Abstract
Modifying the length of the Huntington's disease (HD) CAG repeat, the major determinant of age of disease onset, is an attractive therapeutic approach. To explore this we are investigating mechanisms of intergenerational and somatic HD CAG repeat instability. Here, we have crossed HD CAG knock-in mice onto backgrounds deficient in mismatch repair genes, Msh3 and Msh6, to discern the effects on CAG repeat size and disease pathogenesis. We find that different mechanisms predominate in inherited and somatic instability, with Msh6 protecting against intergenerational contractions and Msh3 required both for increasing CAG length and for enhancing an early disease phenotype in striatum. Therefore, attempts to decrease inherited repeat size may entail a full understanding of Msh6 complexes, while attempts to block the age-dependent increases in CAG size in striatal neurons and to slow the disease process will require a full elucidation of Msh3 complexes and their function in CAG repeat instability.
Keywords: Huntington's disease, trinucleotide, instability, repeat, striatum, repair, mouse, knock-in, pathogenesis
Introduction
Huntington's disease (HD) is a dominantly inherited neurodegenerative disorder caused by the expansion of a CAG repeat within the HD gene encoding huntingtin (Huntington's disease collaborative research group 1993). The HD CAG expansion initiates a protracted cascade of events that results initially in the degeneration of medium-spiny neurons in the striatum, motor, cognitive and psychiatric decline and eventual death (Vonsattel et al. 1985; Harper 1999). The mutant HD CAG repeat is remarkably unstable when transmitted to subsequent generations, with a bias towards expansion in transmissions from fathers (Duyao et al. 1993; MacDonald et al. 1993; Zuhlke et al. 1993; Telenius et al. 1994; Wheeler et al. 2007). The repeat also exhibits somatic instability, undergoing expansion particularly in the striatum (Telenius et al. 1994; Kennedy et al. 2003; Shelbourne et al. 2007; Veitch et al. 2007; Gonitel et al. 2008). Significantly, age of onset and disease severity are highly dependent on CAG length (Duyao et al. 1993; Andrew et al. 1993; Snell et al. 1993; Stine et al. 1993; Gusella et al. 1996). Therefore, as intergenerational instability alters the inherited HD CAG repeat length and somatic instability further alters repeat length in the target tissue, understanding the factors that influence both intergenerational and striatal instability is critical as these factors may modify the disease.
HD homologue (Hdh) CAG knock-in mice (White et al. 1997; Wheeler et al. 1999) that replicate the genetic mutation in HD patients provide ideal models in which to study the instability of the HD CAG repeat in its appropriate genomic context. We have shown that HdhQ111 knock-in mice recapitulate many features of repeat instability seen in patients, namely intergenerational repeat length changes with a paternal expansion bias at frequencies seen in humans, and somatic expansion that is predominant in striatum (Lloret et al. 2006; Wheeler et al. 1999). Importantly, HdhQ111 mice, exhibiting accurate expression of mutant huntingtin, also display early presymptomatic phenotypes that exhibit key features of the human disease mechanism, namely, dominant inheritance, CAG length and time dependence, and striatal specificity, allowing modifiers of the HD CAG pathogenic process to be tested (Fossale et al. 2002; Gines et al. 2006; Gines et al. 2003; Wheeler et al. 2000; Wheeler et al. 2002; Wheeler et al. 2003;). Significantly, the CAG repeat length dependence of knock-in phenotypes provides the possibility of testing directly whether factors that alter repeat length also alter the pathogenic process.
We previously investigated the role of mismatch repair gene Msh2 in HD CAG repeat instability and the HD CAG pathogenic process in HdhQ111 mice (Wheeler et al. 2003). In paternal transmissions Msh2 was required for intergenerational HdhQ111 CAG repeat expansions and protected against contractions. Msh2 was also required for striatal HdhQ111 CAG repeat instability and dramatically enhanced an early histological phenotype, the accumulation or epitope-accessibility of a conformation of full-length mutant huntingtin in striatal neurons, as revealed by nuclear immunostaining with the EM48 anti-huntingtin antibody. This finding strongly suggested that striatal instability contributes to the HdhQ111 pathogenic process.
Here, taking advantage of the opportunity afforded by HdhQ111 mice to dissect both intergenerational and somatic instability as well as HD CAG pathogenesis, we have investigated Msh2's binding partners, Msh3 and Msh6 (Acharya et al. 1996), in these processes, as well as nucleotide excision repair gene Xpc with the aim of exploring the roles of other DNA repair pathways. We have crossed HdhQ111 mice onto backgrounds deficient in either Msh3, Msh6 or Xpc to determine whether: 1. these genes influence HD CAG striatal and intergenerational instability in a precise genetic model of HD, 2. the same or different mechanisms underlie striatal and intergenerational instability, and 3. these genes are modifiers of an early, dominant, CAG length-dependent phenotype, nuclear mutant huntingtin immunostaining in striatal neurons.
Materials and Methods
Mice
HdhQ111 knock-in mice (White et al. 1997; Wheeler et al. 1999) were maintained on a CD1 (Charles River Laboratories) background. Msh3 and Msh6 knockout mice (Edelmann et al. 2000; Edelmann et al. 1997) (C57BL/6J) were obtained from Dr. Winfried Edelmann and Xpc knockout mice (Cheo et al. 1997) (C57BL/6J) were obtained from Dr. Errol Friedberg. Msh2 knockout mice (129Ola/FVB) were from Dr. Hein te Riele (Toft et al. 1999). HdhQ111/+ mice were crossed with each DNA repair knockout strain to generate mice heterozygous for each mutation. These mice were then crossed to generate HdhQ111/+ littermates that were wild-type (+/+), heterozygous (+/-) or homozygous mutant (-/-) for each DNA repair gene. These mice were either used for assessment of somatic effects (instability, nuclear huntingtin immunohistochemistry), or were the transmitting parents for the intergenerational instability experiments. Genetic background can modify instability and nuclear huntingtin accumulation (Lloret et al. 2006). However, assuming that such modifier genes segregate independently from the repair genes being analyzed they will occur equally in the repair gene mutants and their wild-type littermates. Thus, sufficient numbers of mice of each genotype will allow identification of effects of the repair gene mutants over and above those due to genetic background. Therefore, to minimize potential confounding effects of genetic background all genotypic comparisons for intergenerational instability, somatic instability and EM48-immunohistochemistry were performed using littermates. The number of mice we have used in each analysis was based on numbers of mice needed to detect modifier effects of the Msh2 gene on a mixed genetic background (Wheeler et al. 2003). All analyses were with heterozygous HdhQ111/Hdh+ mice. All animal procedures were carried out to minimize pain and discomfort, under an approved Institutional Animal Care and Use Committee protocol.
Genotyping and HD CAG repeat length determination
Genomic DNA was isolated from tail biopsies at weaning (for routine genotyping and intergenerational instability analysis) or from adult tail and striatum dissected from adult mice brains (for somatic instability analysis) using the PureGene DNA isolation kit (Gentra, Minneapolis, MN, USA). Genotyping of the HdhQ111 knock-in allele was carried out using a combination of a human-specific PCR assay that amplifies the HD CAG repeat from the knock-in allele but does not amplify the mouse sequence, and a wild-type mouse-specific assay that generates a PCR product from the wild-type mouse allele but not from the knock-in allele. The human-specific assay has been previously described (Mangiarini et al. 1997). For the wild-type mouse-specific assay primers 5′-CCTGGAAAAGCTGATGAAGG (forward) and 5′-TGGACAGGG AACAGTGTTGGC (reverse) were used in a PCR buffer containing 67 mM Tris-HCl pH 8.8, 16.7 mM (NH4)2SO4, 2 mM MgCl2, 0.17 mg/mg BSA, 10 mM 2-mercaptoethanol, 10% DMSO, 200μM dNTPs, 5 ng/μl primers with 0.5 U Taq polymerase (Perkin Elmer). Cycling conditions were 94°C 90 sec, 35 cycles of 94°C 30 sec, 56°C 30 sec, 72°C 90 sec, followed by 10 minutes at 72°C. Products were resolved in 0.8% agarose gels. DNA repair knockout mice were genotyped as described (Cheo et al. 1997; Edelmann et al. 2000; Edelmann et al. 1997; Toft et al. 1999).
HD CAG repeat size was determined using the human HD CAG repeat-specific PCR assay (Mangiarini et al. 1997). The forward primer was fluorescently labeled with 6-FAM (Perkin Elmer) and products were resolved using either the ABI 377 or AB1 3730xl automated DNA analyzer (Applied Biosystems). GeneScan and Genotyper software packages with GeneScan 500-TAMRA as internal size standard (ABI 377) or GeneMapper v3.7 with GeneScan 500-LIZ as internal size standard (ABI 3730) were used to assign repeat size. Runs included the same control DNAs of known HD CAG repeat size. The HD CAG size was assigned as the highest peak in the GeneScan trace.
Analysis of intergenerational instability
For assessment of intergenerational instability, breeding pairs were established between 2-4 different HdhQ111/Hdh+ mice of each sex and DNA repair gene genotype (+/+, +/- and -/-) and Hdh+/+ wild-type littermates that were either +/+ or +/- for the DNA repair gene mutation. Genomic DNA was extracted from tail biopsies of HdhQ111/+ transmitting parents and HdhQ111/+ progeny at weaning. Amplification of the HD CAG repeat and repeat size analysis were carried out as described above. Intergenerational instability was determined by comparing HD CAG repeat size in the HdhQ111/+ transmitting parent with those in the HdhQ111/+ progeny. Parent and pups were compared in the same ABI automated DNA sequencer run.
Immunohistochemistry
Immunohistochemistry was carried out on 7 μm paraffin-embedded coronal sections of brains perfused with periodate-lysine-paraformaldehyde as described (Wheeler et al. 2002). Immunostaining with polyclonal anti-huntingtin antibody EM48 (amino acids 1-256) (Gutekunst et al. 1999) was as described (Wheeler et al. 2002). All EM48 immunostaining experiments were performed under identical conditions. Diffuse EM48 immunostaining was quantified in striata from 3-5 HdhQ111/+ mice of each DNA repair genotype (+/+, +/-. -/-) to be investigated at 5 months of age. The mean nuclear stain intensity was determined in four 750μm × 500μm regions of dorsal striatum, matched in terms of their anterior/posterior location, using the ‘histogram’ function in Adobe Photoshop to convert the signal intensity in all stained nuclei to arbitrary units. Background signal, the mean value for 10 fields within each 750μm × 500μm region analyzed, was subtracted from the nuclear signal. To quantify the total amount of nuclear stain in a way that reflects both the immunostaining intensity and the number of immunostained nuclei, a staining index (SI) was calculated as the product of the mean nuclear stain intensity and the number of stained nuclei for each 750μm × 500μm region analyzed.
Statistical analyses
Intergenerational instability
Progeny genotype has been found to influence intergenerational instability (Savouret et al. 2003). Therefore we first assessed the influence of progeny genotype (+/+, +/-, -/-) on intergenerational instability in transmissions from heterozygous parents. There was no effect of progeny genotype on intergenerational instability for any of the genes analyzed (data not shown), therefore progeny genotype was not accounted for in subsequent comparisons of parental genotype. Although we attempted to match transmitting parents as closely as possible for CAG length and age, we nevertheless controlled for these variables in our statistical analyses to ensure that these factors did not confound the outcome.
We used four statistical models in order to assess the influence of parental genotype on repeat length changes transmitted to progeny. To compare overall repeat size change distributions we used a multinomial distribution with a cumulative logit function to determine the probability of having a cumulative higher change than the cumulative lower changes. We also compared the overall frequencies of changed versus unchanged alleles using a binomial distribution. As additional tests we used binomial distributions to model the odds of expanded alleles compared to grouped unchanged and contracted alleles (‘Risk’ model) as well as the odds of contracted alleles compared to grouped unchanged and expanded alleles (‘Protective’ model). Analyses in all cases were carried out using Generalized Estimating Equations (GENMOD procedure in SAS v 9.1) (SAS Institute 1999), controlling for parental CAG repeat size, parental age and for the correlation between pups from the same parent. We set the alpha level to 0.01, since a Bonferroni adjustment for multiple comparisons is too conservative given the correlated nature of the tests performed.
Immunohistochemical staining
Statistical analysis to compare quantified immunohistochemical staining index values between different genotypes was carried out using Generalized Estimating Equations (GENMOD procedure in SAS) adjusting for CAG size and for repeated observations from each mouse.
Results
Intergenerational repeat instability
The HdhQ111 CAG repeat exhibits an expansion bias in paternal transmissions and a contraction bias in maternal transmissions (Lloret et al. 2006; Wheeler et al. 1999; Wheeler et al. 2003). We previously showed that the absence of Msh2 abolished paternal HdhQ111 CAG expansions and resulted in an increased frequency of contractions (Wheeler et al. 2003). Msh2's binding partners Msh3 and Msh6 (Acharya et al. 1996) might therefore also play a role in generating paternal repeat expansions and/or protecting against paternal contractions. We were also interested to test whether Xpc, encoding the major DNA recognition protein in global genome nucleotide excision repair (Volker et al. 2001), influenced intergenerational instability, particularly the maternal repeat length changes and paternal repeat contractions that did not depend on Msh2 (Wheeler et al. 2003). To determine the roles of these genes in intergenerational HdhQ111 CAG repeat instability the HdhQ111 allele was crossed onto Msh3-, Msh6- and Xpc-deficient mouse strains and the size of the HdhQ111 CAG repeat was determined in heterozygous HdhQ111/+ male and female parents and their progeny.
To determine the influence of the DNA repair genes on intergenerational instability several statistical methods were used that captured the magnitude, frequency and direction of the repeat length changes (see Materials and Methods for details). First, the overall distribution of repeat length changes was modeled using a multinomial distribution to ascertain the likelihood of having a positive repeat size change. Second, the frequency of changed alleles (irrespective of direction) versus unchanged alleles was modeled using a binomial distribution. Finally, we wished to determine the effect of the DNA repair genes on the relative frequencies of expansions and contractions. As the exclusion of unchanged alleles resulted in loss of data we instead used two alternative binomial distributions: the first modeled the odds of positive change (‘Risk’ model) by comparing the frequency of expansions with the combined frequencies of contractions and unchanged alleles. The second distribution modeled the odds of negative change (‘Protective’ model) by comparing the frequency of contractions with the combined frequencies of expansions and unchanged alleles. In all statistical models odds ratios (OR) were determined for homozygous and heterozygous DNA repair gene knockout mice compared to wild-type controls. Table 1 and Supplementary Table 1 show the results of these analyses for paternal and maternal transmissions, respectively.
Table 1.
Paternal transmissions
| Model | Parental genotype compared to +/+ | Msh 2 | Xpc | Msh 6 | Msh 3 | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR (99% CI) |
p-value | OR (99% CI) |
p-value | OR (99% CI) |
p-value | OR (99% CI) |
p-value | ||
| Multinomial (cumulative higher changes vs. cumulative lower changes) | -/- | 0.20 (0.04, 0.92) |
0.0065 | 1.03 (0.52, 2.02) |
0.91 | 1.18 (0.46, 3.03) |
0.65 | 0.44 (0.15, 1.35) |
0.061 |
| +/- | 0.41 (0.04, 4.54) |
0.34 | ND | ND | 0.43 (0.07, 2.72) |
0.24 | 0.51 (0.14, 1.84) |
0.17 | |
| changed vs. unchanged | -/- | 0.52 (0.18, 1.48) |
0.11 | 0.55 (0.26, 1.20) |
0.05 | 1.43 (0.67, 3.03) |
0.23 | 0.88 (0.32, 2.42) |
0.74 |
| +/- | 0.54 (0.85, 3.45) |
0.39 | ND | ND | 4.98 (2.72, 9.14) |
< 0.0001 | 0.35 (0.12, 1.04) |
0.013 | |
| ‘Risk’ expansions vs contractions+unchanged | -/- | NR | NR | 0.88 (0.36, 2.16) |
0.72 | 1.40 (0.63, 3.13) |
0.28 | 0.59 (0.27, 1.31) |
0.089 |
| +/- | NR | NR | ND | ND | 1.16 (0.24, 5.52) |
0.81 | 0.40 (0.13, 1.22) |
0.034 | |
| ‘protective’ contractions vs expansions+unchanged | -/- | 2.13 (0.36, 12.64) |
0.28 | 0.09 (0.003, 2.59) |
0.07 | 1.15 (0.26, 5.11) |
0.81 | 1.79 (0.31, 10.35) |
0.39 |
| +/- | 1.40 (0.06, 30.71) |
0.78 | ND | ND | 4.28 (1.02, 18.00) |
0.009 | 0.67 (0.09, 4.98) |
0.61 | |
Results of running four statistical models to determine the effect of Msh2, Xpc, Msh6 and Msh3 DNA repair genes on the intergenerational instability of the HdhQ111 CAG repeat in paternal transmissions. Odds ratios (OR) with 99% confidence intervals (CI) and p-values are given for each model for comparisons between fathers that were deficient in either one (+/-) or two (-/-) copies of each of the DNA repair genes and the respective wild-type (+/+) controls. Values in bold are statistically significant. Values in italics indicate trends that did not reach significance. A p-value of 0.01 was used to determine significance (see Materials and Methods). NR: the ‘Risk’ model was not run as there were zero data points in the expansion group. ND: Xpc heterozygotes were not analyzed. Msh2: 116 total transmissions from 9 fathers; Xpc: 139 total transmissions from 12 males; Msh6: 144 total transmissions from 11 males; Msh3: 134 total transmissions from 12 males.
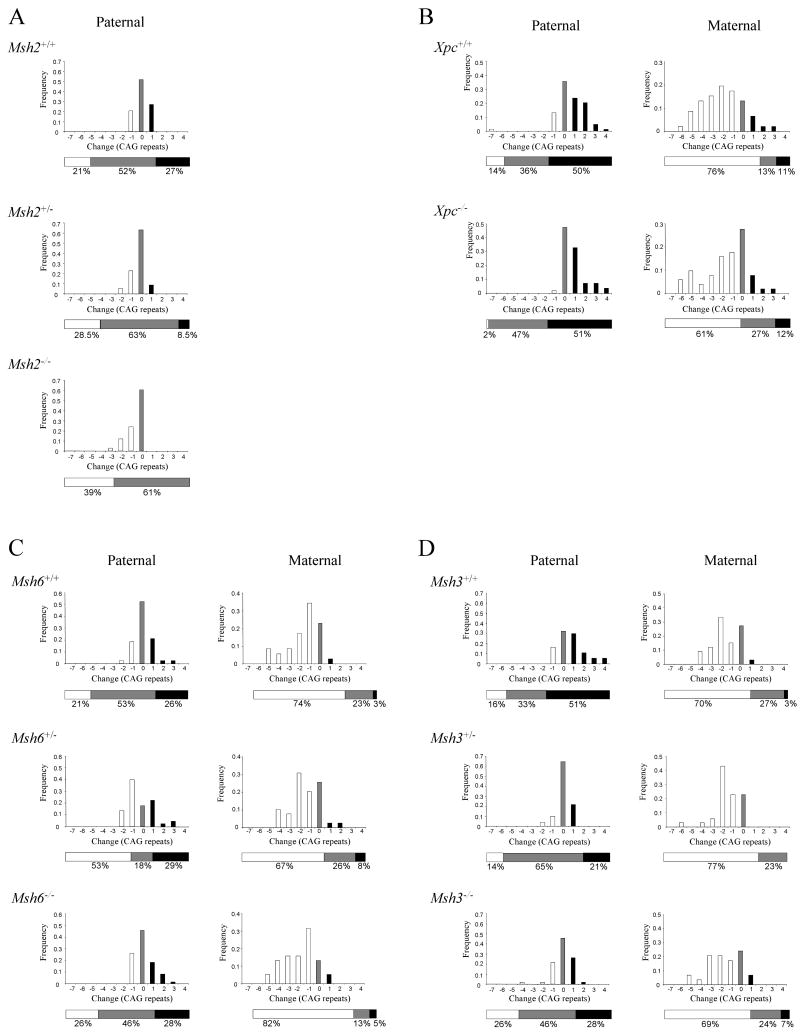
We first quantified repeat length changes in paternal transmissions from Msh2+/+, and Msh2-/- males, as well as Msh2+/- males, which had not previously been evaluated (Wheeler et al. 2003). As shown in Figure 1A and Table 1, expansions were eliminated in transmissions from Msh2-/- males, as previously found (Wheeler et al. 2003), with a statistically significantly lower odds of having a higher repeat size change in transmissions from Msh2-/- compared to Msh2+/+ males (OR=0.2, p=0.0065). In transmissions from Msh2+/- males, the frequency of expansions was also reduced compared to Msh2+/+ males, but this trend did not reach statistical significance. Therefore, the absence of two Msh2 alleles appears necessary to eliminate expansions in paternal transmissions of the HdhQ111 CAG repeat.
Figure 1.
HdhQ111 CAG repeat intergenerational instability.
HdhQ111 mice were crossed onto backgrounds deficient in Msh2 (A), Xpc (B), Msh6 (C) or Msh3 (D). The change in HdhQ111 CAG repeat length upon transmission from heterozygous HdhQ111/+ fathers or mothers to their progeny was determined. The total number of transmissions analyzed for each genotype, the mean age and mean CAG repeat length of the transmitting parents were as follows: (A) Paternal: Msh2+/+ n=48, 5.4 months, 88 CAGs; Msh2+/- n=35, 5.2 months, 87 CAGs; Msh2-/- n=33, 4.4 months, 86 CAGs.
(B) Paternal: Xpc+/+ n= 84, 3.8 months, 103 CAGs; Xpc-/- n=55, 4.0 months, 103 CAGs. Maternal: Xpc+/+ n=55, 4.1 months, 103 CAGs, Xpc-/- n=51, 4.3 months, 102 CAGs).
(C) Paternal: Msh6+/+ n=38, 5.9 months, 101 CAGs; Msh6+/- n=45, 5.6 months, 101 CAGs: Msh6-/- n=61, 5.5 months, 101 CAGs. Maternal: Msh6+/+ n=35, 5.4 months, 100 CAGs; Msh6+/- n=39, 5.7 months, 101 CAGs; Msh6-/- n=38, 5.5 months, 99 CAGs. (D) Paternal: Msh3+/+ n=37, 5.1 months, 101 CAGs; Msh3+/- n=51, 5.0 months, 101 CAGs; Msh3-/- n=46, 4.8 months, 101 CAGs. Maternal: Msh3+/+ n=33, 4.3 months, 101 CAGs; Msh3+/- n=35, 4.4 months, 101 CAGs; Msh3-/- n=29, 4.2 months, 100 CAGs. Bar graphs show frequency distributions of the inherited repeat length changes. The horizontal bars below the graphs depict the total percentage of expansions, contractions and unchanged alleles. Black: expansions; white: contractions; grey: unchanged alleles.
We then investigated Xpc to assess whether the global genome nucleotide excision repair pathway was involved in intergenerational instability. The effect of Xpc on paternal and maternal transmissions of the HdhQ111 CAG repeat is shown in Figure 1B. There was no significant effect of Xpc genotype on maternally inherited HdhQ111 CAG repeat length changes. The absence of Xpc did result in a slight reduction in the frequency of paternal contractions with a corresponding increase in the frequency of unchanged alleles. However, these data did not reach statistical significance (Table 1), and, given the large number of paternal transmissions analyzed (84 Xpc+/+ and 55 Xpc-/-), we propose that Xpc is unlikely to contribute in any significant way to paternal HdhQ111 CAG repeat contractions.
We then analyzed Msh2's binding partners Msh6 and Msh3. The effects of Msh6 and Msh3 genotype on paternal and maternal HdhQ111 transmissions are shown in Figures 1C and 1D respectively. There was no significant effect of Msh6 or Msh3 genotype on maternally inherited HdhQ111 CAG repeat length changes, consistent with the lack of involvement of Msh2 in intergenerational instability in female transmissions (Wheeler et al. 2003). In transmissions from Msh3+/- and Msh3-/- males, there was a shift from expansions to unchanged and contracted alleles compared to transmissions from Msh3+/+ males (Figure 1D). However, this did not reach statistical significance at the pre-specified significance level of 0.01 (Table 1), suggesting that the majority of Msh2-mediated paternal expansions occur independently of Msh3. Msh6 genotype also did not significantly influence the expansion frequency in paternal transmissions (Figure 1C, Table 1). Therefore, Msh2-mediated paternal expansions of the HdhQ111 CAG repeat also occur independently of Msh6. However, in transmissions from Msh6+/- males there was a statistically significant increase in contractions compared to Msh6+/+ males (changed versus unchanged OR=4.98, p<0.0001; ‘Protective’ model OR=4.28, p=0.009), suggesting that Msh6 may protect against paternal contractions (Figure 1C, Table 1). Interestingly, in transmissions from Msh6-/- males, the distribution of repeat length changes was similar to wild-type, suggesting that other mechanisms may compensate for the increase in contractions caused by the absence of Msh6. As Msh2 also protected against paternal repeat contractions (Figure 1A, Table 1), our results suggested that this protective effect is likely to be mediated by Msh2-Msh6 dimers.
Somatic repeat instability
HdhQ111 mice display age-dependent, expansion-biased somatic instability of the HdhQ111 CAG repeat in the striatum that becomes apparent by 5 months of age. In contrast, tail DNA does not exhibit somatic instability at this age (Wheeler et al. 1999). We previously showed that HdhQ111 CAG striatal instability was eliminated in Msh2-/- mice (Wheeler et al. 2003). We have now determined striatal instability of the HdhQ111 CAG repeat in heterozygous Msh2+/- mice to assess the effects of removing a single Msh2 allele. DNA was extracted from dissected striata of HdhQ111/+ mice at 5 months of age and repeat size determined by GeneScan analysis of the PCR-amplified HdhQ111 CAG repeat. As shown in Figure 2A, Msh2+/Msh2+ striatum displayed somatic instability as indicated by a characteristic bimodal repeat size distribution, not seen in somatically stable tail DNA at the same age. Msh2+/- striatum showed a similar bimodal repeat size distribution to Msh2+/Msh2+ striatum, in contrast to Msh2-/- striatum previously shown to have no somatic instability (Wheeler et al. 2003). Therefore, the absence of a single Msh2 allele was not sufficient to eliminate somatic instability in HdhQ111 striatum.
Figure 2.
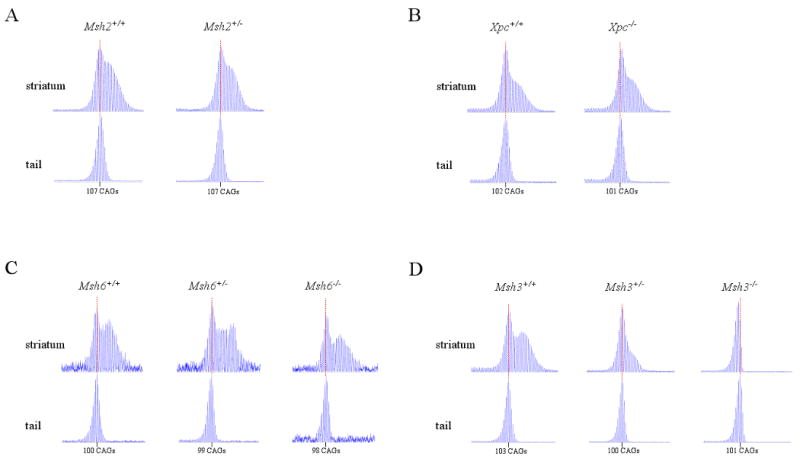
HdhQ111 CAG repeat striatal instability.
HdhQ111 mice were crossed onto backgrounds deficient in Msh2 (A), Xpc (B), Msh6 (C) or Msh3 (D). ABI GeneScan traces are shown of the HdhQ111 CAG repeat PCR-amplified from genomic DNA extracted from striata or tail of heterozygous HdhQ111/Hdh+ mice at 5 months of age. The number of mice of each DNA repair genotype and the constitutive HD CAG repeat size determined from tail in each mouse is as follows: (A) Msh2+/+ n=4, CAG 106, 107, 107, 107; Msh2+/- n=4, CAG 107, 107, 108, 111. (B) Xpc+/+ n=3, CAG 100, 101, 102; Xpc-/- n=3, CAG 95, 99, 101. (C) Msh6+/+ n=3, CAG 98, 100, 100; Msh6+/- n=3, CAG 99, 99, 99; Msh6-/- n=3, CAG 98, 99, 100. (D) Msh3+/+ n=3, CAG 102, 103, 103; Msh3+/- n=4, CAG 100, 100, 101, 102; Msh3-/- n=3, CAG 94, 95, 101, 101. Mice of the same genotype had identical repeat profiles regardless of constitutive CAG repeat number. Representative GeneScan traces are shown. HD CAG repeat size in tail is indicated, the position of which is marked with a dotted red line.
To delve further into the underlying mechanism and to explore an additional DNA repair pathway, we then evaluated the effects of eliminating Xpc, Msh6 and Msh3 on striatal HdhQ111 CAG repeat instability in HdhQ11/+ mice at 5 months of age. As shown in Figure 2B, Xpc+/+ and Xpc-/- striata both displayed a bimodal repeat size distribution characteristic of somatically unstable HdhQ111 CAG repeats, showing that Xpc does not play a major role in HdhQ111 CAG repeat striatal instability. Similarly, as shown in Figure 2C, Msh6+/+, Msh6+/- and Msh6-/- striata all displayed a bimodal repeat size distribution characteristic of somatically unstable HdhQ111 CAG repeats. Therefore, Msh6 does not play a major role in HdhQ111 CAG repeat striatal instability.
In contrast, as shown in Figure 2D, while Msh3+/+ striatum displayed a bimodal repeat size distribution characteristic of somatically unstable HdhQ111 CAG repeats, the GeneScan trace in Msh3-/- striatum was similar to that seen in tail, showing that Msh3 was required for striatal instability. Interestingly, in Msh3+/- striatum the GeneScan trace was not bimodal but was slightly broader than that in Msh3-/- striatum, showing that striatal instability is greatly reduced in the absence of only a single Msh3 allele. These results indicate that HdhQ111 CAG striatal instability is likely to be mediated via Msh2-Msh3 dimers. Furthermore, the finding that striatal instability is significantly reduced in Msh3+/- heterozygotes but not in Msh2+/- heterozygotes indicates that Msh3 levels are a limiting factor in this process.
Nuclear mutant huntingtin immunostaining
The time-dependent immunostaining of mutant huntingtin in the nuclei of striatal neurons is an early, dominant, CAG length-dependent phenotype in HdhQ111 mice that is delayed in the absence of Msh2 (Wheeler et al. 2003). We hypothesized that Msh2 influenced nuclear huntingtin immunostaining as a result of its role in mediating somatic instability in the striatum. The distinct effects of Msh2, Msh3, Msh6 and Xpc genes on striatal instability now afforded the opportunity to explore further the relationship between somatic HdhQ111 CAG instability and the early disease process in HdhQ111 striatum. We asked whether heterozygous loss of Msh2 was sufficient to delay this early phenotype, and explored whether homozygous knockout of Xpc and homozygous and heterozygous knockouts of Msh6 and Msh3 might modify this phenotype. Thus, we examined nuclear mutant huntingtin immunostaining using the EM48 anti-huntingtin polyclonal antibody (Gutekunst et al. 1999) in striata of 5-month heterozygous HdhQ111/+ mice with the same Msh2, Xpc, Msh6 and Msh3 genotypes analyzed above for striatal instability. This antibody specifically detects a conformation of mutant huntingtin in striatal nuclei of HdhQ111 mice (Wheeler et al. 2000).
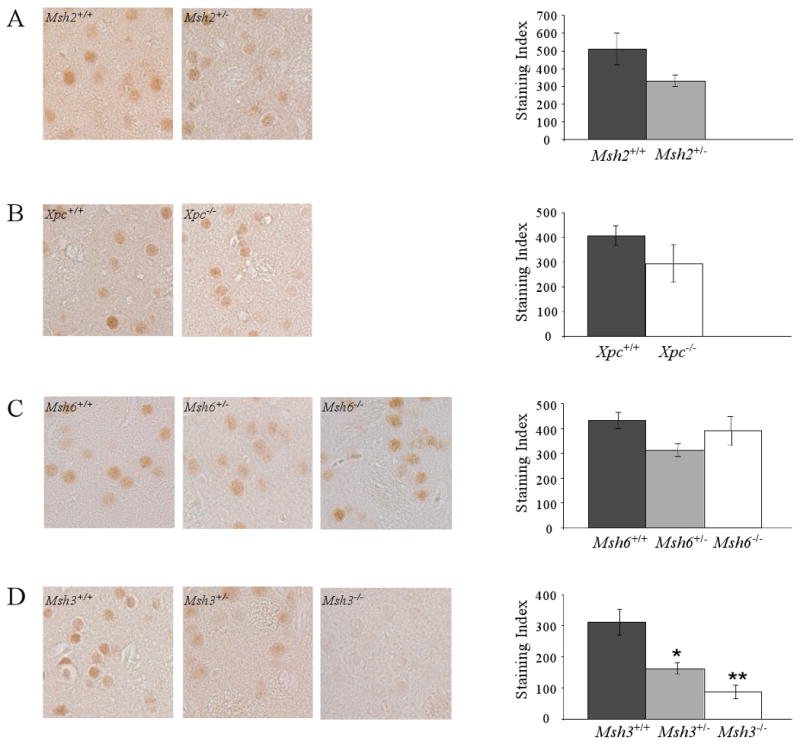
Figure 3 shows the quantified staining indices, reflecting the immunostaining intensity and number of immunostained nuclei, for each DNA repair gene analyzed. The staining index was not significantly altered in the absence of a single Msh2 allele (Figure 3A), in the complete absence of Xpc (Figure 3B), or in the absence of either one or two Msh6 alleles (Figure 3C). By contrast, the staining index was significantly decreased by the loss of either one (p=0.02) or two (p<0.001) Msh3 alleles. In contrast to Msh2, reduction of Msh3 levels, rather than elimination of Msh3, was sufficient to diminish nuclear mutant huntingtin immunostaining. Msh3 genotype did not alter the total amount of huntingtin or the amount of mutant huntingtin in striatum (Supplementary Figure 1). Our findings that only Msh2 and Msh3 were unequivocal contributors to both striatal instability and to nuclear huntingtin immunostaining implicate Msh2-Msh3 dimers in both these events, and provide strong support for the hypothesis that somatic instability in the striatum contributes to the HD CAG pathogenic process.
Figure 3.
Nuclear mutant huntingtin immunostaining in striatal neurons.
HdhQ111 mice were crossed onto backgrounds deficient in Msh2 (A), Xpc (B), Msh6 (C) or Msh3 (D). Striatal sections from HdhQ111/+ mice at 5 months of age were immunostained with anti-huntingtin antibody EM48 and a staining index (SI) calculated for each mouse as the product of the mean staining intensity and the number of immunostained nuclei (see Materials and Methods).
Left: Representative striatal histological sections immunostained with EM48.
Right: Bar graphs showing the mean SI for each genotype +/- standard error.
The number of mice of each DNA repair genotype (+/+, +/-, -/-) and the constitutive HD CAG repeat size determined from tail in each mouse is as follows: (A) Msh2+/+ n=3, CAG 107, 108, 109; Msh2+/- n=4, CAG 106, 106, 107, 107. (B) Xpc+/+ n=4, CAG 99, 101, 101, 101; Xpc-/- n=3, CAG 98, 101, 102. (C) Msh6+/+ n=3, CAG 98, 99, ND#; Msh6+/- n=4, CAG 97, 98, 99, 99; Msh6-/- n=4, CAG 98, 99, 99, 99. (D) Msh3+/+ n=5, CAG 98, 100, 100, 101, 102; Msh3+/- n=5, CAG 97, 97, 100, 101, 105; Msh3-/- n=3, CAG 99, 100, 102. * p=0.02, ** p<0.001. #A repeat value could not be determined for this mouse. Loss of two Msh3 alleles had a slightly greater effect of loss of one Msh3 allele but this effect was not statistically significant (p=0.11).
Discussion
The length of the HD CAG repeat is the single most critical determinant of HD pathogenesis. Therefore, understanding the factors responsible for both intergenerational instability, which alters inherited repeat length, and somatic instability, which further alters repeat length in the striatum, may lead to novel therapeutic targets. Here, using a precise genetic model of HD, HdhQ111 knock-in mice, we have explored mechanisms of intergenerational and striatal instability by performing a detailed analysis of the roles of DNA repair genes Msh3, Msh6 and Xpc, and have asked directly whether these genes are modifiers of an early disease phenotype.
Intergenerational instability
Xpc did not play a significant role in HdhQ111 CAG repeat intergenerational instability. This finding was in agreement with the lack of an effect of XPC on CAG repeat contractions in cultured human cells (Lin and Wilson 2007). In particular, Xpc did not significantly influence HdhQ111 maternal repeat length changes or paternal contractions, neither of which depend on Msh2, implicating pathways other than mismatch repair and global genome nucleotide excision repair in these events.
Msh2 is required for paternal HdhQ111 CAG repeat expansions and protects against paternal HdhQ111 CAG repeat contractions, implicating Msh2's binding partners Msh3 and/or Msh6 in these events. We found that paternal expansions did not require Msh6, and were largely independent of Msh3, strongly contrasting with an absolute requirement for Msh2. These findings suggest that while some paternal expansions of the HdhQ111 CAG repeat may be mediated by Msh2-Msh3 dimers, the majority are mediated by Msh2-dependent mechanisms that are independent of both Msh6 and Msh3. Interestingly a myotonic dystrophy type 1 (DM1) mouse model also exhibited intergenerational CTG expansions that were Msh2-dependent, but independent of both Msh6 and Msh3 (Foiry et al. 2006). However, these only accounted for a minority of the expansion events, with most being dependent on both Msh2 and Msh3. Therefore, it is likely that the effects of the mismatch repair proteins, perhaps at the level of repeat binding (Owen et al. 2005; Pearson et al. 1997), may be modulated by repeat sequence (CAG versus CTG), repeat length and genomic location.
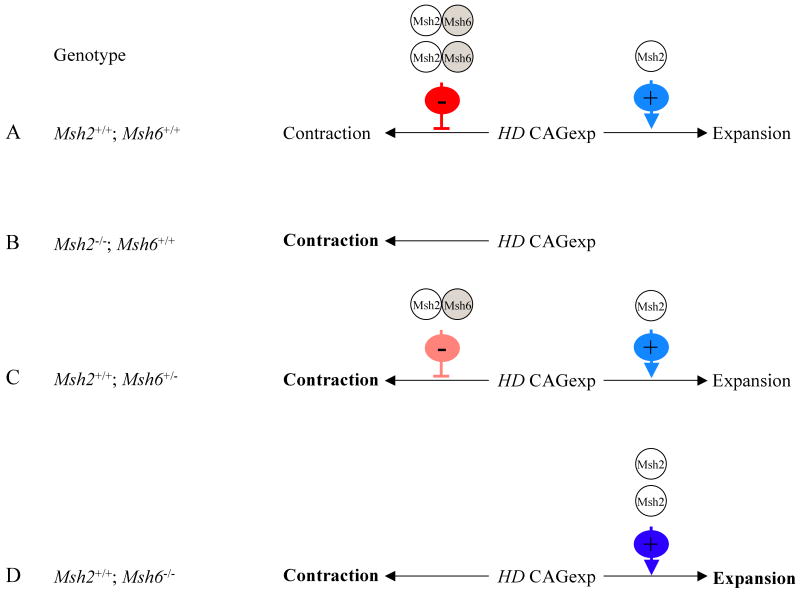
We report a novel role for Msh6 in protecting against paternal intergenerational repeat contractions. As Msh2 also protected against repeat contractions, our results suggests that the protection is afforded by Msh2-Msh6 dimers. However, although the absence of a single Msh6 allele resulted in an increased contraction frequency, this effect was not seen in the absence of two Msh6 alleles. These results were intriguing, and we propose the following model to explain the effects of Msh2 and Msh6 (Figure 4). Two Msh2-dependent mechanisms play a role in paternal intergenerational HdhQ111 CAG repeat instability: Msh2-dependent pathway(s) mediate expansions and Msh2-Msh6 protects against contractions (Figure 4A). The model assumes that repeat length can be influenced sequentially by each mechanism. Thus, the sum of each effect determines whether the repeat ultimately undergoes an expansion or a contraction, and the distribution of inherited repeat length changes depends on the relative influence of each mechanism. In transmissions from Msh2-/- fathers both mechanisms are eliminated, resulting in the loss of expansions and an increase in contraction frequency (Figure 4B). In transmissions from Msh6+/- fathers, the protective mechanism is reduced, resulting in an increased contraction frequency (Figure 4C). In transmissions from Msh6-/- fathers although contractions are increased, we hypothesize that the complete loss of Msh6 shifts the balance of Msh2 complexes in favor of Msh2-dependent pathway(s) that generate expansions sufficient to compensate for the increased number of contractions. Dynamic interactions between different Msh2 complexes have been described previously (Chang et al. 2000; Foiry et al. 2006; van den Broek et al. 2002). The net result of these two effects in Msh6-/- transmissions is a distribution of inherited repeat size changes that is similar to that in Msh6+/+ transmissions (Figure 4D and Figure 1C). Therefore, from a therapeutic perspective, our results suggest that it may be possible to shorten the inherited HD CAG repeat length, the major determinant of onset age in HD, by reducing, but not eliminating, Msh6.
Figure 4.
Proposed model for the roles of Msh2 and Msh6 in HdhQ111 CAG repeat intergenerational instability.
The length of the expanded HD CAG repeat (HD CAGexp) inherited from fathers is determined by the combined influences of two Msh2-dependent mechanisms: Msh2 mediates expansions and Msh2-Msh6 dimers protect against contractions. A) In mice wild-type for both Msh2 and Msh6 (Msh2+/+/; Msh6+/+) expansion (light blue ‘+’ arrow) and inhibition of contraction (red ‘-’ inhibitory symbol) both occur. B) In mice lacking Msh2 (Msh2-/-; Msh6+/+) both mechanisms are abolished, the net result being an increased contraction frequency (bold type). C) In mice lacking one Msh6 allele (Msh2+/+; Msh6+/-) contraction frequency is increased (bold type) as a result of reduced Msh2-Msh6 inhibition (pink‘-’ inhibitory symbol) while expansions are unaffected. D) In mice lacking two Msh6 alleles (Msh2+/+; Msh6-/-) contraction frequency is increased (bold type) due to loss of Msh2-Msh6 inhibition. However, this is compensated by an increase in expansions (dark blue ‘+’ arrow and bold type), as in the absence of Msh6 the balance of Msh2-dependent pathways is shifted in favor of those that mediate expansions.
Striatal instability
Striatal HdhQ111 CAG repeat instability is dependent on Msh2 (Wheeler et al. 2003). We show that striatal instability does not require Xpc or Msh6; however, it is completely dependent on Msh3, indicating that it is mediated via Msh2-Msh3 dimers. These results are consistent with roles of Msh2 and Msh3 in generating somatic CAG and CTG repeat expansions in other tissues in HD and DM1 mouse models (Foiry et al. 2006; Manley et al. 1999; Owen et al. 2005; Savouret et al. 2003; van den Broek et al. 2002). Furthermore, our finding that Msh3 was a limiting factor in somatic instability was also reported in a DM1 mouse model (Foiry et al. 2006). Therefore, similar roles of Msh2-Msh3 in HD and DM1 mouse models suggest a common mechanism of somatic expansion for CAG and CTG repeat diseases. Significantly, we have shown here that Msh3 determines HD CAG repeat length in the striatum, the target of HD pathogenesis.
Msh2-Msh3 dimers have been shown to bind expanded CAG repeat hairpin structures in vitro (Owen et al. 2005), although this has not been demonstrated in vivo. Msh2-Msh3 binding may mediate the formation of such structures (Panigrahi et al. 2005), which are subsequently processed to generate expansions. It has been proposed that Msh2-Msh3 binding subverts the mismatch repair machinery to result in expansion (Owen et al. 2005). However, it is also possible that expansions occur as a consequence of active engagement of the mismatch repair process (Gomes-Pereira et al. 2004). Further experiments are needed to distinguish these possibilities.
A requirement for Msh3 in generating somatic expansions contrasted with the relatively minor role of Msh3 in generating intergenerational expansions. Msh2 homodimers and homomultimers have been identified (Acharya et al. 1996), and purified Msh2 can bind to mismatched bases (Alani et al. 1995), Holliday junctions (Alani et al. 1997) and CAG/CTG repeat structures (Pearson et al. 1997). The different requirements for Msh3 in striatal expansions and intergenerational expansions may reflect the binding affinities of Msh2 and Msh2-Msh3 to different repeat structures that may form in striatal cells and in spermatogenic cells. eg. purified Msh2 binds more strongly to CAG repeat secondary structures than CTG repeat secondary structures (Pearson et al. 1997). Thus, the formation of CTG versus CAG secondary structures in striatal and spermatogenic cells could result in the differential binding of Msh2, and potentially of Msh2-Msh3 dimers. Alternatively, the different requirements for Msh3 could reflect levels of expression of Msh3 in striatal versus sperrmatogenic cells. Interestingly, Msh2 and Msh3 expression patterns differed during mouse spermatogenesis (Richardson et al. 2000), suggesting that, at least for some activities, these two proteins may be functionally uncoupled. Our results imply that drugs that interfere with Msh2 binding to expanded CAG/CTG repeats may inhibit intergenerational expansions, whilst drugs that interfere with Msh2-Msh3 binding to expanded CAG/CTG repeats, or dimerization of Msh2-Msh3 may inhibit striatal expansions.
Nuclear mutant huntingtin immunostaining
Msh3 also modified nuclear mutant huntingtin immunostaining in the striatum, as found previously for Msh2 (Wheeler et al. 2003). As postulated for Msh2, Msh3 could either modify this early phenotype as a consequence of its effect on somatic instability, or via an alternative mechanism, unrelated to its role in somatic instability. In the present study, in which we have examined both somatic instability and nuclear huntingtin immunostaining in striata from mice of many different DNA repair genotypes, we observed a very good correlation between striatal instability and nuclear mutant huntingtin immunostaining. These data provide strong support for a direct contribution of somatic instability to this phenotype. Furthermore, the finding that Msh3 was a limiting factor both in somatic instability and in nuclear mutant huntingtin immunostaining implies a shared underlying mechanism. Thus, we propose that Msh2-Msh3-dependent striatal instability is itself a modifier of the HD CAG pathogenic pathway, accelerating CAG repeat length-dependent striatal phenotypes. Our results, therefore, suggest both Msh2 and Msh3 as potential therapeutic targets. Msh3 is of particular interest as cancer-causing mutations in Msh3 have not been found, and because eliminating only a single allele's worth of Msh3 was sufficient to slow the HdhQ111 CAG pathogenic process.
Conclusions
Reducing HD CAG repeat length is an attractive therapeutic strategy. To determine potential therapeutic targets it is important to understand mechanisms underlying both intergenerational and striatal instability, as both process alter HD CAG repeat length. We have found that distinct Msh2-dependent mechanisms contribute to intergenerational and striatal HD CAG repeat instability. Our studies suggest Msh6 as a target for increasing intergenerational contractions, thus reducing the inherited HD CAG repeat length, and Msh3 as a target for eliminating striatal expansions. Importantly, we show directly that Msh3 is a modifier of the HD CAG pathogenic process, supporting a role of somatic instability in the disease, and suggesting novel routes of intervention in this devastating disorder.
Supplementary Material
A. Immunoblot of protein extracts from striatum of 5-month HdhQ111/+ mice on Msh3+/+ (N=3), Msh3+/- (N=2) and Msh3-/- (N=2) genetic backgrounds, probed with anti-huntingtin antibody mAb2166 (Chemicon International, Temecula, CA, USA), and subsequently with α-tubulin (Sigma) as a load control. Mutant and wild-type huntingtin bands are indicated. Soluble protein was extracted from dissected striata in RIPA buffer with protease inhibitors (Complete protease inhibitor cocktail, Roche, Mannheim, Germany). 30 μg of protein extract were resolved in 3-8 % Tris-acetate gels and proteins detected by immunoblot as previously described (Wheeler et al. 2000, Hum. Mol. Genet., 9, 503–513).
B. Quantification of the immunoblot in (A). Both levels of total huntingin (left) and mutant huntingtin (right) were quantified relative to α-tubulin. Bars represent mean ratios ± s.e. Quantification of immunoblot signals was carried out using a GS-800 Calibrated Densitometer with Quantity One software (BioRad).
Acknowledgments
We would like to thank Dr. Xiao-Jiang Li for generously providing the EM48 polyclonal antibody, Dr. Hein te Riele for the Msh2 knockout mice and Dr. Susan Cotman for discussion and critical reading of the manuscript. This work was funded by the National Institutes of Health NS049206 (V.C.W), NS532167 (M.E.M), NS16367 (M.E.M, K.L.L.); The Huntington's Disease Society of America (V.C.W); The Jerry MacDonald HD Research Fund (K.L.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA, Graham RK, Hayden MR. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- Chang DK, Ricciardiello L, Goel A, Chang CL, Boland CR. Steady-state regulation of the human DNA mismatch repair system. J Biol Chem. 2000;275:18424–18431. doi: 10.1074/jbc.M001140200. [DOI] [PubMed] [Google Scholar]
- Cheo DL, Ruven HJ, Meira LB, Hammer RE, Burns DK, Tappe NJ, van Zeeland AA, Mullenders LH, Friedberg EC. Characterization of defective nucleotide excision repair in XPC mutant mice. Mut Res. 1997;374:1–9. doi: 10.1016/s0027-5107(97)00046-8. [DOI] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Umar A, Yang K, Heyer J, Kucherlapati M, Lia M, Kneitz B, Avdievich E, Fan K, Wong E, Crouse G, Kunkel T, Lipkin M, Kolodner RD, Kucherlapati R. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Can Res. 2000;60:803–807. [PubMed] [Google Scholar]
- Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, Liedtke W, Cohen PE, Kane MF, Lipford JR, Yu N, Crouse GF, Pollard JW, Kunkel T, Lipkin M, Kolodner R, Kucherlapati R. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- Foiry L, Dong L, Savouret C, Hubert L, te Riele H, Junien C, Gourdon G. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- Fossale E, Wheeler VC, Vrbanac V, Lebel LA, Teed A, Mysore JS, Gusella JF, MacDonald ME, Persichetti F. Identification of a presymptomatic molecular phenotype in Hdh CAG knock-in mice. Hum Mol Genet. 2002;11:2233–2241. doi: 10.1093/hmg/11.19.2233. [DOI] [PubMed] [Google Scholar]
- Gines S, Bosch M, Marco S, Gavalda N, Diaz-Hernandez M, Lucas JJ, Canals JM, Alberch J. Reduced expression of the TrkB receptor in Huntington's disease mouse models and in human brain. Eur J Neurosci. 2006;23:649–658. doi: 10.1111/j.1460-9568.2006.04590.x. [DOI] [PubMed] [Google Scholar]
- Gines S, Seong I, Fossale E, Ivanova E, Trettel F, Gusella JF, Wheeler VC, Persichetti F, Macdonald ME. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington's disease knock-in mice. Hum Mol Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- Gonitel R, Moffitt H, Sathasivam K, Woodman B, Detloff PJ, Faull RL, Bates GP. DNA instability in postmitotic neurons. Proc Natl Acad Sci USA. 2008;105:3467–3472. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella JF, McNeil S, Persichetti F, Srinidhi J, Novelletto A, Bird E, Faber P, Vonsattel JP, Myers RH, MacDonald ME. Huntington's disease. Cold Spring Harbor Symposia on Quantitative Biology; 1996. pp. 615–626. [PubMed] [Google Scholar]
- Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li XJ. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper PS. Huntington's disease: a clinical, genetic and molecular model for polyglutamine repeat disorders. Phil Trans Royal Soc London - Series B: Biological Sciences. 1999;354:957–961. doi: 10.1098/rstb.1999.0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington's disease collaborative research group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Kennedy L, Evans E, Chen C, Craven L, Detloff P, Ennis M, Shelbourne P. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret A, Dragileva E, Teed A, Espinola J, Fossale E, Gillis T, Lopez E, Myers RH, MacDonald ME, Wheeler VC. Genetic background modifies nuclear mutant huntingtin accumulation and HD CAG repeat instability in Huntington's disease knock-in mice. Hum Mol Genet. 2006;15:2015–2024. doi: 10.1093/hmg/ddl125. [DOI] [PubMed] [Google Scholar]
- MacDonald ME, Barnes G, Srinidhi J, Duyao MP, Ambrose CM, Myers RH, Gray J, Conneally PM, Young A, Penney J, et al. Gametic but not somatic instability of CAG repeat length in Huntington's disease. J Med Genet. 1993;30:982–986. doi: 10.1136/jmg.30.12.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Mahal A, Mott R, Seller M, Bates GP. Instability of highly expanded CAG repeats in mice transgenic for the Huntington's disease mutation. Nat Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- Owen BA, Yang Z, Lai M, Gajek M, Badger JD, 2nd, Hayes JJ, Edelmann W, Kucherlapati R, Wilson TM, McMurray CT. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Ewel A, Acharya S, Fishel RA, Sinden RR. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum Mol Genet. 1997;6 doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- SAS Institute, I. SAS/STAT Users Guide. SAS Institute Inc.; Cary, NC: 1999. [Google Scholar]
- Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelbourne PF, Keller-McGandy C, Bi WL, Yoon SR, Dubeau L, Veitch NJ, Vonsattel JP, Wexler NS, Arnheim N, Augood SJ. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum Mol Genet. 2007;16:1133–1142. doi: 10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]
- Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, MacDonald ME, Gusella JF, Harper PS, Shaw DJ. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- Stine OC, Pleasant N, Franz ML, Abbott MH, Folstein SE, Ross CA. Correlation between the onset age of Huntington's disease and length of the trinucleotide repeat in IT-15. Hum Mol Genet. 1993;2:1547–1549. doi: 10.1093/hmg/2.10.1547. [DOI] [PubMed] [Google Scholar]
- Telenius H, Kremer B, Goldberg YP, Theilmann J, Andrew SE, Zeisler J, Adam S, Greenberg C, Ives EJ, Clarke LA, et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]
- Toft NJ, Winton DJ, Kelly J, Howard LA, Dekker M, te Riele H, Arends MJ, Wyllie AH, Margison GP, Clarke AR. Msh2 status modulates both apoptosis and mutation frequency in the murine small intestine. Proc Natl Acad Sci USA. 1999;96:3911–3915. doi: 10.1073/pnas.96.7.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJ, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- Veitch NJ, Ennis M, McAbney JP, The U.S.-Venezuela Collaborative Research Project. Shelbourne PF, Monckton DG. Inherited CAG.CTG allele length is a major modifier of somatic mutation length variability in Huntington disease. DNA Repair (Amst) 2007;6:789–796. doi: 10.1016/j.dnarep.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP. Neuropathological classification of Huntington's disease. J Neuropath Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, Persichetti F, McNeil S, Mysore J, Mysore S, Macdonald ME, Myers RH, Gusella JF, Wexler NS, The U.S.-Venezuela Collaborative Research Project Factors associated with HD CAG repeat instability in Huntington's disease. J Med Genet. 2007;44:695–701. doi: 10.1136/jmg.2007.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler VC, Auerbach W, White JK, Srinidhi J, Auerbach A, Ryan A, Duyao MP, Vrbanac V, Weaver M, Gusella JF, Joyner AL, MacDonald ME. Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum Mol Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, Gutekunst CA, Vrbanac V, Lebel LA, Schilling G, Hersch S, Friedlander RM, Gusella JF, Vonsattel JP, Borchelt DR, MacDonald ME. Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum Mol Genet. 2002;11:633–640. doi: 10.1093/hmg/11.6.633. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, Lebel LA, Vrbanac V, Teed A, Te Riele H, MacDonald ME. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum Mol Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, White JK, Gutekunst CA, Vrbanac V, Weaver M, Li XJ, Li SH, Yi H, Vonsattel JP, Gusella JF, Hersch S, Auerbach W, Joyner AL, MacDonald ME. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum Mol Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- White JK, Auerbach W, Duyao MP, Vonsattel JP, Gusella JF, Joyner AL, MacDonald ME. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- Zuhlke C, Riess O, Bockel B, Lange H, Thies U. Mitotic stability and meiotic variability of the (CAG)n repeat in the Huntington disease gene. Hum Mol Genet. 1993;2:2063–2067. doi: 10.1093/hmg/2.12.2063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Immunoblot of protein extracts from striatum of 5-month HdhQ111/+ mice on Msh3+/+ (N=3), Msh3+/- (N=2) and Msh3-/- (N=2) genetic backgrounds, probed with anti-huntingtin antibody mAb2166 (Chemicon International, Temecula, CA, USA), and subsequently with α-tubulin (Sigma) as a load control. Mutant and wild-type huntingtin bands are indicated. Soluble protein was extracted from dissected striata in RIPA buffer with protease inhibitors (Complete protease inhibitor cocktail, Roche, Mannheim, Germany). 30 μg of protein extract were resolved in 3-8 % Tris-acetate gels and proteins detected by immunoblot as previously described (Wheeler et al. 2000, Hum. Mol. Genet., 9, 503–513).
B. Quantification of the immunoblot in (A). Both levels of total huntingin (left) and mutant huntingtin (right) were quantified relative to α-tubulin. Bars represent mean ratios ± s.e. Quantification of immunoblot signals was carried out using a GS-800 Calibrated Densitometer with Quantity One software (BioRad).