Abstract
The F1-V vaccine antigen, protective against Yersinia pestis, exhibits a strong tendency to multimerize that affects larger-scale manufacture and characterization. In this work, the sole F1-V cysteine was replaced with serine by site-directed mutagenesis for characterization of F1-V non-covalent multimer interactions and protective potency without participation by disulfide-linkages. F1-V and F1-VC424S proteins were over-expressed in Escherichia coli, recovered using mechanical lysis/pH-modulation and purified from urea-solubilized soft inclusion bodies, using successive ion-exchange, ceramic hydroxyapatite, and size-exclusion chromatography. This purification method resulted in up to 2 mg per gram of cell paste of 95% pure, mono-disperse protein having ≤ 0.5 endotoxin units per mg by a kinetic chromogenic limulus amoebocyte lysate reactivity assay. Both F1-V and F1-VC424S were monomeric at pH 10.0 and progressively self-associated as pH conditions decreased to pH 6.0. Solution additives were screened for their ability to inhibit F1-V self-association at pH 6.5. An L-arginine buffer provided the greatest stabilizing effect. Conversion to >500-kDa multimers occurred between pH 6.0 and 5.0. Conditions for efficient F1-V adsorption to the cGMP-compatible Alhydrogel® adjuvant were optimized. Side-by-side evaluation for protective potency against subcutaneous plague infection in mice was conducted for F1-VC424S monomer; cysteine-capped F1-V monomer; cysteine-capped F1-V multimer; and a F1-V standard reported previously. After a two-dose vaccination with 2 × 20 µg of F1-V, respectively, 100, 80, 80, and 70% of injected mice survived a subcutaneous lethal plague challenge with 108 LD50 Y. pestis CO92. Thus, vaccination with F1-V monomer and multimeric forms resulted in significant, and essentially equivalent, protection.
INTRODUCTION
The potential use of plague as a biological weapon has necessitated the continued development of effective prophylaxis [1]. A previously licensed human plague vaccine (Plague Vaccine USP), consisting of killed whole-cell Yersinia pestis, protected against plague infection acquired subcutaneously [2]. However, this whole-cell vaccine was later shown to be ineffective against aerosol challenge and to be poorly protective against a virulent strain lacking capsule [3–4]. In an effort to produce a more efficacious vaccine, Heath et al. developed a recombinant vaccine composed of a fusion protein of the Fraction 1 capsular antigen (F1, Caf1) with a second protective immunogen called the V-antigen (LcrV) [5]. F1-V was originally purified using a polyhistidine tag and the his(10)-F1-V vaccine protected experimental mice against pneumonic as well as bubonic plague produced by either F1+ or F1− strains of Y. pestis [5]. By a statistical comparison of potency [6], the recombinant fusion-protein vaccine provided far better protection against the wild-type (F1+) strain than did the former Plague Vaccine USP, and it also showed a significant improvement in protection over a cocktail vaccine composed of the separate F1 and V antigens, as was first indicated after its creation [4,6].
Based on the success of animal protection studies and with the intent to improve the fusion protein for product development, F1-V was subsequently re-engineered to remove the poly-histidine tag and placed under transcriptional control of the IPTG-inducible pET-24a expression system (plasmid pPW731) in Escherichia coli strain BL21 (DE3), and then purified from soft inclusion bodies using 6M urea and a two-column procedure including anion-exchange and hydrophobic interaction chromatography [6]. The untagged fusion protein showed equivalent immunogenicity and protective efficacy, and was less polydisperse in molecular structure than the individual F1 subcomponent, but still showed a tendency to aggregate under certain conditions. The tendency of F1-V to self-associate was revealed by analytical size exclusion chromatography (HPLC-SEC) coupled to multiple angle laser light scattering (called SEC-MALLS in combination), which clearly showed mixtures of monomer, dimer, and multimeric species of higher mass in all standard preparations of F1-V [6]. The prior study did not eliminate a possible contribution to aggregation or stability by the single cysteine in F1-V, which provided an un-paired, solution-accessible free-thiol.
During production, F1-V characteristically formed loose inclusion bodies – insoluble collections of protein – as expressed at high levels in E. coli [6–8]. Subsequently, inclusion body dispersal and re-association of F1-V by on-column refolding embodied a substantial effort for downstream processing, and solution-state heterogeneity (i.e., monomer, self-dimer, and self-multimer forms) persisted throughout chromatographic isolation of the target species. Thus, the prior technology presented risks for large-scale manufacture including: 1) possible entrapment of contaminants within multimeric forms, which may lower process yields and increase process costs to achieve purity; and 2) uncontrolled or premature re-folding that may affect fusion-protein structure and thereby impact product consistency and long-term stability [9].
With a view toward developing current good manufacturing practices-compliant F1-V manufacture, wherein final product purity and target protein structural definition are crucial for regulatory approval, we aimed to devise a robust process for recovering monomeric F1-V preparations that contained minimal self- and hetero-protein associated forms. We further aimed to address any uncertainty as to the comparative level of plague protection achievable using monomeric versus multimeric F1-V preparations. Concerns regarding monomeric F1-V plague vaccine efficacy are based upon prior haptan reports, where monodisperse antigens induced weaker immune responses than did protein assemblies [10], and the disease context in which F1 subunits are encountered as multimeric fiber structures [11–12].
In support of these central objectives, we report herein: 1) the development of a new purification scheme for isolation of monomeric F1-V under reducing conditions (designated ‘F1-VMN’); 2) the use of site-directed mutagenesis to substitute the sole cysteine in F1-V (C424) with serine (designated ‘F1-VC424S-MN’), to prevent disulfide dimer formation and to eliminate in-process reducing agents, oxygen exclusion, and reducing agent clearance; 3) recovery and purification of monomeric F1-VC424S-MN under atmospheric oxygen conditions; 4) characterization of the resulting F1-VMN and F1-VC424S, MN preparation solution states with respect to pH and stabilizing additives; 5) conversion of the F1-VMN form to multimeric form (designated ‘F1-VAG’ under controlled, low pH conditions; and 6) assessment of vaccine protective efficacy against subcutaneous plague infection provided by an Alhydrogel-adsorbed, two-dose vaccination with F1-VC424S-MN, F1-VMN, and F1-VAG forms compared to the previously reported standard F1-V preparation (designated F1-VSTD’) [6]. We report that vaccination with all F1-V forms tested resulted in significant, and essentially equivalent, protection against up to 108 LD50 of wild-type Y. pestis.
MATERIALS AND METHODS
Bacterial strain, plasmid construction, cultivation, and induction
For F1-VMN the E. coli strain BLR130 and the F1-V expression vector, pPW731 (USAMRIID), controlled under a T7 promoter, were used for F1-V expression [6]. A growth medium of soytone, yeast extract, and glucose (J.T. Baker, Phillipsburg, NJ) and the antibiotics kanamycin (30 mg/L) and tetracycline (15 mg/L; Sigma, St. Louis, MO) were used in phosphate buffer, pH ∼7.3. Sterile medium in shaker flasks (300 ml) was inoculated with 1 ml of the strain from a previously made glycerol stock and incubated for ∼13 h at 37 °C with shaking at 220 rpm. Batch cultivations were carried out in a Bioflo 4500 (New England Biolabs, Ipswich, MA) equipped with a 15-L vessel and 10-L working volume. Growth medium (9.7 L) was inoculated with 300 ml of seed culture. The dissolved oxygen concentration was maintained above 15% air saturation at 37 °C by controlling the aeration and agitation rates through BIOCOMMAND software (New England Biolabs). Solution pH was kept between 7.2 and 7.4 by adding 0.1 N HCl or 30% NH4OH. After 3.5 h, the culture was induced with IPTG (1 mM) and harvested 2 h later by centrifugation. Cell paste aliquots were stored below –70 °C.
For F1-VC425S, TOP10, BL21 (DE3), and BL21 Star (DE3) E. coli strains were from Invitrogen (Carlsbad, CA). BL21 Star cells carried a mutated rne gene that encoded a truncated RNase E protein lacking the capacity to degrade mRNA and leading to increased mRNA stability and enhanced protein expression. The F1-V pET24a(+) Cys425→Ser425 expression plasmid (F1-VC424S) was prepared by site-directed mutagenesis of the original cysteine-containing caf1-lcrV gene fusion (expressing F1-VSTD) on source plasmid F1-VSTD pET-24a (pPW731) [6]. Site-directed mutagenesis was performed with the Quick-change site-directed mutagenesis kit (Stratagene, La Jolla, CA). Complementary mutagenic primers F1-V-CS-F (5′ - CT CAC TTT GCC ACC ACC TCC TCG GAT AAG TCC AGG CCG C - 3′) and F1-V-CS-R (5′ – GCG GCC TGG ACT TAT CCG AGG AGG TGG TGG CAA AGT GAG – 3′) were constructed with consideration of primer length (39 bp), % GC (59%), and melting temperature (85.8 °C). Each primer (125 ng) was combined with 50 ng of F1-V pET-24a (pPW731), along with the additional reaction chemistry as recommended by the manufacturer. Cycling parameters for the mutagenesis reaction included one cycle of 95 °C for 30 sec, followed by 12 cycles of 95 °C melting for 30 sec, 53 °C annealing for 1 min, and 68 °C extension for 7 min, concluding with a 4 °C hold. The mutagenesis reaction was then digested with 1 µl of DpnI at 37 °C for 1 h. The DpnI-digested mutagenesis reaction (1 µL) was used to transform chemically competent TOP10 E. coli, and the transformed cells were grown on LB plates containing 50 µg/ml kanamycin. Positive clones were verified by bidirectional DNA sequence analysis on an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA). The F1-VC424S vector was transformed into BL21 Star cells for protein expression under control of the isopropyl-β-D-thiogalacto-pyranoside (IPTG)-inducible T7 promoter. F1-VC424S-BL21 Star E. coli starter cultures were grown overnight in four 4-L shaker-flasks filled with a total of 10-L LB medium at 37 °C, and 250-rpm shaking in the presence of 50 mg/L of kanamycin. Starter cultures were then diluted 1:10 in fresh kanamycin-supplemented LB medium and grown at 37 °C, 250 rpm to an OD600 of 0.5 – 0.8. Protein expression was induced by adding IPTG (0.5 mM). After 3 h at 37 °C with 250-rpm shaking, cells pellets were collected by centrifugation at 10,000 × g for 20 min and stored at –70 °C.
Recovery of F1-VMN
For F1-VMN, combined wet cell paste from two fermentations was re-suspended to 40% w/v with 1.2 L of lysis buffer (50 mM Tris, 50 mM EDTA, 20 mM DTE, pH 9.0), sheared for 10 min with a HAAKE A82 (Thermo-Electron, Waltham, MA), and homogenized by three passages at 12,000 psi through a NS1001-L2K mechanical homogenizer (Niro-Soavi, S.p.A., Parma, Italy). The homogenizer was fitted with a chilled reservoir and cooling coil that was kept below 11 °C. The homogenized paste was adjusted to pH 8.3 +/- 0.2, clarified by centrifugation for 1 h at 10,000 RPM in a JA-10 rotor at 4 °C (Beckman Coulter, Fullerton, CA), and the supernatant was collected. F1-V was precipitated by a slow, well-mixed adjustment of the supernatant to pH 4.8 with 1 M acetic acid (pH 2.25). An off-white, granular pellet, enriched in F1-V, was collected by centrifugation for 1 h. The pellet was washed in an equal volume of 5 mM citric acid, pH 4.8 and then centrifuged, washed again, and the resulting pellet stored below –70 °C. The washed pellet was re-suspended in 2.5 volumes (∼1 L) of solubilization buffer (10 mM Tris, 10 mM ethanolamine, 5 mM L-cysteine, 50 mM EDTA, pH 9.0) and mixed to disperse the pellet at 20 °C for 20 min. The solution was adjusted to pH 11.0 by a slow, drop-wise addition of 10 N NaOH and held for 5 min at 20 °C, then adjusted to pH 8.3 with vigorous mixing and slow addition of 1 M acetic acid. The F1-V-enriched supernatant was separated from a lower density, colorless precipitate, enriched in contaminants (notably 40 kDa E. coli membrane protein I, identified by N-terminal sequencing) by centrifugation. The supernatant was re-precipitated by slow adjustment to pH 4.8 and the F1-V enriched pellet was stored below –70 °C.
For F1-VC424S-MN, cell paste was re-suspended to 20% w/v with 50 mM Tris, 50 mM EDTA, pH 9.0, (without reducing agents) and homogenized by three passages through an EmulsiFlex-C5 MicroFluidizer (Avestin, Canada) at a backpressure of 10,000 to 15,000 psi. The homogenized paste was clarified by centrifugation for 35 min at 15,000 rpm in an SS-34 rotor at 4 °C (Beckman Coulter, Fullerton, CA). Using methods similar to those used for F1-V, except without the addition of reducing agents, F1-VC424S was recovered from the supernatant. The recovered F1-VC424S-enriched pellet was stored below –70 °C.
Initial IEX
Columns and chromatography systems were cleaned and depyrogenated by exposure to 0.05 N NaOH for greater than 12 h or 0.5 N NaOH for 1 h followed by rinsing to neutral pH. For F1-VMN, F1-V-enriched pellet (400-g) was thawed at 20 °C and re-suspended 1:10 into 4 L of IEX-A buffer (10 mM Tris, 10 mM ethanolamine, 4.5 M urea, pH 8.3; then nitrogen sparged; and 5 mM fresh L-cysteine added). The load (∼2.9 mS/cm) was held at 20 °C for ∼3 h for F1-V dispersal and applied onto Q-Sepharose FF resin (BPG100/500, 10 cm D x 20 cm H bed, 90-µm bead size; GE Healthcare, Piscataway, NJ) and developed with one CV rinse and six CV linear gradient elutions at 60 cm/h to 3.5 M urea, 500 mM Gdn HCl in similar buffer (IEX-B). Monomer-enriched fractions, identified by HPLC-SEC analysis, were examined by SDS-PAGE to facilitate selection of the target monomeric F1-V species. The first major F1-V elution peak was collected between the 80- to 130-mM chloride ion (6.0 to 9.7 mS/cm) range. The Q-Sepharose FF elution pool was stored below –70 °C.
For F1-VC424S-MN, the F1-VC424S enriched pellet was re-suspended to 20-ml final volume with 10 mM Tris, 10 mM ethanolamine, 10 mM Gdn HCL, pH 8.3, and then then adjusted to pH 10.3, held for 30 min, and re-adjusted to pH 8.3 with 1 M acetic acid. High-purity solid urea was added to obtain a concentration of 4.5 M urea and the solution was held at 20 °C for 1 to 2 h before being loaded onto Q-Sepharose FF resin (1.6 × 10 cm, 90-µm bead size; GE Healthcare) equilibrated with 10 mM Tris, 10 mM ethanolamine, 4.5 M urea, 10 mM Gdn HCl, pH 8.3, followed by washing and linear gradient elution to 3.5 M urea / 500 mM Gdn HCl at 120 cm/h. The leading shoulder of a complex multi-peak structure was excluded from pooling to eliminate contaminants, identified by SDS-PAGE fraction analysis. F1-VC424S monomer-enriched fractions were collected and pooled from the first major peak eluting between 40 and 80 mM chloride, and stored below –70 °C. The second half (85 mg), was pooled separately and not processed further. The Q-Sepharose FF pool was diluted with high-quality water to 3.4 mS/cm (∼2.5-fold), loaded onto Source 15Q resin (1.6 × 10 cm, 15-µm bead size, GE Healthcare) equilibrated with IEX-A buffer, and eluted with a linear gradient to 40% B over 16 CV at 120 cm/h (4 ml/min). The leading half of the main peak was pooled and stored below –70 °C.
For F1-VMN, buffers IEX-A and IEX-B were made as above except for replacement of L-cysteine with 1 mM DTT. To ensure complete protein reduction, DTT (5 mM) was added to the monomer-enriched pool. After 2.3-fold dilution (from ∼9.5 mS/cm to 4.2 mS/cm, 4.75 L final volume) with IEX-A buffer, the pool was loaded onto Source 15Q resin (BPG100/500, 10 cm D x 20 cm H, 15-µm bead size; GE Healthcare), and eluted with a linear gradient to 40% IEX-B over eight CV at 60 cm/h. The F1-V monomer, eluting below 100 mM chloride ion, was pooled based on HPLC-SEC and SDS-PAGE fraction analysis. Contaminants present in a leading shoulder of a complex multi-peak structure were excluded from pooling. Two trailing shoulders, while also containing F1-V, were pooled separately and not processed further. The Source 15Q Elution Pool was separated into 5 × 200 mL aliquots and stored below –70 °C.
CHT affinity chromatography
For F1-VMN, CHT Type 2 resin (BPG100/500, 10 cm x 12 cm, 20-µm beads, BioRad, Hercules, CA), was charged with high phosphate buffer and equilibrated just before use with CHT-A buffer (10 mM Tris, 150 mM NaCl, 1 mM NaH2PO4, 0.1 mM CaCl2, pH 7.8; argon sparged; 1 mM DTE added, used immediately). For each of five CHT-T2 cycles, a 200 mL Source 15Q Elution Pool aliquot was thawed at ∼20 ºC, adjusted to 1 mM NaH2PO4, 0.1 mM CaCl2, from 100 mM stocks, diluted fivefold into CHT-A buffer, applied to the column at 50 cm/h and eluted with a linear gradient to 50% Buffer CHT-B (CHT-A + 200 mM NaH2PO4) over 16 CV. CHT T2 elution fractions were collected into containers pre-loaded with L-arginine stock (1.3 M L-arginine, pH 10.0) to obtain a final concentration of 200 mM L-arginine in each collected fraction. An early-eluting, sharp, F1-V-containing peak was excluded from pooling. Center fractions within a broader major peak were pooled and concentrated to 8 to 9 mg/ml of total protein by A280 over a 1-ft2 PrepScale-TFF 10-kDa MW cut off spiral tangential flow filtration membrane (regenerated cellulose, Cat# CDUF001LG; Millipore, Billerica, MA). The concentrated (7.4 mg/ml) CHT-T2 pool was divided into 3 × 95 mL aliquots and stored below –70 °C.
For F1-VC424S (MN), CHT-T1 resin (1.6 × 10 cm, 20-µm bead size; BioRad, Hercules, CA) was equilibrated and developed similarly to CHT Type 2 resin above. The Source 15Q pool was thawed and processed through two CHT-T1 column cycles. During the first cycle, performed without trace phosphate added to the load, a portion of F1-VC424S did not bind. For the second cycle, 1 mM phosphate was added to the load, leading to complete F1-VC424S binding. For both cycles a single, notably sharp, concentrated elution peak, was pooled with a minor, extended tail excluded. The fractions were stored chilled.
Size exclusion chromatography formulation of F1-VMN
Each CHT-T2 aliquot (1.2% CV) was adjusted to pH 10.0, held overnight at 4 °C, loaded onto Superdex 200 PG resin (10 cm x 90 cm in a BPG 100/950 column, 34-µm bead size) and eluted with formulation buffer (20 mM L-arginine, 10 mM NaCl, argon, 1 ml of L-cysteine, pH 10.0) at a flow rate of 22 cm/h. The early eluting dimer-enriched fractions were pooled separately (252 mg) and stored below –70 °C. Fractions in the first half of the monomer peak, essentially free of contaminants, were 0.2-µm filtered, aliquoted, and stored below –70 °C. Trailing monomer-peak fractions, enriched in contaminants, were concentrated as above, re-fractionated, and combined with initial monomer-enriched fractions. This final pool was 0.2-µm filtered distributed into sterile cryo-vials; and stored below –70 °C.
For F1-VC424S-MN, main peak fractions were pooled (40 ml) and adjusted to 500 mM L-arginine by adding 3.5 g of solid L-arginine pre-dissolved in 7 ml water. After 10 min at pH 11.0, the pool was adjusted to pH 10.1 by the slow addition of HCl and held overnight at 4 °C. This yielded ∼47 ml at 5.0 mg/mL or 235 mg of total protein. The adjusted CHT-T1 pool was fractionated by size-exclusion chromatography through Superdex 200 PG resin (two tandem columns, 10 cm x 90 cm in BPG 100/950 columns, 34-µm bead size),equilibrated and developed with 20 mM L-arginine, 10 mM NaCl, pH 10.0 (with no L-cysteine) at a flow rate of 22 cm/h. A 43-ml sample (0.3% of CV) was applied. Fractions in the first half of the monomer peak were pooled and stored below –70 °C; thawed; concentrated using YM-10 Centripreps (Millipore, Billerica, MA) at 4 °C; 0.2-µm filtered; distributed into sterile cryo-vials; and stored below –70 °C.
Conversion of monomeric F1-VMN to multimeric F1-VAG
An aliquot of formulated F1-VMN, at pH 10.0, was converted to F1-VAG by slow titration with acetic acid to pH 5.1, incubated overnight at 4 °C, and then stored below – 70 °C.
Optional freeze-drying of F1-VMN
F1-V in formulation buffer was adjusted to 2% w/v low endotoxin D-mannitol (Ferro Phanstiehl Laboratories, Inc., Waukegan, IL) added from a 20% D-mannitol stock dissolved in formulation buffer. The product was distributed into 3-ml glass vials, frozen at a plate temperature of –48 °C, and lyophilized in an AdVantange-ES Benchtop freeze-dryer (VirTis, Gardiner, NY) for 30 h at –45 °C, 8 h at –37 °C, followed by 15 h at +37 °C. Condenser coils were maintained at –80 °C. Vial stoppers were mechanically seated within the chamber while under vacuum and crimped externally. The vials were stored below –70 °C.
Total protein, Endotoxin and SDS-PAGE
Protein concentrations were measured by A280 divided by an absorption co-efficient of E = 0.468 A280, 1cm per (mg of F1-V/ml), calculated using methods [13] automated on the ExPASy Proteomic Server, ProtParm (2005 version, http://www.expasy.org). For solubilized pellets, total protein was estimated with E = 1.0. For endotoxin measurement, the commercially available Charles River (Charleston, SC) kinetic chromogenic limulus amoebocyte lysate reactivity endotoxin kit was used, which had a lower detection limit of 0.005 EU/ml, established versus the provided endotoxin standard. For SDS-PAGE, 4– 12% Bis-Tris NuPAGE gels and reagents, Mark 12 size standards, and Sypro Ruby fluorescent stain were obtained from Invitrogen. Samples were reduced with 5% v/v 2-mercaptoethanol. Destained gels were scanned with a Molecular Dynamics model 595 scanning laser fluorimeter (GE Healthcare) and integrated with ImageMaster 1D Elite software (Version 4.1, GE Healthcare).
SEC-MALLS
Size-exclusion chromatography coupled to multiangle laser light scattering (SEC-MALLS) was applied as previously reported for F1-V [6], but with modifications as published [14, 15]. The HPLC pumps were Rainin HPXL 10 ml/min pumps (Varian, Walnut, CA) run at 0.4 ml/min. Fractionation was performed through two tandem G3000SWxl analytical size-exclusion chromatography columns (7.8 × 250 mm, 5-µM bead size, 250-Å pore size; Tosho Biosciences, Montgomeryville, PA), equilibrated with 0.2-µm filtered, helium-sparged mobile phase (0.1 M KH2PO4, 0.1 M Na2SO4, 0.3 M NaCl, pH 7.0). The light-scattering detector series consisted of a Rainin Dynamax UV-1 A280 detector (Varian); a Dawn EOS multi-angle, static, light-scattering detector (Wyatt Technology Corporation, Santa Barbara, CA); and an Optilab DSP interferometric refractometer (Wyatt). Average molar mass measurements were determined from aligned elution profiles within ASTRA for Windows software (Revision 4.90.07/QELS version 1.00, Wyatt). Bovine serum albumin (2 mg/ml, 10 µL injections) containing a mixture of solution forms (66.3 kDa monomer, 132.6 dimer kDa, and 198.9- kDa trimer; Pierce-Endogen, Rockford, IL) was used to normalize detectors, establish detector train delay times, set software parameters, and confirm system suitability before test sample analysis. According to previously reported methods [15], the standard optical constant was calculated as K* = 1.85 × 10–7 mol cm2 g –2; as derived from (dn/dc) = 0.185 ml g–1, n0 = 1.33; and λ0 = 681 nm; with the form factor set to unity.
Peptide mapping
For each sample, 100-µg aliquots were dried, re-solubilized to 1 M Gdn HCl in 0.1 M Tris, pH 8.0, divided in half and digested (1:30 enzyme-to-substrate ratio) with modified trypsin or chymotrypsin overnight at 37 °C with mixing by vortex at 1,200 rpm. The digest was quenched by acidification and the samples stored at 4 °C until analysis. The samples were injected onto a reverse-phase column (Grace Vydac LC/MS C18, 2.1 × 250 mm, C/N 218MS52, 35 °C; Hesperia, CA) fitted to an HPLC (Thermo Electron, Surveyor LC System, Waltham, MA) followed by a hold at 5% for 5 min and elution over 55 min at 0.2 ml/min using a 1% per min linear gradient of acetonitrile containing 0.08% trifluoroacetic acid and 0.02% formic acid with elution monitored at 214 nm. The effluent was directed into an ion trap mass spectrometer (Thermo Electron, LCQ-Deca MS) for detection by electrospray mass spectrometry (ESI-MS) in positive mode ionization with 250 °C capillary temperature, ∼95 psi sheath gas pressure, ∼5 psi auxiliary gas pressure, source at 5.5 kV with capillary at 44 V, lens offset by 50 V, multipole offset by –5.5 and –10.5V, inter multipole lens at –28V, entrance lens at –88V and a trap DC offset of –10V. MS/MS was performed using 35% collision energy. Sequential scanning, consisting of full-scan ESI-MS from m/z 500 to 2000 and triplicate MS/MS scans of the three most abundant base peak (BP) ions, was employed. Equine skeletal muscle myoglobin (Sigma-Aldrich, M0630, St. Louis, MO) was analyzed as a sample preparation and instrument performance standard. The resulting MS and MS/MS data sets were processed using Bioworks© (Thermo Electron, Version 3.1) and Xcaliber© Software (Thermo Electron, Version 1.3). Except where noted, fragment ion identity assignments were based upon automated software MS/MS analysis of primary-ion peak fragments with software default Xcorr thresholds set for assignment acceptance. The sequence coverage for the mutant myoglobin standard was 100%.
Reagent scouting
For disulfide-linked dimer dispersal scouting, a sub-fraction of purified F1-VMN formulated at ∼0.7 mg/ml in 20 mM L-arginine, 10 mM NaCl, pH 9.9, without added 1 mM L-cysteine, was air oxidized to form ∼22% disulfide-linked dimer. Reagents were added from un-adjusted, acidic, 100-mM stocks of freshly prepared DTE, L-cysteine, and IAA. For the two-reagent conditions, the reductant was added first, followed by a 10-min hold at 25 °C before adding IAA. Adjusted samples were held at 25 °C within the HPLC-SEC auto injector before analysis. Samples were analyzed through HPLC-SEC with two tandem columns (G3000SWxl) on an Agilent 1100 system (Agilent Technologies, Palo Alto, CA) eluted at 0.8 ml/min with 0.1 M KH2PO4, 0.1 M Na2SO4, 0.3 M NaCl, pH 7.0. Column performance was confirmed by running high MW size standards (BioRad). The percentage of integrated A230 eluting in each peak relative to total protein-related integrated absorbance was calculated within Chemstation 2.0 software (Agilent).
For non-covalently-linked multimer dispersal scouting, reagents were prepared as 10-fold stocks in high-quality water and adjusted as needed to ∼pH 6.5. An aliquot of F1-VMN, initially formulated at ∼0.7 mg/ml in 20 mM L-arginine, 10 mM NaCl, 1 mM L-cysteine, pH 9.9, was titrated by micro-addition of HCl to pH 6.5. In less than 5 min, the aliquots were divided and transferred with mixing into containers pre-loaded with 1/10th volume of additive stocks. Samples were held at 4 °C before HPLC-SEC analysis by methods similar to those described for disulfide-linked dimer dispersal scouting above.
Animal vaccinations
Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals, and experiments involving animals were conducted according to the principles set forth in the Guide for the Care and Use of Laboratory Animals [16]. The facility where this research was conducted is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Groups of 10 female, 8- to 10-week-old outbred (Hsd:ND4) Swiss Webster mice were inoculated subcutaneously (s.c.) with purified, recombinant F1-VSTD, F1-VMN, F1-VAG or F1-VC424S-MN preparations. To evaluate the effect of aggregation state on the protective efficacy of F1-V as well as the efficacy of the new F1-VC424S, various F1-V aggregation state formulations were produced. Vaccine candidate formulations included monomeric F1-VC424S-MN, the cysteine-capped, monomeric F1-VMN, and the converted multimer, F1-VAG. F1-VAG was produced by incubating F1-VMN overnight at pH 5.1 and 4 °C to enhance F1-V aggregation. One group of 10 mice was inoculated s.c. with the previously reported mixed solution state F1-VStd as a positive control [6]. In order to maximize immunogenicity, each protein antigen was adsorbed to aluminum hydroxide adjuvant (Alhydrogel. 1.3%; Superfos Biosector, Vedbaek, Denmark; 0.19 mg of aluminum per dose), critically before exposure of adjuvant to injection buffer (1x PBS). Each antigen-adjuvant mixture (200 µL) containing 20 µg of each antigen was administered at a single subcutaneous site on the backs of the animals. After 30 days, the animals were boosted with an identical dose at the same injection site.
Measurement of serum antibody titer using ELISA
Mice were anesthetized with a mixture of 5 mg of xylazine (XYLA-JECT; Phoenix Pharmaceutical, Inc., St. Joseph, MO) per kg, 0.83 mg of acetylpromazine (Fermenta Animal Health Co., Kansas City, MO) per kg, and 67 mg of ketamine hydrochloride (Ketamine; Phoenix Pharmaceutical, Inc.) per kg administered intramuscularly. Blood was collected by retro-orbital sinus puncture for the determination of antibody titers 56 days after the initial injection by standard enzyme-linked immunosorbent assay (ELISA). Briefly, 100 ng of each purified protein in carbonate buffer, pH 9.4, was applied to each well of a 96-well microtiter plate and allowed to incubate overnight at 4 °C. Plates were then washed with 1x PBS + 0.05% Tween 20. Plates were blocked with 100 µl of assay diluent (1 x PBS, 1% bovine serum albumin, 0.05% Tween 20) for 1 h at 37 °C. Plates were washed again and serial dilutions of antiserum in assay diluent ranging from 1:50 to 1:2,048,000 were applied in triplicate. Plates were allowed to incubate at 37 °C for 1 h, washed, and a 1:5000 dilution of horseradish peroxidase-conjugated goat anti-mouse IgG was applied for 1 h at 37 °C. Plates were washed and the chromogenic substrate 3,3′,5,5′ tetramethylbenzidine (TMB; BD Biosciences, Pharmingen, San Diego, CA) was added. After a 30-min incubation at 37 °C in the dark, the reaction was stopped with 25 µl of 2 N sulfuric acid. Plates were read at an optical density of 450 nm (OD450).
Y. pestis lethal challenge
Each of the vaccinated animals designated to receive s.c. challenges was administered 104, 107, 108, or 109 50% lethal doses (LD50) of wild-type Y. pestis CO92, 30 days after the booster dose. The s.c. LD50 for adult mice challenged with CO92 is 1.9 colony-forming units (CFU) as determined by serial dilution and plating. The mice were observed daily for 28 days, at which time the survivors were killed. Fisher’s two-tailed exact tests were used to evaluate animal survival data. Mean time to death after lethal plague challenge was evaluated using Student’s t-tests. Significance in pair-wise comparisons of delayed time to death between groups was computed using Student’s t-tests.
RESULTS AND DISCUSSION
In order to evaluate the effect of super molecular structure (i.e., its state of aggregation) of the F1-V-based plague vaccine antigen on protective efficacy and to facilitate vaccine production, the sole cysteine (C424) in F1-V was replaced with a serine residue by site-directed mutagenesis. Standard F1-VMN and the modified F1-VC424S-MN proteins were independently over-expressed in E. coli, recovered by mechanical lysis/pH-modulation, and purified from urea-solubilized, soft inclusion bodies with successive ion-exchange, ceramic hydroxyapatite, and size-exclusion chromatography stages. Aggregation characteristics for the purified proteins were characterized and compared under variable pH and buffer solution-additive conditions. The biological activities of the two purified proteins in various super molecular states were then evaluated for immunogenicity and efficacy in mice against lethal Y. pestis challenge.
F1-VMN and F1-VC424S-MN expression
F1-VMN and the modified F1-VC424S-MN proteins were expressed as described in the Methods section. The original pET-24a-based F1-V expression vector (pPW731) was modified by site-directed mutagenesis to replace the sole cysteine (Cys424) with a serine residue (Fig. 1). This mutation was performed to eliminate the necessity for reducing conditions during the F1-V protein purification process and to evaluate the effect of the cysteine residue on F1-V protein aggregation. After induction with 0.5 mM IPTG, the F1-VC424S vector over-expressed an insoluble 53-kDa protein as determined by SDS-PAGE (Fig. 2). A pH-based precipitation process was employed to enrich the F1-VC424S protein before solubilization with 5M urea and ion-exchange chromatography.
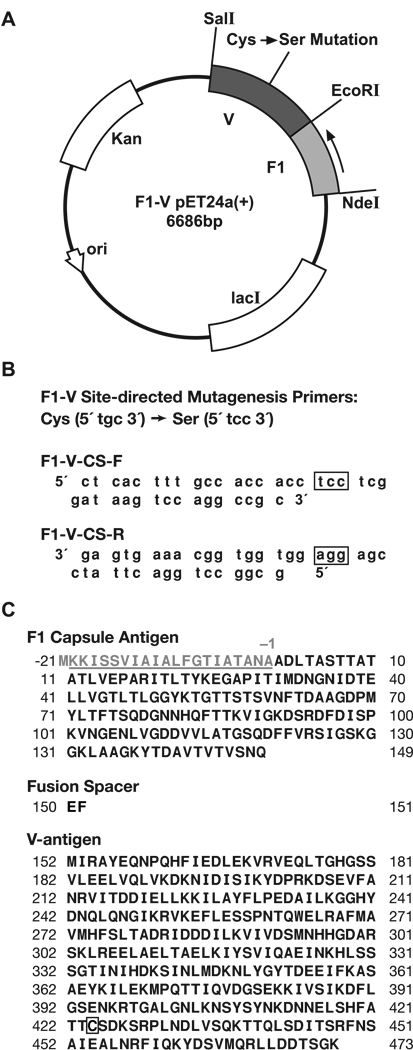
Fig. 1.
(A) Diagram of F1-V pET24a(+) Cys425 -> Ser425 expression plasmid. (B) Site-directed mutagenesis primers F1-V-CS-F and F1-V-CS-R that were used to produce the desired Cys -> Ser mutation at amino acid position 424. (C) The complete F1-V fusion protein amino acid sequence contains three marked points of interest: 1) The F1 capsule signaling/leader sequence is not included in the fusion; 2) An EcoRI restriction site was used to link the F1 protein and V antigen. This restriction recognition site results in a two-amino acid (EF) linker between the F1 protein and the V antigen; and 3) the specific location of the Cys -> Ser mutation within the fusion protein.
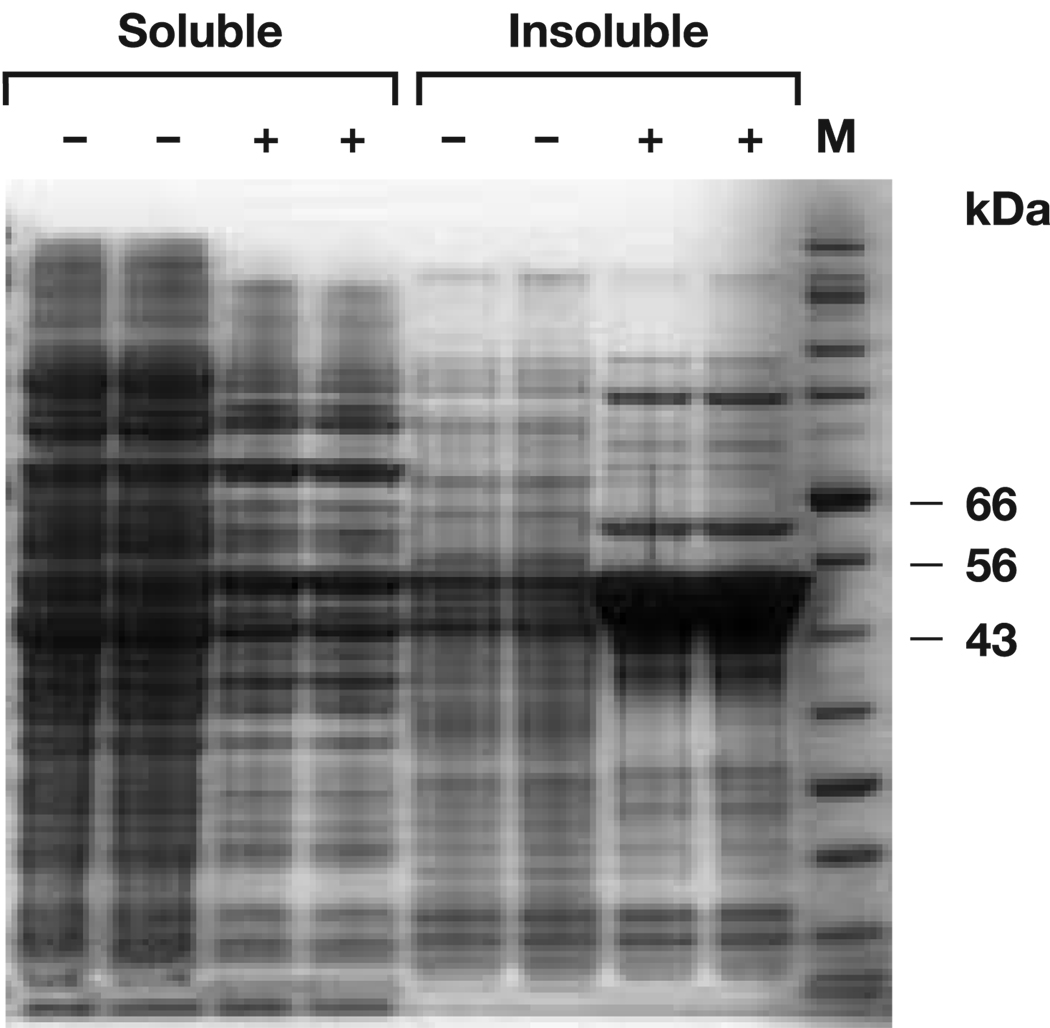
Fig. 2.
Simply Blue-stained (Invitrogen), reduced SDS-PAGE analysis of F1-VC424S expression for whole broth soluble and insoluble fractions before (−) and after (+) IPTG induction. Lane M - protein molecular mass standard. The calculated molecular mass of F1-VC424S monomer is 53 kDa. Cells and supernatants were mixed with 4 x LDS sample buffer and heated at 70 °C for 10 min before electrophoresis through Invitrogen NuPAGE 4–12% Bis-Tris gels, using MOPS SDS running buffer.
At the larger scale, the time course for cultivating E. coli., BLR130 transformed with pPW731 plasmid (containing the coding sequence for the unmodified, cysteine-containing F1-V [6]) had a controlled induction response (Fig. 3A). Resulting F1-V was precipitated by slow pH adjustment to pH 4.8 and additional contaminants were removed (compare lanes 1 vs. 7 in Fig. 3B) by pH modulation before re-solubilization of the enriched F1-V pellet in urea (Fig. 3B).
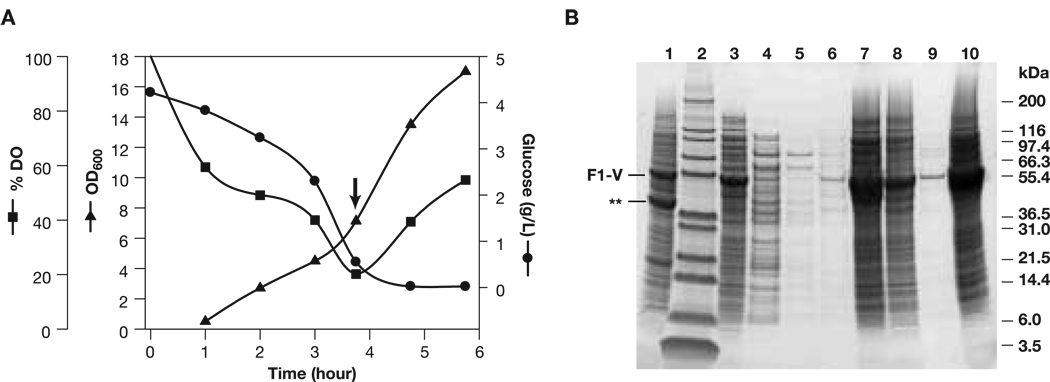
Fig. 3.
(Panel A) Fermentation time course for cultivation of E. coli., BLR130 transformed with plasmid pW731 expressing F1-V. Arrow, induction with 1 mM IPTG, 0.2% arabinose (ADM). (Panel B) Sypro Ruby-stained, reduced SDS-PAGE analysis of F1-VMN recovery (ADM). Pellets were first prepared by ∼25-fold dilution into 2x reducing running buffer. Loadings were 20 µL/well or 40 µL/well (lanes 4, 5, and 6). Lanes: 1) washed initial pellet lysate; 2) Mark 12 molecular mass markers (Invitrogen); 3) lysate supernatant; 4) pH 4.8 supernatant; 5) 1st pH 4.8 rinse; 6) 2nd pH 4.8 rinse; 7) pH 4.8 pellet; 8) post resolubilization and pellet; 9) 2nd pH 4.8 step supernatant; 10) 2nd pH 4.8 step pellet.
Ion-exchange chromatography
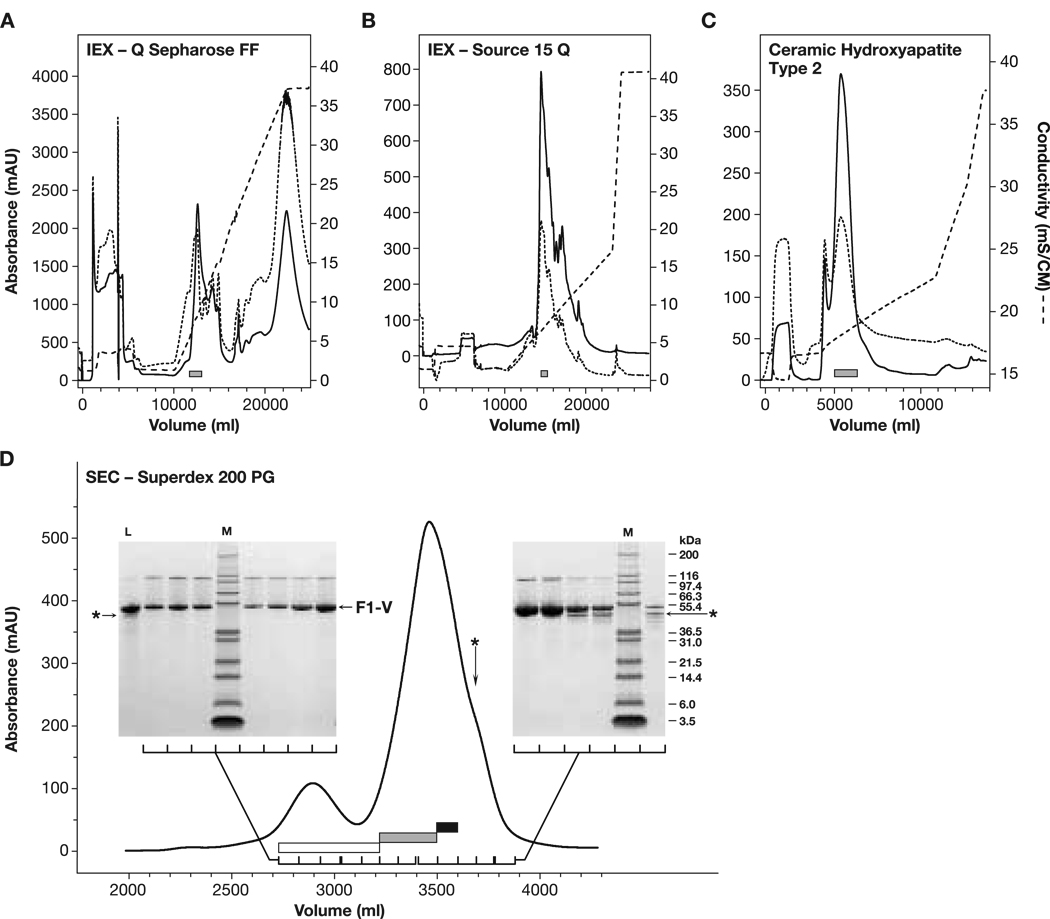
Standard F1-V and the F1-VC424S variant purified similarly through the ion-exchange stages of the improved process. Profiles from purification of the standard (cysteine-containing) F1-V performed at the larger scale are shown (Fig. 4). The Q-Sepharose FF chromatography stage, performed under partially dissociating conditions and eluted with the denaturing Gdn cation, was effective as a charge-based step for isolating F1-V (Fig. 4A). F1-VC424S monomer eluted at Gdn HCl concentrations below 100 mM, similar to what was observed with F1-VSTD-MN. Dimer and trimer forms of F1-V remained intact during reducing SDS-PAGE analysis when samples were prepared by heating to less than 70 °C for 10 min. These same forms were dispersed and ran as apparent F1-V monomers when heated to 100 °C for 10 min (DNS), corroborating prior observations of strong self association F1-V by gel electrophoresis [6]. HPLC-SEC analysis confirmed that the trailing edge of the Q-Sepharose FF F1-V peak (elution volume 1400 ml, Fig. 4A) contained dimer and trimer forms of F1-V between 60 and 120 mM Gdn (DNS). A portion of F1-V remained in the Q-Sepharose FF non-bound fraction. Re-application to the hydroxide-stripped column recovered only a small proportion of the F1-V present, suggesting the F1-V flow- through was not due to column overloading. This phenomenon also concurs with prior findings of an unrecoverable fraction consistently observed during F1-V purification [6]. A subpopulation of F1-V within inclusions, relatively resistant to dissociation in 5 M urea, was likely excluded from the Q-Sepharose FF resin. The Source 15Q elution profile was characteristically jagged and extended with multiple, sharp, minor peaks apparently superimposed on top of a broader three-peak profile (Fig. 4B). The jagged nature of this elution profile was observed in multiple runs during development work and was not related to particular instrumentation or the range of the UV detector.
Fig. 4.
F1-VMN purification (ADM). (grey boxes) Eluate pools. (Panel A) Q-Sepharose FF IEX profile. (dashed trace) Eluate conductivity. A254 (dotted trace) and A280 (solid trace) were monitored across a 2-mm path length cell. The load and wash occurred before the elution started at 6 L. (Panel B) Source 15Q IEX profile with 7 L elution start and 2-mm cell. (Panel C) CHT-T2 chromatography profile with 2 L elution start and 10-mm cell. (Panel D) Preparative scale Superdex 200 PG SEC profile with 2% CV load and 10-mm cell. Three pools were made: a leading, highly-pure, dimer-enriched F1-V pool (open box); a central target F1-VMN pool (light gray box); and a third trailing pool (dark box) containing monomeric F1-VMN and a ∼49 kDa contaminant (asterisk). After re-concentration, the third pool was reprocessed through the Superdex 200 PG stage. (Insets) Sypro-Ruby-stained, reduced 4–12% NuPAGE SDS-PAGE analysis from elution fractions.
Aggregate dissociation
To further understand F1-V losses to non-binding, small-scale F1-V multimer dissociation studies, monitored using HPLC-SEC analysis, showed that F1-V multimers were not fully dispersed by even 7 M urea. Maximum dispersal was observed using 6 M Gdn HCl (DNS). Upon buffer exchange over G-25 resin, from 6M Gdn HCl into Source 15Q Buffer, F1-V remained substantially monomeric (DNS).
Affinity chromatography
Ceramic hydroxyapatite chromatography, being insensitive to high concentrations of Gdn HCl, was used to exchange F1-V into non-denaturing conditions while providing additional purification. Including trace PO42− and Ca2+ ions was critical for efficient F1-V binding and resin stability. Predominantly lower molecular weight contaminants flowed through the CHT-T2 stage. F1-V, processed over CHT Type 2 (T2) resin, eluted primarily in monomeric form (>80%), free of denaturing agents (Fig. 4C). F1-VC424S recovered from the CHT Type 1 (T1) resin contained higher levels of dimer, trimer, and multimer (∼73%). The prior reported method similarly removed denaturants while F1-V was bound to ion-exchange resin [6].
SEC
Superdex 200 PG SEC provided a convenient method for combined final formulation and size classification. F1-V(MN) and F1-VC424S purified similarly by SEC. The mobile phase containing physiologically compatible additives, L-arginine for buffering at pH 10.0, and L-cysteine for thiol capping, maximized the monodispersity of F1-V. Low molecular mass protein trace contaminants in the range of 40 to 49-kDa overlapped with the monomer peak trailing edge (Fig. 4, Panel D, astrix and black bar). The major contaminant at ∼49-kDa was identified by N-terminal sequencing as E. coli serine hydroxymethyl transferase (DNS). Separating and selectively pooling the purest fractions based upon SDS-PAGE analysis minimized these trace contaminants (Fig. 4, Panel D, the three rightmost product lanes were not pooled). Although not used for the vaccination trials, the dimer/multimer pool was essentially 100% pure F1-V with no detectable low molecular mass contaminants by SDS-PAGE (Fig. 4, Panel D, Lanes 2, 3, 4, and 6 from the left). Thus, after initial purification, F1-V and F1-VC424S preferentially self-associated while the 40- and 49-kDa trace contaminants remained as apparently low molecular species. The monomeric and dimeric F1-V forms were well-separated, especially when tandem columns were employed. Thus, the use of a size-based purification method as the last stage critically ensured maximally monodisperse F1-V for use in vaccination trials.
Protein purification process yield
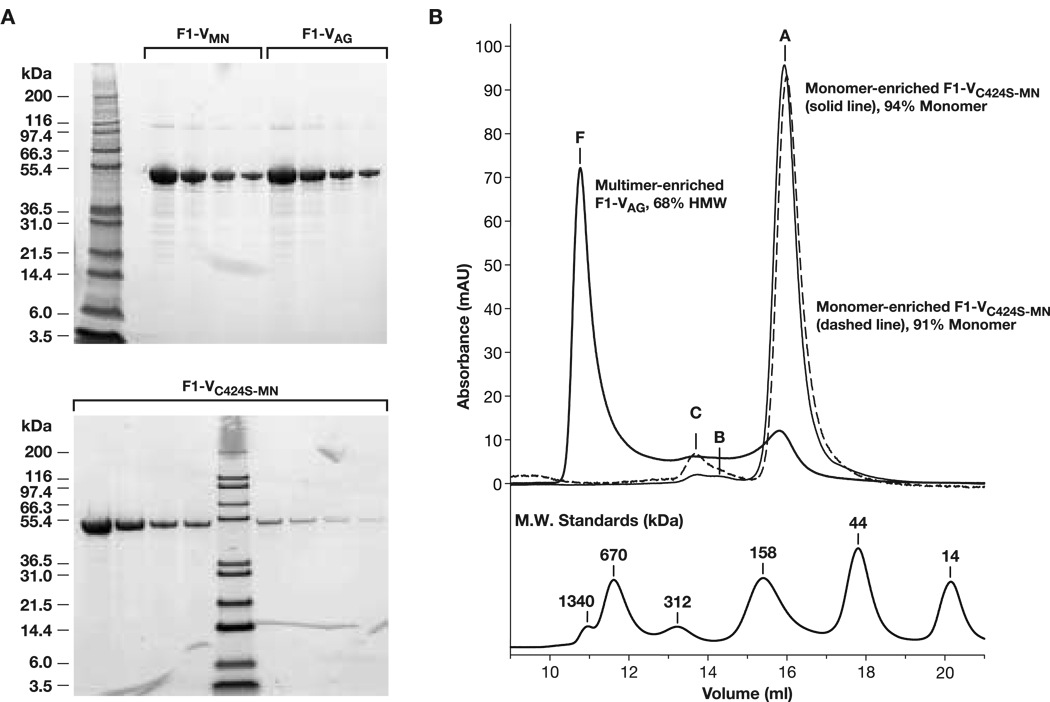
From 765 g of cell paste, 823 mg of monodisperse F1-V was recovered for a final process yield of ∼1.2 mg/g of cell paste (Table 1A). From 23.2 g of F1-VC424S cell paste, 40 mg of F1-VC424S-MN was recovered for a process yield of ∼ 2 mg/g of cell paste (Table 1B). Purity, identity and protective potency testing reported herein were conducted on intermediate bulk materials, prior to final finishing. SDS-PAGE and HPLC-SEC profiles of purified F1-VMN, F1-VAG, and F1-VC424S-MN confirmed greater than 95% purity for the preparations (Fig. 5, Panels A and B). Each preparation was specifically detected in immunoblot analysis as per previously reported methods [6] versus mouse anti-F1 and anti-V antibodies (DNS). Low endotoxin levels (< 0.5 EU/mg) and host cell genomic DNA levels (< 2 pg/mg) were typically observed.
Table 1.
Process summaries for F1-VMN and F1-VC424S (ADM).
| Table 1A-F1V Process Summary for F1-VMN | |||||
|---|---|---|---|---|---|
| Production Stage | Concentration (mg/mL) | Volume (mL) | Total Protein byA280(mg) | Step Yield (%) | Yield (mg TP per g CP) |
| Fermentation (20L) | |||||
| Wet Cell Paste (CP) | − | − | − | − | (765 g CP) |
| Solubilized Pellet | 27.0* | 3,300 | 89,100* | − | 117 |
| Ion Exchange, Q-Sepharose FF | 12.6 | 1,00 | 21,500 | 24 | 28 |
| Ion Exchange, Source 15Q | 5.1 | 1,000 | 5,100 | 24 | 6.6 |
| Affinity, Ceramic Hydroxyapatite Type 2 & Concentration | 7.4 | 285 | 2,115 | 42 | 2.8 |
| Size Exclusion, Superdex 200 PG | 0.78 | 1,055 | 823 | 39 | 1.1 |
| Table 1B Process Summary for F1-Vc424S-mn | |||||
|---|---|---|---|---|---|
| Production Stage | Concentration (mg/mL) | Volume (mL) | Total Protein by A280 (mg) | Step Yield (%) | Yield (mg TP per g CP) |
| Fermentation (10 L) | |||||
| Wet Cell Paste (CP) | − | − | − | − | (23 g CP) |
| Solubilized Pellet | *110 | 35 | 3,850* | − | 167 |
| Ion Exchange, Q-Sepharose FF | 3.5 | 140 | 490 | 13 | 21 |
| Ion Exchange, Source 15Q | 3.2 | 80 | 256 | 52 | 11 |
| Affinity, Ceramic Hydroxyapatite Type 1 & Concentration | 5 | 47 | 235 | 92 | 10 |
| Size Exclusion, Superdex 200 PG & Concentration | 0.77 | 52 | 40 | 20 | 1.7 |
Estimated A280 with E = 1.0.
Fig. 5.
(Panel A) SDS-PAGE analysis of final preparations. (Top) Comparison of F1-VMN before and after conversion to F1-VAG, loaded with two-fold dilution series starting at 9.2 µg/well; (Bottom) F1-VC424S-MN loaded starting at 9.6 µg/well. (Top, Lane 1, Bottom Lane 5) Mark 12 MW marker. (Panel B) Overlaid HPLC-SEC profiles of final F1-VMN (pH 9.9), F1-VC424S-MN (pH 9.9), and F1-VAG (pH 5.0) preparations. F1-V monomer eluted at an anomalous apparent molecular size of 100 kDa relative to high MW size standards (bottom).
Solution stability versus pH
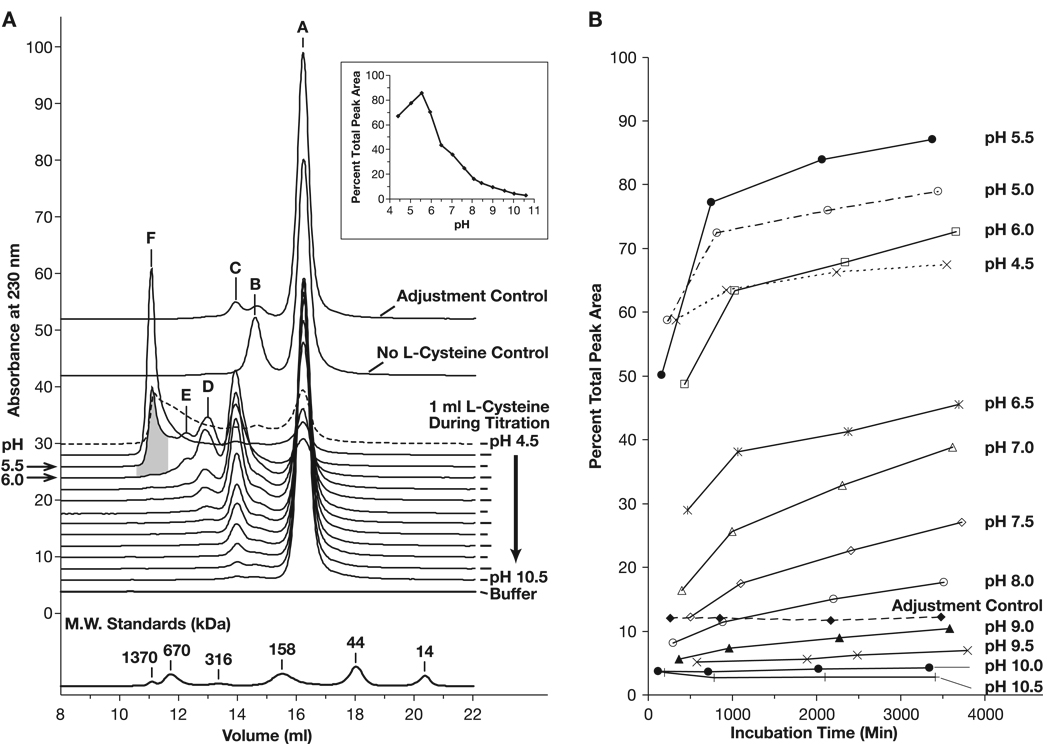
Although the handling of F1-V under neutral to acidic conditions was previously known to be problematic, the details of such effects were not described [6] (B. Powell, unpublished data). As part of an effort to stabilize the monomeric state of F1-V preparations, we further characterized the solution structure of F1-V as a function of diluent pH. Analytical size-exclusion chromatography over a silica-based, wide-pore G3000SWxl column was used to measure the effect of solution composition on the ratio of monomer to dimer/trimer/multimer species as well as the effect on F1-V(NC) and F1-V(S-S) dimer sub-classes. The unique F1-VC424S form enabled us to separately assess the effects of reducing agents and stabilizing additives on structure. A clear trend toward formation of higher molecular mass F1-V associations was observed as a function of lowering solution acidic pH (Fig. 6, Panel A). An identical apparent size profile versus pH trend was observed for F1-VC424S-MN in formulation buffer lacking L-cysteine (DNS) except that the shoulder corresponding to disulfide-linked F1-V dimer (Fig. 6, Panel A, Peak B) was no longer a observed as distinct feature. As shown in the inset to Panel A, the greatest percentage of high molecular mass species appeared between pH 6 and 8. Additionally, the percentage of very high molecular mass species increased as the solution pH dropped from pH 6.0 to 5.5 (Fig. 6, shaded area, Peak F). Aggregation was further exacerbated as solution pH dropped below pH 5.5, observed as a loss of total protein from solution (Fig. 6, Panel A, Inset). For proteins, the histidine imidizole and the N-terminal amine groups become positively charged and thereby decrease the net protein negative charge (calculated F1-V pI = 5.19) below pH 8.0. We speculate that F1-V multimerization at low pH may involve the loss of ionic repulsive forces. A structural re-arrangement exposing hydrophobic patches is also consistent with the pH trend data. The conversion to multimer was time-dependent as shown by the limited conversion to multimer observed in the “adjustment control” sample that was titrated from pH 4.5 back up to pH 9.9 within 10 min of acidification. This time dependence was also clear after plotting the percentage of high molecular mass species versus hold time for each pH condition (Fig. 6, Panel B) where transitions to the stable profiles shown in Panel A were quite slow. This would also be consistent with a relatively slower structural re arrangement during F1-V multimer formation. These results demonstrate that, in the presence of optimized solution additives, moderately basic pH conditions were critical to maintaining a monodisperse F1-V preparation. Thus, preparation under basic conditions and formulation at pH 9.9 maximized recovery of monomer. This concurs with prior empirical findings of optimal F1-V purification at pH 9.5 [6].
Fig. 6.
Analysis of cysteine-stabilized F1-VMN non-covalent self-association (ADM). (Panel A) Stacked HPLC-SEC traces after incubation at 4 °C for 55–64 h at pH 4.5–10.5. The ‘Adjustment Control’ sample was derived from the pH 4.5-sample that was immediately back-titrated to pH 10.0. Peak solution state assignments were based on SEC-MALLS MW determinations (Fig. 8). Peak A, F1-V monomer; Peak B, F1-V(S-S) dimer (DTT-sensitive); Peak C, F1-V(NC) dimer (DTT-insensitive); Peak D, F1-V trimer; Peak E, F1-V multimer less than 0.5 MDa; Peak F, multimer, 0.5 to 6 MDa. The L-cysteine-free control at pH 9.9 established the F1-V(s-s) dimer postion (Peak B). (Panel 1, Inset) Plot of the percentage of integrated peak area contained in dimer and multimer peaks as a function of incubation pH after 55–64 h. incubation (Panel B) Related plots of the percent integrated peak area contained in dimer and multimer peaks as a function of time and pH between 0 and 64 h incubation at 4 °C.
Additive study – thiol reducing and blocking agents
The ability of HPLC-SEC analysis to separate disulfide-bonded F1-V(S-S) dimer (Fig. 6, Panel A; Peak B) from non-covalently associated F1-V dimer, (Peak C) enabled the evaluation of formulation additives intended to minimize disulfide-linkage (Table 2). High DTT concentrations (10 mM) resulted in complete disulfide bond disruption for >20 h at 25 °C. Trace DTT concentrations (0.5 and 1.0 mM) resulted in initial disruption followed by reformation of ∼35% of F1-V(S-S) dimer after 10 h. Intriguingly, the F1-V(S-S) percentage decreased to starting dimer levels after a further hold for 10 h. This observation did not fit a simple disulfide bond exchange/oxidative disulfide bond formation model, and may have been an example of further oxidation [17]. The addition of iodoacetamide alone (2.5 mM), an irreversible free-thiol capping reagent, led to a minor decrease in the F1-V(S-S) dimer over ∼22 h. However, when the F1-V(S-S) dimer was first reduced with DTT (0.5 or 1 mM), held for 10 min at 25 °C, and the resulting free-thiols blocked by excess iodoacetamide (1.25 or 2.5 mM), the level of F1-V(S-S) remained below 5% for more than 20 h.
Table 2.
Percentage F1-V(S-S) dimer (corresponding to Fig 6, Peak C) measured by HPLC-SEC after 25 ºC incubation (in min) at pH 9.9 with thiol-reactive additives (ADM). The percentage of F1-V(NC) dimer (Fig. 6, Peak C) was consistently less than 1.3% (DNS).
| Disulfide Linked Dimer |
||||||
|---|---|---|---|---|---|---|
| Additive Condition | min | % | min | % | min | % |
| No Additions | 68 | 21.4 | 522 | 22.8 | 975 | 23.1 |
| 10 Nm DTE | 417 | 3.1 | 871 | 3.5 | 1385 | 4.0 |
| 0.5 mM DTE | 242 | 30.7 | 696 | 35.8 | 1150 | 23.7 |
| 1 mM DTE | 103 | 3.8 | 556 | 36.8 | 1010 | 21.1 |
| 2.5 mM IAA | 382 | 15.7 | 836 | 13.2 | 1290 | 10.8 |
| 0.5 mM DTE, 1.3 mM IAA | 312 | 3.3 | 766 | 3.9 | 1220 | 3.7 |
| 1 mM DTE, 2.5 mM IAA | 173 | 2.6 | 626 | 3.8 | 1080 | 4.1 |
| 0.5mML-Cys, 1.3 mM IAA | 347 | 4.1 | 801 | 3.7 | 1255 | 3.8 |
| 1 mM L-Cys, 2.5 mM IAA | 208 | 3.8 | 661 | 4.8 | 1115 | 4.4 |
| 0.5 mM L-Cysteine | 277 | 2.8 | 731 | 3.7 | 1185 | 3.3 |
| 1 mM L-Cysteine | 138 | 3.0 | 591 | 3.5 | 1045 | 3.3 |
Unexpectedly, exposing F1-V(S-S) to trace levels of L-cysteine (0.5 mM or 1 mM) alone provided durable disulfide bond disruption. L-cysteine was superior to other additives studied for disulfide disruption, showing an essentially invariant 3.3% F1-V(S-S) content after 20 h at 25 °C. While further experimental elucidation of this interaction remains a possible topic for future work, we hypothesized that L-cysteine formed a relatively more stabile adduct to the F1-V free-thiol than other reducing agents, perhaps stabilized by local ionic or hydrogen bond interactions. This was supported by mass spectrum analysis of tryptic peptides where the uncapped, free-cysteine-containing fragment was not recovered but the cysteine-adduct fragment was isolated and identified with high confidence by MS/MS fragmentation analysis (Supplemental Fig. 9). As a non-toxic, physiological amino acid, L-cysteine (1 mM) was subsequently selected as the agent of choice for suppressing F1-V(S-S) disulfide-bonded dimer levels in native F1-VMN and F1-VAG formulation buffers.
Non-covalent multimer modulation
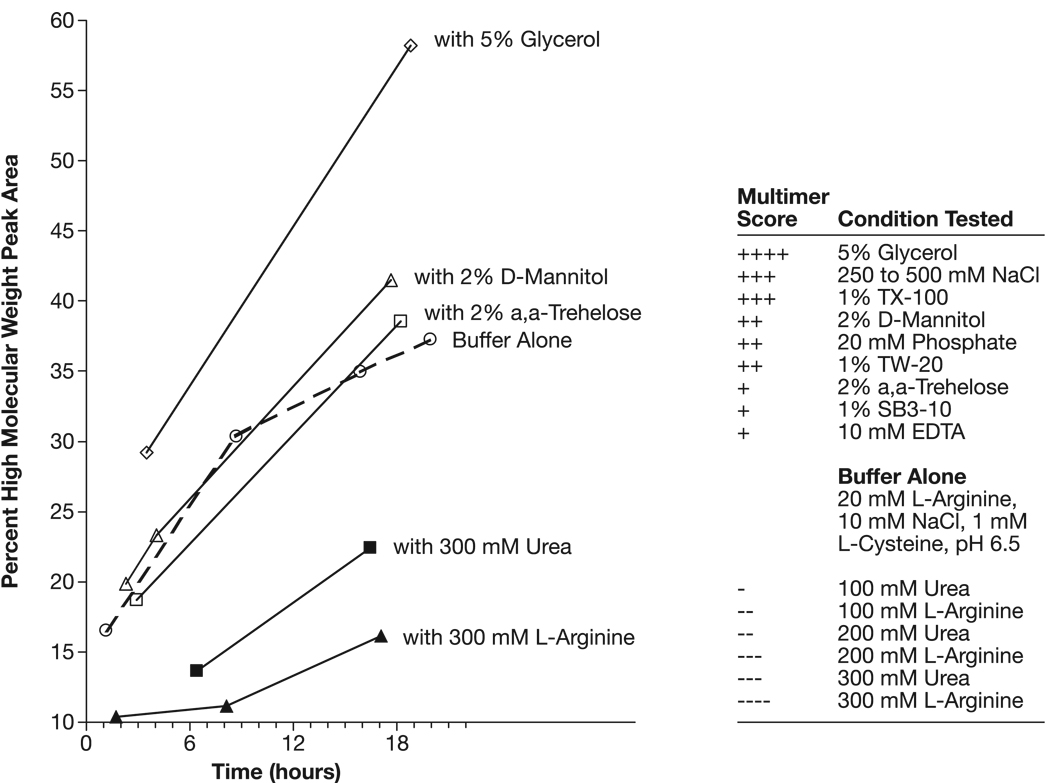
Under conditions of disulfide-bond suppression (1 mM L-cysteine) and non-covalent multimer potentiation (pH 6.5), the effect of solution additives on F1-V monomer solution stability was surveyed. (Fig. 7). Several common formulation additives promoted F1-V self-association, including glycerol, non-reducing sugars, common buffer salts, a non-ionic detergent and a zwitterionic detergent. The multimer-inducing effect of glycerol was surprising in light of its common use to stabilize proteins in solution. These results suggest the existence of a strong, non-covalent (i.e., non-sulfhydryl) self-binding energy within the F1-V protein that drives self-association. Urea and L-arginine suppressed multimer formation with L-arginine (0.3 M) being the most effective F1-V multimer-suppression additive examined. In separate survey, conducted at pH 10.0, L-arginine was more effective than L-lysine for F1-V monomer stabilization (DNS). Thus, the L-arginine guanidinium group may be key to F1-V monomer stabilization. These additive trends informed manufacturing process design and F1-V monomer final formulation.
Fig. 7.
Additive-induced F1-V non-covalent multimer-content modulation at pH 6.5 (ADM).
Freeze drying survey
The materials used in the vaccination portion of this study were not lyophilized. Anticipating the eventual need for a chilled storage product form, lyophilization of F1-V was investigated. Disulfide-linked dimer formation (∼35% dimer, no trimer) was observed in F1-V samples lyophilized without L-cysteine in the formulation buffer (in 20 mM L-arginine, 10 mM NaCl with 2% D-mannitol, pH 9.9) and reconstituted with water. This emergent dimer was dispersed by reconstitution with added 1 mM L-cysteine yielding ∼1.7% non-covalent dimer and ∼2.0% disulfide-linked dimer. We observed minimal F1-V non-covalent dimer formation after lyophilization in formulation buffer supplemented with 2% D-mannitol. This contrasted with the destabilizing effect of 2% D-mannitol observed at pH 6.5 (Fig. 7). Lyophilization with added 1 mM L-cysteine resulted in no discernable increase in dimer content upon rehydration relative to the pre-lyophilization material (DNS). Thus, it may be practical to store F1-V prepared lyophilized in 20 mM L-arginine, 10 mM NaCl, 1 mM L-cysteine, 2% D-mannitol, pH 9.9.
Peptide mapping
F1-V and F1-VC424S identities were determined by peptide mapping (Supplemental Fig. 9). The tryptic-digest sequence coverage for F1-V and F1-VC424S were 73.0 and 85.3%; and for chymotrypic-digest, 58.0 and 61.8%, respectively-confirming target protein expression and recovery. The F1-V tryptic (M+H = 1788.7 Da) and chymotryptic N-terminal peptides were positively identified, and supported the des-Met form of F1-V as reported previously [6]. A modified N-terminus tryptic peptide (M+H = 1831.7 Da, +43.0 Da) was identified. Using high-stringency fragment ion identification criteria (±0.2 Da, Xcorr ≥1.5), searching the centriod data set for carbamylated N-terminal MS/MS ions (+43.0058) identified 4 b-ion identifications versus only 2 b-ion identifications for an acetylation (+42.0105) hypothesis (DNS). Thus, based upon parent and MS/MS ion identifications, N-terminal carbamylation was most strongly supported by the data. Base-peak profiles containing peaks for both the native and modified N-terminus suggested that the proportion of modification was slightly elevated in the F1-VC424S preparation.
The serine mutation in F1-VC424S was confirmed by identification of two tryptic peptides containing serine 424 (residues 398–427, M+2H+2 = 1640.1 Da and residues 406–438, M+2H+2 = 1881.4 Da), and of a single chymotryptic peptide (residues 421– 431, M+H = 1162.5 Da). The corresponding peptides were not found within the F1-V tryptic or chymotryptic MS data sets, confirming assay specificity.
Using automated methods, the F1-V peptides containing cysteine 424 were not identified in tryptic or chymotryptic digests. By visual inspection, a single peak unique to the F1-V tryptic-digest (Supplemental. Fig. 9 at ∼26.2 min within the base-peak profile overlay) remained unassigned. The major ion within this peak corresponded to a 3,411.8 Da peptide that matched the predicted molecular mass for residues 398–427 (3,292.5 Da) if one assumed cysteine 424 was covalently linked to free L-cysteine from the formulation buffer (molecular mass = 121.1 Da, minus 2 H lost upon formation of the disulfide bond). Subsequent examination of MS/MS the fragmentation pattern for this peptide confirmed this assignment. The corresponding peptide was not found within the F1-VC424S tryptic MS data set, further demonstrating assay specificity. Thus, the identities of the native F1-V and F1-VC424S genetic mutant preparations were positively confirmed.
SEC-MALLS
Multiple-angle laser light scattering analysis was performed to assign F1-V solution states to HPLC-SEC assay elution profiles. Based on HPLC-SEC retention volumes alone, the major F1-V peak would have been assigned a molecular weight of ∼100 kDa relative to BioRad high MW size standards (Fig. 6, pH 10 Trace, Peak A). However, by SEC-MALLS the major peak was determined to have an absolute molecular mass between 52.0–55.2 kDa that closely matched the 54 kDa molecular weight expected for F1-V monomer (Fig. 8, Peaks A’ and A). Peak A was thus assigned as monomeric F1-V. This illustrated the known advantage of SEC-MALLS over the conventional methods using reference standards, as SEC protein elution times are known to be affected by differences in molecular radii, molecular shape, and affinities for the column packing.
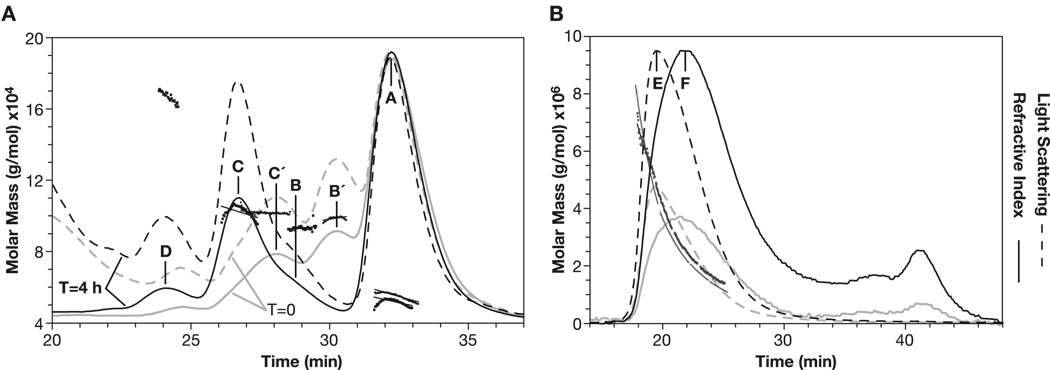
Fig. 8.
SEC-MALLS profiles for titrated F1-VMN (ADM). (Panel A) Calculated molar masses shown as fitted squares. (grey lines, peaks A′, B′, C′) Profiles measured 0.5 h after adjustment to pH 6.5 and, (black lines; peaks A, B, C, D) after 4 h. Calculated peak molar masses with RI-based total protein determination; A’−55.2, A-52.0, B’-98.5, B-93.1, C’-101.8, C-102.2, D-167 kDa. Calculated peak molar masses with A280-based total protein determination; A-44.2., B-83.1, C-86.7, D 137.4 kDa (DNS). (Panel B) Profiles measured <4 h after adjustment to pH 5.1. (grey, 20-µL and black 50- µL injections) of F1-VMN. The light scattering maximum (peak E) preceded the protein content maximum (peak F) resulting in a range of calculated molar masses from 500 to >6,000 kDa.
Upon addition of 1 mM L-cysteine to and adjustment of monomeric F1-V to pH 6.5, a complex transition was observed wherein dimeric F1-V species formed at T=0 (Fig. 8, Panel A, Peaks B′-98.5 kDa and C′-101.8 kDa) and, with time, converted into earlier eluting, apparently more extended, dimeric species (Fig. 8, Panel A, Peaks B-93.1 and C-102.2 kDa). Based upon HPLC-SEC data alone, the ‘F1-V final dimer’ would have been incorrectly assigned as a tetramer (Fig. 6, Panel A, Peak C). Similarly, a well-separated peak with absolute molecular mass of ∼167 kDa was assigned as trimeric F1-V (Fig. 8, Panel A, Peak D). Thus, SEC-MALLS analysis permitted the unequivocal calibration of the SEC-HPLC elution profile for use in establishing that monomeric preparations had been isolated.
After incubation of F1-V monomer at pH 5.0, SEC-MALLS analysis showed conversion to very high molecular mass solution states extending above 1 MDa, with data going off-scale at the beginning of the void peak (Fig. 8, Panel B, Peaks E and F). Thus, SEC-MALLS confirmed that adjustment to pH 5.0 induced formation of an extensively multimerized F1-V population.
ELISA response to F1-V vaccinations
ELISA was performed to determine the anti-F1 and anti-V IgG antibody response against F1-VAG, F1-VC424S, F1-VSTD, and F1-VMN (Table 3). As previously observed [5, 6], the IgG response was dramatically higher against the V antigen compared to the F1 protein for all of the F1-V fusion constructs (Table 3). The average geometric mean anti-V antibody titer was greatest against F1-VC424S but not statistically different than that observed for prior standard preparations of F1-VSTD (119,000 versus 62,000, with a sample size of 30 mice per group). Anti-V antibody titers were statistically equivalent for all of the evaluated F1-V formulations, suggesting that the modified F1-VC424S retains the capacity for recognition by protective anti-V antibodies. Thus, as there is no statistical difference between the anti-F1 and anti-V titers among these antigen groups, these findings support the hypothesis that F1-V aggregation state does not influence the capacity for protective antibodies to recognize the individual component proteins within the F1-V fusion protein.
Table 3.
ELISA titers after vaccination with ALH-alone negative control and with ALH-adsorbed F1-V formulations (ADM).
| Titer Type | Treatment | N | Geometric Mean | Lower 95% CL | Upper 95% CL |
|---|---|---|---|---|---|
| V | ALH | 10 | 300 | 300 | 300 |
| F1-VAG | 30 | 76,000 | 49,000 | 119,000 | |
| F1-Vc424S-MN | 30 | 119,000 | 79,000 | 179,000 | |
| F1-VSTD | 30 | 62,000 | 43,000 | 89,000 | |
| F1-VMN | 29 | 86,000 | 54,000 | 136,000 | |
| F1 | ALH | 10 | 300 | 300 | 300 |
| F1-VAG | 30 | 30,000 | 17,000 | 51,000 | |
| F1-Vc424S-MN | 30 | 30,000 | 17,000 | 54,000 | |
| F1-VSTD | 30 | 19,000 | 12,000 | 30,000 | |
| F1-VMN | 29 | 30,000 | 16,000 | 55,000 |
The anti-F1 titers were substantially lower than anti-V titers for all fusion protein formulations, and these titers did not vary as much as the anti-V titers between the various treatments. A slightly lower, but not statistically significant, anti-F1 titer of 19,000 was observed for the positive control F1-VSTD, while the three additional F1-V formulations demonstrated identical average anti-F1 titers of 30,000. The F1 portion of F1-V was smaller and exhibited a less complex secondary structure than the V protein. Thus, it is not surprising to see less immunogenicity of F1, even after manipulation of the V antigen component.
Protective efficacy and statistical analysis
Purified F1-V formulations (F1-VMN, F1-VAG, F1-VC424S, and F1-VSTD ) were adsorbed to Alhydrogel (ALH) adjuvant in water, diluted into 1x PBS, and used to inoculate mice before s.c. challenge with 107 – 109 LD50 of Y. pestis CO92. The Y. pestis CO92 strain is highly virulent as indicated by 100% fatality among ALH only-vaccinated mice at a much lower challenge dose (104 LD50 compared to 107 – 109 LD50). All of the ALH control animals were dead by day 5 after challenge with an average time to death of 3.2 days. As indicated in Table 4, 100% of F1-VC424S vaccinated mice survived lethal plague challenge with either 107 or 108 LD50 Y. pestis CO92. In comparison, 70% of F1-VSTD animals survived challenge with either 107 or 108 LD50 Y. pestis. Forced monomeric (F1-VMN) and forced multimeric (F1-VAG) forms of F1-V elicited 70–80% survival under the same challenge conditions. The protective efficacy of these F1-V-based vaccines was further demonstrated by 30–50% survival of mice when challenged with 109 LD50 Y. pestis.
Table 4.
(Panel A) Mouse survival after vaccination and lethal plague challenge by group (ADM) (Panel B) Pair-wise survival rate group-comparisons computed with Fisher’s exact test (p < 0.05%, shown bold) (ADM).
| Vaccinated Mouse Survival Data | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | Treatment | Challenge Dose | Alive | Dead | Total | Percent Survival | Mean Survival Time(SE) | Mean Days to Death (SD) | Min | Max |
| 1 | ALH | 104 | 0 | 10 | 10 | 0 | 3.2(0.3) | 3.2 (0.8) | 2 | 5 |
| 2 | FI-Vstd | 107 | 7 | 3 | 10 | 70 | 21.7(3.7) | 7.0 (0.0) | 7 | 7 |
| 3 | F1-VMN | 107 | 7 | 2 | 9 | 78 | 23.2(4.2) | 6.5 (2.1) | 5 | 8 |
| 4 | F1-VAG | 107 | 8 | 2 | 10 | 80 | 23.2(4.3) | 4.0 1.4) | 3 | 5 |
| 5 | F1-Vc424S -MN | 107 | 8 | 0 | 8 | 100 | 28.0(0.0) | − | − | − |
| 6 | F1-VSTD | 108 | 7 | 3 | 10 | 70 | 21.9(3.6) | 7.7 (2.1) | 6 | 10 |
| 7 | F1-VMN | 108 | 8 | 2 | 10 | 80 | 23.1(4.4) | 3.5 (2.1) | 2 | 5 |
| 8 | F1-VAG | 108 | 8 | 2 | 10 | 80 | 23.4(4.1) | 5.0 (2.8) | 3 | 7 |
| 9 | F1-VC424S-MN | 108 | 9 | 0 | 9 | 100 | 28.0(0.0) | − | − | − |
| 10 | FI-Vstd | 109 | 5 | 5 | 10 | 50 | 17.9(3.6) | 7.8 (3.3) | 4 | 13 |
| 11 | F1-VMN | 109 | 4 | 4 | 8 | 50 | 17.1(4.6) | 6.3 (4.g) | 2 | 11 |
| 12 | F1-VAG | 109 | 3 | 7 | 10 | 30 | 13.4(3.8) | 7.1 (3.2) | 3 | 11 |
| 13 | F1-VC424S-MN | 109 | 4 | 6 | 10 | 40 | 16.2(3.4) | 8.3 (3.2) | 3 | 12 |
| Pairwise Comparison p-values by Challenge Dose |
||||||
|---|---|---|---|---|---|---|
| Percent Survival |
Time to Death |
|||||
| Comparison Groups | 107 | 108 | 109 | 107 | 108 | 109 |
| F1-VSTDvs. F1-VMN | 1.0000 | 1.0000 | 1.0000 | 0.5881 | 0.0220 | 0.9349 |
| F1-VSTDvs.F1-VAG | 1.0000 | 1.0000 | 0.8062 | 0.0153 | 0.1360 | 0.9800 |
| F1”VsTDVS- F1-Vc424S-MN | 0.5437 | 0.5118 | 0.0655 | ** | ** | 0.9800 |
| F1-VSTDvs. ALH | 0.0009 | 0.0015 | 0.0076 | 0.0010 | 0.0019 | 0.0126 |
| F1-VMNvs.F1-VAG | 1.0000 | 1.0000 | 0.8948 | 0.0423 | 0.5031 | 0.9798 |
| F1-VMNVS. F1-VC424S-MN | 0.8351 | 0.8044 | 1.0000 | ** | ** | 0.7977 |
| F1-VMNvs.ALH | 0.0003 | 0.0004 | 0.0248 | 0.0032 | 0.3795 | 0.0426 |
| F1-VAGVS. F1-VC424S-MN | 0.8354 | 0.8044 | 1.0000 | ** | ** | 0.9349 |
| F1-VAGvs. ALH | 0.0001 | 0.0004 | 0.1776 | 0.1534 | 0.1296 | 0.0126 |
| F1-Vc424S-MNVS.ALH | <.0001 | <.0001 | 0.0717 | ** | ** | 0.0035 |
Pairwise statistical comparisons were performed for all treatment groups. The statistical results indicate significant differences in “Percent Survival” among the various vaccination groups compared to the ALH control group (Table 4 Panel B). Statistically significant differences in survival were observed for all vaccination groups compared to the ALH control group at the 107 – 108 LD50 dose range. Only F1-VSTD and F1-VMN retained significant survival percentages at 109 LD50. Statistically significant differences in survival were not observed between the various vaccination treatments when compared to each other.
Whether or not those mice vaccinated with a given F1-V preparation, that died, survived longer than the control mice or mice vaccinated with another F1-V preparation, that died, is illustrated in Table 4, Panel B. The statistical comparison designated “Time to Death” highlights significant increases in average time to death among vaccinated mice compared to ALH-only control mice and to other vaccinated mice groups. For example, at 107 LD50, the average time to death for F1-VMN vaccinated mice was 6.5 days, compared to 3.2 days for ALH inoculated mice. The difference in time to death between F1-VMN and ALH groups was statistically significant (p < 0.0032). F1-VC424S statistical comparisons were not performed at the 107 and 108 challenge dose because none of the mice died under those conditions. The analysis indicates that all of the F1-VSTD vaccinated mice that died during the experiment, regardless of the challenge dose, lived significantly longer than the ALH control mice. Most of the other vaccinated mice (F1-VMN / F1-VAG / F1-VC424S) that died, also survived significantly longer than the control mice. Significant differences in survival time between the test groups compared to each other were observed sporadically.
CONCLUSIONS
The 53-kDa F1-V fusion protein was modified by site-directed mutagenesis to replace the sole cysteine with a serine residue, thus producing F1-VC424S. Novel F1-V purification methods were employed to isolate monomeric F1-V and F1-VC424S that resulted in 1 to 2 mg of > 95% pure, mono-disperse protein per gram of cell paste. Standard (cysteine containing) F1-V and F1-VC424S were compared for stability and aggregation characteristics under various conditions of solution pH and buffer additive. Predominately monomeric F1-V forms were observed at pH 10.0 with progressive aggregation occurring as pH conditions were lowered toward pH 5.0. Of the buffer additives that were compared, L-cysteine was found to provide the best disulfide bond disruption, while L-arginine [18] was found to be the most effective additive for disrupting non-covalent multimer associations.
Standard, cysteine-containing F1-V formulations were evaluated side-by-side with the modified F1-VC424S form for protective efficacy against lethal plague challenge in mice. Our results demonstrate that substitution of the cysteine residue with serine did not statistically affect the activity of F1-V to elicit protective immunity against plague. Moreover, the monomeric and multimeric forms of F1-V exhibit equivalent immunogenicity and protective efficacy against subcutaneous infection.
Numerous expression and purification strategies for F1-V have been published ranging from traditional prokaryotic systems [5, 6, 19, 20] to transgenic tomatoes [21] and the tobacco-like Nicotiana benthamiana [22]. Regardless of the ultimate expression strategy employed, the final F1-V fusion protein will retain a tendency to multimerize because of its subunit composition. Although this self-association is due mainly to the F1 subcomponent, the fusion architecture actually reduces polydispersity compared to the individual F1 protein, which is even more aggregative [6]. Thus, the F1-V fusion based plague antigen is at the forefront of plague vaccine development [23–26]. As demonstrated by the findings reported herein, we propose that use of the described F1-VC424S protein form should facilitate the enhanced production and stability of F1-V–based plague vaccines.
Supplementary Material
(Fig. 9a–9f) Peptide mapping analysis of F1-V and F1-VC424S preparations (ADM). (9a) C18 reverse phase-HPLC/MS base-peak profile overlay of tryptic digests for both preparations and a close-up showing elution positions for peptides corresponding to the native N-terminal fragment (residues 1–18, 25.7 min), modified N-terminus (+43 kDa, ∼28.4 min), two F1-VC424S-derived fragments containing serine 424 (residues 398–427, 26.7 min and residues 406–438, 27.0 min), and a derived F1-V fragment containing cysteine 424 disulfide bonded to a single free L-cysteine (residues 398–427, predicted pre-adduct molecular mass = 3,292.5 Da, plus 121.1 Da L-cysteine adduct, minus 2 H from formation of disulfide bond, actual observed molecular mass = 3,411.8 Da). The peak at 26.6 min was identified as an F1-V fragment (residues 306-340) unrelated to the F1-VC424S derived peak at 26.7 min; (9b) MS (Top) and MS/MS spectra for the F1-V derived native N-terminal fragment; (9c) MS (Top) and MS/MS spectra for the F1-V derived modifed N-terminal fragment; (9d) MS (Top) and MS/MS spectra for a F1-VC424S-derived fragment containing serine 424 (residues 398–427]; (9e) MS (Top) and MS/MS spectra for a F1-VC424S-derived chymotryptic fragment containing serine 424 [residues 421–431, retention time = 17.7 min, M+H = 1162.5 Da]; (9f) MS (Top) and MS/MS spectra for the F1-V-derived fragment adducted to L-cysteine [residues 398–427 + L-cysteine].
ACKNOWLEDGMENTS
We thank the persons who provided invaluable support to this work including Steven Tobery and Anthony Bassett for expert assistance with the animal studies; Sarah Norris for statistical consulting; Jennifer Myers for advice in performing the ELISA studies; George Knapp and Terry Sumpter for mass spectral analysis; Samir Shaban for lyophilization; Trevor Broadt for genomic DNA analysis; and Carolyn Whistler, Richard Frederickson, and Maritta Grau for graphics and editorial advice.
This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400; the National Institute for Allergic and Infectious Diseases (NIAID), National Institutes of Health, through Interagency Agreement Y3-CM-3004; and by US Army Medical Research Institute of Infectious Diseases (USAMRIID) research project number 03-4-5A-013 (J. Goodin).
Selected materials, derived in part from the reported work, were deposited in the NIAID Biodefense and Emerging Diseases Research Resources Repository (NR 2561-3).
The content of this publication does not necessarily reflect the views or policies of either the US Army or the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Opinions, interpretations, conclusions, and recommend-ations are those of the authors.
ABBREVIATIONS
- ∼
Approximately
- AX
absorbance at x nm
- ADM
As described in the materials and methods
- ALH
Alhydrogel adjuvant
- CL
confidence limit
- CV
Column volume
- DNS
Data not shown
- DTE
dithioerythritol
- DTT
dithiothreitol
- F1-VAG
multimer-enriched F1-V preparation derived from F1-VMN
- F1-VMN
monomer-enriched F1-V preparation
- F1-VC424S-MN
F1-V with systeine 424 replaced with serine, monomer-enriched
- F1-VSTD
previously reported preparation of F1-V
- F1-V(S-S)
F1-V disulfide linked dimer
- F1-V(NC
F1-V non-covalently associated dimer
- Gdn HCl
guanidine hydrochloride
- IAA
iodoacetamide
- HPLC-SEC
high-performance liquid chromatography size-exclusion chromatography
- IEX
ion exchange chromatography
- L-Cys
L-cysteine
- MW
molecular weight
- RI
refractive index
- SEC-MALLS
size exclusion chromatography - multi-angle laser light scattering
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
WORKS CITED
- 1.Inglesby TV, Dennis DT, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Koerner JF, Layton M, McDade J, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Schoch-Spana M, Tonat K. Plague as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 2000;283:2281–2290. doi: 10.1001/jama.283.17.2281. [DOI] [PubMed] [Google Scholar]
- 2.Titball RW, Williamson ED. Vaccination against bubonic and pneumonic plague. Vaccine. 2001;19:4175–4184. doi: 10.1016/s0264-410x(01)00163-3. [DOI] [PubMed] [Google Scholar]
- 3.Pitt MLM, Estep EJ, Welkos SL, Friedlander AM. Washington, DC. Friedlander, in: Proceedings of the Abstracts of the 94th General Meeting of the American Society for Microbiology; 1994. [Google Scholar]
- 4.Anderson GW, Heath DG, Bolt CR, Welkos SL, Friedlander AM. Short- and long-term efficacy of single-dose subunit vaccines against Yersinia pestis in mice. Am. J. Trop. Med. Hyg. 1998;58:793–799. doi: 10.4269/ajtmh.1998.58.793. [DOI] [PubMed] [Google Scholar]
- 5.Heath DG, Anderson GW, Mauro JM, Welkos SL, Andrews GP, Adamovicz J, Friedlander AM. Protection against experimental bubonic and pneumonic plague by a recombinant capsular F1-V antigen fusion protein vaccine. Vaccine. 1998;16:1131–1137. doi: 10.1016/s0264-410x(98)80110-2. [DOI] [PubMed] [Google Scholar]
- 6.Powell BS, Andrews GP, Enama JT, Jendrek S, Bolt C, Worsham P, Pullen JK, Ribot W, Hines H, Smith L, Heath DG, Adamovicz JJ. Design and testing for a nontagged F1-V fusion protein as vaccine antigen against bubonic and pneumonic plague. Biotechnol. Prog. 2005;21:1490–1510. doi: 10.1021/bp050098r. [DOI] [PubMed] [Google Scholar]
- 7.Lee SH, Carpenter JF, Chang BS, Randolph TW, Kim YS. Effects of solutes on solubilization and refolding of proteins from inclusion bodies with high hydrostatic pressure. Protein Sci. 2006;15:304–313. doi: 10.1110/ps.051813506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panda AK. Bioprocessing of therapeutic proteins from the inclusion bodies of Escherichia coli. Adv. Biochem. Eng. Biotechnol. 2003;85:43–93. doi: 10.1007/3-540-36466-8_3. [DOI] [PubMed] [Google Scholar]
- 9.Chi EY, Krishnan S, Randolph TW, Carpenter JF. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm. Res. 2003;20:1325–1336. doi: 10.1023/a:1025771421906. [DOI] [PubMed] [Google Scholar]
- 10.Miller J, Williamson ED, Lakey JH, Pearce MJ, Jones SM, Titball RW. Macromolecular organisation of recombinant Yersinia pestis F1 antigen and the effect of structure on immunogenicity. FEMS Immunol. Med. Microbiol. 1998;21:213–221. doi: 10.1111/j.1574-695X.1998.tb01168.x. [DOI] [PubMed] [Google Scholar]
- 11.Zavialov AV, Berglund J, Pudney AF, Fooks LJ, Ibrahim TM, MacIntyre S, Knight SD. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell. 2003;113:587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]
- 12.Williams RC, Gewurz H, Quie PG. Effects of fraction I from Yersinia pestis on phagocytosis in vitro. J. Infect. Dis. 1972;126:235–241. doi: 10.1093/infdis/126.3.235. [DOI] [PubMed] [Google Scholar]
- 13.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;11:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gidh AV, Decker SR, Vinzant TB, Himmel ME, Williford C. Determination of lignin by size exclusion chromatography using multi-angle laser light scattering. J. Chromatogr. A. 2006;1114:102–110. doi: 10.1016/j.chroma.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 15.Casini GL, Graham D, Heinie D, Garcea RL, Wu DT. In vitro papillomavirus capsid assembly analyzed by light scattering. Virology. 2004;325:320–327. doi: 10.1016/j.virol.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 16.National Research Council, Institute of Laboratory Animal Resources. Guide for the Care and Use of Laboratory Animals. Bethesda: U.S. Department of Health and Human Services; 1986. U.S. Department of Health and Human Services publication no. (NIH) 86-23. [Google Scholar]
- 17.Huxtable RJ. New York: Plenum Press; 1986. Biochemistry of the Sulfur; pp. 207–208. [Google Scholar]
- 18.Tsumoto K, Umetsu M, Kumagai I, Ejima D, Philo JS, Arakawa T. Role of arginine in protein refolding, solubilization, and purification. Biotechnol. Prog. 2004;20:1301–1308. doi: 10.1021/bp0498793. [DOI] [PubMed] [Google Scholar]
- 19.Williamson ED. Plague vaccine research and development. J. Appl. Microbiol. 2001;91:606–608. doi: 10.1046/j.1365-2672.2001.01497.x. [DOI] [PubMed] [Google Scholar]
- 20.Andrews GP, Heath DG, Anderson GW, Welkos SL, Friedlander AM. Fraction 1 capsular antigen (F1) purification from Yersinia pestis CO92 and from an Escherichia coli recombinant strain and efficacy against lethal plague challenge. Infect. Immun. 1996;64:2180–2187. doi: 10.1128/iai.64.6.2180-2187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez ML, Pinyerd HL, Crisantes JD, Rigano MM, Pinkhasov J, Walmsley AM, Mason HS, Cardineau GA. Plant-made subunit vaccine against pneumonic and bubonic plague is orally immunogenic in mice. Vaccine. 2006;24:2477–2490. doi: 10.1016/j.vaccine.2005.12.057. [DOI] [PubMed] [Google Scholar]
- 22.Santi L, Giritch A, Roy CJ, Marillonnet S, Klimyuk V, Gleba Y, Webb R, Arntzen CJ, Mason HS. Protection conferred by recombinant Yersinia pestis antigens produced by a rapid and highly scalable plant expression system. Proc. Natl. Acad. Sci. USA. 2006;103:861–866. doi: 10.1073/pnas.0510014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glynn A, Freytag LC, Clements JD. Effect of homologous and heterologous prime-boost on the immune response to recombinant plague antigens. Vaccine. 2005;23:1957–1965. doi: 10.1016/j.vaccine.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 24.Tripathi V, Chitralekha KT, Bakshi AR, Tomar D, Deshmukh RA, Baig MA, Rao DN. Inducing systemic and mucosal immune responses to B-T construct of F1 antigen of Yersinia pestis in microsphere delivery. Vaccine. 2006;24:3279–3289. doi: 10.1016/j.vaccine.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Titball RW, Williamson ED. Yersinia pestis (plague) vaccines. Expert Opin. Biol. Ther. 2004;4:965–973. doi: 10.1517/14712598.4.6.965. [DOI] [PubMed] [Google Scholar]
- 26.Leary SE, Griffin KF, Garmory HS, Williamson ED, Titball RW. Expression of an F1/V fusion protein in attenuated Salmonella typhimurium and protection of mice against plague. Microb. Pathog. 1997;23:167–179. doi: 10.1006/mpat.1997.0141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Fig. 9a–9f) Peptide mapping analysis of F1-V and F1-VC424S preparations (ADM). (9a) C18 reverse phase-HPLC/MS base-peak profile overlay of tryptic digests for both preparations and a close-up showing elution positions for peptides corresponding to the native N-terminal fragment (residues 1–18, 25.7 min), modified N-terminus (+43 kDa, ∼28.4 min), two F1-VC424S-derived fragments containing serine 424 (residues 398–427, 26.7 min and residues 406–438, 27.0 min), and a derived F1-V fragment containing cysteine 424 disulfide bonded to a single free L-cysteine (residues 398–427, predicted pre-adduct molecular mass = 3,292.5 Da, plus 121.1 Da L-cysteine adduct, minus 2 H from formation of disulfide bond, actual observed molecular mass = 3,411.8 Da). The peak at 26.6 min was identified as an F1-V fragment (residues 306-340) unrelated to the F1-VC424S derived peak at 26.7 min; (9b) MS (Top) and MS/MS spectra for the F1-V derived native N-terminal fragment; (9c) MS (Top) and MS/MS spectra for the F1-V derived modifed N-terminal fragment; (9d) MS (Top) and MS/MS spectra for a F1-VC424S-derived fragment containing serine 424 (residues 398–427]; (9e) MS (Top) and MS/MS spectra for a F1-VC424S-derived chymotryptic fragment containing serine 424 [residues 421–431, retention time = 17.7 min, M+H = 1162.5 Da]; (9f) MS (Top) and MS/MS spectra for the F1-V-derived fragment adducted to L-cysteine [residues 398–427 + L-cysteine].