SUMMARY
Densin is a member of the LRR (leucine-rich repeat) and PDZ domain (LAP) protein family that binds several signaling molecules via its C-terminal domains, including calcium/calmodulin-dependent protein kinase II (CaMKII). Here we identify several novel mRNA splice variants of densin that are differentially expressed during development. The novel variants share the LRR domain but are either prematurely truncated or contain internal deletions relative to mature variants of the protein (180 kDa), thus removing key protein-protein interaction domains. For example, CaMKIIα coimmunoprecipitates with densin splice variants containing an intact C-terminal domain from lysates of transfected HEK293 cells, but not with variants that only contain N-terminal domains. Immunoblot analyses using antibodies to peptide epitopes in the N- and C- terminal domains of densin are consistent with developmental regulation of splice variant expression in brain. Moreover, putative splice variants display different subcellular fractionation patterns in brain extracts. Expression of GFP-fused densin splice variants in HEK293 cells shows that the LRR domain can target densin to a plasma membrane-associated compartment, but that the splice variants are differentially localized and have potentially distinct effects on cell morphology. In combination, these data show that densin splice variants have distinct functional characteristics suggesting multiple roles during neuronal development.
INTRODUCTION
Alternative splicing of primary mRNA transcripts occurs widely in humans and other mammals, particularly in the nervous system, to increase the functional diversity of proteins without concomitant increases in the amount of genetic information (Modrek & Lee 2002, Lee & Irizarry 2003, Sorek et al. 2004). For example, certain adult CaMKIIβ gene splice variants, but not embryonic CaMKIIβ splice variants or CaMKIIα gene products, interact directly with F-actin filaments (Shen & Meyer 1998, O'Leary et al. 2006). In contrast, CaMKIIα, but not CaMKIIβ, interacts with densin (Strack et al. 2000, Walikonis et al. 2001), one of a growing number of neuronal CaMKII associated proteins (CaMKAPs) that exhibit distinct mechanisms for interaction with CaMKII (Robison et al. 2005).
Densin was identified as a 1495 amino acid protein enriched in postsynaptic densities (PSDs) (Apperson et al. 1996)1, becoming a founding member of the LAP protein family. LAP proteins share a similar organization of LRR (leucine-rich repeat) and one or more PDZ domains (Bilder et al. 2000, Borg et al. 2000). Other LAP protein family members modulate signal transduction pathways. For example, erbin was recently shown to regulate MAP kinase (ERK) pathways (Dai et al. 2006, Huang et al. 2003, Rangwala et al. 2005) and CaV1.3 L-type voltage-gated Ca2+ channels (Calin-Jageman et al. 2007). Densin may be an important scaffolding protein because C-terminal domains have been shown to interact with a variety of signaling molecules including α-actinin (Walikonis et al. 2001), SHANK(Quitsch et al. 2005), δ-catenin (Izawa et al. 2002) and MAGUIN1 (Ohtakara et al. 2002). In addition, the C-terminal domain of densin binds to the association domain of the CaMKIIα holoenzyme (Strack et al. 2000; Walikonis et al. 2001; Robison et al. 2005). Recent studies have shown that overexpression of densin induces excessive neurite branching and outgrowth in cultured neurons: competitive binding of SHANK and δ-catenin to the C-terminal domain appears to modulate this function (Quitsch et al. 2005). Although densin was initially characterized as a brain-specific protein (Apperson et al. 1996), recent studies have shown that densin is also expressed in the kidney and other peripheral tissues, where it is often localized to sites of cell-cell adhesion, suggesting key roles in modulating cell adhesion and cell-cell contacts (Heikkila et al. 2007, Ahola et al. 2003, Lassila et al. 2007, Rinta-Valkama et al. 2007).
Two alternative splice variants of the N-terminal region of densin were initially identified in rat brain (Apperson et al. 1996), and four splice variants in the C-terminal domains were shown to be differentially expressed during rat brain development (Strack et al. 2000). Here, we report the identification and initial characterization of several novel alternative splice variants of densin that are variably expressed during brain development and exhibit distinct functional properties, including interactions with CaMKII, subcellular localization, and effects on cell morphology. These novel splice variants dramatically increase the potential diversity of functional roles for densin during neuronal development.
EXPERIMENTAL PROCEDURES
RNA isolation and RT-PCR
Trizol reagent (Life Technologies, Inc) was used to isolate total RNA from rat brains of different ages (in days. E: embryonic, P: postnatal, E15, P1, P7, P14). For initial analysis of 5’ splice variants expressed during early development (Fig. 1B), 3 µg of the pooled RNA samples were used to make a full-length cDNA pool (5’-RACE-ready cDNA) with BD SMART™ RACE cDNA Amplification kit as instructed by the manufacturer (BD Biosciences Clontech). Briefly, reverse transcription was performed for 1.5 hr at 42°C in the presence of dNTP using the SMART II™ A oligo, 5’-CDS (modified oligo-dT) and BD PowerScript reverse transcriptase in 1× reaction buffer provided by the manufacturer. The reaction was heat inactivated at 70°C and then diluted to a final volume of 60 µl. The 5’-RACE-ready cDNA (2.5 µl) was amplified using 1F and 22R primers (Table 1) and BD Advantage 2 Polymerase Mix (BD Biosciences Clontech). For analytical scale studies (Fig. 2B), total RNA (1.5 µg) was mixed with Tfl DNA polymerase and AMV reverse transcriptase and primers (25 pmol each) designed to amplify different regions of the densin mRNA (Table 1) in the presence of dNTP in 1× reaction buffer using the Access one-step RT-PCR kit (Promega) as described previously (Strack et al. 2000). For large-scale analyses to identify new densin splice variants (Fig. 3B), 3 µg of pooled RNA (see above) was incubated at 42°C for 1.5 hr with oligo-dT and MMLV reverse transcriptase (Promega) in the presence of dNTP in 1× reaction buffer (final volume of 25 µl). The resulting cDNA (3 µl) was amplified using the 13F and 46R primers (Table 1) using the GeneAmp High Fidelity PCR system (Applied Biosystems). PCR products were fractionated on an agarose gel and visualized by ethidium bromide staining. After purification using the QIAquick PCR Purification Kit (Qiagen), the PCR product was ligated into pGEMT-Easy vector (Promega) and transformed into E. coli DH5α. Clones isolated from individual bacterial colonies were verified by PCR amplification with multiple primer pairs (Table 1) using Taq polymerase (New England Biolabs) and DNA sequencing after plasmid DNA extraction (Qiagen). Sequences were deposited in the GenBank database with Accession Numbers EU348625–EU348644.
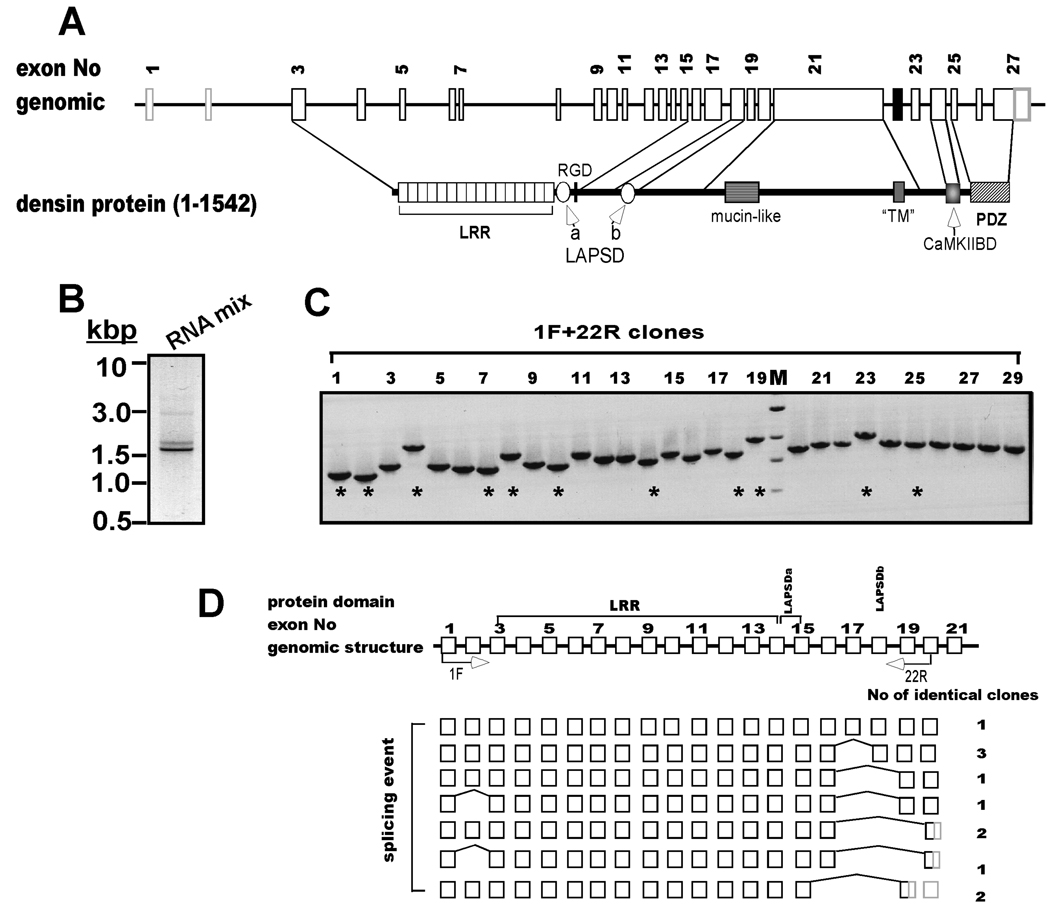
Figure 1. Alternative splicing is detected at the 5’-end of densin.
A. Schematic domain diagram of densin. Structure of densin gene on rat chromosome 2 (GI 34861224) and conserved domains in the full-length form of the protein (1542 amino acids). LRR, leucine-rich repeat; LAPSDa/b, LAP protein specific domains a and b, respectively; RGD: putative integrin interacting motif; “TM”, putative transmembrane domain (see text for discussion); CaMKIIBD, CaMKII binding domain; PDZ, PSD95, dlg1 and ZO1 domain. Sizes of introns, exons and protein domains are depicted to approximate scale. B. Amplification of the 5’ end of the densin mRNA. Full-length cDNA pool was made from a mixture of total mRNAs isolated at different developmental stages by RACE cDNA amplification using primers 1F and 22R (see Experimental Procedures). An aliquot of the product mixture was analyzed on an ethidium bromide-stained gel: an inverted gray-scale image is shown. C. Characterization of individual densin splice variant structures. PCR products from B were inserted into pGEMT Easy vector and 29 isolated bacterial clones containing densin inserts were screened by PCR using 1F and 22R primer set. Products were analyzed on agarose gels stained with ethidium bromide. M: DNA marker: 3.0, 2.0, 1.5 and 1 kbp. D. Summary of densin 5’-splice variant structures. Top: Schematic of the first 21 exons and introns of the densin gene (white boxes and black lines, respectively), not drawn to scale. Labeled arrows below the domain diagram indicate approximate positions of sense and antisense primers used for PCR amplification. DNA sequences of clones (indicated by * in panel C) were aligned to genomic sequence, revealing skipping of 1 or more entire exons (bridging lines). Sequences converted to 3’ untranslated regions (3’ UTRs) by premature stop codons are shaded gray. The number of clones recovered with identical sequences/structures is listed after each variant.
Table 1. Oligonucleotide primers used to identify densin spice variants.
| name | sequence | orientation | position | Exon |
|---|---|---|---|---|
| 1F | TGCTTCTGCGATGATGGAGA | sense | 1-20 | 1 |
| 13F | TTGTGGCTTTCTGACAATCAGTCC | sense | 1386-1409 | 14+15 |
| 22R | AGACAGTGAGTTTTGTCAACTTCAGACTC | antisense | 2233-2205 | 20 |
| 23R | TTTAGTTGTTGGCTGCAAGGACC | antisense | 2342-2320 | 20 |
| 36F | AGAGCAGCCTTTCAATGACTGATC | sense | 3664-3687 | 21 |
| 39F | GACAAGACATCAGATAACAGTG | sense | 3924-3945 | 21 |
| 43R | CTGCTCGGGATAACCGTCCATA | antisense | 4397-4376 | 24 |
| 46R | TCCCCGCGGGACAGTGAGCTCACGTTGAATAA | antisense | 4670-4648 | 27 |
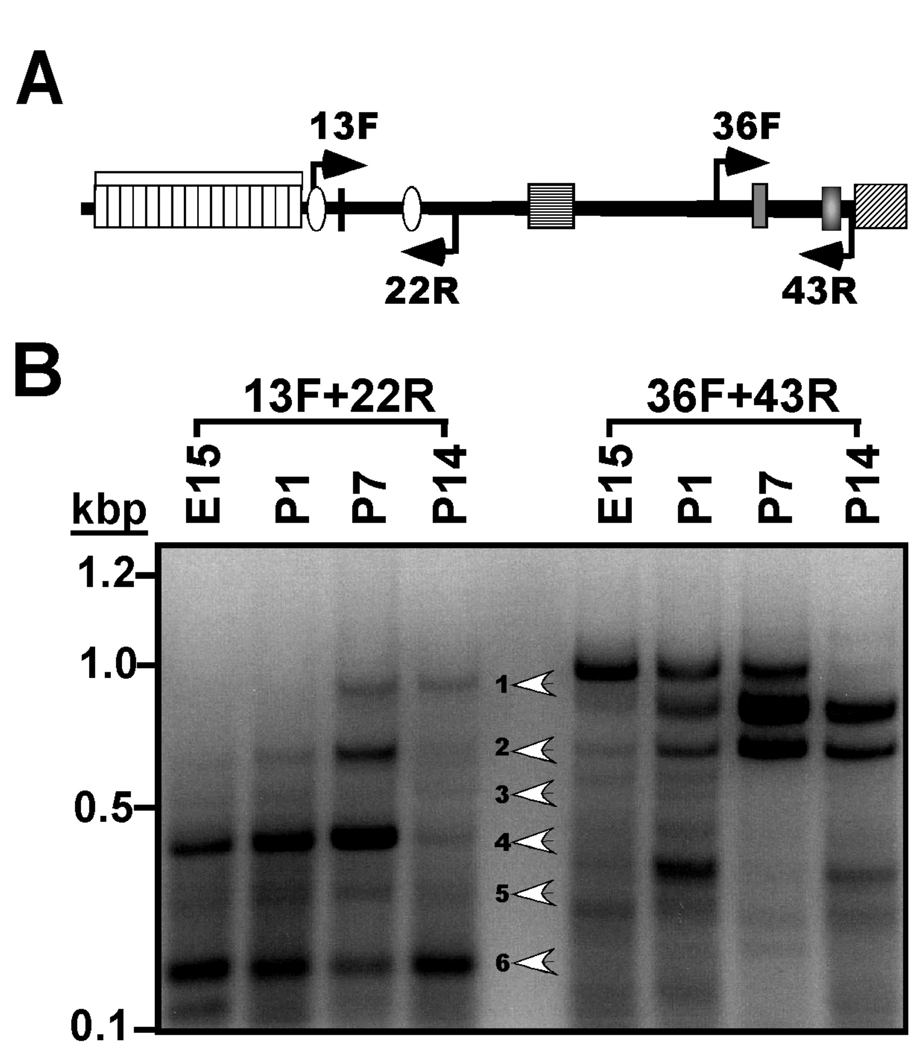
Figure 2. Developmental regulation of the expression of densin 5’ and 3’mRNA splice variants.
A. Schematic domain diagram of densin protein. Labeled arrows indicate approximate positions of primers used in this figure. See Fig. 1A for details of domain structure. B. Analysis of mRNA splicing in the 5’ and 3’ coding regions of the densin mRNA during development. Total RNA isolated from rat brains of the indicated ages (in days; E, embryonic; P, postnatal) was analyzed by RT-PCR using the indicated primers. An ethidium bromide-stained agarose gel is shown with white arrowheads indicating the 6 major products generated using the 13F+22R primer pair.
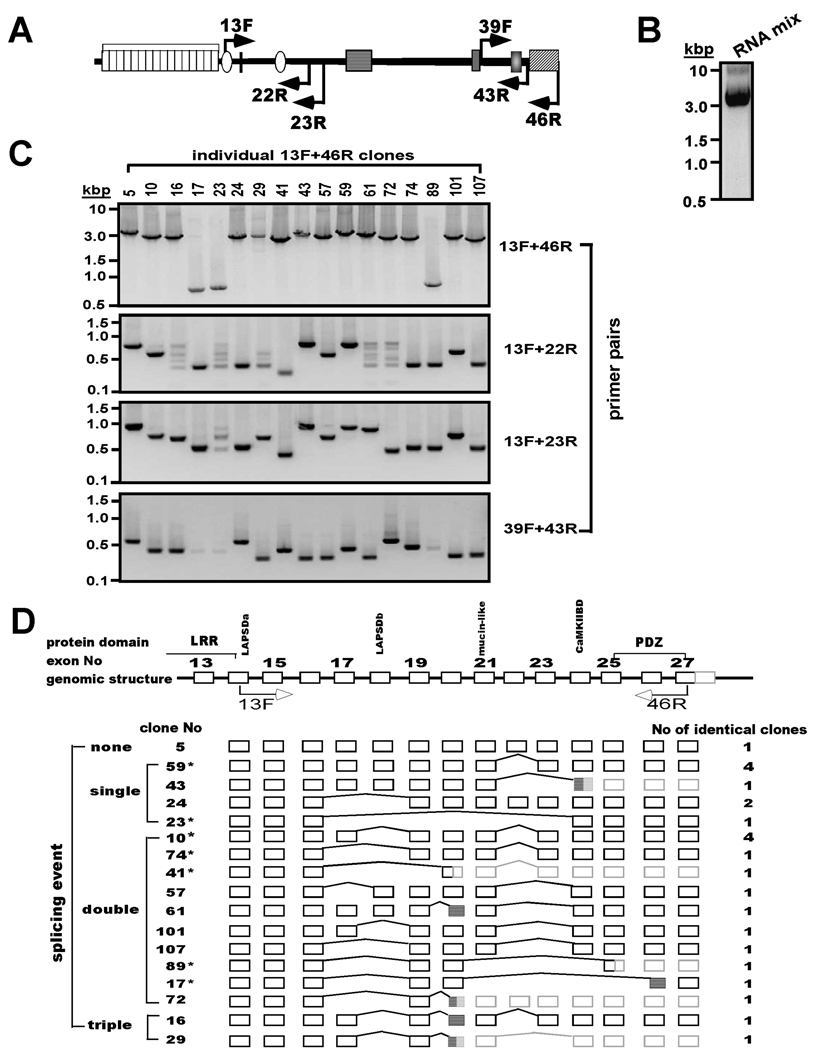
Figure 3. Characterization of novel alternative splice variants of densin.
A. Schematic domain diagram of densin protein. Labeled solid arrows indicate approximate positions of primers used for PCR amplification. See Fig. 1A for details of domain structure. B. Amplification of the 3’ coding region of the densin mRNA. Mixed total RNA samples were amplified by RT-PCR using 13F and 46R primers and an aliquot of the product was analyzed on an ethidium bromide-stained gel. C. Characterization of individual densin splice variant structures. PCR products from B were inserted into pGEMT Easy vector. Isolated bacterial clones containing individual variants (17 representatives shown here) were screened by multiple PCRs using primer sets designed to amplify different segments of the initial 13F/46R products (see panel A). Individual PCR products were analyzed on agarose gels stained with ethidium bromide. D. Summary of densin splice variant structures. Top: Schematic of exon-intron boundaries in part of rat densin gene (GI 34861224) (not drawn to scale). Alignment of sequences of individual densin variants with the genomic structure revealed that length variations could be ascribed to skipping of entire exons (bridging lines) and/or the use of cryptic splice acceptor sites (hatched boxes). Some splicing events introduced premature stop codons and downstream untranslated regions (3’UTRs) in these mRNAs are shaded gray. Clones selected for further studies are indicated by *, and the number of identical clones of each splice variants is listed.
Purification of GST-fusion proteins
GST fusion proteins were generated, expressed in E.coli (BL21-DE3 Gold) and purified using glutathione-agarose (Sigma), essentially as described previously (Strack et al. 2000). GST-DenN contains residues 309–482 and 633–683 of densin, and GST-BΔN contains residues 1247–1290 and 1378–1401 of the densin C-terminal domain. Purified proteins were quantified using Bradford (BioRad) or bicinchonic acid (BCA, Pierce) assays and/or SDS-PAGE analysis.
Generation of densin antibodies
Polyclonal antibodies to two peptide motifs in the N-terminal domain of densin were generated and affinity purified as a custom service by Bethyl Laboratories. Briefly, a single goat was immunized with KLH-conjugated peptides corresponding to residues 451–470 and 652–671 of densin. Antibodies to each peptide were affinity purified from the resulting antiserum (Ab450 and Ab650, respectively). A polyclonal antibody to a C-terminal domain in densin was generated in the Vanderbilt Molecular Recognition Core by immunizing rabbits with the GST-BΔN fusion protein (see above).
Mammalian expression vectors
Entire coding regions of densin splice variants or DNAs encoding desired fragments of densin were amplified by PCR using primers containing BglII (5’) and SacII (3’) restriction enzyme sites and ligated 5' to, and in frame with, GFP in the pEGFP vector (Clontech, BD BioSciences). The densin(Δ1291–1337:P320L), densin-T482(P320L) and densin-T482(R437K/D439E) point mutations were created by QuickChange Site-Directed Mutagenesis (Stratagene). The entire coding region of murine CaMKIIα (GI-31341538) was amplified by PCR and ligated into pcDNA3.1 vector after EcoRI digestion (Invitrogen). pSGL, a synthetic densin fusion protein with myc and GFP tags, was described previously (Strack et al. 2000) and myc-Erbin was a gift from Mei Lin (Medical College of Georgia) (Huang et al. 2001). pEYFP-Mem and pEGFP (Clontech, BD BioSciences) were used as membrane and cytosolic markers, respectively. Plasmid DNA was extracted and purified using Qiagen Maxi-Prep kits and verified by sequencing.
Cell culture and transfection
HEK293 cells were maintained in modified Eagle medium (MEM) containing 10% fetal bovine serum (Gibco), penicillin-streptomycin (Sigma) and 2mM glutamate (Sigma) at 37°C in 5% CO2. Cells were transfected with densin and CaMKII constructs using Fugene® 6 Transfection Reagent (3 µl per µg DNA) (Roche) as instructed by the manufacturer. For cell imaging, cells were grown on 35 mm MatTek plates coated with poly-d-lysine. For immunoprecipitation, cells were grown on 10 cm plates.
HEK293 cell lysis and immunoprecipitation
HEK293 cells (48 hrs after transfection) were rinsed with cold PBS twice and lysed on ice with 1.8 ml per 10 cm plate of Buffer B (50 mM Tris-HCl pH 7.5, 120 mM NaCl, 1 mM EDTA, 0.5% (v/v) Nonidet NP-40, 0.5% (w/v) deoxycholate, 10% (v/v) glycerol, 0.1 mM PMSF, 1 mM benzamidine, 5 mg/l leupeptin, 20 mg/l soybean trypsin inhibitor, 50 mM NaF, 100 mM Na3VO4). After sonication (2×5sec), lysates were centrifuged at 4°C for 15 min at 10,000×g. Equal aliquots of the supernatants were incubated with 10 µg of Densin Ab450 or goat IgG, or 10 µl of rabbit pre-immune serum or AbBΔN antiserum overnight at 4°C. After addition of GammaBind Plus-Sepharose (Amersham Biosciences) (30 µl of 1:1 slurry), incubations were continued for 2 h at 4°C. Beads were collected by microcentrifugation and washed at least three times with 1 ml of Buffer B; beads were transferred to a new microcentrifuge tube during the first wash. Immune complexes were solubilized in SDS-PAGE sample buffer.
Preparation of rat brain extracts for immunoblotting
Whole extracts and subcellular fractions of rat forebrains from different development stages or dissected brain regions were prepared as described previously (McNeill & Colbran 1995). Briefly, tissue was homogenized in 10 ml per g wet weight of Buffer A (10 mM Tris-HCl pH7.5 containing 1mM EGTA, 1mM EDTA, 1mM DTT, 0.1 mM PMSF, 20 mg/l soybean trypsin inhibitor and 5 mg/l leupeptin) using a Polytron (2×15 s bursts separated by 30 sec on ice). After removing an aliquot of the homogenate (whole lysate), remaining homogenate was centrifuged at 100,000×g for 60 min at 4°C. The supernatant (S1: cytosol) was saved and the pellet was rehomogenized in the same volume of Buffer A containing 1% (v/v) Triton X-100. After a second centrifugation as before, the supernatant (S2: Triton-soluble extract) was removed and the pellet (P2: Triton-insoluble cytoskeletal extract) was resuspended in the same volume of Buffer A. Protein concentrations were determined by Bradford assay. Extracts were aliquoted and stored at −80°C until use.
Immunoblotting
Samples were separated by SDS-PAGE under reducing conditions and transferred to nylon reinforced nitrocellulose membranes in 10 mM N-Cyclohexyl-3-aminopropanesulfonic acid (CAPS) buffer. After blocking with TTBS buffer (50 mM Tris-HCl pH 7.5, 0.1% Tween 20, 150 mM NaCl) containing 5% Carnation nonfat milk, membranes were incubated for either 2 hr at room temperature or overnight at 4°C with the specified primary antibodies diluted in TTBS with 5% milk. Densin antibodies Ab450 and Ab650 were diluted 1:1000 while AbBΔN was diluted 1:200. Additional antibodies used in the present study were mouse monoclonal antibodies to CaMKIIΔ (Affinity BioReagents: 1:2000), GFP (Santa Cruz Biotechnology: 1:2000) and Myc (Zymed Laboratories: 1:500). Membranes were washed 3 times in TTBS and incubated with horseradish peroxidase (HRP)-conjugated (American Qualex or Santa Cruz Biotechnology) secondary antibodies diluted in TTBS with 5% milk for 1 hr at room temperature. After extensive washing with TTBS, HRP-conjugated secondary antibodies were detected using enhanced chemiluminescence (Perkin-Elmer).
Immunoflorescence and cell imaging
For live cell imaging shown in Fig. 8, transfected HEK293 cells were imaged directly using a Zeiss LSM510 fluorescence laser scanning confocal microscope about 24 hours after transfection. GFP was excited at 488 nm and images of single optical sections (<1 µm) were taken with a 63×1.4 oil immersion objective with electronic zoom factors of 1.5–2 and exported to Canvas 9 (ACD Systems).
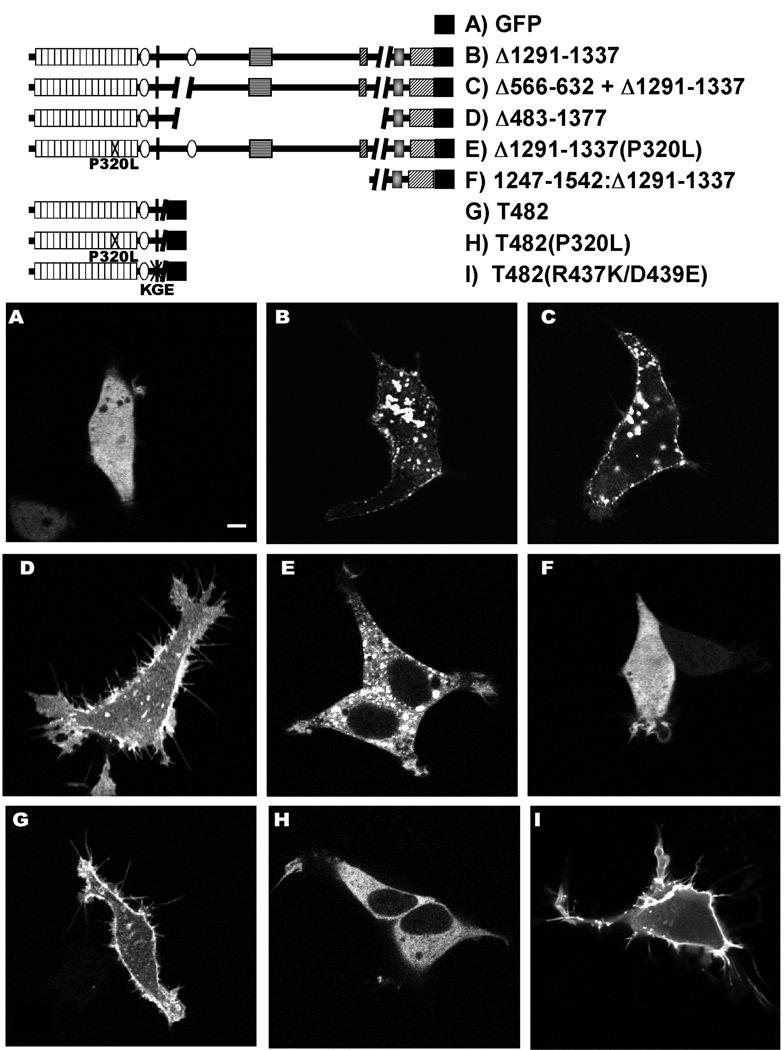
Figure 8. Subcellular distribution of densin variants in transfected HEK293 cells and effects on cell morphology.
The legend (top) shows domain structures of specific densin splice variants with or without the indicated mutations that were expressed in HEK293 cells with a C-terminal GFP. The panels show single optical sections (≈ 1 µm thick) of representative live cells collected using a confocal microscope approximately 24 hours after transfection. The scale bar in Panel A indicates 5 µm and applies to all panels.
For Fig. 9, 24 hr after transfection, cells were rinsed with PBS, fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. After blocking with 10% donkey serum for 30 min, cells were incubated with rhodamine-conjugated phalloidin (1:50, Molecular Probes) diluted in 2% donkey serum for 2 hr at room temperature to label F-actin. Cells were then washed extensively in PBS. Nuclei were stained by incubation with DRAQ5™ (1:250, Axxora LLC) at 37°C for 2 min before taking pictures. Images of labeled cells were collected using a Zeiss LSM510 fluorescence laser scanning confocal microscope with a 63×1.4 oil immersion objective with electronic zoom factors of 1.5–2. GFP, rhodamine and DRAQ5 were excited at 488 nm, 543 nm and 633 nm, respectively and a series of images with a 1 µm z-section were collected to span the entire depth of the cell. The z-section series was stacked together by projection with one angle and separated for individual channels. Images were then exported to Canvas 9 (ACD systems).
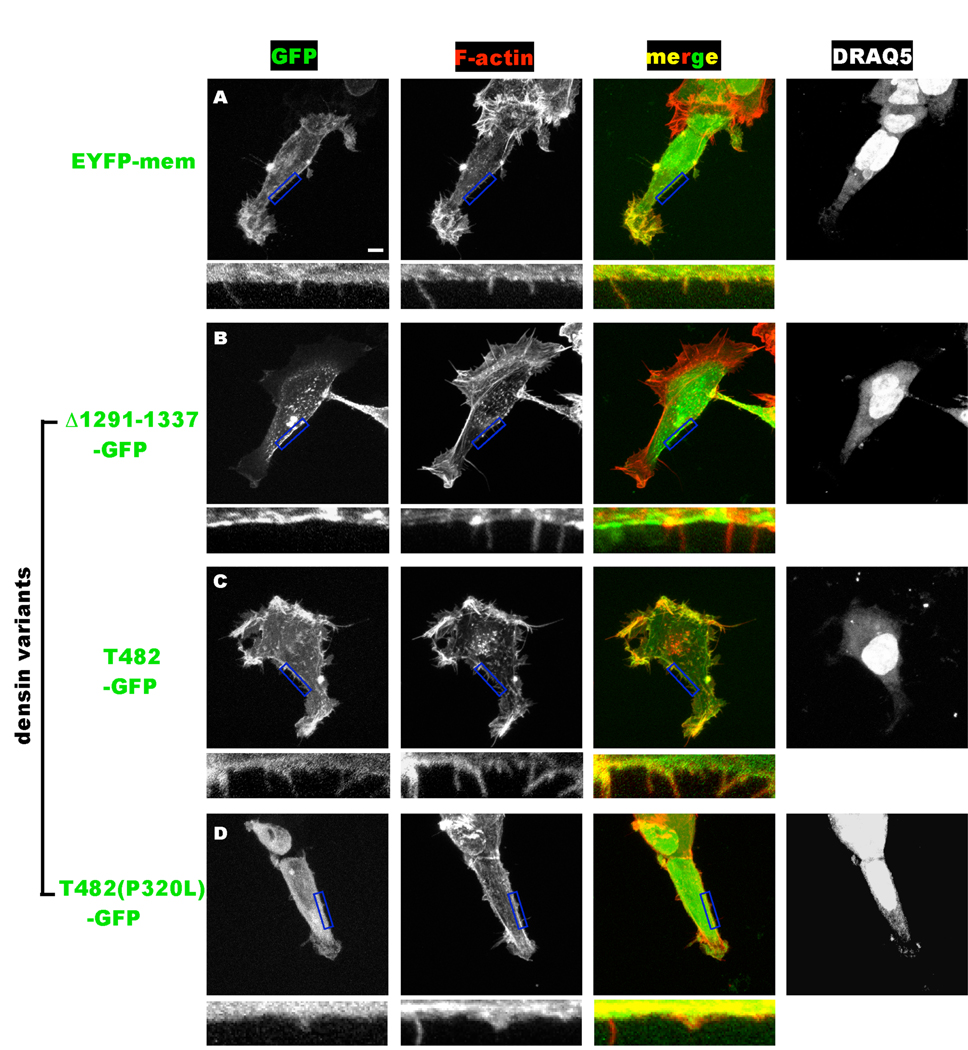
Figure 9. Differential targeting of densin splice variants to F-actin correlates with changes of HEK293 cell morphology.
Cells expressing EYFPmem or the indicated densin-GFP fusion proteins (at left) were fixed ≈24 hours after transfection and stained with rhodamine-phalloidin (F-actin) and DRAQ5 (nuclei). The main panels show Z-stacks of a series of 1 µm optical sections/images of representative cells of each color channel in grayscale. A merged color image of the GFP (green) and F-actin (red) channels is also shown with the color channels intentionally over-saturated to detect fine filopodia-like processes. The scale bar indicates 5 µm, and applies to all of the main panels. Regions of the plasma membrane outlined by the blue box in the main GFP, F-actin and merged panels are magnified below to show filopodia.
RESULTS
Identification of novel alternative splice variants of densin
As a first step toward generating full-length cDNA clones of densin (Fig. 1A), we employed the Rapid Amplification of cDNA ends (RACE) kit to generate a full length cDNA pool from brain total RNA preparations (see Experimental Procedures). To obtain fragments of cDNAs encoding N-terminal domains of densin, 5’-RACE-ready cDNA was used as template for amplification with primer 1F and 22R designed against rat densin cDNA (GenBank Accession Number GI 1657757) (Fig. 1D). Initial PCR experiments amplified 2 major products of 1.5–2.0 kb (Fig 1B). After ligation of the mixed PCR products into pGEMT vector, isolated clones were PCR amplified with the same set of primers in order to identify densin inserts. Notably, the length of the amplified products varied considerably among the 29 clones analyzed (Fig 1C). DNA sequencing of 10 clones (indicated by asterisk in Fig. 1C) revealed variable internal deletions relative to the expected size of 2233 bp (based on GI 1657757). Analysis of genomic sequence GI 34861224 revealed that the densin mRNA (≈5kb) consists of 27 exons within a genomic sequence of around 0.4 Mb (Fig. 1A). Only one of the 10 densin clones isolated contained sequences derived from all the expected exons (Fig. 1D). Shorter variants contained internal deletions of nucleotides derived from one or more entire exons: Δ110–186 (exon 2, E2), Δ1632–1880 (E17), Δ1632–2081(E17 + 18), Δ1632–2193 (E17–19) and Δ1507–2081(E16–18). Interestingly, deletion of E17–19 or E16–18 changes the reading frame of downstream mRNA, creating novel predicted amino acid sequences C-terminal to the splice site and a premature stop codon. In combination, these data show that the primary densin mRNA transcript is subjected to extensive alternative splicing.
These initial data prompted us to systematically explore the possibility that alternative splicing of the densin mRNA was regulated during development. Therefore, we carried out RT-PCR analysis of RNA samples isolated from rat brains at different developmental stages using a primer pair (13F/22R, Fig. 2A) designed to amplify nucleotide 1386–2233 from the 5’ portion of the densin mRNA. These primers were predicted to amplify ≈840 bp product from mRNAs that contain sequences derived from all of the intervening exons. Indeed, the expected ≈840 bp product (band 1) was amplified from postnatal day (P) 7 and P14 RNA. However, several smaller bands were also detected in the P7 sample, while samples derived from embryonic day (E) 15 and P1 brains contained only smaller PCR products (Fig. 2B). The smaller products of 590bp (band 3), 390bp (band4), 280bp and 260bp (band 5) are consistent with predicted sizes that would be amplified from mRNAs lacking specific exon combinations identified in Fig. 1: Δ1632–1880 (E17), Δ1632–2081 (E17 + E18), Δ1632–2193 (E17–19) and Δ1507–2081 (E16–18), respectively, and DNA sequencing confirmed that band 4 lacked exons 17 and 18. The ≈640 bp product (band 2) could result from skipping of exon18 (also see Fig. 3). However, DNA sequencing showed that no specific product was recovered from band 6. Parallel control reactions utilizing primers 36F and 43R revealed a developmentally-regulated pattern of 3' mRNA splicing similar to that reported previously (Fig. 2B) (Strack et al. 2000). In combination, these data show that alternative splicing of both the 5’ and 3' portions of the densin mRNA is developmentally regulated.
We then investigated whether specific combinations of 5’- and 3’-terminal splice variants are preferentially expressed. Primers (13F/46R, Fig. 3A) were chosen to amplify cDNA fragments spanning nucleotide 1386 to the 3’ end of the coding region (nucleotide 4670) from a pool of rat brain RNAs isolated at E15, P1, P7 and P14. The resulting PCR products (Fig. 3B) were ligated into pGEMT-Easy vector and transformed into E. coli. Clones isolated from individual colonies were confirmed by PCR using the same primers (Fig. 3C, top): most contained 2.8–3.2 kb inserts, but some clones contained much shorter (≈0.7 kb) inserts (e.g., clones 17, 23, 89). The individual clones were then analyzed by PCR using additional primer sets (representative analyses of 17 positive clones shown in Fig. 3C), further illustrating the diversity of densin fragments obtained. For example, amplification using 13F/22R primer pair detected the expected ≈840 bp “full length” product in only three clones. Smaller products of approximately 700bp, 400 bp or 250bp were amplified from most of the other clones. Although no specific products were generated from 5 clones using 13F and 22R primers, a combination of the 13F and 23R primers amplified specific products from 4 of these clones, suggesting that the 22R primer failed to anneal to these 4 variants. These studies independently confirmed most of the mRNA variants identified in Fig. 1, with the exception of the variant lacking exons 16–18. Similar PCR analysis to detect 3’ splice variants within these clones (39F/43R primers) detected three different variants that were previously characterized in this lab: the ≈650 bp full-length product (previously termed the D-splice variant: n=3), an ≈ 500 bp product lacking nucleotides encoded by Exon 22 (previously termed the A variant: n=5) and an ≈ 380 bp product lacking nucleotides encoded by Exons 22 and 23 (previously termed the B variant: n=5). Notably, the previously characterized C variant was not detected and no 3’ products were amplified from 3 of these 17 clones. There was no immediately discernible pattern linking 3’- splice variants to specific 5’-splice variants.
DNA sequences of the 24 positive clones obtained from initial PCR amplification using the 13F/46R primer set were compared to the genomic DNA sequence. Most of the variants result from deletion (skipping) of one, two or three entire exons (Fig. 3D) during mRNA splicing, although some variants (clones 16, 61, 17, 29, 72, 43) were apparently derived from the use of cryptic splice acceptor sites within an exon. Table 3 compares the 3' splice acceptor sites used in these clones to the splice acceptor site at the beginning of the corresponding exons and to the consensus splice acceptor sequence. The majority of splicing events produce “in frame” deletions, in agreement with observations that alternative splicing tends to preserve the protein reading frame (Modrek et al. 2001). Proteins produced from these splice variants are predicted to encode proteins with intact N- and C-terminal domains and internal deletions of highly variable size. However, several splicing events produce out of frame deletions that introduce premature stop codons, often quite close to the 3’ splicing sites, such that these variants encode truncated densin products (e.g., clones 43, 41, 89, 29, 72 in Fig. 3C). The complete nucleotide and amino acid sequences of full-length densin and the splice variants identified here are summarized in a supplementary figure available online.
Table 3. Summary of cryptic 3' splice acceptor sites used in densin splice variants.
The splice acceptor sites used in exons 20, 24 and 26 of densin are compared to the canonical splice acceptor sequence. Intron sequences are in capital letters and the first nucleotide of each exon sequence is indicated by a small letter. Y: C or T. N: any nucleotide
| Exon | Splice Site | Densin clone | Actual DNA sequence | “Symbolized” sequence |
|---|---|---|---|---|
| Canonical | -- | …YYYYYYYYNCAGg… | ||
| 20 | Normal | most | …TTTAAATTTCAGa… | …YYYAAAYYYCAGa… |
| Cryptic a | 16, 61 | …GTGTCTCCTCAGg… | …GYGYYYYYYCAGg… | |
| Cryptic b | 29, 72 | …TCTGAATAACAGt… | …YYYGAAYAACAGt… | |
| 24 | Normal | most | …TGCTCTCCACAGg | …YGYYYYYYACAGg |
| Cryptic a | 43 | …CTCCACAGGCAGg | …YYYYAYAGGCAGg | |
| 26 | Normal | most | …TCTGACTTATAGg… | …YYYGAYYYATAGg… |
| Cryptic a | 17 | …CTTTGTTACTAGg… | …YYYYGYYAYTAGg… |
Characterization of densin expression in brain at the protein level
We suggest here a systematic and flexible nomenclature for densin splice variants. We define "full-length" densin (densin-FL) as containing all of the coding exons. Densin-FL encodes a protein of 1542 amino acids by virtue of the inclusion of exon 22, as represented by clone 5, which was previously defined as the 3' D splice variant that was only detected in embryonic brain (Strack et al. 2000). Predicted proteins containing internal deletions are described as densin(Δ###-###), where numbers replacing the # symbols define amino acids missing from that splice variant. For example, clone 59 contains all of the 5' exons but lacks exon 22, and the protein encoded by this mRNA variant is therefore defined as densin(Δ1291–1337). This variant is the original 1495 amino acid densin-180 protein and may be the major variant expressed in adult brain (Strack et al. 2000, Apperson et al. 1996). Clone 10 lacks exon 18 in addition to exon 22, and the predicted protein is therefore designated densin(Δ566–632 + Δ1291–1337). Predicted proteins encoded by splice variants that shift the reading frame to cause premature truncation of the protein are designated as densin-T###, where ### designates the final amino acid, as exemplified by densin-T482, the 482 amino acid protein (≈55 kDa) encoded by clone 41 which contains only the LRR and LAPSDa domains. Fig. 4A summarizes the diverse domain structures of proteins encoded by selected densin splice variants, with predicted molecular weights ranging from 51 kDa to 167 kDa. However, differential posttranslational modification, such as glycosylation, may significantly affect the apparent size of these proteins.
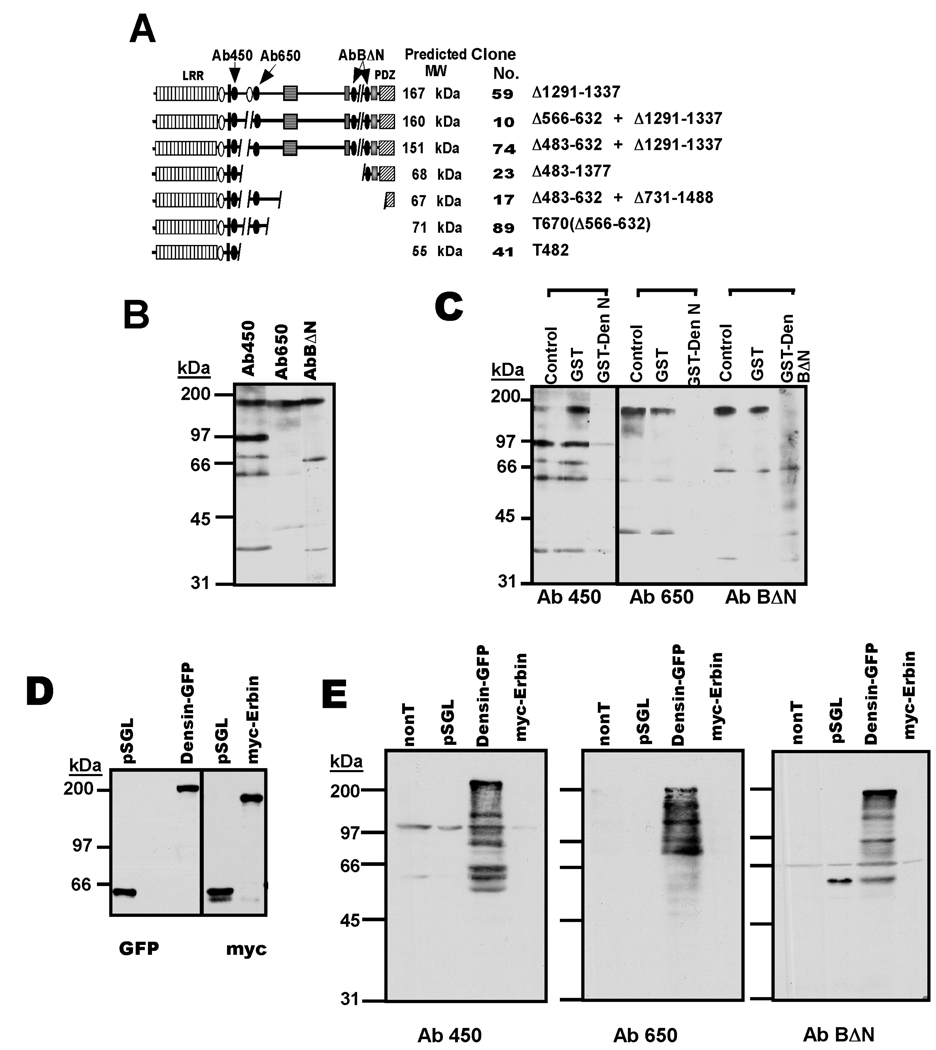
Figure 4. Expression of densin variants in adult brain.
A. Cartoon depicting domain structures of predicted densin spice variants and the locations of epitopes used to generate antibodies. The protein variant at the top is the major adult form (Apperson et al. 1996), with conserved domains and motifs annotated as in Fig. 1A. Structures below were predicted from mRNA variants summarized in Fig. 3C with the corresponding clone number listed afterwards. B. Initial testing of densin antibodies. Rat forebrain whole lysates were immunoblotted using the three antibodies (indicated in Panel A). C. Characterization of antibody specificity for endogenous densin. Antibodies were pre-incubated for 1.5 hours at room temperature with GST, GST-DenN or GST-BΔN (25 µg each in 1 ml), as indicated, and then used to immunoblot adult rat forebrain whole lysates. D. Expression of densin and erbin in transfected HEK293 cells. Myc-Erbin, densin(Δ1291–1337)-GFP, and a control myc/GFP-tagged densin protein (pSGL) were expressed in HEK293 cells and cell lysates were probed for myc or GFP. Using pSGL as a control, amounts of myc-erbin and densin-GFP lysates loaded to the gels were adjusted to contain approximately equal amounts of erbin and densin. E. Characterization of densin antibody specificity in heterologous system. myc-Erbin, densin-GFP and pSGL were expressed in HEK293 cells: nonT, non transfected cells. Gels loaded with similar amounts of erbin and densin lysates (see panel D) were immunoblotted using densin antibodies.
In order to begin to address densin expression at the protein level, three antibodies to different epitopes were generated (see Experimental Procedures): the Ab450 epitope is present in all but one of the proteins predicted by data in Fig. 1 and Fig. 3, whereas the Ab650 and AbBΔN epitopes are differentially incorporated into these proteins (Fig. 4A). Consistent with the original identification of densin, all three antibodies predominantly detected a ≈180 kDa protein in adult rat whole forebrain lysates (Fig. 4B) and careful alignment of multiple parallel immunoblots of different samples showed that these prominent bands co-migrated. All three antibodies also detected several additional smaller proteins in the whole forebrain extracts. Pre-adsorption with GST proteins containing the respective epitopes, but not with GST alone, blocked recognition of the ≈180 kDa protein by all three antibodies (Fig. 4C). GST-DenN, but not GST alone, almost completely blocked the detection of all smaller proteins by Ab450 and Ab650, suggesting that these proteins contain epitopes closely related to densin. In contrast, GST-DenBΔN did not block the detection of proteins at 66 and 40kDa by AbBΔN, implying that these proteins are not related to densin. A protein of ≈60kDa was specifically recognized by both Ab450 and Ab650. Thus, in contrast to the full-length 180 kDa protein, most smaller immunoreactive proteins were recognized by only one or two of the densin antibodies.
The immunoreactive bands specifically recognized by densin antibodies may represent other brain proteins containing similar epitopes. Therefore, we searched rat/mouse genomic databases for other proteins containing similar amino sequences to the epitopes of the three densin antibodies using BlastP (<http://www.ncbi.nlm.nih.gov/blast/Blast.cgi>). The only hit was erbin, another LAP protein family member, which shared homology within the epitopes for Ab450 and AbBΔN. Erbin is the closest relative of densin in the LAP protein family (Santoni et al. 2002), has a similar molecular weight and is also highly enriched in the brain (Huang et al. 2001). Therefore, we tested the selectivity of our antibodies between Erbin and densin. HEK293 cells were transfected to express either densin-GFP, myc-Erbin or pSGL, a synthetic fusion protein containing residues 1378–1451 of densin with both myc and GFP tags (Strack et al. 2000). Cell lysates were immunoblotted in parallel for myc and GFP epitopes, using the pSGL extract to normalize the signals detected. Loading of myc-erbin and densin-GFP extracts was adjusted to yield comparable signals to pSGL with both antibodies (Fig. 4D). Thus, comparable amounts of erbin and densin were loaded on these gels. The normalized densin and erbin extracts were then re-analyzed by probing with the densin antibodies (Fig. 4E). Repeated use of the same extracts for multiple blots was necessary to get appropriately normalized loading, but resulted in significant degradation of densin, such that all three antibodies detected the full length ≈200 kDa densin-GFP protein and multiple proteolytic fragments. (Note: AbBΔN also detected the pSGL protein because this construct contains part of the epitope for this antibody.) However, none of the densin antibodies detected the expressed myc-erbin protein. Thus, under these conditions, all three densin antibodies specifically detected densin over its closest relative, erbin.
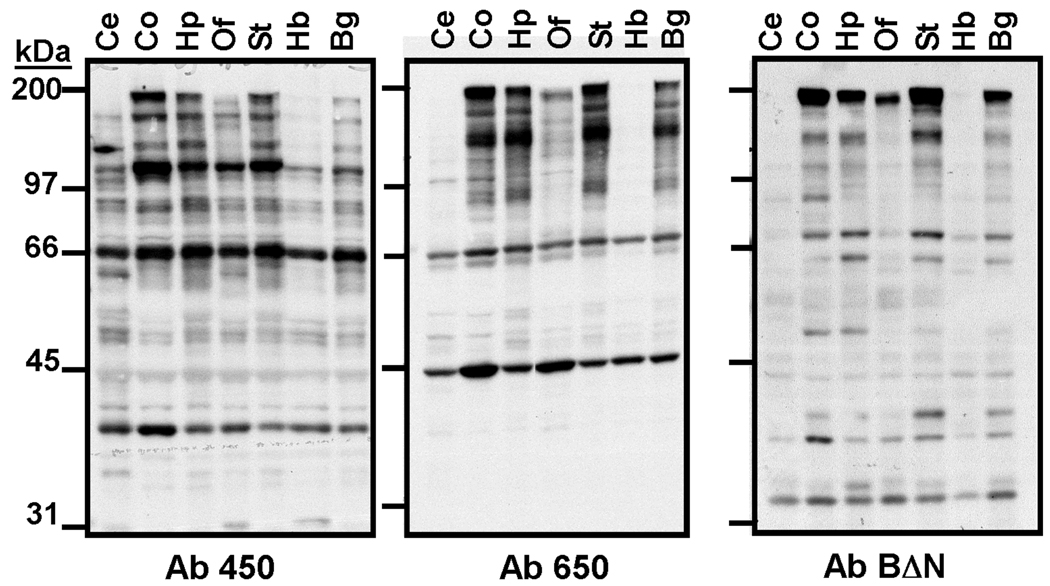
Additional immunoblot studies showed that the ≈180 kDa form of densin was highly expressed in whole extracts of rat cortex, hippocampus, striatum and basal ganglia, with reduced levels in olfactory bulb and hindbrain, and little or no expression in cerebellum (Fig. 5). The smaller proteins detected using Ab450 and Ab650 exhibited a somewhat different distribution between the brain regions from the ≈180 kDa form. For example, Ab450 detects ≈66 kDa and ≈40 kDa proteins in cerebellum and hindbrain, but the ≈180 kDa form is not present. Similarly, Ab650 detects a prominent ≈45 kDa protein in olfactory bulb, where the ≈180 kDa form is less abundant. Notably, recognition of all of these smaller proteins was blocked by pre-adsorption of the antibodies using GST fusion proteins containing the respective epitopes (Fig. 4B and data not shown). In combination, these immunoblotting data are consistent with the expression in distinct brain regions of smaller proteins differentially containing these three densin epitopes. Sizes of these smaller proteins are similar to predicted sizes of densin splice variants that variably include the respective epitopes. However, we cannot rule out the possibility that some of the proteins might be proteolytic fragments derived from longer variants.
Figure 5. Expression of densin in adult rat brain regions.
Whole lysates were immunoblotted using densin antibodies characterized in Fig. 4. Ce: Cerebellum; Co: cortex; Hp: Hippocampus; Of: Olfactory Bulb; St: Striatum; Hb: Hindbrain; Bg: Basal Ganglia.
Interaction of CaMKII with densin is modulated by alternative splicing
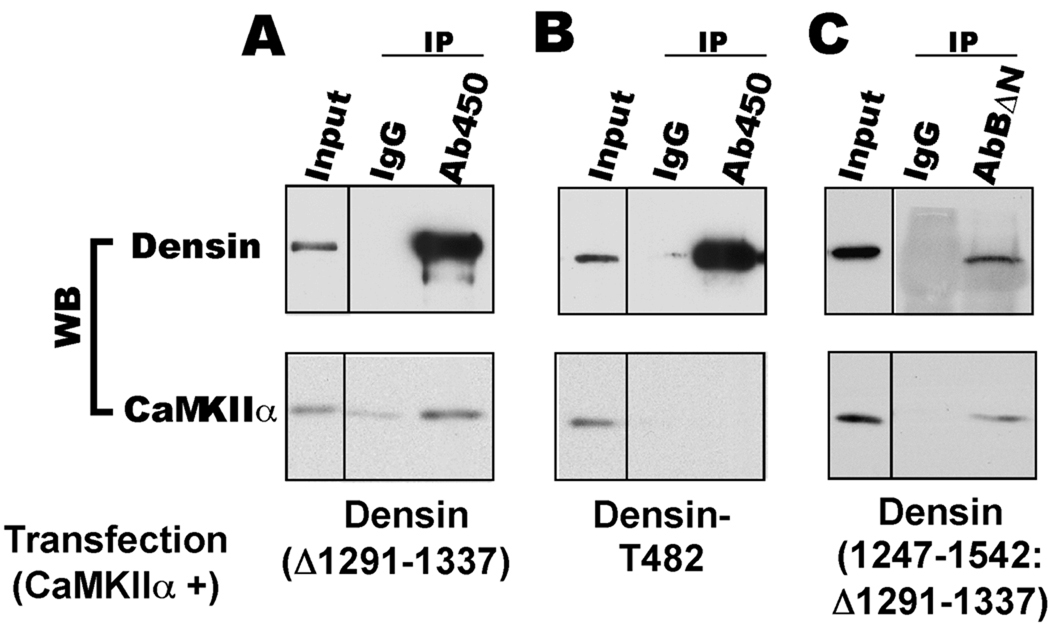
Complexes containing CaMKIIα and densin have been immunoprecipitated from adult rat brain extracts (Walikonis et al. 2001, Robison et al. 2005). In order to further establish that this interaction is due to direct binding of densin to CaMKII and to investigate the effect of alternative splicing on CaMKII binding, we co-expressed different densin splice variants with CaMKIIα in HEK293 cells and analyzed their interactions using co-immunoprecipitation assays. CaMKIIα associated with immunoprecipitated densin(Δ1291–1337) (Fig. 6A), the major form of densin in adult brain. A fragment of the densin C-terminal domain was previously shown to be sufficient to target CaMKIIα in HEK293 cells (Strack et al. 2000). Consistent with this observation, CaMKIIα associated with an immunoprecipitated C-terminal domain fragment of densin(1247–1542: Δ1291–1337) (Fig. 6C). However, CaMKIIα could not be detected in densin-T482 immunoprecipitates (Fig. 6B). In combination, these data show that CaMKII associates with densin in HEK cells via the C-terminal domains and that alternative splicing of the densin mRNA modulates this interaction.
Figure 6. CaMKII associates with the C-terminal domain of densin.
CaMKIIα was expressed with densin(Δ1291–1337), densin-T482 or densin(1247–1542: Δ1291–1337) (all with a C-terminal GFP fusion) in HEK293 cells. Cell lysates were immunoprecipitated using Ab450 (panels A and B) or AbBΔN (panel C), with parallel controls using non-immune IgG. Input samples and immune complexes were probed for densin (using GFP antibody) or CaMKIIα.
Expression of densin variants during development
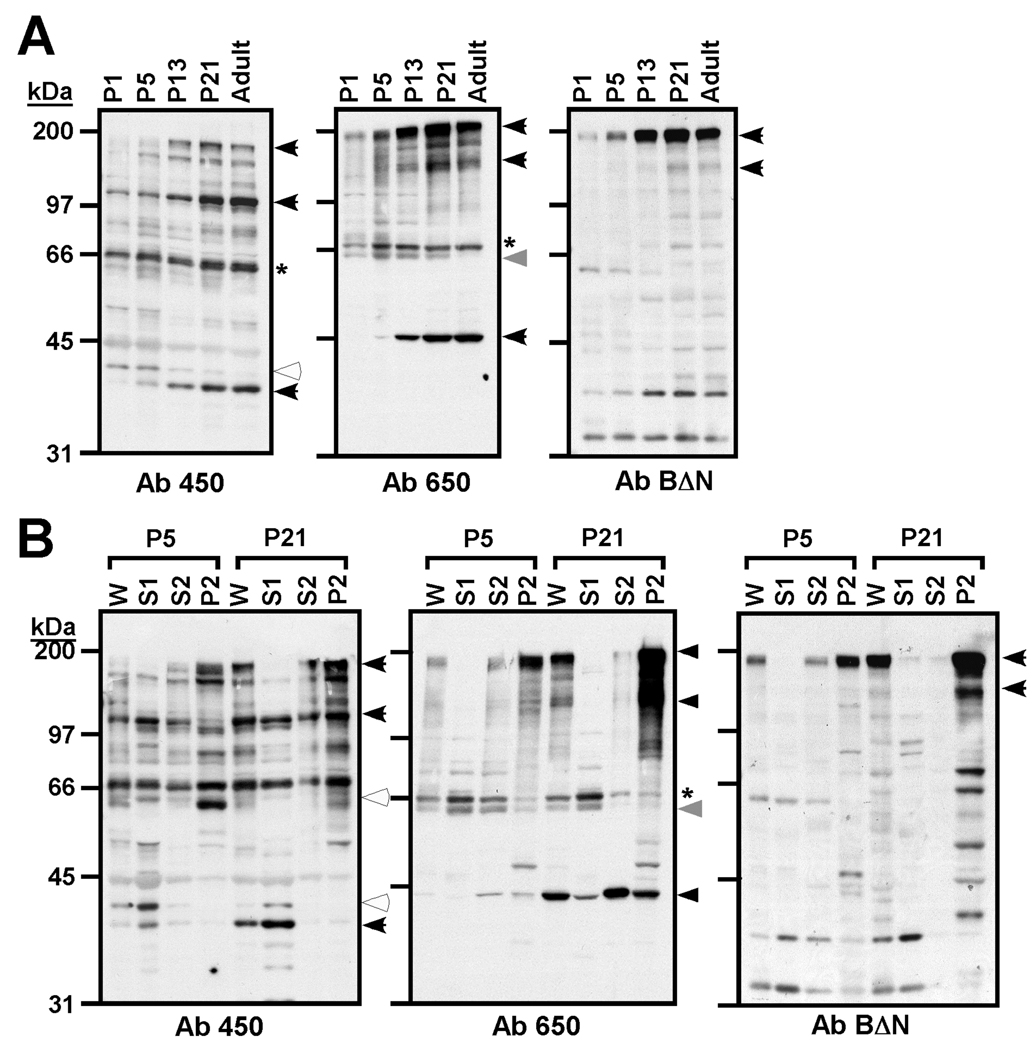
RT-PCR analyses indicated that alternative splicing of densin mRNA is regulated during development (Fig. 2B). Therefore, we used our antibodies to three different domains in densin to immunoblot whole extracts of rat brains dissected at various times during postnatal development. All three antibodies detected a strong up-regulation of the ≈180kDa form of densin between P5 and P13 (Fig 7A). However, smaller immunoreactive proteins exhibited distinct developmental profiles. Proteins of ≈40 kDa and ≈100 kDa detected using Ab450, of ≈120 kDa and ≈45 kDa detected using Ab650, were expressed with a similar developmental profile to the ≈180 kDa protein. On the other hand, expression of proteins of ≈42 kDa detected using Ab450 decreased from P1 to adulthood. Expression of a ≈64 kDa protein (detected by Ab650) appeared to be transiently modulated, peaking at P5–P15 (Fig. 7A). However, expression of a ≈ 66 kDa protein detected using Ab450 and Ab650 was not strongly regulated. In combination these data are consistent with the differential expression of proteins containing different combination of these specific densin epitopes during development.
Figure 7. Expression of densin during development.
A. Densin expression in whole forebrain homogenates. Whole forebrain lysates from rat at the indicated developmental stages were immunoblotted with antibodies Ab450, Ab650 or AbBΔN. B. Subcellular fractionation of densin in forebrain extracts at P5 and P21. Rat forebrain extracts (P5 or P21) were fractionated (see Experimental Procedures) and immunoblotted with Ab450, Ab650 or AbBΔN. W: whole lysates; S1: cytosolic fraction; S2: Triton-soluble fraction; P2: Triton-insoluble cytoskeletal fraction. Black and while arrows indicate proteins with increased and decreased expression, respectively, during development. The grey arrow indicates a protein that was transiently expressed during development. The "*" indicates a protein with relatively constant expression during development.
Subcellular fractionation of putative densin variants expressed in brain
Differential incorporation of protein-protein interaction domains into densin splice variants (Fig. 4A) may affect subcellular localization of the proteins. Therefore, subcellular fractions prepared from rat forebrain dissected at P5 and P21 were immunoblotted for densin and the putative splice variants. The ≈180 kDa form of densin detected using all three antibodies was most abundant in PSD-enriched detergent-insoluble cytoskeletal (P2) fractions isolated at both developmental stages (Fig. 7B), consistent with its original identification as a PSD-enriched protein (Apperson et al. 1996). Notably, smaller immnoreactive proteins that are differentially detected by these antibodies are strongly concentrated in distinct subcellular fractions. S1 cytosolic fractions at P21 are selectively enriched in proteins of ≈40 kDa (Ab450) and ≈66 kDa (Ab650). S2 Triton-soluble fractions are enriched in a ≈45 kDa protein at P21 (Ab650), whereas P2 cytoskeletal fractions are enriched in a ≈60 kDa protein at P5 (Ab450), and proteins of ≈120 kDa (Ab650) at P21 (Fig. 7B). These subcellular fractionation studies also emphasized developmental changes in the expression of proteins recognized by the densin antibodies. For example, expression of the ≈60 kDa protein detected using Ab450 in the P2 fraction at postnatal day 5 was strongly reduced by postnatal day 21, further supporting data showing that expression of proteins containing specific densin epitopes is developmentally regulated, presumably reflecting changes in densin mRNA splice variant expression (Fig. 2). However, the signal for this protein was relatively weak and difficult to resolve in the whole extracts shown in Fig. 7A. Thus, while full-length densin is enriched in fractions containing postsynaptic densities, putative splice variants differentially recognized by the densin antibodies exhibit diverse subcellular fractionation patterns, perhaps reflecting distinct subcellular localizations.
Alternative splicing of densin modulates subcellular localization and cell morphology
In order to more specifically investigate the subcellular localization of densin splice variants, live HEK293 cells expressing selected proteins with GFP fused at the C-terminus were imaged about 24 hours after transfection. Immunoblot analyses of lysates prepared from the transfected cells using antibodies to GFP and densin demonstrated that splice variant proteins are expressed at the expected sizes (data not shown). In images of single optical sections (1 µm), the major adult densin splice variant (densin(Δ1291–1337)) adopted a predominantly punctate staining pattern on or close to the plasma membrane and on intracellular vesicles (Fig. 8B), in contrast to the diffuse localization of GFP alone throughout the cell (Fig. 8A). A truncated densin containing only the carboxy-terminal domain exhibited diffuse cytosolic localization (Fig. 8F), similar to the subcellular distribution of GFP alone, suggesting that domains in the N-terminal portions of densin are important for the association with the plasma membrane. Among densin splice variants, deletion of the LAPSDb domain (densin(Δ566–632 + Δ1291–1337)) had no obvious effect on subcellular localization (Fig. 8C). However, a larger internal deletion in densin(Δ483–1377) resulted in more diffuse localization near the plasma membrane, with weaker intracellular fluorescence (Fig. 8D). The densin-T482 variant that lacks the entire C-terminus also exhibited diffuse membrane association with weaker intracellular fluorescence than densin(Δ1291–1337) (Fig. 8G). Closer inspection of the transfected cells revealed that HEK293 cells expressing densin(Δ483–1377) or densin-T482 typically exhibited an increased number of fine fluorescent processes extending from the plasma membrane that resemble filopodia (Fig. 8D/G). In combination, these data show that densin splice variants are differentially localized within HEK293 cells, and suggest that some may regulate cell morphology.
In order to gain a better understanding of the potential effects of densin splice variants on cell morphology, we fixed HEK293 cells expressing selected GFP-densin splice variants and stained them with rhodamine phalloidin and DRAQ5 to label the F-actin cytoskeleton (red) and nuclei (blue), respectively. We also analyzed cells transfected to express a membrane-targeted YFP protein using the pEYFPmem construct as a positive marker of the plasma membrane and overall cell morphology. Images of thin optical sections of individual cells covering the entire z-dimension were collected, and then merged in Z-stacks. Fig. 9 shows representative cells. As is typical of transfected cells in culture, cell morphologies varied to some extent, even on the same dish/transfection, but these cells typically displayed cortical F-actin staining with evidence of lamellipodia, especially in cells growing in isolation. In control cells, F-actin staining overlapped with YFPmem fluorescence and closer examination revealed overlap of these signals in a few filopodia (Fig. 9A, insets). These data show that imaging under these conditions is unable to clearly distinguish proteins directly associated with the plasma membrane (YFPmem) from those associated with the cortical F-actin cytoskeleton (rhodamine phalloidin). Expression of densin(Δ1291–1337)-GFP (the full length adult variant) had little effect on F-actin stained with rhodamine-phalloidin, and GFP fluorescence was clustered on or near the plasma membrane and in intracellular structures (Fig. 9B), consistent with live cell images (Fig. 8). Higher magnification of the images revealed that these cells typically displayed a few filopodia (stained with rhodamine phalloidin), but that densin(Δ1291–1337)-GFP was not significantly colocalized with rhodamine phalloidin within these structures or the cortical cytoskeleton (Fig. 9B, inserts). In contrast, cells expressing densin-T482-GFP usually displayed more profuse filopodia when compared with cells expressing YFPmem or densin(Δ1291–1337)-GFP. Moreover, densin-T482-GFP fluorescence colocalized with rhodamine-phalloidin within the filopodia and in cortical regions, and was much less abundant in intracellular clusters than densin(Δ1291–1337)-GFP (Fig. 9C), consistent with data shown in Fig. 8G. In combination, these data show that densin-T482, but not densin(Δ1291–1337), is targeted to F-actin both in the cortical cytoskeleton and in filopodia. Moreover, expression of densin-T482 seems to enhance the abundance of filopodia, suggesting that some splice variants may modulate cell morphology.
The LRR domain of densin modulates cell morphology
In order to probe mechanisms underlying the differential subcellular localization of densin variants and their possible effects on cell morphology, we expressed mutated densins as GFP fusion proteins. The N-terminal region of densin contains a canonical RGD motif, representing a possible integrin binding site that may be involved in regulating subcellular localization and or morphological effects (Teti et al. 2002). Mutation of the RGD motif to KGE in the L1 cell adhesion molecule disrupts neurite outgrowth in dorsal root ganglion cells (Yip et al. 1998). However, the corresponding mutation in densin-T482 had no obvious effect on either the subcellular distribution or the induction of filopodia-like projections (Fig. 8I). Two additional conserved domains (LRR and LAPSDa) may play a role in regulating localization and/or cell morphology (Fig. 8G). A Pro to Leu mutation in LRR domains of scribble and erbin disrupts plasma membrane association (Legouis et al. 2003). The corresponding mutation (P320L) in densin-T482 abrogated plasma membrane association and colocalization with the F-actin cytoskeleton (Fig. 8H, Fig. 9D), and appeared to reduce the number of filopodia-like projections stained with rhodamine phalloidin, compared to wild type densin-T482 (Fig 9D). The P320L mutation also reduced plasma membrane clustering of densin(Δ1291–1337), with a concomitant increase in intracellular fluorescent puncta, but had no obvious effect on cell morphology (Fig. 8E). In combination, these data show that the integrity of the LRR region is important for the plasma membrane association of multiple densin variants. Moreover, changes in densin localization may be linked to modulation of cell morphology.
DISCUSSION
Here, we identified new alternative splice variants of densin that are differentially expressed during brain development. Previous studies defined a limited number of alternative splice variants in the 5’ and 3’ portions of the densin mRNA (Strack et al. 2000, Apperson et al. 1996). The present systematic efforts revealed several additional alternative splicing sites, giving rise to novel mRNA variants that are differentially expressed during late embryonic and postnatal development. However, it should be noted that we have not attempted to exhaustively identify all densin splice variants. It seems likely that similar analyses of mRNA isolated at different developmental stages, from specific brain regions or from other tissues using different PCR primers will uncover additional variants. Nevertheless, many of the shorter predicted proteins lack known protein-protein interaction domains, exemplified by the deficiency of CaMKII binding to densin-T482 (Fig. 6), and/or conserved sequence motifs with as yet unknown roles. Moreover, antibodies raised to different domains in densin specifically detect multiple proteins of highly variable sizes that are similar to the predicted sizes of proteins encoded by the mRNA splice variants. The expression of these proteins is developmentally regulated and they exhibit distinct subcellular fractionation patterns. Indeed, we show that alternative splicing results in differential targeting of densin in cells, affects the interaction with CaMKIIα and may modulate cell morphology. Thus, it is likely that densin splice variants have distinct but perhaps overlapping functional roles.
Alternative splicing of densin modulates CaMKII binding
The original studies showed that the C terminal domain of multiple densin splice variants contains a CaMKII-binding domain encoded by exon 24 that interacts with CaMKIIα (Walikonis et al. 2001, Strack et al. 2000, Robison et al. 2005). This interaction is relatively insensitive to kinase activation, suggesting that densin constitutively targets CaMKIIα. Interestingly, exon 24 is absent in some of the novel splice variants identified in the present study, such as densin-T482. Coimmunoprecipitation studies showed that densin(Δ1291–1337) and the C-terminal domain of densin(1247–1542: Δ1291–1337), but not densin-T482, interact with CaMKIIα (Fig. 8), demonstrating that the C-terminal domains of densin are sufficient for CaMKII interaction in mammalian cells. Densin variants that cannot bind CaMKII (and presumably other signaling proteins) likely have distinct roles.
Alternative splice variants of densin are differentially targeted in intact cells
The full-length ≈180 kDa adult form of densin (densin(Δ1291–1337): previously referred to as densin-1801) was identified as an abundant protein in isolated postsynaptic densities and is targeted to excitatory synapses. Initial analyses showed that densin is an O-sialoglycoprotein, containing a weak signal peptide sequence and a single putative transmembrane domain near the C-terminus, suggesting that the LRR domain is extracellular and that the PDZ domain is intracellular (Apperson et al. 1996). Consistent with this model, densin(Δ1291–1337) was associated with the plasma membrane when expressed in HEK293 cells with GFP fused at the C-terminus (Fig. 8B). However, according to this model, truncation of the entire C-terminal domain (including the putative transmembrane domain) should result in secretion of the densin-T482 splice variant. In contrast, densin-T482 remained strongly associated with the plasma membrane (Fig. 8G) and could not be detected in media collected from cells expressing this splice variant, even following immunoprecipitation (Jiao & Colbran, data not shown). Moreover, mutation of the RGD canonical cell adhesion motif had no effect on the membrane localization, arguing that densin-T482 is not associated with integrins outside the plasma membrane (Fig. 8). Our observations suggest that densin is not a transmembrane protein, consistent with a report that cell surface biotinylation failed to label densin, although data supporting this statement were not shown (Izawa et al. 2002).
The LRR domain appears to play an important role in localizing densin variants close to the plasma membrane. This conclusion is supported by the observation that densin-T482 is targeted to the plasma membrane, and that a single point mutation in the LRR domain (P320L) results in diffuse cytosolic localization (Fig. 8G/H). These observations are consistent with recent studies showing that the LRR domain is critical for plasma membrane localization of other intracellular, membrane associated LAP protein family members (Legouis et al. 2003, Saito et al. 2001). Our data cannot distinguish direct association with lipids in the plasma membrane from association with other proteins that are themselves localized to the plasma membrane or to the cortical cytoskeleton. We favor the hypothesis that the LRR domain interacts with other proteins, as suggested previously for LRR domains in other LAP proteins (Legouis et al. 2003, Kallay et al. 2006). However, the identity of protein targets of LRR domains remains unknown.
Densin-T482 is relatively evenly distributed on the plasma membrane and colocalizes with F-actin in cortical subcellular domains and in filopodia (Fig. 8, 9). In contrast, full-length densin has a more punctate plasma membrane association that is not strongly colocalized with F-actin (Fig. 8, Fig. 9) even in more confluent HEK cells (data not shown). Moreover, introduction of the P320L mutation in full length densin results in a punctate intracellular localization quite distinct from that of the wild type protein, rather than the diffuse intracellular localization of densin-T482(P320L) (Fig. 8). In combination, these data suggest that domains outside of the LRR domain also play important roles in controlling the subcellular targeting of densin. This interpretation is further supported by the diffuse plasma membrane localization adopted by the densin(Δ483–1377) (Fig. 8). Interestingly, the PDZ domain does not appear to play a dominant role in subcellular targeting under these conditions because a C-terminal domain fragment adopts a diffuse localization throughout the cell (Fig. 8), which is indistinguishable from GFP alone (Fig. 8). However, neuronal PDZ domain ligands that are absent in HEK293 cells may further modulate the subcellular localization in vivo. Thus, these data suggest that the large poorly characterized central domain in densin has an important role in subcellular targeting, in addition to the critical role of the LRR domain. This likely does not involve the LAPSDb domain since the densin(Δ566–632) variant lacks this domain but adopts a localization that is essentially indistinguishable from the full-length protein (Fig. 8).
Differential targeting of densin variants in HEK293 cells appears to reflect selective localization in the brain because immunoblot analyses of subcellular fractions isolated from rat brain showed that while a 180 kDa form of densin, presumably densin(Δ1291–1337), is concentrated in cytoskeletal PSD-enriched fractions, shorter putative splice variants that are differentially detected by our antibodies have variable subcellular fractionation patterns during brain development (Fig. 6). Additional studies will be required to uncover mechanisms underlying the roles of the LRR domain and the central domain in the differential targeting of densin splice variants in neurons.
Selective effects of densin splice variants on cell morphology
Densin was recently shown to be important for branching of neurites/dendrites during neuronal development (Quitsch et al. 2005). For example, over-expression of a full length densin or an artificial N-terminal fragment containing the LRR domain enhanced dendritic branching in cultured neurons. Interestingly, SHANK interacts with C-terminal domains in densin, and co-expression of SHANK suppressed morphological effects of the full length densin, but not the N-terminal fragment (Quitsch et al. 2005). Another recent study in cultured mouse kidney podocytes showed that a full length densin variant colocalized with F-actin at cell-cell junctions but not with other pools of F-actin, and that the localization of densin was modulated in concert with that of β-catenin (Heikkila et al. 2007).
The present heterologous cell expression studies demonstrated that densin splice variants might also differentially modulate cell morphology. Although expression of full-length densin(Δ1291–1337) had little effect on HEK293 cell morphology, a truncated splice variant lacking the central and C-terminal domains (i.e., densin-T482) seemed to consistently enhance the appearance of “spiky” filopodia-like projections (Fig. 8, 9). Indeed, densin-T482 is strongly localized to the F-actin cytoskeleton in cortical regions of the cell and within the filopodia, in contrast to the longer variants that do not affect HEK293 cell morphology (e.g., densin(Δ1291–1337)). A functional LRR domain is required for this cell phenotype because the P320L mutation in densin-T482 disrupted membrane targeting and the appearance of these projections. Interestingly, densin(Δ483–1377) lacks the large central domain, but retains the C-terminal PDZ domain, yet appears to adopt a similar subcellular localization and has similar morphological effects as densin-T482. More quantitative morphological analyses will be necessary to conclusively establish the effects of densin variants on cell morphology. Nevertheless, these data suggest that the poorly characterized central domain of densin is important in limiting the effects of longer adult variants on HEK293 cell morphology. We propose that the naturally occurring densin splice variants lacking the central or C-terminal domains may have important roles during neuronal morphogenesis in the developing brain and the transition to adult splice variants may be important in stabilizing cell morphology at appropriate times during later development.
The LAP proteins have been subdivided according to the number of PDZ domains. Thus, in mammals, scribble, erbin and Lano have 4, 1 and 0 PDZ domains, respectively (Santoni et al. 2002). While densin was previously placed in the same subclass as erbin, the present studies suggest that densin could function similarly to erbin or to Lano, depending on the specific splice variant expressed. In addition, although there is high sequence and structural similarity between N-terminal LRR and C-terminal PDZ domains in LAP proteins, the length and sequence of the central domain is highly variable (Legouis et al. 2000), implying that this region might confer member-specific protein interactions or functions. Therefore, additional studies will be required to understand the possible role of the central region of densin in these processes. In addition, because expression of these shorter splice variants is regulated during brain development, it will be important to investigate how they interact with the complex signaling pathways that modulate cell morphology and neuronal development.
Overall summary
The present studies identify a tremendous diversity of alternative splice variants generated from the densin gene that are differentially expressed during development. Proteins that are selectively recognized by densin antibodies raised to distinct epitopes, likely representing densin splice variants, are differentially expressed in the brain during development and exhibit distinct subcellular fractionation patterns. It seems likely that complex mechanisms involving several protein-protein interactions control the subcellular localization of densin in multiple cell types. Moreover, densin splice variants identified in the present studies adopt distinct subcellular localizations that could differentially affect cell morphology. Elucidating cellular mechanisms underlying these diverse effects of densin splice variants will be an important future goal. In addition, alternative splicing modulates interaction with CaMKII so that some variants can function as CaMKAPs whereas others cannot. These data suggest that densin splice variants can play multiple distinct roles during neuronal development and in the mature brain.
Supplementary Material
Table 2. Oligonucleotide primers used for Densin-GFP constructs and site directed mutagenesis.
| name | sequence | orientation | position |
|---|---|---|---|
| BglIIheadF | GAAGATCTATGCAGTGCCTGGAGATGACCACC | sense | 186-209 |
| SacIItailR | TCCCCGCGGGACAGTGAGCTCACGTTGAATAAC | antisense | 4670-4647 |
| SacIIT482R | TCCCCGCGGTGACAGTGAGTTTTGTCAACTTCAGACTC | antisense | 2232-2205 |
| BglIID1F | GAAGATCTCGACAAGACATCAGATAAC | sense | 3924-3941 |
| SacIICtailR | TCCCCGCGGTGAGACAGTGAGCTCACGTTGAATAAC | antisense | 4670-4647 |
| P320LmutF | GAGCTGGAGTCCCTCCTTCCCACCATTGGTTACC | sense | 1128-1161 |
| P320LmutR | GGTAACCAATCCTGGGAAGGAGGGACTCCAGCTC | antisense | 1161-1128 |
| RGDmutF | GTTTCCCCAGCAGCCCAAGGGTGAGGAAGATTTCCAGTCAG | sense | 1478-1518 |
| RGDmutR | CTGACTGGAAATCTTCCTCACCCTTGGGCTGCTGGGGAAAC | antisense | 1518-1478 |
ACKNOWLEDGEMENTS
This work was supported by the NIH (RO1-MH63232, PO1-NS44282, F31-MH68129). Fluorescence microscopy was performed in the VUMC Cell Imaging Shared Resource, supported in part by NIH grants CA68485, DK20593, DK58404, HD15052, DK59637 and EY08126. The mammalian CaMKIIα expression construct was a generous gift from Liza Nikandrova.
Footnotes
The original work described “densin-180” as a protein in postsynaptic density fractions isolated from adult rat brains with an apparent size of ≈180 kDa on SDS polyacrylamide gels. We favor the more general name of “densin” to reflect the diverse sizes of densin gene products that are expressed at different stages of development and may be localized to different subcellular compartments.
References
- Ahola H, Heikkila E, Astrom E, Inagaki M, Izawa I, Pavenstadt H, Kerjaschki D, Holthofer H. A novel protein, densin, expressed by glomerular podocytes. J Am Soc Nephrol. 2003;14:1731–1737. doi: 10.1097/01.asn.0000075553.33781.9f. [DOI] [PubMed] [Google Scholar]
- Apperson ML, Moon IS, Kennedy MB. Characterization of densin-180, a new brain-specific synaptic protein of the O-sialoglycoprotein family. J Neurosci. 1996;16:6839–6852. doi: 10.1523/JNEUROSCI.16-21-06839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder D, Birnbaum D, Borg JP, et al. Collective nomenclature for LAP proteins. Nat Cell Biol. 2000;2:E114. doi: 10.1038/35017119. [DOI] [PubMed] [Google Scholar]
- Borg JP, Marchetto S, Le Bivic A, et al. ERBIN: a basolateral PDZ protein that interacts with the mammalian ERBB2/HER2 receptor. Nat Cell Biol. 2000;2:407–414. doi: 10.1038/35017038. [DOI] [PubMed] [Google Scholar]
- Calin-Jageman I, Yu K, Hall RA, Mei L, Lee A. Erbin enhances voltage-dependent facilitation of Ca(v)1.3 Ca2+ channels through relief of an autoinhibitory domain in the Ca(v)1.3 alpha1 subunit. J Neurosci. 2007;27:1374–1385. doi: 10.1523/JNEUROSCI.5191-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P, Xiong WC, Mei L. Erbin inhibits RAF activation by disrupting the sur-8-Ras-Raf complex. J Biol Chem. 2006;281:927–933. doi: 10.1074/jbc.M507360200. [DOI] [PubMed] [Google Scholar]
- Heikkila E, Ristola M, Endlich K, Lehtonen S, Lassila M, Havana M, Endlich N, Holthofer H. Densin and beta-catenin form a complex and co-localize in cultured podocyte cell junctions. Molecular and cellular biochemistry. 2007;305:9–18. doi: 10.1007/s11010-007-9522-6. [DOI] [PubMed] [Google Scholar]
- Huang YZ, Wang Q, Xiong WC, Mei L. Erbin is a protein concentrated at postsynaptic membranes that interacts with PSD-95. J Biol Chem. 2001;276:19318–19326. doi: 10.1074/jbc.M100494200. [DOI] [PubMed] [Google Scholar]
- Huang YZ, Zang M, Xiong WC, Luo Z, Mei L. Erbin suppresses the MAP kinase pathway. J Biol Chem. 2003;278:1108–1114. doi: 10.1074/jbc.M205413200. [DOI] [PubMed] [Google Scholar]
- Izawa I, Nishizawa M, Ohtakara K, Inagaki M. Densin-180 interacts with delta-catenin/neural plakophilin-related armadillo repeat protein at synapses. J Biol Chem. 2002;277:5345–5350. doi: 10.1074/jbc.M110052200. [DOI] [PubMed] [Google Scholar]
- Kallay LM, McNickle A, Brennwald PJ, Hubbard AL, Braiterman LT. Scribble associates with two polarity proteins, Lgl2 and Vangl2, via distinct molecular domains. J Cell Biochem. 2006;99:647–664. doi: 10.1002/jcb.20992. [DOI] [PubMed] [Google Scholar]
- Lassila M, Juhila J, Heikkila E, Holthofer H. Densin is a novel cell membrane protein of sertoli cells in the testis. Mol Reprod Dev. 2007;74:641–645. doi: 10.1002/mrd.20659. [DOI] [PubMed] [Google Scholar]
- Lee CJ, Irizarry K. Alternative splicing in the nervous system: an emerging source of diversity and regulation. Biol Psychiatry. 2003;54:771–776. doi: 10.1016/s0006-3223(03)00375-5. [DOI] [PubMed] [Google Scholar]
- Legouis R, Gansmuller A, Sookhareea S, Bosher JM, Baillie DL, Labouesse M. LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat Cell Biol. 2000;2:415–422. doi: 10.1038/35017046. [DOI] [PubMed] [Google Scholar]
- Legouis R, Jaulin-Bastard F, Schott S, Navarro C, Borg JP, Labouesse M. Basolateral targeting by leucine-rich repeat domains in epithelial cells. EMBO Rep. 2003;4:1096–1102. doi: 10.1038/sj.embor.7400006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill RB, Colbran RJ. Interaction of autophosphorylated Ca2+/calmodulin-dependent protein kinase II with neuronal cytoskeletal proteins. Characterization of binding to a 190-kDa postsynaptic density protein. J Biol Chem. 1995;270:10043–10049. doi: 10.1074/jbc.270.17.10043. [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary H, Lasda E, Bayer KU. CaMKIIbeta association with the actin cytoskeleton is regulated by alternative splicing. Mol Biol Cell. 2006;17:4656–4665. doi: 10.1091/mbc.E06-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtakara K, Nishizawa M, Izawa I, Hata Y, Matsushima S, Taki W, Inada H, Takai Y, Inagaki M. Densin-180, a synaptic protein, links to PSD-95 through its direct interaction with MAGUIN-1. Genes Cells. 2002;7:1149–1160. doi: 10.1046/j.1365-2443.2002.00589.x. [DOI] [PubMed] [Google Scholar]
- Quitsch A, Berhorster K, Liew CW, Richter D, Kreienkamp HJ. Postsynaptic shank antagonizes dendrite branching induced by the leucine-rich repeat protein Densin-180. J Neurosci. 2005;25:479–487. doi: 10.1523/JNEUROSCI.2699-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala R, Banine F, Borg JP, Sherman LS. Erbin regulates mitogen-activated protein (MAP) kinase activation and MAP kinase-dependent interactions between Merlin and adherens junction protein complexes in Schwann cells. J Biol Chem. 2005;280:11790–11797. doi: 10.1074/jbc.M414154200. [DOI] [PubMed] [Google Scholar]
- Rinta-Valkama J, Aaltonen P, Lassila M, Palmen T, Tossavainen P, Knip M, Holthofer H. Densin and filtrin in the pancreas and in the kidney, targets for humoral autoimmunity in patients with type 1 diabetes. Diabetes Metab Res Rev. 2007;23:119–126. doi: 10.1002/dmrr.655. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Bass MA, Jiao Y, MacMillan LB, Carmody LC, Bartlett RK, Colbran RJ. Multivalent interactions of calcium/calmodulin-dependent protein kinase II with the postsynaptic density proteins NR2B, densin-180, and alpha-actinin-2. J Biol Chem. 2005;280:35329–35336. doi: 10.1074/jbc.M502191200. [DOI] [PubMed] [Google Scholar]
- Saito H, Santoni MJ, Arsanto JP, et al. Lano, a novel LAP protein directly connected to MAGUK proteins in epithelial cells. J Biol Chem. 2001;276:32051–32055. doi: 10.1074/jbc.C100330200. [DOI] [PubMed] [Google Scholar]
- Santoni MJ, Pontarotti P, Birnbaum D, Borg JP. The LAP family: a phylogenetic point of view. Trends Genet. 2002;18:494–497. doi: 10.1016/s0168-9525(02)02738-5. [DOI] [PubMed] [Google Scholar]
- Shen K, Meyer T. In vivo and in vitro characterization of the sequence requirement for oligomer formation of Ca2+/calmodulin-dependent protein kinase IIalpha. Journal of neurochemistry. 1998;70:96–104. doi: 10.1046/j.1471-4159.1998.70010096.x. [DOI] [PubMed] [Google Scholar]
- Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Strack S, Robison AJ, Bass MA, Colbran RJ. Association of calcium/calmodulin-dependent kinase II with developmentally regulated splice variants of the postsynaptic density protein densin-180. J Biol Chem. 2000;275:25061–25064. doi: 10.1074/jbc.C000319200. [DOI] [PubMed] [Google Scholar]
- Teti A, Migliaccio S, Baron R. The role of the alphaVbeta3 integrin in the development of osteolytic bone metastases: a pharmacological target for alternative therapy? Calcif Tissue Int. 2002;71:293–299. doi: 10.1007/s00223-001-2071-1. [DOI] [PubMed] [Google Scholar]
- Walikonis RS, Oguni A, Khorosheva EM, Jeng CJ, Asuncion FJ, Kennedy MB. Densin-180 forms a ternary complex with the (alpha)-subunit of Ca2+/calmodulin-dependent protein kinase II and (alpha)-actinin. J Neurosci. 2001;21:423–433. doi: 10.1523/JNEUROSCI.21-02-00423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip PM, Zhao X, Montgomery AM, Siu CH. The Arg-Gly-Asp motif in the cell adhesion molecule L1 promotes neurite outgrowth via interaction with the alphavbeta3 integrin. Mol Biol Cell. 1998;9:277–290. doi: 10.1091/mbc.9.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.