL/RCAR receptor family.
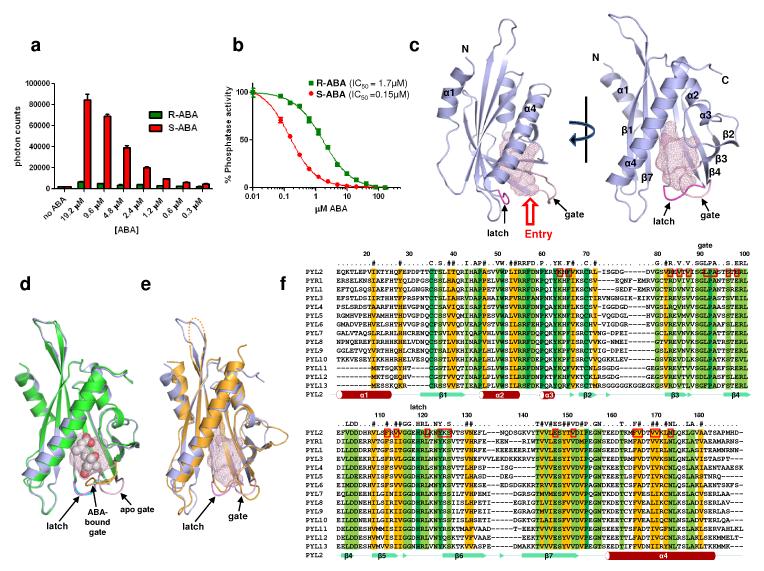
To prepare proteins for crystallization, the entire PYR/PYL family was expressed in E. coli and screened for suitability in crystallization trials. PYR1 and PYL1-6 were soluble and yielded high levels of purified proteins (Supplementary Fig. 1A). Each of these family members was capable of interacting with the ABA-signaling PP2Cs HAB1, ABI1, and ABI2 (Supplementary Fig. 1B). However, as suggested previously by yeast two hybrid assays7,8, the PYR/PYL proteins and PP2Cs showed differential interactions in response to the presence or absence of ABA, the particular stereoisomer utilized, and the particular PP2C probed. These biochemical observations point to possible functional variations between family members in PP2C binding and signaling. Among the proteins expressed, PYL1 and PYL2 produced high-quality crystals of apo receptors, and PYL2 crystals diffracted better than 2.0 Å both in the absence and presence of ABA (Supplementary Fig. 1C). PYL2 interacted with the PP2C HAB1 in an ABA-dependent manner and showed clear preference for the natural S-stereoisomer (Fig. 1A). PYL2 was also able to inhibit the phosphatase activity of HAB1 with an IC50 of 0.15 μM with S-ABA, but with an IC50 of 1.7 μM with the biologically less active R-isomer (Fig. 1B). This 11-fold difference in selectivity is comparable to the 19-fold selectivity shown for PYL5, another member of the PYR/PYL family 9. The PYL1 and PYL2 structures were solved by molecular replacement starting from a model of the birch pollen allergen Bet VI (PDB code 1bv1)14, with the statistics of the structure refinement shown in Supplementary Table 1.
Fig. 1. Structures of apo- and ABA-PYL2, and apo-PYL1.
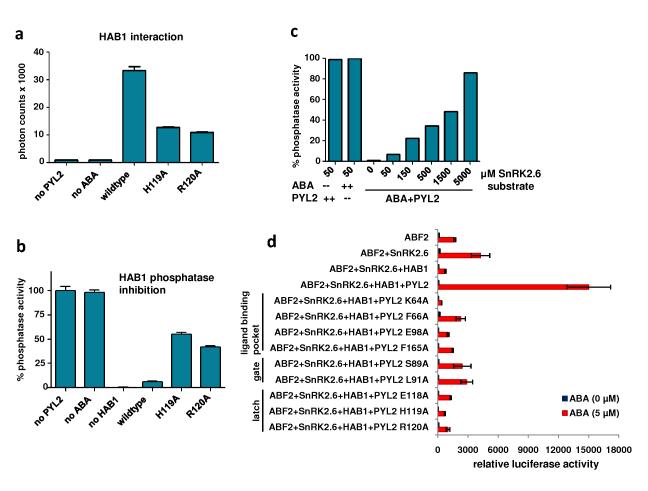
a, ABA-dependent interaction of PYL2 with HAB1. Binding of recombinant H6GST-PYL2 to recombinant biotin-HAB1 at the indicated concentrations of S- and R-ABA was determined by AlphaScreen assays as described in Methods and in Supplementary Fig 4A (n=3, error bars=SD). b, ABA-dependent inhibition of HAB1 phosphatase activity by PYL2. Phosphate release from pS175 of 100μM SnRK2.6 phosphopeptide was determined by colorimetric assay in the presence of recombinant HAB1 and PYL2 (n=3, error bars=SD). The IC50 values were derived from curve fitting based on a competitive inhibitor model for the binding of ABA-bound PYL2 to HAB1 (R2= 0.998 [S-ABA] and 0.997 [R-ABA]). c, Two views of the apo-PYL2 structure with its empty ligand binding pocket shown as mesh. The gate (pink) and latch (magenta) for the ligand entry are indicated by arrows. d, Overlay of the apo (grey) and ABA-bound (green) PYL2 structures. ABA is shown as ball model with its surrounding ligand binding pocket as mesh. The latch (magenta for apo- and cyan for ABA-bound PYL2) and the conformational change in the gate (pink and yellow in apo- and ABA-bound structures, resp.) are indicated by arrows. e, Overlay of the apo structures of PYL2 (grey) and PYL1 (yellow). f, Sequence alignment of the PYR/PYL family with secondary structure elements of PYL2 below the sequences and amino acid position of full length PYL2 above the alignment. Note that PYL2 in the crystal structure starts at position 14 relative to the full length protein. Conserved residues are highlighted and residues within a 5Å range of the ligand are boxed, and the gate and latch residues are noted.
The structures of apo- and ABA-bound PYL2 have an α-β-α2-β6-α topology and exhibit nearly identical helix-grip folds, which is a hallmark of the START protein superfamily15 (Fig. 1C). A dominant feature of the structure is the long C-terminal α-helix enfolded by an anti-parallel β-sheet of seven strands and α2 and α3 helices. The packing interactions of these elements form the hydrophobic ligand-binding cores of START proteins. Between the C-terminal helix and the β-sheet is a large pocket of 543Å3 in the apo structure (Fig. 1C), and 480Å3 in the ABA-bound structure (Fig. 1D). This pocket is surrounded by 23 residues, which are mostly hydrophobic and highly conserved (Fig. 1F). PYL1 and PYL2 share 51% sequence identity (Supplementary Table 2), and the apo-PYL1 structure resembles the apo-PYL2 structure (Fig. 1E and Supplementary Fig. 2B). The major differences between the apo-PYL1 and apo-PYL2 structures are seen in the relative position of their N-terminal helices and their loop structures between β strands 6 and 7, which is invisible in the apo-PYL1 structure. In contrast, the C-terminal α-helix and the β-strands surrounding the ligand binding pocket are highly similar.
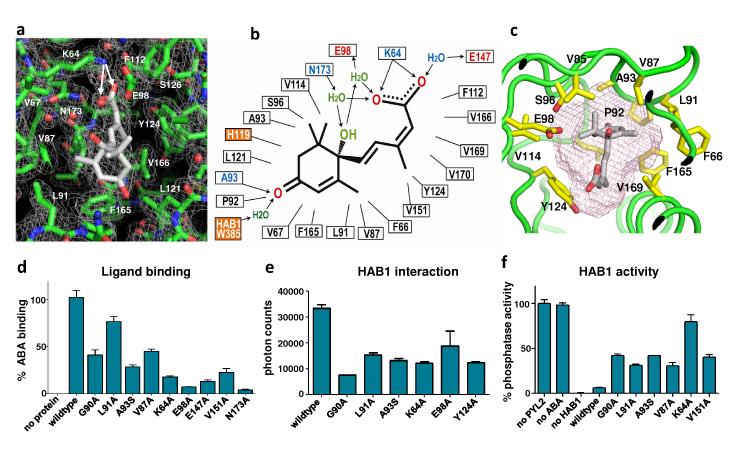
The overall structure of PYL2-ABA complex reveals a monomeric receptor/ligand complex with 1:1 stoichiometry. The binding mode of ABA is clearly defined by a high resolution electron density map (Fig. 2A and Supplementary Fig. 3A). ABA is centered in the pocket with its hydrocarbon chain and cyclohexene ring fitting snugly into the hydrophobic pocket of PYL2. The inter-molecular interactions between ABA and PYL2 are diagrammed in Fig. 2B. Each of the three polar groups of ABA forms direct or water-mediated hydrogen bonds with the receptor. The acid head group forms a charged interaction with the side chain of K64 (Fig. 2A), and a complex network of water-mediated hydrogen bonds with residues E98, N173 and E147 (Fig. 2A-B and Supplementary Fig. 3A). The hydroxyl group from the chiral carbon also forms two water-mediated hydrogen bonds with E98 and N173. Mutations in these polar residues and several other pocket residues in PYL2 reduced its ability to bind to radio-labeled ABA, to interact with HAB1 in response to ABA, and to inhibit HAB1’s PP2C activity (Fig. 2 D-F). The ABA-bound PYL2 structure also reveals the basis for its stereo-selectivity. Fig. 2C shows that the mono-methyl group from the cyclohexene ring fits into a narrow pocket formed by residues F66, V87, L91, P92, F165, and V169, while the dimethyl group fits into a larger pocket formed by A93, S96, and V114. The dimethyl group flipped in the R-isomer would cause stereo collision between the dimethyl group and the narrow pocket that accommodates the mono-methyl group (Fig. 2C). Thus, the size and shape of PYL2’s pocket that accommodates the cyclohexene ring, together with the specific hydrogen bonds between PYL2 and ABA, contribute to its stereo-selectivity.
Fig. 2. Structural, functional, and mutational analysis of the ABA binding pocket.
a, A 2Fo-Fc electron density map of bound ABA and its surrounding residues contoured at 1.0 σ. The charge interaction between the ABA acid group and K64 is indicated by two arrows. b, Schematic presentation of the interactions between PYL2 binding pocket residues and the bound S-ABA. Charged interactions and hydrogen bonds are indicated by arrows, hydrophobic interactions by solid lines with H-bond donors in blue and acceptors in red. The two orange-boxed residues, PYL2 H119 and HAB1 W385, contact ABA only upon formation of the trimeric PYL2-ABA-HAB1 complex. c, The pocket topology of PYL2 surrounding the mono-methyl and the dimethyl groups of ABA shown with key PYL2 residues and the ligand binding pocket (mesh). d-f, Mutations in key ligand binding pocket residues of PYL2 compromise (d) ligand binding as determined by scintillation proximity assay with 3H-labelled ABA (n=3, error bars=SD), (e) HAB1 interaction determined by AlphaScreen assays (n=3, error bars=SD, and (f) inhibition of HAB1 phosphatase activity (n=3, error bars=SD).
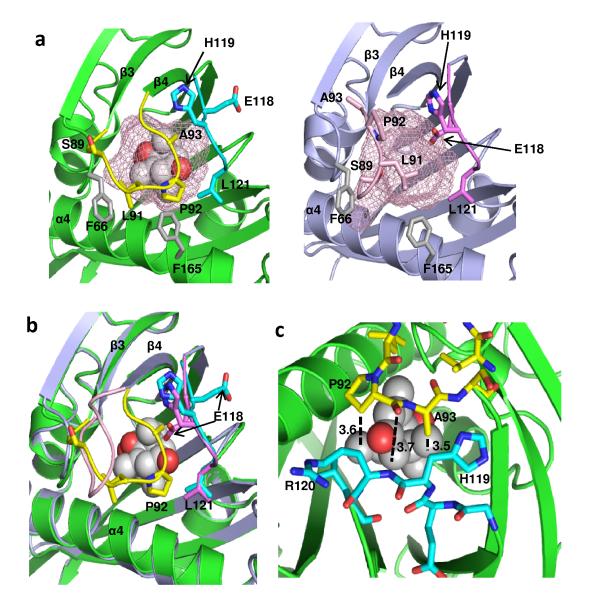
Comparison of the apo- and ABA-bound PYL2 structures suggests that a gate and latch mechanism is employed in hormone binding and signal transduction. The entry of the ligand binding pocket is surrounded by a gate-like loop between β-strands 3 and 4 (residues 89-93: SGLPA) on one side, and on the other side, a latch-like region formed by a loop between β-strands 5 and 6 (residues 119-121: HRL) (Fig. 3A). The positions of H119 and L121 (latch) do not change substantially between apo- and ABA-bound structures. In contrast, ABA binding induces a dramatic conformational change in the SGLPA loop, which shifts 3-9 Å away from the corresponding loop of the apo-structure (Fig. 3A&B), a swing that closes the ligand entry gate and seals the bound ABA molecule from exposure to the solvent.
Fig. 3. A gate and latch mechanism seals ABA in the binding pocket.
a, Close view of the ligand entry site in the apo- and ABA-bound PYL2 structure with the pocket shown as mesh. Side chains in the entry site are marked. The gate and latch are shown in yellow and cyan in the ABA-bound structure and in pink and magenta in the apo structure. b, Overlap of the ligand binding pocket in the ABA-bound and apo form with the gate and latch in the same color code as in (a). c, Interactions between gate (yellow) and latch (cyan) residues in the PYL2-ABA complex. Distances between residues are indicated in Å. ABA is shown as ball model.
Outside of the SGLPA gate, the most dramatic change of conformation occurs at residue E118, which is the residue immediately preceding the HRL latch motif (Fig. 3A and 1F). In the apo-structure, the E118 side chain points into the pocket, a position that would prevent the closure of the SGLPA gate. In the ABA-bound structure, the E118 side chain is flipped by ~150 degrees, so that it points outside of the pocket. This creates the space required for the SGLPA gate to close onto the HRL latch, with the distance between LPA (gate) and HRL (latch) changing from 11-14 Å to 3.5-3.7 Å (Fig. 3A-C). Upon closing the gate, the SGLPA loop together with the HRL residues form a new interface, which we predicted could fit into the active site of the PP2C by using molecular modeling (see methods and Supplementary Fig. 3B).
To validate our prediction and unravel the mechanism of PP2C inhibition by PYR/PYL proteins, we prepared a ternary complex for crystallization by mixing the purified proteins of PYL2 and HAB1 in the presence of S-ABA (Supplementary Fig. 1D). Because of the high binding affinity of PYL2 to HAB1 in the presence of S-ABA, (estimated to be 0.15 μM, see Supplementary Fig. 4), the ternary complex was stable and readily formed crystals (Supplementary Fig. 1C) that diffracted better than 1.95 Å. The structure was determined by molecular replacement with the ABA-PYL2 structure and a HAB1 model built from the PPM1B structure, a human PP2C16. The statistics of the structure refinement are shown in Supplementary Table 1.
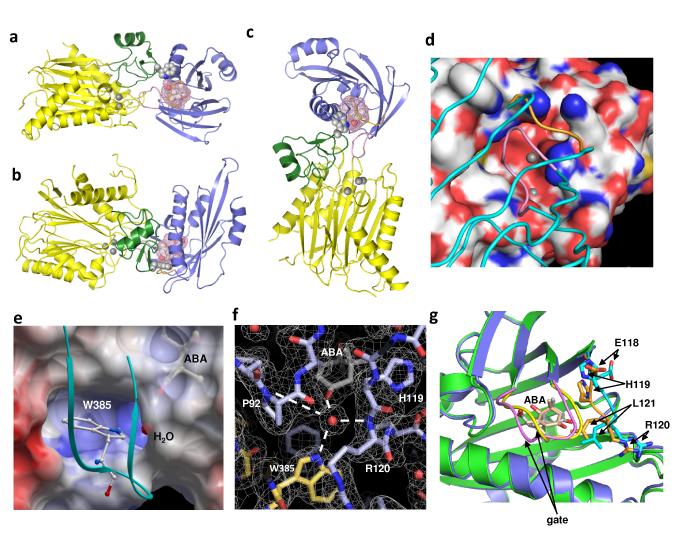
The overall structure of the PYL2-ABA-HAB1 complex reveals a monomeric receptor/ligand/PP2C complex with 1:1:1 stoichiometry (Fig. 4A). The catalytic domain of HAB1 adopts a typical PP2C fold17, containing two five-stranded β-sheets that are sandwiched by two pairs of anti-parallel α-helices. Between the two central β-sheets, there is a small domain of one short α-helix and two anti-parallel β-strands (residues 361-412) that forms the major part of the binding interface with PYL2 (Fig. 4A-C). The catalytic site, which is located at the bottom edge of the two central β-sheets with three Mg2+ ions, is partially occupied by the bound PYL2 (Fig. 4C-D).
Fig. 4. Structure of the PYL2-ABA-HAB1 co-receptor complex.
a-c, Three views of the PYL2-ABA-HAB1 complex. HAB1 is shown with its catalytic domain in yellow (Mg2+ ions as balls) and the ABA-interacting domain in green (W385 lock as ball model). PYL2 is shown in blue and ABA as ball model with its surrounding ligand binding pocket as mesh. d, HAB1 active site as space-filling presentation with Mg2+ ions as balls and the docking PYL2 shown as carbon backbone (gate in pink and latch in gold color). e, HAB1 W385 docking into a PYL2 cavity enables it to make a water-mediated interaction with the ketone group of ABA. The loop containing W385 is shown as ribbon in cyan. f, Network of water-mediated interactions with the ketone group of ABA in the PYL2-ABA-HAB1 complex. ABA and key residues are shown with the surrounding electron density map (2Fo-Fc) contoured at 1σ, W385 of HAB1 is highlighted in yellow. g, HAB1 docking induces conformational changes in the PYL2 gate and latch. Close-up overlay view of PYL2-ABA in the presence (blue) and absence (green) of HAB1. The latch is shown in cyan (−HAB1) and gold (+HAB1), the gate in yellow (−HAB1) and pink (+HAB1), and ABA in grey (−HAB1) and peach (+HAB1).
The complex contains a large complementary interface of 1820 Å2 between the ABA-bound PYL2 and HAB1, whose intermolecular interactions are summarized in Supplementary Table 3. Consistent with our modeling, ABA-bound PYL2 docks into the catalytic active site of HAB1 (Fig. 4D), with the SGLPA gate packing closely with residues D243 to G246, which forms the active site loop of the PP2C. The G246D mutation in HAB1 and the corresponding residues in other group A PP2Cs are well documented to cause dominant ABA-insensitivity18. The distance of G246 to the SGLPA gate is only 2.8 to 4.0 Å, which likely explains why G246D can disrupt the PP2C/PYR1 interaction7-9,19.
Importantly, the ternary complex structure reveals several critical insights of ABA binding and signal transduction that were not predicted by our modeling studies or from the PYL2-ABA complex. The most striking observation is that the indole ring of W385 of HAB1 inserts between SGPLA gate and the HRL latch. In so doing, it points into the ABA-binding pocket and makes a water-mediated hydrogen bond to the ketone group of ABA (Fig. 4E-F). This observation therefore provides direct proof that HAB1 can serve as a co-receptor that senses the binding of ABA into the PYR/PYL receptors.
In response to the insertion of W385 of HAB1 into the ABA-binding pocket of PYL2, the SGPLA gate and the HRL latch undergo several unexpected conformational changes between the PYL2-ABA and ternary PYL2-ABA-HAB1 structures (Fig. 4G). The most drastic changes are seen in the latch residues E118 and H119, whose Cα atoms move 4.5Å and 1.9Å, respectively. In both apo and ABA-bound PYL2, the side chain of H119 is outside of the pocket. However, in the HAB1 complex, PYL2’s H119 side chain points into the pocket and makes contacts with the dimethyl groups of ABA’s cyclohexene ring. Correspondingly, the position of ABA’s cyclohexene ring shifts 1.2-1.4Å, allowing its ketone group to face directly toward the water molecule that makes a hydrogen bond with W385 of HAB1(Fig. 4E-F). In this configuration, W385 of HAB1 functions like a molecular lock that keeps the gate and the latch in the closed positions. This locking mechanism likely explains the higher ABA binding affinities measured for PYR/PYLs in the presence of PP2Cs7,8. Thus, PP2C binding modifies the PYL2-ABA contact surface and creates a water-mediated contact between ABA and PP2C.
The importance of the gate and latch loops is supported by extensive mutational data (Fig. 2D-F). Mutations in both the gate and latch compromised ABA-dependent HAB1 interaction and PP2C activity (H119A and R120A in Fig. 5A-B). Additionally, it has been noted that mutations in the PP2C active site (mutations that cause the dominant ABA-insensitive phenotype, e.g. G180D in ABI1, G168D in ABI2, and G246D in HAB1) disrupt PP2C’s interaction with PYR17-9,19, which is consistent with our model, as the larger side chain of the mutated residues would collide with the SGPLA gate (Supplementary Fig. 3C). Furthermore, the direct docking of PYL2’s SGPLA loop into the active site of HAB1 suggests that the ABA-bound PYL2 functions as a competitive inhibitor of PP2Cs. To test this prediction, we investigated whether increasing concentrations of a PP2C substrate, a SnRK2.6 peptide containing residues 170-180 and a phosphorylated serine at position 175, could overcome the inhibition of HAB1 PP2C activity by ABA-bound PYL2 (Fig. 5C). SnRK2.6 is a natural substrate of PP2C and phosphorylation at S175 is required for SnRK2.6 kinase activation and its role in ABA signaling5. Our results are consistent with a competitive inhibitor mechanism between ABA-bound PYL2 and the PP2C active site (Fig. 4D and 5C). Thus, our structures illuminate both the mode of ligand binding and PP2C inhibition.
Fig. 5. Mutational analysis of PYL2 and HAB1 interface residues.
a+b, Functional analysis of mutations in PYL2 latch residues by (A) HAB1 interaction as determined by AlphaScreen (n=3, error bars=SD) and by (B) HAB1 phosphatase activity (n=3, error bars=SD). c, Increasing concentrations of the SnRK2.6 substrate overcomes HAB1 inhibition by ABA-bound PYL2. Uninhibited HAB1 phosphatase activity (absence of either ABA or PYL2) is shown as control. d, Reconstituted ABA signaling pathway for gene expression in Arabidopsis mesophyll protoplasts. PYL2, HAB1, SnRK2.6 and ABF2 expression vectors were cotransfected into protoplasts together with a luciferase reporter plasmid driven from the promoter of the ABA-responsive RD29B gene. Mutations in the PYL2 ligand binding pocket (K64A, F66A, E98A, F165A), the SGLPA gate (S89A and L91S), and the latch (E118A, H119A, R120A) are defective in the reconstituted signaling pathway (n=3, error bars=SEM).
To validate the functional roles of the PYL2/HAB1 interface in ABA signaling, we mutated key residues that form the gate and latch of PYL2, and analyzed their function in protoplasts20, where ABA-induction of gene expression can be reconstituted by coexpression of a PYR/PYL protein, PP2Cs, SnRK2 kinases, the ABF2 transcription factor and an ABA-responsive promoter reporter construct11. Using this system, ABA induces reporter gene activation in the presence of PYL2 and HAB1 (Fig 5D). However, mutations in PYL2 residues involving ligand binding or formation of the gate and latch compromise the ability of PYL2 to activate the reporter in response to ABA. These mutations also reduced the ability of PYL2 function through ABI1 (Supplementary Fig. 5). Thus, our mutational and structural data suggest that the gate and latch residues involved in conformational changes are critical components of the information transfer mechanism.
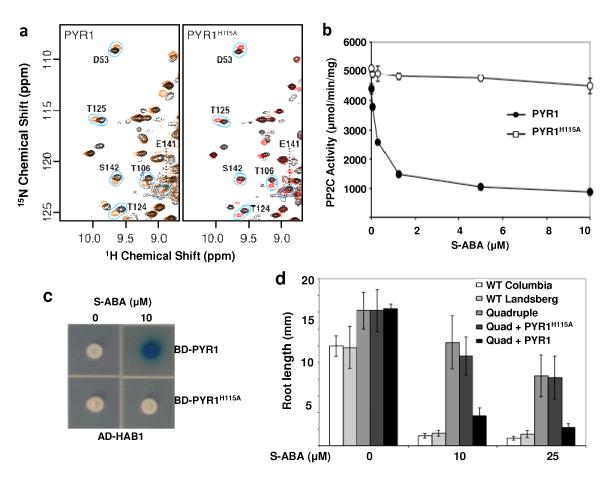
The 14 members of the PYR/PYL family share 38-84% sequence identity over the entire length of the proteins (Supplementary Table 2), but the SGLPA and HRL sequences are invariant among the PYR/PYL family members (Fig. 1F), suggesting that the gate and latch mechanism is likely to be a common feature of these receptors. To further investigate this mechanism and ascertain if it is used by other family members, we studied the consequences of ABA binding on PYR1 using HSQC spectroscopy of NMR, which is a complementary method because it enables measurements of dynamics directly in solution. PYR1 was chosen as a model because its HSQC spectra of NMR have been previously investigated7. ABA binding induced either exchange broadening or chemical shift perturbation in residues surrounding the ligand binding pocket (Supplementary Fig.6A). In contrast, the vast majority (66 of 71, 93%) of unperturbed residues in the PYR1 spectrum are located outside the ABA binding pocket (Supplementary Fig. 6B). Notably, the SGLPA gate and the HRL latch residues exhibit broadening or large shift perturbation, consistent with a large conformational change upon ligand binding (Supplementary Fig. 6C-D). Thus, the chemical shift perturbations induced by ABA binding to PYR1, collected by solution NMR, correlate exceedingly well with the molecular interactions observed in the crystal structures of apo- and ABA-bound PYL2. These observations strongly argue that conformational dynamics accompany ligand binding in multiple PYR/PYL family members and strengthen our conclusion that they are central to the mechanism of signal transduction. We also performed transgenic plant studies of PYR1 with a mutated latch residue (H115A). The homologous residue (H119) in PYL2 is pointed outward to the solvent in both the apo- and ABA-bound PYL2 structures (Fig. 3A-B), thus the H115A mutation is predicted not to interfere with ABA binding in the absence of HAB1. Indeed, NMR spectra of 15N-labelled PYR1H115A show that the mutated protein forms a stably folded protein that responds to ABA similarly to the wild type PYR1 (Fig. 6A), indicating that the H115A PYR1 has comparable ligand binding capability as the wild type in the absence of HAB1. However, consistent with the important role of this latch residue in ABA binding in the PYL2/PP2C complex, the H115A PYR1 protein was defective in ABA mediated PP2C inhibition in vitro (Fig. 6B) and was not able to interact with HAB1 in response to S-ABA (Fig. 6C). Furthermore, H115A PYR1 failed to rescue the ABA-response defect of a quadruple pyr1/pyl1/pyl2/pyl4 mutant in transgenic plants, while the wild type PYR1 did (Fig. 6D). Together, these results demonstrate that the latch residue H115 is not important for ligand binding in the context of PYR1 itself, but is critical for the receptor to relay the ABA binding signal to PP2Cs and downstream signaling effectors.
Fig. 6. Mutations in the PYR1 latch and gate affect ABA signaling in vitro and in vivo.
a, ABA binding is unaltered by the H115A mutation in the PYR1 latch. Comparisons of 1H-15N HSQC spectra for PYR1 (left) and PYR1(H115A) (right) in the presence (red contours) and absence (black contours) of S-ABA indicate that residues surrounding the binding site observed in the ABA-PYL2 crystal structure are similarly perturbed in both proteins. b, HAB1 phosphatase activity is abolished in H115A PYR1 (n=3, error bars=SD). c, The H115A PYR1 mutant is defective in ABA-mediated HAB1 interaction as shown by yeast two-hybrid assay. AD: activation domain, BD: DNA binding domain. d, The H115A PYR1 mutant is defective in inhibiting the root growth of transgenic plants in response to ABA. A quadruple pyr1/pyl1/pyl2/pyl4 mutant (Q) defective in root growth inhibition can be complemented by wildtype PYR1, but not by PYR1 (H115A) (n=6-10, error bars=SD).
In this paper, we present four crystal structures involving two ABA receptors, PYL1 and PYL2, in apo-forms, and the ABA-bound forms of PYL2, either alone or in complex with the HAB1 PP2C. In combination with biochemical, mutagenesis, NMR, and transgenic plant studies, our structures highlight a gate-latch-lock mechanism of ABA binding and signaling by the PYR/PYL family and PP2C. The ternary complex structure reveals that PP2Cs can function as co-receptors to facilitate PYR/PYL proteins to interact with ABA, and establish that these proteins together are bona fide ABA receptors that transduce plant stress signals. Elucidation of the structures and mechanisms of these core components of ABA signaling pathways will help construct a rational framework for understanding plant stress biology as well as for engineering plants that are resistant to salt and drought environments.
The START domain was originally identified in proteins involved in steroidogenesis and metabolism in metazoans21-23. Although members of the START domain family continue to be identified15,24, the cellular functions of most START proteins still remain elusive beyond the established roles in transfer of lipids and cholesterol25. Identification of the gate-latch-lock mechanism for hormone binding and signaling by ABA receptors suggests that other members in the large family of START domain proteins may also function as ligand sensing and signal transduction molecules like ABA receptors.
Methods Summary
PYL2, PYL1, and HAB1 were expressed as H6-GST or H6Sumo fusion proteins in E. coli. Proteins were purified by Ni-NTA chromatography, followed by proteolytic release of tags and size-exclusion chromatography. For formation of PYL2-ABA and HAB1-PYL2-ABA complexes, ABA was mixed with PYL2 and HAB1-PYL2 at 5:1 ratios. Crystals were grown by vapor diffusion and diffraction data were collected from cryo-protected crystals at beamlines 21-ID-D and 21-ID–F at the Advanced Photon Source at Argonne National Laboratories. Structures were solved by molecular replacement in PHASER 26 using the structure of the plant START protein Bet v 1 as model for PYL2 and the structure of the human PP2C PPM1B as model for HAB1. Models were manually fitted using O and Coot 27,28 and further refined using CNS and Refmac5 29,30.
Mutant proteins were expressed as H6GST-fusion proteins and purified by glutathione sepharose chromatography. Protein-protein interactions were determined by luminescence proximity AlphaScreen assay and by yeast two-hybrid assay. Biotinylated HAB1 for the luminescence proximity assay was generated by in vivo biotinylation of an avitag-HAB1 fusion protein. ABA binding was determined by scintillation proximity assay using 3H-labelled ABA. HAB1 phosphatase activity was measured by phosphate release from a SnRK2.6 phosphoprotein (Fig. 1-5) or from a generic pNPP phosphate substrate (Fig. 6b).
For transgenic studies, wildtype and mutant 35S::GFP-PYR1 constructs were transformed by the floral dip method into pyr1/pyl1/pyl2/pyl3 quadruple mutants. Mutant complementation of GFP+ seedlings was assayed by root length measurements. The ABA signal transduction pathway was reconstituted in protoplasts by transient transfection of PYL2, PP2C, SnRK2.6, and ABF2 expression plasmids. Activation of an ABA-inducible CBF3promoter-LUC reporter by PYL2 mutant proteins was determined by luciferase assays normalized for β-glucuronidase activity from a UQ10-GUS reporter. Full Methods accompany this paper at www.nature.com/nature.
Online Methods
Protein Preparation
PYL2 (residues 14–188) was expressed as a 6×His-GST (H6GST) fusion protein from the expression vector pET24a (Novagen). The modified fusion protein contains a H6-tag (MKKGHHHHHHG) at the N terminus and a thrombin protease site between GST and PYL2. BL21(DE3) cells transformed with the expression plasmid were grown in LB broth at 16°C to an OD600 of ~1.0 and induced with 0.1 mM IPTG for 16 hr. Cells were harvested, resuspended in 100 ml extract buffer (20 mM Tris, pH 8.0, 200 mM NaCl, and 10% glycerol) per 6 liters of cells, and passed three times through a French press with pressure set at 1000 Pa. The lysate was centrifuged at 16,000 rpm in a Sorvall SS34 rotor for 30 min, and the supernatant was loaded on a 50 ml Nickel HP column. The column was washed with 10% buffer B (20 mM Tris, pH 8.0, 200 mM NaCl, 500 mM imidazole, and 10% glycerol) for 600 ml and eluted in two steps with 50% buffer B for 200 ml, then 100% buffer B for 100 ml. The eluted H6GST-PYL2 was dialyzed against extract buffer and cleaved overnight with thrombin at a protease/protein ratio of 1:250 in the cold room. The cleaved H6GST tag was removed by passing through a Nickel HP column, and the protein was further purified by chromatography through a HiLoad 26/60 Superdex 200 gel filtration column in 25 mM Tris, pH8.0, 200 mM ammonium acetate, 1mM dithiotreitol and 1mM EDTA. Apo-PYL2 eluted as a sharp single peak with an estimated molecular weight of 23KD, suggesting that apo-PYL2 is a monomer in solution (predicted MW of 21KD).
PYL1 (residues 36–211) was expressed as a H6-SUMO fusion protein from the expression vector pSUMO (LifeSensors). The expression and purification of PYL1 followed the same method as for PYL2 except that the H6SUMO tag was cleaved overnight with SUMO protease at a protease/protein ratio of 1:1000 in the cold room. A typical yield of the purified protein is ~6 mg for PYL1 or ~1 mg for PYL2 from each liter of cells. The apo-PYL1 eluted from the gel filtration column as a broad peak that corresponds to an estimated MW of 31 KD, suggesting the apo-PYL1 may exhibit a monomer-dimer equilibrium in solution. To prepare the protein-ligand complex, we added S-ABA to the purified proteins at a 5:1 molar ratio.
HAB1 (residues 172–511) was expressed as a H6GST fusion protein from the expression vector pET24a (Novagen). Expression and purification of HAB1 followed the same method as for PYL2 except that all buffers contained 5 mM MgCl2. A typical yield of the purified protein was about 5 mg from each liter of cells. To prepare the protein-ligand-PP2C ternary complex, we added S-ABA and purified PYL2 to purified HAB1 at a 5:1:1 molar ratio in the presence of 5mM MgCl2.
Small scale purifications of H6GST-tagged PYL/PYR proteins for expression screening (Fig. S1A) and for binding studies of wildtype and mutant proteins were performed by standard glutathione sepharose chromatography. To generate biotinylated proteins for luminescence proximity assays (AlphaScreen), PP2C open reading frames (orfs) were expressed in E. coli BL21(DE3) cells from a pETDuet (Novagen) derivative vector. The first T7 polymerase-driven expression unit of this vector contains the PP2C orf as H6-thioredoxin-thrombin cleavage site-avitag fusion, the second site the E. coli biotin-ligase gene BirA. The 14 amino acid avitag functions as a defined in vivo biotinylation site in E. coli 31. Cells grown in the presence of 40 μM biotin were lysed and fusion protein purified over Nickel HiTrap columns as above. Following thrombin release of the H6-thioredoxin tag, PP2C proteins with biotinylated avitags were purified over monomeric avidin columns (Pierce) according to the manufacturer’s instructions. Residual biotin was removed by extensive dialysis prior to their use in luminescence proximity assays.
Crystallization
The apo-PYL1 crystals were grown at room temperature in hanging drops containing 1.0 μl of the purified PYL1 protein at 9.1 mg/ml and 1.0 μl of well solution containing 0.1M ammonium sulfate, 0.1M Na-HEPES, pH7.5, 10% w/v PEG 4000, and 30% glycerol. Crystals of about 100 μm in length appeared within 1–2 days. Crystals were flash frozen in liquid nitrogen prior to data collection. The ABA-PYL1 complex did not yield crystals.
Apo-PYL2 crystals were grown at room temperature in hanging drops containing 1.0 μl of the purified PYL2 protein at a concentration of 6.6 mg/ml and 1.0 μl of well solution containing 0.056M sodium phosphate monobasic monohydrate/1.344M potassium phosphate dibasic, pH 8.2, and 5% butanediol. Crystals appeared within 1–2 days and grew to a dimension of 200–250 μm in length over a period of 4 days. Crystals were serially transferred to well buffer with increasing glycerol concentration (20% v/v final) prior to flash freezing in liquid nitrogen.
The PYL2-ABA complex crystals were grown at room temperature in hanging drops containing 1.0 μl of the above protein-ligand solutions at a concentration of 6.6 mg/ml, and 1.0 μl of well solution containing 0.1M HEPES, pH 7.5, 2M ammonium sulfate, and 20% sorbitol. Crystals of about 250 μm in length appeared the following day. Crystals were soaked in well solution with a final concentration of 30% sorbitol prior to flash freezing in liquid nitrogen.
The PYL2-ABA-HAB1 complex crystals were grown at room temperature in hanging drops containing 1.0 μl of the above protein-ligand-PP2C solutions and 1.0 μl of well solution containing 0.2M Ammonium sulfate, 0.1M BIS-Tris pH 6.5, and 15% PEG 3350. Crystals appeared within 1–2 days and grew to a dimension of 450-600 μm in length on the third day. Crystals were serially transferred to well buffer with increasing PEG3350 concentration (35% v/v final) prior to flash freezing in liquid nitrogen.
Data Collection and Structure Determination
The datasets were collected with MAR300 and MAR225 CCD detectors (MAR Research) at the ID-D and ID-F lines of sector-21(LS-CAT) at the Advanced Photon Source at Argonne National Laboratory. The observed reflections were reduced, merged, and scaled with DENZO and SCALEPACK in the HKL2000 package 32. Crystals of apo-PYL1, apo-PYL2, ABA-PYL2, and ABA-PYL2-HAB1 diffracted to resolutions of 2.40Å, 1.85Å, 1.95Å, and 1.95Å, respectively. The apo-PYL1 crystals formed in the P65 space group with two receptors in each asymmetry unit. The weak dimer interface observed in the apo-PYL1 crystals is consistent with monomer-dimer behavior of the protein in solution. The apo-PYL2 crystals formed in the C2221 space group with three receptors in each asymmetry unit. The ABA-bound PYL2 crystals formed in the P6122 space group with one receptor in each asymmetry unit and the ABA-PYL2-HAB1 crystals formed in the P212121 space group with one complex in each asymmetric unit. The intermolecular packing in both apo- and ABA-bound crystals is less extensive than the apo-PYL1 crystals, consistent with the monomeric behavior of the protein in solution.
Molecular replacement was performed by using the Collaborative Computational Project 4 (CCP4) program Phaser 26. Programs O and Coot were used to manually fit the protein model 27,28. Model refinement was performed with CNS and the CCP4 program Refmac5 29,30. The volumes of the ligand binding pocket were calculated with the program Voidoo by using program default parameters and a probe with a radius of 1.4 Å 33. All structure figures were prepared by using PyMOL (DeLano Scientific). The statistics of data collection and the model refinement are summarized in Table S1.
Molecular modeling
The PYL2 model was built with the Prime module (Schrödinger, LLC) using the structure of Bet V I, a birch pollen allergen that contains a START domain (PDB code: 1bv1), and this PYL2 model was used as template for molecular replacement. The HAB1 model was built with the Prime module (Schrödinger, LLC) using the structure of PPM1B, a human PP2C that shares 38% homology with the HAB1 PP2C core domain (PDB code: 2P8E) 16. The HAB1-PYL2/ABA complex model was built with ZDOCK 34 via the protein-protein docking web server (http://zdock.bu.edu/) by uploading the crystal structure of the ABA-bound PYL2 and the homology model of HAB1. The top docking models were retrieved from this server and visually inspected. Structure-based sequence alignments were performed with the ICM program package (Molsoft Inc).
Assays for the interactions between PYR/PYL and PP2Cs
Interactions between PYR/PYL and PP2Cs were assessed by luminescence-based AlphaScreen technology (Perkin Elmer) that our group has used extensively to determine ligand-dependent protein-protein interactions for nuclear receptors 35-37. The scheme of AlphaScreen is illustrated in Supplementary Fig. 4A. Briefly, biotinylated PP2Cs were attached to streptavidin-coated donor beads, and H6-tagged PYR/PYL receptors were attached to nickel-chelated acceptor beads. The donor and acceptor beads were brought into proximity by the interactions between PYR/PYLs and PP2Cs, which were measured with and without abscisic acid, the R-isoform (Sigma) or the S-isoform (A.G. Scientific, Inc). When excited by a laser beam of 680 nm, the donor beam emits singlet oxygen that activates thioxene derivatives in the acceptor beads, which releases photons of 520-620 nm as the binding signal. The experiments were conducted with 100 nM of PP2Cs and PYR/PYL proteins in the presence of 5ug/ml donor and acceptor beads in a buffer of 50 mM MOPS, pH7.4, 50mM NaF, 50mM CHAPS, and 0.1mg/ml bovine serum albumin. The results were based on an average of three experiments with standard errors typically less than 10% of the measurements.
Radio-ligand binding assay
2 μM H6GST-PYL2 were incubated with 250 μg of Ytrium silicate copper-chelating scintillation proximity assay (SPA) beads (GE Healthcare) in a buffer of 50 mM MOPS, pH 7.4, 50 mM NaF, 50 mM CHAPS, and 0.1 mg/ml bovine serum albumin for 40 minutes shaking on ice. H6GST-PYL2 bound to SPA was separated from free H6GST-PYL2 by centrifugation at 5200xg for 30 seconds. Bead pellets were washed with 1 ml of the same buffer, then resuspended in 50 μl of the buffer supplemented with 45 nM 3H-labelled ABA (GE Healthcare; mixture of S-/R- and cis-/trans-isomers), 1 μM unlabelled S-ABA and 10 μM HAB1 and incubated shaking for 1 hour at room temperature. 3H-ABA-PYL2 binding brings the radioactive ABA into the immediate proximity of the scintillant embedded in the SPA beads resulting in the generation of light, which was quantified in a liquid scintillation counter.
Assays of HAB1 phosphatase activity
100 nM biotin-HAB1 and 500 nM Pyl2 were pre-incubated in 50 mM imidazole, pH 7.2, 5 mM MgCl2, 0.1% β-mercaptoethanol and 0.5 μg/ml BSA for 30min at room temperature. Reactions were started by addition of 100 μM of a phosphopeptide corresponding to amino acids 170-180 of SnRK2.6 (HSQPKpSTVGTP). This peptide is phosphorylated at a single residue corresponding to Ser175 in SnRK2.6, whose phosphorylation is required for SnRK2.6 kinase activity2. Phosphate release from pS175 from the phosphopeptide was determined by colorimetric assay (BioVision). Phosphatase inhibition assays in Fig. 6B were performed using the generic pNPP phosphatase substrate, as described previously 7, with the minor modification that the assay buffer utilized 10 mM Mn++ instead of Mg++, which we found to enhance HAB1’s specific activity ~10 fold.
Mutagenesis
Site-directed mutagenesis was carried out using the QuickChange method (Stratagene) or the GeneTailor™ System (Invitrogen). Mutations and all plasmid constructs were confirmed by sequencing prior to both protein expression and yeast hybrid assays. Expression and purification of GST-HAB1, H6-PYR1, and H6-PYR1H115A were performed as described previously 7.
PYR1 transgenic studies
To probe function of mutant PYR1 proteins in planta, we cloned mutant cDNAs into pEGAD38 to create the 35S::GFP-PYRH115A construct, which was then transformed into the pyr1/pyl1/pyl2/pyl4 quadruple mutant line as described 7 using floral dip transformation. For complementation tests, T1 seeds were germinated on MS plates after 4 days stratification and two day old seedlings were screened using a fluorescence-dissecting microscope to identify GFP+ seedlings.6 – 10 GFP+ seedling were transferred to MS plates (at least 6 seedlings per plate) containing differing ABA concentrations, and grown vertically under fluorescent lighting. Root lengths were measured 4 days post transfer. For analysis of 35S::GFP-PYR1, a homozygous single insert line in the quadruple mutant background was used, which was made using a previously described DNA construct 7.
Protoplast transient assays
An ABA-inducible luciferase reporter was constructed by replacing the CBF3 promoter in the CBF3-LUC construct with the promoter region of RD29B, a highly ABA-inducible gene 8. CBF3-LUC, UQ10-GUS and ABI1 protoplast expression plasmid vectors were kindly provided by Jen Sheen (Massachusetts General Hospital, Boston, MA 02114). The coding regions of His-PYR1 and mutants, ABI1-myc, ABF2-HA, and SnRK2.6-Flag were used to replace ABI1 in the ABI1 protoplast expression plasmid vectors.
Protoplasts were isolated from Arabidopsis thaliana ecotype Columbia-0 plants, which were grown on Jiffy7 soil (Jiffy Products Ltd., Canada) in an environment-controlled chamber at 22°C with a photosynthetically active radiation of 75 μmol m−2 s−1 and a day/night cycle of 13 hours light/11 dark. Transient activity assays were performed in Arabidopsis mesophyll protoplasts from Columbia-0 wild-type plants as described by the Sheen laboratory (http://genetics.mgh.harvard.edu/sheenweb ) 20. Transfected protoplasts were incubated for 5 hours in light in the presence of 0 or 5 μM (+) ABA, and then used for measuring LUC and GUS activity. The UQ10- GUS reporter plasmid (β-glucuronidase) was used as internal control to normalize transfection efficiency in protoplast assays.
NMR studies of the wild type and H115A mutated PYR1
The wild type and H115A mutated PYR1 proteins for NMR were expressed in E. coli and purified as described previously 7. ABA-PYR1 complexes were obtained by addition of a two-fold excess of S-ABA to the purified proteins. NMR experiments were performed at 37°C on a Bruker Avance 600MHz spectrometer equipped with a 5 mm TCI CryoProbe and processed with NMRPipe 39. Backbone 1H, 15N and 13C chemical shifts of ABA-PYR1 were obtained using automated assignment software 40,41 and manual analysis of 3D HNCO, HNCOCA, HNCA, HNCOCACB, HNCA, HNCACB, HNCACO, CCONH, HBHACONH, HCCONH, and 15N-edited NOESY-HSQC spectra. For apo-PYR1, only 3D HNCA, 3D HNCO, and 3D 15N NOESY experiments were performed before sample degradation prevented further data collection, but these spectra were sufficient to permit identification of all signals assigned in the ABA-PYR1complex. Assignments were confirmed for 120 of the 183 non-proline residues of PYR1. Portions of the PYR1 sequence could not be assigned due to extreme line broadening in the 2D and 3D spectra, including residues 1–7, 54–64, 81–88, 99–106, 117–123, and 152–166. Many of these residues participate in a dimer interface observed in the crystal packing of apo-PYL2, suggesting that line broadening associated with monomer-dimer exchange is responsible for the majority of missing signals.
Supplementary Material
Acknowledgments
We thank the staff of LS-CAT for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Use of the Advanced Photon Source was supported by the Office of Science of the U. S. Department of Energy. This work was supported by the Jay and Betty Van Andel Foundation (H.E.X.); National Institutes of Health (H.E.X., B.F.V., and J-K. Z.); and National Science Foundation (S.R.C.).
Footnotes
Author Information/ Data deposition: The structure factors and atomic coordinates discussed in this work have been deposited in the Protein Data Bank (www.pdb.org ). The accession codes are: 3KAY for the apo-PYL1; 3KAZ for the apo-PYL2; 3KB0 for the ABA-bound PYL2 complex; and 3KB3 for the ternary complex of PYL2-ABA-HAB1.
Competing interest statement: The authors declare no competing financial interests.
References
- 1.Yoshida T, et al. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinnusamy V, Gong Z, Zhu JK. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol. 2008;50:1187–1195. doi: 10.1111/j.1744-7909.2008.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida R, et al. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 5.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennisi E. Plant biology. Stressed out over a stress hormone. Science. 2009;324:1012–1013. doi: 10.1126/science.324_1012. [DOI] [PubMed] [Google Scholar]
- 7.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 9.Santiago J, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi Y, et al. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- 11.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci U S A. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 14.Gajhede M, et al. X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nat Struct Biol. 1996;3:1040–1045. doi: 10.1038/nsb1296-1040. [DOI] [PubMed] [Google Scholar]
- 15.Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 16.Almo SC, et al. Structural genomics of protein phosphatases. J Struct Funct Genomics. 2007;8:121–140. doi: 10.1007/s10969-007-9036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- 18.Bertauche N, Leung J, Giraudat J. Protein phosphatase activity of abscisic acid insensitive 1 (ABI1) protein from Arabidopsis thaliana. Eur J Biochem. 1996;241:193–200. doi: 10.1111/j.1432-1033.1996.0193t.x. [DOI] [PubMed] [Google Scholar]
- 19.Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci U S A. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 21.Allen-Baume V, Segui B, Cockcroft S. Current thoughts on the phosphatidylinositol transfer protein family. FEBS Lett. 2002;531:74–80. doi: 10.1016/s0014-5793(02)03412-9. [DOI] [PubMed] [Google Scholar]
- 22.Seedorf U, Ellinghaus P, Nofer J. Roch. Sterol carrier protein-2. Biochim Biophys Acta. 2000;1486:45–54. doi: 10.1016/s1388-1981(00)00047-0. [DOI] [PubMed] [Google Scholar]
- 23.Wirtz KW. Phospholipid transfer proteins in perspective. FEBS Lett. 2006;580:5436–5441. doi: 10.1016/j.febslet.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 24.Schrick K, Nguyen D, Karlowski WM, Mayer KF. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004;5:R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olayioye MA, et al. StarD10, a START domain protein overexpressed in breast cancer, functions as a phospholipid transfer protein. J Biol Chem. 2005;280:27436–27442. doi: 10.1074/jbc.M413330200. [DOI] [PubMed] [Google Scholar]
- 26.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Kleywegt GJ, Jones TA. Efficient rebuilding of protein structures. Acta Crystallogr D Biol Crystallogr. 1996;52:829–832. doi: 10.1107/S0907444996001783. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 31.Smith PA, et al. A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res. 1998;26:1414–1420. doi: 10.1093/nar/26.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 33.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 34.Wiehe K, et al. The performance of ZDOCK and ZRANK in rounds 6-11 of CAPRI. Proteins. 2007;69:719–725. doi: 10.1002/prot.21747. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Kovach A, Suino-Powell K, Martynowski D, Xu HE. Structural and biochemical basis for the binding selectivity of peroxisome proliferator-activated receptor gamma to PGC-1alpha. J Biol Chem. 2008;283:19132–19139. doi: 10.1074/jbc.M802040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suino K, et al. The Nuclear Xenobiotic Receptor CAR; Structural Determinants of Constitutive Activation and Heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 37.Xu HE, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 38.Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci U S A. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Bartels C, Günterts P, Billeter M, Wüthrich K. GARANT - a General Algorithm for Resonance Assignment of Multidimensional Nuclear Magnetic Resonance Spectra. J. Comp. Chem. 1997;18:139–149. [Google Scholar]
- 41.Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS computational biology. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.