Abstract
This review summarizes selected studies on galectin-3 (Gal3) as an example of the dynamic behavior of a carbohydrate-binding protein in the cytoplasm and nucleus of cells. Within the 15-member galectin family of proteins, Gal3 (Mr ~30,000) is the sole representative of the chimera subclass in which a proline- and glycine-rich NH2-terminal domain is fused onto a COOH-terminal carbohydrate recognition domain responsible for binding galactose-containing glycoconjugates. The protein shuttles between the cytoplasm and nucleus on the basis of targeting signals that are recognized by importin(s) for nuclear localization and exportin-1 (CRM1) for nuclear export. Depending on the cell type, specific experimental conditions in vitro, or tissue location, Gal3 has been reported to be exclusively cytoplasmic, predominantly nuclear, or distributed between the two compartments. The nuclear versus cytoplasmic distribution of the protein must reflect, then, some balance between nuclear import and export, as well as mechanisms of cytoplasmic anchorage or binding to a nuclear component. Indeed, a number of ligands have been reported for Gal3 in the cytoplasm and in the nucleus. Most of the ligands appear to bind Gal3, however, through protein-protein interactions rather than through protein-carbohydrate recognition. In the cytoplasm, for example, Gal3 interacts with the apoptosis repressor Bcl-2 and this interaction may be involved in Gal3’s anti-apoptotic activity. In the nucleus, Gal3 is a required pre-mRNA splicing factor; the protein is incorporated into spliceosomes via its association with the U1 small nuclear ribonucleoprotein (snRNP) complex. Although the majority of these interactions occur via the carbohydrate recognition domain of Gal3 and saccharide ligands such as lactose can perturb some of these interactions, the significance of the protein’s carbohydrate-binding activity, per se, remains a challenge for future investigations.
Keywords: Carbohydrate-binding proteins, lectins, nucleo-cytoplasmic transport
1. Introduction
Galectin-3 (Gal3) belongs to a family of widely distributed proteins that: (a) bind to β-galactoside-containing glycoconjugates; and (b) contain characteristic amino acid sequences in the carbohydrate recognition domain (CRD) of the polypeptide [1]. The protein consists of a single polypeptide (Mr ~30,000) whose amino acid sequence suggests that it is a chimera of two distinct domains: an NH2-terminal domain containing repeats of a proline- and glycine-rich motif and a COOH-terminal CRD that shows sequence similarity with the corresponding CRDs of other members of the galectin family [1,2]. Each CRD, ~130 amino acids, contains highly conserved amino acid residues that interact with carbohydrate, as revealed by X-ray crystallography [3].
The carbohydrate-binding specificity of purified Gal3 has been studied extensively [2]. The protein binds type 1 or type 2 Galβ1→3(4)GlcNAc chains and the affinity for straight chain polylactosamine structures or complex-type branched glycans is increased over the simple disaccharide. Fucosylation (H-type 1 or H-type 2), or sialylation, or further substitution by (α1→3)—linked galactose or N-acetylgalactosamine on the terminal galactose residue of the lactosamine unit does not affect binding whereas substitution of the penultimate N-acetylglucosamine residue drastically reduces binding. Using isothermal microcalorimetry, thermodynamic analysis of the binding of Gal3 to a series of saccharide ligands yielded the following order of Kd values: (a) lactose (Lac) ~900 μM; (b) thiodigalactoside (TDG) ~550 μM; (c) N-acetyllactosamine (LacNAc) ~150 μM; and (d) blood group A tetrasaccharide, ~60 μM [4]. Other physico-chemical measurements, such as equilibrium dialysis using radioactive Lac as the saccharide ligand or fluorescence polarization assays using the same series of carbohydrates, usually yielded Kd values 5-10 times lower, although the relative affinities were essentially identical [5-8].
Since its initial isolation on the basis of carbohydrate-binding activity, a vast number of phenomenological observations have been reported on Gal3 [9, 10]. At least part of the reason for the large number of studies has to do with the fact that Gal3 exhibits a diverse range of subcellular localizations and molecular interactions. First, Gal3 (along with several other members of the galectin family) exhibits the striking phenomenon of dual localization [11], being found in both the intracellular as well as the extracellular compartments [12, 13]. The mechanism of externalization appears to be unusual inasmuch as the amino acid sequence of Gal3 does not reveal an obvious signal sequence for directing the polypeptide into the classical endomembrane pathway for secretion [13]. Human, rat, and murine Gal3 can be isolated, respectively, from human WiL-2 cells, rat basophilic leukemia cells, and mouse 3T3 fibroblasts in the absence of any reducing agents [14]. The purified recombinant proteins expressed from the rat and mouse cDNA constructs did not require reducing agents such as thiols to maintain carbohydrate-binding activity [14, 15]. Therefore, Gal3 does not appear to be sensitive to oxidative inactivation as might occur in the extracellular compartment. Nevertheless, the NH2-terminus of the polypeptide isolated from cell extracts is blocked by acetylation [16] and Gal3 is found predominantly as an intracellular protein in most cell types studied.
Second, intracellular Gal3 has been localized in both the nucleus and cytoplasm. Depending on the cell type and specific experimental conditions, the protein has been reported to be exclusively/predominantly cytoplasmic [17, 18], predominantly nuclear [17, 19] or distributed between the two subcellular compartments. Finally, even within the nucleus, Gal3 can be found diffusively in the nucleoplasm as well as being associated with a number of discrete punctate structures [20, 21]. These latter may correspond to subnuclear domains characterized by distinct ultrastructural features or by specific marker proteins under light microscopy: interchromatin granule clusters, speckles, Cajal bodies, etc. [22, 23].
On the basis of these observations, there appear to be exquisite mechanisms by which Gal3 localization, transport, and association with distinct subcellular components are regulated. In this special issue on nucleocytoplasmic glycosylation, it seems appropriate to review the studies that have shed new light on the dynamic behavior of a carbohydrate-binding protein in the nucleus and in the cytoplasm. Using Gal3 as a paradigm, the conditions that govern its nuclear versus cytoplasmic distribution, the signals for its nuclear import and export, its association with and dissociation from macromolecular complexes, and subnuclear domains will be discussed.
2. Nuclear versus Cytoplasmic Distribution
2.1 Intracellular distribution under different experimental conditions
A large number of observations on the nuclear versus cytoplasmic distribution of Gal3 have been reported, correlating the presence or absence of the protein in a particular compartment of the cell to various parameters such as source of the cells under study, specific cell type, culture conditions, proliferation status of the cell/culture, or neoplastic transformation. For example, it has been reported that adaptation of murine peritoneal macrophages to in vitro culture reduces the nuclear content of Gal3 [24]. It has also been shown that nuclear Gal3 is elevated in macrophages derived from tumor-bearing hosts, relative to those from normal hosts [25]. Murine resting T lymphocytes express little/no Gal3 but high level expression is induced in mitogen-activated CD4+ and CD8+ T cells. The protein is found predominantly in intracellular compartments: nucleus, cytosol, and a membrane-containing fraction [26].
In baby hamster kidney (BHK) and Madin-Darby canine kidney (MDCK), Gal3 is cytoplasmic as determined by indirect immunofluorescence microscopy [17, 27, 28]. The exclusively cytoplasmic localization of Gal3 in MDCK cells may be hard to interpret inasmuch as saponin was used to permeabilize the cells for antibody staining [28]. Saponin and digitonin belong to the same family of drugs that selectively permeabilizes the cholesterol-rich plasma membrane without allowing antibody access to antigens, including galectins, in the nucleus [29]. On the other hand, the permeabilization agents used in the studies on BHK cells (methanol at -20 °C or triton X-100) do effectively permeabilize the nuclear membrane and therefore, the antibody reagent has full access to Gal3 if present in the nucleus [17, 27]. Moreover, the exclusive cytoplasmic localization was observed both for endogenous Gal3 as well as for the protein overexpressed in the same cells transfected with a cDNA construct encoding the hamster polypeptide.
Overexpression of the same cDNA in Cos-7 cells or rabbit smooth muscle Rb-1 cells, on the other hand, resulted in a predominantly nuclear localization [17]. This cell-type difference in nuclear versus cytoplasmic distribution of Gal3 may reflect the presence or absence of an interacting partner that either has a potent nuclear export signal (NES) or tethers it to a compartment-specific anchor. Indeed, it has been reported that the transcriptional regulator Sufu (Suppressor of fused) interacts with Gal3 and can alter the nuclear versus cytoplasmic distribution of the latter when both proteins are cotransfected into HeLa cells [30].
In fibroblasts, the nuclear versus cytoplasmic distribution of the protein appears to depend on the proliferative state of the cells under analysis. In quiescent cultures (serum-starved or density-inhibited), Gal3 was predominantly cytoplasmic; proliferating cultures of the same cells showed intense nuclear staining [19]. Parallel nuclear run-off transcription assays and Northern blotting for accumulated mRNA levels showed that Gal3 is an immediate-early gene, whose activation upon serum stimulation of quiescent fibroblasts does not depend on de novo protein synthesis [31]. Human diploid fibroblasts have a finite replicative life span when subjected to in vitro culture. While Gal3 could be found in both the nucleus and cytoplasm of young, proliferating cells, the protein was predominantly cytoplasmic in senescent human fibroblasts that have lost replicative competence [18, 32].
2.2 Correlative observations on tumor samples and cell lines
There is a wealth of published data regarding Gal3 expression in cancer, on both tumor samples and cancer cell lines. The phenomenological observations have yielded, however, a wide divergence of results in different malignancies: Gal3 expression is up-regulated in cancers of the thyroid, liver, stomach, and central nervous system but down-regulated in carcinomas of the breast, ovary, uterus, and prostate (see reviews in [33-35]). In the context of the present discussion, attention is focused on those studies reporting an alteration of intracellular distribution in certain types of cancers.
Gal3 expression and its intracellular distribution vary along the crypt-to-surface axis of human colonic epithelia. The protein is concentrated in nuclei of differentiated colonic epithelial cells. The progression from normal mucosa to adenoma to carcinoma is characterized by a striking absence of Gal3 in the nuclei of adenoma and carcinoma cells [36]. The nuclear localization of Gal3 in normal colonic mucosa was confirmed in a subsequent study, but the latter investigators found that the cytoplasmic pool was down-regulated in adenomas and up-regulated in carcinomas [37].
This general trend of shifting the nuclear localization of Gal3 in favor of the cytoplasmic compartment during neoplastic progression has also been reported for tongue and prostate cancer. Using immunohistochemistry, a survey of 77 tongue specimens (54 squamous cell carcinomas and 23 samples of normal mucosa) showed that levels of nuclear Gal3 decreased during the progression from normal to cancerous states. Concomitantly, cytoplasmic expression was increased. On this basis, it was suggested that the observed nuclear-to-cytoplasmic translocation of Gal3 during neoplastic progression may serve as a prognostic factor for tongue cancer patients [38]. A similar survey of 145 prostate carcinoma samples revealed that Gal3 was usually not expressed or was decreased compared to normal glands. When Gal3 was detected in the cancer cells, it was consistently excluded from the nucleus and only present in the cytoplasm [39]. LNCaP, a Gal3-negative human prostate cancer cell line, was transfected with constructs to express Gal3 either in the nucleus or predominantly in the cytosol. Cells exhibiting predominantly cytoplasmic localization of Gal3 showed increased Matrigel invasion, anchorage-independent growth, and in vivo tumor growth and angiogenesis, while cells with nuclear localization of Gal3 affected these parameters in the opposite fashion [40].
In contrast to these results, transformation of a normal rat thyroid cell line with the Ki-ras oncogene resulted in Gal3 localization in the nucleus [41]. Similarly, in a survey of different human cancer cell lines, Gal3 was found maximally expressed in nuclei of papillary cancer cells. Gal3 interacts directly with the thyroid-specific transcription factor TTF-1, up-regulating its transcriptional activity and thus contributing to the proliferation of the thyroid cells. Concomitant expression of nuclear Gal3 and TTF-1 was associated with a worse clinical outcome [42].
3. Nuclear-cytoplasmic Transport
3.1 Gal3 shuttles between the nucleus and the cytoplasm
Nucleo-cytoplasmic shuttling is typically defined as the repeated bidirectional movement of a protein across the nuclear pore complex. By this criterion, Gal3 shuttles between the nucleus and cytoplasm [43]. Human fibroblasts were fused with mouse fibroblasts and the localization of human Gal3 in the heterodikaryon was monitored using the monoclonal antibody NCL-GAL3, which recognizes human Gal3 but not the mouse homolog [44]. The human Gal3 protein localized to both nuclei in a large percentage of heterodikaryons. Addition of the antibiotic leptomycin B (LMB), which inhibits nuclear export of Gal3 (see below), decreased the percentage of heterodikayrons showing human Gal3 in both nuclei. These results suggest that Gal3 can exit one nucleus, travel through the cytoplasm, and enter the second nucleus, matching the definition of shuttling.
Using hydrodynamic methods such as gel filtration [45] as well as thermodynamic methods such as equilibrium sedimentation [46], most measurements of the solution molecular weight of purified Gal3 have yielded a value of ~30,000, suggesting that the polypeptide exists predominantly as a monomer in solution. While this is of a size that could be accommodated by the aqueous channel of the nuclear pore complex (exclusion limit of 40-60 kDa), the studies cited above on the compartmentalization of Gal3 suggest that passive diffusion is an unlikely means of translocation between the compartments. Thus, observations of a predominantly nuclear localization for Gal3 under one set of conditions and a predominantly cytoplasmic localization under a different set of circumstances imply a specific, regulated mode of transport.
Moreover, as measured by fluorescence recovery after photobleaching (FRAP), the rates of both nuclear import and export of Gal3 are similarly rapid, with the half-life dependent upon the cell type (EJA, unpublished observations). These findings are consistent with the nuclear versus cytoplasmic distribution of Gal3, which suggests a steady state of nuclear shuttling in which rapid nuclear import would be required to offset its rapid nuclear export (see below).
3.2 Signals for nuclear import
Early studies provided conflicting information regarding the amino acid residues essential for nuclear localization. Gong et al. [47] reported that the first eleven residues of human Gal3 governed its nuclear import. This conclusion was based on the observation that deletion of the 11 residues at the amino terminus of Gal3 resulted in complete exclusion of the protein from the nucleus. Furthermore, attachment of this sequence of amino acids to the Green Fluorescent Protein (GFP) reporter resulted in nuclear accumulation of the fusion protein. In direct contradiction to these results, Gaudin et al. [17] reported that deletion of residues 1-103 of the hamster homolog had no effect on the nuclear localization of the truncated protein.
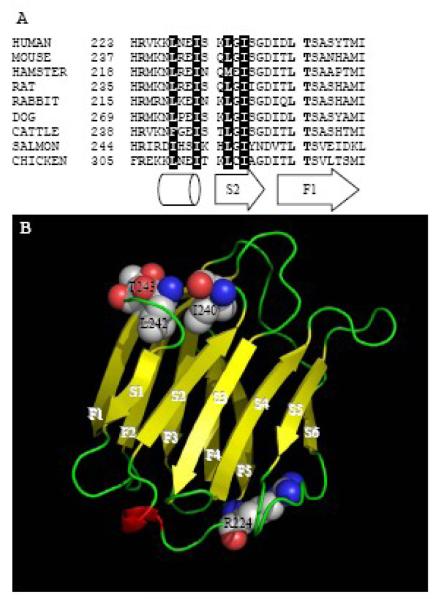
More recent experiments have provided at least some general agreement that the COOH-terminal end (the last 28 amino acids) of the Gal3 polypeptide is important for nuclear localization (see Fig. 1, panel A). First, GFP fusions of murine Gal3 identified a region, encompassing residues 254-257, that is required for nuclear import of Gal3 in mouse 3T3 fibroblasts [48]. Alanine scanning mutagenesis of each of the residues I254, L256, and T257 compromised nuclear import, whereas mutagenesis of T255 had no effect. A triple mutant, I254A;L256A;T257A, showed only cytoplasmic localization; this effect could not be reversed by addition of LMB. This sequence is in agreement with the IXLT motif identified in the nuclear localization activity of the Drosophila protein Dsh [49]. In the context of the entire protein, this sequence lies at the beginning of a single strand of a β-sheet in the CRD of the Gal3 polypeptide (Fig. 1, panel B).
Figure 1.
(A) The amino acid sequence of the carboxyl-terminal 28 amino acids residues of several homologs of the Gal3 polypeptide. The Protein Data Bank accession numbers are as follows: human (NP 002297), mouse (NP 034835), long-tailed hamster (CAA55479), Norway rat (NP 114020), rabbit (NP 001075807), dog (P38486), cattle (NP 001095811), salmon (NP 001134305), chicken (NP 999756). Sequences were retrieved and aligned by Vector NTI (Invitrogen). Residue numbers corresponding to the positions on the polypeptides of each respective species are shown at left. The conserved residues implicated in nuclear import, corresponding to R224 in the human sequence and I-LT in the murine sequence, are highlighted in boldface type, and the conserved hydrophobic-rich NES is shown with white letters on a black background. The cylinder and the arrows beneath the sequences align them with the corresponding single α helix and the S2 and F1 β strands in the crystal structure of the CRD shown in panel B. (B) Ribbon diagram showing the three-dimensional structure of the polypeptide backbone of the human CRD of Gal3 (Protein Data Bank # 2NN8 [92]), which was generated with PyMOL (W.L. DeLano, The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA, USA. http://www.pymol.org). The strands of the two β sheets that comprise the β sandwich are labeled F1-F5 for the five stranded sheet and S1-S6 for the six-stranded sheet. The residues determined to be critical for nuclear import are displayed with space filling representations and are numbered according to the human sequence.
The IXLT motif is conserved in Gal3 sequences from other mammals (Fig. 1, panel A). In the original proposal for naming the galectins [1], it was collectively agreed to restrict the galectin numbering system to mammalian species because, while it is easy to recognize the same galectin in different mammals due to great conservation of amino acid sequence, it was much more difficult to infer the relationship of particular mammalian galectins to those found in lower vertebrates and invertebrates. Nevertheless, we have included two other “galectin-3” sequences in Figure 1A: both the salmon and chicken sequences shown contain the proline-, glycine-rich NH2-terminal domain characteristic of Gal3 and both sequences are identified as “galectin-3” in the Protein Data Bank. Thus, despite differences in other parts of the amino acid sequence, the sequences from salmon and chicken align well in our region of interest (Fig. 1, panel A) and will likely have similar three-dimensional structures. This commonality makes the case even stronger that these amino acid residues are indeed important. Similar considerations apply to the leucine-rich NES that is found in the same region of the polypeptide (see Section 3.3 below).
Second, a subsequent study was carried out using the GFP reporter fused onto the human Gal3 sequence. In agreement with the results obtained with the murine polypeptide, it was reported that truncation of residues 240-250 of the human sequence (the IXLT motif begins at residue 240; see Fig. 1, panel A) resulted in complete abrogation of nuclear localization [50]. Thus, the COOH-terminal end of the Gal3 polypeptide is clearly critical for targeting to the nucleus.
Third, Nakahara et al. [51] identified a sequence in residues 223-228 (HRVKKL) of human Gal3 (Figure 1, panel A) that is similar to the lysine/arginine-rich nuclear localization signal (NLS) found in many nuclear proteins. Deletion of the HRVKKL sequence resulted in the complete loss of nuclear import. Site directed mutagenesis of the sequence in a Gal3-GFP fusion protein demonstrated that only R224, located on the opposite side of the CRD from the IXLT motif (see Fig. 1, panel B), had any role in Gal3 nuclear import. Interestingly, although active transport of Gal3 into the nucleus requires both importin-α and importin-β, Gal3 only binds directly to importin-α [51]. GST pull-down assays demonstrated that Gal3 interacted directly with importin-α1, importin-α3, and importin-α5 (in order of decreasing affinity); the R224A mutation abolished all importin-α interactions. The requirement for importin-β was confirmed with RNAi knockdown experiments, as depletion of importin-β inhibited the nuclear localization of the Gal3-GFP fusion protein.
Despite the general agreement that the COOH-terminal portion of the Gal3 polypeptide is critical for nuclear localization, what remain unclear are the findings that: (a) deletion of the first 11 residues at the NH2-terminus resulted in loss of nuclear localization and fusion of this 11-residue sequence to GFP localized the reporter to the nucleus [47]; and (b) GFP fusion proteins containing residues 1-115 were localized to the nuclear compartment [50]. The results that implicate an NH2-terminal NLS are confounding because: (a) polypeptides derived from truncation from the NH2-terminal end (e.g. deletion of residues 1-73 or of residues 1-127 of the murine homolog) of the Gal3 protein retained nuclear localization like the endogenous polypeptide [17, 48]; and (b) truncation of the COOH-terminal 10 amino acids resulted in loss of nuclear localization [48, 50].
3.3 Identification of a nuclear export signal (NES)
The observation that Gal3 shuttles between the nuclear and cytoplasmic compartments [43] implied that there must be a mechanism of exporting the protein from the nucleus. A digitonin-permeabilized cell assay was used to explore this process [52]. Mouse 3T3 fibroblasts, without fixation, were permeabilized with digitonin which selectively permeabilizes the cholesterol-rich plasma membrane while retaining the integrity of the nuclear membrane. The levels of Gal3 in the nucleus and in the transported fraction were monitored as a function of time by immunofluorescence and by immunoblotting. This analysis revealed that Gal3 was rapidly exported from the nucleus in a temperature-dependent fashion, with a half-life of less than five minutes at both 37°C and 25°C and little to no export at 4° C.
The addition of LMB resulted in the retention of Gal3 in the nucleus; concentrations of LMB as low as 3.8 nM inhibited the export of Gal3 [52]. Because LMB binds to and inhibits the interaction of exportin-1 (CRM1) with leucine-rich NESs, this result implicated the presence of such an NES in the Gal3 polypeptide. Indeed, a leucine-rich sequence can be found in residues 241-256 of murine Gal3, at the COOH-terminus of the protein (Figure 1, panel A). This putative NES fits the spacing of the hydrophobic residues in the canonical nuclear export sequence Lx2-3Lx2-3LxL described by la Cour et al. [53] and is conserved in Gal3 from other mammals as well as in “galectin-3” from salmon and chicken (Figure 1, panel A). When the putative NES was inserted into the pRev(1.4)-GFP vector, an NES-deficient variant that can be used to evaluate potential NESs [54], the protein exhibited LMB-sensitive nuclear export activity [55]. Moreover, site-directed mutagenesis confirmed L248 and I250 as the two critical residues of the NES.
In the context of the entire protein, the NES (residues 241-256) is found in the hydrophobic β-sandwich motif of the CRD (Fig. 1, panel B). Critical hydrophobic residues L242 and I245 lie in the α-helix joining two strands of the β-sheet, and residues L248 and I250 are found in the adjacent β-strand. The side chains of the NES residues are located on the same face of the structure [55]. These properties fit the parameters outlined by a computational study of leucine-rich NESs [53].
In mouse 3T3 fibroblasts, Gal3 exists as two isoelectric variants: (a) a pI 8.7 species corresponding to the non-phosphorylated polypeptide; and (b) a pI 8.2 phosphorylated derivative [56]. The non-phosphorylated form is found exclusively in the nucleus, while phosphorylated Gal3 can be found both in the nucleus and cytoplasm. In nuclear export assays carried out using the digitonin-permeabilized cell system, it was found that only phosphorylated Gal3 is transported out of nuclei [52]. Mass spectrometry of the canine homolog of Gal3 identified the serine at residue 6 as the major site of phosphorylation in vivo, with a minor site at serine-12 [57]. In response to apoptotic insults, phosphorylated Gal3 is exported from the nucleus to the cytoplasm and protects cells from drug-induced apoptosis [58]. In contrast, the S6A mutant, which cannot be phosphorylated, is not exported from the nucleus and failed to protect cells from apoptosis. Thus, phosphorylation of Gal3 and its export from the nucleus to the cytoplasm appear to be important for a critical biological activity of the protein.
4. Activities and Ligands in the Cytoplasm and Nucleus
4.1 General comments on Gal3 ligands
In addition to the transport receptors importin-α and exportin-1, many other proteins that interact with Gal3 have been reported (Table 1). In the context of the present article on dynamics of Gal3 inside cells, several general comments on these ligands might be illuminating. First, although some of these ligands have been documented to be compartment specific (e.g. the cysteine-, histidine-rich protein Chrp is exclusively cytoplasmic [62]), other ligands appear to coexist with Gal3 in both subcellular compartments. Moreover, certain ligands (e.g. the Sufu transcription regulator [30]) may move between the compartments with the shuttling Gal3 molecule itself.
Table 1.
Intracellular Proteins that bind galectin-3
| Ligand | Subcellular Compartment |
Gal3 Domain1 | Carbohydrate Sensitivity2 |
Direct or Indirect |
Reference |
|---|---|---|---|---|---|
| Bcl-2 | cytoplasm | CRD | yes | nk3 | 59 |
| β-catenin | cytoplasm and nucleus |
CRD | yes | direct | 60 |
| CBP70 | nucleus | CRD | yes | nk | 61 |
| Chrp | cytoplasm | CRD | no | direct | 62, 63 |
| Cytokeratin | cytoplasm | CRD | yes | direct | 64 |
| Gemin4 | cytoplasm and nucleus |
CRD | nd4 | direct | 65 |
| hnRNP Q | nucleus | nd | nd | nk | 66 |
| nucling | cytoplasm | nd | nd | nk | 67 |
| OCA-B | cytoplasm | CRD | no | direct | 68 |
| K-Ras | cytoplasm and plasma membrane |
nd | nd | nk | 69 |
| Sufu | cytoplasm and nucleus |
nd | nd | direct | 30 |
| synexin | cytoplasm and mitochondria |
nd | nd | direct | 70 |
| TFII-I | nucleus | CRD | yes | nk | 71 |
| TTF-1 | nucleus | nd | nd | direct | 41 |
The portion of the Gal3 polypeptide required for binding of the ligand.
Sensitivity of Gal3-ligand interaction to saccharide inhibition.
nk, not known because the assays to detect the interaction were carried out using cell lysates or subcellular mixtures rather than purified proteins and thus the possibility of a bridging molecule mediating the interaction cannot be excluded.
nd, not determined.
Second, most of the ligands listed in Table 1 bind Gal3 via protein-protein interactions, rather than lectin-carbohydrate recognition. The lone exception might be the cytokeratins, displaying terminal α1,3 N-acetylgalactosamine residues for Gal3 binding [64]. Despite this fact, however, it should be noted that the domain of Gal3 responsible for interacting with the majority of the listed ligands is the CRD and, in many cases, saccharide ligands such as Lac can perturb the interaction (e.g. the apoptosis suppressor Bcl-2 [59]). Two mechanisms may explain how Lac disrupts these interactions. One possibility is that the binding site(s) for Lac and for the protein ligand overlap and the presence of the saccharide competitively inhibits the binding of the latter. Alternatively, the binding of the saccharide ligand to the carbohydrate-binding site results in a conformational change that disrupts the interaction of the Gal3 polypeptide with other proteins. Conformational changes in the CRD of Gal3 have been reported on the basis of NMR analysis [72] and differential scanning calorimetry studies [15].
Finally, the interaction of many of the ligands with Gal3 has been documented through direct binding assays using purified proteins (e.g. Chrp [62]). On the other hand, there are also other ligands (e.g. the general transcription factor TFII-I [71]) whose association with Gal3 might be indirect because the assays (e.g. co-immunoprecipitation or glutathione S-transferase (GST) pull-down) were carried out with cell lysates or subcellular fractions. The mediation via an intermediary molecule cannot be rigorously ruled out. Therefore, the delineation under the column “Direct or Indirect” in Table 1 may simply reflect the experimental conditions used to document the interaction rather than actual evidence of a bridging molecule.
4.2. Selected ligands of Gal3 in the cytoplasm
Gal3 exhibits anti-apoptotic activity. Human T lymphoma Jurkat cells ectopically expressing Gal3 survive longer than control cells when subjected to a variety of apoptosis-inducing agents [59]. It has also been shown that Gal3 inhibits apoptosis caused by the loss of cell anchorage (anoikis) [73]. Gal3 interacts with the apoptosis repressor Bcl-2, mimicking the ability of the Bcl-2 family members to form heterodimers. In apoptosis of BT549 human breast carcinoma cells induced by administration of cisplatin, Gal3 is translocated to mitochrondrial membranes, where it prevents mitochondrial damage, cytochrome c release, and apoptosome activity [70]. This translocation is dependent on synexin, a calcium- and phospholipid-binding protein. Direct interaction between Gal3 and synexin was documented by a yeast two-hybrid assay, as well as by GST pull-down assays. Gal3 may also negatively regulate apoptosis by interacting with nucling, a novel pro-apoptotic protein found in the cytoplasm [67]. Thus, Bcl-2, nucling and synexin are three ligands of Gal3 related to the latter’s apoptosis inhibiting activity.
Shimura et al. [60] reported that in the human breast cancer cell line BT-549, β-catenin stimulation of both cyclin D1 and myc expression is Gal3 dependent. Gal3 binds β-catenin/Tcf and colocalizes with the complex to the nucleus to induce trancriptional activity in the Wnt signaling pathway. Using purified proteins and GST pull-down assays, direct binding between Gal3 and β-catenin was documented; the CRD of Gal3 and the NH2-terminal portion of β-catenin were required for this interaction [60]. More recent studies have supported a role for Gal3 in stabilizing β-catenin and its nuclear accumulation in Wnt signaling in human colorectal cancer cells [74, 75]. However, the direct interaction between Gal3 and β-catenin has been called into question [75]. It was proposed that Gal3 mediates AKT phosphorylation of glycogen synthase kinase-3β, decreasing the latter’s activity on β-catenin. This, in turn, stabilizes β-catenin in the cytoplasm, allowing for nuclear accumulation and transcriptional activity.
Through interactions with the Gli family of transcription factors, Sufu serves as a negative regulator of the Hedgehog signal transduction pathway. A yeast two-hybrid screen using Sufu as bait identified Gal3 as a potential interacting protein [30]. This direct interaction was confirmed by GST pull-down assays using GST-Sufu fusion protein and radiolabled Gal3. Like Gal3, Sufu shuttles between the cytoplasm and nucleus. The possibility is raised, then, that Gal3 may play a role in the nuclear versus cytoplasmic distribution of Sufu, thereby affecting transcriptional activity.
4.3 Selected ligands of Gal3 in the nucleus
Gal3 can also modulate transcriptional activity more directly. Nuclear thyroid-specific transcription factor TTF-1 interacts via its homeodomain with Gal3 [41]. Gel retardation assays showed that this interaction stimulated the DNA-binding activity of TTF-1. It was proposed that Gal3 can up-regulate the transcriptional activity of TTF-1 through this mechanism, contributing to the proliferation of the thyroid cells.
When nuclear extract (NE) of HeLa cells was subjected to adsorption on GST-Gal3, one protein specifically bound was identified to be the general transcription factor TFII-I [71]. This interaction required the CRD, and Lac as well as other saccharide ligands inhibited GST-Gal3 pull-down of TFII-I while non-binding carbohydrates failed to yield the same effect. Although initially identified as a general transcription factor, TFII-I has actually been studied under a wide variety of contexts, and therefore the same polypeptide has acquired a number of different names (see reference [76] for a review). In the context of activities of Gal3 in pre-mRNA splicing (see below), it was of particular interest that TFII-I had been identified as a spliceosome-associated protein in a large scale proteomic screen of splicing complexes [77].
Gal3 (and another family member galectin-1 (Gal1)) directly interacts with the carboxyl terminal 50 amino acids of Gemin4 [65]. Gemin4 is a member of the SMN (survival of motor neuron) complex that assembles the Sm proteins onto snRNAs in the normal biogenesis of these ribonucleoprotein complexes (RNPs) in the cytoplasm [78, 79]. The SMN complex then enters the nucleus with the snRNPs, delivering them to serve as components of the splicing machinery. Gemin4 is so named because it is a component of the SMN complex that is localized to gemini of Cajal Bodies (either coincident with or immediately adjacent to Cajal Bodies, depending on the cell) [22, 23, 79]. The association of Gal3 with such a complex would suggest that it might also be localized to similar subnuclear domains.
Indeed, early immunofluorescence studies had noted that, in addition to a diffuse distribution in the nucleoplasm, Gal3 appears to be localized with a number of discrete punctate structures designated as speckles [20]. In fact, Gal3 is co-localized in certain of these speckles with two other proteins identified as components of the splicing machinery: the Sm core polypeptides of snRNPs and the serine- and arginine-rich (SR) family of splicing factors [29, 80]. At the ultrastructural level, the speckled structures have been found to correspond to interchromatin granule clusters and perichromatin fibrils [81]. Perichromatin fibrils are readily labeled with short pulses of [3H]uridine [82], suggesting that they represent nascent transcripts at the site of mRNA synthesis and early events of pre-mRNA processing. Immunogold labeling at the ultrastructural level has indeed disclosed Gal3 in perichromatin fibrils [21].
5. Association of Galectin-3 with Ribonucleoprotein Complexes
5.1 Dynamics of Gal3 association with the spliceosome
Several lines of evidence have now been accumulated to document that Gal3 and Gal1 are two of many proteins involved in the splicing of pre-mRNA, as assayed in a cell-free system [80, 83]: (a) NE derived from HeLa cells, capable of carrying out splicing of pre-mRNA, contained both Gal1 and Gal3; (b) depletion of both galectins from NE, either by Lac affinity chromatography or by antibody adsorption, resulted in the concomitant loss of splicing activity; (c) either recombinant Gal1 or recombinant Gal3 was able to reconstitute splicing in a galectin-depleted extract; and (d) saccharides which bind to Gal1 and Gal3 with high affinity, such as Lac and TDG, inhibited the splicing reaction when added to a complete NE, whereas non-binding saccharides such as cellobiose failed to have any effect.
The finding that antibodies directed against Gal3 co-precipitated all mRNA species (pre-mRNA, intermediates, product mRNA and excised lariat) from an in vitro splicing reaction further supports the conclusion that Gal3 is a pre-mRNA splicing factor [84]. In a time course immunoprecipitation experiment, Gal3 was found associated with mRNA intermediates and products as they appeared in the splicing reaction, suggesting that Gal3 is a component of the splicing machinery throughout the entire splicing process. Additionally, there is an interesting observation regarding the apparent redundant function of the Gal3 and Gal1 proteins in splicing. The results of a sequential immunoprecipitation protocol suggest that Gal3 and Gal1 are mutually exclusive in terms of their presence in a spliceosome [84]. Thus, a spliceosome may contain the Gal3 or Gal1 polypeptide but not both. This explains why in vitro splicing reactions devoid of only Gal3 show little to no inhibition and why either galectin can reconstitute splicing in a galectin-depleted NE [80].
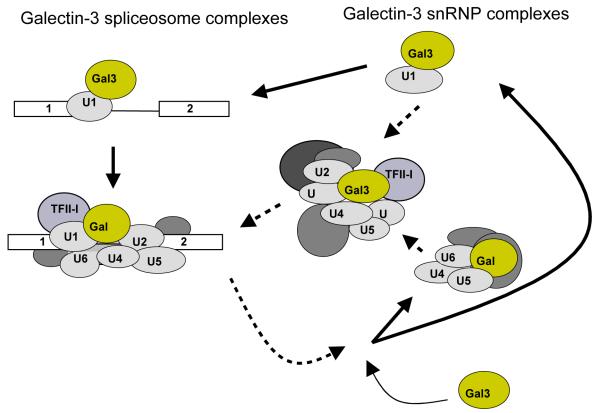
Identifying the actual binding partner of Gal3 on the spliceosome remains an unresolved question. As indicated earlier, one promising candidate for the protein partner of Gal3 on the spliceosome is the general transcription factor TFII-I [71]. Although the Gal3-TFII-I interaction was originally identified in NE devoid of exogenous pre-mRNA, it is possible that this same interaction could occur on the larger scaffold of the spliceosome (Fig. 2, left side).
Figure 2.
Diagram showing the association of Gal3 with snRNPs and the pre-mRNA splicing substrate. The pre-mRNA is shown on the left-side as two rectangular exons joined by a single line intron. Newly identified associations of Gal3 with snRNPs outside the spliceosome are indicated on the right-side. Gal3 can enter the splicing pathway at an early assembly stage via its association with U1 snRNP. In addition, Gal3 also associates with multiple snRNPs in larger complexes outside of the spliceosome. Solid arrows indicate steps supported by accumulated experimental data; dashed arrows represent hypothetical Gal3 involvement that require experimental confirmation.
5.2 Association of Gal3 with RNPs outside of the spliceosome
Aside from a role of Gal3 in pre-mRNA splicing, there are also indications that the protein associates with numerous complexes containing splicing factors in the absence of pre-mRNA. Gal3 was identified through mass spectrometry as a member of a large complex (~60S) termed the PCC (PSF-containing complex) [85]. In addition to containing PSF (pyrimidine track binding protein (PTB)-associated splicing factor), this PCC contained many other proteins identified in complexes with Gal3 such as Gemin4, SMN, and TFII-I [65, 71]. This PCC also co-sedimented on a glycerol gradient in a region with all five snRNAs and contained numerous other spliceosomal proteins, such as the Sm core proteins, snRNP-specific proteins and various SR splicing factors [85]. Interestingly, this PCC could be formed in NE depleted of polyA RNA and did not contain several required second-step splicing factors, suggesting that this complex is distinct from a catalytically active spliceosome. Taken together, these results may point to some role for the PCC in spliceosome assembly and suggest a role for Gal3 in the pre-mRNA splicing pathway outside of the spliceosome itself (Fig. 2, right side).
When HeLa NE was subjected to immunoprecipitation by antibodies against Gal3, all five splicing snRNAs (U1, U2, U4, U5 and U6), but not 5S rRNA, were co-precipitated (Fig. 2, right side) [86]. When the NE was fractionated on a glycerol gradient, immunoprecipitation of pooled gradient fractions with anti-Gal3 show the association of Gal3 with multiple snRNP complexes of varying size and snRNA and protein composition. One such isolated complex showed Gal3 with the U1 mono-snRNP [86] (Fig. 2). This complex was further tested for the ability to bind a pre-mRNA, a property originally assigned to the U1 snRNP on the basis of its recognition of the 5’-splice site [87]. Indeed when the isolated Gal3-U1 complex is combined with exogenous pre-mRNA, antibodies against Gal3 immunoprecipitate the U1 snRNA, snRNP proteins (U1-70K, Sm B/B’) and the pre-mRNA along with the cognate antigen [86]. Using the same Gal3-U1 complex, Gal3 did not associate with exogenous pre-mRNA if it lacked consensus splice sites or if the U1 snRNP was first treated with nuclease to destroy the U1 snRNA of the complex.
It should be noted that one possible explanation for the above observations is that Gal3 directly interacts with RNA. However, attempts to demonstrate direct binding of recombinant Gal3 to in vitro transcribed pre-mRNA substrate [84] or U1 snRNA (KCH and RJP, unpublished observations) via gel mobility shift assays have failed. Therefore it appears that RNA itself is not a nuclear ligand for Gal3.
5.3 Dynamics of Gal3 association with RNPs
One critical question to consider is what regulates the association of Gal3 with nuclear RNPs. Early experiments with Gal3 nuclear localization showed that treating permeablized fibroblasts with ribonuclease A, but not deoxyribonuclease I, abolished Gal3 nuclear staining, a pattern similar to what is observed for the Sm core polypeptides [20]. In addition, when U1 snRNA is first degraded by prior nuclease treatment, immunoprecipitation experiments with antibodies against Gal3 failed to co-precipitate the U1-70K protein and exogenously added pre-mRNA substrate [86]. This suggests that the association of Gal3 with U1 snRNP is a mechanism by which Gal3 can enter the spliceosome (Fig. 2). Thus, the integrity of the RNA in U1 snRNP appears to be critical for Gal3’s participation in pre-mRNA splicing.
Another key consideration for Gal3-RNP association is ionic strength. Indeed, Gal3 has not, to date, been identified in any proteomic study of the spliceosome. However, it appears that the association of Gal3 with splicing complexes is sensitive to disruption by high salt concentrations. Immunoprecipitations carried out at salt concentrations that allow an in vitro splicing reaction to occur (60 mM KCl) show Gal3 associated with the mRNA species contained in a spliceosome. However when this experiment is performed under conditions containing 250 mM KCl, over 90% of the associated spliceosomal mRNA had dissociated from Gal3 [84]. Similarly, the association of Gal3 with snRNPs in the absence of pre-mRNA (Fig. 2, right side) shows the same sensitivity to ionic strength.
Finally, it appears that the association of Gal3 with the nuclear RNPs may be sensitive to carbohydrate ligands as well. Pre-treatment of HeLa NE with lactose before sedimentation on a glycerol gradient shifted the distribution of Gal3 towards the top of the gradient (i.e. towards free protein) [86]. This is not observed if the nuclear extract is pre-treated with cellobiose. These results suggest that carbohydrate ligands may release Gal3 from nuclear snRNP complexes. This release of Gal3 from snRNP complexes may explain the effect of carbohydrates on pre-mRNA splicing, in which Lac and TDG, saccharide ligands of Gal3, inhibit splicing when added to a complete NE while non-binding saccharides such as cellobiose failed to yield the same effect [83]. It should be noted, however, that carbohydrate-binding activity of the Gal1 protein, per se, is not required for pre-mRNA splicing [71]. A site-directed mutant (N46D) of Gal1, devoid of saccharide-binding activity, could still reconstitute splicing activity in a galectin-depleted NE. Due to the redundancy of Gal3 and Gal1 in pre-mRNA splicing and the similarity of the two proteins, it is possible that the above finding may apply to Gal3 as well.
6. Concluding Remarks
The studies reviewed in the present article have documented that Gal3, as an example of an intracellular carbohydrate-binding protein, exhibits dynamic behavior in the nucleus and cytoplasm of cells. A considerable amount of evidence has been accumulated that Gal3 is associated with RNP complexes and participates in splicing of pre-mRNA. Strikingly, it has recently been reported that at least three members of another family of nuclear splicing factors, characterized by an arginine- and serine-rich domain (the SR splicing factors), are similar to the galectins in two respects: they exhibit carbohydrate-binding activity and can be found at the cell surface [88]. Using a glycan array, it was determined that these SR splicing factors bound a series of fucosylated oligosaccharides, including H-type 1, H-type 2, and LeX structures. As detailed in the introduction of this article, Gal3 binds to polylactosamine, as well as to H-type 1 and H-type 2 structures [2].
Based on compositional analysis of the purified protein, Goletz et al. [64] reported that cytokeratins are modified by oligosaccharide(s) containing terminal N-acetylgalactosamine, a possible ligand for Gal3. The major caveat is that the possibility of a contaminating glycoprotein in the cytokeratin preparation needs to be ruled out rigorously. The modification of many nuclear and cytoplasmic proteins with O-linked N-acetylglucosamine has been well documented (for reviews, see [89] and article in present volume). However, neither Gal3 nor the SR splicing factors exhibit appreciable affinity for the monosaccharide. Similarly, the splicing factors do not bind to α1→4-linked glucose moieties on glycogenin, the cytoplasmic glycoprotein that serves as a primer for glycogen synthesis [90]. In the slime mold Dictyostelium, the cytoplasmic/nuclear protein Skp1, a component of the E3 ubiquitin ligase complex, is glycosylated on hydroxyproline to yield the oligosaccharide Galα1→6Galα1→Fucα1→2Galβ1→3 GlcNAc (for reviews, see [91] and article in present volume). This structure could serve as a carbohydrate ligand for both families of splicing factors. The characterization of the oligosaccharide structure found on Skp1 and of the cytosolic (rather than endomembrane pathway) enzymes responsible for its glycosylation have been carried out meticulously in the Dictyostelium system. The same level of rigorous work remains to be extended to higher eukaryotes.
On a number of accounts, therefore, evidence for an endogenous carbohydrate ligand inside cells for the galectin and SR families of splicing factors appears to be less-than-firm at present. It is possible that the nuclear activities of these proteins rely on protein-protein interactions and that protein-carbohydrate binding is reserved for a portion of the protein that localizes to the extracellular compartment. Thus, the significance of the saccharide-binding activity of either Gal3 or SR splicing factors remains as a challenge for future investigations.
Acknowledgements
The work carried out in the authors’ laboratories has been supported by a Calvin Research Fellowship from Calvin College (EJA), by grants Cottrell College Science Award from the Research Corporation (EJA), GM-38740 from the National Institutes of Health (JLW), 06-IRGP-858 from the Michigan State University Intramural Research Grant Program (JLW), and MCB-0092919 from the National Science Foundation (RJP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Barondes SH, Castronovo V, Cooper DW, Cummings RD, Drickamer K, Feizi T, et al. Galectins: a family of animal β-galactoside-binding lectins. Cell. 1994;76:597–598. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- [2].Leffler H, Carlsson S, Hedlund M, Qian Y, Poirier F. Introduction to galectins. Glycoconj. J. 2004;19:433–440. doi: 10.1023/B:GLYC.0000014072.34840.04. [DOI] [PubMed] [Google Scholar]
- [3].Rini JM, Lobsanov YD. New animal lectin structures. Curr. Opin. Struct. Biol. 1999;9:578–584. doi: 10.1016/s0959-440x(99)00008-1. [DOI] [PubMed] [Google Scholar]
- [4].Bachhawat-Sikder K, Thomas CJ, Surolia A. Thermodynamic analysis of the binding of galactose and poly-N-acetyllactosamine derivatives to human galectin-3. FEBS Lett. 2001;500:75–79. doi: 10.1016/s0014-5793(01)02586-8. [DOI] [PubMed] [Google Scholar]
- [5].Hsu DK, Zuberi RI, Liu F-T. Biochemical and biophysical characterization of human recombinant IgE-binding protein, an S-type animal lectin. J. Biol. Chem. 1992;267:14167–14174. [PubMed] [Google Scholar]
- [6].Knibbs RN, Agrwal N, Wang JL, Goldstein IJ. Carbohydrate binding protein 35. II. Analysis of the interaction of the recombinant polypeptide with saccharides. J. Biol. Chem. 1993;268:14940–14947. [PubMed] [Google Scholar]
- [7].Ahmad N, Gabius H-J, Sabesan S, Oscarson S, Brewer CF. Thermodynamic binding studies of bivalent oligosaccharides to galectin-1, galectin-3, and the carbohydrate recognition domain of galectin-3. Glycobiology. 2004;14:817–825. doi: 10.1093/glycob/cwh095. [DOI] [PubMed] [Google Scholar]
- [8].Sorme P, Arnoux P, Kahl-Knutsson B, Leffler H, Rini JM, Nilsson UJ. Structural and thermodynamic studies on cation-π interactions in lectin-ligand complexes: high affinity galectin-3 inhibitors through fine tuning of an arginine-arene interaction. J. Am. Chem. Soc. 2005;127:1737–1743. doi: 10.1021/ja043475p. [DOI] [PubMed] [Google Scholar]
- [9].Dumic J, Dabelic S, Flogel M. Galectin-3: an open-ended story. Biochim. Biophys. Acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- [10].Krzeslak A, Lipinska A. Galectin-3 as a multifunctional protein. Cell. Mol. Biol. Lett. 2004;9:305–328. [PubMed] [Google Scholar]
- [11].Arnoys EJ, Wang JL. Dual localization: proteins in extracellular and intracellular compartments. Acta Histochemica. 2007;109:89–110. doi: 10.1016/j.acthis.2006.10.002. [DOI] [PubMed] [Google Scholar]
- [12].Wang JL, Gray RM, Haudek KC, Patterson RJ. Nucleocytoplasmic lectins. Biochim. Biophys. Acta. 2004;1673:75–93. doi: 10.1016/j.bbagen.2004.03.013. [DOI] [PubMed] [Google Scholar]
- [13].Hughes RC. Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochim. Biophys. Acta. 1999;1473:172–185. doi: 10.1016/s0304-4165(99)00177-4. [DOI] [PubMed] [Google Scholar]
- [14].Frigeri LG, Robertson MW, Liu F-T. Expression of biologically active recombinant rat IgE-binding protein in Escherichia coli. J. Biol. Chem. 1990;265:20763–29769. [PubMed] [Google Scholar]
- [15].Agrwal N, Sun Q, Wang S-Y, Wang JL. Carbohydrate binding protein 35. I. Properties of the recombinant polypeptide and the individuality of the domains. J. Biol. Chem. 1993;268:14932–14939. [PubMed] [Google Scholar]
- [16].Hermann J, Turck CW, Atchison RE, Huflejt ME, Poulter L, Gitt MA, Burlingame AL, Barondes SH, Leffler H. Primary structure of the soluble lactose binding lectin L-29 from rat and dog and interaction of its non-collagenous proline-, glycine-, tyrosine-rich sequence with bacterial and tissue collagenase. J. Biol. Chem. 1993;268:26704–26711. [PubMed] [Google Scholar]
- [17].Gaudin J-C, Mehul B, Hughes RC. Nuclear localisation of wild type and mutant galectin-3 in transfected cells. Biol. Cell. 2000;92:49–58. doi: 10.1016/S0248-4900(00)88763-8. [DOI] [PubMed] [Google Scholar]
- [18].Openo KP, Kadrofske MM, Patterson RJ, Wang JL. Galectin-3 expression and subcellular localization in senescent human fibroblasts. Exp Cell Res. 2000;255:278–290. doi: 10.1006/excr.1999.4782. [DOI] [PubMed] [Google Scholar]
- [19].Moutsatsos IK, Wade M, Schindler M, Wang JL. Endogenous lectins from cultured cells: nuclear localization of carbohydrate-binding protein 35 in proliferating cells. Proc. Natl. Acad. Sci. USA. 1987;84:6452–6456. doi: 10.1073/pnas.84.18.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Laing JG, Wang JL. Identification of carbohydrate binding protein 35 in heterogeneous nuclear ribonucleoprotein complex. Biochemistry. 1988;27:5329–5334. doi: 10.1021/bi00414a057. [DOI] [PubMed] [Google Scholar]
- [21].Hubert M, Wang S-Y, Wang JL, Seve A-P, Hubert J. Intranuclear distribution of galectin-3 in mouse 3T3 fibroblasts: comparative analyses by immunofluorescence and immunoelectron microscopy. Exp. Cell Res. 1995;220:397–406. doi: 10.1006/excr.1995.1331. [DOI] [PubMed] [Google Scholar]
- [22].Spector DL. Nuclear domains. J. Cell Science. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- [23].Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- [24].Dumic J, Lauc G, Hadzija M, Flogel M. Transfer to in vitro conditions influences expression and intracellular distribution of galectin-3 in murine peritoneal macrophages. Z. Naturforsch, C. 2000;55:261–266. doi: 10.1515/znc-2000-3-419. [DOI] [PubMed] [Google Scholar]
- [25].Askew D, Burger CJ, Elgert KD. Tumor growth and adherence change the expression of macrophage Mac-2. Cancer Lett. 1993;69:67–74. doi: 10.1016/0304-3835(93)90034-7. [DOI] [PubMed] [Google Scholar]
- [26].Joo H-G, Goedegebuure PS, Sadanaga N, Nagoshi M, von Bernstorff W, Eberlein TJ. Expression and function of galectin-3, a β-galactoside-binding protein in activated T lymphocytes. J. Leukoc. Biol. 2001;69:555–564. [PubMed] [Google Scholar]
- [27].Sato S, Burdett I, Hughes RC. Secretion of the baby hamster kidney 30-kDa galactose-binding lectin from polarized and nonpolarized cells: a pathway independent of the endoplasmic reticulum-Golgi complex. Exp. Cell Res. 1993;207:8–18. doi: 10.1006/excr.1993.1157. [DOI] [PubMed] [Google Scholar]
- [28].Lindstedt R, Apodaca G, Barondes SH, Mostov KE, Leffler H. Apical secretion of a cytosolic protein by Madin-Darby canine kidney cells. Evidence for polarized release of an endogenous lectin by a nonclassical secretory pathway. J. Biol. Chem. 1993;268:11750–11757. [PubMed] [Google Scholar]
- [29].Vyakarnam A, Lenneman AJ, Lakkides KM, Patterson RJ, Wang JL. A comparative nuclear localization study of galectin-1 with other splicing components. Exp. Cell Res. 1998;242:419–428. doi: 10.1006/excr.1998.4111. [DOI] [PubMed] [Google Scholar]
- [30].Paces-Fessy M, Boucher D, Petit E, Paute-Briand S, Blanchet-Tournier MF. The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, galectin-3 and other nuclear proteins. Biochem. J. 2004;378:353–362. doi: 10.1042/BJ20030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Agrwal N, Wang JL, Voss PG. Carbohydrate-binding protein 35. Levels of transcription and mRNA accumulation in quiescent and proliferating cells. J. Biol. Chem. 1989;264:17236–17242. [PubMed] [Google Scholar]
- [32].Hamann KK, Cowles EA, Wang JL, Anderson RL. Expression of carbohydrate-binding protein 35 in human fibroblasts: variations in the levels of mRNA, protein, and isoelectric species as a function of replicative competence. Exp. Cell Res. 1991;196:82–91. doi: 10.1016/0014-4827(91)90458-7. [DOI] [PubMed] [Google Scholar]
- [33].van den Brule F, Califice S, Castronovo V. Expression of galectins in cancer: a critical review. Glycoconj. J. 2004;19:537–542. doi: 10.1023/B:GLYC.0000014083.48508.6a. [DOI] [PubMed] [Google Scholar]
- [34].Liu F-T, Rabinovich GA. Galectins as modulators of tumour progression. Nat. Rev. Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- [35].Nakahara S, Oka N, Raz A. On the role of galectin-3 in cancer apoptosis. Apoptosis. 2005;10:267–275. doi: 10.1007/s10495-005-0801-y. [DOI] [PubMed] [Google Scholar]
- [36].Lotz MM, Andrews CW, Jr., Korzelius CA, Lee EC, Steele GD, Jr., Clarke A, Mercurio AM. Decreased expression of Mac-2 (carbohydrate binding protein 35) and loss of its nuclear localization are associated with the neoplastic progression of colon carcinoma. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3466–3470. doi: 10.1073/pnas.90.8.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sanjuan X, Fernandez PI, Castells A, Castronovo V, van den Brule FA, Liu F-T, Cardesa A, Campo E. Differential expression of galectin-3 and galectin-1 in colorectal cancer progression. Gastroenterology. 1997;113:1906–1915. doi: 10.1016/s0016-5085(97)70010-6. [DOI] [PubMed] [Google Scholar]
- [38].Honjo Y, Inohara H, Akahani S, Yoshii T, Takenaka Y, Yoshida J, Hattori K, Tomiyama Y, Raz A, Kubo T. Expression of cytoplasmic galectin-3 as a prognostic marker in tongue carcinoma. Clin. Cancer Res. 2000;6:4635–4640. [PubMed] [Google Scholar]
- [39].van den Brule FA, Waltregny D, Liu F-T, Castronovo V. Alteration of the cytoplasmic/nuclear expression pattern of galectin-3 correlates with prostate carcinoma progression. Int. J. Cancer. 2000;89:361–367. doi: 10.1002/1097-0215(20000720)89:4<361::aid-ijc8>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- [40].Califice S, Castronovo V, Bracke M, van den Brule F. Dual activities of galectin-3 in human prostate cancer: tumor suppression of nuclear galectin-3 vs tumor promotion of cytoplasmic galectin-3. Oncogene. 2004;23:7527–7536. doi: 10.1038/sj.onc.1207997. [DOI] [PubMed] [Google Scholar]
- [41].Paron I, Scaloni A, Pines A, Bachi A, Liu F-T, Puppin C, Pandolfi M, Ledda L, Di Loreto C, Damante G, Tell G. Nuclear localization of Galectin-3 in transformed thyroid cells: a role in transcriptional regulation. Biochem. Biophys. Res. Commun. 2003;302:545–553. doi: 10.1016/s0006-291x(03)00151-7. [DOI] [PubMed] [Google Scholar]
- [42].Puglisi F, Minisini AM, Barbone F, Intersimone D, Aprile G, Puppin C, Damante G, Paron I, Tell G, Piga A, Di Loreto C. Galectin-3 expression in non-small cell lung carcinoma. Cancer Lett. 2004;212:233–239. doi: 10.1016/j.canlet.2004.03.006. [DOI] [PubMed] [Google Scholar]
- [43].Davidson PJ, Davis MJ, Patterson RJ, Ripoche M-A, Poirier F, Wang JL. Shuttling of galectin-3 between the nucleus and cytoplasm. Glycobiology. 2002;12:329–337. doi: 10.1093/glycob/12.5.329. [DOI] [PubMed] [Google Scholar]
- [44].Gray RM, Davis MJ, Ruby KM, Voss PG, Patterson RJ, Wang JL. Distinct effects on splicing of two monoclonal antibodies directed against the amino-terminal domain of galectin-3. Arch. Biochem. Biophys. 2008;475:100–108. doi: 10.1016/j.abb.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Roff CF, Wang JL. Endogenous lectins from cultured cells. Isolation and characterization of carbohydrate-binding proteins from 3T3 fibroblasts. J. Biol. Chem. 1983;258:10657–10663. [PubMed] [Google Scholar]
- [46].Morris S, Ahmad N, Andre S, Kaltner H, Gabius HJ, Brenowitx M, Brewer CF. Quarternary solution structures of galectins-1, -3, and -7. Glycobiology. 2004;14:293–300. doi: 10.1093/glycob/cwh029. [DOI] [PubMed] [Google Scholar]
- [47].Gong HC, Honjo Y, Nangia-Makker P, Hogan V, Mazurak N, Bresalier RS, Raz A. The NH2 terminus of galectin-3 governs cellular compartmentalization and functions in cancer cells. Cancer Res. 1999;59:6239–6245. [PubMed] [Google Scholar]
- [48].Davidson PJ, Li S-Y, Lohse AG, Vandergaast R, Verde E, Pearson A, Patterson RJ, Wang JL, Arnoys EJ. Transport of galectin-3 between the nucleus and cytoplasm. I. Conditions and signals for nuclear import. Glycobiology. 2006;16:602–611. doi: 10.1093/glycob/cwj088. [DOI] [PubMed] [Google Scholar]
- [49].Itoh K, Brott BK, Bae GU, Tatcliffe MJ, Sokol SY. Nuclear localization is required for dishevelled function in Wnt/β-catenin signaling. J. Biol. 2005;4:3. doi: 10.1186/jbiol20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nakahara S, Oka N, Wang Y, Hogan V, Inohara H, Raz A. Characterization of the nuclear import pathways of galectin-3. Cancer Res. 2006;66:9995–10006. doi: 10.1158/0008-5472.CAN-06-1772. [DOI] [PubMed] [Google Scholar]
- [51].Nakahara S, Hogan V, Inohara H, Raz A. Importin-mediated nuclear translocation of galectin-3. J. Biol. Chem. 2006;281:39649–39659. doi: 10.1074/jbc.M608069200. [DOI] [PubMed] [Google Scholar]
- [52].Tsay Y-G, Lin NY, Voss PG, Patterson RJ, Wang JL. Export of galectin-3 from nuclei of digitonin-permeabilized mouse 3T3 fibroblasts. Exp. Cell Res. 1999;252:250–261. doi: 10.1006/excr.1999.4643. [DOI] [PubMed] [Google Scholar]
- [53].la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- [54].Henderson BR, Eleftherious A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- [55].Li S-Y, Davidson PJ, Lin NY, Patterson RJ, Wang JL, Arnoys EJ. Transport of galectin-3 between the nucleus and cytoplasm. II. Identification of the signal for nuclear export. Glycobiology. 2006;16:612–622. doi: 10.1093/glycob/cwj089. [DOI] [PubMed] [Google Scholar]
- [56].Cowles EA, Agrwal N, Anderson RL, Wang JL. Carbohydrate- binding protein 35. Isoelectric points of the polypeptide and a phosphorylated derivative. J. Biol. Chem. 1990;265:17706–17712. [PubMed] [Google Scholar]
- [57].Huflejt ME, Turck CW, Lindstedt R, Barondes SH, Leffler H. L-29, a soluble lactose-binding lectin, is phosphorylated on serine 6 and serine 12 in vivo and by casein kinase I. J. Biol. Chem. 1993;268:26712–26718. [PubMed] [Google Scholar]
- [58].Takenaka Y, Fukumori T, Yoshii T, Oka N, Inohara H, Kim HRC, Bresalier RS, Raz A. Nuclear export of phosphorylated galectin-3 regulates its anti-apoptotic activity in response to chemotherapeutic drugs. Mol. Cell. Biol. 2004;24:4395–4406. doi: 10.1128/MCB.24.10.4395-4406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yang RY, Hsu DK, Liu F-T. Expression of galectin-3 modulates T-cell growth and apoptosis. Proc Natl Acad Sci U. S. A. 1996;93:6737–6742. doi: 10.1073/pnas.93.13.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shimura T, Takenaka Y, Tsutsumi S, Hogan V, Kikuchi A, Raz A. Galectin-3, a novel binding partner of β-catenin. Cancer Res. 2004;64:6363–6367. doi: 10.1158/0008-5472.CAN-04-1816. [DOI] [PubMed] [Google Scholar]
- [61].Seve AP, Felin M, Doyennette-Moyne MA, Sahraoui T, Aubery M, Hubert J. Evidence for a lactose-mediated association between two nuclear carbohydrate-binding proteins. Glycobiology. 1993;3:23–30. doi: 10.1093/glycob/3.1.23. [DOI] [PubMed] [Google Scholar]
- [62].Menon RP, Strom M, Hughes RC. Interaction of a novel cysteine and histidine-rich protein with galectin-3 in a carbohydrate-independent manner. FEBS Lett. 2000;470:227–231. doi: 10.1016/s0014-5793(00)01310-7. [DOI] [PubMed] [Google Scholar]
- [63].Bawumia S, Barboni EAM, Menon RP, Hughes RC. Specificity of interactions of galectin-3 with Chrp, a cysteine- and histidine-rich cytoplasmic protein. Biochimie. 2003;85:189–194. doi: 10.1016/s0300-9084(03)00007-5. [DOI] [PubMed] [Google Scholar]
- [64].Goletz S, Hanisch FG, Karsten U. Novel αGalNAc containing glycans on cytokeratins are recognized in vitro by galectins with type II carbohydrate recognition domains. J Cell Sci. 1997;110:1585–1596. doi: 10.1242/jcs.110.14.1585. [DOI] [PubMed] [Google Scholar]
- [65].Park JW, Voss PG, Grabski S, Wang JL, Patterson RJ. Association of galectin-1 and galectin-3 with Gemin4 in complexes containing the SMN protein. Nucleic Acids. Res. 2001;29:3595–3602. doi: 10.1093/nar/29.17.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yoo BC, Hong SH, Ku JL, Kim YH, Shin YK, Jang SG, Kim IJ, Jeong SY, Park JG. Galectin-3 stabilizes heterogeneous nuclear ribonucleoprotein Q to maintain proliferation of human colon cancer cells. Cell Mol Life Sci. 2009;66:350–364. doi: 10.1007/s00018-009-8562-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Liu L, Sadi T, Sano N, Fukui K. Nucling mediates apoptosis by inhibiting expression of galectin-3 through interference with nuclear factor κB signaling. Biochem. J. 2004;380:31–41. doi: 10.1042/BJ20031300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yu X, Siegel R, Roeder RG. Interaction of the B-cell specific transcriptional coactivator OCA-B and galectin-1 and a possible role in regulating BCR-mediated B cell proliferation. J. Biol. Chem. 2006;281:15505–15516. doi: 10.1074/jbc.M509041200. [DOI] [PubMed] [Google Scholar]
- [69].Elad-Sfadia G, Haklai R, Balan E, Kloog Y. Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J. Biol. Chem. 2004;279:34922–24930. doi: 10.1074/jbc.M312697200. [DOI] [PubMed] [Google Scholar]
- [70].Yu F, Finley RL, Jr., Raz A, Kim H-RC. Galectin-3 translocates to the perinuclear membranes and inhibits cytochrome c release from the mitochondria. A role for synexin in galectin-3 translocation. J. Biol. Chem. 2002;277:15819–15827. doi: 10.1074/jbc.M200154200. [DOI] [PubMed] [Google Scholar]
- [71].Voss PG, Gray RM, Dickey SW, Wang W, Park JW, Kasai K, Hirabayashi J, Patterson RJ, Wang JL. Dissociation of the carbohydrate-binding and splicing activities of galectin-1. Arch. Biochem. Biophys. 2008;478:18–25. doi: 10.1016/j.abb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Umemoto K, Leffler H, Venot A, Valafar H, Prestegard JH. Conformational differences in liganded and unliganded states of galectin-3. Biochemistry. 2003;42:3688–3695. doi: 10.1021/bi026671m. [DOI] [PubMed] [Google Scholar]
- [73].Akahani S, Nangia-Makker P, Inohara H, Kim H-R, Raz A. Galectin-3: a novel antiapoptotic molecule with a functional BH1 (NWGR) domain of Bcl-2 family. Cancer Res. 1997;57:5272–5276. [PubMed] [Google Scholar]
- [74].Shi Y, He B, Kuchenbecker KM, You L, Xu X, Mikami I, Yagui-Beltran A, Clement G, Lin YC, Okamoto J, Bravo DT, Jablons DM. Inhibition of Wnt-2 and galectin-3 synergistically destabilizes β-catenin and induces apoptosis in human colorectal cancer cells. Int J Cancer. 2007;121:1175–1181. doi: 10.1002/ijc.22848. [DOI] [PubMed] [Google Scholar]
- [75].Song S, Mazurek N, Liu C, Sun Y, Ding QQ, Liu K, Hung MC, Bresalier RS. Galectin-3 mediates nuclear β-catenin accumulation and Wnt signaling in human colon cancer cells by regulation of glycogen synthase kinase-3β activity. Cancer Res. 2009;69:1343–1349. doi: 10.1158/0008-5472.CAN-08-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Roy AL. Signal-induced functions of the transcription factor TFII-I. Biochim. Biophys. Acta. 2007;1769:613–621. doi: 10.1016/j.bbaexp.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- [79].Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4: a novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Vyakarnam A, Dagher SF, Wang JL, Patterson RJ. Evidence for a role for galectin-1 in pre-mRNA splicing. Mol. Cell. Biol. 1997;17:4730–4737. doi: 10.1128/mcb.17.8.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Fakan S, Leser G, Martin TE. Ultrastructural distribution of nuclear ribonucleoproteins as visualized by immunocytochemistry on thin sections. J. Cell Biol. 1984;98:358–363. doi: 10.1083/jcb.98.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Fakan S, Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int. Rev. Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- [83].Dagher SF, Wang JL, Patterson RJ. Identification of galectin-3 as a factor in pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1213–1217. doi: 10.1073/pnas.92.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wang W, Park JW, Wang JL, Patterson RJ. Immunoprecipitation of spliceosomal RNAs by antisera to galectin-1 and galectin-3. Nucleic Acids Res. 2006;34:5166–5174. doi: 10.1093/nar/gkl673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Peng R, Hawkins I, Link AJ, Patton JG. The splicing factor PSF is part of a large complex that assembles in the absence of pre-mRNA and contains all five snRNPs. RNA Biol. 2006;3:69–76. doi: 10.4161/rna.3.2.3017. [DOI] [PubMed] [Google Scholar]
- [86].Haudek KH, Voss PG, Wang JL, Patterson RJ. A mechanism for galectin-3 incorporation into the spliceosome through its association with U1 snRNP. Biochemistry. 2009;48:7705–7712. doi: 10.1021/bi900071b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA. The U1 small nuclear RNA-protein complex selectively binds a 5’ splice site in vitro. Cell. 1983;33:509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- [88].Hatakeyama S, Sugihara K, Nakayama J, Akama TO, Wong S-MA, Kawashima H, Zhang J, Smith DF, Ohyama C, Fukuda M, Fukuda MN. Identification of mRNA splicing factors as the endothelial receptor for carbohydrate-dependent lung colonization of cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3095–3100. doi: 10.1073/pnas.0810110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- [90].Alonso MD, Lomako J, Lomako WM, Whelan WJ. A new look at the biogenesis of glycogen. FASEB J. 1995;9:1126–1137. doi: 10.1096/fasebj.9.12.7672505. [DOI] [PubMed] [Google Scholar]
- [91].West CM, van der Wel H, Gaucher EA. Complex glycosylation of Skp1 in Dictyostelium: implications for the modification of other eukaryotic cytoplasmic and nuclear proteins. Glycobiology. 2002;12:17R–27R. doi: 10.1093/glycob/12.2.17r. [DOI] [PubMed] [Google Scholar]
- [92].Collins PM, Hidari KI, Blanchard H. Slow diffusion of lactose out of galectin-3 crystals monitored by X-ray crystallography: possible implications for ligand-exchange protocols. Acta Crystallogr. D. Biol. Crystallogr. 2007;63:415–419. doi: 10.1107/S090744490605270X. [DOI] [PubMed] [Google Scholar]