Abstract
In rice (Oryza sativa), brassinosteroids (BRs) induce cell elongation at the adaxial side of the lamina joint to promote leaf bending. We identified a rice mutant (ili1-D) showing an increased lamina inclination phenotype similar to that caused by BR treatment. The ili1-D mutant overexpresses an HLH protein homologous to Arabidopsis thaliana Paclobutrazol Resistance1 (PRE1) and the human Inhibitor of DNA binding proteins. Overexpression and RNA interference suppression of ILI1 increase and reduce, respectively, rice laminar inclination, confirming a positive role of ILI1 in leaf bending. ILI1 and PRE1 interact with basic helix-loop-helix (bHLH) protein IBH1 (ILI1 binding bHLH), whose overexpression causes erect leaf in rice and dwarfism in Arabidopsis. Overexpression of ILI1 or PRE1 increases cell elongation and suppresses dwarf phenotypes caused by overexpression of IBH1 in Arabidopsis. Thus, ILI1 and PRE1 may inactivate inhibitory bHLH transcription factors through heterodimerization. BR increases the RNA levels of ILI1 and PRE1 but represses IBH1 through the transcription factor BZR1. The spatial and temporal expression patterns support roles of ILI1 in laminar joint bending and PRE1/At IBH1 in the transition from growth of young organs to growth arrest. These results demonstrate a conserved mechanism of BR regulation of plant development through a pair of antagonizing HLH/bHLH transcription factors that act downstream of BZR1 in Arabidopsis and rice.
INTRODUCTION
Plant development and morphogenesis is driven by cell elongation and expansion as well as cell division. As such, plant cell elongation is regulated by both environmental signals and multiple endogenous hormones, including brassinosteroid (BR), a plant steroid hormone that plays a major role in promoting cell elongation. In addition, BR regulates multiple developmental and physiological processes, such as vascular differentiation, reproductive development, photomorphogenesis, and stress responses (Clouse and Sasse, 1998; Bishop and Koncz, 2002; Fukuda, 2004). BR-deficient and BR-insensitive mutant plants show dwarfism, dark-green leaves, male sterility, and constitutive photomorphogenesis in the dark (Chory et al., 1991; Szekeres et al., 1996; Li and Chory, 1997; Mori et al., 2002).
Extensive molecular genetic and biochemical studies in Arabidopsis thaliana have illustrated a BR signal transduction pathway from the BRI1 receptor kinase at the cell surface to the BZR transcription factors, which regulate BR-responsive gene expression (Vert et al., 2005; Wang et al., 2006b). In the absence of BR, the BZR transcription factors BZR1 and BZR2 (also named BES1) are phosphorylated by the GSK3-like kinase BIN2 (He et al., 2002; Li and Nam, 2002; Yin et al., 2002) and consequently inactivated due to retention in the cytoplasm through interaction with 14-3-3, degradation by the proteasome, and loss of DNA binding activity (He et al., 2002; Vert and Chory, 2006; Gampala et al., 2007). BR binding to the extracellular domain of the BRI1 receptor kinase activates its kinase activity (He et al., 2000; Kinoshita et al., 2005; Wang et al., 2005), partly by inducing dissociation of the inhibitory BKI1 protein and association/transphosphorylation with the coreceptor kinase BAK1 (Wang and Chory, 2006). Activated BRI1 phosphorylates the BR-signaling kinases (Tang et al., 2008), which bind to the BSU1 phosphatase. BSU1 dephosphorylates BIN2 at a conserved phosphotyrosine residue and inactivates BIN2, leading to nuclear accumulation of unphosphorylated BZR1 and BZR2 (Kim et al., 2009) and BR-responsive genes expression and plant growth.
BZR1 and BZR2 share 88% over all sequence identity and play major roles in BR regulation of gene expression and plant growth (Wang et al., 2002; Yin et al., 2002). Gain-of-function mutation of either of them, bzr1-1D or bes1-D, suppresses BR-deficient and insensitive phenotypes (Wang et al., 2002; Yin et al., 2002). BZR1 specifically binds to the BR response element (CGTGT/CG) and represses gene expression (He et al., 2005). On the other hand, BES1 was shown to interact with a basic helix-loop-helix (bHLH) transcription factors (BIM1) and together bind to the E-box of a BR-induced gene. BES1 also functions together with MYB30 to promote BR target gene expression (Yin et al., 2005; Li et al., 2008).
Compared with the well-defined BR signaling pathway in Arabidopsis, much less is known about BR signaling in monocot. Studies of BRI1 and BZR1 in rice (Oryza sativa) suggest a conserved BR signaling mechanism (Yamamuro et al., 2000; Bai et al., 2007), although a BR-signaling function has not been demonstrated for the rice homologs of other components of the Arabidopsis BR signaling pathway. There is evidence that the rice heterotrimetric G protein α1 plays a role in BR signaling (Wang et al., 2006a; Oki et al., 2008). A few BR-regulated rice genes have been shown to contribute to BR-regulated development, including a direct target gene of BZR1 (Tong et al., 2009; Wang et al., 2009). Furthermore, many studies have shown important roles of BR in specific developmental processes. For example, overexpression of BR biosynthetic gene increases grain filling and rice yield (Wu et al., 2008). One of the most sensitive BR responses in rice is lamina joint inclination (Wada et al., 1981). Weakly BR-deficient or insensitive rice plants have erect leaves, which allow higher growth density and higher grain yield per hectar (Sakamoto et al., 2006), whereas increasing BR biosynthesis or signaling enhances the rice lamina joint bending (Hong et al., 2005; Bai et al., 2007; Nakamura et al., 2009). Thus genetic analysis based on rice leaf bending is likely to lead to insights into the molecular mechanism of growth regulation by BR in monocots.
The bHLH proteins are a group of important transcription factors that carry out diverse functions in both animals and plants. For example, bHLH transcription factors play key roles in phytochrome signal transduction, cell fate determination, stomata differentiation, and BR-responsive gene expression (Bernhardt et al., 2005; Duek and Fankhauser, 2005; Serna, 2007). Typical bHLH proteins contain the basic region that serves as a DNA binding motif. A class of HLH proteins represented by the human Id proteins (Inhibitor of DNA binding) (Sun et al., 1991) lack the basic region and cannot bind DNA but specifically inhibit other bHLH transcription factors by heterodimerization through the HLH region (Ruzinova and Benezra, 2003). Through inhibiting bHLH proteins such as E, the Id proteins play critical roles in cell differentiation and development in mammals (Ruzinova and Benezra, 2003; Kee, 2009). An Arabidopsis HLH protein has been found to interact with and inhibit a bHLH transcription factor involved in light regulation of plant development (Hyun and Lee, 2006).
In this study, we identified a rice mutant (ili1-D) showing phenotypes of increased lamina joint inclination and hypersensitivity to BR, which are caused by overexpression of a HLH transcription factor (ILI1). We further identified another bHLH transcription factor, IBH1, as an ILI1 binding protein. Transgenic experiments indicate that ILI1 and Os IBH1, as well as their Arabidopsis homologs, PRE1 and At IBH1, antagonize each other in regulating cell elongation. BR activates the expression of ILI1 and PRE1 but represses IBH1 as direct target genes of BZR1 in rice and Arabidopsis. Our results demonstrate that ILI1-Os IBH1 and PRE1-At IBH1 are two conserved pairs of antagonistic HLH/bHLH transcription factors that function downstream of BZR1 to mediate BR regulation of cell elongation in rice and Arabidopsis. We further propose that members of the ILI1/PRE1 family integrate multiple signaling pathways to regulate cell elongation and plant development.
RESULTS
Isolation and Characterization of the ili1-D Mutant
One of the most sensitive BR responses in rice is the leaf inclination (Wada et al., 1981). To find new components of the BR pathway in rice, we screened a large collection of T-DNA insertion rice lines (Wan et al., 2009) and identified a mutant with increased leaf inclination (ili1-D) (Figures 1A and 1B). Measurement of the leaf angles showed that most leaves of the wild type bend around 30° and those of the ili1-D mutant bend around 120° to 150° (see Supplemental Figure 1 online). Close-up views by scanning election microscopy show increased growth of cells on the adaxial side of the lamina joint in ili1-D, similar to that of BR treated wild-type rice (Figure 1C). When treated with increasing concentrations of 24-epibrassinolide (eBL; an active form of BRs), lamina joint of ili1-D reached maximum bending (180°) at ∼10 nM eBL, whereas maximum leaf bending of wild-type rice requires about two magnitude higher concentration of eBL (1 μM) (Figure 1D). The coleoptile elongation of ili1-D also showed higher sensitivity to BR treatment than wild-type rice (Figure 1E). These observations suggest that the ili1-D mutant is hypersensitive to BR. Measurement of BR levels did not show significant difference in BR content between ili1-D and wild-type rice (see Supplemental Figure 2 online). These results suggest that ili1-D acts in a branch or downstream of the BR regulatory pathway.
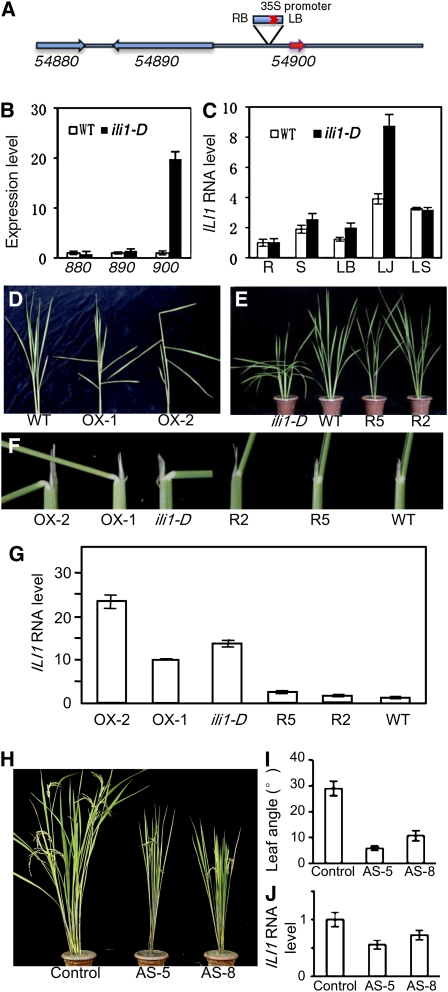
Figure 1.
The ili1-D Mutant Shows Increased Lamina Joint Inclination and BR Sensitivity.
(A) Seven-day-old rice seedlings of wild type, wild type treated with 10 ng/μL eBL for 2 d (WT+BL), and the ili1-D mutant. Arrows point to the lamina joints.
(B) Tillering-stage wild type and the ili1-D mutant plants grown in soil.
(C) Scanning electron microscopy images of the adaxial surface of the lamina joint of wild type, wild type treated with 10 ng/μL eBL, and the ili1-D mutant. Bars = 300 μm.
(D) Lamina inclination angles of the wild type and ili1-D grown on medium containing various concentration of eBL. Bars indicate sd (n = 12).
(E) Coleoptile lengths of the wild type and ili1-D grown in medium containing indicated concentration of eBL. Bars indicate sd (n = 15).
[See online article for color version of this figure.]
The Increased Leaf Inclination of ili1-D Was Caused by Overexpression of an HLH Transcription Factor
Segregation analysis of the heterozygous population showed a 3:1 ratio of ili1-D mutants to wild type, indicating that ili1-D is a dominant mutant. A T-DNA flanking sequence was identified in ili1-D using thermal asymmetric interlaced-PCR (Liu et al., 1995). The BLASTn results showed that the T-DNA was inserted between LOC_Os04g54890 and LOC_Os04g54900 on chromosome 4 (Figure 2A). Quantitative RT-PCR analysis showed that the expression of Os04g54900 was dramatically increased in the ili1-D mutant plants, but the other flanking genes were not significantly affected (Figure 2B). Expression of Os04g54900 is preferentially expressed in the lamina joint section and is increased only in lamina joint in the ili1-D mutant (Figure 2C).
Figure 2.
The Enlarged Leaf Angle of ili1-D Is Caused by Overexpression of a HLH Transcription Factor.
(A) A diagram of the genomic region flanking the T-DNA insertion site in ili1-D.
(B) Quantitative RT-PCR analysis of the expression of the genes surrounding the T-DNA insertion. Tubulin was used as a control.
(C) Tissue-specific expression of ILI1 RNA in the wild type and ili1-D rice analyzed by quantitative RT-PCR in seedlings at trefoil stage. R, root; S, stem; LB, leaf blade; LJ, lamina joint; LS, leaf sheath.
(D) Overexpression of ILI1 in transgenic rice (two independent lines: OX-1 and OX-2) recapitulates the increased laminar inclination phenotype of ili1-D.
(E) Suppressing ILI1 expression using RNAi in the ili1-D mutant background (two independent lines: R2 and R5) rescues the lamina inclination phenotype.
(F) Close-up view of individual lamina joint of ILI1-OX and ili1-D/ILI1-RNAi transgenic plants shown in (C) and (D).
(G) Quantitative RT-PCR analysis of the expression levels of ILI1 in the control, ILI1-OX, and ILI1-RNAi transgenic plants.
(H) to (J) Antisense suppression of ILI1 expression in wild-type rice causes erect leaves.
(H) ILI1-AS transgenic plants (AS5 and AS8) at the heading stage.
(I) Quantitation of the lamina joint angles of the flag leaves.
(J) Quantitative RT-PCR analysis of the expression levels of ILI1 in the control and ILI1-AS plants. The negative transgenic plant was used as control. Error bars indicate sd.
To test whether the high expression of Os04g54900 lead to enlarged leaf angle of the ili1-D mutant, we transformed wild-type rice with a construct of cauliflower mosaic virus 35S promoter driving expression of the Os04g54900 cDNA. A total of 42 transgenic lines were generated, and about two-thirds of them showed no obvious phenotype. The other lines showed phenotypes that range from extremely distorted and retarded growth and infertility (seven plants; see Supplemental Figure 3 online) to semidwarf plants with increased lamina joint bending (approximately eight lines) (Figures 2D, 2F, and 2G). Furthermore, suppressing the expression of Os04g54900 in the ili1-D mutant using RNA interference (RNAi) restored wild-type erect leaves (Figures 2E to 2G). These results indicate that overexpression of Os04g54900 is responsible for the ili1-D phenotype, and we thus named the Os04g54900 gene ILI1 (for Increased Leaf Inclination1). Antisense suppression of ILI1 expression in wild-type background (ILI1-AS) caused an erect leaf phenotype, confirming an essential role of ILI1 in regulating lamina joint bending (Figures 2H to 2J).
ILI1 encodes a 105–amino acid group-D bHLH transcription factor that is characterized by harboring only the HLH domain but lacking the basic region. Protein BLAST search showed that ILI1 has seven homologous genes in the rice genome and is homologous to members of the Arabidopsis PRE (Paclobutrazol resistance) family, tomato (Solanum lycopersicum) Style2.1, and mammalian Id protein (see Supplemental Figures 4 and 5 online). Arabidopsis PRE1 has been implicated in gibberellic acid (GA) responses, as overexpression of PRE1 rescues GA-deficient phenotypes and promotes seed germination and elongation of hypocotyls and petioles (Lee et al., 2006). Tomato Style 2.1 has been shown to promote cell elongation in the style (Chen et al., 2007). Our finding of the role of ILI1 in promoting elongation of lamina joint cells suggests a conserved role of this gene family in promoting plant cell elongation.
ILI1 and PRE1 Interact with Os IBH1 and At IBH1, Two Homologous bHLH Transcription Factors
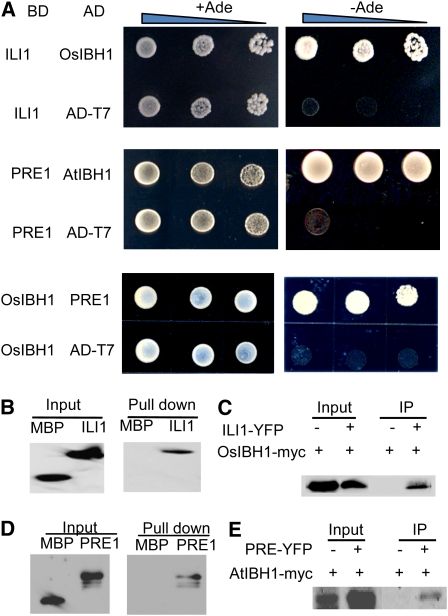
Studies of human Id proteins showed that this type of HLH transcription factors act through interacting with other typical bHLH proteins (Ruzinova and Benezra, 2003). To investigate the functional mechanism of ILI1, we performed a yeast two-hybrid screen for interacting proteins using the full-length ILI1 protein as bait. From three million yeast transformants, we identified 10 clones that grew on the -Ade/-His selection medium. Three of these positive clones contain the same cDNA that encodes a typical bHLH transcription factor that we named IBH1 (for ILI1 Binding bHLH Protein1) (Figure 3A). Sequence analysis of IBH1 showed that the Arabidopsis protein most similar to Os IBH1 is At2g43060 (see Supplemental Figure 6 online). We named At2g43060 as At IBH1. Yeast two-hybrid experiments showed that both At IBH1 and Os IBH1interact with PRE1 in yeast, suggesting evolutionary conservation of the interaction (Figure 3A).
Figure 3.
ILI1 and Its Arabidopsis Homolog PRE1 Interact with Rice and Arabidopsis IBH1.
(A) A yeast two-hybrid screen identified Os IBH1 as an ILI1-interacting protein, and the interaction is conserved between their Arabidopsis homologs PRE1 and At IBH1. Three clones of yeast containing each combination of bait (BD) and prey (AD) vectors were grown on medium with Ade or without Ade. AD-T7, pGAD-T7 empty vector was used as a negative control.
(B) ILI1 interacts with Os IBH1 in vitro. Glutathione agarose beads containing GST-Os IBH1 were incubated with equal amounts of MBP or MBP-ILI1. Proteins bound to GST-OsIBH1 were detected by immunoblotting using anti-MBP antibody.
(C) Co-IP assay showing ILI1 interaction with Os IBH1. Tobacco leaves coexpressing 35S:ILI1-YFP and 35S:OsIBH1-myc or only 35S:OsIBH1-myc were used to immunoprecipitate with anti-GFP antibody, and the immunoblot was probed with anti-myc antibody.
(D) and (E) PRE1 interacts with At IBH1 in vitro and in vivo.
(D) In vitro pull-down assay, performed as in (C), of the interaction between GST-AtIBH1 and MBP-PRE1.
(E) Arabidopsis plants expressing At IBH1-myc only or coexpressing At IBH1-myc and PRE1-YFP were used to immunoprecipitate with anti-GFP antibody, and the immunoblot was probed with anti-myc antibody.
[See online article for color version of this figure.]
To confirm the interaction between ILI1 and IBH1, we performed a glutathione S-transferase (GST) pull-down assay using purified protein expressed from Escherichia coli. As shown in Figure 3B, GST-Os IBH1 can pull down maltose binding protein (MBP)-ILI1 but cannot pull down MBP alone, indicating that ILI1 interacts with IBH1 in vitro. To determine whether ILI1 interacts with IBH1 in vivo, we performed a coimmunoprecipitation (Co-IP) assay. When ILI1-YFP and Os IBH1-Myc fusion proteins were transiently coexpressed in tobacco (Nicotiana tabacum) leaf cells (Figure 3C), the IBH1-Myc fusion protein can be coimmunoprecipitated with ILI1-YFP. In vitro pull-down assay (Figure 3D) and Co-IP assay using transgenic Arabidopsis (Figure 3E) also confirmed that PRE1 binds to At IBH1 in vitro and in vivo. These results demonstrate conserved interactions between ILI1/PRE1 and IBH1 in rice and Arabidopsis.
ILI1/PRE1 and IBH1 Function Antagonistically in Regulating Plant Development
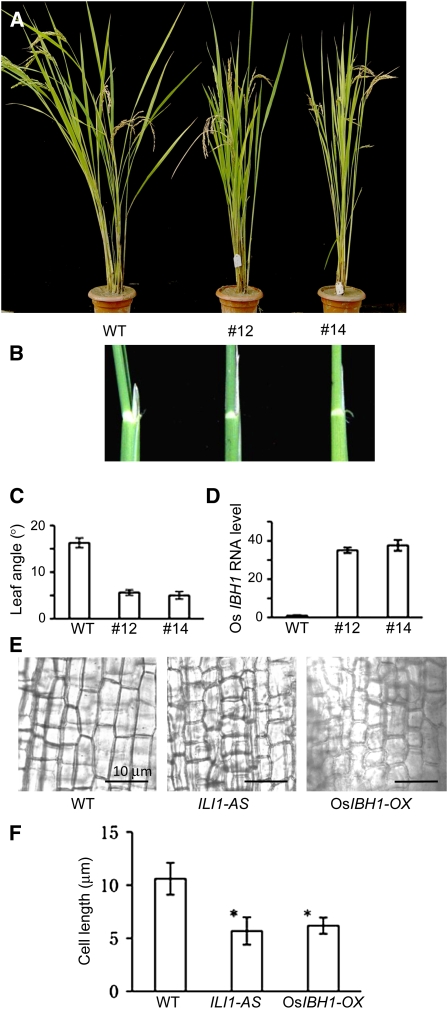
To study the function of IBH1 in plant development, we overexpressed IBH1 in transgenic rice. The 35S:Os IBH1-GFP (green fluorescent protein) transgenic plants showed erect leaves (Figure 4A). The angles of leaf inclination were significantly reduced in the IBH1-overexpressing transgenic plants compared with the wild type (Figures 4B to 4D). The reduced leaf bending in both ILI1-AS and IBH1-OX plants was due to reduced cell elongation in the adaxial side of the lamina joint (Figures 4E and 4F). As such, IBH1 appears to have an opposite action on cell elongation and lamina bending than its interacting protein ILI1.
Figure 4.
Overexpression of IBH1 in Rice Causes an Erect Leaf Phenotype.
(A) The wild-type control plant and 35S:OsIBH1-GFP transgenic rice plants (lines 12 and 14) at heading stage.
(B) The lamina joints of the wild-type and 35S:OsIBH1-GFP transgenic rice plants.
(C) Quantitation of leaf angle of the control and 35S:OsIBH1-GFP transgenic plants. Data from 12 to 15 leaves of each plant. Bars indicate sd.
(D) Os IBH1 RNA expression levels in the wild-type and 35S:OsIBH1-GFP transgenic rice plants. Bars indicate sd.
(E) Cells of lamina joint of wild-type control, ILI1-AS, and OsIBH1-OX plants.
(F) Measurement of the lengths of cells shown in (E). Bars are means ± sd. Asterisks indicate significant difference from the wild type (P < 0.05, Student's t test).
[See online article for color version of this figure.]
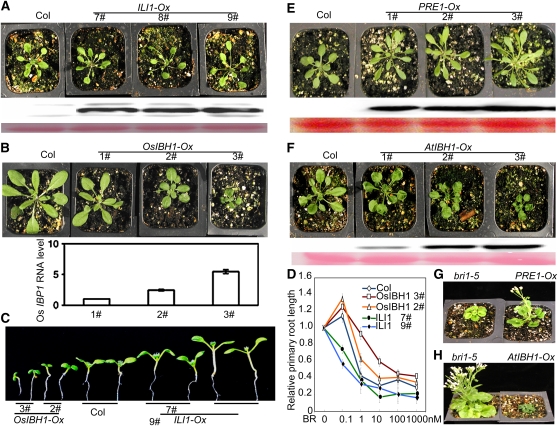
To further study the function of ILI1 and IBH1 and their Arabidopsis homologs, we overexpressed ILI1, PRE1, Os IBH1, and At IBH1 in transgenic Arabidopsis. The ILI1-overexpression (ILI1-OX) transgenic plants displayed elongated petioles and light-green and slightly narrow leaves (Figure 5A), whereas the Os IBH1-OX transgenic plants exhibited shorter petioles and dark-green and round leaves (Figure 5B). The ILI1-OX plants showed increased hypocotyl length and increased BR sensitivity in root inhibition assay, whereas Os IBH1-OX plants showed reduced hypocotyl length and reduced BR sensitivity (Figures 5C and 5D). These results suggest that rice ILI1 and IBH1 can promote and inhibit BR responses in Arabidopsis as they do in rice. Consistent with such conserved roles of ILI1 and IBH1, the PRE1-OX plants displayed similar phenotypes as the ILI1-OX plants (Figure 5E), whereas At IBH1-OX plants showed similar dwarf phenotypes (Figure 5F) as the Os IBH1-OX plants. These results indicated that the function of ILI1/PRE1 and IBH1 is conserved in monocots and dicots, and ILI1/PRE1 promotes cell elongation, whereas IBH1 inhibits cell elongation. Overexpression of PRE1 suppressed the weak allele of bri1-5 (Figure 5G), whereas At IBH1-OX enhanced the dwarf phenotype of bri1-5 (Figure 5H), supporting a role of these interacting HLH/bHLH factors downstream of BRI1 in the BR pathway.
Figure 5.
ILI1/PRE1 and IBH1 Function Antagonistically in Regulating Plant Growth.
(A) Overexpression of ILI1 in Arabidopsis increases petiole elongation. Panels from top to bottom are pictures of wild-type (Columbia) and 35S:ILI1-YFP plants (ILI1-Ox) grown in soil for 3 weeks, immunoblot probed using GFP antibody, and the gel blot stained for protein with Ponceau S.
(B) Overexpression of Os IBH1 in Arabidopsis inhibits petiole elongation. The 35S:OsIBH1-GFP plants (OsIBH1-Ox) were grown in soil for 3 weeks. The bottom panels show RT-PCR analysis of the expression levels of Os IBH1 in the plants with UBQ5 as a loading control.
(C) Seedling of OsIBH1-Ox and ILI1-Ox transgenic lines grown on half-strength Murashige and Skoog (MS) medium in constant light for 7 d.
(D) The ILI1-Ox and OsIBH1-Ox transgenic plants show increased and decreased sensitivity to BR, respectively. Seedlings were grown on medium containing different concentrations of BL for 7 d under constant light. Relative root lengths were average of 30 plants and normalized to the untreated plants. Error bars represent sd.
(E) Overexpression PRE1-YFP (PRE1-Ox) causes longer petioles. The plants were grown in soil for 3 weeks. Bottom panels show immunoblot probed with anti-GFP antibodies and Ponceau S staining for loading control.
(F) The 35S:AtIBH1-Myc (AtIBH1-Ox) transgenic plants show shorter petioles. The plants were grown in soil for 3 weeks. Bottom panels show immunoblot probed with anti-myc antibodies and Ponceau S staining for loading control.
(G) Overexpression of PRE1-YFP partly suppresses the bri1-5 phenotype.
(H) Overexpression of At IBH1-Myc enhances the bri1-5 dwarf phenotype.
The opposite effects on cell elongation, physical interaction between IBH1 and ILI1/PRE1, suggest that ILI1/PRE1 might inhibit IBH1 in a manner similar to the inhibition of the E bHLH factors by Id in humans (Ruzinova and Benezra, 2003). To address this possibility, we crossed the Os IBH1-OX transgenic Arabidopsis plants with ILI1-OX transgenic Arabidopsis plants and At IBH1-OX transgenic plants with PRE1-OX transgenic plants. As shown in Figure 6, all F1 plants exhibited a long petiole phenotype similar to that of the ILI1-OX or PRE1-OX transgenic plants (Figures 6A and 6B). These results suggest that ILI1/PRE1 inhibits the activity of IBH1 in vivo.
Figure 6.
ILI1 and PRE1 Suppress the Activity of IBH1 in Vivo.
(A) Four-week-old plants of ILI1-Ox, OsIBH1-Ox, and their cross progeny (F1). The bottom panels show the PCR genotyping of the transgenes.
(B) Four-week-old PRE1-Ox and AtIBH1-Ox plants and their cross progeny (F1). The bottom panels show immunoblots probed with anti-myc or anti-GFP antibodies.
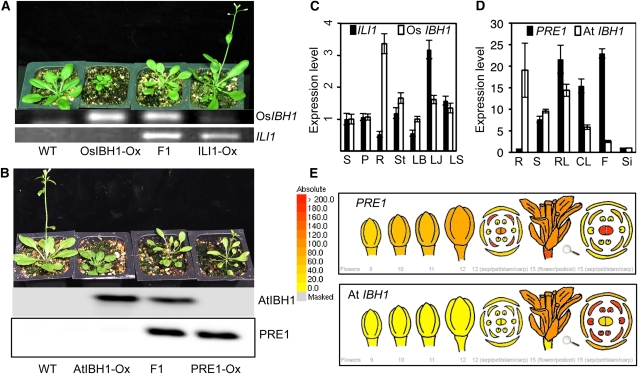
(C) Quantitative RT-PCR analysis of ILI1 and Os IBH1 in various tissues of rice. S, stem; P, panicle; R, root; LB, leaf blade; LJ, lamina joint; LS, leaf sheath. Error bars indicate sd.
(D) Quantitative RT-PCR analysis of PRE1 and At IBH1 in various tissues of Arabidopsis. R, root; S, seedling; RL, rosette leaf; CL, cauline leaf; F, unopen floral buds; Si, silique. Error bars indicate sd.
(E) Relative expression levels of PRE1 and At IBH1 in floral buds (left) and the open flower (right) based on microarray data displayed in the eFP browser (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Color scale shows microarray signal level.
To understand the developmental context in which these HLH/bHLH pairs function, we analyzed their tissue-specific expression. Quantitative RT-PCR analysis showed that rice ILI1 and IBH1 are coexpressed in rice seed, panicle, root, leaf blade, lamina joint, leaf sheath, and stem, while highest expression of ILI1 was observed in lamina joint and IBH1 in roots (Figure 6C). The quantitative RT-PCR analysis on Arabidopsis also showed overlapping expression patterns of PRE1 and IBH1 in various tissues (Figure 6D). Based on Arabidopsis microarray data displayed in the eFP browser, PRE1 is expressed at a high level in young and growing tissues, whereas IBH1 is expressed in mature organs where growth is arrested (Figure 6E; see Supplemental Figure 7 online). In particular, the expression level of PRE1 is high in the floral buds until right before the flower opens, with the highest level observed in petals of opening floral buds, the most rapidly growing organ. In open flowers, PRE1 level is low in the outer floral organs (sepals, petals, and stamens) that have stopped growing, but remains high in carpals, which start rapid growth upon fertilization. By contrast, IBH1 is not expressed in floral buds but is highly expressed in the growth-arrested outer floral organs of open flowers (Figure 6E). PRE1 is also expressed at a higher level than IBH1 in the meristem and at an early stage of seed development, whereas IBH1 is highly expressed in mature or senescing leaves and at later stages of seed development (see Supplemental Figure 7 online). Such sequential and overlapping expression patterns of PRE1 and IBH1 are consistent with their positive and negative roles in cell growth and suggest that downregulation of PRE1 and upregulation of IBH1 plays a role in the transition from growth of young organs to cessation of growth in mature organs.
In Rice, ILI1/IBH1, and in Arabidopsis, PRE1/IBH1, Are Direct Target Genes of BZR1
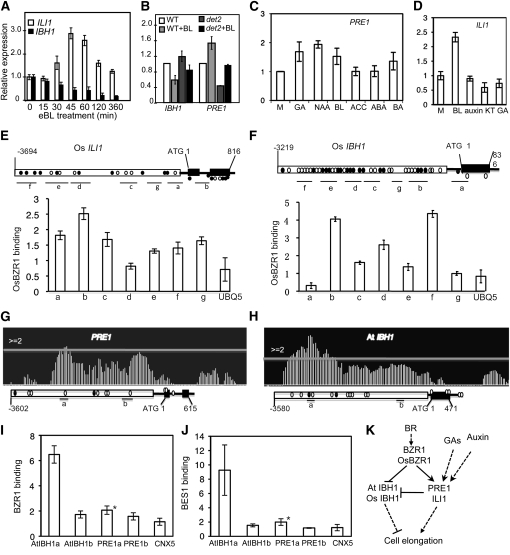
To understand the functional mechanism of ILI1/PRE1 and IBH1 in BR response, we analyzed how BR regulates the expression of these genes. BR treatment of rice seedlings increased the ILI1 RNA level and repressed the expression of IBH1 (Figure 7A). Similarly, BR treatment caused an increase of PRE1 expression but a decrease of IBH1 expression in Arabidopsis (Figure 7B). Expression of PRE1 is also activated by auxin and GA, two other hormones that promote cell elongation (Figure 7C), consistent with a role of PRE1 in cell responses to these hormones in Arabidopsis. Interestingly, auxin and GA did not significantly affect the expression of ILI1 (Figure 7D), consistent with weaker effects and independent pathways of these hormones on lamina joint inclination (Nakamura et al., 2009). It is also possible that auxin and GA act through other homologs of ILI1 to promote cell elongation in their target tissues in rice.
Figure 7.
BR Increases the Transcript Levels of ILI1 and PRE1, but Represses Os IBH1 and At IBH1 Directly through OsBZR1 and BZR1 in Rice and Arabidopsis.
(A) Quantitative RT-PCR analysis of the transcripts of ILI1 and Os IBH1 in wild-type rice treated with 1 μM 24-eBL for different times.
(B) Quantitative RT-PCR analysis of the transcripts of PRE1 and At IBH1 in wild-type Arabidopsis and det2 treated with 100 nM eBL or mock solution for 2 h.
(C) and (D) Quantitative RT-PCR analysis of the effects of different plant hormones on the expression of PRE1 (C) and ILI1 (D).
(C) Seven-day-old Arabidopsis seedling were treated for 1 h with mock solution (M), 50 μM gibberellic acid (GA), 1 μM naphthalenacetic acid (NAA), 100 nM bassinolide (BL), 2 μM 1-aminocyclopropane-1-carboxylic acid (ACC), 100 μM abscisic acid (ABA), or 100 nM 6-benzyl aminopurine (6-BA).
(D) Rice seedlings were soaked in water containing no hormone (M), 10 μM eBL, 10 μM 2,4-D, 100 μM kinetin (KT), or 100 μM GA for 3 h.
(E) to (L) ChIP assays of binding of Os BZR1 ([E] and [F]), BZR1 ([G] to [I]), and BES1 (J) to the promoters of ILI1 (E), Os IBH1 (F), PRE1, and At IBH1 ([G] to [J]). For gene structure diagrams for ILI1 (E), Os IBH1 (F), PRE1 (G), and At IBH1 (H), open boxes show promoter regions, black lines show untranslated regions and introns, black boxes show coding sequences, black circles show BR response element motifs, and white circles show putative E-box motifs. Regions analyzed by quantitative PCR are shown by short lines marked with letters (a to g), and the quantity of binding by Os BZR1, BZR1, or BES1 is shown in the bar graph of (E), (F), (I), and (J) as fold of enrichment over the control sample. Asterisks indicate significant differences (P < 0.05) from UBQ5 and CNX5, which are control genes in rice and Arabidopsis. Error bars indicate sd.
(G) and (H) The BZR1 ChIP-microarray data displayed by Integrated Genome Browser software at selected chromosomal regions marked by the gene structure of PRE1 and At IBH1. The horizontal lines indicate the cutoff of twofold enrichment.
(K) A model for the functions of PRE1/ILI1 and IBH1 in BR response in Arabidopsis and rice. IBH1 negatively regulates cell elongation. BR signaling directly represses the transcription of IBH1 and activates PRE1/ILI1 through BZR1 binding to their promoters. PRE1/ILI1 inhibits IBH1 at the protein level by heterodimerization. Such double repression of IBH1 at the transcriptional and protein levels effectively releases its inhibition of cell elongation, leading to robust growth responses, such as the BR-induced lamina inclination. PRE1/ILI1 or their homologs might also be targets for other signals that regulate cell elongation, such as GA and auxin.
Previous studies have shown that BR signaling primarily regulates the activity of the transcription factor BZR1. We further tested whether BZR1 directly regulates PRE1 and IBH1 in Arabidopsis and whether BZR1 regulates ILI1 and IBH1 in rice using chromatin immunoprecipitation (ChIP) experiments. ChIP was performed using Os BZR1-GFP transgenic rice (Bai et al., 2007) and BZR1-CFP transgenic Arabidopsis (Wang et al., 2002) plants. The ChIP DNA was analyzed for ILI1 and Os IBH1 whole promoter (Figures 7E and 7F) and two different promoter regions of PRE1 and At IBH1 that were chosen based on the BZR1 ChIP microarray data (Figures 7G and 7H). Quantitative PCR analysis showed that the IBH1 promoter DNA was enriched by immunoprecipitation of BZR1-GFP in rice (Figure 7F), and IBH1 promoter was enriched by BZR1-CFP in Arabidopsis (Figures 7H and 7I). ILI1 and PRE1 were also significantly enriched, though to a lesser degree (Figures 7E, 7G, and 7I). These results demonstrate that BR directly represses IBH1 transcription and activates the expression of PRE1 and ILI1 through BZR1 direct binding in Arabidopsis and rice. Similar to BZR1, BES1 also binds strongly to the promoter of IBH1 but only weakly to PRE1 (Figure 7J). Together, our results demonstrate the conserved function of a pair of interacting HLH/bHLH factors in BR regulation of cell elongation in Arabidopsis and rice (Figure 7K).
DISCUSSION
Plant cell elongation is regulated by multiple signals, BR being a major one of them. On the other hand, BR regulates multiple cellular and physiological processes, with cell elongation being the major one. Many BR signaling components have been identified, and the signal transduction pathway from receptor kinase to the BZR transcription factors has been characterized in detail. In this study, we identified ILI1 as an HLH protein involved in BR-promoted elongation of rice lamina joint cells that control leaf bending. We further identified an ILI1-interacting protein in rice, IBH1, and demonstrated that ILI1 and IBH1 regulate cell elongation downstream of the BR signaling pathway. ILI1 and IBH1 are oppositely regulated by BR at the transcriptional level, and they antagonize each other at the protein level similar to the Id and E proteins in humans (Ruzinova and Benezra, 2003). We further provide evidence that the mechanism is conserved in Arabidopsis. Our results indicate that the antagonizing pair of HLH/bHLH factors, ILI1 and IBH1 in rice and PRE1 and IBH1 in Arabidopsis, act downstream of BZR1 to mediate BR regulation of cell elongation. We further propose that such HLH/bHLH pairs are conserved mechanism for cell elongation regulation by various signaling and developmental pathways.
A Pair of HLH/bHLH Factors, ILI1 and IBH1, Mediate BR Regulation of Lamina Inclination in Rice
Different cells and tissues show quantitatively or even qualitatively different responses to BR, as well as other hormones. Lamina joint bending is one of the most sensitive BR responses in rice. BR deficiency or insensitivity causes erect leaves, and increasing BR biosynthetic or signaling results in bending leaves in transgenic rice (Sakamoto et al., 2006; Bai et al., 2007). Both loss-of-function and gain-of-function experiments demonstrated that BR regulation of lamina joint bending is mediated by BZR1 (Bai et al., 2007).
In this study, we demonstrate that BZR1 regulates lamina joint inclination through the ILI1/IBH1 pair of antagonizing HLH/bHLH factors. First, both gain-of-function and loss-of-function experiments confirmed an essential role of ILI1 in lamina joint bending. Overexpression of ILI1 in the ili1-D mutant or in 35S-ILI1 transgenic plants caused severe leaf bending due to growth of the adaxial cells, similar to that caused by BR treatment, whereas suppression of ILI1 expression by RNAi or antisense rescued the ili1-D phenotype and caused erect leaves in wild-type background (Figures 2D, 2E, and 2H). Second, the ili1-D mutant showed increased response to BR in both lamina joint bending assay and coleoptile elongation assay, supporting a role of ILI1 in BR response (Figures 1D and 1E). Third, ILI1 expression level is increased by BR treatment (Figures 7A and 7D). Fourth, ILI1 interacts with a bHLH protein IBH1 (Figures 3A to 3C). Overexpression of IBH1 causes erect leaf and dwarf phenotypes, indicating that IBH1 is a negative regulator of BR response (Figures 4A and 4B). Overexpression of ILI1 is epistastic to IBH1 overexpression in Arabidopsis, supporting the notion that ILI1 inhibits IBH1 at the protein level, similar to the inhibition of bHLH proteins by the human HLH protein Id (Figure 6A). In this context, ILI can also stand for Id-like protein. Finally, we showed that IBH1 is a direct target gene of BZR1, whose expression is repressed by BR (Figures 7A and 7F). Although the precise role of IBH1 in rice development is yet to be evaluated by loss-of-function mutations, these results support a model that ILI1 and IBH1 function downstream of BZR1 to mediate BR regulation of cell elongation in rice. By activating ILI1 and repressing IBH1 at the transcriptional level, BR signaling effectively tips the balance between two counteracting factors, triggering robust downstream gene expression responses, which would contribute to the extreme BR sensitivity of the lamina joint cells. By contrast, ILI1 expression is not induced by GA and auxin (Figure 7D), consistent with their milder effect than BR on lamina joint inclination (Nakamura et al., 2009). A recent study presents evidence for auxin regulation of lamina joint bending through a novel BRI1-independent pathway (Nakamura et al., 2009).
Antagonizing HLH/bHLH Factors Function as a Conserved Central Mechanism for Regulating Cell Elongation by Multiple Signaling and Developmental Pathways
The functions of ILI1 and IBH1 in promoting and repressing cell elongation downstream of the BR pathway are conserved in Arabidopsis. Overexpression of ILI1 and its Arabidopsis homolog PRE1 increases hypocotyl and petiole elongation (Figures 5A and 5E), whereas overexpression of rice IBH1 and its Arabidopsis homolog causes dwarf phenotypes in transgenic Arabidopsis (Figures 5B and 5F). Overexpression of PRE1 suppressed the Arabidopsis bri1 (Figure 5G) and bin2 (see Supplemental Figure 8A online) mutants, whereas overexpression of its antagonizing partner IBH1 enhanced the dwarf phenotypes of these BR mutants (Figure 5H; see Supplemental Figure 8B online), suggesting a positive role of ILI1 and negative role of IBH downstream of BIN2 in the BR pathway. Consistent with the genetic interactions, RNA expression and ChIP experiments demonstrate that Arabidopsis IBH1 is a BR-repressed direct target gene of BZR1, similar to rice IBH1. Like ILI1, PRE1 RNA level is induced by BR, likely through a direct mechanism as BZR1 binds weakly but significantly to the ILI1 and PRE1 promoters, respectively (Figure 7). These results provide evidence that not only primary BR perception and signal transduction mechanisms, but also downstream transcription network of the BR pathway is conserved between dicots and monocots (Figure 7K).
Rice ILI1 and Arabidopsis PRE1 have similar structure features to the human Id proteins (see Supplemental Figure 4 online), containing a HLH domain but lacking the basic domain present in DNA binding bHLH transcription factors. Without the basic domain required for DNA binding, Id not only cannot bind DNA but also inhibits DNA binding of its interacting bHLH proteins, such as the E proteins. Id proteins play crucial roles in cell differentiation and proliferation by acting downstream of many signaling pathways, such as transforming growth factor β and bone morphogenic protein signaling pathways (Ruzinova and Benezra, 2003). Like Id, ILI1 and PRE1 seem to function as non-DNA binding HLH proteins that specifically inhibit bHLH transcription factors IBH1. Genetic and transgenic experiments indicated opposite actions of ILI1/PRE1 from IBH1 on cell elongation, and overexpression of ILI1 or PRE1 suppressed the dwarf phenotype caused by overexprssion of IBH1. As such, the activity of IBH1 is negatively regulated at the protein level by ILI1/PRE1 and at the transcriptional level by BZR1. By activating BZR1 proteins and increasing the expression of ILI1/PRE1, BR triggers double inhibition of IBH1 at both transcript and protein levels, which potentially amplifies the signal and causes robust response of downstream genes (targets of IBH1). Mild response of early BR response genes and robust response of late BR response genes have been observed in microarray analysis in Arabidopsis (Goda et al., 2004). Taken together, our results demonstrate a conserved key mechanism for BR regulation of cell elongation through a pair of antagonizing HLH/bHLH transcription factors.
BRs regulate many common developmental processes with other hormonal and light signals. Many developmental responses induced by BR are similar to those induced by GAs and auxin. In particular, both BR and GA promote seed germination, cell elongation, and flowering. BRs are also required for inhibiting photomorphogenesis in the dark and thus have opposite effects than light on seedling growth. Previous studies indicated that PRE1 is also involved in GA response. Expression of PRE1 is induced by GA, and overexpression of PRE1 can rescue the phenotype of the GA-deficient mutant ga1-3 and GA-insensitive mutant gai-1 (Lee et al., 2006). Our results demonstrate that BRs also induce the expression of PRE1, and PRE1 and its rice homolog ILI1 play an important role in BR response. Such similar regulation of PRE1 expression could explain the similar developmental and physiological effects of BR and GA. We also found that PRE1 expression is increased by auxin treatment (Figure 7C). Therefore, it appears that PRE1 is a common node for regulation of cell elongation by multiple growth-promoting hormones (Figure 7K).
There are six members of the PRE gene family in Arabidopsis and seven members of the ILI family in rice. Interestingly, the Arabidopsis PRE family and rice ILI1 family appear to have evolved after separation of monocots and dicots (see Supplemental Figure 5 online), suggesting functional specification related to specific developmental processes. The function of ILI1 in the lamina joint, a specialized organ in monocots, supports a role of the expanded PRE/ILI1 family in evolution of specific developmental processes. While both ILI1 and PRE1 are activated by BR, only Arabidopsis PRE1 but not rice ILI1 is induced by GA and auxin (Figures 7C and 7D). Whether GA and auxin regulate other members of the ILI1 family in rice remains to be tested. Another member of the Arabidopsis PRE family, PRE6/KIDARI was shown to interact with the bHLH transcription factor HFR1 (Hyun and Lee, 2006), which is involved in light signaling by phytochrome and cryptochrome. Overexpression of KIDARI reduces light sensitivity and causes a long hypocotyl phenotype. Expression of KIDARI shows circadian oscillation, and KIDARI is proposed to mediate circadian regulation of light-responsive cell elongation by inhibiting HFR1 (Hyun and Lee, 2006). A homolog of PRE1 in tomato, Style2.1, regulates cell elongation in developing styles. A mutation in the Style2.1 promoter that results in a downregulation of Style2.1 expression is responsible for short styles and autogamy in cultivated tomatoes (Chen et al., 2007). Interestingly, the spatial and temporal expression patterns of PRE1 and IBH1 during flower development are consistent with a role of PRE1 in promoting organ elongation/expansion before the flower opens and IBH1 in cessation of organ growth after flower opens. The general trend of PRE1 expression in young and growing organs and IBH1 in mature and senescing organs is consistent with higher BRI1 level and BR sensitivity of young organs than mature organs (Friedrichsen et al., 2000). These results thus support a hypothesis that growth hormones, such as BR, GA, and auxin, maintain a high level of PRE1 and low level of IBH1 in developing organs. During the transition from growing to mature nongrowing stage, PRE1 level drops and IBH1 level increases, likely as a result of reduced hormonal signaling. Such a dynamic change of two antagonizing factors would provide an effective mechanism for growth arrest as an organ matures.
It is possible that different members of the ILI1/PRE family mediate regulation of cell elongation in a different developmental or signaling context or play a redundant role in the same developmental process (Tanaka et al., 2009). We propose that the ILI1/PRE gene family, encoding key regulators of cell growth, has expanded during recent evolution to provide regulation of cell elongation by various signaling and developmental pathways, which is essential for diverse shapes and forms of different plant species under different environmental conditions. The ILI1/PRE proteins promote cell elongation by binding to and inhibiting bHLH factors, including but most likely not limited to IBH and HFR1, which inhibit cell elongation. These HLH/bHLH pairs may regulate diverse developmental processes or mediate coregulation of common developmental processes by multiple signals such as the regulation of seedling photomorphogenesis by BR, GA, auxin, and light signals. Further studies of the regulation of PRE/ILI1 factors and identification of their specific interacting bHLH factors will shed new light on the molecular mechanism of plant growth and development.
METHODS
Plant Materials and Growth Conditions
Rice plants (Oryza sativa) were grown in the field or in the greenhouse at 30°C/25°C day/night cycles. For the analysis of BR induction in coleoptile elongation and root elongation, rice seeds were sterilized with 1% NaClO and grown in half-strength MS medium with the indicated concentration of eBL (Sigma-Aldrich) at 30°C under continuous light. Seedlings were examined 7 d after germination. Wild-type Arabidopsis thaliana (Columbia-0) and BR-related mutants were grown in a growth chamber at 24°C/22°C (day/night). For hypocotyl and root growth assays, seedlings were grown on vertical phytoagar plate containing half-strength MS medium with or without indicated concentration of BL under continuous light for 7 d.
Scanning Election Microscopy
Germinated seeds with uniform coleoptile length were transplanted into soil and grown at 30°C for 7 d, and then the second lamina joint (0.5 cm long) was excised for imaging the adaxial surface. For BR treatment, 1 μL of ethanol containing 10 ng/μL eBL and 0.1% Triton 100 was spotted at the adaxial surface of the lamina joint 2 d before fixing tissues in formalin:glacial acetic acid:50% ethanol (2:1:17, v:v:v). After dehydration in a graded ethanol series, the sections were dried for 3 h (Hitachi critical point dryer, HCP-2; Hitachi Koki), surface-sprayed with gold powder, and examined under a scanning electron microscope (Hitachi S3000N).
Measurement of Endogenous BRs
Shoots of the wild type and ili1-D were harvested after growth in a greenhouse for 2 months. About 30 g of fresh tissues was frozen in liquid nitrogen and then lyophilized. The BR content was analyzed by gas chromatography–mass spectrometry as described previously (Noguchi et al., 1999; Fujioka et al., 2002).
Lamina Joint Inclination Assay
Sterilized seeds were grown for 8 d in a dark chamber. Uniform seedlings were then sampled by excising ∼2-cm segments that contained the second-leaf lamina joint under dim light condition. These were floated on distilled water containing various concentrations of 24-eBL. After incubation in a dark chamber at 30°C for 72 h, the angle between the lamina and the sheath was measured (Wada et al., 1981).
Phylogenic and Sequence Analyses
The amino acid sequences were aligned using MUSCLE 3.6 with the default settings (Edgar, 2004) and then adjusted manually using GeneDoc (version 2.6.002) software (Pittsburgh Supercomputing Center; http://www.psc.edu/biomed/genedoc/). Using the MEGA software (version 3.1; http://www.megasoftware.net/index.html) (Kumar et al., 2004) and based on full-length protein sequences, midpoint-rooted neighbor-joining trees were constructed with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed).
Total RNA Isolation and Quantitative RT-PCR Analysis
Total RNA was extracted from 2-week-old rice seedlings of the wide type, the ili1-D mutant, and transgenic rice or from 1-week-old Arabidopsis seedlings using the Trizaol RNA extraction kit (Invitrogen). The first-strand cDNAs were synthesized using AMV reverse transcriptase (Takara) and used as RT-PCR templates. Quantitative real-time PCR analyses were performed on a Mx3000P (Stratagene) using a SYBR green detection protocol according to the manufacturer's instructions. The RT-PCR was repeated at least three times for separately harvested samples. RT-PCR was performed using gene-specific primers (see Supplemental Table 1 online).
Vector Construction and Transformation
The cDNA of ILI1 was amplified by PCR from a rice cDNA library and ligated into PSN1301 or 35S-LNY binary vectors for overexpression. The ILI1 RNAi and antisense plasmids were constructed according to the method described previously (Wang et al., 2004). Full-length cDNAs of Os IBH1, PRE1, and At IBH1 without the stop codon were amplified by PCR from rice cDNA or Arabidopsis cDNA and cloned into pENTR/SD/D-TOPO vectors (Invitrogen) and then subcloned into destination vector pMDC83 (35S:C-GFP), pEarlygate101 (35S:C-YFP), p1390-Myc (35S:C-Myc), pDEST15 (N-GST), pMAL2CGW (N-MBP), pCY86 (N-GAL4BD), and pGAL4ADGW (N-GAL4AD). All binary vector constructs were transformed into Agrobacterium tumefaciens strain GV3101 or strain EHA105 and then transformed into Arabidopsis using the floral dip method and transformed into rice calli using the A. tumefaciens–mediated transformation method (Clough and Bent, 1998; Wang et al., 2004). Primers used for the construction are listed in Supplement Table 1 online.
Yeast Two-Hybrid Screening
The full-length cDNA of ILI1 was cloned into the pBD-GAL4 vector (Stratagene) and transformed into yeast strain AH109. The resulting yeast cells were then transformed with plasmid DNA derived from a rice cDNA library constructed from 3-week-old seedlings of the Japonica cultivar “Taipei309” (Bai et al., 2007). Transformants (∼3.86 × 106) were screened for growth on medium lacking histidine but containing 5 mM 3-aminotriazol. Recovered clones were then assayed for LacZ activity using a filter lift assay. The HIS3+LacZ+ yeast clones were cultured, and the prey plasmids were isolated and then transformed into Escherichia coli for sequencing.
Pull-Down Assay
Os IBH1 and At IBH1 fused to GST were purified using glutathione beads (GE Healthcare). ILI1 and PRE1 fused to maltose binding protein (MBP) were purified using amylose resin (NEB). Glutathione beads containing 1 g of GST-Os IBH1 or GST-At IBH1 were incubated with 1 g MBP, MBP-ILI1, or MBP-PRE1 in pull-down buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, and 1 mM DTT). The mixture was rotated in a cold room for 1 h, and the beads were washed five times with wash buffer (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.1% Nonidet P-40, 1 mM EDTA, and 1 mM DTT). The proteins were eluted from the beads by boiling in 50 μL 2× SDS buffer and loaded onto a SDS-PAGE gel. Gel blots were analyzed using an anti-MBP antibody (NEB).
Co-IP Assay
All procedures were performed at 4°C. Tobacco (Nicotiana tabacum) leaves expressing ILI1-YFP and Os IBH1-myc or Arabidopsis rosette leaves of 3-week-old transgenic plants were ground in IP buffer (20 mM HEPES, pH 7.5, 40 mM KCl, 1 mM EDTA, and 1% Triton X-100), filtered, and centrifuged at 20,000g for 10 min. Supernatant (1 mL) was incubated with anti-GFP coupled to Protein A sepharose beads for 30 min. Beads were washed four times with wash buffer (20 mM HEPES, pH 7.5, 40 mM KCl, and 0.1% Triton X-100), and bound proteins were eluted with 2× SDS buffer.
ChIP
ChIP was performed following the protocol described previously (He et al., 2005), using 3-week-old rice wild-type and 35S:OsBZR1-GFP transgenic rice plants or using Arabidopsis wild-type, pBZR1:BZR1-CFP transgenic Arabidopsis plants and 35S:BES1-GFP transgenic Arabidopsis plants. An affinity-purified anti-GFP polyclonal antibody was used for immunoprecipitation. The ChIP products were analyzed by quantitative real-time PCR (primer sequences are listed in Supplemental Table1 online), and enrichment was calculated as ratio between the transgenic sample and wild-type control sample. The ChIP experiments were performed with three biological replicates, from which the means and standard deviations were calculated. The ChIP microarray experiment (Figures 7G and 7H) was performed using the pBZR1:BZR1-CFP line with three biological repeats. ChIP DNAs were hybridized to the Affymetrix GeneChip Arabidopsis Tiling 1.0R array, which contains a 25-nucleotide oligo for every 35 bp of the Arabidopsis genome.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ILI1, Os04g54900; ILI2, Os11g39000; ILI3, Os03g07540; ILI4, Os06g12210; ILI5, Os02g51320; ILI6, Os03g07510; ILI7, Os10g26460. PRE1, At5g39860; PRE2, At5g15160; PRE3, At1g74500; PRE4, At3g47710; PRE5, At3g28857; PRE6, At1g26945; Style2.1, ABX82931; Id1, CAI20171; Os IBH1, Os04g0660100; and At IBH1, At2g43060.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Lamina Joint Angles of the Flag Leaf.
Supplemental Figure 2. There Is No Obvious Change of Endogenous Levels of Active BRs in the ili1-D Mutant.
Supplemental Figure 3. Some 35S:ILI1 Rice Lines Show Severe Growth Defects Compared with Wild-Type Control.
Supplemental Figure 4. Phylogenic Analyses of ILI1 in Higher Plants.
Supplemental Figure 5. The Phylogenic Tree of Rice ILIs, Arabidopsis PREs, and Related Proteins in Other Higher Plants with Sequenced Genomes.
Supplemental Figure 6. The Phylogenic Tree of Rice IBH1 (Os04g0660100), Arabidopsis IBH1 (At2g43060), and Related Proteins in Other Higher Plants.
Supplemental Figure 7. Gene Expression Pattern of At IBH1 and PRE1 Displayed by the eFP Brower.
Supplemental Figure 8. PRE1 and At IBH1 Function Downstream of BIN2 in the BR Pathway.
Supplemental Table 1. Sequences of PCR Oligos Used in This Study.
Supplemental Data Set 1. Text File of Alignment Corresponding to the Phylogenetic Tree in Supplemental Figure 5.
Supplemental Data Set 2. Text File of Alignment Corresponding to the Phylogenetic Tree in Supplemental Figure 6.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National High-Tech Research and Development (863) Project (project numbers 2006AA10A101 and 2007AA10Z104), the National Science Foundation of China (30870207 and 30328004), the National Institutes of Health (R01GM066258), and a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to S.F. (Grant 19380069). We thank Suguru Takatsuto (Joetsu University of Education) for supplying deuterium-labeled internal standards.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhi-Yong Wang (zywang24@stanford.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bai, M.Y., Zhang, L.Y., Gampala, S.S., Zhu, S.W., Song, W.Y., Chong, K., and Wang, Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132 291–298. [DOI] [PubMed] [Google Scholar]
- Bishop, G.J., and Koncz, C. (2002). Brassinosteroids and plant steroid hormone signaling. Plant Cell 14(Suppl): S97–S110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K.Y., Cong, B., Wing, R., Vrebalov, J., and Tanksley, S.D. (2007). Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318 643–645. [DOI] [PubMed] [Google Scholar]
- Chory, J., Nagpal, P., and Peto, C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D., and Sasse, J.M. (1998). BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 427–451. [DOI] [PubMed] [Google Scholar]
- Duek, P.D., and Fankhauser, C. (2005). bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 10 51–54. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen, D.M., Joazeiro, C.A., Li, J., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, S., Takatsuto, S., and Yoshida, S. (2002). An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 130 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5 379–391. [DOI] [PubMed] [Google Scholar]
- Gampala, S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., and Yoshida, S. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134 1555–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J.X., Gendron, J.M., Sun, Y., Gampala, S.S., Gendron, N., Sun, C.Q., and Wang, Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J.X., Gendron, J.M., Yang, Y., Li, J., and Wang, Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z., Wang, Z.Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., and Chory, J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288 2360–2363. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Ueguchi-Tanaka, M., Fujioka, S., Takatsuto, S., Yoshida, S., Hasegawa, Y., Ashikari, M., Kitano, H., and Matsuoka, M. (2005). The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17 2243–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun, Y., and Lee, I. (2006). KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61 283–296. [DOI] [PubMed] [Google Scholar]
- Kee, B.L. (2009). E and ID proteins branch out. Nat. Rev. Immunol. 9 175–184. [DOI] [PubMed] [Google Scholar]
- Kim, T.-W., Guan, S., Sun, Y., Deng, Z., Tang, W., Shang, J., Sun, Y., Burlingame, A.L., and Wang, Z.-Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11 1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Cano-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., and Chory, J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433 167–171. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lee, S., Yang, K.Y., Kim, Y.M., Park, S.Y., Kim, S.Y., and Soh, M.S. (2006). Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 47 591–600. [DOI] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., and Nam, K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li, L., Yu, X., Thompson, A., Guo, M., Yoshida, S., Asami, T., Chory, J., and Yin, Y. (2008). Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Mori, M., Nomura, T., Ooka, H., Ishizaka, M., Yokota, T., Sugimoto, K., Okabe, K., Kajiwara, H., Satoh, K., Yamamoto, K., Hirochika, H., and Kikuchi, S. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A., Fujioka, S., Takatsuto, S., Tsujimoto, M., Kitano, H., Yoshida, S., Asami, T., and Nakano, T. (2009). Involvement of C-22-hydroxylated brassinosteroids in auxin-induced lamina joint bending in rice. Plant Cell Physiol. 50 1627–1635. [DOI] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., Feldmann, K.A., and Tax, F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki, K., Inaba, N., Kitagawa, K., Fujioka, S., Kitano, H., Fujisawa, Y., Kato, H., and Iwasaki, Y. (2008). Function of the alpha subunit of rice heterotrimeric G protein in brassinosteroid signaling. Plant Cell Physiol. 50 161–172. [DOI] [PubMed] [Google Scholar]
- Ruzinova, M.B., and Benezra, R. (2003). Id proteins in development, cell cycle and cancer. Trends Cell Biol. 13 410–418. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24 105–109. [DOI] [PubMed] [Google Scholar]
- Serna, L. (2007). bHLH proteins know when to make a stoma. Trends Plant Sci. 12 483–485. [DOI] [PubMed] [Google Scholar]
- Sun, X.H., Copeland, N.G., Jenkins, N.A., and Baltimore, D. (1991). Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol. Cell. Biol. 11 5603–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M., Nemeth, K., Koncz-Kalman, Z., Mathur, J., Kauschmann, A., Altmann, T., Redei, G.P., Nagy, F., Schell, J., and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85 171–182. [DOI] [PubMed] [Google Scholar]
- Tanaka, A., et al. (2009). BRASSINOSTEROID UPREGULATED 1, Encoding a Helix-Loop-Helix Protein, is a Novel Gene Involved in Brassinosteroid Signaling and Controls Bending of the Lamina Joint in Rice. Plant Physiol. 151 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W., Kim, T.W., Oses-Prieto, J.A., Sun, Y., Deng, Z., Zhu, S., Wang, R., Burlingame, A.L., and Wang, Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, H., Jin, Y., Liu, W., Li, F., Fang, J., Yin, Y., Qian, Q., Zhu, L., and Chu, C. (2009). DWARF AND LOW-TILLERING, a new member of GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58 803–816. [DOI] [PubMed] [Google Scholar]
- Vert, G., and Chory, J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441 96–100. [DOI] [PubMed] [Google Scholar]
- Vert, G., Nemhauser, J.L., Geldner, N., Hong, F., and Chory, J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21 177–201. [DOI] [PubMed] [Google Scholar]
- Wada, K., Marumo, S., Ikekawa, N., Morisaki, M., and Mori, K. (1981). Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol. 22 323–325. [Google Scholar]
- Wan, S., et al. (2009). Activation tagging, an efficient tool for functional analysis of the rice genome. Plant Mol. Biol. 69 69–80. [DOI] [PubMed] [Google Scholar]
- Wang, L., Wang, Z., Xu, Y., Joo, S.H., Kim, S.K., Xue, Z., Xu, Z., and Chong, K. (2009). OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 57 498–510. [DOI] [PubMed] [Google Scholar]
- Wang, L., Xu, Y.Y., Ma, Q.B., Li, D., Xu, Z.H., and Chong, K. (2006. a). Heterotrimeric G protein alpha subunit is involved in rice brassinosteroid response. Cell Res. 16 916–922. [DOI] [PubMed] [Google Scholar]
- Wang, X., and Chory, J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313 1118–1122. [DOI] [PubMed] [Google Scholar]
- Wang, X., Goshe, M.B., Soderblom, E.J., Phinney, B.S., Kuchar, J.A., Li, J., Asami, T., Yoshida, S., Huber, S.C., and Clouse, S.D. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17 1685–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Chen, C., Xu, Y., Jiang, R., Han, Y., Xu, Z., and Chong, K. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22 409–417. [Google Scholar]
- Wang, Z.Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., Yang, Y., Fujioka, S., Yoshida, S., Asami, T., and Chory, J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2 505–513. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Wang, Q., Chong, K., Wang, F., Wang, L., Bai, M., and Jia, C. (2006. b). The brassinosteroid signal transduction pathway. Cell Res. 16 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.Y., et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell 20 2130–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120 249–259. [DOI] [PubMed] [Google Scholar]
- Yin, Y., Wang, Z.Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., and Chory, J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109 181–191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.