Abstract
In the sarcomeric M-band, the giant ruler proteins titin and obscurin, its small homologue obscurin-like-1 (obsl1), and the myosin cross-linking protein myomesin form a ternary complex that is crucial for the function of the M-band as a mechanical link. Mutations in the last titin immunoglobulin (Ig) domain M10, which interacts with the N-terminal Ig-domains of obscurin and obsl1, lead to hereditary muscle diseases. The M10 domain is unusual not only in that it is a frequent target of disease-linked mutations, but also in that it is the only currently known muscle Ig-domain that interacts with two ligands—obscurin and obsl1—in different sarcomeric subregions. Using x-ray crystallography, we show the structural basis for titin M10 interaction with obsl1 in a novel antiparallel Ig-Ig architecture and unravel the molecular basis of titin-M10 linked myopathies. The severity of these pathologies correlates with the disruption of the titin-obsl1/obscurin complex. Conserved signature residues at the interface account for differences in affinity that direct the cellular sorting in cardiomyocytes. By engineering the interface signature residues of obsl1 to obscurin, and vice versa, their affinity for titin can be modulated similar to the native proteins. In single-molecule force-spectroscopy experiments, both complexes yield at forces of around 30 pN, much lower than those observed for the mechanically stable Z-disk complex of titin and telethonin, suggesting why even moderate weakening of the obsl1/obscurin-titin links has severe consequences for normal muscle functions.
Keywords: immunoglobulin domain, protein complex, x-ray crystallography, mechanosensor, myopathy
Sarcomeres are the smallest contractile units of striated muscles. They are highly ordered assemblies of precisely tailored actin and myosin filaments that are crosslinked at Z-disks and M-bands, respectively. The assembly of hundreds of protein subunits into ordered sarcomeres forms the structural basis for striated muscle contraction. The global layout of sarcomere assembly requires a giant ruler protein, titin, a 3000 kDa modular protein with a length of over 1.2 μm, displaying binding sites for proteins along the entire distance from Z-disk to M-band (1–3). Titin is composed of hundreds of immunoglobulin- and fibronectin-3-like (Ig and Fn3) domains that are arranged in specific patterns in the subcompartments of the sarcomere (3). The Ig-domains in sarcomeric proteins like titin, myomesin, or obscurin present a functionally versatile surface around a highly stable structural scaffold (4).
At the M-band, the centers of myosin filaments are crosslinked into a mechanically stable network that involves titin, the cytoskeletal protein myomesin (5), and the giant obscurin (6), the latter acting as a linker to the sarcoplasmic reticulum (SR) (7). Obscurin, with a mass of around 800 kDa, was initially discovered as a ligand of Z-disk titin (3). Like titin, it is a modular protein composed of Ig and Fn3 domains. Intriguingly, obscurin localizes predominantly to the M-band in mature myofibrils (8), a behavior unexpected for a giant sarcomeric protein. This multicompartment localization is explained by the presence of a further, M-band-specific binding site for obscurin at the most C-terminal titin domain M10. This domain interacts with the N-terminal domain of both obscurin and its small homologue, obscurin-like-1 (obsl1), which lacks the signaling domains and ankyrin-binding sites that link the obscurin C terminus to the SR. In agreement with the SR interaction, obscurin is localized to the myofibril periphery, whereas obsl1 is an integral component of the myofibril (6).
Titin’s C terminus plays a critical role in muscle function, as multiple mutations at this position cause hereditary myopathies. Tibial muscular dystrophy (TMD) and limb girdle muscular dystrophy 2J (LGMD2J) are caused by several missense or deletion/substitution mutations in titin M10 (9, 10), while larger deletions of the titin C terminus cause Salih myopathy (11). Most of these mutations weaken or abrogate binding of obscurin/obsl1 to the M-band (6) and cause gradual failure of sarcomere maintenance. Recently, mutations in obsl1 were identified as the cause of the primordial growth disorder 3-M syndrome (12, 13). Here, using an integrated structural, biophysical, and cell biological approach we illustrate how the titin M10 domain interacts with the N terminus of obs|1, suggesting a molecular explanation for the differential affinity of obscurin and obsl1 to titin M10. We also provide a basis for a mechanistic understanding of the titin mutations linked to LGMD2J and TMD myopathies. Differences in affinity determine the sorting of obscurin and obs|1 in cardiac myofibrils but not the mechanical stability of the titin-obsl1/obscurin complexes as probed by single-molecule force spectroscopy.
Results
Overall Architecture of the M10-OL1 Complex.
To understand at the atomic level the sarcomeric M-band interaction between titin and obsl1, we reconstituted the minimal interacting complex in vitro and solved its x-ray crystal structure in two different crystal forms (Form I, trigonal and Form II, triclinic—PDB codes 2wp3 and 2wwm, respectively). Titin M10 and the N-terminal Ig module of human obsl1 form a heterodimer in solution (see SI Methods and Fig. S1) and in the crystal. No major structural differences are seen in the two different crystal forms, suggesting that the complex observed crystallographically faithfully recapitulates that in solution. After restrained refinement, the best M10-OL1 model solved at the 1.48 Å resolution is characterized by an Rfree value of 21.7% and excellent quality indicators (Table S1).
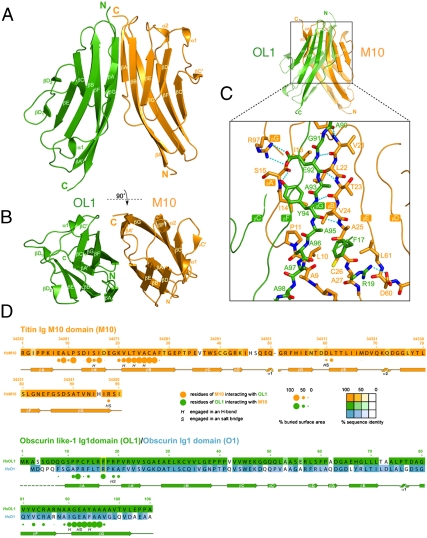
The M10-OL1 heterodimer is shown in Fig. 1A and B. Both M10 and OL1 protomers belong to the intermediate set (I-set) of the Ig family (4, 14) and share the typical barrel-like architecture in which a total of nine strands are arranged into two distinct β-sheets folded into a β-sandwich (15). The two β-sheets are formed by strands βA-βB-βE-βD and βA′-βG-βF-βC-βC′, respectively. In OL1, a proline residue breaks the βD strand, yielding two shorter βD1 and βD2 strands. Overall, the M10 and OL1 are structurally very similar. They display a root mean square (rms) deviation of 1.12 Å for 95 equivalent Ca atoms in spite of their limited (22.1%) sequence identity. The Ig-domains in the M10-OL1 complex are arranged in a head-to-tail fashion with the initial portion of the OL1 βG strand paired parallel to that of the M10 βB strand giving rise to an extended intermolecular β-sheet between OL1 βC′-βC-βF-βG-βA′ and M10 βA-βB-βE-βD β-sheets.
Fig. 1.
(A and B) Cartoon representation of the M10-OL1 heterodimer. As indicated, the view in (B) is rotated by 90 degrees around the x axis compared to the view in (A). M10 and OL1 are shown in orange and green, respectively. (C) Enlarged view of the boxed overall representation shown in the top panel highlighting important M10-OL1 interactions. Some structural elements in the overall representation are shown partly transparent to highlight the mixed intermolecular β-sheet. Color coding for carbon atoms is orange and green for M10 and OL1, respectively; nitrogen, blue; oxygen, red; sulfur, yellow. Hydrogen bonds are dotted cyan lines. (D) Sequence and secondary structure of the M10 and OL1 domains. The sequence of the O1 domain is also given. Amino acids are color-coded according to sequence conservation as highlighted by the conservation bar. Colored circles show residues of one domain contacting the other as indicated in the inset. The radius of the circles is proportional to the buried area percentage as calculated by PISA (34). The letter codes H and S indicate residues involved hydrogen bonds and salt bridges, respectively. Figures were generated with Pymol (www.pymol.org) and Adobe Illustrator.
The M10-OL1 Binding Interface.
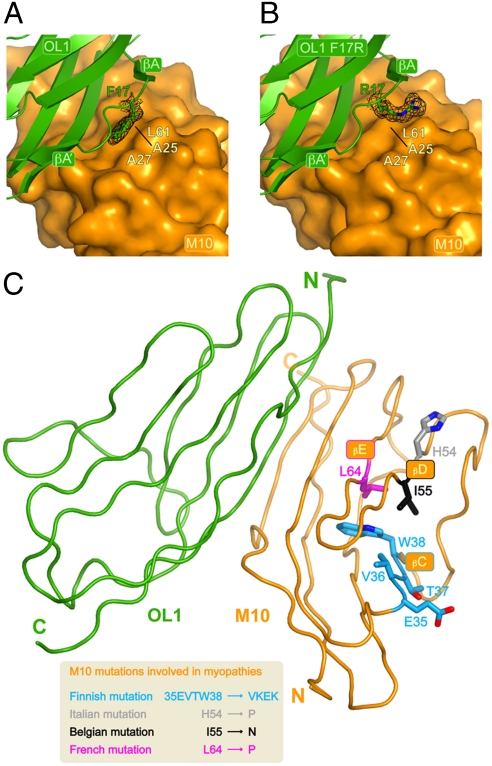
A search in the SCOPPI database (16) of protein-protein interfaces reveals that the M10-OL1 complex represents a mode of interaction previously unobserved for Ig-domains. M10 and OL1 interact with each other using different structural elements of their common Ig-fold (Fig. 1D), with the N terminus of M10 interacting with the C terminus of OL1. M10 contacts the OL1 C terminus encompassing the stretch ranging from the segment connecting M10 βA to βA′ to the first half of the βB strand. Conversely, OL1 interacts with the M10 N terminus between OL1 βF and the first half of βG as well as via its N-terminal stretch preceding the βA′ strand (Fig. 1C and D). The elements involved in establishing the interface are amongst the most structurally divergent (Fig. S2). A mixture of hydrogen bonds, salt bridges and hydrophobic interactions holds the complex together (Fig. 1C). The intermolecular mixed β-sheet connecting the OL1 and M10 β-sheets (see above) is engendered by five hydrogen bonds between residues A90, E92 and Y94 of the OL1 βG strand and residues V21, T23, A25 of the M10 βB strand. Residues E92 and Y94 of the OL1 βG strand also utilize their side-chain to stabilize the complex. The carboxylate group of E92 interacts with the guanidinium group of OL1 R97, whilst the hydroxyl group of Y94 is H-bonded to the main-chain carbonyl oxygen of M10 S15. A second salt bridge connects the side chain of OL1 R19 with that of M10 D60. A number of hydrophobic residues are buried upon complex formation. For example, the side chain of M10 A9 residue points its methyl group toward the main chain of the triple alanine stretch (96AAA98) of the OL1 βG strand. The structure predicts that a large amino acid at this position would impair complex formation. Another hydrophobic residue involved in establishing the M10-OL1 interface is the first residue of the four-amino acid OL1 linker connecting βA to βA′, F17. This residue fits its aromatic side chain in between the methyl groups M10 A25 and A27, resting partly on the aliphatic side chain of M10 L61 (Fig. 2A). Overall, the M10-OL1 interaction buries approximately 670 Å2.
Fig. 2.
(A) Close-up on the OL1 F17 residue with M10 shown as surface representation. The OL1 F17 side chain is buried and stabilized by M10 residues A25, A27 and L61. 2mFo-DFc electron density for OL1 F17 is shown in black at the 1.3σ contour level. (B) Closeup of the OL1 R17 residue in the M10-OL1 F17R complex engineered to partly mimic the M10-O1 complex. OL1 R17 points its charged side-chain towards the solvent. 2mFo-DFc electron density for OL1 R17 is shown in black at the 1.3σ contour level. The orientation is the same as in A. (C) View of the M10-OL1 complex highlighting the position of the M10 residues involved human cardiomyopathies as a result of four different (Finnish, Belgian, French and Italian) genetic mutations (see main text). Amino acid changes resulting from the different mutations are shown in the inset.
Modulation of M10/OL1 and M10/O1 Complex Stability.
At the M-band, titin interacts not only with obsl1, but also with obscurin (6). Pull-down experiments confirm the interaction of titin M10 with the first Ig-domain of obscurin and obsl1 (O1 and OL1, Fig. S3). The OL1 and O1 domains are 38.5% identical and display a strong conservation of the residues involved in forming the M10-OL1 interface (Fig. 1D) suggesting that O1 binds to M10 similarly to OL1. A striking difference, however, is the replacement of OL1 F17 with an arginine residue (R15) in O1 (Yellow Box in Fig. 1D, Fig. S4). As described, OL1 F17 interacts with a hydrophobic pocket on M10 using its aromatic side chain. F17 and R15 are totally conserved within the vertebrate obs|1 and obscurin families, respectively, effectively representing a signature of their Ig1 domains (Fig. S4).
We investigated the importance of this particular side chain in both OL1 and O1 by isothermal titration calorimetry (ITC) and pull-down assays. ITC measurements (Table 1) indicate that the formation of both M10-OL1 and M10-O1 complexes is driven by entropic gain rather than by enthalpic factors. The M10-OL1 complex is more stable than the M10-O1 complex, with a 3.4-fold higher affinity than the latter. Quantification of GFP-OL1 and GFP-O1 pull-down using GST-tagged M10 suggests an even larger difference in stability in favor of the M10-OL1 complex (Fig. S3A). Replacement of residue OL1 F17 with an arginine residue (OL1 F17R) diminished the affinity of this domain for M10 compared to wild-type OL1 (Table 1). Conversely, the R15F variation in O1 (O1 R15F, “obsl1-ized” O1) resulted in a higher affinity for M10 compared to both wild-type O1 and OL1. This trend is confirmed in the pull-down experiment (Fig. S3B and C). Overall, we can therefore conclude that the presence of a phenylalanine residue at the beginning of the βA-βA′ linker favors the stability of both M10-OL1 and M10-O1 complexes over that of an arginine residue. To understand in more detail the effect of an arginine residue at this position, we solved the crystal structure of the M10-OL1 F17R complex (PDB code 2wwk, Table S1). The structure reveals that “obscurin-ization” of OL1 only affects the complex locally, resulting in the positively charged arginine side chain pointing toward the solvent (Fig. 2B). A cluster of three ordered water molecules fills the volume occupied by the aromatic side chain of F17 in the M10-OL1 complex. In agreement with the thermodynamic data, the decrease in stability due to the OL1 F17R replacement can be linked to a loss of entropic gain.
Table 1.
Determination of binding affinities for different M10-OL1 and M10-O1 complexes by ITC (see SI Methods). The acronym Bel M10 indicates the M10 I55N variant of the Belgian mutation. The parameters N, KA, ΔH, T, ΔS, ΔG indicate binding stoichiometry, binding affinity, binding enthalphy, temperature, binding enthropy, and Gibbs’ free energy of binding, respectively
| Interaction | N | KA (105 M-1) | ΔH (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
| M10-OL1 | 1.04 ± 0.008 | 12.6 ± 1.5 | 3.87 ± 0.34 | 12.0 | -8.1 |
| M10-O1 | 1.05 ± 0.005 | 3.7 ± 0.2 | 11.16 ± 0.68 | 18.6 | -7.4 |
| M10-OL1 F17R | 0.99 ± 0.014 | 7.8 ± 1.3 | 0.66 ± 0.11 | 8.6 | -7.9 |
| M10-O1 R15F | 1.04 ± 0.003 | 24.2 ± 1.4 | 5.64 ± 0.18 | 14.2 | -8.6 |
| Bel M10-OL1 | 1.05 ± 0.005 | 11.7 ± 0.9 | 3.66 ± 0.20 | 11.8 | -8.1 |
| Bel M10-O1 | 1.07 ± 0.010 | 6.4 ± 0.7 | 9.6 ± 0.98 | 17.3 | -7.7 |
M10 Residues Involved in Human Myopathies.
The M10 domain of titin has emerged as a hotspot for autosomal dominant and recessive mutations, causing at least three distinct human myopathies. An 11-bp deletion/insertion at position 293,269-293,279 of the titin gene (FINmaj) leads to the exchange of four sequential amino acids in M10 in Finnish patients (17), resulting in the M10 35EVTW38 to VKEK variation; a point mutation at position 293,357 leads to the exchange of M10 Leu64 to Pro in French patients (17), and a point mutation at position 293,329 leads to the exchange of I55 to N in Belgian patients. Very recently, the variation M10 H54 to P has been found in Italian patients (9). The structure of the M10-OL1 complex reveals that these disease-linked residues are not directly involved in the protein–protein interface (Fig. 2C). The predicted outcome of the amino acid variations induced by the Finnish and French mutations is a severe disruption of the M10 fold. The Finnish mutation replaces, among others, W33 with a lysine residue within the βC strand. A tryptophan residue at this position is strictly conserved in the Ig fold. Replacement to a lysine suggests a collapse of the βD/βE strands, which are supported by the bulky hydrophobic tryptophan side chain. Similarly, the introduction by the French mutation of a proline in place of the conserved L64 in the βE strand probably destabilizes this structural element with global repercussions on the general fold. A similar effect can be suggested for the Italian mutation H54P in βD. In contrast, the M10-OL1 structure suggests that the Belgian mutation, which causes the replacement within the βD strand of I55 with asparagine, should have no major detrimental effects on the stability of M10. The side chain of I55 faces a solvent accessible region lined by the heterocycle of W38 side chain and the carbonyl groups of I45 and V36. The side chain of an asparagine residue can easily be accommodated at this position. Consistently, ITC measurements show that substitution of M10 I55 for an asparagine has virtually no effect in the case of this interaction (compare M10-OL1 and Bel M10-OL1 in Table 1). Instead, M10 I55N replacement increases M10 affinity for O1 approximately 1.73-fold. Pull-down data are also consistent with a stronger effect of this amino acid replacement on M10-O1 than on M10-OL1 (Fig. S3B and C). To check whether this differential response could be due to radically different interfaces and to further validate the M10-OL1 complex observed crystallographically, we tested binding of both OL1 and O1 to M10 A9Y. As noted, the structure of M10-OL1 is not compatible with a large residue at this position. Both OL1 and O1 binding to M10 A9Y is completely abrogated (Fig. S3B and C).
Cellular Sorting of Obscurin.
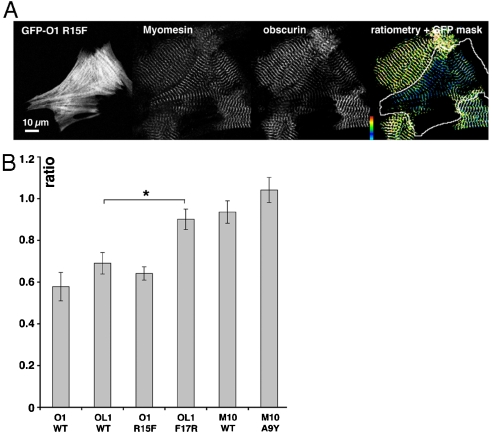
Ratiometric image analysis in neonatal rat cardiomyocytes (6) was used to assess the role of the signature residues in the complex context of the native M-band. Competition of the interactions of obsl1 or obscurin with titin by overexpressed GFP-fused OL1 or O1 leads to the loss of the endogenous obscurin from the M-band (6). Because our currently available obsl1 antibodies are directed against OL1, similar competition assays for obsl1 were not possible. When we overexpressed wild-type O1 or OL1, both domains targeted to M-bands, in addition to other subcellular localizations, and displaced endogenous obscurin (Fig. 3A and Fig. S5), suggesting that the M10-OL1 and M10-O1 interactions are mutually exclusive. The competitive effect is constant within the observable range of GFP expression levels, suggesting that the cellular assay works near saturation (Fig. S6). This confirms that the differential competitive effects observed are due to the nature of the competing protein rather than cellular concentration differences. The mutation OL1 F17R strongly weakens the cellular competition with obscurin, whereas the increase of affinity in O1 R15F had no additional effect under the saturation conditions used (Fig. 3B). The competitive effect of overexpressed titin M10 on obscurin is abrogated by the M10 A9Y mutation (Fig. 3B). These observations support the notion that the crystallographically identified signature residues and key side-chain contacts at the interface play important roles in cellular sorting.
Fig. 3.
(A) Representative example of the competitive effect of overexpressed GFP-fused obscurin/obs|1 (here: GFP-O1 R15F) on endogenous obscurin. The separate channels for GFP, endogenous myomesin and obscurin, and the ratiometric image with overlaid GFP mask for the outline of the transfected cell are shown. The false-color scale range indicator shows a range of 0 (Black) to 3 (White). Scale bar: 10 μm. (B) Average ratio of endogenous obscurin to myomesin in NRC expressing various O1, OL1, and M10 constructs. Whereas the high affinity interactions of O1, O1 R15F and OL1 show no significant differences (p > 0.17), weakening the OL1 interaction leads to a significant loss in M-band affinity (p = 0.003). The weaker competing effect of M10 is completely abrogated in the mutant M10 A9Y. Error bars: SEM.
Mechanical Stability of the Titin-obsl1 and Obscurin Complex.
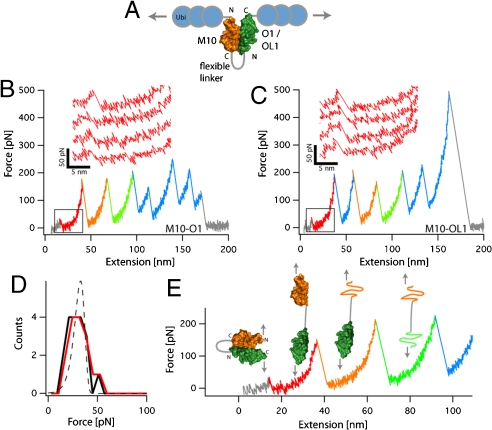
Currently available information on M-band structure suggests that the quaternary complex of titin, obsl1 or obscurin, myomesin, and myosin is crucial for the mechanical integrity of the M-band. However, which of the individual components, and which of their manifold interactions, is most important in bearing mechanical strain is unknown. We therefore tested the mechanical stability of the titin-OL1 and O1 complexes by single-molecule force spectroscopy. If probed directly, the dissociation of the complex, which is held together by noncovalent interactions, will give rise to an unbinding fingerprint that is indistinguishable from the detachment of the polypeptide chain from the AFM cantilever. Guided by the crystal structure, we therefore fused the C terminus of the titin M10 domain to the N terminus of either O1 or OL1 by means of an unstructured, 33 aa linker composed of glycine and serine residues. This single-chain M10-O(L)1 complex was then sandwiched between three concatenated ubiquitin domains (scheme in Fig. 4A), which serve as handles for attachment to the cantilever and the surface of the AFM. It is important to note that this geometry reflects the direction of the force acting in vivo. When load is applied to this chimaeric protein, the sawtooth-like shaped unfolding signature typical for single-molecule mechanical unfolding can be observed (Fig. 4B). At the beginning of the unfolding trace, we can identify two longer unfolding events (orange and green events, contour length increase ΔL = 30.1 ± 0.3 nm, n = 72) followed by the repetitive, ∼23 nm ubiquitin unfolding events (blue). We attribute the orange and green events, whose increase in contour length corresponds well to the length increase expected for a 93 aa protein, to the unfolding of the M10 or O1 domains (expected value 30.2 nm). With an unfolding force of 181 ± 6 pN (n = 72), the stability of the M10 or O1 Ig-domains falls within the normal range of Ig-domains from titin (18, 19). Closer inspection of the unfolding traces reveals that these unfolding events are regularly preceded by a low-force peak of around 30 pN (red). This peak occurs consistently at the same position and force (inset Fig. 4B and C), and reflects the unbinding of the two interacting Ig-domains (ΔL = 17.8 ± 0.7 nm, expected value 17.8 nm). Applying this unbinding assay to the titin-OL1 complex results in comparable unfolding traces (Fig. 4C ΔL = 19.1 ± 0.6 nm, expected value 18.6 nm) and a comparison of the unfolding forces (Fig. 4D) of the unbinding event demonstrates that the mechanical stability of the two complexes is very similar (titin-O1, 29 ± 2 pN, n = 18, titin-OL1 32 ± 3 pN, n = 16). Our results thus reveal the titin-O1/OL1 complex as a mechanically rather labile structure, particularly if compared to the very rigid complex formed between titin and telethonin in the Z-disk at the other end of the sarcomere (20). Moreover, dissociation of the titin-O1/OL1 complex occurs at forces much lower than those necessary to unfold the individual Ig-domains (Fig. 4E).
Fig. 4.
(A) Schematic representation of the chimaeric single-chain titin-obscurin/obsl1 constructs fused to ubiquitin used in AFM. (B and C), representative single-molecule force-extension traces for O1 (B) and OL1 (C) showing the unbinding of the complex at low forces (Boxed) and the subsequent unfolding of the M10 and O1/OL1 and ubiquitin domains. It is important to note that in these traces the unfolding events of M10 and O1/OL1 are indistinguishable in both contour length increase and unfolding force. The orange/green coloring of the unfolding traces is hence merely for illustration. (D) Frequency analysis of the unbinding forces for titin O1 (Black Distribution) and titin-OL1 (Red Distribution) with a force distribution (Dashed Line) calculated with a potential width Δx = 8 Å , an off-rate koff = 0.7 s-1 and a loading rate of 2500 pN s-1. (E) Schematic interpretation of the observed single-molecule experiments where unbinding of the titin-O1/OL1 complex precedes the sequential unfolding of Ig and ubiquitin domains.
Discussion
The complex of titin M10 and OL1 reveals the canonical Ig I-frame as their underlying building principle (4), similar to the M-band domains M5 (PDB 1tnm) (21) and M1 (PDB 2bk8). However, the mode of their antiparallel interaction is unlike the only other known M-band Ig complex, the myomesin My13 homodimer. The two Ig-domains of titin and obsl1 interact in a side-on antiparallel fashion over an area of 670 Å2, with both main-chain as well as side-chain interactions contributing to the interface. Although the arrangement is globally reminiscent of the antiparallel My13 dimer (22) with an interface area of 710 Å2 or other antiparallel cytoskeletal complexes (23), there are unique differences in the complex formation. Myomesin My13 strictly homodimerises; other protein interactions could not be identified (24). In contrast, titin M10 interacts with at least two different proteins in different sarcomeric subcompartments (6). The signature side-chain interactions we reveal here as hallmarks of obscurin or obsl1 are not observed in myomesin, and similarly, the titin-telethonin heterotrimer is bonded exclusively by main-chain interactions (25). The highly conserved, discriminating surfaces in M10-OL1 and M10-O1 account for specific differences in affinity that are reflected in the cellular specificity of their sarcomeric interactions (Fig. 3B). That the key residue differences are biologically significant is underscored by their remarkable cross-species conservation (Fig. 1D, Fig. S4). The high affinity interaction of titin with obsl1 is involved in formation of core myofibrils, whereas lower-affinity interaction directs the peripheral integration of obscurin. Our assays in cardiomyocytes suggests that additional elements of specificity might be involved, possibly by ternary interactions of the M10-O1 complex, as the competition of M10 seems lower in the cellular context. However, it is important to stress that protein interaction dynamics in the solid-state system of the cytoskeleton cannot be simply reduced to individual binary interactions and their measured affinities in solution. Given these constraints, our data lend excellent support that the structurally determined titin-obscurin/obsl1 interface is used in vivo and that the signature residues govern affinity in the cellular context. The antiparallel orientation of titin and obsl1/obscurin will need to be considered in future revised M-band models, but our lack of knowledge of the C-terminal interactions of obsl1 and the overall layout of titin preclude a more detailed model.
Our AFM experiments reveal low unbinding forces of 30 pN for both the obscurin and obsl1/titin complexes when compared to the mechanically stable Z-disk complex of titin and telethonin (20). At first sight, our finding that the mechanical stability of titin-O1 and titin-OL1 is only slightly different might seem at odds with the lower affinity of the titin-OL1 complex measured by ITC. However, a change in Kd of 3.4 corresponds to an energy difference of only 1.2kBT (see SI Text for explanation). The width of our measured distribution allows us to estimate the transition state position of the unbinding process to Δx = 8 Å (see Dashed Line in Fig. 4D). Hence, the expected difference in force between O1 and OL1 is 1.2kBT/8 Å = 6 pN. It is important to note that this estimate is an upper boundary of the expected shift in force, assuming that the difference in binding free energy can be fully attributed to a change in dissociation rates of the two complexes. In agreement with our results, previous experiments on the mechanical unbinding of antibody-antigen complexes, which covered a wide range of dissociation constants, have also shown that the difference in unbinding force for a fourfold change in dissociation constant is only on the order of a few pN (26).
M-band assembly involves multiple interactions: those of myomesin with myosin, of titin with myomesin and obscurin/obsl1, of obscurin/obsl1 with myomesin (3, 5, 6) and possibly a novel splice variant of MyBP-C (27). The genetic diseases disrupting the titin-obsl1/obscurin interaction are late-onset diseases, and although the Finnish and French titin mutations lead to a total loss of obscurin and obsl1 binding, they are not embryonically lethal, possibly because the myomesin interactions will be retained. Furthermore, knockout mice of obscurin develop normal myofibrils but show misaligned SR and late-onset myopathic features (28), supporting the notion that obscurin is not required for primary myofibril assembly. Of note, the M-band is the sarcomeric structure that ruptures first under isometric tetanic contractions (29) and shows lowest transverse stiffness (30), suggesting that its strain-bearing structures are less stable than those of the Z-disk, even though some M-band interactions are probably mechanically stronger than titin-obscurin/obsl1. Our observations therefore suggest that M-band mechanical stability resides in a cooperative interaction network rather than in any one particular interaction, and weakening of these individual interactions caused by titin mutations may therefore play a major role in various myopathies.
Materials and Methods
Detailed experimental procedures for all techniques used are given in SI Methods.
Protein expression and purification.
All Ig-domains used in this study were expressed as N-terminal His6-tag fusion protein in E.coli and purified initially by immobilized metal affinity chromatography (IMAC). Following TEV cleavage the Ig-domains were further purified by IMAC and size-exclusion chromatography (SEC) using a Superdex S75 column (GE Healthcare). Formation of the M10-OL1 complex was achieved by mixing stoichimetric amounts of the individual components followed by SEC.
Crystallization, structure solution and refinement.
Crystals of the M10-OL1 complex were grown by the hanging drop method at 18 °C either in the presence of 0.10 M Bis-Tris pH 5.5, 2.0 M ammonium sulphate (Form I, trigonal) or in the presence of 0.10 M MIB buffer pH 5.0, 25% (w/v) PEG 1500 (Form II, triclinic). Monoclinic crystals of the M10-OL1 F17R complex were obtained from 0.1 M Bis-Tris pH5.5, 0.2 M Ammonium sulphate, 25% (w/v) PEG 3350. Diffraction data for the M10-OL1 complex (Form I) were initially collected in-house on an Oxford Diffraction Nova system and the structure solved by molecular replacement using the program MOLREP (31) using telokin (PDB code 1FHG) as search model. High-resolution data were later collected at the Diamond Light Source. All structures were refined with the program REFMAC5 (32, 33). Data collection and refinement statistics are shown in Table S1.
Isothermal Titration Calorimetry (ITC).
Samples used for ITC experiments were freshly purifed as described above and extensively dialyzed against ITC buffer (20 mM Tris/HCl pH 7.5, 50 mM NaCl, 2 mM TCEP-HCl). ITC measurements were performed on a MicroCal VP-ITC instrument (MicroCal Inc.) at 20 °C. Binding constants and other thermodynamic parameters were calculated by fitting the integrated titration data assuming a single set of binding sites using the Origin software package (OriginLab).
Confocal Microscopy Competition Assay in Neonatal Rat Cardiomyocytes (NRCs) and Image Ratiometry.
NRCs were isolated from rats, transfected with GFP-fused O1, OL1 and M10 and their mutants and cultured essentially as described in (6). Ratiometric image analysis was performed as in (6).
Atomic Force Microscopy (AFM).
Expression of chimaeric titin-obscurin/obsl1 constructs and single-molecule force spectroscopy was performed as described (20) on a custom-built atomic force microscope using freshly activated Nickel-NTA surfaces for protein immobilization via their C-terminal poly-histidine-tags.
Supplementary Material
Acknowledgments.
We thank Birgit Brandmeier for excellent technical assistance and the colleagues of Diamond Light Source beamlines I03 and I04 for their help during data collections. We are indebted to Mark Pfuhl for generous help with the ITC measurements and to Elisabeth Ehler for cardiomyocyte preparations. This work was supported by King’s College London British Heart Foundation (BHF) Centre of Research Excellence and the Medical Research Council of Great Britain. M.G. holds the BHF Chair of Molecular Cardiology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913736107/DCSupplemental.
References
- 1.Tskhovrebova L, Trinick J. Titin: Properties and family relationships. Nat Rev Mol Cell Biol. 2003;4:679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 2.Ehler E, Gautel M. Sarcomere and sarcomerogenesis. Exp Med Biol. 2008;642:1–14. doi: 10.1007/978-0-387-84847-1_1. [DOI] [PubMed] [Google Scholar]
- 3.Lange S, Ehler E, Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16:11–18. doi: 10.1016/j.tcb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Otey CA, Dixon R, Stack C, Goicoechea SM. Cytoplasmic Ig-domain proteins: Cytoskeletal regulators with a role in human disease. Cell Motil Cytoskeleton. 2009;66:618–634. doi: 10.1002/cm.20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agarkova I, Perriard JC. The M-band: An elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–485. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Fukuzawa A, et al. Interactions with titin and myomesin target obscurin and its small homologue, obscurin-like 1, to the sarcomeric M-band: Implications for hereditary myopathies. J Cell Sci. 2008;121:1841–1851. doi: 10.1242/jcs.028019. [DOI] [PubMed] [Google Scholar]
- 7.Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol Rev. 2009;89:1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. J Cell Biol. 2001;154:123–136. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollazzon M, et al. The first Italian family with tibial muscular dystrophy caused by a novel titin mutation. J Neurol. 2009 doi: 10.1007/s00415-009-5372-3. doi: 10.1007/s00415-009-5372-3. [DOI] [PubMed] [Google Scholar]
- 10.Udd B. Third filament diseases. Adv Exp Med Biol. 2008;642:99–115. doi: 10.1007/978-0-387-84847-1_9. [DOI] [PubMed] [Google Scholar]
- 11.Carmignac V, et al. C-terminal titin deletions cause a novel early-onset myopathy with fatal cardiomyopathy. Ann Neurol. 2007;61:340–351. doi: 10.1002/ana.21089. [DOI] [PubMed] [Google Scholar]
- 12.Huber C, et al. OBSL1 mutations in 3-M syndrome are associated with a modulation of IGFBP2 and IGFBP5 expression levels. Hum Mutat. 2010;31:20–26. doi: 10.1002/humu.21150. [DOI] [PubMed] [Google Scholar]
- 13.Hanson D, et al. The primordial growth disorder 3-M syndrome connects ubiquitination to the cytoskeletal adaptor OBSL1. Am J Hum Genet. 2009;84:801–806. doi: 10.1016/j.ajhg.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 15.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 16.Winter C, Henschel A, Kim WK, Schroeder M. SCOPPI: A structural classification of protein-protein interfaces. Nucleic Acids Res. 2006;34:D310–314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackman P, et al. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am J Hum Genet. 2002;71:492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rief M, Gautel M, Schemmel A, Gaub HE. The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by atomic force microscopy. Biophys J. 1998;75:3008–3014. doi: 10.1016/S0006-3495(98)77741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, et al. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 20.Bertz M, Wilmanns M, Rief M. The titin-telethonin complex is a directed, superstable molecular bond in the muscle Z-disk. Proc Natl Acad Sci USA. 2009;106:13307–133310. doi: 10.1073/pnas.0902312106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfuhl M, Pastore A. Tertiary structure of an immunoglobulin-like domain from the giant muscle protein titin: A new member of the I-set. Structure. 1995;3:391–401. doi: 10.1016/s0969-2126(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 22.Pinotsis N, Lange S, Perriard JC, Svergun DI, Wilmanns M. Molecular basis of the C-terminal tail-to-tail assembly of the sarcomeric filament protein myomesin. Embo J. 2008;27:253–264. doi: 10.1038/sj.emboj.7601944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinotsis N, Abrusci P, Djinovic-Carugo K, Wilmanns M. Terminal assembly of sarcomeric filaments by intermolecular beta-sheet formation. Trends Biochem Sci. 2009;34:33–39. doi: 10.1016/j.tibs.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Lange S, et al. Dimerisation of myomesin: Implications for the structure of the sarcomeric M-band. J Mol Biol. 2005;345:289–298. doi: 10.1016/j.jmb.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 25.Zou P, et al. Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature. 2006;439:229–233. doi: 10.1038/nature04343. [DOI] [PubMed] [Google Scholar]
- 26.Schwesinger F, et al. Unbinding forces of single antibody-antigen complexes correlate with their thermal dissociation rates. Proc Natl Acad Sci USA. 2000;97:9972–9977. doi: 10.1073/pnas.97.18.9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackermann MA, Hu LY, Bowman AL, Bloch RJ, Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol Biol Cell. 2009;20:2963–2978. doi: 10.1091/mbc.E08-12-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange S, et al. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J Cell Sci. 2009;122:2640–2650. doi: 10.1242/jcs.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horowits R, Podolsky RJ. The positional stability of thick filaments in activated skeletal muscle depends on sarcomere length: Evidence for the role of titin filaments. J Cell Biol. 1987;105:2217–2223. doi: 10.1083/jcb.105.5.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshikawa Y, Yasuike T, Yagi A, Yamada T. Transverse elasticity of myofibrils of rabbit skeletal muscle studied by atomic force microscopy. Biochem Biophys Res Commun. 1999;256:13–19. doi: 10.1006/bbrc.1999.0279. [DOI] [PubMed] [Google Scholar]
- 31.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Steiner RA, Lebedev AA, Murshudov GN. Fisher’s information in maximum-likelihood macromolecular crystallographic refinement. Acta Crystallogr D. 2003;59:2114–2124. doi: 10.1107/s0907444903018675. [DOI] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.