Abstract
Panic disorder (PD) and social anxiety disorder (SAD) are moderately heritable anxiety disorders. We analyzed five genes, derived from pharmacological or translational mouse models, in a new case-control study of PD and SAD in European Americans: (1) the serotonin transporter (SLC6A4), (2) the serotonin receptor 1A (HTR1A), (3) catechol-o-methyltransferase (COMT), (4) a regulator of g-protein signalling, RGS2, and (5) the gastrin releasing peptide receptor (GRPR). Cases were interviewed using the Schedule for Affective disorders and Schizophrenia (SADS-LA-IV) and were required to have a probable or definite lifetime diagnosis of PD (N = 179), SAD (161) or both (140), with first onset by age 31 and a family history of anxiety. Final diagnoses were determined using the best estimate procedure, blind to genotyping data. Controls were obtained from the NIMH Human Genetics Initiative; only subjects above 25 years of age who screened negative for all psychiatric symptoms were included (N = 470). A total of 45 SNPs were successfully genotyped over the 5 selected genes using Applied Biosystems SNPlex protocol. SLC6A4 provided strong and consistent evidence of association with the PD and PD+SAD groups, with the most significant association in both groups being at rs140701 (χ2=10.72, p=0.001 with PD and χ2=8.59, p=0.003 in the PD+SAD group). This association remained significant after multiple test correction. Those carrying at least one copy of the haplotype A-A-G constructed from rs3794808, rs140701 and rs4583306 have 1.7 times the odds of PD than those without the haplotype (90%CI 1.2-2.3). The SAD only group did not provide evidence of association, suggesting a PD driven association. The findings remained after adjustment for age and sex, and there was no evidence that the association was due to population stratification. The promoter region of the gene, 5-HTTLPR, did not provide any evidence of association, regardless of whether analyzed as a triallelic or biallelic locus, nor did any of the other four candidate genes tested. Our findings suggest that the serotonin transporter gene may play a role in PD; however, the findings require replication. Future studies should attend to the entire genetic region rather than the promoter.
Keywords: anxiety disorder, social phobia, association, SLC6A4, 5-HTTLPR, serotonin receptor 1A (HTR1A), catechol-o-methyltransferase (COMT), regulator of g-protein signalling, gastrin releasing peptide receptor (GRPR)
Introduction
We report on a candidate gene study for panic disorder and social anxiety disorder. Anxiety disorders as a group are the most prevalent psychiatric disorders in the community. Although they are classified as discrete disorders, comorbidity among them is high and they often respond to the same treatments. Anxiety disorders are of interest to study genetically because they are moderately heritable; furthermore, the neural circuitry required for learned and innate fear is well worked out in both human and animal models (1-3). This neural circuitry can suggest candidate genes that can be tested with disease related variants in clinical samples.
There are no clear guides as to which of the anxiety disorders are more likely than others to share common genetic etiology. We elected to study panic disorder (PD) and social anxiety disorder (SAD) because they can be clinically well defined and are moderately heritable. Panic disorder is characterized by discrete and recurrent episodes of rapid sudden onset of uncontrollable fear and accompanying cardiac, respiratory and other symptoms. At least some of the symptoms appear to arrive “out of the blue” and are not related to consistent cues. The lifetime prevalence ranges from 2-5%, age of first onset is in the early 20's, PD is more common in women, and there is consistency in clinical presentation cross-nationally (4, 5). Family studies indicate that PD is highly familial (e.g, (6-9)) with the risk to first-degree relatives of PD probands with onset under age 20 being 17-fold, and under 30, six-fold, as compared to risk in relatives of controls (6, 7). A meta-analysis of twin and family studies suggests a heritability of about 48% (8).
Phobias are also a clinically heterogeneous group of conditions with a common feature of avoidance behaviour secondary to irrational fears of a specific activity, object or situation. The most consistent data suggesting genetic heritability among the phobias concern social phobias, termed interchangeably as social anxiety disorder (SAD), in DSM IV. SAD includes a concern about appearing shameful or stupid in the presence of others, resulting in a persistent fear of public performance and of situations in which there is possible eating, bathing in public, or use of public lavatories. Exposure to a phobic stimulus pushes an immediate anxiety response, and the phobic situation is avoided or is endured with intense anxiety. The lifetime prevalence varies with ranges for generalized social phobia of 3-4% in the U.S., mean age of onset in the early teens, and higher prevalence in women than men.(1-3). While family studies for SAD are fewer than for PD, they indicate over 3-fold increase of SAD, especially generalized SAD, in first-degree relatives of probands with SAD (4-6). Heritability of SAD based on twin studies is similar to that for PD (1). While other anxiety disorders likely have a genetic component in their etiology, their evidence is limited or more equivocal(7, 8).
Linkage and association studies have identified genes potentially related to the etiology of PD (9-11) and SAD (6, 12, 13). We had a large and well-defined sample of PD and SAD cases, and we used this sample to follow up on genes that have been reported in the literature to be associated with PD or SAD phenotypes. We choose five genes, derived from pharmacological or translational mouse models, to analyze in a case-control association study: (1) the serotonin transporter, SLC6A4, (2) the serotonin receptor 1A (HTR1A), (3) catechol-o-methyltransferase (COMT), (4) a regulator of g-protein signalling, RGS2, and (5) the gastrin releasing peptide receptor (GRPR). We choose 45 single nucleotide polymorphisms (SNPs) for typing in these 5 genes, as shown in Table 1.
Table 1.
The 45 SNPs analyzed in the five candidate genes for this case-control association study. Information on bp position and the extent of the coding region 5′ to 3′
| db SNP Build 128 | ||||
|---|---|---|---|---|
| Gene | 5′ | 3′ | Chromosomal location (bp) | SNP |
| RGS2 | 191044794 | 191048026 | 191044782 | rs12130714 |
| 191045850 | rs2746073 | |||
| 191047121 | rs17647363 | |||
| 191047795 | rs4606 | |||
| 191048262 | rs3767488 | |||
| 5HTR1A | 63292034 | 63293302 | 63292485 | rs1800042 |
| 63292752 | rs1800043 | |||
| 63293009 | rs6294 | |||
| 63293256 | rs1800041 | |||
| 63294321 | rs6295 | |||
| SLC6A4 | 25549032 | 25586831 | 25549137 | rs1042173 |
| 25554071 | rs2054848 | |||
| 25555919 | rs3794808 | |||
| 25562658 | rs140701 | |||
| 25562841 | rs4583306 | |||
| 25566514 | rs717742 | |||
| 25567515 | rs140700 | |||
| 25571040 | rs2020942 | |||
| 25572936 | rs6355 | |||
| 25574940 | rs2020936 | |||
| 25575791 | rs2066713 | |||
| 25583308 | rs8073965 | |||
| 25585881 | rs2020933 | |||
| 25585881 | rs2020933 | |||
| 5- | ||||
| 25588134 | HTTLPR_HAPTYPE | |||
| MB-COMT | 18309309 | 18336530 | 18308092 | rs2097603a |
| S-COMT | 18330070 | 18336528 | 18310109 | rs737866 |
| 18311407 | rs933271 | |||
| 18311668 | rs1544325 | |||
| 18314051 | rs174675 | |||
| 18317638 | rs5993883 | |||
| 18321947 | rs5992500 | |||
| 18325177 | rs740603 | |||
| 18330235 | rs4633 | |||
| 18330268 | rs740602 | |||
| 18331201 | rs769223 | |||
| 18331271 | rs4680 | |||
| 18332132 | rs4646316 | |||
| 18333176 | rs174696 | |||
| 18336781 | rs165599 | |||
| GRPR | 16051345 | 16081562 | 16049496 | rs12850070 |
| 16057042 | rs10218163 | |||
| 16064270 | rs7876221 | |||
| 16080998 | rs3747411 | |||
| 16081881 | rs2353576 | |||
| 16084478 | rs6527664 |
rs2097603 is the same as rs2075507. The two SNPs that reportedly interact with rs4684 to affect a working memory phenotype (rs2097603 and rs165599) (Meyer-Lindenberg et al., 2006) map approximately 2,000 bp proximal to rs737866 and 3,600 bp distal to rs174696, respectively.
Substantial evidence points to the involvement of the serotonin system in PD and SAD, notably the effectiveness of drugs such as monoamine oxidase (MAO) inhibitors and selective-serotonin reuptake inhibitors (SSRI's) in treatment (14). SLC6A4, which is the primary target of SSRIs, and HTR1A have been studied extensively in these disorders, with reports of both positive and negative findings(15). However, the majority of these studies assayed only one or two select polymorphisms in case-control studies of relatively small sample size. Most of these studies focused on the functional serotonin transporter promoter polymorphism, 5-HTTLPR, again with conflicting results (15), although a recent meta-analysis concludes that there is no relationship with PD (16). While these discrepancies in the promoter may be attributable to genetic heterogeneity, phenotype definition, or variable power, it is also possible that the association is with another region of the gene, or that the standard subdivision of this allele into long (L) and short (S) may be oversimplified. A recent report identified 5-HTTLPR as functionally triallelic, with a common A/G substitution within the long (L) allele creating a functional AP2 transcription-factor binding site and resulting in reduced gene expression comparable to that of the short (S) allele (17). We have incorporated this finding in our evaluation of 5-HTTLPR.
The catechol-O-methyltransferase (COMT) gene, which codes for one of the major methylation enzymes metabolizing monoaminergic neurotransmitters including dopamine has repeatedly been suggested as a promising candidate gene in the pathogenesis of panic disorder (18-20). The COMT gene maps to chromosome 22q11.2 where it is expressed as a larger, membrane-bound protein or a smaller, soluble form. Both forms encode a functional polymorphism (val158met) wherein individuals harboring the valine allele show significantly higher COMT activity relative to those harboring the methionine allele (21, 22). A number of independent case-control and/or family-based association studies have been reported comparing the functional COMT val158met polymorphism with anxiety disorder; some showing no evidence of association (23, 24) while others report association with the valine allele (25-27), or conversely the methionine allele (28, 29). A recent meta-analysis of six studies with total sample size comparable to the present study reported significant association of the COMT 158val allele with panic disorder in Caucasian samples (30). In a recent study of prefrontal working memory response, it was reported that the val158met variant exerts its phenotypic effect via interaction with two additional COMT variants located just upstream and downstream of the gene-coding region (31).
The final two genes, RGS2 and GRPR, have been shown to be involved in mouse models of anxiety and fear responses, respectively (32-34). Furthermore, significant evidence for association to variants in RGS2 has been reported in a sample of 173 patients with PD and 173 controls (35).
Our goal was to determine whether SNPs in any of the 5 candidate genes were associated with any or all of the three diagnostic categories in our sample: PD, SAD, or the combined diagnosis of PD and SAD, and to determine whether that phenotype could be further refined by comorbidities or age-of-onset. Comorbid conditions, such as agoraphobia and specific phobia, are common among these PD and SAD diagnostic categories. In addition, there is evidence that PD with onset before age 20 may represent a more genetically heritable form of the disease (36, 37), with pre-pubescent patients alone (onset before 12 years of age) studied only minimally. We thus further assessed (1) whether any association between the PD and SAD diagnostic groups and the candidate genes could be refined using the available sample information on the comorbid conditions of agoraphobia, specific phobia, or early-onset recurrent depression (recurrent MDD), as well as (2) whether there was an age-of-onset component to the association evidence; that is, whether the association was stronger in those with an earlier age of onset (less than 20 or 12) than in those with a later onset.
Materials and Methods
Subjects
Sample recruitment and characteristics have been previously detailed (38). Briefly, PD and SAD subjects in the age range of 18 to 65 were recruited between May 2004 and December 2006, with New York State Psychiatric Institute's Institutional Review Board approval. Requirements for inclusion included: a probable or definite diagnosis of PD with or without agoraphobia, SAD, or both; a family history of anxiety in at least one first-degree relative; and first onset of the full syndrome prior to age 31. Agoraphobia was not required, as our family study data showed no increase in familial aggregation of panic by whether or not the proband with PD had agoraphobia (39). Patients with SAD were required to have the generalized form, including at least three situations that elicit extreme social anxiety(6). Subjects with a history of bipolar disorder, schizophrenia, or anti-social personality; those for whom a medical or neurological disorder or treatment might explain the anxiety symptoms; and those who had participated in any of our other genetic studies were excluded prior to DNA collection. For the primary analyses we included only European American cases and NIMH controls. A sample of African American cases and controls were also analyzed in an exploratory manner for association at SLC6A4.
Psychiatric diagnoses were ascertained using the Schedule for Affective Disorders and Schizophrenia-Lifetime Version modified for the study of anxiety disorders and updated for DSM IV (SADS-LA IV) (40). Information on first-degree relatives was obtained using the Family History Screen (FHS), with the proband as the informant (41). All diagnostic assessments were administered by trained interviewers who were doctoral- and master's-level mental health professionals. Interviewers were required to complete 3-4 page narrative summaries following a common format. The narrative included historical summary of all symptoms, descriptions of mental status, and explanation for all uncertain ratings. In addition basic demographic and medical history was obtained. Final psychiatric diagnoses were made by MMW blinded to subjects' genetic data, using the Best Estimate Procedure (42). Any cases of diagnostic uncertainty were reviewed by AJF and were reinterviewed or eliminated if the diagnosis could not be made with certainty.
DNA for control subjects was obtained from the National Institute of Mental Health (NIMH) Human Genetic Initiative (http://www.nimhgenetics.org). Two thousand nine hundred fifty-nine samples were provided to us at the time of analyses. All subjects were European American or African American. Of these, we excluded subjects who displayed any symptoms of schizophrenia, schizoaffective disorder, bipolar disorder, any anxiety or depressive disorder, or substance abuse or dependence, resulting in a sample of 838 (28%) subjects. We then further excluded females under age 25 and males under age 30 to minimize the likelihood that we were including subjects who were still at risk for developing PD. The final sample consisted of 551 European Americans (470) and African Americans, 282 were females and 269 were males (with the lower cut-off for females selected to better match controls to the predominantly female cases).
Psychiatric symptoms in the controls were assessed at the time of their original recruitment by the NIMH via an online self-report that included demographic and medical information, and a psychiatric history based on the Composite International Diagnostic Interview-Short Form (CIDI-SF) (43) (see (44) for detailed description). We did not assess the control subjects in the present study.
Lab Methods
A total of 45 SNPs were successfully genotyped over the 5 selected candidate genes using Applied Biosystem's SNPlex protocol (45). SNPs were selected for genotyping across each gene based on the following procedure: 1) LD patterns across the gene were determined using Haploview (46) with HapMap CEPH genotype data (www.hapmap.org). Then the tagger algorithm, with an r2 threshold of 0.8, was used to choose a set of tagSNPs over each gene. 2) The literature was searched for variants in the candidate genes that had been specifically implicated in any of the phenotypes under investigation.
5-HTTLPR genotyping was carried out in two stages, first discriminating between the L and S alleles and secondly determining the status of the A/G SNP within the long allele (LA/LG). Stage one genotyping was carried out as described in Yonan et al. (47). In the second stage of genotyping, the remaining PCR product of those samples with an L allele was digested with 10 units of HpaII for 4 hours at 37°C, followed by 20 minutes at 65°C, and products resolved on the ABI3730XL as before. The presence of the G nucleotide creates a restriction enzyme cut site, resulting in either a 155 or 329 base pair product corresponding to the LG or LAallele, respectively.
Statistical Methods
All SNP association analyses were conducted using Splus (7.0 for Windows, Insightful Corp.) and STATA (Stata/SE 9.0 for Windows, College Station, TX). To assess the association evidence between markers in the candidate genes and the diagnostic groups and sub-types, we used a trend test, adjusted for gender and age using logistic regression, at each of the 45 SNPs typed. The 5-HTTLPR triallelic marker in SLC6A4 was assessed for association as both a trialleic and biallelic variant, and independently based upon the A/G SNP variant within the 5-HTTLPR long allele. We corrected for multiple hypothesis tests by using a Bonferroni correction, since it provides the most conservative adjustment and our SNPs were chosen using a tagging approach. D' was calculated to measure the degree of linkage disequilibrium in our control sample. Haplotypes were constructed using Phase 2.1.1 (48). To estimate the haplotypes we used 5000 iterations, 1 thinning interval and 5000 burn-ins. The positions of the markers were not specified. Multiple runs varying the seed were used to determine whether the Phase assignments were consistent. We tested for differences in haplotype and haplo-genotype frequencies between cases and controls using chi-square statistics. We also computed odds ratios with 95% confidence intervals.
An additional 90 neutral SNPs were genotyped in the sample to test for the existence of population stratification. We selected SNPs from the http://rosenberglab.bioinformatics.med.umich.edu/datasets.html website (49, 50). The site represents the allele frequencies for 8,700 SNPs derived from genotypic analysis of 42 East Asian, 42 African American, and 42 European-American samples. For each SNP, the site then lists four “I_n statistic” values which measure ability to infer ancestry from each population comparison, and thereby assesses the utility of the marker for detecting population stratification. Employing the most general statistic (I125), the average I_n for the 90 markers is 0.23, and the minimum value is 0.19. Only 1.5% of the 8,700 SNPs characterized in Rosenberg et al., (49) were more informative. These 90 SNPs are provided in Table 4.
Table 4.
The 90 Neutral SNPs typed for Population Stratification Analysis
| SNP | Chromosome | Position (bp) |
|---|---|---|
| rs716924 | 1 | 36196924 |
| rs2038026 | 1 | 23720557 |
| rs15864 | 1 | 110094378 |
| rs1554615 | 1 | 198544703 |
| rs1040501 | 1 | 167354937 |
| rs1015140 | 1 | 198545381 |
| rs729253 | 2 | 117648706 |
| rs714649 | 2 | 157849337 |
| rs163077 | 2 | 38139108 |
| rs878172 | 3 | 71705138 |
| rs592275 | 3 | 40787980 |
| rs317575 | 3 | 4063808 |
| rs1983273 | 3 | 190066320 |
| rs1568598 | 3 | 190012861 |
| rs1443529 | 3 | 64374558 |
| rs1399272 | 3 | 103294465 |
| rs1108718 | 3 | 65779622 |
| rs1107043 | 3 | 61813448 |
| rs1588041 | 4 | 117366454 |
| rs1525760 | 4 | 117354828 |
| rs1506739 | 4 | 117373198 |
| rs1485768 | 4 | 177877416 |
| rs1395433 | 4 | 117330553 |
| rs1385737 | 4 | 177901866 |
| rs871722 | 5 | 17490493 |
| rs729800 | 5 | 133769779 |
| rs434363 | 5 | 117000414 |
| rs430952 | 5 | 117005715 |
| rs217776 | 5 | 37386146 |
| rs216377 | 5 | 37372467 |
| rs2059849 | 5 | 66646665 |
| rs1156387 | 5 | 103953546 |
| rs1078703 | 5 | 153545632 |
| rs860751 | 6 | 14741351 |
| rs714389 | 6 | 83638065 |
| rs276497 | 6 | 137388819 |
| rs276477 | 6 | 137383666 |
| rs22662 | 6 | 5128534 |
| rs2180052 | 6 | 170431913 |
| rs2078265 | 6 | 131803369 |
| rs1455201 | 6 | 104797083 |
| rs1358716 | 6 | 139487696 |
| rs1322393 | 6 | 137376638 |
| rs1076782 | 6 | 134774571 |
| rs880028 | 7 | 50537629 |
| rs739611 | 7 | 24350306 |
| rs722103 | 8 | 40618275 |
| rs2001433 | 8 | 10940884 |
| rs2001329 | 8 | 11024268 |
| rs1455640 | 8 | 2306616 |
| rs998599 | 9 | 17690365 |
| rs1888952 | 9 | 16248117 |
| rs947603 | 10 | 95239594 |
| rs1904649 | 10 | 68626830 |
| rs1904648 | 10 | 68626960 |
| rs757080 | 11 | 60598809 |
| rs729404 | 11 | 60977693 |
| rs725192 | 11 | 83013213 |
| rs620778 | 11 | 120515009 |
| rs612415 | 11 | 60616461 |
| rs1806995 | 11 | 131689089 |
| rs917587 | 12 | 3412935 |
| rs1548837 | 12 | 12945583 |
| rs751531 | 13 | 59020753 |
| rs748144 | 13 | 112149884 |
| rs1337038 | 13 | 73624564 |
| rs741272 | 14 | 88935704 |
| rs730570 | 14 | 100212642 |
| rs716873 | 14 | 91116456 |
| rs1951033 | 14 | 100205070 |
| rs1872234 | 15 | 78877137 |
| rs1863459 | 15 | 24443768 |
| rs1426208 | 15 | 24392346 |
| rs764551 | 16 | 73761782 |
| rs67302 | 16 | 64379299 |
| rs1019800 | 16 | 75906702 |
| rs717742 | 17 | 25566513 |
| rs4583306 | 17 | 25562840 |
| rs3794808 | 17 | 25555919 |
| rs2054848 | 17 | 25554070 |
| rs140701 | 17 | 25562657 |
| rs1042173 | 17 | 25549136 |
| rs1833422 | 18 | 24936256 |
| rs753842 | 19 | 4928743 |
| rs1806931 | 19 | 15700364 |
| rs733578 | 20 | 20901309 |
| rs293554 | 20 | 30549517 |
| rs1001519 | 20 | 58223865 |
| rs1008552 | 21 | 24599352 |
| rs739200 | 22 | 34762282 |
We used both STRUCTURE (51) and the DC method (52, 53) to test for the existence of population stratification. We used STRUCTURE to determine whether our 90 SNPs could detect any sub-structure in our combined sample of cases and controls of reported European American ancestry. We then used the DC method to assess whether any sub-structure in the data differed between cases and controls and could result in population stratification. Using this DC approach one can estimate the amount of population stratification in one's sample and test whether this estimate differs significantly from zero. If so a correction factor is employed in the same way (although the correction factor differs) as one would use the genomic control λ (54).
Results
Sample Characteristics
The analyzed sample comprised 409 cases (PD, N = 163; SAD, 130; PD + SAD, 119) and 470 controls. Table 2 shows the distribution of cases and controls by age and gender. All study participants, unless noted otherwise, were European American. (Note that the controls were chosen to be older, so as to minimize their likelihood of still being at risk for PD or SAD). Comorbid conditions were common among the PD and SAD cases, as shown in Table 2. For example, 142 (31%) of all cases also had early-onset recurrent MDD (defined as age of onset < 30 yrs), and 139 (34%) had specific phobia. Among the panic cases, 19.5% had had their first onset by age 12, and over half (52%) by age 20. A small sample of African American cases with PD or PD+SAD (N=24) and NIMH controls (N=81) were analyzed in an exploratory manner for association with SLC6A4.
Table 2.
Demographic characteristics and comorbidity among European American panic and/or social anxiety disorder cases, and controls
| Panic N = 163 | Social Anxiety N = 130 | Panic + Social N = 119 | Controls N = 470 | |
|---|---|---|---|---|
| Demographics | ||||
| % female | 81 | 77 | 80 | 49 |
| median age (range), yrs | 35 (19-66) | 35 (19-64) | 36 (18-64) | 56 (26-75) |
| Comorbid Conditions | ||||
| Recurrent MDDa | 53 (33) | 34 (26) | 54 (45) | N/A |
| Specific Phobia | 56 (34) | 33 (25) | 50 (42) | N/A |
| Agoraphobia | 130 (80) | 3 (2) | 80 (67) | N/A |
| Age of Onset | ||||
| Panic Onset ≤ 12 yrs | 29 (16) | N/A | 25(21) | N/A |
| Panic Onset ≤ 20 yrs | 79 (44) | N/A | 72 (61) | N/A |
Early onset recurrent major depressive disorder, with first onset by age 30
PD and SAD Candidate Gene Association Study
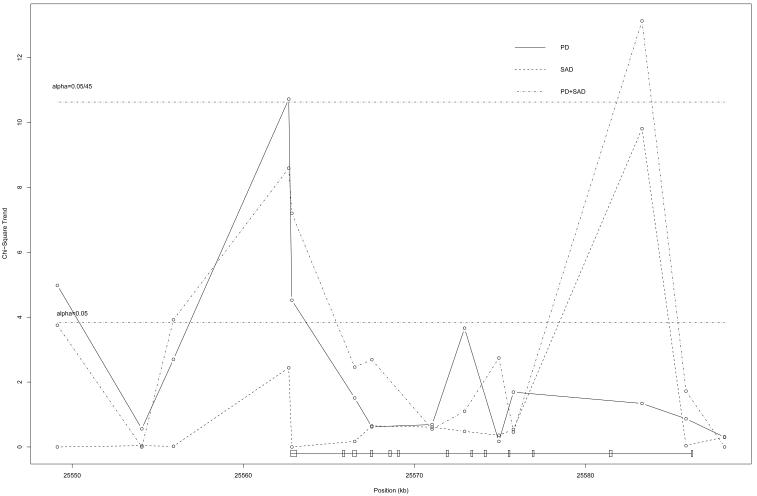
Of all the 45 SNPs tested in the 5 candidate genes, only SNPs in SLC6A4 provided evidence of association with PD and PD+SAD, at the 5% level. This gene continued to provide significant evidence of association, even after Bonferroni correction for the 45 tests of association, and after adjustment for age and sex. The estimated odds ratios remained similar after age and sex adjustment. The promoter region of the gene, 5-HTTLPR, did not provide significant evidence of association with PD or PD+SAD, regardless of whether it was analyzed as a triallelic, biallelic or a straight analysis of the A/G SNP. There was little evidence of association between the SNPs in SLC6A4 and SAD alone, which indicates that the association is restricted to PD or PD+SAD; that is, it appears to be a PD association. The association evidence in this gene with PD and with the PD+SAD diagnostic group was strong and consistent. rs140701 provided the largest χ12 statistics for both diagnostic groups, with χ12=10.72 (p=0.001) and χ12=8.59 (p=0.003) for PD and PD+SAD, respectively, and both diagnostic groups followed a similar pattern of association. One SNP near 5-HTTLPR provides some significant association evidence with SAD (rs8073965); however the flanking SNPs provide little corroborating evidence, the minor allele frequency at this SNP is low (see Table 3), and there is a great deal of linkage disequilibrium between this SNP and rs140701 as measured by D'. Figure 1 plots the chi-square statistics (1 df test of trend) for SNPs typed in this gene against the SNP location in kb on the xaxis. The solid black line represents the association at each SNP with PD, while the dotted line represents the association with the phenotype of PD+SAD. The horizontal line at χCRIT2=10.63 represents the required critical value for significance at the 5% level, after taking the 45 SNP tests into account using a Bonferroni approach. The similar shapes of the PD and PD+SAD lines provide support for combining the two groups into one, and interpreting the association as one with PD, regardless of SAD status as a comorbid condition; in this case, χ12=15.81 (p=0.0001) at rs140701. Table 3 lists the minor allele frequencies in the control sample.
Table 3.
Minor Allele Frequencies (European American Controls) for SNPs in SLC6A4. The minor allele frequency for 5-HTTLPR is presented when treated as biallelic
| SNPs in SLC6A4 | Alleles (major/minor) | Minor Allele Frequency in Controls |
|---|---|---|
| rs1042173 | G/T | 0.436 |
| rs2054848 | G/A | 0.003 |
| rs3794808 | A/G | 0.427 |
| rs140701 | A/G | 0.408 |
| rs4583306 | G/A | 0.408 |
| rs717742 | T/A | 0.203 |
| rs140700 | A/G | 0.091 |
| rs2020942 | A/G | 0.380 |
| rs6355 | C/G | 0.030 |
| rs2020936 | C/T | 0.197 |
| rs2066713 | T/C | 0.383 |
| rs8073965 | T/G | 0.025 |
| rs2020933 | A/T | 0.073 |
| 5-HTTLPR_HAPTYPE | n/a | 0.484 |
Figure 1.
Association between SNPs in SLC6A4 and PD, SAD or PD+SAD; Chi-square critical value corresponding to Bonferroni adjustment for 45 SNPs studied in 5 candidate genes, is provided by the top dotted line. The location of the genotyped SNPs relative to the SLC6A4 gene is depicted below the x-axis.
Population Stratification
To determine whether this association could be due to population stratification, we typed 90 neutral loci, looked for population sub-structure using the program STRUCTURE (51) and then tested, using the DC method (52,53), whether any observed structure could result in population stratification providing a spurious association result with SLC6A4.
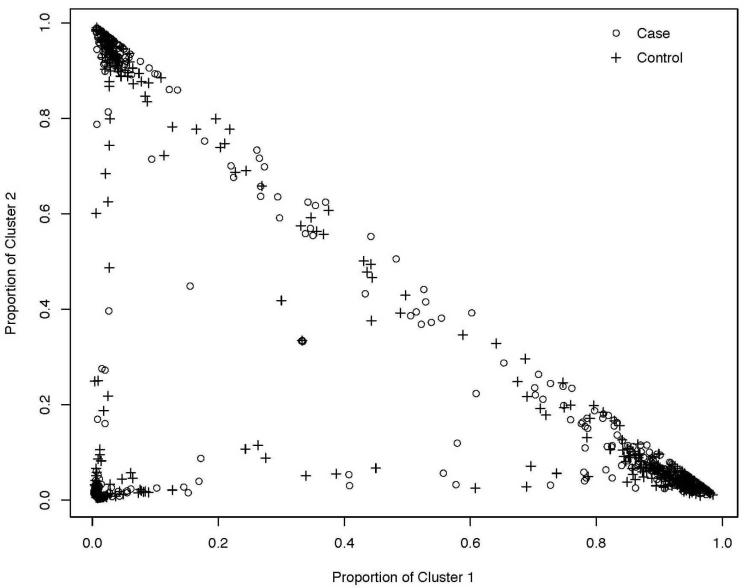
We used 300,000 burnins and 300,000 replications in STRUCTURE with the 90 neutral SNPs (Table 4) for four runs each of k=2,3,4,5,6 on our sample of PD and PD+SAD cases and controls. At k=3 there appeared to be a plateau in the likelihood providing evidence of three clusters in our sample, with relatively consistent information across the four runs at k=3. One of the runs provided evidence of as many as 86% of the sample loading greater than 80% in one of the three clusters (see Figure 2). Thus the 90 neutral SNPs appear to be able to detect structure in the combined sample of European American cases and controls. Yet, Figure 2 also illustrates that the cases and controls are well-distributed across the three clusters. In fact, when the average loadings across the three clusters were calculated, these averages were similar between cases and controls. This indicates that although there may be structure in the data, the cases and controls do not appear to differ in frequency across the clusters; an indication that the cases and controls are comparable with respect to ancestry. The DC method allows us to formally test whether there is a difference between the cases and controls that could result in population stratification.
Figure 2.
Clustering results from STRUCTURE, with k=3 in the European American sample of cases and controls. The proportional membership in cluster 1 is plotted against the proportional membership in cluster 2. Each + represents the proportional membership for a control and the circle represents the proportional membership in the three clusters for each case.
In the DC method one first tests for the existence of population stratification due to allele frequency differences between cases and controls using a t-test, which tests that the DC statistics differs from zero. From our 90 neutral loci we calculated an estimated ; a value which can be interpreted as very small (53). This resulted in an observed t-statistic of −0.3980 (p=0.694), indicating that even though there is some structure in the combined sample of European American cases and controls as uncovered in the STRUCTURE analysis, the cases and controls do not appear to differ from each other significantly based on the set of 90 SNPs used. Thus, there is no evidence that the observed association between PD (PD+SAD) and SLC6A4 is due to population stratification.
Association of SLC6A4 Haplotypes with PD
The LD in SLC6A4 was calculated from the control sample using D'. The region containing our association peak (from 25550 to 25570 kb) is characterized by a large amount of linkage disequilibrium (D'>0.9). However, the region from 25570 kb on appears to have lower pair wise LD. It is interesting to note that, although rs140701 and rs8073965 are 20kb apart, D'> 0.8, however the pair wise linkage disequilibrium between rs140701 and other SNPs surrounding rs8073985 is negligible.
Haplotype analysis using Phase 2.1.1, allowed us to determine whether there were haplotypes or haplo-genotypes in SLC6A4 that were significant predictors of PD. Haplotype construction of SNPs across the gene indicated that a three-SNP haplotype and haplogenotype constructed from rs3794808, rs140701 and rs4583306 was associated with PD (the PD phenotype included those with PD or PD+SAD). The odds of PD in those with at least one copy of the haplotype A-A-G was 1.7 times the odds of those without at least one copy of the haplotype (95% CI 1.2-2.3, p=0.0017). Whereas those who had two copies of the G-G-A haplotype where 1.8 times more likely to not have PD (95% CI 1.2-2.5, p=0.0009) than those with any other genotype. There was no evidence of a haplotype in the region of 5-HTTLPR and/or rs8073965 that differed significantly in frequency between cases and controls.
Possible Confounders
To avoid confounding due to ethnicity, we removed a sample of African American cases and controls from the primary analysis. This African American sample was analyzed separately at SLC6A4 to assess heterogeneity. We had 24 African Americans with PD or PD+SAD, and 81 NIMH African American controls. In this small sample there does appear to be some evidence of association at rs140701 in the African American sample (χ2(1)= 3.22, p=0.07; OR(95% CI) = 0.535 (0.27-1.07)). This indicates that HTTLPR might be associated with PD in both European and African Americans, however, this requires further follow-up.
It is also of interest to determine whether other comorbid disorders, such as agoraphobia, recurrent MDD, and specific phobia, are associated with SLC6A4. Since patients were ascertained on the basis of whether they had PD or SAD, and only PD was associated with SNPs in SLC6A4, it is possible that PD status may be confounding the relationship between SLC6A4 and an alternative phenotype. We used logistic regression in the sub-sample of those with PD (omitting all NIMH controls from these analyses), determining the relationship between SNPs in SLC6A4 and those with (1) specific phobia, (2) recurrent MDD, and (3) agoraphobia. As before, adjusted and unadjusted (for age and sex) analyses were conducted.
The breakdown in the data indicated that those with agoraphobia had primarily PD or PD+SAD diagnoses, as expected (Table 2), while specific phobia and recurrent MDD were more evenly distributed across the PD and SAD diagnostic groups. There was no evidence of association between any of the SNPs in SLC6A4 and recurrent MDD, specific phobia or agoraphobia in the sub-sample of PD cases, with rs140701 resulting in p-values of 0.431, 0.411 and 0.074, respectively, for the three groups. Thus, none of these sub-types could further refine the PD-SLC6A4 association.
Age-of-Onset Analysis
The association evidence in SLC6A4 appears to be pointing to a relationship with PD, regardless of the age-of-onset of the PD (Figure 1). Yet there is evidence that early-onset PD (< age 20) may represent a more genetic, or at least more familial, form of the disease (36). Since prepubertal onset PD is uncommon, we also examined the association with PD onset less than 12 years of age. We thus repeated our case-control analysis restricting our sample to those with PD (in the PD or PD+SAD diagnostic groups), and treating those with onset of PD less than 20 (12) as the cases, and those with onset greater than or equal to 20 (12) the controls. We did not find any evidence that the relationship to SNPs in SLC6A4 differed between the cases and controls, for either age cut-off (p=0.77 and 0.17 for age 20 and age 12, respectively at rs140701). Thus, we were not able to further refine the PD-SLC6A4 association by age-of-onset either.
Discussion
We conducted a candidate gene association study and observed that of the 5 candidates we studied, only SLC6A4 appears to be significantly associated with the phenotype of PD. This PD association could not be further refined by age-of-onset or comorbidity with other psychiatric disorders, does not appear to be due to population stratification, and may also be present in African Americans although further analysis in this group is required. In the complex system of neural communication, SLC6A4 is involved in the transport of serotonin from synaptic spaces to presynaptic neurons, thereby maintaining the pool of available serotonin for subsequent release (55). SLC6A4 knockout mice consistently show increased anxiety-like behaviour and inhibited exploratory locomotion (56). Furthermore, significantly lower binding of SLC6A4 in the midbrain raphe, temporal lobes and hypothalamus of patients with PD has been observed, as has a significant inverse correlation of binding to the severity of PD in the former two brain regions (15).
Importantly, even though we found a significant association with the SLC6A4 gene, we did not find any significant association evidence within the promoter region (5-HTTLPR) of the gene, which is consistent with a recent meta-analysis demonstrating that the promoter region was not associated with the disorder (16). This meta-analysis, however, considered only the biallelic locus; we here further find there is no association with the triallelic variant either. Finally, we also did not find any association with any of the other candidate genes tested.
Recent studies suggest that regulation of SLC6A4 gene expression is determined at the 5-HTTLPR promoter region in concert with regulatory effects exerted from other locations throughout the gene unit. Martin et al. (57) report that at least two SNPs located in intron 1 are more highly correlated with allelic expression differences in SLC6A4 than 5-HTTLPR promoter variants, and that together the upstream promoter and intronic variants account for significantly more variance in gene expression than the promoter variants alone. One of the putative regulatory variants reported by Martin et al. (57), rs2020933, was evaluated in our study and does not show significant evidence of association with the anxiety phenotypes. It is possible that rs140701, located in intron 9 of SLC6A4 represents a new regulatory variant, or that it resides in linkage disequilibrium with such a variant.
Interestingly, a number of studies have found PD to be highly familial [e.g., (39, 58)]. The absolute rate varies by methods but the findings are consistent in diverse countries, with about an 8-fold median relative risk of PD in the first-degree relatives of PD subjects compared to relatives of controls. When combordity in probands was controlled for in the above study, the aggregation in first-degree relatives of panic probands was highly specific to panic disorder, suggesting that panic is a distinct disorder (36). The family studies showed that the risk of PD in first-degree relatives of probands whose PD began before age 20 was increased 17-fold as compared to the risk in relatives of not ill controls (37), and early onset may be transmitted (59). However, these family studies also showed that onset of PD prior to age 31 conferred a six fold increased risk of PD to relatives. Based on these family study findings we selected only cases with onset age prior to 31 for the new study. Thus patients with early onset PD might be the most promising for genetic studies. However, neither age of onset group appeared to refine our PD association.
Finally, it should be noted that the NIMH controls used in this study were not directly interviewed, and information on their first-degree relatives was not available. However a separate study by our group (44) comparing the NIMH controls to a clinically interviewed control sample found that the subset of subjects who did not report any psychiatric symptoms-i.e., those selected as controls for this analysis- were indistinguishable from the clinically interviewed controls on neuroticism and extraversion traits, and may be representative of healthy, non-ill populations. Regardless, under-reporting of psychiatric symptoms among controls would reduce the association evidence; thus, using these controls results in a conservative assessment of the relationship between the candidate genes and the anxiety disorders.
Given the efficacy of selective serotonin reuptake inhibitors for panic disorder, the serotonin transporter likely plays an important mechanistic role in panic disorder. In this report, we do find association evidence for the transporter, but not within the promoter region. Further studies examining the entire genetic region rather than the promoter are therefore warranted. These findings require replication, and a genome-wide association study of PD is currently underway.
Acknowledgements
This research was supported by a clinical studies project (Myrna M. Weissman, P.I) of NIMH Program Project NIMH PO1 MH60970-04 (Rene Hen, P.I.). We would also like to acknowledge the support of MH-48858 (Susan E. Hodge, P.I.).
The NIMH sample was collected by ‘Molecular Genetics of Schizophrenia II’ collaboration, and included the following investigators: ENH/Northwestern University, Evanston, IL, MH059571 - Pablo V. Gejman, MD (Collaboration Coordinator; PI), Alan R. Sanders, MD; Emory University School of Medicine, Atlanta, GA, MH59587 - Farooq Amin, MD (PI); Louisiana State University Health Sciences Center; New Orleans, LA, MH067257 - Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA, MH60870 - William Byerley, MD (PI); Washington University, St Louis, MO, U01, MH060879 - C. Robert Cloninger, MD (PI); University of Iowa, Iowa, IA, MH59566 - Raymond Crowe, MD (PI), Donald Black, MD; University of Colorado, Denver, CO, MH059565 - Robert Freedman, MD (PI); University of Pennsylvania, Philadelphia, PA, MH061675 - Douglas Levinson, MD (PI); University of Queensland, QLD, Australia, MH059588 - Bryan Mowry, MD (PI); Mt Sinai School of Medicine, New York, NY, MH59586 - Jeremy Silverman, PhD (PI).
References
- 1.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. The genetic epidemiology of phobias in women. The interrelationship of agoraphobia, social phobia, situational phobia, and simple phobia. Arch Gen Psychiatry. 1992;49(4):273–81. doi: 10.1001/archpsyc.1992.01820040025003. [DOI] [PubMed] [Google Scholar]
- 2.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. Major depression and phobias: the genetic and environmental sources of comorbidity. Psychol Med. 1993;23(2):361–71. doi: 10.1017/s0033291700028464. [DOI] [PubMed] [Google Scholar]
- 3.Weissman MM, Bland RC, Canino GJ, Greenwald S, Lee CK, Newman SC, et al. The cross-national epidemiology of social phobia: a preliminary report. Int Clin Psychopharmacol. 1996;11(Suppl 3):9–14. doi: 10.1097/00004850-199606003-00003. [DOI] [PubMed] [Google Scholar]
- 4.Coelho HF, Cooper PJ, Murray L. A family study of co-morbidity between generalized social phobia and generalized anxiety disorder in a non-clinic sample. J Affect Disord. 2007;100(13):103–13. doi: 10.1016/j.jad.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Fyer AJ. Heritability of social anxiety: a brief review. J Clin Psychiatry. 1993;54(Suppl):10–2. [PubMed] [Google Scholar]
- 6.Stein MB, Chartier MJ, Hazen AL, Kozak MV, Tancer ME, Lander S, et al. A direct-interview family study of generalized social phobia. Am J Psychiatry. 1998;155(1):90–7. doi: 10.1176/ajp.155.1.90. [DOI] [PubMed] [Google Scholar]
- 7.Kendler KS, Walters EE, Neale MC, Kessler RC, Heath AC, Eaves LJ. The structure of the genetic and environmental risk factors for six major psychiatric disorders in women. Phobia, generalized anxiety disorder, panic disorder, bulimia, major depression, and alcoholism. Arch Gen Psychiatry. 1995;52(5):374–83. doi: 10.1001/archpsyc.1995.03950170048007. [DOI] [PubMed] [Google Scholar]
- 8.Neale MC, Walters EE, Eaves LJ, Kessler RC, Heath AC, Kendler KS. Genetics of blood-injury fears and phobias: a population-based twin study. Am J Med Genet. 1994;54(4):326–34. doi: 10.1002/ajmg.1320540411. [DOI] [PubMed] [Google Scholar]
- 9.Crowe RR, Goedken R, Samuelson S, Wilson R, Nelson J, Noyes R., Jr. Genomewide survey of panic disorder. Am J Med Genet. 2001;105(1):105–9. [PubMed] [Google Scholar]
- 10.Fyer AJ, Hamilton SP, Durner M, Haghighi F, Heiman GA, Costa R, et al. A thirdpass genome scan in panic disorder: evidence for multiple susceptibility loci. Biol Psychiatry. 2006;60(4):388–401. doi: 10.1016/j.biopsych.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Smoller JW, Acierno JS, Jr., Rosenbaum JF, Biederman J, Pollack MH, Meminger S, et al. Targeted genome screen of panic disorder and anxiety disorder proneness using homology to murine QTL regions. Am J Med Genet. 2001;105(2):195–206. doi: 10.1002/ajmg.1209. [DOI] [PubMed] [Google Scholar]
- 12.Blum K, Braverman ER, Wu S, Cull JG, Chen TJ, Gill J, et al. Association of polymorphisms of dopamine D2 receptor (DRD2), and dopamine transporter (DAT1) genes with schizoid/avoidant behaviors (SAB) Mol Psychiatry. 1997;2(3):239–46. doi: 10.1038/sj.mp.4000261. [DOI] [PubMed] [Google Scholar]
- 13.Lochner C, Hemmings S, Seedat S, Kinnear C, Schoeman R, Annerbrink K, et al. Genetics and personality traits in patients with social anxiety disorder: a case-control study in South Africa. Eur Neuropsychopharmacol. 2007;17(5):321–7. doi: 10.1016/j.euroneuro.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Kent JM, Coplan JD, Gorman JM. Clinical utility of the selective serotonin reuptake inhibitors in the spectrum of anxiety. Biol Psychiatry. 1998;44(9):812–24. doi: 10.1016/s0006-3223(98)00210-8. [DOI] [PubMed] [Google Scholar]
- 15.Maron E, Shlik J. Serotonin function in panic disorder: important, but why? Neuropsychopharmacology. 2006;31(1):1–11. doi: 10.1038/sj.npp.1300880. [DOI] [PubMed] [Google Scholar]
- 16.Blaya C, Salum GA, Lima MS, Leistner-Segal S, Manfro GG. Lack of association between the Serotonin Transporter Promoter Polymorphism (5-HTTLPR) and Panic Disorder: a systematic review and meta-analysis. Behav Brain Funct. 2007;3:41. doi: 10.1186/1744-9081-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78(5):815–26. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richard IH, Schiffer RB, Kurlan R. Anxiety and Parkinson's disease. J Neuropsychiatry Clin Neurosci. 1996;8(4):383–92. doi: 10.1176/jnp.8.4.383. [DOI] [PubMed] [Google Scholar]
- 19.Shulman R, Griffiths J, Diewold P. Catechol-O-methyl transferase activity in patients with depressive illness and anxiety states. Br J Psychiatry. 1978;132:133–8. doi: 10.1192/bjp.132.2.133. [DOI] [PubMed] [Google Scholar]
- 20.Simon NM, Emmanuel N, Ballenger J, Worthington JJ, Kinrys G, Korbly NB, et al. Bupropion sustained release for panic disorder. Psychopharmacol Bull. 2003;37(4):66–72. [PubMed] [Google Scholar]
- 21.Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S, et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004;75(5):807–21. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lachman HM, Morrow B, Shprintzen R, Veit S, Parsia SS, Faedda G, et al. Association of codon 108/158 catechol-O-methyltransferase gene polymorphism with the psychiatric manifestations of velo-cardio-facial syndrome. Am J Med Genet. 1996;67(5):468–72. doi: 10.1002/(SICI)1096-8628(19960920)67:5<468::AID-AJMG5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 23.Ohara K, Nagai M, Suzuki Y, Ochiai M, Ohara K. No association between anxiety disorders and catechol-O-methyltransferase polymorphism. Psychiatry Res. 1998;80(2):145–8. doi: 10.1016/s0165-1781(98)00062-6. [DOI] [PubMed] [Google Scholar]
- 24.Samochowiec J, Hajduk A, Samochowiec A, Horodnicki J, Stepien G, Grzywacz A, et al. Association studies of MAO-A, COMT, and 5-HTT genes polymorphisms in patients with anxiety disorders of the phobic spectrum. Psychiatry Res. 2004;128(1):21–6. doi: 10.1016/j.psychres.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Domschke K, Freitag CM, Kuhlenbaumer G, Schirmacher A, Sand P, Nyhuis P, et al. Association of the functional V158M catechol-O-methyl-transferase polymorphism with panic disorder in women. Int J Neuropsychopharmacol. 2004;7(2):183–8. doi: 10.1017/S146114570400416X. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton SP, Fyer AJ, Durner M, Heiman GA, Baisre de Leon A, Hodge SE, et al. Further genetic evidence for a panic disorder syndrome mapping to chromosome 13q. Proc Natl Acad Sci U S A. 2003;100(5):2550–5. doi: 10.1073/pnas.0335669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothe C, Koszycki D, Bradwejn J, King N, Deluca V, Tharmalingam S, et al. Association of the Val158Met catechol O-methyltransferase genetic polymorphism with panic disorder. Neuropsychopharmacology. 2006;31(10):2237–42. doi: 10.1038/sj.npp.1301048. [DOI] [PubMed] [Google Scholar]
- 28.Woo JM, Yoon KS, Choi YH, Oh KS, Lee YS, Yu BH. The association between panic disorder and the L/L genotype of catechol-O-methyltransferase. J Psychiatr Res. 2004;38(4):365–70. doi: 10.1016/j.jpsychires.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Woo JM, Yoon KS, Yu BH. Catechol O-methyltransferase genetic polymorphism in panic disorder. Am J Psychiatry. 2002;159(10):1785–7. doi: 10.1176/appi.ajp.159.10.1785. [DOI] [PubMed] [Google Scholar]
- 30.Domschke K, Deckert J, O'Donovan MC, Glatt SJ. Meta-analysis of COMT vall58met in panic disorder: ethnic heterogeneity and gender specificity. Am J Med Genet B Neuropsychiatr Genet. 2007;144(5):667–73. doi: 10.1002/ajmg.b.30494. [DOI] [PubMed] [Google Scholar]
- 31.Meyer-Lindenberg A, Nichols T, Callicott JH, Ding J, Kolachana B, Buckholtz J, et al. Impact of complex genetic variation in COMT on human brain function. Mol Psychiatry. 2006;11(9):867–77. 797. doi: 10.1038/sj.mp.4001860. [DOI] [PubMed] [Google Scholar]
- 32.Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, Whishaw IQ, et al. Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc Natl Acad Sci U S A. 2000;97(22):12272–7. doi: 10.1073/pnas.220414397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shumyatsky GP, Tsvetkov E, Malleret G, Vronskaya S, Hatton M, Hampton L, et al. Identification of a signaling network in lateral nucleus of amygdala important for inhibiting memory specifically related to learned fear. Cell. 2002;111(6):905–18. doi: 10.1016/s0092-8674(02)01116-9. [DOI] [PubMed] [Google Scholar]
- 34.Yalcin B, Willis-Owen SA, Fullerton J, Meesaq A, Deacon RM, Rawlins JN, et al. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat Genet. 2004;36(11):1197–202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
- 35.Leygraf A, Hohoff C, Freitag C, Willis-Owen SA, Krakowitzky P, Fritze J, et al. Rgs 2 gene polymorphisms as modulators of anxiety in humans? J Neural Transm. 2006;113(12):1921–5. doi: 10.1007/s00702-006-0484-8. [DOI] [PubMed] [Google Scholar]
- 36.Goldstein RB, Weissman MM, Adams PB, Horwath E, Lish JD, Charney D, et al. Psychiatric disorders in relatives of probands with panic disorder and/or major depression. Arch Gen Psychiatry. 1994;51(5):383–94. doi: 10.1001/archpsyc.1994.03950050043005. [DOI] [PubMed] [Google Scholar]
- 37.Goldstein RB, Wickramaratne PJ, Horwath E, Weissman MM. Familial aggregation and phenomenology of ‘early’-onset (at or before age 20 years) panic disorder. Arch Gen Psychiatry. 1997;54(3):271–8. doi: 10.1001/archpsyc.1997.01830150097014. [DOI] [PubMed] [Google Scholar]
- 38.Talati A, Ponniah K, Strug LJ, Hodge SE, Fyer AJ, Weissman MM. Panic Disorder, Social Anxiety Disorder, and a Possible Medical Syndrome Previously Linked to Chromosome 13. Biol Psychiatry. 2007;63(6):594–601. doi: 10.1016/j.biopsych.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weissman MM. Family genetic studies of panic disorder. J Psychiatr Res. 1993;27(Suppl 1):69–78. doi: 10.1016/0022-3956(93)90018-w. [DOI] [PubMed] [Google Scholar]
- 40.Fyer A, Endicott J, Mannuzza S, Klein DF. Schedule for affective disorders andschizophrenia-lifetime version, modified for the study of anxiety disorders (SADS-LA) New York: 1985. [DOI] [PubMed] [Google Scholar]
- 41.Weissman MM, Wickramaratne P, Adams P, Wolk S, Verdeli H, Olfson M. Brief screening for family psychiatric history: the family history screen. Arch Gen Psychiatry. 2000;57(7):675–82. doi: 10.1001/archpsyc.57.7.675. [DOI] [PubMed] [Google Scholar]
- 42.Leckman JF, Sholomskas D, Thompson WD, Belanger A, Weissman MM. Best estimate of lifetime psychiatric diagnosis: a methodological study. Arch Gen Psychiatry. 1982;39(8):879–83. doi: 10.1001/archpsyc.1982.04290080001001. [DOI] [PubMed] [Google Scholar]
- 43.Walters E, Kessler RC, Nelson CB, Mroczek DC. Scoring the World Health Organization's Composite International Diagnostic Interview Short Form (CIDI-SF); revised December 2002. [Google Scholar]
- 44.Talati A, Fyer AJ, Weissman MM. A Comparision between NIMH screened an clinically interviewed control samples on Neuroticism and Extraversion. Molecular Psychiatry. 2008;13(2):122–30. doi: 10.1038/sj.mp.4002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM, et al. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J Biomol Tech. 2005;16(4):398–406. [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 47.Yonan AL, Palmer AA, Gilliam TC. Hardy-Weinberg disequilibrium identified genotyping error of the serotonin transporter (SLC6A4) promoter polymorphism. Psychiatr Genet. 2006;16(1):31–4. doi: 10.1097/01.ypg.0000174393.79883.05. [DOI] [PubMed] [Google Scholar]
- 48.Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73(6):1402–22. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akey JM, Zhang G, Zhang K, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–14. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pritchard JK, Stephens M, Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gorroochurn P, Hodge SE, Heiman G, Greenberg DA. Effect of population stratification on case-control association studies. II. False-positive rates and their limiting behavior as number of subpopulations increases. Hum Hered. 2004;58(1):40–8. doi: 10.1159/000081455. [DOI] [PubMed] [Google Scholar]
- 53.Gorroochurn P, Hodge SE, Heiman GA, Greenberg DA. A unified approach for quantifying, testing and correcting population stratification in case-control association studies. Hum Hered. 2007;64(3):149–59. doi: 10.1159/000102988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 55.Lesch KP, Gutknecht L. Pharmacogenetics of the serotonin transporter. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29(6):1062–73. doi: 10.1016/j.pnpbp.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 56.Holmes A, Murphy DL, Crawley JN. Reduced aggression in mice lacking the serotonin transporter. Psychopharmacology (Berl) 2002;161(2):160–7. doi: 10.1007/s00213-002-1024-3. [DOI] [PubMed] [Google Scholar]
- 57.Martin J, Cleak J, Willis-Owen SA, Flint J, Shifman S. Mapping regulatory variants for the serotonin transporter gene based on allelic expression imbalance. Mol Psychiatry. 2007;12(9):881. doi: 10.1038/sj.mp.4001952. [DOI] [PubMed] [Google Scholar]
- 58.Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 2001;158(10):1568–78. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- 59.Battaglia M, Bertella S, Politi E, Bernardeschi L, Perna G, Gabriele A, et al. Age at onset of panic disorder: influence of familial liability to the disease and of childhood separation anxiety disorder. Am J Psychiatry. 1995;152(9):1362–4. doi: 10.1176/ajp.152.9.1362. [DOI] [PubMed] [Google Scholar]