Abstract
The goal of this study was to elucidate the control mechanisms by which exogenous proteins regulate keratinocyte growth factor (KGF) expression in fibroblasts adhered to differing substrates and thereby provide insights into both fundamental in vitro cell signaling and cell-biomaterial interaction research. A serum-free culture system in which cells maintained their proliferative capacity was established and employed. The addition of transforming growth factor- α (TGF-α), interleukin-1β (IL-1β) and platelet-derived growth factor-BB (PDGF-BB) individually showed no effect on KGF protein release, however, IL-1β addition led to increased KGF mRNA transcription, intracellular KGF protein synthesis, and granulocyte-macrophage colony-stimulating factor (GM-CSF) release. Intracellular KGF protein synthesis and extracellular release were enhanced when fibroblasts were treated with a combination of IL-1β and PDGF-BB which suggests KGF synthesis and release are largely regulated by synergistic mechanisms. Surface-bound fibronectin-derived ligands and individual exogenous proteins promoted fibroblast adhesion to semi-interpenetrating polymer networks (sIPNs) but did not stimulate KGF release despite enhancement of KGF mRNA transcription. Additionally, serum conditioning was found to have a significant impact on KGF synthesis and the subsequent mechanisms controlling KGF release. This study demonstrates that KGF release from fibroblasts is likely regulated by multiple mechanisms involving post-transcriptional and exocytic controls which may be impacted by the presence of serum and how serum is removed from the in vitro cell environment.
Keywords: Keratinocyte growth factor, granulocyte-macrophage colony-stimulating factor, interpenetrating network, Interleukin-1β, Transforming growth factor-α, platelet-derived growth factor-BB
INTRODUCTION
Mammalian fibroblast growth factors (FGFs) form a large family of polypeptide growth factors which function in cell signaling, proliferation and differentiation.[1–3] Keratinocyte growth factor/fibroblast growth factor-7 (KGF/FGF-7) has been shown to be secreted by mesenchymal cells with mitogenic activity on keratinocytes.[1] In a wound healing environment, KGF is produced via fibroblast-keratinocyte paracrine interaction[4–6] and subsequently enhances the proliferation and migration of dermal keratinocytes.[7,8] KGF also influences epithelial cell differentiation and regulates the expression of wound remodeling proteases.[9,10] A wide range of growth factors and cytokines able to induce KGF expression, such as transforming growth factor- α (TGF-α), interleukin-1β (IL-1β), platelet-derived growth factor-BB (PDGF-BB) and tumor necrosis factor-α (TNF-α), have been reported.[1,11–13] Most of these studies, however, are based on conditions using serum-containing media and a tissue culture polystyrene (TCPS) substrate, thereby reflect the combined consequences of the various serum components and the likely substrate-specific effects of a single type of surface. Moreover, the majority of published reports on the effects of growth factors on KGF expression involve cells transferred from serum-containing media directly into serum-free media without any type of gradual transition period.[11–13] Although viability of the cells used in these studies is unclear, such culture conditions result in a sudden change of environment which may lead to stress-induced functional differences. Furthermore, previous work has not drawn a connection between KGF gene transcription and subsequent protein synthesis and/or release. Additionally, while serial transitioning of cells from serum-containing into serum-free media has been shown to be selectively accompanied by up- or down-regulation of certain genes, demonstrating a fine-tuning of signaling pathways, [14,15] there is an overall deficiency in studies comparing the cell behavior in abruptly versus serially transitioned serum-free cultures.
In this study, human dermal fibroblasts successfully adapted into serum-free media were seeded onto differing surfaces with or without modification with adhesive peptide motifs. Fibronectin-derived peptides arginine-glycine-aspartic acid (RGD) and proline-histidine-serine-arginine-asparagine (PHSRN) were employed as they have shown synergistic effects on cell adhesion, migration, proliferation and protein expression, and have previouly been utilized to modify biomaterials.[16–18] These peptides were conjugated to a semi-interpenetrating polymer network (sIPN) based on poly(ethylene glycol) (PEG) and gelatin to gain mechanistic insights into our previous studies, where we had shown that the sIPN modified with ECM-derived ligands enhanced cell adhesion and influenced cytokine release in fibroblast-keratinocyte and fibroblast-monocyte coculture systems as well as promoted ECM remodeling in rodent full-thickness dermal defect and porcine partial-thickness defect models in vivo.[6,19–22] The use of dissimilar substrates also allowed examination of whether KGF control mechanisms were present when fibroblasts were adhered to different surfaces. The effects of exogenous proteins on adhesion and protein release were analyzed in cells which were serially transitioned into serum-free media. The resulting variations in KGF transcription and protein release depending on exogenous protein or serum stimulation provide insights into the control mechanisms governing KGF release by fibroblasts and the development of tissue engineering strategies for wound regeneration.
MATERIALS AND METHODS
Synthesis of sIPNs containing PEGylated fibronectin-derived peptides
RGD and PHSRN were prepared by solid phase peptide synthesis utilizing Fmoc chemistry. Peptide formation was confirmed by 1H-NMR and matrix assisted laser desorption/ionization (MALDI, University of Wisconsin School of Pharmacy) with 95% purity confirmed with HPLC. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Synthesis and characterization of PEGylated-peptide modified gelatin has been described previously.[23] Briefly, terminal hydroxyl groups of PEG diol (2 kDa, Fluka) were converted to carboxylic acid with the addition of t-butyl bromoacetate and dioxane, and then reacted with N-hydroxysuccinimide to synthesize PEG-bis-N-hydroxysuccinimide (PEG-bis-NSu). Glycine-glycine-glycine (GGG, Fluka), RGD, and PHSRN were grafted onto one of the two terminal groups of PEG-bis-NSu by the addition of N,N-diisopropyl ethylamine to form N-hydroxysuccinimide-PEG-ligand. Gelatin was grafted to the other terminal group by adding N-hydroxysuccinimide-PEG-ligand to 1% gelatin in phosphate buffered saline (PBS) solution (pH=8) with N,N-diisopropyl ethylamine. The final product was purified by a pressurized ultrafiltration system (Millipore, Bedford, MA) with a 30 kDa membrane followed by lyophilization. The degree of peptide-PEG grafting onto gelatin was analyzed by trinitrobenzenesulfonic acid-based spectrophotometry.[19,20] The percent modification of gelatin was approximately 70 percent for GGG modified gelatin, 66 percent for RGD modified gelatin and 71 percent for PHSRN modified gelatin.
sIPNs were prepared by combining 575 Da poly(ethylene glycol) diacrylate (PEGdA) and PEGylated-peptide grafted gelatin or unmodified gelatin dissolved in ddH2O to create a 13 w/w% PEGdA, 9 w/w% gelatin solution. The initiator, 2,2-dimethoxy-2-phenyl-acetophenone, was incorporated into the PEGdA and gelatin solution to a final concentration of 0.1 wt%. The solution was poured into 9.5 mm diameter Teflon® molds and crosslinked with UV light (CF 1000, Clearstone Technologies, Inc., Minneapolis, MN) at 365nm for 3 minutes. sIPNs were sterilized with 70% ethanol for 30 minutes and washed three times with PBS before treatment with antibiotics (Penicillin 300 IU/mL, Streptomycin 300 μg/mL and Amphotericin B 0.75 μg/mL, Mediatech, Inc., Herndon, VA) overnight.[24] sIPNs were then washed three times with PBS for 15 minutes each rinse before being fitted into 48-well plates and equilibrated in serum-free fibroblast basal media (FBM, Lonza, Walkersville, MD) at 37°C for 2 hours. The FBM was then replaced for cell seeding.
Cell culture and transitioning to serum-free media
Neonatal human dermal fibroblasts (NHDF) were obtained from Lonza and cultured in fibroblast growth medium-2 (FGM-2, Lonza) containing FBM, 0.1% insulin, 0.1% recombinant human fibroblast growth factor-B, 0.1% GA-1000, and 2% fetal bovine serum (FBS). Fibroblasts for all experiments were derived from the same cell stock at passage 6 to 8 and cells were harvested and split into each experimental group at confluence.
To successfully study the efficacy of serially transitioning cells to serum-free media, cells were split into two groups. One group contained cells which were passaged directly into serum-free media [FBM or Dulbecco’s Minimum Essential Media:Ham’s Nutrient Mixture F-12, 1:1 Mix (DMEM/F-12, Mediatech)].[12] In the other group, over the course of three passages, the serum concentration was slowly reduced by serially adjusting the ratio of FGM-2 to the new media at each passage (from 1:1 to 1:3 to 1:9 and then to serum-free media). DMEM/F12 supplemented with 5% FBS (DMEM/F12+5%FBS) was also used as a serum-containing media control. Cells from both groups were seeded onto 48-well tissue culture polystyrene (TCPS, Becton Dickinson Labware, NJ) at a seeding density of 10,000 cells/cm2. Adherent cell density was quantified and cell viability was observed at 24, 72, 120, and 168 hours using the method described below.
Cell adhesion and viability studies by LIVE/DEAD viability/cytotoxicity assay
Adherent cell density was quantified by LIVE/DEAD viability/cytotoxicity assay for mammalian cells (Invitrogen, Eugen, OR). Cells were washed twice with Dulbecco’s PBS (D-PBS, Mediatech) followed by addition of 2μM calcein-AM and 2μM ethidium homodimer-1 and then incubated at 37 °C for 30 minutes. Cell morphology and viability were then observed using an inverted microscope (Nikon Eclipse TE 300) and images were recorded for cell counting. Five images per sample were taken at random fields of view.
KGF and GM-CSF expression and release studies by ELISA
To study the effect of serum on KGF protein release, cells were cultured on TCPS in FBM, FBM with 2% FBS, FBM with 5% FBS or FGM-2 at a cell seeding density of 50,000 cell/cm2. Cell supernatant was assayed for KGF at 24, 72, 120, and 168 hours using an enzyme-linked immunosorbent assay (ELISA, Quantikine KGF immunoassay colorimetric kit, R&D Systems, Minneapolis, MN).
In order to study the effects of individual exogenous proteins on cell adhesion as well as KGF and granulocyte-macrophage colony-stimulating factor (GM-CSF) release, serum-free FBM was supplemented with one of the following human recombinant proteins: TGF-α (BioVision, Mountain View, CA), IL-1β (BioVision), PDGF-BB (Sigma-Aldrich) or a combination of IL-1β and PDGF-BB. A concentration of 10 ng/mL of each growth factor or cytokine was used based on literature showing enhanced fibroblast KGF mRNA expression at this concentration.[11–13] After serial media transition, fibroblasts were seeded onto TCPS and sIPNs at a seeding density of 50,000 cells/cm2. FBM supplemented with 5% FBS served as a positive control. Cell culture supernatant was collected for ELISA (Quantikine KGF and GM-CSF immunoassay colorimetric kits, R&D Systems) and adherent cell densities were quantified with LIVE/DEAD assay at 24, 72, 120 and 168 hours.
Cell lysate studies were performed to gain insights into protein release and extracellular transport. Cells were seeded onto 6-well TCPS plates with or without sIPNs at a density of 50,000 cells/cm2 in serum-free FBM or FBM supplemented with 10ng/mL IL-1β, 10ng/mL IL-1β and 10ng/mL PDGF-BB, or 5% FBS. At 48 and 120 hours, cells were washed with cold D-PBS and lysed in mammalian protein extract reagent (M-PER, Pierce, Rockford, IL) supplemented with EDTA-free Halt™ protease inhibitor cocktail (Pierce). After centrifugation at 7,000 RPM for 5 minutes, the KGF concentration in the supernatant was analyzed via ELISA. The total protein of cells cultured on TCPS was determined by BCA™ protein assay (Pierce) and the KGF content in cell lysate was presented as a ratio of KGF to total protein concentration. An accurate measure of total protein from cells adhered to sIPNs could not be obtained due to the presence of gelatin in the sIPN.
Reverse transcription-polymerase chain reaction (RT-PCR)
Semi-quantification of KGF mRNA was achieved with RT-PCR. All chemicals and reagents used for RT-PCR were obtained from Invitrogen (Carlsbad, CA) unless otherwise stated. After serial media transition, fibroblasts were seeded onto sIPNs and TCPS at a density of 50,000 cells/cm2 in FBM, FBM plus 10ng/ml IL-1β, or FBM supplemented with 5% FBS. At 24, 72 and 120 hours, Cells were washed with D-PBS twice and total RNA was collected using TRIzol® reagent according to the manufacturer’s recommendations. The RNA concentration and purity was determined by NanoDrop ND-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE). 1 μg of total RNA from each TCPS sample and 600 ng of total RNA from each sIPN sample were treated with DNase I (amplification grade). cDNA was produced using the Moloney murine leukemia virus (M-MLV) reverse transcriptase system and 200 ng per 1 μg total RNA of 15-mer random primers (University of Wisconsin Biotechnology Center) was used in place of random hexamers. KGF and RNA polymerase II (RPII) primers (University of Wisconsin Biotechnology Center) were designed to bind to human cDNA using Primer3Plus software [25] based on DNA sequences obtained from the NCBI database (Bethesda, MD) (Table 1). RPII has been shown to have stable RNA transcription in various tissues and experimental conditions making it an appropriate choice for a reference gene for this study.[26] PCR was accomplished by using the Platinum Blue PCR SuperMix system and was optimized to measure amplicon levels in the linear phase of the amplification reaction. All samples were run in duplicate. Briefly, 2μl cDNA from the reverse transcriptase reaction or the appropriate dilution was added to 200nM of sense primers, 200nM antisense primers and the SuperMix. The mixture was loaded into a thermal cycler (Bio-Rad, Hercules, CA), hot-started for 3 minutes at 94°C, and then cycled 35x as follows: denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 60 seconds. Amplified DNA samples were maintained at 4°C prior to gel electrophoresis. KGF and RPII amplicons for all samples at one time point were run on a 1.5% agarose gel with 0.005% ethidium bromide (Bio-Rad). Gel electrophoresis results were observed and recorded by FOTO/Analyst® Investigator System (Fotodyne Inc., Hartland, WI) and the band intensities were measured by ImageJ software version 1.32j [27] to calculate the intensity ratio between KGF and the reference gene RPII.
Table 1.
KGF and polymerase (RNA) II primers
| Gene | Oligo | Sequence | Accession No. | Amplicon size (bp) |
|---|---|---|---|---|
| FGF-7 (KGF) | KGF-sense | 5′-GGCCTCCATCCCTCTTACTC -3′ | NM_002009 | 242 |
| KGF-antisense | 5′-GAAGTGCATTGCCAAGACATAG -3′ ACCEPTED | |||
| Polymerase (RNA) II Polypeptide A (RPII) | RPII-sense | 5′-ACTGGAGACAGCAAGGTCGT -3′ | NM_000937 | 222 |
| RPII-antisense | 5′-CAGGTGGATGTTGAAGAGCA -3′ |
Statistical Analysis
All data is shown as mean ± standard deviation of samples in independent experiments. Cell density data, ELISA data and RT-PCR data were analyzed by unpaired Student-t test. A value of p< 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
Serum-free media transitioning
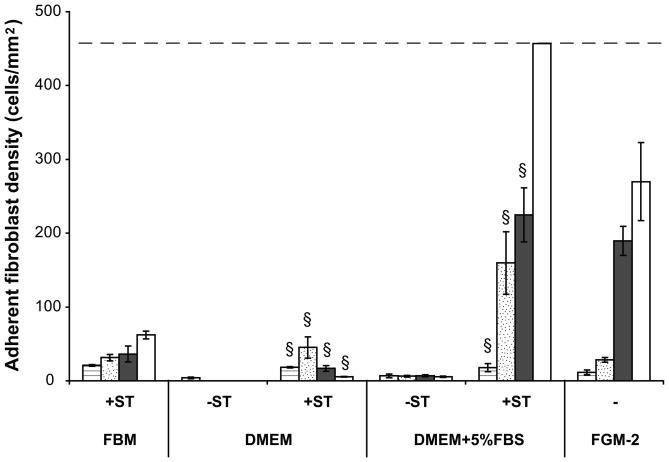
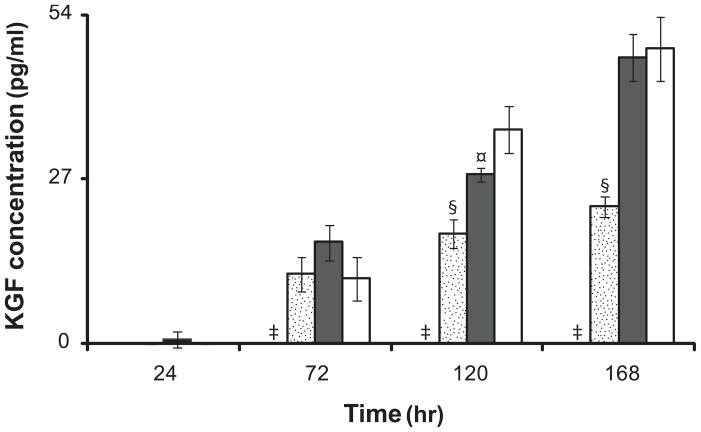
The transitioning of fibroblasts from serum-containing into serum-free media is crucial in maintaining cell viability and productivity. Results demonstrated that neither DMEM/F12 (Figure 1) nor FBM (data not shown) supported fibroblast proliferation when cells were transferred directly from complete growth medium, FGM-2, to serum-free media. Serially transitioning cells into DMEM/F12 and DMEM/F12+5%FBS was successful in increasing adherent cell density over 168 hours as compared to non-transitioned cells. Additionally, cells transitioned into FBM and DMEM/F12+5%FBS demonstrated proliferative capacity while cells transitioned into DMEM/F12 showed decreasing adherent density from 72 to 168 hours (Figure 1). Furthermore, fibroblasts transitioned into FBM showed 0±0 pg/mL KGF release from 24 to 168 hours (Figure 2). As serial transitioning to FBM allowed the fibroblasts to maintain their viability and proliferative capacity while not inducing KGF release, subsequent studies were conducted in this manner.
Figure 1.
Adherent human dermal fibroblast density on TCPS with or without serial transitioning (ST). Cells were split into two groups. One group of cells were passaged directly into DMEM/F12 with or without 5% FBS. The other group of cells were serially passaged into FBM or DMEM/F12 with or without 5% FBS by gradually reducing serum concentration. Cell seeding density was 10,000 cells/cm2. Adherent cell density was quantified by LIVE/DEAD assay at 24 ( ), 72 (
), 72 ( ), 120 (
), 120 ( ) and 168 (□) hours. Complete growth medium FGM-2 served as a control. All data presented as average ± standard deviation (n=3). Dashed line represents the cell confluency.
) and 168 (□) hours. Complete growth medium FGM-2 served as a control. All data presented as average ± standard deviation (n=3). Dashed line represents the cell confluency.
§: Significantly different compared to cells without transitioning at the same time point, p<0.05.
Figure 2.
KGF release from fibroblasts cultured on TCPS in FBM ( ), FBM+2% FBS (
), FBM+2% FBS ( ), FBM+5%FBS (
), FBM+5%FBS ( ), and FGM-2 (□). Serially transitioned cells were cultured on TCPS in serum-free and serum-containing media. FGM-2 served as a positive control using cells which did not undergo serial transitioning. KGF concentrations in cell culture supernatant from 24 to 168 hours were measured by ELISA. All data presented as average ± standard deviation (n=3).
), and FGM-2 (□). Serially transitioned cells were cultured on TCPS in serum-free and serum-containing media. FGM-2 served as a positive control using cells which did not undergo serial transitioning. KGF concentrations in cell culture supernatant from 24 to 168 hours were measured by ELISA. All data presented as average ± standard deviation (n=3).
‡: Significantly different compared to FBM+2%FBS, FBM+5%FBS, and FGM-2 at the same time point, p < 0.05.
§: Significantly different compared to FBM+5%FBS, and FGM-2 at the same time point, p < 0.05.
¤: Significantly different compared to FGM-2 at the same time point, p < 0.05.
Effect of FBS supplementation on KGF release
The effect of FBS on KGF release was studied in serially transitioned fibroblasts cultured in FBM with or without supplementation of FBS. The addition of FBS significantly increased KGF release in a time- and dose-dependent manner. Fibroblasts cultured with 2% FBS increased KGF secretion from 0±0 to 52.2±1.70 pg/mL as compared to fibroblasts cultured with 5% FBS which showed a greater increase from 0.74±1.28 to 92.22±3.90 pg/mL at 24 and 168 hours, respectively (Figure 2).
Fibronectin-derived peptide modified substrates and exogenous proteins influence fibroblast attachment in the absence of serum
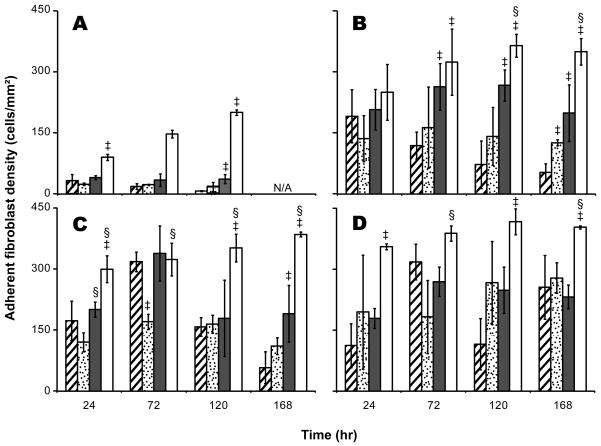
The addition of fibronectin-derived peptide motifs, RGD (37±11 cells/mm2) and PHSRN (36±9 cells/mm2), to the sIPN substrate enhanced cell attachment at 120 hours (p<0.01) as compared to unmodified sIPNs (7±1 cells/mm2). Additionally, the adherent cell number on RGD-modified sIPNs was stable over time as compared to cell adhesion to unmodified and GGG-modified sIPNs (Figure 3A).
Figure 3.
Adherent cell density of fibroblasts cultured on unmodified sIPN ( ), GGG-modified sIPN (
), GGG-modified sIPN ( ), RGD-modified sIPN (
), RGD-modified sIPN ( ) and TCPS (□) at 24, 72, 120 and 168 hours in FBM supplemented with no exogenous protein (A), 10ng/mL TGF-α (B), 10ng/mL IL-1β (C), and 10ng/mL PDGF-BB (D). Cells were stained with LIVE/DEAD fluorescent assay kit and observed under microscope at 10× magnification. Images were recorded for cell counting. Five images per sample were taken at random fields of view. All data presented as average ± standard deviation (n=3).
) and TCPS (□) at 24, 72, 120 and 168 hours in FBM supplemented with no exogenous protein (A), 10ng/mL TGF-α (B), 10ng/mL IL-1β (C), and 10ng/mL PDGF-BB (D). Cells were stained with LIVE/DEAD fluorescent assay kit and observed under microscope at 10× magnification. Images were recorded for cell counting. Five images per sample were taken at random fields of view. All data presented as average ± standard deviation (n=3).
‡: Significantly different compared to unmodified-sIPN with treatment by the same exogenous protein, p < 0.05.
§: Significantly different compared to GGG-sIPN with treatment by the same exogenous protein, p < 0.05.
N/A: Data not collected for that time point.
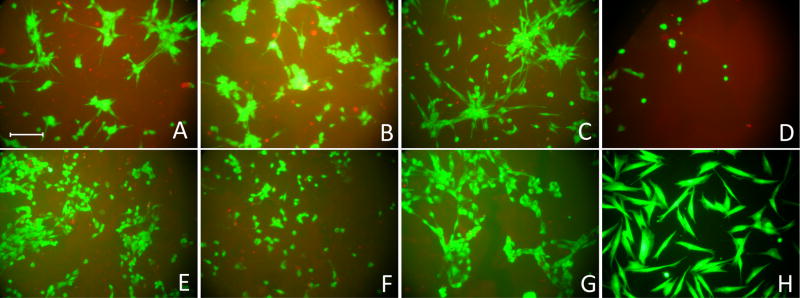
A serum-free culture system provides a platform to study the effect of individual proteins on fibroblast adhesion to various surfaces without interference from other serum components. In this study, FBM supplemented with TGF-α, IL-1β or PDGF-BB was employed to elucidate the effect of these proteins on cell adhesion. Results revealed that all these factors enhanced cell attachment dramatically on both sIPN and TCPS surfaces (Figure 3). At 168 hours, higher cell densities were observed on unmodified and GGG-modified sIPNs with PDGF-BB compared to that with TGF-α and IL-1β (p<0.01). At 24 hours, cells on sIPN surfaces with the addition of TGF-α, IL-1β, and PDGF-BB adapted a more spindle-like morphology (Figure 4A-C) which indicates the cells were more viable and proliferative than cells with a round shape (Figure 4D). At later time points, however, cells on sIPNs treated with TGF-α or IL-1β (Figure 4E–F) qualitatively lost most of the spindle-like morphology in contrast to cells treated with PDGF-BB (Figure 4G). This observation is consistent with the fact that PDGF-BB is a mitogen which not only stimulates cell proliferation, but also serves as a stronger chemoattractant than TGF-α and IL-1β.[28,29]
Figure 4.
Inverted microscope images of human dermal fibroblasts adherent to RGD-modified (A–G) and TCPS (H) surfaces at 24 (A–D, H) and 168 hours (E–G). Cells were cultured in FBM (D, H) or FBM supplemented with TGF-α (A, E), IL-1β (B, F) and PDGF-BB (C, G). Cells were stained with LIVE/DEAD fluorescent assay kit and incubated at 37°C for 30 minutes followed by observation under inverted microscope. Live and dead cells were stained by calcium-AM (green) and ethidium homodimer (red) respectively. The sIPN was slightly stained red in the background. All images were taken at 10× magnification. Scale bar represents 100 μm.
The significant increase in adherent cell density and the change in cell morphology may be attributed to several factors. First, the synthesis of extracellular matrix proteins was likely enhanced by the addition of exogenous proteins. In serum-free conditions, IL-1β has been observed to induce more dermatan sulfate and hyaluronate release, and also stimulated the production of high molecular weight glycosaminoglycan hyaluronate. [30–32] PDGF-BB has been shown to upregulate collagen I and III mRNA expression in dermal fibroblasts and also increased the release of fibronectin, laminin, type IV and V collagens in rat testicular myoid cells under serum-free conditions.[33, 34] Such changes in extracellular matrix synthesis in serum-free media and the subsequent adsorption of these proteins to TCPS and sIPN surfaces may have provided cells with more sites for adhesion. Second, adhesion may have been modulated by the regulation of integrin expression.[35] IL-1β and PDGF-BB have been found to affect integrin expression and function in serum-containing media. IL-1β stimulated expression of αv and β3 integrins when interacting with fibronectin/collagen materials.[36,37] PDGF-BB acted to upregulate αvβ3 and αvβ5 integrins and modulate membrane motility of β1 integrin.[38] Taken together, these studies suggest that the exogenous protein-induced expression of integrins may also contribute to enhanced cell attachment in serum-free media, however, studies on IL-1β and PDGF-BB stimulated integrin expression in serum-free conditions are still lacking.
Effects of exogenous TGF-α, IL-1β, and PDGF-BB on GM-CSF and KGF expression in serum-free media
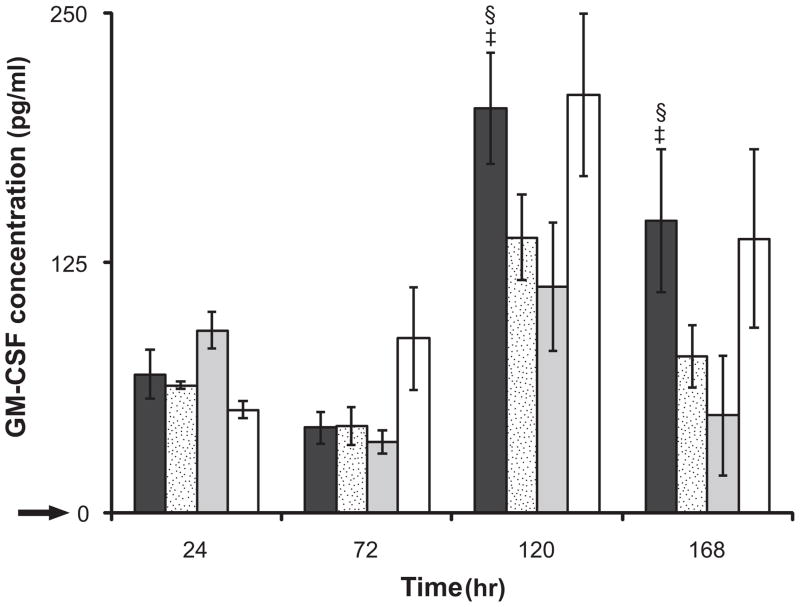
In this study, we observed significantly increased GM-CSF release when IL-1β (Figure 5), but not TGF-α or PDGF-BB, was supplemented. Fibroblasts showed marginal cumulative GM-CSF release of less than 10 pg/mL (p<0.01) in TGF-α or PDGF-BB supplemented media. However, GM-CSF release from cells stimulated with IL-1β was significantly enhanced, particularly at 120 hours. There was no significant difference observed in the GM-CSF concentration among cells adhered to modified or unmodified sIPNs at 24 and 72 hours. Cells adhered to GGG-modified sIPNs, however, showed a greater increase in GM-CSF release than RGD-modified and unmodified sIPNs at 120 and 168 hours (p<0.05). The GGG peptide has been shown to be involved in stimulating the secretion of GM-CSF in mouse bone marrow cells which may correlate with the response seen in this study.[39]
Figure 5.
GM-CSF release in IL-1β (10ng/mL) supplemented FBM from human dermal fibroblasts adhered to GGG-modified sIPN ( ), RGD-modified sIPN (
), RGD-modified sIPN ( ), unmodified gelatin sIPN (
), unmodified gelatin sIPN ( ) and TCPS (□).
) and TCPS (□).  represents the GM-CSF release from fibroblasts cultured on TCPS in FBM without supplemented IL-1β. All data presented as average ± standard deviation (n=3).
represents the GM-CSF release from fibroblasts cultured on TCPS in FBM without supplemented IL-1β. All data presented as average ± standard deviation (n=3).
§: Significantly different compared to unmodified sIPN, p<0.05.
‡: Significantly different compared to RGD modified sIPN, p<0.05;
Individual exogenous proteins, TGF-α, IL-1β, or PDGF-BB, however, had no effect on KGF release from confluent fibroblasts over time. Since individual growth factors were insufficient to replicate the stimulatory effects of FBS and complete fibroblast growth media on KGF release (Figure 2), IL-1β and PDGF-BB were combined to test whether the secretion of KGF was regulated by synergistic effects of growth factors/cytokines. Result showed a detectable KGF concentration (11.79±2.35 pg/mL) in cell culture supernatant at 72 hours which was comparable to released KGF from cells cultured with 2% FBS at the same time point.
Previous research has shown that stimulation of fibroblasts with individual growth factors or cytokines such as IL-1β and PDGF-BB increased KGF mRNA expression.[11–13] Our data, however, showed that these exogenous proteins induced no KGF protein release. Based on our results, it is possible that IL-1β and PDGF-BB were acting synergistically with serum proteins present in the media since, in some studies, cells were made quiescent with media supplemented with 0.5% FBS.[11,13] To further elucidate the relationship between IL-1 β induced KGF mRNA transcription and KGF protein release in serum-free media, RT-PCR and cell lysate analysis were conducted.
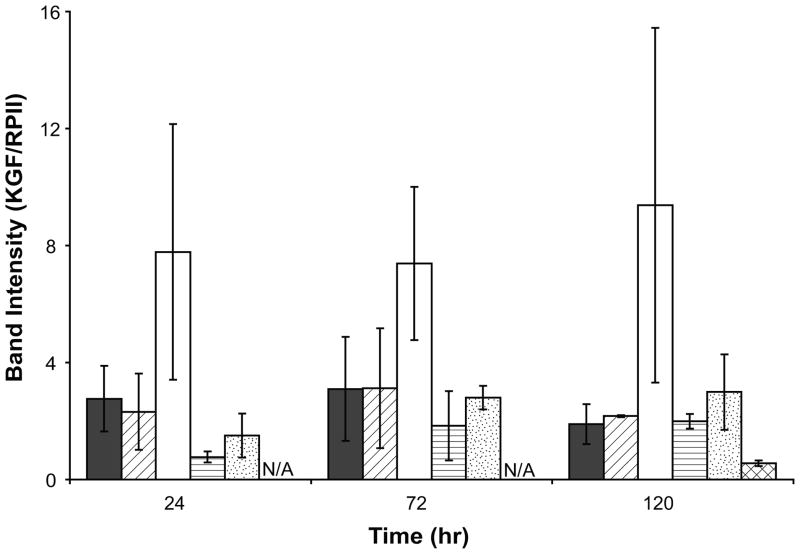
KGF mRNA induction by serum and IL-1β on TCPS and sIPNs
To study the regulation of KGF gene transcription, RT-PCR was performed with serially transitioned fibroblasts which were cultured in FBM, FBM with IL-1β and FBM with 5% FBS on TCPS and sIPN substrates. Transitioned fibroblasts in FBM showed similar KGF mRNA level to cells cultured in FBM supplemented with 5% serum on TCPS (Figure 6). The ratio of KGF/RPII for cells gradually transitioned into FBM (2.8±1.1 and 3.1±1.8 at 24 and 72 hours respectively) was generally higher than that of cells directly passaged into FBM without transitioning (1.1±0.11 at 48 hours). This result suggests that the abrupt media change down-regulated KGF gene transcription and possibly also changed the expression of other genes in dermal fibroblasts. It has been reported that in fibroblasts without media transition, the induction of KGF mRNA decreased with time in both serum and IL-1β supplemented media.[11] In this study, however, the addition of IL-1β to FBM increased KGF mRNA levels from 2.8±1.1 to 7.8±4.4 at 24 hours on TCPS, and the level of KGF mRNA did not change significantly over time (Figure 6). A stimulatory effect of IL-1β on KGF mRNA expression was also observed in cells cultured on unmodified sIPN, although the mRNA level and the extent of IL-1β stimulation were less than that seen in cells adhered to TCPS (Figure 6). Furthermore, the KGF mRNA expression in fibroblasts adhered to RGD-modified sIPNs was significantly lower than that on unmodified sIPN at 120 hours (p<0.01).
Figure 6.
KGF mRNA levels of human dermal fibroblasts cultured in FBM ( ), FBM+5%FBS (
), FBM+5%FBS ( ) and FBM+10ng/mL IL-1β (□) on TCPS; in FBM (
) and FBM+10ng/mL IL-1β (□) on TCPS; in FBM ( ) and FBM+10ng/mL IL-1β (
) and FBM+10ng/mL IL-1β ( ) on unmodified sIPN as well as in FBM on RGD-modified sIPN (
) on unmodified sIPN as well as in FBM on RGD-modified sIPN ( ) at 24, 72 and 120 hours presented as the ratio between KGF and RPII band intensities. KGF and RPII band intensities were measured by ImageJ software version 1.32j. FBM+5%FBS served as a control. All data presented as average ± standard deviation (n=3 for TCPS and RGD-modified sIPN samples; n=2 for unmodified sIPN samples).
) at 24, 72 and 120 hours presented as the ratio between KGF and RPII band intensities. KGF and RPII band intensities were measured by ImageJ software version 1.32j. FBM+5%FBS served as a control. All data presented as average ± standard deviation (n=3 for TCPS and RGD-modified sIPN samples; n=2 for unmodified sIPN samples).
N/A: data not collected for that time point.
Changes between serum-free and serum-containing conditions have been discussed in terms of cell cycle control, autocrine expression of growth factors, and gene expression regulation. [14,40,41] The cell cycle is expected to be arrested in G1 or G0 phase in serum-deprived media and therefore cells cease growing if they are transitioned from serum-containing to serum-free media directly.[40] However, in this study, the growth of serially adapted cells in FBM was not inhibited by such cycle control and cells still proliferated into confluence gradually. Moreover, serum supplemented media had significantly higher KGF protein release which was not observed in serum-free media despite similar mRNA induction. Thus, the results suggest that the intracellular signaling pathways may be differentially regulated under serum-free conditions. One possibility is that KGF mRNA translation in human dermal fibroblasts might be regulated by various post-transcriptional controls which resulted in no KGF protein expression. An alternative is that KGF mRNA was translated, but extracellular transport of the protein was inhibited and the protein was stored intracellularly.
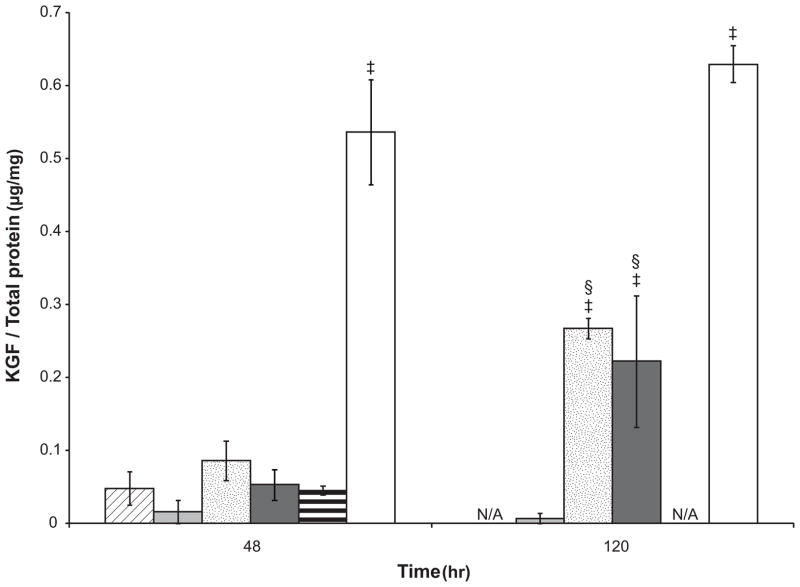
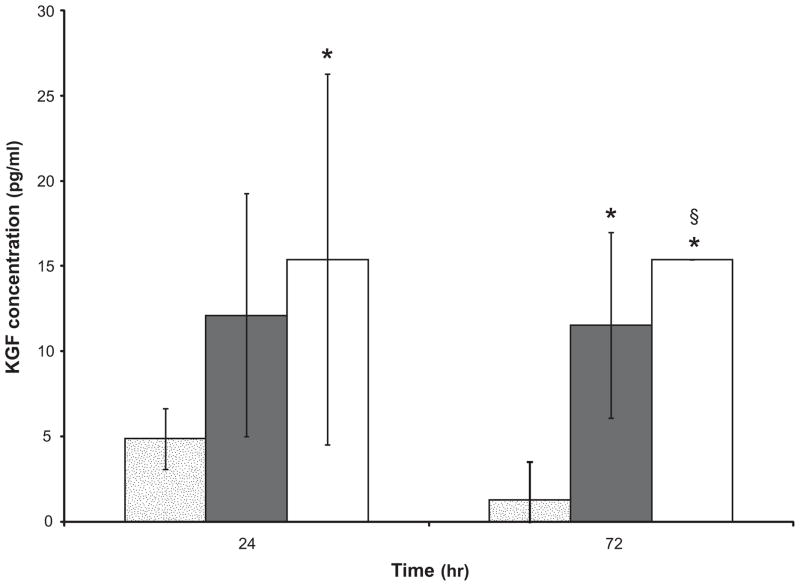
To understand the regulatory mechanisms of KGF protein production in serum-free media, lysates of cells cultured on TCPS (Figure 7) and sIPN (Figure 8) surfaces were analyzed via ELISA. Supplementation with both 5% FBS and IL-1β led to higher amounts of intracellular KGF protein at 48 hours as compared to cells in FBM on TCPS and this stimulatory effect was significantly enhanced over time (48 vs. 120 hours, p<0.05). There was a small amount of KGF protein stored in cells serially transitioned into serum-free FBM, however, abrupt transition from FGM-2 into FBM led to generally higher KGF protein stored in cells (Figure 7). Considering the lower level of KGF gene transcription in cells without media transition as discussed above, the KGF present was likely translated prior to cells being passaged into FBM or within the first 24 hours after cell seeding. The low levels of intracellular KGF protein in fibroblasts serially transitioned into FBM at both 48 and 120 hours suggests a possible inhibitory post-transcriptional control that leads to a decrease of KGF protein synthesis. Interestingly, the KGF mRNA expression on RGD-modified sIPN was significantly lower than unmodified sIPN (Figure 6, p<0.01), however, the KGF protein production in cells on RGD-modified sIPN was significantly higher than that on unmodified sIPN (Figure 8, p<0.05). This substrate effect is probably due to an inverse post-transcriptional regulation of KGF expression which is similar to what was reported by Shaoul et al. with human gastric tissue: KGF protein production was increased in gastric diseases but KGF mRNA level remained at low level.[42] The addition of IL-1β to FBM significantly increased intracellular KGF protein levels (Figure 7 and 8) as well as mRNA levels (Figure 6) on both TCPS and unmodified sIPN surfaces, however, there was still no KGF protein release detected. This implies that IL-1β regulates KGF expression in a manner likely related to protein release inhibition. The dramatic increase of intracellular KGF protein in cells treated with a combination of IL-1β and PDGF-BB on TCPS was consistent to the synergistic effect observed in KGF protein release in cell supernatant.
Figure 7.
KGF protein levels in the lysate of human dermal fibroblasts cultured on TCPS in different media: FBM without serial transitioning ( ), FBM (
), FBM ( ), FBM+5% FBS (
), FBM+5% FBS ( ), FBM+10ng/mL IL-1β (
), FBM+10ng/mL IL-1β ( ), FGM-2 (
), FGM-2 ( ) and FBM+10ng/mL IL-1β+10ng/mL PDGF-BB (□). FBM+5%FBS and FGM-2 served as controls. Cells were seeded in 6-well plates and lysed by M-PER with protease inhibitor at 48 and 120 hours. The KGF concentration in cell lysate was measured by ELISA and the total protein concentration was quantified by a BCA assay. The KGF protein expression was presented as the ratio of KGF to total cell protein. All data presented as average ± standard deviation (n=3).
) and FBM+10ng/mL IL-1β+10ng/mL PDGF-BB (□). FBM+5%FBS and FGM-2 served as controls. Cells were seeded in 6-well plates and lysed by M-PER with protease inhibitor at 48 and 120 hours. The KGF concentration in cell lysate was measured by ELISA and the total protein concentration was quantified by a BCA assay. The KGF protein expression was presented as the ratio of KGF to total cell protein. All data presented as average ± standard deviation (n=3).
‡: compared to FBM with adaptation at the same time point, p<0.05;
§: compared to the same culture medium at 48 hours, p<0.05.
N/A: Data not collected for that media.
Figure 8.
KGF protein concentrations in the lysate of human dermal fibroblasts cultured on unmodified sIPN with FBM ( ) and with FBM+10ng/mL IL-1β (
) and with FBM+10ng/mL IL-1β ( ) as well as on RGD-modified sIPN with FBM (□). Cells were seeded onto sIPNs which were fitted into 48-well plates. Cells were then lysed by M-PER with protease inhibitor at 24 and 72 hours. KGF concentrations were measured by ELISA. All data presented as average ± standard deviation.
) as well as on RGD-modified sIPN with FBM (□). Cells were seeded onto sIPNs which were fitted into 48-well plates. Cells were then lysed by M-PER with protease inhibitor at 24 and 72 hours. KGF concentrations were measured by ELISA. All data presented as average ± standard deviation.
§: significantly different compared to unmodified sIPN when cells were cultured in FBM, p<0.05.
*: sample size n=2, otherwise n=3.
CONCLUSION
A serum-free model was established to study the effects of specific serum proteins and different substrates on human dermal fibroblast behavior. Exogenous TGF-α, IL-1β and PDGF-BB significantly increased adherent cell number on both TCPS and sIPNs, but individually had no effect on KGF protein release. IL-1β and PDGF-BB, however, synergistically acted to increase protein expression and release. Increased KGF mRNA expression and intracellular KGF protein synthesis in IL-1β treated fibroblasts suggest IL-1β stimulated KGF production but not release of the protein, which implies there is some manner of extracellular transport regulation. Results also demonstrated significant effects of both the presence of serum and the manner by which serum is removed from the cell environment on protein synthesis and the subsequent post-transcriptional and post-translational control mechanisms. Results of this study will further our knowledge of cell signaling and wound regeneration-related cell-biomaterial interaction research based on a serum-free environment.
Acknowledgments
The authors thank Amy Chung for her guidance in the polymer synthesis and RT-PCR and for her kind review of this manuscript. This work was supported in part by NIH Grant R01 EB6613.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finch PW, Rubin JS. Keratinocyte growth factor/fibroblast growth factor 7, a homeostatic factor with therapeutic potential for epithelial protection and repair. Adv Cancer Res. 2004;91:69–136. doi: 10.1016/S0065-230X(04)91003-2. [DOI] [PubMed] [Google Scholar]
- 2.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-reviews3005. reviews 3005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werner S, Krieg T, Smola H. Keratinocyte-fibroblast interactions in wound healing. J Invest Dermatol. 2007;127:998–1008. doi: 10.1038/sj.jid.5700786. [DOI] [PubMed] [Google Scholar]
- 5.Mass-Szabowski N, Shimotoyodome A, Fusenig NE. Keratinocyte growth regulation in fibroblast cocultures via a double paracrine mechanism. J Cell Sci. 1999;112:1843–1853. doi: 10.1242/jcs.112.12.1843. [DOI] [PubMed] [Google Scholar]
- 6.Witte RP, Kao WJ. Keratinocyte-fibroblast paracrine interaction: the effect of substrate and culture condition. Biomaterials. 2005;26:3673–3682. doi: 10.1016/j.biomaterials.2004.09.054. [DOI] [PubMed] [Google Scholar]
- 7.Werner S. Keratinocyte growth factor: a unique player in epithelial repair processes. Cytokine Growth Factor Rev. 1998;9:153–165. doi: 10.1016/s1359-6101(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 8.Niu J, Chang Z, Peng B, Xia Q, Lu W, Huang P, et al. Keratinocyte growth factor/fibroblast growth factor-7-regulated cell migration and invasion through activation of NF-κB transcription factors. J Biol Chem. 2007;282:6001–6011. doi: 10.1074/jbc.M606878200. [DOI] [PubMed] [Google Scholar]
- 9.Putnins EE, Firth JD, Uitto VJ. Stimulation of collagenase (MMP-1) synthesis in histiotypic epithelial cell culture by heparin and heparan sulfate with further stimulation by KGF. Matrix Biol. 1996;15:21–29. doi: 10.1016/s0945-053x(96)90123-7. [DOI] [PubMed] [Google Scholar]
- 10.Putnins EE, Firth JD, Uitto VJ. Keratinocyte growth factor stimulation of gelatinase (MMP-9) and plasminogen activator (uPA) in histiotypic epithelial cell culture. J Invest Dermatol. 1995;104:989–994. doi: 10.1111/1523-1747.ep12606233. [DOI] [PubMed] [Google Scholar]
- 11.Brauchle M, Angermeyer K, Hübner G, Werner S. Large induction of keratinocyte growth factor expression by serum growth factors and pro-inflammatory cytokines in cultured fibroblasts. Oncogene. 1994;9:3199–3204. [PubMed] [Google Scholar]
- 12.Chedid M, Rubin JS, Csaky KG, Aaronson SA. Regulation of keratinocye growth factor gene expression by Interleukin 1. J Biol Chem. 1994;269:10753–10757. [PubMed] [Google Scholar]
- 13.Weng J, Mohan RR, Li Q, Wilson SE. IL-1 upregulates keratinocyte growth factor and hepatocyte growth factor mRNA and protein production by cultured stromal fibroblast cells: Interleukin-1β expression in the cornea. Cornea. 1997;16:465–471. [PubMed] [Google Scholar]
- 14.Zander L, Bemark M. Identification of genes deregulated during serum-free medium adaptation of a Burkitt’s lymphoma cell line. Cell Prolif. 2008;41:136–155. doi: 10.1111/j.1365-2184.2007.00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaluria P, Konstantopoulos K, Betenbaugh M, Shiloach J. Egr1 and Gas6 facilitate the adaptation of HEK-293 cells to serum-free media by conferring enhanced viability and higher growth rates. Biotechnol Bioeng. 2008;99:1443–1452. doi: 10.1002/bit.21707. [DOI] [PubMed] [Google Scholar]
- 16.Massia SP, Hubbell JA. An RGD spacing of 440 nm is sufficient for integrin αvβ3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. J Cell Biol. 1991;114:1089–1100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao WJ, Lee D, Schense JC, Hubbell JA. Fibronectin modulates macrophage adhesion and FBGC formation: The role of RGD, PHSRN, and PRRARV domains. J Biomed Mater Res. 2001;55:79–88. doi: 10.1002/1097-4636(200104)55:1<79::aid-jbm110>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 18.Mann BK, West JL. Cell adhesion peptides alter smooth muscle cell adhesion, proliferation, migration, and matrix protein synthesis on modified surfaces and in polymer scaffolds. J Biomed Mater Res. 2002;60:86–93. doi: 10.1002/jbm.10042. [DOI] [PubMed] [Google Scholar]
- 19.Waldeck H, Chung AS, Kao WJ. Interpenetrating polymer networks containing gelatin modified with PEGylated RGD and soluble KGF: synthesis, characterization, and application in in vivo critical dermal wound. J Biomed Mater Res A. 2007;82:861–871. doi: 10.1002/jbm.a.31054. [DOI] [PubMed] [Google Scholar]
- 20.Chung AS, Gao Q, Kao WJ. Either integrin subunit β1 or β3 is involved in Mediating monocyte adhesion, IL-1β protein and mRNA expression in response to surfaces functionalized with fibronectin-derived peptides. J Biomater Sci Polym Ed. 2007;18:713–729. doi: 10.1163/156856207781034179. [DOI] [PubMed] [Google Scholar]
- 21.Gao Q, Chung AS, Kao WJ. Monocytic U937 adhesion, tumor necrosis factor-alpha and interleukin-1beta expression in response to gelatin-based networks grafted with arginine-glycine-aspartic acid and proline-histidine-serine-arginine-asparagine oligopeptides. Tissue Eng. 2007;13:179–185. doi: 10.1089/ten.2006.0007. [DOI] [PubMed] [Google Scholar]
- 22.Kleinbeck KR, Faucher L, Kao WJ. Multifunctional in situ photopolymerized semi-interpenetrating network system is an effective donor site dressing: a cross comparison study in a swine model. J Burn Care Res. 2009;30:37–45. doi: 10.1097/BCR.0b013e3181921f98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burmania JA, Martinez-Diaz GJ, Kao WJ. Synthesis and physicochemical analysis of interpenetrating networks containing modified gelatin and poly(ethylene glycol) diacrylate. J Biomed Mater Res A. 2003;67:224–234. doi: 10.1002/jbm.a.10106. [DOI] [PubMed] [Google Scholar]
- 24.Shearer H, Ellis MJ, Perera SP, Chaudhuri JB. Effects of common sterilization methods on the structure and properties of poly(d,l lactic-coglycolic acid) scaffolds. Tissue Eng. 2006;12:2717–2727. doi: 10.1089/ten.2006.12.2717. [DOI] [PubMed] [Google Scholar]
- 25.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 26.Radoni A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 27.Rasband WS. ImageJ. U.S. National Institutes of Health; Bethesda, MD: Online. 1997–2008. Available from URL: http://rsb.info.nih.gov/ij. [Google Scholar]
- 28.Sasaki M, Kashima M, Ito T, Watanabe A, Izumiyama N, Sano M, et al. Differential regulation of metalloproteinase production, proliferation and chemotaxis of human lung fibroblasts by PDGF, interleukine-1β and TNF-α. Mediators Inflamm. 2000;9:155–160. doi: 10.1080/09629350020002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto M, Aoyagi M, Fukai N, Matsushima Y, Yamamoto K. Differences in cellular responses to mitogens in arterial smooth muscle cells derived from patients with moyamoya disease. Stroke. 1998;29:1188–1193. doi: 10.1161/01.str.29.6.1188. [DOI] [PubMed] [Google Scholar]
- 30.Bronson RE, Argenta JG, Bertolami CN. Interleukin-1-induced changes in extracellular glycosaminoglycan composition of cutaneous scar-derived fibroblasts in culture. Coll Relat Res. 1988;8:199–208. doi: 10.1016/s0174-173x(88)80040-2. [DOI] [PubMed] [Google Scholar]
- 31.Bronson RE, Bertolami CN, Siebert EP. Modulation of fibroblast growth and glycosaminoglycan synthesis by interleukin-1. Coll Relat Res. 1987;7:323–332. doi: 10.1016/s0174-173x(87)80025-0. [DOI] [PubMed] [Google Scholar]
- 32.Denk PO, Roth-Eichhorn S, Gressner AM, Knorr M. Cytokine regulation of hyaluronate production by human tenon’s capsule fibroblasts. Curr Eye Res. 2000;20:77–80. [PubMed] [Google Scholar]
- 33.Brink HE, Stalling SS, Nicoll SB. Influence of serum on adult and fetal dermal firbroblast migration, adhesion and collagen expression. In Vitro Cell Dev Biol Anim. 2005;41:252–257. doi: 10.1290/0503023R.1. [DOI] [PubMed] [Google Scholar]
- 34.Gnessi L, Emidi A, Scarpa S, Palleschi S, Ragano-Caracciolo M, Silvestroni L, et al. Platelet-derived growth factor effects on purified testicular peritubular myoid cells: binding, cytosolic Ca2+ increase, mitogenic activity, and extracellular matrix production enhancement. Endocrinology. 1993;133:1880–1890. doi: 10.1210/endo.133.4.8404630. [DOI] [PubMed] [Google Scholar]
- 35.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 36.Milam SB, Magnuson VL, Steffensen B, Chen DL, Klebe RJ. IL-1β and prostaglandins regulate integrin mRNA expression. J Cell Physiol. 1991;149:173–183. doi: 10.1002/jcp.1041490202. [DOI] [PubMed] [Google Scholar]
- 37.Rinaldi N, Weis D, Brado B, Schwarz-Eywill M, Lukoschek M, Pezzutto A, et al. Differential expression and functional behaviour of the αv and β3 integrin subunits in cytokine stimulated fibroblast-like cells derived from synovial tissue of rheumatoid arthritis and osteoarthritis in vitro. Ann Rheum Dis. 1997;56:729–736. doi: 10.1136/ard.56.12.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman S, He S, Jin M, Ehren M, Wiedemann P, Ryan SJ, et al. A selective cyclic integrin antagonist blocks the integrin receptors αvβ3 and αvβ5 and inhibits retinal pigment epithelium cell attachment, migration and invation. BMC Ophthalmol. 2005;5:16. doi: 10.1186/1471-2415-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takizawa M, Hida T, Horiguchi T, Hiramoto A, Harada S, Tanida S. TAN-1511 A, B and C, microbial lipopeptides with G-CSF and GM-CSF inducing activity. J Antibiot (Tokyo) 1995;48:579–588. doi: 10.7164/antibiotics.48.579. [DOI] [PubMed] [Google Scholar]
- 40.Kamely D, Rudland PS. Induction of DNA synthesis and cell division in human diploid skin fibroblasts by fibroblast growth factor. Exp Cell Res. 1976;97:120–126. doi: 10.1016/0014-4827(76)90661-3. [DOI] [PubMed] [Google Scholar]
- 41.Kaplan PL, Anderson M, Ozanne B. Transforming growth factor(s) production enables cells to grow in the absence of serum: an autocrine system. Proc Natl Acad Sci USA. 1982;79:485–489. doi: 10.1073/pnas.79.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaoul R, Eliahu L, Sher I, Hamlet Y, Miselevich I, Goldshmidt O, et al. Elevated expression of FGF7 protein is common in human gastric diseases. Biochem Biophys Res Commun. 2006;350:825–833. doi: 10.1016/j.bbrc.2006.08.198. [DOI] [PubMed] [Google Scholar]