In unicellular fungi, Mob1 facilitates mitotic exit and cytokinesis through its interaction with NDR family kinases. However, its role in regulating cell division in metazoans is less well understood. In this report, we find that Mob1 is required for the recruitment of the chromosomal passenger complex to the spindle midzone during early anaphase.
Abstract
The spatial and temporal coordination of chromosome segregation with cytokinesis is essential to ensure that each daughter cell receives the correct complement of chromosomal and cytoplasmic material. In yeast, mitotic exit and cytokinesis are coordinated by signaling cascades whose terminal components include a nuclear Dbf2-related family kinase and a noncatalytic subunit, Mps one binding (Mob) 1. There are five human Mob1 isoforms, all of which display redundant localization patterns at the spindle poles and kinetochores in early mitosis, and the spindle midzone during cytokinesis. Mob1 shares similar localization patterns to Polo-like kinase (Plk1) and the chromosomal passenger complex (CPC), and although depletion of Plk1 resulted in a loss of Mob1 from the spindle poles, Mob1 recruitment to kinetochores was unaffected. Conversely, disruption of CPC signaling resulted in a loss of Mob1 from kinetochores without disrupting recruitment to the spindle poles. In Mob1-depleted cells, the relocalization of the CPC and mitotic kinesin-like protein (MKLP) 2 to the spindle midzone was delayed during early anaphase, and as a consequence, the midzone recruitment of MKLP1 also was affected. Together, these results suggest that Mob1 and the other mammalian orthologues of the mitotic exit network regulate mitotic progression by facilitating the timely mobilization of the CPC to the spindle midzone.
INTRODUCTION
Accurate execution of cell division is essential for proper development and survival of the organism. Errors in either chromosome segregation or cytokinesis can be lethal during development and have a destabilizing effect on genomic stability (Kops et al., 2004, 2005; Weaver and Cleveland, 2007). Much of our current understanding of how the cell cycle orchestrates the final stages of cell division in higher eukaryotes is based on genetic studies in unicellular fungi. And although nearly all cell cycle processes defined in yeast have been shown to be conserved in metazoans, it remains to be determined whether the mechanisms by which mitotic exit and cytokinesis are coordinated in yeast are similarly conserved in animal cells.
Studies in yeast have identified homologous cascades termed the septation initiation network (SIN) in Schizosaccharomyces pombe and the mitotic exit network (MEN) in Saccharomyces cerevisiae that regulate the temporal coordination of mitosis and cytokinesis (Jaspersen et al., 1998; Bardin and Amon, 2001; McCollum and Gould, 2001; Krapp and Simanis, 2008). Despite their similarities, some differences do exist in the mechanisms by which these pathways are activated as well their downstream targets (Bardin and Amon, 2001). In both organisms, a monomeric GTPase localizes on the spindle poles during anaphase and initiates a kinase cascade that terminates with the activation of a nuclear Dbf2-related (NDR) family kinase (Sid2/Dbf2) bound to its regulatory subunit Mps one binding (Mob) 1 (Schmidt et al., 1997; Bardin et al., 2000). In fission yeast, the SIN pathway triggers the initiation of contraction of a centrally placed actomyosin ring and formation of the division septum (Krapp et al., 2004). In contrast, the MEN pathway maintains the cytoplasmic localization cell division cycle (Cdc) 14 phosphatase, preventing its sequestration in the nucleolus during late anaphase and allowing Cdc14 to antagonize CDK phosphorylation of key substrates (Jaspersen et al., 1998; Visintin et al., 1998; Stegmeier and Amon, 2004). In fission yeast, the SIN also regulates a phosphatase (Clp1), but in contrast to budding yeast, Clp1 is not required for mitotic exit but instead antagonizes mitotic cyclin-dependent kinase (Cdk) function to ensure the timely onset of cytokinesis (Guertin et al., 2000; Chen et al., 2008).
Mob1 acts as a regulatory subunit of Sid2 (S. pombe) and Dbf2 (S. cerevisiae) and is essential for the late mitotic progression in both species. Cytokinetic failure, multinucleation, and failure to septate have been reported in temperature sensitive mob1 alleles in S. pombe (Hou et al., 2000; Salimova et al., 2000). In budding yeast, Mob1 is essential for mitotic exit, with mob1 mutants arresting in mitosis as large-budded cells with long, anaphase spindles (Luca and Winey, 1998; Luca et al., 2001). Examination of Mob1 function in genetic backgrounds where the mitotic exit defect was overridden revealed that loss of Mob1 function results in cellular chains with segregated nuclei but no cytokinesis (Luca et al., 2001). During mid-anaphase, Mob1 relocalizes from the spindle pole bodies to the bud-neck just before cytokinesis in a manner similar to that observed in fission yeast (Hou et al., 2000; Salimova et al., 2000; Luca et al., 2001). And last, conditional alleles of mob1 demonstrated that the Mob1–Dbf2 complex is required for dissociating Ipl1/Aurora B from the kinetochore and for maintaining chromosomal passenger proteins on the anaphase spindle (Stoepel et al., 2005), suggesting that in addition to its function in regulating Cdc14, the SIN/MEN pathway also may be involved in the regulation of the chromosomal passenger complex (CPC).
Although the MEN/SIN pathways are well described in fungi, full characterization of an analogous cascade in other phyla remains incomplete. This is due in part because functional homologues of upstream MEN/SIN components such as Spg1/Tem1 have yet to be identified. In contrast to Tem1, Mob1 is highly conserved throughout the eukaryotic kingdom, and there are indications that its function during mitosis is conserved. In plants, Mob1 is localized to the forming division site (phragmoplast) in a manner analogous to that seen in yeast (Van Damme et al., 2004; Citterio et al., 2006). Similarly, Mob1 down-regulation in Trypanosoma alters the timing of cytokinesis and placement of the cleavage furrow (Hammarton et al., 2005). In animal cells, orthologues of the MEN/SIN seem to function in pathways associated with the negative regulation of cell proliferation and the promotion of apoptosis (Lai et al., 2005). The Drosophila Mob1 orthologue Mats and the Sid2/Dbf2 orthologue Warts are essential in the Hippo/Salvador/Warts pathway, which regulates cell proliferation and organ development downstream of the proto-cadherin Fat (Hariharan, 2006; Harvey and Tapon, 2007; Matallanas et al., 2008). And although mitotic exit defects have been reported in cells depleted of the Sid2/Dbf2 orthologues large tumor suppressor (Lats) 1 and Lats2 (McPherson et al., 2004; Bothos et al., 2005), there have been relatively few studies examining the localization or functional roles of Mob1 in animal cell division.
Nearly all aspects of the eukaryotic cell cycle are conserved from yeast to humans. However, fundamental differences exist among fungi, plant, and animal cells in the mechanics by which these cells execute nuclear and cytoplasmic division. Moreover, there has been considerable expansion of MEN/SIN orthologues over the course of evolution with five isoforms of Mob1 and four Sid2/Dbf2 orthologues in the human genome (Hergovich et al., 2006b). In an effort to understand how MEN/SIN function is either conserved or has diverged in animal cells, we studied the localization dynamics of Mob1 and its function during mitosis in human cells. We report the different Mob1 isoforms displayed redundant localization patterns on the spindle poles and kinetochores early in mitosis, and the spindle midzone and midbody during anaphase and cytokinesis. Using a combination of pharmacological inhibitors and RNA interference, we have determined that Mob1 has a functional interaction with the CPC and suggest that Mob1 plays a role in coordination the late events of cell division in a manner similar to that reported in yeast.
MATERIALS AND METHODS
Reagents and Antibodies
Unless specified otherwise, all chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO). Nocodazole was purchased from Calbiochem (San Diego, CA), and ZM447439 was purchased from Tocris Bioscience (Ellisville, MO). Mitotic kinesin-like protein (MKLP) 1 antibody was purchased from Cytoskeleton (Denver, CO); Mob1A (Mob4A) antibodies specific for either the N or C termini were purchased from Abgent (San Diego, CA); Aurora B, Hec1, RacGAP, MKLP2, and BubR1 antibodies were purchased from Abcam (Cambridge, MA); inner centromeric protein (INCENP) and Polo-like kinase (Plk1) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); and phosphohistone H3 (Ser10) antibody was purchased from Cell Signaling Technology (Danvers, MA). AlexaFluor-labeled secondary antibodies and the transfection reagent Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA). The transfection reagent DharmaFECT I was purchased from Dharmacon RNA Technologies (Lafayette, CO).
Mammalian Cell Culture, Transfections, and Cell Synchronization
HeLa cells (American Type Culture Collection, Manassas, VA) were maintained in minimal essential medium with Earle's balanced salt solution (Lonza Walkersville, Walkersville, MD), supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, fungicide, 1.5 g/l sodium bicarbonate, and 1.0 mM sodium pyruvate (Sigma-Aldrich). RPE1-hTERT cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium and Ham's F-12 medium (1:1) with 2.5 mM l-glutamine, and 15 mM HEPES (Invitrogen and Lonza Walkerville), supplemented with 10% fetal bovine serum, 0.5 mM sodium pyruvate, 1.2 g/l sodium bicarbonate, and 10 μg/ml hygromycin B. HeLa cells were subcultured at a concentration of 2 × 105 cells/ml, where as RPE1-hTERT cells were subcultured at concentration of 4 × 104 cells/cm2 onto 12-well plates 18 h before transfection. Green fluorescent protein (GFP) plasmids were transfected at a final concentration of 1 μg/ml by using Lipofectamine 2000 (Invitrogen) according to manufacturer's recommendations in media without antibiotics, and they were imaged 24 h after transfection. Small interfering RNA (siRNA) transfection was carried out overnight at a final concentration of 100 nM by using DharmaFECT transfection reagent according to manufacturer's recommendations. After siRNA transfection, cells were released into regular media without antibiotics for an additional 24 h. To synchronize cells at G1/S, cells were treated with thymidine at 2 mM for 16 h. Cells were transfected with plasmid and/or siRNA during an 8-h release from thymidine, and followed by a second 16-h thymidine treatment. For some of the Mob1A depletions, cells were released for an additional 24 h after second thymidine treatment. For early mitotic arrest, cells were treated with nocodazole at of 0.2 μM for 3 h. The aurora kinase inhibitor ZM447439 was used at a final concentration of 5 μM.
Expression Constructs
The full-length coding sequence for Mob1A, Mob1C, Mob1D, and Mob1E were subcloned into the LoxP vector lp-pEGFP-C1 mammalian expression vector (BD Biosciences, San Jose, CA). SMARTselection designed siRNAs were synthesized by Thermo Fisher Scientific (Waltham, MA) (Supplemental Table 1).
Quantitative Polymerase Chain Reaction (PCR)
RNA was harvested from control and Mob1-depleted cultures (in triplicate) 48 h after transfection using RNAeasy Mini kit (QIAGEN, Valencia, CA) following the manufacturer's specifications. RNA samples were treated with DNase (Applied Biosystems, Foster City, CA), and 1 μg of RNA from each sample was reverse transcribed into cDNA by using random primers and SuperScript reverse transcriptase (Invitrogen). Quantitative PCR (qPCR) was performed in an ICycler IQ multicolored real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) using 2 μl of cDNA template (in triplicate) and SYBR Green Supermix (Bio-Rad Laboratories) as a detection dye. Reactions lacking cDNA template were used as nontemplate controls. Data were collected and real-time analysis was carried out using iCycler iQ real-time detection system software version 3.1(Bio-Rad Laboratories). The 2−ΔΔCt (Livak and Schmittgen, 2001) method was used for calculating -fold change in transcript levels in controls and Mob1-depleted cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin were used as internal controls to normalize Mob1 expression. The sequences for gene-specific primers used in the qPCR were designed using Primer3Plus (www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and are listed in Supplemental Table 2.
Immunofluorescence and Image Acquisition
Cells grown on coverslips were fixed either by immersion in −20°C methanol or with 3.7% formaldehyde/phosphate-buffered saline (PBS) followed by permeabilization with 0.5% Triton X-100/PBS. Coverslips were blocked with 3% bovine serum albumin/PBS for 1 h and then incubated for 1–4 h with primary antibodies diluted in blocking buffer and 1 μg/ml Hoechst 33342 (Invitrogen). Primary antibodies were detected with the appropriate AlexaFluor-labeled secondary antibodies (Invitrogen) and mounted in 1× PBS/90% glycerol. Cells were imaged using a 63× Plan-aprochromat 1.4 numerical aperture objective mounted on an Axiovert 200M inverted microscope (Carl Zeiss, Thornwood, NY) equipped for standard epifluorescence as well as an Apotome structured illumination module. For imaging of GFP-Mob1 constructs, only low-level–expressing cells were selected for imaging. Images were collected with a 12-bit AxioCam MrM charge-coupled device camera driven by Axiovision 4.5 software (Carl Zeiss). Then, 8-bit images were exported, and figures were prepared using Photoshop version CS2 software (Adobe Systems, Mountain View, CA).
Western Blot Analyses
RNA interference-mediated depletion of target proteins was confirmed by Western blotting. Cell lysates were generated by extracting cells with radioimmunoprecipitation assay buffer (120 mM NaCl, 0.5% NP-40 detergent, 0.5% SDS, 5 mM EGTA, and 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, and supplemented with 220 μM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and protease inhibitors; Calbiochem, Gibbstown, NJ). Proteins were resolved on 4–15% gradient SDS-polyacrylamide gel electrophoresis gels and then transferred to Immobilon membranes (Millipore, Billerica, MA). The blots were blocked in either 5% bovine serum albumin or 3% milk and then probed for the targeted proteins. Blots were stripped and reprobed for actin or tubulin as loading controls. Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Pittsburgh, PA) and chemiluminescence (Bio-Rad Laboratories) using a ChemiDoc XRS molecular imaging system (Bio-Rad Laboratories).
Measurement of MKLP1 Localization at the Spindle Midzone
To determine whether there was a difference in the localization of MKLP1 between control and Mob1-depleted cells, the width of the MKLP1 signal on anaphase spindles was measured as a function of total spindle length. Control- or Mob1 siRNA-transfected cells were fixed and processed for MKLP1 and tubulin localization, and for 200 cells per condition, the width of the MKLP1 zone was measured at three different locations within the spindle. In addition, the distance between spindle poles was measured to determine total spindle length. A scatter plot of MKLP1's average width was generated as a function of spindle length, and the data were displayed as a best fit using Excel (Microsoft, Redmond, WA). Analysis of covariance (ANCOVA) was performed to determine whether the differences in MKLP1 localization between control- and Mob1-depleted cells were statistically significant (http://faculty.vassar.edu/lowry/VassarStats.html).
RESULTS
Mob1 Family Members Localize to Key Mitotic Structures during Cell Division
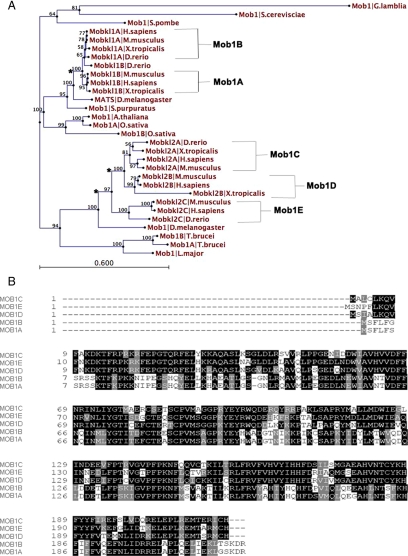
In budding and fission yeast, Mob1 facilitates mitotic exit and cytokinesis by acting as a regulatory subunit for Dbf2/Sid2 kinase (Komarnitsky et al., 1998; Luca et al., 2001; Devroe et al., 2004; Hou et al., 2004). Mob1 and its effector kinase localize first to the spindle pole bodies until anaphase onset, at which time the complex relocalizes to the bud neck/actin ring (Frenz et al., 2000; Salimova et al., 2000; Luca et al., 2001; Yoshida and Toh-e, 2001; Stoepel et al., 2005). As an initial effort toward understanding how the MEN might function in animal cells, we sought to follow Mob1 localization dynamics through mitosis in cultured human cells. In contrast to unicellular fungi where there is a single Mob1 gene, there are five Mob1 genes in the human genome (Figure 1). Moreover, the nomenclature for these genes varies greatly (Supplemental Table 3), creating confusion in both the literature and in the National Center for Biotechnology Information Database. A dendogram of Mob1 sequences from multiple model organisms rooted to the basal eukaryote Giardia lamblia (Figure 1A) resolved Mob1 into two monophyletic groups. The first clade includes a single lineage of plant Mob1 proteins, single copy Drosophila- and sea urchin Mob1 proteins, and two vertebrate Mob1 proteins (MobKL1B and MobKL1A), arising from a likely gene duplication. Following the nomenclature used by Stavridi et al. (2003), we designated these proteins as Mob1A and Mob1B, respectively. Mob1A and B are 96% identical to each other at the amino acid level (Figure 1B) and are known to bind the NDR1/2- and Lats1/2 kinases (Bichsel et al., 2004; Devroe et al., 2004; Bothos et al., 2005; Hergovich et al., 2005, 2006a; Chow et al., 2009). The other major cluster includes protozoan Mob1's, a single Drosophila Mob1 isoform and three vertebrate Mob1 isoforms arising from two gene duplication events (denoted by asterisks, Figure 1A). These three human Mob1's share ∼50% identity with Mob1A and Mob1B, and do not interact with Lats1 or Lats2 (Chow et al., 2009). Otherwise, little is known of their biological activity. In keeping with the nomenclature used for the other isoforms, we therefore assigned- and hereafter refer to these noncanonical isoforms as Mob1C–E.
Figure 1.
Phylogenetic analysis of the Mob1 family. (A) A dendogram was constructed using Mob1 protein sequences from across multiple phyla (Supplemental Table 3), rooted to the basal eukaryote Giardia lamblia, with bootstrap values provided for each node. Based on this analysis and the nomenclature of Stavridi (Stavridi et al., 2003), vertebrate Mob1's were given the assignations Mob1A–E. Branches noted by the asterisks indicate possible gene duplications. (B) Amino acid alignment of the five human Mob1 isoforms. Identical residues are shaded in black, similar residues are shaded in gray and nonconserved residues are not shaded.
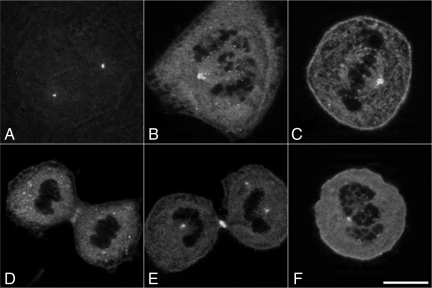
Given the high degree of similarity between the Mob1 proteins (and lack of discriminating antibodies), we tagged four of the human isoforms (Mob1A, Mob1C, Mob1D, and Mob1E) with enhanced green fluorescent protein (EGFP) at their N termini to follow their localization dynamics during mitosis and cytokinesis. Transient expression in HeLa cells revealed redundant localization patterns for all isoforms examined, with Mob1E displaying the least consistent expression and localization patterns, and was discontinued for all subsequent experiments. GFP-tagged Mob1's localized to the centrosomes in late G2 and remained associated with the spindle poles throughout mitosis and cytokinesis (96% of 544 cells scored) (Figure 2, A–E, and Supplemental Figure 1, A–H). Mob1 could also be observed at kinetochores beginning in early prophase before nuclear envelope breakdown (Supplemental Figure 1, A and E) with signal intensity peaking at prometaphase (Figure 2B and Supplemental Figure 1, A and E) (90%; n = 170 cells). However, by metaphase (Figure 2C), only 69% of Hec1-positive kinetochores were Mob1 positive (n = 55), and by anaphase no kinetochore localization could be detected (n = 45). Treatment of cells with nocodazole failed to alter GFP-Mob1 recruitment to spindle poles or kinetochores, suggesting that Mob1 recruitment to these structures was microtubule-independent (Figure 2F). After anaphase onset, Mob1 localized weakly to the spindle midzone (Figure 2D and Supplemental Figures 1, C and G, and 2A, g and h; 64% of 45 cells counted) but was enriched at the midbody at the end of cytokinesis (Figure 2E and Supplemental Figure 1, D and H, and 2J) (99%; n = 107). Although localization at kinetochores was only poorly visible using antibodies specific for Mob1A (Supplemental Figure 2, B and M), GFP-Mob1 localization was otherwise superimposable with the endogenous protein. Kinetochore localization could not be detected if EGFP was expressed alone (Supplemental Figure 1C, p–s), nor did expression of GFP-Mob1 effect the centrosomal localization of the Mob1-binding protein Lats1 or the MEN effector Cdc14A (data not shown), which together suggested that the observed localization patterns of the GFP-chimera were not an artifact of overexpression of the GFP-Mob1 chimeras or were disruptive to endogenous Mob1 function.
Figure 2.
GFP-Mob1 localization in dividing HeLa cells. Optical sections of HeLa cells transfected with GFP-Mob1A revealed that Mob1 recruitment to centrosomes began in late G2 phase and was maintained through cytokinesis (A–F). Kinetochore localization was most prominent during prometaphase (B) but lost during anaphase and cytokinesis (D and E). Mob1 localized to the spindle midzone during anaphase and was enriched in the midbody late in cytokinesis (D and E). Treatment with 250 nM nocodazole failed to disrupt either spindle pole- or kinetochore localization (F). Bar, 10 μm.
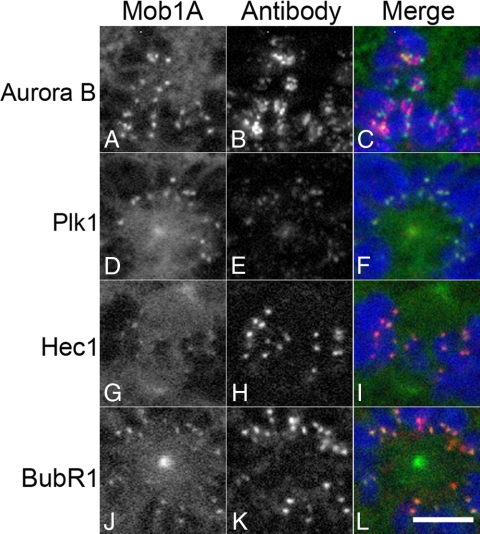
To more closely examine Mob1 localization at the kinetochore, we processed GFP-Mob1A–transfected HeLa cells for colocalization with key kinetochore markers. Aurora B has a broad signal, spanning the inner centromere (Carmena and Earnshaw, 2003) and when colocalized with Mob1A, Mob1A could be detected at both ends of the Aurora B signal (Figure 3, A–C). Compared with Plk1, Hec1 and BubR1, all of which reside within the outer two layers of the kinetochore, the Mob1A signal was superimposable with each marker (Figure 3, D–L). Examination of GFP-Mob1C and Mob1D revealed similar patterns, suggesting that the observed kinetochore localization was not isoform specific.
Figure 3.
GFP-Mob1 colocalization with prominent kinetochore components. GFP-Mob1A was transiently transfected into HeLa cells, processed for Aurora B (A–C), Plk1 (D–F), Hec1 (G–I), and BubR1 (J–L) localization. Note that although Mob1 could be found on either side of the Aurora B centromeric signal (C), Mob1 localization overlapped with Plk1, Hec1, and BubR1 (D–L). Bar, 5 μm.
Mob1 Recruitment to Kinetochores Requires the Chromosomal Passenger Complex
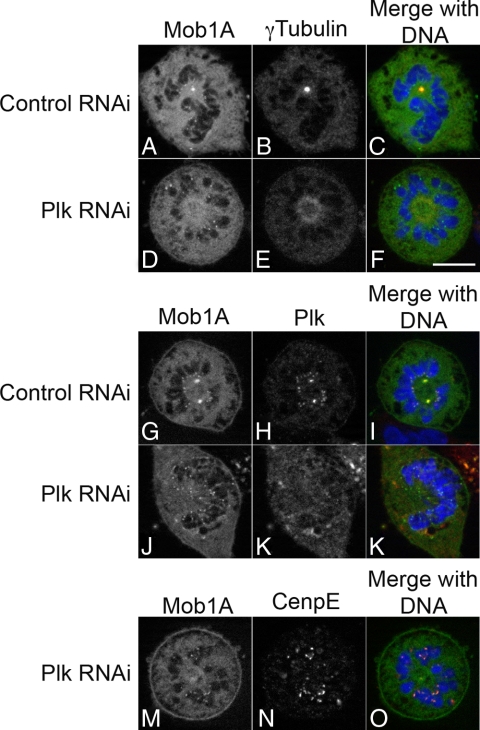
The localization of Mob1 during mitosis was highly reminiscent of the key mitotic kinases Plk1 (Golsteyn et al., 1995; van de Weerdt and Medema, 2006) and Aurora B kinase (Schumacher et al., 1998; Adams et al., 2001b; Giet and Glover, 2001; Murata-Hori et al., 2002), and we sought to determine whether Mob1's localization dynamics was dependent on these kinases. Plk1 is a major regulator throughout mitosis, playing roles in the transition G2/M, centrosomal maturation, spindle bipolarity and cytokinesis (Glover et al., 1998; van de Weerdt and Medema, 2006). Plk1 genetically interacts with Mob1 in yeast (Luca and Winey, 1998; Lee et al., 2001; Tanaka et al., 2001) and in animal cells is recruited to the spindle poles, kinetochores and spindle midzone (Golsteyn et al., 1995; Glover et al., 1998; Barr et al., 2004; van de Weerdt and Medema, 2006; Petronczki et al., 2008). Mob1 and Plk1 colocalized in mitotic cells (Figure 3, D–F), and to determine whether Plk1 influenced Mob1 localization in dividing cells, HeLa cells transiently expressing GFP-Mob1A, -C, and -D were depleted of Plk1 by RNA interference. Consistent with previous reports, Plk1-depleted cells arrested in mitosis with a characteristic monopolar spindle with diffuse γ-tubulin localization (Figure 4E; Lane and Nigg, 1996; Donaldson et al., 2001). In Plk1-depeleted cells, Mob1A was lost from the spindle poles (66%; n = 372 cells) but kinetochore localization was unaffected (Figure 4D, J and M). Identical results were obtained when GFP-Mob1C and Mob1D were expressed in Plk1-depleted cells (Supplemental Figure 3, D, J, P, and V).
Figure 4.
Plk1 is required for Mob1 recruitment to the spindle poles. GFP-Mob1A–expressing HeLa cells were transfected with nontargeting control and Plk1 siRNA (for Western blot confirmation, see Supplemental Figure 4). Twenty-four hours post-siRNA transfection, cells were processed for localization of γ tubulin (A–F) and Plk1 (G–L). In the absence of Plk1, cells formed a monopolar spindle, and although Mob1 was lost from the centrally located spindle pole (D–F), kinetochore localization was unaffected (D–F, J–L, and M–O). Bar, 10 μm.
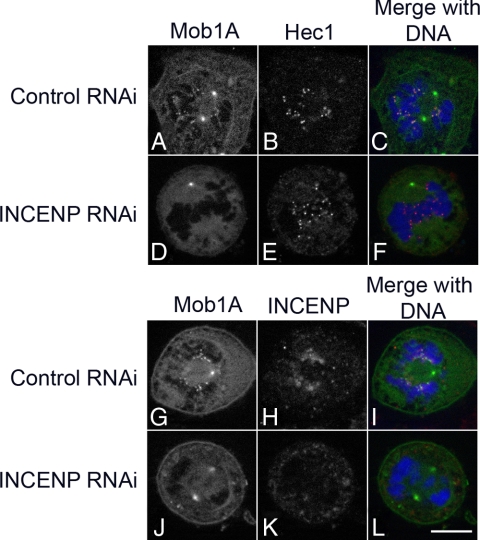
The CPC localizes to the inner centromere early in mitosis and functions in organizing the spindle as well as monitoring tension at the kinetochore (May and Hardwick, 2006; Ruchaud et al., 2007b). In addition, upon anaphase onset the CPC plays a critical role in organizing the spindle midzone and facilitating cytokinesis (Vader et al., 2008; Glotzer, 2009). Because Mob1 was also found at the kinetochores and spindle midzone (Figure 2 and Supplemental Figures 1 and 2), we wanted to determine whether Mob1 localization at the kinetochores was dependent on the CPC in mammalian cells. When INCENP was depleted in HeLa cells expressing GFP-Mob1A, -C, or -D, kinetochore localization of Mob1A was partially lost in 42% and completely lost 43% in cells scored (n = 834; Figure 5 and Supplemental Figure 5). In contrast, spindle pole localization was unperturbed in all cells observed.
Figure 5.
The chromosomal passenger complex is required for Mob1A recruitment to the kinetochore. GFP-Mob1A–expressing HeLa cells were transfected with nontargeting control (A–C, G–I) and INCENP (D–F, J–L) siRNA (for Western blot confirmation, see Supplemental Figure 4), and 24 h post-siRNA transfection, cells were processed for colocalization of Hec1 (B and E) and INCENP (H and K). When INCENP was depleted, Mob1A no longer localized on the kinetochore, but spindle pole localization was unaffected (D and J) (only a single pole was visible in the optical sections shown in D and G). Bar, 10 μm.
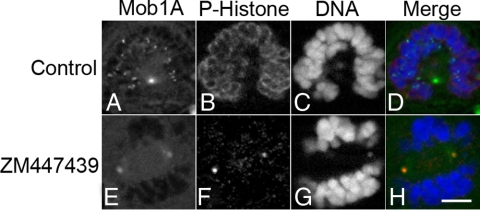
INCENP is required for Aurora B activation and function (Adams et al., 2000; Terada, 2001; Bishop and Schumacher, 2002; Honda et al., 2003) and acts as a binding scaffold for the entire CPC complex (Sessa et al., 2005; Jeyaprakash et al., 2007). To determine whether the catalytic activity of the CPC (Aurora B) is required for Mob1 recruitment to kinetochores, GFP-Mob1A–, -C–, and -D–expressing HeLa cells were synchronized in prometaphase with nocodazole and then released into either dimethyl sulfoxide (DMSO) carrier control (Figure 6, A–D, and Supplemental Figure 6, A–D and I–L) or the Aurora B inhibitor ZM477439 (Figure 6, E–H, and Supplemental Figure 6, E–H and M–P). Kinetochore localization of Mob1A, -C, and -D was lost (93%; n = 148 cells) in the absence of Aurora B activity (Figure 6E and Supplemental Figure 6, E and M) as was Ser10 phosphorylation of histone H3 (Figure 6F and Supplemental Figure 6, F and N), an established marker for Aurora B activity (Hsu et al., 2000; Adams et al., 2001a; Giet and Glover, 2001; George et al., 2006). Thus, either siRNA depletion or pharmacologic inhibition of the CPC blocked Mob1 recruitment to the kinetochores.
Figure 6.
Mob1A localization at kinetochore is lost in the absence of Aurora B activity. GFP-Mob1A–expressing HeLa cells were treated with 200 nM nocodazole for 3 h and then released into DMSO (A–D) or 5 μM ZM477439 (E–H) for an additional 30 min. Cells were fixed and counterstained with phospho- (Ser10) histone H3 (B and F). Note that although spindle pole localization was maintained in cells with compromised Aurora B activity (A and E), kinetochore localization was lost (E). Bar, 10 μm.
Depletion of Mob1 Alters CPC Localization and Function
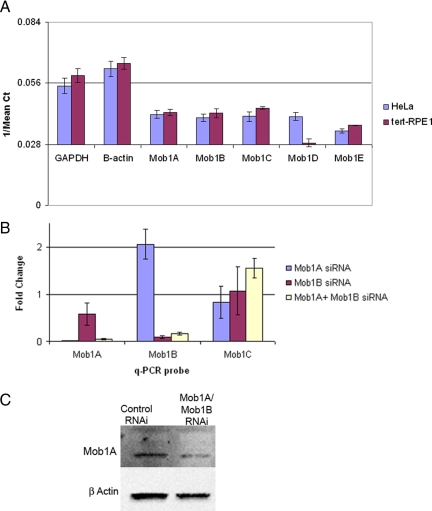
Alteration of Mob1 activity in yeast compromises Dbf2/Sid2- as well as Cdc14/Clp1 function (Mah et al., 2001) and alters CPC relocation to the spindle midzone (Stoepel et al., 2005). To identify the possible functions of Mob1 at the kinetochore or spindle midzone, we depleted Mob1 isoforms in cultured human cells. For these experiments, we used hTERT-immortalized retinal pigmented (RPE1) cells, which in comparison with HeLa cells display low rates of mitotic errors (Kigasawa et al., 1994; Jiang et al., 1999; Thompson and Compton, 2008) and express only Mob1A–D as detected by quantitative PCR (Figure 7A). Because Mob1C and -D do not bind the mammalian NDR family kinases (Chow et al., 2009), Mob1A and -B were depleted by transfection with siRNAs targeting each isoform and analyzed by quantitative PCR and Western blotting (Figure 7, B and C). Although simultaneous depletion of Mob1A and -B drastically reduced both transcript levels without affecting Mob1C expression (Figure 7B), transfection with siRNA targeting Mob1A and Mob1B isoforms resulted in only a partial reduction (30–50%) in Mob1A/B protein levels 48 h after transfection (Figure 7C), suggesting that Mob1 may be a relatively long-lived protein.
Figure 7.
Expression profile and depletion of Mob1 in HeLa and hTERT-RPE1 cells. (A) Quantitative PCR was performed using cDNA from HeLa and RPE1 cells. The graph shows the reciprocal values of Ct on y-axis to represent relative expression of Mob1 isoforms in HeLa and RPE1 cells. Error bars, SD. (B) RPE1 cells were transfected with Mob1A and Mob1B siRNA singly or in combination, and RNA was harvested 48h after transfection. qPCR was performed to determine the transcript levels for Mob1A, -B, and -C (which was not targeted). The -fold changes shown are the means of at least two different experiments done in triplicate. Values between 0.5 and 2.0 were not considered as significant change in gene expression. Error bars, SD. (C) Confirmation of protein depletion was determined by Western blot using an antibody specific for both Mob1A and Mob1B.
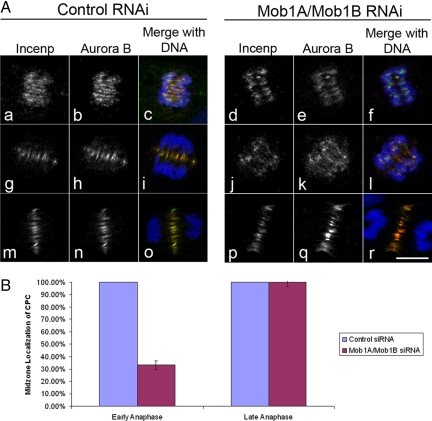
In addition to its role in regulating mitotic exit, functional interactions between Mob1 and the CPC have been described in yeast (Stoepel et al., 2005). Our own examination of Mob1 in human cells indicated that Mob1A localization to the kinetochore required INCENP and Aurora B (Figures 5 and 6 and Supplemental Figures 5 and 6) and to determine whether Mob1A was important for the CPC localization, we examined the organization of Aurora B and its downstream effectors in the presence or absence of Mob1A and 1B. In control cells, Aurora B displayed a well-defined, broad centromeric distribution early in mitosis up until anaphase onset at which time Aurora B and the CPC localize to the spindle midzone during anaphase (Figure 8A, g–h and m–o), where the CPC influences the organization of the spindle midzone and factors required for the execution of cytokinesis (Glotzer, 2009). However in Mob1A/B-depleted cells, we observed Aurora B and INCENP spread throughout the spindle midzone during early anaphase in 66.8% of cells scored (n = 482) with a fraction of the CPC remaining associated with the segregating sister chromatids (Figure 8A, j–l). This stood in sharp contrast to controls where Aurora B and INCENP were cleared from the centromeres by early anaphase and were confined within a narrow zone at the spindle midzone by mid-anaphase (Figure 8A, g–i). However, later in anaphase, no significant difference could be seen in the CPC localization between Mob1-depleted cells (Figure 8A, p–r; and B) and controls (Figure 8A, m–o; and B), suggesting that CPC localization recovered as mitotic exit progressed. Because all four components of the CPC function as a single functional unit (Jeyaprakash et al., 2007), the fact that both Aurora B and INCENP displayed this phenotype suggested that the mobilization of the entire CPC was being affected.
Figure 8.
Depletion of Mob1 disrupts CPC localization to the spindle midzone during early anaphase. (A) RPE1 cells were synchronized by double thymidine block and transfected with either nontargeting siRNA (control, left) or siRNA targeting both Mob1A and Mob1B (Mob1A/1B, right). In contrast to controls where the CPC rapidly relocated to the forming spindle midzone (a–c and g–i), Aurora B and INCENP was found throughout the spindle as associated with chromatin in Mob1A-depleted cells (d–f and j–l). However, by late anaphase, the CPC could be seen in a narrow band at the cell equator in both controls (m-o) and Mob1A-depleted cells (p–r). Bar, 10 μm. (B) Quantification of the CPC phenotype indicated that during early anaphase, 66.8% of cells depleted of Mob1A and Mob1B in early anaphase (n = 482) displayed diffuse Aurora B and INCENP localization along the spindle, whereas by late anaphase, CPC localization to the midzone was normal in 100% Mob1A/B-depleted cells (n = 270).
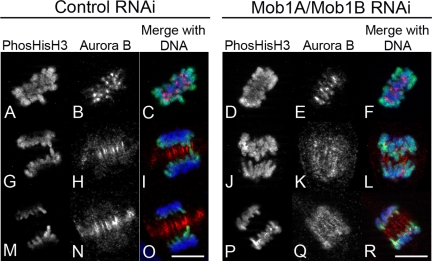
In Mob1A/Mob1B-depleted cells, Aurora B was spread out throughout the early anaphase spindle (Figure 8A, j–l), but there were none of the dramatic phenotypes normally associated with compromised Aurora B function (Kallio et al., 2002; Hauf et al., 2003). To determine whether there are any functional consequences to this altered localization we probed control- and Mob1A/Mob1B-depleted cells for histone H3 phosphorylation, a convenient marker for Aurora B activity. In control cells, phosphohistone reactivity during anaphase is normally restricted to the trailing arms of segregating chromatids (Figure 9, G and M). However, in 55% of Mob1A/Mob1B-depleted cells (n = 70), phosphohistone reactivity was maintained along the entire length of the chromatid arms (Figure 9, J and P), suggesting that although the CPC was not localizing normally to a tight zone at the cell equator, its catalytic activity was normal.
Figure 9.
Histone H3 phosphorylation is maintained on chromatid arms during anaphase in Mob1A/B-depleted cells. Thymidine-synchronized RPE1 cells were transfected with either nontargeting control (left) or Mob1A/Mob1B siRNAs (right), and probed for phospho-Ser10 histone H3 and Aurora B localization. Note that in contrast to cells in metaphase (A–F), anaphase phosphohistone reactivity in controls was limited to those regions of trailing chromatin arms closest to the spindle midzone (G–I and M–O). In Mob1A/Mob1B-depleted cells where Aurora B localization was spread throughout the anaphase spindle, phosphohistone reactivity was found along the entire length of the sister chromatid arms (J–L and P–R). Bar, 10 μm.
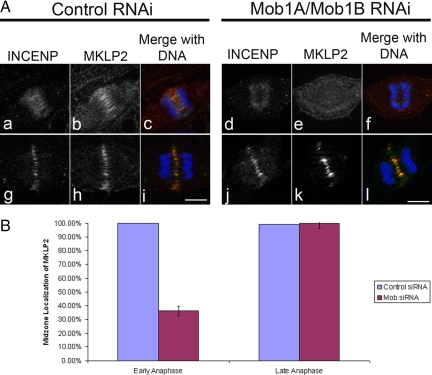
Later in anaphase, Aurora B and INCENP (Figure 8A, m–r) could be found enriched at the spindle midzone in both control- and Mob1A/Mob1B-depleted cells, suggesting that CPC recruitment to the midzone recovered in the absence of Mob1A/Mob1B. The relocalization of the CPC from the centromeres to the spindle midzone requires the action of the kinesin-like motor MKLP2 (Gruneberg et al., 2004), and to determine whether the effect of Mob1A/Mob1B on CPC localization involved this motor, cells were depleted of Mob1A/Mob1B and probed for MKLP2. In early anaphase, MKLP2 begins to localize at the spindle midzone (Figure 10A, a–c), but in Mob1A/Mob1B-depleted cells, MKLP2 recruitment was reduced or absent in >63% of cells observed (n = 92) (Figure 10A, d–f; and B). However, as was the case with the CPC, by late anaphase MKLP2 recruitment to the midzone in Mob1-depleted cells (Figure 10A, j–l; and B) was indistinguishable from controls (Figure 10A, g–i; and B).
Figure 10.
MKLP2 localization is altered in Mob1A/Mob1B-depleted cells. (A) Thymidine-synchronized RPE1 cells were transfected with either nontargeting control (left) or Mob1A/Mob1B siRNA's (right), and probed for INCENP and MKLP2 localization. In contrast to controls (a–c), MKLP2 recruitment during early anaphase was disrupted in Mob1A/B-depleted cells (d–f), but by mid-late anaphase (j–l), MKLP2 recruitment to the midzone resembled controls (g–i). Bar, 10 μm. (B) Disruption of MKLP2 localization to the central spindle occurred in 63.7% of cell depleted of Mob1A/1B during early anaphase (n = 92 cells, SE = 0.0358), but by mid-late anaphase, MKLP2 localization matched controls (n = 102).
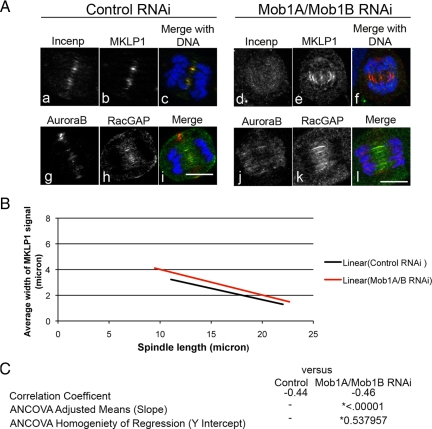
The principle target of the CPC during anaphase is the centralspindlin complex of MKLP1 and male germ cell Rac GTPase activating protein (MgcRacGap), which helps organize the spindle midzone and recruit the RhoGEF Ect2 to the cell equator (D'Avino et al., 2005; Glotzer, 2005). Examination of both centralspindlin components in Mob1A/Mob1B-depleted cells revealed a similar phenotype in cells in early anaphase. In contrast to controls (Figure 11A, b and h), the zone of both MKLP1 and MgcRacGap was slightly wider in Mob1-depleted cells (Figure 11A, e and k). However, because the zone of MKLP1 at the midzone gradually narrows as anaphase and cytokinesis progresses in control cells, we needed to compare the MKLP1 zones between control- and Mob1A/Mob1B-depleted cells as a function of spindle length to determine whether the observed differences were consistent throughout anaphase (Figure 11A). Measurements of the MKLP1 zone and spindle length were made on cells (stained for tubulin and MKLP1) for 200 cells per condition, and examination of the two data sets revealed that the MKLP1 zone narrowed as anaphase progressed in both control (Figure 11B, black line)- and Mob1A/Mob1B (Figure 11A, red line)-depleted cells. However, in Mob1A/Mob1B-depleted cells the MKLP1 zone was wider than controls throughout anaphase (Figure 11B, red line). ANCOVA determined that there was no difference between the slopes of the two curves (p = 0.537957), indicating that the rate by which the MKLP1 zone narrowed was not significantly different between control- and Mob1A/Mob1B-depleted cells (Figure 11C). However, the difference in the width of the MKLP1 zone between control- and Mob1A/Mob1B-depleted cells was significant (p = 0.00001) (Figure 11C), with the MKLP1 zone in Mob1A/Mob1B-depleted cells slightly wider both early and late in anaphase. However, cells in both conditions were capable of initiating cytokinesis, suggesting that the broaden zone of centralspindlin was not a strong enough defect to inhibit RhoA activation and contractile ring assembly (data not shown). Thus, although the absence of Mob1A/Mob1B resulted in only subtle defects in spindle midzone organization, Mob1A and Mob1B did seem to play a role in restricting the zone by which the CPC defined the cell equator in dividing cells.
Figure 11.
Depletion of Mob1A affects the organization of centralspindlin during anaphase. (A) RPE1 cells transfected with control- or Mob1A/1B siRNAs were probed for MKLP1 or MgcRacGAP and counterstained with INCENP or Aurora B, respectively. Bar, 10 μm. (B) Control- or Mob1A/1B-depleted cells were processed for tubulin and MKLP1 localization, and the zone of MKLP1 was measured as a function of spindle length for 200 cells per condition. For each cell, the width of the MKLP1 signal at the midzone was measured, along with the length of the spindle and width of central spindle. Lines of correlation that best fit the scatter plot data are depicted for control (black line) and Mob1A/1B-depleted cells (red line). As shown in the graph, the MKLP1 zone narrowed as anaphase progressed for both control- and Mob1-depleted cells. However, in contrast to controls, the zone of MKLP1 was wider in Mob1-depleted cells both early and late in anaphase. (C) Analysis of covariance was performed on the two data sets, and although there was no statistical significance in the rate of MKLP1 narrowing between control and Mob1-depleted cells (p = 0.537957) (measured by comparing the 2 slopes), there was a significant difference between the widths of the two zones as cells progressed through anaphase (p = 0.00001).
DISCUSSION
Nearly every facet of cell cycle control described in yeast is conserved in metazoans, with increasing layers of complexity added as a result of gene duplications and the addition of intrinsic- and extrinsic regulatory inputs. Given the destabilizing effects of aneuploidy to multicellular organisms, the mechanisms in place to ensure the integrity of the genome have also increased in complexity. The spatiotemporal coordination of nuclear- and cytoplasmic division represents one such area where the core features (cyclin degradation, CDK inactivation, and activation of the cytokinetic machinery) are conserved between yeast and animal cells, with variations in the manner of nuclear division (open or closed) and the mechanisms by which cells execute cytokinesis. The septation initiation and mitotic exit networks, which coordinate mitosis and cytokinesis in yeast, are well conserved in unicellular fungi but poorly described in animal cells. However, most reports in the literature ascribe roles for MEN/SIN orthologues in tumor suppressor pathways regulating organ size and cell proliferation (Pan, 2007; Saucedo and Edgar, 2007). Here, we demonstrate that in human cells, Mob1 associates with mitotic structures in a manner highly reminiscent of the localization dynamics observed for Mob1 in yeast and higher plants, and functions during anaphase to regulate the distribution of the chromosomal passenger complex on the spindle midzone. And although the spindle midzone differs in both organization and function between yeast and animal cells, the dependence of the CPC on elements of the MEN/SIN pathway in animal cells suggests that many of the core functions of the MEN/SIN pathway are conserved throughout evolution.
Evolutionary Conservation of Mob1 Localization Dynamics
Mammals have five Mob1 genes (Figure 1), and given the high degree of similarity between the different isoforms and lack of discriminating antibodies, we elected to express GFP fusions to follow isoform-specific localization dynamics. To our surprise, we found almost superimposable localization dynamics between the isoforms, beginning in late G2/prophase, where Mob1 could be detected at the forming spindle poles and kinetochores (Figure 2 and Supplemental Figure 1). Whereas spindle pole-associated Mob1 was maintained throughout mitosis and cytokinesis, kinetochore recruitment peaked during prometaphase and was lost by anaphase (Figure 2D and Supplemental Figure 1, C and G). Last, Mob1 could be detected at the spindle midzone and midbody (Figure 2, D and E, and Supplemental Figure 1, D and H). And although localization of Mob1A to spindle poles and midbodies in mammalian cells has been reported previously (Bothos et al., 2005), the kinetochore localization of Mob1 isoforms has not been reported to date, and represents a novel and exciting finding. Drosophila Mob4, which most resembles Mob3/phocein in humans, localizes to the spindle poles and weakly to kinetochores, and seems to play a role in spindle pole integrity (Trammell et al., 2008).
In all cases, Mob1 localization in animal cells seems highly reminiscent to that observed in yeast, where Mob1 binds with the spindle pole bodies in G2 (Hou et al., 2000; Salimova et al., 2000; Luca et al., 2001), enters the nucleus and colocalizes with Cdc14 at the kinetochores (Stoepel et al., 2005). After anaphase onset, Mob1 and Dbf2/Sid2 relocalize to the forming actomyosin ring during cytokinesis, where in fission yeast the complex plays a role in initiating constriction of the actin ring and formation of the septum (Hou et al., 2000; Salimova et al., 2000). In plants, Mob1 localizes to microtubule-organizing centers in early prophase and in anaphase to the phragmoplast and forming cell plate (Citterio et al., 2006) suggesting that although there are fundamental differences in the manner by which plants, fungi, and animal cells execute cytokinesis, Mob1 and its binding partners may be playing a conserved role in all of these organisms.
Examination of four of the five mammalian isoforms of Mob1 revealed redundant patterns (Figures 2 and 3 and Supplemental Figures 1 and 2), raising the issue of whether these isoforms are functionally redundant. Quantitative PCR analyses of HeLa- and RPE1 cells indicated that different cell lines express slightly different Mob1 isoform profiles, and differential expression patterns have been described for the Mob1 isoforms in human tissues (Chow et al., 2009). Mob1's C–E do not bind the Lats1/2 kinases (Chow et al., 2009), suggesting that these isoforms perform nonredundant functions. To date, this is the first report to describe the localization patterns of the different Mob1 isoforms, but at this juncture we cannot say whether Mob1C–E can compensate for the loss of other Mob1's during mitosis. Further experimentation will determine whether this is, indeed, the case.
Functional Interaction between the Mitotic Exit Network and the Chromosomal Passenger Complex at the Spindle Midzone
The CPC functions in chromosomal organization and alignment, kinetochore assembly, microtubule attachment, spindle midzone assembly, and cytokinesis (Ruchaud et al., 2007a; Vader et al., 2007). Work in budding yeast found that along with Cdc14, Mob1/Dbf2 partially localizes with kinetochores and is required for mobilization of the CPC from the centromere to the anaphase spindle (Stoepel et al., 2005). We found that in Mob1A/Mob1B-depleted RPE1 cells, the CPC was spread throughout the spindle, with some residual localization on the chromosomes, suggesting that Mob1A was important for the correct redistribution of the CPC to the spindle midzone in a manner analogous for what has been reported in yeast (Figures 8–10). However, there were no indications that CPC function was compromised. Indeed, when we probed cells for known targets of Aurora B (such as phosphohistone H3 and MKLP1), we saw that histone H3 was phosphorylated normally early in mitosis (Figure 9) and that MKLP1 was recruited properly to the midzone during anaphase (Figures 11A). However, in both cases, the diffuse Aurora B localization along the early anaphase spindle seemed to affect the normal dynamics of histone phosphorylation and MKLP1 localization. In Mob1A/Mob1B-depleted cells, the phosphohistone reactivity could be found along the entire length of the segregating sister chromatids (Figure 9, J and P), in contrast to controls where phosphohistone reactivity was limited to those regions of chromatin most proximal to the midzone (Figure 9, G and M). Similarly, although MKLP1 could be found at the midzone of both control- and Mob1A/Mob1B-depleted cells, the zone of MKLP1 was broader in those cells lacking Mob1A/1B (Figure 11). It has been proposed that during anaphase there is a gradient of Aurora B activity at the spindle midzone that acts to influence the late events of mitosis (Fuller et al., 2008). It is possible then that in the Mob1-depleted cells, the diffuse distribution of Aurora B along the entire spindle effectively broadens the activity gradient and thus maintains phosphohistone reactivity and the widened distribution of MKLP1 at the midzone.
We found that in Mob1A/Mob1B-depleted cells, the mitotic kinesin MKLP2 was also late to arrive at the spindle midzone (Figure 10). MKLP2 is required for the CPC to correctly load onto the spindle midzone during anaphase, as well as for the midzone targeting of the MEN effector Cdc14A (Gruneberg et al., 2004). For both the CPC and MKLP2, the absence of Mob1A/B only affected midzone localization early in anaphase (Figures 8 and 10), and that as anaphase and mitotic exit progressed MKLP2/CPC recruitment to the midzone recovered. MKLP2 is antagonized by Cdk1 (Hummer and Mayer, 2009), raising the possibility that MKLP2 is a principle target by which the mitotic exit network regulates the CPC. If Mob1 functions through Cdc14A during mitotic progression (as it does in yeast), then one might predict as CDK1 activity continued to decline through anaphase, those factors negatively regulated by CDK1 (such as MKLP2 and the CPC) would eventually become fully active and function normally. Identifying the responsible NDR kinase (among the four human orthologues of Dbf2) through which Mob1 modulates Cdc14A activity, as well as the direct targets of Cdc14A will reveal exactly how mammalian homologues of the mitotic exit network modulate the final events of cell division.
Supplementary Material
ACKNOWLEDGMENTS
We thank Donovan Bailey for assistance in the phylogenetic analysis of Mob1, Angus Dawe and Ivy Yu for assistance in quantitative PCR, and Lynne Cassimeris and Ken Kaplan for helpful suggestions and encouragement. This work is supported by National Institute of Health grants HD-063917, RR-16480, GM-08136, and GM-61222.
Abbreviations used:
- Cdc
cell division cycle
- CDK
cyclin-dependent kinase
- CPC
chromosomal passenger complex
- INCENP
inner centromeric protein
- Lats
large tumor suppressor
- MEN
mitotic exit network
- MgcRacGAP
male germ cell Rac GTPase activating protein
- MKLP
mitotic kinesin-like protein
- Mob
Mps one binding
- Plk1
polo-like kinase-1
- SIN
septation initiation network.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-06-0471) on December 2, 2009.
REFERENCES
- Adams R. R., Carmena M., Earnshaw W. C. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001a;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- Adams R. R., Maiato H., Earnshaw W. C., Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 2001b;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R. R., Wheatley S. P., Gouldsworthy A. M., Kandels-Lewis S. E., Carmena M., Smythe C., Gerloff D. L., Earnshaw W. C. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Bardin A. J., Amon A. Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Bardin A. J., Visintin R., Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Barr F. A., Sillje H. H., Nigg E. A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Bichsel S. J., Tamaskovic R., Stegert M. R., Hemmings B. A. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J. Biol. Chem. 2004;279:35228–35235. doi: 10.1074/jbc.M404542200. [DOI] [PubMed] [Google Scholar]
- Bishop J. D., Schumacher J. M. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J. Biol. Chem. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothos J., Tuttle R. L., Ottey M., Luca F. C., Halazonetis T. D. Human LATS1 is a mitotic exit network kinase. Cancer Res. 2005;65:6568–6575. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- Carmena M., Earnshaw W. C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- Chen C. T., Feoktistova A., Chen J. S., Shim Y. S., Clifford D. M., Gould K. L., McCollum D. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase. Clp1. Curr. Biol. 2008;18:1594–1599. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A., Hao Y., Yang X. Molecular characterization of human homologs of yeast MOB1. Int. J. Cancer. 2009 doi: 10.1002/ijc.24878. in press. [DOI] [PubMed] [Google Scholar]
- Citterio S., Piatti S., Albertini E., Aina R., Varotto S., Barcaccia G. Alfalfa Mob1-like proteins are involved in cell proliferation and are localized in the cell division plane during cytokinesis. Exp. Cell Res. 2006;312:1050–1064. doi: 10.1016/j.yexcr.2005.12.032. [DOI] [PubMed] [Google Scholar]
- D'Avino P. P., Savoian M. S., Glover D. M. Cleavage furrow formation and ingression during animal cytokinesis: a microtubule legacy. J. Cell Sci. 2005;118:1549–1558. doi: 10.1242/jcs.02335. [DOI] [PubMed] [Google Scholar]
- Devroe E., Erdjument-Bromage H., Tempst P., Silver P. A. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J. Biol. Chem. 2004;279:24444–24451. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- Donaldson M. M., Tavares A. A., Hagan I. M., Nigg E. A., Glover D. M. The mitotic roles of Polo-like kinase. J. Cell Sci. 2001;114:2357–2358. doi: 10.1242/jcs.114.13.2357. [DOI] [PubMed] [Google Scholar]
- Frenz L. M., Lee S. E., Fesquet D., Johnston L. H. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J. Cell Sci. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- Fuller B. G., Lampson M. A., Foley E. A., Rosasco-Nitcher S., Le K. V., Tobelmann P., Brautigan D. L., Stukenberg P. T., Kapoor T. M. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George O., Johnston M. A., Shuster C. B. Aurora B kinase maintains chromatin organization during the MI to MII transition in surf clam oocytes. Cell Cycle. 2006;5:2648–2656. doi: 10.4161/cc.5.22.3444. [DOI] [PubMed] [Google Scholar]
- Giet R., Glover D. M. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. M., Hagan I. M., Tavares A. A. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- Golsteyn R. M., Mundt K. E., Fry A. M., Nigg E. A. Cell cycle regulation of the activity and subcellular localization of PLK1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Honda R., Nigg E. A., Barr F. A. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D. A., Chang L., Irshad F., Gould K. L., McCollum D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 2000;19:1803–1815. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarton T. C., Lillico S. G., Welburn S. C., Mottram J. C. Trypanosoma brucei MOB1 is required for accurate and efficient cytokinesis but not for exit from mitosis. Mol. Microbiol. 2005;56:104–116. doi: 10.1111/j.1365-2958.2005.04542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan I. K. Growth regulation: a beginning for the hippo pathway. Curr. Biol. 2006;16:R1037–R1039. doi: 10.1016/j.cub.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Harvey K., Tapon N. The Salvador-Warts-Hippo pathway—an emerging tumour-suppressor network. Nat. Rev. Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., Peters J. M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Bichsel S. J., Hemmings B. A. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol. Cell. Biol. 2005;25:8259–8272. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Schmitz D., Hemmings B. A. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem. Biophys. Res. Commun. 2006a;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- Hergovich A., Stegert M. R., Schmitz D., Hemmings B. A. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 2006b;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Honda R., Korner R., Nigg E. A. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 2003;14:3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou M. C., Guertin D. A., McCollum D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol. Cell. Biol. 2004;24:3262–3276. doi: 10.1128/MCB.24.8.3262-3276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou M. C., Salek J., McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- Hsu J. Y., et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Hummer S., Mayer T. U. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr. Biol. 2009;19:607–612. doi: 10.1016/j.cub.2009.02.046. [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Charles J. F., Tinker-Kulberg R. L., Morgan D. O. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash A. A., Klein U. R., Lindner D., Ebert J., Nigg E. A., Conti E. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- Jiang X. R., et al. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat. Genet. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- Kallio M. J., McCleland M. L., Stukenberg P. T., Gorbsky G. J. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 2002;12:900–905. doi: 10.1016/s0960-9822(02)00887-4. [DOI] [PubMed] [Google Scholar]
- Kigasawa K., Soushi S., Tanaka Y., Obazawa H. Morphologic and chromosomal study of a human retinal pigment epithelial cell line. Jpn. J. Ophthalmol. 1994;38:10–15. [PubMed] [Google Scholar]
- Komarnitsky S. I., Chiang Y. C., Luca F. C., Chen J., Toyn J. H., Winey M., Johnston L. H., Denis C. L. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G. J., Foltz D. R., Cleveland D. W. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G. J., Weaver B. A., Cleveland D. W. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Krapp A., Gulli M. P., Simanis V. SIN and the art of splitting the fission yeast cell. Curr. Biol. 2004;14:R722–R730. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- Krapp A., Simanis V. An overview of the fission yeast septation initiation network (SIN) Biochem. Soc. Trans. 2008;36:411–415. doi: 10.1042/BST0360411. [DOI] [PubMed] [Google Scholar]
- Lai Z. C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L. L., Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lane H. A., Nigg E. A. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. E., Frenz L. M., Wells N. J., Johnson A. L., Johnston L. H. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 2001;11:784–788. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luca F. C., Mody M., Kurischko C., Roof D. M., Giddings T. H., Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol. Cell. Biol. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F. C., Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A. S., Jang J., Deshaies R. J. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA. 2001;98:7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas D., Romano D., Hamilton G., Kolch W., O'Neill E. A Hippo in the ointment: MST signalling beyond the fly. Cell Cycle. 2008;7:879–884. doi: 10.4161/cc.7.7.5630. [DOI] [PubMed] [Google Scholar]
- May K. M., Hardwick K. G. The spindle checkpoint. J. Cell Sci. 2006;119:4139–4142. doi: 10.1242/jcs.03165. [DOI] [PubMed] [Google Scholar]
- McCollum D., Gould K. L. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- McPherson J. P., et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004;23:3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Hori M., Tatsuka M., Wang Y. L. Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol. Biol. Cell. 2002;13:1099–1108. doi: 10.1091/mbc.01-09-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Petronczki M., Lenart P., Peters J. M. Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev. Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W. C. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 2007a;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W. C. The chromosomal passenger complex: one for all and all for one. Cell. 2007b;131:230–231. doi: 10.1016/j.cell.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Salimova E., Sohrmann M., Fournier N., Simanis V. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J. Cell Sci. 2000;113:1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- Saucedo L. J., Edgar B. A. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- Schmidt S., Sohrmann M., Hofmann K., Woollard A., Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- Schumacher J. M., Golden A., Donovan P. J. AIR-2, an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., Schneider T. R., Stukenberg P. T., Musacchio A. Mechanism of Aurora B activation by INCENP and inhibition by Hesperadin. Mol. Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Stavridi E. S., Harris K. G., Huyen Y., Bothos J., Verwoerd P. M., Stayrook S. E., Pavletich N. P., Jeffrey P. D., Luca F. C. Crystal structure of a human Mob1 protein: toward understanding Mob-regulated cell cycle pathways. Structure. 2003;11:1163–1170. doi: 10.1016/s0969-2126(03)00182-5. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stoepel J., Ottey M. A., Kurischko C., Hieter P., Luca F. C. The mitotic exit network Mob1p-Dbf2p kinase complex localizes to the nucleus and regulates passenger protein localization. Mol. Biol. Cell. 2005;16:5465–5479. doi: 10.1091/mbc.E05-04-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Petersen J., MacIver F., Mulvihill D. P., Glover D. M., Hagan I. M. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 2001;20:1259–1270. doi: 10.1093/emboj/20.6.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y. Role of chromosomal passenger complex in chromosome segregation and cytokinesis. Cell Struct. Funct. 2001;26:653–657. doi: 10.1247/csf.26.653. [DOI] [PubMed] [Google Scholar]
- Thompson S. L., Compton D. A. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trammell M. A., Mahoney N. M., Agard D. A., Vale R. D. Mob4 plays a role in spindle focusing in Drosophila S2 cells. J. Cell Sci. 2008;121:1284–1292. doi: 10.1242/jcs.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G., Cruijsen C. W., van Harn T., Vromans M. J., Medema R. H., Lens S. M. The chromosomal passenger complex controls spindle checkpoint function independent from its role in correcting microtubule kinetochore interactions. Mol. Biol. Cell. 2007;18:4553–4564. doi: 10.1091/mbc.E07-04-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G., Maia A. F., Lens S. M. The chromosomal passenger complex and the spindle assembly checkpoint: kinetochore-microtubule error correction and beyond. Cell Div. 2008;3:10. doi: 10.1186/1747-1028-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D., Bouget F. Y., Van Poucke K., Inze D., Geelen D. Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J. 2004;40:386–398. doi: 10.1111/j.1365-313X.2004.02222.x. [DOI] [PubMed] [Google Scholar]
- van de Weerdt B. C., Medema R. H. Polo-like kinases: a team in control of the division. Cell Cycle. 2006;5:853–864. doi: 10.4161/cc.5.8.2692. [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Weaver B. A., Cleveland D. W. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Toh-e A. Regulation of the localization of Dbf2 and mob1 during cell division of Saccharomyces cerevisiae. Genes Genet. Syst. 2001;76:141–147. doi: 10.1266/ggs.76.141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.