Abstract
E2fs 1-3, also known as activating E2fs, are viewed broadly as critical positive cell cycle regulators. They induce transcription and can drive cells out of quiescence. In flies and mammalian fibroblasts removing activating E2fs causes cell cycle arrest, suggesting an obligate proliferative role 1, 2. However, arrest is indirect as it is alleviated by removing the repressive E2f, dE2f2, in flies, or the tumor suppressor p53 in fibroblasts 3–5. Whether activating E2fs are required for division in vivo is thus an area of lively debate 6. Activating E2fs are also well known pro-apoptotic factors, a defense against oncogenesis 7. In some contexts E2f1 limits irradiation-induced apoptosis 8, 9, but in flies this occurs through repression of hid and the mammalian equivalent, Smac/Diablo is induced not repressed by E2f1 10, and in keratinoctyes it occurs indirectly through induction of DNA repair targets 11. Thus, a direct pro-survival function for activating E2fs in mammals has not been established. To address E2f1-3 function in vivo we focused on the mouse retina, a relatively simple CNS component that can be manipulated without compromising viability and has provided considerable insight into development and cancer 12–14. Here, we show that E2f1-3-deficient retinal progenitor cells or activated Muller glia divide. In the absence of activating E2fs, the Myc family drives proliferation. However, down-regulation of Sirt1, a p53 deacetylase, leads to hyperacetylation of p53 and cell death. Thus, activating E2fs are not universally required for mammalian cell division, but have an unexpected prosurvival role in development.
Keywords: E2f, Neurogenesis, p21Cip1, p57Kip2, Histone deacetylase, Sirtuin, p53, Resveratrol
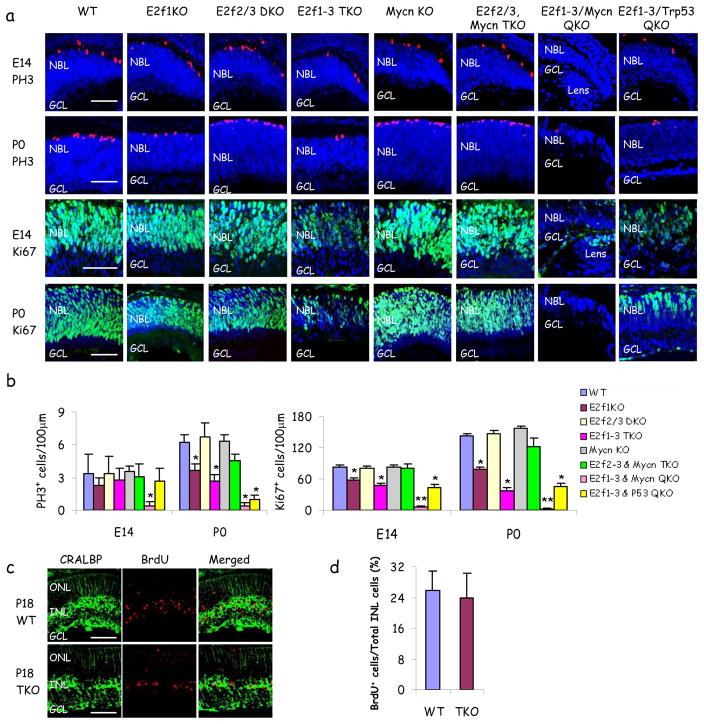
A thin neuroblastic layer (NBL) of progenitors undergoes extensive expansion from mouse embryonic day 11 (E11) to ~ post natal day 8 (P8), generating post-mitotic differentiating cells that develop into the six major retinal neurons and Muller glia. To study the triple knockout (TKO) retina, floxed (f) E2f3 mice were crossed with E2f1−/−, E2f2−/− and α-Cre mice which deletes E2f3 in peripheral retinal progenitors at embryonic day 10 (E10) 15. We assessed Ki67, phosphohistone H3 (PH3), and BrdU incorporation to mark all dividing cells, mitosis and S-phase, respectively (Fig. 1). Some reduction was detected in the E2f1−/− retina, but the E2f2/3 double knockout (DKO) was unaffected, and E2f2 or E2f3 loss did not affect E2f1−/− progenitors (Fig. 1a, b and data not shown). One activating E2f is essential for fibroblast division 2, so we expected a drastic phenotype in the TKO retina where E2f3 is deleted prior to progenitor expansion. Remarkably, we observed many Ki67+ and PH3+ cells in both the E14 and P0 TKO retina (Fig 1a, b). The slightly larger effect at P0 than at E14 is explained by data below. BrdU-labeling was weaker consistent with slower incorporation, but many BrdU+ cells were visible at higher magnification (Supplementary Fig. 1a). Broad Cre-IRES-GFP transgene expression, and analysis of DNA from manually or laser-dissected peripheral retina, mRNA from GFP-sorted E14 retinal cells, and protein at E14 and P0 all proved robust E2f3 targeting (Supplementary Fig. 1b–e). At E14 and P0 cell cycle distribution was the same in WT, E2f1KO or TKO retinas (Supplementary Fig. 2), suggesting a similar effect on all phases. Arrest in TKO fibroblasts is also not phase specific 2. Surprisingly, therefore, although activating E2fs contribute to progenitor expansion, robust division continues in their absence. This partial dispensability in normal dividing progenitors contrasts with the absolute requirement for E2f1 to drive the abnormal cell cycle in Rb-deficient differentiating retinal neurons 15.
Figure 1. Division without activating E2fs.
a, Retinal sections of indicated genotypes and ages were stained for nuclei (DAPI, blue), mitoses (top two panels, PH3, red) or all dividing cells (bottom two panels, Ki67, green). Due to the small size of the E14 E2f1-3/Mycn QKO retina, the lens is also visible in these panels. b, Quantification of PH3+ and Ki67+ cells. c, Domoic Acid/Ouabain/BrdU was injected into the vitreous of P16 mice. Retinas were harvested 48 hrs later, and stained for Muller glia (CRALBP, green) and S-phase (Brdu, red). d, Proportion of BrdU+/CRALBP+cells in the INL from (c) was plotted. Scale bar in (a, c) is 50μm. NBL: neuroblastic layer. GCL: ganglion cell layer.ONL: outer nuclear layer. INL: inner nuclear layer. Data in (b, d) are mean ± SD and asterisks indicate significant difference from WT (* P<0.05, ** P<0.01. Students t-test).
Mature neurons do not divide, but retinal damage triggers reactive gliosis and Muller glia mitosis. We damaged retinas either genetically (homozygous rd that causes photoreceptor degeneration) or by injecting toxins. GFAP induction and loss of p27Kip1 and Cyclin D3 which mark reactive gliosis 16 were observed independent of E2f1-3 (Supplementary Fig. 3 and data not shown). Finally, the proportion of BrdU+/CRALBP+ Muller nuclei in the inner nuclear layer (INL) was identical in control or TKO toxin-treated retina (Fig. 1c, d). Thus, progenitors and mature Muller glia proliferate without activating E2fs.
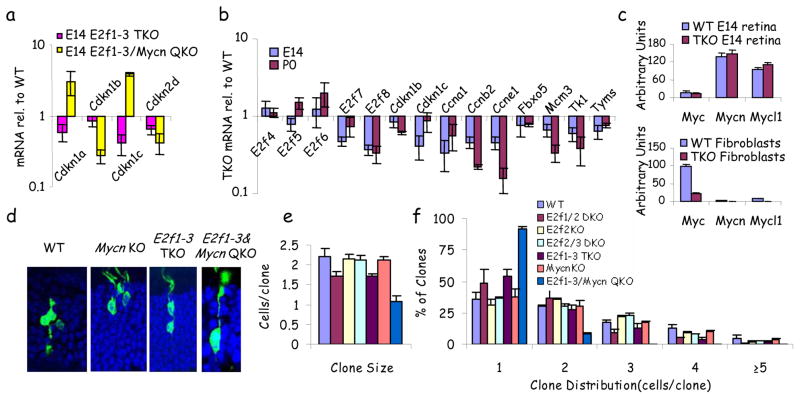
Next, we asked how TKO progenitors divide. In TKO fibroblasts arrest is linked to p53-mediated induction of Cdkn1a, which encodes p21Cip1 [Ref. 2, 3]. Strikingly, both Cdkn1a and 1c (p57Kip2) were reduced in TKO retina (Fig. 2a). Theoretically, induction of positive cell cycle regulators may indirectly interfere with Cdkn1a activation, but as expected, several E2f targets (e.g. Cyclins/Mcm3/Tk) were reduced in TKO progenitors (Fig. 2b). Reducing repressive E2fs might maintain division, but while E2f7/8 were slightly down in the TKO retina, E2f4-6 were unaffected at E14 and two were induced at P0 (Fig. 2b). Myc represses the Cdkn1a promoter in vitro 17 and its over-expression bypasses arrest caused by inhibiting E2f 18, 19. E2f-inhibition in these studies targeted all E2fs, so it is unclear if Myc would override loss of activating E2fs alone, or whether physiological levels of Myc can compensate for E2fs in vivo. Notably, Mycn and Mycl1 were higher than Myc in E14 retina, but the reverse applied in fibroblasts (Fig. 2c). E2f1-3 loss did not reduce Myc family expression in progenitors even though Myc was down-regulated in fibroblasts (Fig. 2c). To test Mycn function, the floxed locus was introduced to the TKO background and deletion of conditional alleles confirmed (Supplementary Fig. 4). Deleting Mycn with a Chx10-Cre driver, expressed across the entire retina, affects eye size 20, but α-Cre mediated deletion in the peripheral retina, alone or with E2f2/3 loss, did not affect eye size, proliferation or survival (Fig. 1a, b, Fig. 3a, b, and data not shown). Nestin-Cre deletion of retinal Mycn also had no effect (T. Reh, unpublished data), thus eye size phenotypes may be Cre-specific. Here, in the E2f1-3/Mycn QKO peripheral retina, eye size was also unaffected, but strikingly progenitor division was dramatically reduced with 12-fold or 67-fold fewer Ki67+ cells than WT retina at E14 and P0, respectively and the NBL was very thin (Fig. 1a). PH3 or BrdU analysis confirmed this dramatic effect (Fig. 1a, b, and data not shown). In agreement, there was a general reduction in E2f target expression, but strikingly, while Cdkn1a and Cdkn1c were reduced in the E2f1-3 TKO retina, they were up-regulated in E2f1-3/Mycn QKO retina (Fig 2a, Supplementary Fig. 5). Thus, Mycn represses Cdk inhibitors and maintains division of E2f1-3 null progenitors, providing the first indication that physiological Myc levels drive division in the absence of activating E2fs.
Figure 2. Mycn drives progenitor division in the absence of E2f1-3.
a, qRT-PCR for indicated genes in E14 WT, E2f1-3 TKO and E2f1-3/Mycn QKO retinas. Levels are plotted relative to WT. b, qRT-PCR for the indicated genes in E14 and P0 WT and E2f1-3 TKO retinas. TKO expression levels are shown relative to WT. c, qRT-PCR for Myc relatives in indicated cells. E2f1-3 TKO cells were GFP-sorted from α-Cre;E2f1−/−;E2f2−/−;E2f3f/f retinas or E2f1−/−;E2f2−/−;E2f3f/f fibroblasts transduced with MXIE-Cre virus. d–f, Control or Cre retrovirus was injected into the subretinal space of P0 mice to generate clones of the indicated genotypes (see text for details). Examples of retinal cell clones (d) along with graphs plotting the average clone size (e) and clone size distribution (f) are shown. Data (a–c, e, f) are mean ± SD.
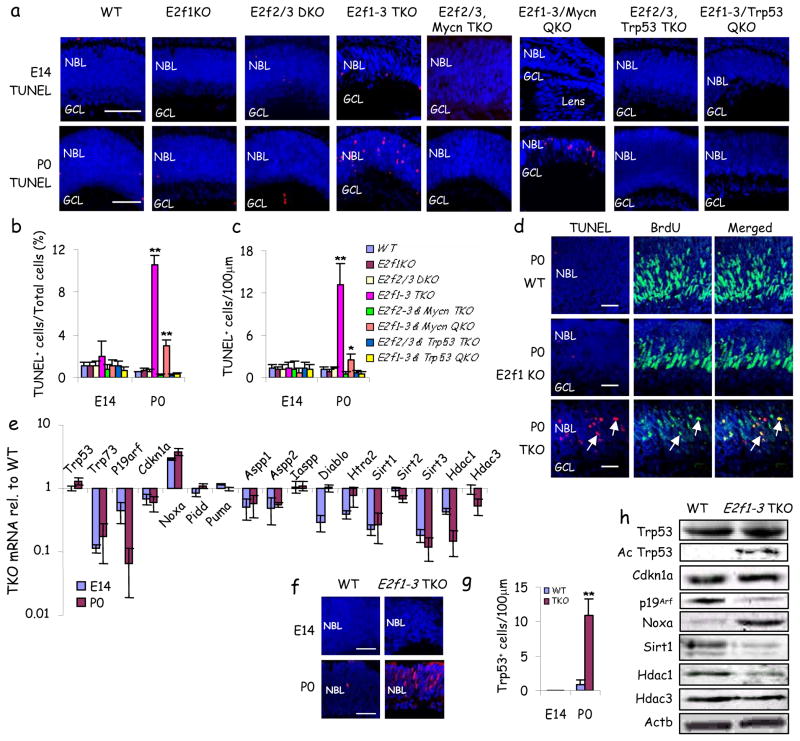
Figure 3. A pro-survival role for activating E2fs.
a, Retinal sections of indicated genotypes and ages were stained for nuclei (DAPI, blue) and apoptosis (TUNEL, red). b–c, Quantification of TUNEL+ cells: b, proportion; c, number per 100μm. d, P0 retinal sections of the indicated genotypes were labeled for 2 hr with BrdU, and stained for nuclei (DAPI, blue), S phase (BrdU, green) and apoptosis (TUNEL, red). Arrows highlight two examples of BrdU+/TUNEL+ cells. e, qRT-PCR analysis of indicated genes in E14 and P0 WT and E2f1-3 TKO retinas. Expression levels in TKO are shown relative to those in WT cells. f, Retinal sections of the indicated genotypes and ages were stained for nuclei (DAPI, blue) and p53 (red). g, Quantification of p53+ cells. h, Cell lysates of P0 WT and E2f1-3 TKO retinas were probed with the indicated antibodies on Western Blots. E2f1-3 TKO cells were dissected from α-Cre; E2f1−/−; E2f2−/−; E2f3f/f peripheral retina. Scale bars in (a,d,f) are 50μm. NBL: neuroblastic layer. GCL: ganglion cell layer. Data in (b, c, e, g) are mean ± SD. Asterisks in (b,c) indicate significant difference from WT (* P<0.05, ** P<0.01. Students t-test).
To further address Mycn/E2f redundancy, Cre retrovirus co-expressing GFP (Supplementary Fig. 6) was injected subretinally into newborn mice and clone size analyzed at P21. The average size of WT, E2f2 KO, E2f2/3 DKO and Mycn KO clones was similar (~ 2.1 cells/clone), but E2f1/2 DKO and E2f1-3 TKO clones were smaller (~ 1.7 cells/clone), but E2f1-3/Mycn QKO clones were almost always single cells (1.07 cells/clone) (Fig. 2d–f). Thus, Mycn promotes division of TKO progenitors cell autonomously.
We considered that E2fs may have another role in retinal development. Activating E2fs are pro-apoptotic 21, but at E14/E17 E2f1-3 loss had no effect on low background apoptosis (Fig. 3a–c, Supplementary Fig. 7). Remarkably, apoptosis was elevated in post-natal TKO retina, returning to normal by P8 when progenitor division is ending (Fig. 3a–c, Supplementary Fig. 7a–c and data not shown). TUNEL+ cells in the TKO retina were also BrdU+, confirming their progenitor status (Fig. 3d). This finding may explain greater reduction in progenitor number at P0 versus E14 (Fig 1a, b). Apoptosis was suppressed by any single activating E2f (Fig. 3a–c, Supplementary Fig. 7).
In flies, dE2f1 protects some irradiated cells by repressing pro-apoptotic hid 8, but in the TKO retina the mammalian equivalents Smac/Diablo and Omi/Htra2 were down-regulated (Fig. 3e). The E2f target p73 was also down-regulated, but immunoreactivity for its relative p53 was elevated, and the p53 pro-apoptotic target NoxA was induced (Fig 3e–g) so we tested whether p53 might drive apoptosis. The floxed locus was introduced into the TKO background and recombination of conditional alleles confirmed (Supplementary Fig. 8). There was no difference in Ki67+, PH3+ or BrdU+ cells in E2f1-3 TKO or E2f1-3/p53 QKO progenitors at E14 or P0 (Fig. 1a–b, and data not shown), which is consistent with the lack of Cdkn1a induction in the E2f1-3 TKO retina (Fig. 2a). In stark contrast, apoptosis was suppressed in E2f1-3/p53 QKO P0 progenitors (Fig. 3a–c). Unlike p53, Mycn loss did not reduce apoptosis: consistent with reduced progenitor numbers the E2f1-3/Mycn QKO P0 retina had fewer total apoptotic cells (Fig 3c), but the proportion of apoptotic (TUNEL+) dividing (Ki67+) progenitors at P0 was not reduced, rather it was slightly elevated (Supplementary Fig. 9) consistent with slightly elevated Noxa expression relative to the TKO retina (Supplementary Fig 5). Thus, unexpectedly, p53 does not reduce division in E2f1-3 TKO progenitors, as in fibroblasts, but drives apoptosis, providing and explaining the first prosurvival role for activating E2fs in normal cells.
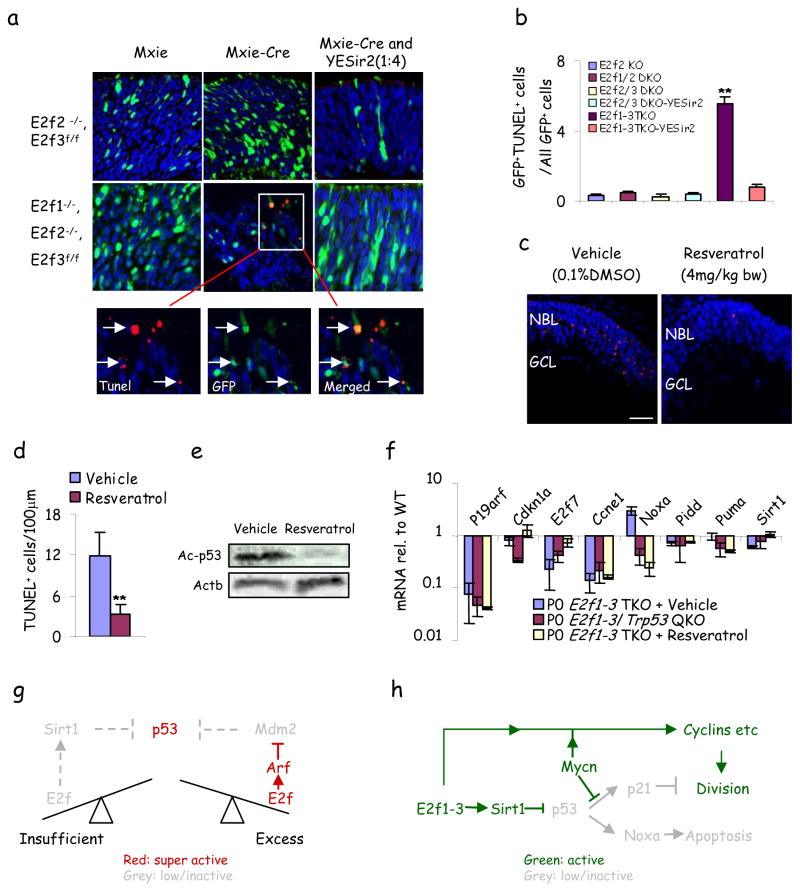
Next, we examined how E2fs constrain p53. Aspp proteins promote p53 pro-apoptotic function but they are E2f-induced 22, 23 and were down-regulated in TKO progenitors, and the Aspp inhibitor, Iaspp, was unaffected (Fig. 3e). p19Arf stabilizes p53 by inhibiting Mdm2, and E2f3 loss derepresses p19Arf transcription in fibroblasts 24, but we observed less p19Arf mRNA and protein in TKO progenitors (Fig. 3e, h). E2f1-3 loss did not affect p53 mRNA, and protein increased only ~1.5 fold (Fig. e, h), suggesting that the 13-fold increase in immunoreactivity may reflect epitope exposure (Fig. 3f, g), perhaps through posttranslational modification 25. E2f1-3 loss did not induce p53 phosphorylation (data not shown) but markedly increased acetylation (Fig. 3h), which is essential for p53 activity 26 is mediated by Tip60, Pcaf and Cbp/p300 and reversed by Hdacs and Sirt1 25. Strikingly, Sirt1/2/3 and Hdac1/3 mRNA were reduced in TKO retina, as were Sirt1/Hdac1 proteins (Fig. 3e, h). Hdac1/3 down-regulation was greater at P0 than E14, correlating with p53 immunodetection specifically at P0 (Fig. 3e). Sirt1 is an E2f1 target 27, but whether other activating E2fs sustain its expression is unknown, and a functional E2f-Sirt1-p53 axis has not been established. To test whether Sirt1 blocks TKO progenitor cell death, we electroporated Cre and Sirt1 expression plasmids 28 into E2f1−/−;E2f2−/−;E2f3f/f P0 retinas and analyzed them two days later. Cre expression induced apoptosis, thus the pro-survival role of E2fs is cell autonomous, and Sirt1 rescued cell death (Fig. 4a, b). Moreover, treatment of pregnant dams from E16 with the Sirt1 agonist resveratrol blocked > 70% of TKO progenitor apoptosis at P0, correlating with p53 deacetylation and blockade of p53-dependent NoxA induction, with no effect on p19Arf expression (Fig. 4c–f). Just as p53 loss did not influence division (Fig. 1a–b), resveratrol did not alter Cyclin levels or proliferation (Fig. 4f, Supplementary Fig 10). In summary, activating E2fs interchangeably induce Sirt1 to block p53 acetylation and apoptosis. These findings explain a novel survival role for activating E2fs in vivo, connecting low E2f to p53 activation (Fig. 4g). As noted above, E2f1-3 redundantly regulated other deacetylases (Fig. 3e) which may also inhibit p53. Integrating the Sirt1 data with our prior findings exposes an E2f/Myc/Sirt1 network that coordinates progenitor proliferation and survival (Fig. 4h).
Figure 4. E2fs promote survival through Sirt1-mediated p53-deacetylation.
a, Mxie plasmid, Mxie-Cre plasmid, or Mxie-Cre plasmid plus YESir2 plasmid expressing Sirt1 were injected subretinally and electroporated into the P0 retinas of the indicated genotypes and analyzed at P2. Retinal sections were stained for nuclei (DAPI, blue) and apoptosis (TUNEL, red). Arrows show examples of GFP+/TUNEL+ cells. b, Quantification of GFP+/TUNEL+ cells as a fraction of all GFP+ cells in clones of the indicated genotype. c, Vehicle (0.1%DMSO) or 4mg/kg body weight of Resveratrol were injected daily into E16 pregnant mice intraperitoneally and newborn E2f1-3 TKO retinas were stained for nuclei (DAPI, blue) and apoptosis (TUNEL, red). NBL: neuroblastic layer, GCL: ganglion cell layer. Scale bar is 50μm. d, Quantification of TUNEL+ cells in (c). e, Cell lysates of dissected P0 E2f1-3 TKO peripheral retina treated with vehicle or resveratrol were probed with indicated antibodies. f, Quantitative real time RT-PCR analysis of the indicated genes in P0 peripheral retina of the indicated genotypes/treatments. Levels are relative to those in WT. Actb mRNA was used to normalize for loading. g, Insufficient or excess E2f activate p53, but by distinct mechanisms. Red, high/active; Grey, low/inactive. h, An integrated E2f/Myc/Sirt1 network promotes division and survival in retinal progenitors. Green and red show active positive and negative functions, respectively. Inactive factors/functions are in grey.
Activating E2fs are critical for division in flies and mammalian fibroblasts, where they counteract repressive E2f or p53, respectively 1–5. We provide the first in vivo evidence that E2f1-3 are not indispensable in every context. Mycn drives E2f-independent division, preventing p53-mediated Cdkn1a induction. In vitro over-expression studies suggested that Myc overcomes E2f repression 18, 19 but did not reveal if normal Myc levels substitute for activating E2fs in vivo. Our genetic strategies reveal that physiological levels compensate for the activating E2fs cell autonomously. Other studies highlight redundancy among related cell cycle regulators 29. Our results indicate redundancy between unrelated cell cycle regulators. We also describe an unexpected prosurvival activity for E2f1-3 in vivo. Loss of any two was harmless, but removing all three impaired progenitor survival. Thus, activating E2fs have interchangeable prosurvival roles, not only E2f1 and not only in irradiated cells 8, 9, 11. We defined the first direct mechanism linking activating E2fs to mammalian cell survival. Thus, E2f1-3 maintain expression of deacetylases, including Sirt1, a proven E2f1 target 27. Our analyses revealed a novel cell autonomous E2f-Sirt1-p53 axis that maintains p53 in a poised state, promoting progenitor survival. This axis could also be important in p53+ tumor cells. Irrespective, its importance for survival in normal cells underscores the need for care in applying E2f inhibitors to treat any disease in vivo.
METHODS SUMMARY
For reactive gliosis 1.0 μl of 2mM domoic acid and 7mM ouabain plus 1mM BrdU was injected into the eyes of P16 ketamine/xylazine anesthetized mice 16. For resveratrol, time pregnant female mice were injected daily with 4 mg/kg of body weight (Cat. 60512A, AKSci) or 0.1%DMSO intraperitoneally from E16. Virus injection and electroporation were as described 30. Fixation, antibody labeling, TUNEL, and quantification of sections, or dissociation and quantification of cells, and RNA and protein analysis were essentially as described 15, 31. The PALM microlaser system was used for LCM following the manufacturer’s recommendations. For flow cytometry E2f3f/f and E2f1−/−, E2f2−/−, E2f3f/f fibroblasts were infected with MXIE or MXIRE-Cre retrovirus. E14 retinas were dissected and dissociated. GFP+ cells were sorted using a FACSAria (Becton Dickinson). For cell cycle analysis, cells were fixed in ice-cold 80% ethanol, counterstained with propidium iodide and analyzed with a BD FACSCaliburTM system. Data were collected using CELLFIT software.
Full Methods and associated references are available in the online version of the paper.
Supplementary Material
Acknowledgments
We thank L. van Parijs, D and R. Weinberg for plasmids, R. Eisenman for mice, and M. Cayouette and J. Wrana for comments. This work was supported by a grant from the Canadian Institutes for Health Research to R.B.
References
- 1.van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–24. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 2.Wu L, et al. The E2F1–3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–62. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 3.Timmers C, et al. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol. 2007;27:65–78. doi: 10.1128/MCB.02147-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma N, et al. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J Biol Chem. 2006;281:36124–31. doi: 10.1074/jbc.M604152200. [DOI] [PubMed] [Google Scholar]
- 5.Frolov MV, et al. Functional antagonism between E2F family members. Genes Dev. 2001;15:2146–60. doi: 10.1101/gad.903901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowland BD, Bernards R. Re-evaluating cell-cycle regulation by E2Fs. Cell. 2006;127:871–4. doi: 10.1016/j.cell.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–57. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moon NS, et al. Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev Cell. 2005;9:463–75. doi: 10.1016/j.devcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Wikonkal NM, et al. Inactivating E2f1 reverts apoptosis resistance and cancer sensitivity in Trp53-deficient mice. Nat Cell Biol. 2003;5:655–60. doi: 10.1038/ncb1001. [DOI] [PubMed] [Google Scholar]
- 10.Xie W, et al. Novel link between E2F1 and Smac/DIABLO: proapoptotic Smac/DIABLO is transcriptionally upregulated by E2F1. Nucleic Acids Res. 2006;34:2046–55. doi: 10.1093/nar/gkl150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berton TR, Mitchell DL, Guo R, Johnson DG. Regulation of epidermal apoptosis and DNA repair by E2F1 in response to ultraviolet B radiation. Oncogene. 2005;24:2449–60. doi: 10.1038/sj.onc.1208462. [DOI] [PubMed] [Google Scholar]
- 12.Dyer MA, Bremner R. The search for the retinoblastoma cell of origin. Nat Rev Cancer. 2005;5:91–101. doi: 10.1038/nrc1545. [DOI] [PubMed] [Google Scholar]
- 13.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–18. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 14.Pacal M, Bremner R. Insights from animal models on the origins and progression of retinoblastoma. Curr Mol Med. 2006;6:759–81. doi: 10.2174/1566524010606070759. [DOI] [PubMed] [Google Scholar]
- 15.Chen D, et al. Rb-Mediated Neuronal Differentiation through Cell-Cycle-Independent Regulation of E2f3a. PLoS Biol. 2007;5:e179. doi: 10.1371/journal.pbio.0050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyer MA, Cepko CL. Control of Muller glial cell proliferation and activation following retinal injury. Nat Neurosci. 2000;3:873–80. doi: 10.1038/78774. [DOI] [PubMed] [Google Scholar]
- 17.Claassen GF, Hann SR. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta -induced cell-cycle arrest. Proc Natl Acad Sci U S A. 2000;97:9498–503. doi: 10.1073/pnas.150006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alevizopoulos K, Vlach J, Hennecke S, Amati B. Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. Embo J. 1997;16:5322–33. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santoni-Rugiu E, Falck J, Mailand N, Bartek J, Lukas J. Involvement of Myc activity in a G(1)/S-promoting mechanism parallel to the pRb/E2F pathway. Mol Cell Biol. 2000;20:3497–509. doi: 10.1128/mcb.20.10.3497-3509.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martins RA, et al. N-myc coordinates retinal growth with eye size during mouse development. Genes Dev. 2008;22:179–93. doi: 10.1101/gad.1608008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 22.Chen D, Padiernos E, Ding F, Lossos IS, Lopez CD. Apoptosis-stimulating protein of p53–2 (ASPP2/53BP2L) is an E2F target gene. Cell Death Differ. 2005;12:358–68. doi: 10.1038/sj.cdd.4401536. [DOI] [PubMed] [Google Scholar]
- 23.Fogal V, et al. ASPP1 and ASPP2 are new transcriptional targets of E2F. Cell Death Differ. 2005;12:369–76. doi: 10.1038/sj.cdd.4401562. [DOI] [PubMed] [Google Scholar]
- 24.Aslanian A, Iaquinta PJ, Verona R, Lees JA. Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev. 2004;18:1413–22. doi: 10.1101/gad.1196704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 26.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–26. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–31. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 28.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 29.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 30.Livne-Bar I, et al. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0600083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D, et al. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5:539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.