Abstract
MicroRNAs (miRNAs) play key roles in gene expression regulation by guiding Argonaute (AGO)-containing microribonucleoprotein (miRNP) effector complexes to target polynucleotides. There are still uncertainties about how miRNAs interact with mRNAs. Here we employed a biochemical approach to isolate AGO-containing miRNPs from human H4 tumor cells by co-immunoprecipitation (co-IP) with a previously described anti-AGO antibody. Co-immunoprecipitated (co-IPed) RNAs were subjected to downstream Affymetrix Human Gene 1.0 ST microarray analysis. During rigorous validation, the “RIP-Chip” assay identified target mRNAs specifically associated with AGO complexes. RIP-Chip was performed after transfecting brain-enriched miRNAs (miR-107, miR-124, miR-128, and miR-320) and nonphysiologic control miRNA to identify miRNA targets. As expected, the miRNA transfections altered the mRNA content of the miRNPs. Specific mRNA species recruited to miRNPs after miRNA transfections were moderately in agreement with computational target predictions. In addition to recruiting mRNA targets into miRNPs, miR-107 and to a lesser extent miR-128, but not miR-124 or miR-320, caused apparent exclusion of some mRNAs that are normally associated with miRNPs. MiR-107 and miR-128 transfections also result in decreased AGO mRNA and protein levels. However, AGO mRNAs were not recruited to miRNPs after either miR-107 or miR-128 transfection, confirming that miRNAs may alter gene expression without stable association between particular mRNAs and miRNPs. In summary, RIP-Chip assays constitute an optimized, validated, direct, and high-throughput biochemical assay that provides data about specific miRNA:mRNA interactions, as well as global patterns of regulation by miRNAs.
Keywords: brain, TXNIP, DBI, neurons, astrocytes
INTRODUCTION
MicroRNAs (miRNAs) are important regulators of gene expression in plants and animals (Ambros 2004; Liu et al. 2008). The mechanisms of miRNA action are incompletely understood. One function of miRNAs is to guide microribonucleoproteins (miRNPs) to target mRNAs for post-transcriptional gene expression regulation. As the name suggests, miRNPs are composed of RNA and proteins. Argonaute (AGO) proteins, which bind directly to mature miRNAs, are a central component of most mammalian miRNPs (Hammond 2005; Liu et al. 2008). AGO proteins help to coordinate the miRNA-related gene expression changes through direct and indirect mechanisms (Hutvagner and Simard 2008; Liu et al. 2008). Four paralogous AGO genes are expressed in humans (AGO 1-4, also known as EIF2C 1-4) (Carmell et al. 2002).
In combination with AGO proteins, a single miRNA species has the potential to target thousands of different mRNAs (Farh et al. 2005; Baek et al. 2008), and each mRNA may also be targeted by multiple miRNAs. It is a challenge to predict which miRNAs will bind to which individual mRNA targets, because most metazoan miRNAs bind to mRNA targets through partial sequence complementarity. Much has been learned about the principles that govern these interactions. Computational algorithms, based on prior experimental studies, can help to predict miRNA:mRNA binding (Enright et al. 2003; Krek et al. 2005; Lewis et al. 2005; Miranda et al. 2006). However, computational methods are imperfect and disparate from each other, and the biochemistry underlying miRNA:mRNA interactions still needs to be more fully elucidated. New high-throughput techniques are required to test current hypotheses and to move the field forward.
A promising method for characterizing miRNPs is co-immunoprecipitation (co-IP) that pulls down AGO proteins along with associated molecules (Mourelatos et al. 2002; Nelson et al. 2004). Using AGO co-IP assays, researchers have isolated multiple proteins, miRNAs, and mRNA targets from miRNPs (Beitzinger et al. 2007; Easow et al. 2007; Karginov et al. 2007; Andachi 2008; Hendrickson et al. 2008; Landthaler et al. 2008; Chi et al. 2009). AGO-associated RNA co-IP experiments with downstream application to high-throughput microarrays, which have been given the designation “RIP-Chip” (Keene et al. 2006), have high potential to study mRNA targeting.
Here we describe a RIP-Chip procedure that uses anti-AGO to co-IP mRNA targets. The monoclonal antibody (“anti-AGO”, which was also termed “2A8”) was raised against human AGO2, and recognizes a C-terminal epitope on AGO2 (Nelson et al. 2007). Multiple human and mouse AGO paralogs are also recognized by anti-AGO, both in co-IPs and on Western blots. Anti-AGO was also used to co-IP miRNAs from human brain, blood, and cultured cells (Nelson et al. 2007) and has also been used in previous co-IPs for mRNA targets (Chi et al. 2009). In the present study, RIP-Chip was applied to H4 glioneuronal cell line (Arnstein et al. 1974), which enabled us to evaluate how individual miRNA transfections can change the mRNA composition of miRNPs.
RESULTS
MiRNA transfections
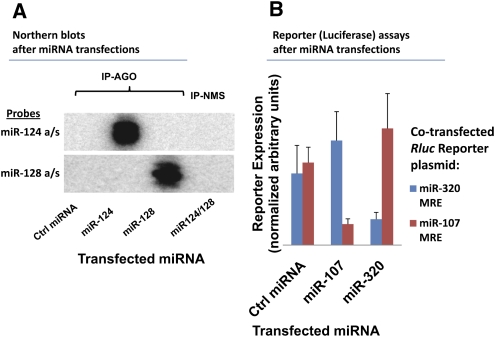
The effectiveness and specificity of miRNA transfections were evaluated using Northern blots and Renilla luciferase (Rluc) reporter assays (Fig. 1). Northern blots (Fig. 1A) were performed using RNA isolated from H4 glioneuronal cells 48 h after transfection with 25 nM of miRNAs. These experiments demonstrate that transfected miRNAs (miR-124 and miR-128) were incorporated appropriately into cells. To test whether the miRNAs were also biologically active, reporter assays were used (Fig. 1B). In these experiments, Rluc pRLTK plasmids with miRNA recognition sequences subcloned into the 3′ untranslated region (UTR) were co-transfected followed by miRNA transfection at 25 nM. Dual luciferase activity assays, including PGL-3 as internal control, were performed as previously described (Kiriakidou et al. 2004). In contrast to the negative control miRNA, specific knockdown of reporter expressions were clearly evident after miR-107 and miR-320 transfections.
FIGURE 1.
MicroRNA (miRNA) transfections are effective and lead to specific targeting by miRNAs. All transfections (25 nM) were performed in H4 glioneuronal cells and cells were harvested 48 h after transfection. (A) Northern blot analysis of miRNPs isolated after miRNA transfections with miR-124 and miR-128. The Northern blot analysis showed that miRNAs were specifically recruited to miRNPs after miRNA transfections. (B) Renilla luciferase (Rluc) reporter assays showed that miRNAs are functionally incorporated after miRNA transfections. MiR-107 and miR-320 specifically target reporter constructs bearing 3′-untranslated region recognition sequences for those miRNAs.
RIP-Chip assay
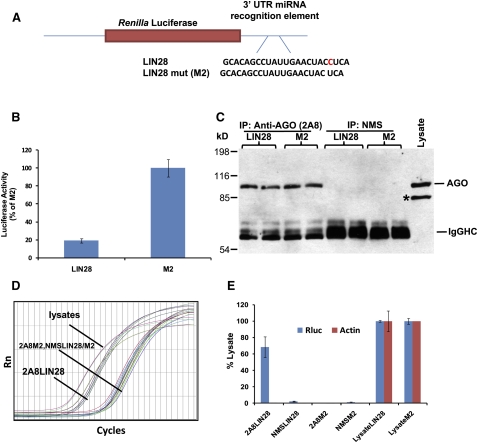
Anti-AGO antibody (Nelson et al. 2007; Kirino and Mourelatos 2008) was used to pull down endogenous AGO-containing miRNP complexes and their associated mRNAs (experimental design shown in Fig. 2). The protocol was first optimized using known positive (miRNA target) control and closely matched negative controls encoded by transfected Rluc pRLTK plasmids (Fig. 3). Reporter plasmids were designed to carry Let-7 miRNA recognition element (MRE) of LIN28 (designated as Rluc-LIN28) or its mutated sequence (Rluc-M2) in the 3′UTR (Fig. 3A). As expected, endogenous Let-7 miRNA inhibited the appropriate Rluc reporter expression (Rluc-LIN28), but not the closely related control reporter (Rluc-M2) (Fig. 3B).
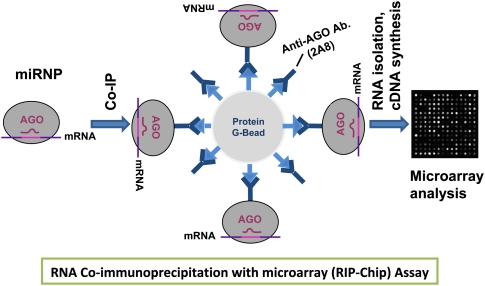
FIGURE 2.
Schematic illustration of biochemical identification of miRNA targets using RIP-Chip. miRNPs are co-IPed with anti-AGO (2A8) antibodies (Nelson et al. 2007) bound to protein G-agarose beads. RNAs that associated with AGO protein are processed for Affymetrix Gene 1.0 ST microarrays and this high-throughput assay can resolve miRNA targets.
FIGURE 3.
RIP-Chip was validated in H4 cells using positive miRNA target control and closely matched negative controls with transfected Renilla luciferase (Rluc) reporter plasmids. (A) Let-7 miRNA recognition element (MRE) of LIN28 or its closely related mutated sequence (designated “M2”) in the 3′-untranslated region (UTR) of the Rluc gene as previously described (Kiriakidou et al. 2004). (B) Dual luciferase activity assay shows that LIN28 reporter activity was decreased as expected by endogenous Let-7 miRNA. (C) Western blots analysis of co-IPed products using anti-AGO antibody. As expected, AGO proteins were co-IPed with anti-AGO (2A8), but not with nonimmunized mouse serum (NMS). Co-IPs were performed on cells transfected with reporter plasmid Rluc-LIN28 or Rluc-M2. An asterisk (*) shows a band that is Radixin, a protein recognized on Western blots, but not via co-IP by anti-AGO 2A8. (D) Although AGO protein was co-IPed regardless of the transfected plasmids, RT-qPCR detected Rluc mRNA only in the anti-AGO co-IP from the cell lysates transfected with Rluc-LIN28. (E) Quantification of qPCR shows that Rluc mRNA was highly enriched only in co-IPed RNAs from cells transfected with Rluc-LIN28, demonstrating a highly specific association of Rluc mRNA with AGO proteins via miRNA binding sites.
The co-IP assay was optimized and validated by examining the specific “pull down” of the Rluc-LIN-28 mRNA relative to Rluc-M2 mRNA. Results using the optimized conditions are shown (Fig. 3C–E). Western blots from co-IPs (Fig. 3C) show that AGO is specifically isolated with the anti-AGO antibody, but not with nonimmunized mouse serum (NMS). Rluc transcripts were detected by real-time quantitative polymerase chain reaction (RT-qPCR) using specific primers. Although the level of AGO proteins co-immunoprecipitated (co-IPed) with 2A8 antibody was the same from all the lysates, Rluc mRNA was highly enriched only in the 2A8 co-IP from the cell lysates transfected with Rluc-LIN28. In contrast, very low levels (less than 1%) of Rluc were associated with AGO co-IP in cells transfected with Rluc-M2 plasmid that carries a mutated Let7 MRE sequence in the 3′UTR. Co-IP with NMS did not result in any enrichment of Rluc mRNA from lysate of H4 cells transfected with either Rluc-LIN28 or Rluc-M2. Moreover, RT-qPCR detected negligible levels of β-actin mRNA from all co-IPs (Fig. 3D,E). These experiments revealed a highly specific association of Rluc mRNA with co-IPed AGO proteins via miRNA-mediated interactions.
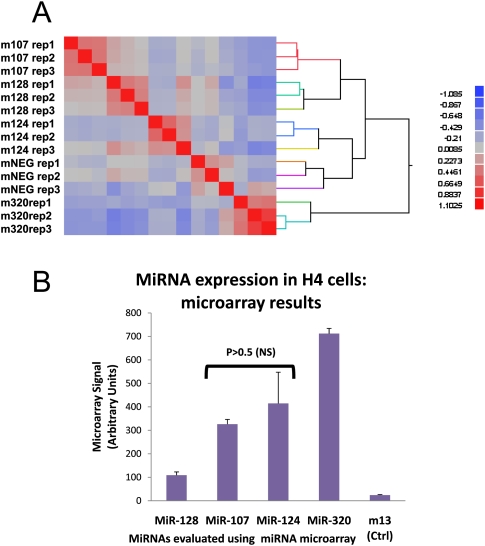
We next analyzed AGO-associated mRNA species by microarray hybridization. For these RIP-Chip experiments, RNAs from 2A8 or NMS co-IPs of H4 cells were isolated, processed, and applied to an Affymetrix 1.0 ST Gene microarray. Microarray results were obtained from H4 cells transfected with five different miRNAs: a nonphysiological negative control miRNA sequence and human miR-107, miR-124, miR-128, and miR-320. Figure 4A shows the results of 15 different biological replicates, three each from the five different miRNA transfections. A cluster analyses shows the ∼7500 most highly expressed genes on the arrays following AGO Co-IP in 15 different biological replicates. Note that all the individual members of the biological replicates cluster together, indicating that between-group variability was higher than within-group variability in these biological replicates.
FIGURE 4.
(A) Clustering analysis of RIP-Chip microarray expression data indicating that upstream miRNA transfections cause consistent changes in the mRNAs recruited into the miRNP. Anti-AGO co-IPs of H4 cells were processed for Affymetrix Gene 1.0 ST microarrays. This heat-map representation of the microarray expression profiling after transfection with various miRNAs and RIP-Chip comprises 15 different biological replicates. The abscissa refers to the same 15 biological replicates in the same order. The analysis indicates that the transfections produced consistent results with between-group variability exceeding within-group variability. (B) Endogenous expression levels of selected miRNAs in H4 cells. Levels of selected brain enriched miRNAs were evaluated as described previously (Wang et al. 2008b) by miRNA microarray analysis of H4 cell RNAs.
Figure 4B shows the results of miRNA microarray study of H4 glioneuronal cells showing relative expression level of miR-107, miR-124, miR-128, and miR-320 under normal culture conditions. These arrays were performed in triplicate on biological replicates exactly as previously described (Wang et al. 2008b). MiR-320 is highest expressed, miR-128 is lowest expressed, and the difference between miR-107 and miR-124 expression is not statistically significant in the cells.
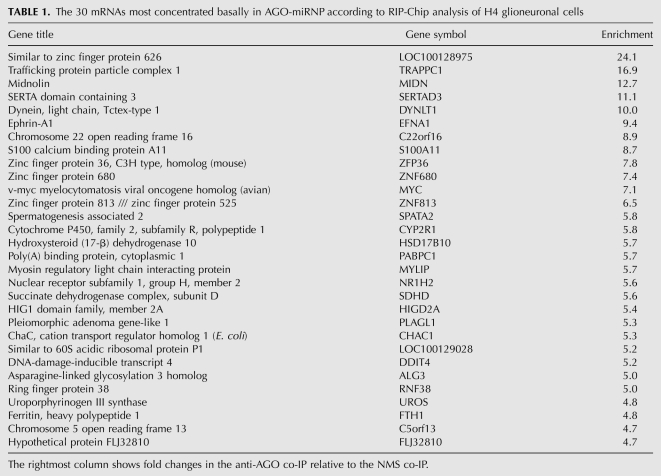
Table 1 shows the 30 mRNAs that had the highest relative enrichment in AGO-miRNP under normal culture conditions, as determined by the relative amount of those mRNAs in the anti-AGO RIP-Chip versus the NMS RIP-Chip. Additional experiments were performed to validate the microarray data. Separate AGO and NMS co-IPs were performed and the RNA again was isolated and this time gene-specific primers were used to assess the enrichment of specific targets that RIP-Chip had flagged. RT-qPCR on four randomly selected AGO-miRNP enriched mRNAs confirmed that they were substantially enriched in the anti-AGO co-IP relative to the NMS co-IP (Supplemental Fig. 1).
TABLE 1.
The 30 mRNAs most concentrated basally in AGO-miRNP according to RIP-Chip analysis of H4 glioneuronal cells
Identification of miRNA targets
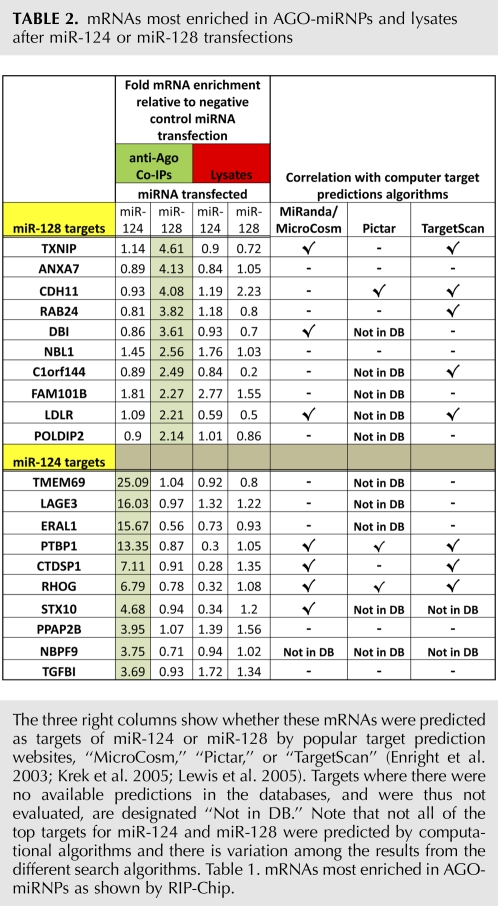
Following transfections with miR-107, miR-124, miR-128, and miR-320, RIP-Chip revealed enrichment of mRNA species that are presumably targets of corresponding miRNAs. Table 2 lists the top 10 targets for brain-enriched miRNAs, miR-124, and miR-128. See Supplemental Table 1 for more data; complete results for miR-107 and miR-320 will be published elsewhere. We compared the RIP-Chip data with computational target prediction programs “miRanda/MicroCosm,” TargetScan, and PicTar (Enright et al. 2003; Krek et al. 2005; Lewis et al. 2005) because these are the three databases that are directly linked to the Rfam miRNA Registry. There is variation among the different prediction algorithms, but in general, these predictions are moderately correlated to our RIP-Chip target identification in the H4 cells.
TABLE 2.
mRNAs most enriched in AGO-miRNPs and lysates after miR-124 or miR-128 transfections
We also analyzed total mRNA expression by microarray analysis using the cell lysates (sampled after cell lysis but prior to the co-IP procedure) used for RIP-Chip. We noted that many mRNA targets that were enriched in miRNPs were down-regulated in the cell lysates (Table 2). However, there are also some miRNP-enriched mRNAs that were not changed, or even up-regulated at mRNA levels. See Supplemental Table 1 for comprehensive microarray data from RIP-Chip and lysates following transfections with miR-124, miR-128, and a negative control miRNA.
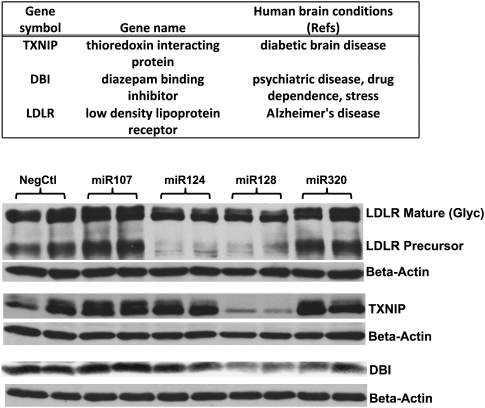
Validating RIP-Chip assay with human disease-relevant miR-128 targets
To test the validity of the RIP-Chip data, we evaluated the expression of a subset of predicted miR-128 targets following miRNA transfections. We chose a subset of miR-128 targets because three of the top 10 miR-128 targets have potential relevance to human neurological disease as shown (Fig. 5, top). The three miR-128 targeted proteins LDLR, TXNIP, and DBI (Fig. 5) were effectively knocked-down by miR-128 transfections. LDLR was also strongly reduced by miR-124 transfection. Note that microarray analysis of cell lysate RNA showed that LDLR mRNA was decreased following miR-124 transfection (Table 2). Several top targets of miR-124 identified by our RIP-Chip (e.g., PTBP1, CTDSP1, and RHOG) have been reported as top miR-124 targets by other laboratories (Karginov et al. 2007; Chi et al. 2009). These data further support the effectiveness of our RIP-Chip in identifying miRNA targets.
FIGURE 5.
Western blot analysis after miRNA transfections helped validate RIP-Chip results. H4 cells were first transfected with full-length cDNA plasmids: pCMV6-LDLR, pCMV6-TXNIP, and pCMV6-DBI for 24 h, and were then transfected with 25 nM of miR-107, miR-124, miR-128, miR-320 precursors, or a negative control miRNA for additional 48 h. Cells were harvested, and Western blot analyses were performed using antibodies against each expressing proteins. Anti-β-actin antibody was used to probe each blot to monitor the total protein loadings. The three miR-128 targeted proteins shown were effectively knocked down by miR-128 transfection. LDLR was also strongly reduced by miR-124 transfection. RIP-Chip did not identify LDLR as miR-124 target, however, microarray analysis of total cell RNA showed that LDLR mRNA was significant decreased upon miR-124 transfection.
MiR-107 and miR-128 transfections lead to shift of endogenous miRNA targets from the miRNP and down-regulation of AGO proteins
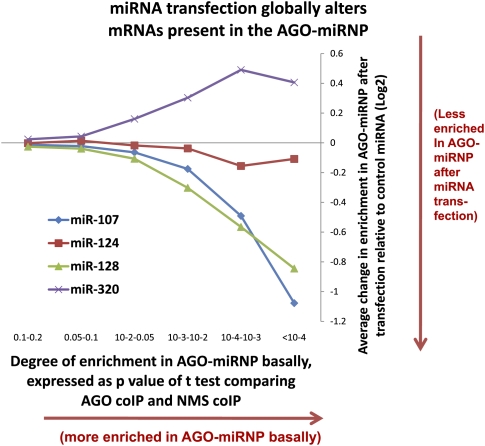
Transfections with individual miRNAs caused some mRNAs to be recruited into miRNPs—these were considered to be likely miRNA targets. By contrast, some miRNA transfections led to a decrease in the amount of mRNAs in the miRNP relative to transfection with a negative control miRNA. These changes differed according to the miRNA transfected. mRNAs that are basally enriched in miRNPs are disproportionately removed from those miRNPs following transfections by miR-107 and miR-128, but not miR-320 or miR-124 (Fig. 6). These data indicate that the presence of miR-107 or miR-128 appears to lessen the association of endogenous miRNPs with their targets.
FIGURE 6.
MiRNA transfections lead to systematic changes in the mRNAs that are incorporated into AGO-miRNPs as determined by RIP-Chip. The abscissa shows six different groups of mRNAs. Enrichment in the AGO-miRNP increases from left to right according to the P-value of t-tests comparing the mRNA amount in anti-AGO co-IP versus anti-NMS co-IP. mRNA groups were P = 0.1–0.2 (N = 3503 mRNAs), P = 0.05–0.1 (N = 1786), P = 0.01–0.05 (N = 1594), P = 0.001–0.01 (N = 531), P = 0.00001–0.0001 (N = 82), and P < 0.00001 (N = 10). Note that for the group with the greatest enrichment in the AGO-miRNPs at baseline (right-most data point), transfections with miR-107 and miR-128, but not miR-124 or miR-320, cause a decrease in these mRNAs in the AGO-miRNP.
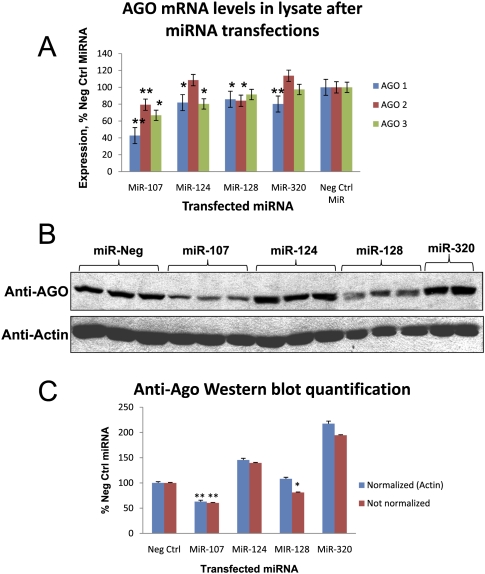
We sought to test how miR-107 and miR-128 decrease the recruitment of mRNAs that are normally enriched in the miRNP. One way this could happen is that miR-107 and miR-128 transfection could cause a decrease in AGO protein, which would decrease mRNA access to miRNPs globally. AGO expression was tested following miRNA transfections in H4 cells (Fig. 7). In microarray data of co-IPed for AGOs, there was no detectable enrichment of any AGO mRNA in miRNPs. However, AGO2 mRNA levels were significantly decreased in cell lysates following miR-107 and miR-128 transfection. AGO1 and AGO3 mRNA was also decreased (Fig. 7A) in miR-107 transfection. Note, only AGO1-3 are shown in Figure 7, since AGO4 is expressed at levels less than one-half of the AGO1-3 in H4 cells (data not shown). At the protein level (Fig. 7B,C), AGO proteins on Western blots following transfections with miR-107 and miR-128 were decreased compared to miR-124, miR-320, and a miRNA negative control transfection (Fig. 7B). This effect was stronger for miR-107 than miR-128. miR-128 did not show decreased AGO protein level on Western blot when β-actin was used as normalization control. The effect of miRNA transfection on AGO was similar in cell lines SH-SY5Y and HeLa (see Supplemental Fig. 2). Together, these data show that miR-107, and perhaps miR-128, target AGO proteins despite the AGO mRNAs not being enriched in the AGO-miRNP.
FIGURE 7.
MiR-107 and miR-128 transfections lead to reduced AGO expression. (A) Microarray analysis of the lysates prior to co-IP shows that different miRNA transfections differentially alter the level of AGO mRNAs. Note that miR-107 and miR-128 transfections cause AGO2 mRNAs to decrease. “*,” P < 0.05; “**,” P < 0.01; using Student's t-test, n = 3 replicates each. (B) Western blot analysis of AGO protein expression after H4 cells were transfected with 25 nM of various miRNAs. Total cell proteins were subjected to SDS-PAGE followed by Western blot analysis using anti-AGO antibody, or anti-β-actin antibody. (C) In a separate experiment, three biological replicates of miRNA transfections were performed for each miRNA and Western blot analysis using anti-AGO and anti-actin. AGO and β-actin protein levels of each individual band were quantified blindly on the Western blot and then normalized to levels of β-actin. Note that AGO protein was knocked down by miR-107 and miR-128. Although equal amounts of total protein were loaded in each well, β-actin levels seemed to be reduced in cells following miR-128 transfections.
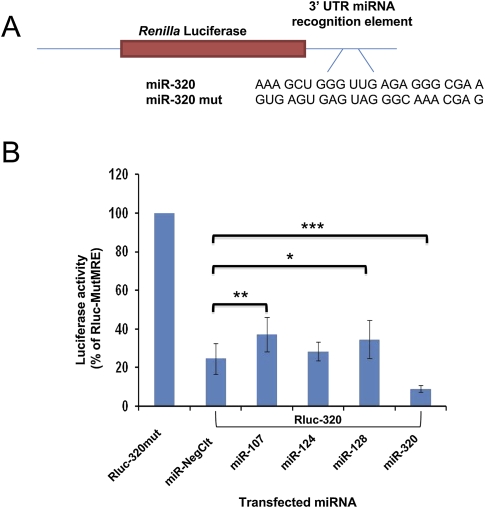
We next employed pRLTK-Rluc reporter system to examine more thoroughly the functional effects of miRNA transfections. H4 cells were first transfected with miRNAs (miR-107, miR-124, miR-128, and miR320, and a negative control miRNA), and the miRNA effects were evaluated by determining reporter activities. pRLTK-Rluc reporter constructs bearing a miR-320 MRE, or a mutated MRE sequence, in the 3′UTR of Rluc, were co-transfected with control plasmid pGL3 48 h after miRNA transfections (Fig. 8). Rluc activity for the negative control miRNA is considered to reflect endogenous miR-320 binding to the MRE with resulting inhibition of translation. Relative increases in Rluc for the miR-320 MRE plasmid indicates less inhibition of reporter gene expression by miR-320. Compared to other miRNA transfections, dual luciferase activity assays demonstrated that miR-107 and miR-128 transfections produced higher luciferase activities in the cells, meaning less inhibition through the miRNP. Hence, miRNA/miRNP function was suppressed by elevated levels of miR-107 and miR-128. Together, the pRLTK/Rluc results, Western blots, and RIP-Chip data suggest that miR-107 and miR-128 decrease the strength of endogenous miRNA activity in association with decreased AGO proteins (Fig. 9).
FIGURE 8.
Reporter assays are used to test how endogenous miRNA/miRNP function is affected by exogenous miRNA transfections. H4 cells were first transfected with various miRNA precursors (negative control, miR-107, miR-124, miR-128, and miR-320) for 48 h A. Reporter plasmid constructs pRL-TK-Rluc carrying miR-320 MRE (Rluc-320), or its mutated sequence (Rluc-320mut) were co-transfected with calibrating plasmid pGL3 for an additional 24 h B. Luciferase activities were determined as previously described (Wang et al. 2008a,b). Luciferase activities, normalized to PGL-3 values, from each transfection were compared to that of negative control miRNA which represents endogenous miR-320 activity. Represented are three independent experiments each with three replicates. Student's t-tests were performed on the data set. “*,” P < 0.05; “**,” P < 0.01; “***,” P < 0.001.
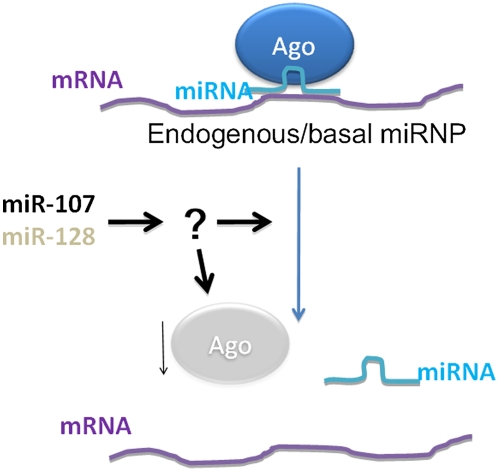
FIGURE 9.
Cartoon schematic to show that miR-107 and miR-128 transfections decrease the enrichment of endogenous mRNA targets in AGO-miRNPs. MiR-107 and miR-128 also lead to decreased miRNA/miRNP activity by reporter assays, correlated with decreased AGO mRNA and protein in the cells. However, since AGO mRNAs are not enriched in miRNPs, the mechanism for the down-regulation of AGO is as yet unknown.
DISCUSSION
Current assumptions about miRNA-related biochemistry must be evaluated critically, especially since the field is so new. Two major challenges in the field are predicting miRNA targets, and profiling the miRNAs that are expressed across different tissues and/or experimental conditions. There has been much progress in these areas. Experiments have included analyses of specific miRNA:mRNA interactions and also high-throughput analyses of downstream effects on mRNAs and proteins. However, there is still a need for more direct experimental techniques including high-throughput assays designed specifically to provide data about miRNA:mRNA interactions.
With regard to predicting miRNA targets, experimental results have focused on the 5′ “seed” region of miRNAs and the 3′UTR of mRNAs, whose complementarity is considered to be the fundamental sequence determinant of miRNA:mRNA binding (Kiriakidou et al. 2004; Baek et al. 2008). These assumptions have helped to guide computer algorithms for predicting miRNA targets. However, these methods frequently disagree with one another, and a number of studies have called the validity of current assumptions into question (Didiano and Hobert 2006; Rigoutsos 2009). Using high-throughput experimental techniques for characterizing miRNA targets, we can perhaps refine and expand the expectations for the “binding rules” that govern miRNA:mRNA interaction. In the current study, microarray analysis of mRNAs isolated from AGO complexes identified thousands of mRNAs associated with AGO-miRNPs. The overall correlations between our results and the computer algorithms suggest that sequence determinants outside of the 5′ seed of miRNAs, and the 3′UTR of mRNAs, should also be considered.
For tissue-level RNA profiling, a common assumption used in normalizing mRNA expression analyses is that the “global expression” levels hold constant. Whether or not this assumption is actually true in mRNA studies, it almost certainly is not valid for miRNA studies, since global miRNA expression appears to be relatively labile. For example, most (Kumar et al. 2007; Sun et al. 2009), but not all (Chiosea et al. 2006), studies show that cancer cells and cancer cell lines have relatively low global expression of miRNAs. Furthermore, prior studies have shown that intracellular signaling mechanisms can have widespread global impact on miRNA processing machinery (Lugli et al. 2005; Adams et al. 2009). Our results are also compatible with a recent study analyzing data from miRNA transfections, which showed that transfecting miRNAs affects the levels of mRNAs predicted to be under endogenous miRNA regulation (Khan et al. 2009). In the current study, transfections with miR-107 and miR-128, but not miR-124, or miR-320 are correlated with decreased AGO mRNA and protein levels. This finding correlates with an altered global pattern of target mRNAs associated with AGO-containing miRNPs and also functional decrease in miRNA/miRNP activity as shown by reporter assays.
Aside from the theoretical implications, specific data about individual miRNA targets were obtained. miR-124 has been studied in a number of contexts because of its extraordinary enrichment in metazoan nervous systems through more than 500 million years of evolution (Peterson et al. 2009). RIP-Chip target identification data for miR-124 generally agree with a prior study in which miR-124 targets were identified in a c-myc-tagged AGO2-293S cell line (Karginov et al. 2007). Three of the top 10 miR-124 targets were identified in both studies. This confirms that co-IP assays are effective in identifying miRNA targets. In our experience, however, tagged AGO constructs are less effective than anti-AGO in pulling down miRNA or mRNA targets (unpublished observations). Our data are also in agreement with prior research on the impact of miR-124 transfections on the transcriptome as assessed by microarrays (Lim et al. 2005; Karginov et al. 2007). RIP-Chip provides an important new layer of information in target identification. Specifically, RIP-Chip can distinguish primary (AGO-miRNP-related) versus secondary changes. For example, RIP-Chip shows that miR-124 targets PTBP1 (polypyrimidine tract binding protein 1) mRNA as previously described (Makeyev et al. 2007). PTBP1 is a protein that affects RNA splicing and the decrease in PTBP1 protein has been shown to down-regulate mRNA levels of PTBP2 (Makeyev et al. 2007). Compatible with that hypothesis, PTBP2 mRNA levels are significantly down-regulated in miR-124 transfected cells, although PTBP2 is not associated with the AGO-miRNP. We observe literally thousands of additional examples of mRNA changes that might be flagged errantly using methods depending on mRNA levels or protein levels alone to indicate miRNA targeting (see Supplemental Table 1). RIP-Chip also confirms a prior report that miR-124 specifically targets SLC16A1 (Li et al. 2009) and leads to recruitment of this mRNA into the AGO-miRNP. By contrast, the level of CDK6 mRNA, which has been shown to be negatively regulated by miR-124 (Pierson et al. 2008), is indeed decreased considerably in the lysate of miR-124 treated cells, but is not apparently incorporated in the AGO-miRNP (see Supplemental Material). This regulation may be another example of “downstream” miR-124 related mechanisms.
miR-128 is another miRNA strongly concentrated in the brain relative to other organs (Lagos-Quintana et al. 2002; Baskerville and Bartel 2005) and enriched in cultured primary rat neurons relative to primary rat astrocytes (Smirnova et al. 2005; Wang et al. 2008b). Interestingly, there is an increase in miR-128 expression in the Alzheimer's disease hippocampus relative to control hippocampus (Lukiw 2007). Prior studies have provided evidence that miR-128 targets E2F3A, BMI-1, NTRK3, and SNAP25 mRNAs (Eletto et al. 2008; Godlewski et al. 2008; Muinos-Gimeno et al. 2009; Zhang et al. 2009). Our results do not provide direct confirmation for those interactions in these cultured cells; however, most of these genes are expressed only at low levels in cultured H4 cells. Although miR-128 transfection decreased the association of many mRNAs with the AGO-miRNP, there were some apparent miR-128 targets. mRNAs that were incorporated consistently into the miRNP after miR-128 transfections included intriguing transcripts, such as TXNIP (thioredoxin interacting protein), DBI (diazepam binding inhibitor, or acyl-Coenzyme A binding protein), and LDLR (low density lipoprotein receptor), each of which may be relevant to specific human diseases (see, for example, Ohkuma et al. 2001; Yamawaki et al. 2003; Gopalraj et al. 2005).
There are some limitations to the current study. Our anti-AGO antibody recognizes a C-terminal epitope of AGO2 (Nelson et al. 2007). AGO-miRNPs have been shown to exert gene expression regulation via multiple mechanisms (Liu et al. 2008). It is posible that the RIP-Chip approach, which requires a target mRNA to be stably bound to the miRNP, only “captures” a subset of the targets according to the specific targeting mechanism. For example, when AGO2 works as a “slicer” to enzymatically cleave target mRNAs (Liu et al. 2004), one would expect a much less stable miRNA:mRNA association and hence a far lower yield of mRNA targets via RIP-Chip.
There were important differences in washing procedures, cells and tissues evaluated, and other methodology between our RIP-Chip method in comparison to experiments using the recently published “HITS-CLIP” method (Chi et al. 2009). In HITS-CLIP experiments the same antibody (Nelson et al. 2007) was used for co-IP experiments with downstream biochemical crosslinking and microarray analyses (Chi et al. 2009). Future experiments can help to understand the biochemical significance of the large overlap and the smaller discrepancies in the data from these two methods.
It is important to note that the shifts in the mRNA targets in the miRNPs probably are not only due to changes such as the AGO protein down-regulation following miR-107 and miR-128 transfections. The changes in RNAs in the miRNP are probably also partly due to competition between the transfected miRNAs and endogenous ones. However, given the miRNA controls we used, that competition cannot explain why some miRNAs, but not others, appear to alter the miRNA targeting at the global level.
Aside from an inability to pinpoint specific mechanisms of mRNA expression changes, these data derive mostly from only a single cancer cell line, H4 cells. H4 cells were isolated from a brain tumor (Arnstein et al. 1974) and express a miRNA profile that more closely resembles cultured astrocytes than cultured neurons (Wang et al. 2008b). These cells have various genetic anomalies (Krex et al. 2001). Also, as with any type of cell, there are presumably many genes that are not expressed in H4 cells that we cannot evaluate for miRNA binding. We used a single cell line in order to rigorously validate microarray studies across many different conditions, including different antibodies, lysates, and different transfections. Different cell lines also were used to show AGO knockdown after miR-107 transfection. Moreover, data from dozens of different microarrays, qPCR, and Western blots, indicate that H4 cells have robust human AGO and miRNA functionality. However, in the future we will perform additional experiments using other cells and tissues to further validate our results.
MATERIALS AND METHODS
Plasmids and antibodies
Full-length cDNA (all UTR-containing) cloned in pCMV6-XL5 plasmid vectors were obtained that express human low density lipoprotein receptor (LDLR, NM_000527.2), thioredoxin interacting protein (TXNIP, NM_006472), and diazepam binding inhibitor (DBI, NM_020548.4) (OriGene Technologies, Inc.). Antibodies used in this work are: Monoclonal anti-AGO (2A8) (Nelson et al. 2007); Rabbit anti-human LDLR antibody (JM-3839-100) and TXINP monoclonal antibody (K0205-3) from MBL; mouse anti-human DBI (MAB0725) from Abnova; Rabbit anti-β-actin antibody (600401886) was purchased from Rockland.
Transfections and dual luciferase reporter assays
H4 cells, Hela cells, and SH-SY5Y cells (American Type Culture Collection), cultured under the vendor's recommended conditions, were plated in 10-cm culture plates at a density of 2.5 × 106/plate day before transfections. Cells were transfected with 25 nM of “Pre-miRNA” (Ambion) referent to hsa-miR-107, hsa-miR-320, hsa-miR-128, hsa-miR-124, and the Negative Control miRNA #1 (sense sequence AGUACUGCUUACGAUACGG) using siRMAX (Invitrogen) according to the manufacturer's instructions. H4 cell transfections and Luciferase reporter activity analyses were performed following the protocol described previously (Wang et al. 2008a). For validation of RIP-Chip, H4 cells plated as above were transfected with 2.5 μg of plasmid pRL-TK (Promega) carrying subcloned hsa-Let-7 MRE of LIN28 (LIN28), or its mutated sequence (M2) in 3′UTR of the Rluc gene as described previously (Kiriakidou et al. 2004; Nelson et al. 2004). Cells were harvested 48 h after transfection. For the miR-320 MRE reporter assays, three different experiments were run with N = 3 each, and the highest and lowest of the nine values were excluded. Student's two-tailed t-tests were performed. Transfection for target validation: Cells were first transfected with 100 ng of LDLR, TXNIP, and DBI plasmids for 24 h, followed by transfection with 25 nM of Pre-miRNAs (negative control miRNA, miR-107, miR-124, miR-128, and miR-320) for an additional 48 h.
Co-IP of miRNPs with anti-AGO antibodies
Protein G agarose beads (Invitrogen) were rinsed four times with PBS, and incubated with monoclonal anti-AGO (Nelson et al. 2007), or NMS (Pierce Biotechnology), or without any IgG in PBS for at least 30 min. After the initial binding, antibody-Protein G beads were blocked with 0.5 mg/mL yeast RNA and 1 mg/mL BSA (Sigma) for an additional 30 min or more. Beads were then washed three times in PBS to remove the unbound IgGs, and washed twice in lysis buffer (25 mM Tris-HCl at pH 8.0, 150 mM NaCl, 2 mM MgCl2, 0.5% NP-40, and 5 mM DTT). Some time (48 h) after the final transfection(s), the cells were first rinsed twice in PBS, and were then lysed on ice for 10 min in a fresh lysis buffer with protease inhibitors (1 tablet/10 mL lysis buffer, Complete Protease Inhibitor Cocktail Tablets, EDTA-free, Roche Applied Science) and RNasin (250 U/mL; Promega). The cell lysates were centrifuged at 10,000g at 4°C for 10 min, and the supernatants saved. The lysates were subjected to preclearance by incubation with pre-blocked Protein G beads at 4°C for 60 min. An aliquot of lysate after preclearance, but before any co-IP, was removed for total RNA and protein analysis. The remaining lysates proceeded to co-IP with either 2A8-Protein G beads or NMS-Protein G beads at 4°C for 90 min. After co-IP, the beads were washed as follows: Twice with lysis buffer; three times with lysis buffer containing 900 mM NaCl and 1% NP-40; twice more with lysis buffer, and then the beads were transferred to a fresh tube and there was one final wash with lysis buffer containing 0.05% NP-40. Following the washes, beads were subjected to DNase treatment by incubating with 250 μL of DNA digestion solution containing 40 mM Tris-HCl at pH 8.0, 10 mM MgSO4, 1 mM CaCl2, 200 U/mL RNasin, and 0.04 U/mL DNase I (Promega). DNase I treatment was carried out at 37°C for 20 min with gentle shaking. An aliquot of beads was removed from each sample and mixed with 2X Laemmli sample buffer for Western blot analysis. RNAs that co-immunoprecipitated with anti-AGO antibodies were extracted using TRIzol LS (Invitrogen) as described in a previous publication (Wang et al. 2008a). Total RNA from cell lysates was isolated using the same procedure, and was also subjected to DNA digestion as described above.
Microarray analysis and RT-qPCR
Microarray analyses of RNAs isolated from co-IP or from total cell lysates were performed using Affymetrix Human Gene 1.0 ST chip at University of Kentucky Microarray Core Facility. At least three biological replicates from at least two individual experiments were carried out in each treatment. For qPCR analysis, RNAs were first reverse-transcribed with qScript cDNA SuperMix (Quanta Biosciences), amplified with specific gene primers (Supplemental Table 1) by TaqMan 2X PCR Master Mix (Applied Biosystems) and SYBR as detector. qPCR was performed using ABI Prism 7000 Sequence Detection System (Applied Biosystems). Each data point represents the results of three technical replicates. Primers were as follows:
pRLTK, forward: TGTATATCCAAGTCACTCAGGAAC;
pRLTK, reverse: GGGAAGGGAAGAGTCAGCTTGTG;
β-actin, forward: GCGAGCATCCCCCAAAGTTCACA; and
β-actin, reverse: GGGCACGAAGGCTCATCATTCA.
Northern blot and Western blot analyses
Northern blot and Western blot analyses were performed as described previously (Wang et al. 2008a).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Ms. Willa Huang for technical and collegial assistance in the project, and Dr. Zissimos Mourelatos for advice and insights. This study was supported by grants R01 NS061933 and K08 NS050110 from the National Institutes of Health, Bethesda, MD.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1905910.
REFERENCES
- Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Andachi Y. A novel biochemical method to identify target genes of individual microRNAs: Identification of a new Caenorhabditis elegans let-7 target. RNA. 2008;14:2440–2451. doi: 10.1261/rna.1139508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnstein P, Taylor DO, Nelson-Rees WA, Huebner RJ, Lennette EH. Propagation of human tumors in antithymocyte serum-treated mice. J Natl Cancer Inst. 1974;52:71–84. doi: 10.1093/jnci/52.1.71. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the microRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA–target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletto D, Russo G, Passiatore G, Del Valle L, Giordano A, Khalili K, Gualco E, Peruzzi F. Inhibition of SNAP25 expression by HIV-1 Tat involves the activity of mir-128a. J Cell Physiol. 2008;216:764–770. doi: 10.1002/jcp.21452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- Gopalraj RK, Zhu H, Kelly JF, Mendiondo M, Pulliam JF, Bennett DA, Estus S. Genetic association of low density lipoprotein receptor and Alzheimer's disease. Neurobiol Aging. 2005;26:1–7. doi: 10.1016/j.neurobiolaging.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Hammond SM. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS One. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes & Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. Site-specific crosslinking of human microRNPs to RNA targets. RNA. 2008;14:2254–2259. doi: 10.1261/rna.1133808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krex D, Mohr B, Hauses M, Ehninger G, Schackert HK, Schackert G. Identification of uncommon chromosomal aberrations in the neuroglioma cell line H4 by spectral karyotyping. J Neurooncol. 2001;52:119–128. doi: 10.1023/a:1010680920087. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li KK, Pang JC, Ching AK, Wong CK, Kong X, Wang Y, Zhou L, Chen Z, Ng HK. miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum Pathol. 2009;40:1234–1243. doi: 10.1016/j.humpath.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu X, Fortin K, Mourelatos Z. MicroRNAs: Biogenesis and molecular functions. Brain Pathol. 2008;18:113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muinos-Gimeno M, Guidi M, Kagerbauer B, Martin-Santos R, Navines R, Alonso P, Menchon JM, Gratacos M, Estivill X, Espinosa-Parrilla Y. Allele variants in functional microRNA target sites of the neurotrophin-3 receptor gene (NTRK3) as susceptibility factors for anxiety disorders. Hum Mutat. 2009;30:1062–1071. doi: 10.1002/humu.21005. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, De Planell-Saguer M, Lamprinaki S, Kiriakidou M, Zhang P, O'Doherty U, Mourelatos Z. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuma S, Katsura M, Tsujimura A. Alterations in cerebral diazepam binding inhibitor expression in drug dependence: A possible biochemical alteration common to drug dependence. Life Sci. 2001;68:1215–1222. doi: 10.1016/s0024-3205(00)01031-6. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: Insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol. 2008;90:1–7. doi: 10.1007/s11060-008-9624-3. [DOI] [PubMed] [Google Scholar]
- Rigoutsos I. New tricks for animal microRNAS: Targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–3248. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Sun R, Fu X, Li Y, Xie Y, Mao Y. Global gene expression analysis reveals reduced abundance of putative microRNA targets in human prostate tumours. BMC Genomics. 2009;10:93. doi: 10.1186/1471-2164-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008a;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Baldwin DA, Isett RB, Ren N, Stromberg A, Nelson PT. Focus on RNA isolation: Obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008b;1779:749–757. doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki H, Haendeler J, Berk BC. Thioredoxin: A key regulator of cardiovascular homeostasis. Circ Res. 2003;93:1029–1033. doi: 10.1161/01.RES.0000102869.39150.23. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, Gong Y, Yin B, Qiang B, Zhao J, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med. 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]