Abstract
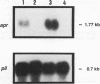
The lasR gene of Pseudomonas aeruginosa is required for transcription of the genes for elastase (lasB) and LasA protease (lasA), two proteases associated with virulence. We report here that the alkaline protease gene (apr) also requires the lasR gene for transcription. Alkaline protease mRNA was absent in the lasR mutant PAO-R1 and present when an intact lasR gene was supplied in trans as determined by Northern (RNA) analysis. The lasR gene also enhances exotoxin A production. Exotoxin A activity in supernatants of PAO-R1 were 30% less than in supernatants of the parental strain, PAO-SR. Multiple copies of lasR in trans in PAO-R1 in increased toxin A activity to twice the parental levels. Analysis of PAO-R1 containing the toxA promoter fused to beta-galactosidase suggests that LasR acts at the toxA promoter or at upstream toxA mRNA sequences. beta-Galactosidase activity was approximately 40% lower in PAO-R1 than in the parental strain, PAO-SR. Furthermore, the effect of LasR on the toxA promoter is not due to the stimulation of transcription of regA, a transcriptional activator of toxA. No difference in chloramphenicol acetyltransferase (CAT) activity was noted between PAO-SR and PAO-R1 containing transcriptional regA promoter-CAT gene fusions. These results broaden the regulatory dominion of lasR and suggest that the lasR gene plays a global role in P. aeruginosa pathogenesis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988 Apr 20;200(4):709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- Bjorn M. J., Sokol P. A., Iglewski B. H. Influence of iron on yields of extracellular products in Pseudomonas aeruginosa cultures. J Bacteriol. 1979 Apr;138(1):193–200. doi: 10.1128/jb.138.1.193-200.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Choi S. H., Greenberg E. P. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J Bacteriol. 1992 Jun;174(12):4064–4069. doi: 10.1128/jb.174.12.4064-4069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D. W., Collier R. J. Enzymatically active peptide from the adenosine diphosphate-ribosylating toxin of Pseudomonas aeruginosa. Infect Immun. 1977 Jun;16(3):832–841. doi: 10.1128/iai.16.3.832-841.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVault J. D., Hendrickson W., Kato J., Chakrabarty A. M. Environmentally regulated algD promoter is responsive to the cAMP receptor protein in Escherichia coli. Mol Microbiol. 1991 Oct;5(10):2503–2509. doi: 10.1111/j.1365-2958.1991.tb02096.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha M. A., Kropinski A. M. Construction of broad-host-range vectors for general cloning and promoter selection in Pseudomonas and Escherichia coli. Gene. 1989 Apr 30;77(2):205–210. doi: 10.1016/0378-1119(89)90068-1. [DOI] [PubMed] [Google Scholar]
- Frank D. W., Storey D. G., Hindahl M. S., Iglewski B. H. Differential regulation by iron of regA and toxA transcript accumulation in Pseudomonas aeruginosa. J Bacteriol. 1989 Oct;171(10):5304–5313. doi: 10.1128/jb.171.10.5304-5313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello M. J., Iglewski B. H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991 May;173(9):3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. C., Vasil M. L. Analysis of transcription of the exotoxin A gene of Pseudomonas aeruginosa. J Bacteriol. 1986 Dec;168(3):1112–1119. doi: 10.1128/jb.168.3.1112-1119.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindahl M. S., Frank D. W., Hamood A., Iglewski B. H. Characterization of a gene that regulates toxin A synthesis in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Jun 24;16(12):5699–5699. doi: 10.1093/nar/16.12.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B. W., Krishnapillai V., Morgan A. F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979 Mar;43(1):73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglewski B. H., Kabat D. NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin,. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2284–2288. doi: 10.1073/pnas.72.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. K., Boese-Marrazzo D. Production and properties of heat-stable extracellular hemolysin from Pseudomonas aeruginosa. Infect Immun. 1980 Sep;29(3):1028–1033. doi: 10.1128/iai.29.3.1028-1033.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. V., Hsieh H. Exotoxins of Pseudomonas aeruginosa. 3. Characteristics of antitoxin A. J Infect Dis. 1973 Oct;128(4):520–526. doi: 10.1093/infdis/128.4.520. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Iglewski B. H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985 Apr;31(4):387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- Ohman D. E., Sadoff J. C., Iglewski B. H. Toxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect Immun. 1980 Jun;28(3):899–908. doi: 10.1128/iai.28.3.899-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K., Morihara K., Atsumi Y., Takeuchi H., Kawamoto S., Kawasaki H., Suzuki K., Fukushima J. Complete nucleotide sequence of the structural gene for alkaline proteinase from Pseudomonas aeruginosa IFO 3455. Infect Immun. 1990 Dec;58(12):4083–4088. doi: 10.1128/iai.58.12.4083-4088.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., DeBusscher G., McCombie W. R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982 Apr;150(1):60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince R. W., Storey D. G., Vasil A. I., Vasil M. L. Regulation of toxA and regA by the Escherichia coli fur gene and identification of a Fur homologue in Pseudomonas aeruginosa PA103 and PA01. Mol Microbiol. 1991 Nov;5(11):2823–2831. doi: 10.1111/j.1365-2958.1991.tb01991.x. [DOI] [PubMed] [Google Scholar]
- Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983 Nov;25(1):71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Shortridge V. D., Pato M. L., Vasil A. I., Vasil M. L. Physical mapping of virulence-associated genes in Pseudomonas aeruginosa by transverse alternating-field electrophoresis. Infect Immun. 1991 Oct;59(10):3596–3603. doi: 10.1128/iai.59.10.3596-3603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol P. A., Cox C. D., Iglewski B. H. Pseudomonas aeruginosa mutants altered in their sensitivity to the effect of iron on toxin A or elastase yields. J Bacteriol. 1982 Aug;151(2):783–787. doi: 10.1128/jb.151.2.783-787.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey D. G., Frank D. W., Farinha M. A., Kropinski A. M., Iglewski B. H. Multiple promoters control the regulation of the Pseudomonas aeruginosa regA gene. Mol Microbiol. 1990 Mar;4(3):499–503. doi: 10.1111/j.1365-2958.1990.tb00616.x. [DOI] [PubMed] [Google Scholar]
- Toder D. S., Gambello M. J., Iglewski B. H. Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol Microbiol. 1991 Aug;5(8):2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- Vasil M. L., Grant C. C., Prince R. W. Regulation of exotoxin A synthesis in Pseudomonas aeruginosa: characterization of toxA-lacZ fusions in wild-type and mutant strains. Mol Microbiol. 1989 Mar;3(3):371–381. doi: 10.1111/j.1365-2958.1989.tb00182.x. [DOI] [PubMed] [Google Scholar]
- Whooley M. A., O'Callaghan J. A., McLoughlin A. J. Effect of substrate on the regulation of exoprotease production by Pseudomonas aeruginosa ATCC 10145. J Gen Microbiol. 1983 Apr;129(4):981–988. doi: 10.1099/00221287-129-4-981. [DOI] [PubMed] [Google Scholar]
- Wick M. J., Frank D. W., Storey D. G., Iglewski B. H. Identification of regB, a gene required for optimal exotoxin A yields in Pseudomonas aeruginosa. Mol Microbiol. 1990 Mar;4(3):489–497. doi: 10.1111/j.1365-2958.1990.tb00615.x. [DOI] [PubMed] [Google Scholar]