Abstract
Disease resistance in plants is often controlled by a gene-for-gene mechanism in which avirulence (avr) gene products encoded by pathogens are specifically recognized, either directly or indirectly, by plant disease resistance (R) gene products. Members of the NBS-LRR class of R genes encode proteins containing a putative nucleotide binding site (NBS) and carboxyl-terminal leucine-rich repeats (LRRs). Generally, NBS-LRR proteins do not contain predicted transmembrane segments or signal peptides, suggesting they are soluble cytoplasmic proteins. RPM1 is an NBS-LRR protein from Arabidopsis thaliana that confers resistance to Pseudomonas syringae expressing either avrRpm1 or avrB. RPM1 protein was localized by using an epitope tag. In contrast to previous suggestions, RPM1 is a peripheral membrane protein that likely resides on the cytoplasmic face of the plasma membrane. Furthermore, RPM1 is degraded coincident with the onset of the hypersensitive response, suggesting a negative feedback loop controlling the extent of cell death and overall resistance response at the site of infection.
Disease resistance in plants often hinges on the ability of the host to recognize specific avirulence (avr) determinants presented by invading microorganisms. This recognition event is controlled by plant disease resistance (R) genes, which are proposed to act as receptors for specific avr-encoded determinants (1). Avr recognition by the plant initiates an elaborate defense response including changes in membrane ion flux, the production of extracellular reactive oxygen intermediates, irreversible plasma membrane (PM) damage, and changes in gene expression and metabolite production (2). This set of responses typically includes localized cell death at the site of pathogen infection, termed the hypersensitive response (HR). In the absence of either R or avr gene, no recognition occurs and the pathogen is able to colonize the host and cause disease.
R genes can be classified by the putative motifs contained in the proteins they encode (1). The largest class encodes a centrally located nucleotide binding site (NBS) and a carboxyl-terminal block of leucine-rich repeats (LRRs). NBS-LRR genes conferring resistance to a number of bacterial, fungal, and viral pathogens have been cloned from a variety of plant species, suggesting that the general NBS-LRR structure is well adapted to recognize a wide range of signals. Subclassification of NBS-LRR sequences is based on the N-terminal domain, which contains either a leucine zipper (LZ) motif or homology with the cytoplasmic domains of the Toll and interleukin-1 receptors (TIR) domain. Computer analysis of predicted NBS-LRR proteins has failed to identify likely transmembrane segments. A potential exception was the prediction of a putative single-pass transmembrane domain in the RPS2 protein. However, site-directed alteration of this sequence to one not predicted to be membrane associated had no effect on RPS2 function (3). Furthermore, with the exception of the predicted flax L6 protein (4), none appear to contain a signal peptide. This led to the hypothesis that NBS-LRR proteins are soluble in the cytoplasm. This predicted localization is consistent with recent findings that suggest that several avr-encoded proteins produced by extracellular pathogens act inside the host cell (3, 5, 6). Delivery of these molecules, in the case of phytopathogenic bacteria such as Pseudomonas syringae and Xanthomonas campestris, requires the evolutionarily conserved type III secretion machinery encoded in plant pathogens by hrp (hypersensitivity and pathogenicity) genes (7).
Whereas there is a growing body of literature regarding the identification of new R gene sequences and the evolution of R gene families, very little is known about the corresponding proteins and the role they play in signal perception and transduction. We are characterizing the product of the Arabidopsis thaliana RPM1 gene. RPM1 encodes an LZ-NBS-LRR protein and confers resistance to the bacterial pathogen P. syringae expressing either of two sequence-unrelated avr genes, avrRpm1 or avrB (8). Using an epitope-tagging strategy, we identified the RPM1 protein in whole plant extracts. In contrast to previous computer-based suggestions for this class of R protein (9), we found RPM1 to be membrane associated and highly enriched in PMfractions. In addition, RPM1 is rapidly degraded at the onset of the HR, perhaps revealing a negative feedback loop that functions to limit the extent of the cell death and the amplitude of the overall disease resistance response.
MATERIALS AND METHODS
Vector Construction and Plant Transformation.
The 3′ end of the RPM1 coding region was PCR amplified with one primer (5′-GCC GCT CGA GAT GAG AGG AAT TAC GG-3′) designed to replace the endogenous translational stop codon with an XhoI site and a second primer (5′-ACA ATG GAG AAT TAC GGT-3′) that annealed upstream of the unique XbaI site in the coding region. The resulting amplification product was cleaved with XbaI and XhoI and used to replace the corresponding portion of the wild-type RPM1 gene. The carboxyl-terminal XhoI site was then ligated to the SalI site of plasmid #CD3–128 (Arabidopsis Biological Resource Center, Ohio State University, Columbus, OH), thus creating a carboxyl-terminal translational fusion with five complete and one partial tandem repeats of the c-Myc epitope (MEQKLISEEDLNE; ref. 10). The chimeric RPM1∷MYC gene, including 1,034 bp of promoter sequence upstream of the translational start codon, was subcloned into a binary vector derived from pGPTV-HPT (11), electroporated into Agrobacterium strain GV3101(pMP90) and transformed by vacuum infiltration (12) into plants.
Plant Growth and P. syringae Infection.
The rps3–1 mutant is a frameshift allele of RPM1 in the Col-0 background referred to henceforth as rpm1-fs (8, 13). Plant growth conditions were described previously (14). DC3000 and the avr-expressing plasmids in pVSP61 have been described (8, 13). P. syringae strains were grown overnight in King’s B medium (14) with appropriate antibiotics, washed once in 10 mM MgCl2 and resuspended to a density of ≈5 × 107 cfu/ml for HR tests and RPM1∷MYC expression analysis. A density of ≈5.5 × 103 cfu/ml was used for analysis of in planta growth. Bacterial suspensions were pressure infiltrated into the abaxial leaf surface by using a needleless 1-ml disposable syringe (14).
Antibody Production, Immunoprecipitation, and Protein Blot Analysis.
RPM1 fragments containing the NBS (aa 177–458) and LRR (aa 536–926) domains (Fig. 1A) were amplified by PCR and cloned into the XhoI/BamHI sites of pET14b (Novagen) to create translational fusions with an N-terminal His6 tag. Fusion proteins were isolated from Escherichia coli as insoluble inclusion bodies, purified by preparative SDS/PAGE and transferred to nitrocellulose filters. Strips of nitrocellulose-bound antigen were solubilized with dimethyl sulfoxide and injected into goats to produce anti-NBS and anti-LRR polyclonal antisera. The anti-c-Myc monoclonal antibody was isolated from hybridoma line 9E10 culture supernatant (University of North Carolina Lineberger Tissue Culture Facility, Chapel Hill, NC).
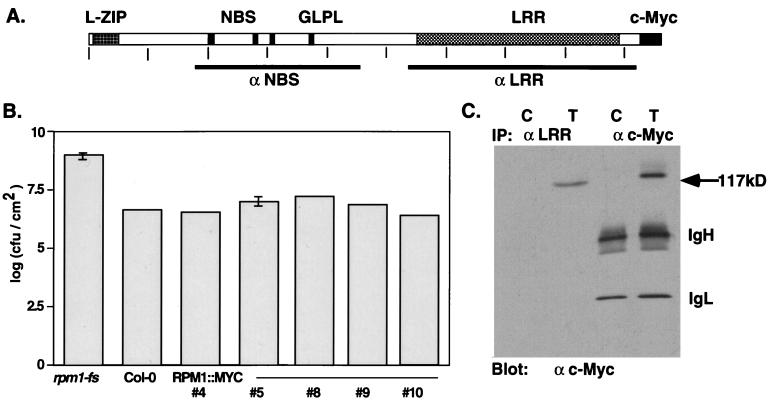
Figure 1.
Structure, function, and immunodetection of RPM1∷MYC. (A) The deduced RPM1 protein indicating the position of the leucine zipper (L-ZIP), putative NBS, and LRR domains. GLPL denotes the core of a conserved sequence motif of unknown function found in all NBS-LRR proteins. Vertical ticks occur every 100 aa. Bars below the schematic define the regions of the protein used to generate the anti-NBS and anti-LRR polyclonal antisera. (B) Complementation of the rpm1-fs mutation with RPM1∷MYC. Bacterial growth in leaves infiltrated with DC3000(avrRpm1) at a density of ≈5.5 × 103 cfu/cm2 was assayed 72 hpi. In planta bacterial titers are given for nontransformed rpm1-fs, wild-type Col-0, and five independent rpm1-fs (RPM1∷MYC) transgenic lines. (C) Immunodetection of RPM1∷MYC by using monoclonal anti-c-Myc antibody after immunoprecipitation with either anti-LRR antisera or monoclonal anti-c-Myc antibody. C is extract from nontransformed control plants, T from transgenic #4 of B. Anti-mouse IgG second step antibody detects the IgG heavy (IgH) and light (IgL) chains of the monoclonal.
Extracts for immunoprecipitation were prepared by grinding ≈200 μg leaf tissue in 500 μl ice-cold extraction buffer (20 mM Tris⋅HCl (pH 7.5)/1 mM EDTA/150 mM NaCl/1% (vol/vol) TX-100). Cell debris was removed by centrifugation at 13,000 × g for 20 min and soluble protein remaining in the supernatant was quantitated by using the DC protein assay (Bio-Rad). For immunoprecipitations, ≈500 μg extract (in a volume of 500 μl) was combined with 5 μl crude anti-RPM1 antisera or 20 μl purified 9E10 anti-myc monoclonal antibody and rotated end-over-end at 4°C for 1 hr. Protein G-agarose beads (30 μl; Boehringer Mannheim) were added and the incubation was continued for 1 hr. Immuno-complexes were washed four times with 1 ml ice-cold extraction buffer, resuspended in 20 μl SDS/PAGE sample buffer, heated to 70°C for 5 min and analyzed by SDS/PAGE, as given below. Extracts for standard protein blot analysis were prepared in the above extraction buffer with the addition of 0.1% (wt/vol) SDS and 5 mM DTT. Approximately 30 μg of protein was loaded per lane of a 7.5% SDS/PAGE gel. Proteins were electroblotted to nitrocellulose membranes and blocked for 1 hr in TBST (20 mM Tris⋅HCl (pH7.6)/130 mM NaCl/0.05% (vol/vol) Tween-20) containing 5% (wt/vol) nonfat dry milk. Reaction with primary antibodies was carried out for 1 hr at room temperature with 1% (wt/vol) nonfat dry milk in TBST. For anti-c-Myc reactions, undiluted 9E10 tissue culture supernatant was used. Horseradish peroxidase-conjugated secondary antibodies (Amersham) were diluted 1:2,000 in TBST containing 1% (wt/vol) nonfat dry milk and reacted with the blots for 1 hr at room temperature. Bands were visualized by using the ECL kit (Amersham).
Membrane Fractionation.
Tissue was homogenized on ice in lysis buffer (0.33 M sucrose/20 mM Tris⋅HCl (pH 7.5)/1 mM EDTA/2 μg/ml aprotinin/2 μg/ml leupeptin/1 μg/ml pepstatin A) by using a Waring blender and polytron. The lysate was cleared of cell-wall debris, nuclei, and intact organelles by filtration through micracloth and centrifugation at 2,000 × g for 10 min. Centrifugation at 100,000 × g for 1 hr generated crude soluble (supernatant) and microsomal membrane (pellet) fractions. Linear and step sucrose gradients were as described (15). Aqueous two-phase partitioning to enrich for PMvesicles was also as described (16), with a polymer concentration of 6.2% (wt/vol).
RESULTS
Identification of the RPM1 Protein in Plant Extracts.
Our strategy to detect RPM1 in plant extracts used polyclonal antibodies raised against two nonoverlapping domains of RPM1, designated NBS and LRR, as well as transgenic plants expressing an RPM1 protein modified by the addition of a carboxyl-terminal c-Myc epitope tag (Fig. 1A). The native RPM1 transcript is present at very low levels (8), suggesting that its protein product may be rare as well. We were therefore concerned that overexpression of the RPM1∷MYC protein in transgenic plants might alter or obscure its normal subcellular localization. Thus, to most closely reflect the endogenous RPM1 expression level, transcription of the RPM1∷MYC construct was driven by ≈1 kb of the native RPM1 promoter.
The carboxyl-terminal epitope-tagged RPM1 protein restored wild-type levels of resistance when transformed into the rpm1-fs mutant line (Fig. 1B), which contains a frame-shift mutation within the RPM1 coding region and is a loss-of-function mutant when challenged with DC3000(avrRpm1) (8, 13). Of 12 transgenic T2 lines assayed, 10 responded to high-dose (5 × 107 cfu/ml) inoculation with DC3000(avrRpm1) by generating a characteristic HR ≈5 hr postinoculation [hours postinoculation (hpi); data not shown]. Furthermore, five of these lines were analyzed for the ability to restrict the growth of DC3000(avrRpm1) in planta. All five displayed an approximate two-log reduction in bacterial growth 3 days postinoculation relative to nontransformed control rpm1-fs plants (Fig. 1B). Both the timing of HR (not shown) and the reduction in growth are comparable to that seen in wild-type RPM1 backgrounds (e.g., Col-0; Fig. 1B). From this functional analysis, we conclude that the subcellular localization of the epitope-tagged RPM1 protein should closely parallel that of the endogenous RPM1 protein.
Immunoprecipitation of RPM1∷MYC protein with the anti-LRR antisera followed by detection on protein blots with the anti-c-Myc antibody revealed a strong band at the predicted size of 117 kDa (Fig. 1C Left). No signal of the expected size was observed in immunoprecipitations from nontransformed control plants (Fig. 1C). Similar results were obtained with the anti-NBS antisera (not shown). We additionally immunoprecipitated and subsequently detected RPM1∷MYC directly using the anti-c-Myc monoclonal antibody (Fig. 1C Right). Thus, the 117-kDa protein is recognized by two distinct anti-RPM1 antibodies and by the anti-c-Myc antibody as well, clearly defining it as the RPM1∷MYC protein. Unexpectedly, neither anti-RPM1 antisera detected endogenous RPM1 or RPM1∷MYC protein on blots of denatured total protein extracts (data not shown). This lack of detection prevented localization of the native RPM1 protein.
RPM1∷MYC Is Peripherally Associated with the Plasma Membrane.
We directly detected RPM1∷MYC routinely on blots of total protein extracts using the anti-c-Myc antibody. Fig. 2A displays a comparison of three independent transgenic lines that express the RPM1∷MYC protein. In each case, the expected band of 117 kDa is present in the total extract (Fig. 2A). Isolation of crude soluble and microsomal membrane fractions by centrifugation at 100,000 × g revealed that RPM1∷MYC was contained completely in the membrane pellet in all three lines examined (Fig. 2A). Two other bands can be seen at ≈98 (variably detected) and ≈150 kDa. These bands are also present in nontransformed control extracts and therefore correspond to endogenous proteins that crossreact with the anti-c-Myc monoclonal antibody (Fig. 2A). In addition, neither of these bands is detected on anti-c-Myc protein blots of immunoprecipitated RPM1∷MYC (Fig. 1).
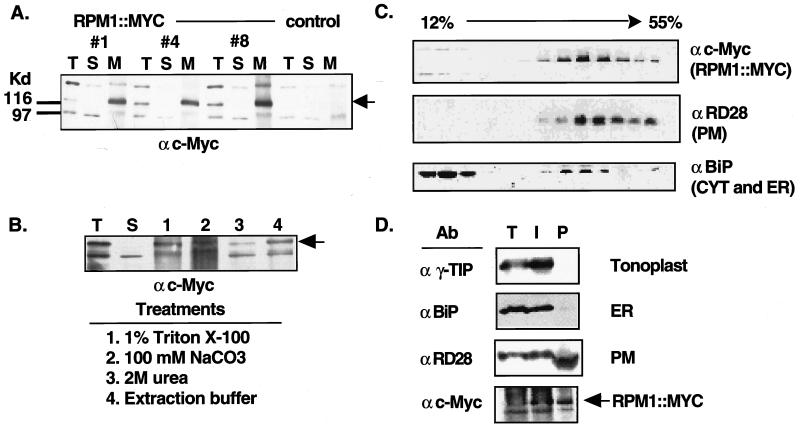
Figure 2.
RPM1∷MYC is a peripheral PM protein. (A) Protein blot of total (T), soluble (S), and microsomal membrane (M) fractions from nontransformed rpm1-fs control plants and three rpm1-fs (RPM1∷MYC) transgenic lines reacted with the anti-c-Myc monoclonal antibody. Equal amounts of protein were loaded in each lane. Arrowhead indicates the position of RPM1∷MYC. Anti-c-Myc crossreacting bands at ≈98 and ≈150 Kd were present in nontransformed control lines. (B) Protein blot reacted with the anti-c-Myc monoclonal antibody demonstrating peripheral association of RPM1∷MYC with the membrane. Total extract (T) was centrifuged at 100,000 × g to generate soluble (S) and microsomal membrane fractions. Membranes were treated as specified to release peripheral membrane proteins. Remaining membranes were pelleted and the newly soluble proteins were analyzed. (C) Fractionation of RPM1∷MYC on sucrose gradients. A 12–55% (wt/vol) linear sucrose gradient was used to fractionate total extract from transgenic plants. Aliquots of each fraction were blotted to nitrocellulose and were analyzed with either anti-c-Myc or the subcellular compartment marker antibodies listed at the right of each panel. (D) Protein blot analysis of membranes fractions obtained by aqueous two-phase partitioning. Total extract (T), intracellular membrane (I), and plasma membrane (P) vesicle fractions were separated by SDS/PAGE, transferred to nitrocellulose and reacted sequentially with antibodies against the c-Myc epitope, RD28, γ-TIP, and BiP.
Because RPM1 lacks an obvious transmembrane segment, we reasoned that peripheral association with the membrane fraction was likely. As expected, treatment of microsomal membranes with detergents such as 1% (vol/vol) Triton X-100 solubilized the microsomal membrane pellet and released RPM∷MYC (Fig. 2B). Treatment of microsomal membranes with 100 mM sodium carbonate or 2 M urea also removed RPM∷MYC from the membrane (Fig. 2B). These treatments do little damage to the integrity of the lipid bilayer and integral proteins, but are known to release many peripheral proteins (17). Indeed, even the physical force of resuspending the membrane pellet in extraction buffer was sufficient to release a portion of the RPM∷MYC protein from the membrane fraction (Fig. 2B). Thus, we conclude that RPM1∷MYC is a peripheral membrane protein.
We conducted a series of subcellular fractionation analyses to determine the membrane compartment in which RPM1∷MYC resides. Numerous attempts to fractionate membranes by linear or step sucrose gradients consistently supported the membrane-association of RPM1∷MYC. RPM1∷MYC was found in fractions containing the endoplasmic reticulum (ER)-localized isoform of BiP (R. Boston, personal communication) and a known PM marker, RD28 (16) (Fig. 2C), but we were unable to define gradient conditions that conclusively resolved PM and ER markers.
Therefore, we used aqueous two-phase partitioning to purify vesicles enriched in PM (16). To verify enrichment of PM vesicles, we sequentially reacted the same protein blot with antibodies demonstrated previously to detect markers for tonoplast (γ-TIP; ref. 18), ER(BiP), and PM (RD28; ref. 16). RD28 dimers (and monomers; not shown) were enriched in the PM lane and γ-TIP and BiP were absent, indicating substantial PM enrichment with undetectable contamination by tonoplast and ER vesicles (Fig. 2D). The continued presence of RD28 in the intracellular membrane fraction is typical of vesicles made with this protocol and reflects the occurrence of “outside-in” PM vesicles partitioning with the intracellular membrane vesicles (19). Consistent with this, γ-TIP and BiP were exclusively detected in the intracellular membrane fraction. RPM1∷MYC, like RD28, is enriched in the PM vesicles (Fig. 2D), strongly suggesting that it is located in the PM.
RPM1∷MYC Is Degraded at a Time Coincident with the Onset of the HR.
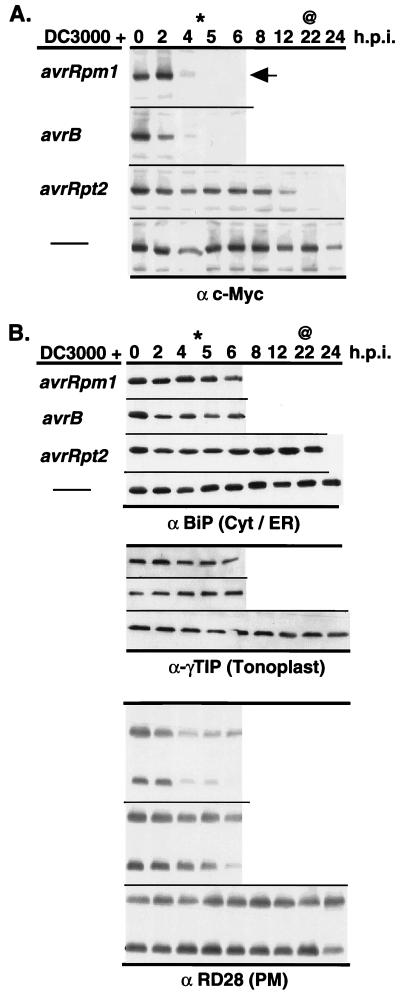
Previous work in our laboratory demonstrated the endogenous RPM1 transcript level is not changed after infection with avirulent P. syringae (M. Grant and J.L.D., unpublished data). Therefore, we wanted to determine whether the expression profile or localization of the RPM1∷MYC protein was altered after infection. We monitored RPM1∷MYC expression over time in total protein extracts from leaves inoculated with the virulent strain DC3000 or with several avirulent DC3000 derivatives at a dose of 5 × 107 cfu/ml. Resistance to strains carrying avrRpm1 and avrB is controlled by RPM1, which triggers a rapid HR with tissue collapse at the infection site occurring ≈5 hpi. In contrast, resistance to the strain carrying avrRpt2 is conferred by a different LZ-NBS-LRR gene, RPS2, which triggers a slower HR with tissue collapse occurring ≈22 hpi. Strikingly, the level of RPM1∷MYC expression declined sharply at a time coincident with the onset of the HR produced by inoculation with DC3000 carrying avrRpm1, avrB, or avrRpt2 (Fig. 3A). In contrast to infection by avirulent strains, inoculation with either virulent DC3000 (Fig. 3A) or MgCl2 buffer alone (not shown) did not result in an HR or degradation of RPM1∷MYC. In fact, RPM1∷MYC persisted even after the initiation of chlorotic disease symptoms caused by DC3000 infection (22 hpi; Fig. 3A). We did not observe the appearance of a low molecular weight carboxyl-terminal RPM1∷MYC fragment (down to ≈20 kDa) in any of our experiments.
Figure 3.
RPM1∷MYC is degraded after inoculation with avirulent P. syringae isolates, which trigger LZ-NBS-LRR resistance genes. (A) Each panel represents infection of RPM1∷MYC transgenics with P. syringae DC3000 expressing a particular avirulence gene or not (thin line) listed at left. Tissue was harvested at the time, in hpi listed at the top, and total protein blots were probed with either anti-c-Myc monoclonal or (B) marker proteins for various subcellular compartments listed at the bottom of each series of blots in B. Extract from control plants (vector) are included for anti-c-Myc and anti-BiP experiments. The arrow marks RPM1∷MYC in A. Star (∗) represent earliest time point where visible RPM1-dependent HR was observed, and ampersand (@) represents the earliest time point where RPS2-dependent HR was observed. This set of data is all from one set of extracts and the experiment was repeated three times.
We addressed whether the RPM1∷MYC disappearance reflected wholesale protein degradation at the onset of HR. Fig. 3B demonstrates that it did not. Marker antigens for cytoplasmic, ER, and tonoplast proteins are intact until well beyond the onset of either RPM1- or RPS2-dependent HR or onset of DC3000 disease symptoms. Whereas monomers and dimers of the PM protein RD28 do begin to disappear at time points when tissue collapse resulting from the HR is complete and the tissue dry, this is later than the time when RPM1∷MYC is completely gone. We observed a similar slow decline in the amounts of a PM-localized ATPase (data not shown), suggesting that subsequent to RPM1∷MYC disappearance, degradation of PM proteins in general begins. The disappearance of RPM1∷MYC is not a peculiarity of the c-Myc epitope, since another protein tagged by a carboxyl-terminal c-Myc epitope, cytosolic AtPEP12p (gift from N. Raikhel), is not lost during this time course (data not shown). Taken together, these results suggest that the loss of RPM1∷MYC signal is not the result of random large-scale protein degradation resulting from HR. We also monitored the localization of RPM1∷MYC over time after inoculation with DC3000(avrRpm1) and found it to remain associated with the membrane fraction as long as it could be detected (data not shown).
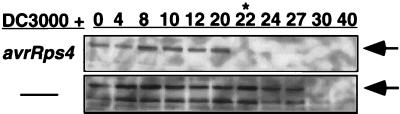
Finally, in light of recent demonstrations that LZ-NBS-LRR class R proteins can differ from TIR-NBS-LRR R proteins in their genetic requirements for function (20), we asked whether RPM1∷MYC disappearance could be triggered by DC3000 carrying avrRps4. This P. syringae gene is recognized by the TIR-NBS-LRR gene, RPS4, resulting in HR at roughly 20 hpi (ref. 21; B. Staskawicz, personal communication). Fig. 4 demonstrates that RPM1∷MYC also disappears at the time of RPS4-mediated HR.
Figure 4.
RPM1∷MYC is degraded after inoculation with an avirulent P. syringae isolate, which triggers a TIR-NBS-LRR resistance gene. Experimental design is as in Fig. 3, but the time course is extended to 40 hpi, at which time all proteins begin to degrade because of disease (DC3000) or complete HR. Star (∗) marks time point of RPS4-dependent HR and arrows denote RPM1∷MYC.
DISCUSSION
Localization of the RPM1∷MYC Protein.
We expressed a fully functional c-Myc-tagged RPM1 protein in transgenic Arabidopsis. The protein could be immunoprecipitated from whole leaf extracts by two antibodies raised against nonoverlapping domains of RPM1 and also identified directly on protein blots by the anti-c-Myc antibody. Contrary to prediction, the RPM1∷MYC protein was not soluble, but rather associated wholly with the microsomal membrane pellet, from which it could be solubilized by treatments known to remove peripheral membrane proteins. Furthermore, membrane fractionation by aqueous two-phase partitioning demonstrated that RPM1∷MYC is enriched in PM vesicles in a manner equivalent to a known PM marker protein. We conclude that RPM1∷MYC is a peripheral membrane protein and that a significant portion of it is localized to the PM. Because the RPM1∷MYC protein was fully functional and expressed from the native RPM1 promoter, its localization should closely parallel that of the endogenous RPM1 protein.
Is the avr Signal Recognized by a Plasma Membrane-Anchored Receptor Complex?
Delivery of avr function to plant cells requires the type III (hrp) secretion system. This system is thought to deliver a variety of molecules, including virulence and avirulence factors, directly into the host cells, as demonstrated for a variety of bacterial pathogens of animals (22, 23). In support of this hypothesis, expression of several avr gene products in the host cell, either stably or transiently, has been shown to be sufficient to trigger a resistance-like response (3, 5, 6). Bacterial type III secretion is thought to proceed through the development of a pilus-like structure connecting the pathogen and host cell (24). Therefore, localization of RPM1 to the cytoplasmic face of the PM may be advantageous in placing the R gene product in a position to intercept incoming pathogen-derived signals.
Whereas RPM1 does not contain a predicted transmembrane segment or signal peptide, our data demonstrate that it is membrane localized and solubilized by treatments known to not disrupt the structure of the lipid bilayer. Therefore, its association with the membrane is likely maintained through interactions with unidentified integral membrane proteins. This finding is not inconsistent with published results using a dog pancreas microsome import assay, which demonstrated that RPS2 protein is not secreted or inserted into the lipid bilayer (3). One candidate RPM1-tethering protein is the product of the recently cloned NDR1 gene. NDR1 is required for the function of a number of Arabidopsis R genes, including RPM1, and is predicted to encode a small novel protein containing two putative transmembrane segments (25). We identified a second potential membrane anchor using the N-terminal portion of RPM1 as bait in a yeast two-hybrid screen. The N terminus of this protein contains six to seven potential transmembrane segments and a novel carboxyl-terminal cytoplasmic domain that interacts with RPM1 (D.C.B., S. Kaufman, and J.L.D., unpublished data).
The genetically defined specificity of avr recognition coupled with recent findings that avr proteins are able to generate R-dependent resistance-like responses when expressed inside plant cells suggests that RPM1 and other NBS-LRR proteins are receptors that directly interact with their cognate avr gene products. Nevertheless, we have not been able to demonstrate an RPM1 interaction with avrRpm1 or avrB through yeast two-hybrid analysis (D.C.B. and J.L.D., unpublished work). In contrast, the product of the tomato R gene Pto, which encodes a serine/threonine protein kinase, has been shown to interact with the avrPto protein of P. syringae (26, 27). Pto function also requires the NBS-LRR protein Prf (28), although the biochemical role that Prf plays in the response is unknown. By analogy, RPM1 may not be sufficient to directly perceive the avr signal, and a Pto-like kinase and/or additional factors may be required to facilitate avr signal recognition. We have additionally demonstrated that avrRpm1 and avrB proteins are localized to plant cell plasma membranes and that this localization is required for avirulence function (Z. Nimchuk, E. Marois, S. Kjemtrup & J.L.D., unpublished work). Based on the PM localization data we have presented for RPM1, we speculate that additional factors required for RPM1 function will be membrane localized as well. In support of this hypothesis, Pto is predicted to have an N-terminal myristolation motif and may therefore be targeted to the membrane. Indeed, the Fen kinase of tomato, which is closely related to Pto and is also Prf dependent, requires a consensus myristolation sequence for function (29).
Degradation of RPM1∷MYC May Define a Negative Feedback Loop That Controls the Extent of Response at the Site of Infection.
RPM1∷MYC protein disappears rapidly at a time coincident with initiation of the HR generated in response to infection by P. syringae DC3000 carrying either avrRpm1, avrB, avrRpt2, or avrRps4 (Figs. 3 and 4). In contrast, little or no loss of RPM1∷MYC is seen at corresponding time points in control treatments that fail to produce an HR or after infection by virulent DC3000. This is true even in leaves exhibiting the onset of disease symptoms, indicating that cell death alone is not sufficient for the initial rapid loss of RPM1∷MYC to occur. The disappearance of RPM1∷MYC signal is most likely the result of increased proteolytic activity. However, the rate of RPM1∷MYC turnover is unknown, and a rapid decrease in the rate of RPM1∷MYC translation could produce a similar result and therefore cannot be ruled out.
At least two other PM proteins degrade subsequent to RPM1∷MYC disappearance and the onset of HR. This suggests that loss of PM integrity is a major cellular event after initial onset of HR. This is consistent with a variety of microscopic analyses (ref. 30) showing that irreversible PM damage is an early sign of HR. In this regard, our results are inconsistent with the canonical early cellular events of animal apoptosis, in which PM integrity is maintained until very late in the cell death process. We also note that at prolonged time points, all proteins assayed, regardless of cellular compartment, disappear as disease develops in DC3000 inoculated tissue (data not shown).
Degradation of RPM1 may be one way in which the cell is able to control HR lesion size and response amplitude at the site of infection. The timing of RPM1∷MYC degradation at the onset of HR is consistent with this idea. The alternative hypothesis, that RPM1∷MYC degradation serves a positive signaling role, is less likely. If this were true, RPM1∷MYC degradation would occur much earlier in the response, before or concurrent with the molecular and physiological changes that promote defense gene activation (31, 32). It is intriguing that RPM1∷MYC is degraded after activation with DC3000(avrRpm1 or avrB) by inoculation of DC3000(avrRpt2), which activates RPS2, another LZ-NBS-LRR protein, and by DC3000(avrRps4), which activates the TIR-NBS-LRR protein RPS4. If these R gene pathways are activated independently, then the mechanism targeting RPM1∷MYC for degradation is likely a general one that degrades engaged as well as unengaged NBS-LRR proteins. This phenomenon is also interesting in light of the recent characterization of the Arabidopsis LSD1 gene (33, 34). Plants carrying a deleted lsd1 allele exhibit spreading necrotic lesions, suggesting that one function of the wild-type gene is to limit the size of the HR. It will be of interest to determine whether the RPM1∷MYC degradation phenomenon will occur in an lsd1 background or in other Arabidopsis mutants that effect disease resistance.
Acknowledgments
We thank Drs. Maarteen Chrispeels (University of California, San Diego, CA), Rebecca Boston (North Carolina State University, Raleigh, NC), and Marc Boutry (Brussels, Belgium) for gifts of antisera and Natasha Raihkel (Michigan State University, East Lansing, MI) for technical advice and the gift of transgenic AtPEP12p-c-Myc tagged plants. We are grateful to Dr. John McDowell and Dr. Susanne Kjemtrup for critical reading of the manuscript and helpful discussions. This work is supported by grants from the National Science Foundation, initially MCB 95056788 and currently IBN 9724075.
ABBREVIATIONS
- avr
avirulence
- hpi
hours postinoculation
- HR
hypersensitive response
- LRR
leucine-rich repeat
- LZ
leucine zipper
- NBS
nucleotide binding site
- ER
endoplasmic reticulum
- PM
plasma membrane
- TIR
Toll/interleukin-1 receptor homology
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Bent A. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leister R T, Ausubel F M, Katagiri F. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence G J, Finnegan E J, Ayliffe M A, Ellis J G. Plant Cell. 1995;7:1195–1206. doi: 10.1105/tpc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopalan S, Bauer D W, Alfano J R, Lonlello A O, He S Y, Collmer A. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Ackerveken G, Marois E, Bonas U. Cell. 1996;87:1307–1316. doi: 10.1016/s0092-8674(00)81825-5. [DOI] [PubMed] [Google Scholar]
- 7.Alfano J R, Collmer A. J Bacteriol. 1997;17779:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant M R, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangl J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 9.Parker J E, Coleman M. Trends Biochem Sci. 1997;22:291–296. doi: 10.1016/s0968-0004(97)01089-x. [DOI] [PubMed] [Google Scholar]
- 10.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker D, Kemper E, Schell J, Masterson R. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- 12.Bechtold N, Ellis J, Pelletier G. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- 13.Bisgrove S R, Simonich M T, Smith N M, Sattler N M, Innes R W. Plant Cell. 1994;6:927–933. doi: 10.1105/tpc.6.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debener T, Lehnackers H, Arnold M, Dangl J L. Plant J. 1991;1:289–302. doi: 10.1046/j.1365-313X.1991.t01-7-00999.x. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed S U, Bar-Peled M, Raikhel N V. Plant Physiol. 1997;114:325–336. doi: 10.1104/pp.114.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniels M J, Mirkov T E, Chrispeels M J. Plant Physiol. 1994;106:1325–1333. doi: 10.1104/pp.106.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludevid D, Hofte H, Himelblau E, Chrispeels M J. Plant Physiol. 1992;100:1633–1639. doi: 10.1104/pp.100.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kjellbom P, Larsson C. Plant Physiol. 1984;62:501–509. [Google Scholar]
- 20.Aarts N, Metz M, Holub E, Staskawicz B J, Daniels M J, Parker J E. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinsch M, Staskawicz B J. Mol Plant–Microbe Interact. 1996;9:55–61. doi: 10.1094/mpmi-9-0055. [DOI] [PubMed] [Google Scholar]
- 22.Michiels T, Wattiau P, Brasseur R, Ruysshaert J-M, Cornelius G R. Infect Immun. 1990;58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galán J. Mol Micobiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 24.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Proc Natl Acad Sci, USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Century K S, Shapiro A D, Repetti P P, Dahlbeck D, Holub E, Staskawicz B J. Science. 1997;278:1963–1965. doi: 10.1126/science.278.5345.1963. [DOI] [PubMed] [Google Scholar]
- 26.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 27.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 28.Salmeron J M, Oldroyd G E D, Rommens C M T, Scofield S R, Kim H- S, Lavelle D T, Dahlbeck D, Staskawicz B J. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 29.Rommens C M T, Salmeron J M, Baulcombe D C, Staskawicz B J. Plant Cell. 1995;7:249–257. doi: 10.1105/tpc.7.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett M, Gallagher M, Fagg J, Bestwick C, Paul T, Beale M, Mansfield J. Plant J. 1996;9:851–866. [Google Scholar]
- 31.Somssich I E, Halbrock K. Trends Plant Sci. 1998;3:86–90. [Google Scholar]
- 32.Yang Y, Shah J, Klessig D F. Genes Dev. 1997;11:1621–1639. doi: 10.1101/gad.11.13.1621. [DOI] [PubMed] [Google Scholar]
- 33.Jabs T, Dietrich R A, Dangl J L. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- 34.Dietrich R A, Richberg M H, Schmidt R, Dean C, Dangl J L. Cell. 1997;88:685–694. doi: 10.1016/s0092-8674(00)81911-x. [DOI] [PubMed] [Google Scholar]