Abstract
Osmotic stresses such as drought, salinity, and cold are major environmental factors that limit agricultural productivity worldwide. Protein phosphorylation/dephosphorylation are major signalling events induced by osmotic stress in higher plants. Sucrose non-fermenting 1-related protein kinase2 family members play essential roles in response to hyperosmotic stresses in Arabidopsis, rice, and maize. In this study, the function of TaSnRK2.4 in drought, salt, and freezing stresses in Arabidopsis was characterized. A translational fusion protein of TaSnRK2.4 with green fluorescent protein showed subcellular localization in the cell membrane, cytoplasm, and nucleus. To examine the role of TaSnRK2.4 under various environmental stresses, transgenic Arabidopsis plants overexpressing wheat TaSnRK2.4 under control of the cauliflower mosaic virus 35S promoter were generated. Overexpression of TaSnRK2.4 resulted in delayed seedling establishment, longer primary roots, and higher yield under normal growing conditions. Transgenic Arabidopsis overexpressing TaSnRK2.4 had enhanced tolerance to drought, salt, and freezing stresses, which were simultaneously supported by physiological results, including decreased rate of water loss, enhanced higher relative water content, strengthened cell membrane stability, improved photosynthesis potential, and significantly increased osmotic potential. The results show that TaSnRK2.4 is involved in the regulation of enhanced osmotic potential, growth, and development under both normal and stress conditions, and imply that TaSnRK2.4 is a multifunctional regulatory factor in Arabidopsis. Since the overexpression of TaSnRK2.4 can significantly strengthen tolerance to drought, salt, and freezing stresses and does not retard the growth of transgenic Arabidopsis plants under well-watered conditions, TaSnRK2.4 could be utilized in transgenic breeding to improve abiotic stresses in crops.
Keywords: Abiotic stress, morphological character, physiological trait, stress responses
Introduction
Drought, salinity, and cold are major environmental stresses that severely reduce agricultural productivity worldwide. To cope with water deficit, plants have developed various mechanisms to protect cellular activities and maintain whole plant integrity. Many stress-induced genes have been identified, including those encoding fundamental enzymes of abscisic acid (ABA) biosynthesis (Bray, 1997), proteins involved in osmotic adaptation and tolerance of cellular dehydration (Shinozaki and Yamaguchi, 1997), cellular protective enzymes (Ingram and Bartels, 1996), and a range of signalling proteins such as transcription factors (Soderman et al., 1996) and protein kinases/protein phosphatases (Hong et al., 1997).
In eukaryotes, reversible protein phosphorylation is central to perception of and response to environmental stresses, and constitutes a major mechanism for the control of cellular functions, such as responses to environmental stimuli and pathogens, and hormonal control of metabolism (Cohen, 1988). Genetic evidence clearly shows that type 2C and 2A protein phosphatases function in the early ABA signalling pathway (Sheen, 1998; Kwak et al., 2002). As a counterpart, various stress-inducible protein kinase families such as mitogen-activated protein kinase (MAPK) (Wrzaczek and Hirt, 2001), calcium-dependent protein kinase (CDPK) (Ludwig et al., 2004), and SNF1-related protein kinase (SnRK), which were first analysed in yeast from where the name originated, are activated by ABA and diverse stress signals.
Yeast SNF1 protein kinase, mammalian AMP-activated protein kinase (AMPK), and plant SnRK protein are highly conserved and play pivotal roles in growth and metabolic responses to cellular stress. In yeast, SNF1 is involved in a variety of functions, including regulation of glucose-responsive genes, control of pseudohyphal growth under nutrient limitations (Cullen and Sprague, 2000), and regulation of meiosis (Honigberg and Lee, 1998). In mammals, AMPK acts as an energy-level sensor to regulate metabolism under low-energy conditions. In plants, SnRKs were grouped into three subfamilies: SnRK1, SnRK2, and SnRK3 (Hrabak et al., 2003). SnRK1 kinase is well characterized at the molecular and biochemical levels, and evidence indicates that SnRK1s have roles in regulating energy metabolism (Hardie et al., 1998). SnRK2 and SnRK3 are unique to plants and are involved in responses to environmental stresses. Several SnRK3 members are extensively characterized, and the well-known SOS2 is required for Na+ and K+ homeostasis in Arabidopsis (Gong et al., 2002).
Current studies indicate that the SnRK2 family is involved in hyperosmotic stress responses and ABA signalling (Boudsocq et al., 2004, 2007; Kobayashi et al., 2004). Ten SnRK2s were identified in Arabidopsis; nine of them were activated by hyperosmotic and salinity stresses, and five of the nine were activated by ABA, whereas none was activated by cold stress (Boudsocq et al., 2004). AtSRK2.6/AtSnRK2E/OST1 and Vicia faba AAPK were activated by ABA and were involved in ABA regulation of stomatal closing and ABA-regulated gene expression (Li et al., 2000; Mustilli et al., 2002; Yoshida et al., 2002). Overexpression of AtSnRK2.8/AtSnRK2C enhances drought tolerance in Arabidopsis (Umezawa et al., 2004). In rice, 10 SnRK2 members were identified; all were activated by hyperosmotic stress, and three were also activated by ABA (Kobayashi et al., 2004). With overexpression of SAPK4, one rice member significantly enhanced salt tolerance of transgenic plants (Diedhiou et al., 2008). In maize, 10 SnRK2 members were cloned, and most ZmSnRK2 genes were induced by one or more abiotic stresses (Huai et al., 2008). In soybean (Glycine max), four SnRK2 members were isolated, and all were activated by hyperosmotic stress (Yoon et al., 1997; Monks et al., 2001). NtOSAK, identified in tobacco, was involved in the response to hyperosmotic stress (Kelner et al., 2004). In wheat, only one SnRK2 member, PKABA1, was induced by ABA and hyperosmotic stress, and it repressed the activities of gibberellic acid-inducible promoters when transiently overexpressed in barley aleurone layers (Gomez-Cadenas et al., 1999, 2001; Shen et al., 2001; Johnson et al., 2002). Previous findings suggest that many SnRK2 members are involved in the response to environmental stimuli, and different members exhibit diverse expression patterns, suggesting they may play different roles in response to abiotic stresses. However, knowledge of specific functions of SnRK2s is fragmentary and their role in stress signalling is still enigmatic. There is a paucity of reports about wheat SnRK2 members.
In this study, TaSnRK2.4, an SnRK2 member from wheat, was cloned and its expression patterns in response to water deficit, high salinity, low temperature, and ABA treatment were determined. Various expression patterns occurred with different stresses. Transgenic experiments indicated that TaSnRK2.4 significantly increased tolerance to drought, salt, and freezing stresses in Arabidopsis. Morphological assays revealed that overexpression of TaSnRK2.4 did not cause negative effects on the growth and yield of transgenic plants. Therefore, TaSnRK2.4 might be utilized to improve abiotic stress tolerance in plants.
Materials and methods
Plant materials and water stress experiments
Wheat (Triticum aestivum L.) genotype ‘Hanxuan 10’ with a conspicuous drought-tolerant phenotype was used in this study. After sterilizing with 75% ethanol and washing with sterilized water, wheat seeds were germinated and cultured with double-distilled water in a growth chamber (20±1 °C, 150 μmol m−2 s−1, 12 h light/12 h dark cycle). Two-leaf seedlings, which showed extreme tolerance to drought stress at this developmental stage in pilot experiments, were treated with polyethylene glycol-6000 (PEG-6000; –0.5 MPa) solution, 250 mM NaCl, low temperature (4 °C), and 50 μM ABA. The treated plants were stressed in the PEG and NaCl solutions, sprayed with ABA, or cultured in low temperature conditions for 1, 3, 6, 12, 24, 48, and 72 h. Untreated control seedlings continued to be grown in the growth chamber. Whole wheat leaves were sampled from the seedlings at different times, frozen immediately with liquid nitrogen, and stored at –80 °C for RNA isolation and other analyses.
To study the expression of target genes at different developmental stages, seedling leaves and roots, the leaf spindle at jointing, and young ears at the heading stage were sampled. Seedlings were grown in the growth chamber as described above, and spindle leaves at jointing and young ears were sampled from plots without environmental stress.
Construction and screening of a full-length cDNA library database
Tissues from wheat seedlings at various stages and from mature plants were collected to extract total RNA with TRIZOL reagent (Invitrogen), and mRNA was isolated with oligo d(T) cellulose (Qiagen). Several full-length cDNA libraries of wheat in λ Zap II (Stratagene) were constructed with the optimized Cap-trapper method (Mao et al., 2005). A full-length wheat cDNA database was generated with the 3′ and 5′ end sequencing data of full-length cDNA clones. To obtain the cDNA sequence of TaSnRK2.4, the amino acid sequence of rice SAPK4 was used as a query probe to screen the wheat full-length cDNA database. Four candidate clones were obtained by blastp, and the full-length cDNA of TaSnRK2.4 was identified by sequencing the ends.
Database searches of the nucleotide and deduced amino acid sequences were performed by NCBI/GenBank/Blasting. Sequence alignment and similarity with other species were determined by the megAlign program in DNAStar. The signal sequence was predicted with SignalP (http://genome.cbs.dtu.dk/services/SignalP). The functional region and activity sites were identified using PROSITE (http://expasy.hcuge.ch/sprot/ prosite.html) and SMART motif search programs (http://coot.embl-heidelberg.de/SMART)
Phylogenetic tree construction of TaSnRK2.4
Phylogenetic analysis was performed to understand the relationship between TaSnRK2.4 and other SnRK2 members from other plant species. A maximum likelihood tree was constructed with the proml program in the PHYLIP (version 3.68) software package, with the putative amino acid sequences. The bootstrap parameter was set at 100.
Gene structure analysis
To analyse the structure of TaSnRK2.4, a pair of primers flanking the open reading frame (ORF) were designed (forward primer, 5′-TGCAGAGTTCCACGATAGGCCG-3′, reverse primer, 5′-CCTACCGACCCAACGAACGAG-3′; LA-Taq (Takara) was utilized to amplify the genome sequence of TaSnRK2.4, and sequences were analysed with the MegAlign program in DNAStar software.
Subcellular localization of TaSnRK2.4 protein
The full-length cDNA clone of TaSnRK2.4 was fused upstream of the green fluorescent protein (GFP) gene and put under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter in the pJIT163-GFP expression vector for construction of a 35S::TaSnRK2.4-GFP fusion protein. Proper restriction sites were added to the 5′ and 3′ ends of the coding region by PCR; the oligonucleotides for fusion GFP subcloning were: forward primer, 5′-GAGAGTCGACATGGAGAAGTACGAGGCGGT-3′ (SalI site in bold italics), reverse primer, 5′-GAGAGGATCCCGAGCTCATGCGGAGCTCT-3′ (BamHI site in bold italics). The PCR product obtained was digested with appropriate restriction endonucleases, and then ligated with the pJIT163-GFP plasmid cut with the corresponding enzyme to create recombinant plasmids for expression of the fusion protein. Positive plasmids were confirmed by restriction analysis, followed by sequencing. The recombinant constructs were transformed into living onion epidermal cells by biolistic bombardment with a GeneGun (Biorad Helios™) according to the instruction manual (helium pressure, 150–300 psi). The subcellular location of TaSnRK2.4 was detected by monitoring the transient expression of GFP in onion epidermal cells. The transformed cells were incubated in Murashige and Skoog (MS) medium at 28 °C for 36–48 h and then observed with a laser scanning confocal microscope (Leika TCS-NT). The images obtained were recorded automatically. The recombinant constructs and the control pJIT163-GFP plasmid were each bombarded into 20 onion epidermal segments.
Quantitative real-time PCR
After treatment with DNase I, the RNA samples were used as templates for cDNA synthesis using the Superscript First-Strand Synthesis System kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) was performed in triplicate with an ABI PRISM® 7000 system using the SYBR Green PCR master mix kit (Applied Biosystems) according to the manufacturer's instructions. A tubulin transcript was used to quantify the relative transcript level. The qRT-PCR primers were: forward primer, 5′-GGTTCATGCAAGCGGAGAGC-3′; reverse primer, 5′-AACCAAAACCAAACAGAAGCAAAC-3′.
The relative level of gene expression was detected using the 2–ΔΔCT method (Livaka and Schmittgen, 2001). ΔΔCT=(CT,Target–CT,Tubulin)Time x–(CT,Target–T,Tubulin)Time 0. The CT (cycle threshold) values for both the target and internal control genes were the means of triplicate independent PCRs. Time x is any treated time point (1, 3, 6, 12, 24, 48, or 72 h) and Time 0 represents the untreated time (0 h). To detect the transcription level of TaSnRK2.4 at different developmental stages, the expression of TaSnRK2.4 in seedling leaves was regarded as standard for its lower level, and the corresponding formula was modified as ΔΔCT=(CT,Target–CT,Tubulin)DST–(CT,Target–CT,Tubulin)SL. DST refers to the developmental stage tissue and SL to the seedling leaf. The tubulin transcript of Arabidopsis was used to quantify the expression levels of TaSnRK2.4 in the transgenic Arabidopsis lines.
Generation of transgenic plants
The coding region of TaSnRK2.4 cDNA was amplified by RT-PCR using primers 5′-GAGAGGATCCGGGATGGAGAAGTACGAGGCG-3′ (the BamHI site is in bold italics) and 5′-GAGAGTCGACGATATGCGTAGCGAGCTCATGC-3′ (the SalI site is in bold italics) and cloned into a pPZP211 vector (Hajdukiewicz et al., 1994) as a GFP-fused fragment driven by the CaMV 35S promoter. The transformation vectors harbouring 35S::GFP and 35S::TaSnRK2.4-GFP were introduced into Agrobacterium, and transferred into wild-type (WT) Arabidopsis (Columbia ecotype) plants by floral infiltration. Positive transgenic lines were firstly screened on kanamycin plates and then identified by fluorescence detection and western blotting. The TaSnRK2.4–GFP for different transgenic lines was detected with a confocal microscope, and the measurement of protein abundance was relative to the intensity of the GFP signal in 2-d-old seedling roots under the same detection conditions.
Morphological characterization of transgenic plant roots and shoots
Transgenic plants were characterized for morphological changes under short-day (12 h light/12 h dark) photoperiods in a growth chamber with a constant temperature of 22 °C. Root morphology was examined on MS medium solidified with 1.0% agar. Briefly, T3 homozygous transgenic and WT seeds were germinated on MS medium and grown vertically for 7 d before measurement of primary root length.
Stomatal aperture measurements
Seedling leaves of the same size were detached; the epidermis was peeled off and immediately placed flat on a glass slide. Guard cells at similar locations were observed and photographed using an Axioplan 2 microscope (Carl Zeiss). The width, length, and area of each stomatal pore in the photographed image were measured, and the ratios of stomata and leaf areas were recorded.
Drought tolerance assays
Drought tolerance assays were performed on seedlings and mature plants. Both WT and transgenic seeds were germinated on MS medium. Four 7-d-old seedlings were planted in identical pots containing a soil mix (1:1 vermiculite:humus) and well watered. The seedlings were cultured in a greenhouse (22 °C, 70% humidity, 150 μmol m−2 s−1, 12 h light/12 h dark cycle) without watering. The difference between the seedling and mature plant treatments was the soil volume in each pot. For the seedling treatment, the soil was half the volume (∼100 ml) of the pot, and for mature plants the soil was three-quarters the volume (∼150 ml).
Salt tolerance assays
Salt tolerance assays were conducted at the seedling stage. Arabidopsis seedlings were cultured as described above. Water was withheld for 3 weeks and plants were then well irrigated with NaCl solution (350 mM) applied at the bottom of the pots. When the soil was completely saturated with salt water, free NaCl solution was removed and the plants were cultured as normal. To make morphological differences under severe salt stress more distinctive, after 1 week the salt-water-logged pots were placed in 2 cm deep fresh water for 24 h to leach the salt completely from the soil. Survival rates were recorded 2 weeks later.
Cold tolerance assays
Cold tolerance assays were carried out on seedlings. Normally cultured Arabidopsis seedlings (4 weeks old) were stressed in a –10 °C freezer for 1.5 h, then cultured at 15 °C for 24 h to facilitate recovery, and finally cultured under normal growing conditions.
Water loss rate determination
Water loss rates were measured using 10 plants each of WT and transgenic plants (including GFP transgenic plants). Four-week-old plants were detached from roots and weighed immediately (fresh weight, FW), then the plants were left on the laboratory bench (humidity, 45–50%, 20–22 °C) and weighed at the designated time intervals. The proportions of fresh weight loss were calculated relative to the initial plant weights. The plants were finally oven dried for 24 h at 80 °C to a constant dry weight (DW). Relative water contents (RWCs) were measured according to the formula: RWC (%)=(desiccated weight–DW)/(FW–DW)×100.
Cell membrane stability
Plant cell membrane stability (CMS) was determined with a conductivity meter (DDS-1, YSI), CMS (%)=(1–initial electrical conductivity/electrical conductivity after boiling)×100. Fifteen 7-d-old seedlings (grown on 1× MS medium, 0.8% agar) were transferred to a horizontal screen; seedling roots were completely submerged in PEG-6000 (25.4%, –1.4 Mpa) or NaCl (250 mM). When signs of stress began to appear on WT plants, seedlings were removed and immediately thoroughly rinsed with double-distilled water (ddH2O) prior to immersion in 20 ml of ddH2O at room temperature. After 2 h the initial conductivities of the solutions were recorded. The samples were then boiled for 30 min, cooled to room temperature, and the final conductivities were measured.
Osmotic potential and free proline determination
Osmotic potential (OP) was measured with a Micro-Osmometer (Fiske® Model 210, Fiske® Associates). Measurements were taken in the freezing point mode at room temperature. Five plants of each line were collected as a sample, which was finely ground using a mortar and pestle before being transferred to a microcentrifuge tube. The supernatant tissue sap was obtained after centrifuging at 12 000 rpm for 10 min at room temperature. Three replications were set for each plant line, and the OP for each sample was measured three times. Free proline was extracted and quantified from fresh tissues of well-watered seedlings (0.5 g) according to the method of Hu et al. (1992).
Chlorophyll fluorescence assays
Chlorophyll florescence was measured with a portable photosynthesis system (LI-COR LI-6400 XTR). Fully expanded leaves were selected for the determination of chlorophyll fluorescence parameters; three measurements were made for each plant, and 20 plants were used for WT and transgenic lines. The maximum efficiency of photosystem II (PSII) photochemistry, Fv/Fm=(Fm–F0)/Fm, was employed to assess changes in the primary photochemical reactions of the photosynthetic potential at an early stage of drought stress.
Protein isolation and western blotting of Arabidopsis seedlings
Total protein was extracted from ∼0.1 g of Arabidopsis seedling tissue using 200 μl of buffer containing HEPES-NaOH (pH 7.5), 5 mM EDTA, 5 mM EGTA, 10 mM Na3VO4, 10 mM NaF, 5% glycerol, 10 mM dithiothreitol (DTT), 1 mM phenylmethylsulphonyl fluoride (PMSF), 10 mg ml−1 leupeptin, 10 mg ml−1 aprotinin, and 10 mg ml−1 antipain. To obtain enough protein, the homogenate was placed on ice for 1–2 h and centrifuged at 20 000 g for 40 min at 4 °C. The supernatant was immediately frozen in liquid nitrogen and further fractionated by centrifugation at 20 000 g for 20 min at 4 °C. The resulting supernatant was assayed for protein quantity according to the Bradford (1976) method. Protein samples were electrophoretically separated on 12.5% polyacrylamide gels with a visible protein marker, and subsequently proteins were transferred to polyvinylidene fluoride (PVDF) membranes (pore size: 0.45 mm) (Amersham) by semi-dry electroblotting (Mini-Protean II system; Bio-Rad) using TBST (2 mM TRIS, 192 mM glycine, 20% methanol complemented with 0.1% Tween-20) as the transfer buffer. The membrane was blocked with 5% skim milk and blotted with commercial GFP-tag rabbit monoclonal antibody diluted in TBST. After extensive washing, the bound primary antibody was detected with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody according to the manufacturer's recommended procedure (Amersham).
Results
Molecular characterization of TaSnRK2.4
TaSnRK2.4 (GQ384359) was obtained by screening wheat full-length cDNA libraries. The TaSnRK2.4 cDNA is 1335 bp in length and consists of 54 bp of 5′-untranslated region (UTR), 1092 bp of ORF, and 173 bp of 3′-UTR. The ORF encodes a polypeptide of 364 amino acid residues (AARs) with a predicted molecular mass of 42.1 kDa and a pI of 6.05. The deduced amino acid sequence shows high homology with counterpart monocot SnRK2 family members, that is Oryza sativa and Zea mays, and relatively lower homology with SnRK2s from dicot species, such as Glycine max and A. thaliana. TaSnRK2.4 has 92.5% identity to O. sativa SAPK4 (Q5N942), 82.2% to ZmSAPK4 (ACG46236), 80.1% to GmPK3 (AAB68961), and 75.5% to ASK1 (NP_172563) and FsPK (Fagus sylvatica) (CAE54075). Scansite analysis indicated that TaSnRK2.4 has the potential for serine/threonine and tyrosine kinase activities and, like other SnRK2s, it has two domains in its N- and C-terminal regions. The N-terminal catalytic domain (4–260 AARs) is highly conserved, containing an ATP-binding site (10–33 AARs) and a protein kinase-activating site (119–131 AARs) (Fig. 1A). The C-terminal region, in which a stretch of acidic amino acids forms a negatively charged domain with the amino acid glutamate, is thought to function in protein–protein interactions and is mainly involved in ABA responsiveness (Kobayashi et al., 2004). The secondary structure prediction revealed that the TaSnRK2.4 sequence formed 11 α-helixes and nine β-pleated sheets. The tertiary structure assay displayed a similar model to the 3D structure of rice SAPK4 (Kobayashi et al., 2004).
Fig. 1.
Sequence alignment of TaSnRK2.4 and SnRK2s in other plant species. (A) Amino acid alignment of TaSnRK2.4 and other SnRK2 family members from selected plant species. The numbers on the left indicate the amino acid position. Identical amino acid residues are shown with a black background. Gaps, indicated by dashed lines, are introduced for optimal alignment. The box indicated by a solid triangle is the ATP-binding region signature. The box indicated by an asterisk is the serine/threonine protein kinase-activating signature. The region underlined indicates the divergent C-terminus. Alignments were performed using the Megalign program of DNAStar. (B) Phylogenetic tree of TaSnRK2.4 and SnRK2 members from other plant species. At, Arabidopsis thaliana; Fs, Fagus sylvatica; Gm, Glycine max; Os, Oryza sativa; Zm, Zea mays. The phylogenetic tree was constructed with the PHYLIP 3.68 package; bootstrap values are in percentages. (This figure is available in colour at JXB online.)
Phylogenetic analysis
Halford and Hardie (1998) and Hardie et al. (1998) divided the SnRK2 family into two groups based on protein size and character of the acidic amino acid-enriched C-terminus, namely SnRK2a and SnRK2b. SnRK2a corresponds to the later defined subclass I, and SnRK2b includes subclasses II and III. A phylogenetic tree was constructed with the putative amino acid sequences of TaSnRK2.4 and some SnRK2 family members of subclass I, with enriched glutamate in the C-terminal region (Fig. 1B). TaSnRK2.4 clustered in the same clade as OsSAPK4 and ZmSAPK4/ZmSnRK2.4. OsSAPK4 enhances tolerance to high salinity in transgenic rice (Diedhiou et al., 2008).
Determination of gene structure
To analyse the gene structure, a pair of primers flanking the ORF was utilized to amplify TaSnRK2.4. The genomic DNA sequence is ∼4.3 kb, consisting of nine exons and eight introns, with all spicing sites following the GT–AG rule. This structure is consistent with counterparts of rice, maize, and Arabidopsis (Huai et al., 2008).
Expression of TaSnRK2.4 in tissues at different developmental stages
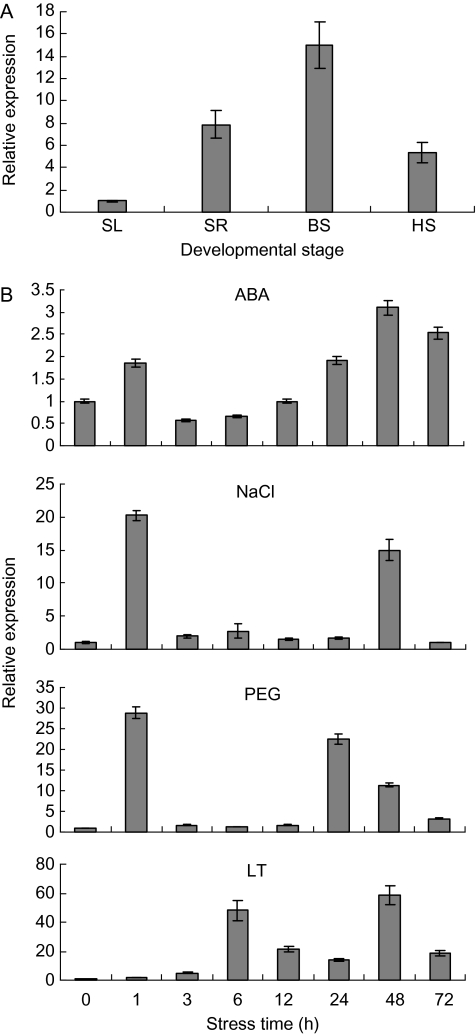
Expression patterns of TaSnRK2.4 in seedling, booting, and heading tissues were analysed by qRT-PCR (Fig. 2A). The highest relative expression occurred in the booting spindle, and the relative expression levels were 15, 7.9, and 5.3 times greater than the control for the booting spindle, seedling root, and heading spike, respectively.
Fig. 2.
Expression patterns of TaSnRK2.4. (A) Expression patterns of TaSnRK2.4 in wheat tissues at different developmental stages. SL, seedling leaf; SR, seedling root; BS, booting spindle; HS, heading spike. The 2–ΔΔCT method was used to measure the relative expression level of the target gene, and the expression of TaSnRK2.4 in seedling leaves was regarded as the standard for its lower level. (B) Expression patterns of TaSnRK2.4 under ABA, salt (NaCl), PEG, and low temperature (LT) treatments. Two-leaf seedlings of common wheat cv. Hanxuan 10 were exposed to abiotic stresses as described in the Materials and methods. The 2–ΔΔCT method was used to measure the relative expression level of the target gene, and the expression of TaSnRK2.4 in non-stressed seedling leaves was regarded as the standard. Means were generated from three independent measurements; bars indicate standard errors.
Early response of TaSnRK2.4 to hyperosmotic stresses
The expression of TaSnRK2.4 was characterized by qRT-PCR in seedling leaves, and various expression patterns were observed under different stresses (Fig. 2B). TaSnRK2.4 was significantly activated by salt, water deficit, and low temperature stresses, and only weakly by ABA. Similar double-peaked expression patterns were apparent for all four treatments; rapid responses for PEG and salt stresses were detected after 1 h (maybe even earlier). The expression levels of TaSnRK2.4 peaked at 1 h for PEG and NaCl, and at 48 h for cold and ABA, and the corresponding maxima were 29, 20, 59, and 2.5 times the normal level, respectively.
Subcelluar localization of TaSnRK2.4
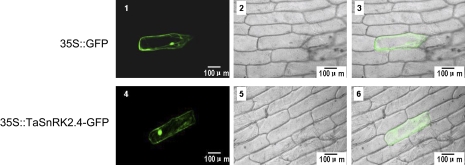
Protein kinases localize to specific cell compartments for proper function, and scanning sequences often specify their intracellular locations. One putative N-myristylation site and a transmembrane region were identified in the TaSnRK2.4 protein by the PlantsP program (http://plantsp.sdsc.edu), suggesting that TaSnRK2.4 might interact with the cell membrane and nuclear system. The subcellular distribution of TaSnRK2.4 in onion epidermis was also examined by transient expression of fusion proteins with GFP under fluorescent microscopy. As predicted, TaSnRK2.4–GFP was present in the cell membrane, cytoplasm, and nucleus (Fig. 3).
Fig. 3.
Subcellular localization of TaSnRK2.4 in onion epidermal cells. Cells were bombarded with constructs carrying GFP or TaSnRK2.4–GFP as described in the Materials and methods. GFP and TaSnRK2.4–GFP fusion proteins were transiently expressed under control of the CaMV 35S promoter in onion epidermal cells and observed with a laser scanning confocal microscope. Images were taken in the dark field for green fluorescence (1, 4), while the outline of the cell (2, 5) and the combination (3, 6) were photographed in a bright field. (This figure is available in colour at JXB online.)
Gene expression level and protein abundance in TaSnRK2.4 transgenic lines
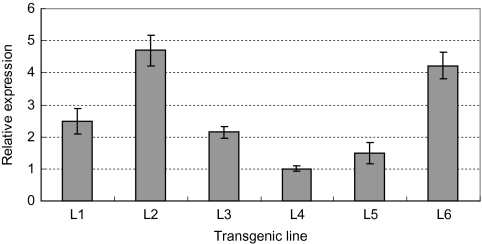
Six transgenic lines were randomly selected for detection of gene expression, and line 4 was used for the quantification of the expression level of TaSnRK2.4 because of its lowest expression. The expression levels of TaSnRK2.4 in different transgenic lines varied greatly. The highest expression occurred in line 2, followed by lines 6, 1, 3, and 5 (Fig. 4). Protein detection revealed that the abundance of TaSnRK2.4 in transgenic lines was quite similar to the gene expression levels, and GFP intensities of lines 4 and 5 were relatively weaker than those of the other four lines (Supplementary Fig. S1 available at JXB online).
Fig. 4.
Expression levels of TaSnRK2.4 in different transgenic Arabidopsis lines. Gene expression level of TaSnRK2.4 in different transgenic Arabidopsis lines. L1–L6, six individual TaSnRK2.4 transgenic lines. The expression of TaSnRK2.4 in L4 was regarded as the standard due to its lower level.
Morphological characteristics of TaSnRK2.4-overexpressing Arabidopsis plants under normal growth conditions
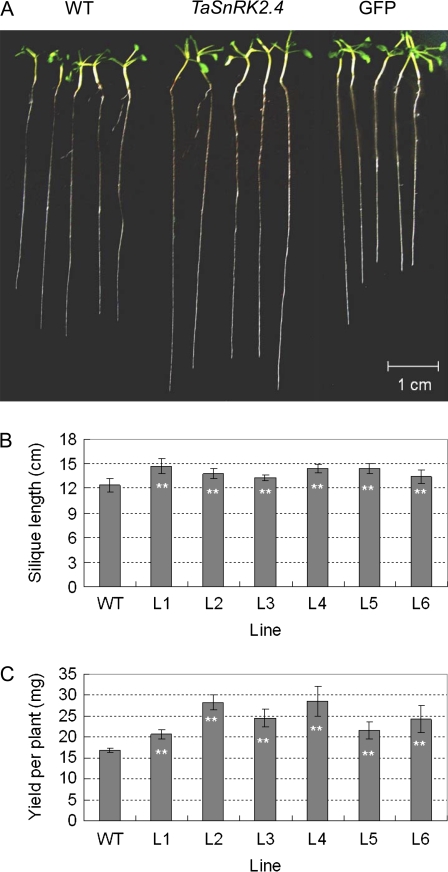
To evaluate the applicability of TaSnRK2.4 in transgenic breeding for abiotic stress tolerance, the phenotypes of TaSnRK2.4 Arabidopsis were characterized at different developmental stages. The seed germination assay indicated that the seedling establishment time (SET) for TaSnRK2.4 (T3 homozygous seeds) was nearly 24 h later than in the WT, whereas the germination rate of transgenic lines was slightly higher than that of the WT, but the difference did not reach a significant level (F-test, P >0.05) (data not show). As a consequence the seedlings of TaSnRK2.4 plants were slightly smaller than those of WT Arabidopsis plants at the very early stages (data not shown), but the difference disappeared after 2 weeks on MS medium (data not shown). The primary root lengths for transgenic lines were significantly greater than those of WT and GFP plants (F-test, *P <0.05) (Fig. 5A; Supplementary Fig. S2 at JXB online). For seedlings (4 weeks old) grown in soil, there was no visible difference between transgenic and WT plants under well-watered conditions (data not shown), but the siliques of transgenic Arabidopsis were significantly longer than those of the WT (Fig. 5B) and the yields of the transgenics were significantly higher than those of the WT (Fig. 5C) (F-test, *P <0.05; **P <0.01)
Fig. 5.
Morphological characterization of TaSnRK2.4 plants. (A) Comparison of primary root lengths. Because of the prolonged SET for transgenic lines, WT seeds were planted 1 d later than the transgenic lines, and root lengths were compared on the seventh day. (B) The silique sizes of TaSnRK2.4 plants were larger than those of the WT under well-watered conditions. Plants of the same size and siliques at the same stem location were selected to measure silique length, and 10 plants were used for each line in triplicate (F-test **P <0.01). (C) TaSnRK2.4 plants had higher yields than the WT. The seeds of transgenic TaSnRK2.4 and WT plants cultured under well-watered conditions were harvested separately, and the yield of each plant was measured after complete dehydration. Thirty plants were used for each line; values are the mean ±SE (F-test **P <0.01). (This figure is available in colour at JXB online.)
TaSnRK2.4-overexpressing plants have a significantly higher osmotic potential
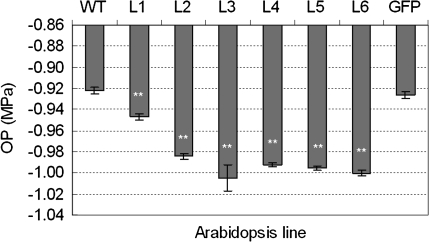
Osmotic stress causes detrimental changes in cellular components, such as reactive oxygen species, various molecular chaperones, and diverse osmoprotectants. The accumulation of osmoprotectants is an effective strategy to enhance plant tolerance to osmotic stresses. To reveal the physiological effects of TaSnRK2.4 overexpression, six transgenic lines under well-watered conditions were selected for an OP assay. The OP of all the transgenic lines was significantly higher than those of WT and GFP plants, which were not significantly different. Thus overexpression of TaSnRK2.4 apparently leads to enhanced OP in transgenic lines (Fig. 6).
Fig. 6.
Transgenic TaSnRK2.4 plants had significantly higher osmotic potential. Six TaSnRK2.4 transgenic lines, as well as WT and GFP plants, cultured under well-watered conditions, were selected to perform osmotic potential assays as described in the Materials and methods. L1–L6, six individual TaSnRK2.4 transgenic lines; WT, wild type; GFP, GFP transgenic line.
Compelling evidence indicates that free proline plays important roles in addressing osmotic stress, including scavenging free radicals, stabilizing subcellular structures, buffering cellular redox, and increasing the OP (Bartels and Sunkar, 2005). To determine the reason for OP augmentation in transgenic plants under normal growing conditions, free proline contents were determined. No differences were identified between the WT and GFP controls and TaSnRK2.4 plants (data not shown).
TaSnRK2.4-overexpressing plants acquire strong water retention ability
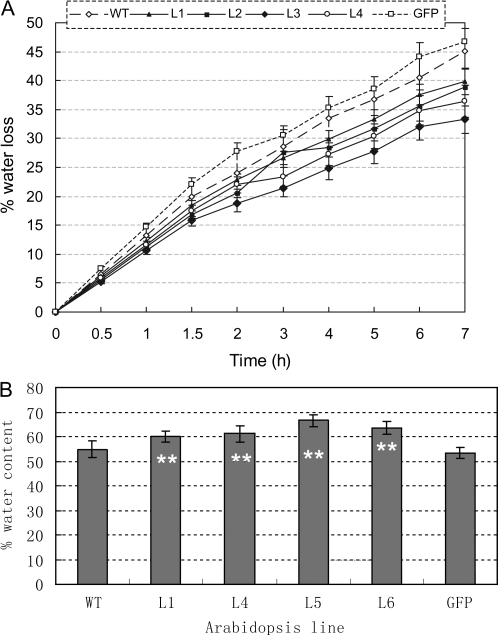
To assess the water retention ability of transgenic Arabidopsis, four transgenic lines were selected for a detached rosette water loss rate assay. Nine time points (7 h period) were selected for measurement of the fresh weight changes in detached rosettes. Compared with WT and GFP plants, the four transgenic lines showed lower water loss rates at each time period (Fig. 7A), and the final relative water contents of TaSnRK2.4 rosettes were significantly higher than those of the two controls (F-test, **P < 0.01) (Fig. 7B).
Fig. 7.
TaSnRK2.4 plants have stronger water retention ability. (A) Comparison of water loss rates for detached rosettes between transgenic plants and WT and GFP controls. Values are the mean ±SE (n=10 plants). (B) Comparison of relative water contents of detached rosettes of transformed plants and controls 7 h after treatment. Values are mean ±SE (n=10 plants).
Overexpression of TaSnRK2.4 increased cell membrane stability of Arabidopsis under adverse stress conditions
To identify the response of TaSnRK2.4 plants to hyperosmotic stress, four homozygous transgenic lines were selected for physiological assays. After germination on MS medium, 7-d-old seedlings were treated with 25.4% (–1.4 MPa) PEG-6000 and NaCl (250 mM) solutions. Signs of PEG stress began to appear on WT and GFP plants 20 h later when samples were collected for CMS measurement. CMS levels in transgenic lines were significantly higher than in the two controls (F-test, *P <0.05) (Supplementary Fig. S3 at JXB online), strongly indicating that PEG stress damage in TaSnRK2.4 plants was much less than in WT plants. In the salinity stress tests, symptoms of salt stress began to appear on WT and GFP plants 4 h after the NaCl treatment was applied; no signs of stress were evident on TaSnRK2.4 plants. CMS levels in TaSnRK2.4 plants were 9–20% higher than in WT and GFP plants; CMS levels in transgenic lines L4 and L5 were significantly increased (F-test, *P <0.05) (Supplementary Fig. S3).
TaSnRK2.4 Arabidopsis has higher photosynthetic potential under moderate drought stress
Growing data indicate that chlorophyll fluorescence is an effective parameter to reveal the early signs of stress, and hence a suitable way to screen for stress tolerance in plants (Chaerle et al., 2007). To evaluate the photosynthetic potential of TaSnRK2.4 plants further, six transgenic lines were used in chlorophyll fluorescence assays. Under well-watered conditions, no differences in the Fv/Fm ratio between TaSnRK2.4 plants and the WT control were evident (data not shown). Under moderate drought stress conditions, the leaf colours of WT and GFP plants were slightly darker than most of the TaSnRK2.4 plants. Chlorophyll fluorescence detection showed that the Fv/Fm ratios of all the TaSnRK2.4 lines were higher than those of the controls, and three of the six lines reached significant levels (F-test, *P <0.05, **P <0.01) (Supplementary Fig. S4 at JXB online).
TaSnRK2.4 plants have pronounced drought tolerance
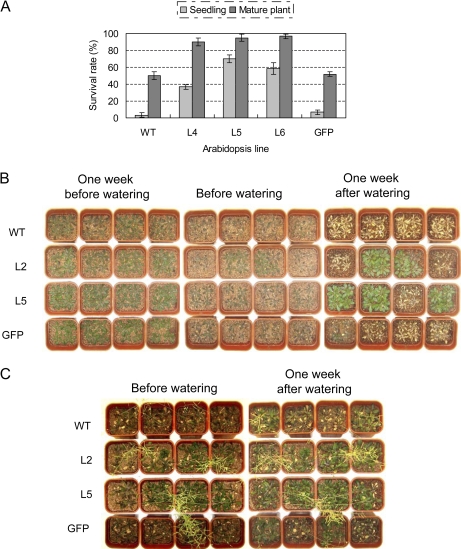
To characterize further the performance of TaSnRK2.4 plants under drought stress in soil, six transgenic lines were selected for drought resistance tests at the seedling and mature growth stages. For the seedling assay, the lower rosette leaves of WT and GFP plants showed slight wilting whereas TaSnRK2.4 plants grew normally after 40 d without watering. On the 49th day (just before watering), WT and GFP plants displayed signs of severe wilting (all rosette leaves were severely curled), whereas only some the TaSnRK2.4 plants showed symptoms of severe drought stress. After watering for 1 week, ∼97% of WT and 94% of GFP plants had died, whereas 37–70% of TaSnRK2.4 plants survived (Fig. 8A, B). For mature plants, after 54 d with no water, WT and GFP plants showed severe wilting (most rosettes were curled), whereas only a few TaSnRK2.4 plants had begun to wilt. To determine if the stressed plants could produce seeds, the drought stress treatments were terminated by re-watering. After watering for 1 week, ∼50% of WT and 51% of GFP plants recovered, whereas 68–97% of TaSnRK2.4 plants survived. Most of the TaSnRK2.4 plants had normal seed development (Fig. 8A, C). The seed-producing abilities of TaSnRK2.4 plants were clearly much higher than those of WT and GFP plants (Fig. 8C).
Fig. 8.
Transgenic TaSnRK2.4 Arabidopsis has enhanced drought tolerance. (A) Survival rates of TaSnRK2.4 transformants and controls following severe and moderate drought stress conditions at two developmental stages. Values are the mean ±SE (n=20 plants). (B) Phenotypes of selected TaSnRK2.4 lines and WT and GFP controls, following severe drought stress at the seedling stage. (C) Phenotypes of selected TaSnRK2.4 lines and WT and GFP controls following moderate drought stress at the mature growth stage. (This figure is available in colour at JXB online.)
TaSnRK2.4 plants have enhanced tolerance to salt stress
Many studies report cross-talk among responses to different stresses. To determine whether TaSnRk2.4 overexpression enhances tolerance to salt stress, Arabidopsis seedlings growing in soil were exposed to 350 mM NaCl solution. About 20 h after initial exposure, leaf tips of all lines began to curl. One week later, signs of salt stress were clear; TaSnRK2.4 plants were much less affected than WT and GFP plants (Fig. 9). Two weeks after salt leaching, only 5.0–6.7% of WT and GFP plants had survived, compared with 15.0–33.3% of transgenic plants. The survival rates of TaSnRk2.4 transgenics were much higher than those of the controls (Fig. 9).
Fig. 9.
Transgenic TaSnRK2.4 Arabidopsis has enhanced salt tolerance. Comparison of survival rates of TaSnRK2.4 lines and WT and GFP controls treated with 350 mM NaCl. Twenty plants of each line were used in each of three experiments. (This figure is available in colour at JXB online.)
TaSnRK2.4 plants exhibit enhanced cold tolerance
To examine TaSnRK2.4 Arabidopsis plants under cold stress, TaSnRK2.4 plants and WT and GFP controls were exposed to severe cold stress. Only 1.7% of WT and 3.3% of GFP plants survived the freezing stress, whereas the survival rate of TaSnRK2.4 plants reached 8.3–31.7%. Three weeks later, the surviving TaSnRK2.4 plants showed normal seed development (Fig. 10).
Fig. 10.
Comparison of freezing tolerance for TaSnRK2.4 and control plants. Normally cultured transgenic seedlings at 4 weeks were stressed at –10 °C for 1.5 h. Twenty plants were used in each of three experiments. Survival rates were determined 2 weeks after freezing. (This figure is available in colour at JXB online.)
Discussion
TaSnRK2.4 possesses the typical features of the SnRK2 subfamily
Hyperosmotic stresses, such as drought, cold, and salinity, severely limit the agricultural yield of wheat. Experimental approaches, including forward and reverse genetics and transcriptome analysis, have been applied to identify key molecular factors that facilitate crop acclimation to environmental stresses. Compelling evidence indicates that protein phosphorylation is one of the central signalling events occurring in response to environmental stress in plants (Ichimura et al., 2000). In this study, an osmotic-stress activated protein kinase gene, TaSnRK2.4, was identified in common wheat, and gene structure analysis indicated similarity to counterparts in Arabidopsis, rice, and maize (Huai et al., 2008), implying SnRK2.4 evolved before separation of monocots and dicots.
The expression pattern can be a direct indication of a gene's involvement in developmental or differential events. In Arabidopsis, AtSRK2.8/AtSRK2C was identified as a root-specific protein kinase, and AtSRK2.6/AtSRK2E/OST1 was confirmed to play a pivotal role in stomatal closure in leaves, suggesting that different SnRK2 members have various roles in different tissues. Higher expression levels of TaSnRK2.4 in both the booting spindle and seedling roots (Fig. 2A) suggested it might act as a fundamental signalling molecule of water and/or nutrient status in soil and play crucial roles in reproductive organ development.
Growing evidence supports a role for the SnRK2 family in response to multienvironmental stress. Kobayashi et al. (2004) observed up-regulation of SAPK4 under ABA or NaCl treatment in rice. Huai et al. (2008) identified the response of ZmSAPK4/ZmSnRK2.4 to ABA, heat, and NaCl in maize seedlings. In this study, expression of TaSnRK2.4 was detected under diverse environmental stresses, including PEG, salt, cold, and ABA treatments, and similar double-peaked expression patterns were identified under various stresses (Fig. 2B). Significant differences in expression levels and response times indicate that TaSnRK2.4 is very sensitive to NaCl and PEG stresses, and less sensitive to ABA treatment (Fig. 2B). These results suggest that TaSnRK2.4 might be involved in the very rapid response to PEG and NaCl stresses.
Evidence from cultured plant cells shows extremely early activation of AtSRK2C and NtOSAK, suggesting that SnRK2s might be activated almost from the beginning of osmotic stress (Kelner et al., 2004; Umezawa et al., 2004). An original aim of the present study was to detect dynamic responses of TaSnRK2.4. Transcriptional peaks were witnessed as early as 1 h after stress was applied, a result that was quite similar to that found for AtSRK2C reported by Umezawa et al. (2004). In future research, additional time points and shorter time periods need be set for more precise identification of response activation of TaSnRK2.4s following osmotic stress.
TaSnRK2.4 overexpression has no adverse effects in Arabidopsis
To investigate the in vivo role of TaSnRK2.4 in plant abiotic resistance, the fused TaSnRK2.4–GFP was overexpressed in Arabidopsis. Before functional analysis of TaSnRK2.4 in Arabidopsis, overexpression of TaSnRK2.4 was re-identified by western blotting (data not shown). Growth retardation is a common phenomenon, often occurring in transgenic plants and severely restricting the utilization of target genes in plant breeding. To assess the feasibility of using TaSnRK2.4 in transgenic breeding for abiotic stress tolerance, the morphological features of transgenic TaSnRK2.4 plants were closely monitored. Seed germination of transgenic plants was delayed by 24 h relative to WT controls (data not shown). Seed dormancy is a complex trait, influenced by a myriad of genetic and environmental factors, many of which are mediated by hormones, with gibberellin, ethylene, and brassinosteroids known to promote germination and ABA known to promote dormancy (Koornneef et al., 2002; Millar et al., 2006). Recent studies show that AtSnRK2D/AtSnRK2.2, AtSnRK2E/AtSnRK2.6, and AtSnRK2I/AtSnRK2.3 protein kinases involved in ABA signalling are essential for the control of seed development and dormancy through extensive control of gene expression (Fujii et al., 2007; Nakashima et al., 2009). The extended SET of TaSnRK2.4 seed may possibly be due to dormancy. The existing reports and results of this study support the classification of AtSnRK2D/AtSnRK2.2, AtSnRK2E/AtSnRK2.6, and AtSnRK2I/AtSnRK2.3 protein kinases in subclass III and TaSnRK2.4 in subclass I of the SnRK2 family. All are involved in seed dormancy, suggesting that participation in dormancy is a basic function of this kinase family (Boudsocq et al., 2004). Pre-harvest sprouting, which commonly occurs in conditions of prolonged rainfall and high humidity before harvesting, severely affects the yield and quality of wheat. The extended SET of TaSnRK2.4 seed can be regarded as a form of sprouting delay that might be usable to improve pre-harvest sprouting resistance in wheat. Although the current data provide no obvious explanation for the delayed germination, more focus should be given to determining the cause of prolonged SET. This will not only provide an understanding of the delayed germination, but overexpression of TaSnRK2.4 may have a role in pre-harvest sprouting control in wheat.
The visibly smaller seedling size of TaSnRK2.4 plants is probably due to the prolonged SET, a difference that vanished later in development. Root length determination results indicate that TaSnRK2.4 plants have longer primary roots than the WT and GFP control plants (Fig. 5A, Supplementary Fig. S2 at JXB online). A longer root would most probably facilitate water absorption from deeper soils especially when water shortages occur, thus strengthening drought tolerance and increasing biomass and yield. In this study, the silique sizes and yield per plant of TaSnRK2.4 transformants were much larger than those of the WT contols (Fig. 5B, C), further suggesting that use of TaSnRK2.4 might be feasible in enhancing yield. Stomatal dynamics are key in reducing water loss and allowing entry of carbon dioxide for photosynthesis. Compelling evidence in Arabidopsis indicates that AtSnRK2.6/AtSnRK2E/OST1 can integrate ABA signals and osmotic stress, and is involved in ABA-dependent stomatal regulation (Mustilli et al., 2002; Yoshida et al., 2006).The width, length, area, and density of stomata, as well as the area ratio of stomata to leaf were measured, and no obvious differences were identified (data not shown), showing that TaSnRK2.4 does not participate in the regulation of stomatal development and aperture.
Physiological changes in transgenic TaSnRK2.4 plants under various conditions
Environmental stresses often cause physiological changes in plants. Physiological indices, including CMS, OP, RWC, and chlorophyll fluorescence, are typical physiological parameters for evaluating abiotic stress tolerance and resistance in crop plants. In general, plants with higher CMS, RWC, OP, and photosynthetic capacities have enhanced tolerance or resistance to environmental stresses.
RWC and detached-leaf water loss rate are essential parameters of water status in plants and have been proposed as important indicators of water status (Clarke et al., 1989; Dhanda and Sethi, 1998). RWC is closely related to cell volume and may more closely reflect the balance between water supply to leaves and transpiration rate (Farquhar et al., 1989). In the present work, the detached-leaf water loss rate of TaSnRK2.4 Arabidopsis was lower than that of the WT and GFP controls, and the final RWCs for TaSnRK2.4 seedlings were significantly higher than those of the controls (Fig. 7), strongly indicating that the transgenic lines had higher water retention ability.
Plant survival depends on maintaining positive turgor pressure, which is important for cell expansion and stomatal opening. A decrease in water availability induced by osmotic stress might lead to turgor reduction. Osmotic adjustment (OA) is a fundamental cell tolerance response to osmotic stress, and can be realized by the accumulation of diverse osmoprotectants. Generally, a higher capacity for OA means broader adaptation and more tolerance to osmotic stress. OP is a direct reflection of OA capability at the physiological level, and has been used as an effective index to screen crop germplasm for osmotic stress tolerance. Our research indicates the OP of TaSnRK2.4 lines is significantly higher than that of WT and GFP controls under well-watered conditions (Fig. 6). These results strongly indicated that the increased OP in transgenic plants is due to the overexpression of TaSnRK2.4, rather than GFP. Increased OP is primarily attributed to accumulation of osmoprotectants, including amino acids, quaternary amines, and various sugars. It is well documented that proline is the most widely distributed multifunctional osmolyte, occurring not only in plants, but also in many other organisms, and playing important roles in enhancing osmotic stress tolerance (Bartels and Sunkar, 2005). An increase in free proline was not detected in TaSnRK2.4 plants, suggesting that proline was not the main reason for OP augmentation, and that TaSnRK2.4 was not likely to be involved in the pathway of proline metabolism. Higher OP commonly predicts higher water retention capacity and a lower rate of water loss, as well as higher water use efficiency. The results of OP analysis were consistent with the above-mentioned detached-leaf water loss rate and RWC results (Fig. 7), and partially explain the enhanced tolerance to drought, salt, and cold stresses.
Cell membranes are one of the first targets of many plant stresses. It is generally accepted that the maintenance of membrane integrity and stability under water stress conditions is a major component of environmental stress tolerance in plants (Levitt, 1980). CMS has been used for assessing tolerance to frost, heat, and desiccation (Farooq and Azam, 2006). In most of these studies, CMS exhibits a positive correlation with several physiological and biochemical parameters conditioning plant responses to environmental conditions such as water use efficiency (Franca et al., 2000), stomatal resistance, OP, leaf rolling index, K+ concentration, OA, and/or RWC (Munns, 2002). The degree of CMS under environmental stresses can be easily estimated through measurements of electrolyte leakage from cells. In this study, the CMS of TaSnRK2.4 plants under both osmotic and salinity stresses was higher than that of the WT and GFP controls, clearly demonstrating that CMS enhancement is caused by overexpression of TaSnRK2.4. As mentioned above, CMS has a positive relationship with several physiological and biochemical parameters; it predicts that TaSnRK2.4 plants might have a strong capacity to tolerate environmental stresses, as verified by the functional assay results in Arabidopsis (Figs 8–10).
Chlorophyll fluorescence from intact leaves, especially fluorescence induction patterns, is a reliable, non-invasive method for monitoring photosynthetic events and reflects the physiological status of the plant (Strasser et al., 2000). The ratio of variable to maximal fluorescence is an important parameter used to assess the physiological status of the photosynthetic apparatus. It represents the maximum quantum yield of the primary photochemical reaction of PSII. Environmental stresses that affect PSII efficiency are known to provoke decreases in the Fv/Fm ratio (Krause and Weis, 1991). In this research, the slightly darker leaf colour and lower Fv/Fm ratio were evident in WT and GFP plants (Supplementary Fig. S4 at JXB online), undoubtedly suggesting that TaSnRK2.4 plants had more robust photosynthetic capabilities than the controls under moderate drought stress conditions.
Overexpression of TaSnRK2.4 enhanced multienvironmental stress responses in Arabidopsis
It is well established that the unique SnRK2 family plays critical roles in responses to hyperosmotic stress and ABA treatment. Ten SnRK2s were identified in Arabidopsis, rice, and maize (Boudsocq et al., 2004, 2007; Kobayashi et al., 2004). Several research groups have demonstrated that OST1/SnRK2E/SRK2.6 and V. faba AAPK are involved in ABA-dependent stomatal regulation (Li et al., 2000; Mustilli et al., 2002; Yoshida et al., 2002). Shin et al. (2007) and Umezawa et al. (2004) showed that overexpression of AtSnRK2C/AtSnRK2.8 increases the expression of stress-related genes and thus enhances drought tolerance in Arabidopsis. Diedhiou et al. (2008) demonstrated that overexpression of SAPK4 significantly enhanced the salt tolerance of transgenic rice through regulating genes with functions in ion homeostasis and the oxidative stress response. In terms of actual mechanisms, it remains unknown as to how SAPK4 prevents Na+ and Cl– from entering the cell and how the cell maintains ion homeostasis. In the present research, transgenic TaSnRK2.4 plants were exposed to severe drought, salt, and freeze stresses. The morphological and physiological evidence strongly demonstrated that the transgenic lines acquired strengthened tolerance to severe drought, high salinity, and freezing stresses relative to WT plants. Our understanding is that enhanced multistress tolerance is possibly due to increased osmotic potential. Under water-deficient conditions, enhancement of OP leads to reduced water loss and increased RWC in plant cells, possibly facilitating the enhanced water retention ability, benefiting the maintenance of regular cell turgor, and avoiding damage to cell membranes, thus enhancing drought tolerance. Under salt stress, the higher OP probably prevents entry of harmful ions, including Na+ and Cl–, thus relieving ion damage to cell membranes, and therefore increasing salt tolerance. Under cold stress, higher OP commonly means more solutes in the plant sap, resulting in lower freezing points and hence reduced freezing damage. In the current work, gene expression levels and protein abundance for the two transgenic lines (L4 and L5) were relatively lower than for the other four transgenic lines (Fig. 4, Supplementary Fig. S1 at JXB online), whereas their capability of osmotic tolerance was much stronger than that of the latter (Fig. 8–10). This seems to suggest that the overexpression level is not directly proportional to the ability for abiotic stress resistance, which hints that an appropriate overexpression level should be considered when generating transgenic plants with a regulatory gene controlled by a constitutive overexpressing promoter for improvement of abiotic stress tolerance.
This study primarily concerned the morphological and physiological features of TaSnRK2.4 overexpression in Arabidopsis under normal and adverse conditions. Further comprehensive investigations to dissect the actual molecular mechanisms for enhancing OP are ongoing and it is believed that the outcomes may enable tolerance to abiotic stresses levels in crop plats to be strengthened.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Protein abundance of TaSnRK2.4 in different transgenic Arabidopsis lines.
Fig. S2. The primary root of TaSnRK2.4 plant is much longer than that of the two controls.
Fig. S3. Cell membrane stability of TaSnRK2.4 plants under adverse stress conditions.
Fig. S4. Comparison of photosynthetic potential for TaSnRK2.4 plants and controls under moderate drought stress.
Supplementary Material
Acknowledgments
The authors thank Professor Zhensheng Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing) for providing the pJIT163-GFP expression vector. We thank Robert A McIntosh (Plant Breeding Institute, University of Sydney, NSW, Australia), Dr Joe Colasanti (Department of Molecular and Cellular Biology, University of Guelph, Canada), and Miss Cynthia Eden (English Language Preparation Program, University of Guelph, Canada) for critical reading of, and comments on the manuscript. This work was supported by the National Key Technologies R&D Program (2009ZX08002-012B).
Glossary
Abbreviations
- AAR
amino acid residue
- ABA
abscisic acid
- AMPK
AMP-activated protein kinase
- CDPK
calcium-dependent protein kinase
- CMS
cell membrane stability
- GFP
green fluorescent protein
- MAPK
mitogen-activated protein kinase
- OA
osmotic adjustment
- OP
osmotic potential
- PEG
polyethylene glycol
- qRT-PCR
quantitative real-time PCR
- RWC
relative water content
- SET
seedling establishment time
- SNF
sucrose non-fermenting
- SnRK
SNF1-related protein kinase
- WT
wild type
References
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Critical Reviews in Plant Science. 2005;24:23–58. [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. Journal of Biolical Chemistry. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Barbier-Brygoo H, Lauriere C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Molecular Biology. 2007;63:491–503. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bray E. Plant responses to water deficit. Trends in Plant Science. 1997;2:48–54. [Google Scholar]
- Chaerle L, Leinonen I, Jones HG, Van Der Straeten D. Monitoring and screening plant populations with combined thermal and chlorophyll fluorescence imaging. Journal of Experimental Botany. 2007;58:773–784. doi: 10.1093/jxb/erl257. [DOI] [PubMed] [Google Scholar]
- Clarke JM, Romagosa JI, Srivastava JP, McCaig TN. Relationship of excised-leaf water loss rate and yield of durum wheat in diverse environments. Canadian Journal of Plant Science. 1989;69:1075–1081. [Google Scholar]
- Cohen P. Protein phosphorylation and hormone action. Proceedings of the Royal Society B: Biological Sciences. 1988;234:115–144. doi: 10.1098/rspb.1988.0040. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF. Glucose depletion causes haploid invasive growth in yeast. Proceedings of the National Academy of Sciences, USA. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SS, Sethi GS. Inheritance of excised-leaf water loss and relative water content in bread wheat (Triticum aestivum) Euphytica. 1998;104:39–47. [Google Scholar]
- Diedhiou CJ, Popova OV, Dietz KJ, Golldack D. The SNF1-type serine–threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biology. 2008;8:49. doi: 10.1186/1471-2229-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq S, Azam F. The use of cell membrane stability (CMS) technique to screen for salt tolerant wheat varieties. Journal of Plant Physiol. 2006;163:629–637. doi: 10.1016/j.jplph.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Farquhar GD, Wong SC, Evans JR, Hubic KT. Photosynthesis and gas exchange. In: Jones HG, Flowers TJ, Jones MB, editors. Plants under stress. Cambridge: Cambridge University Press; 1989. pp. 47–69. [Google Scholar]
- Franca MGC, Thi ATP, Pimentel C, Rossiello ROP, Zuily FY, Laffray D. Differences in growth and water relations among Phaseolus vulgaris cultivars in response to induced drought stress. Environmental and Experimental Botany. 2000;43:227–237. doi: 10.1016/s0098-8472(99)00060-x. [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. The Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Verhey SD, Holappa LD, Shen Q, Ho TH, Walker-Simmons MK. An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proceedings of the National Academy of Sciences, USA. 1999;96:1767–1772. doi: 10.1073/pnas.96.4.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Zentella R, Walker-Simmons MK, Ho TH. Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. The Plant Cell. 2001;13:667–679. [PMC free article] [PubMed] [Google Scholar]
- Gong DM, Guo Y, Jagendorf AT, Zhu JK. Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiology. 2002;130:256–264. doi: 10.1104/pp.004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Molecular Biology. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Halford NG, Hardie DG. SNF1-related protein kinases: global regulators of carbon metabolism? Plant Molecular Biology. 1998;37:735–748. doi: 10.1023/a:1006024231305. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annual Review of Biochemistry. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- Hong SW, Jon JH, Kwak JM, Nam HG. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopis thaliana. Plant Physiology. 1997;113:1203–1212. doi: 10.1104/pp.113.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg SM, Lee RH. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1998;18:4548–4555. doi: 10.1128/mcb.18.8.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, et al. The Arabidopsis CDPK–SnRK superfamily of protein kinases. Plant Physiology. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CA, Delauney AJ, Verma DP. A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proceedings of the National Academy of Sciences, USA. 1992;89:9354–9358. doi: 10.1073/pnas.89.19.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai J, Wang M, He J, Zheng J, Dong Z, Lv H, Zhao J, Wang G. Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Reports. 2008;27:1861–1818. doi: 10.1007/s00299-008-0608-8. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. The Plant Journal. 2000;24:655–665. doi: 10.1046/j.1365-313x.2000.00913.x. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annual Reviews of Plant Physiology and Plant Molecular Biology. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Johnson R, Wagner R, Verhey S, Walker-Simmons MK. The ABA-responsive kinase PKABA1 interacts with a seed-specific ABA response element binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiology. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelner A, Pekala I, Kaczanowski S, Muszynska G, Hardie DG, Dobrowolska G. Biochemical characterization of the tobacco 42-kD protein kinase activated by osmotic stress. Plant Physiology. 2004;136:3255–3265. doi: 10.1104/pp.104.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. The Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Current Opinion in Plant Biology. 2002;5:33–36. doi: 10.1016/s1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- Krause GH, Weis E. Chlorophyll fluorescence and photosynthesis: the basics. Annual Reviews of Plant Physiology and Plant Molecular Biology. 1991;42:313–349. [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI. Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. The Plant Cell. 2002;14:2849–2861. doi: 10.1105/tpc.003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J. Responses of plants to environmental stresses. Water, radiation, salt and other stresses. vol. II. New York: Academic Press; 1980. [Google Scholar]
- Li J, Wang XQ, Watson MB, Assmann SM. Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science. 2000;287:300–303. doi: 10.1126/science.287.5451.300. [DOI] [PubMed] [Google Scholar]
- Livaka KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ludwig AA, Romeis T, Jones JD. CDPK-mediated signalling pathways: specificity and cross-talk. Journal of Experimental Botany. 2004;55:181–188. doi: 10.1093/jxb/erh008. [DOI] [PubMed] [Google Scholar]
- Mao XG, Kong XY, Zhao GY, Jia JZ. Construction of a full-length cDNA library of Aegilops speltoides Tausch with optimized cap-trapper method. Acta Genetica Sinica. 2005;32:811–817. [PubMed] [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid J, Gubler F. Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. The Plant Journal. 2006;45:42–54. doi: 10.1111/j.1365-313X.2006.02659.x. [DOI] [PubMed] [Google Scholar]
- Monks DE, Aghoram K, Courtney PD, DeWald DB, Dewey RE. Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. ThePlant Cell. 2001;13:1205–1219. doi: 10.1105/tpc.13.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. Comparative physiology of salt and water stress. Plant, Cell and Environment. 2002;25:239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. The Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3 involved in ABA-signaling are essential for the control of seed development and dormancy. Plant and Cell Physiology. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proceedings of the National Academy of Sciences, USA. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Gomez-Cadenas A, Zhang P, Walker-Simmons MK, Sheen J, Ho TH. Dissection of abscisic acid signal transduction pathways in barley aleurone layers. Plant Molecular Biology. 2001;47:437–448. doi: 10.1023/a:1011667312754. [DOI] [PubMed] [Google Scholar]
- Shin R, Alvarez S, Burch AY, Jez JM, Schachtman DP. Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proceedings of the National Academy of Sciences, USA. 2007;104:6460–6465. doi: 10.1073/pnas.0610208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi S. Gene expression and signal transduction in water-stress response. Plant Physiology. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderman E, Mattsson J, Engstrom P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and abscisic acid. The Plant Journal. 1996;10:375–381. doi: 10.1046/j.1365-313x.1996.10020375.x. [DOI] [PubMed] [Google Scholar]
- Strasser A, Srivastava A, Tsimilli-Michael M. The fluorescence transient as a tool to characterize and screen photosynthetic samples. In: Mohanty P, Yunus U, Pathre M, editors. Probing photosynthesis: mechanism, regulation and adaptation. London: Taylor and Francis; 2000. pp. 443–480. [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2004;101:17306–17311. doi: 10.1073/pnas.0407758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Hirt H. Plant MAP kinase pathways: how many and what for? Biology of the Cell. 2001;93:81–87. doi: 10.1016/s0248-4900(01)01121-2. [DOI] [PubMed] [Google Scholar]
- Yoon HW, Kim MC, Shin PG. Differential expression of two functional serine/threonine protein kinases from soybean that have an unusual acidic domain at the carboxy terminus. Molecular and General Genetics. 1997;255:359–371. doi: 10.1007/s004380050507. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant and Cell Physiology. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. Journal of Biological Chemistry. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.