Abstract
The Us3 serine/threonine kinase encoded by all alphaherpesviruses performs several important functions during virus multiplication. For example, expression of pseudorabies virus (PRV) Us3 causes reorganization of the actin cytoskeleton into filamentous processes (FPs) that promote cell-to-cell spread of virus infection. PRV Us3-induced FP formation requires Us3 kinase activity. To determine whether these characteristics were shared by HSV-2 Us3, expression plasmids for wild type (WT) and kinase dead (KD) Us3 variants were constructed. Expression of WT Us3 resulted in robust FP formation whereas expression of the KD Us3 variants did not. In the course of these experiments we noted that KD/K220 mutant Us3s were excluded from the nucleus in comparison to WT or KD/D305A Us3, prompting us to investigate Us3 nuclear shuttling properties. Herein we describe determinants of HSV-2 Us3-induced FP formation and present evidence for the presence of a leucine-rich nuclear export signal within HSV-2 Us3.
Keywords: HSV-2, Us3, serine/threonine kinase, actin cytoskeleton, nuclear export signal
INTRODUCTION
Herpes simplex virus type 2 (HSV-2) is a member of the alphaherpesvirus sub-family of the Herpesviridae and is the primary causative agent of genital herpes. Like all alphaherpesviruses, HSV-2 encodes a serine/threonine kinase in the unique short region of its genome, designated Us3. Us3 has several known functions in alphaherpesvirus biology including prevention of virus-induced apoptosis (Geenen et al., 2005; Hata et al., 1999; Leopardi, Van Sant, and Roizman, 1997; Ogg et al., 2004), virion maturation (Reynolds et al., 2002; Schumacher et al., 2005; Wagenaar et al., 1995) and cell-to-cell spread of virus infection (Demmin et al., 2001; Favoreel et al., 2005). Expression of the Us3 gene from HSV-2, as well as Us3 genes from the related alphaherpesviruses pseudorabies virus (PRV) and Marek’s disease virus (MDV), results in alterations in the actin cytoskeleton (Calton et al., 2004; Murata et al., 2000; Schumacher et al., 2005; Van Minnebruggen et al., 2003; Van Minnebruggen et al., 2002). These cytoskeletal alterations include disassembly of actin stress fibres (Schumacher et al., 2005; Van Minnebruggen et al., 2003), and, in the case of PRV, formation of long filamentous processes (FPs) that are believed to facilitate the spread of virus to non-adjacent cells (Favoreel et al., 2005). Us3 kinase activity has been shown to be required for the cytoskeletal remodeling caused by HSV-2 Us3 (Murata et al., 2000) and PRV Us3 (Van den Broeke et al., 2009a) but is not required in the case of MDV Us3 (Schumacher et al., 2008). Recently, it has been demonstrated that the group I p21-activated kinases PAK1 and PAK2 play an essential role in PRV Us3-induced remodeling of the actin cytoskeleton (Van den Broeke et al., 2009b).
We wished to establish whether HSV-2 Us3 was capable of remodeling the actin cytoskeleton into FPs in transiently transfected cells and, if so, whether disruption of kinase activity in HSV-2 Us3 would correlate with disruption of FP formation. During the course of investigating these properties, we observed a striking difference in the subcellular localization of one of our kinase dead (KD) versions of HSV-2 Us3. This observation prompted us to investigate the nuclear shuttling properties of Us3. In this communication, we describe determinants of HSV-2 Us3-induced FP formation and present evidence for the presence of a leucine-rich nuclear export signal within HSV-2 Us3.
RESULTS
Expression of HSV-2 Us3 in transfected cells results in FP formation
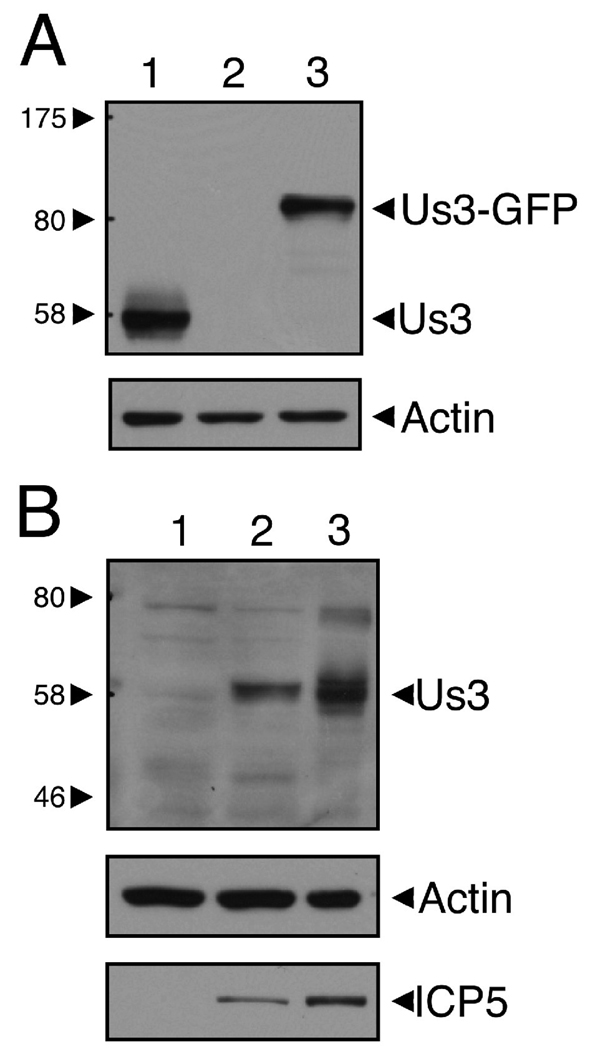
To enable the detection of HSV-2 Us3 in transfected cells, rat polyclonal antiserum was raised against a GST-HSV-2 Us3 fusion protein produced by recombinant baculoviruses. This antiserum detected a protein slightly larger than the predicted molecular weight for HSV-2 Us3 (53 kDa) in extracts prepared from 293T cells transfected with a HSV-2 Us3 expression construct (Figure 1A, lane 1) as well as in extracts prepared from Vero cells infected with HSV-2 (Figure 1B, lane 3). This antiserum was also able to detect HSV-1 Us3 (Figure 1B, lane 2).
Figure 1.
Polyclonal antiserum from rats immunized with GST-HSV-2 Us3 specifically detects HSV-2 Us3. A. Equal volumes of cellular extracts prepared from 293T cells transfected with a plasmid encoding HSV-2 Us3 (lane 1), GFP (lane 2), or HSV-2 Us3-GFP (lane 3) were analyzed by Western blotting. The upper panel was probed with rat polyclonal HSV-2 Us3 antiserum and the loading control in the lower panel was probed with anti-actin monoclonal antibody. No bands were detected in lane 2 of the upper panel (including bands smaller than 58 kDa, which are not shown). The major band detected in lane 1 is slightly larger than the predicted molecular weight of HSV-2 Us3 (53 kDa). Note the shift in molecular weight of the detected band in cells transfected with plasmid encoding HSV-2 Us3-GFP fusion protein. B. Equal volumes of cellular extracts prepared from mock-infected (lane 1), HSV-1 17+-infected (lane 2), or HSV-2 HG52-infected Vero cells (lane 3) were analyzed by Western blotting. The upper panel was probed with rat polyclonal HSV-2 Us3 antiserum, the loading control in the middle panel was probed with anti-actin monoclonal antibody, and the infection control in the lower panel was probed with anti-HSV-2 ICP5. Molecular size markers indicated at the left of each upper panel are in kilodaltons.
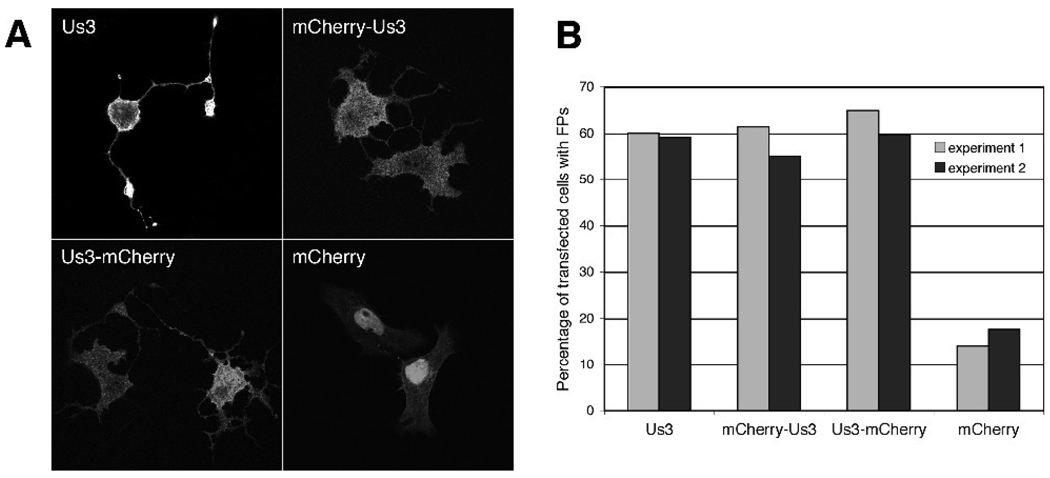
To determine whether HSV-2 Us3 was capable of eliciting FP formation, Vero cells were transfected with a plasmid encoding HSV-2 Us3 along with a plasmid encoding the red fluorescent protein mCherry (Shaner et al., 2004), to aid in identifying transfected cells, then stained with HSV-2 Us3 antiserum and examined by confocal microscopy. HSV-2 Us3 could be detected in both the nucleus and the cytoplasm in transfected cells (Figure 2A; see also Figure 5B and Figure 6). By 24 hours post transfection, the majority of transfected cells had formed long FPs, depicted in Figure 2A, similar to those we have observed previously with PRV Us3 (Calton et al., 2004). The length of the FPs formed was often greater than the length of one cell and elaborate branching of FPs was commonly observed. FPs with similar characteristics were also observed in cells transfected with plasmids encoding HSV-2 Us3 mCherry fusion proteins (denoted mCherry-Us3 and Us3-mCherry in Figure 2A) and in cells transfected with a plasmid encoding HSV-2 Us3 with GFP fused to the C-terminus (data not shown). In addition, comparable numbers of transfected cells with FPs were seen in cells transfected with plasmids encoding mCherry-fused and unfused HSV-2 Us3 (Figure 2B). These data demonstrate that transient expression of HSV-2 Us3 in cells results in FP formation and that fusion of a fluorescent protein tag to either end of HSV-2 Us3 does not impede its ability to induce FP formation.
Figure 2.
Expression of HSV-2 Us3 in transfected cells results in FP formation. A. Representative images of FPs formed in Vero cells transfected with plasmids encoding HSV-2 Us3, mCherry fusions to HSV-2 Us3 or mCherry alone. In the upper left panel, staining with polyclonal antiserum specific for HSV-2 Us3 followed by staining with an Alexa 488 conjugated secondary antibody was used to detect cells expressing HSV-2 Us3. Stained cells were examined by confocal microscopy. B. Percentage of transfected cells with FPs. 50 independent fields of cells containing at least one transfected cell were scored for the presence or absence of FPs. Results from two independent experiments are shown. Numbers of cells scored in experiment 1 and experiment 2 were as follows: for WT Us3 – 115 and 127; for mCherry-Us3 – 145 and 156; for Us3 mCherry – 168 and 154; for mCherry – 206 and 164.
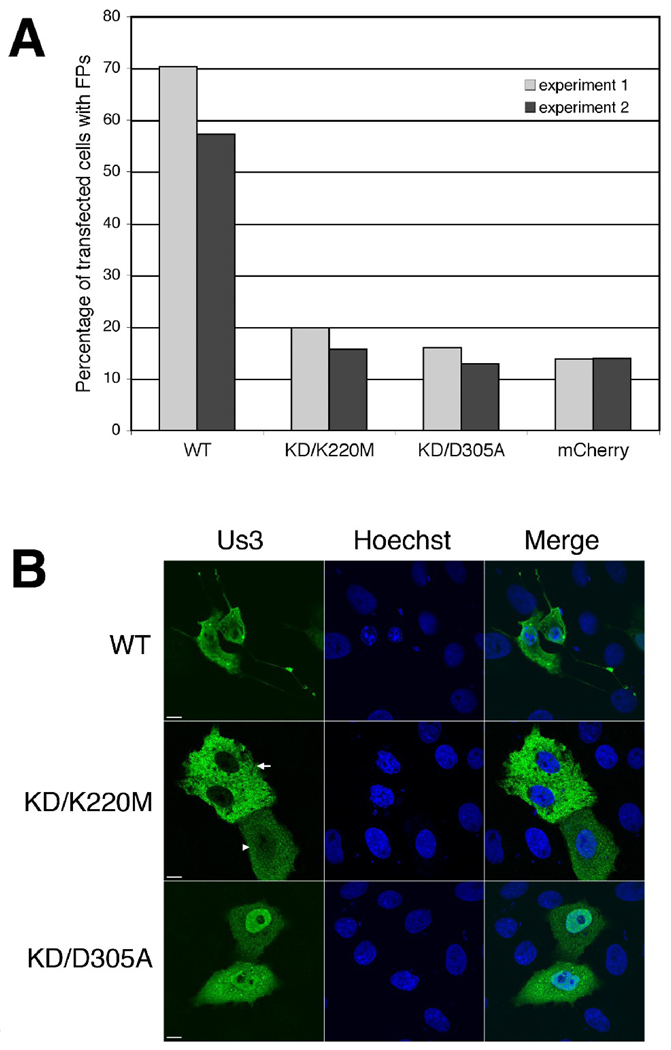
Figure 5.
Us3 kinase activity is required for HSV-2 Us3-induced FP formation. A. Percentage of transfected cells with FPs. Vero cells were transfected with plasmids encoding the indicated proteins. At 24 hours post transfection, cells were fixed and stained with rat polyclonal antiserum specific for HSV-2 Us3 followed by staining with an Alexa 488 conjugated donkey anti-rat secondary antibody. Nuclei were stained with Hoechst 33342. Stained cells were examined by confocal microscopy and 50 independent fields of cells containing at least one transfected cell were scored for the presence or absence of FPs. Results from two independent experiments are shown. Numbers of cells scored in experiment 1 and experiment 2 were as follows: for WT – 111 and 124; for KD/K220M – 151 and 179; for KD/D305A – 138 and 163; for mCherry – 123 and 187. B. Representative images of transfected cells used in this analysis. Arrow and arrowhead indicate the two different subcellular localization patterns of Us3 observed in cells transfected with a plasmid encoding KD/K220M Us3. Scale bar is 10 µm.
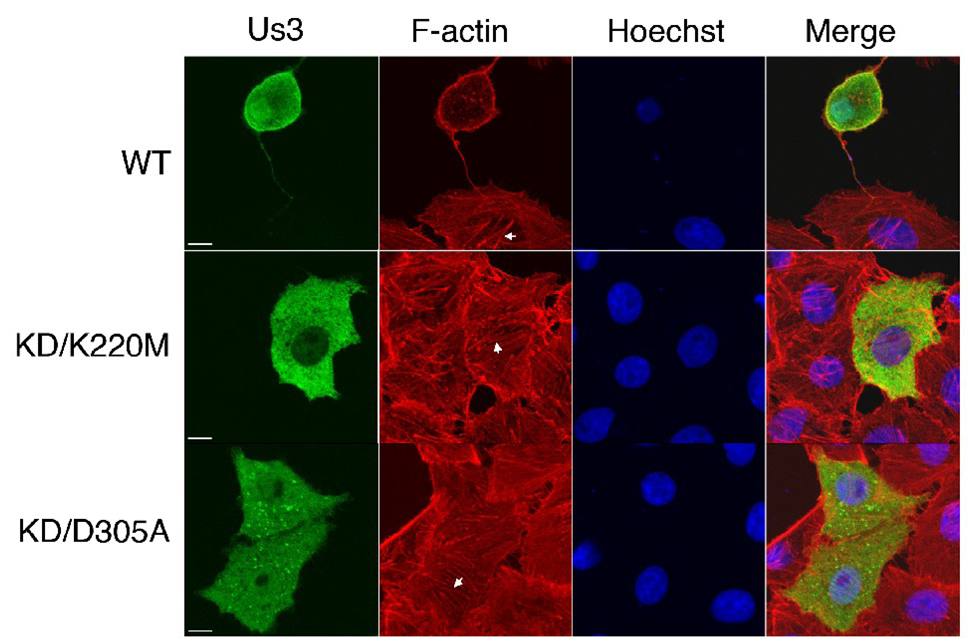
Figure 6.
Us3 kinase activity is required for HSV-2 Us3-induced actin stress fibre breakdown. Vero cells were transfected with plasmids encoding WT, KD/K220M or KD/D305A Us3. At 24 hours post transfection, cells were fixed and stained with rat polyclonal antiserum specific for HSV-2 Us3 followed by staining with an Alexa 488 conjugated donkey anti-rat secondary antibody. Filamentous actin was detected using Texas Red conjugated phalloidin and nuclei were stained with Hoechst 33342. Stained cells were examined by confocal microscopy. Representative images are shown. Arrows indicate actin stress fibres. Scale bar is 10 µm.
Actin polymerization contributes to HSV-2 Us3-induced FP formation
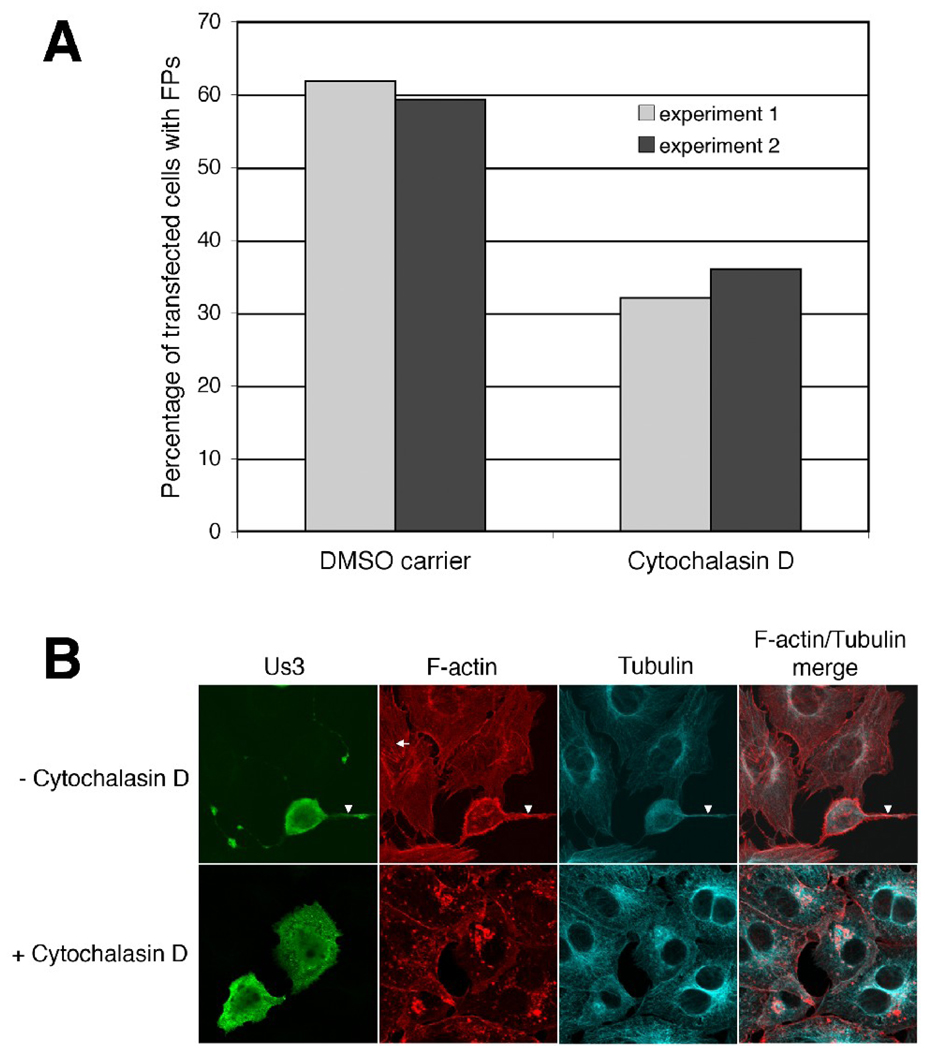
To investigate the specific involvement of actin polymerization in FP formation, Vero cells were transfected with a plasmid encoding HSV-2 Us3 and subsequently incubated with 1 µM cytochalasin D. Filamentous actin was detected using Texas red conjugated phalloidin and microtubules were detected using a monoclonal antibody specific for β-tubulin. The percentage of transfected cells with FPs was diminished in the presence of 1 µM cytochalasin D as compared to cells treated with carrier (0.01% DMSO) (Figure 3A) indicating that the ability to polymerize actin contributes to HSV-2 Us3-induced FP formation. In keeping with previous studies on HSV-2 Us3 (Murata et al., 2000), transfected cells consistently displayed a marked disruption in actin stress fibres; actin stress fibre breakdown occurred regardless of whether the transfected cell had formed FPs (data not shown). By contrast, microtubules in transfected cells remained intact (Figure 3B). FPs induced by HSV-2 Us3 contained both actin and β-tubulin (Figure 3B) as has been observed previously with FPs induced by PRV Us3 (Calton et al., 2004). Thus, the reorganization of the actin cytoskeleton into FPs that contain both actin and microtubules is a common consequence of the expression of PRV and HSV-2 Us3.
Figure 3.
FP formation in the presence of cytochalasin D. A. Percentage of transfected cells with FPs. Vero cells were transfected with a plasmid encoding HSV-2 Us3. At 6 hours post transfection cells were placed in medium containing 1 µM cytochalasin D or 0.01% DMSO (carrier). Cells were incubated in the continuous presence of drug or carrier for 12 hours, then fixed and stained with rat polyclonal antiserum specific for HSV-2 Us3 and mouse monoclonal antibody specific for β-tubulin, followed by staining with Alexa 488 conjugated donkey anti-rat and Alexa 647 conjugated donkey anti-mouse secondary antibodies. Filamentous actin was detected using Texas Red conjugated phalloidin. Stained cells were examined by confocal microscopy and 50 independent fields of cells containing at least one transfected cell were scored for the presence or absence of FPs. Results from two independent experiments are shown. Numbers of cells scored in experiment 1 and experiment 2 were as follows: for Us3-transfected cells treated with DMSO carrier – 110 and 167; for Us3-transfected cells treated with cytochalasin D – 112 and 125. B. Representative images of transfected cells used in this analysis. Arrows indicate actin stress fibres and arrowheads indicate FPs that contain both actin and β-tubulin. Note that actin stress fibres, but not microtubules, are markedly disrupted in cytochalasin D-treated cells.
Us3 kinase activity is required for HSV-2 Us3-induced FP formation and actin stress fibre breakdown
As stated above, the requirement for Us3 kinase activity in Us3-induced cytoskeletal alterations varies between the alphaherpesviruses (Murata et al., 2000; Schumacher et al., 2008; Van den Broeke et al., 2009a). Thus, we wished to establish whether disruption of kinase activity in HSV-2 Us3 would result in a disruption in FP formation and/or actin stress fibre breakdown. Previous work correlating HSV-2 Us3 kinase activity with cell rounding, an indirect read-out of actin stress fibre disruption, had utilized kinase dead (KD) versions of HSV-2 Us3 carrying large deletions (Murata et al., 2000). We wished to construct KD versions of HSV-2 Us3 carrying more precise disruptions in kinase activity. Two different KD versions of HSV-2 Us3 were constructed for our initial investigations. Serine-threonine kinases, including HSV-1 Us3, have been subdivided into eleven subdomains, designated I through XI (Hanks, Quinn, and Hunter, 1988). Subdomain II contains a conserved lysine that is required for catalytic activity (Hanks and Hunter, 1995; Hanks, Quinn, and Hunter, 1988; Kato et al., 2005; Mou, Forest, and Baines, 2007); subdomain VI contains a conserved aspartic acid that is similarly required for catalytic activity (Hanks and Hunter, 1995; Hanks, Quinn, and Hunter, 1988; Knighton et al., 1991). By alignment with HSV-1 Us3, we identified the invariant lysine and aspartic acid in HSV-2 Us3 and changed lysine 220 to methionine (KD/K220M) and aspartic acid 305 to alanine (KD/D305A).
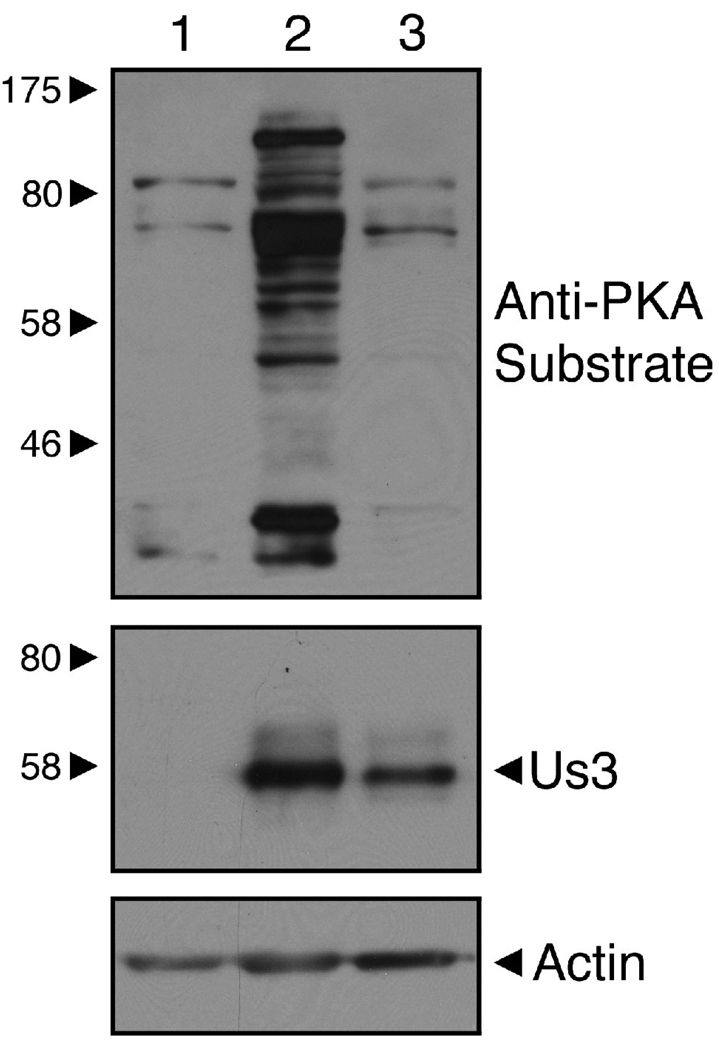
It is well established that mutation of HSV-1 Us3 K220 (K220A or K220M) results in a catalytically inactive enzyme (Kato et al., 2005; Mou, Forest, and Baines, 2007). To confirm that the D305A version of HSV-2 Us3 we constructed lacked kinase activity, whole cell extracts prepared from Vero cells transfected with plasmids encoding WT or KD/D305A were analyzed by Western blotting using antisera reactive against protein kinase A (PKA) substrates (Figure 4). Because of the considerable homology between Us3 and PKA phosphorylation sites, this antisera has been used detect Us3 specific kinase activity (Benetti and Roizman, 2004). Similar levels of phosphorylation were observed in cell lysates prepared from Vero cells transfected with either empty vector or the D305A mutant (Figure 4, lanes 1 and 3). By contrast, expression of WT Us3 resulted in both an increase in the number of phosphorylated proteins detected as well as a marked increase in signal intensity (Figure 4, lane 2). These data indicate that D305A lacks kinase activity.
Figure 4.
HSV-2 Us3 kinase activity is disrupted in a D305A mutant. Equal volumes of cellular extracts prepared from Vero cells transfected with pCINeo vector (lane 1), a plasmid encoding WT HSV-2 Us3 (lane 2), or a plasmid encoding a D305A mutant of HSV-2 Us3 (lane 3) were analyzed by Western blotting. The upper panel was probed with Phospho-(Ser/Thr) PKA substrate antibody, the middle panel was probed with rat polyclonal HSV-2 Us3 antiserum and the loading control in the lower panel was probed with anti-actin monoclonal antibody. Molecular size markers indicated at the left of the panel are in kilodaltons.
Vero cells were transfected with plasmids encoding WT or KD versions of HSV-2 Us3, stained with HSV-2 Us3 antiserum at 24 hours post transfection and then examined by confocal microscopy. The percentage of transfected cells with FPs was diminished in cells transfected with plasmids encoding either KD version of HSV-2 Us3 in comparison to cells transfected with a plasmid encoding WT HSV-2 Us3 (Figure 5A). Similar reductions in FP formation were also observed in cells transfected with KD mCherry-Us3 or GFP fusion proteins (data not shown). The FPs present in cells transfected with plasmids encoding either KD version of HSV-2 Us3 were shorter and tended to be less branched than those observed in cells transfected with a plasmid encoding WT HSV-2 Us3. Consistent with previous work on HSV-2 Us3 (Murata et al., 2000), KD versions did not generally cause cell rounding and breakdown of actin stress fibres (Figure 6). Thus, in terms of the requirement for kinase activity in Us3-induced cytoskeletal alterations, HSV-2 is more similar to PRV than MDV.
Nuclear localization of WT and KD versions of HSV-2 Us3 in transfected cells
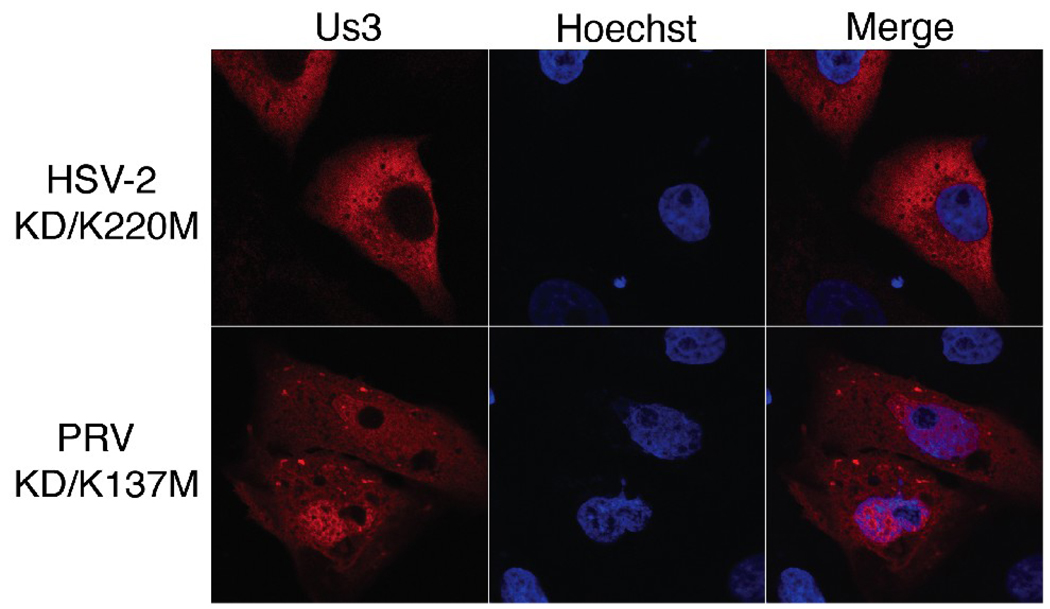
During our characterization of KD versions of HSV-2 Us3, we noted that KD/K220M Us3 often appeared to be excluded from the nucleus of transfected Vero cells, particularly in cells displaying robust expression (Figure 5B). Nuclear exclusion of KD/K220M Us3 was also observed in transfected 293T and HeLa cells (data not shown) and in cells transfected with plasmids encoding KD/K220M Us3-mCherry fusion proteins (Figure 7). To quantify this observation, 50 independent fields of cells transfected with plasmids encoding WT, KD/K220M and KD/D305A HSV-2 Us3 were scored for whether Us3 appeared to be excluded from the nucleus (the staining pattern designated with an arrow in Figure 5B) or whether Us3 could be detected in both nucleus and the cytoplasm (the staining pattern designated with an arrowhead in Figure 5B). In the majority of cells transfected with a plasmid encoding KD/K220M Us3 (93 out of 151 cells in experiment 1 and 117 out of 179 cells in experiment 2), Us3 appeared to be excluded from the nucleus. This was in sharp contrast to cells transfected with either WT or KD/D305A Us3, where nuclear exclusion was rarely observed (3 out of 111 cells or 4 out of 138 cells, respectively in experiment 1 and 0 out of 124 cells or 8 out of 163 cells, respectively in experiment 2). To determine whether the loss of a lysine or conversely, the introduction of a methionine, was responsible for the subcellular localization pattern of K220M, we constructed and analyzed a K220A version of HSV-2 Us3. The subcellular localization pattern of this mutant resembled that of K220M (data not shown). Collectively, these data indicate that mutation of K220, independent of the loss of kinase activity, influences the nuclear localization of HSV-2 Us3. To determine whether this is a common feature of alphaherpesvirus Us3 orthologues, the corresponding mutation was made in PRV Us3 (KD/K137M Us3) and the mutated protein was fused to monomeric red fluorescent protein (mRFP1)(Campbell et al., 2002) to enable its detection in transfected cells. Nuclear exclusion was not observed in cells transfected with a plasmid encoding PRV KD/K137M-mRFP1 Us3 (Figure 7). Thus the involvement of this lysine in nuclear localization may not apply to all alphaherpesvirus Us3 proteins.
Figure 7.
Subcellular localization of PRV KD/K137M Us3. Vero cells were transfected with plasmids encoding mCherry-HSV-2 KD/K220M Us3 or PRV KD/K137M-mRFP1 Us3. At 24 hours post transfection, cells were fixed, nuclei were stained with Hoechst 33342 and examined by confocal microscopy. Representative images are shown.
Evidence for the presence of a leucine-rich nuclear export signal in HSV-2 Us3
The aberrant subcellular localization pattern of Us3 in cells transfected with KD/K220M Us3 prompted us to explore the nuclear shuttling properties of HSV-2 Us3. Conceivably, inhibition of nuclear import of Us3 and/or increase in nuclear export of Us3 could cause Us3 to preferentially accumulate in the cytoplasm. Inspection of the primary amino acid sequence of HSV-2 Us3 did not reveal the presence of lysine/arginine-rich motif conforming to a consensus single or bipartite nuclear localization signal (Dingwall, Sharnick, and Laskey, 1982; Kalderon et al., 1984; Robbins et al., 1991). However, HSV-2 Us3 contains two potential leucine-rich nuclear export signals (NESs)(Bogerd et al., 1996) located within the vicinity of valine 250 (Figure 8A), a residue that is predicted to be part of a leucine-rich NES as determined by the NetNES 1.1 algorithm (la Cour et al., 2004). Both potential NESs are identical in HSV-1 Us3 (Figure 8B). Although the presence of potential leucine-rich NESs in the same region of Us3 orthologues of other alphaherpesviruses was variable, the key hydrophobic residues at the beginning of the first potential NES and the end of the second potential NES were well conserved (Figure 8B).
Figure 8.
Predicted leucine-rich NESs in Us3. A. Predicted leucine-rich NESs in HSV-2 Us3. The top line depicts the most promiscuous consensus sequence encompassing 72% of experimentally verified leucine-rich NESs (la Cour et al., 2004); x’s correspond to any amino acid. Below are two tracts of sequence from HSV-2 Us3, designated 1 and 2, that fit this consensus sequence; amino acids corresponding to the x’s in the consensus sequence are shown in smaller font. Numbers in brackets below each predicted HSV-2 Us3 NES indicate the NES designation along with amino acid numbers that constitute the NES. B. Alignment of Us3 orthologues from other alphaherpesviruses. A portion of a multiple sequence alignment of Us3s from HSV-2, HSV-1, PRV, MDV, varicella zoster virus (VZV), bovine herpesvirus type 1 (BHV-1), and equine herpesvirus type 1 (EHV-1) generated using ClustalW v1.4 multiple sequence alignment software with a blosum similarity matrix is shown. Numbers in brackets correspond to the amino acid numbers of the portion of the protein shown. Identical amino acids are indicated with an asterisk; conserved amino acids are indicated with a period. Arrows above the alignment indicate the amino acids that constitute the two predicted leucine-rich NESs in HSV-2 Us3. The yes (Y) or no (N) designations at the far right indicate whether a sequence conforming to the consensus sequence shown in panel A is present in the portion of the Us3 protein shown.
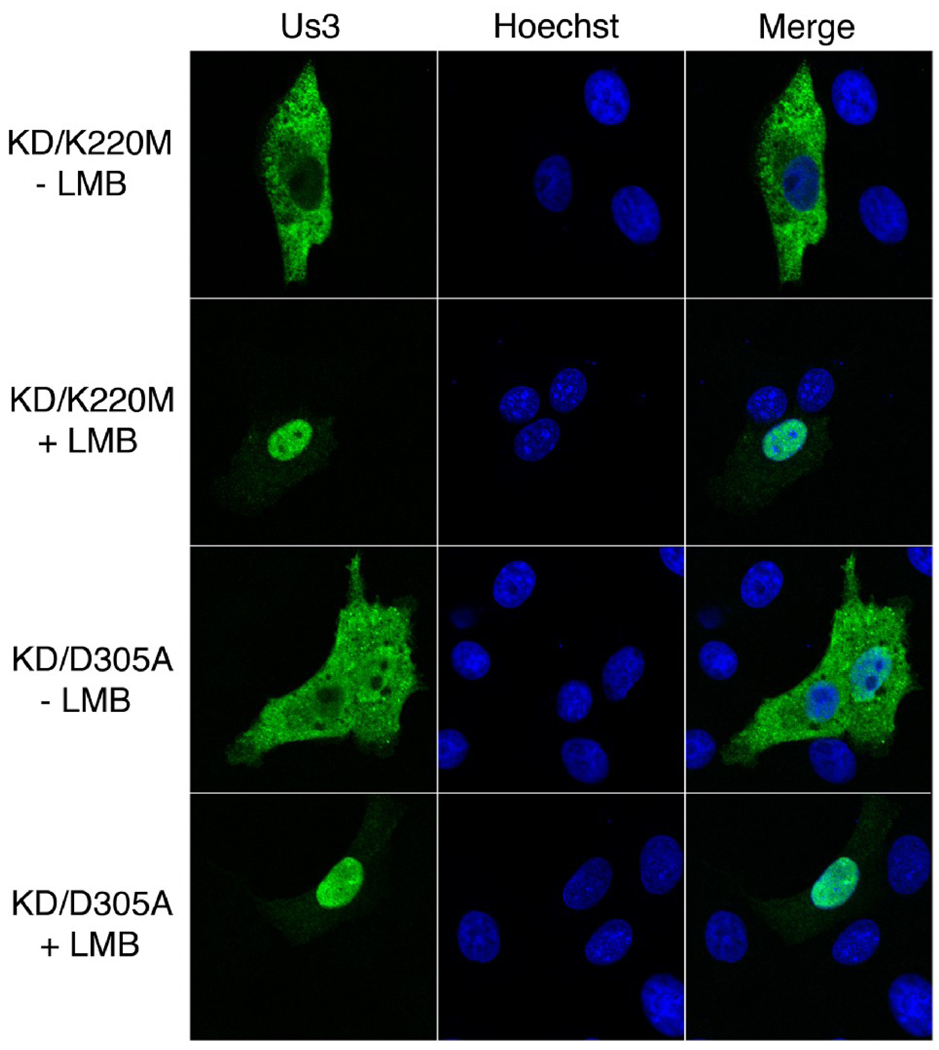
Export of protein from the nucleus via leucine-rich NESs is known to be sensitive to leptomycin B (LMB)(Fornerod et al., 1997; Wolff, Sanglier, and Wang, 1997). To test whether HSV-2 Us3 subcellular localization was affected by LMB, Vero cells were transfected with plasmids encoding KD/K220M or KD/D305A Us3 and cells were subsequently incubated with 10 ηM LMB or 0.07% methanol (carrier) for 16 hours. We utilized KD versions of HSV-2 Us3 for our initial analysis to ensure that transfected cells would remain flat and thus allow better imaging of the subcellular distribution of Us3. In many of the cells treated with LMB (46 out of 100 cells and 56 out of 100 cells for KD/K220M and KD/D305A, respectively), Us3 was located predominately within the nucleus (Figure 9, “+ LMB” panels). This staining pattern was more rarely observed in cells treated with carrier (2 out of 100 cells and 12 out of 100 cells for KD/K220M and KD/D305A, respectively). Thus the subcellular localization pattern of KD HSV-2 Us3 is sensitive to LMB, consistent with the presence of a leucine-rich NES within this protein.
Figure 9.
Subcellular localization of HSV-2 Us3 in the presence of LMB. Vero cells were transfected with plasmids encoding the indicated proteins. At 6 hours post transfection cells were placed in medium containing 10 ηM LMB (+ LMB) or 0.07% methanol carrier (− LMB). Cells were incubated in the continuous presence of drug or carrier for 16 hours, then fixed and stained with rat polyclonal antiserum specific for HSV-2 Us3 followed by staining with an Alexa 488 conjugated secondary antibody. Nuclei were stained with Hoechst 33342. Stained cells were examined by confocal microscopy. A large proportion of transfected cells treated with LMB showed the staining pattern depicted in the “+ LMB” panels.
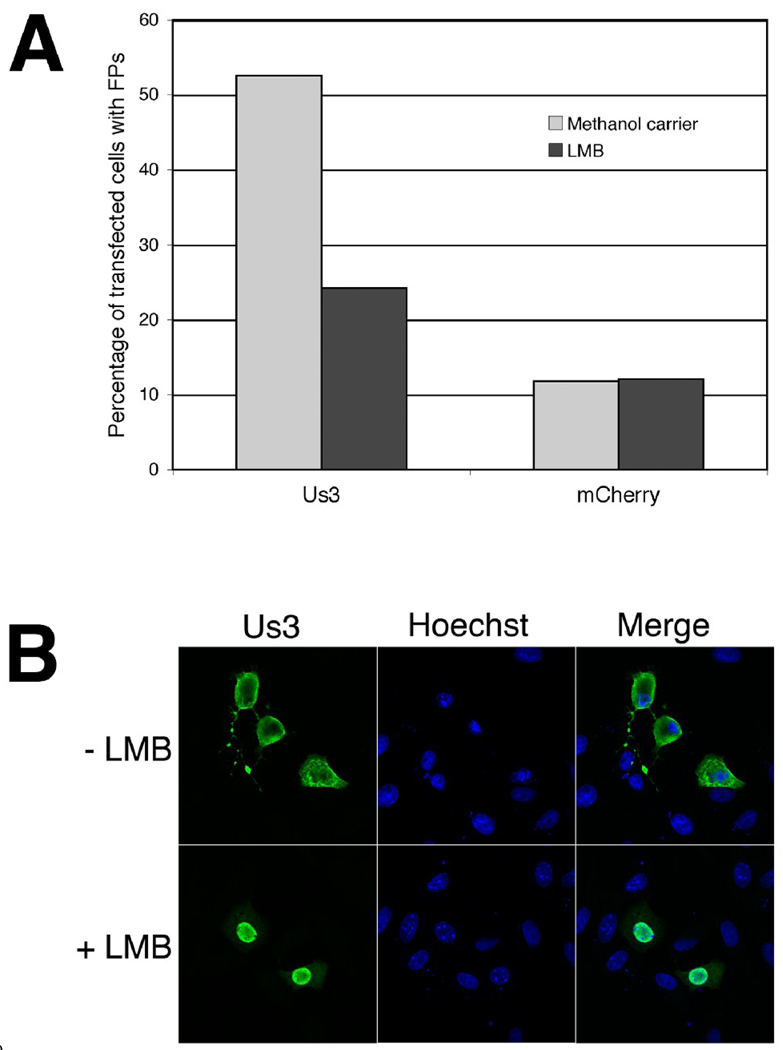
Based on the above findings, we reasoned that LMB treatment might provide a means for distinguishing the roles of nuclear and cytoplasmic HSV-2 Us3 in FP formation. Vero cells were transfected with a plasmid encoding WT HSV-2 Us3 and cells were subsequently incubated with 10 ηM LMB or carrier for 16 hours. Transfected cells were then examined for the subcellular localization of Us3 and for the presence or absence of FPs. The percentage of Us3-transfected cells with FPs was diminished in the presence of 10 ηM LMB as compared to cells treated with carrier (Figure 10A). The percentage of mCherry-transfected cells with FPs did not change in the presence of 10 ηM LMB. Moreover, the FPs formed in the presence of LMB tended to be shorter and much less elaborate than FPs formed in the presence of carrier. In a large proportion of cells treated with LMB (68 out of 101 cells and 71 out of 106 cells in the experiment reported in Figure 10A), Us3 was located predominately within the nucleus (as depicted in Figure 10B, “+ LMB” panel); this staining pattern was more rarely observed in cells treated with carrier (8 out of 137 cells and 9 out of 135 cells in the experiment reported in Figure 10A). Furthermore, the vast majority of cells with Us3 located predominately within the nucleus lacked FPs (55 out of 68 cells and 55 out of 71 cells in the LMB-treated cells in the experiment reported in Figure 10A). These data indicate that LMB treatment has an adverse effect on HSV-2 Us3-induced FP formation and lend support to the notion that cytoplasmic HSV-2 Us3 is specifically involved in actin cytoskeleton remodeling. Thus, subcellular localization can be included along with kinase activity as determinants of HSV-2 Us3-induced FP formation.
Figure 10.
FP formation in the presence of LMB. A. Percentage of transfected cells with FP. Vero cells were transfected in duplicate with plasmids encoding the indicated proteins. At 6 hours post transfection cells were placed in medium containing 10 ηM LMB or 0.07% methanol carrier. Cells were incubated in the continuous presence of drug or carrier for 16 hours, then fixed and stained with rat polyclonal antiserum specific for HSV-2 Us3 followed by staining with an Alexa 488 conjugated secondary antibody. Nuclei were stained with Hoechst 33342. Stained cells were examined by confocal microscopy and transfected cells were scored for the presence or absence of FPs. The average of duplicate samples is shown. Numbers of cells scored are as follows: for Us3-transfected cells treated with carrier – 137 and 135; for Us3-transfected cells treated with LMB – 101 and 106; for mCherry-transfected cells treated with carrier – 122 and 134; for mCherry-transfected cells treated with LMB – 104 and 103. B. Images of transfected cells used in this analysis. A large proportion of transfected cells treated with LMB showed the staining pattern depicted in the “+ LMB” panels.
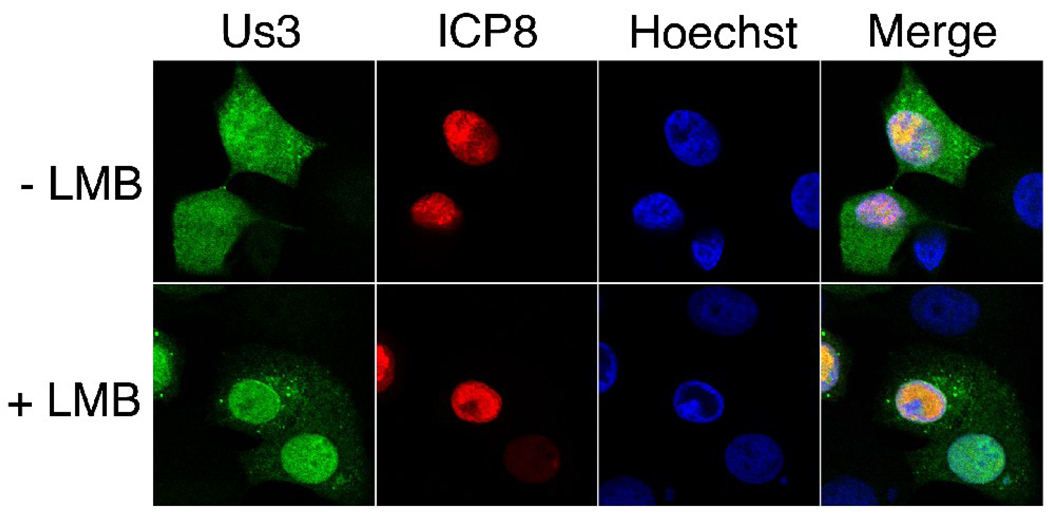
To determine if there was evidence for a functional Us3 NES in virally infected cells, we examined Us3 localization in HSV-2 infected Vero cells in the presence and absence of LMB. Vero cells growing on glass bottom dishes were infected with HSV-2 (HG52) at a multiplicity of infection of 10. At 2 hours post infection either 10 ηM LMB or carrier (0.07% methanol) was added to the cells and the infection was allowed to proceed until 8 hours post infection. Cells were then fixed and stained for Us3 and ICP8, an early viral protein that localizes to the nucleus (Fenwick, Walker, and Petkevich, 1978; Knipe and Spang, 1982; Quinlan, Chen, and Knipe, 1984) (Figure 11). In the absence of LMB, Us3 was distributed throughout the nucleus and cytoplasm as well as to small punctate cytoplasmic structures reminiscent of those seen in PRV infected cells stained with Us3 antiserum (Lyman, Demmin, and Banfield, 2003). In the presence of LMB, Us3 appeared predominantly nuclear in many infected cells. The Us3 localization differences observed in infected cells in the presence of LMB were less dramatic than was observed in Us3-transfected cells in the presence of LMB. Nevertheless, our results with infected cells are in keeping with our results with transfected cells and consistent with the idea that Us3 shuttles between the nucleus and cytoplasm during viral infection.
Figure 11.
Us3 localization in HSV-2 infected cells in the presence or absence of LMB. Vero cells were infected with HSV-2 HG52 at an MOI of 10. At 2 hours post infection cells were placed in medium containing 10 ηM LMB (+ LMB) or 0.07% methanol carrier (− LMB). Cells were incubated in the continuous presence of drug or carrier for 6 hours, then fixed and stained with rat polyclonal antiserum specific for HSV-2 Us3 and mouse monoclonal antibody specific for ICP8, followed by staining with Alexa 488 conjugated donkey anti-rat and Alexa 568 conjugated donkey anti-mouse secondary antibodies. Nuclei were stained with Hoechst 33342. Stained cells were examined by confocal microscopy. Representative images are shown.
DISCUSSION
These investigations were initiated with the goal of establishing whether HSV-2 Us3 was able to induce FP formation as we, and others, have demonstrated for PRV Us3 (Calton et al., 2004; Favoreel et al., 2005). The results of our studies establish that FP formation is a consequence of HSV-2 Us3 expression, and furthermore, that Us3 kinase activity is required for FP formation. During the course of determining the requirement for kinase activity in FP formation, we noted that a KD version of HSV-2 Us3, in which a lysine in catalytic subdomain II (Hanks, Quinn, and Hunter, 1988) was changed to methionine (KD/K220M), was often excluded from the nucleus. Exclusion of Us3 from the nucleus was also apparent in HSV-2 Us3 mutants carrying a deletion encompassing catalytic subdomains I and II (Murata et al., 2000). Nuclear exclusion was not observed with a distinct KD HSV-2 Us3 in which an aspartic acid in catalytic subdomain VI was changed to alanine (KD/D305A). These data indicate that the K220M mutation, rather than loss of kinase activity, is responsible for the aberrant subcellular localization. Furthermore, the localization of the Us3 K220A mutant resembled that of the Us3 K220M mutant, suggesting that it is the loss of a lysine at position 220 rather than the gain of a methionine that is responsible for nuclear exclusion of HSV-2 Us3. Thus, we have identified a single amino acid in HSV-2 Us3 that is involved in nuclear localization. A determinant of nuclear export of HSV-2 Us3 was also identified in these studies. The results of our experiments with LMB, a specific inhibitor of nuclear export via leucine-rich NESs (Wolff, Sanglier, and Wang, 1997), in both transfected and infected cells provide strong evidence for the presence of a leucine-rich NES in HSV-2 Us3.
It is well established that the Us3 kinase plays a variety of roles in alphaherpesvirus multiplication (Demmin et al., 2001; Favoreel et al., 2005; Geenen et al., 2005; Hata et al., 1999; Leopardi, Van Sant, and Roizman, 1997; Ogg et al., 2004; Reynolds et al., 2002; Schumacher et al., 2005; Wagenaar et al., 1995). It follows that Us3 would need to localize to multiple subcellular compartments to carry out its different functions. Indeed, detailed analysis of subcellular localization of the PRV Us3 orthologue has established that Us3 contains intrinsic mitochondrial, nuclear and membrane localization signals (Calton et al., 2004; Van Minnebruggen et al., 2003). It is expected that active transport of HSV-2 Us3 is required in order to move this protein into or out of the nucleus because it has a predicted molecular mass of 53 kDa, which is larger than the limit for passive diffusion of molecules through nuclear pores (Tran and Wente, 2006). It is known that Us3 interacts with substrates in both the nucleus and cytoplasm (Mou, Forest, and Baines, 2007; Munger and Roizman, 2001; Poon, Liang, and Roizman, 2003; Van den Broeke et al., 2009b). Nuclear shuttling would allow Us3 to access substrates that are located exclusively in either the nucleus or the cytoplasm and allow continued access to a substrate if the subcellular localization of that substrate changes. Thus, maintaining the ability to shuttle between the nucleus and cytoplasm is likely vital for Us3 to carry out its different functions.
In our current model for nuclear shuttling of HSV-2 Us3, nuclear import proceeds via an intrinsic, non-canonical NLS or by interaction with a cellular protein carrying an NLS. Export of HSV-2 Us3 proceeds via a leucine-rich NES that may reside within amino acids 242–259 (Figure 8A). This process is sensitive to LMB, implicating the specific involvement of the chromosomal regional maintenance 1 (CRM1) -mediated nuclear export pathway (Fornerod et al., 1997). Our studies do not yet allow us to distinguish whether cytoplasmic accumulation of KD/K220 mutants arises as a result of impaired nuclear import or enhanced nuclear export of Us3. This will be an important issue to resolve in our continuing studies on this protein.
Our previous studies on PRV Us3 demonstrated that determinants of nuclear localization are contained within amino acids 54 through 156 (Calton et al., 2004). The lysine that we have identified as important for proper nuclear localization does fall within the analogous N-terminal region of HSV-2 Us3 (amino acids 64 through 241 using ClustalW v1.4 multiple sequence alignment software with a blosum similarity matrix). Mutation of the analogous lysine in PRV Us3, however, did not result in nuclear exclusion (Figure 7). Also, the potential NESs we have identified in HSV-2 Us3 are not found in the corresponding region of PRV Us3 (Figure 8B) and the NetNES 1.1 algorithm (la Cour et al., 2004) did not predict any amino acids having the potential to be part of a leucine-rich NES in PRV Us3. Thus, the specific requirements and/or mechanisms for nuclear localization of PRV Us3 may be different from those of HSV-2 Us3.
We noted that LMB treatment of HSV-2 Us3-transfected cells correlated with failure to produce FPs (Figure 10A). We ascribed this result to an inability of HSV-2 Us3 to be exported to the cytoplasm where it is required to elicit FPs. However, it is formally possible that LMB inhibits FP formation in a manner that is independent of its effects on Us3 localization. To address this, experiments are underway to define the sequence requirements of the NES of HSV-2 Us3. This will enable the construction of Us3 mutants that are unable to localize to the cytoplasm and allow us to test the idea that cytoplasmic Us3 is required for FP formation without the use of pharmacological inhibitors.
Based on our current understanding of the requirements for proper nuclear localization of HSV-2 Us3, the more pragmatic choice of KD mutant for use in studies where nuclear localization of Us3 is required would be KD/D305A. Our current efforts to map the NES of HSV-2 Us3 as well as to determine whether HSV-2 Us3 contains an intrinsic NLS will be valuable lines of investigation. A more thorough understanding of the nuclear shuttling properties of Us3 will enable specific disruption of nuclear shuttling as a means of studying Us3 functions in distinct cellular compartments.
MATERIALS AND METHODS
Cells and viruses
African green monkey kidney (Vero), 293T and HeLa cells were all maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum in a 5% CO2 environment. HSV-1 strain 17+ and HSV-2 strain HG52 were generously provided by Drs. A. Dolan and D. McGeoch, MRC Virology Unit, University of Glasgow. HSV-1 and HSV-2 strains were propagated and titred on Vero cells.
Construction of Us3 expression plasmids and kinase dead versions of Us3
To construct the HSV-2 Us3 expression plasmid pHSV2Us3, the Us3 gene was amplified by PCR with forward primer 5’CCGAATTCATGGCCTGTCGTAAGTTCTGTGG3’ and reverse primer 5’CCGAATTCTCACTTAGGGTGAAATAGCG3’ from template pATHSV-2HindIIIL (a kind gift from Drs. A. Dolan and D. McGeoch), which contains a HindIII fragment encompassing a portion of the unique short region of the HG52 genome (Dolan et al., 1998), and the amplified fragment was subcloned into pCR-Blunt II-TOPO vector (Invitrogen, Burlington, ON) by using manufacturer’s protocols. All plasmids utilizing PCR in their construction were sequenced to ensure that no spurious mutations were introduced in the process. An EcoRI fragment encompassing the amplified sequences was excised from this intermediate vector, purified, and then ligated to similarly digested pCINeo vector (Promega, Madison, WI). To fuse the red fluorescent protein monomeric Cherry (mCherry)(Shaner et al., 2004) to the N-terminus of HSV-2 Us3, the mCherry gene was amplified by PCR with forward primer 5’CGCGCTAGCATGGTGAGCAAGGGCGAGG3’ and reverse primer 5’CGCCTCGAGTCTTGTACAGCTCGTCCATGC3’ from template pRSETb mCherry (a kind gift from Dr. R. Tsien) and the amplified fragment was subcloned into pCR-Blunt II-TOPO vector. An NheI/XhoI fragment encompassing the amplified sequences was excised from this intermediate vector, purified and ligated to similarly digested pHSV2Us3. To fuse green fluorescent protein (GFP) or mCherry to the C-terminus of HSV-2 Us3, the Us3 gene was amplified by PCR with forward primer 5’ACAAAGCTTATGGCCTGTCGTAAGTTCTGTGG3’ and reverse primer 5’ACAGGTACCGTCTTAGGGTGAAATAGCGG3’ from template pHSV2Us3 and the amplified fragment was subcloned into pCR-Blunt II-TOPO vector. HindIII/KpnI fragments encompassing the amplified sequences were excised from the intermediate vector, purified and ligated to similarly digested pEGFP-N1 (Clontech, Mountain View, CA) or pJR70, a pEGFP-N1-based expression vector in which the EGFP gene was replaced with the mCherry gene (Banfield and Randall, unpublished). To fuse glutathione S-transferase (GST) to the N-terminus of HSV-2 Us3, the Us3 gene was amplified by PCR with forward primer 5’ACAGAATTCGATGGCCTGTCGTAAGTTCTGTGG3’ and reverse primer 5’CCGAATTCTCACTTAGGGTGAAATAGCG3’ from template pHSV2Us3 and the amplified fragment was subcloned into pCR-Blunt II-TOPO vector. An EcoRI fragment encompassing the amplified sequences was excised from the intermediate vector, purified and ligated to similarly digested pGEX-3X vector (GE Healthcare Lifesciences, Piscataway, NJ).
PCR with mutagenic primers using pHSV2Us3 template DNA was used to introduce specific mutations into the HSV-2 Us3 gene: for the K220M mutation, we used forward primer 5’CGGGTAATCGTCATGGCGGGGTGGTACG3’ and reverse primer 5’CGTACCACCCCGCCATGACGATTACCCG3’; for the D305A mutation, we used forward primer 5’GCAAAGGCATCATCCACCGCGCTATTAAGACCGAGAACATCTTC3’ and reverse primer 5’GAAGATGTTCTCGGTCTTAATAGCGCGGTGGATGATGCCTTTGC3’; for the K220A mutation, we used forward primer 5’CATCGGGTAATCGTCGCGGCGGGGTGGTACG3’ and reverse primer 5’CGTACCACCCCGCCGCGACGATTACCCGATG3’. To introduce the KD/K137M mutation into the PRV Us3 gene, we used forward primer 5’CACGGTGGTGCTGATGGTGGGCCAGAAGC3’ and reverse primer 5’GCTTCTGGCCCACCATCAGCACCACCGTG3’ on pPRVUs3bmRFP1 template, a pCINeo-based plasmid containing Us3 sequence derived from the Becker strain of PRV with monomeric red fluorescent protein (mRFP1)(Campbell et al., 2002) fused to the C-terminus. GFP or mCherry was introduced on to the N- or C-terminus of KD versions of HSV-2 Us3 as described above for the wild type (WT) protein or by directly using WT fusion protein expression plasmids as template for mutagenesis.
Preparation of HSV-2 Us3 antiserum
A baculovirus expression system, generously provided by Dr. E. Carstens, Queen’s University, was used to express GST-HSV-2 Us3. The GST-HSV-2 Us3 gene was amplified by PCR with forward primer 5’CGCCTCGAGATGTCCCCTATACTAGG3’ and reverse primer 5’CGCAAGCTTGAATTCTCACTTAGGGTG3’ from pGEX-3X vector containing HSV-2 Us3 sequence (see preceding section) and the amplified fragment was subcloned into pCR-Blunt II TOPO vector. An XhoI and HindIII fragment encompassing the amplified sequences was excised from the intermediate vector, purified and ligated to similarly digested pFast-Bac1 vector to form pFast-Bac-GST-HSV2Us3. DH10Bac E. coli cells, containing bacmid vector, were transformed with pFast-Bac-GST-HSV2Us3. To screen for incorporation of GST-HSV-2 Us3 into the Baculovirus genome, transformants were subjected to blue/white screening on selection plates containing kanamycin (50 µg/ml), gentamycin (7 µg/ml), tetracycline (10 µg/ml), X-gal (100 µg/ml), and IPTG (40 µg/ml). Recombinant bacmid DNA was purified using the Bac-to-Bac Baculovirus Expression System (Invitrogen, Burlington, ON), according to manufacturer’s procedures and then used to transfect Sf21 cells. Cell supernatants containing recombinant viruses were harvested 72 hours post transfection and recombinant virus was plaque purified. Sf21 cells (2 × 106) seeded onto a T25 flask were infected with recombinant virus at a multiplicity of infection (MOI) of 0.1. At 72 hours post infection cells were harvested and centrifuged for 5 min at 3,000 × g. Supernatant was discarded and pelleted cells were lysed with 1 ml of cold Buffer C (50 mM Tris [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 10% glycerol, and 1 mM PMSF) (Kato et al., 2001). Cell lysate was centrifuged at 5,000 × g for 20 min, and insoluble material in the pellet was separated by SDS-PAGE on a 10% acrylamide gel. A band corresponding to the 78 kDa GST-HSV-2 Us3 fusion protein was excised from the gel and used to immunize rats for antibody production (Cedarlane Laboratories, Burlington, ON). Rat polyclonal antiserum against HSV-2 Us3 was used for Western blotting at a dilution of 1:500 and for indirect immunofluorescence microscopy at a dilution of 1:1,000
Other immunological reagents
Anti β-actin monoclonal antibody (Sigma, St. Louis, MO) was used for Western blotting at a dilution of 1:2,000; anti β-tubulin monoclonal antibody (Sigma, St. Louis, MO) was used for indirect immunofluorescence at a dilution of 1:200; anti ICP5 monoclonal antibody (Virusys, Sykesville, MD) was used for Western blotting at a dilution of 1:3,000; anti ICP8 monoclonal antibody (Virusys, Sykesville, MD) was used for indirect immunofluorescence at a dilution of 1:1,000; Phospho-(Ser/Thr) PKA substrate antibody (Cell Signaling Technology, Danvers, MA) was used for Western blotting at a dilution of 1:1,000; horseradish peroxidase conjugated goat anti-mouse, goat anti-rabbit and rabbit anti-rat (Sigma, St. Louis, MO) were used for Western blotting at dilutions of 1:10,000, 1:10,000 and 1:80,000, respectively; Alexa Fluor 488 conjugated donkey anti-rat, Alexa Fluor 568 conjugated donkey anti-mouse, and Alexa Fluor 647 conjugated donkey anti-mouse (Invitrogen, Burlington, ON) were all used for indirect immunofluorescence at a dilution of 1:500.
Transfections
Transfection of 293T cells for the purpose of preparing cellular extracts was carried out using the calcium phosphate co-precipitation technique (Graham and van der Eb, 1973). Transfection of Vero, 293T and HeLa cells for the purpose of microscopic analyses was carried out using FuGene 6 (Roche, Laval, QC) according to manufacturer’s instructions. Transfection of Vero cells for the purpose of preparing whole cell extracts was carried out using the Amaxa Nucleofector transfection system (Lonza, Basel, Switzerland) according to manufacturer’s instructions.
Preparation and analysis of cellular extracts
For preparation of cellular extracts from infected cells, confluent monolayers of Vero cells grown in 6-well plates were infected with HSV-1 or HSV-2 at an MOI of 1. At 24 hours post infection, the medium was removed and the cells washed with phosphate-buffered saline (PBS). Cells were scraped into 200 µl of lysis buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% NP40, 1% Na deoxycholate) containing protease inhibitors (Roche, Laval, QC) and transferred to a 1.5-ml microfuge tube. Lysates were kept on ice for 30 minutes with intermittent mixing and then centrifuged at 10,000 × g for 5 min. Supernatants were collected in 1.5-ml microfuge tubes and stored at −20°C. For preparation of cellular extracts from transfected 293T cells, sub-confluent monolayers of cells grown in 35 mm dishes were transfected as described in the preceding section. At 24 hours post transfection, extracts were prepared and stored as described above. For Western blot analysis, 5 to 10 µl of cellular extract was mixed with SDS-PAGE loading buffer, heated to 100°C for 5 min, and electrophoresed through SDS-12.5% PAGE gels. Separated proteins were transferred to PVDF membranes (Millipore, Billerica, MA) and probed with appropriate dilutions of primary antibody followed by appropriate dilutions of horseradish peroxidase conjugated secondary antibody. For preparation of whole cell extracts from transfected Vero cells, 1 × 106 cells grown in 35 mm dishes were transfected using the Amaxa Nucleofector system. At 24 hours post transfection, cells were scraped off the dish in 100 µl of PBS, transferred to a 1.5-ml microfuge tube and lysed by the addition of 33 µl of 3X SDS-PAGE loading buffer. The lysate was repeatedly passed through a 28 1/2 gauge needle to reduce viscosity and then heated at 100°C for 5 min. Twenty µl of whole cell lysate was used for Western blot analysis as described above.
Inhibition of actin polymerization
To inhibit actin polymerization in Us3-transfected Vero cells, cells were incubated in medium containing either 1 µM cytochalasin D or 0.01% DMSO (carrier) starting at 6 hours post transfection. Cells were maintained in the continuous presence of cytochalasin D or carrier for a total of 12 hours.
Indirect immunofluorescence microscopy
Cells for microscopic analyses were grown either on glass coverslips or on glass bottom dishes (MatTek, Ashland, MA). Cells were fixed by incubation with freshly prepared 4% paraformaldehyde in PBS for 10 min at room temperature. Fixed cells were washed 3 times with PBS containing 1% BSA (PBS/BSA) and permeabilized for 10 min at room temperature with PBS/BSA containing 0.1% Triton X-100. Cells were then washed 3 times with PBS/BSA and 150 µl of primary antiserum diluted appropriately in PBS/BSA was applied for 45 min at room temperature. Cells were washed 3 times with PBS/BSA and 150 µl of Alexa Fluor conjugated secondary antibody (Invitrogen, Burlington, ON) diluted appropriately in PBS/BSA was applied for 30 min at room temperature. Cells were then washed 3 times with PBS/BSA. To visualize nuclei, cells were incubated with Hoechst 33342 (Sigma, St. Louis, MO) diluted to 0.5 µg/ml in PBS for 7 min at room temperature. To visualize filamentous actin, cells were incubated with Texas Red-phalloidin (Invitrogen, Burlington, ON) diluted to 165 ηM in PBS for 7 min at room temperature. Stained cells on coverslips were washed 3 times in PBS and mounted in PBS containing 50% (v/v) glycerol onto glass slides; stained cells in glass bottom dishes were washed 3 times in PBS and stored under PBS/BSA. Images were captured using an Olympus FV1000 laser scanning confocal microscope and Fluoview CS3 software. Images were captured using a 40× or a 60× objective and a digital zoom factor ranging from 2 to 4. Composites of representative images were prepared using Adobe Photoshop software.
ACKNOWLEDGEMENTS
This work was supported in part by NIH grant AI48626, CFI award 16389, and CIHR operating grant 93804 to BWB. We thank Dr. R. Tsien (UCSD/HHMI) and J. Randall and J. Cooper (UCDHSC) for providing plasmids used in this study and Drs. A. Dolan and D. McGeoch (MRC Virology Unit, U. Glasgow) for providing plasmids and viruses used in this study. We thank Dr. E. Carstens (Queen’s U.) for providing reagents and expertise for expressing HSV-2 Us3 in baculovirus and Dr. K. Gee for access to the Amaxa Nucleofector. We acknowledge members of the Banfield lab for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Benetti L, Roizman B. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc Natl Acad Sci U S A. 2004;101(25):9411–9416. doi: 10.1073/pnas.0403160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16(8):4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calton CM, Randall JA, Adkins MW, Banfield BW. The pseudorabies virus serine/threonine kinase Us3 contains mitochondrial, nuclear and membrane localization signals. Virus Genes. 2004;29(1):131–145. doi: 10.1023/B:VIRU.0000032796.27878.7f. [DOI] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99(12):7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmin GL, Clase AC, Randall JA, Enquist LW, Banfield BW. Insertions in the gG gene of pseudorabies virus reduce expression of the upstream Us3 protein and inhibit cell-to-cell spread of virus infection. J Virol. 2001;75(22):10856–10869. doi: 10.1128/JVI.75.22.10856-10869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30(2):449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- Dolan A, Jamieson FE, Cunningham C, Barnett BC, McGeoch DJ. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72(3):2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favoreel HW, Van Minnebruggen G, Adriaensen D, Nauwynck HJ. Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an alphaherpesvirus are associated with enhanced spread. Proc Natl Acad Sci U S A. 2005;102(25):8990–8995. doi: 10.1073/pnas.0409099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick ML, Walker MJ, Petkevich JM. On the association of virus proteins with the nuclei of cells infected with herpes simplex virus. J Gen Virol. 1978;39(3):519–529. doi: 10.1099/0022-1317-39-3-519. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Geenen K, Favoreel HW, Olsen L, Enquist LW, Nauwynck HJ. The pseudorabies virus US3 protein kinase possesses anti-apoptotic activity that protects cells from apoptosis during infection and after treatment with sorbitol or staurosporine. Virology. 2005;331(1):144–150. doi: 10.1016/j.virol.2004.10.027. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9(8):576–596. [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241(4861):42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hata S, Koyama AH, Shiota H, Adachi A, Goshima F, Nishiyama Y. Antiapoptotic activity of herpes simplex virus type 2: the role of US3 protein kinase gene. Microbes Infect. 1999;1(8):601–607. doi: 10.1016/s1286-4579(99)80059-8. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kato A, Yamamoto M, Ohno T, Kodaira H, Nishiyama Y, Kawaguchi Y. Identification of proteins phosphorylated directly by the Us3 protein kinase encoded by herpes simplex virus 1. J Virol. 2005;79(14):9325–9331. doi: 10.1128/JVI.79.14.9325-9331.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Kawaguchi Y, Tanaka M, Igarashi M, Yokoyama A, Matsuda G, Kanamori M, Nakajima K, Nishimura Y, Shimojima M, Phung HT, Takahashi E, Hirai K. Epstein-Barr virus-encoded protein kinase BGLF4 mediates hyperphosphorylation of cellular elongation factor 1delta (EF-1delta): EF-1delta is universally modified by conserved protein kinases of herpesviruses in mammalian cells. J Gen Virol. 2001;82(Pt 6):1457–1463. doi: 10.1099/0022-1317-82-6-1457. [DOI] [PubMed] [Google Scholar]
- Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253(5018):407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- Knipe DM, Spang AE. Definition of a series of stages in the association of two herpesviral proteins with the cell nucleus. J Virol. 1982;43(1):314–324. doi: 10.1128/jvi.43.1.314-324.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17(6):527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Leopardi R, Van Sant C, Roizman B. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc Natl Acad Sci U S A. 1997;94(15):7891–7896. doi: 10.1073/pnas.94.15.7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman MG, Demmin GL, Banfield BW. The attenuated pseudorabies virus strain Bartha fails to package the tegument proteins Us3 and VP22. J Virol. 2003;77(2):1403–1414. doi: 10.1128/JVI.77.2.1403-1414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou F, Forest T, Baines JD. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J Virol. 2007;81(12):6459–6470. doi: 10.1128/JVI.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Roizman B. The US3 protein kinase of herpes simplex virus 1 mediates the posttranslational modification of BAD and prevents BAD-induced programmed cell death in the absence of other viral proteins. Proc Natl Acad Sci U S A. 2001;98(18):10410–10415. doi: 10.1073/pnas.181344498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Goshima F, Daikoku T, Takakuwa H, Nishiyama Y. Expression of herpes simplex virus type 2 US3 affects the Cdc42/Rac pathway and attenuates c-Jun N-terminal kinase activation. Genes Cells. 2000;5(12):1017–1027. doi: 10.1046/j.1365-2443.2000.00383.x. [DOI] [PubMed] [Google Scholar]
- Ogg PD, McDonell PJ, Ryckman BJ, Knudson CM, Roller RJ. The HSV-1 Us3 protein kinase is sufficient to block apoptosis induced by overexpression of a variety of Bcl-2 family members. Virology. 2004;319(2):212–224. doi: 10.1016/j.virol.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Poon AP, Liang Y, Roizman B. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cells infected with a mutant carrying a cDNA copy of the infected-cell protein no. 0. J Virol. 2003;77(23):12671–12678. doi: 10.1128/JVI.77.23.12671-12678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan MP, Chen LB, Knipe DM. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36(4):857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J Virol. 2002;76(17):8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64(3):615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Schumacher D, McKinney C, Kaufer BB, Osterrieder N. Enzymatically inactive U(S)3 protein kinase of Marek's disease virus (MDV) is capable of depolymerizing F-actin but results in accumulation of virions in perinuclear invaginations and reduced virus growth. Virology. 2008;375(1):37–47. doi: 10.1016/j.virol.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher D, Tischer BK, Trapp S, Osterrieder N. The protein encoded by the US3 orthologue of Marek's disease virus is required for efficient de-envelopment of perinuclear virions and involved in actin stress fiber breakdown. J Virol. 2005;79(7):3987–3997. doi: 10.1128/JVI.79.7.3987-3997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125(6):1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Van den Broeke C, Deruelle M, Nauwynck HJ, Coller KE, Smith GA, Van Doorsselaere J, Favoreel HW. The kinase activity of pseudorabies virus US3 is required for modulation of the actin cytoskeleton. Virology. 2009a;385(1):155–160. doi: 10.1016/j.virol.2008.11.050. [DOI] [PubMed] [Google Scholar]
- Van den Broeke C, Radu M, Deruelle M, Nauwynck H, Hofmann C, Jaffer ZM, Chernoff J, Favoreel HW. Alphaherpesvirus US3-mediated reorganization of the actin cytoskeleton is mediated by group A p21-activated kinases. Proc Natl Acad Sci U S A. 2009b;106(21):8707–8712. doi: 10.1073/pnas.0900436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Minnebruggen G, Favoreel HW, Jacobs L, Nauwynck HJ. Pseudorabies virus US3 protein kinase mediates actin stress fiber breakdown. J Virol. 2003;77(16):9074–9080. doi: 10.1128/JVI.77.16.9074-9080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Minnebruggen G, Van de Walle GR, Favoreel HW, Nauwynck HJ, Pensaert MB. Temporary disturbance of actin stress fibers in swine kidney cells during pseudorabies virus infection. Vet Microbiol. 2002;86(1–2):89–94. doi: 10.1016/s0378-1135(01)00493-x. [DOI] [PubMed] [Google Scholar]
- Wagenaar F, Pol JM, Peeters B, Gielkens AL, de Wind N, Kimman TG. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J Gen Virol. 1995;76(Pt 7):1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- Wolff B, Sanglier JJ, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4(2):139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]