Abstract
A total of 565 methicillin-resistant Staphylococcus aureus (MRSA) isolates were collected mostly from Europe and the Americas (2004 to 2007) during a phase IV clinical trial comparing linezolid with vancomycin for the treatment of complicated skin and skin structure infections proven to be due to MRSA. The isolates were tested for their susceptibilities by the broth microdilution method, they were tested for inducible clindamycin resistance by the D-test, and they were screened for heterogeneous resistance to vancomycin (heterogeneously vancomycin-intermediate S. aureus [hVISA]) by the Etest macromethod. The isolates were evaluated for the MRSA genotype by pulsed-field gel electrophoresis, staphylococcal protein A (spa) typing, multilocus sequence typing (MLST), and staphylococcal cassette chromosome mec (SCCmec) typing. All isolates were inhibited by 4 μg/ml of linezolid (MIC50 and MIC90, 2 and 4 μg/ml, respectively). The vast majority of isolates (92.4%) were resistant to erythromycin, and high clindamycin resistance rates were observed (28.5% constitutive and 16.3% inducible). Only 1.0% of the isolates were hVISA. Isolates from the United States were predominantly USA300 sequence type 8 (ST8)-SCCmec type IV (78.5%), followed by a lower prevalence of USA100 ST5-SCCmec type II isolates (14.2%). Strains belonging to the ST5 lineage were widely distributed in Portugal, South American countries, and Mexico. MRSA strains belonging to ST8-SCCmec type IV predominated in Russia (80.0%) and also emerged in Venezuela and Colombia. The epidemic MRSA type 15 clone predominated in the United Kingdom (55.6%) and Spain (100%). In addition, a new MLST profile (ST1071) was observed in South Africa. This study demonstrated the presence of major clones in particular regions (ST8 in the United States, ST5 in Latin America and Portugal, ST22 in Spain and the United Kingdom); however, emerging clones were identified, suggesting that the epidemiology of MRSA continues to evolve.
During the last few decades, methicillin-resistant Staphylococcus aureus (MRSA) has been a worldwide clinical problem and has increasingly been responsible for infections in both the community and hospital settings (1). This remarkable pathogen has acquired resistance to several classes of antimicrobial agents, therefore commonly exhibiting the multidrug resistance (MDR) phenotype, which poses a continuous threat for antimicrobial therapy (11, 14). More worrisome is the fact that the methicillin resistance rates currently exceed 60% among S. aureus isolates recovered from patients in intensive care units in many institutions in the United States (5).
S. aureus is the main pathogen responsible for skin and skin structure infections (SSSIs), which are the most common human bacterial infections observed in clinical practice (10). Although many cases of SSSIs are successfully treated with empirical therapy and local wound care, the increased prevalence of MRSA isolates causing SSSIs has become a significant therapeutic problem (10). This issue has been further complicated by the recent emergence of virulent MRSA strains among healthy individuals from the community in the United States and other countries worldwide (4, 23, 28). In addition, SSSIs can evolve to bloodstream infections, which may lead to metastatic foci of infections, such as endocarditis, osteomyelitis, and deep-seated abscesses (10).
Although a few surveillance studies have shown the continuous predominance of a small number of clones in some geographic regions, several longitudinal studies have also described that the epidemiology of MRSA can evolve rapidly (12, 15). Therefore, continuous epidemiologic surveillance of MRSA is important to obtain a better understanding of the dynamics of these microorganisms. The aim of this study was to assess the molecular epidemiology of MRSA isolates associated with complicated SSSIs (cSSSIs) collected during a randomized, open-label, controlled, multicenter phase IV clinical trial comparing linezolid with vancomycin. In addition, the antimicrobial potencies of linezolid and clinically used parenteral and oral agents were evaluated.
MATERIALS AND METHODS
Bacterial isolates.
The clinical isolates of MRSA included in this study were those obtained during the screening or baseline visit of 565 microbiologically available subjects enrolled in a multicenter clinical trial comparing linezolid to vancomycin for the treatment of cSSSIs (2004 to 2007). Isolates were cultured from surgical and traumatic wound infections, abscesses, infected decubitus and diabetic ulcers, infected burn wounds, and other cSSSIs requiring systemic antimicrobial therapy. Isolates from simple cellulitis infections were excluded. The specimens were processed and the bacterial pathogens were cultured by each medical laboratory site, according to local practices and trial design. Individual investigators sent all bacterial isolates to Covance Central Laboratory Service (Indianapolis, IN) for identification and susceptibility testing. MRSA identification was performed by routine methods (catalase, coagulase, colony morphology, etc.) and automated procedures, when needed (Vitek system; bioMerieux, Hazelwood, MO). Baseline MRSA isolates were forwarded to JMI Laboratories (North Liberty, IA) for further molecular studies.
Antimicrobial susceptibility testing.
MICs were determined by the broth microdilution method by using panels with cation-adjusted Mueller-Hinton medium and the recommendations of the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS; M07-A8, 2009) (6). Testing, incubation, and MIC interpretations were performed by using the recommendations of the CLSI in document M100-S19 (7). Quality control strains included S. aureus ATCC 29213 and Enterococcus faecalis ATCC 29212; all results were within the ranges published by the CLSI (7). Inducible clindamycin resistance was determined by the D-test disk diffusion method (7). Screening for heterogeneous vancomycin-intermediate S. aureus (hVISA) was performed by the Etest macromethod (AB Biodisk, Solna, Sweden), as described previously (32).
Molecular characterization.
All isolates were submitted to staphylococcal cassette chromosome mec (SCCmec) typing (types I through VI) by a multiplex PCR strategy, as described previously (22), as well as to screening for Panton-Valentine leukocidin (PVL)-encoding genes (lukF-PV and lukS-PV) by multiplex PCR with PVL-specific primers, as previously described by Lina et al. (18), along with generic primers targeting the tuf gene for internal control purposes. Bacterial chromosomal DNA was digested with SmaI and subjected to pulsed-field gel electrophoresis (PFGE) (30). PFGE types were assigned according to the origin of the isolates (continent), followed by a capital letter; however, isolates from South Africa, Malaysia, and Singapore were analyzed separately. Isolates were assigned to the same PFGE type when all bands matched. When one to three band differences were observed, the isolates were assigned to subtypes, as previously established by Tenover et al. (29). Representative isolates corresponding to the USA100, USA300, USA400, USA700, and Hungarian/Brazilian clones were used for comparison purposes.
Additionally, the PFGE patterns of representative isolates belonging to the predominant clone from each country were analyzed by the use of GelCompar II software (Applied Maths, Kortrijk, Belgium). Percent similarities were identified on a dendrogram derived from the unweighted-pair group method using average linkages and based on Dice coefficients. Band position tolerance and optimization were set at 1.2% and 0.5%, respectively. Isolates showing similarity coefficients of ≥80% were considered to be genetically related and to belong to the same type (30). These representative isolates were also further characterized by single-locus (spa) sequence typing and multilocus sequence typing (MLST), as described elsewhere (11, 25). spa types were assigned through the Ridom web server (http://www.ridom.de/spaserver/), and MLST alleles and sequence types (STs) were identified by using the MLST database (http://www.mlst.net).
RESULTS
Bacterial isolates.
A total of 565 baseline MRSA isolates from the cSSSI trial were evaluated. The majority of isolates were collected from subjects in the United States (62.5%; 43 sites), followed by Portugal (8.5%; 5 sites), Venezuela (7.3%; 5 sites), United Kingdom (6.4%; 2 sites), Russia (3.5%; 3 sites), South Africa (2.3%; 3 sites), Chile (2.1%; 2 sites), Italy (1.8%; 5 sites), Mexico (1.6%; 3 sites), Colombia (1.2%; 2 sites), Argentina (0.7%; 3 sites), Brazil (0.7%; 1 site), Spain (0.5%; 2 sites), Malaysia (0.4%; 1 site), Belgium (0.4%; 2 sites), and Singapore (0.2%; 1 site). The isolates were predominantly cultured from abscesses (50.3%) and postoperative infections (22.2%), followed by diabetic ulcers (8.7%), infected skin ulcers (7.1%), wound infections (3.9%), cellulitis (3.2%), decubitus ulcers (2.3%), and other cSSSIs (2.3%).
Antimicrobial susceptibility testing.
Linezolid was active against all isolates (MIC50 and MIC90, 2 and 4 μg/ml, respectively; 100% susceptible), as were vancomycin (MIC50 and MIC90, 1 and 1 μg/ml, respectively), teicoplanin (MIC50 and MIC90, 0.5 and 1 μg/ml, respectively), and daptomycin (MIC50 and MIC90, 0.25 and 0.5 μg/ml, respectively). In addition, quinupristin-dalfopristin inhibited 99.8% of the isolates at ≤1 μg/ml; one isolate from the United States had a MIC of 16 μg/ml (Table 1). The vast majority of MRSA isolates were resistant to erythromycin (92.4%), and high clindamycin resistance rates were observed (28.5% of the isolates showed constitutive resistance and 16.3% showed inducible resistance). The clindamycin resistance rate was the highest in Latin America (88.3%), followed by Europe (74.8%) and the United States (22.7%). Overall, among the erythromycin-resistant, clindamycin-susceptible isolates, 16.3% showed inducible clindamycin resistance, and this rate was the highest in Europe (40.3%), followed by the United States (10.2%) and Latin America (3.9%).
TABLE 1.
Antimicrobial susceptibility testing results for baseline MRSA clinical isolates (2004 to 2007) recovered during a phase IV clinical trial of complicated skin and skin structure infections
| Antimicrobial agent | % Susceptible/% resistant,a by continentb |
Overall (n = 565) |
||||||
|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) |
% Susceptible/% resistanta | |||||||
| Europe (nc = 119) | Latin America (n = 77) | North America (n = 353) | Others (n = 16) | 50% | 90% | Range | ||
| Linezolid | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 2 | 4 | ≤0.25-4 | 100.0/0.0 |
| Erythromycin | 19.3/79.9 | 5.2/82.2 | 3.4/96.3 | 0.0/100.0 | >16 | >16 | 0.25->16 | 6.9/92.4 |
| Clindamycin | 25.2/74.8d | 11.7/88.3e | 77.3/22.7f | 0.0/100.0g | 0.25 | >4 | 0.06->4 | 55.2/44.8h |
| Gatifloxacin | 4.2/95.0 | 13.0/85.7 | 46.7/53.0 | 0.0/100.0 | 2 | 8 | 0.06->8 | 32.0/67.6 |
| Tetracycline | 77.3/22.7 | 81.8/14.3 | 92.9/7.1 | 43.8/56.3 | 0.25 | >16 | ≤0.12->16 | 86.7/12.8 |
| TMP/SMX | 89.9/10.1 | 88.3/11.7 | 99.2/0.8 | 43.8/56.3 | ≤0.5 | ≤0.5 | ≤0.5->4 | 94.2/5.9 |
| Q/Di | 100.0/0.0 | 100.0/0.0 | 99.7/0.3 | 100.0/0.0 | 0.25 | 0.5 | ≤0.12-16 | 99.8/0.2 |
| Daptomycin | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 0.25 | 0.5 | 0.06-1 | 100.0/0.0 |
| Teicoplanin | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 0.5 | 1 | ≤0.25-8 | 100.0/0.0 |
| Vancomycin | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 1 | 1 | ≤0.25-2 | 100.0/0.0 |
MICs were interpreted according to the guidelines in CLSI document M100-S19 (7).
Europe and Latin America are represented by six countries each; North America is represented by the United States; and others is represented by Malaysia, Singapore, and South Africa.
n, number of MRSA isolates.
Includes isolates with constitutive (34.5%) and inducible (40.3%) resistance phenotypes.
Includes isolates with constitutive (84.4%) and inducible (3.9%) resistance phenotypes.
Includes isolates with constitutive (12.5%) and inducible (10.2%) resistance phenotypes.
Includes isolates with constitutive (68.8%) and inducible (31.3%) resistance phenotypes.
Includes isolates with constitutive (28.5%) and inducible (16.3%) resistance phenotypes.
Q/D, quinupristin-dalfopristin.
Gatifloxacin (a fluoroquinolone surrogate) showed limited activity against this collection; and susceptibility rates were 4.2% in Europe, 13.0% in Latin America, and 46.7% in the United States. Isolates from other countries were resistant to gatifloxacin. Trimethoprim-sulfamethoxazole (TMP/SMX) demonstrated a high level of activity against MRSA strains (MIC50 and MIC90, ≤0.5 and ≤0.5 μg/ml, respectively), mainly in the United States (99.2% susceptible); however, lower TMP/SMX susceptibility rates were noted in isolates recovered from Europe and Latin America (89.9 and 88.3%, respectively) and from Malaysia, Singapore and South Africa (43.8% susceptible). The highest tetracycline susceptibility rates were noted in the United States (92.9%), whereas in the other countries the rates ranged from 43.8 to 81.8% (Table 1).
The antimicrobial susceptibility profiles of MRSA isolates of the USA100 and USA300 clones are shown in Table 2. Isolates belonging to these clones were very susceptible to linezolid (100.0%), vancomycin (100.0%), teicoplanin (100.0%), daptomycin (100.0%), quinupristin-dalfopristin (≥99.6%), and TMP/SMX (≥98.0%). USA100 isolates were resistant to erythromycin (96.0%), clindamycin (92.0%; 52.0% inducible), and gatifloxacin (100.0%), whereas USA300 isolates were resistant to erythromycin (97.8%) and gatifloxacin (42.6%). The clindamycin resistance rate was 6.5% (1.8% inducible) when the activity of clindamycin against isolates of USA300 was tested. Tetracycline was slightly more active against USA100 strains (100.0%) than against S. aureus strains of the USA300 clone (91.3%).
TABLE 2.
Antimicrobial susceptibility testing results for baseline MRSA clinical isolates of USA100 and USA300
| Antimicrobial agent | USA100 (n = 50) |
USA300 (n = 277) |
||||||
|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) |
% Susceptible/% resistanta | MIC (μg/ml) |
% Susceptible/% resistanta | |||||
| 50% | 90% | Range | 50% | 90% | Range | |||
| Linezolid | 2 | 4 | 1-4 | 100.0/0.0 | 2 | 4 | 0.5-4 | 100.0/0.0 |
| Erythromycin | >16 | >16 | 0.5->16 | 4.0/96.0 | >16 | >16 | 0.5->16 | 1.8/97.8 |
| Clindamycin | 0.25 | >4 | 0.12->4 | 8.0/92.0b | 0.12 | 0.25 | 0.06->4 | 93.5/6.5c |
| Gatifloxacin | 8 | >8 | 2->8 | 0.0/100.0 | 0.12 | 4 | 0.06->8 | 57.4/42.6 |
| Tetracycline | 0.5 | 0.5 | 0.25-2 | 100.0/0.0 | 0.25 | 0.5 | ≤0.12->16 | 91.3/8.7 |
| TMP/SMX | ≤0.5 | ≤0.5 | ≤0.5->4 | 98.0/2.0 | ≤0.5 | ≤0.5 | ≤0.5->4 | 99.6/0.4 |
| Q/Dd | 0.25 | 0.5 | ≤0.12-1 | 100.0/0.0 | 0.25 | 0.25 | ≤0.12-16 | 99.6/0.4 |
| Daptomycin | 0.25 | 0.5 | 0.12-1 | 100.0/0.0 | 0.25 | 0.5 | 0.12-1 | 100.0/0.0 |
| Teicoplanin | 0.5 | 1 | ≤0.25-2 | 100.0/0.0 | 0.5 | 0.5 | ≤0.25-1 | 100.0/0.0 |
| Vancomycin | 1 | 1 | 0.5-2 | 100.0/0.0 | 1 | 1 | 0.5-1 | 100.0/0.0 |
MICs were interpreted according to the guidelines in CLSI document M100-S19 (7).
Includes isolates with constitutive (40.0%) and inducible (52.0%) resistance phenotypes.
Includes isolates with constitutive (4.7%) and inducible (1.8%) resistance phenotypes.
Q/D, quinupristin-dalfopristin.
The overall vancomycin modal MIC was 1 μg/ml (81.4% of isolates), and S. aureus isolates displaying MICs of 2 μg/ml were rare (3.2%). Moreover, 7.8, 4.2, and 0.8% of the isolates recovered from Latin America, Europe, and the United States showed a vancomycin MIC result of 2 μg/ml, respectively. Vancomycin inhibited all USA300 strains (49.0% of all MRSA strains; data not shown) at ≤1 μg/ml. Only 1.0% (6/565) of the isolates were characterized as hVISA, and these isolates showed vancomycin MICs of 1 (four strains) or 2 μg/ml (two strains). These strains were collected from the United Kingdom (1 of 36 isolates; 2.8%), Venezuela (1 of 41 isolates; 2.4%), Brazil (1 of 4 isolates; 25.0%), and Italy (3 of 10 isolates; 30.0%). Two hVISA isolates from Italy and one from Venezuela were clustered within the EUR-C and LAT-C PFGE types, respectively.
Molecular characterization.
Table 3 shows the epidemiologic data obtained from the collections of organisms evaluated. PFGE analysis revealed the presence of a major cluster (78.5%; 17 subtypes) among isolates from the United States, designated USA-A, which matched the USA300 pulsotype previously described by McDougal et al. (21). Further typing analysis of one representative isolate of this cluster showed spa type YHGFMBQBLO, ST8-SCCmec type IV (SCCmec IV) and a PCR-positive result for PVL, consistent with the characteristics of the USA300 genotype (Fig. 1). The second major PFGE type among U.S. MRSA strains was USA-B, and these isolates harbored SCCmec II and were negative for PVL (14.2%; 14 subtypes). These typing results matched those for USA100 (ST5-SCCmec II; New York/Japan clone) (28). Three isolates displayed identical PFGE patterns, which corresponded to that of USA400 ST1-SCCmec IV (0.8%). The remaining 23 (6.5%) MRSA strains clustered within 14 PFGE types, which showed SCCmec IV and were variable for the presence of PVL determinants or were SCCmec II and PVL negative.
TABLE 3.
Epidemiologic data for MRSA baseline isolates characterized during this study in each country
| Region and country (no. of isolates tested) | PFGE pattern | No. (%) of isolates | No. of subtypes | SCCmec type | PVLa | Type by MLSTb |
|---|---|---|---|---|---|---|
| North America | ||||||
| United States (353) | USA-A/USA300 | 277 (78.5) | 17 | IV | Pos | ST8 |
| USA-B/USA100 | 50 (14.2) | 14 | II | Neg | ST5 | |
| USA-C/USA400 | 03 (0.8) | 1 | IV | Pos | ST1 | |
| USA-D to -Q | 23 (6.5) | 16 | II or IV | Neg/Pos | ||
| Latin America | ||||||
| Argentina (4) | LAT-A | 1 (25.0) | 1 | III | Neg | ST239 |
| LAT-B | 1 (25.0) | 1 | II | Neg | ||
| LAT-C | 2 (50.0) | 2 | I | Neg | ST5 | |
| Brazil (4) | LAT-A | 1 (25.0) | 1 | III | Neg | ST239 |
| LAT-N | 1 (25.0) | 1 | III | Neg | ||
| LAT-O | 2 (50.0) | 2 | III | Neg | ||
| Chile (12) | LAT-A | 1 (8.3) | 1 | III | Neg | ST239 |
| LAT-C | 10 (83.3) | 5 | I | Neg | ST5 | |
| LAT-E | 1 (8.3) | 1 | II | Neg | ||
| Columbia (7) | LAT-C | 2 (28.6) | 2 | I | Neg | ST5 |
| LAT-G | 4 (57.1) | 2 | IV or IVE/F | Pos | ST8 | |
| LAT-H | 1 (14.3) | 1 | I | Neg | ||
| Mexico (9) | LAT-I | 9 (100) | 4 | II | Neg | ST5 |
| Venezuela (41) | LAT-C | 31 (75.6) | 8 | I | Neg | ST5 |
| LAT-F | 3 (7.3) | 1 | IV | Neg | ||
| LAT-G | 6 (14.6) | 3 | IV | Pos | ST8 | |
| LAT-K | 1 (2.4) | 1 | IV | Pos | ||
| Europe | ||||||
| Belgium (2) | EUR-A | 1 (50.0) | 1 | IV | Neg | |
| EUR-B | 1 (50.0) | 1 | IV | Neg | ||
| Italy (10) | EUR-A | 1 (10.0) | 1 | IV | Neg | |
| EUR-B | 1 (10.0) | 1 | IV | Neg | ||
| EUR-C | 2 (20.0) | 1 | I | Neg | ST228 | |
| EUR-D | 1 (10.0) | 1 | III | Neg | ST239 | |
| EUR-E | 1 (10.0) | 1 | I | Neg | ||
| EUR-F | 1 (10.0) | 1 | IV | Neg | ||
| EUR-G | 2 (20.0) | 1 | IV | Neg | ||
| EUR-O | 1 (10.0) | 1 | IV | Neg | ST22 | |
| Portugal (49) | EUR-D | 7 (14.6) | 3 | III | Neg | ST239 |
| EUR-G | 3 (6.3) | 2 | III | Neg | ||
| EUR-H | 1 (2.0) | 1 | III | Neg | ||
| EUR-I | 1 (2.0) | 1 | III | Neg | ||
| EUR-J | 5 (10.4) | 3 | IV | Neg | ||
| EUR-K | 14 (29.2) | 3 | II or IVE/F | Neg | ST5 | |
| EUR-L | 5 (10.4) | 2 | II/IV | Neg | ||
| EUR-O | 9 (18.8) | 1 | IV | Neg | ST22 | |
| EUR-S | 2 (4.0) | 1 | II | Neg | ||
| EUR-U | 1 (2.0) | 1 | III | Neg | ||
| Russia (20) | EUR-M | 4 (20.0) | 3 | III | Neg | |
| EUR-N | 16 (80.0) | 5 | IV | Neg | ST8 | |
| Spain (3) | EUR-O | 3 (100.0) | 1 | IV | Neg | ST22 |
| UK (37) | EUR-O | 20 (55.6) | 6 | IV | Neg | ST22 |
| EUR-Q | 1 (2.7) | 1 | II | Neg | ||
| EUR-R | 12 (33.3) | 6 | IV | Neg | ||
| EUR-T | 3 (8.3) | 3 | IV | Neg | ||
| Other | ||||||
| Malaysia (2) | ASI-A | 1 (50.0) | 1 | III | Neg | ST239 |
| ASI-B | 1 (50.0) | 1 | III | Neg | ||
| Singapore (1) | ASI-C | 1 (100.0) | 1 | III | Neg | |
| South Africa (13) | AFR-A | 1 (7.7) | 1 | IV | Neg | |
| AFR-B | 5 (38.5) | 1 | III | Neg | ||
| AFR-C | 7 (53.8) | 2 | II | Neg | ST1071 |
Pos, positive; Neg, negative.
MLST was performed with isolates selected from the predominant lineage(s) in each country.
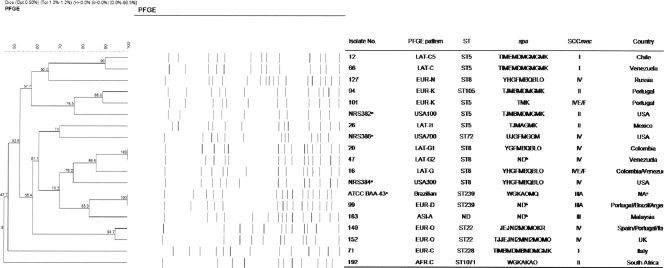
FIG. 1.
PFGE pattern analysis, MLST, and spa and SCCmec typing results for representative isolates belonging to the dominant MRSA clone from each country. The PFGE patterns of the USA100, USA300, USA700, and Hungarian/Brazilian clones are also shown for comparison purposes. Percent similarities were identified on a dendrogram derived from the unweighted-pair group method using average linkages and based on Dice coefficients. Band position tolerance and optimization were set at 1.2% and 0.5%, respectively. Isolates showing similarity coefficients of ≥80% were considered to be genetically related. a, strains NRS384 and NRS382 also correspond to the representative isolates belonging to the two dominant clones found in the United States (USA-A and USA-B, respectively) during this study; b, ND, not determined; c, NA, not applicable.
The majority of isolates from Latin American countries clustered within the LAT-C PFGE type (58.4%; 11 subtypes); and isolates belonging to this clone were found in Argentina (50.0%), Chile (83.3%), Colombia (28.6%), and Venezuela (75.6%). The LAT-C representative isolate was spa type TIMEMDMGMGMK, ST5-SCCmec I, and PVL negative, which corresponded to the Cordobes/Chilean clone (Fig. 1). Most isolates from Colombia clustered within the LAT-G PFGE type (57.1%; two subtypes), and isolates belonging to this clone were also found in Venezuela (14.6%; three subtypes). One representative isolate of LAT-G from Colombia and one from Venezuela were spa type YHGFMBQBLO, ST8-SCCmec IV, and PCR positive for PVL. These results matched those for USA300; however, isolates belonging to LAT-G and USA300 clustered into separate PFGE types (similarity coefficient, 76.2%; Fig. 1). All baseline isolates recovered from Mexico belonged to LAT-I, and the representative isolate was spa type TJMAGMK, ST5-SCCmec II, and PVL negative (Fig. 1), which matched the characteristics of the New York/Japan clone (31).
The EUR-O PFGE type was the predominant clone found in the United Kingdom (55.6%); and the representative isolate was spa type TJJEJNI2MNI2MOMO, ST22-SCCmec IV, and PVL negative, which corresponded to the internationally disseminated United Kingdom epidemic MRSA type 15 (EMRSA-15) clone (Fig. 1). This clone was also noted in Italy (10.0%) and Portugal (18.8%). Furthermore, all MRSA isolates collected from two medical centers located in Spain belonged to EUR-O (United Kingdom EMRSA-15; Fig. 1). Isolates from Russia also showed the presence of a predominant clone (EUR-N; 80.0%). The representative isolate was spa type YHGFMBQBLO, ST8-SCCmec IV, and PVL negative (Fig. 1). Except for the PVL genes, these typing results matched those for USA300, but representative isolates of these clones clustered into distinct PFGE types (Fig. 1).
Isolates from Italy were scattered within eight clones showing variable SCCmec types, all were PCR negative for PVL, and none of these clones predominated. Similar to the isolates from Italy, baseline isolates from Portugal demonstrated greater genetic diversity and the isolates clustered within 10 PFGE types; however, three dominant clones were noted among five Portuguese hospitals: EUR-K (29.2%), EUR-O (18.8%), and EUR-D (14.6%). Two representative isolates of EUR-K showed spa type TMK or TJMBMDMGMK and belonged to ST5-SCCmec IV and ST105-SCCmec II, respectively. The latter was a single-locus variant of ST5. EUR-D was the third most prevalent clone in Portugal, and this PFGE type corresponded to the pandemic Hungarian/Brazilian clone (similarity coefficient, 100.0%; Fig. 1).
The majority of isolates from South Africa clustered within the AFR-C PFGE type (53.8%; two subtypes; Table 1); and the representative isolate showed SCCmec II, was PVL negative and spa type WGKAKAO, and showed a new MLST profile, named ST1071 (Fig. 1). Only two baseline MRSA isolates from Malaysia were evaluated, and they showed unique PFGE types (types ASI-A and -B). The three MRSA isolates from Malaysia and Singapore harbored SCCmec type III and were PVL negative.
Overall, 296 (52.4%) strains were PCR positive for PVL genes, and except for those isolates belonging to LAT-G (Colombia and Venezuela) and one isolate belonging to LAT-K (Venezuela), the remaining PVL-positive isolates clustered mostly within USA-A (USA300). Two LAT-G isolates from Colombia showed a distinct subtype of SCCmec IV (subtype IVE or -F), according to the typing method employed. Similar findings were observed for isolates of PFGE types EUR-K and EUR-L from Portugal (Table 2).
DISCUSSION
All MRSA isolates evaluated during this study showed in vitro susceptibility to linezolid and glycopeptides; however, high rates of resistance (≥44.8%) to erythromycin, clindamycin, and fluoroquinolones were observed overall. The rates of resistance to all these agents except erythromycin among MRSA isolates recovered from the United States were lower than those for isolates recovered from the other regions. These results were likely due to the high prevalence of clone USA300 isolates (78.5%) in the United States, which are more susceptible to clindamycin and fluoroquinolones but which are resistant to erythromycin, likely due to the presence of msrA (15, 21). A worrisome finding was that the rate of resistance to fluoroquinolones (42.6%) found among USA300 isolates causing cSSSIs in this study was similar to that reported among S. aureus isolates of USA300 (recovered in 2005 and 2006) causing invasive disease in U.S. patients (54.6%) (17).
Overall, TMP/SMX showed good activity against the MRSA isolates (MIC50 and MIC90, ≤0.5 and ≤0.5 μg/ml, respectively), mainly against those organisms recovered from the United States (99.2% susceptible; only two isolates displayed MICs of 4 μg/ml). Tetracycline demonstrated variable results, and in addition to linezolid and glycopeptides, tetracycline also showed acceptable in vitro coverage (>90.0% susceptible) against isolates from the United States, regardless of their clonal type (USA300 or USA100). However, isolates of USA100 were slightly more susceptible than isolates of USA300, a finding previously reported for MRSA isolates implicated in invasive infections in the United States (17).
A literature review reported a general frequency of hVISA isolates among MRSA strains of 2.2% (19), which is higher than that found in the present study (1.0%). It is worthwhile to note that vancomycin inhibited 96.8% of isolates at ≤1 μg/ml, which may explain the low prevalence of hVISA isolates observed. In addition, these phenotypes were not confirmed by population analysis, and this rate could become even lower. Two hVISA isolates from Italy were recovered from the same medical site (data not shown) and clustered within the same PFGE type (EUR-C). Interestingly, 50.0% of the hVISA isolates were associated with ST5 (LAT-C [Venezuela]) or ST228 (EUR-C [Italy]), which is a two-locus variant of ST5. The molecular typing results for the hVISA and VISA isolates detected to date revealed that these are mostly derived from hospital-acquired clones, primarily ST5, and it has been speculated that this clone could be predisposed to emerge as VISA (13).
Overall, analysis of the molecular epidemiology of MRSA strains showed a predominant PFGE type in most countries evaluated. In the United States, the predominance of the USA300 clone as a cause of cSSSIs (78.5%) was observed, and this clone was detected in 83.7% of U.S. medical centers and was the predominant clone in 72.1% of them (data not shown). These results are in agreement with those presented in previous reports describing this clone as being the most prevalent cause of staphylococcal SSSIs acquired in the community (28). Moran et al. (23) reported that virtually all MRSA isolates causing SSSIs in U.S. emergency departments (59.0% overall) were community-associated MRSA strains, and 97.0% of these strains were USA300. This rate was higher than that observed in the present study (78.5%), in which 22.2% of the MRSA isolates were recovered from postoperative infections, which are more likely caused by health care-associated strains (17).
Previous reports have described the predominance of the Brazilian clone (ST239-SCCmec IIIA, renamed the South American clone) in hospitals in Brazil, Uruguay, Chile, and Argentina (27); however, recent studies have shown the replacement of this clone by the Cordobes/Chilean clone (ST5-SCCmec I), which has predominantly been isolated in Argentina, Chile, and Paraguay (20, 26). In this study, MRSA strains from South American countries largely clustered within the Cordobes/Chilean clone, being detected in four countries and prevailing in three of them. These results show a continuous predominance of this clone in this region, apart from Colombia, where most isolates clustered within PFGE type LAT-G (57.1%). Although representative isolates of LAT-G and USA300 did not cluster within the same PFGE type (similarity coefficient, 76.2%), they belonged to the same clonal lineage, clonal complex 8, as determined by MLST and spa typing. Further surveillance studies should be performed to monitor the evolution of LAT-G, since these clones (LAT-G and USA300) may also share the determinants responsible for the pathogenicity displayed by USA300 (9).
All MRSA isolates recovered from three medical centers in Mexico were characterized as ST5-SCCmec II, which is in agreement with the findings presented in a previous report describing the displacement of ST30-SCCmec IV by ST5-SCCmec II between 2001 and 2002 (31). In addition, only one of four isolates from Brazil clustered within the Brazilian clone. This may indicate that this clone has also been progressively displaced in this country or that it has genetically evolved, thus displaying greater variability by PFGE. The latter assumption may be supported by the fact that the remaining isolates also harbored SCCmec III, suggesting that they may be derived from ST239, but displayed different PFGE types. However, greater genetic variability is not surprising, since this clone has a long history of causing health care-associated infections.
This study revealed that EMRSA-15 dominated among English isolates; and this clone was also detected in Portugal, Italy, and Spain. EMRSA-15 was first reported in 1991 in southeast England and the Midlands, rapidly spread through numerous hospitals in the United Kingdom, and became one of the two most dominant clones (3). This pandemic clone has been identified in several European countries, but it still comprised less than 2% of the nosocomial MRSA isolates recovered in Spanish hospitals (24). More recently, a higher prevalence of EMRSA-15 (32%) was noted in Majorcan islands hospitals (2). In the present study, all isolates recovered from Spain (Seville and Cordoba) belonged to EMRSA-15, indicating that this clone may have become more prevalent in that country; however, only three isolates were evaluated.
A recent molecular epidemiology evaluation has revealed major shifts in the background of MRSA isolates in a Portuguese tertiary-care hospital (3), where the commonly found Hungarian/Brazilian clone (ST239-SCCmec IIIA) was replaced by EMRSA-15 (80% of the MRSA isolates). This report was later confirmed by Aires de Sousa et al. (1), who described that the majority of isolates (54%) recovered from 11 Portuguese hospitals belonged to EMRSA-15. Interestingly, in the same report, the New York/Japan clone (ST5-SCCmec II), first detected in a Portuguese hospital in 2005 (a single isolate), and the Hungarian/Brazilian clone shared the same overall prevalence (17%), but the New York/Japan clone prevailed among hospitals in Lisbon, Portugal (1). In the present study, a similar coexistence of these three clones was observed among Portuguese MRSA isolates; however, the New York/Japan clone was the dominant clone, consisting of 29.2% of all isolates (18.8% of EMRSA-15 isolates). It is worthwhile to mention that most MRSA isolates (72.9%) from Portugal were recovered from Lisbon or the city of Almada. Furthermore, EMRSA-15 was barely detected in these cities, and most of the isolates belonging to this clone were recovered from Coimbra, Portugal.
Interestingly, the predominant clone found in Russia showed the same MLST profile and spa type as the USA300 clone. Although representative isolates of these clones clustered within different PFGE types, these findings suggest that these isolates may have derived from a common ancestor. Additionally, several reports from Europe have described the dissemination of a new community-acquired MRSA clone associated with ST80 and SCCmec type IV (16). Isolates belonging to this clone display the tetracycline resistance phenotype; however, MRSA isolates with these characteristics from European countries were not observed in this study.
The limitations of this study were the small numbers of MRSA isolates obtained from certain countries, which jeopardized comparisons with the findings presented in previous reports and which limited the epidemiologic conclusions that could be drawn for those regions. However, this study provides some insights into the geographic distribution of MRSA isolates in most regions evaluated. The data showed the dominance of the USA300 clone as a cause of cSSSIs in the United States and the emergence and dissemination of strains associated with ST8 in South American countries and Russia. The major EMRSA-15 clone has usually been considered epidemic in many European hospitals (8) and still predominated in that region (27.7%; data not shown). In addition, the massive dissemination of isolates associated with ST5 was also noted in various Latin American countries (Mexico, Chile, Venezuela, Colombia, and Argentina), which should be a great concern due to its MDR phenotype.
Acknowledgments
We thank Herminia de Lencastre and Keiichi Hiramatsu for providing S. aureus strains HDE288 and WIS, respectively, used in this study as positive controls (SCCmec types V and VI, respectively) during the SCCmec typing procedures.
This study was supported by a grant from Pfizer Inc., New York, NY.
Footnotes
Published ahead of print on 25 November 2009.
REFERENCES
- 1.Aires-de-Sousa, M., B. Correia, and H. de Lencastre. 2008. Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J. Clin. Microbiol. 46:2912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcoceba, E., A. Mena, M. Cruz Perez, E. Ruiz de Gopegui, E. Padilla, J. Gil, A. Ramirez, C. Gallegos, A. Serra, J. L. Perez, and A. Oliver. 2007. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Majorcan hospitals: high prevalence of the epidemic clone EMRSA-15. Clin. Microbiol. Infect. 13:599-605. [DOI] [PubMed] [Google Scholar]
- 3.Amorim, M. L., N. A. Faria, D. C. Oliveira, C. Vasconcelos, J. C. Cabeda, A. C. Mendes, E. Calado, A. P. Castro, M. H. Ramos, J. M. Amorim, and H. de Lencastre. 2007. Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J. Clin. Microbiol. 45:2881-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanc, D. S., C. Petignat, A. Wenger, G. Kuhn, Y. Vallet, D. Fracheboud, S. Trachsel, M. Reymond, N. Troillet, H. H. Siegrist, S. Oeuvray, M. Bes, J. Etienne, J. Bille, P. Francioli, and G. Zanetti. 2007. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus in a small geographic area over an eight-year period. J. Clin. Microbiol. 45:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher, H. W., and G. R. Corey. 2008. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46(Suppl. 5):S344-S349. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2009. M07-A8. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Clinical and Laboratory Standards Institute. 2009. M100-S19. Performance standards for antimicrobial susceptibility testing, 19th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Cuevas, O., E. Cercenado, E. Bouza, C. Castellares, P. Trincado, R. Cabrera, and A. Vindel. 2007. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Spain: a multicentre prevalence study (2002). Clin. Microbiol. Infect. 13:250-256. [DOI] [PubMed] [Google Scholar]
- 9.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 10.Eisenstein, B. I. 2008. Treatment challenges in the management of complicated skin and soft-tissue infections. Clin. Microbiol. Infect. 14(Suppl. 2):17-25. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. U. S. A. 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graber, C. J., M. K. Wong, H. A. Carleton, F. Perdreau-Remington, B. L. Haller, and H. F. Chambers. 2007. Intermediate vancomycin susceptibility in a community-associated MRSA clone. Emerg. Infect. Dis. 13:491-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, C. H., M. Tuckman, A. Y. Howe, M. Orlowski, S. Mullen, K. Chan, and P. A. Bradford. 2006. Diagnostic PCR analysis of the occurrence of methicillin and tetracycline resistance genes among Staphylococcus aureus isolates from phase 3 clinical trials of tigecycline for complicated skin and skin structure infections. Antimicrob. Agents Chemother. 50:505-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, R. N. 2008. Key considerations in the treatment of complicated staphylococcal infections. Clin. Microbiol. Infect. 14(Suppl. 2):3-9. [DOI] [PubMed] [Google Scholar]
- 16.Larsen, A. R., S. Bocher, M. Stegger, R. Goering, L. V. Pallesen, and R. Skov. 2008. Epidemiology of European community-associated methicillin-resistant Staphylococcus aureus clonal complex 80 type IV strains isolated in Denmark from 1993 to 2004. J. Clin. Microbiol. 46:62-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limbago, B., G. E. Fosheim, V. Schoonover, C. E. Crane, J. Nadle, S. Petit, D. Heltzel, S. M. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, W. Schaffner, Y. Mu, and S. K. Fridkin. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J. Clin. Microbiol. 47:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 19.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayor, L., J. Ortellado, C. Menacho, G. Lird, C. Courtier, C. Gardon, H. Meugnier, M. Bes, F. Vandenesch, and J. Etienne. 2007. Molecular characterization of methicillin-resistant Staphylococcus aureus isolates collected in Asuncion, Paraguay. J. Clin. Microbiol. 45:2298-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran, G. J., A. Krishnadasan, R. J. Gorwitz, G. E. Fosheim, L. K. McDougal, R. B. Carey, and D. A. Talan. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666-674. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Roth, E., F. Lorenzo-Diaz, N. Batista, A. Moreno, and S. Mendez-Alvarez. 2004. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J. Clin. Microbiol. 42:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sola, C., P. Cortes, H. A. Saka, A. Vindel, and J. L. Bocco. 2006. Evolution and molecular characterization of methicillin-resistant Staphylococcus aureus epidemic and sporadic clones in Cordoba, Argentina. J. Clin. Microbiol. 44:192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sola, C., G. Gribaudo, A. Vindel, L. Patrito, and J. L. Bocco. 2002. Identification of a novel methicillin-resistant Staphylococcus aureus epidemic clone in Cordoba, Argentina, involved in nosocomial infections. J. Clin. Microbiol. 40:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stryjewski, M. E., and H. F. Chambers. 2008. Skin and soft-tissue infections caused by community-acquired methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46(Suppl. 5):S368-S377. [DOI] [PubMed] [Google Scholar]
- 29.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velazquez-Meza, M. E., M. Aires de Sousa, G. Echaniz-Aviles, F. Solorzano-Santos, G. Miranda-Novales, J. Silva-Sanchez, and H. de Lencastre. 2004. Surveillance of methicillin-resistant Staphylococcus aureus in a pediatric hospital in Mexico City during a 7-year period (1997 to 2003): clonal evolution and impact of infection control. J. Clin. Microbiol. 42:3877-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wootton, M., A. P. MacGowan, T. R. Walsh, and R. A. Howe. 2007. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 45:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]