Abstract
Vibrio cholerae strains of the O1 serogroup that typically cause epidemic cholera can be classified into two biotypes, classical and El Tor. The El Tor biotype emerged in 1961 and subsequently displaced the classical biotype as a cause of cholera throughout the world. In this study we demonstrate that when strains of the El Tor and classical biotypes were cocultured in standard LB medium, the El Tor strains clearly had a competitive growth advantage over the classical biotype starting from the late stationary phase and could eventually take over the population. The classical biotype produces extracellular protease(s) in the stationary phase, and the amounts of amino acids and small peptides in the late stationary and death phase culture filtrates of the classical biotype were higher than those in the corresponding culture filtrates of the El Tor biotype. The El Tor biotype cells could utilize the amino acids more efficiently than the classical biotype under the alkaline pH of the stationary phase cultures but not in medium buffered to neutral pH. The growth advantage of the El Tor biotype was also observed in vivo using the ligated rabbit ileal loop and infant mouse animal models.
Cholera is an ancient human affliction described in Indian, Arabian, Greek, and Roman texts more than 2,500 years ago. In modern times there have been seven recorded pandemics of cholera, all of which have been caused by strains of the O1 serogroup of Vibrio cholerae. The O1 strains can be classified into two biotypes, classical and El Tor. The first six pandemics of cholera occurred between 1817 and 1961 and were caused by classical biotype strains that were in general extremely virulent and could cause devastating epidemics. In 1961, the El Tor biotype emerged in Indonesia to cause the ongoing seventh cholera pandemic. The seventh-pandemic El Tor is a highly adapted organism that within a decade of its emergence displaced the classical biotype as a cause of epidemic cholera (11). Most recently, in 1992 another serogroup, O139, emerged to cause serious cholera epidemics in the Indian subcontinent (22). It has been demonstrated that these strains have been derived from the seventh-pandemic O1 El Tor clone (5, 25).
Several biochemical tests, including hemolysis (2, 23), hemagglutination of chicken erythrocytes (14), resistance to polymyxin B (13), bacteriophage-mediated lysis (4, 6, 27), and acetoin synthesis (15), are used to distinguish strains of the El Tor from the classical biotype. Many studies have revealed dramatic differences in regulation of virulence gene expression between the El Tor and classical biotypes (1, 8, 16, 19). A comparative genomic study revealed that 22 genes are missing in the classical biotype strains compared to seventh-pandemic El Tor strains, with most of these genes located on two chromosomal islands (9). Furthermore, transcriptional profiling experiments indicated that 524 genes were differentially expressed between the two biotypes under conditions that induce virulence expression in the classical biotype. Expression of genes encoding virulence factors was higher in the classical biotype while expression of those encoding proteins required for transport of amino acids and peptides, chemotaxis, and biofilm formation was higher in the El Tor biotype (3). Recently, a unique difference in carbohydrate metabolism between the two biotypes has been reported (29). Classical biotype strains grown in the presence of exogenous sugars produced a sharp decrease in medium pH due to formation of organic acids, resulting in loss of viability. However, growth of El Tor biotype strains in the presence of sugars produced 2,3-butanediol, a neutral fermentation end product that did not inhibit bacterial growth. These results suggest that the ability to metabolize sugars without production of growth-inhibitory acidic end products might account for the improved evolutionary fitness of the V. cholerae El Tor biotype compared to the classical biotype. However, whether this ability could provide a competitive growth advantage to the El Tor biotype when it is grown in cocultures with the classical biotype has not been investigated.
In their natural environment microbial populations compete continuously for limited resources. Different offensive strategies have been adopted by different bacteria for competitive exclusion of related species or even subpopulations of the same species to facilitate their own survival. In Escherichia coli, specific mutations in the rpoS gene encoding the stationary-phase-specific sigma factor and the enhanced ability to catabolize amino acids in an rpoS background under basic conditions have been shown to confer a fitness advantage referred to as growth advantage in the stationary phase (GASP) (10, 31, 32). On the other hand, stationary-phase-specific competition defect (SPCD) has been demonstrated in E. coli mutants defective in SOS DNA polymerase and penicillin binding protein 1b (21, 28).
In this study the relative fitness of El Tor and classical biotype strains has been examined in cocultures. The results obtained indicated that the El Tor biotype had a competitive fitness advantage in the stationary phase when grown with the classical biotype in cocultures. The nature of the fitness gain has been investigated.
MATERIALS AND METHODS
Bacterial strains and growth media.
The V. cholerae strains used in this study are listed in Table 1. Luria-Bertani (LB) medium contained 1% tryptone (Difco), 0.5% yeast extract (Difco), and 0.5% NaCl. When required, the LB medium was buffered to pH 7.2 with 100 mM HEPES buffer. A synthetic salt solution containing 30 mM KH2PO4, 2 mM NH4Cl, 85 mM NaCl, 1 mM MgSO4, and trace amounts of FeCl3 in 50 mM Tris-HCl (pH 7.2 or 8.5) supplemented with 0.2% Casamino Acids or individual amino acids (0.5%) was used in some experiments.
TABLE 1.
Bacterial strains
| V. cholerae strain | Relevant characteristics | Reference or sourcea |
|---|---|---|
| O395 Smr | O1 classical; derivative of wild-type O395 | J.J. Mekalanos, Harvard Medical School, Boston, MA |
| O395 Smr Nalr | O1 classical; derivative of O395 Smr | This study |
| 569B | O1 classical; wild type | Laboratory collection |
| N16961 Smr | O1 El Tor; derivative of wild-type N16961 | J.J. Mekalanos, Harvard Medical School, Boston, MA |
| N16961 Smr Nalr | O1 El Tor; derivative of N16961 Smr | This study |
| C6709 Smr | O1 El Tor; derivative of wild-type C6709 | NICED, Kolkata, India |
| SG24 | O139; wild type | NICED, Kolkata, India |
| E7946 Smr | O1 El Tor; derivative of wild-type E7946 | NICED, Kolkata, India |
NICED, National Institute of Cholera and Enteric Diseases.
Growth conditions.
The V. cholerae strains were grown in LB medium and maintained at −70°C in 20% (vol/vol) glycerol. V. cholerae strains from glycerol stock cultures were streaked on LB agar plates and incubated overnight at 37°C. A loopful of cells from the plate was inoculated into 5 ml of LB medium and grown overnight (16 h) at 37°C with shaking. Cultures were then inoculated (1:100, vol/vol) into fresh LB medium, either individually or mixed together, and incubated at 37°C as above. At regular intervals aliquots were removed, and the number of CFU of each population was determined using appropriate antibiotics, as described previously (20). All experiments were performed at least thrice. In some experiments, individual cultures of the two biotypes were grown for 24 h and then mixed in appropriate volumes so that the number of CFU of the two strains in the mixed culture was approximately 1:1, and incubation was continued. Streptomycin (50 μg/ml) and nalidixic acid (10 μg/ml) were used when required.
Preparation of culture filtrates.
V. cholerae strains were grown in LB medium as described above for 20 or 24 h and centrifuged at 6,000 rpm for 5 min at room temperature. The supernatant was collected and passed through 0.22-μm-pore-size filters (Millipore). A total of 100 μl of the filtered supernatant was plated on the LB agar plates to confirm the absence of bacteria in the filtrates.
Protease assay in culture filtrates.
Protease activity in culture filtrates was assayed as described by Schneider et. al (26). Briefly, 2 μg of bovine serum albumin (BSA) in borate buffer (pH 9) was incubated with 20-h culture filtrates (4 μl, unless stated otherwise in the text) for different time periods. The reaction was stopped by the addition of gel loading buffer and heating in a boiling water bath for 5 min; filtrates were then analyzed by SDS-PAGE. To examine the effect of pH on protease activity in culture filtrates, 4-μl culture filtrates were incubated with BSA dissolved in 50 mM Tris buffer and pH was adjusted to 7 or 8.5.
Ligated rabbit ileal loop assay.
In vivo growth of the El Tor biotype strain N16961 and classical biotype strain O395 in individual and mixed cultures was examined using the ligated rabbit ileal loop model essentially as described by De and Chatterjee (7). Briefly, rabbits were starved overnight with free access to water and anesthetized, and the small intestine was tied into consecutive 6- and 2-cm segments proximally to the mesoappendix. V. cholerae strain N16961 or O395 was grown to the logarithmic phase in LB medium and diluted in normal saline to a concentration of about 107 CFU per ml. An inoculum of 0.5 ml of strain N16961 or O395 or a 1:1 mixture of N16961 and O395 containing about 106 CFU was introduced into each 6-cm segment, and one loop was inoculated with saline as a negative control. The intestine was returned to the peritoneal cavity, and the incision was closed. After 18 h, the animals were sacrificed, and the small intestine was removed. Fluid accumulated in each loop was separately collected and measured, after which the loops were slit open and scraped. The fluid and scrapings were centrifuged (at 8,000 × g for 5 min) to collect the bacteria, washed twice with normal saline, and finally resuspended in saline. Bacterial count in the saline suspension was determined by plating on LB agar as well as on thiosulfate-citrate-bile salt-sucrose (TCBS) agar plates containing appropriate antibiotics. Each sample was inoculated into at least two loops in each animal, and the experiment was performed in three individual animals.
Infant mouse assays.
V. cholerae strains (approximately 105 CFU of individual or mixed cultures) were inoculated intragastrically into 4- to 5-day-old BALB/c suckling infant mice (n = 4), and the infection was allowed to proceed for 20 h. The mice were sacrificed, their small intestines were homogenized, and the total number of CFU obtained from the intestines of four mice was determined for each strain by plating dilutions of intestinal homogenates on LB agar containing appropriate antibiotics.
RESULTS
V. cholerae O1 biotype El Tor strain N16961 has a competitive growth advantage over the biotype classical strain O395.
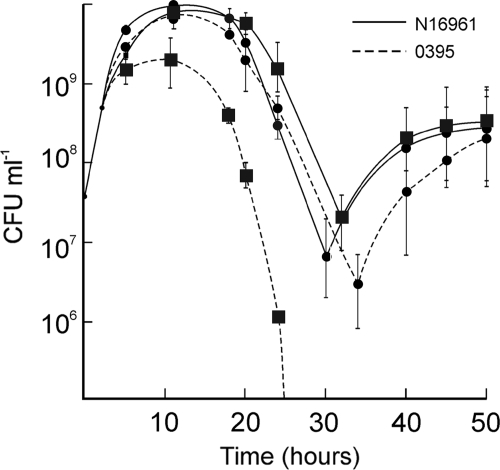
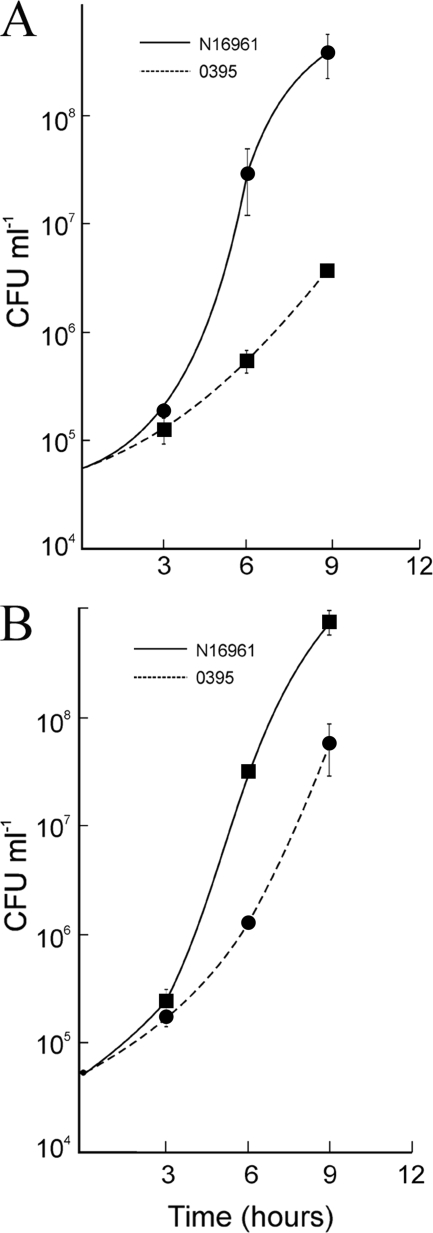
Representative strains N16961 and O395 of the V. cholerae El Tor and classical biotypes, respectively, were grown individually or in mixed cultures in LB medium, and the number of viable cells in each culture was assayed at regular intervals. When grown separately, El Tor strain N16961 and the classical strain O395 had similar growth rates in the logarithmic phase (Fig. 1). In the stationary and death phases small but consistent differences in the viable cell counts and kinetics were observed between the two biotypes; especially, recovery from the death phase was slower in O395 cultures (Fig. 1). Next, the strains N16961 and O395 were cocultured in LB medium, and the growth of each strain in the coculture was monitored (see Materials and Methods). The two populations in the mixed culture could be distinguished by a neutral antibiotic resistance marker carried on one of the strains. The marker was switched between the strains, and cocultures were performed reciprocally to confirm that the marker did not affect fitness. The growth rates of N16961 and O395 in the coculture were comparable in the logarithmic phase (Fig. 1). However, from the onset of the stationary phase (about 5 h), the relative proportion of N16961 in the coculture started increasing. The difference was more noticeable in the death phase, and by 20 (± 3) h the proportion of El Tor N16961 was almost 100-fold higher than the proportion of classical O395 (Fig. 1). By 38 (± 3) h the culture consisted almost exclusively of El Tor N16961 cells (data not shown). Similar results were obtained when the antibiotic resistance marker was present in either strain.
FIG. 1.
Growth of V. cholerae El Tor N16961 and classical O395 strains in individual cultures and mixed cultures. The strains N16961 and O395 were grown individually in monocultures or cocultured together (1:1), and the number of CFU of each strain was determined at regular intervals. Circles and squares represent the number of CFU of each strain in individual cultures and mixed cultures, respectively.
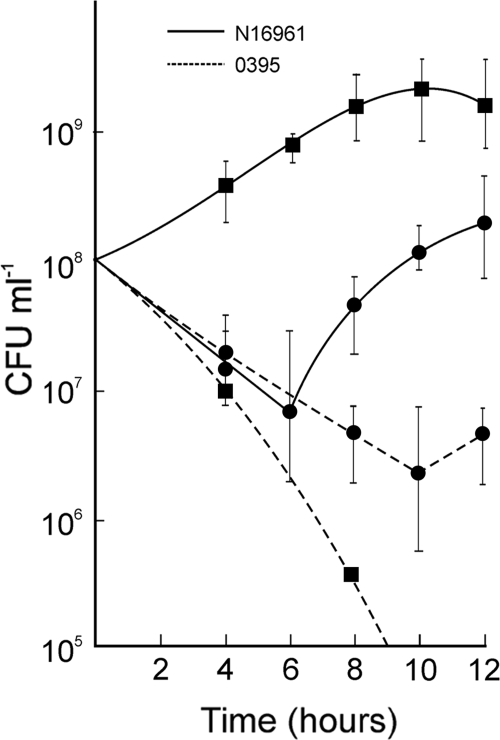
In another set of experiments, the strains N16961 and O395 were grown separately for 20 to 24 h in LB medium; the cultures were then mixed in an appropriate volume so that the cell count of the two strains in the mixed culture was in a ratio of about 1:1, and incubation was continued at 37°C. After 6 h, the viable cell count of El Tor N16961 was about 100-fold higher and that of classical O395 was about 5-fold lower in the mixed cultures than in the individual cultures (Fig. 2).
FIG. 2.
Competition between El Tor and classical biotype strains in the death phase. The El Tor N16961 and classical O395 strains were grown separately for 24 h and then mixed at a ratio of 1:1. The individual cultures and the mixed culture were incubated at 37°C with aeration for a further 12 h, and the numbers of CFU of each strain in the individual and mixed cultures were determined at regular intervals. Circles and squares represent the number of CFU per ml of each strain in individual cultures and mixed cultures, respectively.
In view of the fact that the El Tor biotype strain N16961 exhibited a striking growth advantage during the late stationary and death phases when it was cocultured with the classical biotype strain O395, the molecular basis of the growth advantage phenotype was studied in greater detail.
Growth of El Tor strain N16961 in stationary-phase culture filtrate of the classical strain O395.
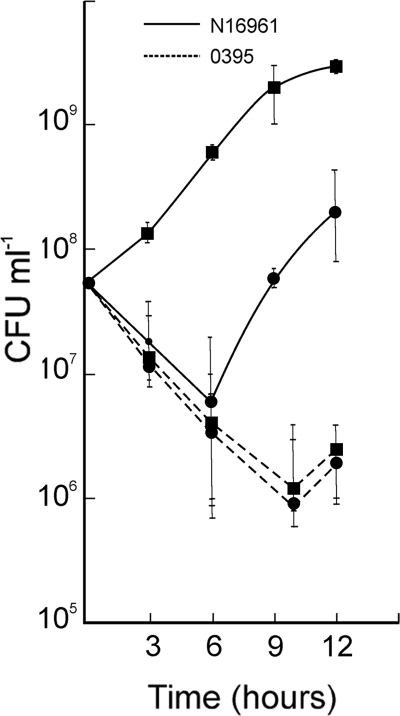
During the bacterial death phase, nutrients are released from dead cells into the medium and may be utilized by cells that are still alive. We considered the possibility that nutrients are present in the classical strain O395 stationary/death-phase cultures that are more efficiently utilized by the El Tor N16961 cells than by O395 cells to account for the competitive growth advantage of the former in cocultures. Hence, the growth of N16961 was examined in conditioned medium prepared from death phase cultures of O395. After 6 h, the viable cell count of N16961 was about 100-fold higher in the O395 culture filtrate than in its own culture (Fig. 3). In a reciprocal experiment when O395 was suspended in its own or in N16961 culture filtrates, the viability of O395 was comparable (Fig. 3).
FIG. 3.
Growth of El Tor strain N16961 and classical strain O395 in 24-h conditioned medium. V. cholerae El Tor strain N16961 and classical strain O395 were grown separately in LB medium for 24 h. Five milliliters of each culture was centrifuged, washed, and suspended in equal volumes of 24-h culture filtrates of N16961 (circles) or O395 (squares), and the number of CFU was determined at regular intervals.
These results indicated that N16961 cells might obtain more nutrients from the death phase culture filtrates of strain O395 than from their own corresponding culture filtrate. In stationary-phase cultures, since the primary nutrients available are amino acids and peptides (32), the amount of amino acids/peptides in 24-h culture filtrates of the two biotypes was estimated using an o-phthaldialdehyde assay (24). It was consistently found that the amount of amino acids/peptides in the culture filtrates of the classical strain O395 was 2- to 3-fold higher than amounts in El Tor N16961 culture filtrates.
Protease activity in culture filtrates of the classical strain O395.
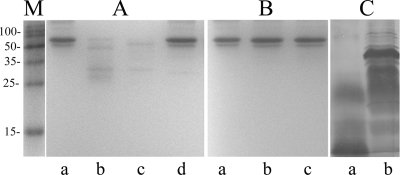
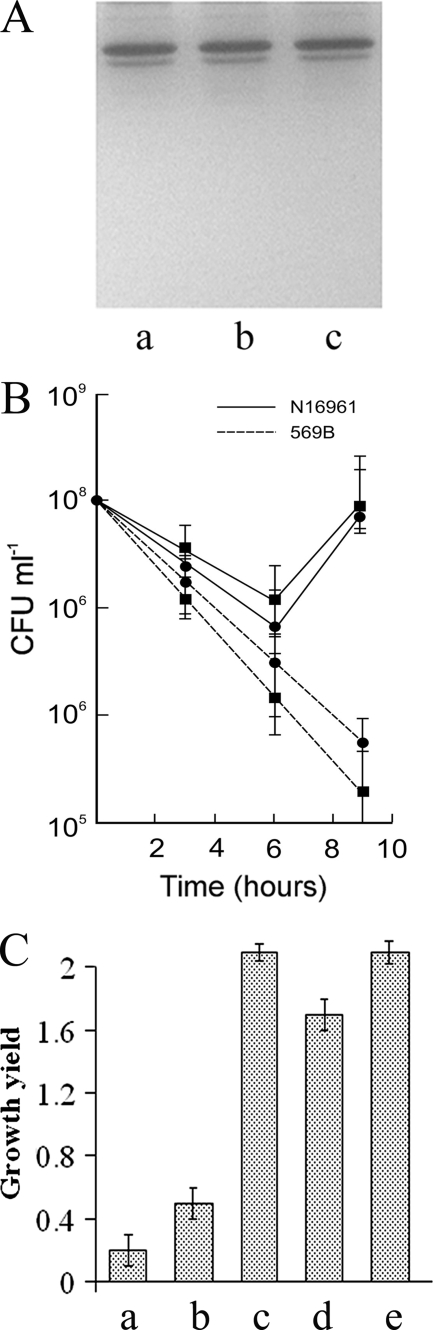
In view of the fact that larger amounts of amino acids and peptides were detected in the death phase (20 to 24 h) culture filtrates of the classical O395 than in the filtrates of the El Tor N16961 strain, we examined if extracellular protease(s) were present in the supernatants that could cleave proteins released by dead cells into amino acids/peptides. BSA was incubated with 20-h culture filtrates of the two biotypes, and aliquots were removed at different time intervals and examined by SDS-PAGE. Extensive proteolysis of BSA was observed within 5 min of incubation with O395 culture filtrate (Fig. 4A). However, when incubated with N16961 culture filtrate, practically no proteolysis of BSA was observed even after 60 min (Fig. 4B). These results clearly indicated the presence of significant protease activity in the culture filtrate of the classical O395 but not in the filtrate of El Tor N16961. Consistent with this observation, intact proteins were, in general, not detected in the 20-h culture filtrate of the classical biotype (Fig. 4C). No protease activity was detected in logarithmic-phase cultures of O395 or N16961 (data not shown).
FIG. 4.
Protease activity in culture filtrates of the El Tor and classical biotypes. (A and B) BSA (2 μg; lanes a) was incubated with 4 μl of 20-h culture filtrates of classical strain O395 (A) or El Tor strain N16961 (B) for 5 min (lanes b) or 60 min (lanes c) or for 60 min in the presence of 1 mM EDTA (lanes d) and analyzed by SDS-PAGE. (C) Proteins from 500 μl of 20-h culture filtrates of strain O395 (lane a) and N16961 (lane b). Lane M, standard molecular size markers. The numbers on the left indicate the sizes of the markers (kDa).
The effect of several protease inhibitors on the proteolytic activity of the 20-h O395 culture filtrate was examined. The protease activity was completely inhibited by protease inhibitor cocktail (Roche). However, when all the individual components of the cocktail were tested, only EDTA could effectively inhibit O395 extracellular protease activity (Fig. 4A, lane d). None of the other components, including bestatin, phosphoramidon, leupeptin, and phenylmethylsulfonyl fluoride (PMSF), had any effect.
Utilization of amino acids by stationary phase cultures of the El Tor N16961 and the classical O395 strains.
Exogenous amino acids and small peptides can be transported into bacterial cells and can serve as carbon and energy sources as well as protein building blocks (18). Minimal medium that contained only inorganic salts and Casamino Acids as the sole source of carbon and energy was tested for the ability to support growth of the El Tor N16961 and classical O395 strains. The medium was inoculated with aged (20- to 24-h grown) N16961 or O395 cells at about 5 × 104 CFU/ml and incubated for up to 9 h. In minimal medium buffered to pH 9 (but not pH 7.2; see below), the growth yield of N16961 was about 100-fold more than that of O395 (Fig. 5A). Practically no growth of either N16961 or O395 was observed in minimal medium with no added Casamino Acids or any other source of carbon (data not shown). This directly demonstrated that the aged N16961 cells were more efficient than those of O395 in taking up and utilizing exogenous amino acids and short peptides as carbon sources. In a similar experiment, minimal medium containing individual amino acids was tested for the ability to support growth of N16961 and O395. No significant growth of either strain was observed in the presence of the amino acids Gly, Val, Ala, Leu, Ile, Tyr, Trp, Phe, Thr, Met, or Cys as the sole carbon source. All other amino acids examined supported growth of both strains although growth of N16961 was in general significantly higher than that of O395 (Table 2). The most striking difference, about 45-fold higher growth yield of N16961 than of O395, was observed with Glu. A considerable growth advantage of N16961 over O395 (4- to 8-fold) was also observed in the presence of Pro, Asp, Lys, Gln, and Arg. Ser, His, and Asn supported the growth of both strains to comparable levels.
FIG. 5.
Growth of the El Tor and classical biotype in minimal medium supplemented with Casamino Acids. The El Tor N16961 and classical O395 strains were grown in LB medium monocultures for 20 h; cells were collected by centrifugation, washed, and suspended in minimal medium supplemented with 0.2% Casamino Acids and grown with aeration at 37°C. The minimal medium was buffered to pH 8.5 (A) or 7.2 (B).
TABLE 2.
Growth of V. cholerae classical O395 and El Tor N16961 strains in minimal medium supplemented with individual amino acidsa
| Amino acid | Individual growth yield of the indicated strainb |
Relative growth yieldc | |
|---|---|---|---|
| O395 | N16961 | ||
| Glutamic acid | 38.33 ± 5 | 1750 ± 200 | 45.65 ± 2.5 |
| Proline | 11.5 ± 3 | 88 ± 10 | 7.65 ± 2.3 |
| Aspartic acid | 28.16 ± 4 | 200 ± 20 | 7.10 ± 0.43 |
| Lysine | 5.8 ± 1.5 | 40.66 ± 2 | 7.01 ± 2.2 |
| Glutamine | 5 ± 2 | 23.33 ± 5.4 | 4.66 ± 1.2 |
| Arginine | 11.6 ± 2 | 41.83 ± 0.5 | 3.6 ± 0.9 |
| Serine | 34 ± 3 | 57.5 ± 2 | 1.69 ± 0.13 |
| Histidine | 16.66 ± 3 | 25.16 ± 5 | 1.5 ± 0.04 |
| Asparagine | 35 ± 5 | 52.5 ± 3 | 1.5 ± 0.19 |
Twenty-hour cultures of the El Tor N16961 and classical O395 strains were suspended in minimal medium (pH 9) supplemented with the indicated amino acid, and the number of CFU was determined after 9 h.
Individual growth yield was calculated as follows: number of CFU of each strain after 9 h/initial number of CFU.
Relative growth yield was calculated as follows: individual growth yield of N16961/individual growth yield of O395.
Taken together, these results (Fig. 4 and 5) suggest that when the classical O395 and the El Tor N16961 strains are grown together in cocultures to the late stationary phase, extracellular protease(s) produced by the classical O395 strain degrades proteins released from dead cells to amino acids/peptides. These amino acids/peptides are taken up and utilized by the El Tor N16961 cells in the coculture with greater efficiency than the classical O395 cells, resulting in the survival and growth advantage of the El Tor N16961 strain in the cocultures. To examine the importance of extracellular protease(s) produced by the classical biotype in providing a growth advantage to the El Tor biotype in cocultures, the following experiments were performed using the classical strain 569B that did not produce extracellular protease(s).
The classical biotype strain 569B does not produce extracellular proteases and confers a lower growth advantage to El Tor N16961 in cocultures.
V. cholerae 569B is a toxigenic strain of the classical biotype. Production of extracellular protease(s) by 569B was examined by incubating BSA with 20-h culture filtrate of 569B for 60 min. Practically no proteolysis of BSA was observed (Fig. 6A), suggesting that, unlike strain O395, strain 569B did not produce extracellular protease(s) although both strains belong to the classical biotype. Whether the El Tor strain N16961 had a growth advantage when cocultured with strain 569B was next examined. Twenty-four-hour cultures of N16961 and 569B were mixed (1:1), and the number of CFU of each strain in the coculture was determined at regular intervals up to 9 h. The number of CFU of N16961 in the cocultures was only slightly higher than that in the individual culture (Fig. 6B). In a similar experiment, when N16961 was grown with the classical strain O395, the growth of N16961 was about 100-fold higher in the mixed culture than in the individual culture (Fig. 2). The viable cell count of 569B was comparable to that of O395 in cocultures and in individual cultures (Fig. 2 and 5). When the classical strains O395 and 569B were grown together, neither strain derived a growth advantage, and growth of each strain in the mixed cultures was similar to that in the individual cultures (data not shown). Next, strain N16961 was incubated in 20-h 569B culture filtrates. After 6 h of incubation, the viable cell count of the El Tor N16961 was 2-fold higher in 569B culture filtrates than in its own culture (Fig. 6C). Under identical conditions, the number of CFU of N16961 increased 10-fold when growth occurred in culture filtrates from O395 (Fig. 6C). To examine if this difference was due to the absence of extracellular protease activity in 569B cultures, 20-h culture filtrate of 569B was treated with proteinase K, and growth of N16961 was examined. The growth of N16961 in proteinase K-treated culture filtrates of 569B was significantly higher than in the untreated filtrate (Fig. 6C). Taken together, these results suggested a role for extracellular protease(s) in conferring a growth advantage to the El Tor N16961 cells.
FIG. 6.
V. cholerae classical strain 569B does not produce extracellular proteases and does not provide a growth advantage to El tor N16961. (A) BSA (2 μg; lane a) was incubated with 4 μl (lane b) or 8 μl (lane c) of 20-h culture filtrate of classical strain 569B for 60 min and analyzed by SDS-PAGE. (B) The El Tor N16961 and classical 569B strains were grown separately for 24 h and then mixed at a ratio of 1:1. The individual cultures and the mixed culture were incubated for a further 9 h, and the numbers of CFU of each strain in the individual and mixed cultures were determined at regular intervals. Circles and squares represent the number of CFU per ml of each strain in individual cultures and mixed cultures, respectively. (C) Growth of 20-h El Tor N16961 cells in 20-h culture filtrates from strain 569B. The number of CFU of N16961 suspended in N16961 (a), 569B (b), or O395 (c) culture filtrate or proteinase K (20 μg per ml)-treated culture filtrate of strain N16961 (d) or 569B (e) was determined after 6 h. Growth yields were calculated by dividing the number of cells after 6 h by the number of cells at inoculation. Data represent the average of three independent experiments; error bars indicate standard error of the means.
Effect of pH on the growth advantage of El Tor strain N16961 in cocultures.
In unbuffered LB medium, the pH of the medium is initially 7.2 but increases in the stationary phase to 8.5 or higher (data not shown). The increase in pH occurs primarily due to the use of amino acids as the major source of carbon and energy after exhaustion of the carbohydrates in the medium (10). To examine if the pH of the growth medium had any role in conferring a competitive growth advantage to El Tor N16961 in cocultures with classical O395, the two strains were grown together in LB medium buffered to pH 7.2, and the survival of N16961 was monitored and compared to that in unbuffered LB medium. In LB medium buffered to neutral pH, the growth advantage of the El Tor biotype N16961 strain was significantly reduced although not totally abolished (data not shown).
The effect of pH on the production and activity of the extracellular proteases by strain O395 was also examined. Protease activity levels in culture filtrates of O395 grown for 20 h in unbuffered LB medium and in LB medium buffered to pH 7.2 were comparable (data not shown). To examine the effect of pH on the activity of the extracellular proteases, culture filtrates of strain O395 grown in unbuffered LB medium for 20 h was incubated with BSA in borate buffer, pH 7.2 or pH 9. Under both conditions, BSA was completely degraded, and the degradation could be inhibited by EDTA. These results suggested that neither production nor activity of the O395 extracellular protease(s) was dependent on pH. Practically no proteolytic activity was detected in the N16961 culture filtrates under any of the above conditions (data not shown).
The effect of pH on the uptake and utilization of amino acids/peptides in minimal medium by the El Tor N16961 and classical O395 strains was next examined. Growth of each strain in minimal medium supplemented with Casamino Acids and buffered to pH 7.2 was compared. Under these conditions, the growth of El Tor N16961 was only 15-fold higher than that of classical O395 (Fig. 5B), whereas in basic medium (pH 9), growth of N16961 was 100-fold higher than that of O395 (Fig. 5A). It may be noted that growth of O395 was significantly lower in basic medium than under neutral conditions, suggesting that O395 is less efficient in uptake and utilization of amino acids under basic conditions than strain N16961.
Competitive growth advantage of other V. cholerae strains.
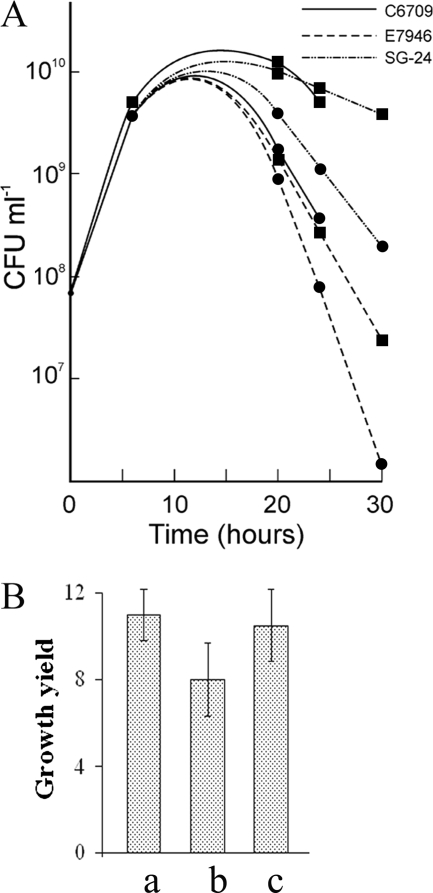
In view of the fact that considerable differences exist among individual strains of the El Tor biotype (9, 11), we have examined if El Tor strains other than N16961 also derive a growth advantage when cocultured with classical strains. When the strains C6709 and E7946 belonging to the El Tor biotype (Table 1) were cocultured with the classical strain O395, both of the El Tor strains exhibited a growth advantage in the cocultures (Fig. 7A). Also the strain SG24 belonging to serogroup O139, which is closely related to the El Tor biotype (5), gave similar results. At 24 h, the growth of strains C6709 and SG24 was about 7-fold higher while that of strain E7946 was about 3-fold higher in the cocultures than in the individual cultures. It may be noted that strain N16961 had about a 3-fold growth advantage in the cocultures. Furthermore, the growth of aged cells of strains C6709, E7946, and SG24 was significantly higher when they were suspended in 24-h culture filtrates of the classical strain O395 than when they were grown in their own culture filtrates although quantitative differences exists between the strains (Fig. 7B).
FIG. 7.
Growth advantage of other V. cholerae strains. (A) The V. cholerae El Tor strains E7946 and C6709 and O139 strain SG24 were grown in monocultures or cocultured with classical O395 (1:1), and the number of CFU of each strain was determined at regular intervals. Circles and squares represent the numbers of CFU of each strain in individual cultures and mixed cultures, respectively. (B) The El Tor strains C6709 (a) and E7946 (b) and O139 strain SG24 (c) were grown for 24 h in LB medium monocultures, centrifuged, washed, and suspended in equal volumes of 24-h culture filtrates of classical strain O395 or their own culture filtrates; the number of CFU per ml of each strain was then determined after 6 h. Growth yields were determined by dividing the number of cells of each strain in O395 culture filtrates by the number of cells in their own culture filtrates. All experiments were performed three times, and results are represented as means ± standard deviations.
In vivo competition between El Tor N16961 and classical O395.
In view of the fact that under standard laboratory conditions the El Tor biotype can out-compete the classical biotype in cocultures, it was important to examine whether a similar population takeover occurs in vivo. The ligated rabbit ileal loop and the infant mouse models were used to assess the in vivo growth of El Tor N16961 and classical O395 in individual and mixed cultures. When inoculated separately in rabbit ileal loops or intragastrically to infant mice, O395 and N16961 grew at comparable rates; however, when the strains were inoculated together at a ratio of 1:1, N16961 had a considerable growth advantage over O395, and the relative ratio of the two strains changed to about 11:1 and 28:1, respectively, in rabbit intestine and infant mice (Table 3). These results clearly indicated that the El Tor biotype has a competitive growth advantage over the classical biotype even in the animal intestine.
TABLE 3.
Competitive growth of El Tor N16961 and classical O395 in rabbit intestine and infant mice
| Strain | Growth in ligated rabbit ileal loop model |
Growth in infant mouse model |
||||
|---|---|---|---|---|---|---|
| No. of CFU in cocultures |
Competitive indexb | No. of CFU in cocultures |
Competitive indexb | |||
| Input | Outputa | Input | Output | |||
| O395 | 1.5 × 106 | 3.9 × 108 | 11.53 | 1.33 × 105 | 4.9 × 105 | 28.3 |
| N16961 | 1.5 × 106 | 4.5 × 109 | 1.06 × 105 | 1.38 × 107 | ||
Output is the total number of CFU in fluid accumulated in the loops plus the total number of CFU in mucosal scrapings. The total number of CFU of each strain in the mucosal scrapings was about 1/10 of the numbers in the fluid.
Competitive index was calculated as follows: (output of N16961/input of N16961)/(output of O395/input of O395), where input and output are numbers of CFU.
DISCUSSION
A unique event in bacterial epidemiology was the displacement of the epidemic-causing classical biotype of V. cholerae by the El Tor biotype within a decade of the emergence of the latter in 1961, marking the beginning of the seventh cholera pandemic (11). To the best of our knowledge, such a rapid competitive exclusion of an existing epidemic-causing strain by a newly emerged strain has not been observed for any other bacterial pathogen. The genetic basis of this phenomenon is still unknown, despite the accumulation of many facts about the molecular differences between the El Tor and the classical biotypes (3, 9, 17). It has been demonstrated in this study that the competitive exclusion of the classical biotype by the El Tor biotype could be reproduced under standard laboratory conditions, and the molecular basis of the fitness gain of the El Tor biotype when the strain is cocultured with the classical biotype has been investigated.
The growth advantage of the El Tor biotype when it is cocultured with the classical biotype was reminiscent of the GASP phenotype exhibited by certain rpoS mutants of E. coli. The GASP mutants have a competitive growth advantage in the stationary and death phases and can finally take over the population when cocultured with the wild-type strains. When grown separately, the GASP mutants and the wild-type strains exhibit similar growth patterns (12, 30). In this respect the E. coli GASP mutant strongly resembled the El Tor biotype that could grow and eventually take over the population when it was cocultured with the classical biotype. Since the GASP phenotype is generally attributed to mutations in the rpoS gene encoding a stationary-phase-specific sigma factor, the rpoS gene sequence and upstream promoter regions of the strains El Tor N16961 and classical O395 were compared (www.tigr.org). No difference in rpoS sequences was detected between the two biotypes (data not shown), indicating that RpoS might have no role in the observed competitive growth advantage of the El Tor biotype.
The observation that the aged (20- to 24-h grown) El Tor N16961 cells derived a pronounced growth advantage from aged O395 culture filtrates compared to growth in their own culture filtrate (Fig. 3) suggested that more nutrients might be present in the former, and aged N16961 cells are more proficient than O395 cells in utilizing the nutrients. Indeed, the amino acids and short-peptide content of the O395 culture filtrates was higher than that of N16961. What could be the source of the higher nutrient content in the classical O395 culture filtrates? One possibility is that a greater number of O395 cells had lysed in the 24-h cultures used in this study, releasing nutrients from the dead cells. However, an examination of the growth curves of N16961 and O395 showed that the numbers of CFU of the two strains in the 24-h cultures were comparable. We next considered the possibility that extracellular proteases might be present in O395 cultures that degrade intact proteins released by dead cells into short peptides and amino acids. This was suggested by the observation that intact proteins were more abundant in death phase culture filtrates of N16961 than in filtrates of O395 (Fig. 4C) though amino acids and peptides were more abundant in the O395 culture filtrates. Indeed, we confirmed high extracellular protease activity starting from the stationary phase (16 h) in culture filtrates of O395 that was not detected in N16961 cultures (Fig. 4). The protease activity was inhibited by only EDTA, indicating the presence of metalloprotease(s). Analysis of the V. cholerae genome database (www.ebi.ac.uk) indicated the presence of at least 19 almost identical metalloprotease genes in strains N16961 and O395. It is not yet clear why protease activity was detected in the culture filtrates of O395 but not in those of N16961 in spite of the presence of almost identical metalloprotease genes in the two biotypes.
How important is the extracellular protease(s) produced by the classical O395 cells in providing a growth advantage to the El Tor N16961 cells in cocultures? Since the protease activity was inhibited by only EDTA, which had a detrimental effect on the survival of V. cholerae, we could not directly inhibit protease(s) to assess its role in providing a growth advantage to the El Tor strains. However, the classical biotype strain 569B that did not produce extracellular proteases failed to provide a growth advantage to the El Tor strain N16961 although 569B culture filtrates treated with exogenously added proteinase K provided a significant growth advantage (Fig. 6), strongly emphasizing the importance of extracellular protease(s) present in aged cultures of the classical strain O395 in conferring a growth advantage to the El Tor cells.
The observation that aged N16961 cells grew more efficiently than cells of O395 in conditioned medium from O395 death phase cultures suggested that the pool of amino acids/peptides produced by the action of the classical O395 proteases on proteins released by dead cells is more efficiently utilized by El Tor N16961 than by the classical O395. Direct evidence for this hypothesis was obtained by comparing the growth rates of N16961 and O395 in minimal medium that contained Casamino Acids or each individual amino acid as the sole source of carbon and energy. Glutamic acid, in particular, conferred a striking growth advantage to the El Tor biotype. It may be mentioned in this context that a microarray-based whole-genome expression analysis had earlier revealed that when the two strains were grown under virulence-inducing conditions (in LB medium, pH 6.6, at 30°C), expression levels of at least eight genes encoding proteins required for transport of amino acids and peptides were significantly higher in the El Tor biotype than in the classical biotype (3). Thus, the growth advantage of El Tor strains in mixed cultures with classical strain O395 or in conditioned medium from aged O395 cultures might be due to a combination of two factors, the extracellular protease(s) released by O395 that increased the available amino acid/peptides in culture supernatants and the more efficient catabolism of these nutrients by El Tor strains than by the classical strain O395. It may be noted that pH of the medium had an important effect on the latter but not the former process (Fig. 5). Although El Tor strains were equally efficient in utilizing amino acids and peptides under neutral and basic conditions, the classical strain O395 could utilize these nutrients only in neutral medium. It is attractive to hypothesize that the sequence of events that might occur when El Tor N16961 and classical O395 strains were cocultured in unbuffered LB medium is as follows: when the culture enters stationary phase, O395 produces extracellular protease(s); in the subsequent death phase the protease(s) degrades proteins released from dead cells into amino acids and short peptides. At this time, the pH of the unbuffered LB medium increases to about 8.5, a change that reduces uptake and utilization of amino acids/peptides by the classical cells but allows the El Tor biotype to utilize the amino acid/peptide pool for its own growth, resulting in a growth advantage of the El Tor biotype. This hypothesis is supported by the observation that the growth advantage of the El Tor biotype was much reduced in medium buffered to neutral pH conditions. It may be mentioned in this context that the GASP phenotype of an E. coli rpoS mutant was also apparent under basic conditions (10) and was due to enhanced amino acid catabolism (32).
From the results presented in this report, it becomes immediately apparent that the intestinal environment might provide excellent conditions for enrichment of the El Tor biotype. V. cholerae colonizes the small intestine, where the milieu is alkaline due to secretion of bicarbonate through the common bile duct into the lumen of the duodenum. Furthermore, the small intestine is thought to contain a pool of amino acids due to the cleavage of dietary proteins by pepsin in the stomach and subsequently trypsin in the duodenum. Thus, it is reasonable to assume that in the intestinal milieu, the El Tor biotype would derive a significantly larger growth advantage than the classical biotype and, in the case of mixed infections, might ultimately exclude the classical biotype from the culture. Indeed, when the El Tor N16961 and classical O395 strains were inoculated into rabbit ileal loops or infant mice at a ratio of 1:1, after 18 h the relative ratio of the two biotypes changed significantly in favor of the El Tor biotype (Table 2).
Acknowledgments
We thank all members of the Biophysics Division for cooperation, encouragement, and helpful discussions during the study.
The work was supported by a research grant [5/8-1(196)/D/2004-ECD-II] from the Indian Council for Medical Research, Government of India. S.P. and A.K.B. are grateful to the Council of Scientific and Industrial Research and University Grants Commission for research fellowships.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Abuaita, B. H., and J. H. Withey. 2009. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect. Immun. 77:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, T. J., and P. A. Blake. 1981. Epidemiological usefulness of changes in hemolytic activity of Vibrio cholerae biotype El Tor during the seventh pandemic. J. Clin. Microbiol. 13:126-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyhan, S., A. D., Tischler, A. Camilli, and F. H. Yildiz. 2006. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect. Immun. 74:3633-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, S. K., R. Chowdhury, and J. Das. 1992. A 14-kilodalton inner membrane protein of Vibrio cholerae biotype El Tor confers resistance to group IV choleraphage infection to classical vibrios. J. Bacteriol. 174:6221-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calia, K. E., M. Murtagh, M. J. Ferraro, and S. B. Calderwood. 1994. Comparison of Vibrio cholerae O139 with V. cholerae O1 classical and El Tor biotypes. Infect. Immun. 62:1504-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury, R., S. K. Biswas, and J. Das. 1989. Abortive replication of choleraphage φ149 in Vibrio cholerae biotype El Tor. J. Virol. 63:392-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De, S. N., and S. N. Chatterjee. 1953. An experimental study of the mechanisms of action of Vibrio cholerae on the intestinal mucous membrane. J. Pathol. Bacteriol. 46:559-562. [DOI] [PubMed] [Google Scholar]
- 8.DiRita, V. J., M. Neely, R. K. Taylor, and P. M. Bruss. 1996. Differential expression of the ToxR regulon in classical and E1 Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc. Natl. Acad. Sci. U. S. A. 93:7991-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. U. S. A. 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell, M. J., and S. E. Finkel. 2003. The growth advantage in stationary phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 185:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkel, S. E. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4:113-120. [DOI] [PubMed] [Google Scholar]
- 13.Han, G. K., and T. S. Khie. 1963. A new method for differentiation of Vibrio comma and Vibrio El Tor. Am. J. Hyg. 77:184-186. [DOI] [PubMed] [Google Scholar]
- 14.Jonson, G., J. Sanchez, and A. M. Svennerholm. 1989. Expression and detection of different biotype-associated cell-bound haemagglutinins of Vibrio cholerae O1. J. Gen. Microbiol. 135:111-120. [DOI] [PubMed] [Google Scholar]
- 15.Kovacikova, G., W. Lin, and K. Skorupski. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol. Microbiol. 57:420-433. [DOI] [PubMed] [Google Scholar]
- 16.Kovacikova, G., and K. Skorupski. 2000. Differential activation of the tcpPH promoter by AphB determines biotype specificity of virulence gene expression in Vibrio cholerae. J. Bacteriol. 182:3228-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matson J. S., J. H. Withey, and V. J. DiRita. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75:5542-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 19.Murley, Y. M., J. Behari, R. Griffin, and S. B. Calderwood. 2000. Classical and El Tor biotypes of Vibrio cholerae differ in timing of transcription of tcpPH during growth in inducing conditions. Infect. Immun. 68:3010-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul, K., A. Ghosh, N. Sengupta, and R. Chowdhury. 2004. Competitive growth advantage of non-toxigenic mutants in the stationery phase in archival cultures of pathogenic Vibrio cholerae strains. Infect. Immun. 72:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepper, E. D., M. J. Farrell, and S. E. Finkel. 2006. Role of penicillin-binding protein 1b in competitive stationary phase survival of Escherichia coli. FEMS Microbiol. Lett. 263:61-67. [DOI] [PubMed] [Google Scholar]
- 22.Ramamurthy, T., S. Garg, R. Sharma, S. K. Bhattacharya, G. B. Nair, T. Shimada, T. Takeda, T. Karasawa, H. Kurazano, A. Pal, and Y. Takeda. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703-704. [DOI] [PubMed] [Google Scholar]
- 23.Richardson, K., J. Michalski, and J. B. Kaper. 1986. Hemolysin production and cloning of two hemolysin determinants from classical Vibrio cholerae. Infect. Immun. 54:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth, M. 1971. Fluorescence reaction for amino acids. Anal. Chem. 43:880-882. [DOI] [PubMed] [Google Scholar]
- 25.Sack, D. A., R. B. Sack, G. B. Nair, and A. K. Siddique. 2004. Cholera. Lancet 363:223-233. [DOI] [PubMed] [Google Scholar]
- 26.Schneider, D. R., S. P. Sigel, and C. D. Parker. 1981. Characterization of Vibrio cholerae protease activities with peptide digest analysis. J. Clin. Microbiol. 13:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeya, K., T. Otohuji, and H. Tokiwa. 1981. FK phage for differentiating the classical and El Tor groups of Vibrio cholerae. J. Clin. Microbiol. 14:222-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. U. S. A. 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon, S. S., and J. J. Mekalanos. 2006. 2,3-Butanediol synthesis and the emergence of the Vibrio cholerae El Tor biotype. Infect. Immun. 74:6547-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zambrano, M. M., and R. Kolter. 1996. GASPing for life in stationary phase. Cell 86:181-184. [DOI] [PubMed] [Google Scholar]
- 31.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 32.Zinser, E. R., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]