Abstract
Few antivirals are effective against positive-strand RNA viruses, primarily because the high error rate during replication of these viruses leads to the rapid development of drug resistance. One of the favored current targets for the development of antiviral compounds is the active site of viral RNA-dependent RNA polymerases. However, like many subcellular processes, replication of the genomes of all positive-strand RNA viruses occurs in highly oligomeric complexes on the cytosolic surfaces of the intracellular membranes of infected host cells. In this study, catalytically inactive polymerases were shown to participate productively in functional oligomer formation and catalysis, as assayed by RNA template elongation. Direct protein transduction to introduce either active or inactive polymerases into cells infected with mutant virus confirmed the structural role for polymerase molecules during infection. Therefore, we suggest that targeting the active sites of polymerase molecules is not likely to be the best antiviral strategy, as inactivated polymerases do not inhibit replication of other viruses in the same cell and can, in fact, be useful in RNA replication complexes. On the other hand, polymerases that could not participate in functional RNA replication complexes were those that contained mutations in the amino terminus, leading to altered contacts in the folded polymerase and mutations in a known polymerase–polymerase interaction in the two-dimensional protein lattice. Thus, the functional nature of multimeric arrays of RNA-dependent RNA polymerase supplies a novel target for antiviral compounds and provides a new appreciation for enzymatic catalysis on membranous surfaces within cells.
Keywords: poliovirus, oligomer, polymerase, complementation, antiviral
INTRODUCTION
Polymerases and other enzymes are often assumed to function as isolated, soluble entities. However, within cells most macromolecular processes occur in large complexes that are immobilized on subcellular structures. For positive-strand viruses such as poliovirus, hepatitis C, and SARS coronavirus, the synthesis of viral RNA occurs on the cytoplasmic surfaces of membranes within the cytoplasm of the infected cells. For poliovirus, the viral RNA polymerases in such complexes can form flat lattices comprised of many copies of the enzyme, geometric structures that are ideally suited to form planar arrays on membrane surfaces. Here, we show that the polymerases within these oligomeric complexes play both enzymatic and structural roles. In fact, inactive polymerases can readily perform the structural roles so long as they are suitably shaped. This principle both illustrates the power of protein oligomerization for a small genome that is to take over a host cell and the importance of understanding the architecture of oligomeric proteins in the design of antimicrobial compounds that target them. Specifically, compounds that inactivate the catalytic sites of enzymes that function as oligomers may allow continued function of the drug-bound proteins in their structural roles, whereas misshaping their surface should destroy all their functions.
Nucleic acid synthesis is often associated with large, immobilized complexes (Cook 1999). For positive-strand RNA viruses such as poliovirus, foot-and-mouth disease virus, hepatitis C virus, and many others, the RNA replication complexes form on the cytosolic surfaces of membranes (for review, see Ahlquist et al. 2005; Mackenzie 2005; Salonen et al. 2005; Ortin and Parra 2006; Wileman 2006). The molecular details by which proteins arrayed in two dimensions on an intracellular surface function is at the heart of many problems at the interface of cell biology and biochemistry.
The RNA-dependent RNA polymerase of poliovirus, termed 3D polymerase, is a soluble enzyme targeted to intracellular membranes by its interaction with other components of the viral RNA replication complex; binding to 3AB, a membrane-associated viral protein has been shown to be sufficient to tether 3D polymerase to membranes (Lama et al. 1994; Towner et al. 1996; Lyle et al. 2002b; Fujita et al. 2007; Strauss and Wuttke 2007). All other nonstructural poliovirus proteins and their precursors (2A, 2B, 2C, 2BC, 3A, 3AB, 3C, and 3CD) are also found in the membrane-associated RNA replication complexes (Egger et al. 1996; Jurgens et al. 2006), as are several host proteins.
Specific binding interactions between many viral proteins in the RNA replication complex have been demonstrated. Homo-oligomerization has been reported for membrane-associated 3A (Xiang et al. 1998), the soluble domain of which can form dimers in solution (Strauss et al. 2003). Interactions between 3C and 3D have been observed by glutaraldehyde cross-linking and enzymatic function (Pathak et al. 2007); two different potential 3C–3D interaction surfaces have been observed crystallographically (Marcotte et al. 2007). Oligomerization of purified 3D RNA-dependent RNA polymerase in solution has been demonstrated by turbidity assays, electron microscopy (Lyle et al. 2002a), glutaraldehyde cross-linking (Pathak et al. 2007), and two-hybrid experiments (Hope et al. 1997; Xiang et al. 1998). The functional nature of such polymerase–polymerase interactions has been supported by observed hallmarks of oligomeric activity such as intermolecular enzymatic uridylylation (Richards et al. 2006), cooperative RNA binding (Pata et al. 1995; Beckman and Kirkegaard 1998), cooperative RNA elongation (Pata et al. 1995; Hobson et al. 2001), defective phenotypes of mutant viruses with mutations at protein–protein interfaces (Hobson et al. 2001; Pathak et al. 2002), and dominant phenotypes of several polymerase alleles (Crowder and Kirkegaard 2005). Purified poliovirus 3D polymerase forms two-dimensional arrays (Lyle et al. 2002a), as has been observed for the polymerase-associated protein p1 of phage F29 (Bravo and Salas 1998). For poliovirus RNA replication, we have suggested (Lyle et al. 2002a) that such planar arrays of polymerase are of an appropriate geometry to function as high-order multimeric patches on the surfaces of intracellular membranes, whether as homo-oligomers of 3D polymerase or in complex with larger precursors (Marcotte et al. 2007; Pathak et al. 2007).
The RNA-dependent RNA polymerases of several other positive-strand RNA viruses also undergo homo-oligomeric complex formation. The RNA-dependent RNA polymerase protein A of flock house virus has been shown to oligomerize both biochemically and via FRET analysis within infected cells. Multimeric polymerase protein A was shown to be sufficient to recruit genomic RNA to cellular membranes (Dye et al. 2005). Oligomerization of the polymerases from hepatitis C virus (Qin et al. 2002; Wang et al. 2002), tobacco mosaic virus (Goregaoker and Culver 2003), and brome mosaic virus (O'Reilly et al. 1997) has also been reported.
To investigate further the functional significance of poliovirus RNA-dependent RNA polymerase oligomerization, wild-type enzyme was allowed to form oligomeric structures with mutant enzymes that contained inactivated catalytic residues, misshapen oligomerization surfaces, or malfolded structures. As shown in Figure 1A, the three-dimensional structure of the full-length RNA-dependent RNA polymerase of poliovirus (Thompson and Peersen 2004) is shaped like a right hand, with the active site residues Asp328 and Asp329 indicated in yellow. In solution, we found that added polymerase molecules that contained mutated active sites could supplement the activity of wild-type enzyme, presumably by performing a structural role. Although this rescuing activity did not require a catalytic active site, it was highly sensitive to mutations that affected polymerase oligomerization or folding. In infected cells, the function of a temperature-sensitive polymerase could be supplemented by the direct intracellular introduction of purified polymerase protein, even when added protein had a defective active site. Thus, both catalytically inactive and active enzymes can function within oligomeric arrays, arguing for architectural functions of the nonenzymatic participants.
FIGURE 1.
Oligomerization properties of poliovirus 3D polymerase in solution and effects of titration of wild-type and YGAA mutant polymerases into subthreshold concentrations of wild-type polymerase. (A) A ribbon diagram of the three-dimensional structure of the 461-amino acid poliovirus RNA-dependent RNA polymerase is shown with the canonical “thumb,” “fingers,” and “palm” polymerase domains indicated. Asp328 and Asp329 at the active site are shown in yellow; both of these residues were mutated to Ala residues in the YGAA mutant polymerase. Residues Leu446, Arg455, and Arg456, involved in the “thumb” surface contact of a polymerase–polymerase interaction termed Interface I are shown in blue. Residues Asp339, Ser341, and Asp349, involved in the “palm” surface of Interface I are shown in pink; these are difficult to see in this view (see also Fig. 3A). (B) Concentrated wild-type poliovirus polymerase stored in a solution that contained 50% glycerol and 140 mM NaCl was diluted to the concentrations indicated in a solution containing 15% glycerol and 40 mM NaCl. Absorbance measurements were taken every 30 sec over a 10-min time period at 350 nm. (C) Concentrated wild-type and YGAA mutant poliovirus polymerase were diluted to concentrations of 5.6 μM and turbidity was monitored. (D) Electrophoretic mobility of HP1 RNA, a 110-nt RNA derived from the 3′ end of the poliovirus genome with several U residues at its 3′ end for self-priming, is shown following incubation for 30 min with increasing amounts of YGAA mutant polymerase (lanes 2–9, containing 70, 130, 250, 500, 750, 1000, 1500, or 1750 nM YGAA mutant polymerase, respectively). (E) To a preparation of 32P-labeled HP1 RNA (lane 1), various concentrations and mixtures of wild-type and YGAA mutant poliovirus polymerase were added, incubated for 30 min, and the products were displayed (lanes 2–13). Each of lanes 2–13 contained 7.5 nM wild-type polymerase, a concentration too low to support detectable template utilization under these conditions (lanes 2,8). This was supplemented with increasing amounts of wild-type 3D polymerase (lanes 3–7, containing an additional 7.5, 22.5, 55, 117.5, or 242.5 nM wild-type polymerase, respectively) or YGAA mutant polymerase (lanes 9–13) at the same concentrations. (F) The percentage of template elongation is shown as a function of the ratio of added:basal polymerase for both added wild-type and added YGAA mutant polymerase. “WT + WT” and “YGAA + YGAA” show the activities when these polymerases were present without mixing. Data points for turbidity and elongation assays are taken from one representative experiment; at least five experiments were performed for each of the proteins with comparable results. (G) Model for the elongation of RNA within a polymerase lattice. Lateral contacts between polymerase molecules along crystallographically defined Interface I are shown via blue (thumb) and pink (palm) contacts. The contacts (Interface II) that align the Interface I fibers into two-dimensional sheets are not yet known. The RNA template (red) is shown binding to several polymerases along an Interface I fiber, and the nascent RNA (yellow) is shown with its 3′ end in the active site (yellow dots) of a single polymerase molecule. Inactive polymerases mixed into the array are depicted as black molecules.
RESULTS
Mutation of the active site of 3D polymerase does not impede its ability to oligomerize or to participate in functional complexes
If some polymerases in an oligomeric structure perform predominately structural roles, they should be able to do so in the absence of catalytic activity. To determine the threshold concentration at which the oligomerization of wild-type and mutant 3D polymerase molecules could be observed using a turbidity assay, several concentrations of wild-type 3D polymerase were assayed. After purification (see Supplemental Material), polymerase preparations were stored in high concentrations of glycerol to prevent oligomerization. Upon dilution, solutions of polymerase at 4.5 and 5.6 μM, but not at 2.8 μM, could be seen to acquire detectable turbidity at 350 nm upon incubation for 10 min (Fig. 1B); longer incubation periods did not result in significant changes for wild-type enzymes (data not shown). Structures that scatter 350 nm light are likely to be large; although turbidity has been shown previously to correlate with the formation of higher-order, functional oligomers of poliovirus polymerase (Lyle et al. 2002a), it is likely that this is a much less-sensitive assay than functional oligomer formation. For example, these concentrations of polymerase are comparable to the concentrations of tubulin required to observe microtubule formation by turbidity assay; however, functional microtubules can be formed in solution at 10-fold lower concentrations (Lee et al. 1975).
To test whether inactive polymerases that contained mutations in active-site residues could also oligomerize, we monitored the acquisition of turbidity by solutions of mutant polymerase in which both Asp328 and Asp329 (Fig. 1A) were changed to Ala (termed 3D-D328A/D329A, or YGAA). Such mutations have been shown to abolish viability of the virus (Diamond and Kirkegaard 1994) and catalytic activity of the polymerase (Jablonski and Morrow 1995). While both wild-type and YGAA polymerases acquired turbidity to comparable extents, the kinetics differed in detail (Fig. 1C). Although the YGAA mutations are distant from any surface residues that might be involved in protein–protein interactions, allosteric interactions between the active site and the protein–protein interaction surfaces have been observed previously for 3D polymerase (Boerner et al. 2005). Nevertheless, mutations in the active site, although they completely abolished catalytic activity (Fig. 1D), did not preclude polymerase oligomerization.
To determine whether functional polymerase oligomers could be created by supplementing low concentrations of wild-type 3D polymerase with catalytically inactive enzyme, we monitored the elongation activity of purified 3D polymerase on self-priming RNA molecules. The RNA template used in these experiments, termed HP1 RNA, is a 32P-labeled, 110-nucleotide (nt) RNA molecule that comprises the 3′-terminal 85 heteropolymeric nucleotides, 20 A residues, and 5 U residues, which form a self-priming hairpin at the 3′ end (Pata et al. 1995; Lyle et al. 2002a). As shown in Figure 1, E and F, the addition of increasing amounts of wild-type polymerase gave rise to a cooperative increase in elongation of this primed RNA template (Pata et al. 1995; Lyle et al. 2002a). This cooperativity can be readily seen by comparing the complete lack of elongation activity at 7.5 and 15 nM polymerase (Fig. 1E, lanes 2,3), with the 14% elongation at 30 nM (Fig. 1E, lane 4), and 75% elongation at 62 nM (Fig. 1E, lane 5). No detectable elongation was observed in this experiment at polymerase concentrations of 15 nM or lower. Therefore, we selected the concentration of 7.5 nM wild-type polymerase as being a basal amount at which no elongation activity could be seen. However, when increasing amounts of catalytically inactive polymerase were added to 7.5 nM of basal, wild-type polymerase, template elongation was readily observed (Fig. 1E, lanes 9–12). These data are quantified in Figure 1F, where it is clear that even at a 1:1 ratio of mutant:wild-type enzyme, the YGAA mutant polymerase stimulated RNA elongation significantly. As the amount of inactive polymerase was increased, the percentage of template utilization reached 12% at ratios of inactive to active polymerase as high as 7:1, then began to decline. The stimulation observed by the addition of inactive enzyme did not result simply from the addition of excess protein, because as will be shown below, other mutant polymerases added at equivalent or higher concentrations did not display this effect. That the YGAA mutant polymerase was more stimulatory at low concentrations than wild-type polymerase (Fig. 1E, cf. lanes 3 and 9), is consistent with its increased oligomerization in the turbidity assay (Fig. 1C). Once the polymerase array contained many inactive enzymes; however, it lost catalytic function; at a 32:1 mutant:wild-type polymerase ratio, no elongation was observed (Fig. 1E, lane 13).
These data support the hypothesis that oligomers of polymerase are required for efficient RNA elongation under these conditions, but that not all of the polymerases within the oligomeric complex need be catalytically active (Fig. 1G). Even when an oligomeric structure is “more dead than alive,” containing three times more “zombie” polymerases than active enzyme, the extent of elongation was identical to that observed for uniformly wild-type enzyme (Fig. 1F). The working model shown in Figure 1G illustrates the hypothesis that polymerase molecules in two-dimensional arrays perform both enzymatic and structural roles. The horizontal polymerase–polymerase contacts are depicted as forming along Interface I, in which the thumb contacts (blue) and the palm contacts (pink) form a fiber, and fiber–fiber contacts form the two-dimensional lattices observed by electron microscopy (Lyle et al. 2002a), with randomly inserted inactive polymerase molecules depicted in black.
Physical mixing of wild-type and mutant polymerases
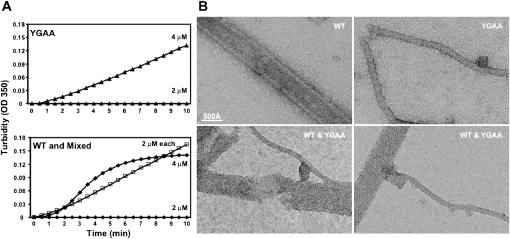
The interpretation of the “zombie” experiment shown in Figure 1 presumes that wild-type and YGAA active-site mutant polymerases can mix in solution. To test this assumption, we monitored the acquisition of turbidity by wild-type and YGAA mutant enzyme separately and together. As shown in Figure 2A, at concentrations of 2 μM, neither wild-type nor YGAA mutant polymerase acquired significant turbidity after a 10-min incubation. However, when mixed together at concentrations of 2 μM each, turbidity was acquired comparable in extent to that of either polymerase alone at 4 μM. Interestingly, the kinetics of turbidity development for the mixed oligomers appeared more similar to that of the YGAA mutant polymerase than of the wild-type polymerase at 4 μM, indicating that the cooperativity of assembly was diminished in the mixture.
FIGURE 2.
Formation and visualization of mixed poliovirus 3D polymerase oligomers. (A) Concentrated YGAA mutant poliovirus polymerase (top) or wild-type poliovirus polymerase (bottom) were diluted to 2 μM or 4 μM as indicated, and turbidity was monitored. A mixture of 2 μM of each polymerase was also monitored for the ability to oligomerize (bottom, □). (B) Negative stained electron micrographs of 1 μM purified wild-type polymerase (top, left), 1 μM purified YGAA mutant polymerase (top, right), and mixtures of 1 μM each (bottom). WT tubes are stain-filled, as seen by the high-protein density visible at the edges of the tube and the lower density in the tube center (protein is white). YGAA tubes are stain-excluding, suggesting tubes without a hollow center. Mixed samples show transition regions, as for example, the YGAA-like tube (bottom, left) that extends from a WT-like region of a thick tube. Scale bar, 500 Å.
To investigate the ultrastructure of the mixed oligomers, electron microscopy was performed on the structures formed by wild-type polymerase, YGAA mutant polymerase, and equimolar mixtures of the two. Both wild-type and YGAA mutant polymerases formed lattices whose uniformity allowed them to form tubular structures, albeit of different diameters (Fig. 2B). Wild-type structures were seen as buffer- and stain-filled tubes and also as “flattened” tubes, due to locally variable drying conditions of negatively stained samples. YGAA structures were thinner tubes that had stain-excluding centers and did not appear flattened. The mixed oligomers showed clear junctional regions between thick wild-type-like, and thinner YGAA-like oligomers of polymerase. Electron microscopy thus confirmed the physical mixing of wild-type and YGAA mutant polymerase. These EM data also support the turbidity data (Fig. 1C), which showed that the rate of formation and the final structures assembled by wild-type and YGAA mutant polymerases differ in detail, even though the YGAA mutations are deep within the folded structure of the polymerase, and therefore were not expected to effect polymerase–polymerase interactions.
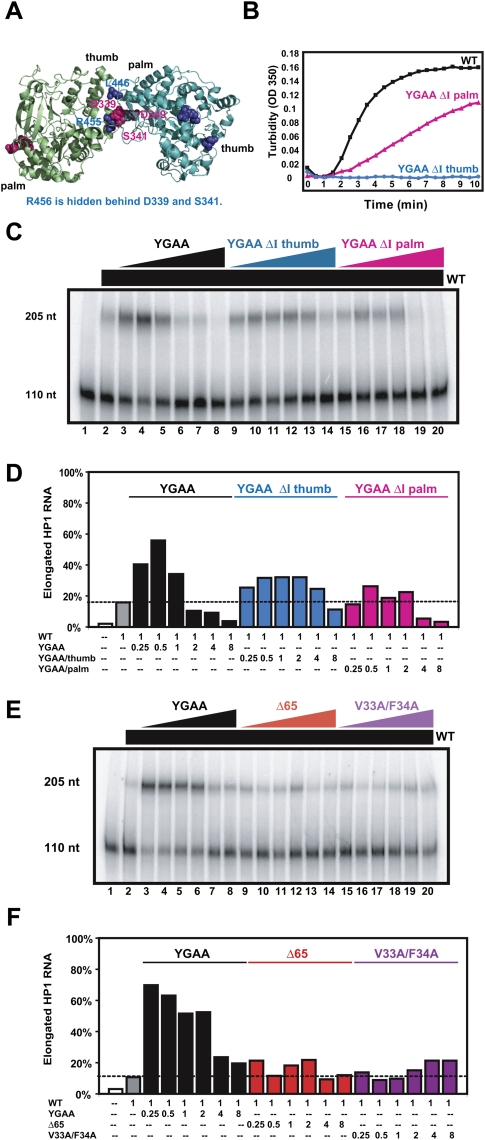
Oligomerization and biochemical rescue activities of polymerases that contain mutations in Interface I
To investigate the structural requirements for biochemical supplementation of active polymerase by catalytically inactive polymerases, we tested the effect of mutations on the two surfaces of Interface I. Previous studies showed that when three of the residues on the “palm” side of Interface I (D339, S341, and L349) were mutated to Ala (Fig. 3A, ΔI palm, shown in pink), the resulting virus displayed a small-plaque phenotype at 37°C (Pathak et al. 2002) and a strong temperature-sensitive phenotype at 39.5°C (Burgon et al. 2009). Mutation of residues on the “thumb” side of Interface I (L446, R455, R456) to Ala (Figs. 1A, 3A, ΔI thumb, highlighted in blue) resulted in viral lethality (Diamond and Kirkegaard 1994; Hobson et al. 2001; Pathak et al. 2002) and reduced cooperativity of RNA binding and elongation (Lyle et al. 2002a). Both the ΔI thumb and ΔI palm mutations reduced oligomerization of purified polymerase using a filter assay that retained structures >450 nm in diameter (Pathak et al. 2002).
FIGURE 3.
Ability of mutant 3D polymerases bearing active-site mutations and mutations at predicted interfaces to oligomerize and to biochemically complement wild-type polymerase. (A) Rendering of two full-length 3D polymerase molecules (Thompson and Peersen 2004) interacting at predicted Interface I (Hansen et al. 1997). Residues Leu446, Arg455, and Arg456 on the “thumb” side are shown in blue; the “ΔI thumb” mutant polymerase contains L446N, R455A, and R456A mutations. Residues Asp339, Ser341, and Asp349 on the “palm” side of Interface I are shown in pink; the “ΔI palm” mutant polymerase contains D339A, S341A, and D349A mutations. (B) Concentrated wild-type, YGAA ΔI thumb, and YGAA ΔI palm polymerases were diluted to concentrations of 5.6 μM and turbidity was monitored as in Figure 1. (C) The elongation of 32P-labeled HP1 RNA (lane 1) by 500 nM of wild-type polymerase resulted in 15% template utilization under the conditions of this experiment (lane 2). To this basal level of wild-type polymerase, increasing concentrations of YGAA, YGAA ΔI thumb, and YGAA ΔI palm mutant polymerases were added: 125 nM (lanes 3,9,15), 250 nM (lanes 4,10,16), 500 nM (lanes 4,11,17), 1 μM (lanes 5,12,18), 2 μM (lanes 6,13,19), and 4 μM (lanes 7,14,20). The RNA species present after a 30-min incubation were displayed by gel electrophoresis. (D) The percentage of template elongation from the experiment in C is shown as a function of the ratio of mutant:wild-type polymerase. These experiments were repeated several times; typical results are shown. (E) The elongation of 32P-labeled HP1 RNA (lane 1) by 500 nM of wild-type polymerase resulted in 14% template utilization under the conditions of this experiment (lane 2). To this basal level of wild-type polymerase, increasing concentrations of YGAA, Δ65, and V33A/F34A mutant polymerases were added: 125 nM (lanes 3,9,15), 250 nM (lanes 4,10,16), 500 nM (lanes 4,11,17), 1 μM (lanes 5,12,18), 2 μM (lanes 6,13,19), and 4 μM (lanes 7,14,20). The RNA species present after a 30-min incubation were displayed by gel electrophoresis. (F) The percentage of template elongation from the experiment in E is shown as a function of the ratio of mutant:wild-type polymerase.
For this study, each of these Interface I mutations was engineered into the YGAA mutant polymerase. Figure 3B shows the effect of the Interface I mutations on the acquisition of turbidity by the YGAA ΔI palm and YGAA ΔI thumb mutant polymerases. Although both mutant polymerases showed reduced rates of turbidity acquisition compared with wild-type (Fig. 3B) or YGAA mutant polymerases (Fig. 1C), quantitative differences were observed. Specifically, the ΔI thumb mutations introduced a nearly complete loss-of-function phenotype with respect to oligomerization, consistent with the lethality of these mutations to the virus, whereas the ΔI palm mutations introduced a partial loss-of-function phenotype to oligomerization, consistent with the small-plaque, temperature-sensitive phenotype of virus that contains them (Diamond and Kirkegaard 1994; Hobson et al. 2001; Pathak et al. 2002; Burgon et al. 2009). Similar data were obtained with purified ΔI palm and ΔI thumb mutant polymerases that lacked the additional YGAA mutation (data not shown). Therefore, despite the previously published, presumed equivalence of the YGAA ΔI palm and YGAA ΔI thumb mutations (Pathak et al. 2002), the actual effects of the mutations are quantitatively different and consistent with the viral phenotypes.
To determine the effect of these mutations of Interface I residues on the ability of mixed oligomers to function in RNA elongation, we titrated increasing amounts of YGAA, YGAA ΔI thumb, and YGAA ΔI palm mutant polymerases into a solution that contained a concentration of wild-type polymerase sufficient to give ∼15% elongation of a labeled HP1 template. This intermediate polymerase concentration was chosen so that both stimulatory and inhibitory effects of the mutant polymerases would be observable. Addition of increasing amounts of YGAA mutant polymerase stimulated RNA elongation activity (Fig. 3C, lanes 2–5) to almost 60%; however, higher ratios of inactive:active polymerase caused a reduction in RNA elongation activity (Fig. 3C, lanes 6–8). Both the YGAA ΔI thumb (Fig. 3C, lanes 9–11) and the YGAA ΔI palm mutant polymerase (Fig. 3C, lanes 15–17) were markedly less stimulatory than the YGAA mutant polymerase. However, high concentrations of the YGAA ΔI palm polymerase (Fig. 3C, lanes 18–20) proved to be more inhibitory than YGAA ΔI thumb polymerase (Fig. 3C, lanes 12–14). Quantitative interpretation of the observed stimulation of polymerase activity is difficult, as it should be a combination of the stimulatory and inhibitory effects of any given mutant polymerase. However, the extent of inhibition of wild-type polymerase activity at high ratios of mutant:wild-type enzyme should be a simple function of the participation of the mutant enzyme in the lattice. Therefore, the increased inhibition of wild-type polymerase observed with the YGAA ΔI palm mutant polymerase (Fig. 3C,D) is consistent with its intermediate ability to oligomerize (Fig. 3B).
To test whether more extreme forms of polymerase malfolding also affected the ability of inactive polymerases to supplement the activity of wild-type polymerase, we tested the effect of mutations in the amino-terminal region of the polymerase on biochemical rescue. The amino-terminal residues of the poliovirus RNA-dependent RNA polymerase constitute the “fingers” domain (Fig. 1A), which docks onto the “thumb” domain via insertion of hydrophobic residues Val33 and Phe34 (Thompson and Peersen 2004; Thompson et al. 2007). Specifically, we tested the ability of Δ65 mutant polymerase, missing the first 65 amino acids at the amino terminus (Hobson et al. 2001) and V33A/F34A mutant polymerase to supplement wild-type polymerase. Both of these mutant polymerases are catalytically inactive (Supplemental Fig. 1). As shown in Figure 3, E and F, Δ65 and V33A/F34A mutant polymerases were titrated into a basal concentration of wild-type polymerase sufficient to extend 13% of the labeled HP1 RNA under the conditions of this experiment (Fig. 3E, lane 2). As before, addition of inactive YGAA mutant polymerase conferred a stimulatory effect (Fig. 3E, lanes 3–8) in this experiment. For both the Δ65 and V33A/F34A mutant polymerases, little to no stimulation was observed. Therefore, misfolding of the polymerase either by mutation of Interface I or by disrupting folding of the amino terminus prevented biochemical complementation.
Exogenously added poliovirus polymerase rescues the defect in V391L mutant virus in tissue culture, regardless of its enzymatic activity
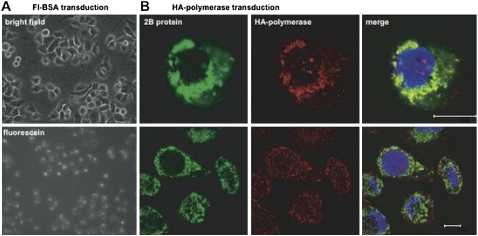
To test whether catalytically inactive, but structurally intact viral polymerases could supplement the function of RNA replication complexes in infected cells, we wanted to demonstrate that purified polymerase could be added exogenously to infected cells. As shown in Figure 4A, fluoresceinated BSA transfected into HeLa cells using a commercial protein transfection reagent could be seen in almost all of the cells of a typical microscopic field. To test whether transfected poliovirus 3D polymerase could participate in preformed viral RNA replication complexes, purified polymerase that contained an HA epitope tag at its C terminus was transfected into cells that had been infected with wild-type poliovirus. Indirect immunofluorescent confocal microscopy (Fig. 4B) revealed that the membrane-associated RNA replication protein 2B encoded by the infecting virus showed significant colocalization with the transfected HA-tagged 3D polymerase protein. Therefore, transfected polymerase could be targeted to the right place to function in RNA replication.
FIGURE 4.
Visualization of transduced proteins in HeLa cells. (A) Epifluorescent light microscopy of HeLa cells transduced with BodipyFL-conjugated BSA. (Top) Visualization of a field of transduced cells under phase contrast; (bottom) visualization of the same field under a fluorescein filter set after the addition of trypan blue to quench extracellular fluorescence. (B) Confocal micrographs from V391L-infected, HA-polymerase-transduced HeLa cells. Visualization of the poliovirus membrane-associated RNA replication protein 2B (green), transduced HA-tagged 3D polymerase (red), and merged fields are shown. Scale bar, 10 microns.
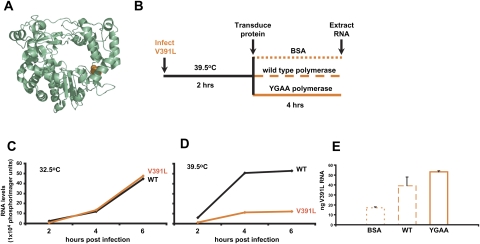
To test the ability of transfected, purified polymerase to rescue RNA replication defects in cells, we monitored viral RNA synthesis during infection with a well-characterized temperature-sensitive virus, 3D-V391L. The V391L mutation (Fig. 5A) was originally identified in a two-hybrid screen for polymerase mutations that interfered with the binding of 3D polymerase to 3AB, its membrane tether (Hope et al. 1997; Lyle et al. 2002b). The temperature sensitivity of the V391L mutant virus is consistent with the hypothesis that, at the nonpermissive temperature, the defective 3D–3AB interaction allows the mutant polymerase to be released from its membrane tether, thus reducing its effective concentration in the RNA replication complex. The experimental design (Fig. 5B) was to supplement infections with 3D-V391L mutant virus with transfected polymerase protein and to monitor the effect on viral RNA synthesis. Figure 5, C and D, document the defect in viral positive-strand RNA accumulation at 39.5°C, the nonpermissive temperature, as determined by quantitative dot blot analysis. As can be seen in Figure 5E, the amount of viral positive-strand RNA within cells infected with 3D-V391L virus at 39.5°C and transduced with bovine serum albumin (BSA) was increased more than twofold by the introduction of purified wild-type polymerase. Therefore, exogenously added polymerase could supplement the function of the poorly tethered V391L mutant polymerase during infection. This function was also provided, even more efficiently, by the same amount of purified, transduced polymerase that was catalytically inactive. Thus, some function of V391L polymerase other than its catalytic activity can be supplemented at the nonpermissive temperature, most likely via the formation of oligomeric structures on the surface of membranous vesicles that facilitate the tethering of the active enzyme.
FIGURE 5.
Complementation of V391L polymerase in infected cells by exogenous poliovirus polymerase protein. (A) Ribbon drawing of poliovirus 3D polymerase with Val391 shown in orange. (B) Experimental design of polymerase transduction into V391L mutant virus-infected cells. At 2-h post-infection with V391L mutant virus at the nonpermissive temperature, the indicated proteins were transduced into the infected cells. (C) Accumulation of positive-strand poliovirus RNA during HeLa cell infection with wild-type or V391L mutant virus at the permissive temperature and (D) at the nonpermissive temperature. (E) Positive-strand viral RNA was quantified 6-h post-infection by dot-blot analysis of serial dilutions of total cellular RNA from the experiment depicted in B, after the transfection of 7 μg of BSA, purified wild-type poliovirus 3D polymerase, or purified YGAA mutant poliovirus 3D polymerase. The amount of positive-strand RNA in a sample of 10 μg of total cellular RNA was determined by comparison to a standard curve of poliovirus RNA diluted in cellular extracts. Mean and standard error of duplicate experiments are shown.
DISCUSSION
It is now appreciated that most polymerases do not move along their DNA or RNA templates, but are immobilized in complexes through which their templates translocate (for review, see Giaever et al. 1988; Droge 1994; Faro-Trindade and Cook 2006). Large oligomeric structures formed by purified poliovirus 3D polymerase have been shown to be two-dimensional arrays, one polymerase in depth and hundreds of molecules in length and width (Lyle et al. 2002a). The geometry of such two-dimensional sheets is well-suited to orient polymerases and associated proteins on the cytoplasmic surfaces of intracellular membranes, but also brings up many questions. What are the molecular contacts that form the two-dimensional arrays? Array formation was shown to be destroyed by deletion of amino-terminal residues and by disruption of a protein–protein interaction termed Interface I (Hansen et al. 1997; Hobson et al. 2001; Lyle et al. 2002a). The molecular contacts observed in Interface I are shown in Figure 3A. The “Interface II” contacts that must exist in order to align the parallel fibers into the observed two-dimensional arrays have not yet been identified; they were originally thought to be due to intermolecular donation of N-terminal residues (Hansen et al. 1997; Hobson et al. 2001), but the intramolecular folding of the N-terminal residues in the full-length structure (Thompson and Peersen 2004) makes this interpretation less likely. The actual disposition of the N-terminal residues of the polymerase in solution is still unclear due to the very low transition temperature of their folded structure (Thompson et al. 2007).
In this study, we have shown that suboptimal polymerase activity in solution (Fig. 1) and in infected cells (Fig. 5) can be rescued by the addition of catalytically inactive polymerases. Therefore, during replication of viral RNA in infected cells and in the elongation of long heteropolymeric RNAs by purified polymerase, enzymatically inactive polymerases can play a crucial role. We argue that this role is architectural, to create a two-dimensional lattice with cooperative RNA-binding properties. Whether this activity is required for RNA binding, the reorganization of RNA structural motifs, or the configuration of the active site of the polymerase with respect to the RNA primer is not yet known.
If every polymerase in an oligomeric array does not have to be enzymatically active, how does elongation proceed? Is one active site used iteratively, or is the elongating chain passed between active sites? RNA-binding experiments with poliovirus polymerase (Beckman and Kirkegaard 1998) and co-crystals of short RNA templates bound to the related foot-and-mouth-disease virus polymerase (Ferrer-Orta et al. 2004) have suggested that each polymerase molecule can bind to ∼12 nt of single-stranded RNA. RNA-binding affinity was reduced by mutation of Interface I residues, but not by deletion of amino-terminal residues (Hobson et al. 2001). Therefore, we propose that cooperative RNA binding is accomplished by fibers formed along Interface I (Fig. 1G). The 110-nt RNA substrate used here contains 95 nt of single-stranded RNA, and should therefore bind to multiple polymerases. If the same active site were used throughout elongation, then the probability of elongating the nascent strand would depend simply on the ability of the primed template RNA to bind to an active polymerase. If, on the other hand, after the nascent strand had elongated significantly when bound to the first polymerase, it were to transfer to the next active site in the fiber, multiple transfers would be required to elongate the entire RNA. As shown in Figure 1F, whether the ratio of added mutant:basal wild-type polymerase was three, seven, or 15, ∼10%–11% of the template RNA was elongated, much more than would be expected for the active-site transfer model. Furthermore, inspection of the gel in Figure 1E and other similar gels (data not shown) has revealed none of the prematurely terminated products expected if nascent strand transfer to nonfunctional active sites occurred. Therefore, we conclude that when RNA is elongated in an oligomeric array, one active site is used iteratively, and most of the polymerases in the oligomer serve as cooperative RNA-binding proteins. This is consistent with the very high processivity of RNA-dependent RNA polymerization by poliovirus polymerase on long RNA templates (Rodriguez-Wells et al. 2001).
What is required for an inactive polymerase to serve its function within an oligomeric array? When additional mutations were introduced into either the “thumb” or “palm” surface of Interface I, the ability of the inactive polymerases to supplement wild-type activity was markedly reduced (Fig. 3). The severity of these alleles correlates with their phenotypes during infection: the “thumb” mutations, which are lethal to the virus (Diamond and Kirkegaard 1994; Hobson et al. 2001; Pathak et al. 2002), abrogated oligomerization as measured by turbidity, whereas the “palm” mutations, which give rise to a small-plaque, temperature-sensitive viruses (Pathak et al. 2002; Burgon et al. 2009), showed some residual oligomerization as measured by turbidity (Fig. 3B). Thus, we consider it likely that intermolecular contacts at Interface I play roles during infection. Other mutant polymerases that failed to supplement low concentrations of wild-type polymerase were polymerase with the amino-terminal 65 amino acids deleted (Δ65) or with V33A/F34A mutations that should interfere with the docking of the polymerase “fingers” to the “thumb.” Whether this failure to complement reflected the failure of the Δ65 and V33A/F34A mutant polymerases to form contacts at Interface II, or due to some other aspect of their malfolding, is not yet known.
The biological significance of two-dimensional arrays of poliovirus polymerase observed by electron microscopy (Lyle et al. 2002a) and discussed herein has been disputed in the literature. In particular, the significance of Interface I has been questioned because sets of mutations that were thought to have equivalent effects on the stability of Interface I (R455A, R456A mutations on the “thumb” face and D339A, S341A, D349A on the “palm” face) displayed lethal and small-plaque phenotypes, respectively (Pathak et al. 2002). In these experiments (Pathak et al. 2002), extents of oligomerization were quantified using filter-binding assays, which impose a threshold size for retention of oligomers. However, as can be seen in Figure 3B, more sensitive quantitation of the unlimited oligomerization of poliovirus polymerase revealed that the L446N/R455A/R456A mutations caused a complete loss of detectable oligomerization, as did the R455A/R456A mutations alone (data not shown). On the other hand, the D339A, S341A, D349A “ΔI palm” mutations caused only a partial loss of oligomerization, which is in fact consistent with the less severe viral phenotype they cause. Another argument that polymerase–polymerase interactions at Interface I residues may not be biologically important, or at least not conserved among picornaviruses, has been that the identity of individual residues is not highly conserved among picornavirus polymerases (Marcotte et al. 2007; Pathak et al. 2007). However, covariation between complementary interfaces is, of course, a better indicator of conserved function, and this possibility has not yet been addressed.
The importance of polymerase oligomerization in RNA elongation in solution has also been (Thompson and Peersen 2004) called into question. Specifically, in assays that measure polymerase activity using high concentrations of either short, synthetic RNAs or homopolymeric templates and primers, the integrity of Interface I has been shown to have no effect (Pathak et al. 2002; Thompson and Peersen 2004; Marcotte et al. 2007). In these experimental conditions, under which RNA binding is not expected to be rate limiting, it is not surprising that the formation of a cooperative RNA-binding surface would be unnecessary. Furthermore, the incorporation of labeled nucleotides gives no information about the efficiency of template utilization, only the function of the active site. However, under the assay conditions reported in the present study and in previous studies from these laboratories (Pata et al. 1995; Lyle et al. 2002b), the elongation efficiency of prelabeled, relatively long (110-nt) heteropolymeric RNA substrate has been monitored. Under these conditions, cooperative polymerase activity and complete template utilization have been repeatedly observed (Figs. 1, 3; Pata et al. 1995; Lyle et al. 2002a) and shown to be sensitive to mutation of Interface I (Lyle et al. 2002b). As has been discussed previously (Lyle et al. 2002b), a consistent interpretation of all of these findings is that polymerase oligomerization is required for high-affinity RNA binding, but not for polymerase activity per se. Both steps in an RNA elongation reaction are expected to be crucial for RNA replication within infected cells, especially at the beginning of an infection, when a single 7500-nt viral RNA molecule must be efficiently translated and replicated.
In an infected cell, it is likely that many 3D polymerase and 3D polymerase-containing molecules form contacts in addition to those formed with other polymerases. For example, studies of the uridylation of the poliovirus protein primer, VPg, support the existence of a functional interaction between the viral 3D polymerase and the 3C moiety of 3CD. 3CD stably accumulates in infected cells and is possibly a precursor of the 3C proteinase and the 3D polymerase. A recent three-dimensional structure of 3CD determined by X-ray crystallography has revealed two interesting 3D–3C interaction surfaces (Marcotte et al. 2007); it was noted that the formation of one of these 3C–3D interactions would not preclude the polymerase–polymerase interactions at Interface I discussed herein. Mutagenesis experiments have also suggested a set of residues of 3D polymerase involved in interaction with a dimer of 3C proteins; however, these proposed contacts (Shen et al. 2008) do not correspond to those described structurally (Marcotte et al. 2007). In any case, it is perfectly possible for the same surface of a protein to be involved in multiple functional interactions because, as exemplified by the present study, it is not likely that each multifunctional polymerase molecule in the cell is involved in an identical, static complex.
Large oligomeric complexes are excellent drug targets for several biological reasons. First, the constituents of large complexes must often serve multiple functions, so their ability to form and reform oligomers with different compositions can be crucial. For this reason, stabilization of a complex can often be as deleterious as its destabilization, and can be more readily accomplished by small molecules (Chardin and McCormick 1999; Zeghouf et al. 2005). Second, the presence of defective subunits, such as those bound to an inhibitory drug, can dominantly inhibit the function of complexes, even if some of those subunits are functional or even drug resistant. In a genomic screen of the poliovirus genome designed to identify proteins that, when defective, could interfere with the growth of wild-type viruses, several dominant polymerase alleles were identified (Crowder and Kirkegaard 2005). The principle of targeting oligomeric proteins to ensure that drug-sensitive genomes are dominant will be especially important in the design and implementation of pharmaceuticals against RNA viruses, whose high error rate makes drug resistance a formidable obstacle. However, as demonstrated here, targeting the active sites of oligomeric enzymes may not be as productive a strategy as targeting structural features that will affect the function of the oligomeric complex.
MATERIALS AND METHODS
Mutagenesis and polymerase purification
SOE PCR (splicing by overlapping extension PCR) was used to generate all mutant 3D polymerase plasmids (Lefebvre et al. 1995). The wild-type 3D expression vector (T5T3D) (Pata et al. 1995) was used as a PCR template. The YGAA mutations (D328A/D329A) replaced the Asp328 and Asp329 codons for Ala codons (GATGAT to GCTGCT). The YGAA ΔI thumb mutant polymerase-encoding plasmid (L446N/R455A/R456A/D328A/D329A) was generated by inserting the YGAA mutations into the T5T3D-ΔInt I plasmid (Lyle et al. 2002b). T5T3D-Δ65 contained a deletion of the codons for the amino-terminal 65 amino acids (Hobson et al. 2001). V33A/F34A mutant polymerase was encoded by T5T3D-V33A/F34A, which substitutes the Val33 and Phe34 codons for Ala codons (GTGTTT to GCCGCG). YGAA ΔI palm polymerase was encoded by T5T3D-ΔI palm, in which the codons for Asp339, S341, and D349 were substituted for Ala codons as described previously (Pathak et al. 2002).
Poliovirus 3D polymerase was purified as described previously (Hobson et al. 2001; Lyle et al. 2002a) with minor modifications. A full description of this protocol is available in the Supplemental Information.
Electron microscopy
To prepare samples for electron microscopy, reaction mixes containing the indicated concentrations of wild-type or YGAA mutant polymerase, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 10% glycerol, 30 mM NaCl, and 0.01% n-octyl glucoside were incubated at 30°C for 30 min, followed by 1 h on ice. Five microliters of each reaction were placed on carbon-coated, copper grids and incubated at room temperature for 5 min. The grids were washed with 12 drops of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and negatively stained with 1% uranyl acetate. Images were recorded at 45,000× magnfication on a Philips CM-12 electron microscope operated at 120 kV. The negatives were digitized using a Nikon 9000 scanner using a pixel size corresponding to 1.41 Å.
RNA synthesis and elongation reactions
HP1 template was generated as previously described (Pata et al. 1995) and purified by electroelution using an IBI electroeluter (IBI Technology). To transcribe HP1 RNA, 1 μg of HP1 template was incubated with 250 U of T7 polymerase (NEB), 1X T7 transcription buffer, 50 μCi [α32P]UTP (3000 Ci/mmol, NEN), 0.5 mM NTP, and 100 U of RNasin (Promega). The reaction was incubated at 37°C for 3 h and treated with 347 U of DNase I (Promega) for 15 min at 37°C. HP1 RNA was purified over a P30 RNase-free Micro Biospin column (Bio-Rad), and then subjected to phenol:chloroform extraction and ammonium acetate precipitation with a 70% ethanol wash. Pelleted HP1 RNA was resuspended in 200 μL 10 mM Tris (pH 8.0) and quantified by spectrophotometer reading (Hitachi U2000).
RNA elongation reactions were performed by incubating 1 nM HP1 RNA with variable concentrations of added polymerase in a solution containing 25 mM MES (pH 5.5), 5 mM MgCl2, 10 mM glycerol, 30 mM NaCl, 0.01% n-octyl glucoside, 5 mM DTT on ice for 10 min. In mixed oligomer experiments, wild-type and mutant 3D polymerases in the high-glycerol storage buffer were pre-incubated together for 10 min on ice prior to inclusion in the elongation reactions. Reactions were transferred to 37°C and equilibrated for 10 min before adding 250 μM ribonucleotide triphosphates. Reactions proceeded for 30 min at 37°C; at low concentrations of polymerase, this was within initial rate conditions (data not shown). Seven and a half microliters of a solution containing 250 μg/mL proteinase K (Sigma), 375 mM Hepes-NaOH (pH 8.0), 0.5% SDS, and 25 mM EDTA was added to terminate the reactions, and the samples were incubated at 37°C for 10 min. Thirty microliters of 80% formamide was added and the samples incubated at 95°C for 4 min, then placed immediately on ice. The RNA species were separated by electrophoresis on 8% acrylamide (19:1 acrylamide:bis-acrylamide), 7 M urea, 0.5X TBE gels run at 1850 V, and maintained at 50°C during the entire run. Gels were subsequently dried and quantified by phosphorimaging (Molecular Dynamics Storm System). The specific activity of polymerase preparations can differ, as can be seen by comparing Figure 1E and Figure 3C; polymerase preparations can lose activity with storage.
Turbidity assays
Turbidity assays were performed in a Hitachi U2000 spectrophotometer, measuring absorbance at 350 nm (Lyle et al. 2002a). Wild-type or mutant polymerase was diluted to a final concentration of 5.6 μM polymerase (except where noted), 40 mM NaCl, and 15% glycerol in a total volume of 50 μL, mixed with 150 μL of 13.5 mM Tris-HCl (pH 7.5) and placed in a 200-μL, 1-cm quartz cuvette (Sigma) at room temperature. Absorbance at OD350 was recorded at 30-sec intervals.
Protein transduction and dot blot assays
HeLa cells were seeded at a density of 7.5 × 105 cells per 6-cm plate 24 h prior to infection. Cells were infected with wild-type or V391L mutant poliovirus as described (Hope et al. 1997) at a multiplicity of infection of 20 plaque-forming units/cell. For RNA accumulation assays, after 30 min adsorbtion at 37°C, cells were incubated at 32.5°C or 39.5°C for the indicated times. When protein transduction was included in the protocol, V391L mutant-infected cells were incubated for 2 h at 39.5°C. Then, transduction of 7 μg of BSA or poliovirus polymerase proteins into cells was performed using Pulsin delivery reagent (Polyplus Transfection) according to the manufacturer's recommendations. The transduction mixture was incubated with cells for 4 h at 39.5°C. At the times indicated, total cellular RNA was extracted using TRIzol reagent (Invitrogen). RNA pellets were resuspended in 50 μL 10 mM Tris (pH 8.0) and quantified on a Nanodrop 1000 spectrophotometer (Thermo Scientific).
Ten micrograms of each RNA sample, along with serial 10-fold dilutions of each sample in mock-treated RNA diluent, were assembled in 20 μL volumes in sterile, nontissue culture-treated 96-well plates (Nunc, Thermo Fisher). Samples were denatured in three volumes of denaturing solution (500 μL formamide, 162 μL 37% formaldehyde [12.3 M] and 100 μL of a solution containing 0.2 M MOPS [pH 7.0], 0.5 M sodium acetate, and 0.01 M EDTA) for 15 min at 65°C. Following denaturation, samples were mixed with 2 vol of ice-cold 20X SSC and immediately spotted onto a Hybond XL membrane (GE Biosciences) using an S&S Minifold I Dot Blot (Schleicher & Schuell). Samples were washed with 2 mL 10X SSC, and the blot was air dried. RNA was UV cross-linked to the membrane using the “Autolink” function on a UV Stratalinker (Stratagene). The blot was rehydrated in 6X SSC and prehybridized with 20 mL of Ultrahyb solution (Ambion) for 30 min at 68°C prior to the addition of a 32P-labeled probe complementary to positive-strand poliovirus RNA. After overnight incubation the blot was washed, exposed to a phosphor screen, and quantitated on a Storm PhosphorImager (Molecular Dynamics).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Scott Crowder, Bill Jackson, Matthew Taylor, and Andres Tellez for helpful advice; Eric Freundt for confocal microscopy lessons; Jing Wang for initial electron microscopy experiments; and Peter Sarnow, Jennifer Ptacek, and Anthony Vecchiarelli for critical reading of the manuscript. This work was supported by ACS postdoctoral fellowship no. PF-04-040-01-GMC to J.F.S., and by NIH grant no. AI-25166 and an NIH Director's Pioneer Award to K.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1955410.
REFERENCES
- Ahlquist P, Schwartz M, Chen J, Kushner D, Hao L, Dye BT. Viral and host determinants of RNA virus vector replication and expression. Vaccine. 2005;23:1784–1787. doi: 10.1016/j.vaccine.2004.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman MT, Kirkegaard K. Site size of cooperative single-stranded RNA binding by poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1998;273:6724–6730. doi: 10.1074/jbc.273.12.6724. [DOI] [PubMed] [Google Scholar]
- Boerner JE, Lyle JM, Daijogo S, Semler BL, Schultz SC, Kirkegaard K, Richards OC. Allosteric effects of ligands and mutations on poliovirus RNA-dependent RNA polymerase. J Virol. 2005;79:7803–7811. doi: 10.1128/JVI.79.12.7803-7811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Salas M. Polymerization of bacteriophage ϕ 29 replication protein p1 into protofilament sheets. EMBO J. 1998;17:6096–6105. doi: 10.1093/emboj/17.20.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgon TB, Jenkins JA, Deitz SB, Spagnolo JF, Kirkegaard K. Bypass suppression of small-plaque phenotypes by a mutation in poliovirus 2A that enhances apoptosis. J Virol. 2009;83:10129–10139. doi: 10.1128/JVI.00642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin P, McCormick F. Brefeldin A: The advantage of being uncompetitive. Cell. 1999;97:153–155. doi: 10.1016/s0092-8674(00)80724-2. [DOI] [PubMed] [Google Scholar]
- Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- Crowder S, Kirkegaard K. Trans-dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nat Genet. 2005;37:701–709. doi: 10.1038/ng1583. [DOI] [PubMed] [Google Scholar]
- Diamond SE, Kirkegaard K. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J Virol. 1994;68:863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droge P. Protein tracking-induced supercoiling of DNA: A tool to regulate DNA transactions in vivo? Bioessays. 1994;16:91–99. doi: 10.1002/bies.950160205. [DOI] [PubMed] [Google Scholar]
- Dye BT, Miller DJ, Ahlquist P. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J Virol. 2005;79:8909–8919. doi: 10.1128/JVI.79.14.8909-8919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D, Pasamontes L, Bolten R, Boyko V, Bienz K. Reversible dissociation of the poliovirus replication complex: Functions and interactions of its components in viral RNA synthesis. J Virol. 1996;70:8675–8683. doi: 10.1128/jvi.70.12.8675-8683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faro-Trindade I, Cook PR. Transcription factories: Structures conserved during differentiation and evolution. Biochem Soc Trans. 2006;34:1133–1137. doi: 10.1042/BST0341133. [DOI] [PubMed] [Google Scholar]
- Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J Biol Chem. 2004;279:47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- Fujita K, Krishnakumar SS, Franco D, Paul AV, London E, Wimmer E. Membrane topography of the hydrophobic anchor sequence of poliovirus 3A and 3AB proteins and the functional effect of 3A/3AB membrane association upon RNA replication. Biochemistry. 2007;46:5185–5199. doi: 10.1021/bi6024758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever GN, Snyder L, Wang JC. DNA supercoiling in vivo. Biophys Chem. 1988;29:7–15. doi: 10.1016/0301-4622(88)87020-0. [DOI] [PubMed] [Google Scholar]
- Goregaoker SP, Culver JN. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J Virol. 2003;77:3549–3556. doi: 10.1128/JVI.77.6.3549-3556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JL, Long AM, Schultz SC. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- Hobson SD, Rosenblum ES, Richards OC, Richmond K, Kirkegaard K, Schultz SC. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 2001;20:1153–1163. doi: 10.1093/emboj/20.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope DA, Diamond SE, Kirkegaard K. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J Virol. 1997;71:9490–9498. doi: 10.1128/jvi.71.12.9490-9498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski SA, Morrow CD. Mutation of the aspartic acid residues of the GDD sequence motif of poliovirus RNA-dependent RNA polymerase results in enzymes with altered metal ion requirements for activity. J Virol. 1995;69:1532–1539. doi: 10.1128/jvi.69.3.1532-1539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens CK, Barton DJ, Sharma N, Morasco BJ, Ogram SA, Flanegan JB. 2Apro is a multifunctional protein that regulates the stability, translation, and replication of poliovirus RNA. Virology. 2006;345:346–357. doi: 10.1016/j.virol.2005.09.067. [DOI] [PubMed] [Google Scholar]
- Lama J, Paul AV, Harris KS, Wimmer E. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J Biol Chem. 1994;269:66–70. [PubMed] [Google Scholar]
- Lee JC, Hirsh J, Timasheff SN. In vitro formation of filaments from calf brain microtubule protein. Arch Biochem Biophys. 1975;168:726–729. doi: 10.1016/0003-9861(75)90308-2. [DOI] [PubMed] [Google Scholar]
- Lefebvre B, Formstecher P, Lefebvre P. Improvement of the gene splicing overlap (SOE) method. Biotechniques. 1995;19:186–188. [PubMed] [Google Scholar]
- Lyle JM, Bullitt E, Bienz K, Kirkegaard K. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science. 2002a;296:2218–2222. doi: 10.1126/science.1070585. [DOI] [PubMed] [Google Scholar]
- Lyle JM, Clewell A, Richmond K, Richards OC, Hope DA, Schultz SC, Kirkegaard K. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J Biol Chem. 2002b;277:16324–16331. doi: 10.1074/jbc.M112429200. [DOI] [PubMed] [Google Scholar]
- Mackenzie J. Wrapping things up about virus RNA replication. Traffic. 2005;6:967–977. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte LL, Wass AB, Gohara DW, Pathak HB, Arnold JJ, Filman DJ, Cameron CE, Hogle JM. Crystal structure of poliovirus 3CD protein: Virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol. 2007;81:3583–3596. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly EK, Paul JD, Kao CC. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J Virol. 1997;71:7526–7532. doi: 10.1128/jvi.71.10.7526-7532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortin J, Parra F. Structure and function of RNA replication. Annu Rev Microbiol. 2006;60:305–326. doi: 10.1146/annurev.micro.60.080805.142248. [DOI] [PubMed] [Google Scholar]
- Pata JD, Schultz SC, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- Pathak HB, Ghosh SK, Roberts AW, Sharma SD, Yoder JD, Arnold JJ, Gohara DW, Barton DJ, Paul AV, Cameron CE. Structure–function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J Biol Chem. 2002;277:31551–31562. doi: 10.1074/jbc.M204408200. [DOI] [PubMed] [Google Scholar]
- Pathak HB, Arnold JJ, Wiegand PN, Hargittai MR, Cameron CE. Picornavirus genome replication: Assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex. J Biol Chem. 2007;282:16202–16213. doi: 10.1074/jbc.M610608200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Luo H, Nomura T, Hayashi N, Yamashita T, Murakami S. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J Biol Chem. 2002;277:2132–2137. doi: 10.1074/jbc.M106880200. [DOI] [PubMed] [Google Scholar]
- Richards OC, Spagnolo JF, Lyle JM, Vleck SE, Kuchta RD, Kirkegaard K. Intramolecular and intermolecular uridylylation by poliovirus RNA-dependent RNA polymerase. J Virol. 2006;80:7405–7415. doi: 10.1128/JVI.02533-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Wells V, Plotch SJ, DeStefano JJ. Primer-dependent synthesis by poliovirus RNA-dependent RNA polymerase (3Dpol) Nucleic Acids Res. 2001;29:2715–2724. doi: 10.1093/nar/29.13.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Reitman ZJ, Zhao Y, Moustafa I, Wang Q, Arnold JJ, Pathak HB, Cameron CE. Picornavirus genome replication. Identification of the surface of the poliovirus (PV) 3C dimer that interacts with PV 3Dpol during VPg uridylylation and construction of a structural model for the PV 3C2-3Dpol complex. J Biol Chem. 2008;283:875–888. doi: 10.1074/jbc.M707907200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss DM, Wuttke DS. Characterization of protein-protein interactions critical for poliovirus replication: analysis of 3AB and VPg binding to the RNA-dependent RNA polymerase. J Virol. 2007;81:6369–6378. doi: 10.1128/JVI.02252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss DM, Glustrom LW, Wuttke DS. Toward an understanding of the poliovirus replication complex: The solution structure of the soluble domain of the poliovirus 3A protein. J Mol Biol. 2003;330:225–234. doi: 10.1016/s0022-2836(03)00577-1. [DOI] [PubMed] [Google Scholar]
- Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004;23:3462–3471. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AA, Albertini RA, Peersen OB. Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J Mol Biol. 2007;366:1459–1474. doi: 10.1016/j.jmb.2006.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Ho TV, Semler BL. Determinants of membrane association for poliovirus protein 3AB. J Biol Chem. 1996;271:26810–26818. doi: 10.1074/jbc.271.43.26810. [DOI] [PubMed] [Google Scholar]
- Wang QM, Hockman MA, Staschke K, Johnson RB, Case KA, Lu J, Parsons S, Zhang F, Rathnachalam R, Kirkegaard K, et al. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2002;76:3865–3872. doi: 10.1128/JVI.76.8.3865-3872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006;312:875–878. doi: 10.1126/science.1126766. [DOI] [PubMed] [Google Scholar]
- Xiang W, Cuconati A, Hope D, Kirkegaard K, Wimmer E. Complete protein linkage map of poliovirus P3 proteins: Interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J Virol. 1998;72:6732–6741. doi: 10.1128/jvi.72.8.6732-6741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeghouf M, Guibert B, Zeeh JC, Cherfils J. Arf, Sec7 and Brefeldin A: A model towards the therapeutic inhibition of guanine nucleotide-exchange factors. Biochem Soc Trans. 2005;33:1265–1268. doi: 10.1042/BST0331265. [DOI] [PubMed] [Google Scholar]