Abstract
Geranyl diphosphate (GPP), the precursor of many monoterpene end products, is synthesized in plastids by a condensation of dimethylallyl diphosphate and isopentenyl diphosphate (IPP) in a reaction catalyzed by homodimeric or heterodimeric GPP synthase (GPPS). In the heterodimeric enzymes, a noncatalytic small subunit (GPPS.SSU) determines the product specificity of the catalytic large subunit, which may be either an active geranylgeranyl diphosphate synthase (GGPPS) or an inactive GGPPS-like protein. Here, we show that expression of snapdragon (Antirrhinum majus) GPPS.SSU in tobacco (Nicotiana tabacum) plants increased the total GPPS activity and monoterpene emission from leaves and flowers, indicating that the introduced catalytically inactive GPPS.SSU found endogenous large subunit partner(s) and formed an active snapdragon/tobacco GPPS in planta. Bimolecular fluorescence complementation and in vitro enzyme analysis of individual and hybrid proteins revealed that two of four GGPPS-like candidates from tobacco EST databases encode bona fide GGPPS that can interact with snapdragon GPPS.SSU and form a functional GPPS enzyme in plastids. The formation of chimeric GPPS in transgenic plants also resulted in leaf chlorosis, increased light sensitivity, and dwarfism due to decreased levels of chlorophylls, carotenoids, and gibberellins. In addition, these transgenic plants had reduced levels of sesquiterpene emission, suggesting that the export of isoprenoid intermediates from the plastids into the cytosol was decreased. These results provide genetic evidence that GPPS.SSU modifies the chain length specificity of phylogenetically distant GGPPS and can modulate IPP flux distribution between GPP and GGPP synthesis in planta.
INTRODUCTION
Terpenoids, the largest class of plant metabolites, play numerous vital roles in basic plant processes, such as respiration, photosynthesis, growth, development, reproduction, and adaptation to environmental conditions (Gershenzon and Kreis, 1999; Rodríguez-Concepción and Boronat, 2002). In addition to their roles as hormones and structural components of the photosynthetic apparatus, membranes, and cell walls, specialized terpenoids synthesized in different plant lineages mediate antagonistic and beneficial interactions among organisms and defend the plant against predators and pathogens (Gershenzon and Dudareva, 2007). Plant-produced terpenoids are essential nutrients in human diets and are used as natural flavor and aroma compounds or as chemotherapeutic agents with antitumor activities (Wagner and Elmadfa, 2003).
Terpenoid biosynthesis in plants occurs in different subcellular compartments, including the cytosol, plastid, and mitochondria. Two compartmentally separated metabolic pathways provide the universal five-carbon isoprenoid precursors, isopentenyl diphosphate (IPP) and its allylic isomer dimethylallyl diphosphate (DMAPP), for terpenoid formation (McGarvey and Croteau, 1995). IPP, derived from the classical mevalonic acid (MVA) pathway, localized mostly in the peroxisomes (Sapir-Mir et al., 2008), serves as a precursor for farnesyl diphosphate (FPP; C15) and, ultimately, the cytosolically synthesized sesquiterpenes and triterpenes, including sterols (Newman and Chappell, 1999). IPP and DMAPP formed via the plastidic methyl-erythritol-phosphate (MEP) pathway provide the precursors for geranyl diphosphate (GPP; C10), geranylgeranyl diphosphate (GGPP; C20), and, ultimately, the monoterpenes, diterpenes, and tetraterpenes (Lichtenthaler, 1999; Rohmer, 1999). Metabolic flux through these pathways can differ dramatically, and the flux through the MEP pathway often exceeds that of the MVA (McCaskill and Croteau, 1995; Dudareva et al., 2005; Wu et al., 2006). It was reported that IPP and perhaps DMAPP could be exchanged between different compartments, particularly in the direction from plastids to cytosol (Hemmerlin et al., 2003; Laule et al., 2003; Schuhr et al., 2003; Dudareva et al., 2005; Phillips et al., 2008), raising a question about the relative contribution of each pathway to the biosynthesis of the various classes of terpenoids. Trafficking of isoprenoid intermediates between organelles, likely mediated by specific metabolite transporters (Bick and Lange, 2003), depends on the plant species, tissue, and physiological state of the plant (Steliopoulos et al., 2002; Dudareva et al., 2005; Hampel et al., 2005a, 2005b).
GPP is the entry point leading to the biosynthesis of many monoterpene end products. It is synthesized in a reaction catalyzed by geranyl diphosphate synthase (GPPS), for which both homodimeric and heterodimeric architecture has been found, depending on the plant species (Burke et al., 1999, Burke and Croteau, 2002b; Tholl et al., 2004; Schmidt and Gershenzon, 2008; Wang and Dixon, 2009). The first heterodimeric GPPS containing large and small subunits was isolated from peppermint (Mentha piperita) glandular trichomes via enzyme purification and protein sequencing (Burke et al., 1999). While both proteins belong to the trans-prenyltransfrease family, the large subunit was similar to bona fide geranylgeranyl diphopsphate synthases (GGPPSs) (Burke et al., 1999; Burke and Croteau, 2002a), and the small subunit exhibited only a low level of homology to existing prenyltransferases (Burke et al., 1999). Both subunits were catalytically inactive when expressed alone in Escherichia coli but formed an active GPPS when coexpressed together (Burke et al., 1999). Subsequently, genes encoding proteins with high similarity to the mint GPPS small subunit (GPPS.SSU) were found in several plant species, including snapdragon (Antirrhinum majus), Clarkia breweri, and hop (Humulus lupulus), which are known to produce large amounts of monoterpenes in specific organs (Tholl et al., 2004; Wang and Dixon, 2009).
With the exception of mint GPPS, protein sequences have not been obtained from plant-purified GPPS. Attempts to identify GPPS in a given plant species have relied primarily on mining DNA databases, expressing candidate genes in a bacterial system, and testing the resulting proteins in vitro (Tholl et al., 2004; Wang and Dixon, 2009). In plants where no genes were found to be orthologous to GPPS.SSU, sequences with homology to trans-prenyltransferases were studied and shown to encode proteins capable of forming homodimers with GPPS activity in E. coli (Bouvier et al., 2000; Burke and Croteau, 2002b, Schmidt and Gershenzon, 2008).
The GPPS.SSU proteins, which are always inactive when produced in E. coli by themselves, constitute a distinct clade of trans-prenyltransferases (Wang and Dixon, 2009). By contrast, there is uncertainty about the identity of GPPS.LSU proteins, since the mint GPPS.LSU sequence was found to be similar to bona fide GGPPS enzymes, and typically plant genomes have multiple genes encoding GGPPS (Okada et al., 2000; Ament et al., 2006; Pateraki and Kanellis, 2008). While the mint large subunit purified from plant is inactive by itself, potential GPPS.LSU in snapdragon and hop represent active GGPPS and change their product specificity to GPP formation upon interaction with GPPS.SSU (Tholl et al., 2004; Wang and Dixon, 2009). Moreover, the ability of GPPS.SSU to modify the chain length specificity of phylogenetically distant bona fide GGPPSs in vitro (Burke and Croteau, 2002a; Tholl et al., 2004; Wang and Dixon, 2009) left the question of whether plants in general have specific genes for GPPS large subunit (GPPS.LSU) or whether GGPPS genes are recruited for this purpose. In both snapdragon and hop, the expression of GPPS.SSU, but not the potential GPPS.LSU, closely correlated with monoterpene emission, suggesting that the small subunit plays a key role in regulating GPP formation (Tholl et al., 2004; Wang and Dixon, 2009).
It is therefore possible that GPPS enzymatic activity and thus the modulation of flux to monoterpene biosynthesis is achieved through the regulation of GPPS.SSU gene expression and the recruitment of bona fide GGPPS proteins. However, genetic evidence for this scenario in planta is lacking. Information on such a mechanism would not only be of interest in understanding terpene biosynthesis, but would also be beneficial in attempts to increase or otherwise modify the synthesis of important terpene products in transgenic plants (Croteau et al., 2005; Wildung and Croteau, 2005). Here, we tested whether snapdragon GPPS.SSU, when overexpressed in tobacco (Nicotiana tabacum), is capable of finding an endogenous GGPPS partner(s), modifying its product specificity, and shifting the metabolic flux toward GPP formation, thus affecting terpenoid biosynthesis in planta.
RESULTS
Generation of Transgenic Tobacco Plants Expressing the Small Subunit of Snapdragon GPPS
To determine whether snapdragon (Am) GPPS.SSU could form a functional heterodimer with phylogenetically distant GGPPS in planta and increase the flux toward GPP formation, we generated transgenic tobacco plants overexpressing Am GPPS.SSU under the control of the petal-specific C. breweri linalool synthase (LIS) promoter (Cseke et al., 1998). This promoter was chosen to limit Am GPPS.SSU expression to flowers in order to avoid potentially deleterious effects on plant growth and development. Seven independent lines with different Am GPPS.SSU levels, as determined by protein gel blot analysis with antibodies generated against the snapdragon GPPS.SSU (Tholl et al., 2004), were obtained and adapted to greenhouse conditions. The Am GPPS.SSU protein level in these transgenics was highest in the GPPS.SSU-5 to GPPS.SSU-7 lines, intermediate in the GPPS.SSU-1 line (68% of that in GPPS.SSU-7), low in the GPPS.SSU-3 line (8% of GPPS.SSU-7; 12% of that in GPPS.SSU-1), and undetectable in the GPPS.SSU-2 and GPPS.SSU-4 lines (Figure 1A). The four transgenic lines with the highest and moderate levels of Am GPPS.SSU protein (GPPS.SSU-5 to GPPS.SSU-7 and GPPS.SSU-1 lines, respectively) showed strong chlorosis and a reduction in stature (Figures 1A to 1C), while lines with low and undetectable Am GPPS.SSU protein levels (GPPS.SSU-3 and GPPS.SSU-2/GPPS.SSU-4 lines, respectively) showed no visible differences from control untransformed tobacco plants. Only plants with low (GPPS.SSU-3) and moderate (GPPS.SSU-1) Am GPPS.SSU protein levels were able to mature and produce viable seeds. Their T1 progenies showed 3:1 segregation for hygromycin resistance, indicating that only one copy of the Am GPPS.SSU was incorporated.
Figure 1.
Effect of Am GPPS.SSU Expression on the Phenotype of Transgenic Tobacco Plants.
(A) Am GPPS.SSU protein levels in different transgenic tobacco lines. The representative protein gel blot shows the 30-kD protein recognized by anti-Am GPPS.SSU antibodies. Proteins were extracted from tobacco leaves of untransformed control and seven independent transgenic lines (shown on the top), and 20 μg of protein were loaded in each lane. The blot shown represents a typical result of three independent experiments.
(B) Effect of high levels of Am GPPS.SSU protein on the phenotype of transgenic tobacco plants. Three transgenic lines with the highest levels of Am GPPS.SSU protein (GPPS.SSU-5 to GPPS.SSU-7) show strong chlorosis and dwarfism relative to the control plant shown in (C).
(C) Phenotype of GPPS.SSU-1 line, which in contrast with GPPS.SSU-5 to GPPS.SSU-7, was able to produce mature plants. The inset shows leaves from control (left) and GPPS.SSU-1 transgenic plants (right). All plants in (B) and (C) are 6 to 7 weeks old.
The GPPS activities in the partially purified chloroplasts from transgenic lines GPPS.SSU-7 (1.75 pkat/mg protein), GPPS.SSU-1 (1.41 pkat/mg protein), and GPPS.SSU-3 (0.40 pkat/mg protein) were ∼8.3-, ∼7-, and ∼2-fold higher, respectively, than the activity detected in chloroplasts of control plants (0.21 pkat/mg protein; Table 1) and positively correlated with the corresponding Am GPPS.SSU protein levels (see Supplemental Figure 1A online).
Table 1.
GPPS and GGPPS Activities in the Isolated Chloroplasts from Control and Transgenic Leaves
| Activity | Control | GPPS.SSU-1 | GPPS.SSU-3 | GPPS.SSU-7 |
|---|---|---|---|---|
| GPPS | 0.21 ± 0.12 | 1.41 ± 0.13** | 0.40 ± 0.03* | 1.75 ± 0.18** |
| GGPPS | 0.42 ± 0.08 | 0.23 ± 0.06** | 0.28 ± 0.08* | 0.12 ± 0.01** |
Values are pkat/mg protein (means ± se, n = 3). Confidence levels were tested by a Student's t test; *, P < 0.01; **, P < 0.003.
Effect of Snapdragon GPPS Small Subunit Expression on the Profile of Emitted Terpenoids
GPPS.SSU-1 and GPPS.SSU-3 tobacco lines producing mature plants were chosen for further analyses. RNA gel blot analysis of Am GPPS.SSU transcript levels in both lines revealed that the transgene was expressed in flowers and leaves despite the use of the petal-specific LIS promoter (see Supplemental Figure 2 online). The level of Am GPPS.SSU transcripts in the GPPS.SSU-3 leaves, determined by quantitative real-time PCR, was 20% of that in the GPPS.SSU-1 line, consistent with the amount of Am GPPS.SSU protein (Figure 1A) and GPPS activities detected in these lines (Table 1).
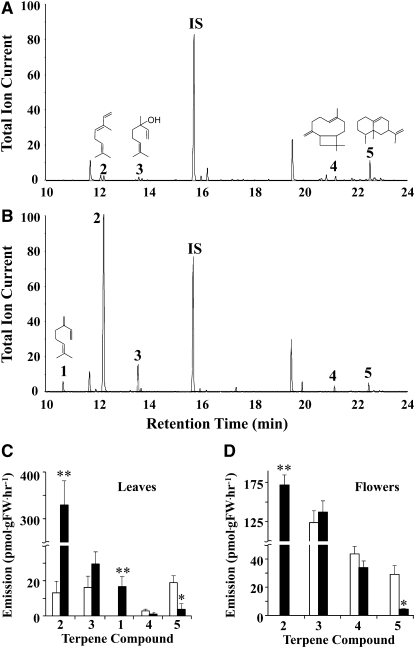
To examine the effect of Am GPPS.SSU overexpression on the level of emitted terpenoids, volatiles were collected from flowers and leaves after 24 h for two consecutive days and analyzed by gas chromatography–mass spectrometry (GC-MS). Leaves of control tobacco plants emit small amounts of the monoterpenes linalool and (E)-β-ocimene at similar levels, as well as the sesquiterpenes β-caryophyllene and 5-epi-aristolochene, with the latter representing the major sesquiterpene constituent (Figure 2A). Leaves of GPPS.SSU-1 transgenic plants showed a 25-fold increase in (E)-β-ocimene emission relative to the control and also began to emit a new monoterpene compound, myrcene (Figures 2B and 2C). No statistically significant changes were observed in the level of emitted linalool (Figures 2A to 2C). The overall increase in monoterpene emission by almost 12-fold (4.3 and 51.8 ng/gFW·h−1 in control and GPPS.SSU-1 leaves, respectively) was accompanied by a decrease in the total emission of sesquiterpenes including 2.4- and 5-fold decreases in β-caryophyllene and 5-epi-aristolochene, respectively (Figure 2C).
Figure 2.
Effect of Am GPPS.SSU Expression on Terpenoid Emission from Leaves and Flowers of Transgenic Tobacco Plants.
(A) and (B) Metabolic profiling of volatiles emitted from leaves of untransformed control (A) and transgenic GPPS.SSU-1 (B) tobacco plants. Volatiles collected from detached leaves were analyzed by electron ionization GC-MS, and total ion currents are plotted. Compounds were identified based on their mass spectra and retention time: 1, myrcene; 2, (E)-β-ocimene; 3, linalool: IS, internal standard (naphthalene); 4, β-caryophyllene; 5, 5-epi-aristolochene.
(C) and (D) Quantitative changes in terpenoids emitted from leaves (C) and flowers (D) of GPPS.SSU-1 tobacco plants relative to the control. White and black bars represent the amount of terpenoids in the control and transgenic line, respectively. Data are means ± sd (n = 3 to 10). Asterisks indicate values that are significantly different from the control. Confidence levels were tested by a Student's t test; *, P < 0.05; **, P < 0.001.
Similar to leaves, flowers of control tobacco plants produced the same spectrum of terpenoid compounds, with the exception of (E)-β-ocimene, which was absent in the floral scent profile (Figure 2D). The total amount of emitted terpenoids was almost 4-fold higher in flowers (33.9 ng·gFW·h−1) than in leaves (8.7 ng·gFW·h−1), with linalool being the major scent compound in flowers. Both tissues produce similar levels of 5-epi-aristolochene (18.9 ± 4 and 29.1 ± 6 pmol·gFW·h−1 in leaves and flowers, respectively), while the amount of linalool and β-caryophyllene were 7.7- and 15.3-fold higher in flowers than leaves (Figures 2C and 2D).
Expression of Am GPPS.SSU in the GPPS.SSU-1 transgenic line did not significantly affect the level of emitted linalool and β-caryophyllene but resulted in significant emission of (E)-β-ocimene, which was not detected in control flowers but became the major scent compound in this transgenic line (Figure 2D). Similar to leaves, the level of 5-epi-aristolochene emitted from flowers was decreased by 6.7-fold relative to the control. The leaves and flowers of GPPS.SSU-1 transgenic tobacco line produced equal levels of total terpenoid volatiles (52.8 versus 52.3 ng·gFW·h−1). Terpenoid emission from the leaves and flowers of the GPPS.SSU-3 transgenic line were not statistically different from that of the control plants, although GPPS activity was increased by almost 2-fold (Table 1).
Methyl jasmonate (MeJA) is known to simulate insect or pathogen attack and induce production of terpenoid compounds involved in plant defense (Martin et al., 2003; Phillips et al., 2007). Indeed, MeJA treatment increased the level of emitted (E)-β-ocimene by 620-fold from leaves of control tobacco plants (Table 2). However, linalool emission was almost unaffected by MeJA treatment despite equal levels of these two monoterpenes in untreated leaves. To examine the impact of increased flux to GPP formation on a plant's response to MeJA, leaves of GPPS.SSU-1 tobacco plants were also treated with MeJA and emitted terpenoids were analyzed. Emission of (E)-β-ocimene and linalool in MeJA-treated transgenic leaves was increased 160- and 3.5-fold, respectively (Table 2). These transgenic leaves emitted 6.6- and 3.6-fold more (E)-β-ocimene and linalool, respectively, than control plants under the same treatment. In addition, MeJA induced the emission of a new monoterpene, limonene, in transgenic leaves but not in control and increased the levels of emitted myrcene by 5.9-fold relative to untreated ones.
Table 2.
Effect of MeJA and Fosmidomycin Treatments on Monoterpene Emission from Control and GPPS.SSU-1 Leaves
| Control (pmol/gFW·h−1) |
GPPS.SSU-1 (pmol/gFW·h−1) |
|||||
|---|---|---|---|---|---|---|
| Compound | Means ± se No Treatment | MeJA | MeJA+ Fa | Means ± se No Treatment | MeJA | MeJA+ Fa |
| Myrcene | 0 | 0 | 0 | 16.7 ± 5.6 | 98.7 ± 26.7 | 0 |
| Limonene | 0 | 0 | 0 | 0 | 10.3 ± 4.3 | 0 |
| Ocimene | 13.1 ± 6.6 | 8,081 ± 2,498 | 95.4 | 330 ± 52 | 53,050 ± 3,800 | 1,020 |
| Linalool | 16.2 ± 6.5 | 29.2 ± 5.3 | 1.5 | 29.5 ± 6.9 | 104 ± 29 | 10.4 |
F, fosmidomycin.
Fosmidomycin, a specific inhibitor of the first committed enzyme of the plastidial MEP pathway, 1-deoxy-d-xylulose-5-phosphate reductoisomerase (DXR) (Kuzuyama et al., 1998), completely eliminated myrcene and limonene emission in transgenic leaves and decreased emitted ocimene by 98 to 99% in both control and transgenics treated with MeJA. Although the linalool level was very similar to that of myrcene (98.7 and 104 pmol/gFW·h−1, respectively) in transgenic leaves after MeJA treatment, fosmidomycin had less effect on linalool emission relative to myrcene, decreasing it by 90 and 95% in transgenic and control leaves, respectively (Table 2).
Effect of Am GPPS.SSU Expression in Tobacco Plants on GGPP-Derived Isoprenoids
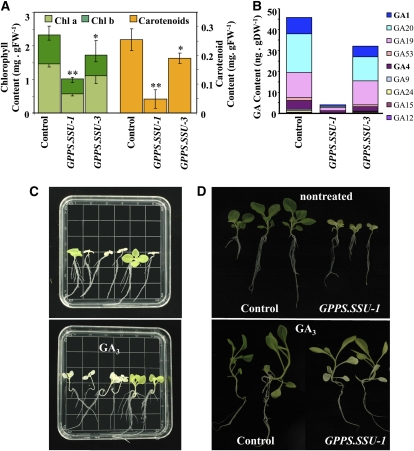
Similar to transgenic plants with the highest Am GPPS.SSU levels (GPPS.SSU-5 to GPPS.SSU-7), the GPPS.SSU-1 tobacco plants exhibited leaf chlorosis, elevated light sensitivity, and delay in their growth when compared with control plants (Figure 1C). Their increased light sensitivity required growing these plants under decreased light intensity (30 to 40 μmol·m−2.s−1), which was accomplished by using shade cloth. Analysis of various plastid-derived isoprenoids revealed that the amount of total chlorophyll in the leaves of these transgenic plants, as well as chlorophyll a and b, was ∼2.5-fold lower than that in the control leaves (Figure 3A). Moreover, the Am GPPS.SSU expression in tobacco affected the total carotenoid level, which was reduced by 5.2-fold when compared with control leaves. Similar to GPPS.SSU-1, the GPPS.SSU-3 transgenic tobacco plants contained 30% less total chlorophylls and carotenoids than control plants (P < 0.05, Student's t test). Notably, the total chlorophyll levels showed negative correlation with GPPS activities in transgenic lines when the amount of chlorophyll detected in line GPPS.SSU-7 was also included in this analysis (see Supplemental Figure 1B online).
Figure 3.
Effect of Am GPPS.SSU Expression on GGPP-Derived Isoprenoids in Transgenic Tobacco Plants.
(A) Chlorophyll and carotenoid contents in the leaves of control and transgenic lines GPPS.SSU-1 and GPPS.SSU-3. Data are means ± sd (n = 3 to 10). Asterisks indicate values that are significantly different from the control. Confidence levels were tested by a Student's t test; * P < 0.05; ** P < 0.001.
(B) Levels of GAs in the leaves of control and transgenic lines GPPS.SSU-1 and GPPS.SSU-3.
(C) Germination of T1 seeds of GPPS.SSU-1 line without (top panel) or with (bottom panel) 30 μM GA3. Resulting 10-d-old T1 seedlings show the expected segregation of the wild type and bleached/reduced growth phenotype. Addition of GA3 rescued the growth of transgenic seedlings.
(D) Continued treatment of transgenic GPPS.SSU-1 seedlings with GA3 recovered their growth to almost that of control plants. Fourteen-day-old plants grown without (top panel) or with (bottom panel) 30 μM GA3 are shown.
Since a reduction in stature of the GPPS.SSU-1 transgenic tobacco plants could be the result of changes in the level of gibberellins (GAs), growth regulators that are formed via the plastidic MEP pathway, endogenous GA levels were analyzed in leaves of control and transgenic plants. The total GA levels in the GPPS.SSU-1 and GPPS.SSU-3 transgenics were 9 and 70% of that in the control leaves (Figure 3B) and were negatively correlated with the Am GPPS.SSU gene expression in these transgenic plants (see Supplemental Figure 2 online). Both bioactive GA4 and GA1, as well as their inactive precursors, contributed to the reduction in the total amount of GAs (Table 3). Addition of GA3 to the growth medium partially rescued leaf size and hypocotyl length of 10-d-old GPPS.SSU-1 transgenic tobacco seedlings, which showed significant improvement in their development relative to untreated plants, with some even becoming similar in size to control seedlings despite their chlorosis (Figure 3C). Continuous growth of these transgenics in the presence of GA3 resulted in 1-month-old GPPS.SSU-1 plants that exhibited similar growth when compared with untransformed control (Figure 3D). To test whether the growth retardation in transgenic tobacco seedlings is due to the reduction in flux toward GA formation, transgenic seedlings were grown in the presence of the volatile ent-kaurene (5 μg/plate), a diterpene precursor of GAs. The Arabidopsis thaliana mutant gal3, deficient in ent-kaurene biosynthesis (Otsuka et al., 2004), was used as a positive control in these experiments. Ent-kaurene had no effect on control tobacco or wild-type Arabidopsis seedlings, but completely rescued the gal3 phenotype. GPPS.SSU-1 seedlings were able to take up ent-kaurene from the air and showed an improved rosette growth with leaf area 40% ± 1.1% larger (P < 0.001) than that in untreated GPPS.SSU-1 seedlings (see Supplemental Figure 3 online). In the GPPS.SSU-3 transgenic plants, the 1.5- to 2-fold decrease in the levels of bioactive GA4 and GA1 (Table 3) had no visible effect on their growth.
Table 3.
Endogenous GA Levels in Leaves of Control and Transgenic Plants
| GA | Control (ng/gDW) | GPPS.SSU-1 (ng/gDW) | GPPSSSU-3 (ng/gDW) |
|---|---|---|---|
| GA4a | 7.88/6.76 | 1.2/0.77 | 1.81/2.71 |
| GA12 | 0.34/0.12 | ND/0.07 | 0.18/0.1 |
| GA15 | 0.69/0.1 | ND/ND | 0.02/0.08 |
| GA24 | 0.71/0.38 | 0.15/0.11 | 0.48/0.33 |
| GA9 | 1.38/– | 0.16/– | 0.48/0.31 |
| GA1a | 9.62/9.14 | 0.71/4.11 | 5.93/4.57 |
| GA53 | 5.0/1.52 | ND/ND | 0.99/0.84 |
| GA19 | 15.9/9.29 | 2.12/1.01 | 17.9/9.66 |
| GA20 | 25.3/17.8 | 0.7/0.39 | 8.57/15.6 |
GA measurements were performed twice using independently prepared plant material and are shown on the left and right, respectively, for each compound. –, Amount was detectable but was not quantified due to comigration of impurities. ND, not detected.
Bioactive GAs are shown in bold.
The reduction of chlorophyll and GA levels in transgenic tobacco plants could be the result of either reduced flux through the MEP pathway, GGPPS elimination from its active enzyme pool via the interaction with Am GPPS.SSU, or the reduction in flux to GGPP due to the redirection of IPP and DMAPP toward GPP formation. The expression levels of 1-deoxy-d-xylulose-5-phosphate synthase (DXS) and DXR, the first two genes that control the flux through the MEP pathway (Takahashi et al., 1998; Estévez et al., 2001), were analyzed in control and transgenic tobacco plants. Quantitative real-time PCR analysis revealed no differences in DXS or DXR mRNA levels in leaves of GPPS.SSU-1 plants relative to control (see Supplemental Figure 4 online). When the GGPPS activity was analyzed in partially purified chloroplasts from leaves of control and transgenic plants, 47 and 35% reductions were observed in GPPS.SSU-1 (0.23 pkat/mg protein) and GPPS.SSU-3 (0.28 pkat/mg protein) chloroplasts, respectively, relative to control (0.43 pkat/mg protein) (Table 1).
Identification of Endogenous Tobacco Partner(s) for Snapdragon GPPS.SSU
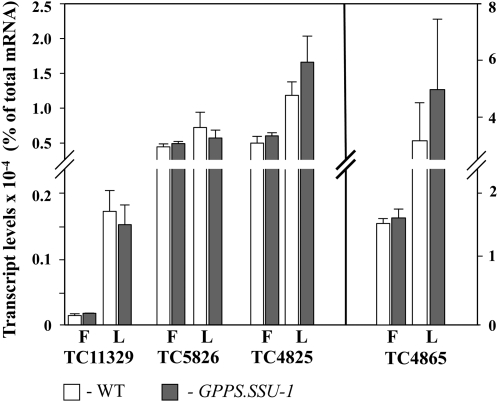
Snapdragon GPPS.SSU is inactive as a homodimer (or as a monomer) (Tholl et al., 2004), and in order to contribute to GPP formation in tobacco, it has to find a partner(s) to form a functional heterodimer. Potential partners could include inactive GGPPS-like proteins (Burke et al., 1999) or active GGPPS that change their chain length specificity upon the interaction with GPPS.SSU (Burke and Croteau, 2002a; Tholl et al., 2004). Thus, we searched the publicly available tobacco EST database for sequences with similarity to Am GPPS.LSU, which itself is a functional GGPPS in the homodimeric state (Tholl et al., 2004). This search revealed four tentative consensus (TC) cDNA sequences, TC4825, TC4865, TC5826, and TC11329, with amino acid identities ranging from 34 to 65% to snapdragon GPPS.LSU (see Supplemental Figure 5 online), with highest values for TC11329 (65% identity; 79% similarity) and TC5826 (61% identity; 76% similarity). TC4865 was a full-length clone, which allowed us to amplify its open reading frame by RT-PCR using mRNA derived from tobacco leaves and clone-specific primers. TC11329 initially lacked its 5′ region, which was obtained based on its high homology to Nicotiana benthamiana and Nicotiana attenuata GGPPSs (see Methods). The missing 3′-translated regions of TC5826 and TC4825 were recovered by 3′ rapid amplification of cDNA ends (RACE). The lengths of predicted TC5826 and TC11329 proteins (365 and 366 amino acids, respectively) were the closest to snapdragon GPPS.LSU (372 amino acids), while the other two, TC4825 and TC4865, were 332 and 402 amino acids in size, respectively. TC4825, TC5826, and TC11329 were annotated as GGPPS-like genes and TC4865 as solanesyl diphosphate synthase. All proteins contain characteristic N-terminal transit peptides, suggesting their plastidic localization. Genes encoding these proteins were expressed in both leaves and flowers of tobacco plants with the highest transcript abundance for TC4865 and the lowest for TC11329 (Figure 4). Expressions of all genes, with the exception of TC5826, were higher in leaves than in flowers, and their levels in transgenic plants were not statistically different from those in the controls (Figure 4).
Figure 4.
Expression Levels of Genes Encoding Potential Endogenous Tobacco Partners for Am GPPS.SSU in the Control and GPPS.SSU-1 Transgenic Line.
Quantitative RT-PCR analysis of expression of tobacco candidates for GPPS.LSU in leaves (L) and flowers (F) of control (white) and GPPS.SSU-1 (gray) is shown. Expression is represented as a percentage of the large subunit candidate mRNA to total mRNA. Each point is the average of three independent experiments. Standard error values are indicated by vertical bars.
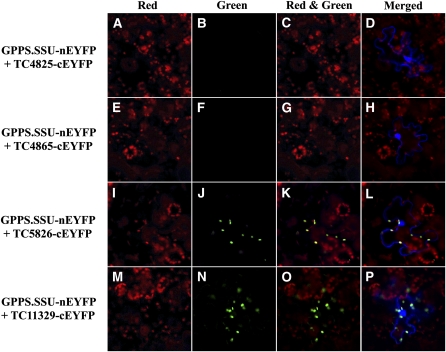
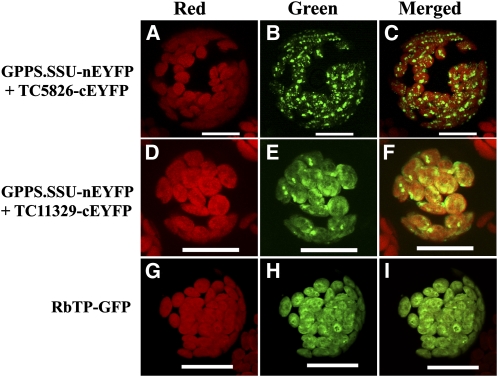
To determine which of the four tobacco candidate genes encode a protein that can interact with Am GPPS.SSU, we employed bimolecular fluorescence complementation (BiFC), which allows in vivo detection of protein–protein interactions as well as subcellular localization of interacting proteins (Citovsky et al., 2008). Of the four potential partners, only TC5826 and TC11329, the two proteins with the highest similarity to snapdragon GPPS.LSU (see Supplemental Figure 5B online), interacted with Am GPPS.SSU, and the resulting heterodimers were localized to plastids similar to snapdragon heterodimeric GPPS, which is confined to the leucoplasts of the conical cell of the inner epidermis layer of flower petals (Tholl et al., 2004). The coexpression of Am GPPS.SSU tagged with the N-terminal part of enhanced yellow fluorescent protein (nEYFP) and tobacco TC5826 or TC11329 tagged with C-terminal part of EYFP (cEYFP) resulted in fluorescence complementation within the plastids of both N. benthamiana epidermal cells (Figures 5I to 5P) and Arabidopsis leaf protoplasts (Figure 6). No signal was observed when TC4825 and TC4865 proteins tagged with cEYFP were coexpressed with Am GPPS.SSU tagged with nEYFP (Figures 5A to 5H).
Figure 5.
BiFC Detection of Protein–Protein Interactions of Snapdragon Am GPPS.SSU with Tobacco GGPPS-Like Proteins in N. benthamiana Leaf Epidermal Cells.
(A) to (D) Coexpression of GPPS.SSU-nEYFP and TC4825-cEYFP.
(E) to (H) Coexpression of GPPS.SSU-nEYFP and TC4865-cEYFP.
(I) to (L) Coexpression of GPPS.SSU-nEYFP and TC5826-cEYFP.
(M) to (P) Coexpression of GPPS.SSU-nEYFP and TC11329-cEYFP.
Fluorescence of reconstructed YFP was detected in the green channel and is shown in the “Green” panel. Chlorophyll autofluorescence was detected in the red channel and is shown in the “Red” panel. The “Red and Green” panel shows merged chlorophyll autoflourescence and YFP signals. Cytosol and nuclear-localized cyan fluorescent protein (CFP) was used to demarcate the boundary of the cell. The “Merged” panel shows combined “Red and Green” and CFP signals.
Figure 6.
BiFC Detection of Protein–Protein Interactions of Am GPPS.SSU with Tobacco GGPPS-Like Proteins in Arabidopsis Leaf Protoplasts.
(A) to (C) Coexpression of Am GPPS.SSU-nEYFP and TC5826-cEYFP.
(D) to (F) Coexpression of Am GPPS.SSU-nEYFP and TC11329-cEYFP.
(G) to (I) Expression of the RbTP-GFP chloroplast marker.
Reconstructed YFP ([B] and [E]) and GFP (H) fluorescence was detected in the green channel and is shown in the “Green” panel; chlorophyll autofluorescence was detected in the red channel and is shown in the “Red” panel; the “Merged” panel shows combined green and red channels. Bars = 50 μm.
Functional Characterization of Am GPPS.SSU Interacting Partners and Chimeric Snapdragon/Tobacco GPPS
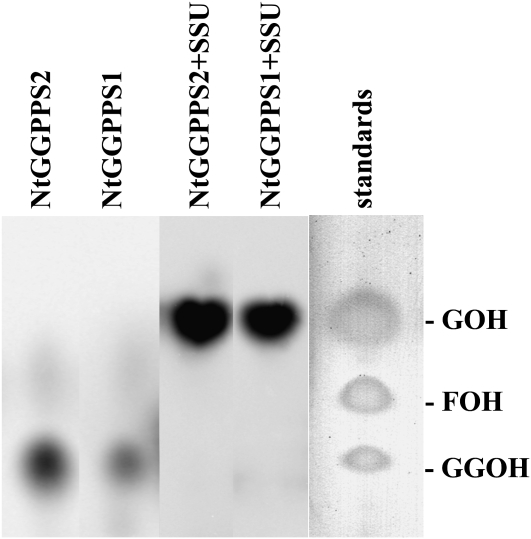
For the functional characterization of proteins showing interactions with Am GPPS.SSU, the coding regions of TC5826 and TC11329 cDNAs without their N-terminal transit peptides were expressed in E. coli as inducible fusion proteins containing a hexahistidine (His)6 tag. Both purified recombinant proteins were active GGPPSs and catalyzed the formation of GGPP from [1-14C]-IPP and its allylic cosubstrate DMAPP (Figure 7) as a major product with minor production of FPP (5 and 10% of total products for TC5826 and TC11329, respectively). These proteins were also able to use GPP and FPP as allylic cosubstrates with IPP, producing GGPP as the predominant product. Kinetic analysis revealed that, while the Km of TC5826 for FPP was almost 2-fold higher than that of TC11329, the recombinant proteins have very similar apparent Km values for DMAPP (15.5 and 22.8 μM for TC5826 and TC11329 proteins, respectively) and GPP (2.4 and 3.6 μM for TC5826 and TC11329, respectively) (Table 4). These values are within the range previously reported for other GGPPSs (Burke and Croteau, 2002a; Wang and Dixon, 2009). Based on this functional evaluation, the TC11329 and TC5826 proteins were designated as Nt GGPPS1 and GGPPS2, respectively.
Figure 7.
Products Generated by Tobacco GGPPS1, GGPPS2, and Chimeric Snapdragon/Tobacco Heterodimeric GPPSs from DMAPP and [1-14C]-IPP in Vitro.
Reaction products were hydrolyzed to their corresponding alcohols, extracted with hexane, and separated by reverse-phase thin layer chromatography. Authentic standards (GOH, geranol; FOH, farnesol; GGOH, geranylgeranol) were visualized by exposing thin layer chromatography plates to iodine vapor. Assays contained 5 μg of recombinant Nt GGPPS1 or Nt GGPPS2 proteins or 40 μg of hybrid heterodimers.
Table 4.
Kinetic Parameters of Tobacco GGPPSs, Recombinant, and Plant Snapdragon/Tobacco Chimeric GPPSs
| Enzyme K | Km IPP (μM)a | Km DMAPP (μM)b | Km MgCl2 (mM)c | Km GPP (μM)b | Km IPP (+GPP) (μM)d | Km FPP (μM)b | Km IPP (+FPP) (μM)e |
|---|---|---|---|---|---|---|---|
| Nt GGPPS1 (TC11329)f | 16.91 ± 0.51 | 22.81 ± 0.83 | 0.51 ± 0.03 | 3.63 ± 0.45 | 19.74 ± 1.89 | 28.82 ± 7.87 | 27.74 ± 7.71 |
| Nt GGPPS2 (TC5826)f | 11.29 ± 2.18 | 15.53 ± 5.03 | 0.19 ± 0.07 | 2.36 ± 0.51 | 19.53 ± 4.64 | 53.2 ± 12.52 | 20.06 ± 2.97 |
| SSU/NtGGPPS1f | 9.25 ± 1.9 | 58.86 ± 6.51 | 0.38 ± 0.09 | NDg | ND | ND | ND |
| SSU/NtGGPPS2f | 20.56 ± 2.54 | 29.04 ± 4.31 | 0.58 ± 0.14 | ND | ND | ND | ND |
| Tobacco hybrid GPPS | 10.56 ± 1.81 | 27.15 ± 8.9 | 0.41 ± 0.13 | ND | ND | ND | ND |
At 66 μM DMAPP.
At 53 μM IPP.
At 53 μM IPP and 66 μM DMAPP.
At 28 μM GPP.
At 42 μM GPP.
Truncated version of GGPPS corresponding to mature protein was used.
ND, activity not detected.
To determine whether the interactions between Am GPPS.SSU and these two tobacco GGPPSs lead to the formation of active functional heterodimers that produce GPP, Am GPPS.SSU cDNA with a (His6) tag extension was coexpressed with either Nt GGPPS1 or GGPPS2 lacking the transit peptides (both without a His tag), followed by affinity purification of recombinant heterodimers. The interaction of Am GPPS.SSU with tobacco GGPPSs changed their chain length product specificity and resulted in the exclusive production of GPP in the presence of IPP and DMAPP (Figure 7). The coexpression of Am GPPS.SSU with tobacco proteins encoded by TC4825 and TC4865, which did not show BiFC interactions with Am GPPS.SSU, resulted in no formation of active GPPS. Similar to recombinant heterodimeric GPPSs from mint and snapdragon (Burke et al., 1999; Tholl et al., 2004), the chimeric snapdragon/tobacco GPPSs were unable to use GPP or FPP as allylic cosubstrates with IPP. Kinetic characterization of chimeric proteins revealed that the GPPS.SSU-GGPPS2 heterodimer had very similar Km values for IPP and DMAPP (20.6 ± 2.5 μM and 29 ± 4.3 μM, respectively). By contrast, the GPPS.SSU-GGPPS1 heterodimer had an apparent Km value for IPP 6.4-fold lower than that for DMAPP (9.3 ± 1.9 and 58.9 ± 6.5 μM, respectively) (Table 4).
Chimeric GPPS was also partially purified from leaves of transgenic tobacco GPPS.SSU-1 plants using a weak anion-exchange DEAE-cellulose column followed by Mono-Q anion-exchange and gel filtration chromatography (see Supplemental Figure 6A online). Protein gel blot analysis confirmed that the purified protein fraction with GPPS activity contained Am GPPS.SSU. It had a molecular mass of 68.4 kD based on gel filtration chromatography, suggesting that it was composed of the mature 29-kD snapdragon small and the mature tobacco 36-kD large subunits (see Supplemental Figure 6B online) (molecular mass predicted by the iPSORT program [http://hc.ims.u-tokyo.ac.jp/iPSORT/] for mature Nt GGPPS1 was 36.2 kD and for mature Nt GGPPS2 was 36.6 kD). Similar to the recombinant snapdragon/tobacco GPPSs purified from E. coli (Figure 7, Table 4), the purified chimeric plant protein produced exclusively GPP from [1-14C]-IPP and DMAPP and was free from other prenyltransferase activities (see Supplemental Figure 6C online). Comparison of the kinetic behavior of this plant protein with that of recombinant snapdragon/tobacco enzymes revealed that it possesses characteristics of both hybrids with apparent Km values for IPP of 10.6 ± 1.8 μM and DMAPP of 27.2 ± 8.9 μM (Table 4), which were very close to that of the native snapdragon plant GPPS (15 ± 2.6 μM and 37.3 ± 2.1 μM for IPP and DMAPP, respectively; Tholl et al., 2004). Unfortunately, we were not able to obtain protein sequences from these plant-purified GPPS enzymes due to their insufficient purity.
DISCUSSION
Snapdragon GPP.SSU Modifies the Chain Length Specificity of Tobacco GGPPS and Regulates GPP Formation in Planta
Despite the ubiquity of monoterpenes, GPPSs have been investigated only in a limited number of species (Croteau and Purkett, 1989; Burke et al., 1999; Narita et al., 1999; Bouvier et al., 2000; Tholl et al., 2001, 2004; Burke and Croteau, 2002a; van Schie et al., 2007; Schmidt and Gershenzon, 2008). Two fundamentally different dimeric GPPSs were discovered using protein- and homology-based cloning strategies. However, very little is known about the function of GPPS enzymes in planta and the mechanisms that regulate the flux to GPP. For heterodimeric GPPSs, it has been proposed that the small subunit plays a regulatory role in controlling the flux to GPP based on its gene expression in several species specialized in high production of monoterpenes (Tholl et al., 2004; Wang and Dixon, 2009) and biochemical studies showing that the small subunit protein can interact with GGPPS from phylogenetically distant plant species, thus changing their GGPPS activity to efficient GPP production in vitro (Burke and Croteau, 2002a; Tholl et al., 2004; Wang and Dixon, 2009). Whether the formation of such a chimeric enzyme can occur in planta and be used as a new approach for enhancing flux toward GPP has not been determined.
Currently, there is almost no information available about endogenous GPPS(s) enzymes and their corresponding genes in N. tabacum, which produces only small amounts of volatile terpenoids. By expressing snapdragon GPPS.SSU in tobacco and detecting an increase in both GPPS activity (Table 1) and the amount of monoterpenes produced in transgenic plants (Figures 2C and 2D, Table 2), we showed that catalytically inactive Am GPPS.SSU can find an interacting partner in planta and form active chimeric GPPS. Moreover, BiFC studies (Figures 5 and 6) and in vitro protein coexpression analysis (Table 4) confirmed that Am GPPS.SSU can interact with two [GGPPS1 (TC11329) and GGPPS2 (TC5826)] of the four identified potential tobacco candidates for the GPPS large subunit (see Supplemental Figure 5B online). Both Nt GGPPS1 and GGPPS2 appeared to be active GGPPSs (Figure 7, Table 4), with the highest similarity to snapdragon GPPS.LSU (see Supplemental Figure 5 online). In addition, both contain one conserved CxxxC motif (where “x” can be Ala, Leu, Ile, Val, Gly, or Ser) (see Supplemental Figure 5A online), which was recently shown to be critical for interaction with the small subunit (Wang and Dixon, 2009). Interestingly, GGPPS2 contained a Met after the first Cys of the CxxxC motif but was still able to interact with the small subunit (Figures 5 and 6), indicating that “x” in the CxxxC motif could be extended to Met as well. The interaction of Am GPPS.SSU with tobacco GGPPS1 and GGPPS2 modified chain length specificities of the GGPPSs, leading to the exclusive formation of GPP from IPP and DMAPP (Figure 7, Table 4). The formation of chimeric snapdragon/tobacco GPPS in planta suggests that Am GPPS.SSU has strong affinity to tobacco GGPPS and can prevent or compete with GGPPS homodimerization, at least under Am GPPS.SSU levels detected in transgenic plants. Moreover, the kinetic properties of the chimeric snapdragon/tobacco GPPSs isolated from transgenic GPPS.SSU-1 leaves were very similar to that of native snapdragon GPPS (Table 4; Tholl et al., 2004).
Of the two noninteracting tobacco candidates, TC4865, annotated as solanesyl diphosphate synthase, lacks the small subunit binding CxxxC motif (see Supplemental Figure 5A online), but has two conserved Asp-rich motifs DD(X)2-4D (where X can be any amino acid), an essential substrate binding element in all short-chain prenyltransferases (Koyama and Ogura, 1999; Wang and Ohnuma, 1999). By contrast, TC4825 contains two CxxxC motifs, a characteristic feature of GPPS.SSU, and appears to belong to the recently defined SSU II subfamily (see Supplemental Figures 5A and 5B online; Wang and Dixon, 2009), likely representing a tobacco GPPS.SSU.
In plants, both GGPPS and GPPS are localized in plastids (Okada et al., 2000; Tholl et al., 2004; Ament et al., 2006; Pateraki and Kanellis, 2008), suggesting that at any given time, GPPS.SSU can interact with GGPPS and modify its product specificity for efficient GPP production. To prevent uncontrolled/undesired heterodimer formation in plastids potentially leading to deleterious effects on GGPP-derived metabolism, the level of GPPS.SSU has to be tightly regulated. Such regulation in plants appears to occur at the transcriptional level, as was demonstrated in snapdragon and hop (Tholl et al., 2004; Wang and Dixon, 2009). In snapdragon flowers, expression of GPPS.SSU, but not GPPS.LSU, displays a tissue-specific profile, which is developmentally regulated and changes rhythmically during the daily light/dark cycle, positively correlating with terpenoid volatile emission (Tholl et al., 2004). Similarly, in hop, GPPS.SSU is highly and specifically expressed in trichomes where myrcene production takes place, while the GPPS.LSU is expressed in all tissues examined (Wang and Dixon, 2009).
In this study, the expression of Am GPPS.SSU under the control of the heterologous LIS promoter resulted in drastic effects on GGPP-derived essential isoprenoids, including gibberellins, carotenoids, and chlorophylls (Figure 3, Table 3). Very high levels of Am GPPS.SSU expression appear to be lethal, as only a small number of transformants was obtained from 1200 transformation events, and three lines highly expressing Am GPPS.SSU (GPPS.SSU-5, -6, and -7) were unable to mature under greenhouse conditions (Figures 1A and 1B). Only plants with low and moderate Am GPPS.SSU levels (GPPS.SSU-3 and GPPS.SSU-1, respectively) were able to produce mature plants, although the latter still developed growth retardation and chlorosis (Figures 1A and 1C). Overall, these results suggest that tight regulation of GPPS.SSU is crucial for plant survival.
Flux Redirection toward GPP Has a Profound Effect on Monoterpenes, Sesquiterpenes, and GGPP-Derived Metabolites
GPPS is the key branchpoint enzyme leading to monoterpene biosynthesis, which generally occurs in plastids (Aharoni et al., 2005). Tobacco plants used in this study produce low levels of volatile terpenoids under normal growth conditions (8.7 and 33.9 ng·gFW h−1 from leaves and flowers, respectively). However, Am GPPS.SSU overexpression in GPPS.SSU-1 transgenic tobacco resulted in a large increase in emitted monoterpenes in both leaves and flowers (Figure 2), providing evidence that the level of GPP for endogenous tobacco monoterpene synthases was limiting in control tobacco plants. Interestingly, the effect of Am GPPS.SSU expression was unevenly distributed between different monoterpene compounds. The leaves of transgenic plants emitted 25-fold more (E)-β-ocimene than controls and produced a new monoterpene, myrcene (Figure 2C). Transgenic flowers produced (E)-β-ocimene, which was not emitted from control flowers (Figure 2D). Unexpectedly, linalool emission was not significantly altered by Am GPPS.SSU expression (Figure 2). Moreover, MeJA treatment, known to induce monoterpene production, drastically increased the levels of emitted (E)-β-ocimene in both control and GPPS.SSU-1 leaves (by 620- and 160-fold, respectively) but had a comparatively small effect on linalool emission, increasing it by 1.8- and 3.5-fold in controls and transgenics, respectively (Table 2). Such a subtle effect of Am GPPS.SSU overexpression or MeJA treatment on linalool emission could be due to its formation in the cytosol by a bifunctional monoterpene/sesquiterpene synthase, as was shown in strawberry (Fragaria spp; Aharoni et al., 2004) and snapdragon (Nagegowda et al., 2008). However, a treatment with fosmidomycin, a specific DXR inhibitor, reduced linalool emission by 90 and 95% in transgenic and control leaves, respectively (Table 2), suggesting that the majority of linalool is made from the MEP pathway–derived isoprenoid precursors synthesized in plastids. Thus, linalool could be formed in the cytosol from GPP exported from plastids by specific transporters (Bick and Lange, 2003), which control the GPP flux to cytosol and subsequently linalool formation. Alternatively, the unequal use of the increased GPP pool in plastids of transgenic plants by monoterpene synthases responsible for (E)-β-ocimene and linalool formation could be due to a low affinity of linalool synthase toward GPP or a high affinity combined with a saturation of enzyme with substrate, sequestration of synthesized linalool within the cell (Lücker et al., 2001), or channeling of GPP to (E)-β-ocimene production. Another explanation is that linalool is not produced from GPP but from another precursor, as was recently shown for other monoterpenes in Solanaceae (Schilmiller et al., 2009).
The increase in monoterpene emission in transgenic plants was accompanied by a decrease in the emission of sesquiterpenes by 2.4- and ∼5-fold for β-caryophyllene and 5-epi-aristolochene in leaves, respectively, and by 6.7-fold for 5-epi-aristolochene in flowers (Figures 2A and 2D). These results suggest that there is metabolic crosstalk between the plastidic MEP and cytosolic MVA pathways in tobacco and that the introduction of Am GPPS.SSU in transgenic plants leads to an increase in flux toward GPP formation in plastids, subsequently reducing the IPP pool and its transport to the cytosol resulting in reduced sesquiterpene formation. Although the GPP transport from plastids to the cytosol could be increased in transgenic plants, our results show that this exported GPP is not available for FPP biosynthesis directed to sesquiterpene formation. Prior studies also observed that the overexpression of three lemon monoterpene synthases in tobacco plants decreased β-caryophyllene levels by 2- to 3-fold (Lücker et al., 2004). Taken together, these results suggest that the plastidic MEP pathway–derived precursors are used for both plastidic monoterpene and cytosolic sesquiterpene biosynthesis in tobacco plants.
GPP is not only a direct precursor for monoterpene formation but also an allylic cosubstrate with IPP for GGPP production by GGPPS. Kinetic evaluation of tobacco GGPPS1 and GGPPS2 revealed that these enzymes have high affinity toward GPP with Km values of 2.4 and 3.6 μM, respectively (Table 4), which are within the range (low μM) reported for monoterpene synthases (Wise and Croteau, 1999), allowing them to compete with monoterpene synthases for GPP use. Increased monoterpene emission, especially (E)-β-ocimene (Figure 2), in the tobacco GPPS.SSU-1 line suggests that monoterpene synthases efficiently use an increased GPP pool in transgenic plants. On the other hand, the levels of GGPP-derived metabolites, including chlorophyll, carotenoids, and GAs, were drastically reduced in these transgenic plants (Figure 3, Table 3). The reduced GGPP production could be due to IPP limitation, GPP unavailability for GGPPS due to channeling toward (E)-β-ocimene formation, and/or GGPPS elimination from its active pool via the interactions with Am GPPS.SSU. Analysis of GGPPS activity in the GPPS.SSU-1 transgenic plants showed a 45% decrease relatively to the control (Table 1). Interestingly, the low level of Am GPPS.SSU expression in GPPS.SSU-3 transgenic plants also led to a 33% decrease in GGPPS activity and 30% reduction of GGPP-derived metabolites (Figures 3A and 3B) without affecting plant phenotype. Although GPPS activity was increased almost 2-fold in the GPPS.SSU-3 line (Table 1), it was not sufficient enough to alter monoterpene emission. These results suggest that when IPP and GPP are not redirected to monoterpene formation and are available for GGPP formation, as occurs in GPPS.SSU-3 transgenic plants, the level of GGPPS enzyme activity determines the level of GGPP produced. By contrast, in GPPS.SSU-1 transgenic plants with the severe phenotype and high levels of emitted monoterpenes, a combination of different factors affects the GGPP formation, among which the limitation of IPP caused by flux redirection toward GPP formation likely plays a major role in the observed effects on GGPP-derived metabolites.
Leaf chlorosis and reduced stature were also observed in transgenic tobacco, producing high levels of patchoulol as a result of the expression of patchoulol systhase coupled with FPP synthase, both targeted to the plastids (Wu et al., 2006), and in transgenic Arabidopsis and potato (Solanum tuberosum) plants expressing strawberry linalool/nerolidol systhase (NES1) targeted to chloroplasts (Aharoni et al., 2003, 2006). In all cases, the overexpression of the target genes led to an increase in the production of the corresponding terpenoids at the expense of the depletion of isoprenoid precursors required for metabolites essential for plant growth and development, although a potential toxic effect of the introduced compounds was not excluded. The observed phenotype in GPPS.SSU-1 tobacco was also typical for plants deficient in the MEP pathway, which supplies IPP and DMAPP for all plastidic isoprenoids (Estévez et al., 2001). The expression levels of the key genes, DXS and DXR, regulating the flux through the MEP pathway remained unaffected in these plants (see Supplemental Figure 4 online), providing further evidence that the flux redirection of isoprenoid precursors toward monoterpene formation is mainly responsible for the reduction in the GGPP-derived metabolites in transgenic plants, although the possible changes at the DXS and/or DXR protein levels despite the unaltered transcript levels cannot be excluded (Cordoba et al., 2009).
METHODS
Plant Material and Transformation
Tobacco (Nicotiana tabacum cv Xantii nc) was used for the generation of transgenic plants. Plants were grown under a 16-h photoperiod in standard greenhouse conditions unless specified. Transgenic plants were obtained via Agrobacterium tumefaciens (strain EHA105 carrying plasmid pEF1.LIS-AmGPPS.SSU) leaf disc transformation using the standard transformation protocol (Curtis et al., 1995). Plants rooted on hygromycin selection (10 mg/L) were screened for the presence of the LIS promoter by PCR with the specific primers: LIS-F (5′-GGCACCCACTTCTTAATGATC-3′) and LIS-R (5′-CTGGGATATGATAGGATGTGG-3′). T0 and T1 transformants were self-pollinated manually, and the seeds obtained were analyzed for segregation by germinating on half-strength Murashige and Skoog medium supplemented with hygromycin (10 mg/L). Untrasformed tobacco plants were used as a control in all described experiments. Wild-type Arabidopsis thaliana ecotype Columbia and ent-kaurene–deficient gal 3 mutant (Otsuka et al., 2004) were used in ent-kaurene treatment experiments.
Cloning of Snapdragon GPPS.SSU in Plant Expression Vector
The binary vector pCAMBIA 1303 (http://www.cambia.org/daisy/cambia/585) was modified by replacing the cauliflower mosaic virus 35S promoter, gusA, and monomeric green fluorescent protein (bases 11,600 to 2564) with the Clarkia breweri LIS promoter (1038 bp) (Orlova et al., 2006; Koeduka et al., 2009) containing the introduced NcoI site at its 3′ end. The coding region of snapdragon (Antirrhinum majus) Am GPPS.SSU of 891 bp in size was then inserted into the NcoI-PstI site of the modified binary vector (plasmid pEF1.LIS-AmGPPS.SSU).
Analysis of Plant Volatiles
Leaves and flowers volatiles were collected from control and transgenic tobacco plants using a closed-loop stripping method under growth chamber conditions (21°C, 50% relative humidity, 150 μmol·m−2·s−1 light intensity, and 12 h photoperiod) (Donath and Boland, 1995; Dudareva et al., 2005). Eleven “ready to open” cut flower buds at stage 10 (Goldberg, 1988; Koltunow et al., 1990) were used for floral scent collections, while 10 to 12 g of detached leaves were used for collection of leaf volatiles. Volatiles were sampled twice during a 48-h period by changing Porapak Q traps (80/100 mesh size; Alltech Associates) every 24 h. Volatiles were eluted with 200 μL of dichloromethane and analyzed by GC-MS (5975 inert mass spectrometer combined with 6890 GC; Agilent) using naphthalene as an internal standard as described previously (Dudareva et al., 2005).
Total Chlorophyll and Carotenoid Measurements
One hundred milligrams of N2-frozen tobacco young leaf tissues were extracted three times with 1 mL of 96% ethanol, and extracted pigments were analyzed spectrophotometrically as described previously (Lichtenthaler, 1987).
GA Analysis
GA levels were determined by liquid chromatography-selected reaction monitoring on a quadrupole/time-of-flight tandem mass spectrometer (Q-Tof Premier; Waters) connected to an Acquity Ultra Performance liquid chromatograph equipped with a reverse phase column (Acquity UPLC BEH-C18; Waters) as described previously (Varbanova et al., 2007). We used 2H-labeled GAs as internal standards.
MeJA, GA3, and ent-Kaurene Treatments
For MeJA treatments, 95% pure MeJA (Sigma-Aldrich) was added to a 5% (w/v) sucrose solution to final concentration of 436 μM. Ten grams of excised young leaves from control and GPPS.SSU-1 transgenic tobacco plants were placed into desiccators containing the MeJA/sucrose solution. For inhibition experiments, the MEP pathway–specific inhibitor fosmidomycin (Invitrogen) was added at a final concentration of 100 μM. Emitted volatiles were collected every 24 h for 48 h as described above. The GA treatment was achieved by growing control and GPPS.SSU-1 tobacco seeds on solid half-strength Murashige and Skoog medium supplemented with 30 μM GA3. For ent-kaurene treatment, Arabidopsis wild-type, gal3 mutant, tobacco control, and GPPS.SSU-1 seeds were grown on solid half-strength Murashige and Skoog medium in the presence of 5 μg of the volatile ent-kaurene on filter paper placed in the Petri plates (Otsuka et al., 2004). After 10 d, seedling growth was analyzed by measuring leaf area using ImageJ software (http://rsbweb.nih.gov/ij/). Each treatment included two biological repetitions with 30 seedlings.
RNA Isolation, RNA Gel Blot Analysis, and Quantitative RT-PCR
Total RNA was isolated from wild-type and transgenic tobacco leaves and 1-d-old flowers (stage 11 according to Goldberg, 1988; Koltunow et al., 1990) using the RNeasy Plant Mini Kit (Qiagen) and analyzed as previously described (Boatright et al., 2004). A 1-kb EcoRI fragment containing the coding region of GPPS.SSU (Tholl et al., 2004) was used as a probe in RNA gel blot analysis. Five micrograms of total RNA were loaded in each lane. The blots were rehybridized with 18S rDNA (Eckenrode et al., 1985) for loading control.
For quantitative RT-PCR (qRT-PCR), total RNA was treated with DNase I to eliminate genomic DNA using the TURBO DNA-free kit (Ambion), and 1 μg of RNA was subsequently reverse transcribed to cDNA in a total volume of 100 μL using the High Capacity cDNA reverse transcription kit (Applied Biosystems). Gene-specific primers, TC4825 forward 5′-ATCCGGTGCTCGGCTTCT-3′ and reverse 5′-CTGGTTGATATCACGAATTAGAGTTGT-3′; TC5826 forward 5′-TGGAAAACCTCCACAGAGACATT-3′ and reverse 5′-TCAAAGTCAAACTTAGGCAAGATGA-3′; TC11329 forward 5′-CTTGTACTGGTAACCCTAATGTTGGA-3′ and reverse 5′-TCCGAGAACTACGGAAGCTTCTA-3′; TC4865 forward 5′-GGAGGATTCAATGCTCGGAAA-3′ and reverse 5′-TGGCGCCACCCCACTA-3′, were designed using the PrimerExpress software (Applied Biosystems). All primers showed 90 to 100% efficiency at a final concentration of 300 nM. For absolute quantification of individual mRNA transcripts, the pCR4-TOPO (Invitrogen) plasmids carrying truncated TC4825, TC5826, TC11329, and TC4865 (see below) were digested with EcoRI, and the resulting fragments were purified from agarose gels using a gel extraction kit (Qiagen). After determination of their DNA concentrations using the NanoDrop 1000 spectrophotometer (Thermo Scientific), the purified DNA fragments were diluted from 2 ng/mL to 3.2 pg/mL and used as templates to obtain standard curves in qRT-PCR with gene-specific primers. DXS, DXR, and Am GPPS.SSU transcript levels were normalized to actin. Gene-specific primers were as follows: 5′-AGTGTTTAACATTCTTGGACAAACC-3′ and 5′-ACAACTATTCTCACCTTAGCTGGTG-3′ forward and reverse for DXS, 5′-CTTAGGCGCATTATATTAACTGCAT-3′ and 5′-GTGGCAGAATCAACAGTAATCTTTT-3′ forward and reverse for DXR, 5′-GATCCGAATATCGAACTTTTGAC-3′ and 5′-AGTTCATGGTATTGCCCTTCTA-3′ forward and reverse for Am GPPS.SSU, and 5′-CTGAAGTCCTTTTCCAACCTTC-3′ and 5′-GCAGTAATTTCCTTACTCATCC-3′ forward and reverse for actin. Individual qRT-PCR reactions were performed in duplicate, each containing 5 μL of the SYBR Green PCR Master Mix (Applied Biosystems), 2 μL of 50 times diluted cDNA, and 1.5 μL of 2 μM forward and reverse primers. Two-step qRT-PCR amplification (40 cycles of 95°C for 3 s followed by 60°C for 30 s) was performed using the StepOn Real-Time PCR system (Applied Biosystems). Based on standard curves, absolute quantities of individual transcripts were calculated and expressed as a percentage of the total cDNA. Each data point represents an average of three independent biological samples.
Purification of Chimeric Snapdragon/Tobacco GPPS from Transgenic Tobacco
Freshly excised GPPS.SSU-1 transgenic tobacco leaves were frozen in liquid N2 and ground to a fine powder using a mortar and pestle. The frozen powder was immediately slurried with freshly prepared extraction buffer (3:1 [v/w], buffer:tissue) containing 25 mM MOPSO, pH 7.0, 10 mM MgCl2, 1 mM DTT, 1% (w/v) polyvinylpyrrolidone (PVP-40), 1 mM phenylmethanesulfonyl fluoride, and 10% (v/v) glycerol. The slurry was homogenized in a chilled glass homogenizer (Wheaton, VWR Scientific Products), passed through two layers of Miracloth, and centrifuged for 20 min at 12,000g. The pellet was discarded and the supernatant was loaded onto a DEAE-cellulose column (20 mL of DE53; Whatman) preequilibrated with 25 mM MOPSO, pH 7.0, 10 mM MgCl2, 1 mM DTT, and 1% (w/v) and 10% (v/v) glycerol (buffer A) at a flow rate of 0.2 mL min−1. The column was washed with 100 mL of buffer A, and the enzyme was eluted with a linear gradient (400 mL) from 0 to 1000 mM KCl in buffer A. Fractions (9 mL each) were collected and assayed for prenyltransferase activity. Fractions with the highest activity in the 200 to 267 mM KCl range were combined (total of 27 mL) and desalted on Econo-Pac10DG columns equilibrated with buffer A.
The desalted eluent (36 mL) was loaded onto a MonoQ anion exchange column (gel bed volume 8 mL) (Amersham Pharmacia Biotech) preequilibrated with buffer A at a flow rate of 0.5 mL min−1. After washing the column with 50 mL of buffer A, the protein was eluted with a linear gradient of 0 to 1000 mM KCl (100 mL, flow rate 0.5 mL min−1) in buffer A. Fractions (1 mL) were tested for prenyltransferase activity, and the highest prenyltransferase activity was eluted from the column at 228 to 397 mM KCl. For size exclusion gel filtration chromatography, 200 μg of protein with the highest prenyltransferase activity after MonoQ-column purification were loaded on a HiLoad 16/60 Superdex 200 column (GE Healthcare) preequilibrated with buffer A, and elution was performed using with the same buffer at a flow rate of 0.2 mL/min. Fractions (0.5 mL each) were analyzed for prenyltransferase activity, product specificity, and by protein blot analysis with polyclonal antibodies against Am GPPS.SSU for the presence of Am GPPS.SSU. Fractions that contained the highest levels of Am GPPS.SSU and exclusively produced GPP as a product were used for kinetic analysis.
Immunoblot Analysis
Crude extracts were prepared from the control and transgenic tobacco leaves and flowers as described previously (Dudareva et al., 2000). Immunodetection was performed using rabbit anti-GPPS.SSU polyclonal antibodies (Tholl et al., 2004; 1:2500 dilution), with goat anti-rabbit IgG horseradish peroxidase conjugate (Sigma-Aldrich; 1:30,000 dilution) as a secondary antibody. Antigen bands were visualized using chemiluminescence reagents (DuPont–New England Nuclear Life Science Products) for protein gel blots according to the manufacturer's protocols and exposed on Eastman Kodak X-OMAT AR film.
BiFC
The pSAT vectors (Lee and Gelvin, 2008) used for BiFC were kindly provided by Stanton Gelvin (Purdue University). The open reading frames of snapdragon GPPS.SSU and tobacco GGPPS-like proteins were fused in frame and upstream of the N-terminal half of EYFP in pSAT4-nEYFP-N1 and the C-terminal half of EYFP in pSAT6-cEYFP-C1, respectively, in the NcoI-BamHI cloning sites of both vectors. BiFC constructs were generated using the following primers: GPPS.SSU forward 5′-CCATGGCTATGGCCCACGGCCTCACCCATTTC-3′ and reverse 5′-GGATCCCAACACCAGCAAGACTATGCTCTAG-3′; TC4865 forward 5′-CCATGGCTATGATGTCTGTGACTTGCCATAATCTTGAG-3′ and reverse 5′-GGATCCCTTCAATTCTCTCCAGATTATACTTCACAAT-3′; TC11329 forward 5′-CCATGGCTATGAGATCTATGAATCTTGTCGATTC-3′ and reverse 5′-GGATCCCATTTTCACGATAAGCAATGTAATCC-3′; TC5826 forward 5′-CCATGGCTATGGCATTTTTGGCTACCATTTCTGGCCATG-3′ and reverse 5′-GGATCCCATTCTGGCGATGTGCAATATAATTAGCAAAATG-3′; TC4825 forward 5′-CCATGGCTATGGTGTTCTCTATGGTGATGAGTTTTTCAC-3′ and reverse 5′-GGATCCCGTTATCAAGGCTAAAACCTCTATCTG-3′. Sequencing confirmed the accuracy of fusions. The resulting constructs were used for transient expression in Arabidopsis protoplasts and leaf epidermal cells of Nicotiana benthamiana. Arabidopsis protoplasts were prepared and transformed as described by Sheen (2002). Arabidopsis leaves (∼20) were cut into 0.5- to 1.0-cm strips and incubated for 3 h without shaking in the dark in a 10 mL of solution containing 1% cellulase (SERVA) and 0.25% macerozyme (SERVA) in 0.4 M mannitol, 20 mM KCl, 20 mM MES, pH 5.7, 10 mM CaCl2, and 0.1% BSA. Protoplasts were filtered through a 105-μm nylon mesh and centrifuged at 100g for 2 min. The pellet was washed with an ice-cold solution containing 154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 7.5, and 5 mM glucose and resuspended in the same solution at a concentration of 2 × 105 cells per mL. Ten micrograms of each plasmid encoding cEYFP and nEYFP fusion proteins were mixed at a 1:1 (w/w) ratio and used for polyethylene glycol–mediated transformation of 100 μL of the ice-cold protoplasts. The plasmid p326-RbTP-SGFP (Lee et al., 2002; Nagegowda et al., 2008) was used as a marker for plastidial localization. Transient expression of the YFP fusion proteins was observed 16 to 20 h after transformation. Images were acquired using a Radiance 2100 MP Rainbow (Bio-Rad) on a TE2000 (Nikon) inverted microscope with a ×60 oil 1.4–numerical aperture lens. The YFP was excited with the 514-nm line of the four-line argon laser, and the emission was collected with a 530LP, 560SP filter combination. Chlorophyll was excited by the 637-nm red diode laser, and emission >660 nm in wavelength was collected.
Transient expression in N. bentamiana leaf epidermal cells was achieved by microbombardment. Plasmids encoding cEYFP and nEYFP fusion proteins were mixed at a 1:1 (w/w) ratio, and a total of 50 μg DNA was adsorbed onto 10 μg of 1-μm gold particles according to the manufacturer's instructions (Bio-Rad; 165-2263). The particles were microbombarded at a pressure of 150 p.s.i. (1 p.s.i. ≈ 6.9 kPa) using a portable Helios Gene Gun (Bio-Rad; 165-2411) into the epidermis of N. benthamiana leaves. Sixteen to twenty-four hours after bombardment, plant tissues were analyzed for EYFP transient expression under a Zeiss LSM 5 Pascal confocal laser scanning microscope equipped with two laser lines and a set of filters capable of distinguishing between the cyan (ECFP) and yellow (EYFP) fluorescence proteins and plastid autofluorescence.
Cloning and Expression of Tobacco Prenyltransferases in Escherichia coli
The publicly available tobacco EST database (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=tobacco) was used to identify cDNA clones with homology to snapdragon GPPS.LSU. TC4865 was full-length, which allowed us to amplify its open reading frame by RT-PCR using mRNA derived from tobacco leaves and clone-specific primers: forward 5′-CATATGATGATGTCTGTGACTTGCCATAATCTTGAG-3′ and reverse 5′-GGATCCTTCAATTCTCTCCAGATTATACTTCAC-3′. Both TC5826 and TC4825 lacked 3′ translated regions, which were obtained by 3′ RACE using the GeneRacer Core kit (Invitrogen) and the following primers: 5′-CCGATCCTATGCATGGCTTCTTGTG-3′ for TC5826 and 5′-CTTGACCTTGAGGGTGGACCAAATG-3′ for TC4825. 3′ RACE of TC4825 resulted in amplification of two 3′ regions with 98% identity, which were used to obtain two cDNA clones with 96% identity at the nucleotide level. The coding region of TC5826 was further amplified by RT-PCR using the forward primer 5′-CATATGATGGCATTTTTGGCTACCATTTCTGGCCATG-3′ in combination with the reverse primer 5′-GGATCCTCAATTCTGGCGATGTGCAATATAATTAGCAAAATG-3′. The forward and reverse primers for the TC4825 coding region were 5′-CATATGGTGTTCTCTATGGTGATGAGTTTTTCAC-3′ and 5′-GGATCCTTAGCTATCAAGGCTAAAACCTCTATCTG-3′, respectively. The TC11329 lacked the 5′ region and showed high nucleotide identity (95%) to N. benthamiana (Nb) and Nicotiana attenuata (Na) GGPPSs. Thus, the forward primer for TC11329 was designed based on Nb GGPPS and Na GGPPS sequences, which were 100% identical in the first 23 nucleotides from the start codon. The forward and reverse primers for TC11329 were 5′-CATATGAGATCTATGAATCTTGTCGA-3′ and 5′-GGATCCCTAATTTTCACGATAAGCAATGTA-3′, respectively. For biochemical characterization, truncated versions of tobacco prenyltransferases with deletions of their N-terminal plastidial targeting sequences were subcloned into the expression vectors pET11a and pET28a (Novagen). In all cases, forward primers introduced an NdeI site at the initiating ATG codon. Truncated versions for TC4865 and TC5826 were obtained using the forward primers 5′-CATATGAATGTAAGTAGAGGAGGATTCA-3′ and 5′-CATATGAGACATTCTTATAGTTTCCTCC-3′, respectively, which introduced a starting Met in place of Asn-32 (TC4865) and Arg-32 (TC5826), in combination with above-mentioned clone-specific reverse primers with BamHI restriction site after the stop codon. Truncated versions of TC4825 and TC11329 clones that started at Met-22 and Met-31, respectively, were obtained using the corresponding forward primers 5′-CATATGCAGAAAACAATCCGGTGCTC-3′ and 5′-CATATGAAAATTCCTGTGAAAAACCC-3′ with the above mentioned clone-specific reverse primers containing BamHI sites after the stop codon. Sequencing revealed that no errors had been introduced during PCR amplifications.
For functional expression, E. coli BL21 (DE3) cells were transformed with the resulting recombinant plasmids as well as pET vectors without inserts as controls. For coexpression, E. coli BL21 (DE3) cells were cotransformed with two plasmids (e.g., small subunit and tobacco prenyltransferases) in a single transformation event. Single positive bacterial colonies were used to inoculate 25 mL Luria-Bertani medium with 37 μg/mL chloramphenicol and 50 μg/mL ampicillin (for pET11a constructs) or 50 μg/mL kanamycin (for pET 28a constructs) or with both antibiotics (for coexpression of both pET11a/pET28a constructs), which were grown overnight at 37°C. Two milliliters of these cultures were transferred to 100 mL of the same medium and continued to grow at 37°C until an OD600 of 0.5 was reached. Cultures were then induced by the addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 0.4 mM and grown for an additional 18 h at 18°C. Protein purification by affinity chromatography on nickel-nitrilotriacetic acid-agarose (Qiagen) was performed as described previously (Tholl et al., 2004). Protein concentration was determined using the Bradford method (Bradford, 1976).
Prenyltransferase Assays and Product Identification
Prenyltransferase assays were performed as described previously (Tholl et al., 2004). The reaction was performed in a final volume of 100 μL containing 40 μM [1-14C]-IPP (50 mCi/mmol) and 40 μM DMAPP in assay buffer (25 mM MOPSO, pH 7.0, 10% [v/v] glycerol, 2 mM DTT, and 10 mM MgCl2). After the reaction was initiated by the addition of an aliquot of the enzyme, it was immediately overlaid with 1 mL of hexane and incubated for 40 min at 30°C. Assays were stopped by adding 10 μL of 3 n HCl and then incubated for an additional 20 min at 30°C to hydrolyze the acid-labile allylic diphosphates formed during the assay to their corresponding alcohols. Hydrolysis products (and alcohols produced by endogenous phosphohydrolase activity during the assay) were extracted into the hexane phase by vigorous mixing for 15 s, and 600 μL of the hexane phase were counted in a liquid scintillation counter. Controls included assays with boiled protein extracts and without substrate. Background radioactivity produced in the controls was subtracted from all results.
In order to determine endogenous phosphohydrolase activity, assays were performed as described above but with [1-14C]-IPP as the only substrate and without acid treatment. After incubation at 30°C, samples were cooled on ice, and the radiolabeled isopentenol was directly extracted into the hexane layer.
For the identification of reaction products, larger-scale assays were performed in a final volume of 200 μL. Assays were performed for 6 h at 30°C. To stop the assay and hydrolyze all diphosphate esters (including unreacted substrate as well as products), 200 μL of a solution containing 2 units of bovine intestine alkaline phosphatase (18 units/mg; Sigma-Aldrich) and 2 units of potato apyrase (25.2 units/mg; Sigma-Aldrich) in 0.2 M Tris-HCl, pH 9.5, were added to samples followed by overnight incubation at 30°C. After enzymatic hydrolysis, the resulting prenyl alcohols were extracted with 1 mL hexane. One hundred nanograms of geraniol (GOH), farnesol (FOH), and geraylgeraniol (GGOH) were added as internal standards and carriers, and the hexane fraction was concentrated to 25 μL. The products were separated on reversed phase TLC plates (C18 silica TLC plates; Sorbent Technologies). Chromatography was performed using an acetone:water (6:1, v/v) mobile phase (Ling et al., 2007), and radioactive spots were identified using a phosphor imager (Molecular Dynamics Typhoon 8600). Nonradioactive standards were visualized by exposure of TLC plates to iodine vapor.
Chloroplast Isolation
For chloroplast isolation, 5 g of young leaf tissue of control, GPPS.SSU-1, GPPS.SSU-3, and GPPS.SSU-7 tobacco plants were homogenized in a blender three times for 10 s each in 20 mL of PIM buffer (0.5M sorbitol, 20 mM HEPES, pH 7.4, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, 10% ethanediol, 5 mM DTT, and 1% BSA) (Kang and Rawsthorne, 1994). The homogenate was filtered through a double layer of Miracloth and centrifuged at 4°C for 5 min at 120g. The pellet was washed in 2 mL of PIM buffer followed by centrifugation. The pellet was then resuspended in 0.5 mL of prenyltransferase extraction buffer and centrifuged at 13,000 rpm for 10 min at 4°C to remove thylakoid membranes using Hermle Z 180M Eppendorf centrifuge, and the supernatant was used for detection of prenyltransferase activity.
Determination of Kinetic Properties of Prenyltranspherases
For kinetic analysis, an appropriate enzyme concentration was chosen so that the reaction velocity was proportional to the enzyme concentration and was linear with respect to incubation. For chimeric snapdragon/tobacco recombinant and plant-purified GPPSs, Km values for IPP were determined at saturating concentrations of DMAPP (66 μM), whereas Km values for DMAPP were determined at saturating concentrations of IPP (53 μM). The apparent Km values for MgCl2 were determined at saturating concentrations of both IPP and DMAPP. For recombinant tobacco GGPPSs, Km values for GPP were determined at saturating concentrations of IPP (53 μM), and Km values for IPP analyzed at saturating concentrations of GPP (28 μM). Kinetic data were evaluated by hyperbolic regression analysis (Hyper.exe version 1.01; J.S. Easterby). Triplicate assays were performed for all analyses.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are as follows: Am GPPS.LSU, AAS82860; Am GPPS.SSU, AAS82859; At GGPS1, AT1g49530; At GGPS2, AT2g18620; At GGPS3, AT2g18640; At GGPS4, AT2g23800; At GGPS5, AT3g14510; At GGPS6, AT3g14530; At GGPS7, AT3g14550; At GGPS8, AT3g20160; At GGPS9, AT3g29430; At GGPS10, AT3g32040; At GGPS11, AT4g36810; At GGPS12, AT4g38460; Hb SPS, DQ437520; Hl GPPS.LSU, FJ455407; Hl GPPS.SSU, FJ455406; Mp GPPS.LSU, AF182828; Mp GPPS.SSU, AF182827; Nt GGPPS1, GQ911583; Nt GGPPS2, GQ911584; Os SPS, NM_001062973; Sl GGPS1, DQ267902; Sl GGPPS2, DQ267903; St GGPPS, TC171867.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Correlation between Am GPPS.SSU Expression, GPPS Activity, and Total Chlorophyll Levels in Transgenic Tobacco Plants.
Supplemental Figure 2. Am GPPS.SSU mRNA Levels in Leaves and Flowers of Control and Transgenic Tobacco Lines GPPS.SSU-1 and GPPS.SSU-3.
Supplemental Figure 3. Effect of ent-Kaurene Treatment on Growth of Seedlings of Arabidopsis thaliana (Columbia), GA-Deficient Arabidopsis Mutant gal3, Nicotiana tabacum cv Xantii, and GPPS.SSU-1 Transgenic Tobacco.
Supplemental Figure 4. DXS and DXR mRNA Expression in Leaves of the Control and GPPS.SSU-1 Transgenic Tobacco.
Supplemental Figure 5. Relatedness of Am GPPS.SSU and Tobacco Clones to Other Plant GPPSs, GGPPSs, and Solanesyl Diphosphate Synthases (SPSs).
Supplemental Figure 6. Purification of Chimeric Snapdragon/Tobacco GPPS from Leaves of GPPS.SSU-1 Tobacco Plants.
Supplemental Data Set 1. Text File of the Alignment Used to Create the Phylogenetic Tree Shown in Supplemental Figure 5.
Supplementary Material
Acknowledgments
This work is supported by National Science Foundation/USDA-National Research Initiative (NRI) Interagency Metabolic Engineering Program, Grants MCB 0331333 (N.D.) and MCB 0331353 (E.P.), and by a grant from the Fred Gloeckner Foundation to N.D. We thank Miyuki Satoh (RIKEN) for technical assistance in GA measurements, Yasuhisa Kaminaga for help with protein purification, and Jennifer Sturgis for confocal microscopy assistance. Confocal microscopy with Arabidopsis protoplasts was performed at the Purdue Cancer Center Analytical Cytometry Laboratories supported by the National Cancer Institutes's core Grant NIH NCI-2P30CA23168.
The author responsible for distribution of material integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Natalia Dudareva (dudareva@purdue.edu).
Online version contains Web-only data.
References
- Aharoni, A., Giri, A.P., Deuerlein, S., Griepink, F., de Kogel, W.J., Verstappen, F.W., Verhoeven, H.A., Jongsma, M.A., Schwab, W., and Bouwmeester, H.J. (2003). Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15 2866–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni, A., Giri, A.P., Verstappen, F.W., Bertea, C.M., Sevenier, R., Sun, Z., Jongsma, M.A., Schwab, W., and Bouwmeester, H.J. (2004). Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16 3110–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni, A., Jongsma, M.A., and Bouwmeester, H.J. (2005). Volatile science? Metabolic engineering of terpenoids in plants. Trends Plant Sci. 10 594–602. [DOI] [PubMed] [Google Scholar]
- Aharoni, A., Jongsma, M.A., Kim, T., Ri, M., Giri, A.P., Verstappen, F.W., Schwab, W., and Bouwmeester, H.J. (2006). Metabolic engineering of terpenoid biosynthesis in plants. Phytochem. Rev. 5 49–58. [Google Scholar]
- Ament, K., Van Schie, C.C., Bouwmeester, H.J., Haring, M.A., and Schuurink, R.C. (2006). Induction of a leaf specific geranylgeranyl pyrophosphate synthase and emission of (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene in tomato are dependent on both jasmonic acid and salicylic acid signaling pathways. Planta 224 1197–1208. [DOI] [PubMed] [Google Scholar]
- Bick, J.A., and Lange, B.M. (2003). Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch. Biochem. Biophys. 415 146–154. [DOI] [PubMed] [Google Scholar]
- Boatright, J., Negre, F., Chen, X., Kish, C.M., Wood, B., Peel, G., Orlova, I., Gang, D., Rhodes, D., and Dudareva, N. (2004). Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135 1993–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier, F., Suire, C., d'Harlingue, A., Backhaus, R.A., and Camara, B. (2000). Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 24 241–252. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Burke, C.C., and Croteau, R. (2002. a). Interactions with the small subunit of geranyl diphosphate synthase modifies the chain length specificity of geranylgeranyl diphosphate synthase to produce geranyl diphosphate. J. Biol. Chem. 277 3141–3149. [DOI] [PubMed] [Google Scholar]
- Burke, C.C., and Croteau, R. (2002. b). Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Arch. Biochem. Biophys. 405 130–136. [DOI] [PubMed] [Google Scholar]
- Burke, C.C., Wildung, M.R., and Croteau, R. (1999). Geranyl diphosphate synthase: Cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc. Natl. Acad. Sci. USA 96 13062–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky, V., Gafni, Y., and Tzfira, T. (2008). Localizing protein-protein interactions by bimolecular fluorescence complementation in planta. Methods 45 196–206. [DOI] [PubMed] [Google Scholar]
- Cordoba, E., Salmi, M., and León, P. (2009). Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 60 2933–2943. [DOI] [PubMed] [Google Scholar]
- Croteau, R.B., Davis, E.M., Ringer, K.L., and Wildung, M.R. (2005). (-)-Menthol biosynthesis and molecular genetics. Naturwissenschaften 92 562–577. [DOI] [PubMed] [Google Scholar]
- Croteau, R., and Purkett, P.T. (1989). Geranyl pyrophosphate synthase: Characterization of the enzyme and evidence that this chain-length specific prenyltransferase is associated with monoterpene biosynthesis in sage (Salvia officinalis). Arch. Biochem. Biophys. 271 524–535. [DOI] [PubMed] [Google Scholar]
- Cseke, L., Dudareva, N., and Pichersky, E. (1998). Structure and evolution of linalool synthase. Mol. Biol. Evol. 15 1491–1498. [DOI] [PubMed] [Google Scholar]
- Curtis, I.S., Davey, M.R., and Power, J.B. (1995). Leaf disk transformation. Methods Mol. Biol. 44 59–70. [DOI] [PubMed] [Google Scholar]
- Donath, J., and Boland, W. (1995). Biosynthesis of acyclic homoterpenes: Enzyme selectivity and absolute configuration of the nerolidol precursor. Phytochemistry 39 785–790. [Google Scholar]
- Dudareva, N., Andersson, S., Orlova, I., Gatto, N., Reichelt, M., Rhodes, D., Boland, W., and Gershenzon, J. (2005). The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 102 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva, N., Murfitt, L.M., Mann, C.J., Gorenstein, N., Kolosova, N., Kish, C.M., Bonham, C., and Wood, K. (2000). Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell 12 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckenrode, V.K., Arnold, J., and Meagher, R.B. (1985). Comparison of the nucleotide sequence of soybean 18S rRNA with the sequences of the other small-subunit rRNA. J. Mol. Evol. 21 259–269. [DOI] [PubMed] [Google Scholar]
- Estévez, J.M., Cantero, A., Reindl, A., Reichler, S., and León, P. (2001). 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 276 22901–22909. [DOI] [PubMed] [Google Scholar]
- Gershenzon, J., and Dudareva, N. (2007). The function of terpene natural products in the natural world. Nat. Chem. Biol. 3 408–414. [DOI] [PubMed] [Google Scholar]
- Gershenzon, J., and Kreis, W. (1999). Biochemistry of terpenoids: Monoterpenes, sesquiterpenes, diterpenes, sterols, cardiac glycosides and steroid saponins. In Biochemistry of Plant Secondary Metabolism, M. Wink, ed (Boca Raton, FL: CRC Press), pp. 222–299.
- Goldberg, R.B. (1988). Plants: Novel developmental processes. Science 240 1460–1467. [DOI] [PubMed] [Google Scholar]
- Hampel, D., Mosandl, A., and Wüst, M. (2005. a). Biosynthesis of mono- and sesquiterpenes in carrot roots and leaves (Daucus carota L.): Metabolic cross talk of cytosolic mevalonate and plastidial methylerythritol phosphate pathways. Phytochemistry 66 305–311. [DOI] [PubMed] [Google Scholar]
- Hampel, D., Mosandl, A., and Wüst, M. (2005. b). Induction of de novo volatile terpene biosynthesis via cytosolic and plastidial pathways by methyl jasmonate in foliage of Vitis vinifera L. J. Agric. Food Chem. 53 2652–2657. [DOI] [PubMed] [Google Scholar]
- Hemmerlin, A., Hoeffler, J.F., Meyer, O., Tritsch, D., Kagan, I.A., Grosdemange-Billiard, C., Rohmer, M., and Bach, T.J. (2003). Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J. Biol. Chem. 278 26666–26676. [DOI] [PubMed] [Google Scholar]
- Kang, F., and Rawsthorne, S. (1994). Starch and fatty acid synthesis in plastids from developing embryos of oilseed rape (Brassica napus L.). Plant J. 6 795–805. [Google Scholar]
- Koeduka, T., Orlova, I., Baiga, T.J., Noel, J.P., Dudareva, N., and Pichersky, E. (2009). The lack of floral synthesis and emission of isoeugenol in Petunia axillaris subsp. parodii is due to a mutation in the isoeugenol synthase gene. Plant J. 58 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow, A.M., Truettner, J., Cox, K.H., Wallroth, M., and Goldberg, R.B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2 1201–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, T., and Ogura, K. (1999). Isopentenyl diphosphate isomerase and prenyltransferases. In Comprehensive Natural Product Chemistry: Isoprenoids Including Carotenoids and Steroids, Vol. 2, D.E. Cane, ed (Oxford, UK: Pergamon Press), pp. 69–96.
- Kuzuyama, T., Shimizu, T., Takahashi, S., and Seto, H. (1998). Fosmidomycin, a specific inhibitor of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett. 39 7913–7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laule, O., Furholz, A., Chang, H.-S., Zhu, T., Wang, X., Heifetz, P.B., Gruissem, W., and Lange, B.M. (2003). Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100 6866–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.H., Kim, D.H., Lee, S.W., Kim, Z.H., and Hwang, I. (2002). In vivo import experiments in protoplasts reveal the importance of the overall context but not specific amino acid residues of the transit peptide during import into chloroplasts. Mol. Cells 14 388–397. [PubMed] [Google Scholar]
- Lee, L.-Y., and Gelvin, S.B. (2008). T-DNA binary vectors and systems. Plant Physiol. 146 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1999). The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 47–65. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148 350–382. [Google Scholar]
- Ling, Y., Li, Z.H., Miranda, K., Oldfield, E., and Moreno, S.N. (2007). The farnesyl-diphosphate/geranylgeranyl-diphosphate synthase of Toxoplasma gondii is a bifunctional enzyme and a molecular target of bisphosphonates. J. Biol. Chem. 282 30804–30816. [DOI] [PubMed] [Google Scholar]
- Lücker, J., Bouwmeester, H.J., Schwab, W., Blaas, J., van der Plas, L.H., and Verhoeven, H.A. (2001). Expression of Clarkia S-linalool synthase in transgenic petunia plants results in the accumulation of S-linalyl-beta-D-glucopyranoside. Plant J. 27 315–324. [DOI] [PubMed] [Google Scholar]
- Lücker, J., Schwab, W., van Hautum, B., Blaas, J., van der Plas, L.H., Bouwmeester, H.J., and Verhoeven, H.A. (2004). Increased and altered fragrance of tobacco plants after metabolic engineering using three monoterpene synthases from lemon. Plant Physiol. 134 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D.M., Gershenzon, J., and Bohlmann, J. (2003). Induction of volatile terpene biosynthesis and diurnal emission by methyl jasmonate in foliage of Norway spruce. Plant Physiol. 132 1586–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill, D., and Croteau, R. (1995). Isoprenoid synthesis in peppermint (Mentha x piperita): Development of a model system for measuring flux of intermediates through the mevalonic acid pathway in plants. Biochem. Soc. Trans. 23 290S. [DOI] [PubMed] [Google Scholar]
- McGarvey, D.J., and Croteau, R. (1995). Terpenoid metabolism. Plant Cell 7 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagegowda, D.A., Gutensohn, M., Wilkerson, C.G., and Dudareva, N. (2008). Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 55 224–239. [DOI] [PubMed] [Google Scholar]
- Narita, K., Ohnuma, S., and Nishino, T. (1999). Protein design of geranyl diphosphate synthase. Structural features that define the product specificities of prenyltransferases. J. Biochem. 126 566–571. [DOI] [PubMed] [Google Scholar]
- Newman, J.D., and Chappell, J. (1999). Isoprenoid biosynthesis in plants: Carbon partitioning within the cytoplasmic pathway. Crit. Rev. Biochem. Mol. Biol. 34 95–106. [DOI] [PubMed] [Google Scholar]
- Okada, K., Saito, T., Nakagawa, T., Kawamukai, M., and Kamiya, Y. (2000). Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 122 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova, I., Marshall-Colón, A., Schnepp, J., Wood, B., Varbanova, M., Fridman, E., Blakeslee, J.J., Peer, W.A., Murphy, A.S., Rhodes, D., Pichersky, E., and Dudareva, N. (2006). Reduction of benzenoid synthesis in petunia flowers reveals multiple pathways to benzoic acid and enhancement in auxin transport. Plant Cell 18 3458–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, M., Kenmoku, H., Ogawa, M., Okada, K., Mitsuhashi, W., Sassa, T., Kamiya, Y., Toyomasu, T., and Yamaguchi, S. (2004). Emission of ent-kaurene, a diterpenoid hydrocarbon precursor for gibberellins, into the headspace from plants. Plant Cell Physiol. 45 1129–1138. [DOI] [PubMed] [Google Scholar]
- Pateraki, I., and Kanellis, A.K. (2008). Isolation and functional analysis of two Cistus creticus cDNAs encoding geranylgeranyl diphosphate synthase. Phytochemistry 69 1641–1652. [DOI] [PubMed] [Google Scholar]
- Phillips, M.A., D'Auria, J.C., Gershenzon, J., and Pichersky, E. (2008). The Arabidopsis thaliana type 1 isopentenyl diphosphate isomerase are targeted to multiple subcellular compartments and gave overlapping functions in isoprenoid biosynthesis. Plant Cell 20 677–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, M.A., Walter, M.H., Ralph, S.G., Dabrowska, P., Luck, K., Urós, E.M., Boland, W., Strack, D., Rodríguez-Concepción, M., Bohlmann, J., and Gershenzon, J. (2007). Functional identification and differential expression of 1-deoxy-D-xylulose 5-phosphate synthase in induced terpenoid resin formation of Norway spruce (Picea abies). Plant Mol. Biol. 65 243–257. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción, M., and Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer, M. (1999). The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 16 565–574. [DOI] [PubMed] [Google Scholar]
- Sapir-Mir, M., Mett, A., Belausov, E., Tal-Meshulam, S., Frydman, A., Gidoni, D., and Eyal, Y. (2008). Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiol. 148 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller, A.L., Schauvinhold, I., Larson, M., Xu, R., Charbonneau, A.L., Schmidt, A., Wilkerson, C., Last, R.L., and Pichersky, E. (2009). Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. USA 106 10865–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., and Gershenzon, J. (2008). Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry 96 49–57. [DOI] [PubMed] [Google Scholar]
- Schuhr, C.A., Radykewicz, T., Sagner, S., Latzel, C., Zenk, M.H., Arigoni, D., Bacher, A., Rohdich, F., and Eisenreich, W. (2003). Quantitative assessment of crosstalk between the two isoprenoid biosynthesis pathways in plants by NMR spectroscopy. Phytochem. Rev. 2 3–16. [Google Scholar]
- Sheen, J. (2002). A transient expression assay using Arabidopsis mesophyll protoplasts. Available at http://genetics.mgh.harvard.edu/sheenweb/.
- Steliopoulos, P., Wüst, M., Adam, K.-P., and Mosandl, A. (2002). Biosynthesis of sesquiterpene germacrene D in Solidago canadensis: 13C and 2H labeling studies. Phytochemistry 60 13–20. [DOI] [PubMed] [Google Scholar]
- Takahashi, S., Kuzuyama, T., Watanabe, H., and Seto, H. (1998). A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. USA 95 9879–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl, D., Croteau, R., and Gershenzon, J. (2001). Partial purification and characterization of the short-chain prenyltransferases, geranyl diphosphate synthase and farnesyl diphosphate synthase, from Abies grandis (grand fir). Arch. Biochem. Biophys. 386 233–242. [DOI] [PubMed] [Google Scholar]
- Tholl, D., Kish, C.M., Orlova, I., Sherman, D., Gershenzon, J., Pichersky, E., and Dudareva, N. (2004). Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 16 977–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie, C.C.N., Ament, K., Schmidt, A., Lange, T., Haring, M.A., and Schuurink, R.C. (2007). Geranyl diphosphate synthase is required for biosynthesis of gibberellins. Plant J. 52 752–762. [DOI] [PubMed] [Google Scholar]
- Varbanova, M., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, K.H., and Elmadfa, I. (2003). Biological relevance of terpenoids: Overview focusing on mono-, di and tetraterpenes. Ann. Nutr. Metab. 47 95–106. [DOI] [PubMed] [Google Scholar]
- Wang, G., and Dixon, R.A. (2009). Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. USA 106 9914–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K., and Ohnuma, S.-I. (1999). Chain-length determination mechanism of isoprenyl diphosphate synthases and implications for molecular evolution. Trends Biochem. Sci. 24 445–451. [DOI] [PubMed] [Google Scholar]
- Wildung, M.R., and Croteau, R.B. (2005). Genetic engineering of peppermint for improved essential oil composition and yield. Transgenic Res. 14 365–372. [DOI] [PubMed] [Google Scholar]
- Wise, M.L., and Croteau, R. (1999). Monoterpene biosynthesis. In Comprehensive Natural Product Chemistry: Isoprenoids Including Carotenoids and Steroids, Vol. 2, D.E. Cane, ed (Oxford, UK: Pergamon Press), pp. 97–153.
- Wu, S., Schalk, M., Clark, A., Miles, R.B., Coates, R., and Chappell, J. (2006). Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat. Biotechnol. 24 1441–1447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.