Abstract
Mechanical force modulates a wide array of cell physiological processes. Cells sense and respond to mechanical stimuli using a hierarchy of structural complexes spanning multiple length scales, including force-sensitive molecules and cytoskeletal networks. Understanding mechanotransduction, i.e., the process by which cells convert mechanical inputs into biochemical signals, has required the development of novel biophysical tools that allow for probing of cellular and subcellular components at requisite time, length and force scales and technologies that track the spatio-temporal dynamics of relevant biomolecules. In this review, we begin by discussing the underlying principles and recent applications of atomic force microscopy, magnetic twisting cytometry, and traction force microscopy, three tools that have been widely used for measuring the mechanical properties of cells and for probing the molecular basis of cellular mechanotransduction. We then discuss how such tools can be combined with advanced fluorescence methods for imaging biochemical processes in living cells in the context of three specific problem spaces. We first focus on fluorescence resonance energy transfer, which has enabled imaging of intra- and intermolecular interactions and enzymatic activity in real time based on conformational changes in sensor molecules. Next, we examine the use of fluorescence methods to probe force-dependent dynamics of focal adhesion proteins. Finally, we discuss the use of calcium ratiometric signaling to track fast mechanotransductive signaling dynamics. Together, these studies demonstrate how single-cell biomechanical tools can be effectively combined with molecular imaging technologies for elucidating mechanotransduction processes and identifying mechanosensitive proteins.
Keywords: mechanotransduction, AFM, MTC, TFM, FRET, focal adhesions, calcium signaling
Introduction
While cells have long been understood to sense and respond to external biochemical stimuli, life scientists are increasingly beginning to appreciate that mechanical inputs can also powerfully regulate cell behavior, thus giving rise to the rapidly growing field of cellular mechanobiology. Mechanosensitivity, the ability of individual cells to sense mechanical stimuli from their surroundings, plays a central role in development, wound healing, tissue homeostasis, and many other critical physiological processes. Mechanotransduction refers to the process through which a cell converts a mechanical stimulus into a biochemical signal and may involve force-induced conformational changes in proteins that lead to opening of membrane channels, expose cryptic binding sites, or alter binding affinities, with eventual activation of signaling cascades [1-3]. Identification of mechanosensory proteins therefore forms a critical piece of our understanding of normal mechanotransduction and may lend insight into diseases that involve abnormal mechanotransduction. This in turn requires methods capable of applying localized forces to living cells under physiological conditions and tracking protein and signaling dynamics at the micro- and nanoscale.
In this review, we describe recent methodological advances in the study of mechanotransduction, with an emphasis on the dynamic tracking of mechanosensitive and mechanotransductive biomolecules in living cells. Key to these advances have been the creation of novel optical probes that enable high-resolution measurement of protein conformation and enzymatic activity in living cells in the setting of applied mechanical force. We begin by reviewing methods for the mechanical interrogation of single living cells and then conclude by discussing how some of these methods have been integrated with optical tools to study mechanotransduction.
Tools for Measuring Mechanical Properties of Single Cells
Over the past few decades, a wide range of methods has been developed for mapping the mechanical properties of living cells and/or subcellular components. These include micropipet aspiration (MPA) [4-6], traction force microscopy (TFM) [7-9], atomic force microscopy (AFM) [10-12], magnetic twisting cytometry (MTC) [13, 14], subcellular laser ablation (SLA) [15, 16], micropost array detectors (MPADs) [17-20], laser particle tracking microrheology (LPTM) [21, 22], and many other MEMS-based devices. While an exhaustive discussion of all these techniques is beyond the scope of this review, we begin by briefly exploring three of the most widely-used approaches: AFM, MTC, and TFM.
Atomic Force Microscopy (AFM)
AFM is a scanning probe microscopy technique that permits imaging and mechanical manipulation of biological materials at the micro- and nanoscale (Fig. 1a). Since its introduction in 1986 [23], AFM has become one of the most widely used biophysical tools in cell biology because of its ability to image biomolecules at nanometer-scale resolution, apply forces to cells over an extremely wide dynamic range (10 pN – 106 pN), and process samples in physiologic media and aqueous buffers [12, 24]. AFM operates by tracking physical interactions between a sample and a nanometer-sized tip attached to the end of a highly flexible cantilever, with the magnitude of the cantilever's deflection reflecting interaction forces between tip and sample. AFM images can be obtained in two types of feedback modes: An “AC” mode in which the tip is oscillated at its resonant frequency and a constant amplitude is maintained as the sample is scanned, or in a “DC” (contact) mode in which the tip is brought into direct contact with the surface and the cantilever deflection is kept constant. AFM imaging has been widely used for studying the structure and mechanics of isolated biomolecules [25-27], components of the cell nucleus [28, 29], and subcellular cytoskeletal structures [10, 30]. In addition to imaging, AFM has been successfully used in a force mode in which the tip is held in a fixed horizontal position and used to indent the sample. This approach has been applied with great success to measure the viscoelastic properties of many different cell types [31-36] and tissue specimens [37, 38], and alterations in stiffness associated with cell differentiation [39] and disease progression [37, 40, 41] (Fig. 1b). The imaging and force modes of AFM can be used in combination to estimate the forces associated with specific cellular processes, including migration [42], lamellipodial protrusion [43, 10], and cell division [45]. The AFM has also been utilized as a rheometer by driving the cell into the cantilever in an oscillatory fashion and using the cantilever to measure the amplitude and phase of the response [46]. In addition to mechanical interrogation of cells and tissues, AFM has emerged as a particularly powerful probe of single-molecule conformational and binding properties, including unfolding of proteins [47-50] and nucleic acids [51]. Using AFM, receptor-ligand interactions can also be probed by measuring unbinding forces between receptors and ligands covalently immobilized on the AFM tip and substrate, respectively (or vice versa). This approach has been successfully employed to estimate molecular binding strengths of various receptor-ligand systems such as integrin-collagen [52] and cadherin-cadherin [53]. In recent studies, AFM has been used in conjunction with total internal fluorescence (TIRF) microscopy and magnetic tweezers for studying force-dependent binding interactions between talin and vinculin [54]. In addition to using purified proteins, living cells can also be used as probes for studying protein-cell and cell-cell adhesion, which has the added advantage of ensuring that proteins are in their native state. For example, Moy and coworkers demonstrated that leukocyte activation induces changes in the interaction between integrins and ICAM-I [55]. Cellular processes such as protein clustering and adhesion strengthening can also be studied by modulating the tip-sample contact time [56].
Figure 1. Mechanobiology tools and their applications.
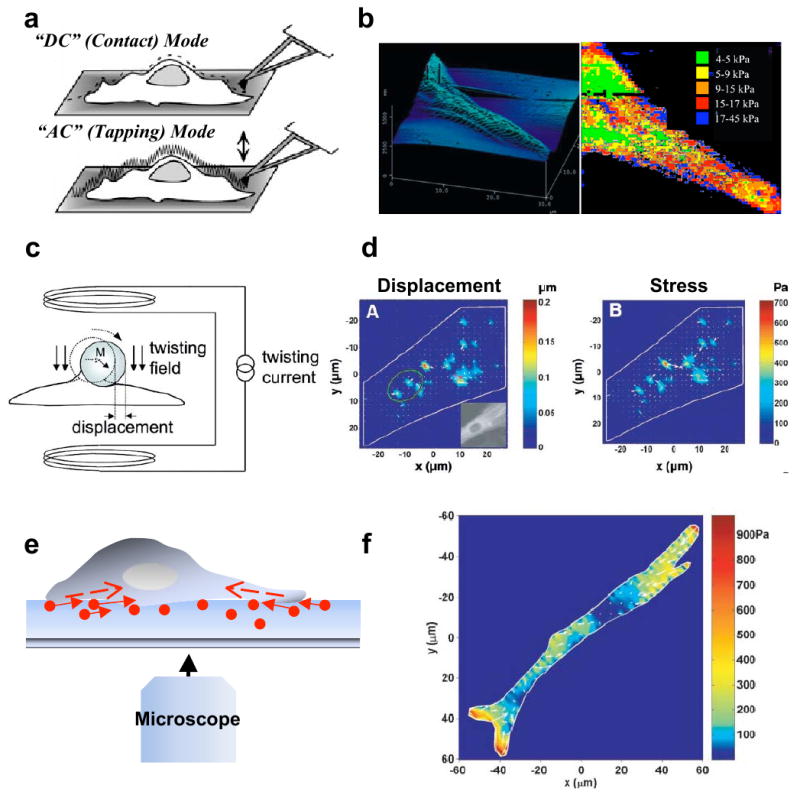
a. Atomic force microscopy (AFM). AFM can be operated either in DC (contact mode) in which the tip is brought into direct contact with the surface and the cantilever deflection is kept constant, or in AC (tapping) mode in which the tip is oscillated at its resonant frequency and a constant amplitude is maintained as the sample is scanned. Figure reprinted from [24] with permission. b. Topographic map and stiffness map of the edge of a fibroblast adhering to a fibronectin-coated glass substrate. Figure reprinted from [31] with permission. c. Magnetic twisting cytometry (MTC). In MTC, a homogeneous magnetic twisting field applied to a cell surface-adhered bead causes the bead to rotate and to displace thereby inducing shear stresses on the cell surface. Figure reprinted from [58] with permission. d. Displacement and stress maps computed using MTC for a human airway smooth muscle (HASM) cell expressing YFP-actin. Figure reprinted from [61] with permission. e. Traction force microscopy (TFM). Schematic of a cell on a hydrogel with embedded beads (circles) used in TFM. Arrows are indicative of the direction of bead motion induced by cell contractility (dotted arrows inside the cell). f. Traction field computed using TFM for a HASM cell on a 1300 Pa gel. Arrows show the direction and the relative magnitude of the tractions. Figure reprinted from [14] with permission.
Magnetic Twisting Cytometry (MTC)
In AFM, localized forces are exerted on biological samples by indentation. In contrast, MTC, which was first applied to cell-surface mechanotransduction by Wang et al. in 1993 [13], exerts forces on cells through magnetic manipulation of surface-bound particles (Fig. 1c). Specifically, cells are incubated with ferromagnetic or superparamagnetic beads coated with either full-length ECM proteins or cell-adhesive peptides (e.g. RGD), often at a multiplicity of ∼20 beads per cell [57]. Application of a strong magnetic field over a very short time orients and induces the magnetic dipoles of the beads to align along the horizontal direction. The application of a subsequent weak but sustained force along an orthogonal direction induces the beads to realign along this new direction, thereby twisting the surface-attached beads. The shear stiffness of the cell is calculated by measuring the bead rotation and the angular bead strain using a magnetometer. Some of the modifications and/or advances that have been made in the area of MTC include oscillatory MTC and 3D MTC. In oscillatory MTC, application of an oscillatory magnetic force allows the simultaneous tracking of both the elastic component and the viscous component of cell stiffness over the duration of force application [58]. In 3D MTC, the transmission of mechanical forces applied through magnetic beads can be mapped to determine force-transduction pathways and probe the mechanical anisotropy of adherent cells [59]. In addition to measuring cell mechanical properties, MTC can be used to probe how strongly specific cell-surface receptors are coupled to the cytoskeleton and to identify proteins involved in various force transduction pathways. For example, the limited rotation of RGD-bound beads in comparison to beads coated with antibodies to metabolic receptors provided one of the first direct demonstrations that ECM-bound integrins engage the cytoskeleton and therefore mechanically couple the cytoskeleton and ECM [13]. The role of the actomyosin network in modulating cell prestress and stiffness have been demonstrated in studies where myosin activators stiffen cells, and inhibitors soften them [14]. MTC has also been used to compare the capacity of various transmembrane proteins to transmit mechanical force [60]. In human airway smooth muscle cells, oscillatory MTC has been used to demonstrate the heterogeneous nature of long-distance force transmission within the cell, together with buildup of displacements and stresses at discrete sites [61] (Fig. 1d).
Traction Force Microscopy (TFM)
In contrast to AFM and MTC, where cell properties are quantified by locally applying forces to the cell surface, TFM measures contractile (traction) forces exerted by cells against the ECM (Fig. 1e). An early demonstration of traction forces between cells and the ECM was provided by Harris and coworkers [62, 63], who visualized cells wrinkling thin, silicone membranes during migration. However, the inherently nonlinear nature of wrinkling complicated quantitative extraction of traction forces from these observations. That problem was overcome with the introduction of traction force microscopy (TFM), which infers tractional stresses and strains from the motions of fiduciary beads embedded in a hydrogel-based ECM [64]. The emergence of polyacrylamide gels [64, 65], which are linearly elastic, optically clear, and amenable to protein conjugation, as a common platform for studying the effects of ECM stiffness on cell behavior has helped made TFM a widely used tool for measuring cell generated traction forces. In a TFM experiment, cells are cultured on ECM-coated polyacrylamide hydrogels studded with fluorescent fiduciary beads whose spatial distribution tracks strains within the gel. Traction forces are then determined by recording, comparing, and analyzing bead displacements prior to (“stressed”) and following (“unstressed”) chemical detachment of the cell. A variety of continuum approaches have been developed for solving the inverse problem of calculating traction forces from bead displacements [8, 66, 67]. For example, Dembo and Wang used a computationally-intense mesh-based approach in their pioneering measurements of traction forces exerted by fibroblasts during locomotion [66, 68]. Later, Butler and colleagues developed a different approach based on transformation of the ECM stress-strain relationship into Fourier space [8]. Since then, TFM has been used to measure traction forces in a variety of different cell-types [14, 69-72] (Fig. 1f), to elucidate the contributions of actomyosin contractility to the generation of these forces [70, 73, 74], and to correlate altered traction forces with cellular pathology. For example, the linear relationship between traction force and substrate stiffness observed in normal cells is lost in transformed cells and may explain these cells' unregulated growth [75]. Such alterations in traction forces have also been observed in transformed sarcoma cells where aggressive invasion into the surrounding matrix has been attributed to increased Rho/ROCK signaling [76]. As the resolution of TFM has increased, it has become possible to measure traction forces generated by individual, micron-sized adhesions. For example, in GFP-zyxin transfected fibroblasts, large traction forces have been observed at transient adhesions at the leading edge, whereas small tractions have been observed at mature focal adhesions [7]. Recently, TFM has been combined with fluorescence speckle microscopy (FSM) to reveal a biphasic dependence of traction forces on actin retrograde speed, with F-actin speed related inversely to traction force at transient adhesions and more directly so at large, mature focal adhesions [78]. TFM has also helped inspire the development of microfabricated post array detectors (MPADs), which greatly simplify the quantification of cell-ECM tractional forces by placing cells atop a bed of elastic pillars whose deformations offer a direct readout of local cell-induced stresses. [20].
Direct Visualization of Mechanotransduction in Living Cells: Selected Examples
Having reviewed several methods for probing mechanotransduction, we now discuss specific, recently-reported cases in which some of these tools have been combined with traditional optical imaging technologies to directly visualize the interconversion of mechanical and biochemical information in living cells.
Measuring Mechanotransduction With Optical Probes
Incorporation of a fluorescent label represents one of the simplest ways to study the localization dynamics of specific molecules in living cells. Due to the discovery of a range of fluorescent proteins with diverse spectral properties that permit imaging with high spatial and temporal resolution, optical probes can be used for studying the interactions between different proteins involved in mechanotransduction [79]. But perhaps the most exciting recent use of fluorescence imaging in the context of mechanotransductive signaling has been the combination of mechanobiology tools with advanced molecular imaging technologies, which include TIRF [70, 80], fluorescence recovery after photobleaching (FRAP) [15, 81], fluorescence correlation spectroscopy (FCS) [82], chromophore-assisted laser inactivation (CALI) [83, 84], fluorescence lifetime imaging microscopy (FLIM) [85], and fluorescence resonance energy transfer (FRET). These novel approaches are beginning to enable the application of a range of mechanical forces for perturbing living cells while following the temporal and spatial dynamics of specific mechanotransductive molecules. Of all of these technologies, FRET has emerged as a particularly powerful tool for visualizing signal transduction in response to mechanical stimulation. FRET is a form of non-radiative energy transfer observed between donor and acceptor fluorophores when the emission spectrum of the donor overlaps with the excitation spectrum of the acceptor. Critically, FRET efficiency between a donor and acceptor decays as the separation distance to the sixth power, which in practical terms means that a FRET signal is observed only when the donor and the acceptor are less than 10 nm apart. FRET-based reporters have been developed to probe the dynamics of a wide variety of focal adhesion proteins and signaling molecules [86-90], and can easily be combined with force measurements to study mechanotransduction. For example, FRET has been utilized for studying the interaction between focal adhesion kinase (FAK), paxillin and Cas and their phosphorylation at focal adhesions by expressing different combinations of these proteins fused to cyan and yellow fluorescent proteins as FRET donor and acceptor pairs [91]. The development of FRET sensors based on fusion of donor/acceptor fluorphores to the GTPases Rho, Rac and Cdc42 and their binding partners has significantly added to our understanding of the activation and spatio-temporal dynamics of these GTPases in various cellular processes [86, 87, 89]. For example, in endothelial cells, FRET was used to demonstrate conformational changes in a G protein coupled receptor (GPCR) in response to mechanical stimulation, independent of ligand engagement [92].
Src activation in response to externally applied force was visualized using a FRET-based sensor in an elegant and influential study by Wang et al. [90]. In this study, the authors developed a genetically encoded Src reporter for imaging and quantification of Src activation in HUVEC cells. The Src reporter was composed of (from N to C terminus) CFP, an SH2 domain, a flexible linker, a p130Cas-derived Src substrate peptide and YFP. When the Src phosphorylation domain is non-phosphorylated, the CFP and the YFP molecules are in close spatial proximity and therefore undergo strong FRET. In contrast, Src phosphorylation produces a conformational change that separates the CFP and YFP moieties and dramatically weakens the FRET signal (Fig. 2a, b). The authors then used OT to apply tensile forces to the cell surface via fibronectin-coated beads, which remarkably led to waves of Src activation in a spatially-directed and temporally-coordinated fashion away from the site of force application (Fig. 2c). Such activation was absent when similar forces were applied through polylysine-coated beads or when cells were treated with cytoskeletal poisons. These results directly demonstrate connections between the activation and propagation of Src and mechanical stimulation and illustrate the importance of an intact cytoskeleton to those connections.
Figure 2. Application of FRET for studying the mechanical activation of Src in HUVECs.
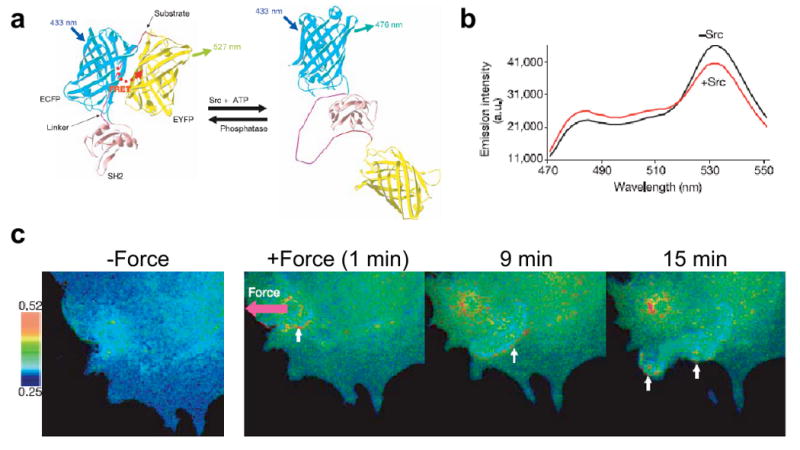
a. Schematic of mechanism of FRET sensor for Src activity. b. Effect of Src phosphorylation on the FRET emission ratio. c. Local application of force led to long range propagation of Src in a direction opposite to the direction of the applied force. Figure reprinted from [90] with permission.
In another creative combination of FRET-based probes with mechanobiological tools, FRET was used in conjunction with TFM for tracking traction-dependent adhesion clustering [93]. Here, MC3T3-E1 preosteoblasts were allowed to spread on alginate hydrogels of defined stiffnesses. As a first step, G4RGDASSK oligopeptides were chemically conjugated to sodium alginate gels and then labeled with either Alexa Fluor 488 or Alexa Fluor 546, with Alexa Fluor 488 as the donor and Alexa Fluor 546 as the acceptor. The hydrogels were then prepared by mixing equal volumes of the labeled polymer solutions with calcium sulfate. FRET signals were only observed in regions of the gel were cells were adhering to the substrate and actively pulling on the substrates, thereby bringing the donor and acceptor in close proximity. Increasing the density of adhesion ligands led to corresponding increase in FRET signal at the peripheries of adherent cells indicating local compression. Nocodazole treatment, which is known to stimulate contractility, led to an increase in FRET intensity because of increased traction forces and focal adhesion formation at the cell periphery. While an increase in hydrogel stiffness from 20 kPa to 110 kPa led to a corresponding increase in traction forces, the FRET signal was biphasic in nature with maximal traction forces observed on 60 kPa gels. In a subsequent study, the same investigators extended this technique to quantify the number of ligand-receptor bonds formed by cells in a three-dimensional ECM gel [94].
Force-Induced Biophysical Changes In Focal Adhesion Proteins
The need for many tissue cells to adhere to a solid matrix and their inability to survive in suspension underscores the importance of adhesions in cellular survival function [95]. In addition to physically coupling cells to the surrounding ECM, focal adhesions are increasingly understood to play a critical role in mechanosensing and mechanotransduction. The sensory function of focal adhesions is made possible by the complex interaction between the vast number (>50) of structural and signaling proteins which modulate focal adhesion assembly, maturation and turnover [96]. Some of these molecules bind transmembrane integrins, thereby linking the cytoskeleton to the ECM. For example, the cytoplasmic domain of β-integrin subunits directly interact with talin [97-99], α-actinin [100, 101], and filamin [97, 102, 103], all of which directly bind F-actin. Key focal adhesion-associated signaling molecules include protein tyrosine kinases (e.g., FAK [104, 105] and Src [90, 106]), protein tyrosine phosphatases (e.g., RPTP-α [107]), serine-threonine kinases (e.g., integrin-linked kinase [108]), and proteases (e.g., calpain [109]). Other important focal adhesion proteins include vinculin [110, 111], zyxin [15, 112] and paxillin [91, 113]. The collective signaling of these molecules determines the dynamics of focal adhesion and mechanotransduction [3, 114, 115]. The force-dependent dynamics of focal adhesions have been demonstrated in a range of different studies across multiple cell types. While application of forces (either internal contractile forces or externally applied forces) is necessary for the formation, maintenance, and growth of stable focal adhesions [116, 117], removal of force either by myosin inhibition [118] or caldesmon expression [119, 120] can produce focal adhesion disassembly. Focal adhesions also play an important role in sensing ECM rigidity, as observed in the preferential spreading of cells and formation of stable adhesions on stiff substrates [64], migration in the direction of increased rigidity [121], and reinforcement of integrin-cytoskeleton bonds upon application of force [106, 122]. Though the role of focal adhesions in mechanosensing is clear from the abovementioned studies, the underlying molecular mechanisms remain unclear.
Using stretchable substrates, Sheetz and coworkers [123] demonstrated the role of p130Cas in adhesion-dependent mechanotransduction. Earlier, the same group had shown that stretching of triton-insoluble cytoskeletons leads to increased binding of paxillin, FAK and p130Cas to cytoplasmic proteins, and that increased tyrosine phosphorylation of p130Cas is involved in the force-dependent activation of the small GTPase Rap1 [124, 125]. In this study, the authors first demonstrated that such stretch-dependent increase in p130Cas phosphorylation was also present in intact cells. Since no increase in c-Src activity was found, the authors hypothesized that increased p130Cas phosphorylation may be due to stretch-dependent increased susceptibility of p130Cas to baseline phosphorylation by c-Src rather than increased c-Src activity per se. This hypothesis was tested by creating a recombinant p130Cas substrate domain that was biotinylated at both the amino and carboxyl terminals, and bound to avidin on a stretchable substrate. Consistent with the hypothesis, p130Cas phosphorylation increased in proportion to stretch though no increase in c-Src activity was measured. Further, using an antibody that recognizes the extended form of p130Cas, the authors demonstrated that p130Cas was localized at sites of high traction forces in living cells. Together, these results support a role for p130Cas in force sensing, and suggest increased susceptibility to phosphorylation as a general mechanism for cell signaling.
In addition to altered susceptibility to phosphorylation, force-induced exposure of binding sites represents another important mode of mechanotransduction. Talin and vinculin are two important focal adhesion proteins implicated in cell adhesion and migration, with talin linking integrins to the cytoskeleton, and talin-vinculin interactions promoting cytoskeletal reorganization. In a recent study, the effect of force on this interaction was studied using a combination of magnetic tweezers, TIRF microscopy and AFM [54]. The talin rod (TR) domain contains up to 11 vinculin binding sites, with activation of these sites leading to conformational change in the vinculin head (Vh) domain. Of the 11 binding sites, 4 are inactive and are hypothesized to be activated by force-induced stretching of the talin rod domain. While magnetic tweezers were used to stretch talin rod domain chemically attached to a glass surface, binding-induced conformational change in the fluorescently-tagged Vh domain was monitored in TIRF, with a drop in the fluorescence intensity observed upon bleaching. The number of bleaching events was proportional to the magnitude of the applied force, suggesting that vinculin-talin binding was force-dependent. AFM was then used for studying the force-dependent stretching of the TR domain and determine the unfolding forces, contour lengths, and unfolding rate for the domain. Together, these results establish that force-induced stretching can expose cryptic binding sites for vinculin in the TR domain and that force-induced exposure of binding sites may represent a common pathway for mechanochemical signaling.
Calcium Signaling
The importance of calcium signaling in biology is widely appreciated, and control of intracellular calcium concentrations plays an important role in regulating a wide range of cellular processes in essentially all cell types [126-128]. Many mechanosensory and mechanotransductive proteins, such as α-actinin [100], gelsolin [129], and myosin light chain kinase [130], are strongly regulated by calcium binding. Cell contractility and adhesion turnover are tightly regulated by intracellular calcium, and transient increases in intracellular calcium are frequently observed in highly motile cells [69, 131]. In fibroblasts, substrate stretch leads to increases in intracellular calcium and traction forces, whereas depletion of extracellular calcium reduces traction forces [132]. Activation of calcium ion channels in HUVECs in response to local stretching of actin stress fibers with laser tweezers revealed a new function of the actin cytoskeleton as a force-transmitting device [133]. While migrating cells have long been known to exhibit a global calcium gradient with lowest concentration at the leading edge [134], the existence of high-calcium microdomains at the leading edge observed in recent studies points to a role of calcium in modulating the turning behavior of migrating cells [135]. In contrast to global calcium gradients in migrating cells, recent reports have documented the existence of oscillatory calcium levels in mesenchymal stem cells (MSCs) cultured on compliant matrices. In earlier studies, substrate rigidity was shown to play a key role in directing MSC differentiation with neuronal differentiation observed on soft substrates and osteogenic differentiation observed on stiff substrates [136]. Interestingly, substrate rigidity also influenced the amplitude and the frequency of calcium oscillations, with negligible oscillations observed on soft 1 kPa substrates, and significantly higher levels of oscillations on stiffer substrates [137].
In a particularly interesting use of calcium ratiometric imaging to study mechanotransduction, AFM was used to determine the strains required to elicit calcium transients in osteoblasts [138]. In this study, calcium transients were tracked after the application of localized indentational forces on cultured osteoblasts with an AFM tip (Fig. 3a, b). In response to mechanical stimulation, an increase in intracellular calcium was observed in 48% of the cells. Further, these transients were observed ∼13% during indentation, ∼13% during retraction, and ∼22% during both indentation and retraction, with propagation of transients to neighboring cells observed in some instances. Also, calcium stimulation was dependent on the amount of strain imparted during indentation, and followed a sigmoidal profile (Fig. 3c). Interestingly, although pharmacologic disruption of the actin cytoskeleton led to cell softening, no changes in calcium transients were observed. In contrast, disruption of microtubules or intermediate filaments markedly reduced calcium transients. These results indicate the existence of stretch-activated calcium channels and voltage-sensitive calcium channels as two distinct pathways for processing mechanical stimuli.
Figure 3. Application of AFM for studying calcium signaling in osteoblasts.
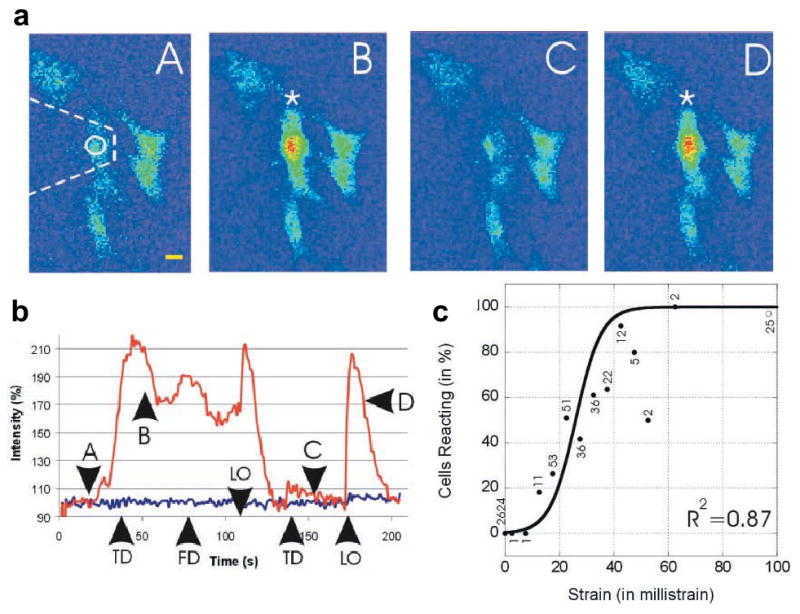
a. Sequence of images (A-D) showing alterations in intracellular calcium during two cycles of touch-down (TD) and lift-off (LO) by the AFM tip. b. Time-course of calcium intensity fluctuations (upper trace), with the time points of touch-down (TD), lift-off (LO), and acquisition of force-distance curves (FD) indicated. The letters A-D on the curves correspond to the sequence of images in (a). c. Plot of percentage of cells reacting as a function of applied strain fitted to a sigmoidal curve, with the numbers corresponding to the number of reactive cells at each strain value. Figure reprinted from [138] with permission.
Stretch-activated calcium channels have also been implicated in the motility of keratocytes. The spatio-temporal regulation of traction forces by calcium in keratocytes was demonstrated by combining calcium imaging and traction force microscopy on gelatin substrates [131]. In this study, the correlation between traction forces and calcium influx was studied by monitoring the changes in traction forces during individual calcium transients over a period of 5-30 seconds. An increase in calcium sensor fluorescence led to a corresponding increase in traction forces with a delay of ∼30 seconds. The strong correlation between the duration of the calcium transient with both the magnitude and duration of traction force increase highlighted the strong coupling between the increase in calcium levels and the increase in traction forces. Taken together, these observations reveal the role of calcium in modulating contractility and maintaining a highly directed cell movement.
Conclusions
In this review, we have discussed three mechanobiology tools (AFM, MTC, and TFM) that are widely used for measuring the mechanical properties of living cells and have been combined with optical technologies for understanding mechanochemical signaling. The versatility of these methods can be gauged from the myriad of different experimental conditions where these tools have been employed, and the range of length and force scales these methods encompass. The capabilities of these methods have been greatly extended by combining them with imaging technologies that permit live-cell imaging and tracking of force-dependent dynamic processes. While we chose here to highlight only a few specific combinations of mechanobiological and optical tools, we anticipate that this kind of multimodal approach will represent an enormous growth area for the field of cellular mechanobiology. Potential examples include the use of TIRF imaging to track the conformation and assembly of specific molecules at the cell-ECM interface and the application of super-resolution imaging technologies to follow dynamic processes in living cells below the diffraction limit [139]. It is also important to note that almost all of the examples discussed in this review feature 2D ECMs that differ in important ways from 3D ECMs encountered in tissues, where the effects of soluble factors, matrix metalloproteases, matrix composition and stiffness are likely to be far more complicated. Hence, one of the major challenges in the field of mechanobiology involves the development of new tools for quantifying the mechanical phenotype of cells in 3D. Importantly, few of the methods currently used to probe single-cell mechanics and mechanotransduction, including those described in the first part of this review, are readily applicable to 3D culture. In principle, the optical modalities described in the second half of the review do not suffer from this limitation, which leads us to believe that these methods will play an increasingly important role in advancing the field over the next decade. This, in turn, may create the need for the development of new molecular probes and/or imaging modalities capable of interrogating living cells and tissues at the high spatial and temporal resolution needed to precisely follow mechanotransductive signaling. It may also spur the development of creative ways to extract mechanical information from preexisting dynamic processes. One such recently developed method is based on single-cell tracking in 3D for mapping out local matrix deformations and studying the interplay between contractile forces and matrix degradation [140]. Another is the recent use of image correlation spectroscopy to predict the elastic properties of collagen ECMs by tracking the constituent fibers [141].
An inherent challenge in elucidating connections between mechanical force and signaling lies in the rich diversity of mechanotransductive pathways in living cells and the many ways in which force could potentially influence these pathways. The redundancy of different mechanotransduction pathways suggests that many of the mechanosensitive proteins may have multiple roles depending on their localization. For example, zyxin, paxillin, and other proteins have been shown to translocate from focal adhesions to the nucleus, and an important avenue of future research is to identify cues that could trigger such translocation [142]. Similar to unfolding of cytoplasmic proteins like talin discussed here, force-dependent unfolding of ECM proteins such as fibronectin are also likely to play an important role in mechanotransduction [143]. In addition to focal adhesions and ion channels, recent literature also shows the role of cell-cell junctions in mechanosensing [144]. In conclusion, the development of new mechanobiology tools together with rapid advances in molecular biology holds great promise in contributing to our understanding of mechanotransduction and opening up new avenues for research.
Acknowledgments
S.K. gratefully acknowledges grant support from the Arnold and Mabel Beckman Foundation, the American Heart Association (0765128Y), NSF (0727420) and the NIH Director's New Innovator Award (1DP2OD004213), a part of the NIH Roadmap for Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10(1):34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10(1):75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 3.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 4.Evans EA, Hochmuth RM. Membrane viscoelasticity. Biophys J. 1976;16(1):1. doi: 10.1016/S0006-3495(76)85658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hochmuth RM. Micropipette aspiration of living cells. J Biomech. 2000;33(1):15–22. doi: 10.1016/s0021-9290(99)00175-x. [DOI] [PubMed] [Google Scholar]
- 6.Jones WR, Ting-Beall HP, Lee GM, Kelley SS, Hochmuth RM, Guilak F. Alterations in the Young's modulus and volumetric properties of chondrocytes isolated from normal and osteoarthritic human cartilage. J Biomech. 1999;32(2):119–127. doi: 10.1016/s0021-9290(98)00166-3. [DOI] [PubMed] [Google Scholar]
- 7.Beningo KA, Dembo M, Kaverina I, Small JV, Wang Yl. Nascent Focal Adhesions Are Responsible for the Generation of Strong Propulsive Forces in Migrating Fibroblasts. J Cell Biol. 2001;153(4):881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol. 2002;282(3):C595–605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 9.Munevar S, Wang Y, Dembo M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J. 2001;80(4):1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotsch C, Jacobson K, Radmacher M. Dimensional and mechanical dynamics of active and stable edges in motile fibroblasts investigated by using atomic force microscopy. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(3):921–926. doi: 10.1073/pnas.96.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Meth. 2008;5(6):491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radmacher M, Wang YL, Discher DE. Methods in Cell Biology. Academic Press; 2007. Studying the Mechanics of Cellular Processes by Atomic Force Microscopy; pp. 347–372. [DOI] [PubMed] [Google Scholar]
- 13.Wang N, Butler J, Ingber D. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260(5111):1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 14.Wang N, Tolic-Norrelykke IM, Chen J, Mijailovich SM, Butler JP, Fredberg JJ, Stamenovic D. Cell prestress. I. Stiffness and prestress are closely associated in adherent contractile cells. Am J Physiol Cell Physiol. 2002;282(3):C606–616. doi: 10.1152/ajpcell.00269.2001. [DOI] [PubMed] [Google Scholar]
- 15.Lele TP, Pendse J, Kumar S, Salanga M, Karavitis J, Ingber DE. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J Cell Physiol. 2006;207(1):187–194. doi: 10.1002/jcp.20550. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S, Maxwell IZ, Heisterkamp A, Polte TR, Lele TP, Salanga M, Mazur E, Ingber DE. Viscoelastic Retraction of Single Living Stress Fibers and Its Impact on Cell Shape, Cytoskeletal Organization, and Extracellular Matrix Mechanics. Biophysical Journal. 2006;90(10):3762–3773. doi: 10.1529/biophysj.105.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci U S A. 2005;102(33):11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan JL, Liu W, Nelson CM, Raghavan S, Chen CS. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 2004;10(56):865–872. doi: 10.1089/1076327041348365. [DOI] [PubMed] [Google Scholar]
- 19.Nelson CM, Pirone DM, Tan JL, Chen CS. Vascular endothelial-cadherin regulates cytoskeletal tension, cell spreading, and focal adhesions by stimulating RhoA. Mol Biol Cell. 2004;15(6):2943–2953. doi: 10.1091/mbc.E03-10-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100(4):1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng Y, Kole TP, Wirtz D. Micromechanical mapping of live cells by multiple-particle-tracking microrheology. Biophys J. 2002;83(6):3162–3176. doi: 10.1016/S0006-3495(02)75319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada S, Wirtz D, Kuo SC. Mechanics of living cells measured by laser tracking microrheology. Biophys J. 2000;78(4):1736–1747. doi: 10.1016/S0006-3495(00)76725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986;56(9):930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Hoh JH. Probing the machinery of intracellular trafficking with the atomic force microscope. Traffic. 2001;2(11):746–756. doi: 10.1034/j.1600-0854.2001.21102.x. [DOI] [PubMed] [Google Scholar]
- 25.Silva LP. Imaging proteins with atomic force microscopy: an overview. Curr Protein Pept Sci. 2005;6(4):387–395. doi: 10.2174/1389203054546389. [DOI] [PubMed] [Google Scholar]
- 26.Muller DJ. AFM: a nanotool in membrane biology. Biochemistry. 2008;47(31):7986–7998. doi: 10.1021/bi800753x. [DOI] [PubMed] [Google Scholar]
- 27.Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu Rev Biochem. 2008;77:127–148. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- 28.Hansma HG, Kasuya K, Oroudjev E. Atomic force microscopy imaging and pulling of nucleic acids. Curr Opin Struct Biol. 2004;14(3):380–385. doi: 10.1016/j.sbi.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Hirano Y, Takahashi H, Kumeta M, Hizume K, Hirai Y, Otsuka S, Yoshimura SH, Takeyasu K. Nuclear architecture and chromatin dynamics revealed by atomic force microscopy in combination with biochemistry and cell biology. Pflugers Arch. 2008;456(1):139–153. doi: 10.1007/s00424-007-0431-z. [DOI] [PubMed] [Google Scholar]
- 30.Pesen D, Hoh JH. Micromechanical architecture of the endothelial cell cortex. Biophys J. 2005;88(1):670–679. doi: 10.1529/biophysj.104.049965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Fibroblast Adaptation and Stiffness Matching to Soft Elastic Substrates. Biophysical Journal. 2007;93(12):4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rotsch C, Radmacher M. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study. Biophys J. 2000;78(1):520–535. doi: 10.1016/S0006-3495(00)76614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rotsch C, Braet F, Wisse E, Radmacher M. AFM imaging and elasticity measurements on living rat liver macrophages. Cell Biol Int. 1997;21(11):685–696. doi: 10.1006/cbir.1997.0213. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann UG, Rotsch C, Parak WJ, Radmacher M. Investigating the cytoskeleton of chicken cardiocytes with the atomic force microscope. J Struct Biol. 1997;119(2):84–91. doi: 10.1006/jsbi.1997.3868. [DOI] [PubMed] [Google Scholar]
- 35.Takai E, Costa KD, Shaheen A, Hung CT, Guo XE. Osteoblast elastic modulus measured by atomic force microscopy is substrate dependent. Ann Biomed Eng. 2005;33(7):963–971. doi: 10.1007/s10439-005-3555-3. [DOI] [PubMed] [Google Scholar]
- 36.Sen S, Kumar S. Cell-matrix de-adhesion dynamics reflect contractile mechanics. Cellular and Molecular Bioengineering. 2009;2(2):218–230. doi: 10.1007/s12195-009-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166(6):877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matyka K, Matyka M, Mroz I, Zalewska-Rejdak J, Ciszewski A. An AFM study on mechanical properties of native and dimethyl suberimidate cross-linked pericardium tissue. J Mol Recognit. 2007;20(6):524–530. doi: 10.1002/jmr.855. [DOI] [PubMed] [Google Scholar]
- 39.Collinsworth AM, Zhang S, Kraus WE, Truskey GA. Apparent elastic modulus and hysteresis of skeletal muscle cells throughout differentiation. Am J Physiol Cell Physiol. 2002;283(4):C1219–1227. doi: 10.1152/ajpcell.00502.2001. [DOI] [PubMed] [Google Scholar]
- 40.Lam WA, Rosenbluth MJ, Fletcher DA. Chemotherapy exposure increases leukemia cell stiffness. Blood. 2007;109(8):3505–3508. doi: 10.1182/blood-2006-08-043570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenbluth MJ, Lam WA, Fletcher DA. Force Microscopy of Nonadherent Cells: A Comparison of Leukemia Cell Deformability. Biophysical Journal. 2006;90(8):2994–3003. doi: 10.1529/biophysj.105.067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laurent VM, Kasas S, Yersin A, Schaffer TE, Catsicas S, Dietler G, Verkhovsky AB, Meister JJ. Gradient of rigidity in the lamellipodia of migrating cells revealed by atomic force microscopy. Biophys J. 2005;89(1):667–675. doi: 10.1529/biophysj.104.052316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotsch C, Jacobson K, Condeelis J, Radmacher M. EGF-stimulated lamellipod extension in adenocarcinoma cells. Ultramicroscopy. 2001;86(12):97–106. doi: 10.1016/s0304-3991(00)00102-9. [DOI] [PubMed] [Google Scholar]
- 45.Matzke R, Jacobson K, Radmacher M. Direct, high-resolution measurement of furrow stiffening during division of adherent cells. Nat Cell Biol. 2001;3(6):607–610. doi: 10.1038/35078583. [DOI] [PubMed] [Google Scholar]
- 46.Mahaffy RE, Shih CK, MacKintosh FC, Kas J. Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Phys Rev Lett. 2000;85(4):880–883. doi: 10.1103/PhysRevLett.85.880. [DOI] [PubMed] [Google Scholar]
- 47.Bhasin N, Carl P, Harper S, Feng G, Lu H, Speicher DW, Discher DE. Chemistry on a single protein, vascular cell adhesion molecule-1, during forced unfolding. J Biol Chem. 2004;279(44):45865–45874. doi: 10.1074/jbc.M404103200. [DOI] [PubMed] [Google Scholar]
- 48.Law R, Carl P, Harper S, Dalhaimer P, Speicher DW, Discher DE. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys J. 2003;84(1):533–544. doi: 10.1016/S0006-3495(03)74872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carl P, Kwok CH, Manderson G, Speicher DW, Discher DE. Forced unfolding modulated by disulfide bonds in the Ig domains of a cell adhesion molecule. Proc Natl Acad Sci U S A. 2001;98(4):1565–1570. doi: 10.1073/pnas.031409698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muller DJ, Sapra KT, Scheuring S, Kedrov A, Frederix PL, Fotiadis D, Engel A. Single-molecule studies of membrane proteins. Curr Opin Struct Biol. 2006;16(4):489–495. doi: 10.1016/j.sbi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Lee GU, Chrisey LA, Colton RJ. Direct measurement of the forces between complementary strands of DNA. Science. 1994;266(5186):771–773. doi: 10.1126/science.7973628. [DOI] [PubMed] [Google Scholar]
- 52.Taubenberger A, Cisneros DA, Friedrichs J, Puech PH, Muller DJ, Franz CM. Revealing early steps of alpha2beta1 integrin-mediated adhesion to collagen type I by using single-cell force spectroscopy. Mol Biol Cell. 2007;18(5):1634–1644. doi: 10.1091/mbc.E06-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panorchan P, Thompson MS, Davis KJ, Tseng Y, Konstantopoulos K, Wirtz D. Single-molecule analysis of cadherin-mediated cell-cell adhesion. J Cell Sci. 2006;119(Pt 1):66–74. doi: 10.1242/jcs.02719. [DOI] [PubMed] [Google Scholar]
- 54.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323(5914):638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen A, Moy VT. Cross-linking of cell surface receptors enhances cooperativity of molecular adhesion. Biophys J. 2000;78(6):2814–2820. doi: 10.1016/S0006-3495(00)76824-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahimou F, Mok LP, Bardot B, Wesley C. The adhesion force of Notch with Delta and the rate of Notch signaling. J Cell Biol. 2004;167(6):1217–1229. doi: 10.1083/jcb.200407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lele TP, Sero JE, Matthews BD, Kumar S, Xia S, Montoya-Zavala M, Polte T, Overby D, Wang N, Ingber DE. Tools to study cell mechanics and mechanotransduction. Methods Cell Biol. 2007;83:443–472. doi: 10.1016/S0091-679X(07)83019-6. [DOI] [PubMed] [Google Scholar]
- 58.Fabry B, Maksym GN, Shore SA, Moore PE, Panettieri RA, Jr, Butler JP, Fredberg JJ. Selected contribution: time course and heterogeneity of contractile responses in cultured human airway smooth muscle cells. J Appl Physiol. 2001;91(2):986–994. doi: 10.1152/jappl.2001.91.2.986. [DOI] [PubMed] [Google Scholar]
- 59.Hu S, Eberhard L, Chen J, Love JC, Butler JP, Fredberg JJ, Whitesides GM, Wang N. Mechanical anisotropy of adherent cells probed by a three-dimensional magnetic twisting device. Am J Physiol Cell Physiol. 2004;287(5):C1184–1191. doi: 10.1152/ajpcell.00224.2004. [DOI] [PubMed] [Google Scholar]
- 60.Wang N, Ingber DE. Probing transmembrane mechanical coupling and cytomechanics using magnetic twisting cytometry. Biochem Cell Biol. 1995;73:327–335. doi: 10.1139/o95-041. [DOI] [PubMed] [Google Scholar]
- 61.Hu S, Chen J, Fabry B, Numaguchi Y, Gouldstone A, Ingber DE, Fredberg JJ, Butler JP, Wang N. Intracellular stress tomography reveals stress focusing and structural anisotropy in cytoskeleton of living cells. Am J Physiol Cell Physiol. 2003;285(5):C1082–1090. doi: 10.1152/ajpcell.00159.2003. [DOI] [PubMed] [Google Scholar]
- 62.Harris AK, Stopak D, Wild P. Fibroblast traction as a mechanism for collagen morphogenesis. Nature. 1981;290(5803):249–251. doi: 10.1038/290249a0. [DOI] [PubMed] [Google Scholar]
- 63.Harris AK, Wild P, Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208(4440):177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 64.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94(25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kandow CE, Georges PC, Janmey PA, Beningo KA. Polyacrylamide hydrogels for cell mechanics: steps toward optimization and alternative uses. Methods Cell Biol. 2007;83:29–46. doi: 10.1016/S0091-679X(07)83002-0. [DOI] [PubMed] [Google Scholar]
- 66.Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76(4):2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z, Lin JS, Chen J, Wang JH. Determining substrate displacement and cell traction fields--a new approach. J Theor Biol. 2006;242(3):607–616. doi: 10.1016/j.jtbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 68.Dembo M, Oliver T, Ishihara A, Jacobson K. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys J. 1996;70(4):2008–2022. doi: 10.1016/S0006-3495(96)79767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doyle AD, Lee J. Cyclic changes in keratocyte speed and traction stress arise from Ca2+-dependent regulation of cell adhesiveness. J Cell Sci. 2005;118(Pt 2):369–379. doi: 10.1242/jcs.01590. [DOI] [PubMed] [Google Scholar]
- 70.Iwadate Y, Yumura S. Actin-based propulsive forces and myosin-II-based contractile forces in migrating Dictyostelium cells. J Cell Sci. 2008;121(Pt 8):1314–1324. doi: 10.1242/jcs.021576. [DOI] [PubMed] [Google Scholar]
- 71.Curtze S, Dembo M, Miron M, Jones DB. Dynamic changes in traction forces with DC electric field in osteoblast-like cells. J Cell Sci. 2004;117(Pt 13):2721–2729. doi: 10.1242/jcs.01119. [DOI] [PubMed] [Google Scholar]
- 72.Tolic-Norrelykke IM, Wang N. Traction in smooth muscle cells varies with cell spreading. J Biomech. 2005;38(7):1405–1412. doi: 10.1016/j.jbiomech.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 73.Lombardi ML, Knecht DA, Dembo M, Lee J. Traction force microscopy in Dictyostelium reveals distinct roles for myosin II motor and actin-crosslinking activity in polarized cell movement. J Cell Sci. 2007;120(Pt 9):1624–1634. doi: 10.1242/jcs.002527. [DOI] [PubMed] [Google Scholar]
- 74.Chen J, Li H, SundarRaj N, Wang JH. Alpha-smooth muscle actin expression enhances cell traction force. Cell Motil Cytoskeleton. 2007;64(4):248–257. doi: 10.1002/cm.20178. [DOI] [PubMed] [Google Scholar]
- 75.Wang HB, Dembo M, Wang YL. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am J Physiol Cell Physiol. 2000;279(5):C1345–1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 76.Rosel D, Brabek J, Tolde O, Mierke CT, Zitterbart DP, Raupach C, Bicanova K, Kollmannsberger P, Pankova D, Vesely P, Folk P, Fabry B. Up-regulation of Rho/ROCK signaling in sarcoma cells drives invasion and increased generation of protrusive forces. Mol Cancer Res. 2008;6(9):1410–1420. doi: 10.1158/1541-7786.MCR-07-2174. [DOI] [PubMed] [Google Scholar]
- 78.Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183(6):999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Shyy J, Y J, Chien S. Fluorescence Proteins, Live-Cell Imaging, and Mechanobiology: Seeing Is Believing. Annual Review of Biomedical Engineering. 2008;10(1):1–38. doi: 10.1146/annurev.bioeng.010308.161731. [DOI] [PubMed] [Google Scholar]
- 80.Ueda M, Sako Y, Tanaka T, Devreotes P, Yanagida T. Single-molecule analysis of chemotactic signaling in Dictyostelium cells. Science. 2001;294(5543):864–867. doi: 10.1126/science.1063951. [DOI] [PubMed] [Google Scholar]
- 81.Lele TP, Thodeti CK, Ingber DE. Force meets chemistry: analysis of mechanochemical conversion in focal adhesions using fluorescence recovery after photobleaching. J Cell Biochem. 2006;97(6):1175–1183. doi: 10.1002/jcb.20761. [DOI] [PubMed] [Google Scholar]
- 82.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311(5767):1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 83.Tour O, Meijer RM, Zacharias DA, Adams SR, Tsien RY. Genetically targeted chromophore-assisted light inactivation. Nat Biotechnol. 2003;21(12):1505–1508. doi: 10.1038/nbt914. [DOI] [PubMed] [Google Scholar]
- 84.Rajfur Z, Roy P, Otey C, Romer L, Jacobson K. Dissecting the link between stress fibres and focal adhesions by CALI with EGFP fusion proteins. Nat Cell Biol. 2002;4(4):286–293. doi: 10.1038/ncb772. [DOI] [PubMed] [Google Scholar]
- 85.Verveer PJ, Wouters FS, Reynolds AR, Bastiaens PI. Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane. Science. 2000;290(5496):1567–1570. doi: 10.1126/science.290.5496.1567. [DOI] [PubMed] [Google Scholar]
- 86.Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290(5490):333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 87.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305(5690):1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 88.Cai X, Lietha D, Ceccarelli DF, Karginov AV, Rajfur Z, Jacobson K, Hahn KM, Eck MJ, Schaller MD. Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol Cell Biol. 2008;28(1):201–214. doi: 10.1128/MCB.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440(7087):1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 90.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434(7036):1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 91.Ballestrem C, Erez N, Kirchner J, Kam Z, Bershadsky A, Geiger B. Molecular mapping of tyrosine-phosphorylated proteins in focal adhesions using fluorescence resonance energy transfer. J Cell Sci. 2006;119(Pt 5):866–875. doi: 10.1242/jcs.02794. [DOI] [PubMed] [Google Scholar]
- 92.Chachisvilis M, Zhang YL, Frangos JA. G protein-coupled receptors sense fluid shear stress in endothelial cells. Proc Natl Acad Sci U S A. 2006;103(42):15463–15468. doi: 10.1073/pnas.0607224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(12):4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kong HJ, Boontheekul T, Mooney DJ. Quantifying the relation between adhesion ligand-receptor bond formation and cell phenotype. Proceedings of the National Academy of Sciences. 2006;103(49):18534–18539. doi: 10.1073/pnas.0605960103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 96.Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114(Pt 20):3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 97.Legate KR, Fassler R. Mechanisms that regulate adaptor binding to beta-integrin cytoplasmic tails. J Cell Sci. 2009;122(Pt 2):187–198. doi: 10.1242/jcs.041624. [DOI] [PubMed] [Google Scholar]
- 98.Lele TP, Thodeti CK, Pendse J, Ingber DE. Investigating complexity of protein-protein interactions in focal adhesions. Biochem Biophys Res Commun. 2008;369(3):929–934. doi: 10.1016/j.bbrc.2008.02.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Giannone G, Jiang G, Sutton DH, Critchley DR, Sheetz MP. Talin1 is critical for force-dependent reinforcement of initial integrin-cytoskeleton bonds but not tyrosine kinase activation. J Cell Biol. 2003;163(2):409–419. doi: 10.1083/jcb.200302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Otey CA, Carpen O. alpha-actinin revisited: A fresh look at an old player. Cell Motility and the Cytoskeleton. 2004;58(2):104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 101.Otey CA, Pavalko FM, Burridge K. An interaction between alpha-actinin and the beta 1 integrin subunit in vitro. J Cell Biol. 1990;111(2):721–729. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3(12):1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- 103.Critchley DR, Holt MR, Barry ST, Priddle H, Hemmings L, Norman J. Integrin-mediated cell adhesion: the cytoskeletal connection. Biochem Soc Symp. 1999;65:79–99. [PubMed] [Google Scholar]
- 104.Frey MT, Tsai IY, Russell TP, Hanks SK, Wang YL. Cellular responses to substrate topography: role of myosin II and focal adhesion kinase. Biophys J. 2006;90(10):3774–3782. doi: 10.1529/biophysj.105.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang HB, Dembo M, Hanks SK, Wang Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci U S A. 2001;98(20):11295–11300. doi: 10.1073/pnas.201201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159(4):695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.von Wichert G, Jiang G, Kostic A, De Vos K, Sap J, Sheetz MP. RPTP-alpha acts as a transducer of mechanical force on alphav/beta3-integrin-cytoskeleton linkages. J Cell Biol. 2003;161(1):143–153. doi: 10.1083/jcb.200211061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sakai T, Li S, Docheva D, Grashoff C, Sakai K, Kostka G, Braun A, Pfeifer A, Yurchenco PD, Fassler R. Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev. 2003;17(7):926–940. doi: 10.1101/gad.255603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Glading A, Lauffenburger DA, Wells A. Cutting to the chase: calpain proteases in cell motility. Trends Cell Biol. 2002;12(1):46–54. doi: 10.1016/s0962-8924(01)02179-1. [DOI] [PubMed] [Google Scholar]
- 110.Coll J, Ben-Ze'ev A, Ezzell RM, Fernandez JLR, Baribault H, Oshima RG, Adamson ED. Targeted Disruption of Vinculin Genes in F9 and Embryonic Stem Cells Changes Cell Morphology, Adhesion, and Locomotion. Proceedings of the National Academy of Sciences. 1995;92(20):9161–9165. doi: 10.1073/pnas.92.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends in Cell Biology. 2006;16(9):453. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 112.Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J Cell Biol. 2005;171(2):209–215. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Katoh K, Kano Y, Ookawara S. Rho-kinase dependent organization of stress fibers and focal adhesions in cultured fibroblasts. Genes to Cells. 2007;12:623–638. doi: 10.1111/j.1365-2443.2007.01073.x. 5 %R. [DOI] [PubMed] [Google Scholar]
- 114.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix and the cytoskeleton. Nat Rev Mol Cell Biol. 2001;2(11):793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 115.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10(1):21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 116.Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153(6):1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133(6):1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Totsukawa G, Wu Y, Sasaki Y, Hartshorne DJ, Yamakita Y, Yamashiro S, Matsumura F. Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J Cell Biol. 2004;164(3):427–439. doi: 10.1083/jcb.200306172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Helfman DM, Levy ET, Berthier C, Shtutman M, Riveline D, Grosheva I, Lachish-Zalait A, Elbaum M, Bershadsky AD. Caldesmon inhibits nonmuscle cell contractility and interferes with the formation of focal adhesions. Mol Biol Cell. 1999;10(10):3097–3112. doi: 10.1091/mbc.10.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grosheva I, Vittitow JL, Goichberg P, Gabelt BT, Kaufman PL, Borras T, Geiger B, Bershadsky AD. Caldesmon effects on the actin cytoskeleton and cell adhesion in cultured HTM cells. Exp Eye Res. 2006;82(6):945–958. doi: 10.1016/j.exer.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 121.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79(1):144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88(1):39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 123.Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127(5):1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tamada M, Sheetz MP, Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev Cell. 2004;7(5):709–718. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 125.Sawada Y, Sheetz MP. Force transduction by Triton cytoskeletons. J Cell Biol. 2002;156(4):609–615. doi: 10.1083/jcb.200110068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stossel TP. On the crawling of animal cells. Science. 1993;260(5111):1086–1094. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 127.Evans JH, Falke JJ. Ca2+ influx is an essential component of the positive-feedback loop that maintains leading-edge structure and activity in macrophages. Proc Natl Acad Sci U S A. 2007;104(41):16176–16181. [Google Scholar]
- 128.Sun S, Liu Y, Lipsky S, Cho M. Physical manipulation of calcium oscillations facilitates osteodifferentiation of human mesenchymal stem cells. FASEB J. 2007;21(7):1472–1480. doi: 10.1096/fj.06-7153com. [DOI] [PubMed] [Google Scholar]
- 129.Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274(47):33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 130.Takashima S. Phosphorylation of myosin regulatory light chain by myosin light chain kinase, and muscle contraction. Circ J. 2009;73(2):208–213. doi: 10.1253/circj.cj-08-1041. [DOI] [PubMed] [Google Scholar]
- 131.Doyle A, Marganski W, Lee J. Calcium transients induce spatially coordinated increases in traction force during the movement of fish keratocytes. J Cell Sci. 2004;117(11):2203–2214. doi: 10.1242/jcs.01087. [DOI] [PubMed] [Google Scholar]
- 132.Munevar S, Wang YL, Dembo M. Regulation of mechanical interactions between fibroblasts and the substratum by stretch-activated Ca2+ entry. J Cell Sci. 2004;117(Pt 1):85–92. doi: 10.1242/jcs.00795. [DOI] [PubMed] [Google Scholar]
- 133.Hayakawa K, Tatsumi H, Sokabe M. Actin stress fibers transmit and focus force to activate mechanosensitive channels. J Cell Sci. 2008;121(4):496–503. doi: 10.1242/jcs.022053. [DOI] [PubMed] [Google Scholar]
- 134.Brundage RA, Fogarty KE, Tuft RA, Fay FS. Calcium gradients underlying polarization and chemotaxis of eosinophils. Science. 1991;254(5032):703–706. doi: 10.1126/science.1948048. [DOI] [PubMed] [Google Scholar]
- 135.Wei C, Wang X, Chen M, Ouyang K, Song LS, Cheng H. Calcium flickers steer cell migration. Nature. 2009;457(7231):901–905. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 137.Kim TJ, Seong J, Ouyang M, Sun J, Lu S, Hong JP, Wang N, Wang Y. Substrate rigidity regulates Ca2+ oscillation via RhoA pathway in stem cells. J Cell Physiol. 2009;218(2):285–293. doi: 10.1002/jcp.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Charras GT, Horton MA. Single Cell Mechanotransduction and Its Modulation Analyzed by Atomic Force Microscope Indentation. Biophysical Journal. 2002;82(6):2970–2981. doi: 10.1016/S0006-3495(02)75638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9(12):929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 140.Bloom RJ, George JP, Celedon A, Sun SX, Wirtz D. Mapping local matrix remodeling induced by a migrating tumor cell using three-dimensional multiple-particle tracking. Biophys J. 2008;95(8):4077–4088. doi: 10.1529/biophysj.108.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Raub CB, Unruh J, Suresh V, Krasieva T, Lindmo T, Gratton E, Tromberg BJ, George SC. Image correlation spectroscopy of multiphoton images correlates with collagen mechanical properties. Biophys J. 2008;94(6):2361–2373. doi: 10.1529/biophysj.107.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr Opin Cell Biol. 2006;18(5):524–532. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 143.Vogel V, Sheetz MP. Cell fate regulation by coupling mechanical cycles to biochemical signaling pathways. Curr Opin Cell Biol. 2009;21(1):38–46. doi: 10.1016/j.ceb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu WF, Nelson CM, Tan JL, Chen CS. Cadherins, RhoA, and Rac1 are differentially required for stretch-mediated proliferation in endothelial versus smooth muscle cells. Circ Res. 2007;101(5):e44–52. doi: 10.1161/CIRCRESAHA.107.158329. [DOI] [PubMed] [Google Scholar]


