Abstract
CD36 is a membrane glycoprotein present on platelets, mononuclear phagocytes, adipocytes, hepatocytes, myocytes, and some epithelia. On microvascular endothelial cells, CD36 is a receptor for thrombospondin-1 and related proteins and functions as a negative regulator of angiogenesis. On phagocytes, through its functions as a scavenger receptor recognizing specific oxidized phospholipids and lipoproteins, CD36 participates in internalization of apoptotic cells, certain bacterial and fungal pathogens, and modified low-density lipoproteins, thus contributing to inflammatory responses and atherothrombotic diseases. CD36 also binds long-chain fatty acids and facilitates their transport into cells, thus participating in muscle lipid utilization, adipose energy storage, and gut fat absorption and possibly contributing to the pathogenesis of metabolic disorders, such as diabetes and obesity. On sensory cells, CD36 is involved in insect pheromone signaling and rodent fatty food preference. The signaling pathways downstream of CD36 involve ligand-dependent recruitment and activation of nonreceptor tyrosine kinases, specific mitogen-activated protein kinases, and the Vav family of guanine nucleotide exchange factors; modulation of focal adhesion constituents; and generation of intracellular reactive oxygen species. CD36 in many cells is localized in specialized cholesterol-rich membrane microdomains and may also interact with other membrane receptors, such as tetraspanins and integrins. Identification of the precise CD36 signaling pathways in specific cells elicited in response to specific ligands may yield novel targets for drug development.
Overview
CD36 was described nearly 30 years ago as “glycoprotein IV,” the fourth major band (of ~88 kD) observed on SDS–polyacrylamide gel electrophoresis gels of platelet membranes (1). It was later found to be identical to the antigen recognized by the monoclonal antibody OKM5, a marker for monocytes and macrophages (2). CD36 is present on many mammalian cell types: microvascular endothelium; “professional” phagocytes including macrophages, dendritic cells, microglia, and retinal pigment epithelium; erythroid precursors; hepatocyes; adipocytes; cardiac and skeletal myocytes; and specialized epithelia of the breast, kidney, and gut (3). CD36 is the defining member of a gene family (4, 5) that includes two other members in vertebrates, LIMP-2 (lysosomal integral membrane protein–2) and CLA-1 (CD36 and LIMP-2 analogous), which is also known as SRB-1 (scavenger receptor B-1). The primary structure of CD36 family members is conserved in mammals, and multiple orthologs have been identified in most orders of insect (Diptera, Hymenoptera, Coleoptera, and Lepidoptera) (6) as well as in nematodes, sponges (7), and slime mold, suggesting that the common ancestral gene appeared more than 300 million years ago.
CD36 in Angiogenesis
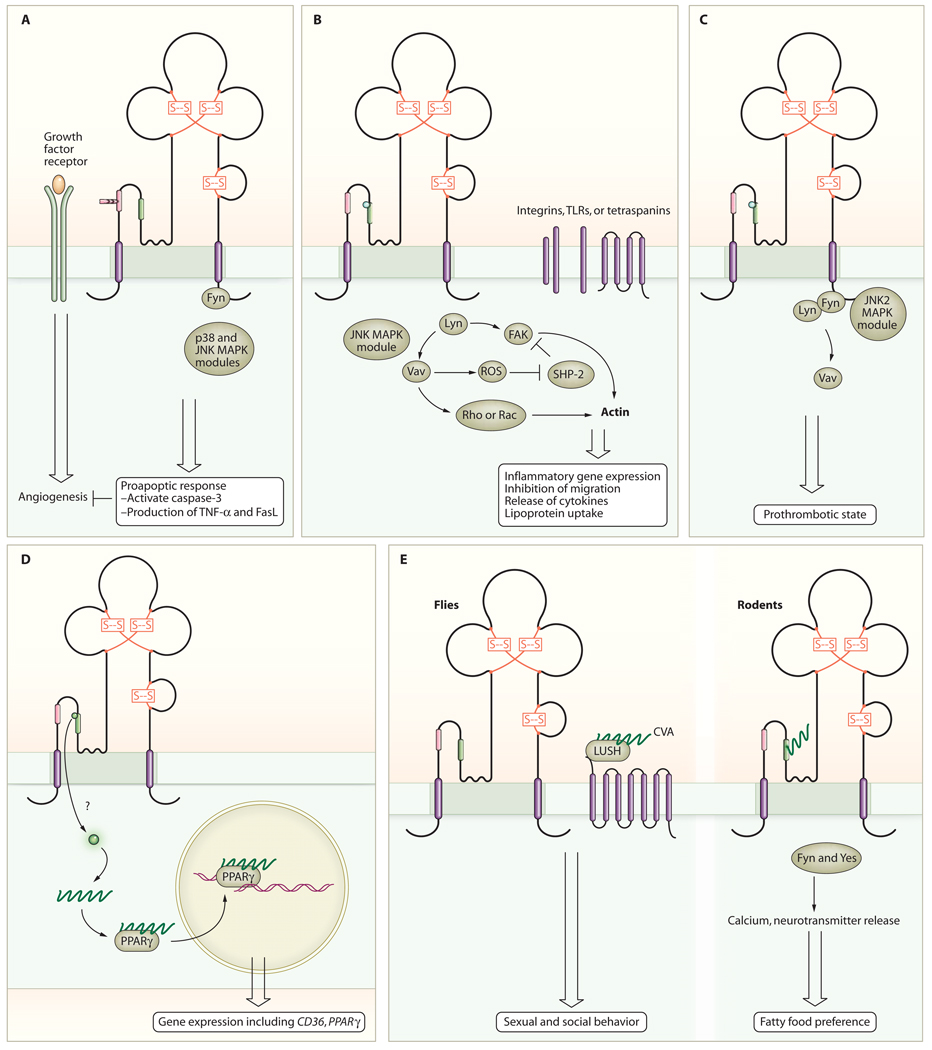
The diverse expression pattern of CD36 is reflective of its multiple cellular functions. It is a receptor for thrombospondin-1 (8, 9) and several other proteins containing peptide domains known as thrombospondin type I repeats (TSRs) (10). In this capacity on microvascular endothelial cells, it functions as an endogenous negative regulator of angiogenesis (11) and, therefore, plays a role in tumor growth, inflammation, wound healing, and other pathological processes requiring neovascularization. CD36 accomplishes this function by inhibiting growth factor–induced proangiogenic signals that mediate endothelial cell proliferation, migration, and tube formation and instead generating anti-angiogenic signals that lead to apoptosis (11, 12). Pharmacologic and genetic knockout experiments identified the signaling pathway downstream of CD36 in this setting as one involving the nonreceptor protein tyrosine kinase Fyn, the mitogen-activated protein kinases (MAPKs) p38 and c-Jun N-terminal kinase (JNK), and caspase-3 (12) (Fig. 1A). Downstream induction of expression of additional proapoptotic effectors, including Fas ligand and tumor necrosis factor–α (TNF-α), is also involved (13, 14).
Fig. 1.
CD36 mediates cell-specific responses. (A) In endothelial cells, CD36 inhibits angiogenesis induced by growth factors, such as fibroblast growth factor 2 (FGF2) and vascular endothelial growth factor (VEGF), and promotes apoptosis. (B) In macrophages and monocytes, CD36 promotes inflammatory responses and phagocytosis. CD36 may interact with other receptors, such as integrins, TLRs, or tetraspanins, to mediate some of the responses. (C) In platelets, CD36 promotes activation, aggregation, and secretion. (D) In monocytes and macrophages, CD36 promotes the uptake of bioactive lipids, leading to activation of PPARγ transcriptional pathways. (E) In sensory cells, CD36 contributes to cellular responses in the mouth and gut to dietary fats in mammals (right) and to pheromone responses in Drosophila (left).
CD36 as a Pattern Recognition Receptor
CD36 functions as a scavenger receptor— that is, a pattern-recognition receptor—on phagocytic cells (15). In addition to CD36, scavenger receptors include the structurally unrelated proteins SRA, CLA-1, lectinlike oxidized low-density lipoprotein receptor (LOX)–1, macrophage receptor with collagenous structure (Marco), Toll-like receptors (TLRs), and CD68, among others. These proteins evolved with the innate immune system as primitive receptors that recognized and helped the organism eliminate foreign agents (for example, bacteria, parasites, and viruses) during an infection (16, 17). The hallmark of these receptors is their ability to recognize specific classes of molecular patterns presented by pathogens or by pathogen-infected cells. CD36 recognizes specific lipid and lipoprotein components of bacterial cell walls (18)—particularly those of staphylococcal and mycobacterial organisms (19), β-glucans on fungal species (20), and erythrocytes infected with falciparum malaria (4, 21)—and thereby triggers a reaction that results in opsonin-independent pathogen internalization. As with many other scavenger receptors, CD36 also recognizes endogenously derived ligands, including apoptotic cells (22–24), shed photoreceptor outer segments (25, 26), oxidatively modified lipoproteins (15, 27), glycated proteins, and amyloid-forming peptides (28, 29). Studies of model organisms have shown that scavenging is an evolutionarily ancient function of CD36. For example, the protein products of the Drosophila croquemort (30, 31) and peste (19) genes are CD36 orthologs present on blood cells and are involved in recognition and clearance of apoptotic cells and mycobacteria, respectively. CO3F11.3, an ortholog on cells of the worm Caenorhabditis elegans, participates in the innate immune response to fungi (20).
Because of the potential importance of CD36 ligands in disease pathogenesis, including atherosclerosis and Alzheimer’s disease, the scavenger receptor functions of CD36 have been extensively studied. For example, recognition and internalization of oxidized forms of low-density lipoprotein (oxLDL) by macrophage CD36 promotes the formation of lipid-laden “foam cells” and atherosclerotic plaque (27, 32–34); triggers proinflammatory reactions, such as activation of the transcription factor nuclear factor κB (NF-κB) (35, 36), release of cytokines, and production of reactive oxygen species (ROS) (37, 38); and inhibits macrophage migration (38, 39) [thus trapping cells in an inflammatory milieu (40)] (Fig. 1B). Similarly, CD36-dependent interaction of β-amyloid (Aβ) peptide with microglial cells leads to internalization of the peptides and a proinflammatory response (29, 41, 42) and may contribute to the pathogenesis of Alzheimer’s disease. On blood platelets, the interaction of CD36 with oxLDL (43) or anionic phospholipids presented on the surface of cell-derived microparticles (44) renders the platelets more sensitive to activation by low doses of agonists and may represent a mechanistic link between oxidative stress, hyperlipidemia, inflammation, and pathological thrombosis (Fig. 1C). On dendritic cells of the immune system, CD36 mediates uptake of apoptotic cells and provides a mechanism for cross-presentation of antigens to cytotoxic T cells (24). CD36 may also play a role in cytokine-mediated macrophage fusion and granuloma formation (45).
Studies of mice with targeted deletion of the cd36 gene show that loss of CD36 confers protection from diet-induced atherosclerosis (32) and thrombosis (44) and limits inflammation and tissue infarction associated with acute cerebrovascular occlusion (37), but may increase susceptibility to certain infections (18). The athero-protective role of CD36 deficiency is controversial (46–48), but the preponderance of evidence supports an important role for CD36 in mediating macrophage responses described above that are proatherogenic. Studies in several in vivo model systems show smaller plaque lesions (32, 49–51) or less-complex lesions (48), less aortic cholesterol, or all three in the absence of CD36 (48). The reason for discrepancy among murine studies may relate to environmental factors that induce stress or inflammation and the type of analysis performed. We hypothesize that CD36, because of its unique specificity for ligands generated during host responses to atheroinflammatory diseases (that is, oxidized phospholipids and apoptotic cells), may function as a “disease sensor” capable of triggering a signaling cascade that could influence macrophage and platelet responses and contribute to disease pathology.
The mechanisms by which CD36 signaling in response to scavenger ligands leads to multiple outcomes, such as providing the trigger for internalization or phagocytosis of the bound ligand [a so-called “eat me signal” (21–28, 52)], induction of leukocyte proinflammatory responses (29, 35–42, 53), and promotion of platelet granule secretion and integrin activation (43, 44, 54), have been partially characterized. In all cases, the intracellular signals involve recruitment and activation of Src family nonreceptor protein tyrosine kinases and serine/threonine kinases of the MAPK family, analogous but not identical, to kinases downstream of CD36 in endothelial cells (12, 42, 52, 55). Lyn and JNK2 are critical effectors of both macrophage and platelet responses, whereas Fyn and p38 are the primary mediators of endothelial cell responses, showing that cellular context is critical to understanding the signaling components engaged by CD36 (Fig. 1). The signaling partners downstream of the Src family kinases and MAPKs are not fully elucidated, although studies have implicated focal adhesion components, including the tyrosine kinases Pyk 2 and FAK (focal adhesion kinase) and the adaptor proteins p130cas and paxillin in some responses (38, 56). For example, after macrophage exposure to oxLDL, FAK undergoes prolonged phosphorylation and activation (38) because of direct activation by Src family kinases coupled with inactivation of a specific phosphatase, SHP-2. The latter was the result of intracellular ROS generation and subsequent oxidative inactivation of a critical cysteine residue in the enzymatic active site. The net results of these intracellular events were enhanced actin polymerization, increased cellular spreading, and inhibition of migration. The Vav family of proteins may also link CD36 to downstream events (57). These proteins are known substrates for Fyn and Lyn and, when activated by phosphorylation, function as guanine nucleotide exchange factors (GEFs) for Rho and Rac guanosine triphosphatases (GTPases). Vavs are large multidomain proteins that contain two SH3 domains flanking a single SH2 domain, and, thus, they function as scaffolds as well as GEFs. Vavs are phosphorylated in macrophages, microglial cells, and platelets in a CD36-dependent manner and may be an important link between CD36 and responses requiring small molecular weight guanine nucleotide–binding proteins (G proteins).
CD36-Mediated Lipid Uptake
A ligand-specific aspect of CD36 signaling involves its capacity to deliver biologically active lipids to cells (58) (Fig. 1D). Incubation of monocytes with oxLDL increases transcription of several genes, including the one encoding CD36, through activation of the nuclear hormone receptor peroxisome proliferator–activated receptor γ (PPARγ) (59). The activating ligand(s) for PPARγ are oxidized phospholipids, such as 9- and 13-HODE (hydroxyoctadecadienoic acid), and these, their precursor lipids, or both are part of the cargo delivered to the cell within the oxLDL particle. Although PPARγ is generally thought to promote anti-inflammatory responses, the presence of macrophages with abundant CD36 expression in the setting of oxLDL is proinflammatory and proatherogenic. It is likely that, before the advent of the fat- and calorie-rich Western diet, uptake of modified LDL was rare and localized and up-regulation of CD36 was a mechanism for sequestration of this aberrant ligand. Continuous exposure to oxLDL, however, leads to development and accumulation of lipid-laden macrophages and contributes to a dysfunctional response. Although PPARγ activation is clearly an important component of CD36 signaling in monocytes and macrophages, most of the responses to CD36 ligands cannot be accounted for by a transcriptional mechanism. For example, internalization of large particulate ligands requires rapid induction of intracellular signals to effect cytoskeletal reorganization and direct internalized ligands to specific intracellular compartments but does not require new protein synthesis. Similarly, the rapid proinflammatory, prothrombotic responses are mostly nontranscriptional.
CD36 also functions on adipocytes, muscle cells, enterocytes, and hepatocytes as a facilitator of long-chain fatty acid transport (60–64). There is evidence for CD36 binding to fatty acids and oxidized fatty acids, but the mechanism by which it facilitates fatty acid uptake remains vague (60, 65). Expression of the gene encoding CD36 in this context is controlled by the lipogenic transcription factors PPARγ, liver X receptor (LXR), and pregnane X receptor (PXR) (59, 61), and in skeletal muscle its abundance correlates with oxidative potential and is regulated by exercise and insulin (66, 67). In muscle, there is evidence for an intracellular pool of CD36 that can be translocated to the plasma membrane to increase fatty acid uptake (68). Cd36-null mice show abnormal plasma lipid and lipoprotein profiles and resting hypoglycemia (69), attributable in part to a marked impairment of fatty acid utilization in cardiac and striated muscle and of fatty acid uptake by adipose tissues (62). Studies in rodents and humans suggest that CD36–fatty acid interactions may contribute to the pathogenesis of metabolic disorders, such as insulin resistance, obesity, and nonalcoholic hepatic steatosis (70–75). The mechanistic role of CD36 in metabolic disease is likely to be complex and context dependent. CD36 contributes to intracellular lipid accumulation and might, therefore, be expected to promote lipotoxicity and, hence, insulin resistance. In several animal models, ablation of CD36-mediated lipid uptake in muscle or liver prevented lipotoxicity (76–78). In models in which CD36 was specifically induced in the liver by pharmacologic means or cDNA transduction, CD36 contributed to steatosis, which can also contribute to metabolic disorders (61, 68). Alternatively, a cd36-null mutation in the spontaneously hypertensive rat strain has been linked to insulin resistance (79, 80); however, the complete absence of CD36 in that model has come into question (81). Insulin and glucose infusion studies of a small number of human participants lacking CD36 showed insulin resistance (71), but other similar studies have not (82–84). CD36 deficiency in human participants is associated with multiple different compound mutations that may not only affect CD36 but also expression of other genes that could affect phenotype (85, 86). Although it may be counterintuitive, ablation of CD36 may also lead to insulin resistance. Mice null for cd36 are not whole-body insulin resistant, but they demonstrate liver insulin resistance (87). The phenotype is masked by the essential utilization of glucose by muscle because fatty acid uptake is impaired. One can imagine that differential organ or cell-specific CD36 expression may differentially affect insulin resistance and that this may underlie the discrepancies among the human studies.
In the gut, CD36 promotes absorption of long-chain fatty acids (63, 64) and participates in carotenoid uptake for vitamin A metabolism (88, 89). The mammalian CD36 homolog, SRB1, is present on steroidogenic tissues and hepatocytes, where it mediates selective cellular cholesterol uptake from high-density lipoprotein (HDL) particles (90). This provides cells with essential precursors for steroid hormone synthesis and also provides a mechanism to “unload” excess cholesterol from peripheral tissues through so-called “reverse cholesterol transport.” The mechanisms by which CD36 and related family members affect these lipid uptake functions are topics of ongoing study and controversy.
CD36 in Sensory Perception
As with the scavenging functions of CD36, study of model organisms has shed light on biological implications of CD36–fatty acid interactions (Fig. 1D). Many insect species have a CD36 ortholog, SNMP (sensory neuron membrane protein), on dendrites of the specialized neural cells in antennae involved in pheromone detection (91, 92). Drosophila SNMP functions as part of a multiprotein signaling complex, along with specific G protein–coupled odorant receptors (including Or67d and Or83b), that recognizes a fatty acid–derived pheromone, cis-vaccenyl acetate (CVA) (93, 94). CVA binds to the soluble odorant binding protein LUSH, inducing a conformational change in LUSH and allowing it to bind to the odorant receptor complex and initiate sexual and social aggregation behavior (95, 96). SNMP is required for electrophysiologic responses to CVA; snmp-null cells show increased spontaneous electrical activity but totally lack induced activity in response to CVA-LUSH, suggesting perhaps that SNMP serves a regulatory function in signal transduction. Interestingly, more than a dozen CD36 orthologs have been identified in the genomes of insect species with functions related to pheromone signaling, innate immunity, phagocytic clearance of apoptotic cells, and various aspects of fatty acid metabolism. The ninaD and santa maria genes in Drosophila, for example, encode proteins involved in gut uptake of carotenoids and activation of β-carotene monooxygenase in the eye (97).
CD36 also has a role in chemical sensory responses in mammalian species. CD36 is abundant on the apical surface of about 12 to 15% of circumvallate taste bud cells in the lingular papillae (98). Exposure to long-chain fatty acids, such as linoleic acid, causes a CD36-dependent rise in intracellular calcium concentration in these cells associated with neurotransmitter release, activation of gustatory neurons in the brain, and a rapid and sustained increase in the flux and protein content of pancreatobiliary secretions (98–101). In rodents, this series of reactions results in preferential attraction to lipid-rich food and at the same time readies the gut for a fatty meal (98, 99). These traits are lost in cd36-null animals. Similar to the signaling pathways identified in endothelial cells, macrophages, and platelets, the CD36-mediated neurosensory response is associated with phosphorylation of specific Src family kinases (in this case, Fyn and Yes), and inhibition of these kinases blocks neurotransmitter release in response to linoleic acid (99). Interestingly, duodenal infusion of fat (bypassing the tongue) in rodents leads to mobilization of oleoylethanolamide (OEA), a fatty acid derivative that suppresses feeding behavior. OEA synthesis requires dietary oleic acid and is deficient in cd36-null animals (102). Thus, CD36 sensing of fatty acids in the gut provides negative feedback to the fat feeding– promoting activity of CD36 sensing in the tongue.
CD36 Organization, Membranes, Partners
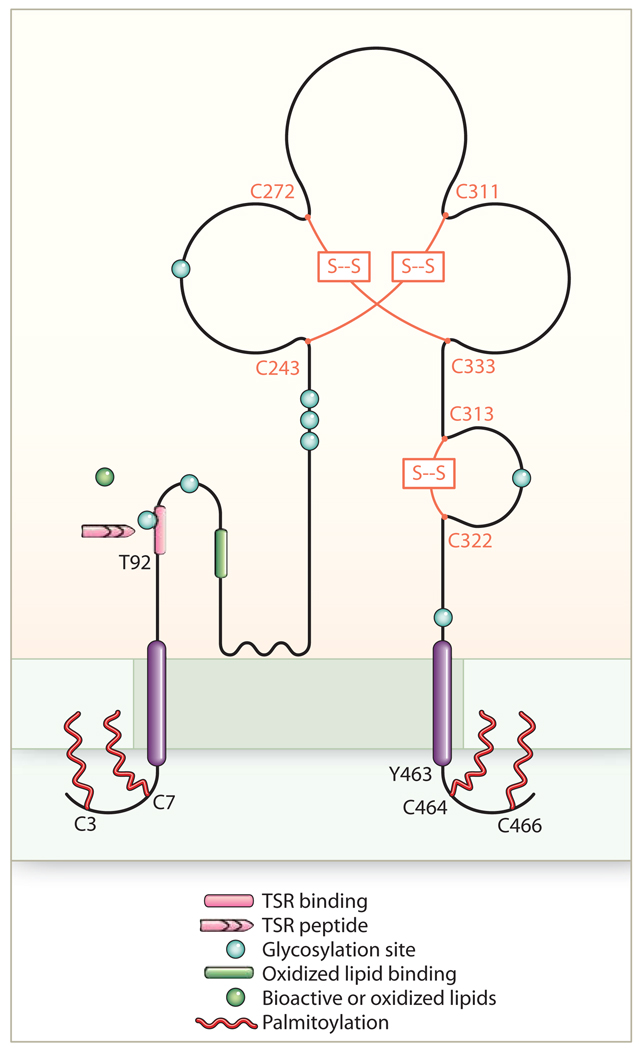
As summarized above, CD36-mediated biological responses to its varied array of ligands in numerous cells and tissues have relevance to multiple homeostatic, developmental, and pathological processes, including eating preferences and sexual behavior. Considerable interest is currently focused on the mechanisms by which CD36 transmits intracellular signals and thereby affects its many functions. CD36 has two transmembrane domains, short intracytoplasmic domains of five to seven and 11 to 13 amino acids and a large extracellular domain with six conserved cysteines linked in three disulfide bridges (3, 86) (Fig. 2). The extracellular domain includes a hydrophobic region between amino acids 184 and 204 that may potentially interact with the plasma membrane. The extracellular domain is heavily glycosylated, accounting for the observed molecular weight that is 30 to 40,000 daltons greater than the weight predicted from the cDNA. Studies of recombinant proteins expressed in insect cells suggest that 9 of the 10 asparagine residues in the extracellular domain are glycosylated and that glycosylation is necessary for correct trafficking to the plasma membrane (103). Both of the intracellular domains contain paired cysteine residues that are lipid acylated and thus probably tightly associated with the inner leaflet of the plasma membrane (104) (Fig. 2). CD36 can be ubiquitinated on lysines 469 and 472 of the C-terminal domain (105), and this can target it to lysosomes, thus regulating its abundance. Interestingly, CD36 ubiquitination was stimulated by increasing fatty acid concentrations and inhibited by insulin, suggesting potential physiologic regulation of CD36 activity by this pathway (105).
Fig. 2.
Topology and domains of CD36. This ditopic transmembrane receptor resides in lipid raft membrane domains (shown shaded) and may interact with other cell surface receptors, such as integrins, tetraspanins, and TLRs (not shown). Ligand recognition may be modulated by phosphorylation of CD36 at Thr (92) (T). The extracellular domain contains three disulfide bridges (Cys243- Cys311, Cys272-Cys333, and Cys313-Cys322), multiple glycosylation sites, and at least two separate ligand binding domains, one for proteins with thrombospondin repeat (TSR) domains and one for oxidized lipids. The N- and C-terminal tails contain paired palmitoylated cysteine residues. Tyr463 and Cys464 in the C-terminal tail are important for ligand binding and for interaction with downstream signaling molecules.
The extracellular domain mediates ligand recognition and contains independent binding sites for TSR peptides (106, 107) and oxidized phospholipids (108, 109); the structural basis for fatty acid binding remains largely unknown (5, 60). The mechanisms by which a receptor with minimal intracellular presence, no intrinsic kinase or phosphatase activity, no known intracellular scaffolding domain(s), and no direct link to GTPases activates multiple signaling pathways remain poorly understood but are under intense study. A common theme in CD36 signal transduction is activation of Src family kinases (110) and MAPKs. Antibodies to CD36 coprecipitate specific Src kinases and upstream MAPK kinases (MAPKKs) from lysates of different cell types, and incubation of cells with CD36 ligands, such as oxLDL, increases the amount of activated (phosphorylated) Src kinases in the precipitates (52, 54). These studies suggest that CD36 associates with and participates in assembly of a dynamic signaling complex essential to downstream functions. It is highly likely that the C-terminal cytoplasmic domain of CD36 directs these associations. Point mutations of specific tyrosine or cysteine residues (Y463 or C464) in this domain (53) result in loss of response to ligands, and a recombinant protein containing this cytoplasmic domain precipitated a multiprotein complex from monocytes that contained Lyn, a MAPKK, and several as-yet-unidentified proteins (52). Because CD36 resides in cholesterol-rich, detergent-insoluble lipid raft domains and copurifies with caveolae from some tissues (111–114), it is possible that CD36 signaling relates to its localization in these membrane regions in which signaling molecules, such as Src, accumulate. It is also possible that residence in these domains facilitates lipid transport through a docking mechanism and aid from associated membrane and cytoplasmic proteins. CD36 is phosphorylated on an extracellular threonine residue (115), which influences binding of several ligands (115, 116) and may effect internalization and signaling.
It is also likely that CD36 may effect signal transduction, in part, by interacting with other membrane receptors, such as integrins, tetraspanins (117), and TLRs. The latter was elegantly demonstrated in studies showing cooperation between CD36 and TLR2 or TLR6 in macrophage recognition and response to bacteria and bacterial cell wall components, such as Staphylococcus-derived lipoteichoic acid (LTA) and diacylated lipoproteins (18, 118, 119). In some studies, however, some components of the proinflammatory response and bacteria uptake were shown to be dependent on CD36-JNK signaling (120, 121) and did not require TLR-mediated activation of NF-κB. TLRs do not appear to be required for CD36-dependent uptake of oxLDL (52) or apoptotic cells. Several CD36 functions require integrins, and both β2 (28, 57) and β3 (117) integrins coimmunoprecipitated with CD36. Internalization of apoptotic cells and photoreceptor outer segments requires integrins–αvβ3 in macrophages (22) and αvβ5 in dendritic cells (24) and retinal pigment epithelia (122). Microglial responses to Aβ require β2 integrins, and the spreading of brain tumor cells on thrombospondin- 1 seems controlled by a functional interaction between β1 integrins and CD36 (123).
In summary, CD36-mediated signaling pathways are conserved, defined by certain common themes, and involved in many critical cellular processes, but still relatively poorly understood. Figure 1 outlines our current model. Careful dissection of pathways in the context of specific cells and ligands may yield novel insights for drug development of multiple disorders.
References and Notes
- 1.Clemetson KJ, Pfueller ST, Luscher EF, Jenkins CSP. Isolation of the membrane lycoproteins of human blood platelets by lection affinity chromatography. Biochim. Biophys. Acta. 1977;464:493–508. doi: 10.1016/0005-2736(77)90025-6. [DOI] [PubMed] [Google Scholar]
- 2.Knowles DM, Tolidijian B, Marboe C, Agati VD, Grimes M, Chess L. Monoclonal anti-human monocyte antibodies OKM1 and OKM5 possess distinctive tissue distributions including differential reactivity with vascular endothelium. J. Immunol. 1984;132:2170–2173. [PubMed] [Google Scholar]
- 3.Febbraio M, Hajjar DP, Silverstein RL. CD36: A class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Invest. 2001;108:785–791. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oquendo P, Hundt E, Lawler J, Seed B. CD36 directly mediates cytoadherence of Plasmodium falciparum parasitized erythrocytes. Cell. 1989;58:95–101. doi: 10.1016/0092-8674(89)90406-6. [DOI] [PubMed] [Google Scholar]
- 5.Calvo D, Dopazo J, Vega MA. The CD36, CLA-1 (CD36L1), and LIMPII (CD36L2) gene family: Cellular distribution, chromosomal location, and genetic evolution. Genomics. 1995;25:100–106. doi: 10.1016/0888-7543(95)80114-2. [DOI] [PubMed] [Google Scholar]
- 6.Hart K, Wilcox MA. Drosophila gene encoding an epithelial membrane protein with homology to CD36/LIMP II. J. Mol. Biol. 1993;234:249–253. doi: 10.1006/jmbi.1993.1580. [DOI] [PubMed] [Google Scholar]
- 7.Müller WEG, Thakur NL, Ushijima H, Thakur AN, Krasko A, Pennec GL, Indap MM, Perovi-Ottstadt P, Schröder HC, Lang G, Bringmann G. Matrix-mediated canal formation in primmorphs from the sponge Suberites domuncula involves the expression of a CD36 receptor-ligand system. J. Cell Sci. 2004;117:2579–2590. doi: 10.1242/jcs.01083. [DOI] [PubMed] [Google Scholar]
- 8.Asch AS, Barnwell J, Silverstein RL, Nachman RL. Isolation of the thrombospondin membrane receptor. J. Clin. Invest. 1987;79:1054–1061. doi: 10.1172/JCI112918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverstein RL, Baird M, Yesner L. Sense and anti-sense cDNA transfection of glycoprotein IV (CD36) in melanoma cells: Role of CD36 as a thrombospondin receptor. J. Biol. Chem. 1992;267:16607–16612. [PubMed] [Google Scholar]
- 10.Kaur B, Sandberg EM, Devi NS, Zhang S, Shim H, Mao H, Febbraio M, Klenotic P, Cork S, Silverstein RL, Crat DJ, Olson JJ, Van Meir EG. Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res. 2009;69:1212–1220. doi: 10.1158/0008-5472.CAN-08-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson DW, Pearce SFA, Zhong R, Silverstein RL, Frazier WA, Bouck NP. CD36 mediates the inhibitory effects of thrombospondin-1 on endothelial cells. J. Cell Biol. 1997;138:707–717. doi: 10.1083/jcb.138.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jimenez B, Volpert OV, Crawford SE, Febbraio M, Silverstein RL, Bouck NP. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat. Med. 2000;6:41–48. doi: 10.1038/71517. [DOI] [PubMed] [Google Scholar]
- 13.Volpert OV, Zaichuk T, Zhou W, Reiher F, Ferguson TA, Stuart PM, Amin M, Bouck MNP. Inducer-stimulated Fas targets activated endothelium for destruction by anti-angiogenic thrombospondin-1 and pigment epithelium-derived factor. Nat. Med. 2002;8:349–357. doi: 10.1038/nm0402-349. [DOI] [PubMed] [Google Scholar]
- 14.Rege TA, Stewart J, Jr., Dranka B, Benveniste EN, Silverstein RL, Gladson CL. Thrombospondin-1-induced apoptosis of brain microvascular endothelial cells can be mediated by TNF-R1. J. Cell. Physiol. 2009;218:94–103. doi: 10.1002/jcp.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endemann G, Stanton LW, Madden KS, Bryant CM, White RT, Protter AA. CD36 is a receptor for oxidised low density lipoprotein. J. Biol. Chem. 1993;268:11811–11816. [PubMed] [Google Scholar]
- 16.van Berkel TJ, Out R, Hoekstra M, Kuiper J, Biessen E, van Eck M. Scavenger receptors: Friend or foe in atherosclerosis? Curr Opin. Lipidol. 2005;16:525–535. doi: 10.1097/01.mol.0000183943.20277.26. [DOI] [PubMed] [Google Scholar]
- 17.Akira S, Takeda K. Toll-like receptor signaling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 18.Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, Crozat K, Sovath S, Shamel L, Hartung T, Zahringer U, Beutler B. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 19.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–1253. doi: 10.1126/science.1116006. published online 14 July 2005 (10.1126/science. 1116006) [DOI] [PubMed] [Google Scholar]
- 20.Means TK, Mylonakis E, Tampakakis E, Colvin RA, Seung E, Puckett L, Tai MF, Stewart CR, Pukkila-Worley R, Hickman SE, Moore KJ, Calderwood SB, Hacohen N, Luster AD, El Khoury J. Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARF1 and CD36. J. Exp. Med. 2009;206:637–653. doi: 10.1084/jem.20082109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith TG, Serghides L, Patel S, Febbraio M, Silverstein RL, Kain KC. CD36-mediated non-opsonic phagocytosis of erythrocytes infected with stage I and IIA gametocyes of Plasmodium falciparum. Infect. Immun. 2003;71:393–400. doi: 10.1128/IAI.71.1.393-400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savill J, Hogg N. Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J. Clin. Invest. 1992;90:1513–1522. doi: 10.1172/JCI116019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren Y, Silverstein RL, Allen J, Savill J. CD36 gene transfer confers capacity for phagocytosis of cells undergoing apoptosis. J. Exp. Med. 1995;181:1857–1862. doi: 10.1084/jem.181.5.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albert ML, Pearce SFA, Francisco L, Sauter B, Silverstein RL, Bhardwaj N. Immature dendritic cells phagocytose apoptotic cells via αvβ5 and CD36, and cross-present antigens to CTLs. J. Exp. Med. 1998;188:1359–1368. doi: 10.1084/jem.188.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryeom SW, Sparrow JR, Silverstein RL. CD36 participates in the phagocytosis of rod outer segments on retinal pigment epithelium. J. Cell Sci. 1996;109:387–395. doi: 10.1242/jcs.109.2.387. [DOI] [PubMed] [Google Scholar]
- 26.Sun M, Finnemann SC, Febbraio M, Shan L, Annangudi SP, Podrez EA, Hoppe G, Darrow R, Organisciak DT, Salomon RG, Silverstein RL, Hazen SL. Light-induced oxidation of photoreceptor outer segment phospholipids generates ligands for CD36-mediated phagocytosis by retinal pigment epithelium: A potential mechanism for modulating outer segment phagocytosis under oxidant stress condition. J. Biol. Chem. 2006;281:4222–4230. doi: 10.1074/jbc.M509769200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podrez EA, Febbraio M, Sheibani N, Schmitt D, Silvertein RL, Hajjar DP, Cohen PA, Frazier WA, Hoff HF, Hazen SL. The macrophage scavenger receptor CD36 is the major receptor for LDL modified by monocyte-generated reactive nitrogen species. J. Clin. Invest. 2000;105:1095–1108. doi: 10.1172/JCI8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bamberger ME, Harris ME, McDonald DR, Husemann J, Landreth GE. A cell surface receptor complex for fibrillar β-amyloid mediates microglial activation. J. Neurosci. 2003;23:2665–2674. doi: 10.1523/JNEUROSCI.23-07-02665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Khoury JB, Moore KJ, Means TK, Leung J, Terada K, Toft M, Freeman MW, Luster AD. CD36 mediates the innate host response to β-amyloid. J. Exp. Med. 2003;197:1657–1666. doi: 10.1084/jem.20021546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity. 1996;4:431–443. doi: 10.1016/s1074-7613(00)80410-0. [DOI] [PubMed] [Google Scholar]
- 31.Franc NC, Heitzler P, Ezekowitz RAB, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284:1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- 32.Febbraio M, Podrez EA, Smith JD, Hazen SL, Hoff HF, Sharma K, Hajjar DP, Silverstein RL. Targeted disruption of the class B scavenger receptor, CD36, protects against atherosclerotic lesion development in mice. J. Clin. Invest. 2000;105:1049–1056. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunjathoor VV, Febbraio M, Podrez EA, Moore KJ, Andersson L, Koehn S, Rhee JS, Silverstein R, Hoff HH, Freeman MV. Scavenger receptors class A-I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J. Biol. Chem. 2002;277:49982–49988. doi: 10.1074/jbc.M209649200. [DOI] [PubMed] [Google Scholar]
- 34.Podrez EA, Batyreva E, Shen Z, Zang R, Deng Y, Sun M, Finton PJ, Shen L, Febbraio M, Hajjar DP, Silverstein RL, Hoff HF, Salomon RG, Hazen SL. A novel family of atherogenic oxidized phospholipids promotes macrophage foam cell formation via the scavenger receptor CD36 and is enriched in atherosclerotic lesions. J. Biol. Chem. 2002;277:38517–38523. doi: 10.1074/jbc.M205924200. [DOI] [PubMed] [Google Scholar]
- 35.Janabi M, Yamashita S, Hirano K, Sakai N, Hiraoka H, Matsumoto K, Zhang Z, Nozaki S, Matsuzawa Y. Oxidized LDL-induced NF-κB activation and subsequent expression of proinflammatory genes are defective in monocyte-derived macrophages from CD36-deficient patients. Arterioscler. Thromb. Vasc. Biol. 2000;20:1953–1960. doi: 10.1161/01.atv.20.8.1953. [DOI] [PubMed] [Google Scholar]
- 36.Martin-Fuentes P, Civeira F, Recalde D, Garcia-Otin AL, Jarauta E, Marzo I, Cenarro A. Individual variation of scavenger receptor expression in human macrophages with oxidized low-density lipoprotein is associated with a differential inflammatory response. J. Immunol. 2007;179:3242–3248. doi: 10.4049/jimmunol.179.5.3242. [DOI] [PubMed] [Google Scholar]
- 37.Cho S, Park EM, Febbraio M, Anrather J, Park L, Racchumi G, Silverstein RL, Iadecola C. The class B scavenger receptor CD36 mediates free radical production and tissue injury in cerebral ischemia. J. Neurosci. 2005;25:2504–2512. doi: 10.1523/JNEUROSCI.0035-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park YM, Febbraio M, Silverstein RL. CD36 modulates migration of mouse and human macrophages in response to oxidized LDL and contributes to macrophage trapping in the arterial intima. J. Clin. Invest. 2009;119:136–145. doi: 10.1172/JCI35535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harb D, Bujold K, Febbraio M, Sirois MG, Ong H, Marleau S. The role of the scavenger receptor CD36 in regulating mononuclear phagocyte trafficking to atherosclerotic lesions and vascular inflammation. Cardiovasc. Res. doi: 10.1093/cvr/cvp081. published online 26 March 2009 (10.1093/ cvr/cvp081) [DOI] [PubMed] [Google Scholar]
- 40.Curtiss LK. Reversing atherosclerosis? N. Engl. J. Med. 2009;360:1144–1146. doi: 10.1056/NEJMcibr0810383. [DOI] [PubMed] [Google Scholar]
- 41.Coraci IS, Husemann J, Berman JW, Hulette C, Dufour JH, Campanella GK, Luster AD, Silverstein SC, El-Khoury JB. CD36, a class B scavenger receptor, is expressed on microglia in Alzheimer’s disease brains and can mediate production of reactive oxygen species in response to β-amyloid fibrils. Am. J. Pathol. 2002;160:101–112. doi: 10.1016/s0002-9440(10)64354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore KJ, El Khoury J, Medeiros LA, Terada K, Geula C, Luster AD, Freeman MW. A CD36-initiated signaling cascade mediates inflammatory effects of β-amyloid. J. Biol. Chem. 2002;277:47373–47379. doi: 10.1074/jbc.M208788200. [DOI] [PubMed] [Google Scholar]
- 43.Podrez EA, Byzova TV, Febbraio M, Salomon RG, Ma Y, Valiyaveettil M, Poliakov E, Sun M, Finton PJ, Curtis BR, Chen J, Zhang R, Silverstein RL, Hazen SL. Platelet CD36 links hyperlipidemia, oxidant stress and a pro-thrombotic phenotype. Nat. Med. 2007;13:1086–1095. doi: 10.1038/nm1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh A, Li W, Febbraio M, Espinola RG, McCrae K, Silverstein RL. Platelet CD36 mediates interactions with endothelial cell-derived microparticles and contributes to thrombosis in vivo. J. Clin. Investig. 2008;118:1934–1943. doi: 10.1172/JCI34904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helming L, Winter J, Gordon S. The scavenger receptor CD36 plays a role in cytokine-induced macrophage fusion. J. Cell Sci. 2009;22:453–459. doi: 10.1242/jcs.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore KJ, Kunjathoor VV, Koehn SL, Manning JJ, Tseng AA, Silver JM, Mc-Kee M, Freeman MW. Loss of receptor-mediated lipid uptake via scavenger receptor A or CD36 pathways does not ameliorate atherosclerosis in hyperlipidemic mice. J. Clin. Invest. 2005;115:2192–2201. doi: 10.1172/JCI24061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore KJ, Freeman MW. Scavenger receptors in atherosclerosis: Beyond lipid uptake. Arterioscler. Thromb. Vasc. Biol. 2006;26:1702–1711. doi: 10.1161/01.ATV.0000229218.97976.43. [DOI] [PubMed] [Google Scholar]
- 48.Manning-Tobin JJ, Moore KJ, Seimon TA, Bell SA, Sharuk M, Alvarez-Leite JI, de Winther MP, Tabas I, Freeman MW. Loss of SR-A and CD36 activity reduces atherosclerotic lesion complexity without abrogating foam cell formation in hyperlipidemic mice. Arterioscler. Thromb. Vasc. Biol. 2009;29:19–26. doi: 10.1161/ATVBAHA.108.176644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Febbraio M, Guy E, Silverstein RL. Stem cell transplantation reveals that absence of macrophage CD36 is protective against atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2004;24:2333–2338. doi: 10.1161/01.ATV.0000148007.06370.68. [DOI] [PubMed] [Google Scholar]
- 50.Guy E, Kuchibhotla S, Silverstein R, Febbraio M. Continued inhibition of atherosclerotic lesion development in long term Western diet fed CD36o /apoEo mice. Atherosclerosis. 2007;192:123–130. doi: 10.1016/j.atherosclerosis.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 51.Kuchibhotla S, Vanegas D, Kennedy DJ, Guy E, Nimako G, Morton RE, Febbraio M. Absence of CD36 protects against atherosclerosis in ApoE knock-out mice with no additional protection provided by absence of scavenger receptor A I/II. Cardiovasc. Res. 2008;78:185–196. doi: 10.1093/cvr/cvm093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rahaman SO, Lennon DJ, Febbraio M, Podrez EA, Hazen SL, Silverstein RL. CD36-dependent activation of c-Jun N-terminal kinase by oxidized LDL is required for macrophage foam cell formation. Cell Metab. 2006;4:211–221. doi: 10.1016/j.cmet.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart LM, Deng J, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, Ezekowitz RA, Moore KJ. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J. Cell Biol. 2005;170:477–485. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen K, Febbraio M, Li W, Silverstein RL. A specific CD36-dependent signaling pathway is required for platelet activation by oxidized LDL. Circ. Res. 2008;102:1512–1519. doi: 10.1161/CIRCRESAHA.108.172064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDonald DR, Brunden KR, Landreth GE. Amyloid fibrils activate tyrosine kinase-dependent signaling and superoxide production in microglia. J. Neurosci. 1997;17:2284–2294. doi: 10.1523/JNEUROSCI.17-07-02284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart LM, Bell SA, Stewart CR, Silver JM, Richard J, Goss JL, Tseng AA, Zhang A, El Khoury JB, Moore KJ. CD36 signals to the actin cytoskeleton and regulates microglial migration via a p130Cas complex. J. Biol. Chem. 2007;282:27392–27401. doi: 10.1074/jbc.M702887200. [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson B, Koenigsknecht-Talboo J, Grommes C, Lee CY, Landreth G. Fibrillar β-amyloid-stimulated intracellular signaling cascades require Vav for induction of respiratory burst and phagocytosis in monocytes and microglia. J. Biol. Chem. 2006;281:20842–20850. doi: 10.1074/jbc.M600627200. [DOI] [PubMed] [Google Scholar]
- 58.Han J, Hajjar DP, Febbraio M, Nicholson AC. Native and modified low density lipoproteins increase the functional expression of the macrophage class B scavenger receptor, CD36. J. Biol. Chem. 1997;272:21654–21659. doi: 10.1074/jbc.272.34.21654. [DOI] [PubMed] [Google Scholar]
- 59.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 60.Abumrad NA, el-Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J. Biol. Chem. 1993;268:17665–17668. [PubMed] [Google Scholar]
- 61.Zhou J, Febbraio M, Zhai Y, Kuruba R, Wada T, Khadem S, Ren S, Li S, Silverstein RL, Xie W. LXR, PXR, and PPARγ cooperate in regulating fatty acid transporter CD36 and promoting hepatic lipogenesis. Gastroenterology. 2008;134:556–567. doi: 10.1053/j.gastro.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 62.Coburn CT, Knapp FF, Jr., Febbraio M, Beets AL, Silverstein RL, Abumrad NA. Defective uptake and utilization of long-chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J. Biol. Chem. 2000;275:32523–32529. doi: 10.1074/jbc.M003826200. [DOI] [PubMed] [Google Scholar]
- 63.Drover VA, Nguyen DV, Bastie CC, Darlington YF, Abumrad NA, Pessin JE, London E, Sahoo D, Phillips MC. CD36 mediates both cellular uptake of very long chain fatty acids and their intestinal absorption in mice. J. Biol. Chem. 2008;283:13108–13115. doi: 10.1074/jbc.M708086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nassir F, Wilson B, Han X, Gross RW, Abumrad NA. CD36 is important for fatty acid and cholesterol uptake by the proximal but not distal intestine. J. Biol. Chem. 2007;282:19493–19501. doi: 10.1074/jbc.M703330200. [DOI] [PubMed] [Google Scholar]
- 65.Baillie AG, Coburn CT, Abumrad NA. Reversible binding of long-chain fatty acids to purified FAT, the adipose CD36 homolog. J. Membr. Biol. 1996;153:75–81. doi: 10.1007/s002329900111. [DOI] [PubMed] [Google Scholar]
- 66.Bonen A, Dyck DJ, Ibrahimi A, Abumrad NA. Muscle contractile activity increases fatty acid metabolism and transport and FAT/CD36. Am. J. Physiol. 1999;276:E642–E649. doi: 10.1152/ajpendo.1999.276.4.E642. [DOI] [PubMed] [Google Scholar]
- 67.Su X, Abumrad NA. Cellular fatty acid uptake: A pathway under construction. Trends Endocrinol. Metab. 2009;20:72–77. doi: 10.1016/j.tem.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nickerson JG, Momken I, Benton CR, Lally J, Holloway GP, Han XX, Glatz JF, Chabowski A, Luiken JJ, Bonen A. Protein-mediated fatty acid uptake: Regulation by contraction, AMP-activated protein kinase, and endocrine signals. Appl. Physiol. Nutr. Metab. 2007;32:865–873. doi: 10.1139/H07-084. [DOI] [PubMed] [Google Scholar]
- 69.Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, Pearce SFA, Silverstein RL. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J. Biol. Chem. 1999;274:19055–19062. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 70.Glazier AM, Scott J, Aitman TJ. Molecular basis of the CD36 chromosomal deletion underlying SHR defects in insulin action and fatty acid metabolism. Mamm. Genome. 2002;13:108–113. doi: 10.1007/s00335-001-2132-9. [DOI] [PubMed] [Google Scholar]
- 71.Miyaoka K, Kuwasako T, Hirano K, Nozaki S, Yamashita S, Matsuzawa Y. CD36 deficiency associated with insulin resistance. Lancet. 2001;357:686–687. doi: 10.1016/s0140-6736(00)04138-6. [DOI] [PubMed] [Google Scholar]
- 72.Corpeleijn E, van der Kallen CJ, Kruijshoop M, Magagnin MG, de Bruin TW, Feskens EJ, Saris WH, Blaak EE. Direct association of a promoter polymorphism in the CD36/FAT fatty acid transporter gene with type 2 diabetes mellitus and insulin resistance. Diabet. Med. 2006;23:907–911. doi: 10.1111/j.1464-5491.2006.01888.x. [DOI] [PubMed] [Google Scholar]
- 73.Griffin E, Re A, Hamel N, Fu C, Bush H, McCaffrey T, Asch AS. A link between diabetes and atherosclerosis: Glucose regulates expression of CD36 at the level of translation. Nat. Med. 2001;7:840–846. doi: 10.1038/89969. [DOI] [PubMed] [Google Scholar]
- 74.Liang CP, Han S, Okamoto H, Carnemolla R, Tabas I, Accili D, Tall AR. Increased CD36 protein as a response to defective insulin signaling in macrophages. J. Clin. Invest. 2004;113:764–773. doi: 10.1172/JCI19528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greco D, Kotronen A, Westerbacka J, Puig O, Arkkila P, Kiviluoto T, Laitinen S, Kolak M, Fisher RM, Hamsten A, Auvinen P, Yki-Järvinen H. Gene expression in human NAFLD. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;294:G1281–G1287. doi: 10.1152/ajpgi.00074.2008. [DOI] [PubMed] [Google Scholar]
- 76.Koonen DP, Febbraio M, Bonnet S, Nagendran J, Young ME, Michelakis ED, Dyck JR. CD36 expression contributes to age-induced cardiomyopathy in mice. Circulation. 2007;116:2139–2147. doi: 10.1161/CIRCULATIONAHA.107.712901. [DOI] [PubMed] [Google Scholar]
- 77.Yang J, Sambandam N, Han X, Gross RW, Courtois M, Kovacs A, Febbraio M, Finck BN, Kelly DP. CD36 deficiency rescues lipotoxic cardiomyopathy. Circ. Res. 2007;100:1208–1217. doi: 10.1161/01.RES.0000264104.25265.b6. [DOI] [PubMed] [Google Scholar]
- 78.Koonen DP, Jacobs RL, Febbraio M, Young ME, Soltys CL, Ong H, Vance DE, Dyck JR. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes. 2007;56:2863–2871. doi: 10.2337/db07-0907. [DOI] [PubMed] [Google Scholar]
- 79.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, St. Lezin E, Kurtz RW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat. Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 80.Pravenec M, Landa V, Zídek V, Musilová A, Kazdová L, Qi N, Wang J, St Lezin E, Kurtz TW. Transgenic expression of CD36 in the spontaneously hypertensive rat is associated with amelioration of metabolic disturbances but has no effect on hypertension. Physiol. Res. 2003;52:681–688. [PubMed] [Google Scholar]
- 81.Bonen A, Han XX, Tandon NN, Glatz JF, Lally J, Snook L, Luiken JJ. FAT/CD36 expression is not ablated in spontaneously hypertensive rats. J. Lipid Res. 2009;50:740–748. doi: 10.1194/jlr.M800237-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kajihara S, Hisatomi A, Ogawa Y, Yasutake T, Yoshimura T, Hara T, Mizuta T, Ozaki I, Iwamoto N, Yamamoto K. Association of the Pro90Ser CD36 mutation with elevated free fatty acid concentrations but not with insulin resistance syndrome in Japanese. Clin. Chim. Acta. 2001;314:125–130. doi: 10.1016/s0009-8981(01)00658-1. [DOI] [PubMed] [Google Scholar]
- 83.Furuhashi M, Ura N, Nakata T, Shimamoto K. Insulin sensitivity and lipid metabolism in human CD36 deficiency. Diabetes Care. 2003;26:471–474. doi: 10.2337/diacare.26.2.471. [DOI] [PubMed] [Google Scholar]
- 84.Yanai H, Chiba H, Morimoto M, Jamieson GA, Matsuno K. Type I CD36 deficiency in humans is not associated with insulin resistance syndrome. Thromb. Haemost. 2000;83:786. [PubMed] [Google Scholar]
- 85.Love-Gregory L, Sherva R, Sun L, Wasson J, Schappe T, Doria A, Rao DC, Hunt SC, Klein S, Neuman RJ, Permutt MA, Abumrad NA. Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum. Mol. Genet. 2008;17:1695–1704. doi: 10.1093/hmg/ddn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rać ME, Safranow K, Poncyljusz W. Molecular basis of human CD36 gene mutations. Mol. Med. 2007;13:288–296. doi: 10.2119/2006-00088.Rac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goudriaan JR, Dahlmans VE, Teusink B, Ouwens DM, Febbraio M, Maassen JA, Romijn JA, Havekes LM, Voshol PJ. CD36 deficiency increases insulin sensitivity in muscle, but induces insulin resistance in the liver in mice. J. Lipid Res. 2003;44:2270–2277. doi: 10.1194/jlr.M300143-JLR200. [DOI] [PubMed] [Google Scholar]
- 88.Voolstra O, Kiefer C, Hoehne M, Welsch R, Vogt K, von Lintig J. The Drosophila class B scavenger receptor NinaD-I is a cell surface receptor mediating carotenoid transport for visual chromophore synthesis. Biochemistry. 2006;45:13429–13437. doi: 10.1021/bi060701u. [DOI] [PubMed] [Google Scholar]
- 89.van Bennekum A, Werder M, Thuahnai ST, Han CH, Duong P, Williams DL, Wettstein P, Schulthess G, Phillips MC, Hauser H. Class B scavenger receptor-mediated intestinal absorption of dietary beta-carotene and cholesterol. Biochemistry. 2005;44:4517–4525. doi: 10.1021/bi0484320. [DOI] [PubMed] [Google Scholar]
- 90.Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-B1 as a high density lipoprotein receptor. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 91.Nichols Z, Vogt RG. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 2008;38:398–415. doi: 10.1016/j.ibmb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 92.Forstner M, Gohl T, Gondesen I, Raming K, Breer H, Krieger J. Differential expression of SNMP-1 and SNMP-2 proteins in pheromone-sensitive hairs of moths. Chem. Senses. 2008;33:291–299. doi: 10.1093/chemse/bjm087. [DOI] [PubMed] [Google Scholar]
- 93.Jin X, Ha TS, Smith DP. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10996–11001. doi: 10.1073/pnas.0803309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- 95.Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stowers L, Logan DW. LUSH shapes up for a starring role in olfaction. Cell. 2008;133:1137–1139. doi: 10.1016/j.cell.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 97.Wang T, Jiao Y, Montell C. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J. Cell Biol. 2007;177:305–316. doi: 10.1083/jcb.200610081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Laugerette F, Passilly-Degrace P, Patris B, Niot I, Febbraio M, Montmayeur JP, Besnard P. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J. Clin. Invest. 2005;115:3177–3184. doi: 10.1172/JCI25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.El-Yassimi A, Hichami A, Besnard P, Khan NA. Linoleic acid induces calcium signaling, Src kinase phosphorylation, and neurotransmitter release in mouse CD36-positive gustatory cells. J. Biol. Chem. 2008;283:12949–12959. doi: 10.1074/jbc.M707478200. [DOI] [PubMed] [Google Scholar]
- 100.Sclafani A, Ackroff K, Abumrad NA. CD36 gene deletion reduces fat preference and intake but not post-oral fat conditioning in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;293:R1823–R1832. doi: 10.1152/ajpregu.00211.2007. [DOI] [PubMed] [Google Scholar]
- 101.Gaillard D, Laugerette F, Darcel N, El-Yassimi A, Passilly-Degrace P, Hichami A, Khan NA, Montmayeur JP, Besnard P. The gustatory pathway is involved in CD36-mediated orosensory perception of long-chain fatty acids in the mouse. FASEB J. 2008;22:1458–1468. doi: 10.1096/fj.07-8415com. [DOI] [PubMed] [Google Scholar]
- 102.Schwartz GJ, Fu J, Astarita G, Li X, Gaetani S, Campolongo P, Cuomo V, Piomelli D. The lipid messenger OEA links dietary fat intake to satiety. Cell Metab. 2008;8:281–288. doi: 10.1016/j.cmet.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoosdally SJ, Andress EJ, Wooding C, Martin CA, Linton KJ. The human scavenger receptor CD36: Glycosylation status and its role in trafficking and function. J. Biol. Chem. 2009 April 15; doi: 10.1074/jbc.M109.007849. published online (10.1074/jbc.M109.007849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tao N, Wagner SJ, Lublin DM. CD36 is palmitoylated on both N- and C-terminal cytoplasmic tails. J. Biol. Chem. 1996;271:22315–22320. doi: 10.1074/jbc.271.37.22315. [DOI] [PubMed] [Google Scholar]
- 105.Smith J, Su X, El-Maghrabi R, Stahl PD, Abumrad NA. Opposite regulation of CD36 ubiquitination by fatty acids and insulin: Effects on fatty acid uptake. J. Biol. Chem. 2008;283:13578–13585. doi: 10.1074/jbc.M800008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pearce SFA, Wu J, Silverstein RL. Recombinant GST/CD36 fusion proteins define a thrombospondin binding domain: Evidence for a single calcium-dependent binding site on CD36. J. Biol. Chem. 1995;270:2981–2986. doi: 10.1074/jbc.270.7.2981. [DOI] [PubMed] [Google Scholar]
- 107.Leung LL, Li WX, McGregor JL, Albrecht G, Howard RJ. CD36 peptides enhance or inhibit CD36-thrombospondin binding. A two-step process of ligand-receptor interaction. J. Biol. Chem. 1992;267:18244–18250. [PubMed] [Google Scholar]
- 108.Pearce SFA, Roy P, Febbraio M, Nicholson AC, Hajjar DP, Silverstein RL. Recombinant GST/CD36 fusion proteins define an oxidized LDL binding domain. J. Biol. Chem. 1998;273:34875–34881. doi: 10.1074/jbc.273.52.34875. [DOI] [PubMed] [Google Scholar]
- 109.Kar NS, Ashraf MZ, Valiyaveettil M, Podrez EA. Mapping and characterization of the binding site for specific oxidized phospholipids and oxidized low density lipoprotein of scavenger receptor CD36. J. Biol. Chem. 2008;283:8765–8771. doi: 10.1074/jbc.M709195200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang MM, Bolen JB, Barnwell JW, Shattil SJ, Brugge JS. Membrane glycoprotein IV (CD36) is physically associated with the Fyn, Lyn, and Yes protein-tyrosine kinases in human platelets. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7844–7848. doi: 10.1073/pnas.88.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lisanti MP, Scherer PE, Vidugiriene J, Tang Z, Hermanowski-Vosatka A, Tu YH, Cook RF, Sargiacomo M. Characterization of caveolin-rich membrane domains isolated from an endothelial-rich source: Implications for human disease. J. Cell Biol. 1994;126:111–126. doi: 10.1083/jcb.126.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pohl J, Ring A, Korkmaz U, Ehehalt R, Stremmel W. FAT/CD36-mediated long-chain fatty acid uptake in adipocytes requires plasma membrane rafts. Mol. Biol. Cell. 2005;16:24–31. doi: 10.1091/mbc.E04-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zeng Y, Tao N, Chung KN, Heuser JE, Lublin DM. Endocytosis of oxidized low density lipoprotein through scavenger receptor CD36 utilizes a lipid raft pathway that does not require caveolin-1. J. Biol. Chem. 2003;278:45931–45936. doi: 10.1074/jbc.M307722200. [DOI] [PubMed] [Google Scholar]
- 114.Kincer JF, Uittenbogaard A, Dressman J, Guerin TM, Febbraio M, Guo L, Smart EJ. Hypercholesterolemia promotes a CD36-dependent and endothelial nitric-oxide synthase-mediated vascular dysfunction. J. Biol. Chem. 2002;277:23525–23533. doi: 10.1074/jbc.M202465200. [DOI] [PubMed] [Google Scholar]
- 115.Asch AS, Liu I, Briccetti FM, Barnwell JW, Kwakye-Berko F, Dokun A, Goldberger J, Pernambuco M. Analysis of CD36 binding domains: Ligand specificity controlled by dephosphorylation of an ectodomain. Science. 1993;262:1436–1440. doi: 10.1126/science.7504322. [DOI] [PubMed] [Google Scholar]
- 116.Ho M, Hoang HL, Lee KM, Liu N, MacRae T, Montes L, Flatt CL, Yipp BG, Berger BJ, Looareesuwan S, Robbins SM. Ectophosphorylation of CD36 regulates cytoadherence of Plasmodium falciparum to microvascular endothelium under flow conditions. Infect. Immun. 2005;73:8179–8187. doi: 10.1128/IAI.73.12.8179-8187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miao WM, Vasile E, Lane WS, Lawler J. CD36 associates with CD9 and integrins on human blood platelets. Blood. 2001;97:1689–1696. doi: 10.1182/blood.v97.6.1689. [DOI] [PubMed] [Google Scholar]
- 118.Hashimoto M, Tawaratsumida K, Kariya H, Kiyohara A, Suda Y, Krikae F, Kirikae T, Gotz F. Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J. Immunol. 2006;177:3162–3169. doi: 10.4049/jimmunol.177.5.3162. [DOI] [PubMed] [Google Scholar]
- 119.Triantafilou M, Gamper FG, Haston RM, Mouratis MA, Morath S, Hartung T, Triantafilou K. Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J. Biol. Chem. 2006;281:31002–31011. doi: 10.1074/jbc.M602794200. [DOI] [PubMed] [Google Scholar]
- 120.Nilsen NJ, Deininger S, Nonstad U, Skjeldal F, Husebye H, Rodionov D, von Aulock S, Hartung T, Lien E, Bakke O, Espevik T. Cellular trafficking of lipoteichoic acid and Toll-like receptor 2 in relation to signaling: Role of CD14 and CD36. J. Leukoc. Biol. 2008;84:280–291. doi: 10.1189/jlb.0907656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baranova IN, Kurlander R, Bocharov AV, Vishnyakova TG, Chen Z, Remaley AT, Csako G, Patterson AP, Eggerman TL. Role of human CD36 in bacterial recognition, phagocytosis, and pathogen-induced JNK-mediated signaling. J. Immunol. 2008;181:7147–7156. doi: 10.4049/jimmunol.181.10.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Finnemann SC, Silverstein RL. Differential roles of CD36 and αvβ5 integrin in photoreceptor phagocytosis by the retinal pigment epithelium. J. Exp. Med. 2001;194:1289–1298. doi: 10.1084/jem.194.9.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pijuan-Thompson V, Grammar JR, Stewart J, Silverstein RL, Pearce SF, Tuszynski G, Murphy-Ullrich JE, Gladson CL. Retenoic acid alters the mechanism of attachment of malignant astrocytoma and neuroblastoma cells to thrombospondin-1. Exp. Cell Res. 1999;249:86–101. doi: 10.1006/excr.1999.4458. [DOI] [PubMed] [Google Scholar]
- 124.This work was supported in part by NIH grants HL81011, HL87018, and HL72942 to R.L.S. and M.F.; HL85718 to R.L.S.; and HL082511 and HL70083 to M.F.; and by an American Heart Association Grant in Aid to M.F.