Abstract
NAD-dependent butanediol dehydrogenase (Bdh1p) from Saccharomyces cerevisiae reversibly transforms acetoin to 2,3-butanediol in a stereospecific manner. Deletion of BDH1 resulted in an accumulation of acetoin and a diminution of 2,3-butanediol in two S. cerevisiae strains under two different growth conditions. The concentrations of (2R,3R)-2,3-butanediol are mostly dependent on Bdh1p activity, while those of (meso)-2,3-butanediol are also influenced by the activity of NADP(H)-dependent oxidoreductases. One of them has been purified and shown to be d-arabinose dehydrogenase (Ara1p), which converts (R/S)-acetoin to meso-2,3-butanediol and (2S,3S)-2,3-butanediol. Deletion of BDH2, a gene adjacent to BDH1, whose encoded protein is 51% identical to Bdh1p, does not significantly alter the levels of acetoin or 2,3-butanediol in comparison to the wild-type strain. Furthermore, we have expressed Bdh2p with a histidine tag and have shown it to be inactive toward 2,3-butanediol. A whole-genome expression analysis with microarrays demonstrates that BDH1 and BDH2 are reciprocally regulated.
Acetoin and 2,3-butanediol are minor products generated by Saccharomyces cerevisiae during alcohol fermentation. Their sensory impacts on wine are poorly documented. Acetoin may affect the wine bouquet, although its perception threshold in wine is relatively high, around 150 mg/liter (21, 31). On the other hand, 2,3-butanediol is odorless (33) and cannot be expected to appreciably affect the sensory quality of wine. However, the compound may contribute to the wine body (28).
Acetaldehyde, pyruvate, and α-acetolactate are the main precursors of acetoin in S. cerevisiae. Acetoin can be formed from acetaldehyde and/or pyruvate through an anomalous reaction of pyruvate decarboxylase. Thus, although its main activity is to irreversibly decarboxylate pyruvate to acetaldehyde, it can also catalyze carbon-carbon bond formation, yielding acetoin from pyruvate and/or acetaldehyde (2, 4). In addition, α-acetolactate would produce acetoin through its nonenzymatic decarboxylation to diacetyl and subsequent reduction to acetoin through the action of several NADH- and NADPH-dependent oxidoreductases (12). However, the situation is more complex in wine fermentation, where other yeasts and bacteria display supplementary enzymatic activities capable of producing both acetoin and 2,3-butanediol (1, 27).
We have previously characterized a butanediol dehydrogenase (Bdh1p) as a medium-chain dehydrogenase/reductase (MDR) that can reversibly transform R-acetoin and S-acetoin to (2R,3R)-2,3-butanediol and meso-2,3-butanediol, respectively, in a NAD(H)-dependent reaction (10). BDH2 is a gene adjacent to BDH1 whose uncharacterized protein product (Bdh2p) shares 51% sequence identity with Bdh1p. To evaluate the in vivo roles of Bdh1p and Bdh2p, we compared the levels of several extracellular metabolites in cultures of wild-type and deficient strains. The results show that, although Bdh1p is the main enzyme in 2,3-butanediol production [essentially the (2R,3R)-2,3-butanediol stereoisomer], some meso-2,3-butanediol is still produced by the bdh1Δ strains. We have characterized Ara1p as an oxidoreductase that can reduce racemic acetoin to meso-2,3-butanediol and (2S,3S)-2,3-butanediol in the presence of NADPH.
Furthermore, we have overexpressed Bdh2p with a histidine tag at its carboxyl terminus and have shown it to be inactive toward acetoin and 2,3-butanediol. A microarray study indicated that BDH1 and BDH2 are reciprocally regulated under the conditions studied.
MATERIALS AND METHODS
Materials.
Restriction enzymes and T4 DNA ligase were from Boehringer Mannheim (Mannheim, Germany). Vent polymerase was from New England Biolabs, Inc. (Beverly, MA). DNA oligomers were synthesized and purified by Sigma-Genosys (Haverhill, United Kingdom). Chemicals were purchased from Fluka, Aldrich, or Sigma (Saint Louis, MO) and were of the highest available quality. Formate dehydrogenase and glucose-6-phosphate dehydrogenase were from Sigma.
Yeast and bacterial strains.
Escherichia coli XL-1 blue (Stratagene, La Jolla, CA) or DH5α was used for cloning experiments. The S. cerevisiae organisms used were derived from four laboratory strains with different genetic backgrounds: FY834α (MATα his3Δ200 ura3-52 leu2Δ1 lys2Δ202 trp1Δ63) (38); CEN.PK2-1C (MATa ura3-52 leu2-3,112 trp1-289 his3Δ1 MAL2-8c SUC2) (36); WCG4-11/22a (MATa pre1-1 pre2-2 ura2 leu2-3,112 his3-115), a yeast strain with an impaired proteasome (23); and WV36-405 (MATa ura3-52 trp1 adh1Δ adh2Δ adh3 adh4::TRP1), constructed by Wolfgang Vogel (Neuherberg, Germany), an Adh− strain. The mutant strains FY bdh1Δ, FY bdh2Δ, FY bdh1Δ bdh2Δ, CEN bdh1Δ, CEN bdh2Δ, CEN bdh1Δ bdh2Δ, WCG4-11/22a bdh1Δ, and WV36-405 bdh1Δ were constructed by disrupting the BDH1 and BDH2 genes from the parental strains by PCR-based gene targeting with the kanMX4 (19) and natMX4 (9) markers, respectively (see below).
Plasmids, DNA manipulations, cloning techniques, and transformation methods.
All DNA manipulations were performed under standard conditions, as described previously (30). E. coli plasmid DNA was obtained by using a commercial kit provided by Sigma. The disruption of the BDH1, BDH2, and ARA1 genes was done by the one-step gene replacement method (29) using three DNA fragments containing kanMX4 and natMX4 flanked by 40 nucleotides identical to those of the coding regions of BDH1, BDH2,and ARA1. These DNA fragments were obtained from three PCRs by using the oligonucleotides 5′ GGA ACT AAA AAA AGT TTT AAT TAA TTA TGA GAG CTT TGG CCG TAC GCT GCA GGT CGA C 3′ and 5′ CGC GAG GGG CCC CAA ATA TTA TTT TGT CAT TAC TTC ATT TTC GAT GAA TTC GAG CTC G 3′ with the plasmid pUG6 (11) as a template (to delete BDH1), the oligonucleotides 5′ GCA ATA AGA ATA ACA ATA AAT TCA TTG AAC ATA TTT CAG ACG TAC GCT GCA GGT CGA CGG 3′ and 5′ ACC GCG GGA TTA ACA CGA GAA CGT GAG TAC TCA ATC ACA AAT ACG ACT CAC TAT AGG GAG 3′ with plasmid pAG25 (9) (to delete BDH2), and the oligonucleotides 5′ TCA ATT GAT AAA AGC GTC TTG ATT TTA ATC AAC TGC TAT CAG CTT GCC TTG TCC CC 3′ and 5′ AAG AAG CGA ACT AAA TAA AGT GAA AAT AAA GTC GTT GTC GCT CGT TTT CGA CAC T 3′ with plasmid pAG25 (9) (to delete ARA1). The conditions used for the PCRs were those described for the amplification of the kanMX4 and natMX4 genes (9, 37). The linear fragments were introduced into the yeast strains by the lithium acetate method (13), and the transformants were selected on YPD agar medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose-containing 2% agar) supplemented with 200 μg/ml G418 from Gibco (Minnesota) or 100 μg/ml clonNAT (Hans-Knöll Institute für NaturstonForschung, Jena, Germany). The deletions of the BDH1, BDH2, and ARA1 genes were checked by appropriate PCRs of the genomic DNA obtained from several transformants. BDH1 and BDH2 (tagged with His6 at their C termini) were amplified by PCR by using genomic DNA from the strain FY834α as a template and cloned in the pYES2 vector from Stratagene (La Jolla, CA). The primers 5′ GCG GGA TCC ATG AGA GCT TTG GCA TAT TTC AAG AAG GG 3′ and 5′ GCG GAA TTC TTA ATG ATG ATG ATG ATG ATG CTT CAT TTC ACC GTG ATT GTT AGG 3′ were used to clone BDH1, and the primers 5′ GCG GGA TCC ATG AGA GCC TTA GCG TAT TTC GGT AAA GG 3′ and 5′ CGG GAA TTC TCA ATG ATG ATG ATG ATG ATG TGT GTG ACG CAG TTT AGC CTC G 3′ were used to clone BDH2. The amplified fragments and the pYES2 vector were digested with BamHI and EcoRI (to clone BDH1 and BDH2). Both constructs (pYES2-BDH1-6His and pYES2-BDH2-6His) were sequenced to verify that no mutations had been introduced by the Vent polymerase enzyme. Both constructs, together with constructs that did not contain the His tag, were used to transform the yeast strains by the lithium acetate method. ARA1 (tagged with His6 at its C terminus) was amplified by PCR by using genomic DNA from the strain FY834α as a template and cloned in the pYES2 vector. The primers 5′ CGG CGG ATC CAT CAT GTC TTC TTC AGT AGC CTC AAC C 3′ and 5′ GCG TCT AGA TTA ATG ATG ATG ATG ATG ATG ATA CTT TAA ATT GTC CAA GTT TGG 3′ were used to histidine tag and clone ARA1, with BamHI and XbaI digestion in pYES2.
Media and growth conditions.
Aerobic cultures were grown at 28°C on a rotary shaker (250 rpm) in YPD medium containing 20% glucose, starting with an inoculum at an optical density at 600 nm (OD600) of 0.05. Anaerobic growth was performed in 200-ml fermentors equipped with fermentation locks in YPD containing 15% glucose at 28°C with continuous stirring (230 rpm). The expression of Bdh1p (see below) was followed under aerobic conditions in YPD and in 1% yeast extract, 2% peptone, and 2% galactose or 3% ethanol. The FY834α(pYES2-BDH1-6His), FY834α(pYES2-BDH2-6His), WCG4-11/22a(pYES2-BDH1-6His), and WCG4-11/22a(pYES2-BDH2-6His) strains were grown in synthetic complete medium lacking uracil (SC-Ura) in the presence of 2% galactose.
Analytical methods.
The levels of glucose, succinate, glycerol, pyruvate, ethanol, acetate, acetaldehyde, acetoin, and 2,3-butanediol were measured in the growth media at different times after inoculation of yeast cells by previously described methods (22). To determine the levels of the different acetoin and 2,3-butanediol stereoisomers, the supernatants of the yeast cultures (together with 1-hexanol as an internal standard) were extracted by chloroform, as reported previously (17), and analyzed by gas chromatography (GC). The different acetoin and 2,3-butanediol stereoisomers were resolved on a chiral column (Supelco β-DEX 120; 30 m in length; 0.25-mm inner diameter) coupled to a Hewlett-Packard gas chromatograph equipped with a mass spectrophotometer as a detector, under the conditions reported previously (10).
MALDI-TOF MS analyses.
The Coomassie-stained protein spots were excised from the acrylamide gel, destained, and digested with 25 ng sequencing-grade trypsin (Promega) for 3 h at 37°C. All mass spectrometry (MS) samples were prepared by mixing 1 μl of sample with the same volume of a solution of α-cyano-4-hydroxycinnamic acid matrix (10 mg/ml in 30% acetonitrile, 60% water plus 0.1% trifluoroacetic acid) and were spotted onto a ground-steel plate (Bruker) and allowed to air dry at room temperature. Matrix-assisted laser desorption ionization (MALDI) mass spectra were recorded in the positive ion mode on an Ultraflex time of flight (TOF) instrument (Bruker). Ion acceleration was set to 20 kV. All mass spectra were externally calibrated for each matrix using a standard peptide mixture. For peptide mass fingerprint analysis, a Mascot search engine (Matrix Science) was used with the following parameters: mass spectrometry protein sequence database, a maximum of 2 missed trypsin cleavages, cysteine carbamidomethylation and methionine oxidation as variable modifications, and 50-ppm tolerance. Positive identifications with P values higher than 0.05 were accepted.
Ara1p purification.
The yeast extract from strain WV36-405 bdh1Δ (pYES2-ARA1-6His) was used to purify Ara1p-6His. The extract was loaded on a Ni-Sepharose column (GE Healthcare, Sant Cugat, Spain) that was extensively washed with 50 mM sodium phosphate, pH 7.4, containing 150 mM NaCl and 20 mM imidazol. Ara1p-6His was eluted in the presence of the same buffer, but with 0.5 M imidazol. The active fractions were recovered and loaded on a PD10 column (GE Healthcare, Sant Cugat, Spain) to change the buffer to 20 mM KP, pH 7. The desalted fraction was then loaded on a hydroxyapatite column and eluted with a gradient of 20 mM KPi, pH 7 (50 ml), and 600 mM KPi, pH 7 (50 ml). The active fractions were pooled and loaded on a PD10 column to change the buffer composition to 100 mM HEPES, pH 7, 50 mM NaCl.
Enzymatic activities and coenzyme-regenerating systems.
Butanediol dehydrogenase activity was determined spectrophotometrically by measuring the change of absorbance at 340 nm and 25°C, corresponding to the oxidation of NADH (ɛ340 = 6,220 M−1·cm−1). One unit of activity (U) corresponded to 1 μmol of NAD formed per minute. The standard specific activity was measured in 33 mM sodium phosphate buffer at pH 7 in the presence of 50 mM (R/S)-acetoin and 0.2 mM NADH. For the characterization of the products generated by the acetoin reductase activity from yeast extracts, cells were homogenized by agitation with glass beads in 33 mM sodium phosphate, pH 7. Assay mixtures were prepared with 50 mM racemic acetoin and 0.2 mM NADH (or NADPH), together with the following coenzyme-regenerating systems: 100 mM sodium formate (at pH 7) and 3 U of formate dehydrogenase were used for NADH regeneration, and 50 mM glucose-6-phosphate and 10 U of glucose-6-phosphate dehydrogenase were used for NADPH regeneration.
Western blots and isoelectric focusing (IEF) gels.
Bdh1-6His and Bdh2-6His were detected by Western blotting by using a mouse monoclonal anti-His6 antibody (Roche). Briefly, FY834, FY834 bdh1Δ, and WCG4-11/22a transformed with plasmids pYES2-BDH1-6His and pYES2-BDH2-6His were grown in SC-Ura and galactose. Cells at an OD600 of 2.5 were collected at the early logarithmic (OD600 = 0.5) and stationary (OD600 = approximately 5) phases and treated with 0.2 M NaOH as described previously (16). The extracts were loaded in a 10% SDS-PAGE gel that was blotted onto a polyvinylidene difluoride (PVDF) membrane (Millipore) and treated with the primary anti-His6 antibody. To visualize the relevant bands, the blot was treated with a secondary goat anti-mouse antibody (Bio-Rad) conjugated to horseradish peroxidase (HRP), and the bands were detected by chemiluminescence with luminol and peroxide with the aid of a Bio-Rad Chemidoc XRS.
To visualize the butanediol dehydrogenase activities of crude extracts, we used precast isoelectric focusing (pH 3 to 9) gels from Bio-Rad (Criterion). After the samples were loaded, the voltage was set at 100 V for 1 h, followed by 250 V for 1 h and 500 V for half an hour. BDH activities were visualized on the gel by activity staining with 0.5 M 2,3-butanediol (a mixture of isomers), 2 mM NAD, 0.08 mg/ml of phenazine methosulfate (PMS), and 0.8 mg/ml of nitroblue tetrazolium (NBT) and quantified by densitometry (25).
Microarray analysis of global gene expression.
Yeast cells from wild-type (FY834) and mutant (FY bdh1Δ, FY bdh2Δ, and FY bdh1Δ bdh2Δ) strains were grown for 40 h under respirofermentative conditions at 20% glucose, and total RNA was extracted by using the RiboPure Yeast kit from Ambion, following the manufacturer's instructions.
Five hundred nanograms of total RNA was reverse transcribed, amplified, labeled by in vitro transcription with Cy3 or Cy5 using a Low Input Linear Amplification Kit (Agilent 5184-3523), and hybridized following the manufacturer's instructions (Agilent Gene expression two-color 60-mer oligonucleotide microarray-processing protocol, version 5.7, March 2008) on yeast whole-genome microarrays (G2519-AMADID-015072). The oligonucleotide probes used to measure the expression of the BDH1 and BDH2 genes were 5′ CAC AAG GAA TCC AAC GTT AAG ATT CTA TTG ACG CCT AAC AAT CAC GGT GAA ATG AAG TAA 3′ and 5′ CGG AAA GAT CAA GAA AGA CTA CGA GAA TCA ATA AAC GAG GCT AAA CTG CGT CAC ACA TGA 3′.
For comparison of single-deletion mutants to the wild-type strain, four-biological-replicate experiments were performed, whereas two biological replicates were used for the comparison of the double-deletion mutant to the wild type. For all biological-replicate pairs, samples labeled with two different dyes were cohybridized on two separate microarrays with dye swapping to correct for dye bias effects. Thus, eight microarray hybridizations were processed for each of the two single-mutant-versus-wild-type comparisons and four for the double-mutant-versus-wild-type comparison, totaling 20 array data sets.
Fluorescence images were obtained using an Agilent G2565BA scanner at 100% photomultiplier tube intensity and 100% laser power settings and quantified with GenePix 6.0 software (Axon, Molecular Devices, Sunnyvale, CA) using the irregular-feature-finding option. The extracted raw data were processed using the Limma package developed within the Bioconductor project in the R statistical programming environment (8). After normexp background subtraction was applied (24), the two channels on each array were balanced by lowess normalization using 0.3 as the span parameter with reduced weights in control and poor-quality spots, followed by a scaling step to make all arrays comparable, using the Limma package in the R environment (32). For statistical assessment of differential gene expression, the Statistical Analysis of Microarrays (SAM) algorithm was used by applying the samr library in R (34). All software was run in a Web interface implementation of the R-based software (J. Lozano, L. Opatowski, G. Canton, E. Gonzalez, B. Minana, M. Vilardell, X. Pastor, M. Hummel, and L. Sumoy, unpublished data).
RESULTS
Expression of Bdh1p during yeast growth.
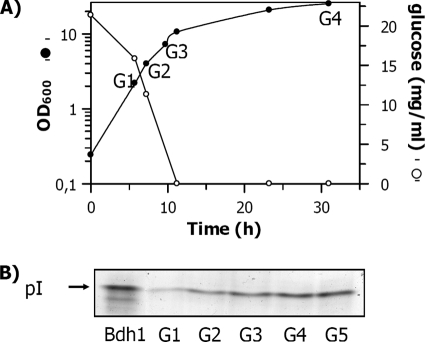
The FY834 strain was grown in 2% glucose, and the Bdh1p activity was determined by a zymogram (Fig. 1). The intensities of the bands revealed a 3-fold increase of the Bdh1p activity in the stationary phase, when glucose was not detectable in the medium, compared to the activity in the exponential phase. The Bdh1p specific activity of the crude extract at an OD of 2 (G1 in Fig. 1) was 0.05 U/mg, while the samples at an OD of 20 (G4) and at an OD of 34 (G5) yielded a specific activity of 0.17 U/mg. The densitometries in the zymogram analysis of samples grown in galactose and ethanol media, in both exponential and stationary phases, were similar to the intensities of the Bdh1p activity bands from the sample grown in 2% glucose in the stationary phase, when no glucose was detectable in the medium (data not shown).
FIG. 1.
Yeast growth and zymogram showing the Bdh1p activity of the FY834 strain extracts at different times under respirofermentative conditions. (A) Growth curve of the FY834 yeast strain (in rich medium at 2% glucose) under respirofermentative conditions superimposed on the glucose concentrations of the supernatants. (B) Zymogram of the yeast extracts prepared at the times indicated in the growth curve. Sample G5 was taken at an OD of 34. Bdh1p bands on the gel were visualized by activity staining with (2R,3R)-2,3-butanediol and NAD.
Metabolite levels under anaerobic and respirofermentative conditions.
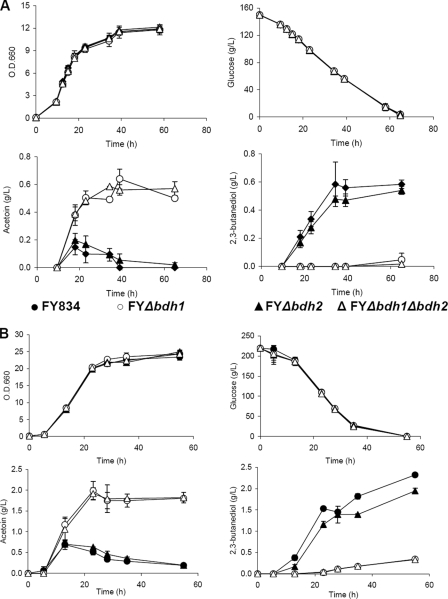
Metabolite levels were analyzed in strains FY834, FY bdh1Δ, FY bdh2Δ, FY bdh1Δ bdh2Δ, CEN.PK2-1C, CEN bdh1Δ, CEN bdh2Δ, and CEN bdh1Δ bdh2Δ. In addition to the OD600 and glucose concentration (Fig. 2), the levels of succinate, pyruvate, ethanol, acetate, acetaldehyde, and glycerol were also similar between the strains, under both anaerobic and respirofermentative conditions (results not shown). However, clear differences between strains were observed for acetoin and 2,3-butanediol (Fig. 2). An inverse relationship between acetoin and 2,3-butanediol accumulation for all the strains and growth conditions was clearly shown. Thus, at the end of both respirofermentative and anaerobic growth curves, an accumulation of acetoin by the FY bdh1Δ and FY bdh1Δ bdh2Δ strains paralleled a small amount of 2,3-butanediol. Moreover, the yeast strains FY834 and FY bdh2Δ accumulated 2,3-diols, while only traces of acetoin could be measured in the supernatants of their growth media. While deletion of BDH1 impaired the conversion of acetoin to 2,3-butanediol, deletion of BDH2 seemed to alter the 2,3-butanediol levels only marginally (Fig. 2B). The levels of acetoin and 2,3-butanediol (per mmol glucose) were approximately 2-fold higher under respirofermentative than under anaerobic conditions.
FIG. 2.
Concentrations of glucose, acetoin, and 2,3-butanediol in the growth media under anaerobic and respirofermentative conditions for the FY834 and derived bdh1Δ and bdh2Δ yeast strains. The FY834 (wild type), FY bdh1Δ, FY bdh2Δ, and FY bdh1Δ bdh2Δ strains were incubated in rich media containing 15% glucose at 28°C in 200-ml fermentors equipped with fermentation locks with continuous stirring (A) and in 20% glucose at 28°C in an orbital shaker at 250 rpm (B). The growth and compositions of the media were determined at different times. The error bars correspond to the standard deviations of three experiments.
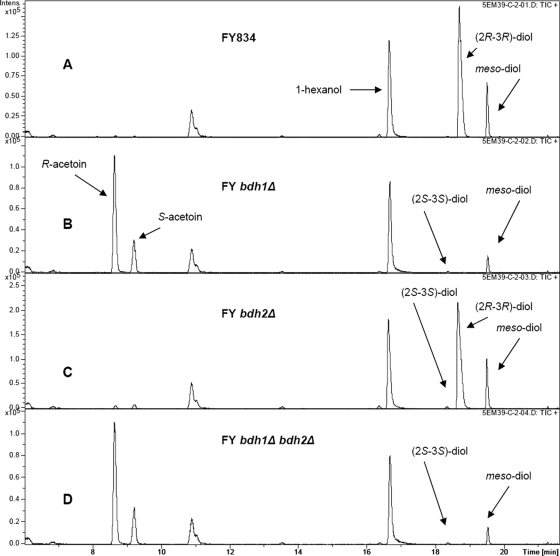
A chiral column in a gas chromatograph, coupled with a mass spectrometer, was used to identify the acetoin and 2,3-butanediol stereoisomers accumulated in the growth media of different strains. Figure 3 shows the levels of the stereoisomers at the end of the growth curve (71 h after inoculation) for the FY strains. Two different patterns could be discerned from the four yeast strains. The first pattern, represented by the FY834 and FY bdh2Δ strains (Fig. 3A and C), showed accumulation of (2R,3R)-2,3-butanediol (2/3 of the total 2,3-butanediol) and meso-2,3-butanediol (1/3), with traces of (2S,3S)-2,3-butanediol and acetoin. The second pattern, represented by the FY bdh1Δ and FY bdh1Δ bdh2Δ strains (Fig. 3B and D), shows accumulation of R-acetoin (2/3 of the total acetoin level), S-acetoin (1/3), and a minor amount of meso-2,3-butanediol, with traces of (2S,3S)-2,3-butanediol. The stereoisomer composition for the CEN.PK2-1C strains was similar in all cases to that of the corresponding FY strains (results not shown). Thus, the deletion of BDH1 resulted in the accumulation of acetoin and the disappearance of (2R,3R)-2,3-butanediol, while a small amount of meso-2,3-butanediol remained.
FIG. 3.
Gas chromatography analysis of acetoin and 2,3-butanediol stereoisomers in the growth media under respirofermentative conditions. Rich media containing 20% glucose were inoculated with the FY834 (wild type) (A), FY bdh1Δ (B), FY bdh2Δ (C), and FY bdh1Δ bdh2Δ (D) strains. After 71 h of growth in an orbital shaker at 250 rpm and 28°C, the growth media were centrifuged and the supernatants were analyzed by gas chromatography.
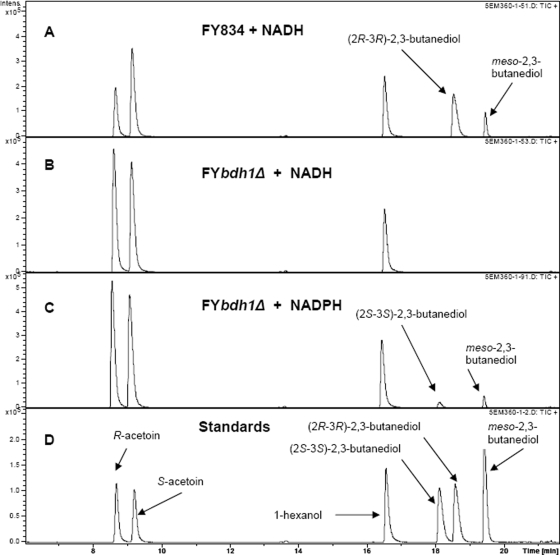
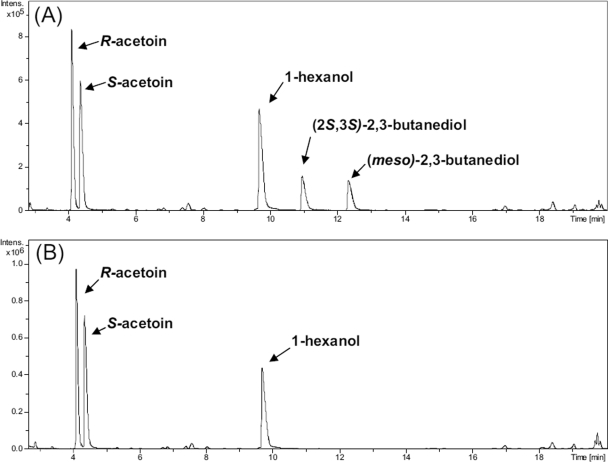
The production of the meso-isomer in the absence of Bdh1p demonstrated the existence of a different enzyme activity that was able to synthesize the compound. To further characterize this activity, we studied the NADH- and NADPH-dependent acetoin reductase activities of the FY834 and FY bdh1Δ strains. Thus, crude extracts of those strains (after 40 h of growth under respirofermentative conditions) were assayed for their acetoin reductase activities (Fig. 4). The FY bdh1Δ strain did not produce any 2,3-butanediol when assayed with NADH (Fig. 4B) but produced meso-2,3-butanediol and (2S,3S)-2,3-butanediol when assayed with NADPH (Fig. 4C). These results indicated the presence of an NADPH-dependent acetoin reductase activity with S specificity for acetoin. This putative activity would produce meso-2,3-butanediol from R-acetoin and (2S,3S)-2,3-butanediol from S-acetoin.
FIG. 4.
Analysis by gas chromatography of NADH- and NADPH-dependent acetoin reductase activities of the FY834 (wild type) and FY bdh1Δ strain extracts. Yeast extracts were prepared from the corresponding cells 40 h after they were inoculated in rich medium containing 20% glucose under respirofermentative conditions. The extracts were incubated with 50 mM (R/S)-acetoin (racemic mixture) and 0.2 mM NADH (or NADPH), together with the corresponding coenzyme-regenerating system (see Materials and Methods). After 20 h of incubation, the reaction mixtures were analyzed by gas chromatography. (A) Analysis of the reaction mixture with the FY834 strain (with an NADH-dependent acetoin reductase specific activity of 0.20 U/mg), (R/S)-acetoin, NADH, and the NADH-regenerating system. (B) Analysis of the reaction mixture with the FY bdh1Δ strain, (R/S)-acetoin, NADH, and the NADH-regenerating system. (C) Analysis of the reaction mixture with the FY bdh1Δ strain (with an NADPH-dependent acetoin reductase specific activity of 0.030 U/mg), (R/S)-acetoin, NADPH, and the NADPH-regenerating system. (D) Analysis of the chloroform extract of a mixture composed of 10 mM R-acetoin, 10 mM S-acetoin, 20 mM (2S,3S)-2,3-butanediol, 20 mM (2R,3R)-2,3-butanediol, and 20 mM meso-2,3-butanediol, together with 1-hexanol as an internal standard.
Partial purification and characterization of Ara1p as the NADPH-dependent oxidoreductase producing meso-2,3-butanediol.
To characterize the enzyme(s) responsible for the production of meso-2,3-butanediol in the bdh1Δ strains, we used an extract of WV36-405 bdh1Δ yeast cells and followed their NADPH reductase activities through different purification steps (DEAE-Sepharose and red Sepharose chromatography). The active fractions were pooled, concentrated, and loaded on an SDS gel. The more abundant bands were excised from the gel, digested with trypsin, and analyzed by MALDI-TOF, yielding Zwf1p, Oye2p, Erg19p, Ara1p, and Trr1p with a high probability score (results not shown). All of them except Erg19p are known to require NADP(H) for their activities, an expected result because of the use of red Sepharose [a resin used to purify NADP(H)-binding proteins] in the enrichment process. Since it is known that d-arabinose dehydrogenase (Ara1p) displays NADPH-dependent diacetyl reductase activity (14, 35), we decided to purify the enzyme and determine its acetoin reductase activity. We His tagged and purified Ara1p-6His from WV36-405 bdh1Δ(pYES2-ARA1-6His) through a Ni-nitrilotriacetic acid (NTA)-Sepharose column and a hydroxyapatite column. We characterized the products of the reaction between (R/S)-acetoin and Ara1p-6his in the presence of a NADPH-regenerating system by the use of a chiral column in a GC-MS system. Figure 5 shows that the reaction yielded meso-2,3-butanediol and (2S,3S)-2,3-butanediol, while no 2,3-butanediol was obtained in the control reaction mixture containing all the ingredients except Ara1p-6His.
FIG. 5.
The NADPH-dependent acetoin reductase reaction catalyzed by purified Ara1p-6His analyzed by gas chromatography. (A) Profile of reaction products obtained from Ara1p-6his. Ara1p-6His (0.07 U measured as NADPH-dependent acetoin reductase activity at 50 mM acetoin and 0.2 mM NADPH) was incubated for 24 h with 50 mM (R/S)-acetoin, 1 mM NADPH, 50 mM glucose-6-phosphate, and 10 U of glucose-6-phosphate dehydrogenase. The reaction mixture, together with 1-hexanol as an internal standard, was extracted with chloroform as described previously (13). (B) Profile of reaction products obtained in the control reaction. The reaction mixture contained the same reagents as in panel A, except for Ara1p-6His.
2,3-Butanediol levels in the FY bdh1Δ and FY bdh1Δ ara1Δ strains.
To ascertain the role of ARA1 in the in vivo production of 2,3-butanediol, cultures of the strains FY834, FY bdh1Δ, and FY bdh1Δ ara1Δ were grown in triplicate under respirofermentative conditions on 20% glucose. After 70 h of growth, aliquots of the cultures were taken, extracted with chloroform (see Materials and Methods), and analyzed with a chiral column coupled to a gas chromatograph. The levels of (2S,3S)-2,3-butanediol and meso-2,3-butanediol were 0.053 ± 0.005 g/liter and 0.18 ± 0.03 g/liter, respectively, for the FY bdh1Δ strain and 0.038 ± 0.001 g/liter and 0.120 ± 0.006 g/liter for the FY bdh1Δ ara1Δ strain. The concentrations of (2R,3R)-2,3-butanediol were almost undetectable for both strains. The levels of R-acetoin and S-acetoin were 1.305 ± 0.045 g/liter and 0.352 ± 0.013 g/liter for the FY bdh1Δ strain and 1.230 ± 0.045 g/liter and 0.345 ± 0.019 g/liter for the FY bdh1Δ ara1Δ strain. By comparing the mean values for the metabolites by Student's t test (P < 0.05), significant differences were found between the concentrations of (2S,3S)-2,3-butanediol (t = 5.09 and P = 0.036) and meso-2,3-butanediol (t = 3.39 and P = 0.027) in the FY bdh1Δ and FY bdh1Δ ara1Δ strains, but the differences were not significant for the acetoin levels.
Expression and enzymatic activities of Bdh2p.
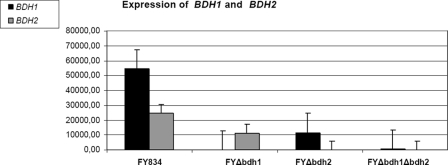
Since Fig. 2 suggests a mild effect on the 2,3-butanediol levels in the FY bdh2Δ strain compared to the FY strain, we decided to study the role of BDH2 more directly in this respect. First, we measured the expression levels of BDH1 and BDH2 under respirofermentative conditions at 40 h after the inoculation (a time within the stationary phase, where clear differences in acetoin and 2,3-butanediol levels were found between strains). As Fig. 6 shows, both genes are reciprocally regulated: BDH2 is expressed in the FY bdh1Δ strain (at approximately a 50% of its level in the FY strain). The data also indicate clear repression of BDH1 in the FY bdh2Δ strain (the level of BDH1 expression was approximately 20% of that in the FY strain). However, the levels of acetoin and 2,3-butanediol were much more affected in the FY bdh1Δ strain than in the FY bdh2Δ strain. We decided to determine the putative butanediol dehydrogenase activity of Bdh2p in crude extracts of the FY bdh1Δ(pYES2-BDH2) and WV36-405 bdh1Δ(pYES2-BDH2) strains. The NADH-dependent acetoin reductase activity of the WV36-405 bdh1Δ(pYES2-BDH1) extract was 14.4 U/mg, while the corresponding cellular extracts of the WV36-405 bdh1Δ(pYES2-BDH2) and WV36-405 bdh1Δ(pYES2) strains were devoid of NADH-dependent acetoin reductase activity.
FIG. 6.
Expression levels of BDH1 and BDH2 in the FY834 (wild type) and derived bdh1Δ and bdh2Δ yeast strains. The FY834, FY bdh1Δ, FY bdh2Δ, and FY bdh1Δ bdh2Δ strains were incubated in rich media containing 20% glucose at 28°C in an orbital shaker at 250 rpm for 40 h. Total RNA was extracted, and the expression levels of BDH1 and BDH2 were monitored from the intensities of the respective signals, as indicated in Materials and Methods. The means of the expression levels are given in arbitrary units, together with their standard deviations.
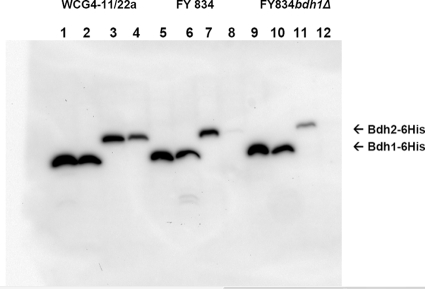
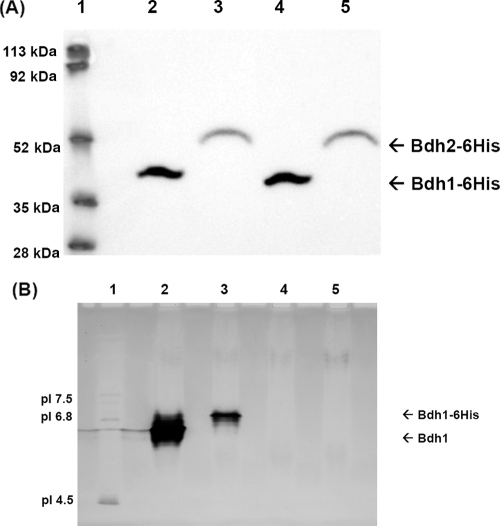
To explore whether Bdh2p was really expressed, we tagged Bdh2p with 6 histidines (in two constructs: N terminus, 6His-Bdh2, and C terminus, Bdh2-6His) and studied the presence of the tagged proteins by means of an anti-His antibody in different yeast genetic backgrounds. Positive results were obtained when Bdh2p was tagged with 6 histidines in its C terminus (Bdh1p was also tagged at its C terminus as a control). Figure 7 shows the expression patterns of Bdh1-6His and Bdh2-6His in the yeast strains FY834 (lanes 5 to 8) and FY834 bdh1Δ (lanes 9 to 12) and in a proteasome-impaired yeast strain (WCG4-11/22a [lanes 1 to 4]). The expression of both proteins was followed in the early logarithmic (lanes 1, 3, 5, 7, 9, and 11) and stationary (lanes 2, 4, 6, 8, 10, and 12) phases. Bdh2-6His was more stable in the proteasome-impaired strain (compare lanes 3 and 4 with 7 and 8 and with 11 and 12) and in the early logarithmic phase (compare lanes 3 and 4). We could not detect the expression of 6His-Bdh2p (Bdh2p tagged at its N terminus) in any of the strains checked (results not shown). Therefore, we decided to overexpress Bdh2-6His (and Bdh1-6His as control) in a WCG4-11/22a bdh1Δ yeast strain. The yeast strains were grown in galactose to the early logarithmic phase, and after the cells were broken, the crude extracts were subjected to Western blotting, BDH activity determination, and IEF analysis. Although both Bdh2p-6His and Bdh1p-6His were detected by Western blotting (Fig. 8A, lanes 4 and 5; the higher Mr of Bdh2p-6His was due to its 34-amino-acid extension at the C terminus in respect to Bdh1p-6His), only Bdh1-6His showed butanediol dehydrogenase activity in the IEF gel and in the spectrophotometer. The IEF gel visualized by BDH activity showed that Bdh1p-6His had a higher pI than Bdh1p (the experimental value was close to the predicted pI value of 6.2), probably due to the His6 tag (Fig. 8B, lanes 1 and 2). Lanes 1 and 2 (Fig. 8B) also show a multiplicity of bands, probably due to different oxidation states of the proteins. Bdh2 and Bdh2-6His did not show activity on the IEF gels (Fig. 8B, lanes 4 and 5). The specific activity of the extract overexpressing Bdh1p was 25 U/mg, while the one overexpressing Bdh1-6His yielded 1.6 U/mg. No acetoin reductase activity (and no 2,3-butanediol dehydrogenase) was found in the extracts overexpressing neither Bdh2 nor Bdh2-6His. The lack of activity of Bdh2p toward 2,3-butanediol (from the IEF and spectrophotometer determinations) and acetoin (from the spectrophotometer) was correlated with the acetoin content in the supernatants of the FY, FY bdh1Δ, FY bdh2Δ, and FY bdh1Δ bdh2Δ strains, where no difference could be attributed to the deletion of BDH2 under the experimental conditions studied. Since the deletion of BDH2 has been found to affect the expression of BDH1, the small effect on 2,3-butanediol concentrations observed in the FY bdh2Δ strain (Fig. 2B) could be an indirect effect due to the lower level of BDH1 expression, although the repression of BDH1 does not necessarily result in less Bdh1p.
FIG. 7.
Western blot analysis of the expression of Bdh1p-6His and Bdh2p-6His in different yeast strains and growth phases. Crude extracts from yeast cells at an OD600 of 2.5 (strains FY834, FY834 bdh1Δ, and WCG4-11/22a transformed with plasmids pYES2-BDH1-6His and pYES2-BDH2-6His) were grown in SC-Ura and galactose. The cells were collected in the early logarithmic (OD600 = 0.5; lanes 1, 3, 5, 7, 9, and 11) and stationary (OD600 = approximately 5; lanes 2, 4, 6, 8, 10, and 12) phases and treated with 0.2 M NaOH as described previously (34). Lanes 1, 2, 5, 6, 9, and 10 show the expression levels of Bdh1p-6His in the cell extracts of strains WCG4-11/22a (lanes 1 and 2), FY834 (lanes 5 and 6), and FY834 bdh1Δ (lanes 9 and 10). Lanes 3, 4, 7, 8, 11, and 12 show the expression of Bdh2p-6His in the cell extracts of strains WCG4-11/22a (lanes 3 and 4), FY834 (lanes 7 and 8), and FY834 bdh1Δ (lanes 11 and 12).
FIG. 8.
Western blot and isoelectric focusing analyses of the expression of Bdh1p-6His and Bdh2p-6His. (A) Western blot analysis. Yeast cells at an OD of ∼10 from strains WCG4-11/22a bdh1Δ(pYES2-BDH1-6His) (lane 2) and WCG4-11/22a bdh1Δ(pYES2-BDH2-6His) (lane 3) were treated as described previously (34) for Western blot analysis with an anti-His antibody. Lane 4, 54 μg of protein from extract WCG4-11/22a bdh1Δ(]pYES2-BDH1-6His); lane 5, 57 μg of protein from extract WCG4-11/22a bdh1Δ(pYES2-BDH2-6His); lane 1, molecular mass standards. (B) Isoelectric focusing analysis. Shown is an isoelectric focusing gel (pH 3 to 9) of Bdh1p, Bdh1p-6His, Bdh2p, and Bdh2p-6His visualized by butanediol dehydrogenase activity. Lane 1, pI standards; lane 2, 75 μg of protein from extract WCG4-11/22a bdh1Δ(pYES2-BDH1) containing 1.9 U of BDH activity; lane 3, 81 μg of protein from extract WCG4-11/22a bdh1Δ(pYES2-BDH1-6His) containing 0.15 U of BDH activity; lane 4, 81 μg of protein from extract WCG4-11/22a bdh1Δ(pYES2-BDH2) with no BDH activity; lane 5, 86 μg of protein from extract WCG4-11/22a bdh1Δ(pYES2-BDH2-6His) with no BDH activity.
DISCUSSION
We showed above that Bdh1p is induced upon glucose depletion. This augmentation of Bdh1p activity is supported by microarray data reporting upregulation of BDH1 during stationary phase and upon the diauxic shift (5, 7). On the other hand, we observed that the production of acetoin and 2,3-butanediol catalyzed by Bdh1p increased under respirofermentative conditions compared to anaerobic conditions. A similar effect of the oxygen supply during yeast fermentation on the formation of 2,3-butanediol through acetoin was previously observed (15). This change was attributed to reduction of the NADH availability by oxygen, decreasing the rate of glycerol and ethanol formation and resulting in the accumulation of acetaldehyde and derivative compounds. As a whole, these data emphasize the role of Bdh1p in the metabolic remodeling associated with the transition from fermentation to respiration.
We have previously shown the enantioselectivity of Bdh1p toward the reduction of the carbonyl groups from acetoin, leading to an R-alcohol (10). Thus, (2R,3R)-2,3-butanediol and meso-2,3-butanediol are formed by Bdh1p from R-acetoin and S-acetoin, respectively. We now demonstrate that Bdh1p accounts for most of the (2R,3R)-2,3-butanediol produced under fermentative and respirofermentative conditions in S. cerevisiae growing in rich media containing 15 to 20% glucose. Thus, almost no (2R,3R)-2,3-butanediol was detected in the culture medium of the FY bdh1Δ or CEN bdh1Δ strain. The fact that the levels of meso-2,3-butanediol did not decrease to the same degree can be attributed to an independent oxidoreductase activity that was shown to be NADPH dependent (Fig. 4). This finding agrees with the fact that certain strains have been found to produce meso-2,3-diol without producing (2R,3R)-2,3-butanediol (20), suggesting that a different enzymatic activity produces meso-2,3-diol without the need to accumulate the (2R,3R) isomer. The MALDI-TOF analysis of a partially purified active fraction suggested that Ara1p could be involved. Purified His-tagged Ara1p was able to produce meso-2,3-butanediol and (2S,3S)-2,3-butanediol from (R/S)-acetoin (Fig. 5). Ara1p would be selective toward the acetoin carbonyl group, leading to an S-alcohol. This is the first report of acetoin reductase activity by Ara1p with characterization of the enantiomeric form of the products. Thus, Ara1p could contribute to the production of meso-2,3-butanediol and (2S,3S)-2,3-butanediol from acetoin by the bdh1Δ strains. We tested this hypothesis directly by comparing the in vivo levels of acetoin and 2,3-butanediol in a FY bdh1Δ strain and a FY bdh1Δ ara1Δ strain. While the differences between the acetoin concentrations were not significant, the levels of both isomers, (2S,3S)-2,3-butanediol and meso-2,3-butanediol, were significantly lower in the FY bdh1Δ ara1Δ strain than in the FY bdh1Δ strain. However, given the fact that the differences were small, the data suggest a minor role of Ara1p in the production of 2,3-butanediol under these conditions. Another relevant conclusion from the work is that the double-mutant strain still produces (2S,3S)-2,3-butanediol and meso-2,3-butanediol. Since five more aldoketoreductases have been described in yeast (3), it is possible that some of them could play additional roles in the production of 2,3-butanediol. However, these would be minor pathways, since Bdh1p accounts for most of the 2,3-butanediol production (Fig. 3). Interestingly, the [R-acetoin]/[S-acetoin] ratio of 3.1 found in the present work for the bdh1Δ strains closely corresponds to the ratio of 3.3 resulting from the pyruvate decarboxylase activity from pyruvate (4). Moreover, the [(2R,3R)-2,3-butanediol]/[meso-2,3-butanediol] ratio of 3.1 found here for the wild-type strain is close to the ratio of 2.1 found in an early report of yeast grown anaerobically (18).
The levels of extracellular metabolites (especially acetoin) measured in the bdh2Δ strains are close to the levels found in their wild-type counterparts. Moreover, the levels of acetoin isomers and 2,3-butanediol are similar in the bdh1Δ and bdh1Δ bdh2Δ strains (Fig. 3). Although these facts could imply that Bdh2p is not involved in the production of 2,3-butanediol (and is devoid of acetoin reductase activity), we decided to address this issue more directly. First, we showed that the mRNA of BDH2 was expressed in the FY strain and repressed in the FY834 bdh1Δ strain, although no butanediol dehydrogenase activity was present in a FY834 bdh1Δ(pYES2-BDH2) strain. We also could demonstrate the presence of the protein Bdh2-6His in a Western blot that did not show BDH activity in an IEF gel or in the spectrophotometer. Bdh2-6His seems to be a labile protein that could be expressed in a proteasome-deficient yeast strain, although at a lower level than Bdh1-6His expressed under similar experimental conditions.
The final conclusion about Bdh2p that can be deduced from the present work is that it is devoid of BDH activity. In effect, Fig. 8 shows expression of Bdh2p-6His, but no BDH activity (as opposed to Bdh1p-6His). Although it could be argued that the His tail impairs its putative BDH activity, we do not favor this explanation for two reasons: Bdh1p-6His displays BDH activity, and the IEF gel loaded with an extract of the strain WCG4-11/22a bdh1Δ(pYES2-BDH2) does not show BDH activity (Fig. 8B, lane 4). Since an extract of the strain WCG4-11/22a bdh1Δ(pYES2-BDH1) grown under the same conditions showed high BDH activity (lane 2), Bdh2p, if active, should be visible in lane 4 (unless, of course, it was extremely labile). Thus, although YAL061W affects the expression of BDH1 (under our experimental conditions, at least), its designation as BDH2 (currently in the databases) should be reconsidered, since the results from this work do not corroborate its presumed BDH activity.
An interesting application of the present results is that the control of BDH1 expression could affect the balance between acetoin and 2,3-butanediol and could have an impact on the flavor and aroma of wines produced by industrial yeast strains. In fact, some strains reported as non-2,3-butanediol producers (26) could have a nonfunctional or a repressed BDH1 gene. Alternatively, BDH1 could be lost in those strains, given its subtelomeric position in chromosome I. We have recently shown the utility of overexpressing BDH1 in a wine yeast strain to reduce its acetoin content (6). Another application of Bdh1p overproduction is in the production of enantiopure 2,3-butanediol isomers.
Acknowledgments
This work was supported by grants from the Ministry of Education and Science (BFU-2006-10401, BIO2007-64659, and BFU2008-02945) and Generalitat de Catalunya (2005 SGR 00112), Spain.
We thank Danièle Urban-Grimal (Institut Jacques Monod, Paris, France) for sending us the WCG4-11/22a strain. The MALDI-TOF MS analyses were carried out by Sílvia Bronsoms in the proteomics and bioinformatics facility (SePBio) of UAB, a member of the Proteored network. The technical assistance of Brigitte Cambon (UMR 1083 Sciences pour l'Oenologie, INRA, Montpellier, France) is acknowledged.
Footnotes
Published ahead of print on 4 December 2009.
REFERENCES
- 1.Bartowsky, E. J., and P. A. Henschke. 2004. The buttery attribute of wine-diacetyl-desirability, spoilage and beyond. Int. J. Food Microbiol. 96:235-252. [DOI] [PubMed] [Google Scholar]
- 2.Bornemann, S., D. H. G. Crout, H. Dalton, D. Hutchinson, G. Dean, N. Thomson, and M. M. Turner. 1993. Stereochemistry of the formation of lactaldehyde and acetoin produced by the pyruvate decarboxylases of yeast (Saccharomyces sp.) and Zymomonas mobilis: different Boltzmann distributions between bound forms of the electrophile, acetaldehyde, in the two enzymatic reactions. J. Chem. Soc. Perkin 1:309-311. [Google Scholar]
- 3.Chang, Q., T. A. Griest, T. M. Harter, and J. M. Petrash. 2007. Functional studies of aldo-keto reductases in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, G. C., and F. Jordan. 1984. Brewers' yeast pyruvate decarboxylase produces acetoin from acetaldehyde: a novel tool to study the mechanism of steps subsequent to carbon dioxide loss. Biochemistry 23:3576-3582. [DOI] [PubMed] [Google Scholar]
- 5.deRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 6.Ehsani, M., M. R. Fernández, J. A. Biosca, A. Julien, and S. Dequin. 2009. Engineering of 2,3-butanediol dehydrogenase to reduce acetoin formation by glycerol-overproducing, low-alcohol Saccharomyces cerevisiae. Appl. Environ. Microbiol. 75:3196-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasch, A., P. Spellman, C. Kao, O. Carmel-Harel, M. Eisen, G. Storz, D. Botstein, and P. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 10.González, E., M. R. Fernández, C. Larroy, L. Solá, M. A. Pericás, X. Parés, and J. A. Biosca. 2000. Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene product. Disruption and induction of the gene. J. Biol. Chem. 275:35876-35885. [DOI] [PubMed] [Google Scholar]
- 11.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidlas, J., and R. Tressl. 1990. Purification and properties of two oxidoreductases catalyzing the enantioselective reduction of diacetyl and other diketones from baker's yeast. Eur. J. Biochem. 188:165-174. [DOI] [PubMed] [Google Scholar]
- 13.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, S. T., W. K. Huh, B. H. Lee, and S. O. Kang. 1998. d-Arabinose dehydrogenase and its gene from Saccharomyces cerevisiae. Biochim. Biophys. Acta 1429:29-39. [DOI] [PubMed] [Google Scholar]
- 15.Kuriyama, H., and H. Kobayashi. 1993. Effects of oxygen supply on yeast growth and metabolism in continuous fermentation. J. Ferment. Bioeng. 75:364-367. [Google Scholar]
- 16.Kushnirov, V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857-860. [DOI] [PubMed] [Google Scholar]
- 17.Michnick, S., J. L. Roustan, F. Remize, P. Barre, and S. Dequin. 1997. Modulation of glycerol and ethanol yields during alcoholic fermentation in Saccharomyces cerevisiae strains overexpressed or disrupted for GPD1 encoding glycerol 3-phosphate dehydrogenase. Yeast 13:783-793. [DOI] [PubMed] [Google Scholar]
- 18.Neish, A. C. 1950. Stereoisomers of 2,3-butanediol produced by yeast. Can. J. Res. 28B:660-661. [Google Scholar]
- 19.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 20.Patel, S., and T. Shibamoto. 2002. Effect of different strains of Saccharomyces cerevisiae on production of volatiles in Napa Gamay wine and Petite Sirah wine. J. Agric. Food Chem. 50:5649-5653. [DOI] [PubMed] [Google Scholar]
- 21.Peinado, R. A., J. Moreno, M. Medina, and J. C. Mauricio. 2004. Changes in volatile compounds and aromatic series in sherry wine with high gluconic acid levels subjected to aging by submerged flor yeast cultures. Biotechnol. Lett. 26:757-762. [DOI] [PubMed] [Google Scholar]
- 22.Remize, F., J. L. Roustan, J. M. Sablayrolles, P. Barre, and S. Dequin. 1999. Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in by-product formation and to a stimulation of fermentation rate in stationary phase. Appl. Environ. Microbiol. 65:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter-Ruoff, B., D. H. Wolf, and M. Hochstrasser. 1994. Degradation of the yeast MAT alpha 2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 354:50-52. [DOI] [PubMed] [Google Scholar]
- 24.Ritchie, M. E., J. Silver, A. Oshlack, M. Holmes, D. Diyagama, A. Holloway, and G. K. Smyth. 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23:2700-2707. [DOI] [PubMed] [Google Scholar]
- 25.Robertson, E. F., H. K. Dannelly, P. J. Malloy, and H. C. Reeves. 1987. Rapid isoelectric focusing in a vertical polyacrylamide minigel system. Anal. Biochem. 167:290-294. [DOI] [PubMed] [Google Scholar]
- 26.Romano, P., L. Granchi, M. Caruso, G. Borra, G. Palla, C. Fiore, D. Ganucci, A. Caligiani, and V. Brandolini. 2003. The species-specific ratios of 2,3-butanediol and acetoin isomers as a tool to evaluate wine yeast performance. Int. J. Food Microbiol. 86:163-168. [DOI] [PubMed] [Google Scholar]
- 27.Romano, P., and G. Suzzi. 1996. Origin and production of acetoin during wine yeast fermentation. Appl. Environ. Microbiol. 62:309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romano, P., G. Suzzi, V. Brandolini, E. Menziani, and P. Domizio. 1996. Determination of 2,3-butanediol in high and low acetoin producers of Saccharomyces cerevisiae wine yeasts by automated multiple development (AMD). Lett. Appl. Microbiol. 22:299-302. [DOI] [PubMed] [Google Scholar]
- 29.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Shinohara, T., Y. Shimazu, and M. Watanabe. 1979. Dosage de l'acétoine et du lactate d'éthyle dans les vins par chromatographie en phase gazeuse et étude de leur formation dans les vins. Agric. Biol. Chem. 43:2569-2577. [Google Scholar]
- 32.Smyth, G. K., and T. P. Speed. 2003. Normalization of cDNA microarray data. Methods 31:265-273. [DOI] [PubMed] [Google Scholar]
- 33.Syu, M. J. 2001. Biological production of 2,3-butanediol. Appl. Microbiol. Biotechnol. 55:10-28. [DOI] [PubMed] [Google Scholar]
- 34.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Bergen, B., R. Strasser, N. Cyr, J. D. Sheppard, and A. Jardim. 2006. Alpha,beta-dicarbonyl reduction by Saccharomyces D-arabinose dehydrogenase. Biochim. Biophys. Acta 1760:1636-1645. [DOI] [PubMed] [Google Scholar]
- 36.van Dijken, J. P., J. Bauer, L. Brambilla, P. Duboc, J. M. Francois, C. Gancedo, M. L. Giuseppin, J. J. Heijnen, M. Hoare, H. C. Lange, E. A. Madden, P. Niederberger, J. Nielsen, J. L. Parrou, T. Petit, D. Porro, M. Reuss, N. van Riel, M. Rizzi, H. Y. Steensma, C. T. Verrips, J. Vindelov, and J. T. Pronk. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 26:706-714. [DOI] [PubMed] [Google Scholar]
- 37.Wach, A., A. Brachat, C. Rebischung, S. Steiner, K. Pokorni, S. te Heesen, and P. Philippsen. 1998. PCR-based gene targeting in Saccharomyces cerevisiae. Methods Microbiol. 26:67-81. [Google Scholar]
- 38.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]