SUMMARY
The translational machinery of the cell re-localizes to focal adhesions following the activation of integrin receptors. This response allows for rapid, local production of components needed for adhesion complex assembly and signaling. Vinculin links focal adhesions to the actin cytoskeleton following its activation by integrin signaling, which severs intramolecular interactions of the vinculin head and tail (Vt) domains. Our vinculin:raver1 crystal structures and binding studies show that activated Vt selectively interacts with one of the three RNA recognition motifs (RRM) of raver1, that the vinculin:raver1 complex binds to F-actin, and that raver1 binds selectively to RNA, including a sequence found in vinculin mRNA. Further, mutation of residues that mediate interaction of raver1 with vinculin abolish their co-localization in cells. These findings suggest a feed-forward model where vinculin activation at focal adhesions provides a scaffold for recruitment of raver1 and its mRNA cargo to facilitate the production of components of adhesion complexes.
Keywords: focal adhesion, actin cytoskeleton, crystallography, RNP motif, RNA binding
INTRODUCTION
Vinculin links integrin-mediated focal adhesion junctions to the actin cytoskeleton and is essential for the formation of proper cell-matrix contacts as well as for directed cell migration (DeMali and Burridge, 2003; Ziegler et al., 2006). Vinculin is held in an inactive, closed conformation through extensive hydrophobic interactions of its N-terminal seven-helix bundle domain (Vh1; Figure 1) in its globular “head” to a five-helix bundle domain present in its tail (Vt) domain (Bakolitsa et al., 2004; Borgon et al., 2004; Izard et al., 2004). Severing this interaction is sufficient to activate vinculin (Johnson and Craig, 1995; Gilmore and Burridge, 1996; Izard et al., 2004; Izard and Vonrhein, 2004; Bois et al., 2005; Fillingham et al., 2005; Bois et al., 2006; Nhieu and Izard, 2007), and can be triggered by the binding of the α-helical vinculin binding sites (VBS) of talin or α-actinin, which displace Vt from a distance by inducing profound conformational changes in the structure of Vh1 (Izard et al., 2004; Bois et al., 2005). This then allows interactions with other partners, including binding of Arp2/3, VASP, and vinexin to the proline-rich loop region that connects the head and tail domains of vinculin, and the binding of F-actin, paxillin, phosphatidylinositol (4,5)-bisphosphate (PIP2), and the hnRNP protein raver1 to the Vt domain (Johnson and Craig, 1995; Gilmore and Burridge, 1996; Hüttelmaier et al., 2001; Chandrasekar et al., 2005; Bois et al., 2006; Ziegler et al., 2006).
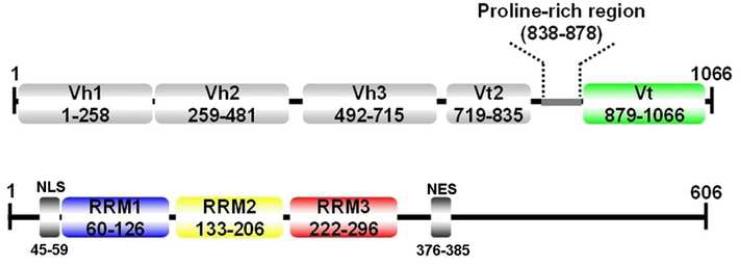
Figure 1.
Domain Organization of human Vinculin (top) and human Raver1 (bottom). Vinculin domains Vh1, Vh2, Vh3, and Vt2 together comprise the vinculin head domain VH. NES, nuclear export sequence; NLS, nuclear localization signal.
The interaction of raver1 with vinculin that occurs at adhesion sites (Hüttelmaier et al., 2001) is particularly intriguing since raver1 harbors three tandem N-terminal RNA recognition motifs (RRM1-3; Figure 1) and has an alternative role as a regulator of mRNA processing, where it interacts with the splicing regulator polypyrmidine-tract binding protein PTB (Gromak et al., 2003; Spellman et al., 2005; Rideau et al., 2006). RRM domains generally harbor two ribonucleoprotein consensus sequences on their central β-strands (β3 and β1, coined RNP1 and RNP2, respectively) that direct interactions with RNA, yet affinity and specificity can also be affected by adjacent N- or C-terminal extensions and loops present in these and adjacent RRM motifs. However, RRM domains also serve as docking sites for protein-protein interaction (reviewed by Kielkopf et al., 2004; Maris et al., 2005; Lunde et al., 2007; Cléry et al., 2008), and we therefore reasoned that one or more of the RRM motifs of raver1 might mediate its interaction with the Vt domain of activated vinculin (Hüttelmaier et al., 2001).
Here we present the first crystal structure of the key functional C-terminal domain of activated vinculin in complex with one of its partners, raver1. Our vinculin:raver1 crystal structures explain why vinculin activation is necessary for this interaction, identify a new mechanism for vinculin interaction with its partners, as well as a new mode for protein-protein interactions directed by an RRM motif. Further, we show that residues of raver1 that mediate its contact with vinculin are also required for raver1 and vinculin co-localization in cells. Finally, our studies establish that the RRM1 motif of raver1 binds to a select RNA sequence that is found in vinculin mRNA, and that the vinculin-raver1 complex is permissive for binding to F-actin. Collectively these findings suggest that activated vinculin functions as a scaffold for the recruitment of raver1 and its mRNAs cargo to focal adhesions, a scenario that would promote localized synthesis of adhesion complexes by the translational machinery.
RESULTS
The RRM1 Domain of Raver1 Directs its Interactions with Vinculin
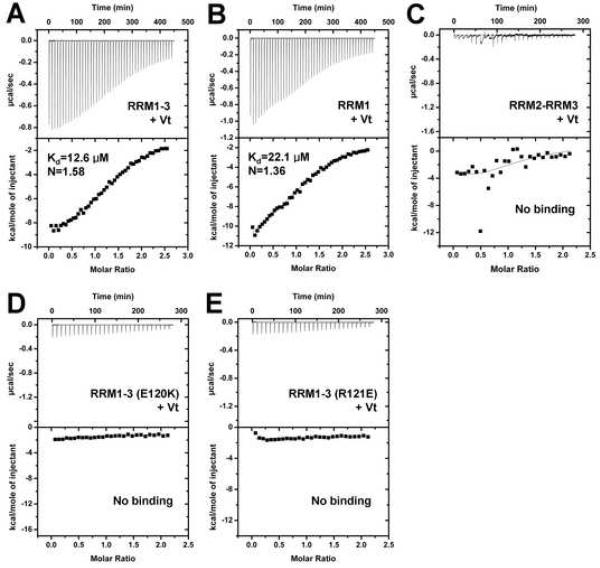
RRM motifs direct both RNA-protein and protein-protein interactions (Kielkopf et al., 2004; Maris et al., 2005; Lunde et al., 2007; Cléry et al., 2008). The raver1-vinculin interaction was known to be mediated by binding of the N-terminus of raver1 to Vt (Hüttelmaier et al., 2001). We reasoned that this was mediated by the RRM motifs present in the N-terminus of raver1 (Figure 1) and to test this we performed isothermal titration calorimetry (ITC). Indeed, Vt bound to the RRM1-3 region (residues 39-321) and also to RRM1 alone (residues 59-130) with Kd values of 12.6 μM and 22.1 μM, respectively, but failed to bind to the tandem RRM2-RRM3 (residues 129-321) motifs (Figure 2). Therefore, the RRM1 domain of raver1 directs its interactions with vinculin. As expected (Hüttelmaier et al., 2001), the interactions of raver1 with vinculin required its activation, as full length, inactive human vinculin failed to bind to raver1 (Supplementary Figure S1A, S1B). Similar findings were evident in size exclusion chromatography, where RRM1-3 bound to Vt but not to full-length human vinculin (Supplementary Figure S1C, S1D) and in native gel analyses, where RRM1-3 bound to Vt but not to full-length vinculin (data not shown).
Figure 2. The RRM1 Domain of Raver1 Directs its Interactions with Vinculin.
(A) Binding of Vt to RRM1-3 (ΔH, -10.5±0.2 kcal/mol; ΔS, -12.9 cal/mol) as determined by ITC. Representative analyses are shown. The Kd of the interactions is provided in the lower inset. N represents the stoichiometry of the binding reactions.
(B) Binding of Vt to RRM1 (ΔH, -15.1±0.5 kcal/mol; ΔS, -29.7 cal/mol).
(C) Binding of Vt to RRM2-RRM3.
(D), (E) Mutations of raver1 predicted to disrupt electrostatic interactions with vinculin. Glu120Lys (D) or Arg121Glu (E) substitution mutations abolish binding to Vt. Binding of raver1 mutant E120K was also not detected by size exclusion chromatography (data not shown).
Crystal Structures of Human Vinculin in Complex with Raver1
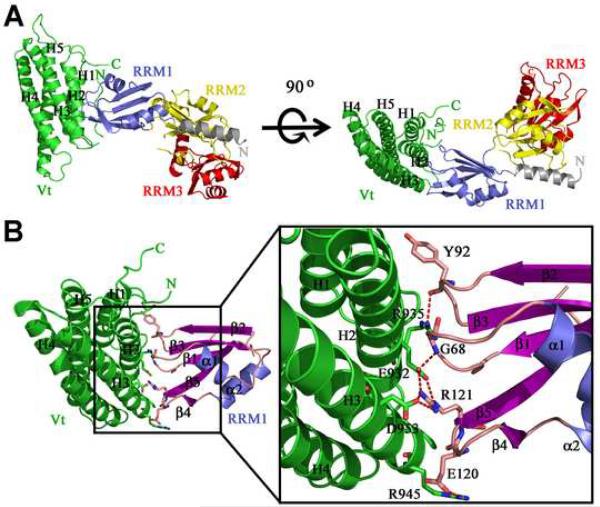
To define the interaction of raver1 with vinculin we solved the crystal structures of human Vt in complex with RRM1 or with the RRM1-3 domains of human raver1 at 2.9 Å and 2.75 Å resolution, respectively (Figure 3; Supplementary Figure S2; Tables 1 and 2). The conformations of Vt and RRM1 as seen in the Vt:RRM1 and the Vt:RRM1-3 crystal structures are very similar, where the Cα traces superimpose with a root-mean-square deviation of 0.9 Å for 250 equivalent residues (Supplementary Figure S3), despite the fact that these crystals were obtained using different conditions (see Experimental Procedures), eliminating crystallization artifacts. As for other RRM domains, these crystal structures showed that the RRM1, RRM2, and RRM3 domains of raver1 have a canonical βαββαββ topology with five-stranded anti-parallel β-strands (β1-β5) and two α-helices (α1, α2) (Figure 3; Supplementary Figure S2). Surprisingly, the interaction was largely directed by the binding of the H2 and H3 α-helices of Vt with three connecting loop regions (the β1-α1, β2-β3, and β4-β5 loops) of the RRM1 domain (Figure 3; Supplementary Figure S2, S3). To our knowledge, this mode of RRM-mediated protein-protein interaction differs from all known examples involving RRM domains (Kielkopf et al., 2001; Fribourg et al., 2003; Lau et al., 2003; Shi and Xu, 2003; Maris et al., 2005; Ngo et al., 2008). Specifically, in the U2B/U2A structure (Price et al., 1998), the α-helix of the U2B RRM domain interacts with the leucine rich region of U2A, whereas the interactions of human Y14 with Magoh (Fribourg et al., 2003; Lau et al., 2003; Shi and Xu, 2003), UPF2 with UPF3 (Kadlec et al., 2004), and P14 with SF3b (Schellenberg et al., 2006; Spadaccini et al., 2006) occur via the surface of the β-sheet that is involved in RNA binding of canonical RRMs. Further, in the RRM protein interactions in the U2AF35/U2AF65 (Kielkopf et al., 2001), U2AF65/SF1 (Selenko et al., 2003), and SPF45/SF3b155 (Corsini et al., 2007) complexes the UHM (U2AF-homology motif) signature sequences present in U2AF35, U2AF65, and SPF45 (which include a Arg-X-Phe loop sequence and acidic residues) all recognize a UHM-ligand motif (a Trp and positively charged residue) of the ligand. Finally, in the interaction between PTB with raver1, the raver1 peptide (496-PGVSLLGAPPKD-507) interacts with the hydrophobic groove of the PTB RRM2 motif and this complex is compatible with a simultaneous RNA binding (Rideau et al., 2006). The Vt-raver1 interaction has little in common with these examples of RRM-directed protein-protein interactions.
Figure 3. Structure of the Vinculin:Raver1 Complex.
(A) The Vt:RRM1-3 complex structure is shown in ribbon representation with the five α-helices (H1-H5) of the tail domain of human vinculin in green, the N-terminal helix of raver1 in gray, RRM1 in blue, RRM2 in yellow, and RRM3 in red. Left, head-on view, and right, rotated by 90°, showing the interaction of the loops connecting the β3-β2, β1-α1, and β4-β5 strands of raver1 with H2 and H3 α-helices of vinculin's tail domain.
(B) The Vt:RRM1 structure showing the β-sheets and α-helices of RRM1 in purple and light blue, respectively. Residues involved in the Vt-RRM1 interaction are shown on the right panel. The intermolecular hydrogen-bonding network is indicated by dotted lines. The side chain atoms, NE and NH2 of raver1-Arg121 engage in electrostatic interactions with vinculin-Glu932 atoms OE1 and OE2, respectively, while raver1-Arg121 (NH1) is held in place through interactions with the main chain oxygen of raver1-Arg119 as well as OD2 of vinculin-Asp953. An additional electrostatic interaction was also found between side chains of raver1-Glu120 and vinculin-Arg945, which spatially orients the side chain of vinculin-Arg945 such that it is parallel to the side chain of raver1-Arg117 and provides a stable surface for hydrophobic interaction.
Table 1.
Data Reduction Statistics of the Vt:RRM1 and Vt:RRM1-3 Data Sets
| Vt:RRMl | Vt:RRM1-3 | |
|---|---|---|
| Resolution | 50 Å-2.9 Å | 50 Å-2.75 Å |
| last shell | 2.95 Å-2.9 Å | 2.8 Å- 2.75 Å |
| Total Measurements | 40,086 | 248,668 |
| Number of Unique Reflections | 23,190 | 40,684 |
| last shell | 1,094 | 1,974 |
| Space Group | P1 | P212121 |
| Unit Cell Dimensions | ||
| a | 41.8 Å | 86.8 Å |
| b | 70.9 Å | 94.9 Å |
| c | 99.3 Å | 186 Å |
| α | 89.9° | 90° |
| β | 90° | 90° |
| γ | 104° | 90° |
| Wavelength | 1.04 Å | 1 Å |
| Rsyma | 0.093 | 0.058 |
| last shell | 0.26 | 0.387 |
| I/σ(I) | 12.18 | 30.49 |
| last shell | 3.25 | 3.41 |
| Completeness | 0.941 | 0.998 |
| last shell | 0.857 | 0.977 |
Table 2.
Crystallographic Refinement Statistics of the Vt:RRM1 and Vt:RRM1-3 Structures
| Vt:RRMl | Vt:RRMl-3 | |
|---|---|---|
| Resolution | 20 Å-2.9 Å | 20 Å-2.75 Å |
| last shell | 3.07 Å-2.9 Å | 2.92 Å- 2.75 Å |
| No. of Reflections (working set) | 23,051 | 40,500 |
| No. of Reflections (test set) | 1,182 | 2,024 |
| R-factora | 0.21 | 0.225 |
| last shell | 0.26 | 0.27 |
| Rfreeb | 0.277 | 0.274 |
| last shell | 0.348 | 0.289 |
| No. of amino acid residues | 977 | 915 |
| No. of protein atoms | 7,689 | 7,126 |
| No. of solvent molecules | 4 | 41 |
| Average B-factor (protein) | 55.74 Å2 | 63.91 Å2 |
| Average B-factor (solvent) | 41.93 Å2 | 49 Å2 |
| R.m.s.d. from ideal geometry: | ||
| Covalent bond lengths | 0.011 Å | 0.011 Å |
| Bond angles | 1.203° | 1.335° |
The free R-factor is a cross-validation residual calculated by using 5% of reflections, which were randomly chosen and excluded from the refinement.
The intermolecular interaction of activated vinculin and raver1 is mediated through electrostatic interactions (vinculin-Asp953, -Arg945, and -Glu932 residues with raver1-Arg121, -Glu120, and -Arg121, respectively), a hydrogen bond network (side chains of vinculin-Arg935 and -Glu932 with the main chain carboxyl groups of raver1-Tyr92 or the main chain amide group of -Gly68, respectively), and hydrophobic interactions (raver1-Tyr92 with vinculin residues Phe885, Pro886, Met899, and Ala931; Figure 3B; Supplementary Figure S2). Indeed, mutating raver1 residue Glu120 to a Lysine, or Arg121 to a Glutamate, prevents the electrostatic interactions with Vt residues Arg945 or Asp953 and Glu932, respectively, and abolished binding to Vt in solution (Figure 2D, 2E) while still preserving micromolar affinity of raver1 for RNA (data not shown).
The interaction of vinculin with the RRM1 domain of raver1 occurs on the opposite surface of its putative RNA-binding β-sheet (β1 and β3 strand) and at a considerable distance from those present in RRM2 and RRM3. Thus, the structures of the RNP motifs that could direct binding with RNA by raver1 are not affected by its interaction with vinculin. Structure-based sequence alignment shows that the residues in the Vt-RRM1 interface are strictly conserved in the RRM1 domain of raver1 of several species, and that they are not present in the RRM2 and RRM3 domains (Supplementary Figure S4A), explaining the selectivity of the interaction of vinculin for the RRM1 motif of raver1. Finally, the interaction of Vt with raver1 buries 669 Å2 of the surface area of Vt and 748.1 Å2 of the surface area of RRM1-3. This relatively small interface is consistent with the ~10 μM Kd of this interaction. Nonetheless, the ability of raver1 to crystallize with vinculin, and to readily interact with vinculin in solution and in cells (Hüttelmaier et al., 2001) suggests that once bound to activated vinculin raver1 has a low off rate.
Raver1 Residues that Direct Contact with Vinculin are Required for Raver1 and Vinculin Co-localization
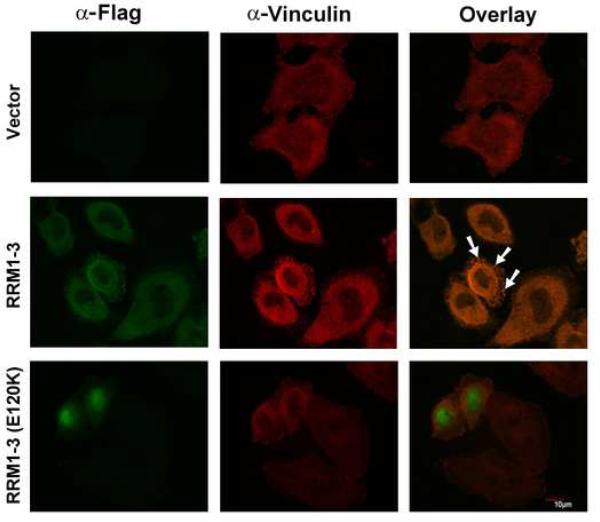
Raver1 interacts with the splicing regulator PTB in the nucleus, but with vinculin in cytoplasm and at focal adhesions (Hüttelmaier et al., 2001). To test whether residues of raver1 that were required for raver1 and vinculin binding by ITC (Figure 2D-E) were also required for their colocalization in cells, HeLa cells were transfected with an vector driving the expression of Flag epitope-tagged RRM1-3 domain of raver1, or expressing RRM1-3 domain harboring the E120K mutation. As expected, immunofluorescence analyses demonstrated that wild type raver1 RRM1-3 co-localized with endogenous vinculin, whereas mutant RRM1-3 (E120K) did not. Rather, this mutant was predominately nuclear in its localization (Figure 4). Therefore, interactions with vinculin appear to be important for cytoplasmic and adhesion complex localization of raver1.
Figure 4. Raver1 Residues that Direct Interactions with Vinculin are Required for Vinculin and Raver1 Co-Localization in Cells.
HeLa cells were transfected with p3xFLAG-CMV-7.1 alone (Vector) or with this vector expressing raver1 RRM1-3 or RRM1-3 (E120K). After 48 hours, cells were stained with anti-Flag antibody (green, left) and anti-vinculin antibody (red, center). The co-localization of vinculin and raver1 RRM1-3 (white arrows) is seen as yellow (right) from superposition of green and red channels. By contrast, raver1 RRM1-3 (E120K) is predominately nuclear and does not co-localize with endogenous vinculin.
Vinculin Activation is Necessary for its Interaction with Raver1
Full length, inactive vinculin does not bind to raver1 either in solution (Supplementary Figure S1A, S1B, S1D) or in solid phase binding assays (Hüttelmaier et al., 2001). In full-length, inactive vinculin, a portion of the raver1 binding site is occluded by the 882-890 loop region, which likely hinders its interactions with the raver1. Specifically, superposition of the 882-890 loop region of inactive versus raver1-bound vinculin shows significant alterations in its structure, and that the binding sites for raver1 are masked in inactive vinculin. Therefore, although RRM1 binds to the face of Vt that is opposite that which directs its interactions with the Vh1 seven-helix bundle domain (Borgon et al., 2004; Izard et al., 2004), the interactions of the loops of RRM1 with helices H2 and H3 of Vt are nonetheless compromised by the 882-890 loop of vinculin (Figure 5A; Supplementary Figure S5).
Figure 5. Structural Constraints and Alterations Provoked by the Vinculin-Raver1 Interaction.
(A) Superposition of the Vt:RRM1-3 complex (color coding is the same as in Figure 3) with full-length, inactive human vinculin (shown in pink). The raver1 binding site is occluded by the 882-890 loop region of inactive vinculin. Asp882 of Vt in its raver1 bound state is indicated as well as Pro877 of inactive vinculin (dashed arrows).
(B) Interaction with vinculin alters the structure of the binding loops of raver1. Superposition of the native mouse RRM1 domain (PDBID 1WI6, yellow) onto human RRM1 (blue) when bound to vinculin (green). Upon binding to vinculin, β-strand β4 moves as indicated by red arrows. In the unbound raver1 structure, only two interactions are found between the β-hairpin and the loop region situated between strand β1 and helix α1 (Arg127 and Asp77 as well as Arg129 and Leu74; not shown for clarity). However, in the Vt:raver1 complex the side chain of Arg121 interacts with main chain oxygen of Arg119; the side chain of Arg119 engages in electrostatic interactions with the side chains of Asp69 and Glu74; and the side chain of Arg117 is in contact with the main chain oxygen of Glu120. The hydrophobic cavity observed on the surface near the β-hairpin is rather large in the unbound conformation, which almost completely disappears in the complex.
Structural Alterations in Raver1 Provoked by Vinculin Binding
Comparison of the native NMR structure of the mouse RRM1 domain (PDB ID 1WI6) with that of the crystal structure of human RRM1 bound to vinculin also revealed significant alterations in the structure of the β-hairpin residing on the β4-β5 loop, which shifts β-strand (β4) by about 15° when bound to vinculin (Figure 5B). This region is well defined in the NMR structure ensemble of 20 best structures, where two intramolecular interactions are found between the β-hairpin and the loop region that is positioned between strand β1 and helix α1, namely between Arg127 and Asp77 as well as Arg129 and Leu74. By contrast, in Vt:raver1 complex the intramolecular interactions of raver1 are transformed, where the side chains of Arg119 and Arg121 (corresponding to murine residues Arg127 and Arg129) now engage in electrostatic interactions with the side chains of Asp69 and Glu74, or interact with main chain oxygen of Arg119, and where the side chain of Arg117 interacts with the main chain oxygen of Glu120. Further, the Vt-induced movement directs the strong interactions of Glu120 and Arg121 of raver1 with Arg945 and Asp953 of vinculin (Figure 3B). Interestingly, there are some parallels of the raver1 interaction with vinculin with that of nearly equivalent loop regions of the RRM2 domain of the spliceosome factor ASF/SF2 with SRPK1 protein kinase, where residues of these loops of ASF/SF2 are also involved in electrostatic interactions with SRPK1 (Supplementary Figure S6); however, here the β4 strand appears to unfold following substrate binding (Ngo et al., 2008).
Raver1 Selectively Binds to RNA
The RNP1 and RNP2 motifs present in the RRM1-3 domains of raver1 direct interactions with RNA in other RRM-containing proteins (Kielkopf et al., 2004; Maris et al., 2005; Lunde et al., 2007; Cléry et al., 2008). We therefore reasoned that raver1 bound to RNA. To identify possible RNA targets of raver1, we screened raver1 for its ability to bind to twenty different RNA sequences that have been identified in RRM:RNA complex structures. RNA oligonucleotides harboring these sequences were tested for their ability to bind to raver1 by isothermal titration calorimetry under physiological conditions. Ultimately, these analyses established that the RRM1 domain of raver1 selectively binds to a 12-nucleotide RNA having the sequence UCAUGCAGUCUG with micromolar affinity (Supplementary Figure S7). Interestingly, the sequence of the RNA bound by the RRM1 motif of raver1 is an exact match with a portion of the coding region of human vinculin mRNA (nucleotides 3089-3100). Therefore, raver1 is indeed an RNA binding protein, and its RRM1 motif directs specific binding to RNA and additionally directs interactions with vinculin.
The Raver1-Vinculin Interaction is Permissive for Binding to F-actin
A hallmark of vinculin activation is its ability to bind to F-actin (Johnson and Craig, 1995). To test whether the vinculin-raver1 interaction was also permissive for binding to F-actin, we performed F-actin co-sedimentation assays by incubating Vt and/or raver1 at equimolar ratios with F-actin. As expected, Vt avidly bound to F-actin, whereas raver1 did not (Figure 6). Further, pre-formed Vt-raver1 complexes also bound to F-actin (Figure 6). Thus, the vinculin-raver1 interaction is also permissive for binding to F-actin.
Figure 6. The Vinculin:Raver1 Complex is Permissive for Binding to F-actin.
While raver1 does not bind F-actin, the Vt:raver1 complex co-sediments with F-actin. After F-actin co-sedimentation, supernatants (S) were removed and pellets (P, having polymerized F-actin) were washed twice.
DISCUSSION
In the nucleus, raver1 functions as a co-repressor of PTB-directed splicing of select mRNAs, for example of α-tropomyosin transcripts in muscle cells (Gromak et al., 2003; Spellman et al., 2005; Rideau et al., 2006). Raver1 binds to PTB through the agency of its C-terminus (Rideau et al., 2006), a scenario that leaves the RRM domains present in its N-terminus free to bind to other partners, which we have shown includes RNA having the sequence UCAUGCAGUCUG, a sequence that is found in vinculin mRNA. In muscle cells and other cell types raver1 re-localizes from the nucleus to adhesion complexes where it binds to vinculin (Hüttelmaier et al., 2001; Jockusch et al., 2003; Zieseniss et al., 2007) and here our studies indicate that the raver1-vinculin interaction appears to be important for cytoplasmic and focal adhesion localization of raver1, as a form of raver1 that cannot bind to vinculin is predominately nuclear (Figure 4). Importantly, we have shown that the RRM1 domain directs the interactions of raver1 with the tail domain of activated vinculin and that this interaction is permissive for vinculin binding to F-actin (Figure 6). Importantly, our studies have shown that raver1 binds to the vinculin mRNA cargo (Supplementary Figure S7).
The translational machinery rapidly re-localizes to nascent focal adhesions and here vinculin also co-localizes with mRNA (Chicurel et al., 1998; Hüttelmaier et al., 2005; Rodriguez et al., 2006). We hypothesize that raver1 might transport mRNA cargo to these microcompartments via its interaction with vinculin, and that this feed-forward delivery includes transcripts encoding components of adhesion complexes, including vinculin mRNA itself (Figure 7). Similar interactions of vinculin with other RRM-containing proteins could also contribute to this response and allow for the production of other components, along with other specialized mRNA delivery systems, such as the hnRNP K homology domain-containing zipcode binding proteins ZBP1 and ZBP2 that deliver β-actin mRNAs to the adhesion complex (Hüttelmaier et al., 2005; Czaplinski and Singer, 2006; Pan et al., 2007). Collectively, these systems would allow for the rapid and coordinated in situ production of components required for the expanding adhesion complex by the translational machinery, akin to the scenario observed for the localization of a distinct cast of RNAs at protruding pseudopodia (Weis, 2004; Mili et al., 2008).
Figure 7. Vinculin Functions as a Scaffold for the Production of Adhesion Components by the Translational Machinery at Nascent Focal Adhesions.
The activation of vinculin (blue), by talin and perhaps other signals (Izard et al., 2004; Weis, 2004; del Rio et al., 2009), following the engagement of integrin receptors (α and β subunits) provokes alterations in the structure of its head (VH) domain that disrupt the head-tail interaction that holds vinculin in its inactive conformation. Activated vinculin then binds to the RRM1 domain of raver1 (red) through the agency of its tail (Vt) domain. This interaction is permissive for the interactions of the three tandem RRM domains of raver1 with their mRNA cargo (combs), which might include vinculin mRNA and transcripts encoding other components of adhesion complexes. The vinculin-raver1 interaction is also permissive for binding to actin filaments (white spheres). This feed-forward delivery system would ensure rapid and coordinated production of components of adhesion complexes by the translational machinery at these adhesion microcompartments, as seen in vivo (Chicurel et al., 1998; Rodriguez et al., 2006).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The C-terminal tail domain of human Vinculin (Vt, residues 879-1066), and the RRM1 (residues 59-130), RRM2-RRM3 (residues 129-321), and RRM1-3 (residues 39-321) domains of human raver1 were cloned into the pET-28a vector (Novagen) and expressed in Escherichia coli strain BL21-DE3 (Invitrogen) to produce N-terminal (or C-terminal for RRM1-3) hexahistidine-tagged proteins having a thrombin cleavage site for removal of the His-tag. The vectors carrying the Glu120Lys or Arg121Glu mutants were cloned by PCR amplification using mutagenic primers designed against wild-type RRM1-3 vector. The PCR products were treated with DpnI (New England BioLabs) to degrade template plasmids. All constructs were verified by sequencing. Bacteria were grown at 37°C in Luria-Bertan medium containing 20 mg/l kanamycin. Bacterial cultures were induced at OD600=0.8 by adding IPTG to 0.5 mM and incubating at 20°C for 24 hours. The fusion proteins were purified by immobilized metal affinity chromatography on Co2+-TALON beads (BD Biosciences). After cleavage of the hexahistidine-tag at 4°C overnight, the proteins were purified by gel filtration using a Superdex-75 column (Amersham) equilibrated in 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl. Protein samples were concentrated to 14.2 mg/ml (Vt), 9.4 mg/ml (raver1 RRM1), 16.4 mg/ml (RRM2-RRM3), or 39.6 mg/ml (RRM1-3). For isothermal titration calorimetry (ITC) experiments, the Vt:RRM1-3 complex was further purified by gel filtration using a Superdex-200 column.
Purified Vt (545 μM) or full length vinculin (320 μM) and RRM1-3 (529 μM) were mixed and loaded on a Superdex-200 26/60 column (Amersham Biosciences) equilibrated with 20 mM Tris-HCl pH 8 and 150 mM NaCl. Elution was performed at 4°C, and the flow-rate was 1 ml/min. Protein detection was performed on SDS-polyacrylamide PhastGels stained with Coomassie blue.
Crystallization of Vinculin:Raver1 Complexes
We originally obtained needle shaped crystals of the Vt:RRM1 complex using C-terminal hexahistidine-tagged Vt and un-tagged RRM1 proteins. To improve the crystal quality, we removed the C-terminal hexahistidine-tag of Vt, which allowed crystallization of this complex. Crystals of the Vt:RRM1 complex were obtained after mixing Vt (402 mM) and RRM1 (440 mM) in an equal volume in a solution containing 100 mM Tris (pH 7.5), 200 mM sodium nitrate, and 16% PEG-3,350. Crystals were grown at 4°C by sitting drop vapor diffusion. Crystals appeared after two days. The crystals were transferred to Paratone-N solution (Molecular Dimensions Ltd.) before flash-freezing in liquid nitrogen. The crystals belonged to space group P1 with unit cell dimensions a = 41.8 Å, b = 70.9 Å, c = 99.3 Å, a= 89.8°, b= 90.0°, and g= 104.0°. The four Vt:RRM1 heterodimers in the asymmetric unit result in a solvent content of 49.4% (VM = 2.4 Å3/Da) (Matthews, 1968).
After testing several different RRM1-3 boundaries for co-crystallization, we eventually obtained Vt:RRM1-3 crystals using raver1 residues 39-321. These Vt:RRM1-3 crystals were obtained after mixing Vt (545 μM) and RRM1-3 (529 μM) in an equal volume in a solution containing 10% PEG-8,000, 8% ethylene glycol, and 100 mM Na-HEPES (pH 7.5) by sitting drop vapor diffusion method. Glycerol (28%) and ethylene glycol (13%) were used as the cryoprotectant. The crystals belonged to space group P212121 with unit cell dimensions a = 86.8 Å, b = 94.9 Å, and c = 186.0 Å. There were two Vt:RRM1-3 heterodimers in the asymmetric unit, giving a solvent content of 66.3% (VM = 3.7 Å3/Da) (Matthews, 1968). All X-ray data were collected at the Advanced Photon Source (SER-CAT 22BM beamline) and processed using HKL2000 (Otwinowski and Minor, 1997) (Table 1).
Determination of Vt:RRM1 and Vt:RRM1-3 Structures
Molecular replacement using the program MOLREP (Vagin and Teplyakov, 1997) and the Vt structure (PDBID 1RKE) as the search model yielded four Vt solutions for the Vt:RRM1 complex. The four Vt models were then fixed and the four positions of RRM1 were also determined by molecular replacement using mouse raver1 RRM1 structure as the search model (PDBID 1WI6) and the program MOLREP. The entire structure of Vt (four molecules) and RRM1 (four molecules) complex was checked and manually rebuilt into the composite omit 2Fobs-Fcalc and Fobs-Fcalc electron density maps by using O (Jones et al., 1991) in several iterative rounds. CNS (Brünger et al., 1998) was used to calculate omit maps. Crystallographic refinement was further performed using REFMAC (Murshudov et al. 1997) and BUSTER/TNT (Tronrud et al 1987; Bricogne 1997) and resulted in a final crystallographic R-value of 0.210 (Rfree=0.277; Table 2).
Vt amino acids 879-883 and 1064-1066 of subunit A; 879-883, 888-891, 1046-1054 and 1062-1066 of subunit B; 879-883, 888-892, 1047-1054, and 1064-1066 of subunit C; and 879-883, 888-892, and 1064-1066 of 839 subunit D are not present in our Vt:RRM1 model due to disorder. In the Ramachandran plot, 91.8% and 8.2% of the residues fell in the most favored and additionally allowed regions, respectively, with no residues in the generally allowed or disallowed regions.
The Vt:RRM1-3 complex structure was determined by molecular replacement using the program MOLREP (Vagin and Teplyakov, 1997). The refined Vt and RRM1 models of our Vt:RRM1 complex structure were used as search models. The two Vt models and two RRM1 models were then fixed and the two positions of RRM2 and RRM3 were determined by molecular replacement using the first RRM domain structure of HuD protein (PDBID 1FXL) as the search model. CNS (Brünger et al., 1998) was used to calculate omit maps. Crystallographic refinement was further performed using REFMAC (Murshudov et al. 1997) and BUSTER/TNT(Tronrud et al 1987; Bricogne 1997). Side chains were successively rebuilt according to 2Fobs-Fcalc and Fobs-Fcalc electron density maps followed by iterative cycles of model rebuilding with O (Jones et al., 1991). At the very last step, positional and B-factor refinement was carried out using BUSTER/TNT (Bricogne, 1997). The final R-factor was 0.225 with a free R of 0.274.
Vt amino acids 879-881, and 1064-1066 (subunit A); 879-882, 889-891, and 1064-1066 (subunit C) and raver1 residues 318-321 (subunit B) and 319-321 (subunit D) are not present in the final Vt:RRM1-3 model due to disorder. In the Ramachandran plot, 89.3%, 90.4%, 9.4% and 0.01% of the residues fell in the most favored, additionally allowed regions, and in the generally allowed or disallowed regions, respectively, with no outliers.
Isothermal Titration Calorimetry (ITC)
High-sensitivity ITC experiments were used to assess the interaction between raver1 and Vt. Assays were performed at 23°C in 20 mM Tris-HCl pH 8, 150 mM NaCl using a VP-ITC calorimeter (Microcal, Inc.). For each experiment, the sample cell (volume 1.4 ml) was filled with 10-45 μM raver1 protein solution. A 300 μl syringe was filled with a 150-500 μM Vt protein solution or 90-300 μM RNA solution. The reference cell contained 20 μM Tris-HCl pH 8, 150 μM NaCl buffer. Titration experiments were typically consisted of 25-45 injections, each of 6-10 μl volume and 20 seconds duration, with 10 minutes interval between additions. The stirring rate was 300 r.p.m. The binding isotherms were fitted via a nonlinear least squares minimization method to determine the binding stoichiometry (n), the equilibrium binding constant (KD), the change in enthalpy (ΔH) and entropy (ΔS).
To screen RNAs for their ability to bind to raver1, we concatenated RNA sequences described in the twenty RNA:RRM structures in the literature into five concatenated oligonucleotides; i.e., each oligonucleotide had four known RNAs that bind to RRM motifs. Of these five concatenates, only one 12-mer showed micromolar affinity. We then truncated this RNA from each end and tested these new RNAs in binding assays (data not shown). We eventually found that a 6-mer (UGCAGU) bound to raver1 (data not shown). Finally, having found this sequence present in human vinculin mRNA, we then tested whether a 12-mer matching the vinculin sequence surrounding the UGCAGU core bound to raver1 by ITC and showed that this was the case (Supplementary Figure S7).
Raver1-Vt-F-actin Binding Assays
The preformed Vt:raver1 complex (1:1.3 ratio) was obtained by mixing Vt and raver1 and incubating for 20 minutes. F-actin co-sedimentation assays were performed as described (Bois et al., 2006) using 12 μl of polymerized F-actin, and equimolar amounts of Vt and RRM1-3 raver1 protein, alone or pre-incubated together for 1 hour (at room temperature, in a final total reaction volume of 100 μl). Following incubation the samples were sedimented at 95,000 × g at 25°C in a Beckman ultracentrifuge for 15 minutes and pellet and supernatant fractions were collected. Equal volumes of supernatants and pellets were resolved on 20% SDS-polyacrylamide gels and the proteins were visualized by staining with Coomassie Blue.
Transfections and Immunofluorescence Microscopy
For immunofluorescence experiments, HeLa cells were plated onto glass coverslips overnight and then transfected with a total of 0.8 μg p3xFLAG-CMV-7.1 vector, FLAG-RRM1-3, or FLAG-RRM1-3 (E120K) DNA using Lipofectamine 2000 (Invitrogen). After 48 hours, the cells were fixed with 4% paraformaldehyde (20 minutes at room temperature), permeabilized with PBS and 0.25% Triton X-100 for 5 minutes, and blocked with PBS and 2% BSA for 30 minutes. Endogenous vinculin was detected by staining with anti-vinculin mouse monoclonal antibody (Sigma) and followed by donkey anti-mouse Alexa 568 (Invitrogen) conjugated secondary antibody. After washing 3 times the cells were stained for raver1 using Anti-FLAG M2 mouse monoclonal FITC-conjugated antibody (Sigma). Cells were then washed and mounted using Vectorshield mounting medium (Vector laboratories Inc.,) and indirect fluorescence images were recorded either with an Olympus FluoView 1000 Confocal Microscope and processed using FV10-ASW.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to John Cleveland (Scripps-Florida) for discussions and critical review of the manuscript, Philippe Bois (Scripps-Florida) for many helpful discussions and substantial help with our RNA experiments, Zhen Wu (Scripps-Florida) for sequencing, and Oliver Smart and Gerard Bricogne (GlobalPhasing Ltd.) for crystallographic refinement using autoBUSTER. We also thank HaJeung Park (Scripps-Florida) for assistance at the synchrotron. We are grateful to the staff at the Advanced Photon Source, SER-CAT, for synchrotron support. TI is supported by grants from the National Institutes of Health (GM071596, AI055894, and AI067949) and by start-up funds provided to Scripps Florida from the State of Florida. This is publication no. 19546 from the Scripps Research Institute.
abbrevsiations footnote
- BSA
bovine serum albumin
- CNS
crystallography & NMR system
- ITC
isothermal titration calorimetry
- FITC
fluorescein isothiocyanate
- hnRNP
heterogeneous nuclear ribonucleoprotein
- IPTG
isopropyl-β-D-thiogalactopyranoside
- mRNA
messenger ribonucleic acid
- NMR
nuclear magnetic resonance
- PCR
polymerase chain reaction
- PIP2
phosphatidylinositol-4,5-bisphosphate
- PEG
polyethylene glycol
- PTB
polypyrimidine tract binding protein
- RRM
RNA recognition motif
- SDS
sodium dodecyl sulfate
- VBS
vinculin binding site
- Vh1
vinculin's N-terminal seven-helical bundle
- Vh2
vinculin's 2nd seven-helical bundle domain
- Vh3
vinculin's 3rd seven-helical bundle domain
- Vt
vinculin's C-terminal five-helical bundle domain
ACCESSION NUMBERS
The coordinates and structure factors for the complexes have been deposited to the Protein Data Bank with accession codes 3H2V (Vt:RRM1) and 3H2U (Vt:RRM1-3).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- Bois PRJ, Borgon RA, Vonrhein C, Izard T. Structural dynamics of α-actininvinculin interactions. Mol. Cell Biol. 2005;25:6112–6122. doi: 10.1128/MCB.25.14.6112-6122.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Bois PRJ, O'Hara BP, Nietlispach D, Kirkpatrick J, Izard T. The vinculin binding sites of talin and α-actinin are sufficient to activate vinculin. J. Biol. Chem. 2006;281:7228–7236. doi: 10.1074/jbc.M510397200. [DOI] [PubMed] [Google Scholar]
- Borgon RA, Vonrhein C, Bricogne G, Bois PRJ, Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Bricogne G. The Bayesian viewpoint in crystallography: Basic concepts and applications. Methods Enzymol. 1997;276:361–423. doi: 10.1016/S0076-6879(97)76069-5. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chandrasekar I, Stradal TE, Holt MR, Entschladen F, Jockusch BM, Ziegler WH. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 2005;118:1461–1472. doi: 10.1242/jcs.01734. [DOI] [PubMed] [Google Scholar]
- Chicurel ME, Singer RH, Meyer CJ, Ingber DE. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature. 1998;392:730–733. doi: 10.1038/33719. [DOI] [PubMed] [Google Scholar]
- Cléry A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcárcel J, Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007;14:620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem. Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- DeMali KA, Burridge K. Coupling membrane protrusion and cell adhesion. J. Cell Sci. 2003;116:2389–2397. doi: 10.1242/jcs.00605. [DOI] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham I, Gingras AR, Papagrigoriou E, Patel B, Emsley J, Critchley DR, Roberts GC, Barsukov IL. A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure. 2005;13:65–74. doi: 10.1016/j.str.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat. Struct. Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidylinositol-4-5-bisphosphate. Nature. 1996;381:531–535. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- Gromak N, Rideau A, Southby J, Scadden AD, Gooding C, Hüttelmaier S, Singer RH, Smith CW. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttelmaier S, Illenberger S, Grosheva I, Rüdiger M, Singer RH, Jockusch BM. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J. Cell Biol. 2001;155:775–786. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Izard T, Evans G, Borgon RA, Rush CL, Bricogne G, Bois PR. Vinculin activation by talin through helical bundle conversion. Nature. 2004;427:171–175. doi: 10.1038/nature02281. [DOI] [PubMed] [Google Scholar]
- Izard T, Vonrhein C. Structural basis for amplifying vinculin activation by talin. J. Biol. Chem. 2004;279:27667–27678. doi: 10.1074/jbc.M403076200. [DOI] [PubMed] [Google Scholar]
- Jockusch BM, Hüttelmaier S, Illenberger S. From the nucleus toward the cell periphery: a guided tour for mRNAs. News Physiol. Sci. 2003;18:7–11. doi: 10.1152/nips.01413.2002. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models into electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat. Struct. Mol. Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, Lücke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Diem MD, Dreyfuss G, Van Duyne GD. Structure of the Y14-Magoh core of the exon junction complex. Curr. Biol. 2003;13:933–941. doi: 10.1016/s0960-9822(03)00328-2. [DOI] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystal. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Ngo JC, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu XD, Adams JA, Ghosh G. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol. Cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nhieu GT, Izard T. Vinculin binding in its closed conformation by a helix addition mechanism. EMBO J. 2007;26:4588–4596. doi: 10.1038/sj.emboj.7601863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pan F, Hüttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol. Cell. Biol. 2007;27:8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B“-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Hüttelmaier S, Singer RH, Matthews S, Curry S, Smith CW. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nature Struct. Mol. Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J. Cell Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Lührmann R, Glover JN, MacMillan AM. Crystal structure of a core spliceosomal protein interface. Proc. Natl. Acad. Sci. USA. 2006;103:1266–1271. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xu RM. Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev. 2003;17:971–976. doi: 10.1101/gad.260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Krämer A, Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- Spadaccini R, Reidt U, Dybkov O, Will C, Frank R, Stier G, Corsini L, Wahl MC, Lührmann R, Sattler M. Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA. 2006;12:410–425. doi: 10.1261/rna.2271406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- Tronrud DE, Eyck LFT, Matthews BW. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta Crystallogr. 1987;A43:489–501. [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- Weis WI. Cell biology: how to build a cell junction. Nature. 2004;430:513–515. doi: 10.1038/430513a. [DOI] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Zieseniss A, Schroeder U, Buchmeier S, Schoenenberger CA, van den Heuvel J, Jockusch BM, Illenberger S. Raver1 is an integral component of muscle contractile elements. Cell Tissue Res. 2007;327:583–594. doi: 10.1007/s00441-006-0322-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.