The enzyme Psd2 catalyzes endosomal synthesis of the phospholipid PE. While this pool of PE represents a minority of total cellular PE, function of Psd2 is required for normal activity of the vacuolar ABC transporter Ycf1. Psd2 controls vacuolar PE levels by acting at the level of the endosome.
Abstract
Control of lipid composition of membranes is crucial to ensure normal cellular functions. Saccharomyces cerevisiae has two different phosphatidylserine decarboxylase enzymes (Psd1 and Psd2) that catalyze formation of phosphatidylethanolamine. The mitochondrial Psd1 provides roughly 70% of the phosphatidylethanolamine (PE) biosynthesis in the cell with Psd2 carrying out the remainder. Here, we demonstrate that loss of Psd2 causes cells to acquire sensitivity to cadmium even though Psd1 remains intact. This cadmium sensitivity results from loss of normal activity of a vacuolar ATP-binding cassette transporter protein called Ycf1. Measurement of phospholipid levels indicates that loss of Psd2 causes a specific reduction in vacuolar membrane PE levels, whereas total PE levels are not significantly affected. The presence of a phosphatidylinositol transfer protein called Pdr17 is required for Psd2 function and normal cadmium tolerance. We demonstrate that Pdr17 and Psd2 form a complex in vivo that seems essential for maintenance of vacuolar PE levels. Finally, we refine the localization of Psd2 to the endosome arguing that this enzyme controls vacuolar membrane phospholipid content by regulating phospholipids in compartments that will eventually give rise to the vacuole. Disturbance of this regulation of intracellular phospholipid balance leads to selective loss of membrane protein function in the vacuole.
INTRODUCTION
The biochemical nature of integral membrane proteins demands that their polypeptide backbones are in direct contact with the membrane bilayer. This physical proximity supports the view that changes in the lipid content of membranes would naturally influence the activity of these integral membrane proteins. However, testing this idea is complicated by the multiple roles played by membrane lipids in terms of cell physiology. This problem is exacerbated in eukaryotic organisms with multiple membrane-defined compartments, each with a unique lipid composition (reviewed in Maxfield and Tabas, 2005; van Meer, 2005). To approach the contribution of particular lipids to membrane function in eukaryotic cells, mutant strains of Saccharomyces cerevisiae have been used with specific defects in biosynthesis of phospholipids (recently reviewed in Carman and Han, 2009) or sterols (reviewed in Sturley, 2000). These types of experiments have been quite useful in determining the importance of sterols in permease association with lipid rafts (Bagnat et al., 2000; Dupre and Haguenauer-Tsapis, 2003; Hearn et al., 2003; Umebayashi and Nakano, 2003) and in implicating the essential nature of the phospholipid phosphatidylethanolamine (PE) in delivery of nutrient transporters to the plasma membrane (Opekarova et al., 2005).
The primary route of PE production in S. cerevisiae, like most eukaryotic cells, is via the mitochondrially localized phosphatidylserine (PS) decarboxylase enzyme (Psd) 1 (Achleitner et al., 1995). However, cells that lack the PSD1 are still capable of growth in the absence of exogenous ethanolamine due to the presence of a second phosphatidylserine decarboxylase called Psd2 (Trotter et al., 1995; Trotter and Voelker, 1995). Psd2 has been argued to enrich in fractions corresponding to the Golgi/vacuolar region of the cell and to contain a C2 domain associated with membrane binding (Kitamura et al., 2002). Other than its role in maintaining ethanolamine prototrophy in a psd1Δ cell, no specific phenotypes have been associated with loss of Psd2.
In this report, we provide evidence that cells lacking Psd2 exhibit a cadmium hypersensitive defect. Importantly, although these psd2Δ cells are cadmium sensitive, psd1Δ strains are no more cadmium sensitive than a wild-type strain. Our earlier work determined that overproduction of Psd1 (Gulshan et al., 2008), even in a catalytically inactive form, was able to elevate expression of a plasma membrane multidrug transporter called Pdr5 (Balzi et al., 1994; Bissinger and Kuchler, 1994; Hirata et al., 1994). Overexpression of Psd2 elevated cadmium tolerance but only if the protein was proteolytically matured. The action of Psd2 on cadmium tolerance is mediated by the vacuolar membrane-localized ATP-binding cassette (ABC) transporter protein Ycf1, a well-known determinant of cadmium resistance (Szczypka et al., 1994). Fluorescence microscopy and biochemical fractionation experiments demonstrate that Psd2 is localized to the endosomal system of S. cerevisiae. Together, these data indicate that Psd2 influences PE content in the endocytic system and the vacuole. Changes in vacuolar PE content influence the activity of the Ycf1 ABC transporter protein.
MATERIALS AND METHODS
Yeast Strains and Media
Yeast strains used in this study are listed in Table 1. Cells were grown in cultures containing YPD (2% yeast extract, 1% peptone, and 2% glucose) under nonselective conditions or appropriate CSM media (Bio 101, Vista, CA) under selective conditions (Sherman et al., 1979). Drug resistance was measured by the spot test assay on plates with either a single concentration of drug or gradient plates (Katzmann et al., 1999). Transformation was performed using the lithium acetate technique (Ito et al., 1983).
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα leu2-3, 112ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel− | S. Emr (Cornell University,Ithaca, NY) |
| BY4742 | MATα his3-Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Open Biosystems |
| YPH499 | MATα ade2-101 ura3-52 his-3Δ200, trp1-Δ63 leu2Δ1Lys2-801 | A. K. Bachhawat (IMTECH,Chandigarh, India) |
| PSY4 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 psd2Δ::natR-MX4 | Gulshan et al. (2008) |
| KG21 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2–801 psd2Δ::HIS3MX6 | Gulshan et al. (2008) |
| KGS22 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 psd1Δ::kanMX2 psd2Δ::HIS3MX6 | Gulshan et al. (2008) |
| KGS25 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 cho2Δ::kanMX2 | Gulshan et al. (2008) |
| KGS26 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 opi3Δ::kanMX2 | Gulshan et al. (2008) |
| KGS27 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 cho2Δ:: natR-MX4 | Gulshan et al. (2008) |
| KGS28 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 cho2Δ:: natR-MX4 opi3Δ::kanMX2 | Gulshan et al. (2008) |
| KGS50 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 pdr17Δ::kanMX2 | This study |
| KGS51 | MATα leu2-3, 112ura3-52 his3-Δ200 trp1-Δ901 lys2-801 pdr16Δ::kanMX2 | This study |
| KGS52 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 ycf1Δ::kanMX2 | This study |
| KGS53 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 ycf1Δ::natR-MX4 bpt1Δ::kanMX2 | This study |
| KGS54 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 PSD2-3 × HA::TRP1 | This study |
| KGS55 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 psd1Δ::kanMX2 PSD2-3XHA::TRP1 | This study |
| KGS56 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 PSD2-GFP::TRP1 | This study |
| KGS57 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 psd1Δ::kanMX2 PSD2-GFP::TRP1 | This study |
| KGS58 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 kanMX2::TDH3p-PSD2-GFP::TRP1 | This study |
| KGS59 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 Pdr17-TAP::HIS3MX6 | This study |
| KGS60 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 Pdr16-TAP::HIS3MX6 | This study |
| KGS61 | MATα leu2-3, 112ura3-52 his3-Δ200 trp1-Δ901 lys2-801 psd2Δ::natR-MX4PDR17-TAP::HIS3MX6 | This study |
| KGS62 | MATα leu2-3, 112ura3-52 his3-Δ200 trp1-Δ901 lys2-801 PDR17-TAP::HIS3MX6PSD2-3XHA::TRP1 | This study |
| KGS63 | MATα leu2-3, 112ura3-52 his3-Δ200 trp1-Δ901 lys2-801 YCF1-GFP::HIS3MX6 | This study |
| KGS64 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 psd2Δ::natR-MX4YCF1-GFP::HIS3MX6 | This study |
| KGS65 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 YCF1-TAP::HIS3MX6 | This study |
| KGS66 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 psd2Δ::natR-MX4YCF1-TAP::HIS3MX6 | This study |
| KGS67 | MATα ade2-101 ura3-52 his-3Δ200 trp1-Δ63 leu2-Δ1 lys2-801 psd2Δ::natR-MX4 | This study |
| KGS68 | MATα ade2-101 ura3-52 his-3Δ200 trp1-Δ63 leu2-Δ1 lys2-801 pdr17Δ::kanMX2 | This study |
Strain Construction
The open reading frames (ORFs) PDR17, PDR16, and YCF1 were disrupted in SEY6210 by polymerase chain reaction (PCR)-mediated gene disruption using the KanMX4 cassette, yielding KGS50, KGS51, and KGS52, respectively. The following primers were used for amplification of KanMX4 deletion cassettes of YCF1, PDR17, and PDR16, YCF1-Del1, YCF1-Del2, Pdr17-For, Pdr17-Rev, Pdr16-For, and Pdr16-Rev. The PSD2 ORF and PDR17 ORF were also disrupted in the YPH499 background by using PCR-mediated gene disruption with the psd2Δ::natMX4 and pdr17::KanMX4 cassettes to yield KGS67 and KGS68. The primers used to amplify the deletion cassettes for psd2 natR::MX4 and pdr17::KanMX4 were Psd2 Del-For, Psd2 Del-Rev, and Pdr17-For, Pdr17-Rev, respectively. A strain carrying deletions in the YCF1 and BPT1 genes was constructed by transforming the bpt1::KanMX4 cassette into a strain carrying the ycf1::natMX4, allele, yielding KGS53. The bpt1::KanMX4 deletion cassette was amplified by using primers Bpt1-Del-For and Bpt1-Del-Rev. A C-terminal 1X hemagglutinin (HA)-tagged allele of Psd2 (obtained from YEp352-PSD2–1XHA (Kitamura et al., 2002) was first generated in psd1Δ psd2Δ background by recombining in the PSD2-1XHA allele to restore ethanolamine prototrophy. The 1X HA-tagged version was then exchanged with 3XHA-TRP1 and GFP-TRP1 cassettes amplified from plasmids pFA6a-3XHA and pFA6a-GFP (Longtine et al., 1998), yielding KGS 55 and KGS 57.
The C-terminal 3X HA tag and green fluorescent protein (GFP) tag fusions of PSD2 were also constructed in SEY6210 by transforming this strain with a 3XHA-TRP1 or GFP-TRP1 cassettes amplified from plasmids strains KGS55 and KGS57, yielding KGS54 and KGS56. To generate strains carrying tandem affinity purification (TAP) tag fusions of PDR16 and PDR17 in wild-type cells, SEY6210 strain was transformed with a TAP-HIS3MX6 cassettes, amplified by using primers Pdr16 TAP-For, Pdr16 TAP-rev, Pdr17 TAP-For and Pdr17 TAP-Rev, from the TAP tag strain collection (Open Biosystems, Huntsville, AL), yielding KGS59 and KGS60. The TAP-HIS3MX6 cassette for PDR17 for making C-terminal TAP-tagged fusions was also transformed in strains PSY4 and KGS54, yielding KGS61 and KGS62, respectively. Strains carrying C-terminal GFP and TAP tag fusions of YCF1 were constructed in SEY 6210 and psd2Δ (PSY4) by transforming these strains with a GFP-HIS3MX6 and TAP-HIS3MX6 cassette amplified from GFP tag strain collection (Open Biosystems), yielding KGS63, KGS 64, KGS 65, and KGS66. All the strains were confirmed by PCR. Primer sequences are available on request.
Plasmids
A high-copy-number plasmid carrying PSD2 with a single HA epitope at its C terminus was provided by Dennis Voelker (Kitamura et al., 2002). A pBG1805 (2-μm URA3) clone carrying a GAL1-PSD2 fusion gene was purchased from Open Biosystems. A 1.053-kb PCR fragment containing the PDR17 ORF was amplified as a BamHI fragment from SEY6210 (wild-type) genomic DNA. This fragment was then cloned under control of the strong PGK1 promoter in pXTZ138 (Zhang and Moye-Rowley, 2001) to generate pKGE38. The plasmid pBG1805 carrying the PSD2 gene with a C-terminal HA tag was cut with SacI restriction enzyme to release the GAL1 promoter as well as 250 base pairs of N-terminal PSD2 ORF sequences. A 850-base pair PCR fragment carrying 600 base pairs of the PSD2 promoter and 250 base pairs of the PSD2 ORF starting from ATG was amplified as SacI fragment from wild-type genomic DNA and cloned in plasmid pBG1805-GAL1-PSD2-HA cut with SacI, to generate pKGE36. The plasmid pKGE36 was used to construct a catalytically inactive mutant of GGST motif of PSD2 ORF. A 230-base pair fragment was amplified from C-terminal region of PSD2 ORF by using primers PSD2 GGS-AAA-For (TTTGccGcggCTACTATAATAATCATTATCCCG) (mutant residues in lowercase) and PSD2-3351-Rev. Another 416 base pairs were amplified from the C-terminal region of PSD2 ORF by using primers Psd2-ClaI-200-For and PSD2-GGST-Rev. Both fragments were then used as template to amplify 646 bp of mutated C-terminal PSD2 ORF by using primers Psd2-ClaI-200-For and PSD-GGST-Rev. Primer sequences are available on request. The 646-base pair fragment was then transformed into a psd1Δ psd2Δ strain along with plasmid KGE36 cut with ClaI and AatII that removed 12 amino acids from the extreme C-terminal coding region of PSD2 ORF while leaving the GGST motif intact. The transformants were recovered on CSM-URA plates containing 1 mM ethanolamine. Multiple transformants were then streaked on media lacking ethanolamine to identify the mutated GGST motif. The plasmid was recovered from a colony that exhibited the expected ethanolamine auxotrophy and named pKGE37. This plasmid was then sequenced as well as retested for defective ethanolamine complementation of psd1Δ psd2Δ strain. The plasmid carrying the PSD2-3X HA C-terminal tag was generated by transforming psd1Δ psd2Δ cells with plasmid YEp352 2 μm URA3 PSD2-HA with PSD2-3X HA::TRP1 cassette amplified from plasmid pFA6a-3X HA::TRP1 (Longtine et al., 1998). TRP1 transformants were recovered and the ability of the 3X HA-tagged Psd2 to normally confer ethanolamine prototrophy verified. The physical structures of all plasmids were confirmed by restriction digestions and DNA sequencing. Multicopy plasmid vectors expressing Tlg1-mCherry and Tlg2-mCherry were obtained from David Katzmann (Mayo Medical School, Rochester, MN) and the clone expressing Snc1-RFP was from Kazuma Tanaka (Hokkaido University, Sapporo, Japan).
Monochlorobimane (MCB) Transport
This assay was adapted from previous work (Li et al., 1996). Yeast cultures were grown in rich YPD medium until saturation. Cells were reinoculated in fresh YPD media with a starting OD600 nm of 0.1. After allowing cell growth for 2 h at 30°C, 50 μM 1-(chloromethyl)-2,6,7-trimethylpyrazolo[1,2-a]pyrazole-3,5-dione (MCB) was added to each culture. After incubation for either 2 or 4 h, the cells were centrifuged for 2 min at 14K. The cells were washed three times with YPD to remove excess MCB dye. These cells were then viewed under a fluorescence microscope equipped with UV excitation filter.
Adenine (Ade) Pigment Transport
Vacuolar ABC transporters have been demonstrated previously to mediate ade pigment conjugate transport to vacuoles (Chaudhuri et al., 1997; Sharma et al., 2002). Qualitative measurement of ade pigment transport was accomplished by growing cells to saturation in YPD media containing excess adenine. Cells were transferred to CSM media containing excess adenine after 6 h of growth to prevent any pigment formation. After allowing them to reach early log phase, cells were extensively washed with CSM media without adenine. After washing, these cells were transferred to CSM media containing only 5 μg/ml adenine for 2 or 4 h. Cells were washed with sterile water and viewed under a fluorescent microscope. For quantification of ade pigment, cells were grown overnight in YPD containing 0.5% yeast extract (0.5% YPD) and then reinoculated in fresh 0.5% YPD media to an OD600 nm of 0.2. These cells were then grown at 30°C until an OD of 0.8 was reached. Equal numbers of cells were then harvested from each sample, suspended in 5% sulfosalicylic acid, and extracts were made by breaking these cells with glass beads. After clearing this extract by centrifugation, ade pigment accumulation was quantified by measuring absorbance at 530 nm.
Sucrose Density Gradient
Cells carrying integrated PSD2-3X HA::TRP1 and PDR17-TAP::HIS3 fusion genes were grown in 100 ml of YPD to an OD600 nm of 0.6. Cells were incubated in ice for 15 min after addition of 10 mM sodium azide and potassium fluoride, harvested, and broken with glass beads in cold STE10 buffer (10 mM Tris, 10 mM EDTA, and 10% sucrose) in the presence of protease inhibitors. Cell extracts were loaded on sucrose density gradients prepared by sequentially pouring STE solutions containing 60, 35, 20, and 10% sucrose to form a 5-ml step gradient. The gradient was allowed to diffuse for 6 h. Gradients were subjected to 14 h of centrifugation at 50,000 rpm at 4°C in a SW55Ti rotor (Beckman Coulter, Fullerton, CA). Fractions were collected from the top of the gradient and precipitated with 50% trichloroacetic acid (TCA). These fractions were subjected to Western blot analysis using monoclonal anti-HA (1:5000), polyclonal anti-TAP (1:5000), monoclonal anti-Pma1 (1:1000; Abcam, Cambridge, MA), polyclonal anti-Kar2 (1:12,000), and monoclonal anti-Pep12 (1:5000). Primary antibodies were detected with horseradish peroxide-conjugated anti-rabbit secondary antibody (1:12,000) and anti-mouse antibody (1:12,000), followed by measurement of chemiluminescence (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Lipid Extraction and Analysis
For whole cell lipid extractions, cells were grown in YPD to an OD600 nm of 0.6 and washed twice with ice-cold water. These cells were then resuspended in water and 2 volumes of ethanol added. The samples were boiled for 1 h. Lipids were extracted by the Bligh Dyer method (Bligh and Dyer, 1959). To prepare vacuolar membrane lipids, vacuoles were isolated from spheroplasts by a Ficoll gradient flotation method using DEAE-dextran lysis as described previously (Wemmie and Moye-Rowley, 1997). Vph1 was blotted from vacuolar fractions to check enrichment of vacuolar protein. To prepare endosomal enriched fractions, the differential centrifugation technique of Singer-Kruger et al. (1993) was used. Two independent spheroplast preparations were assayed three times. Phospholipids were extracted from vacuolar and endosomal membranes as described above. The chloroform phases from all lipid samples were dried under nitrogen gas and dissolved in an appropriate volume of chloroform/methanol (9:1, vol/vol). The samples were subjected to one-dimensional thin layer chromatography (TLC), and phospholipids were visualized by exposure to iodine vapors. Individual phospholipids were scratched from TLC silica plate along with blank from areas of plate where no lipid is present. The percentage of phospholipids was calculated by comparison with standards made from 1 mM Na2HPO4. In brief, dried lipids were suspended in 0.3 ml of 70% perchloric acid and heated to 180°C for 30 min. Samples were allowed to cool, and 1.67 ml of distilled water was added. To all samples, 0.33 ml of ammonium molybdate (2.5%) solution was added, vortexed briefly, and 0.33 ml of ascorbic acid (10%) was added. Samples were kept at 50°C for 15 min to develop color. Samples were spun down and OD of supernatant was taken at 820 nm.
Fluorescence Microscopy
Strains expressing different GFP- or red fluorescent protein (RFP)-tagged proteins were grown to saturation. These cultures were then reinoculated to a starting OD600 nm of 0.1. Cells were allowed to grow for 3–4 h to an approximate OD600 nm of 0.4–0.6 and then visualized by fluorescence microscopy. For the N-[3-triethylammoniumpropyl]-4-[p-diethylaminophenylhexatrienyl] pyridinium dibromide (FM4-64) chase experiment, cells were labeled with the dye at 0°C for 12 min, washed twice with YPD, and then chased in prewarmed medium at 30°C for the indicated times. Fluorescence microscopic images were produced by placing a few microliters of a culture of interest on a glass slide, overlaying with a coverslip, and examining under the 100× oil objective of an Olympus BX60 microscope. Indirect immunofluorescence was carried out as described previously (Katzmann et al., 1999) using anti-HA (Covance Research Products, Princeton, NJ) or anti-Vma2 (Invitrogen, Carlsbad, CA) mouse monoclonal antibodies. Texas Red-conjugated goat anti-mouse antibodies (Invitrogen) were used to detect binding of the primary antibodies. Fluorescent micrographs were captured on a digital camera (Hamamatsu, Bridgewater, NJ).
Western Analysis
For Western analysis, cells were grown in 100 ml of YPD or selective medium. Cells were harvested at OD600 nm of ∼1.0, washed, and lysed by glass bead lysis in a buffer containing 300 mM sorbitol, 100 mM NaCl, 5 mM MgCl2, 10 mM Tris, pH7.4, and Complete protease inhibitors (Roche Diagnostics, Indianapolis, IN). The Bradford assay (Bradford, 1976) was used to determine the concentration of proteins in different samples. Equal amounts of proteins were loaded on 10% polyacrylamide gel. The proteins were transferred to nitrocellulose membrane, blocked with 5% nonfat dry milk in phosphate-buffered saline, and then probed with either anti-HA or anti-TAP antibodies. Horseradish peroxidase-conjugated secondary antibody and the ECL kit (Pierce Chemical, Rockford, IL) were used to visualize immunoreactive proteins.
Coimmunoprecipitation
All immunoprecipitation assays were performed using lysed spheroplasts. In brief, cells growing in early log phase were washed with spheroplast solution I (1 M sorbitol, 10 mM MgCl2, 30 mM dithiothreitol, 100 μg/ml phenylmethylsulfonyl fluoride, 50 mM K2HPO4), resuspended in spheroplast solution II (1 M sorbitol, 10 mM MgCl2, 30 mM dithiothreitol, 100 μg/ml phenylmethylsulfonyl fluoride, 50 mM K2HPO4, and 25 mM sodium succinate, pH 5.5) containing oxylyticase and incubated at 30°C for 30 min. Spheroplasts were collected by centrifuging at 5000 rpm for 20 min at 4°C in a JA-20 rotor (Beckman Coulter). These spheroplasts were lysed using NP-40 lysis buffer (1% Nonidet P-40/Triton X-100, 0.15 M NaCl, and 50 mM Tris-HCl, pH 7.2). Glass beads were added to cells suspended in NP-40 lysis buffer, followed by addition of 2 mM EDTA, 200 μM sodium vanadate, and 50 mM sodium fluoride. Lysis was accomplished by shaking cell suspensions on a Tomy shaker for 30 min at 4°C. Protein extracts were clarified by centrifuging lysates at 14,000 rpm for 5 min in an Eppendorf microcentrifuge. For immunoprecipitation, extracts were incubated with anti-TAP antibody for 4 h at 4°C. Prewashed protein A-agarose beads were added to these samples and incubated for 2 h. Beads were washed and immunoprecipitated proteins along with input proteins were then loaded on a 10% polyacrylamide gel and analyzed by Western blotting with anti-TAP and anti-HA antibodies.
RESULTS
Psd2 Is Required for Tolerance to Heavy Metals
In the model eukaryote S. cerevisiae, three different routes exist to support the biosynthesis of the phospholipid PE (recently reviewed in Carman and Han, 2009). The major source of PE biosynthesis comes through the decarboxylation of PS in the mitochondria catalyzed by the Psd1 phosphatidylserine decarboxylase enzyme (Clancey et al., 1993; Trotter et al., 1993). A second nonmitochondrial phosphatidylserine decarboxylase enzyme called Psd2 is sufficient to retain ethanolamine independent growth in psd1Δ cells (Trotter et al., 1995). The final avenue for PE biosynthesis, the Kennedy pathway, requires the presence of exogenous ethanolamine to synthesize adequate PE to support viability (Birner et al., 2001).
Previous work from our laboratory has established that changes in the dosage of PSD1 influence expression of the multidrug resistance gene PDR5 (Gulshan et al., 2008). This gene encodes a plasma membrane ABC transporter protein that is thought to act as a broad specificity drug pump (Kolaczkowski et al., 1996). Although overproduction of Psd1 in ρ+ cells led to a large increase in PDR5 transcription, loss of Psd1 from cells had insignificant effects on resistance to either cycloheximide (Pdr5 substrate) or cadmium (independent of Pdr5). Surprisingly, although loss of Psd2 also had negligible consequences to cycloheximide resistance, a large decrease in cadmium tolerance was seen (Figure 1). An isogenic ycf1Δ strain was also used to compare the relative increase in cadmium sensitivity seen in psd2Δ cells. YCF1 encodes an ABC transporter protein localized to the limiting membrane of the vacuole that is a key determinant of cadmium resistance in S. cerevisiae (Wemmie and Moye-Rowley, 1997). A psd2Δ strain exhibited cadmium sensitivity that was nearly as severe as a ycf1Δ mutant, indicative of a pronounced defect in the ability to tolerate this heavy metal.
Figure 1.
Genetic analysis of Psd2 interaction with Ycf1 substrates. (A) The indicated mutant strains and isogenic wild-type were grown to mid-log phase, and then 1000 cells of each placed on solid medium containing a gradient of the compound listed at the bottom of the panel (Cyh, cycloheximide). Gradient plates were prepared as described previously (Katzmann et al., 1999), and the relative increase in drug concentration was indicated by the bar of increasing width. Plates were incubated at 30°C and photographed once growth was visible. (B) Isogenic wild-type or ycf1Δ bptΔ cells were transformed with the high-copy-number vector plasmid carrying wild-type PSD2 (2 μm PSD2) or the empty vector (Vector). Transformants were grown to mid-log phase with selection for the plasmid and tested for resistance as described above.
Because Psd2 and Ycf1 seem to cause similar effects on cadmium resistance, we wanted to determine the relationship between the two genes encoding these proteins. A high-copy-number plasmid containing the wild-type PSD2 gene was introduced into isogenic wild-type and ycf1Δ bpt1Δ cells. Bpt1 is a homologue of Ycf1 (Klein et al., 2002; Sharma et al., 2002), and the double mutant was used to eliminate the background activity of cadmium resistance otherwise supported by Bpt1. Similar results were obtained when using ycf1Δ single mutants (data not shown). Transformants were placed on media containing a gradient of either HgBr or cadmium. After several days' growth, these plates were photographed (Figure 1).
Two important findings emerged from this experiment. First, overproduction of Psd2 led to an increase in resistance to both heavy metals. These heavy metals have previously both been established as substrates of Ycf1 but Bpt1 observed to influence cadmium tolerance only (Gueldry et al., 2003). Second, this resistance is not seen in ycf1Δ bpt1Δ cells. Together, these data support the view that Ycf1 function responds in a dose-dependent manner to levels of Psd2.
Catalytic Function of Psd2 Is Required to Support Cadmium Resistance
Our previous work with the mitochondrial Psd1 enzyme produced the unexpected finding that the catalytic function of this protein was not required to exert the effect of this protein on drug resistance (Gulshan et al., 2008). To determine whether catalytic activity of Psd2 was required for the enzyme to influence cadmium tolerance, a mutant form of this gene was constructed. Earlier work in Escherichia coli (Li and Dowhan, 1990), and our more recent studies in S. cerevisiae (Gulshan et al., 2008), demonstrated the necessity for C-terminal processing of phosphatidylserine decarboxylase enzymes. Detailed biochemical analyses in E. coli and mammalian cells revealed that this processing is required to generate the active site of the enzyme (Li and Dowhan, 1990; Kuge et al., 1996). The serine residue that is exposed upon cleavage between the glycine and serine residues is required for phosphatidylserine decarboxylase activity (recently reviewed in Schuiki and Daum, 2009). Similarly, mutation of an LGST sequence corresponding to the presumptive processing site in the mitochondrial Psd1 of S. cerevisiae eliminated the ability of the resulting mutant protein to confer ethanolamine prototrophy on cells (Gulshan et al., 2008). Psd2 contains a related sequence (GGST) (Trotter et al., 1993) that is located at an analogous position in the polypeptide chain. A site-directed mutant from of Psd2 was prepared in which the GGST was changed to AAAT. Wild-type and GGS1041AAA mutant forms of Psd2 were expressed as epitope-tagged proteins in a psd2Δ background. Transformants were analyzed for the expression level of Psd2 by Western blotting and tested for their ability to complement the cadmium hypersensitivity of the psd2Δ strain (Figure 2).
Figure 2.
Catalytic activity of Psd2 is required for heavy metal resistance. (A) Loss of residues required for Psd2 catalysis eliminates the ability of the resulting protein to stimulate cadmium resistance. A high-copy-number plasmid containing wild-type or the GGS1041AAA allele of PSD2 or the empty vector plasmid was transformed into a psd2Δ strain. These plasmids were designed to produce full-length wild-type or GGS1041 Psd2 with a single HA epitope tag fused at the C terminus. Transformants were analyzed for the ability to support growth in the presence of a concentration gradient of cadmium as described above. (B) Site-directed mutation in PSD2 blocks C-terminal processing. Whole cell protein extracts were produced from the transformants in A and analyzed by Western blotting with anti-HA antibody. Molecular mass standards are indicated on the left-hand side of the panel in kilodaltons. The position of the properly processed Psd2 C-terminal fragment is indicated by the arrow. (C) Addition of ethanolamine suppresses the cadmium defect in a psd2Δ mutant strain. An isogenic series of strains of the indicated relevant genotype denoted on the right hand side of the panel were grown to mid-log phase. Serial dilutions of these cultures were plated on medium containing a single cadmium concentration. Where indicated, 15 mM ethanolamine was added to the medium. Plates were incubated at 30°C and photographed.
The mutant GGS1041AAA Psd2 protein failed to complement cadmium hypersensitivity of the psd2Δ strain, whereas the wild-type protein restored tolerance to this heavy metal. Western blot analysis indicated that although the expected C-terminal product was seen in cells expressing the wild-type protein, the mutant GGS1041AAA Psd2 did not generate any C-terminal fragment. Similarly, the GGS1041AAA Psd2 was unable to support ethanolamine independent growth when present in a psd1Δ psd2Δ cell (data not shown). These observations argue that normal processing and catalytic activity of Psd2 are required for this protein to carry out its biological activities, both at the level of PE production and cadmium resistance. These findings suggest that levels of Psd2-produced PE are critical for normal cadmium tolerance. An attractive model to explain these data are that Psd2 plays a major role in controlling levels of PE in the vacuole and Ycf1 is influenced by changes in PE content in this organelle.
To test this idea, the ability of exogenous ethanolamine to suppress the cadmium sensitivity of a psd2Δ strain was examined. We hypothesized that a specific pool of PE, normally produced by Psd2, is depleted, and then cadmium resistance is lowered. If ethanolamine is added to the medium, then PE levels in the cell can be elevated through function of the Kennedy pathway (reviewed in Daum et al., 1998), which may in turn suppress the cadmium sensitivity of the psd2Δ strain. Because this PE pool acts via influencing Ycf1, then ethanolamine supplementation would be unable to suppress the cadmium sensitivity caused by loss of the YCF1 gene. We also tested the ability of the Kennedy pathway to suppress the cadmium sensitivity of a strain lacking the phosphatidylinositol transfer protein (PITP) homologue Pdr17. Pdr17 was previously shown to be required for PE biosynthesis by Psd2 (Wu et al., 2000), suggesting the possibility that loss of this PITP might similarly cause a reduction in cadmium resistance. An isogenic series of disruption strains was tested for cadmium tolerance on plates containing or lacking supplementation with ethanolamine (Figure 2).
Loss of either Psd2 or Pdr17 causes a large reduction in cadmium resistance, but this sensitivity can be suppressed with the addition of ethanolamine to cadmium-containing medium. This behavior was not reproduced by a ycf1Δ strain that was cadmium sensitive irrespective of the presence of ethanolamine. These data support the view that both Psd2 and Pdr17 are required for normal cadmium tolerance through their roles in PE biosynthesis. Furthermore, both Psd2 and Pdr17 act through the vacuolar ABC transporter Ycf1.
Unique Contributions of Psd2 and Pdr17 to Cadmium Tolerance
Because loss of either Psd2 or Pdr17 caused cadmium sensitivity, we tested the ability of increased dosage of each protein to suppress the cadmium defect caused by loss of the other. A high-copy-number plasmid that overproduced Psd2 or a fusion gene between the strong PGK1 promoter and PDR17 was used to drive elevated levels of each protein. These plasmids were introduced, along with a vector control, into isogenic psd2Δ and pdr17Δ strains. Transformants were then analyzed for the ability to confer cadmium resistance as well as the steady-state level and the membrane association of each protein (Figure 3).
Figure 3.
Both Psd2 and Pdr17 are required for wild-type cadmium resistance. (A) Isogenic strains with the relevant phenotype indicated at the right were transformed with an empty vector plasmid (Vector) or with plasmids conferring overexpression of either HA epitope-tagged Psd2 (2 μm PSD2) or Pdr17 (PGK-PDR17). Transformants were grown to mid-log phase and then tested for the ability to grow on YPD medium containing a gradient of cadmium. (B) Whole cell protein extracts were prepared from the transformants above and analyzed by Western blotting with anti-HA antibody. The molecular masses of the immunoreactive species are indicated. (C) Whole cell protein extracts were prepared from isogenic PDR17 or pdr17Δ strains containing an integrated PSD2-3X HA fusion gene (top) or isogenic PSD2 or psd2Δ strains containing an integrated PDR17-TAP fusion gene (bottom). Aliquots of the whole cell protein extract (T) were retained and the remainder of the sample centrifuged at 10,000 × g to produce a 10K pellet (P) and 10K supernatant (S). Equal amounts of protein were then subjected to Western analysis using anti-HA or anti-TAP antibodies to detect Psd2-3X HA or Pdr17-TAP, respectively. When Psd2-3X HA is expressed at normal chromosomal levels, only the C-terminal processing product (17 kDa) can be seen, and this is indicated at the right.
Overproduction of Psd2 was unable to suppress the cadmium sensitivity of a pdr17Δ strain, although it was able to complement the same phenotype in a psd2Δ strain. Similarly, PGK-driven expression of Pdr17 complemented the cadmium sensitivity of a pdr17Δ strain but not of the isogenic psd2Δ cell. These data argue that, as seen previously for PE biosynthesis (Wu et al., 2000), Pdr17 and Psd2 carry out unique, essential roles in conferring normal cadmium resistance on cells.
The mutual dependence of cadmium tolerance shown here and PE biosynthesis shown previously on the presence of both Psd2 and Pdr17 suggested that these proteins might exhibit biochemical phenotypes in the absence of the other. To determine the basis of the defects caused by loss of either one of these partner proteins, steady-state levels of both Psd2 and Pdr17 were compared in the presence and absence of the other subunit. The transformants described above were grown to mid-log phase and levels of both proteins determined by Western blot analysis (Figure 3).
Steady-state levels and processing of Psd2 were independent of the presence of Pdr17. Similarly, Pdr17 was equally expressed in isogenic wild-type and psd2Δ cells. These data demonstrate that the steady-state levels of these two proteins are controlled independently.
Previous experiments determined that the membrane association of Psd2 requires the presence of a C2 domain present in the amino terminus of this protein (Kitamura et al., 2002). Removal of the C2 domain retained catalytic activity but eliminated the ability of the resulting mutant to function in vivo. To determine whether membrane association of either Psd2 or Pdr17 might require the presence of the other, the association of these two phospholipid biosynthetic enzymes was assessed in isogenic wild-type or mutant strain lacking potential partner protein. This analysis was carried out using integrated epitope-tagged alleles of each gene in order to avoid possible complications from overproduction. Whole cell protein extracts were prepared under native conditions and then centrifuged at 10,000 × g to generate a 10K pellet and supernatant fraction. Equal amounts of protein were analyzed by Western blotting.
Both Psd2 and Pdr17 were found in the 10K pellet fractions exclusively, consistent with membrane localization. Extraction of these pellet fractions with Na2CO3 but not Triton X-100 released Psd2 from the pellet fractions. Pdr17 exhibited very similar fractionation properties (data not shown). This biochemical behavior is consistent with peripheral membrane association via these factors. However, association of these phospholipid biosynthetic enzymes with their target membranes is independent of the presence of the other. These experiments argue that although function of Psd2 and Pdr17 depends on the presence of both proteins, their expression and membrane association do not.
Psd2 and Pdr17 Are Physically Associated
Because both cadmium resistance and PE biosynthesis required the presence of both proteins, we wondered whether these factors might form a heteromeric complex. A large-scale analysis of interacting proteins in S. cerevisiae has also provided support for the idea that Psd2 and Pdr17 interact (Krogan et al., 2006). To directly assess the possibility that these proteins might physically interact, a coimmunoprecipitation assay was carried out. A control PITP (Pdr16) was used to evaluate the specificity of any potential Psd2:Pdr17 interaction. We examined the cadmium and fluconazole resistance profiles of strains lacking the related PITPs Pdr16 and Pdr17. PDR16 and PDR17 were initially identified as downstream targets of the Pdr1/Pdr3 transcription factors (van den Hazel et al., 1999). To evaluate the phenotypes caused by loss of these different PITPs, we constructed isogenic wild-type, pdr16Δ and pdr17Δ mutants. These strains were tested for their ability to tolerate challenge by cadmium or the antifungal drug fluconazole (Figure 4). Loss of Pdr16 was found to cause azole sensitivity, a finding reported previously in a different genetic background (van den Hazel et al., 1999), whereas loss of Pdr17 alone caused cadmium sensitivity as detailed above. These data indicate that, although Pdr16 is related to Pdr17 by sequence conservation (reviewed in Griac, 2007), their functions are distinct.
Figure 4.
Psd2 and Pdr17 associate in vivo. (A) An isogenic series of strains consisting of wild-type (WT), pdr16Δ, or pdr17Δ was tested for the ability to tolerate different levels of cadmium or fluconazole by gradient plate assay. (B) Wild-type strains containing integrated PDR16-TAP or PDR17-TAP fusion genes at the native loci were transformed with a high-copy-number vector (−) or the same plasmid producing Psd2-HA (+). Transformants were grown to midlog phase, whole cell protein extracts prepared and then equal amounts of protein were withdrawn to analyze as control for input proteins (INPUT). The remainder of the protein extracts were subjected to immunoprecipitation (IP) using anti-TAP antibody. Input and immunoprecipitated samples were analyzed by Western blotting using anti-TAP and anti-HA antibodies.
To evaluate interaction of Psd2 with Pdr17, two TAP-tagged strains were generated by integrating the TAP cassette at the C termini of the PDR16 and PDR17 genes. Strains containing these TAP-tagged forms of Pdr16 or Pdr17 were transformed with a high-copy-number plasmid producing Psd2-HA. Transformants were grown to mid-log phase, protein extracts prepared and analyzed by immunoprecipitation using anti-TAP antiserum. Immunoprecipitates were resolved in SDS-polyacrylamide gel electrophoresis (PAGE) and blotted for TAP- and HA-tagged proteins (Figure 4).
Psd2-HA was specifically recovered in the anti-TAP immunoprecipitate prepared from the Pdr17-TAP–expressing strain. No Psd2-HA was seen in immunoprecipitates from the Pdr16-TAP strain. We also carried out a similar analysis using a Pdr17-TAP–expressing strain containing an integrated PSD2-HA fusion gene and again observed coimmunoprecipitation of these two proteins (data not shown). These observations support the view that Psd2 and Pdr17 specifically associate to form a complex that catalyzes the decarboxylation of PS to form PE and that this PE is essential for normal cadmium tolerance. Given the finding that overproduction of Psd2 can increase cadmium resistance in a Ycf1-dependent manner, we set out to determine how changes in PSD2 gene dosage influenced Ycf1-dependent cadmium resistance.
Psd2 Action Enhances Ycf1-dependent Transport Activity
Psd2 function could trigger increased Ycf1 activity in a variety of ways. A YCF1-lacZ plasmid we have characterized previously (Wemmie et al., 1994) was used to allow a facile readout for YCF1 gene expression. Ycf1-TAP and Ycf1-GFP fusion proteins obtained from large-scale collections of tagged proteins (Ghaemmaghami et al., 2003; Huh et al., 2003) were used to examine steady-state levels and vacuolar localization, respectively. These different assays for Ycf1 expression and localization failed to show any significant differences when compared in wild-type and isogenic psd2Δ cells (data not shown). These results suggested that any changes in Ycf1 function were more likely to be due to effects on the transport activity of this protein. To test this idea, we examined vacuolar accumulation of two different Ycf1 substrates, MCB and an endogenous red pigment that accumulates in adenine biosynthetic mutant strains (Chaudhuri et al., 1997).
MCB has been demonstrated previously to accumulate in the vacuole in a Ycf1-dependent manner (Li et al., 1996). Wild-type and psd2Δ cells were incubated with MCB for 0, 2, or 4 h, and then they were examined by fluorescence microscopy. These cells were also labeled with the dye FM4-64 to visualize the vacuolar membranes (Figure 5).
Figure 5.
Psd2 stimulates Ycf1-dependent transport activity. (A) Wild-type (wt) or isogenic psd2Δ cells were grown to mid-log phase and then analyzed for vacuolar accumulation of MCB as described previously (Li et al., 1996). After labeling with MCB, cells were visualized at 2 and 4 h by Nomarski optics (DIC) and fluorescence microscopy. Cells were also stained with FM4-64 (FM) to confirm accumulation of MCB inside the vacuole. (B) Isogenic ade2Δ cells containing (wt) or lacking (psd2Δ) the PSD2 gene were grown on limiting adenine to induce formation of the ade pigment (Ade). Microscopy was as above with the exception that the presence of the autofluorescent ade pigment was detected.
As can be seen in Figure 5, after 2 h of incubation, MCB fluorescence was clearly seen in the lumen of the vacuole of wild-type cells. Conversely, no detectable fluorescence was found in the vacuolar lumen of a psd2Δ strain at this same time point. After extended incubations (4 h), both wild-type and psd2Δ cells showed lumenal fluorescence indicative of MCB accumulation.
This finding supported the view that loss of Psd2 depressed the level of Ycf1 transport activity present in the vacuolar membrane and that this reduction was probably the cause of the cadmium hypersensitivity of the psd2Δ strain. Because the MCB accumulation assay requires the dye to first enter the cells, the reduced vacuolar accumulation seen in psd2Δ strains could potentially be due to defects in MCB uptake. To address this issue, we carried out a second Ycf1 transport assay using the endogenous adenine pigment as the substrate for Ycf1. In some adenine mutants, biosynthetic intermediates accumulate upstream of the enzymatic block in the pathway that are modified and produce an intense red autofluorescence (referred to as the ade pigment) (Smirnov et al., 1967). The red color requires the transport of these intermediates into the lumen of the vacuole, a process catalyzed by Ycf1 and related ABC transporters (Chaudhuri et al., 1997). Because this pigment is produced endogenously, no concerns exist with the ability of the substrate to present equally to Ycf1 in wild-type or psd2Δ cells. This assay was performed using isogenic ade2Δ mutant strains, containing or lacking PSD2. These cells were grown in the presence of high adenine to repress pigment formation; washed to remove adenine; and then incubated for 0, 4, or 6 h. Cells were microscopically imaged at each time point (Figure 5).
The ade pigment was detected inside the vacuoles of wild-type cells within 4 h after adenine withdrawal. Pigment accumulation was only seen in psd2Δ cells after 6 h. As with the exogenously supplied MCB, ade pigment accumulation was delayed in psd2Δ cells, consistent with a reduction in Ycf1 transport activity. This effect was quantitated by extracting ade pigment and measuring its levels in these backgrounds. Wild-type accumulation was set to 100%. The psd2Δ strain only accumulated 60 ± 5% of the wild-type strain while the ycf1Δ mutant accumulated 40 ± 6%. Together, these measurements of ade pigment accumulation support the notion that loss of Psd2 reduces Ycf1-dependent transport of both cadmium and the ade pigment.
To ensure that any defects seen in either MCB accumulation or ade pigment formation could not be explained by a global loss of vacuole function in the absence of Psd2, wild-type and psd2Δ cells were stained with the dye quinacrine, which is accumulated on the basis of the pH gradient of the vacuole and serves as an indicator for function of the vacuolar ATPase activity. No differences in quinacrine staining were detected when comparing wild-type and psd2Δ cells (data not shown).
Psd2 Localizes to the Endosome
Previous experiments have reported localization of Psd2 to a region termed the Golgi/vacuole (Wu and Voelker, 2001). Because the experiments above indicated that Psd2 function had an important impact on both Ycf1 transport activity and PE levels in the vacuole, we revisited the question of Psd2 localization. A more refined identification of the membrane likely to be targeted by Psd2 is important to the understanding of how this enzyme regulates intracellular phospholipid homeostasis.
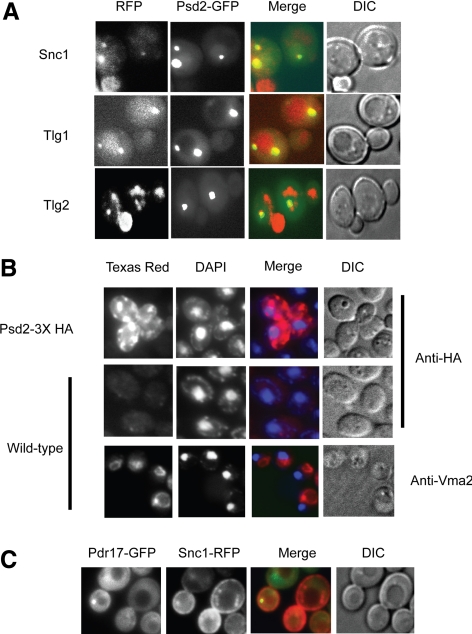
To examine Psd2 localization, we first constructed a strain in which GFP was integrated at the wild-type PSD2 locus to form a Psd2-GFP fusion protein. This strain expressed a fluorescent form of Psd2 at levels modulated by its authentic chromosomal locus. This PSD2-GFP fusion gene was able to confer ethanolamine independent growth when introduced into a psd1Δ background indicating that the fusion protein retained enzymatic function (data not shown). Fluorescent microscopic analysis of the Psd2-GFP–expressing cells exhibited punctate fluorescence at structures within the cytoplasm. These punctate structures did not colocalize with the labeling seen for Ycf1-GFP (vacuolar membrane) or Anp1-RFP (Golgi) (data not shown). To determine whether Psd2-GFP might be localized within the endocytic system, we used several different marker proteins known to be associated with this membrane compartment including a Snc1-RFP fusion protein, known to localize to the plasma membrane and early endosomes (Gurunathan et al., 2000; Lewis et al., 2000), as well as Tlg1- and Tlg2-mCherry fusion proteins as markers for this compartment (Kama et al., 2007). The strain expressing Psd2-GFP was transformed individually with expression plasmids driving accumulation of Snc1-RFP, Tlg1-mCherry, or Tlg2-mCherry. Transformants were grown to mid-log phase and processed for fluorescence microscopy (Figure 6).
Figure 6.
Psd2 localizes to the endosome. (A) Wild-type cells containing an integrated PSD2-GFP fusion gene were transformed with plasmids expressing a Snc1-RFP fusion protein (Furuta et al., 2007) or Tlg1-mCherry or Tlg2-mCherry (obtained from D. Katzmann). Transformants were grown to mid-log phase and analyzed microscopically as described above. (B) Wild-type cells containing or lacking an integrated Psd2-3X HA fusion gene were grown to mid-log phase and processed for indirect immunofluorescence as described previously (Katzmann et al., 1995). Primary antibodies used were either anti-HA or anti-Vma2 that detects a component of the vacuolar ATPase. Bound antibodies were visualized with an anti-mouse Texas Red conjugate. Cells were also stained with 4,6-diamidino-2-phenylindole (DAPI) to provide a constant internal reference. (C) A strain containing an integrated TDH3-PDR17-GFP fusion gene was transformed with the Snc1-RFP expressing plasmid. A transformant was imaged as described above.
We found that Psd2-GFP formed only a few punctuate structures in the cytoplasm and that these structures were also labeled by the fluorescent Snc1, Tlg1, and Tlg2 proteins. To determine if Pdr17 colocalized to the endocytic system as did Psd2, a Pdr17-GFP fusion gene was constructed. The Snc1-RFP plasmid was introduced into cells expressing Pdr17-GFP and distribution of these two fluorescent proteins compared (Figure 6C). As seen for Psd2-GFP and Snc1-RFP, colocalization of Pdr17 and Snc1 was detected. These fluorescent microscopic data supported the conclusion that both Psd2 and Pdr17 are localized to the endocytic system rather than either Golgi network or vacuole as suggested previously (Wu and Voelker, 2001).
To further validate our identification of the endocytic system as the site of Psd2 localization, we performed indirect immunofluorescent localization using a Psd2-3X HA-expressing strain. The presence of Psd2 was detected using a mouse anti-HA antibody. We also visualized a component of the vacuolar ATPase, the Vma2 protein using an antibody directed against this V1 subunit (Yamashiro et al., 1990). Cells were also stained with DAPI to provide a reference for nuclear and mitochondrial DNA (Figure 6B).
As seen when using the Psd2-GFP fusion protein, Psd2-3X HA was found to be present in a limited number of cytoplasmic structures. These structures were clearly resolvable from the location of the vacuolar membrane indicated by anti-Vma2 staining. Together with the colocalization with the fluorescent endosomal marker proteins above, these data argue against vacuolar localization of Psd2 and rather that this phosphatidylserine decarboxylase activity is found in the endocytic system.
Further support for our hypothesis that Psd2 localized to endosomes came from a FM4-64 chase experiment. FM4-64 is a widely used vacuolar membrane dye that reaches the vacuole via the endocytic pathway (Vida and Emr, 1995). At early times after labeling with FM4-64, the early endocytic system can be visualized but this disappears over the time course of labeling as the dye makes it way through the endocytic pathway. We labeled cells expressing the Psd2-GFP fusion protein with FM4-64, washed away excess dye and then visualized both the dye and Psd2-GFP with increasing time (Figure 7).
Figure 7.
Evidence for endosomal location of Psd2 and Pdr17. Wild-type cells containing a PSD2-GFP (A) or a TDH3-PDR17-GFP (B) fusion gene were grown to mid-log phase. Samples were withdrawn, labeled with FM4-64 at 0°C, washed to remove unbound dye, and then chased at 30°C for the indicated times. Aliquots of cells were imaged for FM4-64 and the GFP fusion proteins using fluorescence microscopy.
Immediately after labeling, FM4-64 can be seen along with Psd2-GFP but there was no overlap in these fluorescent signals. After 5 min of incubation, double labeling was detected in this cells indicating that the dye had arrived in the Psd2-GFP–positive compartment. This double labeling was no longer seen as the incubation was continued to 10 min. Note that the limiting membrane of the vacuole can be seen at 10 min and that this structure is clearly distinct from the Psd2-GFP signal. This experiment is consistent with Psd2 being localized to an endosomal compartment that can be labeled with FM4-64. We performed these same analyses with cells expressing Pdr17-GFP. The expression of Pdr17p-GFP was found to be quite low with only few cells showing punctate cytoplasmic fluorescence (data not shown). To circumvent this problem we placed Pdr17-GFP under the strong TDH3 promoter. Using this elevated level of Pdr17-GFP expression, this fusion protein was localized to the plasma membrane, cytoplasm as well as in endosomes (Figure 7B). These data support the view that both Psd2 and Pdr17 are localized to the endosomal system.
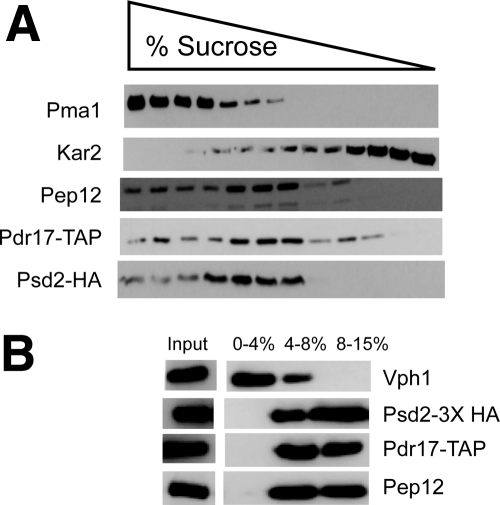
To confirm these microscopic analyses, we carried out biochemical fractionation of intracellular membranes using sucrose gradient analysis (Katzmann et al., 1999). To carry out this experiment, we used a strain containing a version of PSD2 containing an integrated 3X HA tag at its C terminus. Similarly, we integrated a TAP epitope tag at the C terminus of PDR17 in the same strain which allowed detection of both Psd2 and Pdr17 in the same gradient fractions. This dually epitope-tagged strain was grown to mid-log phase, gently lysed and membranes fractionated on a 10–60% sucrose gradient. Aliquots of each fraction were resolved on SDS-PAGE and subjected to Western blot analysis using the antibodies indicated (Figure 8).
Figure 8.
Psd2 cofractionates with an endosomal marker protein. (A) A wild-type strain containing fusion genes expressing Pdr17-TAP and Psd2-3X HA was grown to mid-log phase and whole cell lysates prepared under native conditions. The lysates were loaded onto sucrose density gradient and centrifuged at 50,000 rpm in a SW55.1 rotor (Beckman Coulter) for 14 h at 4°C. Fractions were removed from the gradient, concentrated, and resolved on SDS-PAGE. Proteins were then analyzed by Western blotting with antibodies to the polypeptides indicated at the left-hand side of the figure. Fractions containing increasing concentrations of sucrose are indicated by the bar of increasing width. (B) A parallel culture of the same strain containing the epitope-tagged Psd2 and Pdr17 above was grown to mid-log phase, lysed, and membrane fractions prepared by Ficoll density gradient centrifugation as described previously (Wemmie and Moye-Rowley, 1997). A fraction of the original lysate was reserved as an input control (Input). Samples were recovered from the interface fractions between the different Ficoll concentrations, proteins precipitated by TCA and then separated on SDS-PAGE. The relevant interface fractions are indicated at the top of the panel. Proteins were transferred to nitrocellulose and subjected to Western blotting to detect the indicated proteins. Vph1 corresponds to an integral membrane component of the vacuolar ATPase (Manolson et al., 1992).
Both Psd2-3X HA and Pdr17-TAP were enriched in overlapping aliquots from the center of this sucrose gradient. These proteins were also enriched in the same fractions containing the highest levels of the endosomal marker protein Pep12 (Gerrard et al., 2000) and distinct from either a plasma membrane protein (Pma1) (Serrano et al., 1986) or an endoplasmic reticulum marker (Kar2) (Rose et al., 1989).
We also carried out Ficoll gradient analysis of membrane proteins which clearly resolves vacuolar proteins from other membrane components as previously demonstrated (Vida et al., 1990; Wemmie and Moye-Rowley, 1997). The Psd2-3X HA and Pdr17-TAP–expressing strain was grown to mid-log phase and gently lysed. Lysates were separated by centrifugation in a Ficoll step gradient and membrane fractions collected at each interface. Proteins were recovered by TCA precipitation and subjected to Western blotting analysis (Figure 8B).
Both Psd2 and Pdr17 were found to coenrich with Pep12, a marker of the endocytic system (Becherer et al., 1996). These three proteins were clearly resolved from the Vph1 protein which is found in the V0 complex of the vacuolar ATPase (Manolson et al., 1992). These analyses support the conclusion that Psd2 and Pdr17 localize to the endosomal system where this protein complex is likely to regulate phospholipid synthesis.
Loss of Psd2 Specifically Reduces Endosomal and Vacuolar PE Content
Because loss of Psd2 leads to a decrease in Ycf1-dependent transport activity, we hypothesized that a change in PE content of the vacuolar membrane could explain this reduction. Previous work has demonstrated that Psd1 provides the major route for PE biosynthesis with Psd2 carrying out the remainder (Birner et al., 2001). This finding suggested that loss of Psd2 might cause a decrease in the vacuolar pool of PE. To test this idea, we prepared vacuolar membranes from wild-type, psd1Δ, and psd2Δ cells by using Ficoll gradient centrifugation. Enrichment for vacuoles was confirmed by Western blotting for Vph1 (data not shown). We also analyzed phospholipid content in whole cell lipid extracts. Phospholipids were analyzed using an assay for lipid phosphorous content (Chalvardjian and Rudnicki, 1970) and quantitated (Figure 9).
Figure 9.
Psd2 is required to maintain normal vacuolar phospholipid composition. (A) Vacuolar-enriched membrane fractions (Vacuolar) were prepared by Ficoll density gradient centrifugation as described above. Total cellular phospholipids (Total) were extracted from exponentially growing cells of the indicated relevant genotypes. PE and PS levels were determined in these lipid fractions (Chalvardjian and Rudnicki, 1970). (B) A wild-type strain expressing Psd2-3X HA and Pdr17-TAP was grown to mid-log phase and lysates prepared. Lysates were resolved into soluble (S2 and S3) or membrane-associated (P2 and P3) fractions by differential centrifugation following the protocol of Singer-Kruger et al. (1993). These fractions were assessed for their relative purity by Western blotting to detect the indicated proteins. (C) Membrane-associated fractions described above were prepared from the indicated strains and assayed for their levels of PE and PS. These values represent the averages of two independent determinations.
PE levels measured in vacuole-enriched membrane fractions were reduced from 40% in wild-type or psd1Δ cells to 25% in psd2Δ strains. Consistent with this observed decrease in PE found in psd2Δ cells, levels of PS increased. PS levels were not significantly different when comparing wild-type and psd1Δ cells. Control experiments measuring these same phospholipids present in total cellular lipids reproduced the findings of others (Birner et al., 2001) as loss of Psd1 caused a drop in total PE and an increase in PS, whereas psd2Δ cells showed no significant difference from the isogenic wild-type strain.
Although the reduction in vacuolar PE levels would explain the observed decrease in Ycf1-mediated function, the lowered vacuolar PE was unlikely to result from a direct effect of Psd2 or Pdr17 because these proteins were found on the endosome (Figures 6–8). Together, these data suggest the possibility that the action of the endosomally localized Psd2:Pdr17 complex acts to regulate the PE content of the vacuole. To test this suggestion, we prepared endosome-enriched fractions using differential centrifugation (Singer-Kruger et al., 1993) and assessed phospholipid and protein levels (Figure 9, B and C).
The most endosome-enriched fraction was represented by the P3 samples. P3 was found to have very little Vph1, consistent with this purification separating vacuolar membranes from endosomal fractions. Some endoplasmic reticulum contamination was still present as evidenced by the detection of the luminal Kar2 protein in this fraction. However, when P3 fractions were generated from cells lacking Psd2 and analyzed for phospholipid content, PE levels were found to be reduced. This reduction did not occur when the same fraction was produced from either wild-type or psd1Δ cells. PS levels were not significantly different in any of these samples. P2 fractions exhibited a reduction in PE levels in samples from psd1Δ cells but no other changes of note. These assays are consistent with the interpretation that PE levels are lowered in endosomes from psd2Δ strains and that this reduction is linked to a similar decrease in vacuolar PE levels. This depletion in vacuolar membrane PE is coupled to the observed reduction in Ycf1 activity in cells lacking Psd2.
DISCUSSION
Biosynthesis of phospholipids is of central importance in the normal functioning of biological membranes. S. cerevisiae has served as an outstanding model system allowing the fundamental mechanisms of eukaryotic membrane production to be defined. While genetics have clearly indicated the existence of both a mitochondrial and a extra-mitochondrial route for de novo PE production, clear phenotypes only existed for the mitochondrially localized Psd1 pathway (Birner et al., 2001). Here, we provide evidence that the second route of PE production catalyzed by the phosphatidylserine decarboxylase Psd2 is required for normal function of a vacuolar membrane protein. This functional requirement is mediated by the need for correct PE levels in the vacuolar membrane that support normal activity of Ycf1 and possibly other proteins.
Psd2 represents a relatively unusual type of phosphatidylserine decarboxylase enzyme and is found in plants but not in bacteria or mammals (Voelker, 1997). Psd1 homologues are present in mammalian mitochondria and at the inner membrane of bacteria (reviewed in Vance, 2003). Proteins exhibiting high sequence similarity to Psd2 have been described in plants (Nerlich et al., 2007), suggesting the possibility that a common role for control of internal membrane PE exists in these organisms. Ycf1 homologues have been described in plants that localize to internal membranes and are required for cadmium resistance (Tommasini et al., 1996).
Although Psd1 and Psd2 catalyze the same reaction, the PS substrate is apparently presented to each enzyme quite differently. Psd1 is located close to the site of PS synthesis, closely resembles the E. coli Psd enzyme and has not been shown to require any other enzyme to carry out its role. Conversely, Psd2 is nearly twice the size of Psd1 and not as closely related to the bacterial enzyme (Trotter et al., 1995). Psd2 also requires the presence of the Pdr17 PITP to be able to productively interact with PS as even elevated dosage of PSD2 is unable to bypass a pdr17Δ mutation (Figure 3). Previous work has demonstrated that membrane association of Psd2 requires the presence of an amino-terminally located C2 domain (Kitamura et al., 2002). Together, these data suggest that although Psd2 and Pdr17 form a complex on the membrane, the independent membrane association of both proteins is likely required to permit complex formation.
The finding that Psd2 was localized to the endosomes was unexpected. Previous assignment of Psd2 to a Golgi/vacuole distribution (Trotter and Voelker, 1995) was based strictly on biochemical fractionation experiments which did not provide the resolution to define the endosomal location we show here. We anticipated finding Psd2 on the vacuolar membrane because we found that loss of this enzyme triggered a cadmium-sensitive defect centered on reduction of Ycf1 function. The endosomal enrichment of Psd2/Pdr17, coupled with the demonstration that loss of this complex lowered vacuolar PE levels, indicates that phospholipid levels of the vacuolar membrane may be controlled by the direct action of this enzyme complex on the endosome. This suggestion is directly supported by our measurements of PE content of internal membranes. Modulation of endosomal PE levels is eventually communicated to the vacuolar membrane through vesicular transport. The indirect control of phospholipid content by Psd2 is reminiscent of the effect of the P-type ATPase Drs2 on plasma membrane asymmetry (Chen et al., 2006). Drs2 is localized to the Golgi membranes (Chen et al., 1999), yet its loss causes defects in distribution of phospholipids on the plasma membrane.
Changes in PE levels have been extensively documented to lead to problems in folding of membrane proteins (recently reviewed in Dowhan and Bogdanov, 2009). Most of these studies have been carried out using reconstituted membrane proteins in an in vitro setting. Our experiments provide in vivo demonstration of the importance of phospholipid composition in regulating membrane protein function. An alternative view of the consequences of loss of Psd2 on Ycf1 function is that elevated PS levels act to inhibit transporter activity. Based on the extensive documentation of the stimulatory effect of PE on folding and subsequent activity of membrane proteins (Bogdanov et al., 1999, 2002; Zhang et al., 2003; Hakizimana et al., 2008), we favor the positive effect of PE levels increasing Ycf1 activity rather than high PS levels inhibiting function of this ABC transporter.
Another important distinction between previous work carried out primarily on prokaryotic permeases and stimulation of their folding by PE (Wang et al., 2002), comes from the fundamental differences in the biogenesis of prokaryotic and eukaryotic membrane proteins. The exceptionally well-studied prokaryotic lactose permease, in which the positive effect of PE on folding has been demonstrated (Bogdanov et al., 1999, 2002), is inserted into its final membrane destination as it is being synthesized. This is quite different from Ycf1 that follows the typical itinerary of eukaryotic membrane proteins consisting of initial biosynthesis and insertion into the membrane of the endoplasmic reticulum, transit through the Golgi and only then reaching its vacuolar membrane site of function (Wemmie and Moye-Rowley, 1997). Our data suggest a possible means of regulation of Ycf1 as the unique phospholipid composition of the vacuole compared with other internal membranes is evidently required for full activity of this transporter. This would have the result of keeping Ycf1 activity relatively low as it transits the secretory pathway en route to its functional residence in the vacuolar membrane.
Our previous work on Psd1 demonstrated that this phosphatidylserine decarboxylase protein has another function that in mitochondrial-nuclear signaling in addition to its enzymatic activity (Gulshan et al., 2008). Psd1 also closely resembles its bacterial counterpart Psd (Clancey et al., 1993; Trotter et al., 1993) that has been carefully studied in enzymological terms. Although Psd1 and Psd2 carry out the same conversion of PS to PE, they execute this reaction in dramatically different ways. All available data indicate that Psd1 requires no other protein to function as a phosphatidylserine decarboxylase, whereas Psd2 clearly requires Pdr17 (Wu et al., 2000). Additionally, loss of Psd1 C-terminal processing prevented enzymatic function but had no detectable effect on steady-state level of the resulting mutant protein (Gulshan et al., 2008). Conversely, introducing this same C-terminal processing block into Psd2 led to a reduction in the level of mutant protein. Perhaps the different subcellular distributions of Psd1 and Psd2 explain the differential dependence on normal C-terminal processing for protein expression.
ACKNOWLEDGMENTS
We thank Wayne Riekhof, Dennis Voelker and Rob Piper for providing reagents and many helpful discussions. We also thank Kazuma Tanaka and David Katzmann for sending the endosomal marker constructs. Dan Stringer and Rob Piper provided important assistance with microscopy. This work was supported by National Institutes of Health grant GM-75120.
Abbreviations used:
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- Psd
phosphatidylserine decarboxylase
- TAP
tandem affinity purification
- PITP
phosphatidylinositol transfer protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-06-0519) on December 16, 2009.
REFERENCES
- Achleitner G., Zweytick D., Trotter P. J., Voelker D. R., Daum G. Synthesis and intracellular transport of aminoglycerophospholipids in permeabilized cells of the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:29836–29842. doi: 10.1074/jbc.270.50.29836. [DOI] [PubMed] [Google Scholar]
- Bagnat M., Keränen S., Shevchenko A., Shevchenko A., Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzi E., Wang M., Leterme S., Van Dyck L., Goffeau A. PDR5: a novel yeast multidrug resistance transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- Becherer K. A., Rieder S. E., Emr S. D., Jones E. W. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner R., Burgermeister M., Schneiter R., Daum G. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:997–1007. doi: 10.1091/mbc.12.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissinger P. H., Kuchler K. Molecular cloning and expression of the S. cerevisiae STS1 gene product. J. Biol. Chem. 1994;269:4180–4186. [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bogdanov M., Heacock P. N., Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M., Umeda M., Dowhan W. Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J. Biol. Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Carman G. M., Han G. S. Regulation of phospholipid synthesis in yeast. J Lipid Res. 2009;50(suppl):S69–S73. doi: 10.1194/jlr.R800043-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalvardjian A., Rudnicki E. Determination of lipid phosphorus in the nanomolar range. Anal. Biochem. 1970;36:225–226. doi: 10.1016/0003-2697(70)90352-0. [DOI] [PubMed] [Google Scholar]
- Chaudhuri B., Ingavale S., Bachhawat A. K. apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics. 1997;145:75–83. doi: 10.1093/genetics/145.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Ingram M. F., Rosal P. H., Graham T. R. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 1999;147:1223–1236. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Wang J., Muthusamy B. P., Liu K., Zare S., Andersen R. J., Graham T. R. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic. 2006;7:1503–1517. doi: 10.1111/j.1600-0854.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- Clancey C. J., Chang S. C., Dowhan W. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J. Biol. Chem. 1993;268:24580–24590. [PubMed] [Google Scholar]
- Daum G., Lees N. D., Bard M., Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Dowhan W., Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu. Rev. Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre S., Haguenauer-Tsapis R. Raft partitioning of the yeast uracil permease during trafficking along the endocytic pathway. Traffic. 2003;4:83–96. doi: 10.1034/j.1600-0854.2003.40204.x. [DOI] [PubMed] [Google Scholar]
- Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., Tanaka K. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell. 2007;18:295–312. doi: 10.1091/mbc.E06-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard S. R., Levi B. P., Stevens T. H. Pep12p is a multifunctional yeast syntaxin that controls entry of biosynthetic, endocytic and retrograde traffic into the prevacuolar compartment. Traffic. 2000;1:259–269. doi: 10.1034/j.1600-0854.2000.010308.x. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Griac P. Sec14 related proteins in yeast. Biochim. Biophys. Acta. 2007;1771:737–745. doi: 10.1016/j.bbalip.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Gueldry O., Lazard M., Delort F., Dauplais M., Grigoras I., Blanquet S., Plateau P. Ycf1p-dependent Hg(II) detoxification in Saccharomyces cerevisiae. Eur. J. Biochem. 2003;270:2486–2496. doi: 10.1046/j.1432-1033.2003.03620.x. [DOI] [PubMed] [Google Scholar]
- Gulshan K., Schmidt J., Shahi P., Moye-Rowley W. S. Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression. Mol. Cell. Biol. 2008;28:5851–5864. doi: 10.1128/MCB.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S., Chapman-Shimshoni D., Trajkovic S., Gerst J. E. Yeast exocytic v-SNAREs confer endocytosis. Mol. Biol. Cell. 2000;11:3629–3643. doi: 10.1091/mbc.11.10.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakizimana P., Masureel M., Gbaguidi B., Ruysschaert J. M., Govaerts C. Interactions between phosphatidylethanolamine headgroup and LmrP, a multidrug transporter: a conserved mechanism for proton gradient sensing? J. Biol. Chem. 2008;283:9369–9376. doi: 10.1074/jbc.M708427200. [DOI] [PubMed] [Google Scholar]
- Hearn J. D., Lester R. L., Dickson R. C. The uracil transporter Fur4p associates with lipid rafts. J. Biol. Chem. 2003;278:3679–3686. doi: 10.1074/jbc.M209170200. [DOI] [PubMed] [Google Scholar]
- Hirata D., Yano K., Miyahara K., Miyakawa T. Saccharomyces cerevisiae YDR1, which encodes a member of the ATP-binding cassette (ABC) superfamily, is required for multidrug resistance. Curr. Genet. 1994;26:285–294. doi: 10.1007/BF00310491. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kama R., Robinson M., Gerst J. E. Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol. Cell. Biol. 2007;27:605–621. doi: 10.1128/MCB.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Epping E. A., Moye-Rowley W. S. Mutational disruption of plasma membrane trafficking of Saccharomyces cerevisiae Yor1p, a homologue of mammalian multidrug resistance protein. Mol. Cell. Biol. 1999;19:2998–3009. doi: 10.1128/mcb.19.4.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Hallstrom T. C., Voet M., Wysock W., Golin J., Volckaert G., Moye-Rowley W. S. Expression of an ATP-binding cassette transporter encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura H., Wu W. I., Voelker D. R. The C2 domain of phosphatidylserine decarboxylase 2 is not required for catalysis but is essential for in vivo function. J. Biol. Chem. 2002;277:33720–33726. doi: 10.1074/jbc.M205672200. [DOI] [PubMed] [Google Scholar]
- Klein M., Mamnun Y. M., Eggmann T., Schuller C., Wolfger H., Martinoia E., Kuchler K. The ATP-binding cassette (ABC) transporter Bpt1p mediates vacuolar sequestration of glutathione conjugates in yeast. FEBS Lett. 2002;520:63–67. doi: 10.1016/s0014-5793(02)02767-9. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski M., van der Rest M., Cybularz-Kolaczkowski A., Soumillion J.-P., Konings W. N., Goffeau A. Anticancer drugs, ionophoric peptides and steroids as substrates of the yeast multidrug transporter Pdr5p. J. Biol. Chem. 1996;271:31543–31548. doi: 10.1074/jbc.271.49.31543. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kuge O., Saito K., Kojima M., Akamatsu Y., Nishijima M. Post-translational processing of the phosphatidylserine decarboxylase gene product in Chinese hamster ovary cells. Biochem. J. 1996;319:33–38. doi: 10.1042/bj3190033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. J., Nichols B. J., Prescianotto-Baschong C., Riezman H., Pelham H. R. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. X., Dowhan W. Studies on the mechanism of formation of the pyruvate prosthetic group of phosphatidylserine decarboxylase from Escherichia coli. J. Biol. Chem. 1990;265:4111–4115. [PubMed] [Google Scholar]
- Li Z.-S., Szczypka M., Lu Y.-P., Thiele D. J., Rea P. A. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J. Biol. Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Manolson M. F., Proteau D., Preston R. A., Stenbit A., Roberts B. T., Hoyt M. A., Preuss D., Mulholland J., Botstein D., Jones E. W. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H+-ATPase. J. Biol. Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- Maxfield F. R., Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- Nerlich A., von Orlow M., Rontein D., Hanson A. D., Dormann P. Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria. Plant Physiol. 2007;144:904–914. doi: 10.1104/pp.107.095414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opekarova M., Malinska K., Novakova L., Tanner W. Differential effect of phosphatidylethanolamine depletion on raft proteins: further evidence for diversity of rafts in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1711:87–95. doi: 10.1016/j.bbamem.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Misra L. M., Vogel J. P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Schuiki I., Daum G. Phosphatidylserine decarboxylases, key enzymes of lipid metabolism. IUBMB Life. 2009;61:151–162. doi: 10.1002/iub.159. [DOI] [PubMed] [Google Scholar]
- Serrano R., Keilland-Brandt M. C., Fink G. R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+ and Ca2+-ATPases. Nature. 1986;319:689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- Sharma K. G., Mason D. L., Liu G., Rea P. A., Bachhawat A. K., Michaelis S. Localization, regulation, and substrate transport properties of Bpt1p, a Saccharomyces cerevisiae MRP-type ABC transporter. Eukaryot. Cell. 2002;1:391–400. doi: 10.1128/EC.1.3.391-400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G., Hicks J. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1979. [Google Scholar]
- Singer-Kruger B., Frank R., Crausaz F., Riezman H. Partial purification and characterization of early and late endosomes from yeast. Identification of four novel proteins. J. Biol. Chem. 1993;268:14376–14386. [PubMed] [Google Scholar]
- Smirnov M. N., Smirnov V. N., Budowsky E. I., Inge-Vechtomov S. G., Serebrjakov N. G. Red pigment of adenine-deficient yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1967;27:299–304. doi: 10.1016/s0006-291x(67)80096-2. [DOI] [PubMed] [Google Scholar]
- Sturley S. L. Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim. Biophys. Acta. 2000;1529:155–163. doi: 10.1016/s1388-1981(00)00145-1. [DOI] [PubMed] [Google Scholar]
- Szczypka M. S., Wemmie J. A., Moye-Rowley W. S., Thiele D. J. A yeast metal resistance protein similar to human CFTR and multidrug resistance-associated protein. J. Biol. Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- Tommasini R., Evers R., Vogt E., Mornet C., Zaman G.J.R., Schinkel A. H., Borst P., Martinoia E. The human multidrug resistance-associated protein functionally complements the yeast cadmium factor 1. Proc. Natl. Acad. Sci. USA. 1996;93:6743–6748. doi: 10.1073/pnas.93.13.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter P. J., Pedretti J., Voelker D. R. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 1993;268:21416–21424. [PubMed] [Google Scholar]
- Trotter P. J., Pedretti J., Yates R., Voelker D. R. Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression, and creation of the null allele. J. Biol. Chem. 1995;270:6071–6080. doi: 10.1074/jbc.270.11.6071. [DOI] [PubMed] [Google Scholar]
- Trotter P. J., Voelker D. R. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:6062–6070. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- Umebayashi K., Nakano A. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 2003;161:1117–1131. doi: 10.1083/jcb.200303088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hazel H. B., Pichler H., do Valle Matta M. A., Leitner E., Goffeau A., Daum G. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J. Biol. Chem. 1999;274:1934–1941. doi: 10.1074/jbc.274.4.1934. [DOI] [PubMed] [Google Scholar]
- van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J. E. Molecular and cell biology of phosphatidylserine and phosphatidylethanolamine metabolism. Prog Nucleic Acid Res Mol Biol. 2003;75:69–111. doi: 10.1016/s0079-6603(03)75003-x. [DOI] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T. A., Graham T. R., Emr S. D. In vitro reconstitution of intercompartmental protein transport to the yeast vacuole. J. Cell Biol. 1990;111:2871–2884. doi: 10.1083/jcb.111.6.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker D. R. Phosphatidylserine decarboxylase. Biochim. Biophys. Acta. 1997;1348:236–244. doi: 10.1016/s0005-2760(97)00101-x. [DOI] [PubMed] [Google Scholar]
- Wang X., Bogdanov M., Dowhan W. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmie J. A., Moye-Rowley W. S. Mutational analysis of the Saccharomyces cerevisiae ATP-binding cassette transporter protein Ycf1p. Mol. Microbiol. 1997;25:683–694. doi: 10.1046/j.1365-2958.1997.5061868.x. [DOI] [PubMed] [Google Scholar]
- Wemmie J. A., Szczypka M. S., Thiele D. J., Moye-Rowley W. S. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J. Biol. Chem. 1994;269:32592–32597. [PubMed] [Google Scholar]
- Wu W. I., Routt S., Bankaitis V. A., Voelker D. R. A new gene involved in the transport-dependent metabolism of phosphatidylserine, PSTB2/PDR17, shares sequence similarity with the gene encoding the phosphatidylinositol/phosphatidylcholine transfer protein, SEC14. J. Biol. Chem. 2000;275:14446–14456. doi: 10.1074/jbc.275.19.14446. [DOI] [PubMed] [Google Scholar]
- Wu W. I., Voelker D. R. Characterization of phosphatidylserine transport to the locus of phosphatidylserine decarboxylase 2 in permeabilized yeast. J. Biol. Chem. 2001;276:7114–7121. doi: 10.1074/jbc.M010278200. [DOI] [PubMed] [Google Scholar]
- Yamashiro C. T., Kane P. M., Wolczyk D. F., Preston R. A., Stevens T. H. Role of vacuolar acidification in protein sorting and zymogen activation: a genetic analysis of the yeast vacuolar proton-translocating ATPase. Mol. Cell. Biol. 1990;10:3737–3749. doi: 10.1128/mcb.10.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Bogdanov M., Pi J., Pittard A. J., Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J. Biol. Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- Zhang X., Moye-Rowley W. S. Saccharomyces cerevisiae multidrug resistance gene expression inversely correlates with the status of the Fo component of the mitochondrial ATPase. J. Biol. Chem. 2001;276:47844–47852. doi: 10.1074/jbc.M106285200. [DOI] [PubMed] [Google Scholar]