Abstract
Although over expression and 15N enrichment facilitate the observation of resonances from disordered proteins in Escherichia coli, 15N enrichment alone is insufficient for detecting most globular proteins. Here we explain this dichotomy and overcome the problem while extending the capability of in-cell NMR by using 19F labeled proteins. Resonances from small (~10 kDa) globular proteins containing the amino acid analog 3-fluoro-tyrosine can be observed in cells, but for larger proteins the 19F resonances are broadened beyond detection. Incorporating the amino acid analog trifluoromethyl-L-phenylalanine allows larger proteins (up to 100 kDa) to be observed in cells. We also show that site specific structural and dynamic information about both globular and disordered proteins can be obtained inside cells by using 19F NMR.
Keywords: 19F NMR, in-cell NMR, intrinsically disordered proteins, site specific labeling, protein structure, protein dynamics
Most proteins function inside cells under crowded and complex conditions, where the concentration of macromolecules can reach ~400 g/L.1–3 Studying proteins in the cellular interior, although difficult, is important for understanding how environments affect functions. In-cell NMR provides a means to assess protein structure, function, and interactions with other proteins, DNA and small molecules at atomic resolution in living cells.4–19 Recently, the high-resolution NMR structure of a small, 66 residue, protein in the cytosol of Escherichia coli has been reported.20
The success of in-cell NMR experiments depends on overcoming several obstacles. As currently practiced, in-cell NMR in E. coli requires protein over expression, which may lessen its biological significance. Current practice also requires growth on nutrients enriched in NMR-active nuclei, usually 15N or 13C. Normal metabolism of these nutrients causes a background spectrum that obscures signals from the protein being studied. Proteins that leak from the cell also cause artifacts.21 Furthermore, the crowded intracellular environment broadens resonances from globular proteins, lowering the sensitivity of NMR experiments. For instance, specific methyl labeling, a technique typically used only for large proteins, was required to obtain sufficient long-range structure restraints for the small protein cited above. Augustus et al. showed that the repressor protein MetJ is completely undetectable in E. coli because of weak, non-specific DNA binding,4 and indetectability has been reported for other globular proteins.11
Since natural proteins contain no fluorine, this 100% abundant spin-½ nucleus with its high sensitivity (83% of 1H), spectral simplicity, and large chemical shift range is attractive for protein NMR in vitro and in cells.22–2519F in-cell NMR was first applied to detect protein mobility in the yeast Saccharomyces cerevisiae,26–28 and a preliminary study has been reported in E. coli.29 Here we describe detailed studies using this bacterium.
We examine one disordered and five globular proteins containing 15N and/or 19F. More specifically, we incorporate the fluorinated amino acid analogs, 3-fluoro-tyrosine (3FY) and trifluoromethyl-L-phenylalanine (tfmF) into proteins ranging in size from 7 kDa to 100 kDa.
Materials and Methods
Expression Systems
The ubiquitin (UBQ),6 calmodulin (CAM),30 and α-synuclein (αSYN) expression systems were gifts from Alexander Shekhtman (State University of New York at Albany), Anthony Persechini (University of Missouri, Kansas City), and Peter Lansbury (Harvard), respectively. pET28a plasmids (Novagen) containing the gene for truncated chymotrypsin inhibitor 2 (CI2)31 or PDZ332 domain were a gift from Andrew Lee (University of North Carolina at Chapel Hill). The GFP and histidinol dehydrogensase (HDH) expression system has been described.29,33 For 15N enrichment, the plasmids were transformed into BL-21(DE3-Gold) competent cells. The CAM, CI2, and PDZ3 transformants were spread onto Luria broth agar plates containing 60 μg/mL kanamycin, the others were spread onto plates containing 60 μg/mL ampicillin.
15N Enrichment and 3FY Labeling
The procedure was similar to that described by Khan et al.34 and Li et al.24 Details are given in the Supporting Information.
tfmF Labeling
Amber stop codons (TAG) were incorporated at the sites for tfmF labeling by using site-directed mutagenesis (QuickChange, Stratagene) of the target genes, which are present in the arabinose-inducible expression vector, pBAD. The labeling procedure was similar to that described by Hammill et al.35 Details are given in the Supporting Information.
Preparing for in-cell NMR
Cultures (usually ~100 mL) were centrifuged at 1,200g for 30 min at room temperature. The cell pellets were resuspended in 2 mL of LB media. The samples, comprising 90:10 mixtures of cell slurry:D2O, were placed in 5-mm NMR tubes for data acquisition. Supernatants were collected by centrifugation (Eppendorf, model 5418, 2000g for 10 min) after the experiments to assess leakage.21 The pellets were resuspended in buffer (50-mM Tris, pH 8.0) to a final volume of 1 mL. Lysates were made from the resuspended pellets by sonication (Fisher Scientific, Sonic Dismembrator Model 500) on ice for 10 min with a duty cycle of 2 s on, 5 s off. The lysate was collected after centrifugation at 16,000g for 10 min. Viscosities were measured with a Viscolite 700 viscometer (Hydramotion Ltd., England).
NMR
15N-1H-HSQC spectra were acquired on a cold-probe equipped Varian Inova 500-MHz spectrometer at 25 °C. The 1H dimension had a sweep width of 8401 Hz and comprised 1024 complex points. The 15N dimension had a sweep width of 2200 Hz and comprised 64 complex points. The data were processed with NMRPipe36 and NMRDraw.37 19F spectra were acquired at 37 °C on a Varian Inova 600-MHz spectrometer equipped with a 5 mm 19F {H} z-gradient probe. The spectra comprised 128 to 2048 transients, a 30 kHz sweep width, and a 2 s delay before acquisition. 19F chemical shifts are referenced to trifluoroethanol at 0 ppm.
Results
3FY Labeled, 15N Enriched α–Synuclein (αSYN)
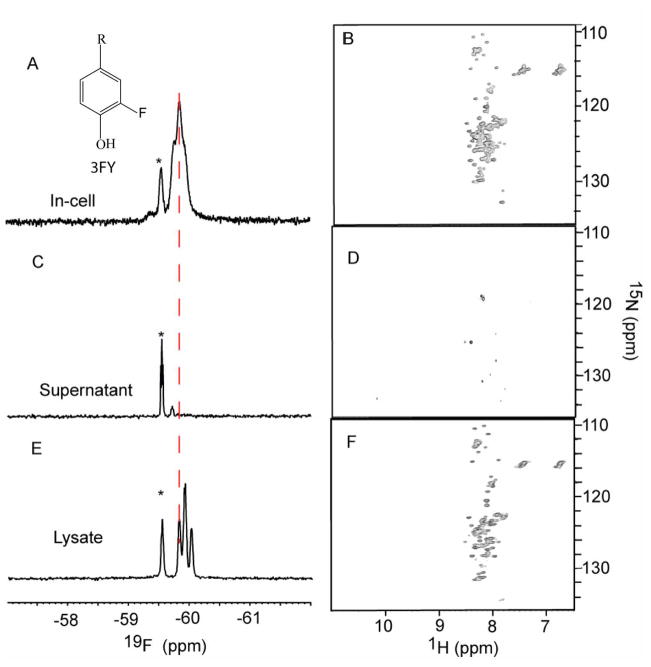
There are four tyrosines in this 140 residue, intrinsically disordered protein, one at position 39 and three near the C-terminus, at positions 125,133 and 136. We labeled all these residues with 3FY. As shown in Figure 1A, the 19F spectrum of the cell slurry shows a broad protein resonance at ~−60 ppm and a sharp resonance from free 3FY at −59.6 ppm, The assignment of the αSYN resonance was confirmed by comparison to the spectrum of the purified protein. The assignment of the 3FY resonance was confirmed by comparison to the supernatant spectrum. The 15N-1H HSQC spectrum of the cell slurry (Figure 1B) shows numerous protein crosspeaks, consistent with previous work.10,38 To check for leakage, we subjected the cell slurry to centrifugation and examined the supernatant. The presence of only the free 3FY resonance in the 19F spectrum (Figure 1C) and the near absence of crosspeaks in the HSQC spectrum (Figure 1D) indicate that little or no αSYN had leaked. The cells were then lysed by sonication, the cellular debris removed by centrifugation, and the clear lysate examined by NMR. The 19F resonances sharpened (Figure 1E), revealing three protein peaks that shifted upfield by ~0.1 ppm. The 19F spectrum has been assigned.24 The middle peak comprises 3FY resonances from residues 39 and 125. The downfield and upfield resonances correspond to residues 136 and 139, respectively. The crosspeaks in the HSQC spectrum of the lysate are sharper than those from the cell slurry, but the spectrum is essentially unchanged. The limited chemical shift dispersion of the 19F and 1H resonances show that αSYN is disordered in cells, consistent with other work.10,38
Figure 1.
19F spectra of 3FY labeled αSYN (left panels) and 1H-15N HSQC spectra of 15N-enriched αSYN (right panels). Panels A and B show in-cell spectra. The inset in panel A shows the structure of 3FY. Panels C and D show spectra of supernatants collected immediately after completing the in-cell spectra. Panels E and F show spectra of supernatants from the clear lysates. The asterisks indicate the free 3FY resonances. The dashed vertical line shows the upfield shift on cell lysis.
tfmF Labeled αSYN
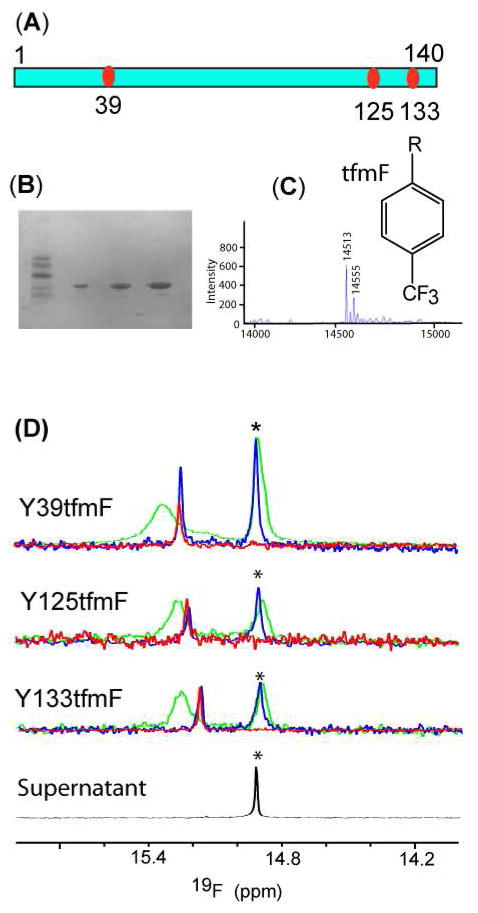
To overcome the incomplete resolution of the four 19F resonances from 3FY labeled tyrosines in cells (Figure 2A), we labeled the protein with tfmF at three of the four tyrosines by using an orthogonal aminoacyl synthase system.29 Before performing in-cell NMR experiments, we assessed the system by purifying the labeled protein (Figure 2B) and using mass spectrometry to confirm the expected 52 Da increase in mass, from 14461 to 14513 Da. The peak at 14555 is labeled and acetylated protein.
Figure 2.
Sites of tfmF incorporation in α-synuclein (A). SDS-PAGE of the three purified tfmF labeled α-synucleins (B). ESI-mass spectrum of tfmF39 labeled α–synuclein (C). The inset shows the structure of tfmF. 19F spectra of labeled synuclein (D). Spectra from cell slurries are shown in green. Spectra from clear lysates are shown in blue. Spectra from purified tfmF labeled proteins are shown in red. Spectra from supernatants collected immediately after the in-cell NMR experiments are shown in black. The asterisks indicate the free tfmF resonances.
The in-cell 19F spectra for proteins labeled at positions 39, 125 and 133 are shown as green traces in Figure 2D. The tfmF 39 resonance is broader than the tfmF 125 and 133 resonances in the cell slurry. The resonances from the lysates and from the purified proteins are narrower and shift upfield by ~0.1 compared to those from the cell slurry. Only a free tmfF resonance is observed in the supernatants, showing that the proteins do not leak.
19F Labeled, 15N Enriched Ubiquitin (UBQ)
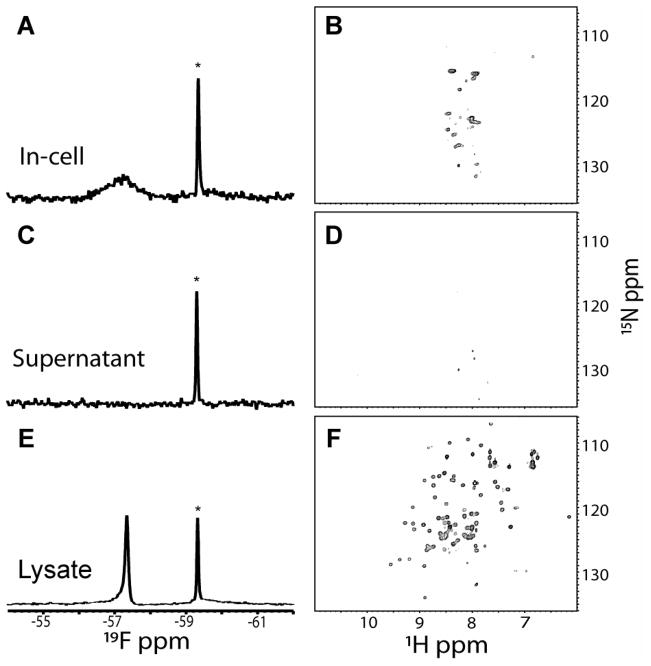
This 8 kDa globular protein has one tyrosine. Figure 3A shows the 19F spectrum of the cell slurry. The spectrum contains a sharp free 3FY resonance and a broad protein resonance. The identity of these resonances was confirmed by comparisons to spectra of the purified protein and the supernatant. Figure 3B shows the HSQC spectrum from the slurry. Only metabolite signals39 are observed. Figures 3C and 3D show the 19F and HSQC spectra from the supernatant collected immediately after the in-cell NMR experiment. Only the free 3FY resonance is present in the 19F spectrum, and the HSQC spectrum is nearly blank. These observations show that UBQ has not leaked from the cells. The cells were then lysed. The 19F lysate spectrum (Figure 3E) shows a single sharp protein resonance and the HSQC spectrum (Figure 3F) closely resembles that of pure UBQ.40 We also collected HSQC data on the globular, 11 kDa PDZ3 domain of PSD9532,41 in cells. Like UBQ, the PDZ3 domain is expressed at mM levels (Figure S1, Table S1), but its HSQC spectrum cannot be obtained from the cell slurry. The protein signals, however, appear upon lysis (Figure S2). We do not understand our inability to reproduce the published results on UBQ, which has been reported to yield high resolution spectra in E. coli.6,42 Our use of a different growth medium is not the reason, because we obtain similar results to those shown in Figure 3 when we use the media described in the publications. We also tried expressing the protein at different temperatures without success. Our studies were conducted on a cold-probe equipped 500 MHz instrument. Lack of sensitivity does not explain our inability to detect UBQ in cells because we obtain the same results with a cold-probe equipped 700 MHz spectrometer.
Figure 3.
19F- and 1H-15N HSQC- spectra of 15N-enriched, 3FY labeled UBQ. The panels are labeled as described in the caption to Figure 1.
Freezing cells prior to in-cell NMR studies has been suggested.6 We prepared another 15N enriched UBQ sample for in-cell experiments, but stored the sample at −20 °C overnight. The sample was thawed and used to collect an in-cell spectrum. The spectrum of native UBQ,40 which is not observed in a fresh sample (Figure 3B), is visible in the previously frozen sample (Figure S3). Storing the sample at −80 °C gives the same result. Adding 10% (v/v) glycerol decreases, but does not always prevent, leakage. We conclude that cells should not be frozen if they are to be studied by using in-cell NMR.
3FY Labeled, 15N Enriched Chymotrypsin Inhibitor 2 (CI2)
This 7 kDa globular protein has one tyrosine. Figure 4A shows the 19F spectrum of 3FY labeled cell slurry. Three resonances are observed. The sharpest resonance is from free 3FY. The other two resonances are from CI2. Both have a chemical shift of −59.2 ppm. One protein resonance is broad, with a width at half height of ~1.5 ppm. The other resonance is sharper and superimposed on the broad resonance. The HSQC spectrum of the cell slurry (Figure 4B) shows a spectrum almost identical to that of purified CI2.31 The spectrum from the supernatant collected immediately after the in-cell experiment contains a resonance from both free 3FY and 3FY labeled CI2 (Figure 4C). The HSQC spectrum of the supernatant (Figure 4D) is almost identical to the spectrum from the cell slurry (Figure 4A). These data show that CI2 has leaked from the cells, consistent with previous work.38 After lysis (Figure 4E), only free 3FY and a single sharp resonance from the labeled protein is observed. The HSQC spectrum of the lysate is identical to the HSQC spectrum from the cell slurry. Comparing the three 19F spectra, suggests that the broad resonance at −59.2 ppm in the cell slurry is intracellular CI2 and the superimposed sharper resonance is from CI2 that has leaked from the cells.
Figure 4.
19F spectra of 3FY labeled CI2 (left panels) and 1H-15N HSQC spectra of 15N-enriched CI2. The panels are labeled as described in the caption to Figure 1.
tfmF Labeled CI2
Figure 5 shows the 19F spectra of CI2 labeled at positions 18 and 42 in cells and lysates. The protein resonances have a width at half height of ~0.20 ppm in cells. They shift upfield by 0.15–0.20 ppm and narrow to ~0.03 ppm upon lysis. There are no protein signals from the supernatants collected after the NMR experiments, indicating that tfmF labeled CI2 does not leak. This result is surprising considering the results from the 3FY labeled protein (Figure 4). As discussed below, a lower expression level may explain the absence of leakage. The small signals near the free tfmF may be a degradation product of labeled CI2 or a tfmF metabolite.
Figure 5.

19F spectra of K18tfmF CI2 in cells (green) and lysates (blue) (A), Y42tfmF CI2 in cells and in lysates (B), and supernatants collected after the in-cell NMR experiments (C). The asterisks indicate the free tfmF resonances.
3FY Labeled, 15N Enriched Calmodulin (CAM)
This 16 kDa two-lobed globular protein has two tyrosine residues. Figure 6A shows the 19F spectrum of the 3FY labeled cell slurry. Three resonances are evident. The sharp resonance is from free 3FY. The other two, one on either side of the 3FY resonance, are from the protein.43 The HSQC spectrum from the slurry (Figure 6B) shows only metabolite signals.38,39 Figure 6C and 6D show the 19F spectrum and the HSQC spectrum from the supernatant collected immediately after the in-cell NMR experiment. Only free 3FY is observed in the 19F spectrum, and the HSQC spectrum is nearly devoid of crosspeaks, indicating that CAM does not leak from the cells. Figure 6E shows the 19F spectrum of the cleared lysate. The broad protein resonances observed in the cell slurry narrow on lysis, but the width at half height for the broadest resonance is still >0.5 ppm. The observation of CAM crosspeaks43,44 in the HSQC spectrum of the lysate (Figure 6F) proves that detectable amounts of the protein are present. The HSQC spectrum of the lysate also shows that CAM is not fully Ca2+ loaded.43
Figure 6.
19F- and 1H-15N HSQC- spectra of 15N-enriched, 3FY labeled CAM. The panels are labeled as described in the caption to Figure 1.
3FY Labeled, 15N Enriched Green Fluorescence Protein (GFP)
This 27 kDa globular protein contains 12 tyrosines. The resonances in the 19F and 15N-1H HSQC spectra are too broad to detect in cells and lysates (Figure S4), but SDS-PAGE analysis and the fluorescence of the samples show that the protein is over expressed (Figure S1, Table S1).
tfmF Labeled GFP
tfmF might be a better label for larger proteins because the trifluromethyl group adds rotational motion that is independent of molecular tumbling. The green traces in Figure 7 show the 19F spectra of GFP labeled at position 39 and position 221 in cells. The 19F resonances from the two proteins in cells are broad, with widths at half height of ~0.4 ppm, but observable. The corresponding resonances from the purified protein are narrower, with widths of <0.1 ppm. The only resonance in the supernatant from the cell slurry is from free tfmF, which shows that labeled GFP does not leak. Lysis caused an upfield shift of 0.10–0.15 ppm.
Figure 7.
19F spectra of tfmF 39 labeled GFP in cells (green) and the purified protein in solution (blue) (A), tfmF 221 labeled GFP in cells and the purified protein in solution (B), and supernatants collected after the in-cell NMR experiments (C). The asterisks indicate the free tfmF resonances.
tfmF Labeled Histidinol Dehydrogensase (HDH)29
We applied the tfmF labeling method to this 98 kDa homodimer. The 19F spectra are presented in Figure 8. As shown by comparisons to spectra for the purified protein and the supernatant, the sharp resonance in the cell slurry spectrum is from free tfmF and the broad resonance (width at half height of ~1.0 ppm) is from HDH. The only resonance in the supernatant is from free tfmF, showing that labeled HDH does not leak.
Figure 8.
19F spectra of L225tfmF HDH. In-cell sample (A), purified protein (B), supernatant collected after the in-cell NMR experiments (C). The asterisks indicate the resonance from free tfmF.
Discussion
We used NMR to study six proteins enriched in 15N and/or labeled with 19F in E. coli cells. The proteins are present in the cytoplasm [although some αSYN33 and CI2 (Figure S5) is periplasmic]. Two 19F labeling strategies were used. One strategy, incorporating 3FY in place of tyrosine, was accomplished by expressing the protein in 15N enriched minimal media containing 3FY, phenylalanine, tryptophan, and N-(phosphomonoethyl) glycine.34 The other strategy involved an orthogonal tRNA synthase system29 to replace residues with tfmF.
A one dimensional 19F spectrum can be acquired in minutes (compared to an hour for 15N-1H HSQC spectra shown here), which allows the study of proteins near their physiological concentrations. We estimate an intracellular concentration of the tfmF labeled proteins of 50 to 100 μM from the areas of the free tfmF and the protein resonances, the tfmF concentration in the media, and the fact that the cells occupy half the slurry volume of NMR samples. This concentration equals that of the most abundant soluble E. coli proteins.45,46 Furthermore, these 19F experiments can be performed as a function of time to obtain data on signal transduction and metabolism.
19F labeling is well suited to assess leaking. Controls must be performed to ensure the protein of interest is inside the cells during the NMR experiment.21 For CI2 (Figure 4), we see a sharp 3FY resonance from leaked protein and a broad resonance from intracellular protein. By comparing the signal intensity in the supernatant (leaked CI2) to that in the lysate (total CI2) we estimate that 5–10% of the protein leaks from the cells. Importantly, this small fraction of leaked CI2 accounts for 100% of the CI2 signal in the HSQC spectrum of the cell slurry.
To assess the effect of the expression system on leakage, we repeated the experiments in the same E. coli strain [BL21(DE3)] with CI2 under control of the araBAD promoter33 rather than the T7 promoter. We did not observe CI2 resonances in the HSQC spectrum from the cell slurry, but we did observe CI2 resonances in the lysate (Figure S6). These experiments confirm the indetectability of CI2 HSQC spectra in cells and suggest that the expression system affects protein leakage. For our pBAD experiment, however, leaked protein might not be detected because CI2 expression was also lower [~0.2 mM compared to 1 mM in BL21(DE3)]. We repeated the experiment using strain DH10B, but expression was so low that CI2 crosspeaks were not observable even in the lysate. Additionally, in opposition to what has been recommended,6 storing cells in the freezer should be avoided. As we have shown for UBQ (Figure S3), the freeze-thaw cycle disrupts a fraction of the cells, spilling the enriched protein into the surrounding dilute solution. We have shown elsewhere that encapsulating the cells controls leakage.38 In summary, although more experiments are required to deconvolute the effects of the expression system, expression level, and strain, 19F provides a straightforward assay for leakage.
19F labeling extends the utility of in-cell NMR for studying intrinsically disordered proteins. For αSYN, we observe both backbone 15N and side chain19F signals in cells, although the resonances from the cells are broader than those from lysates and dilute samples (Figure 1). Due to the limited chemical shift dispersion of disordered proteins, tfmF labeling (Figure 2) is preferred over 3FY labeling (Figure 1) because any natural, ribosomally encoded, amino acid can be replaced with tfmF. Moreover, tfmF labeling provides dynamic information at a single site. For α-synuclein in cells, the tfmF 39 resonance is broader than the C-terminal tfmF resonances. This observation indicates constrained motion at position 39, consistent with reports that position 39 has residual structure while the C-terminal region is completely disordered.24,47,48 By the same reasoning, the increased width of the tfmF 39 resonance in GPF compared to the tfmF 221 resonance (Figure 7) indicates that the side chain at position 221 is more mobile than the side chain at position 39. Such dynamic information is masked in dilute solution studies of purified proteins because the difference in the intrinsic line width is small in dilute solution.
Counter to the utility of 15N enrichment for in-cell studies of disordered proteins, we do not observe 15N signals from globular proteins in cells. Similar problems have been observed elsewhere,4,11 and Sakakibara et al. report the instance of a 7 kDa globular protein that is amenable to in-cell NMR in one E. coli strain, but not another.20 We can exclude several causes for our failure to detect the HSQC spectra of the globular proteins studied here. It is not insufficient expression. The data in Table S1 show that the 15N enriched proteins are expressed at mM levels, which should allow detection. We can also rule out over expression because the low concentration of tfmF labeled proteins in cells (50–100 μM) relative to 15N enriched proteins (mM) still leads to broad 19F resonances. Augustus et al.4 showed that DNA binding explains the absence of an in-cell HSQC spectrum from the MetJ protein, but this is not a reasonable explanation for our results because the proteins are not DNA binders and all have pI values of 6.5 or less. We can rule out insolubility because the proteins are found in the supernatant of the lysates, not in the pellets. Strong membrane binding is also excluded because membranes are found in the pellets. The proteins also appear to be mostly, and perhaps completely, in their native states because HSQC spectra from lysates of cells expressing UBQ, CI2, CAM, apocytochrome b521 and PDZ3 (Figure S2) are like those of the native proteins in dilute solution. Even though the HSQC spectrum of GFP is not observed in cell slurries or lysates, we know that GFP is in its native state because the samples fluoresce.
Increased viscosity in cells is one reason for our inability to observe HSQC spectra of globular proteins in E. coli. High viscosity slows molecular tumbling, increasing the breadth of crosspeaks, which decreases their detectability.38 The measured viscosity of the clear supernatants from the lysates is 2–4 fold times that of water. Since we did not add liquid during lysis, we can use the protein concentration in the cells and the lysate (Table S1) to estimate that the cytoplasm is diluted 1.5–3.0 fold in the lysates. Combining these ranges, and assuming a direct relationship between viscosity and concentration, gives a crude estimated intracellular viscosity of 3–12 times that of water.
The Stokes-Einstein-Debye equation49 predicts a direct linear dependence between viscosity and the apparent molecular size of globular proteins. If we assume this equation is valid inside cells, the apparent molecular weight of UBQ, the smallest protein studied here, would be 24–96 kDa. The lower value is compatible with the detection by the NMR methods we used, but the upper value is too large to yield an HSQC spectrum of the protein. This increase in apparent molecular weight will be even greater for the larger proteins. In summary, if our assumptions are valid, the increased viscosity in cells can explain our inability to detect globular proteins. At least one assumption, however, is suspect.
The Stokes-Einstein-Debye relationship breaks down in cells and in lysates because the definition of the viscosity assumes that the species increasing the viscosity (the viscogen) is infinitely smaller than the test molecule. This definition is valid in systems comprising small viscogens (like glycerol) and a globular test protein, but it is invalid in cells where the viscogens and the test protein are approximately the same size. Such macromolecular crowding can cause negative deviation from the Stokes-Einstein-Debye law, at least when synthetic polymers are used as crowding agents.50 That is, increases in viscosity decrease the tumbling rate by a smaller amount than is predicted by the equation. Given this negative deviation, the apparent molecular sizes will be less than the estimates given above, providing confidence that if viscosity were the only factor, we should have observed at least the smallest protein, UBQ, in cells.
We suggest that nonspecific interactions also contribute to our inability to detect globular protein HSQC spectra in E. coli cells. There is precedence for the idea that weak, nonspecific interactions are a feature of the cellular interior. It was suggested in the 1930s that the cells might be highly organized, and in the 1940s the complete enzymic repertoire of the Krebs cycle was isolated as a whole.51 Recent NMR studies indicate that 50% of the proteins in bacterial cells are completely immobile52 and that nonspecific interactions occur in vitro when proteins are used as crowding agents50 In summary, we suggest that a combination of increased viscosity and nonspecific protein interactions explains our inability to obtain high quality solutions NMR spectra from globular proteins in E. coli.
19F labeling not only facilitates leakage detection, but also overcomes the problem of detecting small globular proteins in cells (e.g., Figures 3, 4, and 6). Even though the 3FY resonances in UBQ, CI2 and CAM are broad, they are detectable in cells. We attribute this detectability to the low background and high sensitivity afforded by 19F and the limited number of labels in the proteins. The width of 3FY resonances in UBQ, CI2, and CAM were used to provide a rough estimate of ~40–100 ns for the rotational correlation times.34 These values represent a ten-fold increase in correlation time compared to dilute solution. Such an increase is also consistent with our inability to detect crosspeaks from 15N enriched globular proteins in cell slurries.
For larger globular proteins like GFP, even the 19F resonances from 3FY are too broad to observe (Figure S4). A clue to overcoming this problem came from our work on disordered proteins. NMR spectra of disordered proteins are observable in cells because the disorder facilitates internal protein motions.38 These motions are almost completely damped by the inherent order of globular proteins, such that global protein motion of globular proteins determines the line width of their resonances,53 and hence their detectability.
We reasoned that the independent internal motion of the trifluoromethyl group of tfmF would sharpen the 19F resonances, thereby facilitating the detection of larger globular proteins. This prediction is borne out. Resonances from tfmF labeled GFP and HDH are observed in cells (Figures 7 and 8). Furthermore, as expected, the 19F peak width of tfmF is molecular weight dependent, increasing from 0.20 ppm for CI2 to 0.50 ppm for GFP, to ~0.6 ppm for the 48 kDa homodimer, nitroreductase,29,35 and ~1 ppm for the 98 kDa homodimer, histidinol dehydrogensase29,35 (Figure 8). This idea of increased sensitivity via increased internal motion is used in the labeling of methyl groups with 13C,18 but tfmF offers the advantage that the labeled compound is less susceptible to metabolic scrambling.
The chemical shift of 19F is sensitive to its environment. This sensitivity is readily seen upon cell lysis. Lysis causes a ~0.1 ppm upfield shift of resonances from the intracellular protein for every protein investigated. An upfield shift was also observed for small 19F containing molecules by Xu et al.54 By comparing the 31P to 19F shifts for compounds containing both nuclei, these authors showed that the difference between extracellular and intracellular compounds arises because of differences in protein hydration inside and outside of cells. These differences in hydration may help explain the changes in protein stability in cells compared to the dilute solution.55,56
Conclusion
The high viscosity and weak interactions in the cytoplasm can make routine 15N enrichment a poor choice for in-cell NMR studies of globular proteins in E. coli. We demonstrated that 19F labeling is a suitable labeling method for studying not only globular proteins, but also disordered proteins in cells with NMR. The 19F chemical shift and line width provides site-specific structural and dynamic information in cells. In addition, we have shown that the increased motion of tfmF expands the application of in-cell NMR to larger globular proteins. Finally, the decreased rotational motion of globular proteins suggests that high resolution magic angle spinning57 might be well suited for in-cell NMR.
Supplementary Material
Acknowledgments
We thank Andrew Lee, Alexander Shekhtman, Anthony Persechini, and Peter Lansbury for supplying the expression systems, Gregory B. Young and Marc ter Horst for NMR training and spectrometer maintenance, and Elizabeth Pielak for comments on the manuscript. This work was supported by the National Science Foundation (MCB-051647) and the National Institutes of Health (5DP1OD783).
Footnotes
Supporting Information Available. Complete ref 22, additional descriptions of sample preparation, NMR parameters and additional in-cell spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Luby-Phelps K. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 2.Malmström J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmerman SB, Trach SO. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 4.Augustus AM, Reardon PN, Spicer LD. Proc Natl Acad Sci USA. 2009;106:5065–5069. doi: 10.1073/pnas.0811130106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brindle KM, Williams SP, Boulton M. FEBS Lett. 1989;255:121–124. [Google Scholar]
- 6.Burz DS, Dutta K, Cowburn D, Shekhtman A. Nat Protoc. 2006;1:146–152. doi: 10.1038/nprot.2006.23. [DOI] [PubMed] [Google Scholar]
- 7.Dedmon MM, Patel CN, Young GB, Pielak GJ. Proc Natl Acad Sci USA. 2002;99:12681–12684. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubbard JA, MacLachlan LK, King GW, Jones JJ, Fosberry AP. Mol Microbiol. 2003;49:1191–1200. doi: 10.1046/j.1365-2958.2003.03628.x. [DOI] [PubMed] [Google Scholar]
- 9.Inomata K, Ohno A, Tochio H, Isogai S, Tenno T, Nakase I, Takeuchi T, Futaki S, Ito Y, Hiroaki H, Shirakawa M. Nature. 2009;458:106–109. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- 10.McNulty BC, Young GB, Pielak GJ. J Mol Biol. 2006;355:893–897. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 11.Pielak GJ, Li C, Miklos AC, Schlesinger AP, Slade KM, Wang G, Zigoneanu IG. Biochemistry. 2008;48:226–234. doi: 10.1021/bi8018948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakai T, Tochio H, Tenno T, Ito Y, Kokubo T, Hiroaki H, Shirakawa M. J Biomol NMR. 2006;36:179–188. doi: 10.1007/s10858-006-9079-9. [DOI] [PubMed] [Google Scholar]
- 13.Selenko P, Serber Z, Gadea B, Ruderman J, Wagner G. Proc Natl Acad Sci USA. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selenko P, Wagner G. Nat Methods. 2006;3:80–81. doi: 10.1038/nmeth0206-80. [DOI] [PubMed] [Google Scholar]
- 15.Serber Z, Dötsch V. Biochemistry. 2001;40:14317–14323. doi: 10.1021/bi011751w. [DOI] [PubMed] [Google Scholar]
- 16.Serber Z, Keatinge-Clay AT, Ledwidge R, Kelly AE, Miller SM, Dötsch V. J Am Chem Soc. 2001;123:2446–2447. doi: 10.1021/ja0057528. [DOI] [PubMed] [Google Scholar]
- 17.Serber Z, Selenko P, Hansel R, Reckel S, Lohr F, Ferrell JE, Jr, Wagner G, Dötsch V. Nat Protoc. 2006;1:2701–2709. doi: 10.1038/nprot.2006.181. [DOI] [PubMed] [Google Scholar]
- 18.Serber Z, Straub W, Corsini L, Nomura AM, Shimba N, Craik CS, Ortiz de Montellano P, Dötsch V. J Am Chem Soc. 2004;126:7119–7125. doi: 10.1021/ja049977k. [DOI] [PubMed] [Google Scholar]
- 19.Xie J, Thapa R, Reverdatto S, Burz DS, Shekhtman A. J Med Chem. 2009;52:3516–3522. doi: 10.1021/jm9000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakakibara D, Sasaki A, Ikeya T, Hamatsu J, Hanashima T, Mishima M, Yoshimasu M, Hayashi N, Mikawa T, Walchli M, Smith BO, Shirakawa M, Guntert P, Ito Y. Nature. 2009;458:102–105. doi: 10.1038/nature07814. [DOI] [PubMed] [Google Scholar]
- 21.Pielak GJ. Biochemistry. 2007;46:8206. doi: 10.1021/bi700744h. [DOI] [PubMed] [Google Scholar]
- 22.Cellitti SE, et al. J Am Chem Soc. 2008;130:9268–9281. doi: 10.1021/ja801602q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danielson MA, Falke JJ. Ann Rev Biophys Biomol Struct. 1996;25:163–195. doi: 10.1146/annurev.bb.25.060196.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Lutz EA, Slade KM, Ruf RA, Wang G, Pielak GJ. Biochemistry. 2009;48:8578–8584. doi: 10.1021/bi900872p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Frieden C. Proc Natl Acad Sci USA. 2007;104:11993–11998. doi: 10.1073/pnas.0705253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haggie PM, Brindle KM. Journal of Biological Chemistry. 1999;274:3941–3945. doi: 10.1074/jbc.274.7.3941. [DOI] [PubMed] [Google Scholar]
- 27.Williams SP, Fulton AM, Brindle KM. Biochemistry. 1993;32:4895–4902. doi: 10.1021/bi00069a026. [DOI] [PubMed] [Google Scholar]
- 28.Williams SP, Haggie PM, Brindle KM. Biophys J. 1997;72:490–498. doi: 10.1016/S0006-3495(97)78690-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson JC, Hammill JT, Mehl RA. J Am Chem Soc. 2007;129:1160–1166. doi: 10.1021/ja064661t. [DOI] [PubMed] [Google Scholar]
- 30.Pervushin KV, Wider G, Riek R, Wüthrich K. Proc Natl Acad Sci USA. 1999;96:9607–9612. doi: 10.1073/pnas.96.17.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charlton LM, Barnes CO, Li C, Orans J, Young GB, Pielak GJ. J Am Chem Soc. 2008;130:6826–6830. doi: 10.1021/ja8005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Proc Natl Acad Sci USA. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slade KM, Baker R, Chua M, Thompson NL, Pielak GJ. Biochemistry. 2009;48:5083–5089. doi: 10.1021/bi9004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan F, Kuprov I, Craggs TD, Hore PJ, Jackson SE. J Am Chem Soc. 2006;128:10729–10737. doi: 10.1021/ja060618u. [DOI] [PubMed] [Google Scholar]
- 35.Hammill JT, Miyake-Stoner S, Hazen JL, Jackson JC, Mehl RA. Nat Protoc. 2007;2:2601–2607. doi: 10.1038/nprot.2007.379. [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Johnson BA, Blevins RA. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 38.Li C, Charlton LM, Lakkavaram A, Seagle C, Wang G, Young GB, Macdonald JM, Pielak GJ. J Am Chem Soc. 2008;130:6310–6311. doi: 10.1021/ja801020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryant JE, Lecomte JT, Lee AL, Young GB, Pielak GJ. Biochemistry. 2005;44:9275–9279. doi: 10.1021/bi050786j. [DOI] [PubMed] [Google Scholar]
- 40.Schneider DM, Dellwo MJ, Wand AJ. Biochemistry. 1992;31:3645–3652. doi: 10.1021/bi00129a013. [DOI] [PubMed] [Google Scholar]
- 41.Law AB, Fuentes EJ, Lee AL. J Am Chem Soc. 2009;131:6322–6323. doi: 10.1021/ja809915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burz DS, Shekhtman A. PLoS ONE. 2008;3:e2571. doi: 10.1371/journal.pone.0002571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitevski-LeBlanc JL, Evanics F, Prosser RS. J Biomol NMR. 2009;45:255–264. doi: 10.1007/s10858-009-9359-2. [DOI] [PubMed] [Google Scholar]
- 44.Jaren OR, Kranz JK, Sorensen BR, Wand AJ, Shea MA. Biochemistry. 2002;41:14158–14166. doi: 10.1021/bi026340+. [DOI] [PubMed] [Google Scholar]
- 45.Pedersen S, Bloch PL, Neidhardt FC. Cell. 1978;14:179–190. doi: 10.1016/0092-8674(78)90312-4. [DOI] [PubMed] [Google Scholar]
- 46.Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner M, Frishman D. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eliezer D, Kutluay E, Bussell R, Jr, Browne G. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 48.Wu KP, Kim S, Fela DA, Baum J. J Mol Biol. 2008;378:1104–1115. doi: 10.1016/j.jmb.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Debye PJ. Polar molecules. Chemical Catalog Company; New York: 1929. [Google Scholar]
- 50.Li C, Wang Y, Pielak GJ. J Phys Chem B. 2009;113:13390–13392. doi: 10.1021/jp907744m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srere PA. Trends Biochem Sci. 2000;25:150–153. doi: 10.1016/s0968-0004(00)01550-4. [DOI] [PubMed] [Google Scholar]
- 52.Persson E, Halle B. Proc Natl Acad Sci U S A. 2008;105:6266–6271. doi: 10.1073/pnas.0709585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jarymowycz VA, Stone MJ. Chem Rev. 2006;106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- 54.Xu ASL, Waldeck R, Kuchel PW. NMR Biomed. 1993;6:136–143. doi: 10.1002/nbm.1940060206. [DOI] [PubMed] [Google Scholar]
- 55.Ghaemmaghami S, Oas TG. Nat Struct Mol Biol. 2001;8:879–882. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 56.Ignatova Z, Gierasch LM. Proc Natl Acad Sci U S A. 2004;101:523–528. doi: 10.1073/pnas.0304533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindon JC, Beckonert OP, Holmes E, Nicholson JK. Prog Nucl Magn Reson Spectrosc. 2009;55:79–100. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.