Abstract
APE1/Ref-1 (APE1), the mammalian ortholog of Escherichia coli Xth, and a multifunctional protein possessing both DNA repair and transcriptional regulatory activities, has a pleiotropic role in controlling cellular response to oxidative stress. APE1 is the main apurinic/apyrimidinic endonuclease in eukaryotic cells, playing a central role in the DNA base excision repair pathway of all DNA lesions (uracil, alkylated and oxidized, and abasic sites), including single-strand breaks, and has also cotranscriptional activity by modulating genes expression directly regulated by either ubiquitous (i.e., AP-1, Egr-1, NF-κB, p53, and HIF) and tissue specific (i.e., PEBP-2, Pax-5 and −8, and TTF-1) transcription factors. In addition, it controls the intracellular redox state by inhibiting the reactive oxygen species (ROS) production. At present, information is still inadequate regarding the molecular mechanisms responsible for the coordinated control of its several activities. Both expression and/or sub-cellular localization are altered in several metabolic and proliferative disorders such as in tumors and aging. Here, we have attempted to coalesce the most relevant information concerning APE1's different functions in order to shed new light and to focus current and future studies to fully understand this unique molecule that is acquiring more and more interest and translational relevance in the field of molecular medicine. Antioxid. Redox Signal. 11, 601–619.
Introduction: An Overall View on APE1. General Considerations on APE1 Biological and Molecular Functions. Involvement of APE1 in Controlling Pro-Apoptotic and Pro-Survival Transcription Factors. Is This a Biological Paradox?
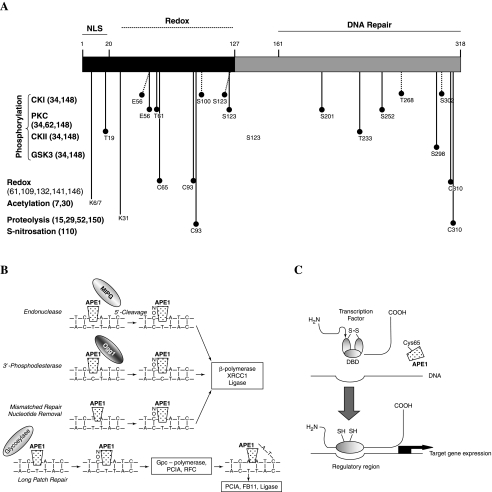
Cellular response to oxidative stress is a highly regulated and complex biological process (42). APE1/Ref-1 (also called HAP1 or APEX, and here referred to APE1), the mammalian ortholog of Escherichia coli Xth (exonuclease III), is a vital protein that acts as an essential master regulator of this response, highly contributing to the maintenance of the genome stability. After cloning by two independent groups in 1991 (21, 114) as a DNA repair enzyme first and as a redox protein the following year (144), APE1 has been described, in ~300 articles, as playing a role in several biological contexts. APE1 is a dual function protein involved both in the base excision repair (BER) pathways of DNA lesions, acting as the major apurinic/apyrimidinic endonuclease, and in eukaryotic transcriptional regulation of gene expression. This effect is obtained as a redox co-activator of different transcription factors such as the early growth response protein-1 (Egr-1), nuclear factor-κB (NF-κB), p53, hypoxia inducible factor-1α (HIF-1α), cAMP response element binding protein (CREB), activator protein-1 (AP-1), and paired box-containing proteins (Pax) in different cell systems (126). These two biological activities are located in two functionally distinct domains. The N-terminus, containing the nuclear localization signal (NLS) region, is principally devoted to the redox activity, through Cys65, while the C-terminus exerts the enzymatic activity on the abasic sites of DNA (146) (Fig. 1A). Homology with ExoIII starts from residue 62. A Blast homology search clearly displays the peculiar distribution of the primary structure conservation of APE1 (http://www.expasy.ch/cgi-bin/blast.pl, accession number: P27695). While the C-terminal part of the protein is highly conserved during phylogeny, the N-terminus is not. Besides in the mammalian proteins, in which the N-terminus is highly conserved (>90%), this region is almost always absent in other organisms, with the exception of Zebrafish, Drosophila, Xenopus, and Dictyostelium, where homology is <40%, suggesting that it may be a recent acquisition of evolution.
FIG. 1.
Representations of APE1 domains structure and functions. (A) Schematic structure, based on functional studies (66, 144) of APE1 structure with critical residues. Schematic diagram of putative or experimental determined post-translational modifications of APE1/Ref-1. CKI, casein kinase I; CKII, casein kinase II; GSK3, glycogen synthase kinase 3; NLS, nuclear localization signal; PKC, protein kinase C. (B) APE1 functions in base excision repair. FEN1, flap endonuclease 1; MPG, methylpurine DNA glycosylase; Ogg1, 8-oxoguanine DNA glycosylase, PCNA, proliferating cell nuclear antigen; RFC, replication factor C; XRCC1, X-ray cross-species complimenting 1. (C) Theoretical molecular model of the redox function of APE1 as a transcriptional coactivator of several transcription factors. Thioredoxin (Trx) is responsible for completing the redox cycle by which APE1 reduced form is restored. DBD, DNA binding domain.
The redox function of APE1 is found only in mammals and not in other vertebrates, as demonstrated by the lack of redox function of the Zebrafish APE1 (zAPE1). The acquisition of the redox function in APE1 proteins is discussed in a recent publication (39). The two major functions of APE1, redox and repair, are completely independent in their actions, as shown by the observation that a mutation of the Cys at position 65 (C65) abolishes the redox function but does not affect the repair function (86), whereas mutation of a variety of amino acids required for DNA repair activity, such as Histidine 309 (H309) and others (87, Vascotto et al., unpublished observations), do not affect the redox function. While the DNA repair active site of APE1 has been clearly delineated (44, 90), the redox domain is much less defined. The only Cys residue required for full redox function is C65, which is buried within the APE1 protein (44, 90), as recently confirmed by Georgiadis who mutated in the zAPE1 a Thr (T58) to a Cys located at the same position of the Cys in the mammalian APE1, resulting in the acquisition of redox activity of the mutated protein (39).
The vital role of APE1 seems to be due to its fundamental activity in the base excision repair (BER) pathway of DNA lesions (35). However, the biological relevance in eukaryotic transcriptional regulation of gene expression as a redox co-activator of different transcription has yet to be fully elucidated (126).
Another interesting function is associated to its ability to indirectly bind to the negative calcium response elements (nCaRE) of some promoters (i.e., parathyroid hormone (PTH) and APE1 promoters) acting as a transcriptional repressor. In particular, binding to nCaRE-B, in the PTH gene promoter, requires Ku70 (Ku86) as an additional factor in the complex (17), while binding to nCaRE-B2 in the APE1 promoter involves hnRNPL interaction (77). This activity seems to be regulated by acetylation of APE1 through the CBP/p300 HAT action (7). A more recent finding has identified APE1's role in mediating production of single-strand DNA (ssDNA) breaks in gene promoters during repair of targeted base oxidation lesions caused by oxygen radicals generated during physiologic signaling (10, 153, 154). Thus, defects in the APE1-mediated step in BER pathway could be linked to altered gene expression besides altering transcription factor state. This is discussed later in this review.
APE1 subcellular distribution, in different mammalian cell types, is mainly nuclear and is critical in controlling cellular proliferative rate (35, 57, 67). Interestingly, the expression of APE1 is strictly connected to different tumorigenic processes (126). The necessity of APE1 for cellular survival and its frequent overexpression in tumor cells strongly suggest a fundamental role of this protein in preventing cell death and in controlling cellular proliferation. However, APE1 abilities to activate transcription factors, such as p53 and Egr-1 (14, 30, 38, 54, 64, 107, 117), mainly involved in controlling cell-cycle arrest and apoptotic programs, leave open the debate concerning the mechanisms responsible for controlling the different functions in several contexts. APE1 protein is also localized within mitochondria in different cell types but, at present, the role in this organelle has not been completely elucidated (15, 88, 122), as well as the molecular mechanism regulating its mitochondrial localization (15).
The fact that APE1 is essential for cell viability was originally demonstrated by genetic studies. Knockout of APE1 in mice causes postimplantation embryonic lethality on days E5 to E9 (85, 147), and attempts to isolate stable APE1-knockout cell lines were totally unsuccessful. For these reasons, a detailed comprehension of molecular targets of APE1 functions has been very difficult. In the last 3 years, conditional knockout and knockdown strategies (35, 67, Vascotto et al., unpublished observations) confirmed the crucial role of this protein for cellular existence and allowed the establishment of cell models to better investigate and characterize the major functions of APE1. However, complete knowledge of molecular effectors regulated by APE1 in determining its biological essentiality is still scanty. The necessity of APE1 for mammalian cells seems to be mainly due to its DNA repair activity in BER pathway. Interestingly, attempts to restore the DNA repair activity in cells not expressing APE1, by using the yeast homologous Apn1 (35) which lacks the redox-transcriptional activation domain, or an APE1 mutant lacking the acetylation sites but not the DNA repair activity (67), or with the redox-defective mutant C65S (Vascotto et al., unpublished observations) did not completely restore the loss of cell viability. Moreover, by specifically blocking the APE1 redox but not DNA activity with an APE1-specific redox inhibitor, E3330, it has been shown that the cytokine-mediated hemangioblast development in vitro was significantly impaired (155). Collectively, these data demonstrate that the exact knowledge of the precise molecular mechanisms for APE1's vital role in mammalian cells is still inadequate. They do, however, indicate that the repair function is crucial for survival and the redox function for cell growth (Vascotto et al., unpublished observations).
DNA Repair Activity of APE1
It is already well established that a plethora of base lesions are induced in mammalian cell genomes by different physical and chemical agents, among which reactive oxygen species (i.e., O2, H2O2 and OH, collectively named ROS) play a dominant role. These lesions, if not adequately repaired, are at the basis of a variety of diseases (including cancer) and of aging.
The BER is the most used pathway to cope with the single base lesion. The BER pathway is also involved in repairing the DNA single-strand breaks (SSB) induced by free radical agents. One of the key enzymes of the BER pathway in mammals is APE1. The basic reactions of the BER pathway have been extensively reviewed (26, 58, 81, 88, 126). They require the coordinated activity of a number of enzymes including: (a) a DNA glycosylase capable of excising a specific modified base; (b) an AP endonuclease, such as APE1, which cleaves the 5′ phosphodiester bond, generating 3′OH and 5′dRP termini; (c) an exonuclease activity (β-polymerase, FEN, APE1); (d) a DNA polymerase (β-polymerase, XRCC1 or δ/ɛ-polymerase with PCNA); and, finally (e) a ligation activity (DNA ligases I and III, XRCC1), (Fig. 1B). The three ways by which APE1 produces the 3′OH terminus for priming and the types of evolutionarily conserved amino acids necessary for these functions have been already reviewed (26, 58 and other articles in this issue of ARS).
All the steps of the BER pathway are finely orchestrated, both from the thermodynamic and the kinetic point of view, to provide an accurate repair of the damaged base and to avoid the generation of intermediate products that are toxic for the cell (58). This implies the coordinated interaction of the various players in the BER process and, when necessary, their further interaction with the DNA replication machinery, as demonstrated by the co-immunoprecipitation of the BER repair proteins with cyclin A and DNA replication proteins (103). In this context, it is noteworthy that RPA proteins are able to suppress the APE1 endonuclease activity in ssDNA of a replicative fork but not in a transcription bubble or in dsDNA (27) and that Cockayne syndrome B protein potentiates the APE1 activity on fully paired AP-DNA but much more on bubble AP-DNA, suggesting a role for this protein in the transcription-repair pathway (143).
Of particular interest in the definition of new functions and targets of APE1 is the finding that, after UVA irradiation, APE1 co-localizes with Ogg1 in the nuclear speckles, organelles associated to transcription and RNA processing. This localization is abolished in the presence of antioxidants (12), suggesting that ROS are the driving force of this localization. Another interesting APE1 function, connected to its capacity to bind to SSB, is that of inhibiting the same binding by PARP1, with consequent inactivation of the poly-ADP-ribosylation and prevention of necrosis (121).
A good example of how APE1 may use its dual nature to produce a cut in the DNA strand and to activate other proteins is that of the VEGF gene which contains a hypoxic response element (HRE) target of HIF-1α. Oxygen deprivation induces formation of an abasic site in the response element followed by HIF-1α binding. The data suggest that the abasic site is the target of APE1 activity with consequent increased flexibility of DNA which causes HIF-1 binding (10) and transcriptional activation of the VEGF gene (46, 153).
Very recently, a role for APE1 (and for APE2) has been suggested also in the antibody class switch recombination, where the abasic site generated by uracil DNA glycosylase, following cytosine deamination, is converted in a single-stranded break by a standard AP-endonuclease procedure (51). Additionally, the recently found inhibitory crosstalk between the oncogene protein Bcl2 and the DNA repair activity of APE1 (152), suggests a novel mechanism which may promote genetic instability and tumorigenesis.
Redox Regulation of Transcription Factors Activities
The intracellular redox status reflects the balance between the activity of antioxidant enzymatic and nonenzymatic cell systems (including GSH/GSSG, superoxide dismutase, catalase, peroxidases, glutathione peroxidases, etc.) and the amount of ROS produced: (a) as byproducts of respiration; (b) as a consequence of external noxious agents such as ionizing radiation; (c) as ‘second messengers' (23, 42); and (d) during pathological states in activated neutrophils (93). Of notice is the observation that variations in the redox state may result in alterations in gene expression profile asset. The molecular mechanism at the basis of this regulation is exerted through the modulation of transcription factors (TFs) activity. In particular, the redox status of reactive Cys residues, located within the DNA-binding domain of some TFs, may control the transcriptional activity of the TFs itself. APE1 has been identified as a protein capable of nuclear redox activity, inducing the DNA binding activity of several transcription factors, such as AP-1 (144), NF-κB (95), Myb (145), PEBP2 (1), HLF (25), NF-Y (94), Egr-1 (14, 64), HIF-1α (63), ATF/CREB family (145), p53 (38, 54, 117), and Pax proteins (13, 128, 129). In each case, this effect was accomplished by maintaining the cysteine residues of the TFs in the reduced state (Fig. 1C), through a redox cycle in which Trx would restore the reduced form of APE1 (61, 109, 132, 141). By using ‘in vitro’ assays, it has been shown that Cys65 is the redox-active site of the protein (39, 139). Since Cys65 appears buried in the 3-D model structure of the protein (44, 90), the redox regulation may implicate the occurrence of unfolding or conformational change of APE1 to allow interaction with redox-sensitive TFs. However, Jayaraman et al. suggested that the stimulatory role played by APE1 on p53 activity may also occur in a redox-independent way (70). This was subsequently corroborated by Ordway et al. (101) who provided the first in vivo evidences that the Cys65 residue of APE1 is unexpectedly not essential for redox regulation of AP-1 DNA binding and, thus, challenging the previous hypotheses. However, the presence of compensatory mechanisms occurring in transgenic mice cannot be excluded, and recent publication by Georgiadis et al. has confirmed the original finding of Cys65 being required for APE1 redox function (39).
Regulation of APE1 at the Gene Expression Level, at the Subcellular Localization Level, and Role of Identified Posttranslational Modifications
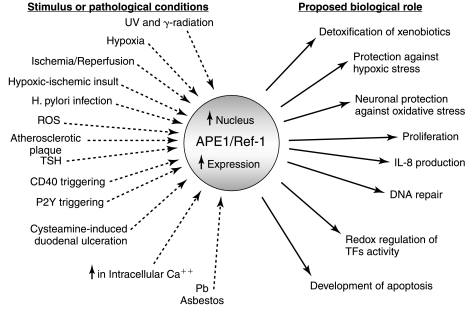
APE1 functional activation is a consequence of different stimuli that may generate both physiological and toxic oxidative stress conditions or increase the intracellular cAMP levels leading to different outcomes (Fig. 2).
FIG. 2.
Schematic representation of some of the stimuli known to activate APE1/Ref-1 expression and/or function. APE1 functional activation, which is associated to nuclear accumulation and upregulation of protein expression, is a consequence of different conditions and implies different outcomes. Thus, APE1 is a central actor in the adaptive cellular response to oxidative stress.
The regulatory functions of the different APE1 activities can be fine-tuned and implemented via three different mechanisms: (a) increase in APE1's level after transcriptional activation (107, 108, 111, 126); (b) relocalization of APE1 from the cytoplasm to the nucleus (126); and (c) modulation of APE1's post-translational modifications (PTM), such as acetylation and phosphorylation. As recently demonstrated, in addition to redox regulation, acetylation appears to have a fine-tuning role in affecting APE1's different activities (7, 30, 67).
Both in vivo and in vitro studies demonstrated that different oxidative or toxic agents and/or intracellular produced ROS efficiently and rapidly (within minutes to hours, depending on the specific ROS-generating stimuli) promote a transient increase in APE1 protein levels, which is inhibited by cycloheximide (111, 129). Different transcription factors, including Sp-1 (36), Egr-1 (107), STAT3 (53), CREB (48), and Jun/ATF4 (37) are involved in the inducible expression of APE1. APE1 itself may inhibit its own expression through the binding to nCa-RE sequences within the APE1 distal promoter, thus constituting an autoregulatory functional loop (68). In addition, APE1 expression is linked in a positive autoregulatory loop with Egr-1 (107), and in a negative inhibitory loop with p53 (151). Interestingly, protein upregulation is always associated with an increase in both redox and AP endonuclease activity, followed by an increase in cell resistance toward oxidative stress and DNA damaging agents (49, 87, 88, 111, 126), strengthening the conclusion that an upregulation of APE1 protein levels has profound biological consequences.
The activation of APE1 is also obtained by a process independent from de novo synthesis and involves cytoplasm to nucleus translocation after exposure of cells to oxidative stress conditions (111, 126, 127) or upon physiologic increase in intracellular ROS production (108). Nuclear localization of APE1 is controlled by the first 20 amino acids at the N-terminal sequence, as determined by Jackson et al. (69) and nuclear import is controlled through a bipartite NLS comprising residues 1–7 and 8–13 with the involvement of an importin system. In fact, the first 20 residues directly bind to karyopherin α1 and α2. Data obtained by treatment of cells with the nuclear export inhibitor leptomycin B suggested the presence of a nuclear export signal (NES) that may reside in a Leu-rich region (L291, L292, L295 residues, which are exposed in the 3-D structure) (69). Recently, it has been shown that the region comprising amino acids 64–80 contains a NES (110). Thus, both nuclear import and export may control subcellular distribution of APE1. In addition, the interaction with specific nuclear proteins could be a means to maintain APE1 within the nucleus. This hypothesis, recently proposed by Jung et al. based on their data showing that nuclear localization of APE1 was dependent on GADD45a nuclear protein expression (71), deserves further experimental support.
During the last few years, several lines of evidence have been accumulating, demonstrating that functional triggering of membrane-bound receptors (such as those for TSH, CD40L, ATP, IL-2, etc.) can lead to APE1 functional activation through intracellular generation of sublethal doses of ROS (126). Noteworthy is the observation that APE1 is also directly responsible for the control of the intracellular ROS levels through its inhibitory effect on Rac1 (2, 52, 102, Vascotto et al., unpublished observations), the regulatory subunit of a membrane nonphagocytic NADPH oxidase system. This enzyme, composed of multiple membrane-associated (Mox and p22phox) and cytosolic components (p67phox, p47phox, and Rac1), catalyzes the transformation of the molecular oxygen to the superoxide anion by transferring an electron from the substrates NADH or NADPH (3). Since we have recently demonstrated that NADPH-mediated ROS production induced by P2Y triggering was able to promote APE1 functional activation (108), we propose the existence of an autoregulatory loop between these two systems. This mechanism may be of therapeutic relevance for endothelial, fibroblastic, and smooth muscle cells, and should be analyzed in diseases of the vascular system where an overactivation of the NADPH oxidase system is involved (19), as well as in the angiogenesis process (133), where an additional autoregulatory loop between APE1 and VEGF may be inferred (10, 153). This observation could be therapeutically relevant in the treatment of tumor progression and cancer metastasis.
Based on the above-mentioned considerations, APE1 seems to act as an intracellular signaling tool involved both in modulating the cellular response to acute and chronic oxidative stress conditions, and also in controlling the endogenous ROS levels during the physiological generation of ROS as intracellular signaling molecules. Since the cell system must be able to discriminate between different ROS-generating stimuli, APE1 behaves as an integrating signaling molecule.
APE1 is an abundant protein (∼104–105 copies/cell) within eukaryotic cells and with a relatively long half-life [~8 h (Vascotto et al., unpublished observations)]. Therefore, the fine-tuning of the multiple functions of this pleiotropic protein may reside in the impact that PTMs have on the function of APE1 and on the modulation of the APE1-interactome under different conditions. Whereas for the former hypothesis some experimental evidences have been obtained [acetylation of K6/K7 residues (7, 30)], very little information is now available on the protein interacting partners of APE1. Pioneering in silico studies discovered that several different phosphorylation sites were scattered throughout the molecule. These potential phosphorylation sites included consensus sequences for casein kinase I and II (CKI and CKII), for protein kinase C (PKC), and for GSK3 (Fig. 1A) (34, 148). Initial in vivo studies confirmed a role for PKC in phosphorylating APE1 in response to PMA or to alkylating agents (i.e., MMS) leading to AP-1 activation (62). However, these studies have not been repeated nor followed up. Therefore, the role of phosphorylation on APE1 is still not clear.
APE1 is a site for redox regulation by the dithiol-reducing enzyme Trx (61, 109, 132, 141), through Cys35 and Cys32 in the catalytic center of Trx, and involving the Cys65 redox sensitive site of APE1 (139, 141, 146). The Trx-mediated redox regulation of APE1 is required for the functional activation of p53 (132) and AP-1 (61). Though the biological relevance of Cys65 residue seems determined, it is not currently known whether this Cys residue undergoes PTM in vivo.
Qu et al. (110) demonstrated that two (Cys93 and Cys310) of the seven Cys residues of APE1 can undergo S-nitrosation in response to nitric oxide stimulation, leading to nucleus to cytoplasm relocalization of the protein in a CRM1-independent process, possibly as a consequence of demasking a putative nuclear export signal (aa 64–80). S-nitrosation may therefore constitute a specific molecular switch to strictly control the intracellular distribution of APE1 between nucleus and cytoplasm, and provides a new working hypothesis for the cytoplasmic accumulation of APE1 observed in more aggressive tumors (126). Unfortunately, no detail is available about the functional implications of S-nitrosation on the different biological functions of APE1. Accordingly, since both NO and APE1 are associated with tumorigenesis and neurodegenerative diseases, future work is needed to address whether nitrosative stress leads to genomic instability, and may be the target for designing new therapeutic strategies.
An interesting post-translational processing that has been recently described is proteolysis occurring at residue Lys31. This PT regulation of APE1 protein is responsible for enhanced cell death mediated by granzyme A (GzmA) (29) and granzyme K (GzmK) (52). APE1 is associated with the endoplasmic reticulum in a macromolecular complex of 270–420 kDa containing evolutionarily conserved proteins called SET, pp32, and HMG2. GzmA cleaves APE1 after Lys31, giving rise to a protein form called NΔ33APE1, and alters its ability to be actively accumulated within nuclei of cells (15, 69, and our unpublished observations) and to interact with XRCC1 (136). However, some authors claimed that truncated APE1 may loose its AP-endonuclease activity (29) and acquire a nonspecific DNAse function (150). This peculiar processing is not limited to immune cells but may constitute a general molecular device for redirecting APE1 to mitochondria (125), as suggested by Chattopadhyay et al. (15) and Mitra et al. (88), in spite of the intriguing finding of a proteolysis occurring at the level of Asn33 rather than Lys31. Again, if the removal of the terminal 31–33 amino acids is responsible for APE1 to move to the mitochondria to function in mitochondrial BER as an AP endonuclease, it is hard to understand this truncated protein having nonspecific DNAse activity (150) unless it is very cell-type specific. Accordingly, previous work by many investigators has never observed a nonspecific nuclease activity with the cleavage of the first 61 amino acids (66) and additional data clearly showed that the truncated APE1 protein has an unaltered AP-endonuclease activity (15, and our unpublished observations), at least in vitro.
While it is known that nuclear accumulation of APE1 triggers the activation of several transcription factors, the functional role of acetylation is barely understood. Acetylation of both histones and regulatory proteins is commonly catalyzed by the histone acetyltransferase (HAT) p300/CBP, and can be reversed by histone deacetylases (HDACs), which in turn control the acetylation level of transcription factors or co-activators (50, 74). Bhakat et al. have reported that the balance between the acetyltransferase activity of p300/CBP and the deacetylase activity of HDAC1 maintains APE1's acetylation at Lys residues 6 and 7 (K6, K7) in response to Ca2+ levels, thus controlling expression of target genes (7). More recently, we found that exposure of HeLa cells to H2O2 and to histone deacetylase inhibitors increases acetylation of APE1 at residues Lys6/Lys7, leading to Egr-1-mediated induction of the tumor suppressor PTEN gene expression (30). Our data open new perspectives in the comprehension of the many functions exerted by APE1 in controlling cell response to oxidative stress and underline the double-face nature of APE1 which plays a role in both pro-survival and in cell cycle arrest mechanisms. Interestingly, despite the very low homology degree in the N-terminal region (<40%), K6 or K6/K7 are much more conserved, thus reinforcing their primary role during phylogenesis.
Altogether, these observations have raised the possibility that subtle PTMs provide a means for channeling the multifunctional APE1 to different activities and interactions and thus could act as a regulatory switch in performing different functions. APE1 subcellular localization is quite variable. Most cell types exhibit only nuclear, others display only cytoplasmic, while others show both nuclear and cytoplasmic localization (126). Such a complex distribution pattern suggests that localization is not random but, on the contrary, is controlled by a strictly regulated process. Though of fundamental interest for a full comprehension of the role of APE1 in different pathological conditions, the clear understanding of the biological relevance of APE1 subcellular compartmentalization still remains elusive. Whereas we can rather easily figure out the role for nuclear localization of APE1 based on its main DNA repair and co-transcriptional activity, a convincing explanation for the extranuclear roles of APE1 is still evanescent. Cytoplasmic localization of APE1, such as that reported for fibroblasts, spermatocytes, thyrocytes, lymphocytes, hepatocytes, and hippocampal cells (20, 22, 24, 72, 73, 112, 126, 129, 142), is associated with high metabolic or proliferative rates and may be related to a cell cycle-dependent expression (36). Possible explanatory hypotheses for cytoplasmic expression of APE1 may come from the mitochondrial role of the protein, as described above. A further functional explanation comes from its association with endoplasmic reticulum membranes, as evidenced by ultrastructural (125) and biochemical (28, 29, 47) analysis. It has been suggested that APE1 redox activity in the cytoplasm may be required to maintain newly synthesized transcription factors in a reduced state during their translocation to the nucleus (24). Therefore, future work is required to shed more light on the extranuclear role(s) of APE1, starting from the explanation of its cytoplasmic function.
Molecular Basis for the Vital Role of APE1 in Mammalian Cells: New Perspectives
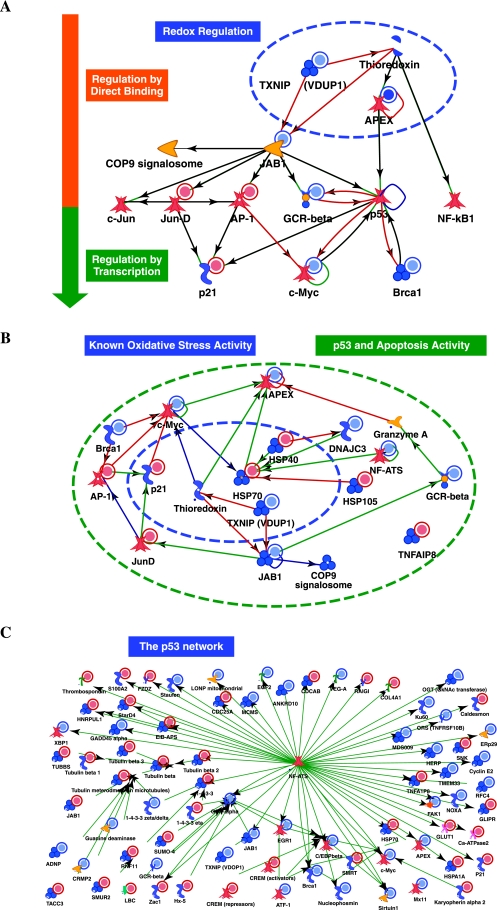
To understand the vital role of APE1 in mammalian cells, we recently combined gene expression array and proteomics analysis to identify the genes directly or indirectly regulated by APE1. By generating an inducible knockdown cell model, in which endogenous APE1 expression could be inhibited by siRNA technology (Vascotto et al., unpublished observations), we built up the molecular networks in which APE1 is involved (see Fig. 3A, B, and C for models network). APE1 silencing induces apoptosis through mitochondrial pathway and a strong inhibitory effect on cell growth. There is evidence of p53 activation and perturbation of the glucocorticoid receptor signaling pathway that could explain an induction of apoptosis. Glucocorticoid receptor NR3C1 (GCR-alpha) itself is downregulated, upon APE1 silencing, whereas p53 is not. Instead, a large number of p53 controlled transcription factors are differentially expressed in our APE1 silencing experiment, further supporting an important role of APE1 in modulating p53 functions (38, 54, 117) (Fig. 3B and C). Differentially expressed genes in our model strongly indicate a significant perturbation of the glucocorticoid receptor signaling pathway. Proteins of the HSP70 complex (i.e., HSPA1A, HSPA1B, and HSPA8) are significantly up-regulated as a consequence of APE1 silencing, and it is known that HSP70 is required for the assembly of the glucocorticoid receptor–HSP90 complex (92). Moreover, other key components and targets of glucocorticoid receptor signaling pathway were also on our gene list (Egr-1, C/EBP-beta, c-Myc, SUMO-4, 14-3-3 proteins, and tubulin proteins).
FIG. 3.
Different model networks for APE1-mediated signaling as derived by gene expression and proteomic data. Direct interaction network for genes dysregulated upon APE1 knock-down, involved in redox control, transcription regulators and chaperones (A) and (B) and in p53-signaling (C). Colors of the symbols indicate inhibition (blue) and activation (red). Here, the name APEX is used to refer to APE1. This model network is accessible at this site: https://portal.genego.com/pub/network/n-860173808.html. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
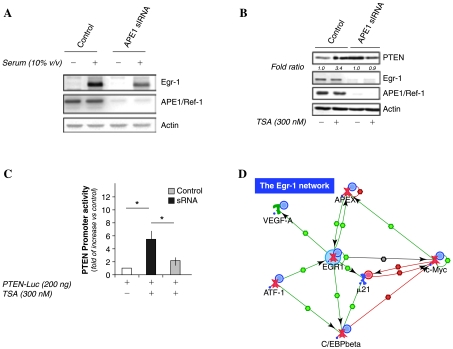
One of the genes that emerged from our analysis and is involved in the control of cell growth processes, as well as in the antioxidant response, is Egr-1. This transcription factor resulted as a good candidate target of APE1, since its expression was significantly reduced in APE1 knocked-down cells both basally and upon serum treatment after starvation (Fig. 4A). We therefore investigated the functional effect of APE1 silencing in controlling the expression of one of the most well-known Egr-1 target genes, the tumor suppressor PTEN, which is induced upon UV- and oxidative stress-cellular damage (5, 137). An impairment of Egr-1 expression upon APE1 silencing affected the inducible expression of PTEN gene with a transcriptional mechanism, confirming the dual nature of APE1 itself (Fig. 4B and C). Thus, the lack of APE1 impairs the adaptive cellular response to damaging agents. These findings further enlighten the double nature of APE1 to be involved in both the processes of cell growth and in growth arrest upon cellular damage. Interestingly, this kind of dual role nature in transcriptional regulation and DNA repair for proteins is consistently present in the five major DNA repair pathways, that is, homologous recombinational repair (HRR), nonhomologous end joining (NHEJ), nucleotide excision repair (NER), BER and mismatch repair (MMR), as in the cases of BRCA1, ATM, and p53 itself (for review, refer to Ref. 6). The existence and the correct regulation of a mechanism shifting cells from DNA repair to apoptosis is central to avoid progression to cancer, preventing clonal expansion of cells in which unrepaired damage would lead to mutation and to carcinogenesis. In this regard, it is interesting to note that a number of DNA repair genes (i.e., GADD45, BRCA1) were downregulated as a consequence of APE1 silencing, suggesting the existence of a cross-regulation of the expression between individual partners of different pathways and underlining the central role of APE1 in DNA repair processes with a different function besides the well-known AP-endonuclease activity. This hypothesis will require further investigation.
FIG. 4.
Functional inactivation of Egr-1 transcriptional activity on PTEN target gene. HDACs inhibitors-induced PTEN upregulation requires APE1 expression. (A) Inducible expression of Egr-1 by 1 h serum treatment after o/n starvation is significantly reduced in APE1-silenced cells. Endogenous APE1 silencing was obtained by inducible RNAi strategy (30). Expression of Egr-1 and APE1 proteins was evaluated by Western blot analysis on 10 μg of nuclear extracts as previously described (30). (B) Induction of endogenous PTEN expression by HDACs inhibitors is inhibited in HeLa cells in which Egr-1 is diminished upon constitutive stable APE1 knock-down. To evaluate the endogenous PTEN levels upon HDACs inhibitors treatment, control and knocked-down HeLa cells were treated with TSA for 30 h and further collected and lysed to obtain total extracts that were subsequently used for Western blot analysis. Ten μg of extracts were separated onto a 10% SDS-PAGE, blotted onto nitrocellulose membranes, and assayed for the presence of APE1 protein by using the monoclonal anti-APE1 antibody, for the presence of PTEN protein by using the monoclonal anti-PTEN antibody and for the presence of Egr-1 protein by using a specific polyclonal antibody. Actin was also measured, as loading control. Values represent the relative amount of PTEN with respect to control untreated cells, after normalization for actin content. The mean value of two independent experiments is reported. (C) Induction of PTEN promoter activity by HDACs inhibitors is prevented in constitutive stable APE1 knocked-down HeLa cells in which Egr-1 is significantly diminished. APE1 silencing was obtained as described in legend to (A). After 10 days of doxycycline treatment, HeLa cells were transfected with plasmid containing the PTEN promoter-Luc sequence and then treated with TSA for 30 h. Twenty-four hours after treatment, cells were harvested, and luciferase and β-galactosidase activities were measured. Bars indicate the mean value ± SD of three independent experiments. (*p < 0.05 by Student's t-test). (D) Model network involving APE1-Egr 1 functional interaction. This model network is accessible at this site: https://portal.genego.com/pub/network/n-839812191.html. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
The interesting finding that, besides its activity as a redox coactivator for Egr-1 transcriptional activity (64, 107), APE1 is also able to regulate Egr-1 expression levels reinforces the previously suggested hypothesis (107) of the existence of a functional loop between APE1 and Egr-1 in reciprocally modulating each other. This finding strengthens the biological complexity typical of this multifunctional protein and could well be a molecular tool by which APE1 may regulate the expression of DNA repair genes such as GADD45 (130).
Egr-1 activates the transcription of genes in response to a variety of mitogenic and nonmitogenic stimuli, including growth factors and hypoxia (75) and it is able to form redox-modulated transcriptional complexes with the AP-1 as well as with APE1 and Trx (76). Accordingly, in line with the data obtained in our cell model, the role of Egr-1 seems to be pivotal. The central effect of APE1 on Egr-1 biological functions is also reinforced by the concomitant downregulation of additional Egr-1 target genes such as C/EBPβ, VEGF and c-Myc (Vascotto et al., unpublished observations; 135). Based on these findings the networking model depicted in Fig. 4D was assembled.
Future Perspectives in Elucidating the Pleiotropic Function of APE1 in Mammalian Cells at the Molecular Level. Looking at the Roles of the Mitochondrial and the Subnuclear Distribution of APE1. Could the N-Terminal Unconserved Domain Make the Difference?
As mentioned above, removal of APE1 NLS through proteolysis controls the amount of APE1 present within the nuclear compartment and would constitute an elegant tool to control APE1 alternative functions in noncanonical subcellular compartments, as mitochondria. Unfortunately, in non-immune cells, neither the identity of the specific protease responsible for this cleavage nor the mitochondrial localization signal (MTS) have been determined yet. The relatively high molecular weight of APE1 is not fully compatible with a passive mechanism of translocation through the outer membrane of mitochondria, and therefore it may require the presence of a specific regulatory transport mechanism. Whereas a large majority of proteins synthesized in the cytoplasm localize into mitochondria by means of an N-terminal MTS, a significant fraction of mitochondrial proteins lack this recognition signal. In the case of APE1, it has been suggested that the MTS may reside in the C-terminal 69 amino residues of the protein (110). Mitochondrial localization of APE1 may be associated to a potential role in DNA repair of oxidized bases in the mitochondrial genome (15, 88, 126). However, since it is not clear whether, in vivo, NΔ33APE1 maintains its DNA repair activity (29) or, as previously suggested, may acquire an aspecific endonuclease activity for dsDNA in vitro (150), at present it is impossible to derive any definite conclusion. Moreover, since generation of truncated NΔ33APE1 form is associated with the occurrence of an apoptotic phenotype (52; Vascotto et al., unpublished observations), it cannot be excluded that this APE1 form may be causatively involved in the cytotoxic effect driving pro-apoptotic triggering directly from mitochondria. Preliminary data obtained in our laboratory in cells expressing a noncleavable mutant APE1 protein (i.e., 31–34A mutant) seem to support this hypothesis. Should this be confirmed, strategies to modulate the proteolytic removal of the APE1 N-terminus will constitute a possible good candidate target for future drug development. On the other side, it is not completely clear whether truncation of APE1 N-terminus may affect APE1 redox activity (29, 52), leaving open the debate on the biological meaning of the process. Since the N-terminal region of the protein seems to be a result of phylogenetic evolution, it is intriguing that this part of the protein could account for the difference in modulating the various functions of APE1.
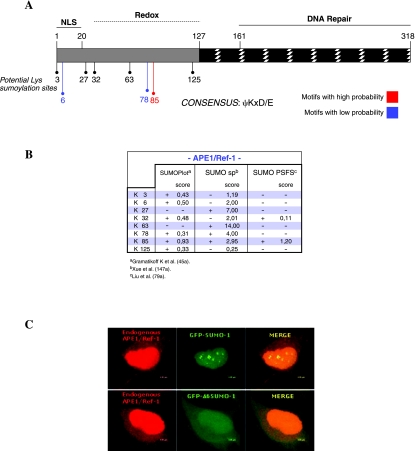
In addition to the canonical 37 kDa APE1 protein, and the truncated NΔ33APE1 form, in some circumstances the appearance of a lower mobility band of ~50 kDa has been detected (our unpublished data). This form would account for a PTM that introduces a substantial modification to the wild-type protein. A recently identified PTM, typically occurring on Lys residues of several nucleoproteins, is sumoylation (40, 56). The addition of a small ubiquitin-like modifier (SUMO) molecule to the target protein accounts for an increase of ~12 kDa of the apparent molecular mass of the target protein itself. Many SUMO-modified proteins function in regulation of transcription, chromatin structure, maintenance of the genome stability, and signal transduction. Upon sumoylation, interactions that are dependent on other post-translational modifications or on unmodified lysine are lost. Sumoylation also promotes novel interactions, in some cases associated with a conformational change in the target protein. The effects of PTM by SUMO to compete for target lysines enhance or inhibit interactions with other proteins (or other binding partners, such as DNA) or induce conformational changes. These effects are not mutually exclusive, but might not all occur on the same substrate. Thus, we investigated the possibility that APE1 may undergo sumoylation. APE1 protein contains 29 Lys residues, 15 of them lie in the N-terminal domain, and 14 residues lie in the C-terminal domain of the protein. Protein sequence analysis using three different softwares for prediction of sumoylation sites (SUMOPlot, SUMO sp, and SUMO PSFS) revealed several “low probability” putative sumoylation sites and a “high probability” putative sumoylation site, which contains a canonical sumoylation ψ-K-X-D/E motif (Fig. 5A and B). Putative sumoylation sites do not distribute homogenously along the whole sequence, but concentrate at the N-terminal domain, which is not present in functionally related proteins from other organisms and is required for the redox activity of APE1. Co-localization experiments on cells transfected with GFP-SUMO-1 clearly demonstrated that endogenous APE1 can be modified by wild-type SUMO-1 but not by a deletion mutant (i.e., Δ6SUMO-1), which looses its ability to modify target proteins (Fig. 5C). In vitro sumoylation experiments also clearly demonstrated that APE1 may undergo sumoylation (unpublished data). Our inability to identify the Lys residue target of sumoylation, by mutagenizing the putative sumoylation sites identified during in silico analysis (unpublished data), may suggest that multiple Lys residues could be the simultaneous target of this PTM. Additional work, which is difficult as sumoylated APE1 represents a very tiny fraction in comparison to the nonsumoylated protein, is required to understand the role of sumoylation in controlling APE1 functions.
FIG. 5.
(A) APE1 protein sequence bears several putative sumoylation sites, as from in silico analysis. APE1 protein contains 29 Lysine residues, 15 of them lie in the N-terminal domain of the protein. Protein sequence analysis using three different softwares for prediction of sumoylation sites (SUMOPlot, SUMO sp, and SUMO PSFS) revealed several “low probability” putative sumoylation sites and a “high probability” putative sumoylation site, which contains a canonical sumoylation ψ-K-X-D/E motif. Putative sumoylation sites do not distribute homogenously along whole sequence, but concentrate at N-terminal domain, which is not present in functionally related proteins from other organisms and is required for the redox activity of APE1. (B) Of the 29 Lys residues, 8 of them are identified as potential sumoylation sites with the three different softwares. Lys85represents a classical consensus sequence, recognized by all progr ams. Lys78 is recognized as low-probability consensus sites by two different programs, while five Lys residues (K3, K6, K27, K63, and K125) are recognized as potential sumoylation sites by only one program. Interestingly, all potential sumoylation sites reside within the Redox transactivation domain. (C) APE1 colocalizes with SUMO-1, but not with Δ6 SUMO-1, into nuclear subdomains, presumably nuclear bodies. HeLa cells were transfected with either pGFPSUMO-1 or pGFPΔ6SUMO-1, as a negative control. The inactive Δ6SUMO-1 is a deletionmutant lacking the 6 C-terminal aminocid residues, including double glycines being the site for the isopeptidic bond with the target protein, whichcannot sumoylate target proteins located in the nuclear bodies. Thus, this mutant relocalizes within nucleus and cytoplasm in a diffuse way. APE1 staining was analyzed using a monoclonal anti-APE1 antibody. Primary antibody and GFPSUMO-1 staining were revealed by incubation with Alexa Fluor 546 conjugated secondary antibody or by intrinsic green fluorescence of GFP, respectively. Merging of the two colors results in a yellow signal, corresponding to co-localized proteins. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
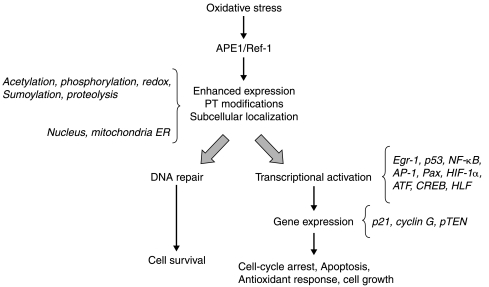
In summary, all of these studies have confirmed the dual role nature of APE1, as a prototypical example of an apparent biological paradox. While a number of reports clearly demonstrated the antiapoptotic roles as well as the positive effect on cell proliferation (for a review, see Ref. 126, and 35, 67, 88), other data underlined its potential role in controlling proapoptotic functions through p53-mediated activation of p21 (38, 54, 117), leading to the arrest of cell cycle by inhibiting cyclin-dependent kinase function (11), and cyclin G, having pro-apoptotic activity (97). However, it is becoming increasingly evident that the antiapoptotic roles of APE1 are ascribable to its DNA repair functions (35) rather than to its activities as a transcriptional co-activator. Thus, it is tempting to speculate that the latter function—activation of transcription factors (i.e., p53 and Egr-1) ensuring efficient cell-cycle arrest—may act in concert with the previous (repair of DNA damage) to protect cells from accumulation of oxidative damage (Fig. 6) and be a later evolutionarily acquired function since only mammals and not just vertebrates appear to have the redox signaling function (39). This may have been an advantageous addition to APE1's interactions and functions that benefited mammalian cells, given the potentially toxic nature of their environment. This may have subsequently led to additional signaling and regulatory interactions and functions. Obviously, for a proper modulation of these two interconnected functions, a fine-tuned regulation of APE1 activities is required. Thus, a better understanding of the processes controlling APE1 subcellular distribution, of the post-translational modifications occurring on the protein itself, of the mechanisms controlling protein half-life, and of the different interacting partners recruited as a function of cellular response, is required to fully address this paradoxical issue.
FIG. 6.
Model of APE1 multifunctional roles in coordinating cell response to oxidative stress. APE1 has dual functions in cellular response tooxidative stress by acting as an AP-endonuclease in DNA repair and as a transcriptional regulator of various transcription factors leading to cell-cycle arrest or to cell survival. APE1's post-translational modifications and regulation of its intracellular trafficking may be critical in in vivo fine-tuning of its activities.
Clinical Perspectives: Altered Expression/Distribution of APE1 and Human Pathology
Accumulating evidence has demonstrated that the heterogeneity of APE1 expression pattern is linked to different pathological conditions ranging from metabolic to differentiative disorders, including cancer and neurodegenerative diseases. Different kinds of human tumors were characterized by alterations in subcellular distribution of APE1 with respect to nontumoral tissue (73; for review, see 126). Generally, APE1 localization is eminently nuclear, while in several carcinomas a nuclear, cytoplasmic, and nuclear/cytoplasmic staining was observed (126). This peculiar distribution correlates well with the aggressiveness and prognosis of the tumor as nuclear localization was always associated with a better prognostic feature together with a higher degree of cellular differentiation, low angiogenesis, and negative lymph node status. As in hepatocellular carcinoma (HCC), a cytoplasmic localization of APE1 was associated with a significant lower degree of differentiation and with a shorter survival time, the localization of APE1 in liver biopsy is of prognostic value (22). Noteworthy, alteration in sub-cellular distribution of APE1 is not functionally related to the ability of cancerous tissue to repair abasic sites, suggesting that DNA repair by BER may not be affected (8, 113, 115). Therefore, it appears that the extranuclear roles of APE1 are responsible for its association with cancer.
Whether the alterations of APE1 subcellular localization are causally responsible or only associated with tumor progression is not clear at present. However, our recent findings in HCC, in which a chronic cellular oxidative stress conditions (91) in a milieu of inflammation (106) plays a central role in tumor progression implying functional activation of pro-survival transcription factors such as NF-κB and STAT3 (43, 84), supports the latter conclusion. To definitely address this point, it is mandatory that the interactome network of APE1 as well as the role of PTM in controlling the cytoplasmic localization of the protein must be unveiled. An interesting working hypothesis for a role of APE1 in the inflammatory process comes from recent works on APE1-mediated IL-8 production in Helicobacter pylori infections (96). These authors demonstrated that APE1 is required for H. pylori infection-mediated induction of IL-8 production by gastric cells through the involvement of NF-κB and AP-1 transcription factors (96). Thus, APE1 seems to play also a leading role in the production of inflammatory cytokines. Future work will be needed to extend these observations and to open new applicative perspectives in molecular medicine.
Other than in proliferative disorders mentioned above, APE1 deregulation has also been demonstrated in other pathologies, in particular degenerative disorders. Neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease, and amyotrophic lateral sclerosis (ALS), are characterized by a condition of chronic oxidative stress that primarily contributes to the pathogenesis through apoptosis of neuronal cells (33). APE1 is highly expressed in selected regions of the central nervous system (99, 100, 142). A reduction in APE1 expression, followed by an increase in the apoptotic rate, occurs in the hippocampus after a hypoxic–ischemic injury (41), in the cortex after compression injury (79), and in the spinal cord after ischemia (116). The hippocampus of patients with Alzheimer shows an increased expression of APE1 levels in senile plaques and plaque-like structures (123). Our recent unpublished data demonstrated an increased nuclear expression of APE1 in neuronal and glial cells of the cerebral cortex in both familial and sporadic Alzheimer (Marcon et al., unpublished data). These findings, together with the observation of an increased DNA repair mechanism in Alzheimer (18), may be associated with the cellular adaptive response to the oxidative stress condition typical of Alzheimer, and may be involved in the pathogenesis of the disease. Despite the abundance of APE1 in neurons and the correlations between alterations in APE1 levels and various neuropathologies, few studies have addressed the role of APE1 in preventing neurotoxicity. Recent data demonstrate that APE1 protects primary cultures of hippocampal and sensory neurons from oxidative damage induced by H2O2 (134). However, despite its fundamental importance for future therapeutic development, the precise mechanism of the protective effect is still poorly understood.
Translational, Clinical Applications of APE1 Redox Inhibition for Cancer or Other Areas
As discussed in the previous sections, APE1 has two primary and major activities: DNA BER and redox signaling of downstream transcription factor targets. While previous studies have demonstrated that altering APE1 levels leads to blockage of cell growth and increased cancer cell sensitivity (8, 9, 16, 26, 31, 32, 55, 59, 65, 78, 80–83, 87, 98, 120, 124, 131, 138, 140, 149), these studies have either used overexpression of APE1, APE1 antisense oligonucleotides, or APE1 siRNA. The dilemma with this approach, while valid, is that each of these procedures changes the total cellular content level of APE1 and removes all of APE1's functions, not just the repair or redox activities. Because APE1 has multiple functions, as well as interactions with many other proteins (belonging to DNA repair, signaling and to the Egr-1 pathway), the increase or decrease of APE1 protein may result in multiple effects in which the APE1's specific role cannot be easily depicted. Furthermore, recent studies tested the hypothesis that APE1 is responsible for mediating production of single-strand DNA (ssDNA) breaks in gene promoters during repair of targeted base oxidation lesions caused by oxygen radicals generated during physiologic signaling (10, 153, 154). Production of ssDNA breaks is believed to play a key role during transcription by imparting substantial flexibility to promoter sequences, enabling them to bend in a manner that establishes the chromatin architecture needed for gene expression. In addition, APE1, besides forming the ssDNA break, is also required for high-fidelity repair of the break (104, 105, 119). Thus, defects in the APE1-mediated step in BER pathway could be linked to altered gene expression besides altering transcription factor state.
Use of specific small molecule inhibitors, such as one that blocks APE1 redox, but not repair, will be important to delineate the distinct roles of APE1 in various cancers, other diseases, and normal cellular functions. Likewise, an APE1 specific repair inhibitor will help to elucidate that role (4). Ultimately, using APE1 redox inhibitors with APE1 specific endonuclease repair inhibitors will give a clearer picture of the multiple activities of APE1.
APE1 was originally identified as the primary target of E3330 (3-[5-(2,3-dimethoxy-6-methyl-1,4-benzoquinoyl)]-2-nonyl-2-propionic acid), a small molecule redox inhibitor (155). E3330 was immobilized on beads and APE1 was identified from a nuclear extract of a leukemia cell line as a protein that specifically bound to E3330. Using surface plasmon resonance (SPR), an equilibrium constant (KD) of 1.6 nM was obtained for the binding of E3330 to APE1, suggesting a specific interaction. E3330 was also shown to block the ability of APE1 to reduce NF-κB, thus interfering with the redox activity of APE1 (45, 60, 89, 118). The proposed binding site on APE1 is somewhat puzzling. The amino acid residues 72–80 form a ridge on the surface of the molecule with no obvious cavities or binding pockets that are large enough to bind E3330. Currently, we are pursuing studies to delineate the binding site or region of E3330 on APE1.
Our recent data have demonstrated that E3330 blocks the redox function of APE1 with AP-1 as the downstream target in vitro (86). Additionally, using a transactivation assay for AP-1 or HIF-1α targets in ovarian cancer cells, increasing amounts of E3330 led to decreased activation of a luciferase reporter downstream of AP-1 and HIF-1α (86). While E3330 blocked APE1's redox function, it had no effect on APE1 repair endonuclease activity nor other members of the BER pathway (86). These studies demonstrate the specificity of E3330 for APE1's redox, but not for its repair function. This is supported by our recently published data in ovarian cancer studies in xenografts demonstrating that the knockdown of APE1 results in the blocking of cell growth and proliferation, but not necessarily cell death (31). This is the first time that cancer cell killing has been reported using a small molecule inhibitor of APE1 redox function.
E3330 was also shown to have single agent inhibition of cell growth using a variety of cancer cell lines including, ovarian, colon, lung, breast, brain, pancreatic, prostate, and multiple myeloma cancers (Kelley et al. unpublished observations). In stark contrast, we do not see significant growth inhibition in our studies with normal cells such as hematopoietic embryonic cells (155) or in human CD34+ progenitor cells (Kelley et al. unpublished observations). These data are novel in that they implicate the redox role of APE1 in cancer, but not “normal”, cell survival.
Recent studies using this APE1 redox inhibitor in angiogenesis model systems have been instructive. Proliferation of endothelial cells (EC) is an important index for angiogenic ability of EC in vitro. E3330 inhibited retinal vascular endothelial cells (RVEC) growth in a dose-dependent manner (86). Additionally, an in vitro angiogenesis assay was also used to determine the effect of E3330 on RVEC formation of capillary-like structures on Matrigel. The inhibitory effect of E3330 was similar to the proliferation assay with the complete loss of tube formation at low micromolar concentrations. These results demonstrate that blocking APE1's redox function attenuates RVEC proliferation and capillary formation in vitro and these findings implicate the use of an APE1 redox inhibitor in antiangiogenic translational studies. Further mechanistic studies as to how this is occurring are in progress, although preliminary studies indicate the redox inhibitory effect is not necessarily related to cell killing, but to a block in cell proliferation or cytostatic effect similar to that observed using APE1 siRNA in vivo (31).
In conclusion, APE1 is a multifunctional protein with both important DNA repair and redox capabilities. However, in order to demarcate the various functions of APE1, small molecule inhibitors of each function will be necessary to ultimately conclude which function is required in normal and cancer cell function. Recent findings with redox inhibition of APE1 have potential clinical translational significance such that a redox inhibitor could be used as a single agent, in combination with current treatments or as a potential antigrowth, cytostatic agent. Furthermore, new APE1 redox analogues could play a role in antiangiogenic therapies.
Acknowledgments
This work was supported by grants from MIUR (FIRB #RBRN07BMCT), Telethon (Grant #GGP05062 and #GGP06268), MAE (Executive Programme of Cooperation in the field of Science and Technology), and Regione Friuli Venezia Giulia to G.T. and C.T., and by the National Institutes of Health, National Cancer Institute CA94025, CA106298, CA114571 and CA121168 and the Riley Children's Foundation to M.R.K.
The authors also thank Dr. Carlo Vascotto, Dr. Milena Romanello, Dr. Damiano Fantini, and Dr. Leo Zeef for gene networks and sumoylation analysis.
Abbreviations
AP, apurinic/apyrimidinic; AP-1, activator protein-1; BER, base excision repair; CKI and CKII, casein kinase I and II; CREB, cAMP response element binding protein; Egr-1, early growth response protein-1; FEN1, flap endonuclease I; GSK3, glycogen synthase kinase 3; GzmA, granzyme A; GzmK, granzyme K; HIF-1α, hypoxia inducible factor-1α; MTS, mitochondrial targeting sequence; MPG, methylpurine DNA glycosylase; nCaRE, negative calcium-responsive regulatory element; NF-κB, nuclear factor-κB; Ogg1, 8-oxoguanosine DNA glycosylase; P2Y, purinergic receptors (G-proteins coupled); Pax, paired box containing genes; PCNA, proliferating cell nuclear antigen; PEBP-2, polyoma virus enhancer-binding protein-2; PKC, protein kinase C; PTH, parathyroid hormone; PTM, post-translational modification; RFC, replication factor C; ROS, reactive oxygen species; SPR, surface plasmon resonance; ssDNA, single-stranded DNA; SUMO, small ubiquitin-like modifier; TFs, transcription factors; Trx, thioredoxin; TTF-1, thyroid transcription factor-1; XRCC1, X-ray cross-species complementing 1.
References
- 1.Akamatsu Y. Ohno T. Hirota K. Kagoshima H. Yodoi J. Shigesada K. Redox regulation of the DNA binding activity in transcription factor PEBP2. The roles of two conserved cysteine residues. J Biol Chem. 1997;272:14497–14500. doi: 10.1074/jbc.272.23.14497. [DOI] [PubMed] [Google Scholar]
- 2.Angkeow P. Deshpande SS. Qi B. Liu YX. Park YC. Jeon BH. Ozaki M. Irani K. Redox factor-1: An extranuclear role in the regulation of endothelial oxidative stress and apoptosis. Cell Death Differ. 2002;9:717–725. doi: 10.1038/sj.cdd.4401025. [DOI] [PubMed] [Google Scholar]
- 3.Babior BM. NADPH oxidase: An update. Blood. 1999;93:1464–1476. [PubMed] [Google Scholar]
- 4.Bapat A. Fishel ML. Georgiadis M. Kelley MR. Going ape as an approach to cancer therapeutics. Antioxid Redox Signal. in press. [DOI] [PMC free article] [PubMed]
- 5.Baron V. Adamson ED. Calogero A. Ragona G. Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13:115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein C. Bernstein H. Payne CM. Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: Fail-safe protection against carcinogenesis. Mutat Res. 2002;511:145–178. doi: 10.1016/s1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 7.Bhakat KK. Izumi T. Yang SH. Hazra TK. Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;1:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobola MS. Blank A. Berger MS. Stevens BA. Silber JR. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin Cancer Res. 2001;7:3510–3518. [PubMed] [Google Scholar]
- 9.Bobola MS. Finn LS. Ellenbogen RG. Geyer JR. Berger MS. Braga JM. Meade EH. Gross ME. Silber JR. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin Cancer Res. 2005;11:7405–7414. doi: 10.1158/1078-0432.CCR-05-1068. [DOI] [PubMed] [Google Scholar]
- 10.Breit JF. Ault–Ziel K. Al–Mehdi AB. Gillespie MN. Nuclear protein-induced bending and flexing of the hypoxic response element of the rat vascular endothelial growth factor promoter. FASEB J. 2008;22:19–29. doi: 10.1096/fj.07-8102com. [DOI] [PubMed] [Google Scholar]
- 11.Brugarolas J. Chandrasekaran C. Gordon JI. Beach D. Jacks T. Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 12.Campalans A. Amouroux R. Bravard A. Epe B. Radicella JP. UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J Cell Sci. 2007;120:23–32. doi: 10.1242/jcs.03312. [DOI] [PubMed] [Google Scholar]
- 13.Cao X. Kambe F. Ohmori S. Seo H. Oxidoreductive modification of two cysteine residues in paired domain by Ref-1 regulates DNA-binding activity of Pax-8. Biochem Biophys Res Commun. 2002;297:288–293. doi: 10.1016/s0006-291x(02)02196-4. [DOI] [PubMed] [Google Scholar]
- 14.Cesaratto L. Calligaris SD. Vascotto C. Deganuto M. Bellarosa C. Quadrifoglio F. Ostrow JD. Tiribelli C. Tell G. Bilirubin-induced cell toxicity involves PTEN activation through an APE1/Ref-1-dependent pathway. J Mol Med. 2007;85:1099–1112. doi: 10.1007/s00109-007-0204-3. [DOI] [PubMed] [Google Scholar]
- 15.Chattopadhyay R. Wiederhold L. Szczesny B. Boldogh I. Hazra TK. Izumi T. Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen DS. Olkowski ZL. Biological responses of human apurinic endonuclease to radiation-induced DNA damage. Ann NY Acad Sci. 1994;726:306–308. doi: 10.1111/j.1749-6632.1994.tb52834.x. [DOI] [PubMed] [Google Scholar]
- 17.Chung U. Igarashi T. Nishishita T. Iwanari H. Iwamatsu A. Suwa A. Mimori T. Hata K. Ebisu S. Ogata E. Fujita T. Okazaki T. The interaction between Ku antigen and REF1 protein mediates negative gene regulation by extracellular calcium. J Biol Chem. 1996;271:8593–8598. doi: 10.1074/jbc.271.15.8593. [DOI] [PubMed] [Google Scholar]
- 18.Davydov V. Hansen LA. Shackelford DA. Is DNA repair compromised in Alzheimer's disease? Neurobiol Aging. 2003;24:953–968. doi: 10.1016/s0197-4580(02)00229-4. [DOI] [PubMed] [Google Scholar]
- 19.De Keulenaer GW. Chappell DC. Ishizaka N. Nerem RM. Alexander RW. Griendling K. K. Oscillatory and steady laminar shear stress differentially affect human endothelial redox state: Role of a superoxide-producing NADPH oxidase. Circ Res. 1998;82:1094–1101. doi: 10.1161/01.res.82.10.1094. [DOI] [PubMed] [Google Scholar]
- 20.Deganuto M. Pittis MG. Pines A. Dominissini S. Kelley MR. Garcia R. Quadrifoglio F. Bembi B. Tell G. Altered intracellular redox status in Gaucher disease fibroblasts and impairment of adaptive response against oxidative stress. J Cell Physiol. 2007;212:223–235. doi: 10.1002/jcp.21023. [DOI] [PubMed] [Google Scholar]
- 21.Demple B. Herman T. Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: Definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Maso V. Avellini C. Crocè LS. Rosso N. Quadrifoglio F. Cesaratto L. Codarin E. Bedogni G. Beltrami CA. Tell G. Tiribelli C. Subcellular localization of APE1/Ref-1 in human hepatocellular carcinoma: possible prognostic significance. Mol Med. 2007;13:89–96. doi: 10.2119/2006-00084.DiMaso. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 24.Duguid JR. Eble JN. Wilson TM. Kelley MR. Differential cellular and subcellular expression of the human multifunctional apurinic/apyrimidinic endonuclease (APE/ref-1) DNA repair enzyme. Cancer Res. 1995;55:6097–6102. [PubMed] [Google Scholar]
- 25.Ema M. Hirota K. Mimura J. Abe H. Yodoi J. Sogawa K. Poellinger L. Fujii–Kuriyama Y. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: Their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans AR. Limp–Foster M. Kelley MR. Going APE over ref-1. Mutat Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 27.Fan J. Matsumoto Y. Wilson DM., 3rd Nucleotide sequence and DNA secondary structure, as well as replication protein A, modulate the single-stranded abasic endonuclease activity of APE1. J Biol Chem. 2006;281:3889–3898. doi: 10.1074/jbc.M511004200. [DOI] [PubMed] [Google Scholar]
- 28.Fan Z. Beresford PJ. Zhang D. Lieberman J. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol Cell Biol. 2002;22:2810–2820. doi: 10.1128/MCB.22.8.2810-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan Z. Beresford PJ. Zhang D. Xu Z. Novina CD. Yoshida A. Pommier Y. Lieberman J. Cleaving the oxidative repair protein APE1 enhances cell death mediated by granzyme A. Nat Immunol. 2003;4:145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- 30.Fantini D. Vascotto C. Deganuto M. Bivi N. Gustincich S. Marcon G. Quadrifoglio F. Damante G. Bhakat KK. Mitra S. Tell G. APE1/Ref-1 regulates PTEN expression mediated by Egr-1. Free Radic Res. 2008;42:20–29. doi: 10.1080/10715760701765616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fishel ML. He Y. Reed AM. Chin–Sinex H. Hutchins GD. Mendonca MS. Kelley MR. Knockdown of the DNA repair and redox signaling protein Ape1/Ref-1 blocks ovarian cancer cell and tumor growth. DNA Repair (Amst) 2008;7:177–186. doi: 10.1016/j.dnarep.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fishel ML. He Y. Smith ML. Kelley MR. Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin Cancer Res. 2007;13:260–267. doi: 10.1158/1078-0432.CCR-06-1920. [DOI] [PubMed] [Google Scholar]
- 33.Fonnum F. Lock EA. The contributions of excitotoxicity, glutathione depletion and DNA repair in chemically induced injury to neurones, exemplified with toxic effects on cerebellar granule cells. J Neurochem. 2004;88:513–531. doi: 10.1046/j.1471-4159.2003.02211.x. [DOI] [PubMed] [Google Scholar]
- 34.Fritz G. Kaina B. Phosphorylation of the DNA repair protein APE/REF-1 by CKII affects redox regulation of AP-1. Oncogene. 1999;18:1033–1040. doi: 10.1038/sj.onc.1202394. [DOI] [PubMed] [Google Scholar]
- 35.Fung H. Demple B. A vital role for APE1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 36.Fung H. Bennett RA. Demple B. Key role of a downstream specificity protein 1 site in cell cycle-regulated transcription of the AP endonuclease gene APE1/APEX in NIH3T3 cells. J Biol Chem. 2001;276:42011–42017. doi: 10.1074/jbc.M106423200. [DOI] [PubMed] [Google Scholar]
- 37.Fung H. Liu P. Demple B. ATF4-dependent oxidative induction of the DNA repair enzyme Ape1 counteracts arsenite cytotoxicity and suppresses arsenite-mediated mutagenesis. Mol Cell Biol. 2007;27:8834–8847. doi: 10.1128/MCB.00974-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaiddon C. Moorthy NC. Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georgiadis M. Luo M. Gaur R. Delaplane S. Li X. Kelley M. Evolution of the redox function in mammalian apurinic/apyrimidinic. Mut Res. 2008;643:54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Gillardon F. Bottiger B. Hossmann KA. Expression of nuclear redox factor ref-1 in the rat hippocampus following global ischemia induced by cardiac arrest. Brain Res Mol Brain Res. 1997;52:194–200. doi: 10.1016/s0169-328x(97)00237-4. [DOI] [PubMed] [Google Scholar]
- 42.Giorgio M. Trinei M. Migliaccio E. Pelicci PG. Hydrogen peroxide: A metabolic by-product or a common mediator of ageing signals? Nat Rev Mol Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 43.Gong G. Waris G. Tanveer R. Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc Natl Acad Sci USA. 2001;98:9599–9604. doi: 10.1073/pnas.171311298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorman MA. Morera S. Rothwell DG. de La Fortelle E. Mol CD. Tainer JA. Hickson ID. Freemont PS. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goto M. Yamada K. Katayama K. Tanaka I. Inhibitory effect of E3330, a novel quinone derivative able to suppress tumor necrosis factor-alpha generation, on activation of nuclear factor-kappa B. Mol Pharmacol. 1996;49:860–873. [PubMed] [Google Scholar]
- 45a.Gramatikoff K. Wu C. Shi X. Fang F. SUMO: The proteome's little prince. In: Kang Z, editor. Frontiers of Biotechnology and Pharmaceuticals. Vol. 4. Science Press USA Inc; 2004. pp. 181–210. [Google Scholar]
- 46.Gray MJ. Zhang J. Ellis LM. Semenza GL. Evans DB. Watowich SS. Gallick GE. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–3120. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- 47.Grillo C. D'Ambrosio C. Scaloni A. Maceroni M. Merluzzi S. Turano C. Altieri F. Cooperative activity of Ref-1/APE and ERp57 in reductive activation of transcription factors. Free Radic Biol Med. 2006;41:1113–1123. doi: 10.1016/j.freeradbiomed.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 48.Grosch S. Kaina B. Transcriptional activation of apurinic/apyrimidinic endonuclease (Ape, Ref-1) by oxidative stress requires CREB. Biochem Biophys Res Commun. 1999;261:859–863. doi: 10.1006/bbrc.1999.1125. [DOI] [PubMed] [Google Scholar]
- 49.Grösch S. Fritz G. Kaina B. Apurinic endonuclease (Ref-1) is induced in mammalian cells by oxidative stress and involved in clastogenic adaptation. Cancer Res. 1998;58:4410–4416. [PubMed] [Google Scholar]
- 50.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 51.Guikema JE. Linehan EK. Tsuchimoto D. Nakabeppu Y. Strauss PR. Stavnezer J. Schrader CE. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Y. Chen J. Zhao T. Fan Z. Granzyme K degrades the redox/DNA repair enzyme Ape1 to trigger oxidative stress of target cells leading to cytotoxicity. Mol Immunol. 2008;45:2225–2235. doi: 10.1016/j.molimm.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 53.Haga S. Terui K. Zhang HQ. Enosawa S. Ogawa W. Inoue H. Okuyama T. Takeda K. Akira S. Ogino T. Irani K. Ozaki M. Stat3 protects against Fas-induced liver injury by redox-dependent and -independent mechanisms. J Clin Invest. 2003;112:989–998. doi: 10.1172/JCI17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanson S. Kim E. Deppert W. Redox factor 1 (Ref-1) enhances specific DNA binding of p53 by promoting p53 tetramerization. Oncogene. 2005;24:1641–1647. doi: 10.1038/sj.onc.1208351. [DOI] [PubMed] [Google Scholar]
- 55.Harrison JF. Rinne ML. Kelley MR. Druzhyna NM. Wilson GL. LeDoux SP. Altering DNA base excision repair: Use of nuclear and mitochondrial-targeted N-methylpurine DNA glycosylase to sensitize astroglia to chemotherapeutic agents. Glia. 2007;55:1416–1425. doi: 10.1002/glia.20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hay RT. SUMO: A history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 57.He T. Weintraub NL. Goswami PC. Chatterjee P. Flaherty DM. Domann FE. Oberley L. W. Redox factor-1 contributes to the regulation of progression from G0/G1 to S by PDGF in vascular smooth muscle cells. Am J Physiol Heart Circ Physiol. 2003;285:H804–812. doi: 10.1152/ajpheart.01080.2002. [DOI] [PubMed] [Google Scholar]
- 58.Hegde ML. Hazra TK. Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herring CJ. West CM. Wilks DP. Davidson SE. Hunter RD. Berry P. Forster G. MacKinnon J. Rafferty JA. Elder RH. Hendry JH. Margison GP. Levels of the DNA repair enzyme human apurinic/apyrimidinic endonuclease (APE1, APEX, Ref-1) are associated with the intrinsic radiosensitivity of cervical cancers. Br J Cancer. 1998;78:1128–1133. doi: 10.1038/bjc.1998.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hiramoto M. Shimizu N. Sugimoto K. Tang J. Kawakami Y. Ito M. Aizawa S. Tanaka H. Makino I. Handa H. Nuclear targeted suppression of NF-kappa B activity by the novel quinone derivative E3330. J Immunol. 1998;160:810–819. [PubMed] [Google Scholar]
- 61.Hirota K. Matsui M. Iwata S. Nishiyama A. Mori K. Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci USA. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsieh MM. Hegde V. Kelley MR. Deutsch WA. Activation of APE/Ref-1 redox activity is mediated by reactive oxygen species and PKC phosphorylation. Nucleic Acids Res. 2001;29:3116–3122. doi: 10.1093/nar/29.14.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang LE. Arany Z. Livingston DM. Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 64.Huang RP. Adamson ED. Characterization of the DNA-binding properties of the early growth response-1 (Egr-1) transcription factor: Evidence for modulation by a redox mechanism. DNA Cell Biol. 1993;12:265–273. doi: 10.1089/dna.1993.12.265. [DOI] [PubMed] [Google Scholar]
- 65.Ide H. Kotera M. Human DNA glycosylases involved in the repair of oxidatively damaged DNA. Biol Pharm Bull. 2004;27:480–485. doi: 10.1248/bpb.27.480. [DOI] [PubMed] [Google Scholar]
- 66.Izumi T. Mitra S. Deletion analysis of human AP-endonuclease: minimum sequence required for the endonuclease activity. Carcinogenesis. 1998;19:525–527. doi: 10.1093/carcin/19.3.525. [DOI] [PubMed] [Google Scholar]
- 67.Izumi T. Brown DB. Naidu CV. Bhakat KK. Macinnes MA. Saito H. Chen DJ. Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Izumi T. Henner WD. Mitra S. Negative regulation of the major human AP-endonuclease, a multifunctional protein. Biochemistry. 1996;35:14679–14683. doi: 10.1021/bi961995u. [DOI] [PubMed] [Google Scholar]
- 69.Jackson EB. Theriot CA. Chattopadhyay R. Mitra S. Izumi T. Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (APE1/Ref1) Nucleic Acids Res. 2005;33:3303–3312. doi: 10.1093/nar/gki641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jayaraman L. Murthy KG. Zhu C. Curran T. Xanthoudakis S. Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 71.Jung HJ. Kim EH. Mun JY. Park S. Smith ML. Han SS. Seo YR. Base excision DNA repair defect in Gadd45a-deficient cells. Oncogene. 2007;26:7517–7525. doi: 10.1038/sj.onc.1210557. [DOI] [PubMed] [Google Scholar]
- 72.Kakolyris S. Kaklamanis L. Engels K. Turley H. Hickson ID. Gatter KC. Harris AL. Human apurinic endonuclease 1 expression in a colorectal adenoma-carcinoma sequence. Cancer Res. 1997;57:1794–1797. [PubMed] [Google Scholar]
- 73.Kakolyris S. Kaklamanis L. Giatromanolaki A. Koukourakis M. Hickson ID. Barzilay G. Turley H. Leek RD. Kanavaros P. Georgoulias V. Gatter KC. Harris AL. Expression and subcellular localization of human AP endonuclease 1 (HAP1/Ref-1) protein: A basis for its role in human disease. Histopathology. 1998;33:561–569. doi: 10.1046/j.1365-2559.1998.00541.x. [DOI] [PubMed] [Google Scholar]
- 74.Katan–Khaykovich Y. Struhl K. Dynamics of global histone acetylation and deacetylation in vivo: Rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 2002;16:743–752. doi: 10.1101/gad.967302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khachigian LM. Lindner V. Williams AJ. Collins T. Egr-1-induced endothelial gene expression: A common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 76.Khomenko T. Deng X. Jadus MR. Szabo S. Effect of cysteamine on redox-sensitive thiol-containing proteins in the duodenal mucosa. Biochem Biophys Res Commun. 2003;309:910–916. doi: 10.1016/j.bbrc.2003.08.092. [DOI] [PubMed] [Google Scholar]
- 77.Kuninger DT. Izumi T. Papaconstantinou J. Mitra S. Human AP-endonuclease 1 and hnRNP-L interact with a nCaRE-like repressor element in the AP-endonuclease 1 promoter. Nucleic Acids Res. 2002;30:823–829. doi: 10.1093/nar/30.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lau JP. Weatherdon KL. Skalski V. Hedley DW. Effects of gemcitabine on APE/ref-1 endonuclease activity in pancreatic cancer cells, and the therapeutic potential of antisense oligonucleotides. Br J Cancer. 2004;91:1166–1173. doi: 10.1038/sj.bjc.6602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lewén A. Sugawara T. Gasche Y. Fujimura M. Chan PH. Oxidative cellular damage and the reduction of APE/Ref-1 expression after experimental traumatic brain injury. Neurobiol Dis. 2001;8:380–390. doi: 10.1006/nbdi.2001.0396. [DOI] [PubMed] [Google Scholar]
- 79a.Liu B, Li S, Wang Y, Lu L, Li Y, and Cai Y. Predicting the protein SUMO modification sites based on properties sequential forward selection (PSFS) Biochem Biophys Res Commun. 2007;358:136–139. doi: 10.1016/j.bbrc.2007.04.097. [DOI] [PubMed] [Google Scholar]
- 80.Liu L. Gerson SL. Therapeutic impact of methoxyamine: Blocking repair of abasic sites in the base excision repair pathway. Curr Opin Investig Drugs. 2004;5:623–627. [PubMed] [Google Scholar]
- 81.Liu L. Nakatsuru Y. Gerson SL. Base excision repair as a therapeutic target in colon cancer. Clin Cancer Res. 2002;8:2985–2991. [PubMed] [Google Scholar]
- 82.Liu L. Yan L. Donze JR. Gerson SL. Blockage of abasic site repair enhances antitumor efficacy of 1,3-bis-(2-chloroethyl)-1-nitrosourea in colon tumor xenografts. Mol Cancer Ther. 2003;2:1061–1066. [PubMed] [Google Scholar]
- 83.Liu L. Yan L. Mahajan V. Donze JR. Gerson SL. Inhibition of AP site repair coupled with action of topoisomerase II poison: A potential strategy to enhance efficacy of chemotherapeutic alkylating agents. Proc Amer Assoc Ca Res. 2003:44. [Google Scholar]
- 84.Liu P. Kimmoun E. Legrand A. Sauvanet A. Degott C. Lardeux B. Bernuau D. Activation of NF-kappa B, AP-1 and STAT transcription factors is a frequent and early event in human hepatocellular carcinomas. J Hepatol. 2002;37:63–71. doi: 10.1016/s0168-8278(02)00064-8. [DOI] [PubMed] [Google Scholar]
- 85.Ludwig DL. MacInnes MA. Takiguchi Y. Purtymun PE. Henrie M. Flannery M. Meneses J. Pedersen RA. Chen DJ. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat Res. 1998;409:17–29. doi: 10.1016/s0921-8777(98)00039-1. [DOI] [PubMed] [Google Scholar]
- 86.Luo M. Delaplane S. Jiang A. Reed A. He Y. Fishel M. Nyland RL., II Borch RF. Qiao X. Georgiadis MM. Kelley MR. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: Small molecule inhibition of Ape1's redox function. Antioxid Redox Signal. 2008 doi: 10.1089/ars.2008.2120. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McNeill DR. Wilson DM., 3rd A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 88.Mitra S. Izumi T. Boldogh I. Bhakat KK. Chattopadhyay R. Szczesny B. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair. 2007;6:461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 89.Miyamoto K. Nagakawa J. Hishinuma I. Hirota K. Yasuda M. Yamanaka T. Katayama K. Yamatsu I. Suppressive effects of E3330, a novel quinone derivative, on tumor necrosis factor-alpha generation from monocytes and macrophages. Agents Actions. 1992;37:297–304. doi: 10.1007/BF02028123. [DOI] [PubMed] [Google Scholar]
- 90.Mol CD. Izumi T. Mitra S. Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 91.Moradpour D. Blum HE. Pathogenesis of hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2005;17:477–483. doi: 10.1097/00042737-200505000-00002. [DOI] [PubMed] [Google Scholar]
- 92.Murphy PJ. Morishima Y. Chen H. Galigniana MD. Mansfield JF. Simons SS., Jr Pratt WB. Visualization and mechanism of assembly of a glucocorticoid receptor.Hsp70 complex that is primed for subsequent Hsp90-dependent opening of the steroid binding cleft. J Biol Chem. 2003;278:34764–34773. doi: 10.1074/jbc.M304469200. [DOI] [PubMed] [Google Scholar]
- 93.Nakamura H. Nakamura K. Yodoi J. Redox regulation of cellular activation. Annu Rev Immunol. 1997;15:351–369. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 94.Nakshatri H. Bhat–Nakshatri P. Currie RA. Subunit association and DNA binding activity of the heterotrimeric transcription factor NF-Y is regulated by cellular redox. J Biol Chem. 1996;271:28784–28791. doi: 10.1074/jbc.271.46.28784. [DOI] [PubMed] [Google Scholar]
- 95.Nishi T. Shimizu N. Hiramoto M. Sato I. Yamaguchi Y. Hasegawa M. Aizawa S. Tanaka H. Kataoka K. Watanabe H. Handa H. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J Biol Chem. 2002;277:44548–44556. doi: 10.1074/jbc.M202970200. [DOI] [PubMed] [Google Scholar]
- 96.O'Hara AM. Bhattacharyya A. Mifflin RC. Smith MF. Ryan KA. Scott KG. Naganuma M. Casola A. Izumi T. Mitra S. Ernst PB. Crowe SE. Interleukin-8 induction by Helicobacter pylori in gastric epithelial cells is dependent on apurinic/apyrimidinic endonuclease-1/redox factor-1. J Immunol. 2006;177:7990–7999. doi: 10.4049/jimmunol.177.11.7990. [DOI] [PubMed] [Google Scholar]
- 97.Okamoto K. Prives C. A role of cyclin G in the process of apoptosis. Oncogene. 1999;18:4606–4615. doi: 10.1038/sj.onc.1202821. [DOI] [PubMed] [Google Scholar]
- 98.Ono Y. Furuta T. Ohmoto T. Akiyama K. Seki S. Stable expression in rat glioma cells of sense and antisense nucleic acids to a human multifunctional DNA repair enzyme, APEX nuclease. Mutat Res. 1994;315:55–63. doi: 10.1016/0921-8777(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 99.Ono Y. Matsumoto K. Furuta T. Ohmoto T. Akiyama K. Seki S. Relationship between expression of a major apurinic/apyrimidinic endonuclease (APEX nuclease) and susceptibility to genotoxic agents in human glioma cell lines. J Neurooncol. 1995;25:183–192. doi: 10.1007/BF01053151. [DOI] [PubMed] [Google Scholar]
- 100.Ono Y. Watanabe M. Inoue Y. Ohmoto T. Akiyama K. Tsutsui K. Seki S. Developmental expression of APEX nuclease, a multifunctional DNA repair enzyme, in mouse brains. Brain Res Dev Brain Res. 1995;86:1–6. doi: 10.1016/0165-3806(94)00212-i. [DOI] [PubMed] [Google Scholar]
- 101.Ordway JM. Eberhart D. Curran T. Cysteine 64 of Ref-1 is not essential for redox regulation of AP-1 DNA binding. Mol Cell Biol. 2003;23:4257–4266. doi: 10.1128/MCB.23.12.4257-4266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ozaki M. Suzuki S. Irani K. Redox factor-1/APE suppresses oxidative stress by inhibiting the rac1 GTPase. FASEB J. 2002;16:889–890. doi: 10.1096/fj.01-0664fje. [DOI] [PubMed] [Google Scholar]
- 103.Parlanti E. Locatelli G. Maga G. Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Parsons JL. Dianova II. Dianov GL. APE1-dependent repair of DNA single-strand breaks containing 3′-end 8-oxoguanine. Nucleic Acids Res. 2005;33:2204–2209. doi: 10.1093/nar/gki518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parsons JL. Dianova II. Dianov GL. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32:3531–3536. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pikarsky E. Porat RM. Stein I. Abramovitch R. Amit S. Kasem S. Gutkovich–Pyest E. Urieli–Shoval S. Galun E. Ben–Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 107.Pines A. Bivi N. Romanello M. Damante G. Kelley MR. Adamson ED. D'Andrea P. Quadrifoglio F. Moro L. Tell G. Cross-regulation between Egr-1 and APE/Ref-1 during early response to oxidative stress in the human osteoblastic HOBIT cell line: Evidence for an autoregulatory loop. Free Radic Res. 2005;39:269–281. doi: 10.1080/10715760400028423. [DOI] [PubMed] [Google Scholar]
- 108.Pines A. Perrone L. Bivi N. Romanello M. Damante G. Gulisano M. Kelley MR. Quadrifoglio F. Tell G. Activation of APE1/Ref-1 is dependent on reactive oxygen species generated after purinergic receptor stimulation by ATP. Nucleic Acids Res. 2005;33:4379–4394. doi: 10.1093/nar/gki751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qin J. Clore GM. Kennedy WP. Kuszewski J. Gronenborn AM. The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal. Structure. 1996;4:613–620. doi: 10.1016/s0969-2126(96)00065-2. [DOI] [PubMed] [Google Scholar]
- 110.Qu J. Liu GH. Huang B. Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res. 2007;35:2522–2532. doi: 10.1093/nar/gkl1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ramana CV. Boldogh I. Izumi T. Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc Natl Acad Sci USA. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rivkees SA. Kelley MR. Expression of a multifunctional DNA repair enzyme gene, apurinic/apyrimidinic endonuclease (APE; Ref-1) in the suprachiasmatic, supraoptic and paraventricular nuclei. Brain Res. 1994;666:137–142. doi: 10.1016/0006-8993(94)90296-8. [DOI] [PubMed] [Google Scholar]
- 113.Robertson KA. Bullock HA. Xu Y. Tritt R. Zimmerman E. Ulbright TM. Foster RS. Einhorn LH. Kelley MR. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 2001;61:2220–2225. [PubMed] [Google Scholar]
- 114.Robson CN. Hickson ID. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rossi O. Carrozzino F. Cappelli E. Carli F. Frosina G. Analysis of repair of abasic sites in early onset breast cancer patients. Int J Cancer. 2000;85:21–26. doi: 10.1002/(sici)1097-0215(20000101)85:1<21::aid-ijc4>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 116.Sakurai M. Nagata T. Abe K. Horinouchi T. Itoyama Y. Tabayashi K. Oxidative damage and reduction of redox factor-1 expression after transient spinal cord ischemia in rabbits. J Vasc Surg. 2003;37:446–452. doi: 10.1067/mva.2003.100. [DOI] [PubMed] [Google Scholar]
- 117.Seo YR. Kelley MR. Smith ML. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc Natl Acad Sci USA. 2002;99:14548–14553. doi: 10.1073/pnas.212319799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shimizu N. Sugimoto K. Tang J. Nishi T. Sato I. Hiramoto M. Aizawa S. Hatakeyama M. Ohba R. Hatori H. Yoshikawa T. Suzuki F. Oomori A. Tanaka H. Kawaguchi H. Watanabe H. Handa H. High-performance affinity beads for identifying drug receptors. Nat Biotechnol. 2000;18:877–881. doi: 10.1038/78496. [DOI] [PubMed] [Google Scholar]
- 119.Sossou M. Flohr–Beckhaus C. Schulz I. Daboussi F. Epe B. Radicella JP. APE1 overexpression in XRCC1-deficient cells complements the defective repair of oxidative single strand breaks but increases genomic instability. Nucleic Acids Res. 2005;33:298–306. doi: 10.1093/nar/gki173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Spitz DR. Azzam EI. Li JJ. Gius D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: A unifying concept in stress response biology. Cancer Metastasis Rev. 2004;23:311–322. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 121.Sukhanova MV. Khodyreva SN. Lebedeva NA. Prasad R. Wilson SH. Lavrik OI. Human base excision repair enzymes apurinic/apyrimidinic endonuclease1 (APE1), DNA polymerase beta and poly(ADP-ribose) polymerase 1: Interplay between strand-displacement DNA synthesis and proofreading exonuclease activity. Nucleic Acids Res. 2005;33:1222–1229. doi: 10.1093/nar/gki266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Szczesny B. Mitra S. Effect of aging on intracellular distribution of abasic (AP) endonuclease 1 in the mouse liver. Mech Ageing Dev. 2005;126:1071–1078. doi: 10.1016/j.mad.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 123.Tan Z. Sun N. Schreiber SS. Immunohistochemical localization of redox factor-1 (Ref-1) in Alzheimer's hippocampus. Neuroreport. 1998;9:2749–2752. doi: 10.1097/00001756-199808240-00012. [DOI] [PubMed] [Google Scholar]
- 124.Taverna P. Liu L. Hwang HS. Hanson AJ. Kinsella TJ. Gerson SL. Methoxyamine potentiates DNA single strand breaks and double strand breaks induced by temozolomide in colon cancer cells. Mutat Res. 2001;485:269–281. doi: 10.1016/s0921-8777(01)00076-3. [DOI] [PubMed] [Google Scholar]
- 125.Tell G. Crivellato E. Pines A. Paron I. Pucillo C. Manzini G. Bandiera A. Kelley MR. Di Loreto C. Damante G. Mitochondrial localization of APE/Ref-1 in thyroid cells. Mutat Res. 2001;485:143–152. doi: 10.1016/s0921-8777(00)00068-9. [DOI] [PubMed] [Google Scholar]
- 126.Tell G. Damante G. Caldwell D. Kelley MR. The intracellular localization of APE1/Ref-1: More than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 127.Tell G. Pellizzari L. Pucillo C. Puglisi F. Cesselli D. Kelley MR. Di Loreto C. Damante G. TSH controls Ref-1 nuclear translocation in thyroid cells. J Mol Endocrinol. 2000;24:383–390. doi: 10.1677/jme.0.0240383. [DOI] [PubMed] [Google Scholar]
- 128.Tell G. Scaloni A. Pellizzari L. Formisano S. Pucillo C. Damante G. Redox potential controls the structure and DNA binding activity of the paired domain. J Biol Chem. 1998;273:25062–25072. doi: 10.1074/jbc.273.39.25062. [DOI] [PubMed] [Google Scholar]
- 129.Tell G. Zecca A. Pellizzari L. Spessotto P. Colombatti A. Kelley MR. Damante G. Pucillo C. An ‘environment to nucleus' signaling system operates in B lymphocytes: redox status modulates BSAP/Pax-5 activation through Ref-1 nuclear translocation. Nucleic Acids Res. 2000;28:1099–1105. doi: 10.1093/nar/28.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thyss R. Virolle V. Imbert V. Peyron JF. Aberdam D. Virolle T. NF-kappaB/Egr-1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death. EMBO J. 2005;24:128–137. doi: 10.1038/sj.emboj.7600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Trzeciak AR. Nyaga SG. Jaruga P. Lohani A. Dizdaroglu M. Evans MK. Cellular repair of oxidatively induced DNA base lesions is defective in prostate cancer cell lines, PC-3 and DU-145. Carcinogenesis. 2004;25:1359–1370. doi: 10.1093/carcin/bgh144. [DOI] [PubMed] [Google Scholar]
- 132.Ueno M. Masutani H. Arai RJ. Yamauchi A. Hirota K. Sakai T. Inamoto T. Yamaoka Y. Yodoi J. Nikaido T. Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J Biol Chem. 1999;274:35809–35815. doi: 10.1074/jbc.274.50.35809. [DOI] [PubMed] [Google Scholar]
- 133.Ushio–Fukai M. Redox signaling in angiogenesis: Role of NADPH oxidase. Cardiovasc Res. 2006;71:226–235. doi: 10.1016/j.cardiores.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 134.Vasko MR. Guo C. Kelley MR. The multifunctional DNA repair/redox enzyme Ape1/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair (Amst) 2005;4:367–379. doi: 10.1016/j.dnarep.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 135.Vidal F. Aragones J. Arantzazu A. de Landazuri MO. Upregulation of vascular endothelial growth factor receptor Flt-1 after endothelial denudation: role of transcription factor Egr-1. Blood. 2000;95:3387–3394. [PubMed] [Google Scholar]
- 136.Vidal AE. Boiteux S. Hickson ID. Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Virolle T. Adamson ED. Baron V. Birle D. Mercola D. Mustelin T. de Belle I. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signaling. Nat Cell Biol. 2001;3:1124–1128. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 138.Walker LJ. Craig RB. Harris AL. Hickson ID. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res. 1994;22:4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Walker LJ. Robson CN. Black E. Gillespie D. Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang D. Luo M. Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 141.Wei SJ. Botero A. Hirota K. Bradbury CM. Markovina S. Laszlo A. Spitz DR. Goswami PC. Yodoi J. Gius D. Thioredoxin nuclear translocation and interaction with redox factor-1 activates the activator protein-1 transcription factor in response to ionizing radiation. Cancer Res. 2000;60:6688–6695. [PubMed] [Google Scholar]
- 142.Wilson TM. Rivkees SA. Deutsch WA. Kelley MR. Differential expression of the apurinic/apyrimidinic endonuclease (APE/ref-1) multifunctional DNA base excision repair gene during fetal development and in adult rat brain and testis. Mutat Res. 1996;362:237–248. doi: 10.1016/0921-8777(95)00053-4. [DOI] [PubMed] [Google Scholar]
- 143.Wong HK. Muftuoglu M. Beck G. Imam SZ. Bohr VA. Wilson DM., 3rd Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xanthoudakis S. Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xanthoudakis S. Miao G. Wang F. Pan YC. Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xanthoudakis S. Miao GG. Curran T. The redox and DNA repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci USA. 1994;91:23–27. doi: 10.1073/pnas.91.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xanthoudakis S. Smeyne RJ. Wallace JD. Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147a.Xue Y, Zhou F, Fu C, Xu Y, and Yao X. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 2006;34:W254–257. doi: 10.1093/nar/gkl207. (web server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yacoub A. Kelley MR. Deutsch WA. The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation. Cancer Res. 1997;57:5457–5459. [PubMed] [Google Scholar]
- 149.Yan L. Bulgar A. Miao Y. Mahajan V. Donze JR. Gerson SL. Liu L. Combined treatment with temozolomide and methoxyamine: Blocking apurininc/pyrimidinic site repair coupled with targeting topoisomerase II{alpha} Clin Cancer Res. 2007;13:1532–1539. doi: 10.1158/1078-0432.CCR-06-1595. [DOI] [PubMed] [Google Scholar]
- 150.Yoshida A. Urasaki Y. Waltham M. Bergman AC. Pourquier P. Rothwell DG. Inuzuka M. Weinstein JN. Ueda T. Appella E. Hickson ID. Pommier Y. Human apurinic/apyrimidinic endonuclease (Ape1) and its N-terminal truncated form (AN34) are involved in DNA fragmentation during apoptosis. J Biol Chem. 2003;278:37768–37776. doi: 10.1074/jbc.M304914200. [DOI] [PubMed] [Google Scholar]
- 151.Zaky A. Busso C. Izumi T. Chattopadhyay R. Bassiouny A. Mitra S. Bhakat KK. Regulation of the human AP-endonuclease (APE1/Ref-1) expression by the tumor suppressor p53 in response to DNA damage. Nucleic Acids Res. 2008;36:1555–1566. doi: 10.1093/nar/gkm1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhao J. Gao F. Zhang Y. Wei K. Liu Y. Deng X. Bcl2 inhibits abasic site repair by downregulating APE1 endonuclease activity. J Biol Chem. 2008;283:9925–9932. doi: 10.1074/jbc.M708345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ziel KA. Campbell CC. Wilson GL. Gillespie MN. Ref-1/Ape is critical for formation of the hypoxia-inducible transcriptional complex on the hypoxic response element of the rat pulmonary artery endothelial cell VEGF gene. FASEB J. 2004;18:986–988. doi: 10.1096/fj.03-1160fje. [DOI] [PubMed] [Google Scholar]
- 154.Ziel KA. Grishko V. Campbell CC. Breit JF. Wilson GL. Gillespie MN. Oxidants in signal transduction: impact on DNA integrity and gene expression. FASEB J. 2005;19:387–394. doi: 10.1096/fj.04-2805com. [DOI] [PubMed] [Google Scholar]
- 155.Zou GM. Luo MH. Reed A. Kelley MR. Yoder MC. APE1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood. 2007;109:1917–1922. doi: 10.1182/blood-2006-08-044172. [DOI] [PubMed] [Google Scholar]