Introduction
The mammalian ribonuclease inhibitor (RI) is a 50-kDa cytosolic protein that binds to pancreatic-type ribonucleases with femtomolar affinity and renders them inactive (for other reviews, see (1–5)). Complexes formed by RI and its target ribonucleases are among the tightest of known biomolecular interactions. The three-dimensional structure of RI is likewise remarkable, being characterized by alternating units of α-helix and β-strand that form a striking horseshoe shape (Fig. 1A) (6). The repeating structural units of RI possess a highly repetitive amino acid sequence that is rich in leucine residues (7, 8). These leucine-rich repeats (LRRs) are present in a large family of proteins that are distinguished by their display of vast surface areas to foster protein protein interactions (9–12). The unique structure and function of RI have resulted in its emergence as the central protein in the study of LRRs, as well as its widespread use as a laboratory reagent to eliminate ribonucleolytic activity (13).
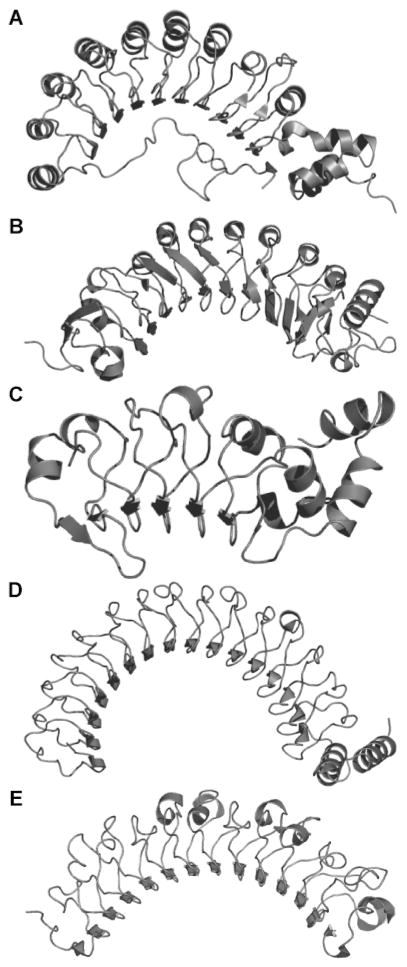
Fig. 1.
Three-dimensional structures of RI and its complexes with ribonucleases. (A) Porcine RI (6) with colors corresponding to exon-encoded modules (40). (B) Porcine RI·RNase A complex (51) (C) Human RI·ANG complex (69).
The biological role of RI is not known in its entirety. The ribonucleases recognized by RI are secreted proteins, whereas RI resides exclusively in the cytosol. Nevertheless, RI affinity has been shown to be the primary determinant of ribonuclease cytotoxicity: only ribonucleases that evade RI can kill a cell (for reviews, see (14–17)). In addition, the complex of RI with human angiogenin (ANG), which stimulates neovascularization by activating transcription in the nucleus (18, 19), is the tightest of known RI·ribonuclease complexes. Yet, a role for RI in angiogenesis is not clear. Also intriguing are the 30–32 cysteine residues of RI, all of which must remain reduced for the protein to retain activity (20). These observations have lead researchers to hypothesize multiple biological roles for RI: (1) to protect cells from invading ribonucleases, (2) to regulate or terminate the activity of ribonucleases with known intracellular functions, and (3) to monitor the oxidation state of the cell in response to factors such as aging and oxidative stress. Here, we review the salient features of RI biochemistry and structure and thereby provide a context for examining the roles of RI in biology.
I. Biochemical Properties
The inhibitory activity of RI in guinea pig liver extracts was discovered in 1952 (21). This activity was inactivated by proteases, heat, or sulfhydryl-group modification, and was sensitive to changes in pH (for a review, see (22)). In addition, the inhibitory activity was isolated in the supernatant fraction during a high-speed centrifugation, indicative of cytoplasmic localization. In the 1970’s, techniques were developed to purify RI to homogeneity, enabling its biochemical characterization (23, 2). Since then, RI has been isolated from numerous mammalian sources, including brain (24–26), liver (27, 28, 26), testis (29), and erythrocytes (30).
A. Purification
RI is particularly abundant in mammalian placenta and liver, which have served as the major source of RI for purification. Human placental RI was first purified to homogeneity using a combination of ion-exchange and ribonuclease-affinity chromatography (23). The tight complex formed by RI and bovine pancreatic ribonuclease (RNase A (31); EC 3.1.27.5) has been exploited to achieve a >103-fold purification of RI in a single chromatographic step using immobilized RNase A. Today, most purification methods rely upon such ribonuclease-affinity chromatography, followed by anion-exchange chromatography (32). Using these purification techniques, approximately 6 mg RI per kg of wet tissue has been isolated from mammalian liver (28) and placenta (33). Human erythrocytes are also rich in RI—the erythrocyte fraction of 100 mL of blood has yielded 430 μg of RI (30).
Several recombinant systems for the production of RI have been reported, three from Escherichia coli and one from Saccharomyces cerevisiae (34–36). Low yields and insolubility have proven to be recurring problems in producing recombinant RI. To date, the most efficient recombinant system utilizes the trp promoter from E. coli to drive expression of porcine RI, and yields approximately 10 mg of RI per liter of culture (37).
B. Characterization
RI is an acidic (pI 4.7) cytosolic protein that binds to pancreatic-type ribonucleases with 1:1 stiochiometry (38). Members of the RNase A superfamily of proteins that are inhibited by RI include RNase A, human pancreatic ribonuclease (RNase 1), ANG, eosinophil-derived neurotoxin (EDN, also known as RNase 2), RNase 4, and monomers of bovine seminal ribonuclease (BS-RNase). When complexed with RI, these ribonucleases are no longer able to bind or degrade RNA (3). RI is ineffective against known non-mammalian homologs of RNase A. The amino acid sequences of human, porcine, mouse, and rat RI share 66% identity (Fig. 2) (7, 8, 39, 40). One third of the residues that differ are conservative substitutions. To date, RI from human and pigs have been characterized most thoroughly and exhibit many identical properties (for reviews, see (4, 5)). Thus, the source of RI will be discussed herein only if a significant divergence occurs with respect to a particular experimental observation.
Fig. 2.
Alignment of the amino acid sequences of RI from human (8), porcine (7), mouse (40), and rat (39). The consensus sequence for the A-type and B-type repeats is indicated, along with the corresponding secondary structure. The initiator methionine residue was not detected in the N-terminal tryptic fragment of human RI and is shown in parentheses. Conserved residues are in boxes. Residues of human RI that contact ANG (69) and residues of porcine RI that contact RNase A (51) are shaded.
The affinity of RI for ribonucleases is extraordinary. Accordingly, substantial effort has been invested in characterizing RI ribonuclease interactions (for a review, see (5)). Techniques to assess binding rely upon the imposition of physical changes or inhibition of catalytic activity. A purely physical method is more convenient to use for ribonucleases with low catalytic activity, such as ANG (41). For example, stopped-flow techniques and the 50% increase in the fluorescence of Trp89 of ANG upon binding to RI have been used to study the association of RI with ANG. They report a two-step binding mechanism that involves formation of a loose enzyme·inhibitor complex (E·I) followed by isomerization to form a tight complex (E·I*), as in Eq. (1):
| (1) |
ANG and RI rapidly form a loose complex (K1 = k−1/k1 = 0.53 μM), which converts slowly (k2 = 97 s−1) to a stable complex. The association rate constant, ka = k1k2/(k−1 + k2) was found to be 1.8× 108 M−1s−1. The dissociation rate constant, kd = k−1k−2/(k−1 + k2), was measured by the monitoring release of ANG from the RI·ANG complex in the presence of excess RNase A as a scavenger, and found to be 1.3 × 10−7 s−1 (35). This value corresponds to a half-life of 62 days for the RI·ANG complex. The resulting value of the equilibrium dissociation constant, Kd = kd/ka = 7.1 × 10−16 M, is exceptionally low, and comparable to the Kd = 6 × 10−16 M of the avidin·biotin complex (42). A competition assay based on fluorescence changes in ANG has been used to measure Kd = 4.4 × 10−14 M for the RI·RNase A complex (41).
RI has only a slight effect on the fluorescence of RNase A, which lacks tryptophan residues. Enzymatic assays in which the value of Ki is determined by the ability of RI to compete with RNA are viable alternatives for this and other ribonucleases that possess high catalytic activity. In general, enzymatic assays require that ribonucleolytic activity can be performed at low enzyme concentrations—no more than 2 orders-of-magnitude greater than the Ki (36). Enzymological methods have been used to assess the affinity of RI for RNase A, RNase 1, and RNase 4 (Table I) (36, 43–45). For examples, the values of ka = 1.7 × 108 M−1s−1, kd = 9.8 × 10−6 s−1, and Ki = 5.9 × 10−14 M were determined by measuring the decrease in ribonucleolytic activity upon addition of RI.
Table I.
Kinetic and thermodynamic parameters for RI ribonuclease interactions
| RI | Ribonuclease | ka (M−1s−1) | kd (s−1) | Ki or Kd (M) | Method | Ref. |
|---|---|---|---|---|---|---|
| Human | ANG | 1.8 × 108 | 1.3 × 10−7 | 7.1 × 10−16 | Physical | (46, 41) |
| ANG | 2.0 × 108 | 1.1 × 10−7 | 5.4 × 10−16 | Physical | (69) | |
| Human | RNase A | 3.4 × 108 | 1.5 × 10−5 | 4.4 × 10−14 | Physical/Enzymological | (46, 41) |
| RNase A | 3.4 × 108 | 1.2 × 10−5 | 3.5 × 10−14 | Physical/Enzymological | (46, 41) | |
| RNase 2 | 1.9 × 108 | 1.8 × 10−7 | 9.4 × 10−16 | Physical/Enzymological | (46, 41) | |
| Porcine | RNase A | 1.7 × 108 | 9.8 × 10−6 | 5.9 × 10−14 | Enzymological | (36) |
| RNase A | 1.3 × 108 | 1.5 × 10−5 | 1.13 × 10−13 | Enzymological | (43) | |
| RNase A | ND | ND | 7.4 × 10−14 | Enzymological | (36) | |
| RNase 4 | 1.5 × 108 | 1.3 × 10−7 | 4.0 × 10−15 | Enzymological | (45) | |
The affinity of RNase A and RNase 2 for RI has also been assessed with a combination of physical and enzymological techniques. The kd value for the RI·RNase A complex was determined by measuring the release of RNase A in the presence of ANG as a scavenger (46, 41). The concentration of free RNase A was detected by high-performance liquid chromatography or by enzymatic activity with RNA substrates that are not cleaved by ANG. Similar assays have been used to determine the kinetic parameters for the RI·RNase 2 interaction (47). The kinetic and thermodynamic parameters determined with a variety of physical and enzymatic methods are in gratifying agreement (Table I).
A fluorescence-based assay has been developed to facilitate rapid measurement of Kd for a wide variety of RI·ribonuclease complexes (48). This assay employs fluorescein-labeled G88R RNase A, which has diminished affinity for RI and exhibits an approximately 20% decrease in fluorescence when bound to RI. Titration of RI with fluorescein-G88R RNase A yielded Kd = 0.55 × 10−9 M for the complex. A competition assay using fluorescein-G88R RNase A was then used to determine the Kd value of unlabeled ribonucleases (Table II). This assay is limited to measuring complexes with Kd values in the nanomolar range or higher, as tighter complexes take too long to reach equilibrium. Nonetheless, this assay has proven to be valuable for determining Kd values of numerous RNase A variants, some of which possess low catalytic activity (49, 50).
Table II.
Properties of Ribonuclease A, its Variants, and Onconase®
| Ribonuclease | kcat/KM (106 M−1s−1) | Kd (nM) | (kcat/KM)cyto (103 M−1s−1) | IC50 (μM) | Ref |
|---|---|---|---|---|---|
| Wild-type RNase A | 43 ± 3 | 6.7 × 10−5 | 0.00072 | >50 | (48–50) |
| G88R RNase A | 14 ± 2 | 0.57 ± 0.05 | 2.0 | 10 ± 1 | (48–50) |
| A4C/G88R/V118C RNase A | 2.6 ± 0.2 | 1.3 ± 0.3 | 0.84 | 4.1 ± 0.6 | (50) |
| K41R/G88R RNase A | 0.6 ± 0.06 | 7.5 ± 1.8 | 1.1 | 5.2 ± 0.7 | (49, 50) |
| A4C/K41R/G88R/V118C RNase A | 0.13 ± 0.03 | 27 ± 3.7 | 0.87 | 7.6 ± 0.9 | (50) |
| K7A/G88R RNase A | 8.8 ± 2.6 | 7.2 ± 0.4 | 15.8 | 1.0 ± 0.1 | (49) |
| ONC | 0.00035 ± 0.00010 | ≥1 × 106 | >0.35 | 0.49 ± 0.065 | (49) |
II. Structure
A. Three-Dimensional Structure
Leucine is the most abundant residue in RI, comprising 18% of its amino acids (23, 28). In 1988, the amino acid sequence of RI from both porcine liver and human placenta was elucidated, revealing that RI is comprised entirely of leucine-rich repeats (LRR) (7, 8). Two types of alternating repeats have been described, A-type (which contains 28 residues) and B-type (which contains 29 residues). Porcine RI is built from 8 A-type and 7 B-type repeats, flanked by short terminal segments (Fig. 2) (10).
RI was the first LRR protein to be crystallized and have its three-dimensional structure determined by X-ray diffraction analysis (6). Its horseshoe shape is one of the most captivating of protein structures. The alternating A- and B-type LRR motifs correspond to structural units, each consisting of an α-helix and β-strand connected by loops (Fig. 2A and 2B). The symmetric and non-globular arrangement of LRRs represents a new protein fold (for reviews, see: (51, 52, 12)). The LRR units of RI are arranged so that the α-helices and β-strands are aligned parallel to a common axis (Fig. 1A). An extended β-sheet defines the inner circumference of the horseshoe and provides a vast surface for interacting with other proteins. Leucines and other aliphatic residues are essential components of the hydrophobic core of the protein, and serve to stabilize the interactions between the LRR units (Fig. 3). The curvature of the RI horseshoe is determined by the difference in distance between neighboring β-strands and α-helices (52, 12). The curvature of RI is quite pronounced, as the addition of only 5 more LRR units to the native 15 would cause the termini of RI to collide (6).
Fig. 3.

(A) Typical A-type of RI (residues 138–165). Typical B-type repeat of RI (residues 223–252). The side chains of conserved aliphatic residues are shown explicitly and numbered within the repeat.
B. A Model Leucine-Rich Repeat Protein
The LRR was first described with respect to the leucine-rich α2-glycoprotein found in human serum (53). RI was the first cytosolic protein discovered to possess LRRs (7, 8). In the past decade, more than a hundred LRR proteins have been identified; these proteins have been found to perform remarkably different functions. In most LRR proteins, however, the LRRs appear to serve as the interface for a protein protein interaction (for reviews, see (54, 52)).
LRR proteins have been classified into subfamilies base on the organism of origin, cellular localization, and LRR consensus sequence (12). To date, seven LRR subfamilies of proteins have been described (Table III), and additional subfamilies could arise with the discovery of more LRR proteins. Members of the RI-like subfamily are intracellular proteins found in animals, and are characterized by repeats of 28/29 amino acids that possess the sequence LXXLXLXX(N/C)XL. Other members of the RI-like subfamily include human MHC class II transactivator (P33076), Ran GTPase activating protein from Saccharomyces pombe (P46060), RNA1 gene product from Saccharomyces cerevisiae (X17376), and the mouse homolog of RNA1 (U20857).
Table III.
Characteristics of LRR Protein Subfamilies
| Subfamily | Source | Location | Representative Protein (organism) | Function | Typical Length of LRR (range) | Secondary Structure of Interstrand Region | PDB code | Ref. |
|---|---|---|---|---|---|---|---|---|
| Typical | Animals, fungi | Extracellular | TSHR (human) | Receptor for thyrotropin | 24 (20 – 27) | α-helix (model) | – | – |
| RI-like | Animals | Intracellular | RI (pig) | Ribonuclease inhibitor | 28–29 (28–29) | α-helix | 1BNH | (6) |
| Cysteine- containing | Animals, plants, fungi | Intracellular | Skp2 (human) | Substrate binding in ubiquitination | 26 (25–27) | α-helix | 1FQV | (57) |
| Plant-specific | Plants, primarily eukaryotes | Extracellular | Pgip (kidney bean) | Pathogen defense | 24 (23 – 25) | 310 helix | 1OGQ | (58) |
| SD22-like | Animals, fungi | Intracellular | U2A′ (human) | RNA Splicing | 22 (21 –23) | 310 helix, α-helix | 1A9N | (55) |
| Bacterial | Gram-negative bacteria | Extracellular | YopM (Yersinia pestis) | Virulence factor | 20 (20 – 22) | PII helix | 1G9U | (56) |
| Small | Mammals | Extracellular | Decorin (human) | Collagen fibrillogenesis | 24 (21 – 30) | 310 helix, PII helix, β-turn, β-strand | 1XKU | (59) |
In general, the β-strand region of the repeat is the most conserved among LRR proteins (12). Subfamilies differ primarily in the secondary structure displayed in the regions between the β-strands (Table III, Fig. 4) (12). Short LRR units result in extended conformations in the interstrand region. For example, members of the bacterial subfamily of LRR proteins are built from repeating units of only 20 amino acid residues. In the SDS22-like family, the α-helix found in RI-like proteins is often replaced by a 310 helix (55). In the structure of YopM, an extracellular protein that confers bacteria with virulence, the α-helix is replaced with a polyproline type-II (PII) helix (Table III) (56). Structures of representative proteins from five subfamilies illustrate the diversity in the size and shape of LRR proteins (Fig. 4) (57–59).
Fig. 4.
Structures of five representative LRR proteins (Table III). (A) cysteine-containing protein Skp2 (57). (B) Plant-specific protein Pgip (58). (C) SDS22-Like protein U2A′ (55). (D) Bacterial protein YopM (56). (E) Decorin (59).
The structure of RI is repetitive and symmetrical, and its surface area is vast and largely concave (Fig. 1A). These unusual attributes make RI a potential platform for the creation of new receptors. Towards this goal, a consensus LRR domain determined from the sequences of rat, pig, and human RI has been used to generate proteins containing 2–12 LRRs (60). Biophysical analyses of the RI-like proteins showed monomeric behavior and circular dichroism spectra characteristic of wild-type RI, suggesting that RI-like proteins are viable templates for engineering.
C. Gene Structure and Evolution
RI homologs have been identified in numerous mammalian species and have been found in nearly every type of organ, tissue, and gland investigated to date. Only one copy of the RI gene exists in the human genome (61), and RIs isolated from different tissues of the same species typically have the same amino acid sequence. Still, subtle divergences exist. For example, alternative splice-site forms have been identified in the 5′ untranslated region of RI from human placenta (61). Yet, Northern blot analysis of RI from both placenta and HeLa cells indicate that RI is expressed as a single transcript (8, 62).
Proteins from all LRR subfamilies are capable of forming horseshoe-like structures similar to that of RI (Fig. 4) (12). Modeling studies suggest that the characteristic LRR of a given LRR subfamily cannot be replaced with the LRR from another subfamily (63). Despite similar tertiary structures, the interstrand segments of LRR proteins exhibit markedly different packing interactions, which are not compatible. These observations suggest that the LRRs from different subfamilies have evolved independently, rather than from a single ancestor.
The human RI gene evolved via gene duplication (40). Structural analysis of the RI gene reveals that the exons of RI correspond directly with the LRR units of RI: each exon codes for two segments of α-helix and β-strand (Fig. 1A). In addition, the exons are exactly the same length (171 bases) and exhibit a high degree of identity (50–60% for the 7 internal exons). Apparently, each module of RI arose from a gene duplication event. Not all of the modules of RI are necessary for RI to bind RNase A (64, 65). In fact, as many as two internal modules (113 residues) of RI can be deleted without abolishing its ability to bind to RNase A or inhibiting its catalytic activity (64). Expansion of the RI gene (and protein) to its current size could have facilitated recognition of additional ribonucleases.
The duplication of RI exons occurred rapidly, perhaps in response to the evolution and divergence of members of the RNase A superfamily (40). The RI gene has continued to diverge slowly over a long period of time. Although there is no direct evidence to support positive selection in the evolution of RI exons, it is probable that RI has co-evolved with its complementary ribonucleases. The binding of RI to members of the RNase A superfamily is class specific. For example, human RI will bind to mammalian ribonucleases, but will not inhibit homologous ribonucleases isolated from chicken liver or frog oocytes (22, 66), consistent with distinct pathways of co-evolution.
III. Complexes with Ribonucleases
A. Three-Dimensional Structures
The three-dimensional structures of porcine RI (6) and the porcine RI·RNase A complex (51) were determined in 1993 and 1995 (Fig. 1B). Approximately 2900 Å2 of surface area is buried at the RI RNase A interface, which is 60% more than in a typical antibody·antigen complex (51). The extensive buried surface likely accounts for its exceptionally high affinity for ribonucleases, producing complexes with a Kd value that is 103-fold lower than that of a typical antibody·antigen complex. The RI RNase A interaction appears to rely on Coulombic forces more than do most protein–protein interactions. The β-sheet lining the inner circumference of the horseshoe contributes only 9 of the residues involved in complex formation. Two contact residues are found in α-helical regions of RI, and the remaining 17 contacts are found in loops connecting the C-termini of the β-strands with the N-termini of the α-helices. Upon binding to RNase A, the structure of RI flexes uniformly, and the distance between the N- and C-termini of RI increases by more than 2 Å.
RNase A is a kidney-shaped molecule (67). The active site of the enzyme is located in a cleft between two lobes of the protein. RI inhibits RNase A by blocking the active site; many of the amino acid residues of RNase A that are important for RNA binding and catalysis also interact with RI (68). Few of the contacts provided by RI mimic the RNase A–RNA interaction, though the phenolic ring of Tyr433 does lie in a nucleoside binding site. Thirteen separate patches of residues (28 amino acids) from dispersed regions of RI interact with 3 clusters of residues (24 amino acids) from RNase A. The C-terminal module of RI forms extensive contacts with RNase A, accounting for approximately 30% of the contacts between the two proteins.
The three-dimensional structure of the human RI·ANG complex was determined in 1997 (69). Although the overall docking of ANG with RI is similar to that of RNase A (Fig. 1C), the flexing of RI in the RI·RNase A complex is not apparent in the RI·ANG complex. As in the RI·RNase A complex, the active site of ANG is blocked by numerous contacts with the C-terminus of RI (69). Yet, both substantial and subtle differences are evident in the two complexes. For example, Lys320 of human RI contacts Asp41 of ANG, whereas the analogous residue in porcine RI, Lys316, interacts with Glu86 of RNase A. Using site-directed mutagenesis, the phenyl group of Tyr434 has been shown to interact with both ANG and RNase A (70). Conversely, the phenolic hydroxyl group of Tyr437 interacts with RNase A, whereas the phenyl group of that residue contacts ANG. The dissimilar binding interactions of the two complexes indicate that the broad specificity of RI for pancreatic-type ribonucleases is derived from a remarkable ability to recognize specific features of each ribonuclease.
B. Biomolecular Analyses
The amino acid sequences of RI vary only slightly between species. Yet, the ribonucleases they inhibit differ significantly, possessing as little as 30% amino acid sequence identity. In addition, the ribonucleases that form tight complexes with RI do not exhibit markedly increased sequence identity with each other than with homologous ribonucleases that do not bind to RI.
Prior to the elucidation of its three-dimensional structure, truncated variants of RI were constructed to examine the requirements of RI binding (64, 65). For example, a library of RI variants was constructed by the deletion of one or more LRR modules (one A-type repeat and one B-type repeat) (64). RI variants missing either modules 3 and 4 or module 6 were found to retain affinity for RNase A, whereas deletion of other modules disrupted binding completely. In addition, deletion of module 6 had a substantially greater affect on the affinity of RI for ANG than for RNase A. In another example, RNase A was found to bind to Δ1–90 RI with only a twofold increase in the value of Ki (65). These data provided the first evidence of the modular structure of RI and demonstrated that RI uses disparate regions of its massive surface area to bind to ribonucleases.
The structure of crystalline RI·RNase A shows Gly88 of RNase A in a hydrophobic pocket formed by three tryptophan residues of RI. To generate an RI-evasive variant of RNase A, Gly88 was replaced with an arginine residue (71). The steric bulk of arginine hinders RI binding, and this single substitution increases the Ki value by 104-fold. A pocket can be created in RI to relieve the steric strain in the RI·RNase A complex imposed by an arginine residue at position 88 of RNase A. Replacing Trp264 in RI with an alanine residue allows RI to accommodate Arg88 of G88R RNase A. Although wild-type RI and the W264A variant inhibit RNase A to a similar extent, only the variant protects 16S- and 23S-rRNA from degradation by G88R RNase A. These data demonstrated that the “knobs-into-holes” concept (72) is applicable to an RI·ribonuclease complex.
Mutagenesis of key binding residues of RI was found to have varying effects on binding energy. Replacing some residues that appear to contact RNase A closely (e.g., Glu287, Lys320, Glu401, or Arg457) have little effect on binding (73). On the other hand, Tyr434, Asp435, Tyr437, and Ser460 of RI were found to constitute a “hot spot“ of binding energy. Only one of those residues, Asp435, is equally important to the binding of ANG. Substitution of any two of these residues has a superadditive effect on ANG binding, but a subadditive effect on RNase A binding (70).
Alterations to a second cluster of RI residues, including Trp261, Trp263, Trp318, and Trp375, have also been shown to display superadditive effects on ANG binding (74). Recent studies have reported superadditive effects in the RI·EDN complex (75); both the C-terminal residues and tryptophan clusters contribute significantly to binding and demonstrate negative cooperativity, as in ANG binding. To date, no such negative cooperativity has been demonstrated for binding to RNase A (70, 74). These results suggest that the binding energy could be more widely distributed in the RI·RNase A complex than in the RI·EDN and RI·ANG complexes.
Structural and biochemical studies have provided significant evidence that the molecular interactions in RI·ribonuclease complexes differ substantially. For example, residues 408–410 in human RI appear to contact RNase A but not ANG. Remodeling these residues to yield C408W/ΔV409/G410W RI decreases the Ki value for RNase A and RNase 1 by >108-fold, but increases that value for ANG by only twofold (76). Thus, the ligand specificity of RI can be altered dramatically by changing only a few residues. It is noteworthy that the C408W/ΔV409/G410W variant of RI could be a useful tool for future studies on the biological function of ANG and the RI·ANG complex.
IV. Cysteine Content and Oxidative Instability
LRR proteins commonly have N- and C-terminal domains that are rich in cysteine residues (12). Still, only proteins from the RI-like and cysteine-containing LRR subfamilies contain cysteine residues in their consensus sequence (12). Human RI and porcine RI contain 32 and 30 cysteine residues, respectively, comprising almost 7% of their amino acid residues (7, 8). Sequence analysis of RI from human, pig, mouse and rat shows that 27 of the cysteine residues are conserved (Fig. 2). Several of the these cysteine residues could play key structural roles: the sulfhydryl group of the cysteine residue at position 10 of the A-type repeat appears to donate a hydrogen bond to the main-chain oxygen of residue 8, whereas the cysteine residue at position 17 of the A-type repeat is part of the hydrophobic core (10) (Fig. 3).
All of its cysteine residues must remain reduced for RI to maintain activity (20). Oxidation of RI is a highly cooperative process (20). Reaction of RI with a substoichiometric amount of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) yields a mixture of completely oxidized, inactive molecules and completely reduced, active molecules. Subsequent to oxidation of only a few cysteines, RI rapidly undergoes a conformational change that results in increasing reactivity of the remaining thiols (20). Several proximal cysteine residues create triggers for the oxidation and denaturation of RI. Replacing Cys328 and Cys329 with alanine residues endows RI with 10- to 15-fold greater resistance to oxidation by hydrogen peroxide with only a minimal effect on its affinity for RNase A (77).
Unlike unbound RI, the RI·RNase A complex can undergo partial oxidation (29). Treatment of the RI·RNase A complex with DTNB oxidizes up to 14 of its 30 cysteine residues and allows the enzyme to express up to 15% of its enzymatic activity. Only after dissociation does RI undergo its typical all-or-none oxidation. Thus, ribonucleases afford RI with some degree of protection from oxidation.
Degradation of RI correlates to its oxidative inactivation. Inducing oxidative damage in LLK-PC1 cells with hydrogen peroxide and diamide results in the degradation of RI (78). Similarly, oxidative stress in human erythrocytes induces decreased levels of glutathione followed by gradual loss of RI activity in the cytosol (30). In contrast to LLK-PC1 cells, inactivated RI is detected in nascent Heinz bodies of human erythrocytes. Oxidation could be a mechanism by which the activity of RI (and thereby its cognate ribonucleases) are regulated in the cytosol.
V. Biological Activities
A. Expression Levels and Tissue Distribution
RI has been found in the cytosol of many cell types. Although it inhibits secretory ribonucleases, RI has not been detected in extracellular fluids, such as plasma, saliva, and urine (26, 79). The expression patterns of RI have been investigated extensively during the previous three decades, with the hope of revealing insight into the biological role of RI. Still, the literature is full of conflicting conclusions. RI biosynthesis seems to correlate positively with anabolic activity, such as cell proliferation; increased RI levels have been found in rat liver after treatment with 2-acetamidofluorene to induce tumors (80) and in developing neonatal rats (81). Yet, RI levels are not elevated in SV-40–transformed hamster embryo fibroblast cells, stimulated HL-60 cells (82), or many hepatocyte lines. The labile nature of RI could have compounded the difficulty of correlating RI levels with physiological relevance. A recent study did, however, find that high RI levels decreased angiogenesis and tumor formation in mouse xenographs (83).
B. Role in Ribonuclease Cytotoxicity
In 1955, RNase A was found to be toxic to carcinomas in mice and rats (84, 85). The antitumor activity of RNase A showed poor promise as a chemotherapeutic because milligram quantities were required to achieve a beneficial effect (86). In 1973, the antitumor activity of dimeric BS-RNase towards Crocker tumor transplants in mice was discovered (87). Further characterization demonstrated, however, that BS-RNase is a poor candidate for cancer chemotherapy, as it has non-specific toxicity; is antispermatogenic (88), hinders embryo development (89) and oocyte maturation (90), and is immunosuppressive (91).
Amphibian ribonucleases from Rana pipiens (92), Rana catesbeiana (93, 94), and Rana japonica (94) were found to contain antitumor activity. Onconase® (ONC) is an RNase A homolog from Rana pipiens and is both cytotoxic and cytostatic towards cultured tumor cells (92, 95). ONC also causes the regression of xenographs in mice (96). ONC has been successful in the treatment of malignant mesothelioma in Phase I (97, 98) and Phase II clinical trials (99). Side effects of ONC are reversible and include renal toxicity and proteinuria. Phase III clinical studies of ONC for the treatment of malignant mesothelioma are in progress.
ONC shares 30% amino acid sequence identity with RNase A (95). Although the key active-site residues of RNase A—His12, Lys41, His119—are conserved in ONC, the amphibian enzyme has ≤0.1% of the ribonucleolytic activity of RNase A (44, 100, 101). The ribonucleolytic activity of ONC is, however, essential for its cytotoxicity (102, 44, 103, 104). The structure of crystalline ONC has been determined, and although ONC is twenty residues shorter than RNase A, the two enzymes share similar secondary and tertiary structure (67, 105). Deletions within ONC are positioned within surface loops and at the N-terminus. ONC contains four disulfide bonds, three of which are present in RNase A. The synapomorphic disulfide bond in ONC secures its C-terminus, and is responsible for endowing ONC with remarkable conformational stability (101, 106). For example, the Tm value of ONC is 90 °C, which is 30 °C higher than that of RNase A.
The mechanism by which a ribonuclease is cytotoxic can be dissected into four steps: (1) cell-surface binding, (2) ribonuclease internalization, (3) translocation into the cytosol, and (4) evasion of RI and degradation of cellular RNA. ONC has low catalytic activity, but is a potent cytotoxin, suggesting that it accomplishes these four steps. In contrast, RNase A is not an efficient toxin. Specifically, RNase A is >103-fold less cytotoxic to cells than is ONC (102). Both RNase A and ONC demonstrate nonspecific binding to the cell surface (K. A. Dickson and R. T. Raines, unpublished results) and no direct measurements of ribonuclease internalization and translocation to the cytosol have been reported to date. The distinguishing attribute of an RNase A homolog with cytotoxic activity is its ability to retain ribonucleolytic activity in the presence of RI. For example, RI does not associate with ONC but binds RNase A with nearly femtomolar affinity (102, 44). As a result, ONC but not RNase A is capable of degrading cellular RNA and causing cell death.
The discovery of ONC in 1988 and its clinical success in subsequent years has intensified the study of other ribonucleases with biological actions. Current studies are focusing on understanding the mechanism of ribonuclease-mediated cytotoxicity with hope to improve potency and specificity. Using the cytotoxicity of ONC as a model, mammalian pancreatic ribonuclease variants have been endowed with toxic activity (for reviews, see (14, 15, 17)). The substantial difference in the binding affinities of ONC and RNase A for RI has proven to be a critical factor in the cytotoxicity of ribonucleases. Variants of pancreatic-type ribonucleases that have been engineered to evade RI possess cytotoxic activity. RI evasion has been achieved by covalently linking other proteins, dimerization, and site-directed mutagenesis.
The most common approach used to generate cytotoxic ribonucleases is to engineer amino acid substitutions that will disrupt contacts in the RI·ribonuclease complex specifically. For example, G88R RNase A is toxic to human leukemia cells (71). Invoking a similar strategy, RNase 1 has been engineered to contain a G88R-like surface loop (107). This variant evades RI and is also toxic to human leukemia cells. Enhanced RI evasion can be attained at the expense of lower ribonucleolytic activity, as in K41R/G88R RNase A and A4C/K41R/G88R/V118C RNase A, without compromising cytotoxicity (Table II) (100, 50).
The ability of a ribonuclease to manifest its catalytic activity in the cytosol is related to its values of kcat/KM and Kd, and the concentration of RI in the cytosol ([RI]cyto = 4 μM (108)). This ability can be described by the parameter (kcat/KM)cyto, which is defined in Eq. (2) (109, 100, 110):
| (2) |
The resulting values of (kcat/KM)cyto for RNase A, its variants, and ONC are listed in Table II. The most toxic RNase A variant reported to date has a double substitution in which Lys7 and Gly88 are replaced with alanine and arginine residues, respectively (49). This variant demonstrates high catalytic activity, evades RI, and is nearly as toxic as ONC to human leukemia cells.
The role of RI in ribonuclease cytotoxicity has been examined directly by modulating intracellular levels of RI. Overexpression of RI in K-562 or HeLa cells diminished the potency of cytotoxic variants of RI without affecting the toxicity of ONC (108). These findings suggest that ONC has no affinity for RI, such that (kcat/KM)cyto= kcat/KM; upon entering a cell, ONC is able to degrade cellular RNA uninhibited. Conversely, the (kcat/KM)cyto values for RNase A variants that maintain affinity for RI are limited by the concentration of cytosolic RI.
Similar results were obtained using RNAi to suppress levels of cytosolic RI. Suppression resulted in increased susceptibility to ribonuclease variants that possess diminished affinity for RI (e.g., G88R RNase A), but did not endow ribonucleases with high affinity for RI with cytotoxic activity (e.g., wild-type RNase A) (111). The amount of intact exogenous ribonuclease that reaches the cytosol of a cell is unknown, but likely to be small. Thus, even trace amounts of cytosolic RI could be sufficient to neutralize an invading ribonuclease with high affinity for RI.
C. Role in Angiogenesis
ANG is a unique ribonuclease (for reviews, see (112–114)). ANG acts on endothelial and smooth muscle cells to induce a wide range of cellular responses including cell proliferation, activation of cell-associated proteases, and cell migration and invasion. ANG binds to a receptor protein and is transported rapidly to the nucleus, where it activates transcription (18, 115–117, 19).
The role of RI in angiogenesis is controversial. The ribonucleolytic activity of ANG is weak (106-fold less than that of RNase A (118, 119)) but essential for its biological activity (120, 121); amino acid substitutions that abolish ribonucleolytic activity also prevent angiogenesis. RI added extracellularly also inhibits angiogenesis (122, 123), most likely by preventing ANG from binding to its receptor. Because the Kd value of the RI·ANG complex is among the lowest of known biomolecular interactions, RI could serve to protect cellular RNA from ANG that leaks inadvertently into the cytosol. On the other hand, RI could serve to control the biological activity of ANG. In one possible scenario, RI negatively regulates ANG that gains access to the cytosol; inactivation of RI reactivates ANG that was sequestered in an RI·ANG complex. Finally, the extraordinary affinity of ANG for RI suggests that the RI·ANG complex itself could have biological activity, though this hypothesis is contradicted by the known angiogenic activity of ANG in chick embryos, which do not possess an RI that binds to mammalian ribonucleases (66, 124).
D. Alternative Biological Roles
The marked oxidation sensitivity of RI in addition to its all-or-none mechanism of oxidative inactivation and denaturation is well documented (20, 77). Yet, the biological significance of these properties remains unclear. One hypothesis suggests that RI is an oxidation sensor in the cell. Overexpression of RI in rat glial cells conferred protection against hydrogen peroxide-induced stress, as indicated by the increased viability of cells, decreased leakage of lactate dehydrogenase, and increased content of reduced glutathione (125). Injection of RI into mice also conferred protection from per-oxidative injuries of the liver induced by exposure to carbon tetrachloride (125). These experiments suggest that RI could protect cells against two distinct onslaughts: invading ribonucleases and oxidative damage.
Surprisingly, significant quantities of RI have been detected in human erythrocytes, which are essentially devoid of ribonucleases and RNA (30). The presence of RI in erythrocytes provides additional evidence that RI serves multiple roles in mammalian cells. Oxidative stress on isolated red blood cells resulted in reduced levels of glutathione followed by gradual loss of RI activity associated with its aggregation in Heinz bodies (30). A similar sequence of inactivation and degradation has been noted for hemoglobin in response to oxidative stress (126) and other proteins (112) associated with aging. Decreases in RI activity have been observed in association with numerous diseases, including cataract formation (127), leukemia (66), and exposure to ionizing radiation (128). Thus, RI in human erythrocytes, as well as nucleated cells, could be a determinant of cellular lifespan or simply a marker of aging.
VI. Conclusions
RI possesses remarkable affinity for pancreatic-type ribonucleases, despite their limited sequence identity. The resulting noncovalent complexes are some of the tightest known in biology. Details of the molecular interactions within RI·ribonuclease complexes have been elucidated from structural and biochemical investigations. Moreover, RI is known to be a sentry, protecting mammalian cells against invading ribonucleases, which abound in extracellular fluids. Still, many questions remain regarding the biological activity of RI: Why have its Ki values evolved to be so low? What is the significance of the oxidation sensitivity of RI? Does the RI·ribonuclease complex itself have a biological role? In addition, the potential of the unique tertiary structure of RI to serve as a scaffold for the design of new receptors is virtually unexplored, but seemingly limitless. Accordingly, future research will likely be directed at elucidating the biological significance of the remarkable biochemical properties of RI, and developing RI as a scaffold for protein engineering. We look forward to learning the results of this effort.
Acknowledgments
Work on the ribonuclease inhibitor protein in the Raines laboratory was supported by grant CA73808 (NIH). K.A.D. was supported by the Louis and Elsa Thomsen Wisconsin Distinguished Fellowship Award from the College of Agricultural and Life Sciences at the University of Wisconsin–Madison.
References
- 1.Roth JS. Some observations on the assay and properties of ribonucleases in normal and tumor tissues. In: Busch H, editor. Methods in Cancer Research. Academic Press; New York: 1967. pp. 153–242. [Google Scholar]
- 2.Blackburn P, Moore S. Pancreatic ribonuclease. The Enzymes. 1982;XV:317–433. [Google Scholar]
- 3.Lee FS, Vallee BL. Structure and action of mammalian ribonuclease (angiogenin) inhibitor. Progress Nucl Acid Res Molec Biol. 1993;44:1–30. doi: 10.1016/s0079-6603(08)60215-9. [DOI] [PubMed] [Google Scholar]
- 4.Hofsteenge J. Ribonuclease inhibitor. In: D’Alessio G, Riordan JF, editors. Ribonucleases: Structures and Functions. Academic Press; New York: 1997. pp. 621–658. [Google Scholar]
- 5.Shapiro R. Cytoplasmic ribonuclease inhibitor. Methods Enzymol. 2001;341:611–628. doi: 10.1016/s0076-6879(01)41180-3. [DOI] [PubMed] [Google Scholar]
- 6.Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 7.Hofsteenge J, Kieffer B, Matthies R, Hemmings BA, Stone SR. Amino acid sequence of the ribonuclease inhibitor from porcine liver reveals the presence of leucine-rich repeats. Biochemistry. 1988;27:8537–8544. doi: 10.1021/bi00423a006. [DOI] [PubMed] [Google Scholar]
- 8.Lee FS, Fox EA, Zhou HM, Strydom DJ, Vallee BL. Primary structure of human placental ribonuclease inhibitor. Biochemistry. 1988;27:8545–8553. doi: 10.1021/bi00423a007. [DOI] [PubMed] [Google Scholar]
- 9.Janin J. Proteins with a ring. Structure. 1994;2:571–573. doi: 10.1016/s0969-2126(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 10.Kobe B, Deisenhofer J. The leucine-rich repeat: A versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 11.Shapiro R, Riordan JF, Vallee BL. LRRning the RIte of springs. Nat Struct Biol. 1995;2:350–354. doi: 10.1038/nsb0595-350. [DOI] [PubMed] [Google Scholar]
- 12.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 13.Pasloske BL. Ribonuclease inhibitors. In: Schein CH, editor. Nuclease Methods and Protocols. Humana Press; Totowa, NJ: 2001. pp. 105–111. [DOI] [PubMed] [Google Scholar]
- 14.Youle RJ, D’Alessio G. Antitumor RNases. In: D’Alessio G, Riordan JF, editors. Ribonucleases: Structures and Functions. Academic Press; New York: 1997. pp. 491–514. [Google Scholar]
- 15.Leland PA, Raines RT. Cancer chemotherapy—ribonucleases to the rescue. Chem Biol. 2001;8:405–413. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matousek J. Ribonucleases and their antitumor activity. Comp Biochem Physiol C Toxicol Pharmacol. 2001;129:175–191. doi: 10.1016/s1532-0456(01)90202-9. [DOI] [PubMed] [Google Scholar]
- 17.Makarov AA, Ilinskaya ON. Cytotoxic ribonucleases: Molecular weapons and their targets. FEBS Letters. 2003;540:15–20. doi: 10.1016/s0014-5793(03)00225-4. [DOI] [PubMed] [Google Scholar]
- 18.Moroianu J, Riordan JF. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc Natl Acad Sci USA. 1994;91:1677–1681. doi: 10.1073/pnas.91.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu ZP, Tsuji T, Riordan JF, Hu GF. The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem Biophys Res Commun. 2003;294:287–292. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]
- 20.Fominaya JM, Hofsteenge J. Inactivation of ribonuclease inhibitor by thiol–disulfide exchange. J Biol Chem. 1992;267:24655–24660. [PubMed] [Google Scholar]
- 21.Pirotte M, Desreux V. Distribution de la ribonuclease dans les extraits de granules cellulaires du foie. Bull Soc Chim Belg. 1952;61:167. [Google Scholar]
- 22.Roth JS. Ribonuclease IX. Further studies on ribonuclease inhibitor. Biochim Biophys Acta. 1962;61:903–915. doi: 10.1016/0926-6550(62)90007-5. [DOI] [PubMed] [Google Scholar]
- 23.Blackburn P, Wilson G, Moore S. Ribonuclease inhibitor from human placenta. Purification and properties. J Biol Chem. 1977;252:5904–5910. [PubMed] [Google Scholar]
- 24.Burton LE, Blackburn P, Moore S. Ribonuclease inhibitor from bovine brain. Int J Peptide Protein Res. 1980;16:359–364. doi: 10.1111/j.1399-3011.1980.tb02959.x. [DOI] [PubMed] [Google Scholar]
- 25.Cho S, Joshi JG. Ribonuclease inhibitor from pig brain: Purification, characterization, and direct spectrophotometric assay. Anal Biochem. 1989;176:175–181. doi: 10.1016/0003-2697(89)90289-3. [DOI] [PubMed] [Google Scholar]
- 26.Nadano D, Yasuda T, Takeshita H, Uchide K, Kishi K. Purification and characterization of human brain ribonuclease inhibitor. Arch Biochem Biophys. 1994;312:421–428. doi: 10.1006/abbi.1994.1328. [DOI] [PubMed] [Google Scholar]
- 27.Gribnau AA, Schoenmakers JG, van Kraaikamp M, Bloemendal H. High purification of the RNase inhibitor from rat liver by affinity chromatography. Biochem Biophys Res Commun. 1970;38:1064–1068. doi: 10.1016/0006-291x(70)90347-5. [DOI] [PubMed] [Google Scholar]
- 28.Burton LE, Fucci NP. Ribonuclease inhibitors from the liver of five mammalian species. Int J Peptide Protein Res. 1982;19:372–379. doi: 10.1111/j.1399-3011.1982.tb02618.x. [DOI] [PubMed] [Google Scholar]
- 29.Ferreras M, Gavilanes JG, Lopez-Otin C, Garcia-Segura JM. Thiol–disulfide exchange of ribonuclease inhibitor bound to ribonuclease A. Evidence of active inhibitor-bound ribonuclease. J Biol Chem. 1995;270:28570–28578. doi: 10.1074/jbc.270.48.28570. [DOI] [PubMed] [Google Scholar]
- 30.Moenner M, Vosoghi M, Ryazantsev S, Glitz DG. Ribonuclease inhibitor protein of human erythrocytes: Characterization, loss of activity in response to oxidative stress, and association with Heinz bodies. Blood Cells Mol Dis. 1998;24:149–164. doi: 10.1006/bcmd.1998.0182. [DOI] [PubMed] [Google Scholar]
- 31.Raines RT. Ribonuclease A. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 32.Garcia MA, Klebe RJ. Affinity chromatography of RNase inhibitor. Mol Biol Rep. 1997;24:231–233. doi: 10.1023/a:1006818400674. [DOI] [PubMed] [Google Scholar]
- 33.Blackburn P. Ribonuclease inhibitor from human placenta: Rapid purification and assay. J Biol Chem. 1979;254:12484–12487. [PubMed] [Google Scholar]
- 34.Vescia S, Tramontano D, Augusti-Tocco G, D’Alessio G. In vitro studies on selective inhibition of tumor cell growth by seminal ribonuclease. Cancer Res. 1980;40:3740–3744. [PubMed] [Google Scholar]
- 35.Lee FS, Vallee BL. Expression of human placental ribonuclease inhibitor in Escherichia coli. Biochem Biophys Res Commun. 1989;160:115–120. doi: 10.1016/0006-291x(89)91628-8. [DOI] [PubMed] [Google Scholar]
- 36.Vicentini AM, Kieffer B, Mathies R, Meyhack B, Hemmings BA, Stone SR, Hofsteenge J. Protein chemical and kinetic characterization of recombinant porcine ribonuclease inhibitor expressed in Saccharomyces cerevisiae. Biochemistry. 1990;29:8827–8834. doi: 10.1021/bi00489a046. [DOI] [PubMed] [Google Scholar]
- 37.Klink TA, Vicentini AM, Hofsteenge J, Raines RT. High-level soluble production and characterization of porcine ribonuclease inhibitor. Protein Expr Purif. 2001;22:174–179. doi: 10.1006/prep.2001.1422. [DOI] [PubMed] [Google Scholar]
- 38.Blackburn P, Jailkhan BL. Ribonuclease inhibitor from human placenta: Interaction with deriviatives of ribonuclease A. J Biol Chem. 1979;254:12488–12493. [PubMed] [Google Scholar]
- 39.Kawanomoto M, Motojima K, Sasaki M, Hattori H, Goto S. cDNA cloning and sequence of rat ribonuclease inhibitor, and tissue distribution of mRNA. Biochim Biophys Acta. 1992;1129:335–338. doi: 10.1016/0167-4781(92)90513-y. [DOI] [PubMed] [Google Scholar]
- 40.Haigis MC, Haag ES, Raines RT. Evolution of ribonuclease inhibitor by exon duplication. Mol Biol Evol. 2002;19:959–963. doi: 10.1093/oxfordjournals.molbev.a004153. [DOI] [PubMed] [Google Scholar]
- 41.Lee FS, Shapiro R, Vallee BL. Tight-binding inhibition of angiogenin and ribonuclease A by placental ribonuclease inhibitor. Biochemistry. 1989;28:225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]
- 42.Green NM. Avidin. Adv Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 43.Zelenko O, Neumann U, Brill W, Pieles U, Moser HE, Hofsteenge J. A novel fluorogenic substrate for ribonucleases. Synthesis and enzymatic characterization. Nucleic Acids Res. 1994;22:2731–2739. doi: 10.1093/nar/22.14.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boix E, Wu Y, Vasandani VM, Saxena SK, Ardelt W, Ladner J, Youle RJ. Role of the N terminus in RNase A homologues: Differences in catalytic activity, ribonuclease inhibitor interaction and cytotoxicity. J Mol Biol. 1996;257:992–1007. doi: 10.1006/jmbi.1996.0218. [DOI] [PubMed] [Google Scholar]
- 45.Hofsteenge J, Vicentini A, Zelenko O. Ribonuclease 4, an evolutionarily highly conserved member of the superfamily. Cell Mol Life Sci. 1998;54:804–810. doi: 10.1007/s000180050209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee FS, Auld DS, Vallee BL. Tryptophan fluorescence as a probe of placental ribonuclease inhibitor binding to angiogenin. Biochemistry. 1989;28:219–224. doi: 10.1021/bi00427a030. [DOI] [PubMed] [Google Scholar]
- 47.Shapiro R, Vallee BL. Interaction of human placental ribonuclease with placental ribonuclease inhibitor. Biochemistry. 1991;30:2246–2255. doi: 10.1021/bi00222a030. [DOI] [PubMed] [Google Scholar]
- 48.Abel RL, Haigis MC, Park C, Raines RT. Fluorescence assay for the binding of ribonuclease A to the ribonuclease inhibitor protein. Anal Biochem. 2001;306:100–107. doi: 10.1006/abio.2002.5678. [DOI] [PubMed] [Google Scholar]
- 49.Haigis MC, Kurten EL, Abel RL, Raines RT. KFERQ sequence in ribonuclease A-mediated cytotoxicity. J Biol Chem. 2002;277:11576–11581. doi: 10.1074/jbc.M112227200. [DOI] [PubMed] [Google Scholar]
- 50.Dickson KA, Dahlberg CL, Raines RT. Compensating effects on the cytotoxicity of ribonuclease A variants. Archives Biochem Biophys. 2003;415:172–177. doi: 10.1016/s0003-9861(03)00214-5. [DOI] [PubMed] [Google Scholar]
- 51.Kobe B, Deisenhofer J. A structural basis of the interactions between leucine- rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 52.Kajava AV. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi N, Takahashi Y, Putnam FW. Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich alpha 2-glycoprotein of human serum. Proc Natl Acad Sci USA. 1985;82:1906–1910. doi: 10.1073/pnas.82.7.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobe B, Deisenhofer J. Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 55.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B″–U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 56.Evdokimov AG, Anderson DE, Routzahn KM, Waugh DS. Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: A leucine-rich repeat protein with the shortest repeating unit. J Mol Biol. 2001;312:807–821. doi: 10.1006/jmbi.2001.4973. [DOI] [PubMed] [Google Scholar]
- 57.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1 Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 58.Matteo AD, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, Lorenzo GD, Tsernoglou D. The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci USA. 2003;100:10124–10128. doi: 10.1073/pnas.1733690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schott PG, McEwan PA, Dodd CM, Bergmann EM, Bishop PN, Bella J. Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan. Proc Natl Acad Sci USA. 2004;101:15633–15638. doi: 10.1073/pnas.0402976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stumpp MT, Forrer P, Binz HK, Pluckthun A. Designing repeat proteins: Modular leucine-rich repeat protein libraries based on mammalian ribonuclease inhibitor family. J Mol Biol. 2003;332:471–487. doi: 10.1016/s0022-2836(03)00897-0. [DOI] [PubMed] [Google Scholar]
- 61.Crawford D, Hagerty K, Beutler B. Multiple splice forms of ribonuclease- inhibitor mRNA differ in the 5′-untranslated region. Gene. 1989;85:525–531. doi: 10.1016/0378-1119(89)90447-2. [DOI] [PubMed] [Google Scholar]
- 62.Schneider R, Schneider-Scherzer E, Thurnher M, Auer B, Schweiger M. The primary structure of human ribonuclease/angiogenin inhibitor (RAI) discloses a novel highly diversified protein superfamily with a common repetitive module. EMBO J. 1988;7:4151–4156. doi: 10.1002/j.1460-2075.1988.tb03310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kajava AV, Kobe B. Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci. 2002;11:1082–1090. doi: 10.1110/ps.4010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee FS, Vallee BL. Modular mutagenesis of human placental ribonuclease inhibitor, a protein with leucine-rich repeats. Proc Natl Acad Sci USA. 1990;87:1879–1883. doi: 10.1073/pnas.87.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hofsteenge J, Vincentini A, Stone SR. Purification and characterization of truncated ribonuclease inhibitor. Biochem J. 1991;275:541–543. doi: 10.1042/bj2750541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kraft N, Shortman K. The phylogeny of the ribonuclease ribonuclease inhibitor system: Its distribution in tissues and its response during leukaemogenesis and aging. Aust J Biol Sci. 1970;23:175–184. [PubMed] [Google Scholar]
- 67.Wlodawer A. Structure of bovine pancreatic ribonuclease by X-ray and neutron diffraction. In: Jurnak FA, McPherson A, editors. Biological Macromolecules and Assemblies, Vol. II, Nucleic Acids and Interactive Proteins. Wiley; New York: 1985. pp. 395–439. [Google Scholar]
- 68.Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with RNase A. J Mol Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- 69.Papageorgiou AC, Shapiro R, Acharya KR. Molecular recognition of human angiogenin by placental ribonuclease inhibitor—an X-ray crystallographic study at 2.0 Å resolution. EMBO J. 1997;16:5162–5177. doi: 10.1093/emboj/16.17.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen CZ, Shapiro R. Superadditive and subadditive effects of “hot spot” mutations within the interfaces of placental ribonuclease inhibitor with angiogenin and ribonuclease A. Biochemistry. 1999;38:9273–9285. doi: 10.1021/bi990762a. [DOI] [PubMed] [Google Scholar]
- 71.Leland PA, Schultz LW, Kim BM, Raines RT. Ribonuclease A variants with potent cytotoxic activity. Proc Natl Acad Sci USA. 1998;98:10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crick FHC. Is α-keratin a coiled coil? Nature. 1952;170:882–883. doi: 10.1038/170882b0. [DOI] [PubMed] [Google Scholar]
- 73.Chen CZ, Shapiro R. Site-specific mutagenesis reveals differences in the structural bases for tight binding of RNase inhibitor to angiogenin and RNase A. Proc Natl Acad Sci USA. 1997;94:1761–1766. doi: 10.1073/pnas.94.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shapiro R, Ruiz-Gutierrez M, Chen CZ. Analysis of the interactions of human ribonuclease inhibitor with angiogenin and ribonuclease A by mutagenesis: Importance of inhibitor residues inside versus outside the C-terminal “hot spot”. J Mol Biol. 2000;302:497–519. doi: 10.1006/jmbi.2000.4075. [DOI] [PubMed] [Google Scholar]
- 75.Teufel DP, Kao RYT, Acharya KR, Shapiro R. Mutational analysis of the complex of human RNase inhibitor and human eosinophil-derived neurotoxin. Biochemistry. 2003;42:1451–1459. doi: 10.1021/bi026852o. [DOI] [PubMed] [Google Scholar]
- 76.Kumar K, Brady M, Shapiro R. Selective abolition of pancreatic RNase binding to its inhibitor protein. Proc Natl Acad Sci USA. 2004;101:53–58. doi: 10.1073/pnas.0307268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim BM, Schultz LW, Raines RT. Variants of ribonuclease inhibitor that resist oxidation. Protein Sci. 1999;8:430–434. doi: 10.1110/ps.8.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blázquez M, Fominaya JM, Hofsteenge J. Oxidation of sulfhydryl groups of ribonuclease inhibitor in epithelial cells is sufficient for its intracellular degradation. J Biol Chem. 1996;271:18638–18642. doi: 10.1074/jbc.271.31.18638. [DOI] [PubMed] [Google Scholar]
- 79.Futami J, Tsushima Y, Murato Y, Tada H, Sasaki J, Seno M, Yamada H. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 1997;16:413–419. doi: 10.1089/dna.1997.16.413. [DOI] [PubMed] [Google Scholar]
- 80.Wojnar RJ, Roth JS. Ribonuclease inhibitor and latent ribonuclease in rat liver during feeding of 2-acetamidofluorene. Cancer Res. 1965;25:1913–1918. [PubMed] [Google Scholar]
- 81.Suzuki Y, Takahashi Y. Developmental and regional variations in ribonuclease inhibitor activity in brain. J Neurochem. 1970;17:1521–1524. doi: 10.1111/j.1471-4159.1970.tb00520.x. [DOI] [PubMed] [Google Scholar]
- 82.Kyner D, Christman JK, Acs G. The effect of 12-O-tetradecanoyl-phorbol 13-acetate on the ribonuclease activity of circulating human lymphocytes. Eur J Biochem. 1979;99:395–399. doi: 10.1111/j.1432-1033.1979.tb13268.x. [DOI] [PubMed] [Google Scholar]
- 83.Botella-Estrada R, Malet G, Revert F, Dasi F, Crespo A, Sanmartin O, Guillen C, Alino SF. Antitumor effect of B16 melanoma cells genetically modified with the angiogenesis inhibitor rnasin. Cancer Gene Ther. 2001;8:278–284. doi: 10.1038/sj.cgt.7700302. [DOI] [PubMed] [Google Scholar]
- 84.Ledoux L. Action of ribonuclease on certain ascites tumours. Nature. 1955;175:258–259. doi: 10.1038/175258b0. [DOI] [PubMed] [Google Scholar]
- 85.Ledoux L. Action of ribonuclease on two solid tumours in vivo. Nature. 1955;176:36–37. doi: 10.1038/176036a0. [DOI] [PubMed] [Google Scholar]
- 86.Roth JS. Ribonuclease activity and cancer: A review. Cancer Res. 1963;23:657–666. [PubMed] [Google Scholar]
- 87.Matousek J. The effect of bovine seminal ribonuclease (AS RNase) on cells of crocker tumour in mice. Experientia. 1973;29:858–859. doi: 10.1007/BF01946329. [DOI] [PubMed] [Google Scholar]
- 88.Matousek J. Aspermatogenic effect of the bull seminal ribonuclease (BS RNase) in the presence of anti BS RNase antibodies in mice. Animal Genet. 1994;25(Suppl 1):45–50. doi: 10.1111/j.1365-2052.1994.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 89.Matousek J. Embryotoxic effect of bull seminal ribonuclease and tissue absorption studies in rats. J Reprod Fertil. 1975;43:171–174. doi: 10.1530/jrf.0.0430171. [DOI] [PubMed] [Google Scholar]
- 90.Slavik T, Matousek J, Fulka J, Raines RT. Effect of bovine seminal ribonuclease and bovine pancreatic ribonuclease A on bovine oocyte maturation. J Exp Zool. 2000;287:394–399. [PubMed] [Google Scholar]
- 91.Matousek J, Soucek J, Riha J, Zankel TR, Benner SA. Immunosuppressive activity of angiogenin in comparison with bovine seminal ribonuclease and pancreatic ribonuclease. Comp Biochem Physiol. 1995;112B:235–241. doi: 10.1016/0305-0491(95)00075-5. [DOI] [PubMed] [Google Scholar]
- 92.Darzynkiewicz Z, Carter SP, Mikulski SM, Ardelt WJ, Shogen K. Cytostatic and cytotoxic effect of Pannon (P-30 Protein), a novel anticancer agent. Cell Tissue Kinet. 1988;21:169–182. doi: 10.1111/j.1365-2184.1988.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 93.Nitta K, Takayanagi G, Kawauchi H, Hakomori S. Isolation and characterization of Rana catesbeiana lectin and demonstration of the lectin-binding glycoprotein of rodent and human tumor cell membranes. Cancer Res. 1987;47:4877–4883. [PubMed] [Google Scholar]
- 94.Nitta K, Ozaki K, Ishikawa M, Furusawa S, Hosono M, Kawauchi H, Sasaki K, Takayanagi Y, Tsuiki S, Hakomori S. Inhibition of cell proliferation by Rana catesbeiana and Rana japonica lectins belonging to the ribonuclease superfamily. Cancer Res. 1994;54:920–927. [PubMed] [Google Scholar]
- 95.Ardelt W, Mikulski SM, Shogen K. Amino acid sequence of an anti-tumor protein from Rana pipiens oocytes and early embryos. J Biol Chem. 1991;266:245–251. [PubMed] [Google Scholar]
- 96.Mikulski SM, Ardelt W, Shogen K, Bernstein EH, Menduke H. Striking increase of survival of mice bearing M109 Madison carcinoma treated with a novel protein from amphibian embryos. J Natl Cancer Inst. 1990;82:151–153. doi: 10.1093/jnci/82.2.151-a. [DOI] [PubMed] [Google Scholar]
- 97.Mikulski SM, Grossman AM, Carter PW, Shogen K, Costanzi JJ. Phase I human clinical trial of ONCONASE (P-30 Protein) administered intravenously on a weekly schedule in cancer patients with solid tumors. Int J Oncol. 1993;3:57–64. doi: 10.3892/ijo.3.1.57. [DOI] [PubMed] [Google Scholar]
- 98.Mikulski SM, Chun HG, Mittelman A, Panella T, Puccio CA, Shogen K, Constanzi JJ. Relationship between response rate and median survival in patients with advanced non-small cell lung cancer: Comparison of ONCONASE® with other anticancer agents. Int J Oncol. 1995;6:889–897. doi: 10.3892/ijo.6.4.889. [DOI] [PubMed] [Google Scholar]
- 99.Mikulski SM, Costanzi JJ, Vogelzang NJ, McCachren S, Taub RN, Chun H, Mittelman A, Panella T, Puccio C, Fine R, Shogen K. Phase II trial of a single weekly intravenous dose of ranpirnase in patients with unresectable malignant mesothelioma. J Clin Oncol. 2002;20:274–281. doi: 10.1200/JCO.2002.20.1.274. [DOI] [PubMed] [Google Scholar]
- 100.Bretscher LE, Abel RL, Raines RT. A ribonuclease A variant with low catalytic activity but high cytotoxicity. J Biol Chem. 2000;275:9893–9896. doi: 10.1074/jbc.275.14.9893. [DOI] [PubMed] [Google Scholar]
- 101.Leland PA, Staniszewski KE, Kim B, Raines RT. A synapomorphic disulfide bond is critical for the conformational stability and cytotoxicity of an amphibian ribonuclease. FEBS Lett. 2000;477:203–207. doi: 10.1016/s0014-5793(00)01804-4. [DOI] [PubMed] [Google Scholar]
- 102.Wu Y, Mikulski SM, Ardelt W, Rybak SM, Youle RJ. A cytotoxic ribonuclease. J Biol Chem. 1993;268:10686–10693. [PubMed] [Google Scholar]
- 103.Newton DL, Xue Y, Boque L, Wlodawer A, Kung HF, Rybak SM. Expression and characterization of a cytotoxic human–frog chimeric ribonuclease: Potential for cancer therapy. Protein Eng. 1997;10:463–470. doi: 10.1093/protein/10.4.463. [DOI] [PubMed] [Google Scholar]
- 104.Newton DL, Boque L, Wlodawer A, Huang CY, Rybak SM. Single amino acid substitutions at the N-terminus of a recombinant cytotoxic ribonuclease markedly influence biochemical and biological properties. Biochemistry. 1998;37:5173–5183. doi: 10.1021/bi972147h. [DOI] [PubMed] [Google Scholar]
- 105.Mosimann SC, Ardelt W, James MNG. Refined 1.7 A X-ray crystallographic structure of P-30 protein, an amphibian ribonuclease with anti-tumor activity. J Mol Biol. 1994;236:1141–1153. doi: 10.1016/0022-2836(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 106.Notomista E, Catanzano F, Graziano G, Di Gaetano S, Barone G, Di Donato A. Contribution of chain termini to the conformational stability and biological activity of onconase. Biochemistry. 2001;40:9097–9103. doi: 10.1021/bi010741s. [DOI] [PubMed] [Google Scholar]
- 107.Leland PA, Staniszewski KE, Kim BM, Raines RT. Endowing human pancreatic ribonuclease with toxicity for cancer cells. J Biol Chem. 2001;276:43095–43102. doi: 10.1074/jbc.M106636200. [DOI] [PubMed] [Google Scholar]
- 108.Haigis MC, Kurten EL, Raines RT. Ribonuclease inhibitor is an intracellular sentry. Nucleic Acids Res. 2002;31:1024–1032. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raines RT. Ribonuclease A: From model system to cancer chemotherapeutic. In: Frey PA, Northrop DB, editors. Enzymatic Mechanisms. IOS Press; Washington, DC: 1999. pp. 235–249. [Google Scholar]
- 110.Futami J, Nukui E, Maeda T, Kosaka M, Tada H, Seno M, Yamada H. Optimum modification for the highest cytotoxicity of cationized ribonuclease. J Biochem (Tokyo) 2002;132:223–228. doi: 10.1093/oxfordjournals.jbchem.a003214. [DOI] [PubMed] [Google Scholar]
- 111.Monti DM, D’Alessio G. Cytosolic RNase inhibitor only affects RNases with intrinsic cytotoxicity. J Biol Chem. 2004;279:39195–39198. doi: 10.1074/jbc.C400311200. [DOI] [PubMed] [Google Scholar]
- 112.Strydom DJ. The angiogenins. Cell Mol Life Sci. 1998;54:811–824. doi: 10.1007/s000180050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pavlov N, Badet J. Angiogenin: Involvement in angiogenesis and tumour growth. Bull Cancer. 2001;88:725–732. [PubMed] [Google Scholar]
- 114.Riordan JF. Angiogenin. Methods Enzymol. 2001;341:263–273. doi: 10.1016/s0076-6879(01)41157-8. [DOI] [PubMed] [Google Scholar]
- 115.Moroianu J, Riordan JF. Identification of the nucleolar targeting signal of human angiogenin. Biochem Biophys Res Commun. 1994;203:1765–1772. doi: 10.1006/bbrc.1994.2391. [DOI] [PubMed] [Google Scholar]
- 116.Hu GF, Riordan JF, Vallee BL. A putative angiogenin receptor in angiogenin-responsive human endothelial cells. Proc Natl Acad Sci USA. 1997;94:2204–2209. doi: 10.1073/pnas.94.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu Z-p, Tsuji T, Riordan JF, Hu G-F. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry. 2003;42:121–128. doi: 10.1021/bi020465x. [DOI] [PubMed] [Google Scholar]
- 118.Harper JW, Vallee BL. A covalent angiogenin/ribonuclease hybrid with a fourth disulfide bond generated by regional mutagenesis. Biochemistry. 1989;28:1875–1884. doi: 10.1021/bi00430a067. [DOI] [PubMed] [Google Scholar]
- 119.Leland PA, Staniszewski KE, Park C, Kelemen BR, Raines RT. The ribonucleolytic activity of angiogenin. Biochemistry. 2002;41:1343–1350. doi: 10.1021/bi0117899. [DOI] [PubMed] [Google Scholar]
- 120.Shapiro R, Fox EA, Riordan JF. Role of lysines in human angiogenin: Chemical modification and site-directed mutagenesis. Biochemistry. 1989;28:1726–1732. doi: 10.1021/bi00430a045. [DOI] [PubMed] [Google Scholar]
- 121.Shapiro R, Riordan JF. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry. 1989;28:7401–7408. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- 122.Shapiro R, Vallee BL. Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc Natl Acad Sci USA. 1987;84:2238–2241. doi: 10.1073/pnas.84.8.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Polakowski IJ, Lewis MK, Muthukkaruppan V, Erdman B, Kubai L, Auerbach R. A ribonuclease inhibitor expresses anti-angiogenic properties and leads to reduced tumor growth in mice. Am J Pathol. 1993;143:507–517. [PMC free article] [PubMed] [Google Scholar]
- 124.Dijkstra J, Touw J, Halsema I, Gruber M, Ab G. Estradiol-induced synthesis of vitellogenin. Biochim Biophys Acta. 1978;521:363–373. doi: 10.1016/0005-2787(78)90278-2. [DOI] [PubMed] [Google Scholar]
- 125.Cui XY, Fu PF, Pan DN, Zhao Y, Zhao J, Zhao BC. The antioxidant effects of ribonuclease inhbitor. Free Radic Res. 2003;37:1079–1085. doi: 10.1080/10715760310001600408. [DOI] [PubMed] [Google Scholar]
- 126.Allen DW, Jandl JH. Oxidative hemolysis and precipitation of hemoglobin. II. Role of thiols in oxidant drug action. J Clin Invest. 1961;40:454–475. doi: 10.1172/JCI104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cavalli L, Galaverni D, Pesando P, Bracchi PG, Campanini G, Maraini G. Control of ribonuclease activity in the human lens during ageing and cataract formation. Ophthalmic Res. 1979;11:416–422. [Google Scholar]
- 128.Kraft N, Shortman K, Jamieson D. The effect of x-irradiation on the balance between alkaline ribonuclease and the ribonuclease inhibitor of mammalian tissues. Radiation Res. 1969;39:655–668. [PubMed] [Google Scholar]