Abstract
WhiD, a member of the WhiB-like (Wbl) family of iron-sulfur proteins found exclusively within the actinomycetes, is required for the late stages of sporulation in Streptomyces coelicolor. Like all other Wbl proteins, WhiD has not so far been purified in a soluble form that contains a significant amount of cluster and characterization has relied on cluster-reconstituted protein. Thus, a major goal in Wbl research is to obtain and characterize native protein containing iron-sulfur clusters. Here we report the analysis of S. coelicolor WhiD purified anaerobically from E. coli as a soluble protein containing a single [4Fe-4S]2+ cluster ligated by four cysteines. Upon exposure to oxygen, spectral features associated with the [4Fe-4S] cluster were lost in a slow reaction that unusually yielded apo-WhiD directly without significant concentrations of cluster intermediates. This process was found to be highly pH dependent with an optimal stability observed between pH 7.0 and 8.0. Low molecular weight thiols, including a mycothiol analogue and thioredoxin, exerted a small but significant protective effect against WhiD cluster loss, an activity that could be of physiological importance. [4Fe-4S]2+ WhiD was found to react much more rapidly with superoxide than with either oxygen or hydrogen peroxide, which may also be of physiological significance. Loss of the [4Fe-4S] cluster to form apo-protein destabilized the protein fold significantly, but did not lead to complete unfolding. Finally, apo-WhiD exhibited negligible activity in an insulin-based disulfide reductase assay demonstrating that it does not function as a general protein disulfide reductase.
WhiB-like (Wbl) proteins are found exclusively in the actinomycetes, a phylum of Gram-positive bacteria that includes Streptomyces, the most abundant source of clinically important antibiotics and other bioactive molecules, and medically important pathogens such as Mycobacterium tuberculosis and Corynebacterium diphtheriae (1, 2). Disruption of wbl genes has shown that Wbl proteins play critical roles in the biology of both Streptomyces and Mycobacterium (1, 3). In Streptomyces coelicolor, WhiB, the founding member of the Wbl family, is required for the initiation of sporulation septation, and WhiD, the subject of this report, is required for the late stages of sporulation (1, 3, 4). In M. tuberculosis, Wbl proteins have been implicated in the ability of the pathogen to persist within its host for long periods of time, despite the low nutrient and oxidative nature of the granuloma, as well as its remarkable tolerance to a wide range of antibiotics (5–9). Recently, WblA has been identified via microarray studies as a pleiotropic regulator of doxorubicin production, as well as morphological differentiation, in Streptomyces peucetius, and Wbl proteins have also been implicated in cell septation/division, stress responses and resistance to antimicrobial drugs (3, 10).
Although genetic analysis has demonstrated the wide biological importance of Wbl proteins, their biochemical function has remained elusive. This issue has been made more intriguing by the demonstration that Wbl proteins bind an iron-sulfur cluster via four highly conserved cysteine residues (see below), and the implicit assumption that this cluster is likely to play a key role in Wbl function (1, 11–13). Consistent with the highly pleiotropic phenotypes of wbl mutants, it has been widely speculated that Wbl proteins function as transcriptional regulators, and there is significant evidence in support of this suggestion (1, 4, 14–16). In the case of M. tuberculosis, yeast two-hybrid studies identified a potential interaction between WhiB3 and a C-terminal fragment (region 4.2) of the principal, essential sigma factor, SigA and, more recently, an E. coli one-hybrid system was used to demonstrate in vivo interaction between WhiB3 and the promoters of several genes whose expression is known to be influenced by whiB3 in vivo (15, 16). This debate has been further stimulated by reports that M. tuberculosis Wbl proteins can function as protein disulfide reductases in their apo-forms (13, 17–19). It was proposed that the Wbl proteins are enzymatically blocked by the binding of an iron-sulfur cluster to the putative catalytic cysteine residues until redox or disulfide stress is encountered, resulting in cluster disassembly and disulfide reductase activity (13). Very recently, a significant step towards understanding the function of Wbl proteins was made by Singh et al (20) who demonstrated strong DNA binding of M. tuberculosis WhiB3 in a cluster-free (apo), oxidized (disulfide bonded) form to the promoter regions of genes involved in polyketide biosynthesis. These observations led the authors to propose a model in which the oxido-reductive stresses encountered by M. tuberculosis during infection lead to cluster loss, with the DNA-binding activity of the resulting apo-protein modulated by its redox state (20).
Wbl proteins are generally small (~10 to 15 kDa) and contain a highly conserved pattern of cysteine residues C(xn)C(x2)C(x5)C (1, 2, 14). We have previously shown that each of the four conserved cysteine residues is essential for the function in vivo of S. coelicolor WhiD, and that, when expressed recombinantly in E. coli under aerobic conditions, colored WhiD inclusion bodies are formed that contain significant amounts of a [2Fe-2S] cluster. Following refolding and anaerobic reconstitution in vitro, WhiD was found to bind a [4Fe-4S] cluster, which was shown to undergo an oxygen-dependent conversion to a [2Fe-2S] form (11). Subsequently, the seven Wbl proteins of M. tuberculosis were also found to contain traces of a [2Fe-2S] cluster following aerobic isolation, and following anaerobic reconstitution, were found to contain an [4Fe-4S] cluster (12, 13). WhiB3, which is important for the virulence of M. tuberculosis in vivo(8), and is the closest homologue of WhiD in M. tuberculosis (see Figure 1), was also found to undergo a [4Fe-4S] to [2Fe-2S] conversion following in vitro reconstitution and exposure to oxygen (8, 12).
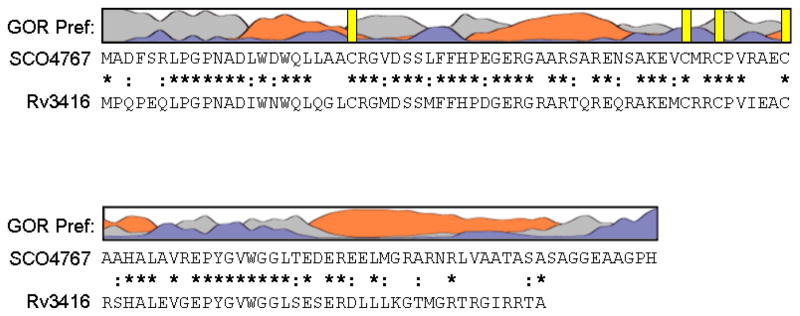
Figure 1. Sequence analysis of WhiD (SCO4767).
The secondary structure of WhiD was predicted using GOR IV (73). The resulting prediction indicated approximately 42% α-helix, 55% coil and 3% beta strand. Preference for α-helix is indicated by orange, β-sheet by blue and coil by grey. Vertical yellow bars indicate the position of cysteine residues. Sequence alignment of WhiD (SCO4767) and WhiB3 (Rv3416) from M. tuberculosis H37Rv is shown. Conserved residues are indicated by a star, conservatively substituted residues are indicated by a colon.
The biological significance of the iron-sulfur cluster found in WhiD and the Wbl proteins of M. tuberculosis is intriguing. In general, iron-sulfur clusters are stable only under anaerobic conditions, and can even assemble in vitro when an appropriate scaffold of ligands is provided (21, 22). However, they are inherently unstable in aerobic conditions (22, 23). In most cases, the polypeptide chain folded around a cluster can control the susceptibility to reactions with oxygen and water in the aerobic environment (21, 23). This opens the way to exploit this susceptibility to sense oxygen, for example (24). Based on the available evidence, it is likely that all Wbl proteins will bind an iron-sulfur cluster and that this cofactor plays a key role in function (1) and recent data indicate that it is the cluster loss reaction together with subsequent oxidation of the coordinating cysteine thiols that is crucial for activity (20). However, to date, all in vitro biochemical and biophysical studies of Wbl proteins, whether from S. coelicolor or M. tuberculosis, have been carried out on proteins produced in E. coli principally as inclusion bodies and/or which have been reconstituted (with cluster) in vitro. Where Wbl proteins have been produced as inclusion bodies, some soluble protein has usually been obtained by varying growth conditions, but the majority of the protein remained in inclusion bodies and cluster incorporation has invariably been to a very low level. Without a functional assay, protein derived from such procedures cannot be authenticated. Therefore, a major goal in Wbl research is to analyse the nature, behavior and reactivities of the iron-sulfur cluster in native protein obtained directly in soluble form with significant cluster incorporation.
Here we report the isolation of a soluble form of WhiD from E. coli, hereafter referred to as native WhiD, which contains a [4Fe-4S]2+ cluster, consistent with our previous study of reconstituted protein. Using UV-visible, CD and resonance Raman spectroscopies, we have investigated in detail the properties of native WhiD and its sensitivity to oxygen, revealing important differences compared with in vitro reconstituted [4Fe-4S] WhiD. An unusual pH sensitivity of oxygen-mediated cluster degradation is reported, along with a significant protecting effect of a physiologically relevant analogue of mycothiol. The sensitivities of native WhiD to hydrogen peroxide and superoxide are also reported, revealing a greatly enhanced rate of reaction with the latter compared to oxygen. Finally, we have investigated the ability of apo-WhiD to function as a protein disulfide reductase. The implications of this work for WhiD function are discussed.
EXPERIMENTAL PROCEDURES
Plasmid Construction
A 0.43 kb fragment carrying an NdeI site overlapping the ATG start codon of whiD, an EcoRI site at the 5′ end, and HindIII site at the 3′ end was generated by PCR using pIJ6626 (3) as template and 5′-GCTACAAGGGAATTCCATATGGCAGATTTCTCCCG-3′ (introducing the NdeI and EcoRI sites), and 5′-GACGCGCTGACCGCGTCGGGAAGCTTGCGGGGCC-3′ (introducing the HindIII site) as primers. The PCR product was digested with EcoRI and HindIII and cloned into EcoRI-HindIII-cut pIJ2925 (25) to create pIJ6630. The resulting allele of whiD was sequenced over its entire length to ensure that only the desired change had been introduced. Finally, the whiD overexpression allele was excised as a 0.43 kb NdeI/BglII fragment and ligated into the expression vectors pET11c and pET15b (Novagen) cut with NdeI and BamHI, generating pIJ6990 and pIJ6631, respectively.
Purification of native-WhiD
Soluble native WhiD was over-produced from plasmid pIJ6631 as a (His)6-tagged protein in aerobic E. coli cultures (BL21 λDE3 Star, Novagen; 37 °C) using LB1 medium supplemented with 100 mg/L ampicillin. Over-production of WhiD was initiated by the addition of 0.4 mM IPTG (50 min, 30 °C) when A600 nm reached ~0.8. To promote the formation of soluble WhiD, cultures were incubated on ice for 18 min prior to induction. To facilitate FeS cluster synthesis, cultures were supplemented with 300 μM ferric ammonium citrate, 75 μM L-methionine and incubated for an additional 3.5 hr at 30 °C. Cells were harvested by centrifugation, washed with lysis buffer (50 mM Tris HCl, 5% Glycerol, 250 mM NaCl, pH 7.3) and stored in an anaerobic freezer (Belle Technology) until needed. Unless otherwise stated, all subsequent purification steps were performed under strict anaerobic conditions in an anaerobic cabinet (Belle Technology), typically operating at ≈2.0 ppm O2 by volume.
Cell pellets were resuspended in lysis buffer with the addition of lysozyme (200 μg/ml), DNaseI (1.3 μg/ml), 1.2 mM PMSF, 3.2 mM benzamidine, 13 mM thymol (26, 27) and 1.7% (v/v) ethanol. The cell suspension was thoroughly homogenised by syringe, passed through a small-bore needle (0.5 mm), transferred to O-ring sealed centrifuge tubes (Nalgene), and centrifuged outside of the cabinet at 40,000 × g for 45 min at 2 °C.
The supernatant was loaded on to a HiTrap chelating column (GE Healthcare) previously charged with NiSO4 and equilibrated with lysis buffer, and then washed with buffer A (50 mM Tris-HCl, 100 mM NaCl, 50 mM imidazole, 5% (v/v) glycerol, pH 7.3). Bound proteins were eluted (1 ml/min) using a linear gradient from 0 to 100% (v/v) buffer B (50 mM Tris-HCl, 100 mM NaCl 500 mM imidazole, 5% (v/v) glycerol, pH 7.3). Fractions (1 ml) containing WhiD were pooled, diluted 10 fold with buffer C (50 mM Tris-HCl, 5% (v/v) Glycerol, pH 7.3) and loaded on to a HiTrap heparin sepharose column (GE Healthcare) previously equilibrated with buffer C. Bound proteins were eluted (0.5 ml/min) using a linear gradient from 0 to 100% (v/v) buffer D (50 mM Tris-HCl, 800 mM NaCl, 5% (v/v) glycerol, pH 7.3). Fractions (0.5 ml) containing WhiD were pooled and stored in an anaerobic freezer until needed.
Purification of reconstituted [4Fe-4S] WhiD
WhiD, lacking a purification tag, was produced in aerobically grown E. coli cultures (BL21 λDE3 Star, Novagen) transformed with pIJ6990. WhiD was isolated from inclusion bodies, refolded and reconstituted, as previously described (11), except that a Q sepharose sepharose column (GE Healthcare) was used in place of a PD10 (GE healthcare) column to remove low molecular weight reactants and to perform buffer exchange (28). Bound protein was eluted with buffer D.
Preparation of apo-WhiD
Native apo-WhiD was prepared from holo-protein using EDTA and potassium ferricyanide, as described by Alam et al. (13) except that a HiTrap Q Sepharose column (GE Healthcare) was used to isolate and concentrate the protein following dialysis. Briefly, the column was equilibrated with 50 mM Tris-HCl, 50 mM KCl, 1 mM DTT, pH 8.0 and bound protein was eluted using a linear gradient between 10 and 100% (v/v) 50 mM Tris-HCl, 800 mM KCl, 1 mM DTT, pH 8.0. Fractions, containing apo-protein were pooled and stored at − 20°C until needed.
Biophysical Characterisation
Sedimentation equilibrium experiments were performed at 25 °C using a Beckman Optima XL-I analytical ultracentrifuge (AUC) fitted with an An50Ti rotor and absorbance optics. Samples (110 μl), in 50 mM Tris-HCl 250 mM NaCl pH 8.0 ±7 mM DTT, containing 12 μM or 24 μM cluster were loaded into 12 mm quartz Epon double sector cells and sealed under anaerobic conditions. Samples were centrifuged at speeds of 22,000 and 25,000 rpm. Absorbance measurements were made at 280 nm and 405 nm once equilibrium was reached, as judged by cessation of changes in scans collected 4 h apart. Buffer served as a reference. Data analysis was performed using Ultrascan II (29). Multiple data sets were simultaneously fitted to a one-component model. The density of the buffer was taken as 1.0099 g/ml and the partial specific volume of (His)6 tagged WhiD was calculated to be 0.7121 ml/g using the program SEDNTERP (30). Gel filtration was carried under anaerobic conditions using the above buffer lacking DTT, a Sephacryl S-100HR HR16/50 column (GE Healthcare), and a flow rate of 1 ml/min.
Equilibrium unfolding experiments were performed anaerobically with apo- and holo-native WhiD (14.1 μM protein) in buffer E (50 mM Tris-HCl, 25 mM NaCl, pH 8.0) using guanidine HCl (GdnHCl), between 0 to 6 M. Samples were incubated anaerobically for ~16 hours at an ambient temperature (~20 °C) prior to far-UV CD measurements. Loss of secondary structure was followed at 222 nm. Tryptophan fluorescence (excitation 280 nm, emission 350 nm) was used to follow thermal denaturation (20 °C to 85 °C) of apo- and holo-proteins (5 μM) in buffer E. The sample was stirred throughout and data was recorded every 1.0 °C with an 8 second averaging time and a 5 nm slit width. Thermodynamic parameters were derived from Van’t Hoff calculations using the Cary Eclipse Bio Software. Changes in the integrity of the iron-sulfur cluster were followed optically over the same temperature range (every 5 °C) by monitoring the ratio of absorbance at 420 and 350 nm.
Stability of the iron-sulfur cluster
To investigate the stability of the iron-sulfur cluster towards oxidative and disulfide stress, aliquots of protein (~7 μM cluster final concentration) and assay buffer (20 mM Mes, 20 mM Tris, 20 mM Bis-Tri-Propane, 100 mM NaCl, 5% (v/v) glycerol, pH 8.0) containing dissolved atmospheric oxygen (234 ± 3 μM) were combined and mixed by inversion in a sealed cuvette outside of the cabinet. Loss of the iron-sulfur cluster was monitored at 406 nm as a function of time. Assays were repeated in the presence of either 2 mM glutathione (GSH), 2 mM glutathiol (oxidised glutathione), 2 mM L-cysteine, 1.8 mM methyl 2(-N-acetyl-L-cysteinyl) amino-2-deoxy-α-D-glucopyrano-side (methyl-mycothiol) (31), 2 mM DL-dithiothreitol (DTT), 2 mM trans-1,2-dithiane-4,5-diol (oxidised DTT) or thioredoxin (360 μM NADPH, 1.5 μM E. coli thioredoxin, 0.2 μM E. coli thioredoxin reductase (Sigma-Aldrich)). Low molecular weight thiols were obtained from Sigma Aldrich and were typically ≥97% purity. Methyl mycothiol was a kind gift (Dr Chris Hamilton, UEA); see Stewart et al (31) for further details. The pH dependence of cluster stability was also investigated using assay buffer containing 2 mM GSH at varying pH values. At pH values ≤5.0 or ≥ 9.0, 20 mM tri-sodium citrate or 20 mM sodium carbonate was added, respectively, to provide additional buffering capacity. Control reactions at pH 8 revealed these components to have no effect on the stability of native WhiD. Experiments to determine the reactivity of the cluster with superoxide ion were carried out using potassium superoxide, KO2 (see other quantitative methods). To confirm the specificity towards superoxide, experiments were repeated in the presence of catalase (1268 units) and/or superoxide dismutase (780 units). Rate constants (ko) were obtained from absorbance data fitted to a single exponential function using Origin (Microcal, Amherst, MA) as previously described (32, 33), In some cases, single exponential fits were not satisfactory and data sets were fitted to a double exponential function and the rate constant of the first phase was taken for the purposes of comparison. Anaerobic assays served as a control.
Protein disulfide reductase assay
To assess whether apo-WhiD can function as a protein disulfide reductase the assay method of Holmgren was used (34). Varying aliquots of apo-WhiD were added to the reaction mixture (63 mM sodium phosphate, 0.13 mM insulin (Sigma-Aldrich), 1 mM EDTA, 1 mM DTT, pH 7.0) to a final concentration between 0 to 25 μM apo-protein. E. coli thioredoxin (Sigma-Aldrich) and E. coli apo-FNR (35) were used as controls. The rate of precipitation of reduced insulin chains was monitored at 650 nm. Enzyme activity was calculated by dividing the slope of the linear portion of the curve by the amount of time required to reach the onset of precipitation (defined as A650nm ≥ 0.05) (13, 17, 34). Buffer alone was used as a control for measuring the intrinsic reduction of insulin by DTT.
Spectroscopy
UV-visible absorbance measurements were made with a Jasco V500 spectrometer. For kinetic measurements with superoxide ions, a fibre optic link (Hellma, Forest Hills, NY) to an anaerobic chamber was utilized to provide necessary rapid mixing and detection. CD measurements were made with a Jasco J810 spectropolarimeter. Far-UV CD was used to characterize the secondary structure of apo- and holo-protein (14.1 μM protein, 1 mm pathlength). Resonance Raman spectra were recorded at 17 K using a Ramanor U1000 spectrometer (Instruments SA, Edison, NJ) and an Innova 10 W-Argon ion laser (Coherent, Santa Clara, CA), with 15 μl frozen droplets of sample mounted on the cold finger of a Displex Model CSA-202E closed cycle refrigerator (Air Products, Allentown, PA). X-band EPR measurements were made with a Bruker EMX spectrometer equipped with a TE-102 microwave cavity and an ESR-900 helium flow cryostat (Oxford Instruments). Spin intensities of paramagnetic samples were estimated by double integration of EPR spectra using 1 mM Cu(II), 10 mM EDTA as the standard. Fluorescence measurements were made using an anaerobic fluorescence cell (1 cm pathlength) on a Varian Cary Eclipse fluorimeter fitted with a peltier controlled thermostatted cell holder.
Other quantitative methods
Protein concentrations were determined using the method of Bradford (BioRad) (36), with bovine serum albumin as the standard. The iron content was determined as previously described (37). Acid-labile sulfide was determined according to the method of Beinert (38). The concentration of dissolved atmospheric oxygen was determined by chemical analysis according to the method of Winkler (39). Hydrogen peroxide solutions were freshly prepared and calibrated as previously described (40). Superoxide (O2−•) solutions (~25 mg/ml potassium superoxide, KO2, Sigma Aldrich) were freshly prepared in a potassium phosphate buffer (50 mM) pH 12.0 (41, 42) and used immediately. Solutions were calibrated using cytochrome c (ε550nm 21,000 M−1 cm−1), and found to contain 2.19 ± 0.17 mM O2−• (41, 43).
RESULTS
Nature of the iron-sulfur cluster
We previously reported that WhiD isolated aerobically from inclusion bodies contained a [2Fe-2S]2+ cluster and that a [4Fe-4S]2+ cluster could be reconstituted in vitro under anaerobic conditions (11). In order to improve the solubility of over-produced WhiD, a hexa-histidine ((His)6) tag was added to the N-terminus. Following optimization of growth conditions, cell lysates were found to contain a soluble, highly colored species, which could be isolated via metal chelate affinity chromatography (see Experimental Procedures).
As isolated, native WhiD had a characteristic straw brown color, displaying an absorbance maximum at 406 nm (ε406 = 16,353 (± 31) M−1 cm−1), together with a broad shoulder at 420 nm, consistent with the presence of a [4Fe-4S] cluster (Figure 2A). Since iron-sulfur proteins derive their optical activity from the fold of the protein to which they are ligated, the CD spectrum provides information about the cluster environment. The CD spectrum of native WhiD contained bands in the 280 to 800 nm region, with three negative features at λmax 325, 398 and 600 nm, and two positive features at λmax 430 and 515 nm. The spectrum is generally similar to that of reconstituted WhiD (inset, Figure 2A), although we note that the negative feature at 325 nm is not clearly resolved in the reconstituted protein spectrum. The Δε values are of the same order of magnitude as in other proteins containing a [4Fe-4S] cluster (44).
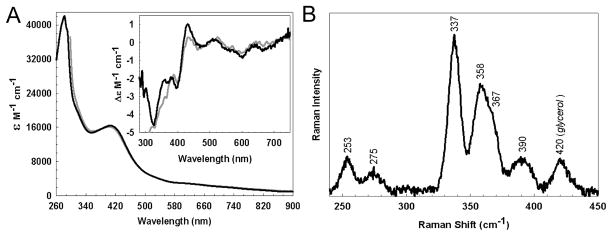
Figure 2. Spectroscopic characterisation of native WhiD.
(A) Optical absorption spectrum and Inset CD spectrum of native WhiD (396 μM [4Fe-4S], 1mm pathlength) in 50 mM Tris 250 mM NaCl 5% (v/v) glycerol, pH 8.0. Extinction coefficients relate to the [4Fe-4S]2+ concentration. The equivalent spectrum arising from in vitro refolded/reconstituted WhiD (dashed gray line) is shown for comparison. (B) Low temperature (17 K) resonance Raman spectrum of native WhiD (0.3 mM) in the Fe-S stretching region using 457.9 nm laser excitation. The spectrum is the sum of 100 scans with each scan involving photon counting for 1 s every 0.5 cm−1 with 6 cm−1 spectral resolution.
The resonance Raman spectrum of native-WhiD (457.9 nm excitation) in the iron-sulfur stretching region (250 to 450 cm−1) is shown in Figure 2B. The Fe-S stretching frequencies and relative resonance enhancements are characteristic of a [4Fe-4S]2+ cluster with complete cysteinyl ligation (45, 46). The bands are readily assigned by analogy with model complexes and simple [4Fe-4S] ferredoxins under idealized Td symmetry (45) (predominantly Fe-Sb stretching modes (cm−1) at 253 (T2), 275 (T1), ~300 (E), 337 (A1), and ~390 (T2), and predominantly Fe-St stretching modes (cm−1) at 358/367 (T2) and ~390 (A1)). Trace amounts of glycerol that remain following buffer exchange are responsible, at least in part, for the 420 cm−1 band.
At 10 K, the EPR spectrum of native WhiD was largely devoid of signals, except for a very weak signal at g = 4.3 indicating a trace component of adventitious ferric ion (not shown). These observations are consistent with a S = 0 [4Fe-4S]2+ cluster that is bound to native-WhiD via four cysteine residues. Attempts to reduce the native-WhiD to the [4Fe-4S]1+ form with sodium ascorbate, sodium dithionite in the presence of methylviologen, and sodium dithionite alone, were unsuccessful. This implies that the cluster, as isolated, has a very low redox potential (below −460 mV at pH 8), and demonstrates a difference between the native and reconstituted protein, for which a dithionite-reduced S = ½ [4Fe-4S]1+ was observed (11).
Biophysical characterization
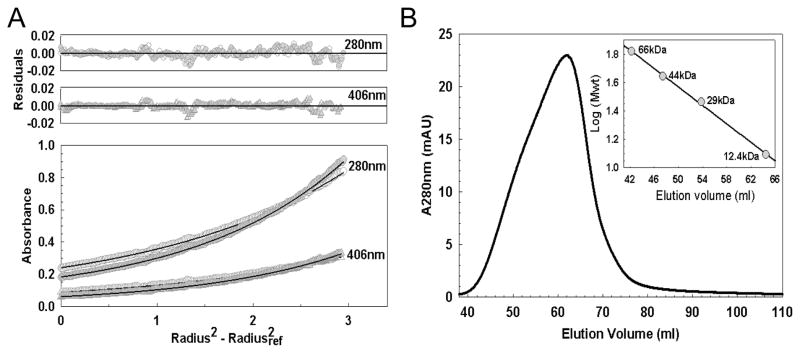
The association state of native WhiD was determined using both equilibrium analytical ultracentrifugation and gel filtration methods. Equilibrium analytical ultracentrifugation data for WhiD at 280 and 405 nm fitted well to a single-component model giving a molecular mass of 14.6 (± 1.0) kDa or 15.3 (± 0.8) kDa, in the presence or absence of 7 mM DTT, respectively (Figure 3A). Using gel filtration, WhiD was found to elute at a volume corresponding to a molecular mass of ~15 kDa (Figure 3B), consistent with the observations above and those made previously with reconstituted material (11). The predicted molecular mass of (His)6-WhiD is 14.1 kDa, clearly demonstrating that the as isolated protein is a monomer. The gel filtration elution peak was somewhat asymmetric, suggesting that a small degree of cysteine side chain oxidation occurred in the absence of a reductant, resulting in a minor component of disulfide bonded dimer. This is consistent with the slight increase in average mass observed by ultracentrifugation in the absence of DTT (Figure 3A). To investigate this further, apo-WhiD and as isolated WhiD were analysed by SDS-PAGE, see Supporting Figure S3A and B. Using a loading buffer lacking DTT a band corresponding to a dimeric form of WhiD was observed at ~29 kDa in addition to the monomeric form at ~15 kDa, and this was significantly more intense in the sample of apo-WhiD. The addition of DTT (250 mM) prior to loading resulted in almost complete loss of the ~29 kDa band, confirming that this was due to an oxidized, disulfide bonded form of the protein. Iron staining of WhiD run on SDS-PAGE in the absence of additional DTT (Supporting Figure S3C) showed that the cluster (or at least iron) was associated with the monomeric form of the protein but not the dimeric form, indicating that the dimeric form is likely to result from disulfide bonded apo-WhiD. Analysis of iron and sulfide contents revealed that WhiD preparations typically contained ≥ 70% holo-protein (based on the assumption that one [4Fe-4S]2+ cluster binds per monomer).
Figure 3. Association state of holo-WhiD.
(A) Equilibrium analytical ultracentrifugation of native-WhiD (21.5 μM) in 50 mM Tris-HCl 250 mM NaCl pH 8.0. The distribution of WhiD was tracked via protein and cofactor absorbancies at 280 nm (circle) and 406 nm (triangle), respectively, in the presence (gray filled) and absence (white filled) of 7 mM DTT, as indicated. Data were fitted to a single component model (black lines, see Experimental procedures) giving a molecular mass of 14.6 kDa and 15.3 kDa in the presence or absence of 7 mM DTT, respectively. Fit residuals are shown above the main plot. (B) Gel filtration chromatogram of native WhiD (450 μM) in 50 mM Tris-HCl 250 mM NaCl pH 8.0. WhiD eluted at a volume corresponding to a molecular mass of ~15 kDa. Inset: Standard calibration curve for the Sephacryl 100HR column.
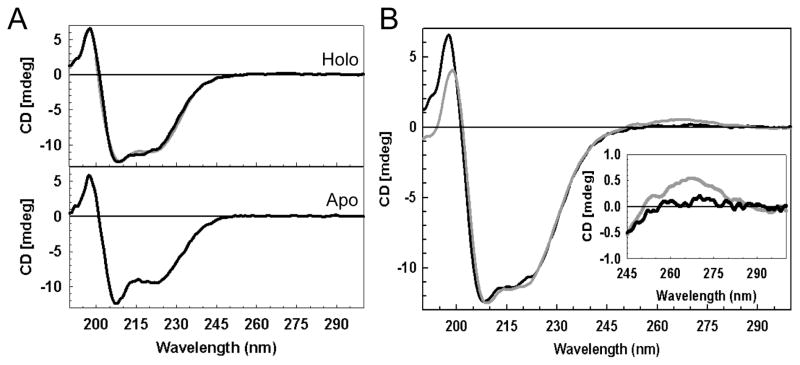
The secondary structure of WhiD, like WhiB3, is predicted to be largely α-helical. Far UV CD spectra for apo- and [4Fe-4S] forms of native WhiD are shown in Figure 4A. The apo-WhiD spectrum was dominated by a negative band at 207 nm, with a weaker feature at 221 nm. We note that this spectrum is similar to that reported by Alam et al. (18) for apo-WhiB3 and is highly reminiscent of CD spectra obtained for 310 helices, which are commonly found in short stretches of secondary structure in proteins (47). In contrast, as isolated [4Fe-4S] WhiD gave a broad negative feature typical of α-helical proteins. An estimation of the secondary structure content was obtained by submitting the far UV CD spectrum to the K2D CD deconvolution server (48). The resulting simulation (Figure 4A, upper panel) indicated approximately 33% α-helix and 17% β-strand. The data indicated that protein conformation is dependent on the iron-sulfur cluster, but also that some secondary structure clearly remained even in the absence of the cluster.
Figure 4. Secondary structure analysis of WhiD.
(A) Far UV CD spectra of native (upper pane) and apo- (lower pane) WhiD (14.1 μM protein) in 50 mM Tris 250 mM NaCl 5% (v/v) glycerol, pH 8.0. A simulation of the native WhiD spectrum generated by the K2D (48) spectral deconvolution programme (gray line, upper pane) is plotted for comparison. (B) Comparison of far UV CD spectra of refolded/reconstituted (gray line) and native (black line) [4Fe-4S] WhiD. Inset is an expanded view of the near-UV CD region.
A comparison of the near UV CD spectra of refolded/reconstituted and native [4Fe-4S] WhiD (see Figure 4B) revealed the presence of a positive band (250 – 300 nm) in reconstituted WhiD which was absent from native [4Fe-4S] WhiD (see Figure 4B, inset) or reconstituted samples prepared from native apo-WhiD (not shown). We note that CD bands in the near-UV (250 – 300 nm) typically reflect the local environment of aromatic amino acid side chains of the protein (49). This implies that in vitro refolded/reconstituted WhiD may have a different conformation in comparison to native WhiD.
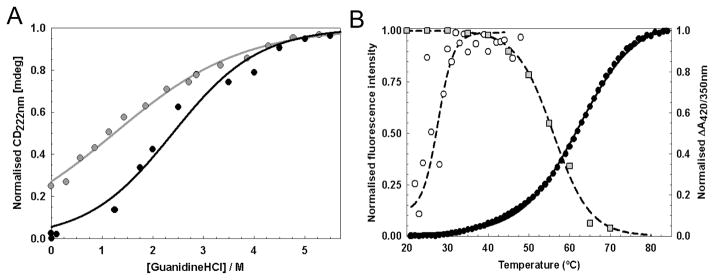
To characterize the relative stabilities of native apo- and holo-WhiD, their chemical (Figure 5A) and thermal (Figure 5B) unfolding properties were investigated (see Table 1). In the case of chemical unfolding, a two-state unfolding model was assumed in order to determine the apparent free energy of unfolding, ΔGunfold, and the transition mid-point, [GdnHCl]½ (50). Holo-WhiD exhibited a sigmoidal transition and fitted well to the two state model with a ΔGunfold, of 7.0 ±0.7 kJ mol−1 and a [GdnHCl]½ of ~2.4 M. For apo-WhiD, a non sigmoidal transition curve was observed, suggesting that the data were associated with the latter portion of a denaturation curve. In order to estimate the stability of apo-WhiD in the absence of denaturant, the initial linear part of the curve was extrapolated back to the intercept with the ordinate axis, which indicated that apo-WhiD is ~25% unfolded at zero denaturant. Based on this, fitting to a two-state unfolding model gave a ΔGunfold of 2.5 ±0.1 kJ mol−1 and a [GdnHCl] ½ of ~1.2 M, indicating that the presence of the cluster significantly stabilizes the protein fold.
Figure 5. Conformational stability of WhiD.
(A) Chemical denaturation of apo- (gray circles) and native-WhiD (black circles) (14.1 μM protein) in 50 mM Tris-HCl, 25 mM NaCl, pH 8.0 with GdnHCl between 0 to 6 M. Fraction of unfolded protein (as normalised intensity at 222 nm) is plotted as a function of GdnHCl concentration. Apo-WhiD displayed a non sigmoidal transition suggesting it is partly unfolded in the absence of denaturant. Assuming apo-WhiD is ~25% unfolded at zero denaturant, both apo- and native-WhiD fitted to a two-state unfolding model (solid lines, see main text). (B) Thermal denaturation of apo- (open circles) and native- WhiD (filled circles) followed by tryptophan fluorescence changes. The thermal stability of the iron-sulfur cluster was followed by plotting the ratio of A420 nm to A350 nm (gray squares, see Table 1). Lines represent fits of the data obtained using the Cary Eclipse Bio Software or Origin (Microcal) to obtain Tm and ΔGunfold values.
Table 1.
Biophysical Characterisation of native WhiD
| [GdnHCl]½ | Tm (°C) | ΔGunfold (kJ.mol−1) | |||
|---|---|---|---|---|---|
| (M) | ΔA406/A350 | ΔFIem350 | Chemical | Thermal | |
| Native WhiD | 2.4 | ~56 | 64.8 ± 2.9 | 7.0 ± 0.7 | 15.1 ± 2.4 |
Thermal denaturation studies were conducted between 20 and 85 °C, in which the folding state of apo- and holo-WhiD was followed by fluorescence. The intrinsic tryptophan flurorescence (FI350 nm) of native holo-WhiD was quenched by the presence of the iron-sulfur cluster and protein unfolding resulted in cluster loss together with a dramatically increased tryptophan emission. Van’t-Hoff analysis of the fluorescence data (see Figure 5B) for holo-WhiD gave a ΔGunfold value of 15.1 ± 2.4 kJ mol−1 and a thermal transition mid-point, Tm, of 64.8 ± 2.9 °C. The thermal stability of the cluster over the same temperature range was monitored by taking the ratio of absorbance at 420 and 350 nm. This gave a Tm of ~56 °C. We note that Leal et al. (50) have also observed a difference in the Tm when comparing tryptophan flurorescence with iron-sulfur absorbance data for the Acidianus ambivalens ferredoxin. We also note that the holo-WhiD sample used here was ~70% replete with cluster, and so the stabilities measured here will be underestimates of the true values. For apo-WhiD, protein unfolding leads to a decrease in the emission intensity of tryptophan and can be followed by plotting the ratio of tryptophan fluorescence emission at 350 nm and 340 nm (50). This revealed a thermal transition between 20 and 40 °C with a Tm of 28.6 ± 0.3 °C see Figure 5B. Further increases in temperature caused a general loss of fluorescence intensity due to precipitation (not shown).
Reaction of [4Fe-4S] WhiD with molecular oxygen, superoxide and hydrogen peroxide
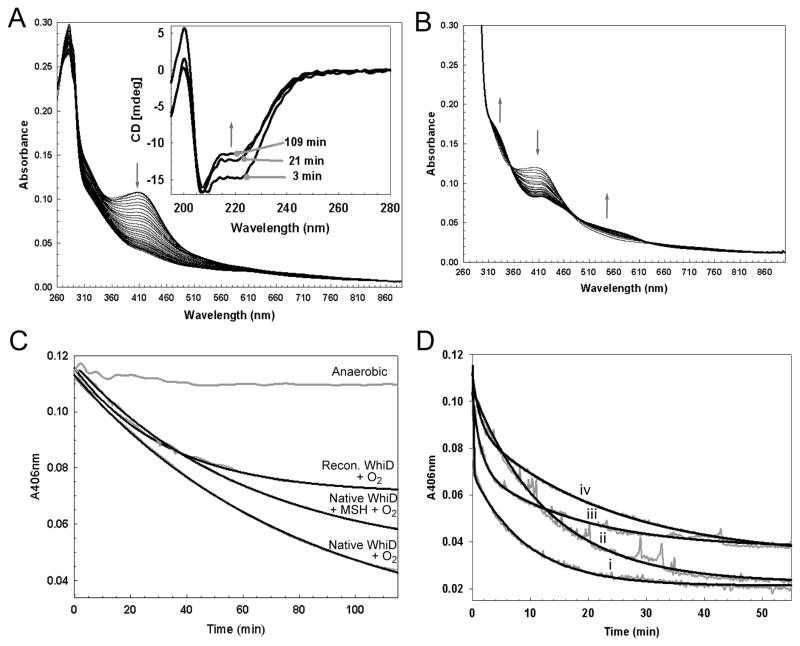
We have previously shown that reconstituted [4Fe-4S] WhiD degrades in the presence of oxygen, first to a [2Fe-2S] cluster, and subsequently to apo-protein (11). Recently, Singh et al (12) reported similar observations for reconstituted WhiB3 from M. tuberculosis. In order to characterize this reaction in more detail, changes in the optical spectrum of native WhiD were monitored on exposure to oxygen. The overlaid time-dependent spectra (Figure 6A), taken at 5 minute intervals, lacked resolved isosbestic points in the 460 nm to 560 nm region and the spectral changes characteristic of a [4Fe-4S] to [2Fe-2S] cluster conversion. Indeed, the final spectrum contained little absorbance in the visible region, indicating that the majority of the protein was in the apo-form. The low intensity residual absorbance is due to unreacted [4Fe-4S] cluster (at 115 min). Reaction of the reconstituted protein (containing between 40 and 70% holo-protein) under identical conditions is shown in Figure 6B; here, the conversion to a [2Fe-2S] form is clearly observed, demonstrating that the reconstituted and native proteins are distinct. A similar gradual loss of the native WhiD [4Fe-4S] signal intensity was observed in the visible CD spectrum (see Supporting Figure S1) and far-UV CD spectra showed a gradual transition from holo- to apo-WhiD (see inset, Figure 6A) consistent with the absence of a stable [2Fe-2S] form of WhiD. The absence of significant levels of [3Fe-4S]+ or [2Fe-2S]2+ cluster intermediates during oxygeninduced degradation of the [4Fe-4S]2+ in native WhiD was also supported by resonance Raman and EPR studies. Oxygen exposure of the Raman sample resulted in decreased intensity of the bands associated with the [4Fe-4S]2+ cluster, relative to the intensity of the ice band at 230 cm−1, without the appearance of the more strongly enhanced Fe-S stretching bands associated with either [3Fe-4S]+ or [2Fe-2S]2+ clusters (data not shown). EPR spectra recorded at increasing times after the addition of O2 revealed the presence of a S = ½ [3Fe-4S]+ signal, but this corresponded to less than 1% of the original cluster concentration, see Supporting Figure S4A. The observed rate constant (ko) of [4Fe-4S] cluster decay was determined by fitting data measured at 406 nm under pseudo first order conditions (in which oxygen was in excess) to a single exponential function (32, 33), see Figure 6C. In the presence of dissolved atmospheric oxygen (~234 μM) native [4Fe-4S] WhiD was converted directly into apo-protein with ko = 12.26 ±0.52 × 10−3 min−1, while reconstituted WhiD was converted into [2Fe-2S] form with ko = 30.38 ±2.57× 10−3 min−1(see Supporting Table S1). To determine whether this was caused by differences in the cluster resulting from in vivo versus in vitro incorporation, or from differences in the protein isolated from soluble or insoluble material, reconstituted [4Fe-4S] WhiD was prepared from apo-native WhiD, resulting in very similar levels of cluster incorporation as observed for WhiD isolated from inclusion bodies. Oxygen reactivity experiments revealed a reactivity very similar to native [4Fe-4S] WhiD (data not shown); therefore, the observed difference in behavior must be derived from differences in the nature of the protein.
Figure 6. Reactivity of the [4Fe-4S] cluster.
(A) Absorption spectra of native-WhiD (~7 μM [4Fe-4S]) following addition to an aerobic buffer (20 mM Tris HCl, 20 mM Mes, 20 mM BisTrisPropane, 100 mM NaCl 5% Glycerol (v/v), pH 8.0); spectra were recorded every 5 min. Inset, Changes in the far UV CD spectra of native-WhiD in response to O2. Spectra were recorded 3, 21 and 109 min after exposure to O2, as indicated. Arrows indicate the direction of movement of spectral features. (B) Absorption spectra of refolded/reconstituted WhiD (~7 μM [4Fe-4S]) following addition to an aerobic buffer; spectra were recorded every 5 min. (C) Plots of A406 nm versus time for anaerobic and aerobic samples of native-WhiD and of refolded/reconstituted WhiD. The data (gray lines) were fitted to a single exponential function (black lines), yielding pseudo first order rate constants. The buffer contained ~234 μM dissolved O2. (D) Plots of ΔA406 nm versus time for the reaction of native WhiD (~7 μM [4Fe-4S]) with i) KO2 (see Supporting Figure S2 for time dependent UV-visible spectra), ii) KO2 plus 780 units superoxide dismutase, iii) KO2 plus 1267 units catalase and iv) KO2 plus superoxide dismutase and catalase, 780 and 1267 units, respectively. The data (gray lines) were fitted to single (ii and iv) or double (i and iii) exponential functions (black lines), yielding pseudo first order rate constants. The buffer contained ~43 μM dissolved superoxide ion (see Experimental procedures). Note that the data are noisy because the experiments were performed with continuous stirring using a spectrometer with a fibre optic link to the anaerobic chamber.
In vivo, it is likely that WhiD will also be exposed to reactive oxygen species during aerobic growth. Therefore, the stability of the cluster towards hydrogen peroxide and superoxide was also investigated. In the presence of hydrogen peroxide (~422 μM) native [4Fe-4S] WhiD was converted directly into apo-protein with ko = 15.47 ± 1.29 × 10−3 min−1, demonstrating that the cluster exhibits a similar reactivity with oxygen and hydrogen peroxide (see Supporting Table S1). EPR studies showed that a higher concentration of [3Fe-4S]1+ was detectable (after 2 mins) but that this corresponded to only ~2 – 4% of the original cluster concentration (depending on the peroxide concentration), and at longer times, the [3Fe-4S]1+ signal became undetectable, see Supporting Figure S4B. Exposure of [4Fe-4S] WhiD to superoxide (added as KO2, see Figure 6D) resulted in data best fitted by a double exponential function with ko1 = 7530 ± 3182 × 10−3 min−1 and ko2 = 93 ± 21 × 10−3 min−1 for the fast and slow phases of the reaction, respectively, indicating a much higher sensitivity to superoxide. However, KO2 yields a mixture of oxygen, superoxide and hydrogen peroxide on dissolution. Therefore, experiments were repeated in the presence of catalase and/or superoxide dismutase (SOD). With catalase present, the rate for the initial rapid phase dropped significantly to give ko1 = 749 ± 36× 10−3 min−1 (with ko2 = 66.2 ± 11.6 × 10−3 min−1). With SOD present, the rate dropped further and the data fitted well to a single exponential with ko = 96 ± 27 × 10−3 min−1. The data confirmed that the rapid reaction was due to superoxide. However, in the absence of hydrogen peroxide but presence of superoxide, the rate was significantly reduced compared to when both were present (ko = ~7500 × 10−3 min−1 compared to ~750 × 10−3 min−1), suggesting that these two reactive oxygen species might exert a cooperative effect. In the presence of both catalase and SOD the data were again well described by a single exponential function with ko = 75 ± 28 × 10−3 min−1. This background reaction, observed in all reactions with KO2, has an associated rate constant significantly greater than that observed for reaction with oxygen alone, suggesting that KO2 in solution likely contains a further reactive component (51). Nevertheless, the effect of this is small compared to that observed for superoxide. Absorbance at 406 nm did not go to or approach zero upon completion of reaction with KO2; UV-visible spectra of [4Fe-4S] WhiD at increasing time points following addition of KO2 (see Supporting Figure S2) were recorded. These revealed that at 5 min intensity had dropped significantly, with a spectrum consistent with the presence of some [2Fe-2S] WhiD. However, this was not stable, as at 25 min, the spectrum was no longer characteristic of an ironsulfur cluster and most likely resulted from iron oxy-hydroxide, iron sulfide or a mixture of these. Consistent with this was the observation of a large EPR signal at g = 4.3 (see Supporting Figure SX), which results from a significant amount of S=5/2 high spin magnetically isolated Fe3+. As for reactions with oxygen and hydrogen peroxide, only very low amounts (~2% of the original cluster) of [3Fe-4S]1+ were detected during the reaction (Supporting Figure S4C).
To investigate further cluster stability to oxygen, absorbance was monitored at 406 nm in the presence of low molecular weight thiol compounds. The pseudo first-order rate constants for cluster loss in the presence of low molecular weight mono-thiols were either unaffected or slightly enhanced in comparison to an untreated sample of native-WhiD (see Supporting Table S1), but the extent of cluster loss was considerably less (≥ 10%) in comparison to untreated samples. In the case of methyl-mycothiol, a synthetic analogue of mycothiol, the natural low molecular weight thiol present in the cytoplasm of actinomycetes, the extent of cluster loss after 115 min was ~45% in comparison to ~62% in an untreated sample (see Figure 6B); somewhat greater protection was obtained with glutathione, cysteine and thioredoxin (see Supporting Table S1). A similar level of protection was observed when glutathione was present in the reaction with hydrogen peroxide. However, no protecting effect was observed with glutathione for reaction with superoxide. The rate constant for reaction was unaffected (see Supporting Table S1) as the extent of cluster degradation (not shown). These observations suggest that mycothiol and/or thioredoxin may play a physiologically relevant role in protecting the cluster in vivo from reaction with oxygen and hydrogen peroxide but not with superoxide.
As observed previously, reactions of [4Fe-4S] WhiD with oxygen carried out in the presence of DTT (2 mM) were found to be biphasic at 505 nm (11). During the reaction time course a gradual shift in λmax, from 406 to 415 nm was observed, together with increased absorbance in the 460 to 600 nm region. This was then followed by further changes in which the spectrum returned to a form closely resembling the starting material. This behavior was not observed in the absence of DTT, or in the presence of low molecular weight mono-thiols such a cysteine or glutathione (not shown), and were not investigated further. The low molecular weight disulfides glutathiol or trans-1,2-dithiane-4,5-diol affected neither the rate nor the extent of cluster loss (see Supporting Table S1).
pH stability
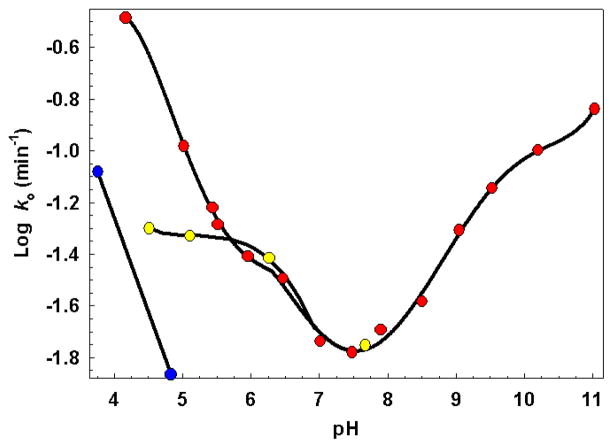
Recently, Geiman et al. (6) reported a 12-fold increase in the expression of WhiB3 in response to an acidic environment. This prompted us to investigate the effect of pH on native holo-WhiD. Figure 7 shows a plot of the pseudo first-order rate constant for oxygen-mediated cluster degradation (monitored at 406 nm) as a function of pH in comparison to equivalent anaerobic samples. In the presence of oxygen the [4Fe-4S] exhibited a remarkably narrow pH stability range of pH 7.0 to 8.0, with the rate constant for cluster degradation increasing ~2 and ~20-fold upon decreasing the pH from 7 to 6 and 7 to 4.2, respectively, and ~7-fold on increasing the pH from 7 to 10.3. In contrast, the addition of methyl-mycothiol (0.5 mM) significantly increased cluster stability, such that the rate constant increased only ~3-fold upon lowering the pH from 7.0 to 4.5, see Figure 7.
Figure 7. pH sensitivity of the [4Fe-4S] cluster.
Pseudo first order rate constants for cluster loss from native-WhiD (7 μM [4Fe-4S]) in aerobic (red circles), aerobic in the presence of methyl mycothiol (yellow circles) and anaerobic (blue circles) buffer are plotted as a function of pH. Solid black lines, derived from polynomial fits, are intended only to indicate overall trends. The buffer was 20 mM Tris HCl, 20 mM Mes, 20 mM BisTrisPropane, 100 mM NaCl 5% Glycerol (v/v), pH as indicated. At pH ≤ 5.5 or ≥ 9.0, 20 mM sodium citrate or 20 mM sodium carbonate, respectively, was added to increase buffer capacity (see Experimental procedures). Solutions contained ~234 μM dissolved O2, 2 mM GSH and 0.5 mM methyl mycothiol as required.
Upon exposure of WhiD to oxygen at pH ≥ 9.5 a new species was formed, which had absorption peaks at 407, 510 and 600 nm. This remained bound to the protein even after returning the sample to pH 7.2, but lacked any CD intensity through the same spectral region (not shown). The far-UV CD spectrum of this species was dominated by a negative band at 207 nm, together with a weaker feature at 221 nm, similar to that observed for apo-WhiD (see Figure 4). EPR analysis of equivalent samples revealed the presence of a signal at g = 4.3, with no obvious low field component (not shown). Although reminiscent of a linear three iron cluster, our observations closely match those of Leal et al. for the formation of iron sulfides bound non-specifically to protein during alkaline cluster degradation (52, 53). Under anaerobic conditions the cluster was stable between pH 6.0 to 11.3, a range comparable to ferredoxin III from C. vinosum (54). Below pH 6, the cluster was unstable, although the rate constant for cluster degradation was always much less than that observed under equivalent aerobic conditions.
Apo-WhiD as a protein disulfide reductase
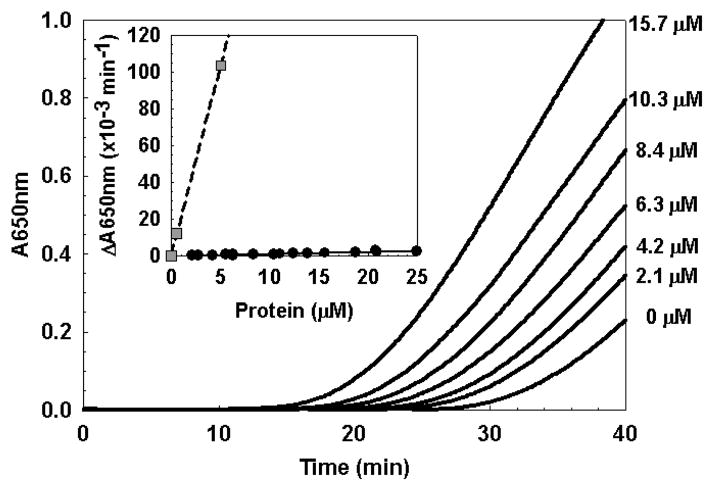
Alam et al. (18) recently provided evidence that apo-WhiB3 can function as general protein disulfide reductase in the insulin disulfide reduction assay originally described by Holmgren (34). Based on the high degree of sequence conservation between WhiD and WhiB3 (see Figure 1), we investigated whether apo-WhiD might also function as a general disulfide reductase. The reduction of insulin was carried out at varying concentrations of apo-WhiD and the apparent reductase activity was calculated according to Garg et al. (17). From Figure 8 it is clear that the rate of insulin reduction was dependent upon the concentration of apo-WhiD. However, the activity of WhiD was minimal in comparison to that of E. coli thioredoxin (see Table 2) and WhiB3 (18). For comparison, assays were also conducted in the presence of apo-FNR, an iron-sulfur cluster containing O2 sensor from E. coli (35, 55), which contains a CxxC motif within its sequence, but has no known function in thiol-disulfide exchange. This showed that apo-FNR has a somewhat higher activity than apo-WhiD. These data indicate that WhiD does not function as a general disulfide reductase.
Figure 8. General protein disulfide reductase activity of apo-WhiD.
Insulin disulfide reduction assay of native apo-WhiD measured as an increase in A650 nm due to the precipitation of insulin caused by the reduction of the soluble oxidized form. The concentration of WhiD in each experiment is indicated. Inset is a plot of the relative reductase activity as a function of protein concentration for native apo-WhiD and E. coli thioredoxin.
Table 2.
Protein disulfide reductase activities of WhiD.
| Sample | Conc. (μM) | Reductase activity (× 10−3) (Ave. ΔA650 nm min−2) |
|---|---|---|
| Apo WhiD | 0.00 | 0.00 (±0.00) |
| 2.10 | 0.09 (±0.04) | |
| 6.26 | 0.41 (±0.06) | |
| 13.82 | 1.19 (±0.42) | |
| 15.64 | 1.62 (±0.12) | |
| 20.74 | 2.17(±0.35) | |
| Apo WhiDa | 3.15 | 0.30 (±0.06) |
| Holo WhiDb | 4.19 | 0.13 (±0.05) |
| Apo-FNR | 2.00 | 0.12 (±0.03) |
| ResA | 20.00 | 7.50c |
| Thioredoxin | 0.53 | 12.31 (±0.45) |
| 5.00 | 104.70 (±1.14) | |
Contained in vitro refolded apo-WhiD isolated from inclusion bodies
Conducted anaerobically.
Calculated from Lewin et. al. (71)
DISCUSSION
In this work we have succeeded in identifying and optimizing growth conditions that allow S. coelicolor WhiD to be expressed in E. coli in a soluble, folded form with ≥70% [4Fe-4S] cluster incorporation following anaerobic purification. This has permitted detailed biophysical and spectroscopic analyses of a cluster bound form of WhiD that has not been resolubilized from inclusion bodies and has not undergone cluster reconstitution in vitro. This is important in the context that, to date, all studies of this class of protein, whether from S. coelicolor or M. tuberculosis, have been carried out on material isolated from E. coli under aerobic conditions, either from insoluble inclusion bodies (which have been solubilized and refolded) (11, 13, 18, 19) or from soluble material that has required in vitro reconstitution in order to generate a significant proportion of cluster containing protein (12, 13, 18, 19). Furthermore, whereas reconstitution has resulted only in [4Fe-4S] cluster formation, aerobic isolation has generated only the [2Fe-2S] form. Because of the lack of an in vitro assay, uncertainty persists about the functional significance of the different forms of Wbl proteins (1). Production of the protein in an E. coli host under conditions that yielded a folded protein with significant in vivo cluster insertion gives some reassurance that the native form of WhiD has been generated here. A detailed biophysical characterization was carried out to define the native state of the protein; these demonstrated that the protein is monomeric, with one [4Fe-4S]2+ cluster ligated by four conserved cysteine residues.
Native WhiD, containing ~70% holo-protein, gave a far-UV CD spectrum characteristic of a protein with significant α-helical content. In equilibrium unfolding experiments, native WhiD exhibited a sigmoidal transition that fitted well to a two-state unfolding model whereas apo-WhiD gave a hyperbolic curve suggesting that the latter is unstable to unfolding following cluster loss. The conformational stabilities (ΔGunfold) of apo- and native-WhiD as determined by chemical denaturation and thermal methods showed that apo-WhiD is much less stable than native WhiD, demonstrating that the [4Fe-4S] cluster contributes significantly to the stability of the WhiD protein fold. Some iron-sulfur cluster proteins undergo partial or complete unfolding upon removal of the cluster (56). Far UV CD data indicated that loss of the cluster did not lead to complete unfolding of WhiD, such that a significant amount of secondary structure was retained. This finding is significant in light of the recent report that M. tuberculosis WhiB3 binds DNA in its apo-form (20).
Following proposals that WhiD may function as an oxygen sensor in S. coelicolor (11), that WhiB3 may be an oxygen sensor involved in the metabolic downturn of M. tuberculosis (12, 57), and that Wbl proteins appear to be involved in stress response (6, 58), we explored the reactivity and chemistry of the [4Fe-4S]2+ cluster in the native state of WhiD with oxygen, hydrogen peroxide, and superoxide ions, as well as thiols, in order to survey reactions with potential biological relevance. All of these reagents had effects ranging from cluster destruction to stabilization. Under anaerobic conditions, the [4Fe-4S]2+ cluster of native WhiD was stable between pH 6.0 to 11.3, a range comparable to that of typical ferredoxins such as FdIII from C. vinosum (54). Upon exposure to oxygen or hydrogen peroxide, the native WhiD [4Fe-4S]2+ cluster reacted slowly, with loss of both iron and sulfide, such that significant concentrations of cluster intermediates (such as [3Fe-4S]1+ or [2Fe-2S]2+) were not observed, yielding apo-WhiD. The initial reaction with oxygen/peroxide may, therefore, represent the rate limiting step, whereby intermediates do not accumulate. The observed rate with oxygen was ~270 times slower than that observed under similar conditions for [4Fe-4S] FNR (59), a known bacterial oxygen sensor, and was more comparable to those reported for other iron-sulfur proteins such as [4Fe-4S] aconitase (60) and FdxH2 from Anabaena variabilis (61) that are not known to play a role in oxygen sensing, even though oxygen may be required to initiate cluster degradation. These results contrast with the oxygen reaction of the [4Fe-4S] cluster in the refolded/reconstituted form of WhiD, which is converted into a stable [2Fe-2S] cluster with spectral features similar to the cluster in WhiD isolated under aerobic conditions (11). Similar observations have been reported for in vitro reconstituted WhiB3 (12). Furthermore, a comparison of CD spectra for refolded/reconstituted and native-WhiD revealed subtle differences in the UV, between 250–350 nm. These reflect differences in the local environment of the aromatic amino acid residues and the [4Fe-4S] cluster, and are likely to be important for the observed differences in reactivity with oxygen (44, 49).
Since inclusion bodies typically contain partially folded protein aggregates and those of WhiD and WhiB3 contain ≤ 30% of a [2Fe-2S] cluster, formation of this cluster may be favored by an incorrectly folded protein (11, 12, 18, 62). Importantly, control reactions using reconstituted material prepared from apo-native WhiD revealed behavior very similar to native [4Fe-4S] WhiD, implying that the [4Fe-4S] to [2Fe-2S] cluster conversion, in this case, is a particular property of in vitro refolded/reconstituted WhiD. Hence the reaction of the WhiD cluster with oxygen is a relatively slow reaction (t½ ~ 57 min) that proceeds via a mechanism that is dependent on the protein fold. Importantly, the data indicate that cluster loss for the native protein involves a direct conversion from [4Fe-4S] to apo-WhiD and, furthermore, that apo-WhiD is susceptible to oxidation, resulting in a disulfide-bonded dimer. These observations are likely to be physiologically important because they indicate that the previously reported [2Fe-2S] form is not a stable intermediate of cluster breakdown, and that the apo-protein can undergo further redox reaction (20).
The oxygen reaction also depended on pH. The [4Fe-4S] cluster of native WhiD exhibited a relatively narrow range of stability from pH 7.0 to 8.0, with the rate constant for cluster degradation increasing ~20-fold upon decreasing the pH from 7.0 to 4.2. Proton dependent redox processes have been noted in a variety of iron-sulfur proteins but the unusually narrow pH profile for WhiD indicates a greater than normal sensitivity to acidic conditions. Geiman et al (6) recently investigated the expression profile of all seven of the M. tuberculosis wbl genes in response to a series of physiological and antimicrobial stresses. Interestingly, a ~12-fold increase in WhiB3 expression in response to an acidic environment (~ pH 4.5) was reported. Survival mechanisms employed by both Gram-positive and Gram-negative bacteria in response to an acid extracellular environment maintain the cytoplasmic pH close to neutrality irrespective of the extracellular pH (63–65). In the case of Streptomyces spp., optimal growth typically occurs between pH 6.5 and 8.0 (66). Remarkably, Mycobacterium tuberculosis can persist in vivo despite phagosomal acidification by activated macrophages (67). This leads to the induction of fatty acid degrading enzymes, DNA repair proteins as well as a remodeling of the cell envelope (68). Therefore, WhiB3 and WhiD may be involved in the cell’s response to pH stress resulting from a significant drop in the pH of the environment.
In the presence of low molecular weight thiols the extent of cluster loss was considerably reduced upon exposure to oxygen. Mycothiol [1-D-myo-inosityl-2-(N-acetylcysteinyl)amido-2-deoxy-α-D-glucopyranoside], an abundant low molecular weight thiol found at millimolar concentrations in most actinomycetes, serves as the major thiol redox buffer for the cell (69). In the presence of methyl-mycothiol (31), a synthetic analogue of mycothiol, and thioredoxin, the extent of cluster loss was significantly reduced compared to an untreated sample. Furthermore, methyl-mycothiol also significantly reduced the sensitivity of native WhiD to low pH, with only a ~3-fold increase in the rate constant upon decreasing the pH from 7.0 to 4.5. These observations suggest mycothiol and/or thioredoxin may play a physiologically important role in protecting the cluster in vivo and may also explain why [4Fe-4S] WhiD can be produced in the glutathione-rich cytoplasm of E. coli grown under aerobic conditions.
The relative stability of [4Fe-4S] WhiD in the presence of both O2 and hydrogen peroxide suggests that this form of WhiD is likely to be found under conditions of mild oxidative stress. However, it should be noted that a sensor that reacts rapidly with its analyte is not always required; the ability to sense prolonged stress over a very long period might be important for metabolic and/or morphological decision making (for example, whether or not to undergo sporulation). Indeed, Geiman et al (6) reported a 3-fold increase in the expression of WhiB3 in response to peroxide stress in M. tuberculosis and the observed rate of oxygen (and perhaps peroxide) induced WhiD cluster loss is comparable to that observed for WhiB3 (12). The much more rapid cluster reaction with the superoxide ion, a by-product of aerobic metabolism, demonstrates a kinetic selectivity of WhiD that may have important physiological consequences. Here, too, conversion from the [4Fe-4S] to apo-form of WhiD was observed, with little or no [3Fe-4S]1+ cluster observed. However, UV-visible absorbance changes suggested the transient formation of a [2Fe-2S] cluster, but this was highly unstable leading to further degradation. We note with interest that, in S. coelicolor, the expression of sodN (SCO5254) is increased in cells under acidic extra-cellular conditions, leading to increased levels of SOD (70). This suggests a possible connection between pH sensitivity and ROS stress.
Recently Alam et al. (18) proposed that apo-WhiB3 can function as a disulfide reductase, in which the WhiB3 catalytic thiols are held inactive by binding of an iron-sulfur cluster until redox or disulfide stress leads to loss of the iron-sulfur cluster, thereby releasing the coordinating cysteines to participate in thiol-disulfide chemistry as a disulfide reductase (13, 18). Our attempts to perturb the stability of native WhiD by simulating oxidative/disulfide stress using a 285-fold molar excess of either GSSG or oxidized DTT in the presence of oxygen were unsuccessful. Using the insulin assay to test for general disulfide reductase activity (34), apo-WhiD was found to exhibit some activity, which was linearly dependent upon the concentration of the protein, but, in comparison to E. coli thioredoxin and M. tuberculosis apo-WhiB3, apo-WhiD was, respectively, ~317 and ~17 fold less efficient (mol for mol). Furthermore, apo-WhiD was found to have a similar activity to other proteins that have no known function in thiol-disulfide exchange, such as E. coli apo-FNR, and was also significantly less active than B. subtilis ResA, a specific thiol-disulfide oxidoreductase involved in cytochrome c maturation, which is believed to require interaction with specific substrates for activation (35, 71). Taken together, the data demonstrate that apo-WhiD does not function as a general disulfide reductase. However, it cannot be discounted that apo-WhiD functions as a target specific reductase, as recently proposed for WhiB1(72).
It was shown recently that M. tuberculosis WhiB3 is active as a DNA binding protein in its cluster-free form (20). If S. coelicolor WhiD functions similarly, then understanding how the protein undergoes cluster loss and its subsequent redox chemistry will be of great physiological importance. The data presented here provide significant new insight into the nature of the WhiD [4Fe-4S] cluster and how a range of physiologically relevant molecules influence (positively or negatively) conversion to apo-WhiD.
Supplementary Material
Acknowledgments
This work was supported by the UK’s Biotechnology and Biological Sciences Research Council grant through the award of grant BB/D00926X/1 to MJB and AJT and Wellcome Trust awards from the Joint Infra-structure Fund for equipment. AJT holds a Leverhulme Emeritus Fellowship. Raman studies at the University of Georgia were supported by grant GM62524 from the NIH to MKJ.
We are grateful to: Christopher J. Hamilton (UEA) and Vishnu Karthik Jothivasan (UEA) for their kind gift of Methyl 2(-N-acetyl-L-cysteinyl) amino-2-deoxy-α-D-glucopyrano-side (methyl-mycothiol); Dennis R. Dean (University of Virginia) for pDB551 encoding NifS; Jeffrey Green (University of Sheffield) for pGS572 encoding FNR; Tom Clarke (UEA) for assistance using the analytical ultracentrifuge and to Maureen Bibb (JIC) and Nick Cull (UEA) for technical assistance. Andrew Thomson holds a Leverhulme Trust Emeritus Fellowship.
Footnotes
The abbreviations used are: BisTrisPropane, 1,3-bis(tris(hydroxymethyl)methylamino)propane; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetate; GdnHCl, guanidine hydrochloride; GSH, glutathione; LB, Luria-Bertani; IPTG, isopropyl-β–D-thiogalactoside; Mes, 2-(N-morpholino) ethanesulfonate;. PMSF, phenylmethanesulphonylfluoride; Tris, 2-amino-2-(hydroxymethyl)-1,3-propanediol.
SUPPORTING INFORMATION AVAILABLE
[4Fe-4S] WhiD cluster reactivities are given in Table S1; The reactivity of the [4Fe-4S] cluster monitored by CD spectroscopy is shown in Figure S1; The reactivity of the [4Fe-4S] cluster with superoxide is shown in Figure S2; Analysis of apo- and as isolated WhiD by SDS-PAGE is shown in Figure S3; EPR spectroscopy data following oxidation of [4Fe-4S] WhiD is shown in Figure S4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.den Hengst CD, Buttner MJ. Redox control in actinobacteria. Biochim Biophys Acta. 2008;1780:1201–1216. doi: 10.1016/j.bbagen.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Soliveri JA, Gomez J, Bishai WR, Chater KF. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology. 2000;146(Pt 2):333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- 3.Molle V, Palframan WJ, Findlay KC, Buttner MJ. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2) J Bacteriol. 2000;182:1286–1295. doi: 10.1128/jb.182.5.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis NK, Chater KF. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol Gen Genet. 1992;232:351–358. doi: 10.1007/BF00266237. [DOI] [PubMed] [Google Scholar]
- 5.Morris RP, Nguyen L, Gatfield J, Visconti K, Nguyen K, Schnappinger D, Ehrt S, Liu Y, Heifets L, Pieters J, Schoolnik G, Thompson CJ. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2005;102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geiman DE, Raghunand TR, Agarwal N, Bishai WR. Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob Agents Chemother. 2006;50:2836–2841. doi: 10.1128/AAC.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 2004;84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Banaiee N, Jacobs WR, Jr, Ernst JD. Regulation of Mycobacterium tuberculosis whiB3 in the mouse lung and macrophages. Infect Immun. 2006;74:6449–6457. doi: 10.1128/IAI.00190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 10.Kang SH, Huang J, Lee HN, Hur YA, Cohen SN, Kim ES. Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. J Bacteriol. 2007;189:4315–4319. doi: 10.1128/JB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakimowicz P, Cheesman MR, Bishai WR, Chater KF, Thomson AJ, Buttner MJ. Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe-4S] cluster. J Biol Chem. 2005;280:8309–8315. doi: 10.1074/jbc.M412622200. [DOI] [PubMed] [Google Scholar]
- 12.Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, Giles GI, Lancaster JR, Jr, Steyn AJ. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc Natl Acad Sci USA. 2007;104:11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alam MS, Garg SK, Agrawal P. Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: a [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol Microbiol. 2007;63:1414–1431. doi: 10.1111/j.1365-2958.2007.05589.x. [DOI] [PubMed] [Google Scholar]
- 14.Green J, Crack JC, Thomson AJ, Le Brun NE. Bacterial sensors of oxygen. Curr Opin Microbiol. 2009;12:145–151. doi: 10.1016/j.mib.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Guo M, Feng H, Zhang J, Wang W, Wang Y, Li Y, Gao C, Chen H, Feng Y, He ZG. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009;19:1301–1308. doi: 10.1101/gr.086595.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steyn AJ, Collins DM, Hondalus MK, Jacobs WR, Jr, Kawakami RP, Bloom BR. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci USA. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg SK, Suhail Alam M, Soni V, Radha Kishan KV, Agrawal P. Characterization of Mycobacterium tuberculosis WhiB1/Rv3219 as a protein disulfide reductase. Protein Expr Purif. 2007;52:422–432. doi: 10.1016/j.pep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Alam MS, Agrawal P. Matrix-assisted refolding and redox properties of WhiB3/Rv3416 of Mycobacterium tuberculosis H37Rv. Protein Expr Purif. 2008;61:83–91. doi: 10.1016/j.pep.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Alam MS, Garg SK, Agrawal P. Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. FEBS J. 2009;276:76–93. doi: 10.1111/j.1742-4658.2008.06755.x. [DOI] [PubMed] [Google Scholar]
- 20.Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibney BR, Mulholland SE, Rabanal F, Dutton PL. Ferredoxin and ferredoxin-heme maquettes. Proc Natl Acad Sci USA. 1996;93:15041–15046. doi: 10.1073/pnas.93.26.15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 23.Tilley GJ, Camba R, Burgess BK, Armstrong FA. Influence of electrochemical properties in determining the sensitivity of [4Fe-4S] clusters in proteins to oxidative damage. Biochem J. 2001;360:717–726. doi: 10.1042/0264-6021:3600717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jervis AJ, Crack JC, White G, Artymiuk PJ, Cheesman MR, Thomson AJ, Le Brun NE, Green J. The O2 sensitivity of the transcription factor FNR is controlled by Ser24 modulating the kinetics of [4Fe-4S] to [2Fe-2S] conversion. Proc Natl Acad Sci USA. 2009;106:4659–4664. doi: 10.1073/pnas.0804943106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen GR, Bibb MJ. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 26.Vasala A, IsomaÌ̂ki R, Myllykoski L, Alatossava T. Thymol-triggered lysis of Escherichia coli expressing Lactobacillus phage LL-H muramidase. Journal of Industrial Microbiology and Biotechnology. 1999;22:39–43. [Google Scholar]
- 27.Bennis S, Chami F, Chami N, Bouchikhi T, Remmal A. Surface alteration of Saccharomyces cerevisiae induced by thymol and eugenol. Lett Appl Microbiol. 2004;38:454–458. doi: 10.1111/j.1472-765X.2004.01511.x. [DOI] [PubMed] [Google Scholar]
- 28.Crack JC, Le Brun NE, Thomson AJ, Green J, Jervis AJ. Reactions of nitric oxide and oxygen with the regulator of fumarate and nitrate reduction, a global transcriptional regulator, during anaerobic growth of Escherichia coli. Methods Enzymol. 2008;437:191–209. doi: 10.1016/S0076-6879(07)37011-0. [DOI] [PubMed] [Google Scholar]
- 29.Cantor CR, Schinmel PR. Ultrascan II Reference. 2. II. WH Freeman and Company; New York: 1980. [Google Scholar]
- 30.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart MJ, Jothivasan VK, Rowan AS, Wagg J, Hamilton CJ. Mycothiol disulfide reductase: solid phase synthesis and evaluation of alternative substrate analogues. Org Biomol Chem. 2008;6:385–390. doi: 10.1039/b716380k. [DOI] [PubMed] [Google Scholar]
- 32.Crack JC, Green J, Cheesman MR, Le Brun NE, Thomson AJ. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc Natl Acad Sci USA. 2007;104:2092–2097. doi: 10.1073/pnas.0609514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton VR, Mettert EL, Beinert H, Kiley PJ. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ Cluster. J Bacteriol. 2004;186:8018–8025. doi: 10.1128/JB.186.23.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 35.Crack JC, Jervis AJ, Gaskell AA, White GF, Green J, Thomson AJ, Le Brun NE. Signal perception by FNR: the role of the iron-sulfur cluster. Biochem Soc Trans. 2008;36:1144–1148. doi: 10.1042/BST0361144. [DOI] [PubMed] [Google Scholar]
- 36.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 37.Crack JC, Green J, Le Brun NE, Thomson AJ. Detection of sulfide release from the oxygen-sensing [4Fe-4S] cluster of FNR. J Biol Chem. 2006;281:18909–8913. doi: 10.1074/jbc.C600042200. [DOI] [PubMed] [Google Scholar]
- 38.Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 39.Vogel AI. Vogel’s Textbook of Quantitative Chemical Analysis. 5. Longman; 1989. [Google Scholar]
- 40.Crack J, Green J, Thomson AJ. Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR) J Biol Chem. 2004;279:9278–9286. doi: 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- 41.Unger F, Westedt U, Hanefeld P, Wombacher R, Zimmermann S, Greiner A, Ausborn M, Kissel T. Poly(ethylene carbonate): A thermoelastic and biodegradable biomaterial for drug eluting stent coatings? J Contr Rel. 2007;117:312–321. doi: 10.1016/j.jconrel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Lokesh BR, Cunningham ML. Further studies on the formation of oxygen radicals by potassium superoxide in aqueous medium for biochemical investigations. Toxicol Lett. 1986;34:75–84. doi: 10.1016/0378-4274(86)90147-5. [DOI] [PubMed] [Google Scholar]
- 43.Vandewalle PL, Petersen NO. Oxidation of reduced cytochrome c by hydrogen peroxide. Implications for superoxide assays. FEBS Lett. 1987;210:195–198. doi: 10.1016/0014-5793(87)81336-4. [DOI] [PubMed] [Google Scholar]
- 44.Stephens PJ, Thomson AJ, Dunn JB, Keiderling TA, Rawlings J, Rao KK, Hall DO. Circular dichroism and magnetic circular dichroism of iron-sulfur proteins. Biochemistry. 1978;17:4770–4778. doi: 10.1021/bi00615a026. [DOI] [PubMed] [Google Scholar]
- 45.Czernuszewicz RS, Macor KA, Johnson MK, Gewirth A, Spiro TG. Vibrational mode structure and symmetry in proteins and analogues containing Fe4S4 clusters: Resonance Raman evidence for different degrees of distortion in HiPIP and ferredoxin. J Am Chem Soc. 1987;109:7178–7187. [Google Scholar]
- 46.Brereton PS, Duderstadt RE, Staples CR, Johnson MK, Adams MWW. Effect of serinate ligation at each of the iron sites of the [Fe4S4] cluster of Pyrococcus furiosus ferredoxin on the redox, spectroscopic, and biological properties. Biochemistry. 1999;38:10594–10605. doi: 10.1021/bi990671d. [DOI] [PubMed] [Google Scholar]
- 47.Yokum TS, Bursavich MG, Gauthier T, Hammer RP, McLaughlin ML. 310-Helix stabilization via side-chain salt bridges. Chem Commun. 1998:1801–1802. [Google Scholar]
- 48.Andrade MA, Chacon P, Merelo JJ, Moran F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Prot Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 49.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Leal SS, Gomes CM. Studies of the molten globule state of ferredoxin: structural characterization and implications on protein folding and iron-sulfur center assembly. Proteins. 2007;68:606–616. doi: 10.1002/prot.21448. [DOI] [PubMed] [Google Scholar]
- 51.Kehrer JP. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology. 2000;149:43–50. doi: 10.1016/s0300-483x(00)00231-6. [DOI] [PubMed] [Google Scholar]
- 52.Leal SS, Teixeira M, Gomes CM. Studies on the degradation pathway of iron-sulfur centers during unfolding of a hyperstable ferredoxin: cluster dissociation, iron release and protein stability. J Biol Inorg Chem. 2004;9:987–996. doi: 10.1007/s00775-004-0599-z. [DOI] [PubMed] [Google Scholar]
- 53.Leal SS, Gomes CM. Linear three-iron centres are unlikely cluster degradation intermediates during unfolding of iron-sulfur proteins. Biol Chem. 2005;386:1295–1300. doi: 10.1515/BC.2005.147. [DOI] [PubMed] [Google Scholar]
- 54.Maskiewicz R, Bruice TC. Dependence of the rates of dissolution of the Fe4S4 clusters of Chromatium vinosum high-potential iron protein and ferredoxin on cluster oxidation state. Proc Natl Acad Sci USA. 1977;74:5231–5234. doi: 10.1073/pnas.74.12.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Outten FW. Iron-sulfur clusters as oxygen-responsive molecular switches. Nat Chem Biol. 2007;3:206–207. doi: 10.1038/nchembio0407-206. [DOI] [PubMed] [Google Scholar]
- 56.Reyda MR, Dippold R, Dotson ME, Jarrett JT. Loss of iron-sulfur clusters from biotin synthase as a result of catalysis promotes unfolding and degradation. Arch Biochem Biophys. 2008;471:32–41. doi: 10.1016/j.abb.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muttucumaru DG, Roberts G, Hinds J, Stabler RA, Parish T. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb) 2004;84:239–46. doi: 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 58.Kim TH, Park JS, Kim HJ, Kim Y, Kim P, Lee HS. The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem Biophys Res Commun. 2005;337:757–764. doi: 10.1016/j.bbrc.2005.09.115. [DOI] [PubMed] [Google Scholar]
- 59.Crack JC, Gaskell AA, Green J, Cheesman MR, Le Brun NE, Thomson AJ. Influence of the environment on the [4Fe-4S]2+ to [2Fe-2S]2+ cluster switch in the transcriptional regulator FNR. J Am Chem Soc. 2008;130:1749–1758. doi: 10.1021/ja077455+. [DOI] [PubMed] [Google Scholar]
- 60.Kennedy MC, Emptage MH, Dreyer JL, Beinert H. The role of iron in the activation-inactivation of aconitase. J Biol Chem. 1983;258:11098–11105. [PubMed] [Google Scholar]
- 61.Singh BB, Curdt I, Jakobs C, Schomburg D, Bisen PS, Bohme H. Identification of amino acids responsible for the oxygen sensitivity of ferredoxins from Anabaena variabilis using site-directed mutagenesis. Biochim Biophys Acta. 1999;1412:288–294. doi: 10.1016/s0005-2728(99)00069-9. [DOI] [PubMed] [Google Scholar]
- 62.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 63.Boot IR, Cash P, O’Byrne C. Sensing and adapting to acid stress. Antonie Van Leeuwenhoek. 2002;81:33–42. doi: 10.1023/a:1020565206835. [DOI] [PubMed] [Google Scholar]
- 64.Booth IR. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. 1985;49:359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foster JW. When protons attack: microbial strategies of acid adaptation. Curr Opin Microbiol. 1999;2:170–174. doi: 10.1016/S1369-5274(99)80030-7. [DOI] [PubMed] [Google Scholar]
- 66.Kontro M, Lignell U, Hirvonen MR, Nevalainen A. pH effects on 10 Streptomyces spp. growth and sporulation depend on nutrients. Lett Appl Microbiol. 2005;41:32–38. doi: 10.1111/j.1472-765X.2005.01727.x. [DOI] [PubMed] [Google Scholar]
- 67.Vandal OH, Nathan CF, Ehrt S. Acid resistance in Mycobacterium tuberculosis. J Bacteriol. 2009;191:4714–4721. doi: 10.1128/JB.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional Adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Newton GL, Fahey RC. Mycothiol biochemistry. Arch Microbiol. 2002;178:388–394. doi: 10.1007/s00203-002-0469-4. [DOI] [PubMed] [Google Scholar]
- 70.Kim YJ, Moon MH, Song JY, Smith CP, Hong SK, Chang YK. Acidic pH shock induces the expressions of a wide range of stress-response genes. BMC Genomics. 2008;9:604. doi: 10.1186/1471-2164-9-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewin A, Crow A, Hodson CT, Hederstedt L, Le Brun NE. Effects of substitutions in the CXXC active-site motif of the extracytoplasmic thioredoxin ResA. Biochem J. 2008;414:81–91. doi: 10.1042/BJ20080356. [DOI] [PubMed] [Google Scholar]
- 72.Garg S, Alam MS, Bajpai R, Kishan KR, Agrawal P. Redox biology of Mycobacterium tuberculosis H37Rv: protein-protein interaction between GlgB and WhiB1 involves exchange of thiol-disulfide. BMC Biochem. 2009;10:1. doi: 10.1186/1471-2091-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.