Abstract
In order to understand the biological importance of naturally occurring sialic acid variations on disialyl structures in nature, we developed an efficient two-step multi-enzyme approach for the synthesis of a series of GD3 ganglioside oligosaccharides and other disialyl glycans containing a terminal Siaα2–8Sia component with different natural and non-natural sialic acids. In the first step, α2–3- or α2–6-linked monosialylated oligosaccharides were obtained using a one-pot three-enzyme approach. These compounds were then used as acceptors for the α2–8-sialyltransferase activity of a recombinant truncated multi-functional Campylobacter jejuni sialyltransferase CstII mutant, CstIIΔ32I53S, to produce disialyl oligosaccharides. The α2–8-sialyltransferase activity of CstIIΔ32I53S has promiscuous donor substrate specificity and can tolerate various substitutions at C-5 or C-9 of the sialic acid in CMP-sialic acid, while its acceptor substrate specificity is relatively restricted. The terminal sialic acid residues in the acceptable monosialylated oligosaccharide acceptors are restricted to Neu5Ac, Neu5Gc, KDN, and some of their C-9 modified forms but not their C-5 derivatives. The disialyl oligosaccharides obtained are valuable probes for their biological studies.
Keywords: carbohydrate, chemoenzymatic synthesis, enzyme, GD3 oligosaccharides, sialic acid, sialosides
Introduction
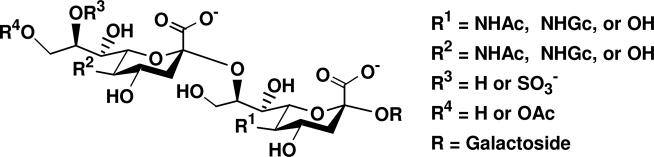
Sialic acids (Sia) are a diverse family of naturally occurring polyhydroxy keto aldonic acids that are broadly distributed in animals and are involved in a wide range of biological processes.1 In most cases, N-acetylneuraminic acid (Neu5Ac), the most abundant sialic acid form, and other common forms such as N-glycolylneuraminic acid (Neu5Gc), keto-deoxy-nonulosonic acid (KDN), and their naturally occurring derivatives, are frequently located on cell surface as the terminal monosialyl residues α2–3- or α2–6-linked to galactosides or 2-acetamino-2-deoxy-galactosides in biologically active glycoconjugates.2 Disialyl structures Siaα2–8Sia containing diverse sialic acid forms (Figure 1) have also been found as constituents of glycans in many glycoproteins and glycolipids including gangliosides which are sialylated glycosphingolipids that are presented on the outer leaflets of plasma membranes.1c
Figure 1.
Structures of common naturally occurring disialyl motifs in glycolipids and glycoproteins.
α2–8-Linked disialyl moiety Neu5Acα2–8Neu5Ac is a common structural unit of GD1c, b-series gangliosides (e.g. GD3, GD2, GD1b, GT1b, GQ1b, and GQ1bα), and of GT1a and GP1c belonging to aand c- series gangliosides respectively.3 The simplest member of this group is ganglioside GD3 which has been shown to be a human melanoma associated antigen.4 Disialyl structures containing other sialic acid forms have also been reported. For example, Neu5Acα2–8Neu5Gc has been found on gangliosides in mouse thymoma and thymocytes,5 cat and sheep erythrocytes,6 and bovine brain.7 Neu5Gcα2–8Neu5Gc has been found on gangliosides in mouse thymoma and thymocytes,5 human gastrointestinal adenocarcinoma, and gastric cancer cell MKN74.8 Neu5Gcα2–8Neu5Ac has been found on gangliosides in mouse5 and rabbit thymus.9 9OAc-Neu5Acα2–8Neu5Ac has been found on GD3 (9OAc-GD3) gangliosides in human melanoma,10 GD1b in bovine brain,11 GT3 in chicken and rat brain,12 and GT2 in Alaskan pollack brain13 and cod brain.14 9OAc-GD3 has been shown to be an important regulatory molecule involved in signal transduction, regulation of cell growth and differentiation, apoptosis, and inflammation, etc.15 Sulfated disialyl structure 8OSO3--Neu5Acα2–8Neu5Ac has been observed in ganglioside GD3 in bovine gastric mucosa16 and sea urchin sperm.17 Although most disialyl sequences are α2–3-linked to a galactose (Gal) moiety in gangliosides, a few Neu5Acα2–8Neu5Ac sequences have also been found to link to N-acetylgalactosamine (GalNAc) or glucose (Glc) through an α2–6-sialyl linkage.17
The α2–8-linked disialyl glycans have also been discovered in glycoproteins.18 For example, Siaα2–8Sia units have been found in both O-linked and N-linked polysialylglycoproteins from trout egg,19 vertebrates/embryonic brain,20 eel/rat brain,21 human tumor,22 fruit fly (Drosophila),23 cicada,24 and rainbow trout ovarian fluid.25 More specifically, Neu5Acα2–8Neu5Ac has been found in O-linked glycoproteins from bovine adrenal medulla26 and human erythrocyte glycophorin,27 as well as N-linked glycoproteins from umbilical cord erythrocyte Band 328 and ovarian fluid of rainbow trout.29 Neu5Gcα2–8Neu5Gc has been found in both O-linked and N-linked glycans in the proteins from bovine adrenal medulla,26 pig spleen,30 rat thymus,31 and recently in mouse serum.32 Although not existing in glycolipids, KDNα2–8KDN has been found in O-linked glycoproteins from rat kidney33 and various rat organs.34 Moreover, polysialic acids with Neu5Acα2–8Neu5Ac repeating units are the major components of capsular polysaccharides of group B Neisseria meningitidis, Escherichia coli K1, Moraxella nonliquifaciens, and Pasteurella haemolytica A2.35
Disialyl structures are believed to play important roles in numerous biological events.15,36 For example, Siglec-7, an inhibitory receptor expressed on natural killer (NK) cells, shows a significant preference for α2–8-linked disialyl ligands37 such as GD3 whose expression on the target cells can suppress NK cell-mediated cytolytic activity.38
Nevertheless, the low availability of pure disialyl glycans and glycoconjugates from natural sources make it difficult to elucidate their biological functions. On the other hand, chemical formation of Siaα2–8Sia-linkage is one of the most challenging tasks in chemical glycosylation due to the sterically hindered tertiary anomeric center, lack of a stereo-directing group adjacent to the anomeric position, the presence of an electron-withdrawing carboxyl group in sialyl donors, and the low reactivity of the C8 hydroxyl group caused by C1 carboxyl and/or the C5 acetamide group in the sialic acid of sialyl acceptors.39 Recently, the introduction of N,N-diacetyl,40 azido,41 N-trifluoroacetyl (N-TFA),42 N-Troc,43 NFmoc,43b,43c N-trichloroacetyl,43b,43c and N-phthalimido group44 at the C5 position in sialyl donors have been reported to exhibit improved donor reactivity towards sialylation. Some of these sialyl donors have been applied in the synthesis of α2–8-linked disialylated oligosaccharides in moderate yields.40b,42b,44 The 1,5-lactam derivative of sialic acids45 and 5-N,4-O-carbonyl protected oxazolidinone sialyl donor46 have also been developed for the synthesis of α2–8-linked disialosides. Despite the advance, current chemical synthesis of sialosides remains to be a time-consuming process and requires skillful expertise.
In comparison, sialyltransferase-catalyzed sialylation with intrinsic high regio- and stereoselectivity, as well as mild and environment-friendly reaction conditions, offers great advantages and is considered an attractive and a practical approach for the synthesis of sialosides including those containing disialyl motifs. Recent identification and cloning of a bi-functional bacterial sialyltransferase CstII from Campylobacter jejuni OH4384 that can catalyze the formation of both α2–8- and α2–3- sialyl linkages47 provide a unique catalyst for efficient synthesis of ganglioside oligosaccharides and their derivatives.48 Nevertheless, both chemical and enzymatic syntheses of α2–8-linked sialosides have been so far limited to Neu5Ac40b, 42b, 44-49 and some Neu5Gc50-containing structures. In order to understand the importance of variations of naturally existing sialic acid forms in α2–8-linked sialosides, herein we report a facile two-step multi-enzyme approach for preparative chemoenzymatic synthesis of α2–8-linked disialyl oligosaccharides containing Neu5Ac, Neu5Gc, KDN, and their derivatives. The success of this method is demonstrated by the production of a series of GD3 ganglioside oligosaccharides and other disialyl glycans containing natural and non-natural sialic acids.
Results and Discussion
Two-step multi-enzyme approach for the synthesis of disialyl oligosaccharides
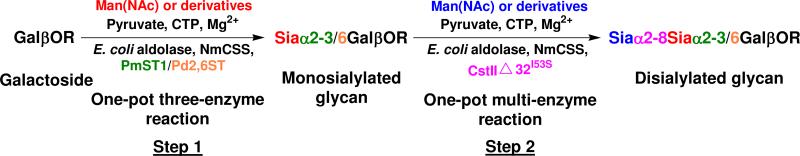
As shown in Figure 2, we used a two-step process to produce disialyl oligosaccharides. In the first step, α2–3- or α2–6-linked monosialyl oligosaccharides containing different sialic acid forms were prepared using the one-pot three-enzyme method and purified as described previously.51 They were then used in the second step as acceptors for the α2–8-sialyltransferase activity of CstIIΔ32I53S for the synthesis of Siaα2–8Siaα2–3/6Gal-terminated disialyl oligosaccharides using a one-pot multi-enzyme approach containing two (with a CMP-sialic acid synthetase and an α2–8-sialyltransferase) or three enzymes (with an additional sialic acid aldolase compared to the two-enzyme approach). CstIIΔ32I53S is a recombinant truncated form of CstII with a single amino acid mutation (I53S mutation was introduced to enhance the α2–8-sialyltransferase activity and to stabilize the enzyme47b) compared to CstII from C. jejuni OH4384.47a It was cloned from a synthetic gene whose codons were optimized for an E. coli expression system.47c Although CstIIΔ32I53S is multifunctional and can catalyze the formation of both Siaα2–3Gal and Siaα2– 8Sia linkages,47,48 its α2–3-sialyltransferase activity is lower than its α2–8-sialyltransferase activity. Therefore, CstIIΔ32I53S was used only for its α2–8-sialyltransferase activity in the two-step process for the synthesis of Siaα2–8Siaα2–3Gal terminated disialyl oligosaccharides to provide a higher efficiency and a better control over the sialylation process.
Figure 2.
Two-step multi-enzyme chemoenzymatic synthesis of diasialyl oligosaccharides containing different sialic acid forms and various sialyl linkages. Enzymes: E. coli aldolase, Escherichia coli K12 sialic acid aldolase; NmCSS, Neisseria meningitidis CMP-sialic acid synthetase; PmST1, Pasteurella multocida sialyltransferase for the formation of α2–3-linked sialosides; Pd2,6ST, Photobacterium damsela α2–6-sialyltransferase for the formation of α2–6-linked sialosides; CstII, Campylobacter jejuni sialyltransferase for the formation of α2–8-linked sialosides. Compounds: Man, mannose; ManNAc, N-acetylmannosamine; CTP, cytidine 5′-triphosphate; Sia, sialic acid.
The preparation of α2–3- or α2–6-linked monosialylated oligosaccharides was carried out using an efficient one-pot three-enzyme chemoenzymatic approach52 developed in our lab. In this system, N-acetylmannosamine (ManNAc), mannose (Man), or their derivatives obtained by chemical or enzymatic modification, was coupled with pyruvate to form sialic acid derivatives by a sialic acid aldolasecatalyzed reaction. The sialic acid derivatives formed were then activated by a CMP-sialic acid synthetase and transferred to a suitable sialyltransferase acceptor for the formation of sialosides. Depending on the specificity of the sialyltransferase, α2–3- or α2–6-linked sialosides could be produced efficiently in a single pot without the purification of intermediates. Sialic acid aldolases from E. coli K1253 and Pasteurella multocida,54 CMP-sialic acid synthetase from N. meningitidis (NmCSS),53 a multifunctional sialyltransferase from Pasteurella multocida (PmST1) for the formation of α2–3-linked sialosides,51a and a sialyltransferase from Photobacterium damsela (Pd2,6ST) for the formation of α2–6-linked sialosides,51b were shown to be excellent catalysts for the synthesis of monosialylated glycans because they were able to be expressed in E. coli in large amounts with high activity and promiscuous substrate specificity.
With α2–3- and α2–6-linked monosialylated oligosaccharides in hands, α2–8-linked disialyl oligosaccharides were synthesized in the second step using the one-pot multi-enzyme process with CstIIΔ32I53S as the α2–8-sialyltransferase and NmCSS with or without E. coli K12 sialic acid aldolase.47c The application of the method in the synthesis of the targeted GD3-type disialyl glycans was explored for two major groups: one group contained a penultimate α2–3-linked Neu5Ac with different terminal α2–8-linked sialic acid forms (Siaα2–8Neu5Acα2–3LacβProN3) and the other contained a terminal α2–8-linked Neu5Ac with different α2–3-linked penultimate sialic acid forms (Neu5Acα2–8Siaα2–3GalβOR). In addition, the synthesis of GD3-type disialyl glycans (Neu5Gc/KDNα2–8Neu5Gc/KDNα2–3LacβProN3) containing the combination of two other common sialic acid forms such as Neu5Gc and KDN was also carried out. The synthesis of Sia2–8Siaα2–6GalβOR-type disialyl glycans was investigated for the compounds containing a terminal α2–8-linked Neu5Ac with different α2–6-linked penultimate sialic acid forms (Neu5Acα2–8Siaα2–6GalβOR) using a one-pot two-enzyme system.
Preparation of GD3-type disialyl oligosaccharides Siaα2–8Neu5Acα2–3LacβProN3 containing a penultimate α2–3-linked Neu5Ac and different terminal α2–8-linked sialic acid forms
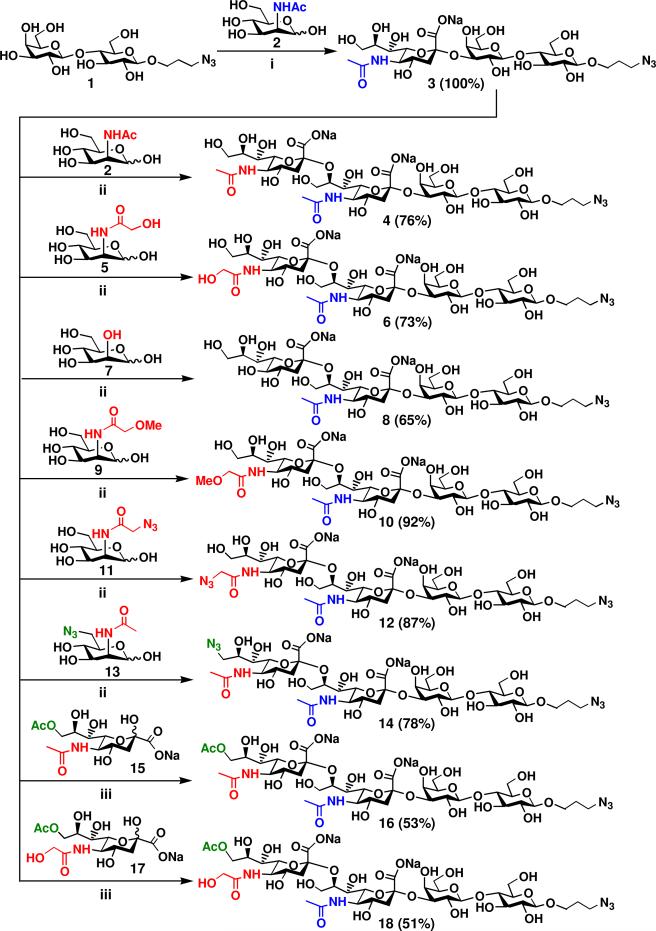
GM3 oligosaccharide with a propyl azide aglycon (Neu5Acα2–3Galβ1–4GlcβProN3 or Neu5Acα2–3LacβProN3) 3 was readily obtained in a quantitative yield by incubating 3-azidopropyl β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (LacβProN3) 1 with N-acetylmannosamine (ManNAc) 2 in the one-pot three-enzyme system as described previously.51a The product Neu5Acα2–3LacβProN3 3 was then used as an acceptor for the α2–8-sialyltransferase activity of CstIIΔ32I53S in the one-pot multiple-enzyme synthesis of GD3-type disialyl oligosaccharides containing different terminal sialic acid forms. As shown in Figure 3, CstIIΔ32I53S has promiscuous donor substrate specificity and can catalyze the transfer of different sialic acids from CMP-sialic acid derivatives synthesized by NmCSS with or without E. coli K12 sialic acid aldolase to form GD3 oligosaccharides 4, 6, 8, 10, 12, 14, 16, and 18 with different terminal sialic acid forms in good to excellent yields (51–92%).
Figure 3.
Synthesis of disialyl GD3 oligosaccharides Siaα2–8Neu5Acα2–3LacβProN3 containing a penultimate α2–3-linked Neu5Ac and different terminal α2–8-linked sialic acid forms. Reagents and conditions: (i) Pyruvate, CTP, Mg2+, Tris-HCl buffer (pH 8.5), E. coli K12 sialic acid aldolase, NmCSS, and PmST1; (ii) Pyruvate, CTP, Mg2+, Tris-HCl buffer (pH 8.5), E. coli K12 sialic acid aldolase, NmCSS, and CstIIΔ32I53S; (iii) CTP, Mg2+, Tris-HCl buffer (pH 7.5), NmCSS, and CstIIΔ32I53S.
The synthesis of GD3 oligosaccharides 4, 6, 8, 10, 12, and 14 was carried out in the one-pot three-enzyme system at pH 8.5. We found that the use of 1.2-fold excess amount of sialic acid precursors was optimum to prevent the multiple α2–8-sialylation by CstIIΔ32I53S.47c, 48 Under these conditions, disialyl GD3 oligosaccharides Neu5Acα2–8Neu5Acα2–3LacβProN3 4 and Neu5Gcα2–8Neu5Acα2–3LacβProN3 6 were obtained in 76% and 73% yields, respectively, from ManNAc 2 and N-glycolyl mannosamine (ManNGc) 5 as sialic acid precursors. The yield (65%) for the formation of KDNα2–8Neu5Acα2–3LacβProN3 8 from mannose 7 was lower due to the formation of by-product with multiple α2–8-linked sialic acids. Interestingly, the synthesis of Neu5GcMeα2–8Neu5Acα2–3LacβProN3 10 containing a terminal modified Neu5Gc with a methyl group at the C5-OH was achieved in high efficiency with a 92% yield from N-methylglycolyl-mannose (ManNGcMe) 9. This may be due to the prevention of additional α2–8-sialylation by the extra methyl group at the C5-OH in the terminal Neu5Gc in the disialyl product 10. Non-natural GD3 oligosaccharides Neu5AcN3α2–8Neu5Acα2–3LacβProN3 12 and Neu5Ac9N3α2–8Neu5Acα2–3LacβProN3 14 containing an azido group at the C-5 or C-9 position of the terminal Neu5Ac were also obtained in good yields (87% and 78%, respectively) from C2- or C6-modified ManNAc derivatives N-azidoacetyl mannoseamine (ManNAz) 11 and N-acetyl-9-azido-mannosamine (9N3ManNAc) 13, respectively.
Gangliosides, including GD3, containing a terminal 9-O-acetyl modified Neu5Ac are common.10,11,55 The biological functions of 9-O-acetylated GD3 are believed to be distinct from its non-acetylated counterpart. For example, it has been found as a marker for neural differentiation and malignant transformation56 and has been suggested to protect tumor cells from apoptosis.57 Both 9-O-acetylated Neu5Ac and Neu5Gc are readily available from their corresponding non-O-acetylated forms by a regioselective chemical acetylation.58 Briefly, treatment of Neu5Ac or Neu5Gc with trimethyl orthoacetate in anhydrous DMSO in the presence of a catalytic amount of p-TsOH gave 9-O-acetyl-N-acetyl neuraminic acid (Neu5,9Ac2) 15 or 9-O-acetyl-N-glycolyl neuraminic acid (Neu5Gc9Ac) 17 in excellent yields (over 90%). GD3 oligosaccharides Neu5,9Ac2α2–8Neu5Acα2–3LacβProN3 16 and Neu5Gc9Acα2–8Neu5Acα2–3LacβProN3 18 were obtained in a one-pot two-enzyme reaction containing NmCSS and CstIIΔ32I53S in 53% and 51% yields from 15 and 17, respectively. A Tris-HCl buffer solution of pH 7.5 was used to prevent the de-acetylation under basic solutions with pH higher than 7.5.
The structures of all purified GD3 oligosaccharide products were confirmed by nuclear magnetic resonance (NMR) spectroscopy and high-resolution mass spectrometry (HRMS). Compairing the 13C NMR spectra of GD3 oligosaccharide Neu5Acα2–8Neu5Acα2–3LacβProN3 4 and GM3 oligosaccharide Neu5Acα2–3LacβProN3 3 indicated a downfield chemical shift of 6.39 ppm for the C-8 of the internal Neu5Ac in Neu5Acα2–8Neu5Acα2–3LacβProN3 4 (78.28 ppm) compared to that for the C-8 of Neu5Ac (71.89 ppm) in Neu5Acα2–3LacβProN3 3. These data confirmed the formation of an α2–8-sialyl linkage by CstIIΔ32I53S-catalyzed sialylation when Neu5Acα2–3LacβProN3 3 was used as an acceptor for CstIIΔ32I53S. Among the GD3 oligosaccharides (4, 6, 8, 10, 12, and 14) synthesized here, only the preparation of Neu5Acα2–8Neu5Ac terminated disialyl oligosaccharides using a similar CstII-catalyzed sialylation of Neu5Ac-containing GM3 oligosaccharides has been reported.47c, 48, 49e–49i
Preparation of GD3-type disialyl oligosaccharides Neu5Acα2–8Siaα2–3GalβOR containing terminalα2–8-linked Neu5Ac and different penultimate α2–3-linked sialic acid forms
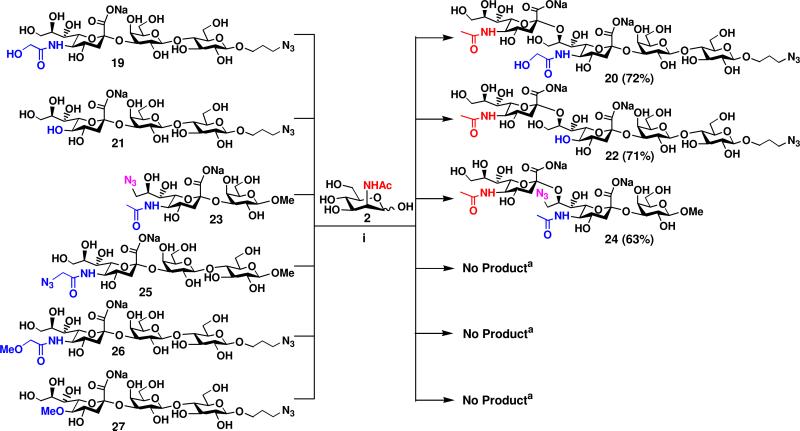
Small scale one-pot three-enzyme reactions were performed first and analyzed by thin-layer chromatography (TLC) to study the acceptor specificity of the α2–8-sialyltransferase (or GD3 synthase) activity of CstIIΔ32I53S. Preparative-scale syntheses were then carried out for suitable acceptors. As summarized in Figure 4, CstIIΔ32I53S exhibited good activity towards monosialylated oligosaccharides 19 or 21 which posses a terminal Neu5Gc or KDN. GD3 oligosaccharides Neu5Acα2–8Neu5Gcα2–3LacβProN3 20 and Neu5Acα2–8KDNα2–3LacβProN3 22 were obtained in 72% and 71% yields, respectively, in the presence of E. coli K12 sialic acid aldolases, NmCSS, and CstIIΔ32I53S using ManNAc 2 as the sialic acid precursor. The yields are comparable to that (76%) for the synthesis of Neu5Acα2–8Neu5Acα2–3LacβProN3 4 (Figure 3) from Neu5Acα2–3LacβProN3 3 (which contains the most abundant sialic acid form Neu5Ac) as an acceptor. Interestingly, substituting the C9-hydroxyl group on the terminal Neu5Ac in sialoside 23 with an azido did not block the α2–8-sialylation reaction catalyzed by CstIIΔ32I53S. Preparative synthesis of disialyl oligosaccharide 24 was achieved in 63% yield. Quite surprisingly, further modification on the C-5 of the terminal Neu5Ac, Neu5Gc, and KDN in α2–3-linked monosialylated oligosaccharides Neu5AcN3α2–3LacOMe 25, Neu5GcMeα2–3LacβProN3 26, and KDN5Meα2–3LacβProN3 27 with either a methyl or an azido group totally blocked the α2–8-sialylation reaction catalyzed by CstIIΔ32I53S. Taken together, these results indicate that the α2–8-sialyltransferase activity of CstIIΔ32I53S can tolerate a limited number of groups (N-acetyl, N-glycol, and hydroxyl) at C-5 and modifications on the C-9 of the terminal sialic acid residue in α2–3-linked monosialylated oligosaccharides as acceptor substrates.
Figure 4.
Synthesis of disialyl GD3 oligosaccharides Neu5Acα2–8Siaα2–3GalβOR containing a terminal α2–8-linked Neu5Ac and different penultimate α2–3-linked sialic acid forms. Reagents and conditions: (i) Pyruvate, CTP, Mg2+, Tris-HCl buffer (pH 8.5), E. coli K12 sialic acid aldolase, NmCSS, and CstIIΔ32I53S. aDetermined by small-scale reaction using TLC analysis.
Among the α2–3-monosialylated oligosaccharides used here (19, 21, 23, 25, 26, and 27) as the acceptors for the α2–8-sialyltransferase activity of CstIIΔ32I53S, the synthesis of 19, 21, 25, and 26 has been reported previously.51a The synthesis of two new α2–3-linked monosialylated oligosaccharides 23 and 27 were carried out in a one-pot three-enzyme system containing an E. coli sialic acid aldolase, NmCSS, and PmST1 similar to that described previously for the synthesis of other α2–3-linked sialosides.51a Compound 23 was obtained from 6-azido-6-deoxy-N-acetyl-d-mannosamine (6-N3-ManNAc) as the precursor of a sialic acid derivative and methyl β-d-galactopyranoside as a sialyltransferase acceptor. Compound 27 was obtained from 2-O-methyl-d-mannose as the precursor of a sialic acid derivative and azidopropyl β-d-lactoside as a sialyltransferase acceptor. The preparation of these starting materials for the one-pot three-enzyme synthesis of α2–3-linked sialosides has been reported previously.51
Preparation of GD3-type disialyl oligosaccharides Neu5Gc/KDNα2–8Neu5Gc/KDNα2–3LacβProN3 containing the combination of Neu5Gc and KDN
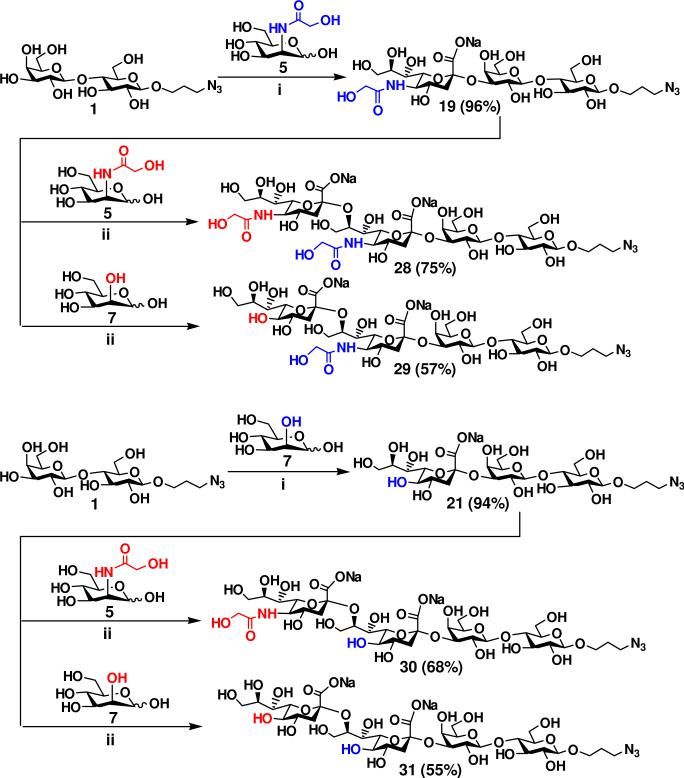
GD3-type disialyl oligosaccharides containing naturally occurring Neu5Gc and KDN sialic acid forms including Neu5Gcα2–8Neu5Gc, KDNα2–8KDN, and hybrid KDNα2–8Neu5Gc and Neu5Gcα2–8KDN units were also synthesized. As shown in Figure 5, monosialylated oligosaccharides Neu5Gcα2–3LacβProN3 19 (96%) and KDNα2–3LacβProN3 21 (94%) were synthesized in the step 1 from LacβProN3 1 and ManNGc 5 or mannose 7 in the presence of E. coli aldolase, NmCSS, and PmST1. Using Neu5Gcα2–3LacβProN3 19 as the acceptor for the α2–8-activity of CstIIΔ32I53S, disialyl oligosaccharides Neu5Gcα2–8Neu5Gcα2–3LacβProN3 28 and KDNα2–8Neu5Gcα2–3LacβProN3 29 were prepared from donor substrates 5 and 7 in the one-pot three-enzyme system in 75% and 57% yields, respectively. Similarly, sialylation of KDNα2–3LacβProN3 21 with the donor substrate 5 and 7 in the presence of E. coli K12 sialic acid aldolase, NmCSS, and CstIIΔ32I53S produced Neu5Gcα2–8KDNα2–3LacβProN3 30 (68%) and KDNα2–8KDNα2–3LacβProN3 31 (55%) in comparable yields. Again, the lower yields (55-57%) for the formation of disialyl oligosaccharides containing a terminal KDN 29 and 31 compared to those (68-75%) for the formation of Neu5Gc-terminated disialyl oligosaccharides 28 and 30 were due to the formation of by-product with multiple α2–8-linked sialic acids for KDN-terminated glycans.
Figure 5.
Enzymatic preparation of GD3-type disialyl oligosaccharides Neu5Gc/KDNα2–8Neu5Gc/KDNα2–3LacβProN3 containing the combination of Neu5Gc and KDN. Reagents and conditions: (i) Pyruvate, CTP, Mg2+, Tris-HCl buffer (pH 8.5), E. coli K12 sialic acid aldolase, NmCSS, and PmST1; (ii) Pyruvate, CTP, Mg2+, Tris-HCl buffer (pH 8.5), E. coli K12 sialic acid aldolase, NmCSS, and CstIIΔ32I53S.
Preparation of Neu5Ac2–8Siaα2–6GalβOR-type disialyl oligosaccharides containing a terminal α2–8-linked Neu5Ac and different penultimate α2–-6-linked sialic acid forms
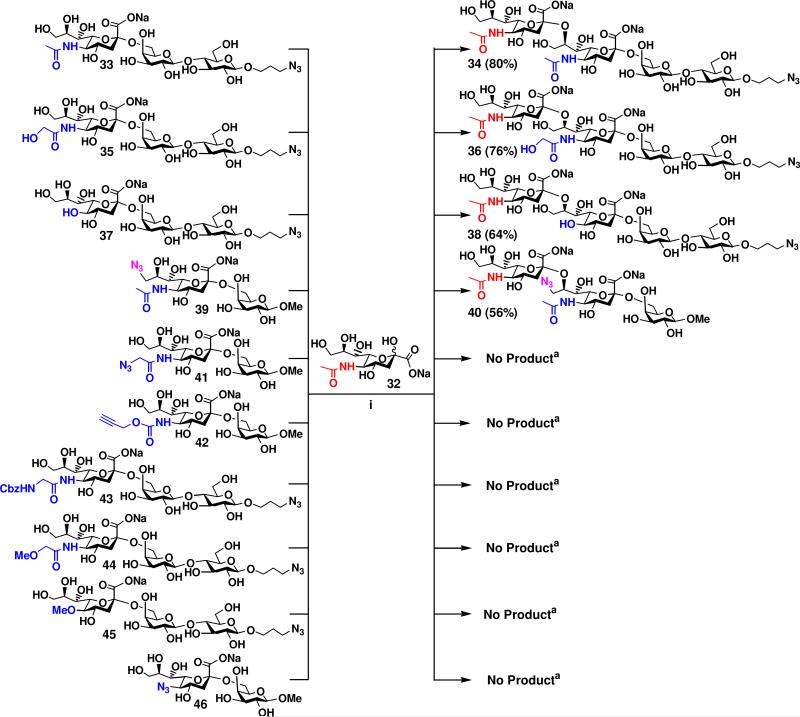
Acceptor specificity of the α2–8-sialyltransferase activity of CstIIΔ32I53S was also explored in a one-pot two-enzyme system with a panel of α2–6-linked monosialylated oligosaccharides containing natural sialic acid forms (Neu5Ac, Neu5Gc, and KDN) and non-natural sialic acids with various modifications at C-9 or C-5. To do this, α2–6-linked monosialylated oligosaccharides Siaα2–6GalβOR were synthesized from GalβOR using Photobacterium damsela α2–6-sialyltransferase (Pd2,6ST) in the one-pot three-enzyme system as described previously.51b Evaluation of Siaα2–6GalβOR as potential acceptors for the α2–8-sialyltransferase activity of CstIIΔ32I53S was carried out in small-scale one-pot two-enzyme reactions from Neu5Ac the reactions and analyzed by thin-layer chromatography (TLC). Similar to human polysialyltransferases ST8SiaII (STX) and ST8SiaIV (PST) reported previously,59 CstII exhibited acceptor specificity towards a list of α2–6-sialosides. α2–6-Linked sialyl lactosides Neu5Acα2–6LacβProN3 33, Neu5Gcα2–6LacβProN3 35, and KDNα2–6LacβProN3 37 containing naturally occurring sialic acid forms including Neu5Ac, Neu5Gc, and KDN served as good acceptor substrates for the α2–8-sialyltransferase activity of CstIIΔ32I53S. As shown in Figure 6, preparative-scale sialylation of 33, 35, and 37 from Neu5Ac 32 as a donor precursor for CstIIΔ32I53S successfully produced the disialylated products Neu5Acα2–8Neu5Acα2–6LacβProN3 34, Neu5Acα2–8Neu5cα2–6LacβProN3 36,and Neu5Acα2–8KDNα2–6LacβProN3 38 in 80%, 76%, and 64% yield, respectively. Similar to its α2–3-monosialylated counterpart Neu5Ac9N3α2–3GalβOMe 23, α2–6-linked sialoside Neu5Ac9N3α2–3GalβOMe 39 containing an azido substitution of the C9-OH of the terminal Neu5Ac was also a suitable acceptor for CstIIΔ32I53S. Sialylation of 39 with Neu5Ac 32 in the one-pot two-enzyme system in preparative scale produced disialyl product Neu5Acα2–8Neu5Ac9N3α2–6GalβOMe 40 in 56% yield. The α2–6-linked monosialylated oligosaccharides containing various substitutions at C-5 of Neu5Ac, Neu5Gc, or KDN, including Neu5AcN3α2–6GalOMe 41, Neu5NPgα2–6LacβProN3 42, Neu5AcCbzα2–6LacβProN3 43, Neu5GcMeα2–6LacβProN3 44, KDN5Meα2–6LacβProN3 45, and KDN5N3α2–6GalOMe 46 did not serve as acceptor substrates for the α2–8-sialyltransferase activity of CstIIΔ32I53S. These data, together with those obtained from the acceptor specificity studies using α2–3-linked monosialylated oligosaccharides, indicate the importance of the C-5 groups on the terminal sialic acid residues, instead of the sialyl linkages, in defining the acceptor specificity of the α2–8-sialyltransferase activity of CstIIΔ32I53S.
Figure 6.
Synthesis of Neu5Ac2–8Siaα2–6GalβOR-type disialyl oligosaccharides containing a terminal α2–8-linked Neu5Ac and different penultimate α2–6-linked sialic acid forms. Reagents and conditions: (i) CTP, Mg2+, Tris-HCl buffer (pH 8.5), NmCSS, and CstIIΔ32I53S. aDetermined by small-scale reactions using TLC analysis.
Conclusions
In conclusion, we have developed an efficient two-step multi-enzyme approach for the synthesis of a series of GD3 ganglioside oligosaccharides and other disialyl glycans containing natural and non-natural sialic acids. The α2–8-sialyltransferase activity of a recombinant multi-functional CstIIΔ32I53S has promiscuous donor substrate specificity and can tolerate various substitutions at C-5 or C-9 of sialic acid residues in the donor. In comparison, the α2–8-sialyltransferase activity of CstIIΔ32I53S has relatively restricted acceptor substrate specificity. While both α2–3- and α2–6-linked monosialyl oligosaccharides are potential acceptors for CstIIΔ32I53S, the terminal sialic acid residues in the acceptable monosialyl oligosaccharide acceptors are limited to Neu5Ac, Neu5Gc, KDN, and some of their C-9 modified forms. Additional modifications at the C-5 of the terminal sialic acid residues in the monosialyl oligosaccharides prevent them to be usable acceptors by the α2–8-sialyltransferase activity of CstIIΔ32I53S. The disialyl oligosaccharides obtained in this work are valuable probes to study the biological importance of naturally occurring sialic acid modifications in disialyl structures in nature.
Experimental Section
Chemicals were purchased and used without further purification. 1H NMR and 13C NMR spectra were recorded on Mercury-300, Varian Inova-400, or Varian VNMRS 600 MHz spectrometer. High resolution electrospray ionization (ESI) mass spectra were obtained at the Mass Spectrometry Facility in the University of California at Davis. Optical rotation was recorded on an Autopol IV Automatic Polarimeter at 589 nm wavelength. Silica gel 60 Å (40–63 m, Sorbent technologies) was used for flash chromatography. Analytical thin-layer chromatography was performed on silica gel plates 60 GF254 (Sorbent technologies) using p-anisaldehyde sugar stain for detection. Gel filtration chromatography was performed using a column (100 cm × 2.5 cm) packed with BioGel P-2 Fine resins (Bio-Rad, Hercules, CA).
Enzymatic Synthesis of Sialosides
Monosialosides
The synthesis of α2–3 and α2–6-linked monosialyl oligosaccharides 3, 19, 21, 25, 26, 33, 35, 37, 39, 41-46 has been reported previously.51 Monosialylated oligosaccharides 23 and 27 were prepared in a one-pot three-enzyme system containing an E. coli sialic acid aldolase, NmCSS, and PmST1 as described previously.51a
Methyl O-(5-acetamido-9-azido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranoside (Neu5Ac9N3α2–3GalβOMe, 23)
Yield, 88%; white foam. 1H NMR (600 MHz, D2O) δ 4.36 (d, 1H, J = 7.8 Hz, H-1), 4.06 (dd, 1H, J = 10.2 and 3.0 Hz), 3.98 (m, 1H), 3.93–3.46 (m, 11H), 3.56 (s, 3H, OMe), 2.74 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq’), 2.03 (s, 3H), 1.78 (t, 1H, J = 12.0 Hz, H-3ax’); 13C NMR (125 MHz, D2O) δ 172.48, 171.32, 101.06, 97.31, 73.36, 72.42, 70.17, 67.91, 66.63, 65.80, 65.01, 58.47, 56.89, 54.59, 50.55, 49.20, 37.20, 19.63. HRMS (ESI) m/z calculated for C18H29N4Na2O13 (M+Na), 532.1629, found 532.1637.
3-Azidopropyl O-(5-O-methyl-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-D-glucopyranoside (KDN5Meα2–3LacβProN3, 27)
Yield, 70%; white form. 1H NMR (600 MHz, D2O) δ: 4.54 (1 H, d, J = 7.8 Hz), 4.51 (1 H, d, J = 8.4 Hz), 4.11–3.59 (19 H, m), 3.59 (3H, s, OMe), 3.50 (2H, t, J = 7.20 Hz), 3.37–3.32 (2 H, m), 2.71 (1 H, dd, J = 12.6 and 4.2 Hz, H-3eq’), 1.94 (2H, m), 1.80 (1H, t, J = 12.0 Hz, H-3ax’); 13C NMR (150 MHz, D2O) δ: 174.17, 102.84, 102.33, 99.86, 79.88, 78.46, 75.63, 75.34, 74.96, 74.53, 73.22, 73.00, 72.35, 70.12, 69.54, 68.15, 67.56, 67.55, 62.82, 61.20, 60.28, 60.24, 48.08, 39.53, 28.42. HRMS (ESI) m/z calculated for C25H42N3Na2O19 (M+Na), 711.2310, found 711.2318.
General procedures for one-pot multi-enzyme preparative synthesis ofα2–8-linked sialosides using CstIIΔ32I53S
A monosialylated oligosaccharide as an acceptor for the α2–8-sialyltransferase activity of CstIIΔ32I53S (2.5–3.0 mg), a sialic acid precursor (mannose, ManNAc, or their derivatives, 1.2 equiv.), sodium pyruvate (7.5 equiv.), and CTP (1.5 equiv.) were dissolved in H2O. Stock solutions of Tris-HCl buffer (1 M, pH 8.5, 1 mL) and MgCl2 (0.5 M, 0.4 mL) were added. After the addition of appropriate amounts of a recombinant E. coli K12 sialic acid aldolase, an N. meningitidis CMP-sialic acid synthetase, and CstIIΔ32I53S, H2O was added to bring the volume of the reaction mixture to 10 mL. The reaction was carried out by incubating the solution in an incubator shaker at 37 °C for 2 h (or at room temperature for overnight) with agitation at 140 rpm. The product formation was monitored by TLC developed with EtOAc:MeOH:H2O:HOAc = 5:3:1.5:0.2 (by volume) and stained with p-anisaldehyde sugar stain. When an optimal yield was achieved, the reaction was quenched by adding the same volume (10 mL) of ice-cold EtOH and incubation at 4 °C for 30 min. The mixture was then centrifuged and the precipitates were removed. The supernatant was concentrated, passed through a BioGel P-2 gel filtration column, and eluted with water to obtain sialoside mixtures. Silica gel flash column was then used to obtain pure disalylated product.
3-Azidopropyl O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (Neu5Acα2–8Neu5Acα2–3LacβProN3, 4)
Yield, 76%; white foam. [α]D22 = -0.23° (c 2.15, H2O); 1H NMR (600 MHz, D2O) δ 4.53 (d, 1H, J = 8.4 Hz, Glc H-1), 4.50 (d, 1H, J = 8.4 Hz, Gal H-1), 4.19–3.99 (m, 6H), 3.94–3.55 (m, 21H), 3.47 (t, 2H, J = 6.6 Hz), 3.33 (t, 1H, J = 8.4 Hz), 2.80 (dd, 1H, J = 12.0 and 4.8 Hz, Neu5Ac H-3eq”), 2.67 (dd, 1H, J = 12.6 and 4.2 Hz, Neu5Ac H-3eq”), 2.08 (s, 3H), 2.05 (s, 3H), 1.93 (m, 2H), 1.77 (t, 1H, J = 12.6 Hz, Neu5Ac H-3ax”), 1.75 (t, 1H, J = 12.0 Hz, Neu5Ac H-3ax”); 13C NMR (125 MHz, D2O) δ 175.18 (2C), 173.87, 173.46, 102.86, 102.33, 100.74, 100.52, 78.28 (Neu5Ac C-8), 78.22, 75.59, 75.32, 74.98, 74.48, 74.25, 73.02, 72.81, 71.96, 69.49, 69.46, 68.64, 68.36, 68.13, 67.89, 67.55, 62.78, 61.73, 61.26, 60.20, 52.44, 51.93, 48.06, 40.68, 39.63, 28.41, 22.48, 22.22. HRMS (ESI) m/z calculated for C37H60N5O27 (M-2Na+H), 1006.3476, found 1006.3476.
3-Azidopropyl O-(5-glycolylamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside (Neu5Gcα2–8Neu5Acα2–3LacβProN3, 6)
Yield, 73%; white foam. [α]D22 = -0.95° (c 1.37, H2O); 1H NMR (600 MHz, D2O) δ 4.49 (d, 1H, J = 8.4 Hz, Glc H-1), 4.45 (d, 1H, J = 7.8 Hz, Gal H-1), 4.18–4.04 (m, 5H), 3.99–3.52 (m, 21H), 3.43 (t, 2H, J = 6.6 Hz), 3.28 (t, 1H, J = 8.4 Hz), 2.77 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.61 (dd, 1H, J = 12.0 and 3.6 Hz, H-3eq”), 2.04 (s, 3H), 1.88 (m, 2H), 1.74 (t, 1H, J = 12.6 Hz, H-3ax”), 1.72 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.86 (2C), 175.15, 173.44, 102.81, 102.28, 100.70, 100.55, 78.21 (Neu5Ac C-8), 78.10, 75.50, 75.24, 74.91, 74.42, 74.26, 72.98, 72.44, 72.00, 69.43, 69.40, 68.31, 68.23, 68.07, 67.95, 67.50, 62.68, 61.70, 61.22, 61.11, 60.14, 52.36, 51.57, 48.00, 40.66, 39.46, 28.37, 22.44. HRMS (ESI) m/z calculated for C37H60N5O28 (M-2Na+H), 1022.3425, found 1022.3433.
3-Azidopropyl O-(3-deoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (KDNα2–8Neu5Acα2–3LacβProN3, 8)
Yield, 65%; white foam. [α]D22 = -10.0° (c 1.64, H2O); 1H NMR (600 MHz, D2O) δ 4.50 (d, 1H, J = 7.8 Hz, Glc H-1), 4.46 (d, 1H, J = 7.8 Hz, Gal H-1), 4.15–4.07 (m, 3H), 4.02–3.49 (m, 21H), 3.44 (t, 2H, J = 6.6 Hz), 3.29 (t, 1H, J = 8.4 Hz), 2.71 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.61 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.04 (s, 3H), 1.89 (m, 2H), 1.76 (t, 1H, J = 12.6 Hz, H-3ax”), 1.67 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.16 (2C), 174.00, 173.57, 102.80, 102.27, 100.72, 100.62, 78.24 (Neu5Ac C-8), 77.97, 75.48, 75.20, 74.90, 74.42, 74.31, 73.73, 72.96, 72.24, 70.52, 69.88, 69.44, 69.35, 68.09, 67.93, 67.49, 62.76, 61.67, 61.21, 60.15, 52.34, 47.99, 40.19, 39.31, 28.36, 22.42. HRMS (ESI) m/z calculated for C35H57N4O27 (M-2Na+H), 965.3210, found 965.3214.
3-Azidopropyl O-(5-methoxyacetamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside (Neu5GcMeα2–8Neu5Acα2–3LacβProN3, 10)
Yield, 92%; white foam. [α]D22 = -0.28° (c 3.2, H2O); 1H NMR (600 MHz, D2O) δ 4.49 (d, 1H, J = 8.4 Hz, Glc H-1), 4.45 (d, 1H, J = 7.8 Hz, Gal H-1), 4.15–4.10 (m, 2H), 4.06–4.01 (m, 3H), 3.98-3.52 (m, 24H), 3.43 (t, 2H, J = 7.2 Hz), 3.39 (s, 3H), 3.28 (t, 1H, J = 8.4 Hz), 2.75 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.64 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.04 (s, 3H), 1.88 (m, 2H), 1.73 (t, 1H, J = 12.0 Hz, H-3ax”), 1.71 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.09 (2C), 173.65, 173.56, 173.54, 102.83, 102.29, 100.70, 100.34, 78.29 (Neu5Ac C-8), 78.16, 75.55, 75.32, 74.92, 74.39, 74.14, 72.99, 72.48, 72.00, 71.03 (OCH3), 69.47, 69.40, 68.34, 68.25, 68.03, 67.60, 67.50, 62.70, 61.72, 61.23, 60.64, 59.17, 52.39, 51.55, 48.01, 40.69, 39.80, 28.38, 22.51. HRMS (ESI) m/z calculated for C38H62N5O28 (M-2Na+H), 1036.3581, found 1036.3579.
3-Azidopropyl O-(5-azidoacetamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside (Neu5AcN3α2–8Neu5Acα2–3LacβProN3, 12)
Yield, 87%; white foam. [α]D22 = +1.37° (c 2.05, H2O); H NMR (600 MHz, D2O) δ 4.49 (d, 1H, J = 7.8 Hz, Glc H-1), 4.46 (d, 1H, J = 7.8 Hz, Gal H-1), 4.16–4.05 (m, 3H), 4.04 (s, 2H), 3.99–3.52 (m, 24H), 3.44 (t, 2H, J = 6.6 Hz), 3.29 (t, 1H, J = 9.0 Hz), 2.76 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.65 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.04 (s, 3H), 1.89 (m, 2H), 1.73 (t, 1H, J = 12.0 Hz, H-3ax”), 1.71 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.11, 173.65, 173.51, 171.27, 102.82, 102.29, 100.68, 100.34, 78.32 (Neu5Ac C-8), 78.15, 75.57, 75.34, 74.93, 74.39, 74.15, 72.99, 72.45, 71.99, 69.52, 69.41, 68.44, 68.21, 68.04, 67.60, 67.50, 62.70, 61.71, 61.23, 60.14, 52.40, 52.04, 51.97, 48.01, 40.62, 39.82, 28.38, 22.50. HRMS (ESI) m/z calculated for C37H59N8O27 (M-2Na+H), 1047.3490, found 1047.3486.
3-Azidopropyl O-(5-acetamido-9-azido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside(Neu5Ac9N3α2–8Neu5Acα2–3LacβProN3, 14)
Yield, 78%; white foam. [α]D22 = +5.11° (c 0.92, H2O); 1H NMR (600 MHz, D2O) δ 4.48 (d, 1H, J = 7.8 Hz, Glc H-1), 4.44 (d, 1H, J = 7.8 Hz, Gal H-1), 4.11 (dd, 1H, J = 12.0 and 3.0 Hz), 4.07-3.94 (m, 6H), 3.81–3.53 (m, 19H), ), 4.47 (dd, 1H, J = 13.2 and 5.4 Hz), 3.42 (t, 2H, J = 6.6 Hz), 3.27 (t, 1H, J = 9.0 Hz), 2.74 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.60 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.02 (s, 3H), 1.99 (s, 3H), 1.87 (m, 2H), 1.72 (t, 1H, J = 12.0 Hz, H-3ax”), 1.69 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.11, 175.06, 173.70, 173.30, 102.79, 102.27, 100.95, 100.39, 78.18 (Neu5Ac C-8), 78.14, 75.52, 75.26, 74.91, 74.41, 73.97, 72.97, 72.59, 72.58, 70.50, 69.40, 69.21, 68.78, 68.59, 68.05, 67.87, 61.56, 61.22, 60.13, 53.09, 52.37, 51.87, 47.99, 40.61, 39.62, 28.36, 22.43, 22.19. HRMS (ESI) m/z calculated for C37H59N8O26 (M-2Na+H), 1031.3540, found 1031.3552.
3-Azidopropyl O-(5-acetamido-9-O-acetyl-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside (Neu5,9Ac2α2–8Neu5Acα2–3LacβProN3, 16)
Yield, 53%; white foam. [α]D22 = -2.36° (c 1.78, H2O); 1H NMR (600 MHz, D2O) δ 4.54 (d, 1H, J = 7.8 Hz, Glc H-1), 4.50 (d, 1H, J = 7.8 Hz, Gal H-1), 4.41 (d, 1H, J = 11.4 Hz), 4.24 (dd, 1H, J = 11.4 and 5.4 Hz), 4.19–4.08 (m, 3H), 4.03-3.57 (m, 22H), 3.48 (t, 2H, J = 6.0 Hz), 3.33 (t, 1H, J = 8.4 Hz), 2.80 (dd, 1H, J = 12.0 and 3.6 Hz, Neu5Ac H-3eq”), 2.67 (dd, 1H, J = 13.2 and 4.8 Hz, Neu5Ac H-3eq”), 2.16 (s, 3H), 2.08 (s, 3H), 2.05 (s, 3H), 1.92 (m, 2H), 1.76 (t, 2H, J = 12.0 Hz, Neu5Ac H-3ax”); 13C NMR (125 MHz, D2O) δ 177.75 (2C), 176.44, 176.03, 105.44, 104.91, 103.33, 103.10, 80.87 (Neu5Ac C-8), 80.80, 78.17, 77.91, 77.56, 77.07, 76.82, 75.61, 75.39, 74.54, 72.06, 72.04, 71.22, 70.94, 70.71, 70.48, 70.12, 65.36, 64.31, 63.84, 62.79, 55.03, 54.51, 50.65, 43.26, 42.22, 30.99, 25.06, 24.80. HRMS (ESI) m/z calculated for C39H62N5O28 (M-2Na+H), 1048.3581, found 1048.3567.
3-Azidopropyl O-(9-O-acetyl-5-glycolylamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside(Neu5Gc9Acα2–8Neu5Acα2–3LacβProN3, 18)
Yield, 51%; white foam. [α]D22 = +3.33° (c 0.63, H2O); 1H NMR (600 MHz, D2O) δ 4.51 (d, 1H, J = 7.8 Hz, Glc H-1), 4.47 (d, 1H, J = 8.4 Hz, Gal H-1), 4.38 (dd, 1H, J = 11.4 and 3.0 Hz), 4.20 (dd, 1H, J = 11.4 and 5.4 Hz), 4.16 (dd, 1H, J = 12.0 and 3.6 Hz), 4.11 (s, 2H), 4.08 (m, 3H), 4.02–3.53 (m, 21H), 3.44 (t, 2H, J = 6.6 Hz), 3.30 (t, 1H, J = 8.4 Hz), 2.78 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.65 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.12 (s, 3H), 2.05 (s, 3H), 1.89 (m, 2H), 1.75 (t, 1H, J = 12.6 Hz, H-3ax”), 1.73 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.81, 175.08, 174.53, 173.62, 173.43, 102.77, 102.26, 100.75, 100.31, 78.32 (Neu5Ac C-8), 78.09, 75.56, 75.31, 74.92, 74.38, 74.01, 72.96, 72.95, 72.27, 69.46, 69.40, 69.38, 69.27, 68.39, 68.03, 67.66, 67.45, 62.59, 61.60, 61.21, 61.08, 52.39, 51.51, 47.96, 40.64, 39.75, 28.34, 22.41, 20.41. HRMS (ESI) m/z C39H62N5O29 (M-2Na+H), 1064.3530, found 1064.3521.
3-Azidopropyl O-(5-acetamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-glycolylamido-3,5-dideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside (Neu5Acα2–8Neu5Gcα2–3LacβProN3, 20)
Yield, 72%; white foam. [α]D22 = -0.78° (c 1.53, H2O); 1H NMR (600 MHz, D2O) δ 4.49 (d, 1H, J = 7.8 Hz, Glc H-1), 4.45 (d, 1H, J = 7.8 Hz, Gal H-1), 4.19–4.06 (m, 5H), 3.99–3.53 (m, 24H), 3.43 (t, 2H, J = 6.6 Hz), 3.29 (t, 1H, J = 8.4 Hz), 2.74 (dd, 1H, J = 12.6 and 4.2 Hz, H-3eq”), 2.66 (dd, 1H, J = 12.6 and 4.2 Hz, H-3eq”), 2.00 (s, 3H), 1.88 (m, 2H), 1.73 (t, 1H, J = 12.0 Hz, H-3ax”), 1.70 (t, 1H, J = 12.6 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 176.24, 175.18, 173.78 (2C), 102.89, 102.33, 100.42 (2C), 78.49 (Neu5Gc C-8), 78.28, 75.64, 75.36, 74.98, 74.47, 73.95, 73.03, 72.79, 71.97, 69.47, 69.25, 68.57, 68.37, 67.81, 67.66, 67.54, 62.79, 61.67, 61.31, 61.26, 60.22, 52.28, 51.93, 48.07, 40.77, 39.77, 28.41, 22.23. HRMS (ESI) m/z calculated for C37H60N5O28 (M-2Na+H), 1022.3425, found 1022.3456.
3-Azidopropyl O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(3-deoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (Neu5Acα2–8KDNα2–3LacβProN3, 22)
Yield, 71%; white foam. [α]D22 = -8.22° (c 3.2, H2O); 1H NMR (600 MHz, D2O) δ 4.48 (d, 1H, J = 7.8 Hz, Glc H-1), 4.45 (d, 1H, J = 7.8 Hz, Gal H-1), 4.16–4.12 (m, 3H), 4.04–3.79 (m, 5H), 3.75–3.51 (m, 18H), 3.43 (t, 2H, J = 6.6 Hz), 3.37 (t, 1H, J = 9.6 Hz), 3.28 (t, 1H, J = 9.0 Hz), 2.73 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.61 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.00 (s, 3H), 1.88 (m, 2H), 1.78 (t, 1H, J = 12.0 Hz, H-3ax”), 1.67 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.13, 173.95, 173.81, 102.85, 102.30, 100.95, 100.29, 78.20 (KDN C-8), 78.08, 75.62, 75.38, 75.20, 74.95, 74.42, 73.00, 72.84, 71.87, 70.74, 69.65, 69.42, 68.67, 68.34, 67.60, 67.51, 62.74, 61.60, 61.22, 61.06, 60.17, 51.89, 48.03, 40.02, 39.43, 28.38, 22.21. HRMS (ESI) m/z calculated for C35H57N4O27 (M-2Na+H), 965.3210, found 965.3211.
Methyl O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-9-azido-3,5,9-trideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-d-galactopyranoside (Neu5Acα2–8Neu5Ac9N3α2–3GalβOMe, 24)
Yield, 63%; white foam. [α]D22 = +10.07° (c 1.47, H2O); 1H NMR (600 MHz, D2O) δ 4.36 (d, 1H, J = 9.6 Hz, Gal H-1), 4.34 (m, 1H), 4.05 (dd, 1H, J = 10.2 and 3.0 Hz), 3.99 (m, 1H), 3.94 (d, 1H, J = 3.0 Hz), 3.90–3.49 (m, 18H), 3.55 (s, 3H), 2.77 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.62 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.05 (s, 3H), 2.01 (s, 3H), 1.72 (t, 1H, J = 12.0 Hz, H-3ax”), 1.71 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.17, 175.15, 174.13, 173.28, 103.66, 100.40, 100.23, 75.84, 75.72, 74.79, 74.30, 72.65, 71.80, 69.38, 69.23, 68.49, 68.10, 68.04, 62.74, 61.12, 60.68, 57.21, 52.25, 51.81, 51.59, 40.40, 39.42, 22.41, 22.23. HRMS (ESI) m/z calculated for C30H50N5O21(M-2Na+H), 816.2998, found 816.2986.
3-Azidopropyl O-(5-glycolylamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-glycolylamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (Neu5Gcα2–8Neu5Gcα2–3LacβProN3, 28)
Yield, 75%; white foam. [α]D22 = -6.90° (c 2.03, H2O); 1H NMR (600 MHz, D2O) δ 4.54 (d, 1H, J = 7.8 Hz, Glc H-1), 4.50 (d, 1H, J = 8.4 Hz, Gal H-1), 4.24–4.10 (m, 7H), 4.04–3.58 (m, 24H), 3.48 (t, 2H, J = 6.6 Hz), 3.33 (t, 1H, J = 9.0 Hz), 2.80 (dd, 1H, J = 12.6 and 4.82 Hz, H-3eq”), 2.72 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 1.93 (m, 2H), 1.77(t, 2H, J = 12.0 Hz, H-3ax”); 13C NMR (100 MHz, D2O) δ 176.24, 175.90, 173.85, 173.59, 102.86, 102.30, 100.34, 100.30, 78.56 (Neu5Gc C-8), 78.15, 75.59, 75.38, 74.95, 74.41, 73.88, 72.99, 72.48, 71.98, 69.41, 69.22, 68.29, 68.20, 67.60, 67.59, 67.50, 62.69, 61.63, 61.27, 61.12, 60.21, 60.15, 52.24, 52.59, 48.01, 40.79, 39.88, 28.37. HRMS (ESI) m/z calculated for C37H60N5O29 (M-2Na+H), 1038.3374, found 1038.3363.
3-Azidopropyl O-(3-deoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-glycolylamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (KDNα2–8Neu5Gcα2–3LacβProN3, 29)
Yield, 57%; white foam. [α]D22 = -7.72° (c 1.84, H2O); 1H NMR (600 MHz, D2O) δ 4.51 (d, 1H, J = 8.4 Hz, Glc H-1), 4.47 (d, 1H, J = 8.4 Hz, Gal H-1), 4.21–4.08 (m, 5H), 4.01–3.52 (m, 24H), 3.45 (t, 2H, J = 6.6 Hz), 3.31 (t, 1H, J = 8.4 Hz), 2.70 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.68 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 1.90 (m, 2H), 1.76 (t, 1H, J = 12.0 Hz, H-3ax”), 1.68 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 178.84, 176.22, 176.20, 105.47, 104.89, 103.23, 102.99, 81.03 (Neu5Gc C-8), 80.85, 78.21, 77.95, 77.55, 77.03, 76.52, 76.38, 75.59, 74.84, 73.07, 72.55, 72.32, 72.02, 71.77, 70.56, 70.21, 70.11, 65.41, 64.19, 63.87, 62.78, 62.07, 54.86, 50.63, 42.96, 42.39, 30.98. HRMS (ESI) m/z calculated for C35H57N4O28 (M-2Na+H), 981.3159, found 981.3143.
3-Azidopropyl O-(5-glycolylamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(3-deoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (Neu5Gcα2–8KDNα2–3LacβProN3, 30)
Yield, 68%; white foam. [α]D22 = -9.90° (c 0.98, H2O); 1H NMR (600 MHz, D2O) δ 4.53 (d, 1H, J = 7.8 Hz, Glc H-1), 4.50 (d, 1H, J = 7.8 Hz, Gal H-1), 4.21–3.56 (m, 27H), 3.48 (t, 2H, J = 6.6 Hz), 3.42 (t, 1H, J = 9.6 Hz), 3.33 (t, 1H, J = 9.0 Hz), 2.80 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.66 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 1.93 (m, 2H), 1.84 (t, 1H, J = 12.0 Hz, H-3ax”), 1.73 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.90, 173.93, 173.71, 102.88, 102.32, 100.99, 100.29, 78.27 (KDN C-8), 78.07, 75.65, 75.39, 75.27, 74.98, 74.47, 73.02, 72.59, 71.95, 70.76, 69.75, 69.67, 69.47, 68.44, 68.33, 67.71, 67.54, 62.74, 61.64, 61.25, 61.16, 60.22, 51.63, 48.06, 40.12, 39.37, 28.41. HRMS (ESI) m/z calculated for C35H57N4O28 (M-2Na+H), 981.3159, found 981.3097.
3-Azidopropyl O-(3-deoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(3-deoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (KDNα2–8KDNα2–3LacβProN3, 31)
Yield, 55%; white foam. [α]D22 = -20.0° (c 0.3, H2O); 1H NMR (600 MHz, D2O) δ 4.37 (d, 1H, J = 7.8 Hz, Glc H-1), 4.35 (d, 1H, J = 7.8 Hz, Gal H-1), 4.06–3.38 (m, 26H), 3.32 (t, 2H, J = 6.6 Hz), 3.26 (t, 1H, J = 9.6 Hz), 3.19 (t, 1H, J = 8.4 Hz), 2.55 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.50 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 1.77 (m, 2H), 1.64 (t, 1H, J = 12.0 Hz, H-3ax”), 1.57 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 173.36, 173.21, 105.46, 104.89, 103.49, 103.03, 80.86 (KDN C-8), 80.58, 78.33, 77.97, 77.85, 77.56, 77.03, 76.45, 75.59, 74.73, 73.31, 73.20, 72.54, 72.32, 72.29, 72.03, 70.62, 70.25, 70.11, 65.41, 64.17, 63.83, 62.79, 50.63, 41.97, 41.96, 30.98. HRMS (ESI) m/z calculated for C33H54N3O27 (M-2Na+H), 924.2945, found 924.2934.
3-Azidopropyl O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (Neu5Acα2–8Neu5Acα2–6LacβProN3, 34)
Yield, 80%; white foam. [α]D22 = -9.14° (c 1.04, H2O); 1H NMR (600 MHz, D2O) δ 4.51 (d, 1H, J = 7.8 Hz, Glc H-1), 4.45 (d, 1H, J = 7.8 Hz, Gal H-1), 4.41 (m, 1H), 4.14 (dd, 1H, J = 12.0 and 3.6 Hz), 4.03–3.54 (m, 25H), 3.48 (t, 2H, J = 6.6 Hz), 3.35 (t, 1H, J = 9.0 Hz), 2.79 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.64 (dd, 1H, J = 12.0 and 4.8 Hz, H-3eq”), 2.08 (s, 3H), 2.05 (s, 3H), 1.94 (m, 2H), 1.76 (t, 1H, J = 12.6 Hz, H-3ax”), 1.70 (t, 1H, J = 12.6 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.19, 175.13, 173.43, 173.30, 103.43, 102.19, 101.00, 100.72, 79.86 (Neu5Ac C-8), 78.49, 74.85, 74.83, 74.29, 73.92, 72.93, 72.88, 72.55, 71.92, 71.01, 69.67, 68.69, 68.60, 68.36, 67.99, 67.51, 63.89, 62.82, 61.70, 60.50, 52.43, 51.95, 48.09, 40.67, 40.12, 28.44, 22.50, 22.26. HRMS (ESI) m/z calculated for C37H60N5O27 (M-2Na+H), 1006.3476, found 1006.3489.
3-Azidopropyl O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-glycolylamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (Neu5Acα2–8Neu5Gcα2–6LacβProN3, 36)
Yield, 76%; white foam. [α]D22 = -5.53° (c 1.03, H2O); 1H NMR (600 MHz, D2O) δ 4.48 (d, 1H, J = 7.8 Hz, Glc H-1), 4.41 (d, 1H, J = 7.8 Hz, Gal H-1), 4.19–4.07 (m, 4H), 4.00–3.50 (m, 25H), 3.44 (t, 2H, J = 6.6 Hz), 3.31 (t, 1H, J = 9.0 Hz), 2.74 (dd, 1H, J = 12.6 and 4.2 Hz, H-3eq”), 2.61 (dd, 1H, J = 12.0 and 4.8 Hz, H-3eq”), 2.00 (s, 3H), 1.89 (m, 2H), 1.71 (t, 1H, J = 12.0 Hz, H-3ax”), 1.68 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 178.20, 176.19, 175.18, 173.88, 103.45, 102.17, 101.16, 100.34, 79.88 (Neu5Gc C-8), 78.85, 74.84, 74.13, 73.92, 72.91, 72.79, 72.53, 71.95, 70.99, 69.80, 69.68, 68.70, 68.56, 68.39, 67.51, 63.92, 62.81, 61.73, 61.30, 60.47, 52.28, 51.94, 48.07, 40.76, 40.28, 28.42, 22.24. HRMS (ESI) m/z calculated for C37H60N5O28 (M-2Na+H), 1022.3425, found 1022.3431.
3-Azidopropyl O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(3-deoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-d-galactopyranosyl-(1→4)-β-d-glucopyranoside (Neu5Acα2–8KDNα2–6LacβProN3, 38)
Yield, 64%; white foam. [α]D22 = -7.45° (c 0.98, H2O); 1H NMR (600 MHz, D2O) δ 4.47 (d, 1H, J = 7.8 Hz, Glc H-1), 4.39 (d, 1H, J = 7.8 Hz, Gal H-1), 4.20 (m, 1H), 4.13–4.08 (m, 2H), 3.99–3.47 (m, 23H), 3.43 (t, 2H, J = 6.6 Hz), 3.38 (t, 1H, J = 9.6 Hz), 3.29 (t, 1H, J = 9.0 Hz), 2.73 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.54 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.00 (s, 3H), 1.88 (m, 2H), 1.77 (t, 1H, J = 12.0 Hz, H-3ax”), 1.62 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.14, 173.98, 173.70, 103.45, 102.17, 101.07, 100.88, 79.95 (KDN C-8), 78.40, 75.34, 74.81, 74.80, 73.94, 72.89, 72.86, 72.51, 71.86, 70.97, 70.75, 69.95, 69.59, 68.68, 68.67, 68.40, 67.50, 63.90, 62.77, 61.66, 60.47, 51.91, 48.06, 40.08, 39.84, 28.40, 22.22. HRMS (ESI) m/z calculated for C35H57N4O27 (M-2Na+H), 965.3210, found 965.3207.
Methyl O-(5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→8)-O-(5-acetamido-9-azido-3,5,9-trideoxy-d-glycero-α-d-galacto-2-nonulopyranosylonic acid)-(2→6)-O-β-d-galactopyranoside (Neu5Acα2–8Neu5Ac9N3α2–6GalβOMe, 40)
Yield, 56%; white foam. [α]D22 = -7.54° (c 1.14, H2O); 1H NMR (600 MHz, D2O) δ 4.37 (m, 1H), 4.27 (d, 1H, J = 7.8 Hz, Gal H-1), 3.97 (m, 1H), 3.89–3.44 (m, 18H), 3.53 (s, 3H), 2.75 (dd, 1H, J = 12.6 and 4.8 Hz, H-3eq”), 2.57 (dd, 1H, J = 12.0 and 4.2 Hz, H-3eq”), 2.03 (s, 3H), 1.99 (s, 3H), 1.72 (t, 1H, J = 12.0 Hz, H-3ax”), 1.58 (t, 1H, J = 12.0 Hz, H-3ax”); 13C NMR (125 MHz, D2O) δ 175.21, 175.13, 173.65, 173.56, 104.04, 101.03, 100.09, 76.29, 74.18, 73.58, 72.73, 72.69, 71.84, 70.77, 69.45, 68.72, 68.59, 68.39, 67.93, 63.65, 62.68, 60.51, 57.53, 52.51, 51.86, 51.37, 40.33, 22.45, 22.18. HRMS (ESI) m/z calculated for C30H50N5O21(M-2Na+H), 816.2998, found 816.2993.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by Award Number R01GM076360 from the National Institute of General Medical Sciences, the Arnold and Mabel Beckman Foundation, and the Alfred P. Sloan Foundation. X.C. is a Beckman Young Investigator, an Alfred P. Sloan Research Fellow, a Camille Dreyfus Teacher-Scholar, and a UC-Davis Chancellor's Fellow.
Footnotes
Supporting Information Available: NMR spectra of monosialylated and disialylated oligosaccharide products. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.a Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Schauer R. Glycoconj. J. 2000;17:485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Angata T, Varki A. Chem. Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 2.a Kelm S, Schauer R, Crocker PR. Glycoconj. J. 1996;13:913–926. doi: 10.1007/BF01053186. [DOI] [PubMed] [Google Scholar]; b Renkonen R. Adv. Exp. Med. Biol. 1998;435:63–73. doi: 10.1007/978-1-4615-5383-0_7. [DOI] [PubMed] [Google Scholar]
- 3.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2nd Ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2008. p. 132. [PubMed] [Google Scholar]
- 4.Nudelman E, Hakomori S, Kannagi R, Levery S, Yeh MY, Hellstrom KE, Hellstrom I. J. Biol. Chem. 1982;257:12752–12756. [PubMed] [Google Scholar]
- 5.Nakamura K, Suzuki M, Taya C, Inagaki F, Yamakawa T, Suzuki A. J. Biochem. 1991;110:832–841. doi: 10.1093/oxfordjournals.jbchem.a123667. [DOI] [PubMed] [Google Scholar]
- 6.Furukawa K, Chait BT, Lloyd KO. J. Biol. Chem. 1988;263:14939–14947. [PubMed] [Google Scholar]
- 7.Nakao T, Kon K, Ando S, Hirabayashi Y. Biochim. Biophys. Acta. 1991;1086:305–309. doi: 10.1016/0005-2760(91)90174-g. [DOI] [PubMed] [Google Scholar]
- 8.Nudelman ED, Levery SB, Stroud MR, Salyan ME, Abe K, Hakomori S. J. Biol. Chem. 1988;263:13942–13951. [PubMed] [Google Scholar]
- 9.Iwamori M, Nagai Y. J. Biol. Chem. 1978;253:8328–8331. [PubMed] [Google Scholar]
- 10.Cheresh DA, Varki AP, Varki NM, Stallcup WB, Levine J, Reisfeld RA. J. Biol. Chem. 1984;259:7453–7459. [PubMed] [Google Scholar]
- 11.Hitoshi S, Kusunoki S, Kon K, Chiba A, Waki H, Ando S, Kanazawa I. J. Neuroimmunol. 1996;66:95–101. doi: 10.1016/0165-5728(96)00024-0. [DOI] [PubMed] [Google Scholar]
- 12.Dubois C, Manuguerra JC, Hauttecoeur B, Maze J. J. Biol. Chem. 1990;265:2797–2803. [PubMed] [Google Scholar]
- 13.Farooqui AA, Horrocks LA. Neurochem. Pathol. 1984;2:189–218. doi: 10.1007/BF02834352. [DOI] [PubMed] [Google Scholar]
- 14.Waki H, Masuzawa A, Kon K, Ando S. J. Biochem. 1993;114:459–462. doi: 10.1093/oxfordjournals.jbchem.a124199. [DOI] [PubMed] [Google Scholar]
- 15.Schauer R. Curr. Opin. Struct. Biol. 2009;19:507–514. doi: 10.1016/j.sbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slomiany A, Kojima K, Banas-Gruszka Z, Slomiany BL. Biochem. Biophys. Res. Commun. 1981;100:778–784. doi: 10.1016/s0006-291x(81)80242-2. [DOI] [PubMed] [Google Scholar]
- 17.Ijuin T, Kitajima K, Song Y, Kitazume S, Inoue S, Haslam SM, Morris HR, Dell A, Inoue Y. Glycoconj. J. 1996;13:401–413. doi: 10.1007/BF00731473. [DOI] [PubMed] [Google Scholar]
- 18.a Sato C, Kitajima K. Trends Glycosci. Glycotechnol. 1999;11:371–390. [Google Scholar]; a Sato C, Fukuoka H, Ohta K, Matsuda T, Koshino R, Kobayashi K, Troy FA, 2nd, Kitajima K. J. Biol. Chem. 2000;275:15422–15431. doi: 10.1074/jbc.275.20.15422. [DOI] [PubMed] [Google Scholar]
- 19.a Sato C, Kitajima K, Tazawa I, Inoue Y, Inoue S, Troy FA., 2nd. J. Biol. Chem. 1993;268:23675–23684. [PubMed] [Google Scholar]; b Inoue S, Iwasaki M. Biochem. Biophys. Res. Commun. 1978;83:1018–1023. doi: 10.1016/0006-291x(78)91497-3. [DOI] [PubMed] [Google Scholar]
- 20.Finne J. J. Biol. Chem. 1982;257:11966–11970. [PubMed] [Google Scholar]
- 21.a James WM, Agnew WS. Biochem. Biophys. Res. Commun. 1987;148:817–826. doi: 10.1016/0006-291x(87)90949-1. [DOI] [PubMed] [Google Scholar]; a Zuber C, Lackie PM, Catterall WA, Roth J. J. Biol. Chem. 1992;267:9965–9971. [PubMed] [Google Scholar]
- 22.a Roth J, Zuber C, Wagner P, Taatjes DJ, Weisgerber C, Heitz PU, Goridis C, Bitter-Suermann D. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2999–3003. doi: 10.1073/pnas.85.9.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Livingston BD, Jacobs JL, Glick MC, Troy FA. J. Biol. Chem. 1988;263:9443–9448. [PubMed] [Google Scholar]; c Martersteck CM, Kedersha NL, Drapp DA, Tsui TG, Colley KJ. Glycobiology. 1996;6:289–301. doi: 10.1093/glycob/6.3.289. [DOI] [PubMed] [Google Scholar]
- 23.Roth J, Kempf A, Reuter G, Schauer R, Gehring WJ. Science. 1992;256:673–675. doi: 10.1126/science.1585182. [DOI] [PubMed] [Google Scholar]
- 24.Malykh YN, Krisch B, Gerardy-Schahn R, Lapina EB, Shaw L, Schauer R. Glycoconj. J. 1999;16:731–739. doi: 10.1023/a:1007115627708. [DOI] [PubMed] [Google Scholar]
- 25.Kanamori A, Inoue S, Iwasaki M, Kitajima K, Kawai G, Yokoyama S, Inoue Y. J. Biol. Chem. 1990;265:21811–21819. [PubMed] [Google Scholar]
- 26.Kiang WL, Krusius T, Finne J, Margolis RU, Margolis RK. J. Biol. Chem. 1982;257:1651–1659. [PubMed] [Google Scholar]
- 27.Fukuda M, Lauffenburger M, Sasaki H, Rogers ME, Dell A. J. Biol. Chem. 1987;262:11952–11957. [PubMed] [Google Scholar]
- 28.Fukuda M, Dell A, Fukuda MN. J. Biol. Chem. 1984;259:4782–4791. [PubMed] [Google Scholar]
- 29.Funakoshi Y, Taguchi T, Sato C, Kitajima K, Inoue S, Morris HR, Dell A, Inoue Y. Glycobiology. 1997;7:195–205. doi: 10.1093/glycob/7.2.195. [DOI] [PubMed] [Google Scholar]
- 30.Sato C, Kitajima K, Inoue S, Inoue Y. J. Biol. Chem. 1998;273:2575–2582. doi: 10.1074/jbc.273.5.2575. [DOI] [PubMed] [Google Scholar]
- 31.Nohara K, Kunimoto M, Fujimaki H. J. Biochem. 1998;124:194–199. doi: 10.1093/oxfordjournals.jbchem.a022079. [DOI] [PubMed] [Google Scholar]
- 32.Yasukawa Z, Sato C, Sano K, Ogawa H, Kitajima K. Glycobiology. 2006;16:651–665. doi: 10.1093/glycob/cwj112. [DOI] [PubMed] [Google Scholar]
- 33.Ziak M, Kerjaschki D, Farquhar MG, Roth J. J. Am. Soc. Nephrol. 1999;10:203–209. doi: 10.1681/ASN.V102203. [DOI] [PubMed] [Google Scholar]
- 34.Ziak M, Meier M, Roth J. Glycoconj. J. 1999;16:185–188. doi: 10.1023/a:1007068102436. [DOI] [PubMed] [Google Scholar]
- 35.a Robbins JB, McCracken GH, Jr., Gotschlich EC, Orskov F, Orskov I, Hanson LA. N. Engl. J. Med. 1974;290:1216–1220. doi: 10.1056/NEJM197405302902202. [DOI] [PubMed] [Google Scholar]; b Rohr TE, Troy FA. J. Biol. Chem. 1980;255:2332–2342. [PubMed] [Google Scholar]; c Aalto J, Pelkonen S, Kalimo H, Finne J. Glycoconj. J. 2001;18:751–758. doi: 10.1023/a:1021147316647. [DOI] [PubMed] [Google Scholar]; d Adlam C, Knight JM, Mugridge A, Williams JM, Lindon JC. FEBS Microbiol. Lett. 1987;42:23–25. [Google Scholar]
- 36.Varki A. Trends Mol. Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaji T, Teranishi T, Alphey MS, Crocker PR, Hashimoto Y. J. Biol. Chem. 2002;277:6324–6332. doi: 10.1074/jbc.M110146200. [DOI] [PubMed] [Google Scholar]
- 38.a Nicoll G, Avril T, Lock K, Furukawa K, Bovin N, Crocker PR. Eur. J. Immunol. 2003;33:1642–1648. doi: 10.1002/eji.200323693. [DOI] [PubMed] [Google Scholar]; b Attrill H, Imamura A, Sharma RS, Kiso M, Crocker PR, van Aalten DM. J. Biol. Chem. 2006;281:32774–32783. doi: 10.1074/jbc.M601714200. [DOI] [PubMed] [Google Scholar]
- 39.Boons GJ, Demchenko AV. Chem. Rev. 2000;100:4539–4566. doi: 10.1021/cr990313g. [DOI] [PubMed] [Google Scholar]
- 40.a Demchenko AV, Boons GJ. Tetrahedron Lett. 1998;39:3065–3068. [Google Scholar]; b Demchenko AV, Boons GJ. Chem.-Eur. J. 1999;5:1278–1283. [Google Scholar]
- 41.Yu CS, Niikura K, Lin CC, Wong CH. Angew. Chem. Int. Ed. Engl. 2001;40:2900–2903. doi: 10.1002/1521-3773(20010803)40:15<2900::AID-ANIE2900>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 42.a Komba S, Galustian C, Ishida H, Feizi T, Kannagi R, Kiso M. Angew. Chem. Int. Ed. Engl. 1999;38:1131–1133. doi: 10.1002/(SICI)1521-3773(19990419)38:8<1131::AID-ANIE1131>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]; b De Meo C, Demchenko AV, Boons GJ. J. Org. Chem. 2001;66:5490–5497. doi: 10.1021/jo010345f. [DOI] [PubMed] [Google Scholar]
- 43.a Ando H, Koike Y, Ishida H, Kiso M. Tetrahedron Lett. 2003;44:6883–6886. [Google Scholar]; b Adachi M, Tanaka H, Takahashi T. Synlett. 2004:609–614. [Google Scholar]; c Tanaka H, Adachi M, Takahashi T. Chem.-Eur. J. 2005;11:849–862. doi: 10.1002/chem.200400840. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka H, Nishiura Y, Adachi M, Takahashi T. Heterocycles. 2006;67:107–112. [Google Scholar]
- 45.a Ando H, Koike Y, Koizumi S, Ishida H, Kiso M. Angew. Chem. Int. Ed. Engl. 2005;44:6759–6763. doi: 10.1002/anie.200501608. [DOI] [PubMed] [Google Scholar]; b Tanaka H, Ando H, Ishida H, Kiso M, Ishihara H, Koketsu M. Tetrahedron Lett. 2009;50:4478–4481. [Google Scholar]
- 46.a Tanaka H, Nishiura Y, Takahashi T. J. Am. Chem. Soc. 2006;128:7124–7125. doi: 10.1021/ja0613613. [DOI] [PubMed] [Google Scholar]; b Tanaka H, Nishiura Y, Takahashi T. J. Am. Chem. Soc. 2008;130:17244–17245. doi: 10.1021/ja807482t. [DOI] [PubMed] [Google Scholar]
- 47.a Gilbert M, Brisson JR, Karwaski MF, Michniewicz J, Cunningham AM, Wu Y, Young NM, Wakarchuk WW. J. Biol. Chem. 2000;275:3896–3906. doi: 10.1074/jbc.275.6.3896. [DOI] [PubMed] [Google Scholar]; b Gilbert M, Karwaski MF, Bernatchez S, Young NM, Taboada E, Michniewicz J, Cunningham AM, Wakarchuk WW. J. Biol. Chem. 2002;277:327–337. doi: 10.1074/jbc.M108452200. [DOI] [PubMed] [Google Scholar]; c Cheng J, Yu H, Lau K, Huang S, Chokhawala HA, Li Y, Tiwari VK, Chen X. Glycobiology. 2008;18:686–697. doi: 10.1093/glycob/cwn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blixt O, Vasiliu D, Allin K, Jacobsen N, Warnock D, Razi N, Paulson JC, Bernatchez S, Gilbert M, Wakarchuk W. Carbohydr. Res. 2005;340:1963–1972. doi: 10.1016/j.carres.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 49.a Ito Y, Numata M, Sugimoto M, Ogawa T. J. Am. Chem. Soc. 1989;111:8508–8510. [Google Scholar]; b Ando H, Ishida H, Kiso M, Hasegawa A. Carbohydr. Res. 1997;300:207–217. doi: 10.1016/s0008-6215(97)00051-7. [DOI] [PubMed] [Google Scholar]; c Ding Y, Fukuda M, Hindsgaul O. Bioorg. Med. Chem. Lett. 1998;8:1903–1908. doi: 10.1016/s0960-894x(98)00332-1. [DOI] [PubMed] [Google Scholar]; d Castro-Palomino JC, Simon B, Speer O, Leist M, Schmidt RR. Chem.- Eur. J. 2001;7:2178–2184. doi: 10.1002/1521-3765(20010518)7:10<2178::aid-chem2178>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]; e Gilbert M, Karwaski M-F, Bernatchez S, Young NM, Taboada E, Michniewicz J, Cunningham A-M, Wakarchuk WW. J. Biol. Chem. 2002;277:327–337. doi: 10.1074/jbc.M108452200. [DOI] [PubMed] [Google Scholar]; f Antoine T, Heyraud A, Bosso C, Samain E. Angew. Chem. Int. Ed. Engl. 2005;44:1350–1352. doi: 10.1002/anie.200461507. [DOI] [PubMed] [Google Scholar]; g Schwardt O, Visekruna T, Zenhausern G, Rabbani S, Ernst B. J. Carbohydr. Chem. 2006;25:543–556. [Google Scholar]; h Rich JR, Wakarchuk WW, Bundle DR. Chem-Eur. J. 2006;2:845–858. doi: 10.1002/chem.200500518. [DOI] [PubMed] [Google Scholar]; i Houliston RS, Yuki N, Hirama T, Khieu NH, Brisson J-R, Gilbert M, Jarrell HC. Biochemistry. 2007;46:36–44. doi: 10.1021/bi062001v. [DOI] [PubMed] [Google Scholar]; j Zhang P, Zuccolo AJ, Li W, Zheng RB, Ling C-C. Chem. Commun. 2009;28:4233–4235. doi: 10.1039/b908933k. [DOI] [PubMed] [Google Scholar]
- 50.Roy R, Pon RA. Glycoconj. J. 1990;7:3–12. [Google Scholar]
- 51.a Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. J. Am. Chem. Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]; b Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Angew. Chem. Int. Ed. Engl. 2006;45:3938–3944. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu H, Chokhawala HA, Huang S, Chen X. Nat. Protoc. 2006;1:2485–2492. doi: 10.1038/nprot.2006.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H, Yu H, Karpel R, Chen X. Bioorg. Med. Chem. 2004;12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Yu H, Cao H, Lau K, Muthana S, Tiwari VK, Son B, Chen X. Appl. Microbiol. Biotechnol. 2008;79:963–970. doi: 10.1007/s00253-008-1506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlosshauer B, Blum AS, Mendez-Otero R, Barnstable CJ, Constantine-Paton M. J. Neurosci. 1988;8:580–592. doi: 10.1523/JNEUROSCI.08-02-00580.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen HY, Challa AK, Varki A. J. Biol. Chem. 2006;281:7825–7833. doi: 10.1074/jbc.M512379200. [DOI] [PubMed] [Google Scholar]
- 57.Kniep B, Kniep E, Ozkucur N, Barz S, Bachmann M, Malisan F, Testi R, Rieber EP. Int. J. Cancer. 2006;119:67–73. doi: 10.1002/ijc.21788. [DOI] [PubMed] [Google Scholar]
- 58.a Ogura H, Furuhata K, Sato S, Anazawa K, Itoh M, Shitori Y. Carbohydr. Res. 1987;167:77–86. doi: 10.1016/0008-6215(87)80269-0. [DOI] [PubMed] [Google Scholar]; b Rauvolfova J, Venot A, Boons GJ. Carbohydr. Res. 2008;343:1605–1611. doi: 10.1016/j.carres.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Angata K, Fukuda M. Biochimie. 2003;85:195–206. doi: 10.1016/s0300-9084(03)00051-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.