Abstract
The Neurospora crassa CYT-18 protein is a mitochondrial tyrosyl-tRNA synthetase that also promotes self-splicing of group I intron RNAs by stabilizing functional structure in the conserved core. CYT-18 binds the core along the same surface as a common peripheral element, P5abc, suggesting that CYT-18 can replace P5abc functionally. In addition to stabilizing structure generally, P5abc stabilizes the native conformation of the Tetrahymena group I intron relative to a globally-similar misfolded conformation, which has only local differences within the core and is populated significantly at equilibrium by a ribozyme variant lacking P5abc (EΔP5abc). Here we show that CYT-18 specifically promotes formation of the native group I intron core from this misfolded conformation. Catalytic activity assays demonstrate that CYT-18 shifts the equilibrium of EΔP5abc toward the native state by at least 35-fold, and binding assays suggest an even larger effect. Thus, like P5abc, CYT-18 preferentially recognizes the native core, despite the global similarity of the misfolded core and despite forming crudely similar complexes, as revealed by DMS footprinting. Interestingly, the effects of CYT-18 and P5abc on folding kinetics differ. Whereas P5abc inhibits refolding of the misfolded conformation by forming peripheral contacts that must break during refolding, CYT-18 does not display analogous inhibition, most likely because it relies to a greater extent on direct interactions with the core. Although CYT-18 does not encounter this RNA in vivo, our results suggest that it stabilizes its cognate group I introns relative to analogous misfolded intermediates. By specifically recognizing native structure, CYT-18 may also interact with earlier folding intermediates to avoid RNA misfolding or to trap native structure as it forms. More generally, our results highlight the ability of a protein cofactor to stabilize a functional RNA structure specifically without incurring associated costs in RNA folding kinetics.
Keywords: Tetrahymena ribozyme, catalytic RNA, RNA folding, RNP assembly, RNA-protein interaction, RNA thermodynamics
INTRODUCTION
RNAs carry out a myriad of cellular functions, many of which require folding to specific structures. Thus, the sequences of these RNAs must encode functional structures that are stable enough to be populated at equilibrium and are kinetically accessible during folding.1 Because structured RNAs typically include base-paired regions that form favorably even as isolated elements, globally unfolded conformations are generally not stable. Therefore, a critical challenge for RNA is to stabilize functional structures relative to alternative conformations that include subsets of the native secondary and tertiary contacts and may also include non-native contacts.1,2 The high stability of local structure also poses challenges for the kinetics of folding, because disruptions of native or non-native contacts that are necessary to allow refolding of misfolded intermediates can be slow enough to limit the overall rate of folding.3,4
Since the discovery of self-splicing more than 25 years ago,5 group I intron RNAs have served as powerful models for studies of RNA folding, structure, and function.6 Particularly valuable have been the intron from Tetrahymena thermophila and its ‘ribozyme’ derivative, which lacks exons and, instead of self-processing, cleaves an oligonucleotide substrate in a guanosine-dependent reaction that mimics the first step of self-splicing.7 The structure consists of two core helical domains, P4–P6 and P3–P8, which are buttressed by peripheral domains encircling the core.8–10 All group I introns share the core domains, but subgroups differ in the identities and abundance of their peripheral domains.11–15
The peripheral domains play key roles in stabilizing the functional structures of many group I introns, including the Tetrahymena intron. One peripheral structure element that is particularly well-studied is P5abc. From comparisons of the wild-type ribozyme with a deletion variant that lacks P5abc (EΔP5abc), it has been shown that P5abc greatly stabilizes the folded structure relative to partially folded intermediates and enhances catalytic activity. 16–18 P5abc also stabilizes the native state relative to a long-lived misfolded conformer that accumulates during Mg2+-induced folding of both the wild-type and EΔP5abc ribozymes19–21 and remains substantially populated at equilibrium in the absence of P5abc.22 Interestingly, although peripheral structure specifically stabilizes the native state, the misfolded conformer appears to be essentially identical to the native state around the periphery, differing locally only within the core in what is suggested to be topological change.23 P5abc can bind the core as a separate molecule,16 and it shifts the equilibrium to strongly favor the native state (Keq = 70,000).22
On the other hand, the presence of P5abc hinders the kinetics of folding, as the misfolded EΔP5abc ribozyme refolds to the native state 100-fold faster than the wild-type ribozyme.21 The folding transition requires extensive unfolding and is greatly accelerated by mutations that abolish native peripheral contacts, including those of P5abc.23 P5abc forms two tertiary contacts to the P4–P6 domain, which packs against P5abc, and a third to the peripheral L2 loop, termed P14. The acceleration of refolding upon deletion of these tertiary contacts suggested that the peripheral ring can form in the context of the misfolded core, and it must be disrupted to allow the core to rearrange to the correct topology. It has been suggested that, while peripheral contacts can stabilize functional RNA structures, they may also increase the likelihood and severity of misfolded intermediates by forming incorrectly or at the wrong time.21
An alternative strategy employed by many group I introns is to stabilize functional structures by interacting with protein cofactors.24 Some of these proteins are encoded within their cognate introns, while others are cellular proteins that have other functions and have apparently been co-opted through evolution.25–28 The Neurospora crassa CYT-18 protein is a prototypical member of this latter group, functioning both as a tyrosyl-tRNA synthetase and a cofactor for several mitochondrial group I introns.25,29 The involvement of CYT-18 in group I intron splicing is thought to have arisen recently in evolution, because functional intron binding depends on insertions in CYT-18 that are present only in one subphylum of fungi.30 These insertions form a critical part of an RNA binding surface that is distinct from the tRNA binding site and allows a CYT-18 dimer to bind to the group I intron core along the face of P4–P6, in roughly the same position occupied in other introns by P5abc.31–35 Importantly, a level of functional equivalency of P5abc and CYT-18 has been demonstrated by the finding that CYT-18 gives substantial rescue of catalytic activity of the EΔP5abc Tetrahymena ribozyme.36
Here, we investigate the extent to which CYT-18 replaces P5abc in folding stability and kinetics of the EΔP5abc ribozyme. We find that CYT-18 stabilizes the native state, in agreement with earlier results, and that it specifically recognizes the native state in preference to the long-lived misfolded conformation. Unlike P5abc, CYT-18 does not slow refolding of the misfolded conformer to the native state. We suggest that this difference in behavior arises from a major difference in the nature of the interactions; whereas P5abc completes a ring of peripheral contacts that encircles the core, CYT-18 forms more extensive contacts directly with the core, and its contacts with the misfolded core can apparently be maintained during refolding. The results have implications for the functions of CYT-18 in vivo and, more generally, for how proteins may evolve to specifically stabilize functional RNA structures.
RESULTS
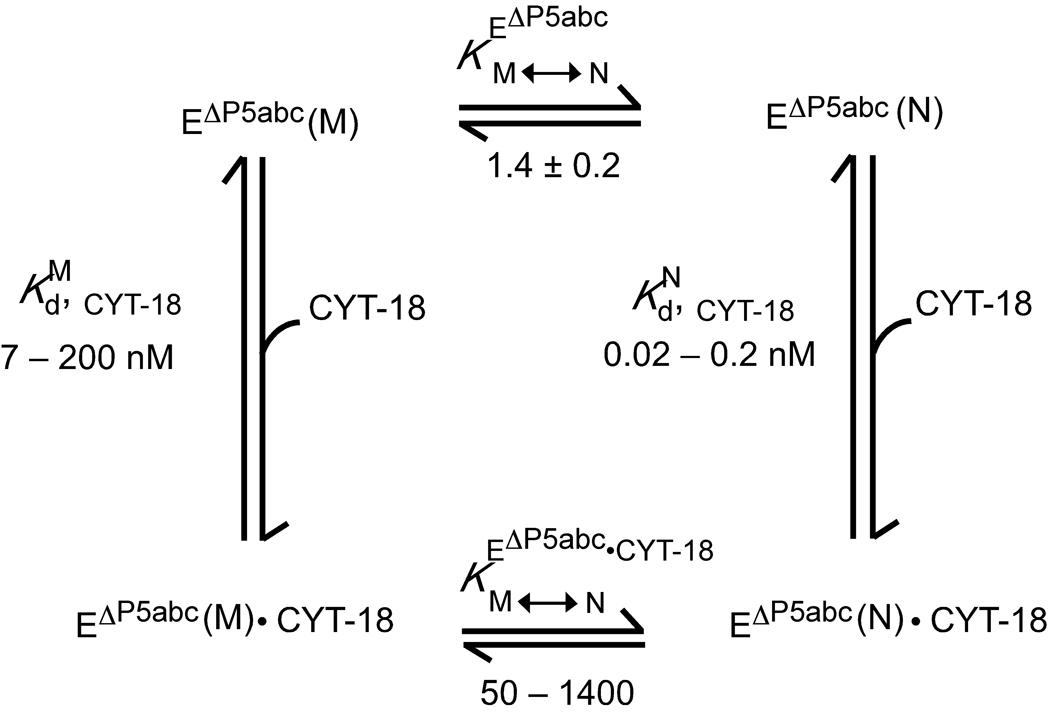
In the sections below, a thermodynamic cycle formalism is used to analyze CYT-18 stabilization of the native structure of the EΔP5abc ribozyme relative to the long-lived misfolded structure (Fig. 1). For the ribozyme alone, the native and misfolded conformations are approximately equal in stability (, top of Fig. 1),22 and we first describe catalytic activity assays that show directly that CYT-18 increases the fraction of native ribozyme by stabilizing it relative to the misfolded form. From Fig. 1 it can be seen that the magnitude of the stabilization conferred by CYT-18 is equal to the difference in its binding affinities for the native and misfolded forms. Therefore, we next describe binding assays and DMS footprinting to confirm that CYT-18 binds preferentially to the native ribozyme core and to probe the magnitude of the difference. In subsequent sections, we probe effects of CYT-18 on the kinetics of folding from the misfolded intermediate to the native state.
Fig. 1.
Thermodynamic cycle depicting binding of CYT-18 to native (N) and misfolded (M) EΔP5abc ribozyme and resulting stabilization of the native state. The equilibrium value of 1.4 between the native and misfolded forms (, top of cycle) was determined previously.22 The equilibrium dissociation constant for binding of the CYT-18 dimer to the native ribozyme (, right) was calculated from kinetics measurements to be 0.2 nM. The range shown arises from the possibility that monomerization of CYT-18 caused the binding rate constant to be underestimated by up to 10-fold. Activity and binding measurements show that CYT-18 binds 35 – 1000-fold weaker to the misfolded ribozyme, leading to the range shown for (calculated from the measured value of for simplicity). The difference between and leads to the calculated range of 50 – 1400 for the equilibrium of the CYT-18 complex between the native and misfolded conformations (, bottom).
CYT-18-mediated stabilization of the native state detected by RNA catalytic activity assay
We used a ribozyme activity assay to determine whether binding of CYT-18 shifts the equilibrium of the EΔP5abc ribozyme toward the native state. Thus, we incubated EΔP5abc to generate an equilibrium mixture of the native and misfolded species.22 Then we added CYT-18 and monitored the fraction of native ribozyme over time by measuring the fraction of trace labeled substrate (S*), added subsequently, that was cleaved in a rapid burst (Fig. 2A; refs. 20–23).
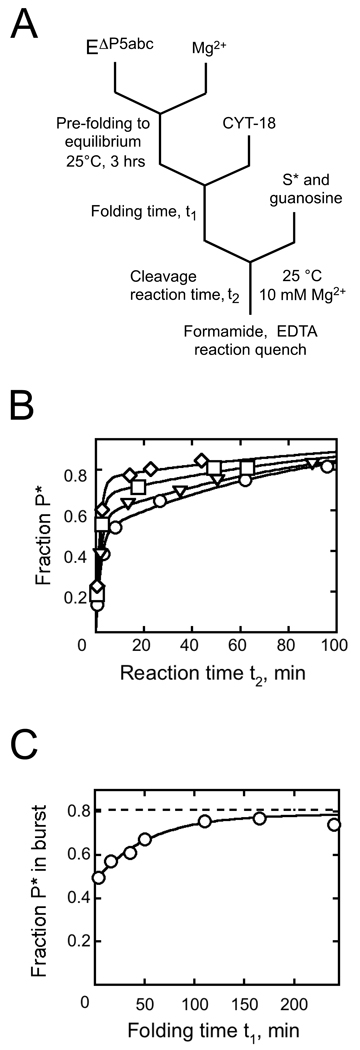
Fig. 2.
Native state stabilization by CYT-18 measured by ribozyme activity. A, Schematic of reaction procedure. B, Substrate cleavage reactions to give oligonucleotide product (P*) after incubation during t1 of 200 nM EΔP5abc with 500 nM CYT-18 for: 3.6 min (circles), 16 min (triangles), 110 min (squares), and 210 min (diamonds). C, The fraction of S* cleaved rapidly to P* (bursts from panel B) plotted against CYT-18 incubation time. Immediately after CYT-18 addition, the fraction of substrate cleaved was 0.48 ± 0.02. The fraction of misfolded ribozyme is inferred from the difference between this value and the maximum burst amplitude after extended incubations of the same ribozyme preparation with P5abc at elevated temperature (0.81 ± 0.02, dashed line; typically 30 min at 50 °C). The ratio of native to misfolded ribozyme gives an equilibrium constant of 1.5 ± 0.1 for folding of the EΔP5abc ribozyme, in good agreement with the value of 1.4 reported previously.22 At later times, the fraction of native ribozyme increased, with kobs = 0.019 ± 0.003 min−1, to a value indistinguishable from that obtained by incubation with P5abc, indicating that CYT-18 shifts the equilibrium to a value of at least 50 (see Results).
For each incubation time with CYT-18 we collected a full time course of the cleavage reaction to unambiguously determine the fraction of native ribozyme over time (Fig. 2B). The substrate cleavage reaction with bound CYT-18 was slower than with P5abc (~0.5 min−1 compared to ~10 min−1). Nevertheless, cleavage was much faster than dissociation of S* and refolding of misfolded ribozyme, such that the amplitude of the cleavage burst provided a good measure of the fraction of native ribozyme (see Materials and Methods). After addition of CYT-18, the fraction of S* that was rapidly cleaved gave a clear time-dependent increase (Fig. 2C). By this measure, the fraction of native ribozyme reached the maximum obtained with P5abc, which indicates the absence of significant misfolded ribozyme (although not all of the substrate is cleaved rapidly because of a small, preparation-specific fraction of ribozyme that is inactive, most likely from damage).22
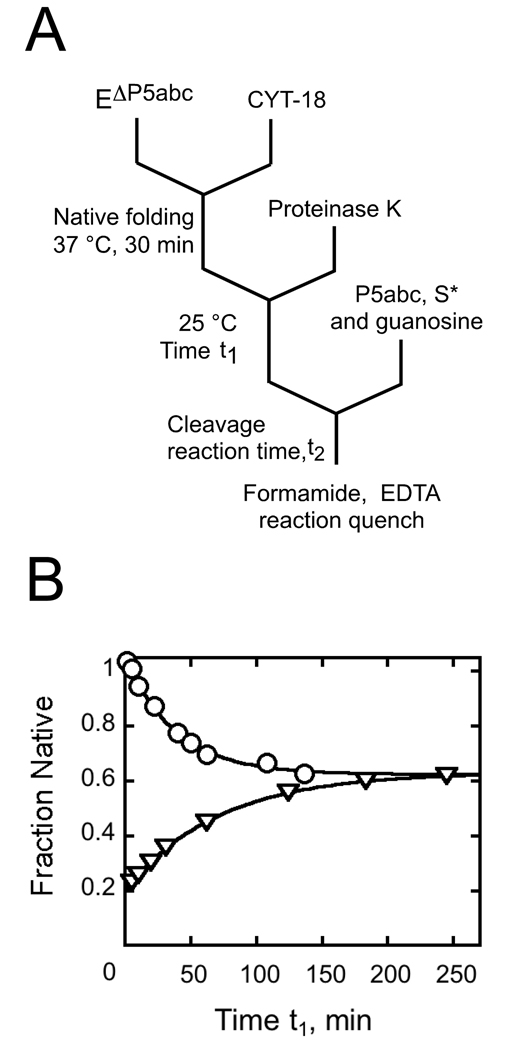
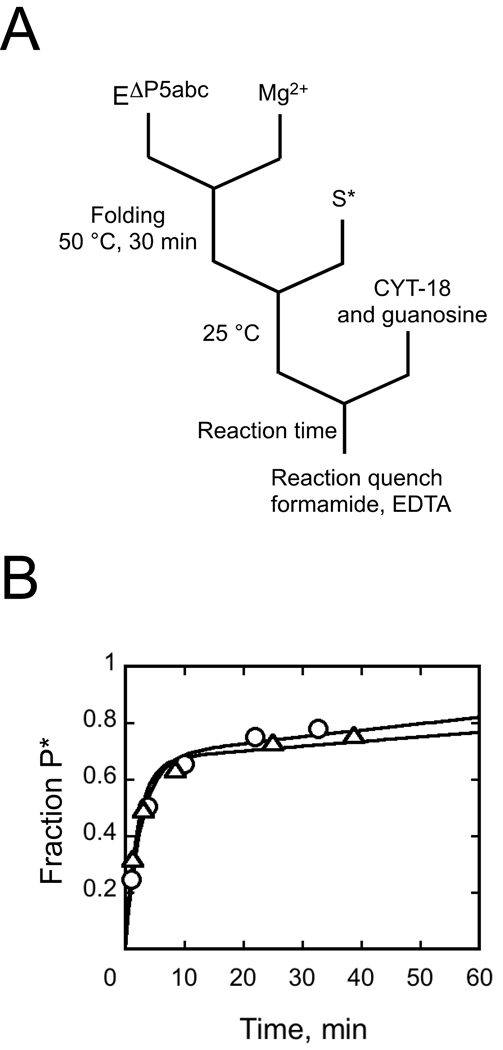
To confirm that CYT-18 shifted the equilibrium toward the native species, we used the same assay to monitor the re-appearance of misfolded ribozyme after incubation with CYT-18 and subsequent inactivation by proteolysis (Fig. 3A). As expected, the fraction of native ribozyme decreased after removal of CYT-18, reaching the same endpoint as a folding reaction in the absence of CYT-18 (Fig. 3B). These two reactions also gave the same rate constant within error, as expected for a two-state equilibrium between the native and misfolded conformations.
Fig. 3.
Return to equilibrium after proteolysis of CYT-18. A, reaction schematic. After allowing formation of native ribozyme by incubating 200 nM EΔP5abc with 500 nM CYT-18, Proteinase K was added (1 mg/ml) to inactivate CYT-18. At various times thereafter (t1), the fraction of native ribozyme was determined by activity. P5abc was added to activate the ribozyme for substrate cleavage without allowing significant redistribution of the native and misfolded forms. B, Upon proteolysis of CYT-18 (circles), the fraction of native ribozyme decreased from a value reflecting essentially 100% native ribozyme (0.86 P* in a cleavage burst) to a value reflecting the expected equilibrium between the native and misfolded species (a burst amplitude of 0.51 ± 0.01, giving an equilibrium constant of 1.5). The decrease gave a rate constant of 0.024 ± 0.003 min−1. A control reaction was performed (triangles) in which EΔP5abc (200 nM) was incubated with 10 mM Mg2+ and the approach to equilibrium was monitored. This reaction gave an identical endpoint (0.52 P*) and rate constant (0.022 min−1), consistent with previous results.22 The plot shows the fraction of functional ribozyme that is present in the native state, obtained by dividing the measured burst amplitude by the maximum level obtained by incubation with P5abc.
Together, these results show that CYT-18 stabilizes the native state of the ribozyme relative to the misfolded form. While it has been shown previously that CYT-18 enhances splicing of group I introns including the P5abc-deleted Tetrahymena intron,36 the increase in the fraction of native ribozyme here indicates that CYT-18 recognizes the native core in preference to the misfolded core (see Fig. 1). The stabilization from this binding preference is substantial, as the misfolded ribozyme is undetectable after extended incubation with CYT-18. Thus, we estimate that CYT-18 generates an equilibrium value of at least 50 (based on 1–2% uncertainty in the amplitude of the cleavage burst), reflecting a stabilization of ≥ 35-fold from the equilibrium value of 1.4 in its absence (≥ 2.1 kcal/mol).
Tight and specific binding of CYT-18 to the native EΔP5abc ribozyme
To determine the equilibrium constants for CYT-18 binding to the native and misfolded ribozyme, we set out to measure the kinetics of binding and dissociation. It is not possible to measure the equilibrium constants directly because CYT-18 binds to the native core of group I introns tightly,29,37 and at the low concentrations of CYT-18 that would be required, the protein could dissociate from functional dimers into monomers.37 Further, the native and misfolded EΔP5abc ribozyme interconvert on the same time scale as equilibrium is reached with subsaturating CYT-18 (data herein), preventing measurement of binding to each conformer separately.
To measure the kinetics of dissociation, we used a pulse-chase procedure, trapping complexes of CYT-18 and radiolabeled EΔP5abc by nitrocellulose filter binding. With an initial pre-incubation to generate largely native ribozyme,22 the major phase of dissociation was easily followed (0.024 ± 0.018 min−1, Fig. 4). This process was considerably faster than dissociation of P5abc (10−6 min−1, ref. 22), moderately faster than dissociation of CYT-18 from its cognate introns (<10−4 – 10−3 min−1, refs. 29,37), and about 5-fold faster than CYT-18 dissociation from the non-cognate yeast intron bI5.38 Nevertheless, the half-life of ~25 minutes indicates a relatively stable complex of CYT-18 and the native EΔP5abc ribozyme.
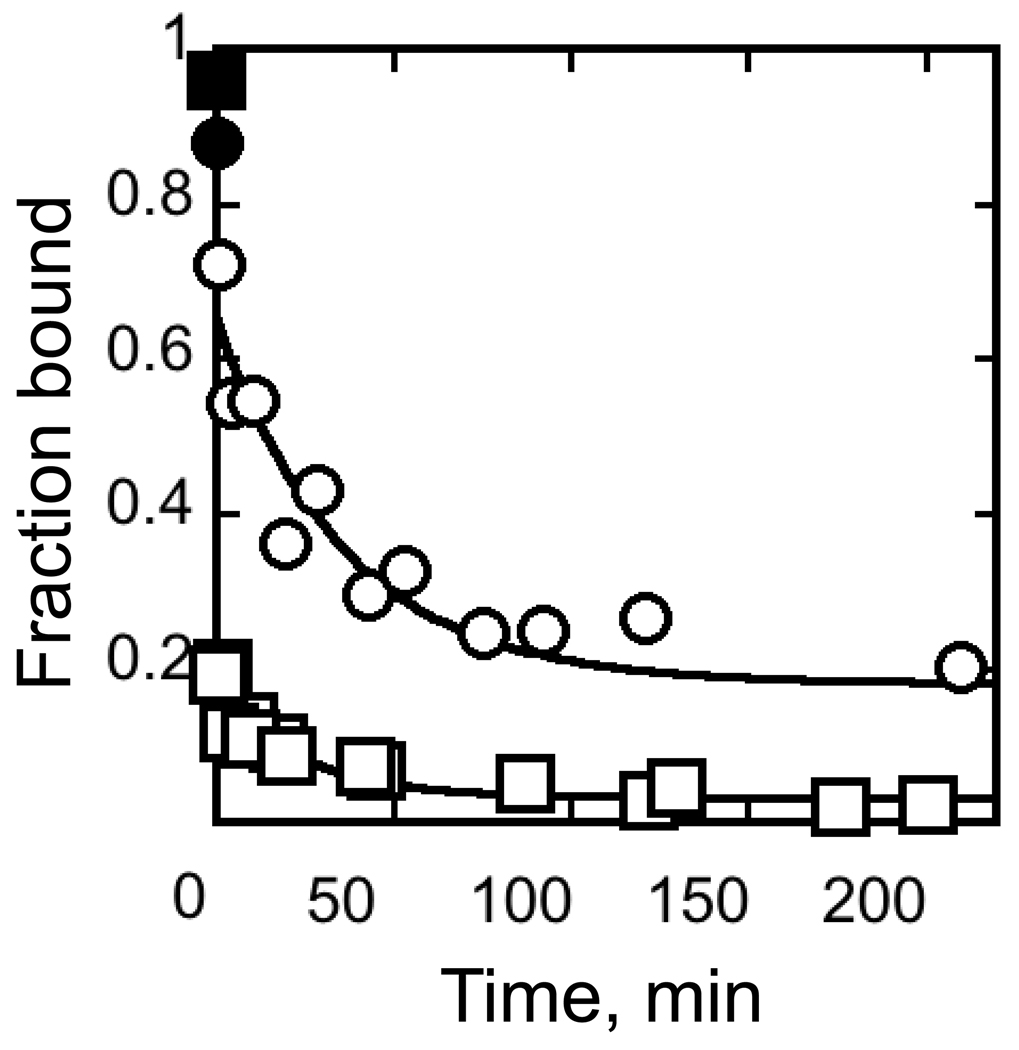
Fig. 4.
CYT-18 dissociation kinetics. CYT-18 (200 nM) was incubated with trace radiolabeled EΔP5abc ribozyme (~3 nM) to give populations of largely native or misfolded ribozyme (circles and squares, respectively; see Methods). Complex dissociation was then followed upon addition of excess unlabeled EΔP5abc (2 µM). Filled symbols show results without unlabeled ribozyme (again, circles and squares represent native and misfolded ribozyme, respectively). For the reaction with native ribozyme, the major phase gave a rate constant of 0.024 ± 0.018 min−1. The endpoint, 0.16 ± 0.05, is in the range expected from the 10-fold excess of unlabeled ribozyme, approximately 60% of which is expected to be native (with most of the remainder misfolded). In experiments monitoring dissociation from misfolded ribozyme, most of the labeled RNA dissociated before the first time point. A minor phase gave a rate constant of 0.042 ± 0.034 with an amplitude of ~0.1. The endpoint, 0.02 ± 0.018, is lower than in reactions with the native ribozyme, despite the same 10-fold excess of unlabeled ribozyme chase, presumably because the labeled misfolded ribozyme competes poorly with the native ribozyme in the chase.
In an analogous experiment with a population of ribozyme that was incubated only briefly with Mg2+ to generate predominantly misfolded ribozyme, the major phase was much more rapid, reaching completion before the first time point (Fig. 4). The observed minor phase gave a rate constant similar to that above (0.042 ± 0.034 min−1), suggesting that it reflected CYT-18 dissociation from the small population of native ribozyme expected under these conditions (~10%, refs. 21,22). Completion of the major phase before the first time point (30 s) indicates a rate constant of ≥ 3 min−1 for dissociation of CYT-18 from the misfolded ribozyme. Increasing Mg2+ concentration to 35 mM or lowering temperature to 5 °C gave modest slowing of CYT-18 dissociation from the native ribozyme (2–3-fold, data not shown) but did not slow dissociation from the misfolded ribozyme to measurable rates, suggesting that dissociation under standard conditions is considerably faster than the lower limit of 3 min−1. CYT-18 does appear to bind the misfolded ribozyme in the ground state under the conditions used, because the level of retention in the absence of chase was substantially greater than the first time point after chase addition (Fig. 4). This result does not reflect CYT-18 free in solution ‘catching’ the ribozyme after being trapped on the filter itself, because pre-spotted CYT-18 gave substantially lower retention of labeled ribozyme applied immediately thereafter (data not shown).
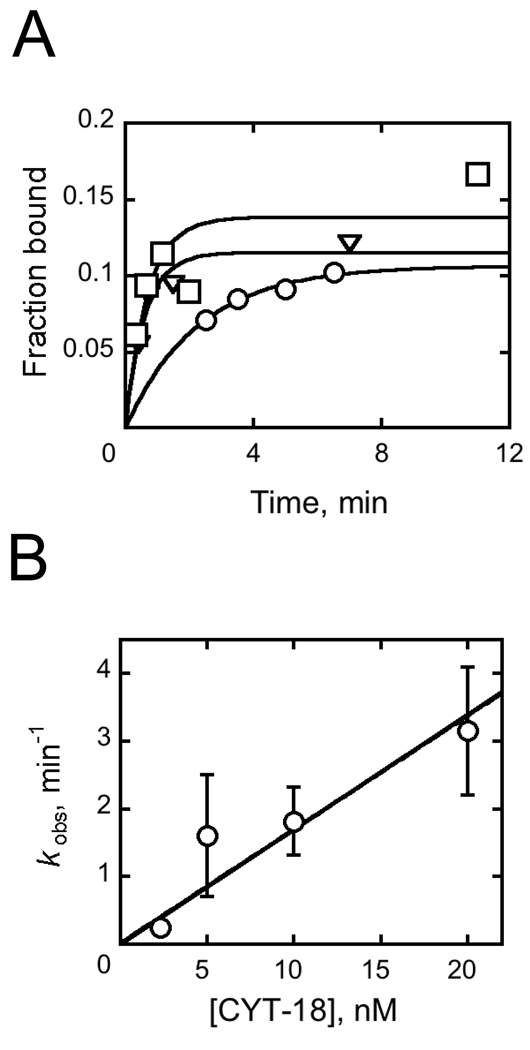
We took advantage of the difference in dissociation kinetics to follow CYT-18 binding to the native ribozyme. We added CYT-18 to an equilibrium mixture of native and misfolded radiolabeled EΔP5abc ribozyme and incubated for various times before adding excess unlabeled ribozyme and applying the solution to a filter. Because the complex formed between CYT-18 and the misfolded ribozyme is expected to dissociate rapidly upon addition of chase, the experiment monitored CYT-18 binding to the native ribozyme. Across a limited range of CYT-18 concentration (5 – 20 nM), the binding process was observable and gave a rate constant of (1.6 ± 0.2) × 108 M−1 min−1 (Fig. 5), ~10-fold lower than for CYT-18 binding to its cognate group I introns and to the yeast bI5 intron.29,37,38 It was not possible to follow CYT-18 binding to the misfolded ribozyme with these methods because the rapid dissociation sets a lower limit on the observed rate constant (kobs = koff + kon [CYT-18]) that is too large to observe by hand.
Fig. 5.
Kinetics of EΔP5abc ribozyme binding by CYT-18. A, Progress curves for radiolabeled EΔP5abc binding to CYT-18 at concentrations of 5 nM (circles), 10 nM (triangles), and 20 nM (squares) CYT-18. Aliquots were quenched with 1 µM unlabeled EΔP5abc ribozyme and then applied to filters. B, Rate constant for CYT-18 binding plotted against CYT-18 concentration. Error bars show standard deviations from two to three replicate measurements.
Weaker binding of CYT-18 than expected from kinetics measurements
In the binding kinetics measurements (Fig. 5A), the final extents of RNA retention were much lower than expected from the equilibrium dissociation constant calculated from koff/kon. The calculated dissociation constant is ~ 0.2 nM, but concentrations of CYT-18 as high as 20 nM gave incomplete binding. Similar observations have been reported for CYT-18 binding to the cognate intron ND129 and for the non-cognate intron bI5.38 The incomplete binding suggested one or more of the following: 1) CYT-18 is largely inactive for group I intron binding, such that the concentrations of active protein are much lower than the total concentrations; 2) a relatively weak complex is formed rapidly and rearranges slowly to a tighter complex;38,39 or 3) CYT-18 dimers dissociate into monomers at low nanomolar concentrations, generating a coupled equilibrium between dimerization and RNA binding and therefore weaker binding than expected.
To test the possibility of inactive CYT-18 in our preparations, we varied the ratio of CYT-18 to ribozyme to determine a stoichiometry (Fig. 6). At each concentration, the fraction of bound RNA was determined by catalytic activity as above. Because one CYT-18 dimer binds and activates one ribozyme molecule,30,37 a greater CYT-18 requirement would suggest the presence of inactive protein. With 100 nM ribozyme, we obtained a stoichiometry of approximately one RNA per CYT-18 dimer, indicating that most of the protein was active. At low ribozyme concentration (1 nM), activation was incomplete even with 10-fold excess CYT-18 (data not shown), confirming the results of Fig. 5 that indicate incomplete binding at these concentrations.
Fig. 6.
Titration of ribozyme with CYT-18. EΔP5abc ribozyme (100 nM) was pre-folded and bound to trace S*, and various concentrations of CYT-18 were added. After a 15 min incubation, the fraction of active ribozyme was determined by measuring the fraction of S* cleaved rapidly upon addition of guanosine. Full activity was observed with ~110 nM CYT-18, as indicated by the intersection of the rising and flat portions of the concentration dependence.
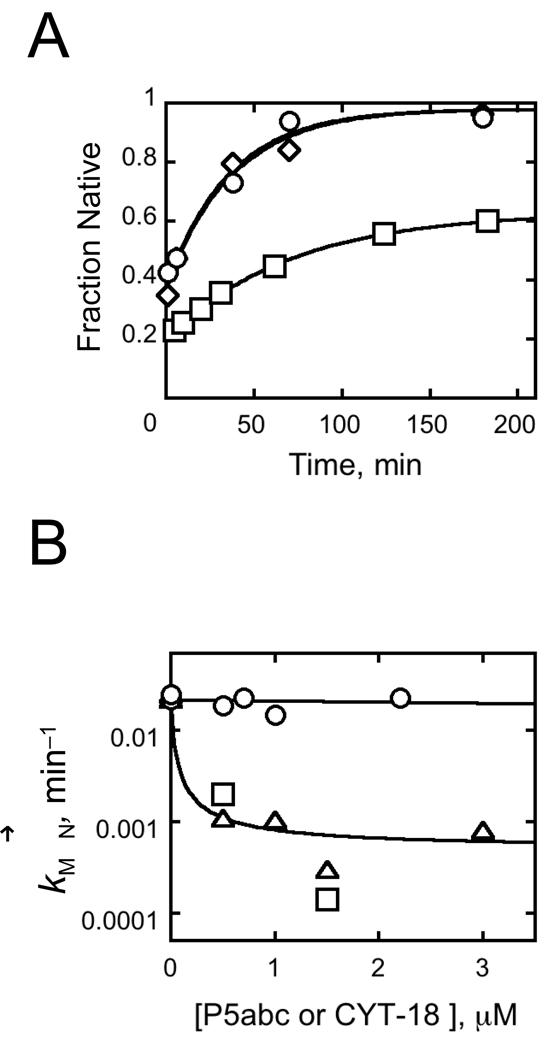
We explored the second possibility, a slow transition from an intermediate to a fully-assembled RNP complex, by adding CYT-18 to the pre-folded, native EΔP5abc ribozyme and measuring substrate cleavage. If a conformational transition were slower than the cleavage step, the observed cleavage reaction would be slower when initiated by CYT-18 than when CYT-18 was pre-incubated with the ribozyme. In contrast to this expectation, the rate constant was the same, 0.4 min−1, whether the reaction was initiated by CYT-18 addition or by addition of guanosine cofactor with CYT-18 pre-bound (Fig. 7). Although CYT-18 binding to this RNA almost certainly involves intermediates,31,37 formation of the functional complex apparently does not involve intermediates that persist on the experimental time scale, ruling out the possibility of an intermediate along this pathway causing the difference between the calculated value and the direct binding measurements.† This result leaves open the possibility of a slow transition to a tight but inactive complex, which could also cause the difference. However, such a model is ruled out by other experiments because it would predict that the fraction of active complex would decrease over time after CYT-18 addition, which is not observed (see Fig. 10 below).
Fig. 7.
Slow conformational changes are not required upon CYT-18 binding to native EΔP5abc ribozyme. A, Reaction schematic. Cleavage reactions were initiated either by adding CYT-18 to pre-folded ribozyme with bound substrate, as shown, or by adding guanosine after a 15 min preincubation with CYT-18. B, Progress curves for reactions initiated by addition of CYT-18 (triangles) or guanosine (circles). The progress curves were the same within error, with rate constants of 0.47 ± 0.07 min−1 and 0.37 ± 0.06 min−1 respectively, and amplitudes of 0.67.
Fig. 10.
CYT-18 does not slow refolding of misfolded ribozyme. A, Time courses of native ribozyme accumulation. After a brief incubation of 200 nM EΔP5abc with 10 mM Mg2+ to allow misfolding, CYT-18 was added (700 nM, circles or 2.2 µM, triangles) and the fraction of native ribozyme was determined by activity at various times thereafter. Values are normalized as described in the Fig. 3 legend. Both reactions gave rate constants of 0.023 min−1. The squares show an equivalent reaction in the absence of CYT-18 (0.015 min−1). B, Refolding rate constants from panel A and additional experiments are plotted against CYT-18 concentration (circles). Results from reactions in which P5abc RNA was added instead of CYT-18 are also shown. Squares show reactions under identical conditions as those with CYT-18 (50 mM Na-MOPS, pH 7.0, 10 mM MgCl2, plus 2.5 mM Tris-Cl, 1% glycerol, and 10 mM KCl), and triangles show reactions under standard RNA folding conditions (50 mM Na-MOPS, pH 7.0, 10 mM Mg2+).
Thus, we are left with the possibility that binding is weakened because of dissociation of CYT-18 dimers to monomers. Simulations showed that the binding data in Fig. 5 and additional analogous experiments are well-described by a model in which CYT-18 dimerizes in solution in the low µM range and the dimer binds tightly to the ribozyme with the measured Kd of 0.2 nM (data not shown).40 To our knowledge, the CYT-18 dimerization constant has not been measured under the conditions of our experiments, but an upper limit of 70 nM was established under higher ionic strength conditions,37 and a related bacterial synthetase dimerizes in the pM range.41 Nevertheless, dimerization is highly sensitive to the details of the interface, as a single point mutation of the bacterial synthetase increases the dimerization constant to 30 µM,41 leaving open the possibility of monomers under the conditions of our binding experiments. Monomerization in solution would also lead to an underestimated binding rate constant, perhaps explaining why the binding rate constant we measured (Fig. 5) is lower than previous results at higher ionic strength with other RNAs.29,37,38 Thus, a pre-formed CYT-18 dimer may bind the ribozyme up to 10-fold faster than indicated by our measured value, approaching the diffusion limit for molecules of this size and complexity.42 This uncertainty is reflected in Fig. 1 as a range for the value of but does not affect the conclusions.
Taken together, results in the preceding sections indicate that CYT-18 binds the native ribozyme core in strong preference to the misfolded core (see Fig. 1). Activity measurements show directly that CYT-18 binds at least 35-fold tighter to the native core. Further, CYT-18 dissociates at least 100-fold faster from the misfolded RNA, suggesting a larger difference than the lower limit established by activity. The affinity difference is unlikely to be more than approximately 1000-fold, because the misfolded RNA is bound by CYT-18 with a Kd of ≤100 nM (Fig. 4). Thus, CYT-18 provides considerable specificity, stabilizing the native state by 2 – 4 kcal/mol relative to the misfolded intermediate.
CYT-18 forms a similar interface with the native and misfolded conformations
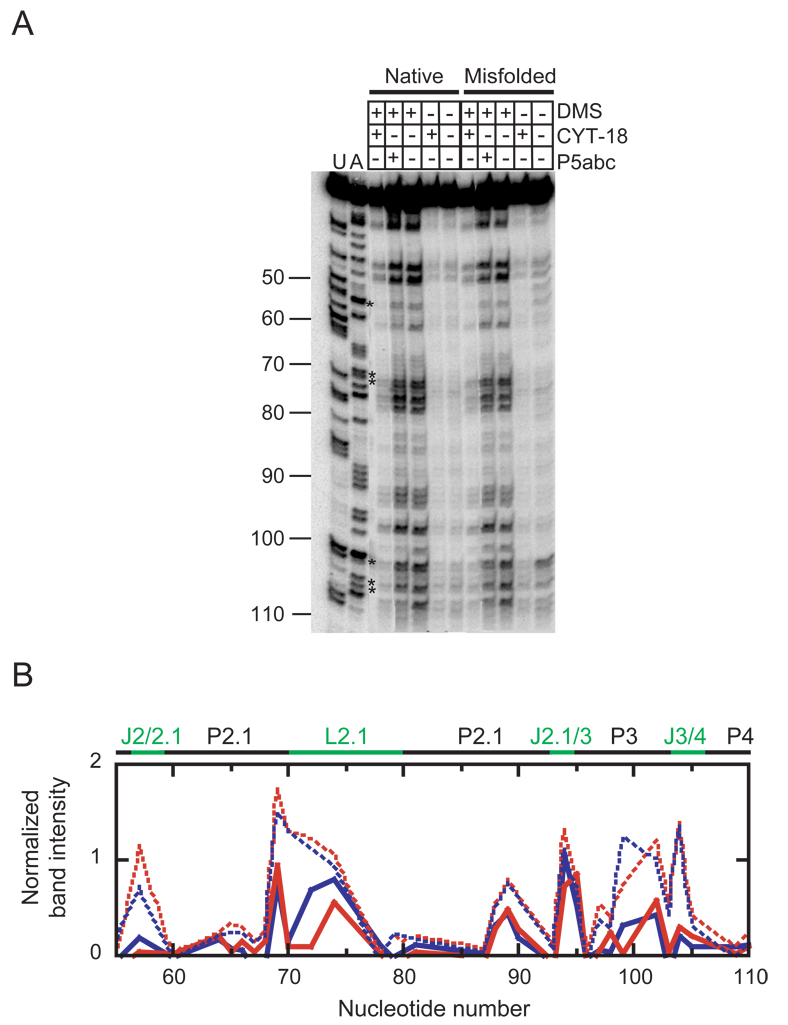
The reduced affinity of CYT-18 for the misfolded ribozyme could reflect binding along the natural P4–P6 interface, but less tightly because of small structural differences relative to the native ribozyme, or it could reflect non-specific binding at other sites.36 To distinguish between these possibilities, we performed footprinting of the native and misfolded forms and their complexes with CYT-18 using dimethyl sulfate (DMS) and reverse transcription, a method that had previously been shown to give a robust signal for CYT-18 binding to the native RNA.36 As expected, CYT-18 gave strong protections of the native ribozyme at certain A and C nucleotides within P4–P6 (A114, A196, A214, A218, A219, A226, A248, C255), and most of these protections also appeared upon addition of P5abc RNA (Fig. 8, Fig. S1, and Fig. S2).18,21 CYT-18 also gave localized changes in other regions including J2/2.1, P2.1, P3, J8/7, P9, and P9.1.
Fig. 8.
DMS footprinting of the native and misfolded ribozyme complexes with CYT-18. A, Gel image of the 5´-region of the ribozyme. B, Quantitation of the region shown in panel A. Data from the native and misfolded ribozyme in complex with CYT-18 are shown as solid blue and red lines, respectively, and data from mixtures of predominantly native and predominantly misfolded ribozyme are shown as dashed blue and red lines, respectively. Asterisks indicate nucleotides that were protected by CYT-18 in both complexes. To correct for differences in loading and for a consistent decrease in labeled material loaded from reactions that included CYT-18, data are shown relative to the fully-extended product and rescaled for simplicity by multiplying these values by 103.
Strikingly, the CYT-18-induced changes in the native ribozyme were largely mirrored with the misfolded ribozyme. All of the strong protections within P4–P6 listed above were also present in the complex with the misfolded ribozyme. Further, all 15 of the most significant protections in other regions of the RNA were also observed in the misfolded complex.
On the other hand, there were also localized differences between the complexes. Within P4–P6, A210 was modestly protected in the native complex but significantly enhanced for modification in the misfolded complex, indicating a local difference in the structural effects of CYT-18. Within J7/3, on the opposite side of the RNA from P4–P6, A269 was significantly enhanced in the native complex but not the misfolded complex. This result was also obtained for P5abc binding,21 suggesting that the exposure of A269 results from a conformational transition specific to the functional structure. There were smaller but significant differences in P9 and P9.1, where protections were generally stronger in the native complex. Last, there were striking local differences in J6/7 (C260, A261, and A262), where the significant protections and enhancements were observed in the misfolded complex with CYT-18 but not in the native complex. Taken together, the results indicate that CYT-18 interacts with the misfolded ribozyme using the same binding interface and forming a globally-similar structure as with the native ribozyme, but with local structural differences that may provide clues as to the physical origin of the preferential binding to the native ribozyme (see Discussion and Fig. 11).
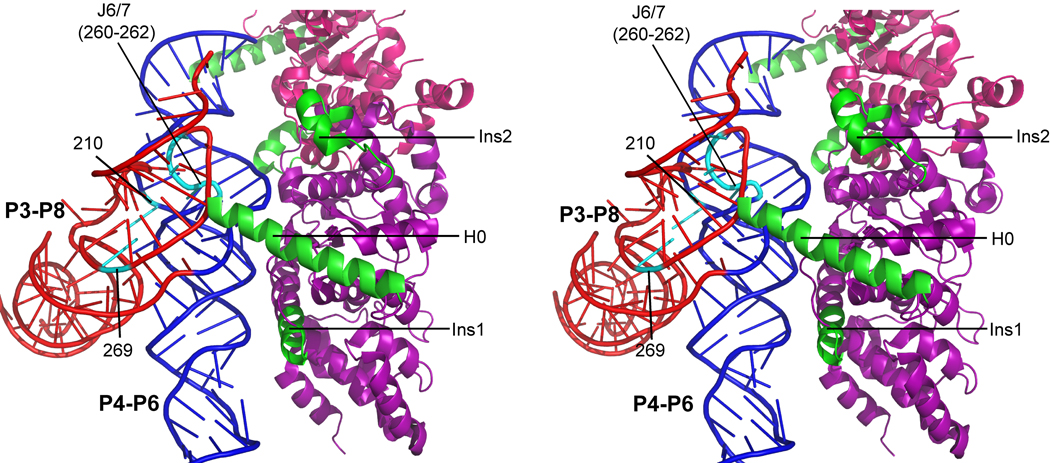
Fig. 11.
Model of CYT-18 in complex with the native Tetrahymena ribozyme core. The P4–P6 domain is blue and P3–P8 is red. Nucleotides that differed in reactivity to DMS between the native and misfolded complexes are shown in cyan and labeled. The subunit of CYT-18 that makes extensive contacts with the core is shown in purple, with inserted regions in green. The other CYT-18 subunit (only partially visible) is magenta. The model was generated by superimposing the structure of the Tetrahymena ribozyme core14 on the Twort intron complex with CYT-18 (see Methods).35
CYT-18 stabilizes native-like conformations relative to less structured conformations
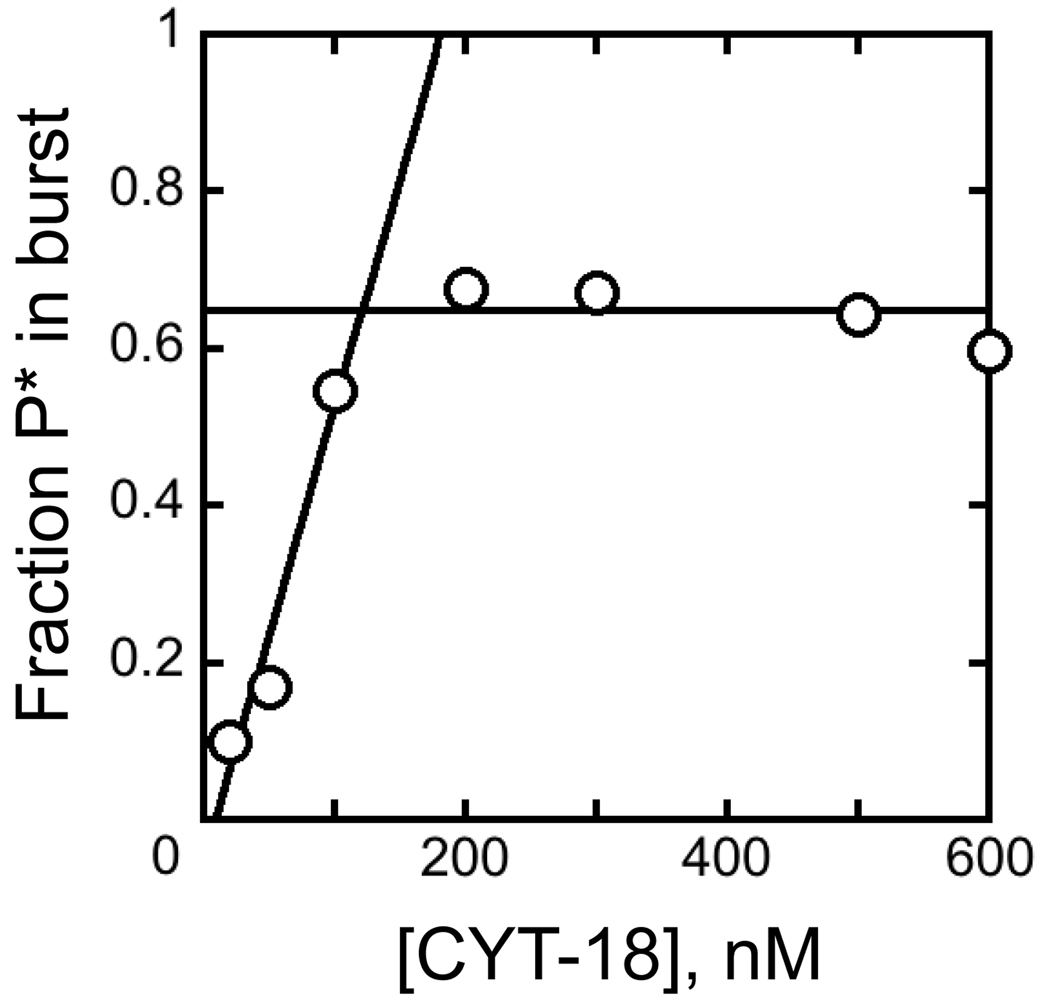
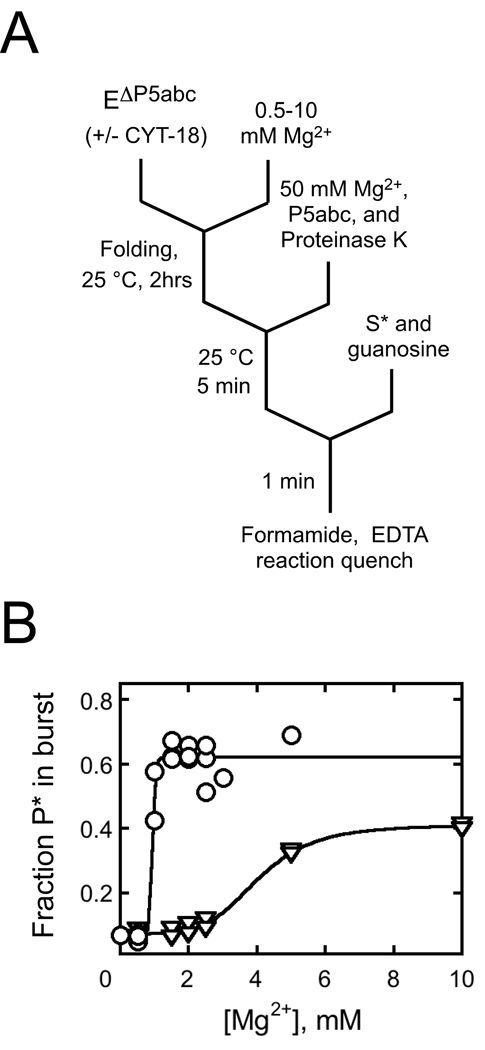
To probe the ability of CYT-18 to favor formation of the native ribozyme in preference to folding intermediates more broadly, we used a previously-developed activity assay.21 In the presence or absence of CYT-18, we varied the Mg2+ concentration across the range required for stable formation of the native state and global structure formation as measured previously by hydroxyl radical footprinting.21,22 After an extended incubation with or without CYT-18, we raised the Mg2+ concentration to support the ribozyme’s catalytic activity and used this activity to determine the fraction of native ribozyme. In the absence of CYT-18, most of the RNA that is non-native during the low Mg2+ incubation misfolds when the Mg2+ concentration is raised, so this method measures the fraction of native ribozyme at the initial Mg2+ concentration.20,21
The presence of CYT-18 substantially reduced the Mg2+ concentration requirement, decreasing the K1/2 value from 4 mM to 0.9 mM Mg2+ (Fig. 9). This result suggests that at low Mg2+ concentrations, CYT-18 stabilizes the native structure relative to the more extended forms that are populated in its absence. It has been shown previously that EΔP5abc ribozyme lacks global tertiary structure at low Mg2+ concentrations,17,21,22 and CYT-18 induces structure formation, as monitored by footprinting, and activates self-splicing of this and other introns.26,36 Thus, the finding here supports and refines the earlier conclusion that CYT-18 enhances splicing by stabilizing native structure.36 Alternatively or in addition, it is possible that CYT-18 favors folding intermediates that subsequently avoid misfolding upon addition of increased Mg2+, an activity that could contribute to group I intron folding in vivo (see Discussion). Such a stabilizing interaction would apparently require greater than 0.5 mM Mg2+ under these conditions and CYT-18 concentration, because reactions with lower Mg2+ concentrations gave only 10% native ribozyme upon subsequent folding, the same value as in the absence of CYT-18.
Fig. 9.
CYT-18 stabilizes the native state relative to less structured folding intermediates. A, reaction schematic. EΔP5abc (200 nM) was incubated at various Mg2+ concentrations to achieve equilibrium between folded and unfolded forms, and then additional Mg2+ was added to trap folding intermediates that subsequently misfolded and to allow determination of the fraction of native ribozyme by activity. B, Data from reactions in the absence (triangles) or presence (circles) of 600 nM CYT-18. The data indicated a cooperative transition, giving K1/2 values of 4 mM Mg2+ and 0.9 mM Mg2+ in the absence and presence of CYT-18, respectively, with Hill coefficients of 5 and >10, respectively. The higher endpoint in the presence of CYT-18 reflects stabilization of the native state relative to the misfolded conformation.
CYT-18 does not slow refolding of the misfolded ribozyme
Previous work has shown that P5abc has a large inhibitory effect on the kinetics of refolding from the misfolded conformation to the native state, as the wild type ribozyme refolds ~80-fold slower than the EΔP5abc ribozyme.21 To determine whether CYT-18 also slows refolding, we added various concentrations of CYT-18 to the misfolded EΔP5abc ribozyme and used the ribozyme activity assay as above to follow native state accumulation. Strikingly, the rate of refolding was not changed by CYT-18 at concentrations up to at least 2.2 µM (Fig. 10). The endpoint increased in the presence of CYT-18, as expected, because CYT-18 shifted the equilibrium toward the native state. Although it did not affect the refolding kinetics, CYT-18 binds to the misfolded ribozyme core at these concentrations (see Fig. 4 and Fig. 8). If it were required to dissociate during refolding, CYT-18 would inhibit refolding at concentrations that support binding in the ground state. Thus, these results suggest that CYT-18 can remain bound to the ribozyme core without interfering with the refolding process. To rule out the possibility that the different behaviors of P5abc and CYT-18` arose because P5abc is covalently linked to the ribozyme but CYT-18 is not, we measured refolding of EΔP5abc with P5abc added as a separate molecule (Fig. S3). As expected, P5abc strongly inhibited refolding even when added in trans (Fig. 10B).
DISCUSSION
Using the EΔP5abc variant of the Tetrahymena ribozyme, we found that CYT-18 can discriminate between the native group I intron core and a misfolded form that is globally similar and of comparable stability. Ribozyme activity assays showed directly that CYT-18 binds more tightly to the native core than the misfolded core, with a difference of at least 35-fold, and binding experiments suggest a larger difference (100–1000-fold). Thus, by binding preferentially to the native form, CYT-18 stabilizes the native structure relative to the misfolded structure. Interestingly, this stabilization is not accompanied by a decreased rate for the folding transition from the misfolded to the native conformation, even under conditions that allow CYT-18 binding to the misfolded conformation. Apparently CYT-18 binds no more tightly to the misfolded intermediate than it does to the transition state ensemble that separates the native and misfolded conformers. This behavior contrasts with that of P5abc, which greatly stabilizes the native state but inhibits the kinetics of the transition because it also binds and stabilizes the misfolded RNA. As described below, this difference, in the context of structural considerations, suggests important differences in the mechanisms of CYT-18 and P5abc in distinguishing the native and misfolded group I intron cores.
Interactions of CYT-18 with the native and misfolded group I intron core
CYT-18 interacts extensively with the P4–P6 domain of the group I intron core, with contacts extending from P5a, at the ‘top’ of the P4–P6 stack, down to J6/6a toward the bottom.10,35,43,44 DMS footprinting in this work showed that the complexes with the native and misfolded species are globally similar, indicating that CYT-18 binds the same surface and makes crudely the same contacts. On the other hand, the local differences may provide clues as to how CYT-18 interacts preferentially with the native intron core.
Fig. 11 shows a model of CYT-18 bound to the native Tetrahymena ribozyme core that highlights nucleotides with significantly different DMS reactivity in the native and misfolded complexes. The model was generated by aligning the Tetrahymena ribozyme core14 with the Twort group I ribozyme from a co-crystal structure with CYT-1835 (see Methods). Interestingly, the nucleotides that give differences in DMS footprinting correspond approximately to the regions that form contacts with CYT-18, as well as to a subset of the regions that have previously been shown to give differences in the native and misfolded forms of the complex with P5abc21 or the full-length ribozyme.23 Most notably, the joining sequence J6/7, which connects the P4–P6 and P3–P8 domains together with J3/4, is positioned very close to the N-terminal helix termed helix H0. Helix H0 inserts directly into the core and forms intimate contacts at the interface between the two domains, perhaps binding to the native state specifically by recognizing the correct geometry in that region.23 CYT-18 has been shown previously to rescue the effects of mutations that weaken the triple helix interactions of J3/4 with the P4–P6 domain and to depend strongly on nucleotide identity within J3/4 for optimal binding,32,45–47 underscoring the importance of this region to specific CYT-18 binding. Local differences in these joining segments could also propagate to give the observed difference DMS reactivity of A210, which is just across the helix from this junction region.
Other differences in DMS footprinting suggest that other regions of CYT-18 also may contribute to the strong discrimination in binding. Within L9, the native complex is more strongly protected (Fig. S1; not shown in Fig. 11). Ins2 of CYT-18 interacts with the minor groove of P9 to stabilize the tetraloop-receptor interaction of L9 and P5. Although the L9-P5 contact likely forms in the misfolded conformation,23 it may be more strongly stabilized by CYT-18 in the native conformation. Last, differences in J2/2.1 and P2 raise the possibility that specificity for the native state is also contributed by the C-terminal domain of CYT-18, which interacts with P3 in close proximity to these regions. This domain is not necessary for CYT-18-enhanced splicing of the EΔP5abc intron,48 but it increases the affinity for the intron (G. Mohr and A. Lambowitz, personal communication).
Although P5abc also stabilizes the native state relative to the misfolded conformation, consideration of the connections of CYT-18 and P5abc to the intron core suggest that they use distinct molecular strategies to confer this stabilization. In addition to contacts with the P4–P6 helix, P5abc contacts the loop of P2 to form the peripheral contact P14, which is necessary to generate a ring of peripheral elements and connections that encircle the core (see Fig. S3).9 Formation of this ring contributes strongly to stabilization of the core and is also critical for stabilization of the native state relative to the long-lived misfolded conformation.22 In contrast, the CYT-18-dependent group I introns either do not include a P2 element or use P2 to contact P8. Thus, CYT-18 may rely to a greater extent on contacts directly with the P4–P6 helix. This difference may also underlie the ability of CYT-18 to perform this stabilization without slowing refolding of the misfolded conformation. Refolding involves substantial unfolding, as the transition is strongly accelerated by urea.21 The intimate association of CYT-18 with the P4–P6 core domain rather than with the periphery may allow unfolding of the periphery, and possibly even the P3–P8 domain, while CYT-18 retains its contacts with the P4–P6 domain.
Implications for interactions of CYT-18 with its cognate group I introns in vivo
It is striking that CYT-18 stabilizes the native state of the Tetrahymena ribozyme relative to the globally-similar misfolded conformer despite not encountering this RNA in vivo. Because all group I introns share the same core structure and topology, they likely have the potential to fold to the same misfolded structure. Indeed, at least two of the three introns that interact with CYT-18 misfold in vitro (ref. 49 and D. Mitchell and R.R., unpublished), although the structures and origins of the misfolding are not known. Thus, it is likely that CYT-18 stabilizes the native forms of its cognate introns relative to analogous misfolded structures, which could be an important part of the function of CYT-18. Below, we consider how this preferential binding may have arisen through evolution and how it may function currently.
Of the mitochondrial tyrosyl tRNA synthetases, only CYT-18 and its close relatives participate in group I intron splicing,30 indicating that this activity arose recently in evolution. At the earliest stages, the protein presumably recognized only a limited structural feature of the RNA, most likely the P4–P6 domain,31,50 and binding was presumably weak and relatively non-specific. With this model, as binding tightened through evolution, CYT-18 acquired the ability to distinguish between the native and misfolded cores. Discrimination by proteins against misfolded group I introns may be common, as two yeast proteins have been shown to stabilize the native bI3 group I intron relative to a misfolded structure with an extensive secondary structure change that accumulates in their absence.51 Discrimination against misfolded conformations with large structural differences could be imagined to arise as a by-product of selective pressure for tight binding to the native RNA, whereas discrimination against closely-related misfolded forms, such as that of the Tetrahymena ribozyme, may require explicit selective pressure against tight binding to the misfolded conformation.
As CYT-18 increased its ability to promote splicing, the group I introns that could interact productively probably continued to lose their inherent abilities to form functional structures. Indeed, the CYT-18-dependent introns that have been studied do not form stable tertiary structure until CYT-18 binds.29,31 Thus, CYT-18 has the potential to first encounter its cognate introns as folding intermediates, and these initial interactions could either help or hinder the folding process. An initial set of interactions with the P4–P6 domain31,50 could stabilize connections to the P3–P8 domain, enforcing the correct geometry and topology within this critical region and guiding folding toward the native structure.35 In this regard, some of the CYT-18-dependent increase in native ribozyme after incubation at low Mg2+ concentrations (Fig. 9) may reflect a stabilization of intermediates that avoid misfolding when the Mg2+ concentration is increased. Nevertheless, any such chaperone activity of CYT-18 in vivo is apparently insufficient to avoid misfolding altogether, because the three introns that bind CYT-18 also require the DEAD-box protein CYT-19.49 Some non-native structures may involve exons,52,53 which would most likely not be affected by CYT-18, or large changes in secondary and/or tertiary structure, rendering the intron core unrecognizable to CYT-18 and requiring a general RNA chaperone.
On the other hand, early association of CYT-18 could hinder folding if it strengthened native contacts that must be disrupted to allow resolution of non-native structure elsewhere in the RNA. Thus, it is interesting that CYT-18 allows facile refolding of the misfolded conformation to the native state, apparently binding no more tightly to the misfolded intermediate than to the less-structured transition state. In contrast, P5abc substantially slows refolding of the misfolded conformation, as well as impeding earlier folding steps.21,54 Thus, recruitment of CYT-18 may have imparted a benefit to its cognate group I introns by allowing them to minimize or delete their peripheral elements and therefore to avoid kinetic traps caused by the peripheral elements while retaining the specific stabilization of the native structure.
MATERIALS AND METHODS
Preparation and labeling of RNA
Wild-type (L-21/ScaI) and EΔP5abc ribozymes and P5abc RNA were prepared by in vitro transcription using T7 RNA polymerase7 and purified using a Qiagen RNeasy column as described previously.55 Ribozymes were 5′ end-labeled by removing 5′-phosphoryl groups with shrimp alkaline phosphatase (MBI Fermentas, Amherst, NY) and then incubating with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). RNA oligonucleotides (Dharmacon Research, Lafayette, CO) were 5′ end-labeled by using [γ-32P]ATP and T4 polynucleotide kinase.7 Labeled RNAs were purified by nondenaturing polyacrylamide gel electrophoresis (PAGE), using a 6% gel for ribozymes and a 20% gel for oligonucleotides.
CYT-18 expression and purification
CYT-18 was expressed and purified essentially as described.34,37 Briefly, CYT-18 was expressed from the plasmid pEX560 in the E. coli strain HMS174 (DE3). Cells were grown in 1 l of LB medium supplemented with 50 µg/ml carbenecillin to an OD600 of 0.6. CYT-18 expression was induced by adding IPTG (1 mM) and growing cells overnight at 25 °C. Cells were pelleted by centrifugation, resuspended in buffer A (25 mM Tris-Cl, pH 7.5, 500 mM KCl, 1 mM EDTA, 10% glycerol), and lysed by incubating with 1 mg/ml lysozyme and sonicating (5 pulses of 20 s). Cellular debris was pelleted by centrifugation (1500 × g, 30 min at 4 °C). All subsequent steps were carried out at 4 °C. Remaining nucleic acids were removed by adding polyethyleneimine (PEI) slowly from a 10% stock to 1%, followed by centrifugation. CYT-18 was precipitated by adding powdered ammonium sulfate to 40% saturation, followed by centrifugation. Ammonium sulfate precipitation was repeated two additional times to remove contaminating ribonucleases. The pellet was resuspended in 30 ml of buffer A containing 500 mM KCl and dialyzed against 1 l of buffer A containing 25 mM KCl for 1 h and then overnight against an additional liter of the same buffer. CYT-18 was further purified using a heparin-sepharose column (16/10, GE Healthcare) previously equilibrated with buffer A containing 25 mM KCl. Bound CYT-18 was eluted with a linear KCl gradient (200–1000 mM), with the bulk of CYT-18 eluting in a peak centered at approximately 400 mM KCl. Fractions containing CYT-18, as determined by SDS-PAGE, were flash-frozen in liquid nitrogen and stored at −80 °C.
Concentration determinations of RNA and CYT-18 protein
CYT-18 concentration was determined using Bradford dye reagent (BioRad, Hercules, CA) with an IgG standard curve. This measurement gave the same values within error as absorbance at 280 nm (not shown). All CYT-18 concentrations refer to the dimer. Ribozyme concentrations were determined spectrophotometrically using the following extinction coefficients (260 nm): wild-type ribozyme, 3.9 × 106 M−1 cm−1; and EΔP5abc, 3.2 × 106 M−1 cm−1.19
Ribozyme catalytic activity assays
Ribozyme activity assays were performed essentially as described previously.20,22,23 EΔP5abc ribozyme (200 nM) was first incubated with Mg2+ for 10 min at 25 °C to generate largely misfolded ribozyme or for 30 min at 37 °C to generate an equilibrium mixture of native and misfolded ribozyme. CYT-18 was then added (500 nM unless otherwise indicated) and a continued incubation was performed under standard conditions (25 °C, 50 mM Na-MOPS, pH 7.0, 10 mM Mg2+). At various times, the fraction of native ribozyme was determined by removing aliquots, adding trace radiolabeled substrate (CCCUCUA5), and quenching time points with two volumes of 90% formamide and separating the substrate and product by 20% denaturing PAGE as described.7
The fraction of S* cleaved rapidly is a good measure of the fraction of native ribozyme because the native and misfolded species bind the substrate with similar rate constants. Once bound to the native ribozyme, S* is rapidly cleaved, whereas S* bound to misfolded ribozyme is not cleaved until the misfolded ribozyme refolds to the native state or S* is released (0.02 min−1) and binds to the native ribozyme.20 For these experiments, the cleavage reaction was carried out by the complex of the EΔP 5abc ribozyme and CYT-18 protein, which gives a lower rate constant for cleavage than the wild-type ribozyme (~0.5 min−1 vs 10 min−1 under these conditions). Instead of collecting only a single time point to indicate the amplitude of the cleavage burst,20,22,23 we carried out complete time courses of S* cleavage and determined the burst amplitude directly by fitting progress curves with an equation that included two exponential terms using Kaleidagraph. Results are given as the average and standard deviation of two to six replicate measurements.
CYT-18 binding and dissociation kinetics
CYT-18 association with the EΔP5abc ribozyme was monitored by retention of 32P-labeled ribozyme on a nitrocellulose filter. For measurements of koff from misfolded ribozyme, trace labeled ribozyme was first incubated at 25 °C in 50 mM Na-MOPS, pH 7.0, 10 mM Mg2+ for 10 min to generate a population of predominantly misfolded ribozyme. CYT-18 was then added to a final concentration of 200 nM and incubated for 5 min to allow binding. For measurements of koff from the native ribozyme, the ribozyme was pre-incubated with CYT-18 under the same solution conditions for 30 min at 37 °C to allow the ribozyme to refold completely to the native state. Excess unlabeled ribozyme was then added (1.6 µM) and at various times, 10 µl aliquots were applied to a nitrocellulose filter that was pre-soaked in reaction buffer and backed with a nylon membrane. Filters were washed immediately with 1 ml of the same buffer and then removed and dried. Labeled RNA on the nitrocellulose and nylon membranes was quantitated by using a phosphorimager. The fraction of labeled material retained on the nitrocellulose membrane was plotted over time and rate constants were determined using Kaleidagraph (Synergy Software).
Experiments to measure binding kinetics to the native ribozyme were performed similarly. Radiolabeled EΔP5abc ribozyme (≤3 nM) was pre-incubated with 10 mM Mg2+ for 30 min at 37 °C to allow equilibration between the native and misfolded species. CYT-18 was added and, at various times thereafter, aliquots were withdrawn, added to a chase of unlabeled ribozyme, and then immediately applied to a filter and processed as described above.
DMS footprinting
Experiments were performed essentially as described.21,56 Conditions were 50 mM Na-MOPS, pH 7.0 and 10 mM Mg2+ at 25 °C. Folding of the EΔP5abc ribozyme (200 nM final concentration) was initiated by adding 10 mM Mg2+ (5 min), and then the ribozyme was further incubated with 500 nM CYT-18 for 30 min at 37 °C to generate the native complex or for 5 min at 25 °C to generate a population dominated by the the misfolded complex. The complex of native ribozyme with P5abc was generated by incubating at 50 °C for 30 min as described previously.21 For all reactions, DMS was then added to 0.67 % by volume and incubated for 5 min, determined empirically to give an optimal level of modification. Reactions were quenched in 30% β-mercaptoethanol, 0.3 M sodium acetate and then phenol extracted and ethanol precipitated. Modifications were detected by reverse transcription (SIII RT, Invitrogen) using 5´-labeled primers 3´-end-complementarity to nucleotides 240, 349 and 390. Reverse transcription products were separated by 8% denaturing gel, visualized using a Phosphorimager, and quantified using SAFA.57 Band intensities were normalized by the fully-extended product after subtracting the corresponding band intensity for a sample that was not treated with DMS, and for convenience these normalized values were multiplied by 103 to produce the y-axis values in Fig. 8 and Fig. S1. Nucleotides were scored as protected or enhanced by CYT-18 if the difference was present in each of three independent determinations and the average intensity differed by at least 1.0 on the scale in Fig. 8 and Fig. S1 (a value determined empirically to capture the bulk of the reproducible changes induced by CYT-18).
Modeling the complex of CYT-18 with the native EΔP5abc ribozyme
The crystal structure model of a thermostable variant of the Tetrahymena ribozyme core (PDB 1X8W)14 was superimposed with the complex of CYT-18 with the Twort ribozyme (PDB-2RKJ)35 using PYMOL. The P4–P6 domains were matched by aligning nucleotides 96–120, 200–226, and 248–268 of the Tetrahymena ribozyme with the corresponding nucleotides in the Twort ribozyme (RMSD = 3.4 Å). The P5abc domain of the Tetrahymena ribozyme was omitted from the model to allow visualization of CYT-18 interaction. The superimposition of P4–P6 resulted in a clash of the Tetrahymena P9 helix with CYT-18; therefore P9 was omitted from the model.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Paukstelis, Roland Saldanha, and Alan Lambowitz for plasmids encoding CYT-18 and for advice on its purification, Yaqi Wan for help preparing Fig. 11, and Georg Mohr, Paul Paukstelis, and members of the Russell lab for comments on the manuscript. This work was supported by grants from the Welch Foundation (F-1563) and the National Institutes of Health (GM070456) to R.R.
Abbreviations
- DMS
dimethyl sulfate
- EDTA
ethylene dinitrilotetraacetic acid
- PAGE
polyacrylamide gel electrophoresis
- P
the oligonucleotide product CCCUCU
- P*
(5′-32P)-labeled product
- S
the oligonucleotide substrate CCCUCUA5
- S*
(5′-32P)-labeled substrate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Two conditions must be met for an intermediate to generate a difference between equilibrium constants determined from kinetics and equilibrium binding experiments.22 First, the intermediate must be sufficiently long-lived to be trapped experimentally in the binding kinetics experiments. With our methodology, the intermediate would have to persist between the addition of chase and application to a filter, approximately 30 s. Second, the intermediate must partition favorably back to dissociation rather than proceeding on to form the final complex. If both conditions are met, the ratio of the rate constants governing these alternative fates will not contribute to the equilibrium constant calculated from kinetics. This is because the binding kinetics experiment will simply measure formation of the intermediate, and the dissociation experiment will not include the ratio of these rate constants because, once formed, the intermediate will go on to dissociate most of the time. However, this ratio will contribute to the true equilibrium binding expression and be present in the direct binding experiment. Because the ratio is <<1, this scenario will give artificially tight binding in kinetics experiments.
Here, our finding that the active complex is formed with an overall rate constant of at least 0.4 min−1 puts an upper limit of 1.7 min on the half-life of any intermediate. This limit alone does not rule out the model, because an intermediate with this lifetime could be trapped experimentally. However, because dissociation of the intermediate must be faster than its forward progression, the lifetime of the intermediate will be decreased substantially by this dissociation. The difference between the equilibrium constant from kinetics and equilibrium measurements is ~100-fold, 0.2 nM vs 20–50 nM, so the dissociation rate constant would have to be ~40 min−1 to account for the difference. This would give any intermediate a lifetime of less than 1 s, which would not allow it to be trapped experimentally.
REFERENCES
- 1.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 2.Sigler PB. An analysis of the structure of tRNA. Annu. Rev. Biophys. Bioeng. 1975;4:477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- 3.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 4.Russell R. RNA misfolding and the action of chaperones. Front. Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 6.Cech TR. Ribozymes, the first 20 years. Biochem. Soc. Trans. 2002;30:1162–1166. doi: 10.1042/bst0301162. [DOI] [PubMed] [Google Scholar]
- 7.Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 8.Michel F, Westhof E. Modeling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 9.Lehnert V, Jaeger L, Michel F, Westhof E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 10.Golden BL, Gooding AR, Podell ER, Cech TR. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science. 1998;282:259–264. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- 11.Tanner MA, Cech TR. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(Ile) of the purple bacterium Azoarcus. RNA. 1996;2:74–83. [PMC free article] [PubMed] [Google Scholar]
- 12.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2–32. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 14.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol. Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 16.van der Horst G, Christian A, Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doherty EA, Herschlag D, Doudna JA. Assembly of an exceptionally stable RNA tertiary interface in a group I ribozyme. Biochemistry. 1999;38:2982–2990. doi: 10.1021/bi982113p. [DOI] [PubMed] [Google Scholar]
- 18.Engelhardt MA, Doherty EA, Knitt DS, Doudna JA, Herschlag D. The P5abc peripheral element facilitates preorganization of the Tetrahymena group I ribozyme for catalysis. Biochemistry. 2000;39:2639–2651. doi: 10.1021/bi992313g. [DOI] [PubMed] [Google Scholar]
- 19.Russell R, Herschlag D. New pathways in folding of the Tetrahymena group I RNA enzyme. J. Mol. Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 20.Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: Commitment to form the native conformation is late in the folding pathway. J. Mol. Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 21.Russell R, Tijerina P, Chadee AB, Bhaskaran H. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry. 2007;46:4951–4961. doi: 10.1021/bi0620149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson TH, Tijerina P, Chadee AB, Herschlag D, Russell R. Structural specificity conferred by a group I RNA peripheral element. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10176–10181. doi: 10.1073/pnas.0501498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell R, Das R, Suh H, Travers K, Laederach A, Engelhardt M, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J. Mol. Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 24.Lambowitz AM, Caprara MG, Zimmerly S, Perlman PS. Group I and Group II ribozymes as RNPs: Clues to the past and guides to the future. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. New York: Cold Spring Harbor; 1999. pp. 451–485. [Google Scholar]
- 25.Akins RA, Lambowitz AM. A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell. 1987;50:331–345. doi: 10.1016/0092-8674(87)90488-0. [DOI] [PubMed] [Google Scholar]
- 26.Lambowitz AM, Perlman PS. Involvement of aminoacyl-tRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem. Sci. 1990;15:440–444. doi: 10.1016/0968-0004(90)90283-h. [DOI] [PubMed] [Google Scholar]
- 27.Labouesse M. The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol. Gen. Genet. 1990;224:209–221. doi: 10.1007/BF00271554. [DOI] [PubMed] [Google Scholar]
- 28.Rho SB, Martinis SA. The bI4 group I intron binds directly to both its protein splicing partners, a tRNA synthetase and maturase, to facilitate RNA splicing activity. RNA. 2000;6:1882–1894. doi: 10.1017/s1355838200001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallweber GJ, Mohr S, Rennard R, Caprara MG, Lambowitz AM. Characterization of Neurospora mitochondrial group I introns reveals different CYT-18 dependent and independent splicing strategies and an alternative 3' splice site for an intron ORF. RNA. 1997;3:114–131. [PMC free article] [PubMed] [Google Scholar]
- 30.Paukstelis PJ, Lambowitz AM. Identification and evolution of fungal mitochondrial tyrosyl-tRNA synthetases with group I intron splicing activity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6010–6015. doi: 10.1073/pnas.0801722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caprara MG, Mohr G, Lambowitz AM. A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- 32.Myers CA, Wallweber GJ, Rennard R, Kemel Y, Caprara MG, Mohr G, Lambowitz AM. A tyrosyl-tRNA synthetase suppresses structural defects in the two major helical domains of the group I intron catalytic core. J. Mol. Biol. 1996;262:87–104. doi: 10.1006/jmbi.1996.0501. [DOI] [PubMed] [Google Scholar]
- 33.Caprara MG, Myers CA, Lambowitz AM. Interaction of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein) with the group I intron P4–P6 domain. Thermodynamic analysis and the role of metal ions. J. Mol. Biol. 2001;308:165–190. doi: 10.1006/jmbi.2001.4581. [DOI] [PubMed] [Google Scholar]
- 34.Paukstelis PJ, Coon R, Madabusi L, Nowakowski J, Monzingo A, Robertus J, Lambowitz AM. A tyrosyl-tRNA synthetase adapted to function in group I intron splicing by acquiring a new RNA binding surface. Mol. Cell. 2005;17:417–428. doi: 10.1016/j.molcel.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 35.Paukstelis PJ, Chen JH, Chase E, Lambowitz AM, Golden BL. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature. 2008;451:94–97. doi: 10.1038/nature06413. [DOI] [PubMed] [Google Scholar]
- 36.Mohr G, Caprara MG, Guo Q, Lambowitz AM. A tyrosyl-tRNA synthetase can function similarly to an RNA structure in the Tetrahymena ribozyme. Nature. 1994;370:147–150. doi: 10.1038/370147a0. [DOI] [PubMed] [Google Scholar]
- 37.Saldanha RJ, Patel SS, Surendran R, Lee JC, Lambowitz AM. Involvement of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing. A new method for purifying the protein and characterization of physical and enzymatic properties pertinent to splicing. Biochemistry. 1995;34:1275–1287. doi: 10.1021/bi00004a022. [DOI] [PubMed] [Google Scholar]
- 38.Webb AE, Rose MA, Westhof E, Weeks KM. Protein-dependent transition states for ribonucleoprotein assembly. J. Mol. Biol. 2001;309:1087–1100. doi: 10.1006/jmbi.2001.4714. [DOI] [PubMed] [Google Scholar]
- 39.Rose MA, Weeks KM. Visualizing induced fit in early assembly of the human signal recognition particle. Nat. Struct. Biol. 2001;8:515–520. doi: 10.1038/88577. [DOI] [PubMed] [Google Scholar]
- 40.Johnson KA, Simpson ZB, Blom T. Global Kinetic Explorer: A new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 41.Park YC, Bedouelle H. Dimeric tyrosyl-tRNA synthetase from Bacillus stearothermophilus unfolds through a monomeric intermediate. A quantitative analysis under equilibrium conditions. J. Biol. Chem. 1998;273:18052–18059. doi: 10.1074/jbc.273.29.18052. [DOI] [PubMed] [Google Scholar]
- 42.Fersht A. Structure and Mechanism In Protein Science: A Guide to Enzyme Catalysis and Protein Folding. U.S.A.: W.H. Freeman and Company; 1999. [Google Scholar]
- 43.Murphy FL, Cech TR. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 44.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 45.Saldanha R, Ellington A, Lambowitz AM. Analysis of the CYT-18 protein binding site at the junction of stacked helices in a group I intron RNA by quantitative binding assays and in vitro selection. J. Mol. Biol. 1996;261:23–42. doi: 10.1006/jmbi.1996.0439. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Gutell RR, Lambowitz AM. Function of tyrosyl-tRNA synthetase in splicing group I introns: an induced-fit model for binding to the P4–P6 domain based on analysis of mutations at the junction of the P4–P6 stacked helices. J. Mol. Biol. 2000;301:265–283. doi: 10.1006/jmbi.2000.3963. [DOI] [PubMed] [Google Scholar]
- 47.Grossberger R, Mayer O, Waldsich C, Semrad K, Urschitz S, Schroeder R. Influence of RNA structural stability on the RNA chaperone activity of the Escherichia coli protein StpA. Nucleic Acids Res. 2005;33:2280–2289. doi: 10.1093/nar/gki515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohr G, Rennard R, Cherniack AD, Stryker J, Lambowitz AM. Function of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase in RNA splicing. Role of the idiosyncratic N-terminal extension and different modes of interaction with different group I introns. J. Mol. Biol. 2001;307:75–92. doi: 10.1006/jmbi.2000.4460. [DOI] [PubMed] [Google Scholar]
- 49.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 50.Caprara MG, Lehnert V, Lambowitz AM, Westhof E. A tyrosyl-tRNA synthetase recognizes a conserved tRNA-like structural motif in the group I intron catalytic core. Cell. 1996;87:1135–1145. doi: 10.1016/s0092-8674(00)81807-3. [DOI] [PubMed] [Google Scholar]
- 51.Duncan CD, Weeks KM. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry. 2008;47:8504–8513. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodson SA, Cech TR. Alternative secondary structures in the 5' exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991;30:2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- 53.Woodson SA. Exon sequences distant from the splice junction are required for efficient self-splicing of the Tetrahymena IVS. Nucleic Acids Res. 1992;20:4027–4032. doi: 10.1093/nar/20.15.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 55.Russell R, Herschlag D. Specificity from steric restrictions in the guanosine binding pocket of a group I ribozyme. RNA. 1999;5:158–166. doi: 10.1017/s1355838299981839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protocols. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.