Abstract
Superinfection exclusion or homologous interference, a phenomenon in which a primary viral infection prevents a secondary infection with the same or closely related virus, has been observed commonly for viruses in various systems, including viruses of bacteria, plants, and animals. With plant viruses, homologous interference initially was used as a test of virus relatedness to define whether two virus isolates were “strains” of the same virus or represented different viruses, and subsequently purposeful infection with a mild isolate was implemented as a protective measure against isolates of the virus causing severe disease. In this study we examined superinfection exclusion of Citrus tristeza virus (CTV), a positive-sense RNA closterovirus. Thirteen naturally occurring isolates of CTV representing five different virus strains and a set of isolates originated from virus constructs engineered based on an infectious cDNA clone of T36 isolate of CTV, including hybrids containing sequences from different isolates, were examined for their ability to prevent superinfection by another isolate of the virus. We show that superinfection exclusion occurred only between isolates of the same strain and not between isolates of different strains. When isolates of the same strain were used for sequential plant inoculation, the primary infection provided complete exclusion of the challenge isolate, whereas isolates from heterologous strains appeared to have no effect on replication, movement or systemic infection by the challenge virus. Surprisingly, substitution of extended cognate sequences from isolates of the T68 or T30 strains into T36 did not confer the ability of resulting hybrid viruses to exclude superinfection by those donor strains. Overall, these results do not appear to be explained by mechanisms proposed previously for other viruses. Moreover, these observations bring an understanding of some previously unexplained fundamental features of CTV biology and, most importantly, build a foundation for the strategy of selecting mild isolates that would efficiently exclude severe virus isolates as a practical means to control CTV diseases.
Superinfection exclusion or homologous interference is a phenomenon in which a preexisting viral infection prevents a secondary infection with the same or a closely related virus, whereas infection by unrelated viruses can be unaffected. The phenomenon was first observed by McKinney (57, 58) between two genotypes of Tobacco mosaic virus (TMV) and later with bacteriophages (21, 94). Since that time, the phenomenon has been observed often for viruses of animals (1, 13, 18, 34, 43, 47, 50, 85, 86-88, 102, 103) and plants (11, 30, 31, 32, 39, 40, 49, 77, 99, 100). In plant virology, homologous interference initially was used as a test of virus relatedness to define whether two virus isolates were “strains” of the same virus or represented different viruses (58, 77). Subsequently, it was developed into a management tool to reduce crop losses by purposely infecting plants with mild isolates of a virus to reduce infection and losses due to more severe isolates, which is referred to as “cross-protection” (reviewed in references 32 and 40).
Homologous superinfection exclusion of animal viruses has been related to several mechanisms acting at various stages of the viral life cycle, including prevention of the incoming virus entry into cells (50, 86, 87), or inhibition of translation or interference with replication (1, 47, 50, 83). Several mechanisms have been postulated for homologous interference of plant viruses, including prevention of the disassembly of the challenge virus as it enters the cell resulting from the expression of the coat protein of the protector virus (67, 84; reviewed in reference 10) and induction of RNA silencing by the protector virus that leads to sequence-specific degradation of the challenge virus RNA (24, 69, 70). However, common mechanisms of superinfection exclusion, expected to be associated with the viruses of plants and animals, have not been elucidated.
Citrus tristeza virus (CTV) is the largest and most complex member of the Closteroviridae family, which contains viruses with mono-, bi-, and tripartite genomes transmitted by a range of insect vectors, including aphids, whiteflies, and mealybugs (3, 6, 19, 20, 46). CTV has long flexuous virions (2,000 nm by 10 to 12 nm) encapsidated by two coat proteins and a single-stranded RNA genome of ∼19.3 kb. The major coat protein (CP) covers ca. 97% of the genomic RNA, and the minor coat protein (CPm) completes encapsidation of the genome at its 5′ end (25, 81). The RNA genome of CTV encodes 12 open reading frames (ORFs) (44, 64) (Fig. 1). ORFs 1a and 1b are expressed from the genomic RNA and encode polyproteins required for virus replication. ORF 1a encodes a 349-kDa polyprotein containing two papainlike protease domains plus methyltransferaselike and helicaselike domains. Translation of the polyprotein is thought to occasionally continue through the polymerase-like domain (ORF 1b) by a +1 frameshift. Ten 3′-end ORFs are expressed by 3′-coterminal subgenomic RNAs (sgRNAs) (37, 45) and encode the following proteins: major (CP) and minor (CPm) coat proteins, p65 (HSP70 homolog), and p61 that are involved in assembly of virions (79); a hydrophobic p6 protein with a proposed role in virus movement (20, 89); p20 and p23, which along with CP are suppressors of RNA silencing (54); and p33, p13, and p18, whose functions remain unknown. Remarkably, citrus trees can be infected with mutants with three genes deleted: p33, p18, and p13 (89).
FIG. 1.
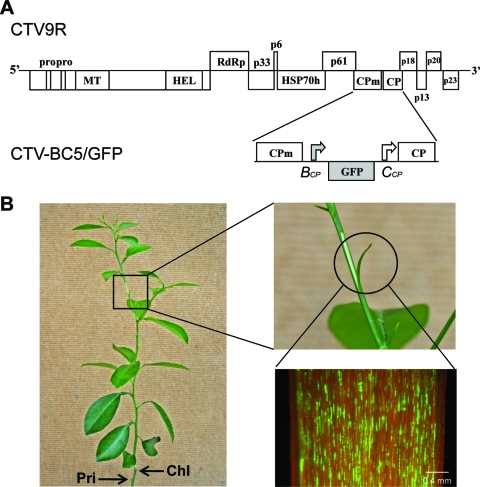
(A) Schematic diagram of the genome organization of wild-type CTV (CTV9R) and its derivative CTV-BC5/GFP encoding GFP. The open boxes represent ORFs and their translation products. PRO, papainlike protease domain; MT, methyltransferase; HEL, helicase; RdRp, an RNA-dependent RNA polymerase; HSP70h, HSP70 homolog; CPm, minor coat protein; CP, major coat protein; GFP, green fluorescent protein. Bent arrows indicate positions of BYV (BCP) or CTV CP (CCP) sgRNA controller elements. Inserted elements are shown in gray. (B) Scheme of the “superinfection exclusion assay.” Young Madam Vinous sweet orange trees were initially inoculated with one of 13 tested CTV isolates. When primary infections were established, the trees were subsequently challenged with CTV-BC5/GFP. All inoculations were done by grafting of the infected tissue into the stem of a tree. The positions of primary (Pri) and challenge (Chl) graft inoculations are shown. The ability of the challenge virus to superinfect trees was determined by visual observation of GFP fluorescence in phloem-associated cells on the internal surface of bark from a young flash starting at about 2 months upon challenge inoculation. Scale bar, 0.4 mm.
The host range of CTV is limited to citrus in which the virus infects only phloem-associated cells. CTV consists of numerous isolates that have distinctive biological and genetic characteristics (38, 48, 56, 72, 74, 75, 95). Recently, a classification strategy for CTV isolates was proposed based on sequence similarity. Analysis of nearly 400 isolates in an international collection revealed five major CTV genotype groups with some isolates undefined (38). For the purposes of the present study, strains are defined as phylogenetically distinct lineages of CTV based upon analysis of nucleotide sequences of the 1a ORF (38). This region of the genome shows high genetic diversity between CTV variants, with levels of sequence identity ranging between 72.3 to 90.3% (38, 48, 52, 74, 75; M. Hilf, unpublished data). Using this definition, T3, T30, T36, VT, and T68 are designated as strains. Individual virus samples are designated as isolates of one of these strains. The ORF 1a nucleotide sequences of isolates of the T36 and T68 strains are equally dissimilar to isolates of the T3, T30, and VT strains, with identities of 72.9, 73, and 72.4% and 77.6, 77.9, and 76.8%, respectively. Identities of ORF 1a range from 89.4 to 90.3% between isolates of the T3, T30, and VT strains. Sequences of ORF1a of isolates belonging to the T36 strain and those from the T68 strain show 72.3% identity. This compares to a range of 89 to 94.8% identity found in the more conserved 3′-half regions of the genomes of isolates from different CTV strains. Each strain is named after a “type isolate” and is composed of isolates with minor sequence divergence (generally less than 5% throughout genome) from the type member. However, isolates of a strain may have significant variations in symptoms and symptoms severity. Remarkably, field trees harbor complex populations of CTV, which are often composed of mixtures of different strains and recombinants between these strains (36, 48, 52, 68, 75, 96, 101). The genetic basis of such frequent coexistence of different strains within the same tree is unknown.
CTV causes economically important diseases of citrus worldwide. One of the most effective management tools has been cross-protection when effective protecting isolates could be found. Preinfection with mild isolates allows commercial production of sweet oranges and limes in Brazil (16) and Peru (9) and grapefruit in South Africa (92). However, identification of protecting isolates has been empirical, difficult, and rare. Cross-protection usually has worked only in certain varieties, and the lack of effective protecting isolates has prevented its use in many varieties and citrus growing areas (15, 41, 61, 73). In general, there has been no understanding why some mild isolates were effective and others failed to protect. Because CTV diseases prevail in citrus growing areas worldwide, elucidation of the mechanisms of exclusion of one CTV variant by another one is an important goal.
In the present study we examined relationships between different genotypes of CTV in terms of their ability to prevent superinfection by another isolate of the virus. We show that superinfection exclusion occurred only between minor genetic variants of the same strain (sequence group) and not between isolates of different strains. When isolates of the same strain were used for sequential plant inoculation, the primary infection provided full exclusion of the challenge isolate. In all combinations of virus isolates belonging to different strains, the primary infection of plants with one strain had no noticeable effect on the establishment of the secondary infection. The results obtained here help elucidate some previously unexplained fundamental features of CTV biology and pose the possibility of an existence of a novel mechanism for superinfection exclusion between virus variants.
MATERIALS AND METHODS
Virus isolates and inoculation of citrus trees.
The Florida CTV virus isolates used in the present study (see Tables 1 and 2) have been maintained in citrus plants under greenhouse conditions. These plants were used as sources of virus for subsequent graft inoculations of young trees. Genomic sequences of five isolates are known: T36 (44, 78), T30-1 (4), T3 and T68 (M. Hilf, unpublished data), and FS577-1-8 (S. Tatineni, unpublished data). The genotype and strain designation of the other isolates have been determined based on genetic marker analysis (38; M. Hilf, unpublished data). In addition to wild-type isolates, the following isolates generated from recombinant virus constructs engineered based on a cDNA clone of T36 (78, 80) and propagated in citrus plants were used in the present study: CTV-BC5/GFP, containing an insertion of the green fluorescent protein (GFP) ORF in the region between the CPm and CP genes in the CTV genome (26); CTV9R-MCA13NR, containing a single nucleotide substitution (U to A) that creates a corresponding amino acid substitution (Phe to Tyr) at position 124 in the CP, removing the epitope recognized by the selective monoclonal antibody MCA13 (82); a set of six T36/T30 hybrid viruses bearing substitutions from different regions of the T30-1 genome into the genome of T36 (3′ nontranslated region (NTR) plus p23 ORF; 3′ NTR plus p23, p20, p13, and part of p18 ORFs; p13 ORF; p18 ORF; p61 ORF; and HSP70h plus p61 ORFs as described by Albiach-Martí et al. in reference 5); and a set of five hybrid viruses engineered from isolates T36 and T68-1 according to the procedures described below.
TABLE 1.
Examination of CTV isolates for their ability to prevent superinfection by T36-based GFP-expressing CTV virus in two citrus hostsa
| Primary inoculation |
Citrus macrophylla |
Madam Vinous sweet orange |
|||
|---|---|---|---|---|---|
| Strain | Isolate | Prechallenge CTV titer ± SD | GFP | Prechallenge CTV titer ± SD | GFP |
| T30 | T30-1 | 3.23 ± 0.015 | Yes | 1.56 ± 0.021 | Yes |
| T55-1 | 2.99 ± 0.019 | Yes | 1.31 ± 0.017 | Yes | |
| T4 | 3.31 ± 0.034 | Yes | 1.81 ± 0.023 | Yes | |
| FL278 | 2.85 ± 0.010 | Yes | 1.52 ± 0.014 | Yes | |
| VT | FL202-1 | 3.30 ± 0.029 | Yes | 1.65 ± 0.031 | Yes |
| FS672 | 3.12 ± 0.040 | Yes | 1.62 ± 0.025 | Yes | |
| FS674HT | 2.90 ± 0.026 | Yes | 1.48 ± 0.015 | Yes | |
| FS701 | 3.09 ± 0.065 | Yes | 1.52 ± 0.011 | Yes | |
| T68 | T68-1 | 3.38 ± 0.055 | Yes | 1.79 ± 0.044 | Yes |
| T3 | T3 | 3.36 ± 0.042 | Yes | 1.89 ± 0.032 | Yes |
| T36 | T36 | 3.33 ± 0.032 | No | 1.57 ± 0.021 | No |
| T66-1 | 3.07 ± 0.043 | No | 1.45 ± 0.010 | No | |
| FS577-1-8 | 3.42 ± 0.065 | No | 1.58 ± 0.038 | No | |
| None | 0.10 ± 0.004 | Yes | 0.09 ± 0.005 | Yes | |
Trees were assayed at 6 weeks after initial inoculation by DAS-I-ELISA using CTV-specific 908 IgG as trapping antibody at 1 μg/ml concentration and ECTV 172 monoclonal antibody as detecting antibody at a 1:50,000 dilution. For the prechallenge CTV titers, the ELISA values (A405) are average values from three plants. The GFP fluorescence was observed in the bark tissue of trees by using a dissecting fluorescence microscope at 2 months after challenge with T36 CTV-BC5/GFP virus.
TABLE 2.
Examination of superinfection exclusion for different combinations of CTV isolates using MCA13 antibodya
| Primary inoculation |
Prechallenge virus titer ± SD | Challenge inoculation |
Postchallenge virus titer ± SD | ||
|---|---|---|---|---|---|
| Strain | Isolate | Strain | Isolate | ||
| None | 0.04 ± 0.003 | T68 | T68-1 | 1.43 ± 0.023 | |
| 0.06 ± 0.001 | T3 | T3 | 1.13 ± 0.014 | ||
| 0.05 ± 0.006 | VT | FL202-1 | 1.66 ± 0.029 | ||
| 0.05 ± 0.005 | T36 | T36 | 1.41 ± 0.018 | ||
| T30 | T30-1 | 0.07 ± 0.005 | T68 | T68-1 | 1.55 ± 0.022 |
| 0.04 ± 0.006 | T3 | T3 | 1.03 ± 0.019 | ||
| 0.06 ± 0.003 | VT | FL202-1 | 1.22 ± 0.020 | ||
| 0.08 ± 0.004 | T36 | T36 | 1.20 ± 0.017 | ||
| T36 | CTV9R-MCA13NR | 0.06 ± 0.008 | T68 | T68-1 | 1.30 ± 0.020 |
| 0.08 ± 0.009 | T3 | T3 | 1.02 ± 0.010 | ||
| 0.05 ± 0.006 | VT | FL202-1 | 1.49 ± 0.015 | ||
| 0.08 ± 0.006 | T36 | T36 | 0.06 ± 0.005 | ||
Prechallenge and postchallenge virus titers were determined using MCA13 antibody as the detecting antibody as follows. For the prechallenge titers, C. macrophylla trees were assayed at 6 weeks after primary inoculation by DAS-I-ELISA using CTV-specific 908 IgG as the trapping antibody at a 1-μg/ml concentration and MCA13 monoclonal antibody as the detecting antibody at a 1:20,000 dilution. For the postchallenge titers, the trees were assayed at 2 months after challenge inoculation using DAS-I-ELISA as described above. The ELISA values (A405) are averages from five plants.
To assess superinfection exclusion, 9- to 12-month-old trees of Madam Vinous sweet orange [C. sinensis (L.) Osbeck] or Citrus macrophylla Wester were initially inoculated by grafting of virus-infected tissue from individual source plants. At 6 weeks after inoculation, systemic tissue was assayed by enzyme-linked immunosorbent assay (ELISA) to confirm the establishment of infection. Secondary (challenge) inoculation of preinfected plants was done by inserting a second graft of bark tissue infected with a challenge virus. When the graft healed, the upper flushes of leaves were trimmed to induce growth of a new flush, which was then evaluated for the ability of the challenge virus to establish systemic infection in plants that were previously infected with a primary virus.
Generation of the CTV hybrid T36/T68 constructs.
The full-length cDNA clone of CTV T36, pCTV9R (78, 80), was the basis of all constructs in the present study. Double-stranded CTV RNA (dsRNA) was extracted from Madam Vinous sweet orange trees infected with isolate T68-1 according to the procedure described by Moreno et al. (60). This dsRNA was used for cDNA synthesis by reverse transcription and subsequent PCR amplification with Pfu DNA polymerase (Stratagene) of an ∼4.0-kb fragment using a pair of primers annealing to the 5′ end of the CPm ORF or the 3′ end of the genomic RNA with incorporated NotI restriction endonuclease cleavage sequence (78). Three restriction endonuclease sites—BglII, PstI, and BssHII—common to the T36 and T68 sequences (nucleotide [nt] positions 15599/15603, 17205/17209, and 18185/18189 in the genome of T36, respectively), along with the NotI site, were used to generate precise in-frame exchanges into the T36 infectious clone. To engineer hybrids H1 to H5, the PCR product amplified from T68-1 cDNA template was digested with BglII and NotI enzymes (H1 hybrid), PstI and NotI (H2), BssHII and NotI (H3), PstI and BssHII (H4), or BglII and BssHII (H5), respectively, and the resulting fragments were substituted for the corresponding regions into the plasmid pUC119 containing a portion of T36 sequence between the PmeI site (nt 11872) and the NotI site at the 3′ end. Resulting plasmids were then digested with PmeI (through incomplete digestion due to the second PmeI site in T68 sequence) and NotI and ligated into PmeI- and NotI-digested pCTV9R. A graphical description of specific hybrid constructs is given in the Results.
Amplification of virions of engineered virus constructs in Nicotiana benthamiana protoplasts for inoculation of citrus trees.
SP6 RNA polymerase-derived transcripts of CTV cDNAs linearized with NotI restriction endonuclease were used for transfection of Nicotiana benthamiana mesophyll protoplasts according to the procedure described by Satyanarayana et al. (78). Protoplasts were harvested at 4 days postinoculation and stored at −70°C for subsequent protoplast passage of virions. Passaging of virions up to 11 successive cycles in protoplasts for amplification of the virus was done as described previously by Satyanarayana et al. (79). Accumulation of virus RNAs was monitored by Northern blot hybridization of the total RNA isolated from protoplasts with a 3′ positive-stranded CTV RNA-specific riboprobe (78). Amplified progeny virions from the final passages in protoplasts were extracted and concentrated by sucrose cushion centrifugation, and the concentrated virions were used for mechanical “bark flap” inoculation of small trees of C. macrophylla as described by Robertson et al. (71). Infected trees were later used as a source of virus for subsequent graft inoculations of young C. macrophylla trees.
Serological assays.
Double antibody sandwich indirect ELISA (DAS-I ELISA) was performed as described previously using antibodies specific to CTV virions (33) to confirm infection in inoculated plants and compare titers of virus accumulation in different citrus species. Purified IgG from rabbit polyclonal antiserum CTV-908 (1 μg/ml) was used as a coating antibody. ECTV172, a broadly reactive CTV monoclonal antibody, or MCA13, selective monoclonal antibody, were used as detecting antibodies.
Examination of fluorescence in citrus plants infected with GFP-tagged CTV.
Samples of bark tissue from CTV-BC5/GFP inoculated trees were examined for GFP fluorescence at different time points beginning at 6 weeks after inoculation using a Zeiss Stemi SV 11 UV-fluorescence dissecting microscope (Carl Zeiss Jena GmbH, Jena, Germany) with an attached camera Olympus Q-color 5 (Olympus America, Inc., Center Valley, PA).
Analysis of virus population accumulated in citrus trees primary infected with T36/T68 or T36/T30 hybrid viruses and challenged with T68-1 or T30-1.
To examine virus population in citrus trees, bark of a young flush was peeled and ground with liquid nitrogen. Total RNA was extracted with the TRIzol Reagent (Invitrogen) and subjected to a reverse transcription-PCR (RT-PCR) procedure using the Titan OneTube RT-PCR System (Roche) and the following sets of primers specific for the 5′ regions of T68-1 or T30-1 genomes: T68-1-5716F and T68-1-7020R annealing to the sequence of T68-1 genome at the nucleotide positions beginning 5716 and 7020, respectively, with the expected size of the amplifying product of 1,304 nt, and T30-1-2F and T30-1-2R annealing to the sequence of T30-1 genome at the nucleotide positions beginning with nt 792 and nt 1635, respectively, with the expected size of the resulting product of 843 nt (Hilf, unpublished). The sequences of the primers are available upon request. The reactions were carried out according to the protocol of the manufacturer. Reaction products were analyzed by electrophoresis in 1% agarose gels containing ethidium bromide at 200 ng/ml.
RESULTS
Ability of different isolates of CTV to prevent superinfection by GFP-tagged CTV T36.
Recently, we developed a GFP-expressing CTV vector (CTV-BC5/GFP) based on an infectious cDNA clone of CTV T36, which is the type isolate of the T36 strain. This virus contains an extra ORF of GFP inserted into the viral genome between the two coat protein ORFs, under the control of the CP sgRNA controller element from Beet yellows virus (26) (Fig. 1A). The biological characteristics of CTV-BC5/GFP in citrus trees were essentially identical to that of wild-type T36. Both viruses exhibited similar time intervals for establishing systemic infections and produced similar symptoms in infected plants and appeared to be equally competitive when inoculated simultaneously into the same tree (26). Multiplication of CTV-BC5/GFP in different citrus varieties produced GFP fluorescence, observation of which allowed visualization of characteristic patterns of virus distribution in phloem-associated cells of these hosts (27). Furthermore, CTV-BC5/GFP has been unusually stable, with continued production of GFP in citrus trees for 6 years thus far. These features made the GFP-expressing CTV a useful tool to examine superinfection.
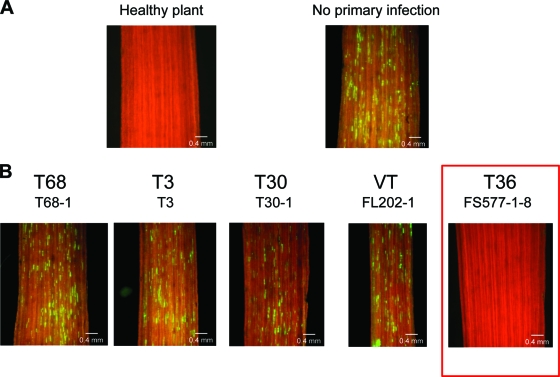
In order to assess the effect of a primary infection of a host plant with an isolate of CTV on the ability of the GFP-tagged T36 to establish superinfection in the same host, small sweet orange trees were first inoculated individually with each of 13 CTV isolates listed in Table 1. (Due to quarantine regulations, only isolates of CTV naturally found in Florida can be propagated in our greenhouses.) These 13 isolates represent five different CTV strains. The primary infections were established by grafting virus-infected tissue into the stems of receptor trees (Fig. 1B, Pri). The upper leaves were trimmed to force the growth of a new set of leaves. At 6 weeks after inoculation, systemic infections of the new leaves were confirmed by ELISA. Similar ELISA values were obtained for all 13 isolates (Table 1), demonstrating similar levels of accumulation of the different isolates in infected plants. The plants were then challenged by putting a second graft of bark tissue containing the T36-based CTV-BC5/GFP (Fig. 1B, Chl). When the graft healed, the upper leaves again were trimmed to induce another new flush of growth. After the development of the second set of new leaves (starting about 6 to 8 weeks after graft-inoculation with the challenge virus) the ability of the challenging virus to superinfect trees was determined by visual observation of GFP fluorescence in the bark tissue of the new flush. As shown in Fig. 2 and Table 1, plants that had primary infections with CTV isolates representing the T30 (T30-1, T55-1, T4, FL278), the VT (FL202-1, FS672, FS674HT, and FS701), the T68 (T68-1) or the T3 (T3) strains all displayed GFP fluorescence similar to that observed in control plants that had no primary infection and were inoculated only with the challenge virus (T36 CTV-BC5/GFP) (Fig. 2). In contrast, no GFP fluorescence was detected in plants primarily infected with isolates of the T36 strain (T36, T66-1, and FS577-1-8) (Fig. 2 and Table 1). These isolates completely prevented superinfection by the T36-based virus, whereas isolates representing other strains of CTV had no interference with infection by the GFP-tagged T36.
FIG. 2.
Observation of GFP fluorescence in phloem-associated cells on the internal surface of bark of sweet orange Madam Vinous trees by using a dissecting fluorescence microscope. (A) Bark from a noninoculated healthy tree (left) and from a tree that was inoculated solely with T36-based CTV-BC5/GFP (right). (B) Bark from trees primarily infected with isolates belonging to five CTV strains and sequentially challenged with CTV-BC5/GFP. Observations were done at 2 months after challenge inoculation. Scale bar, 0.4 mm.
Interaction between CTV isolates is not host dependent.
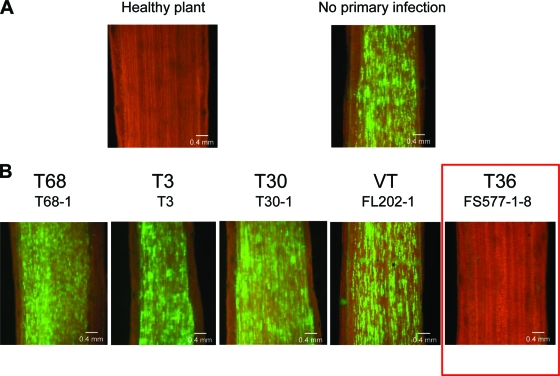
Different varieties of citrus support different levels of accumulation of CTV isolates (14; S. M. Garnsey, unpublished data) and, as demonstrated for T36, the differences in virus titers in different citrus hosts are correlated with the proportion of cells infected (27). To assess the ability of CTV isolates to prevent superinfection in a host in which a significantly higher proportion of cells would become infected with the primary virus and how the increased number of initially infected cells would affect superinfection exclusion, the interactions of the same isolates were examined in C. macrophylla. This variety represents one of the more susceptible citrus hosts to CTV infection and allows accumulation of the virus to high levels. Based on the observation of GFP expression and electron microscopy studies of tissue from T36-infected plants, we estimated that CTV infects about 20 to 30% of phloem-associated cells in this host (27). The primary inoculations of C. macrophylla plants with the 13 isolates resulted in titers higher than those found in sweet orange plants (Table 1). Two months later these plants were challenged with the T36-based CTV-BC5/GFP. Observation of bark tissue of C. macrophylla plants at 2 and 4 months after challenge inoculation revealed that control plants with no primary infection displayed strong GFP fluorescence correlated with the greater number of CTV-BC5/GFP-infected foci than that found in sweet orange plants (Fig. 3). Furthermore, similar to the observations described for sweet orange plants, C. macrophylla trees preinfected with isolates of T68, T3, T30, or VT strains all showed strong GFP fluorescence, indicating that higher levels of primary infections with other isolates did not affect the ability of CTV-BC5/GFP to establish superinfection (Fig. 3 and Table 1). Remarkably, there appeared to be no partial inhibition of the challenging virus by any of these isolates. Even though more cells likely were preoccupied with the primary virus, the levels of GFP fluorescence produced by the challenge virus in these plants were found to be nearly identical to the fluorescence levels in control plants (Fig. 3). At the same time, the T36-based challenge virus was not detected in plants previously infected with isolates of the T36 strain (Fig. 3 and Table 1). Thus, observations made in C. macrophylla confirmed the findings in sweet orange.
FIG. 3.
Observation of GFP fluorescence in phloem-associated cells on the internal surface of bark of C. macrophylla trees by using a dissecting fluorescence microscope. (A) Bark from a noninoculated healthy tree (left) and from a tree that was inoculated solely with T36-based CTV-BC5/GFP (right). (B) Bark from trees primarily infected with isolates belonging to five CTV strains and sequentially challenged with CTV-BC5/GFP. Observations were done at 2 months after challenge inoculation. Scale bar, 0.4 mm.
To assess effect of extended time periods on the ability of the challenging T36 CTV-BC5/GFP virus to maintain infection in combinations with isolates of other strains or to establish infection in trees preinfected with isolates of the T36 strain, the C. macrophylla and sweet orange plants were maintained and monitored for GFP fluorescence during the next 3 years. No change in GFP production was noticed during this period of time (data not shown).
Assessment of superinfection exclusion for different combinations of primary-infecting and challenging isolates of CTV.
In the experiments described above, the only challenge virus was the T36 strain-based GFP-tagged virus CTV-BC5/GFP. To further evaluate the relationships between isolates of different strains of CTV in terms of their ability to interfere with infection by another virus variant, we examined different combinations of primary-infecting and challenging isolates. Because T36 is the only isolate of CTV for which an infectious cDNA clone and GFP-tagged virus constructs have been developed (26, 78), an assay method based on serological differentiation of CTV isolates with monoclonal antibody MCA13 was used to monitor the infection of a challenging virus. This antibody fails to react with isolates of the T30 strain but reacts strongly with isolates of the T68, VT, T3, and T36 strains (66). Furthermore, we recently engineered an MCA13 nonreactive variant of T36, CTV9R-MCA13NR, via mutagenesis of a single nucleotide in the coat protein (82) based on the mapped reactive epitope (63). Thus, two viruses T30-1 and CTV9R-MCA13NR representing the T30 and T36 strains, respectively, were used for primary inoculations of young C. macrophylla trees. At 6 weeks after inoculation, infection of these plants was confirmed with antibody ECTV172 commonly used to detect all CTV strains (data not shown). At the same time plants were assayed with MCA13 antibody to demonstrate the lack of reaction of this antibody with T30-1 and CTV9R-MCA13NR (Table 2). Infected plants then were challenged with the MCA13 reactive T68-1, FL202-1, T3, or T36 isolates of four different strains of CTV, and the establishment of superinfection by those viruses was evaluated at 2 and 4 months postchallenge using the MCA13 antibody. As shown in Table 2, positive MCA13 ELISA values demonstrated a lack of exclusion in any of the combinations in which the primary virus was from a strain different from that of the challenging virus strain. The only incidence of superinfection exclusion occurred when T36 was used as a challenging virus in plants primarily infected with CTV9R-MCA13NR, which represents a virus of the same (T36) strain. The lack of reaction with the MCA13 antibody indicated that CTV9R-MCA13NR completely excluded the wild-type T36 (Table 2).
Effect of homologous sequences on the ability of isolates to prevent superinfection.
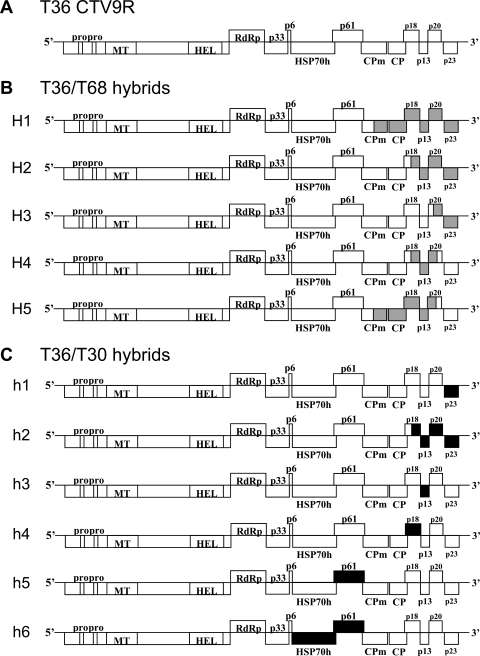
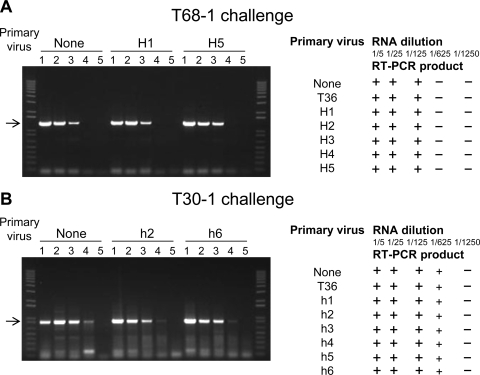
The observation that superinfection exclusion occurred only with isolates of the same strains, which were defined based on the levels of sequence similarity, suggested that RNA silencing might be a component of the exclusion mechanism. Numerous examples from various virus-host systems demonstrated that expression of homologous sequences from a virus genome in a host can trigger sequence-specific degradation of viral RNA, and some examples of superinfection exclusion have been attributed to RNA silencing (42, 70, 98). To examine the role of homologous sequences in superinfection exclusion, we created a series of hybrids in which sequences from one virus were substituted into a virus of a different strain. Five hybrids were generated by substituting different fragments of the T68-1 genome ranging in size between 980 and 3,697 nt and comprising ORFs of p23, p20, p13, p18, CP, and part of CPm ORF into the T36 infectious cDNA clone (Fig. 4A and B, hybrids H1 to H5). These hybrids contained substitutions of genes for proteins involved in virion assembly (CP and CPm), suppression of RNA silencing (p23, p20, and CP) and proteins with unknown functions (p13 and p18). Another set of six T36/T30 hybrid viruses contained substitutions of the following regions of the T30-1 genome into the genome of T36: 3′ NTR plus p23 ORF (hybrid h1); 3′ NTR plus p23, p20, p13, and part of p18 ORFs (h2); p13 ORF (h3); p18 ORF (h4); p61 ORF (h5); and HSP70h plus p61 ORFs (h6) (Fig. 4C) (5). In addition to hybrid viruses with gene substitutions similar to those described for T36/T68 hybrids, this set included hybrids (h5 and h6) with substitutions of genes of the two additional proteins involved in the formation of virions (p61 and HSP70h). Each of these hybrid viruses was used for primary inoculation of C. macrophylla plants. To examine whether introduction of T68-1 or T30-1 sequences into T36 would elicit the ability of these viruses to exclude an isolate of the T68 or the T30 strains, respectively, along with T36, plants preinfected with the T36/T68 hybrids were subsequently challenged with T36 CTV-BC5/GFP or T68-1, and plants preinfected with the T36/T30 hybrids were challenged with T36 CTV-BC5/GFP or T30-1. Potentially, any of the substitutions, particularly substitutions of proteins functioning in virion assembly or suppression of RNA silencing, could have an effect on the exclusion of one virus by another virus that contained introduced homologous regions. At 2 and 4 months after challenge, plants were examined for GFP expression and tested by RT-PCR with T68-1- or T30-1-specific primers for the T36/T68 or T36/T30 sets of plants, respectively (Fig. 5). The RT-PCR primers used in this experiment were designed based on the 5′ end sequences of T68-1 or T30-1 genomes in order to detect replication of the T68-1 or T30-1 challenge isolates but not the respective hybrid viruses bearing fragments from the genomic sequences of those isolates.
FIG. 4.
(A) Schematic diagram of the genome organization of wild-type CTV T36 (T36 CTV9R). (B and C) Schematic representations of the T36/T68 and T36/T30 hybrid constructs, respectively. The open boxes represent ORFs and their translation products. PRO, papainlike protease domain; MT, methyltransferase; HEL, helicase; RdRp, an RNA-dependent RNA polymerase; HSP70h, HSP70 homolog; CPm, minor coat protein; CP, major coat protein. Gray and black boxes indicate the T68 or the T30 sequences substituted within the T36 genome, respectively.
FIG. 5.
Detection of T68-1 (A) and T30-1 (B) multiplication upon challenge inoculation of C. macrophylla trees with no primary infection or preinfected with T36 or T36/T68 (H1 to H5) or T36/T30 (h1 to h6) hybrid viruses, respectively. At 2 months after challenge the total RNA was extracted from tested trees. RT-PCR amplification of viral sequences was carried out by using fivefold serial dilutions of RNA (1/5×, 1/25×, 1/125×, 1/625×, and 1/1,250× [lanes 1, 2, 3, 4, and 5 on the gel pictures, respectively]) and oligonucleotide primers specific for the 5′ regions of the T68-1 or T30-1 genomes, respectively. Reaction products were analyzed by electrophoresis in 1% agarose gel. Images of the agarose gels are shown for RNA samples extracted from T68-1-challenged trees with no primary infection or trees preinfected with T36/T68 hybrids H1 and H5 in panel A and for RNA samples extracted from T30-1 challenged trees with no primary infection or trees preinfected with T36/T30 hybrids h2 and h6 in panel B. Arrows indicate positions of RT-PCR products. The results obtained for other hybrid viruses used for initial inoculations of trees are presented in a table format. +, indicates amplification of the challenge isolate-specific RT-PCR product; -, lack of amplification of the product.
Both sets of hybrid viruses excluded superinfection by the T36 isolate. No GFP fluorescence was observed in plants preinfected with any hybrid viruses and later challenged with CTV-BC5/GFP when plants were examined at different time points (two and 4 months after challenge inoculation and thereafter during the next 2 years) (the results are not shown). Surprisingly, primary infection of plants with any T36/T68 or T36/T30 hybrid viruses did not interfere with the establishment of infection by T68-1 or T30-1, respectively, which was confirmed by obtaining T68-1- or T30-1-specific RT-PCR products identical to the RT-PCR products obtained from control plants that were inoculated only with T68-1 or T30-1 isolates (Fig. 5). Moreover, multiplication of the T68-1 or the T30-1 challenge viruses in trees preinfected with the T36 isolate or the T36/T68 or the T36/T30 hybrids, respectively, was comparable to their multiplication in trees with no primary infection, which was demonstrated by using serial dilutions of RNA extracted from tested trees to amplify challenge isolate-specific RT-PCR fragments (Fig. 5). Thus, sharing of homologous sequences within the 3′ portion of the CTV genome by two viruses did not trigger the ability of one virus to exclude superinfection by another.
DISCUSSION
Results obtained from the experiments in which interactions of several different combinations of primary and challenging viruses were evaluated by observation of GFP expressed from a challenge virus or by ELISA with a differentiating antibody demonstrated that CTV isolates that have established a systemic infection in citrus trees prevent superinfection by an isolate of the same strain, but not by isolates from different strains. Exclusion among isolates of the same strain of CTV was absolute, while isolates from different strains demonstrated a complete lack of exclusion. Furthermore, with the GFP-marked virus used as a challenge virus, we saw no difference in the proportion of cells infected or in the intensity of GFP fluorescence per infected cell in trees infected initially with isolates of heterologous strains compared to inoculation of trees with no primary infection. The isolates of heterologous strains that were established initially appeared to have no effect on infection, movement, and replication of the challenge virus. In addition, when trees were initially infected and later challenged with isolates belonging to the same strain, there was no evidence of infection and replication of the challenge isolate in any of the trees. This contrasts with reports for other viruses, which demonstrate that, in general, depending on the relatedness of the isolates, superinfection exclusion may be partial, resulting in prevention of a secondary isolate infection only in some of the plants (29, 30, 35, 91, 100).
The complete exclusion that occurs between similar isolates is unexpected considering that CTV infects only a portion of the phloem-associated cells (27). The proportion of cells that become infected varies with the different citrus genotypes. Based on previous experiments, C. macrophylla is the most susceptible, with perhaps only one-third or less of the phloem-associated cells infected, whereas significantly fewer cells of sweet orange become infected upon CTV infection (27). However, superinfection exclusion was absolute even in sweet orange in which most of the phloem-associated cells were not infected by the primary isolate. This result suggests that superinfection exclusion was not limited to the infected cells, but the majority of cells that were not infected also were protected. Thus, the exclusion phenomenon must be able to spread beyond the infected cells.
CTV may be somewhat unique among viruses because of the rather large differences in nucleotide sequences among isolates of different strains, which vary in an unusual manner. The 3′ halves of the genomes of the more dissimilar isolates may differ by only 10%, but the 5′ halves progressively become more dissimilar. After examination of many different CTV isolates, it was found, based on sequence analysis of the more diverged 5′ half of the genome, that they generally fall into phylogenetically distinct sequence groups, which have been defined as strains (38). This grouping reflects the pattern of exclusion seen experimentally, suggesting the sequence divergence in this region of the genome may affect intervirus interactions, resulting in the complete lack of superinfection exclusion between isolates of different CTV strains. This result contradicts the premise of one of the original uses for superinfection exclusion as a measure of relatedness of unknown viruses, in which nonexcluded viruses were identified as different viruses (55). Clearly, that is not the case with CTV. Superinfection exclusion defines excluding CTV isolates as members of the same strain and not different strains.
Results presented here provide a possible explanation of some fundamental features of CTV population biology. Isolates of CTV from field trees tend to be complex populations of different strains plus numerous defective RNAs. In addition, some CTV sequences have been shown to have putative recombination sites, suggesting that they were derived from two different strains (36, 48, 52, 68, 75, 96, 101). However, it is paradoxical that the sequences of CTV isolates appear to be unusually stable. For example, two isolates of the T30 strain, one from Spain and the other from Florida, estimated to have a common origin but separated for approximately 100 years, were found to be essentially identical (4). Also, we have found that virus isolates generated from infectious cDNA clones and maintained in trees for up to 10 years had essentially no nucleotide changes (Z. Xiong et al., unpublished data). Furthermore, engineered virus constructs bearing an insertion of a foreign gene (GFP) were stably maintained during 6 years of replication in infected trees (26; unpublished data). Superinfection exclusion has been suggested as a means to maintain stability of viral sequences due to the elimination of multiple related sequences arising through the process of mutation and/or recombination, while reducing the likelihood of the latter event by preventing replication of two or more viral genomes in the same cell (28). It is possible that superinfection exclusion controls the evolution of CTV isolates by fostering extensive genetic changes through recombination between coinfecting isolates of distinct strains while repressing small changes due to replication errors.
Superinfection exclusion, referred to as cross-protection when used as an agricultural practice, has been an effective management procedure that has allowed economic production of citrus in some regions in which endemic severe isolates of CTV made citrus production unprofitable. Stem pitting, a CTV disease syndrome, causes the eventual decline of trees due to a loss of vigor that leads to the production of small unmarketable fruit and may cause tree death. Cross-protection, when possible, has been used to allow commercial viability of citrus plantings. It has been most widely used in Brazil, where more than 50 million Pera orange trees are protected (16). It also is widely used in South Africa, Peru, and Australia (9, 17, 93). However, finding protecting isolates has been empirical and rarely successful. The strategy has been to find mild isolates and test them for the ability to protect against severe isolates in different citrus varieties. This requires years of evaluation, and most isolates fail to protect. However, based on the results presented here, finding protecting isolates now may be relatively straightforward. The first objective is to identify the strain of the severe isolate that needs to be controlled. Then, a mild isolate from that same strain needs to be found. If such an isolate does not occur naturally, it is possible through recombinant DNA methodologies to map the disease determinant(s) of the severe isolate and substitute sequences from a mild isolate of a different strain, as we have done for the decline strain in Florida (5). The resulting mild isolate should exclude the severe isolate.
Alternatively, for some other applications it would be valuable to be able to prevent the exclusion of second virus isolate. For example, in the medical and veterinary fields for repeated applications of vaccines to individuals with persistent infections, for the introduction of multicomponent vaccines, or for gene therapy it would be useful to be able to overcome the exclusion (22). Similar applications could be important for crops protection. The CTV vector used in the present study (26) is sufficiently stable to be considered as a vehicle to deliver any candidate proteins to protect citrus in the field. If exclusion could be prevented, the vector with a different foreign gene could be added again at a later time.
Since homologous superinfection exclusion is a phenomenon of viruses from both the animal and plant kingdoms, it would be expected that there would be some common mechanisms. In animal systems, exclusion has been related to prevention of the secondary virus from entering cells (50, 86, 87), inhibition of translation, or interference with replication (1, 47, 50, 83). Plant viruses do not have specific cell surface receptor recognition processes for entering cells, so an occurrence of a common mechanism at this level of virus infection should not be expected. Alternatively, commonality could exist at the stage of virus replication. Since CTV replicase is translated as a polyprotein that is thought to be processed by viral leader proteases (20, 44, 51, 65), particularly attractive is a hypothesis proposed for Sindbis virus, which relates superinfection exclusion to excess of trans-acting protease produced by the primary virus that fully processes the replicase polyprotein of the secondary virus and thus prevents negative strand synthesis of the challenge virus (47, 88). However, this mechanism would appear unlikely for CTV since uninfected cells, which would not be expected to have the protease of the primary virus, also appear to be protected. Another model of a superinfection exclusion mechanism was described for isolates of tobamoviruses. Exclusion of the incoming virus was shown to be due to excess of the primary virus coat protein preventing uncoating of the secondary virus (8, 53, 84). However, because of the apparent protection of uninfected cells of a citrus host against secondary infection by a related CTV isolate, this “reincapsidation” model also appears to be an unlikely explanation for the phenomenon reported here. Moreover, the substitution of coat protein genes in engineered CTV hybrids did not affect superinfection exclusion as shown for strains of tobamo- and potyviruses (8, 91). Other mechanisms acting at steps of translation or replication also would have to be effective in uninfected cells to be relevant to the process of CTV interference. One such mechanism implicated in superinfection exclusion of several plant viruses and a few animal viruses is RNA silencing (2, 7, 12, 70, 98). RNA silencing, which has been shown to be a major host defense mechanism against viruses in plants and in invertebrate animals, can be induced systemically in both infected and uninfected cells (59, 62, 76, 92, 97). The mechanism targets nearly identical RNA sequences and, thus, introduction of homologous sequences, in some cases as short as 23 nucleotides, into genomes of heterologous viruses has been shown to induce degradation of RNA molecules containing these sequences (42, 70, 90, 98). These characteristics parallel the observations of superinfection exclusion of CTV and could explain the exclusion of closely related isolates and lack of exclusion of isolates of distinct strains. However, this model does not explain how hybrid viruses containing exact cognate sequences up to 3.7 kb in size from isolates of different strains failed to exclude the donor isolates. However, it must be noted that the substitution of homologous sequences only occurred in the 3′ half of the genome.
Prior infection of citrus trees with CTV completely prevented superinfection by an isolate of the same strain, not only in infected cells but also in the uninfected phloem-associated cells. Interestingly, “transmission” of resistance to superinfection from persistently infected to uninfected cells has been shown for an animal virus (23), which also argues for possible common mechanisms driving superinfection exclusion of both animal and plant viruses. Possible mechanisms that involve only cells that are infected, such as inhibition of disassembly, replication, or translation of an incoming virus, appear not to be relevant explanations of this phenomenon. Similarly, superinfection exclusion by CTV appears not to be explained based on an RNA silencing model. Thus, it appears that the superinfection exclusion phenomenon may be due to a biological mechanism that is yet to be determined.
Acknowledgments
We thank Cheryl Graffam and Rachel McCoy for excellent technical assistance.
This research was supported by the Florida Agricultural Experiment Station, an endowment from the J. R. and Addie Graves family, and grants from the Florida Citrus Production Research Advisory Council.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Adams, R. H., and D. T. Brown. 1985. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J. Virol. 54:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adelman, Z. N., C. D. Blair, J. O. Carlson, B. J. Beaty, and K. E. Olson. 2001. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol. Biol. 10:265-273. [DOI] [PubMed] [Google Scholar]
- 3.Agranovsky, A. A. 1996. Principles of molecular organization, expression, and evolution of closteroviruses: over the barriers. Adv. Virus Res. 47:119-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albiach-Martí, M. R., M. Mawassi, S. Gowda, T. Satyanayanana, M. E. Hilf, S. Shanker, E. C. Almira, M. C. Vives, C. López, J. Guerri, R. Flores, P. Moreno, S. M. Garnsey, and W. O. Dawson. 2000. Sequences of Citrus tristeza virus separated in time and space are essentially identical. J. Virol. 74:6856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albiach-Martí, M. R., C. Robertson, S. Gowda, T. Satyanarayana, B. Belluire, S. M. Garnsey, S. Folimonova, P. Moreno, and W. O. Dawson. 2009. The pathogenicity determinant of Citrus tristeza virus causing the seedling yellows syndrome maps at the 3′-terminal region of the viral genome. Mol. Plant Pathol., in press. [DOI] [PMC free article] [PubMed]
- 6.Bar-Joseph, M., S. M. Garnsey, and D. Gonsalves. 1979. The closteroviruses: a distinct group of elongated plant viruses. Adv. Virus. Res. 25:93-168. [DOI] [PubMed] [Google Scholar]
- 7.Baulcombe, D. 2004. RNA silencing in plants. Nature 431:356-363. [DOI] [PubMed] [Google Scholar]
- 8.Beachy, R. N. 1999. Coat-protein-mediated resistance to tobacco mosaic virus: discovery mechanisms and exploitation. Philos. Trans. R. Soc. Lond. 354:659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bederski, K., C. N. Roistacher, and G. W. Müller. 2005. Cross protection against the severe Citrus tristeza virus: stem pitting in Peru, p. 117-126. In Proceedings of the 16th Conference of the IOCV. IOCV, Riverside, CA.
- 10.Bendahmane, M., and R. N. Beachy. 1999. Control of tobamovirus infection via pathogen-derived resistance. Adv. Virus Res. 53:369-386. [DOI] [PubMed] [Google Scholar]
- 11.Bennett, C. W. 1951. Interactions between viruses and virus strains. Adv. Virus Res. 1:39-67. [DOI] [PubMed] [Google Scholar]
- 12.Billecocq, A., M. Vazeille-Falcoz, F. Rodhain, and M. Bouloy. 2000. Pathogen-specific resistance to Rift Valley fever virus infection is induced in mosquito cells by expression of the recombinant nucleoprotein but not NSs nonstructural protein sequences. J. Gen. Virol. 81:2161-2166. [DOI] [PubMed] [Google Scholar]
- 13.Bratt, M. A., and H. Rubin. 1968. Specific interference among strains of Newcastle disease virus 3 mechanism of interference. Virology 35:395-407. [DOI] [PubMed] [Google Scholar]
- 14.Brlansky, R. H., and R. F. Lee. 1990. Numbers of inclusion bodies produced by mild and severe strains of Citrus tristeza virus in seven citrus hosts. Plant Dis. 74:297-299. [Google Scholar]
- 15.Broadbent, P., K. B. Bevington, and B. G. Coote. 1991. Control of stem pitting of grapefruit in Australia by mild strain cross protection, p. 64-70. In Proceedings of the 11th Conference of the IOCV. IOCV, Riverside, CA.
- 16.Costa, A. S., and G. W. Müller. 1980. Tristeza control by cross-protection: a U.S.-Brazil cooperative success. Plant Dis. 64:538-541. [Google Scholar]
- 17.Cox, J. E., L. R. Fraser, and P. Broadbent. 1976. Grapefruit stem pitting: field protection by mild strains, p. 64-70. In Proceedings of the 7th Conference of the IOCV. IOCV, Riverside, CA.
- 18.Delwart, E. L., and A. T. Panganiban. 1989. Role of reticuloendotheliosis virus envelope glycoprotein in superinfection interference. J. Virol. 63:273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolja, V. V., A. V. Karasev, and E. V. Koonin. 1994. Molecular biology and evolution of closteroviruses: sophisticated build-up of large RNA genomes. Annu. Rev. Phytopathol. 32:261-285. [Google Scholar]
- 20.Dolja, V. V., J. F. Kreuze, and J. P. T. Valkonen. 2006. Comparative and functional genomics of closteroviruses. Virus Res. 117:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dulbecco, R. 1952. Mutual exclusion between related phages. J. Bacteriol. 63:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrengruher, M. U., and A. L. Goldin. 2007. Semliki Forest virus vectors with mutations in the nonstructural protein 2 gene permit extended superinfection of neuronal and non-neuronal cells. J. Neurovirol. 13:353-363. [DOI] [PubMed] [Google Scholar]
- 23.Ellenberg, P. M. Edreira, and L. Scolaro. 2004. Resistance to superinfection of Vero cells persistently infected with Junin virus. Arch. Virol. 149:507-522. [DOI] [PubMed] [Google Scholar]
- 24.Fagoaga, C., C. López, A. Hermoso de Mendoza, P. Moreno, L. Navarro, R. Flores, and L. Peña. 2006. Post-transcriptional gene silencing of the p23 silencing suppressor of Citrus tristeza virus confers resistance to the virus in transgenic Mexican lime. Plant Mol. Biol. 66:153-165. [DOI] [PubMed] [Google Scholar]
- 25.Febres, V. J., L. Ashoulin, M. Mawassi, A. Frank, M. Bar-Joseph, K. L. Manjuth, R. F. Lee, and C. L. Niblett. 1996. The p27 protein is present at one end of citrus tristeza particles. Phytopathology 86:1331-1335. [Google Scholar]
- 26.Folimonov, A. S., S. Y. Folimonova, M. Bar-Joseph, and W. O. Dawson. 2007. A stable RNA virus-based vector for citrus trees. Virology 368:205-216. [DOI] [PubMed] [Google Scholar]
- 27.Folimonova, S. Y., A. S. Folimonova, S. Tatineni, and W. O. Dawson. 2008. Citrus tristeza virus: survival at the edge of the movement continuum. J. Virol. 82:6546-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Formella, S., C. Jehle, C. Sauder, P. Staeheli, and M. Schwemmle. 2000. Sequence variability of Borna disease virus: resistance to superinfection may contribute to high genome stability in persistently infected cells. J. Virol. 74:7878-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser, R. S. S. 1998. Introduction to classical cross protection, p.13-24. In D. Foster and S. J. Taylor (ed.), Methods in molecular biology, vol. 81. Plant virus protocols. Humana Press, Inc., Totowa, NJ. [Google Scholar]
- 30.Fulton, R. W. 1978. Superinfection by strains of tobacco streak virus. Virology 85:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Fulton, R. W. 1986. Practices and precautions in the use of cross protection for plant virus disease control. Annu. Rev. Phytopathol. 24:67-81. [Google Scholar]
- 32.Gal-On, A., and Y. M. Shiboleth. 2005. Cross protection, p. 261-288. In G. Loebenstein and J. P. Carr (ed.), Natural resistance mechanisms of plants to viruses. Springer, Dordrecht, The Netherlands.
- 33.Garnsey, S., and M. Cambra. 1991. Enzyme-linked immunosorbent assay (ELISA) for citrus pathogens, p. 193-216. In C. N. Roistacher (ed.), Graft-transmissible diseases of detritus: handbook for detection and diagnosis. FAO, Rome, Italy.
- 34.Geib, T., C. Sauder, S. Venturelli, S. Hassler, P. Staeheli, and M. Schwemmle. 2003. Selective virus resistance conferred by expression of Borna disease virus nucleocapsid components. J. Virol. 77:4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonsalves, D. 1998. Control of papaya ringspot virus in papaya; a case study. Annu. Rev. Phytopathol. 36:415-437. [DOI] [PubMed] [Google Scholar]
- 36.Grant, T. J., and R. P. Higgins. 1957. Occurrence of mixtures of tristeza virus strains in citrus. Phytopathology 47:272-276. [Google Scholar]
- 37.Hilf, M. E., A. V. Karasev, H. R. Pappu, D. J. Gumpf, C. L. Niblett, and S. M. Garnsey. 1995. Characterization of citrus tristeza virus subgenomic RNAs in infected tissue. Virology 208:576-582. [DOI] [PubMed] [Google Scholar]
- 38.Hilf, M. E., V. A. Mavrodieva, and S. M. Garnsey. 2005. Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopathology 95:909-917. [DOI] [PubMed] [Google Scholar]
- 39.Hull, R., and A. Plaskitt. 1970. Electron microscopy on the behavior of two strains of alfalfa mosaic virus in mixed infections. Virology 42:773-776. [DOI] [PubMed] [Google Scholar]
- 40.Hull, R. 2002. Matthews' plant virology. Academic Press, Inc., New York, NY.
- 41.Ieki, H., and A. Yamaguchi. 1988. Protective interference of mild strains of citrus tristeza virus against a severe strain in Morita navel orange, p. 86-90. In Proceedings of the 10th Conference of the IOCV. IOCV, Riverside, CA.
- 42.Jan, F. J., C. Fagoaga, S. Z. Pang, and D. Gonsalves. 2000. A minimum length of N gene sequence in transgenic plants is required for RNA-mediated tospovirus resistance. J. Gen. Virol. 81:235-242. [DOI] [PubMed] [Google Scholar]
- 43.Johnston, R. E. K. Wan, and H. R. Bose. 1974. Homologous interference induced by Sindbis virus. J. Virol. 14:1076-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karasev, A. V., V. P. Boyko, S. Gowda, O. V. Nikolaeva, M. E. Hilf, E. V. Koonin, C. L. Niblett, K. Cline, D. J. Gumpf, R. F. Lee, S. M. Garnsey, and W. O. Dawson. 1995. Complete sequence of the citrus tristeza virus RNA genome. Virology 208:511-520. [DOI] [PubMed] [Google Scholar]
- 45.Karasev, A. V., M. E. Hilf, S. M. Garnsey, and W. O. Dawson. 1997. Transcriptional strategy of closteroviruses: mapping the 5′ termini of the Citrus tristeza virus subgenomic RNAs. J. Virol. 71:6233-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karasev, A. V. 2000. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38:293-324. [DOI] [PubMed] [Google Scholar]
- 47.Karpf, A. R., E. Lenches, E. G. Strauss, J. H. Strauss, and D. T. Brown. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 71:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong, P., L. Rubio, M. Polek, and B. W. Falk. 2000. Population structure and genetic diversity within California Citrus tristeza virus (CTV) field isolates. Virus Genes 21:139-145. [DOI] [PubMed] [Google Scholar]
- 49.Lecoq, H., J. M. Lemaire, and C. Wipf-Scheibel. 1991. Control of zucchini yellow mosaic virus in squash by cross protection. Plant Dis. 75:208-211. [Google Scholar]
- 50.Lee, Y. M., D. M. Tscherne, S. I. Yun, I. Frolov, and C. M. Rice. 2005. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J. Virol. 79:3231-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, Y. P., V. V. Peremyslov, V. Medina, and V. V. Dolja. 2009. Tandem leader proteases of grapevine leafroll-associated virus-2: host-specific functions in the infection cycle. Virology 383:291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.López, C., M. A. Ayllón, J. Navas-Castillo, J. Guerri, P. Moreno, and R. Flores. 1998. Molecular variability of the 5′ and 3′ terminal regions of citrus tristeza virus RNA. Phytopathology 88:685-691. [DOI] [PubMed] [Google Scholar]
- 53.Lu, B., G. Stubbs, and J. N. Culver. 1998. Coat protein interactions involved in tobacco mosaic tobamovirus cross-protection. Virology 248:188-198. [DOI] [PubMed] [Google Scholar]
- 54.Lu, R., A. Folimonov, M. Shintaku, W. X. Li, B. W. Falk, W. O. Dawson, and S. W. Ding. 2004. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. U. S. A. 101:15742-15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matthews, R. E. F. 1991. Plant virology, 3rd ed. Academic Press, London, England.
- 56.Mawassi, M., E. Mietkiewska, R. Gofman, G. Yang, and M. Bar-Joseph. 1996. Unusual sequence relationships between two isolates of citrus tristeza virus. J. Gen. Virol. 77:2359-2364. [DOI] [PubMed] [Google Scholar]
- 57.McKinney, H. H. 1926. Virus mixtures that may not be detected in young tobacco plants. Phytopathology 16:893. [Google Scholar]
- 58.McKinney, H. H. 1929. Mosaic diseases in the Canary Islands, West Africa and Gibraltar. J. Agric. Res. 39:557-578. [Google Scholar]
- 59.Mlotshwa, S., O. Voinnet, M. F. Mette, M. Matzke, H. Vaucheret, S. W. Ding, G. Pruss, and V. B. Vance. 2002. RNA silencing and the mobile silencing signal. Plant Cell S289-301. [DOI] [PMC free article] [PubMed]
- 60.Moreno, P., J. Guerri, and N. Muñoz. 1990. Identification of Spanish strains of citrus tristeza virus (CTV) by analysis of double-stranded RNAs (dsRNA). Phytopathology 80:477-482. [Google Scholar]
- 61.Müller, G. W., A. S. Costa, J. L. Castro, and N. Guirado. 1988. The results from reimmunization tests to control the Capao Bonito strain of tristeza, p. 82-85. In Proceedings of the 10th Conference of the IOCV. IOCV, Riverside, CA.
- 62.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific posttranscriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pappu, H. R., S. S. Pappu, K. L. Manjunath, R. F. Lee, and C. L. Niblett. 1993. Molecular characterization of a structural epitope that is largely conserved among severe isolates of a plant virus. Proc. Natl. Acad. Sci. U. S. A. 90:3641-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pappu, H. R., A. V. Karasev, E. J. Anderson, S. S. Pappu, M. E. Hilf, V. J. Febres, R. M. G. Eckloff, M. McCaffery, V. Boyko, S. Gowda, V. V. Dolja, E. V. Koonin, D. J. Gumpf, K. C. Cline, S. M. Garnsey, W. O. Dawson, R. F. Lee, and C. L. Niblett. 1994. Nucleotide sequence and organization of eight open reading frames of the citrus tristeza cloterovirus genome. Virology 199:35-46. [DOI] [PubMed] [Google Scholar]
- 65.Peng, C. W., V. V. Peremyslov, A. R. Mushegian, W. O. Dawson, and V. V. Dolja. 2001. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 75:12153-12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Permar, T. A., S. M. Garnsey, D. J. Gumpf, and R. F. Lee. 1990. A monoclonal antibody which discriminates strains of citrus tristeza virus. Phytopathology 80:224-228. [Google Scholar]
- 67.Powell-Abel, P. R. S. Nelson, B. De, N. Hoffman, S. G. Rogers, R. T. Fraley, and R. N. Beachy. 1986. Delay of disease development in transgenic plants that express the tobacco virus coat protein gene. Science 232:738-743. [DOI] [PubMed] [Google Scholar]
- 68.Raccah, B., G. Loebenstein, and S. Singer. 1980. Aphid-transmissibility variants of citrus tristeza virus in infected citrus trees. Phytopathology 70:89-93. [Google Scholar]
- 69.Ratcliff, F., B. D. Harrison, and D. C. Baulcombe. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558-1560. [DOI] [PubMed] [Google Scholar]
- 70.Ratcliff, F., S. MacFarlane, and D. C. Baulcombe. 1999. Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robertson, C., S. M. Garnsey, T. Satyanarayana, S. Folimonova, and W. O. Dawson. 2005. Efficient infection of citrus plants with different cloned constructs of Citrus tristeza virus amplified in Nicotiana benthamiana protoplasts, p. 187-195. In Proceedings of the 16th Conference of the IOCV. IOCV, Riverside, CA.
- 72.Rocha-Peña, M. A., R. F. Lee, R. Lastra, C. L. Niblett, F. M. Ochoa-Corona, S. M. Garnsey, and R. K. Yokomi. 1995. Citrus tristeza virus and its vector Toxoptera citricida. Plant Dis. 79:437-445. [Google Scholar]
- 73.Roistacher, C. N., and J. A. Dodds. 1993. Failure of 100 mild Citrus tristeza virus isolates from California to cross protect against a challenge by severe sweet orange stem pitting isolates, p. 100-107. In Proceedings of the 12th Conference of the IOCV. IOCV, Riverside, CA.
- 74.Roy, A., K. L. Manjunath, and R. H. Brlansky. 2005. Assessment of sequence diversity in the 5′-terminal region of Citrus tristeza virus from India. Virus Res. 113:132-142. [DOI] [PubMed] [Google Scholar]
- 75.Rubio, L., M. A. Ayllón, P. Kong, A. Fernández, M. L. Polek, J. Guerri, P. Moreno, and B. W. Falk. 2001. Genetic variation of Citrus tristeza virus isolates from California and Spain: evidence for mixed infections and recombination. J. Virol. 75:8054-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruiz-Medrano, R., B. Xoconostle-Cazares, and F. Kragler. 2004. The plasmodesmatal transport pathway for homeotic proteins, silencing signals and viruses. Curr. Opin. Plant Biol. 7:641-650. [DOI] [PubMed] [Google Scholar]
- 77.Salaman, R. N. 1933. Protective inoculation against a plant virus. Nature 131:468. [Google Scholar]
- 78.Satyanarayana, T., S. Gowda, V. P. Boyko, M. R. Albiach-Martí, M. Mawassi, J. Navas-Castillo, A. V. Karasev, V. Dolja, M. E. Hilf, D. J. Lewandowsky, P. Moreno, M. Bar-Joseph, S. M. Garnsey, and W. O. Dawson. 1999. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc. Natl. Acad. Sci. U. S. A. 96:7433-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Satyanarayana, T., S. Gowda, M. Mawassi, M. R. Albiach-Martí, M. A. Ayllon, C. Robertson, S. M. Garnsey, and W. O. Dawson. 2000. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology 1278:253-265. [DOI] [PubMed] [Google Scholar]
- 80.Satyanarayana, T., S. Gowda, M. A. Ayllon, and W. O. Dawson. 2003. Frameshift mutations in infectious cDNA clones of Citrus tristeza virus: a strategy to minimize the toxicity of viral sequences to Escherichia coli. Virology 313:481-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Satyanayanana, T., S. Gowda, M. A. Ayllón, and W. O. Dawson. 2004. Closterovirus bipolar virion: evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5′ region. Proc. Natl. Acad. Sci. U. S. A. 101:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Satyanarayana, T., C. J. Robertson, S. M. Garnsey, and W. O. Dawson. 2005. Generation of a genetically engineered MCA13 nonreactive variant of the T36 decline isolate of citrus tristeza virus, p. 34-43. In Proceedings of the 16th Conference of the IOCV. IOCV, Riverside, CA.
- 83.Schaller, T., N. Appel, G. Koutsoudakis, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81:4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sherwood, J. L., and R. W. Fulton. 1982. The specific involvement of coat protein in tobacco mosaic virus cross-protection. Virology 119:150-158. [DOI] [PubMed] [Google Scholar]
- 85.Singh, I. R., M. Suomalainen, S. Varadarajan, H. Garoff, and A. Helenius. 1997. Mechanisms for the inhibition of entry and uncoating of superinfecting Semliki Forest virus. Virology 231:59-71. [DOI] [PubMed] [Google Scholar]
- 86.Steck, F. T., and H. Rubin. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology 29:628-641. [DOI] [PubMed] [Google Scholar]
- 87.Steck, F. T., and H. Rubin. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology 29:642-653. [DOI] [PubMed] [Google Scholar]
- 88.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tatineni, S., C. J. Robertson, S. M. Garnsey, M. Bar-Joseph, S. Gowda, and W. O. Dawson. 2008. Three genes of Citrus tristeza virus are dispensable for infection and movement throughout some varieties of citrus trees. Virology 376:297-307. [DOI] [PubMed] [Google Scholar]
- 90.Thomas, C. L., L. D. C. Jones, and A. J. Maule. 2001. Size constraints for targeting posttranscriptional gene silencing and for using RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 25:417-425. [DOI] [PubMed] [Google Scholar]
- 91.Valkonen, J. P., M. L. Rajamäki, and T. Kekarainen. 2002. Mapping of viral genomic regions important in cross-protection between strains of a potyvirus. Mol. Plant-Microbe Interact. 15:683-692. [DOI] [PubMed] [Google Scholar]
- 92.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants: defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 93.Van Vuuren, S. P., R. P. Collins, and J. V. da Graça. 1993. Evaluation of citrus tristeza virus isolates for cross protection of grapefruit in South Africa. Plant Dis. 77:24-28. [Google Scholar]
- 94.Visconti, N. 1953. Resistance to lysis from without in bacteria infected with T2 bacteriophage. J. Bacteriol. 66:247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vives, M. C., L. Rubio, C. López, J. Navas-Castillo, M. R. Albiach-Martí, W. O. Dawson, J. Guerri, R. Flores, and P. Moreno. 1999. The complete genome sequence of the major component of a mild citrus tristeza virus isolate. J. Gen. Virol. 80:811-816. [DOI] [PubMed] [Google Scholar]
- 96.Vives, M. C., L. Rubio, A. Sambade, T. E. Mirkov, P. Moreno, and J. Guerri. 2005. Evidence of multiple recombination events between two RNA sequence variants within a Citrus tristeza virus isolate. Virology 331:232-237. [DOI] [PubMed] [Google Scholar]
- 97.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 98.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 99.Walkey, D. G. A., H. Lecoq, R. Collier, and S. Dobson. 1992. Studies on the control of zucchini yellow mosaic virus in courgettes by mild strain protection. Plant Pathol. 41:762-771. [Google Scholar]
- 100.Wen, F., M. Lister, and F. A. Fattouh. 1991. Cross-protection among strains of barley yellow dwarf virus. J. Gen. Virol. 72:791-799. [DOI] [PubMed] [Google Scholar]
- 101.Weng, Z., R. Barthelson, S. Gowda, M. E. Hilf, W. O. Dawson, D. W. Galbraith, and Z. Xiong. 2007. Persistent infection and promiscuous recombination of multiple genotypes of an RNA virus within a single host generate extensive diversity. PLoS One 2:e917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Whitaker-Dowling, P. A., J. S. Youngner, C. C. Widnell, and D. K. Wilcox. 1983. Superinfection exclusion by vesicular stomatitis virus I. Virology 131:137-143. [DOI] [PubMed] [Google Scholar]
- 103.Wildum, S. M., M. Schindler, J. Munch, and F. Kirchhoff. 2006. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type I-infected T cells to superinfection. J. Virol. 80:8047-8059. [DOI] [PMC free article] [PubMed] [Google Scholar]