Abstract
Gastritis, peptic ulcer disease, and gastric cancer are a few of the diverse disease manifestations that have been shown to be associated with infection by Helicobacter pylori. Why some individuals develop more severe forms of disease remains largely unknown. In this study, 225 South Korean strains were genotyped for vacA and then analyzed to determine if particular genotypes varied across disease state, sex, or cagA allele. Of these strains, 206 strains carried an s1/i1/m1 allele, 11 strains carried an s1/i1/m2 allele, and 8 strains carried an s1/i2/m2 allele. By using Fisher's exact test, a statistical association between variations in the cagA and vacA alleles was identified (P = 0.0007), and by using log linear modeling, this variation was shown to affect the severity of disease outcome (P = 0.027). Additionally, we present evidence that variation within the middle region of VacA contributes significantly to the distribution of vacA alleles across gender (P = 0.008) as well as the association with disease outcome (P = 0.011). In this South Korean population, the majority of H. pylori strains carry the vacA s1/i1/m1 allele and the CagA EPIYA-ABD allele. These facts may contribute to the high incidence of gastric maladies, including gastric cancer.
Helicobacter pylori is a Gram-negative bacterium (28) that chronically infects the gastric mucosa of over half of the world's population (15, 29) and is associated with the development of chronic gastritis, gastric and duodenal ulcers, gastric adenocarcinoma, and mucosa-associated lymphoid tissue (MALT) lymphoma (6, 10, 14, 41). Given H. pylori's high prevalence, chronic persistence, and link to gastric cancer, it is no wonder that gastric cancer is the second most common cause of cancer-associated death (36), with the mortality rate being especially high in East Asian countries such as China, Japan, and South Korea (19).
H. pylori strains express various toxins that enable the bacteria to cause host cell damage. Included among these toxins are cytotoxin-associated gene A (CagA) and vacuolating cytotoxin (VacA) (34). CagA has emerged as a major contributor to disease severity, and there is a direct link between the presence of CagA and an increased cancer risk (7, 17). CagA induces various pathological changes by modulating host cell signaling pathways, primarily after tyrosine phosphorylation at the EPIYA motif (18-22, 35, 45, 54). Interestingly, CagA is polymorphic, and the distribution of EPIYA motif combinations differs geographically.
VacA is another important toxin that is produced and secreted by all H. pylori strains (4, 11) and was previously shown to have various modes of action (12, 16, 27, 39, 49, 50, 52, 56). Like CagA, VacA has been shown to contain a number of polymorphisms. Currently, three polymorphic regions of vacA have been identified: the signal (s), intermediate (i), and middle (m) regions. Each of these polymorphic regions has two main types that divide them further into type 1 and type 2 (3, 43). The s region encodes the N-terminal signal sequence (30, 42), and polymorphisms in the s region affect the anion channel-forming efficiency of the toxin (30); the s1 type has an increased ability to form membrane channels (30). Polymorphisms in the m region affect the cell tropism of the toxin (23); the m1 type of VacA shows toxicity toward a broader range of cells than the m2 type (1, 38). The i region, located between the s and m regions, also displays two main polymorphisms (43). The i1 type of VacA has stronger vacuolating activity than the i2 type (43). Individually, the s1, i1, and m1 types have been shown to be associated with more severe forms of H. pylori-induced disease (5, 43).
Recently, we presented molecular epidemiological evidence that there is a significant association between the development of gastric cancer and infection with H. pylori strains carrying the EPIYA-ABD cagA genotype in South Korea, which has one of the highest rates of H. pylori colonization (51) and one of the highest rates of gastric cancer in the world (17, 47). Given the mounting body of evidence that indicates that cagA and vacA interact, herein we assess vacA polymorphisms across various cagA alleles and in relation to disease development, and we show a significant 3-way association between vacA, cagA, and disease.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The South Korean H. pylori clinical isolates used in this study were previously described by Jones et al. (24) and included 115 gastritis isolates, 55 gastric ulcer isolates, 54 duodenal ulcer isolates, and 30 gastric cancer isolates with epidemiological data on age and gender. H. pylori stocks preserved at −80°C were grown and expanded on antibiotic-supplemented horse blood agar plates under microaerophilic conditions created by an Anoxomat evacuation/replacement system (Spiral Biotech, Norwood, MA) as previously described (9, 24).
vacA genotyping.
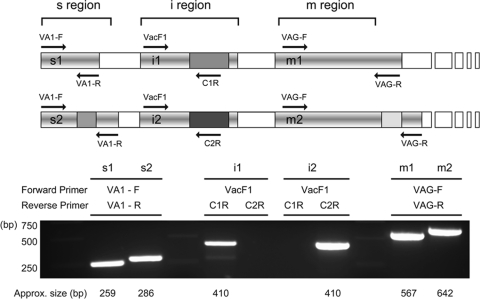
The primers used for vacA genotyping and sequencing of the i region are listed in Table 1. Chromosomal DNA of all 254 H. pylori strains was isolated by using the Easy-DNA kit (Invitrogen, Carlsbad, CA). Four individual PCRs were performed to identify the vacA genotype of each strain (Fig. 1). The s region was identified by amplification with primers VA1-F and VA1-R. The s1 region produced a 259-bp amplicon, whereas the s2 region produced a 286-bp amplicon (3). The m1 and m2 regions were determined by amplification with primers VAG-F and VAG-R, yielding 567-bp and 642-bp products, respectively (4). The i region was genotyped by using two independent PCRs with a universal forward primer (VacF1) and different i region type-specific reverse primers (C1R and C2R), as described previously by Rhead et al. (43). C1R and C2R specifically anneal with the i1 and i2 vacA alleles, respectively (43).
TABLE 1.
Primer sequences
| Primer | Sequence (5′-3′) | Reference |
|---|---|---|
| VA1-Fa | ATGGAAATACAACAAACACAC | 3 |
| VA1-Ra | CTGCTTGAATGCGCCAAAC | 3 |
| VAG-Fb | CAATCTGTCCAATCAAGCGAG | 4 |
| VAG-Rb | GCGTCAAAATAATTCCAAGG | 4 |
| C1Rc | TTAATTTAACGCTGTTTGAAG | 43 |
| C2Rc | GATCAACGCTCTGATTTGA | 43 |
| VacF1c,d | GTTGGGATTGGGGGAATGCCG | 43 |
| VacR9d | TGTTTATCGTGCTGTATGAAGG | 43 |
Part of the primer pair used to genotype the s region.
Part of the primer pair used to genotype the m region.
Part of the primer pair used to genotype the i region.
Part of the primer pair used for i region sequencing.
FIG. 1.
Genotyping of vacA polymorphic regions. (Top) Schematic representation of the vacA alleles, where an s1/i1/m1 allele is shown on the top and an s2/i2/m2 allele is depicted on the bottom. The annealing positions (arrows) and names of the primers used in this study are shown. (Bottom) PCR amplicons of each polymorphic region using the primers listed above the gel are depicted. The approximate size of the amplicon is listed below each band.
Sequencing of 60 isolates from patients suffering from gastritis (24 isolates), duodenal ulcers (10 isolates), gastric ulcers (8 isolates), and cancer (18 isolates) was conducted to identify specific amino acid changes in the i1 allele. Sanger dideoxy sequencing was performed at the Uniformed Services University Health Science Biomedical Instrumentation Center (Bethesda, MD). The resulting DNA sequences were analyzed by using Vector NTI, version 9.1 (Invitrogen, Carlsbad, CA), and Sequencher 4.5 (Gene Codes Corp., Ann Arbor, MI).
Statistical analysis.
Fisher's exact test was used to analyze the association between the vacA allele and disease state or cagA allele (based on the EPIYA motif). Log linear modeling was used to assess higher-order associations. We fit a saturated model using categorical variables representing vacA genotype, cagA genotype, disease state, gender, and age categories. A backward-selection algorithm identified higher-order associations among these variables, which were statistically significant at the 5% level. Data were analyzed by using SPSS software, version 14 or 16 (SPSS Inc., Chicago, IL), or SAS software, version 9.1 (SAS Institute Inc., Cary, NC).
Nucleotide sequence accession numbers.
The sequences for the i1 regions of vacA from 60 strains have been deposited in GenBank under accession numbers GQ338184 to GQ338243 (see Table S1 in the supplemental material).
RESULTS
Sample acquisition and vacA genotyping.
The strains used for this study were previously used for the characterization of the distribution of cagA alleles (24) and represent 260 strains obtained from patients presenting with gastric maladies; 254 of those strains have complete epidemiological data (see Table S1 in the supplemental material). The mean patient age was 51 years, with an age range of 14 to 86 years (Table 2). Within this population, 49.6% (126 patients) were female, with an age range of 21 to 86 years and a mean age of 52 years, and 50.4% (128 patients) were male, with a mean age of 50 years and an age range of 14 to 82 years. Of these samples, 11.8% were from patients with cancer, 42.9% were from patients with peptic ulcer disease (21.7% gastric ulcers and 21.3% duodenal ulcers), and 45.3% were from patients with gastritis (24).
TABLE 2.
vacA-genotyped isolates and disease stateb
| Parameter | Value |
|||||
|---|---|---|---|---|---|---|
| Total | vacA-genotyped isolates | Disease state of vacA-genotyped isolates |
||||
| Gastritis | Gastric ulcer | Duodenal ulcer | Gastric cancer | |||
| Overall total | ||||||
| No. of patients | 254a | 225 | 103 | 43 | 49 | 30 |
| Age range (yr) | 14-86 | 14-86 | 19-82 | 34-84 | 14-72 | 37-86 |
| Mean age (yr) | 51 | 50 | 49 | 55 | 45 | 58 |
| No. of females | 126 | 111 | 68 | 10 | 19 | 14 |
| Age range (yr) | 21-86 | 21-86 | 21-82 | 46-84 | 31-72 | 37-86 |
| Mean age (yr) | 52 | 52 | 49 | 57 | 51 | 61 |
| No. of males | 128 | 114 | 35 | 33 | 30 | 16 |
| Age range (yr) | 14-82 | 14-82 | 19-78 | 34-82 | 14-70 | 38-70 |
| Mean age (yr) | 50 | 49 | 47 | 54 | 41 | 55 |
| Strains carrying s1/i1/m1 | ||||||
| No. of patients | 206 | 95 | 41 | 42 | 28 | |
| Age range (yr) | 14-86 | 19-78 | 34-84 | 14-70 | 37-86 | |
| Mean age (yr) | 50 | 48 | 55 | 43 | 58 | |
| No. of females | 96 | 61 | 8 | 14 | 13 | |
| Age range (yr) | 21-86 | 21-75 | 46-84 | 31-61 | 37-86 | |
| Mean age (yr) | 51 | 49 | 57 | 48 | 60 | |
| No. of males | 110 | 34 | 33 | 28 | 15 | |
| Age range (yr) | 14-82 | 19-78 | 34-82 | 14-70 | 38-70 | |
| Mean age (yr) | 49 | 47 | 54 | 41 | 56 | |
| Strains carrying s1/i1/m2 | ||||||
| No. of patients | 11 | 4 | 1 | 4 | 2 | |
| Age range (yr) | 38-82 | 38-82 | 63 | 41-57 | 46-68 | |
| Mean age (yr) | 54 | 54 | NA | 51 | 56 | |
| No. of females | 9 | 4 | 1 | 3 | 1 | |
| Age range (yr) | 38-82 | 38-82 | 63 | 48-57 | 68 | |
| Mean age (yr) | 56 | 54 | NA | 54 | NA | |
| No. of males | 2 | 0 | 0 | 1 | 1 | |
| Age range (yr) | 41-44 | 0 | 0 | 41 | 46 | |
| Mean age (yr) | 43 | 0 | 0 | NA | NA | |
| Strains carrying s1/i2/m2 | ||||||
| No. of patients | 8 | 4 | 1 | 3 | 0 | |
| Age range (yr) | 38-72 | 38-68 | 56 | 61-72 | 0 | |
| Mean age (yr) | 57 | 51 | NA | 65 | 0 | |
| No. of females | 6 | 3 | 1 | 2 | 0 | |
| Age range (yr) | 38-72 | 38-68 | 56 | 61-72 | 0 | |
| Mean age (yr) | 58 | 53 | NA | 67 | 0 | |
| No. of males | 2 | 1 | 0 | 1 | 0 | |
| Age range (yr) | 43-61 | 43 | 0 | 61 | 0 | |
| Mean age (yr) | 52 | NA | 0 | NA | 0 | |
The total number of samples includes only the 254 that had complete epidemiological data for age and gender.
There was a statistical association between the m allele and gender (P = 0.0233). NA, not applicable.
Four different PCRs were conducted for each strain in order to genotype vacA (Fig. 1 and see Table S1 in the supplemental material). The s region was identified and differentiated by amplification with primers VA1-F and VA1-R (3), and the m region was determined by amplification with primers VAG-F and VAG-R (4). The i region was genotyped by using two independent PCRs using a universal forward primer (VacF1) and one of two i region type-specific reverse primers (C1R for the i1 type and C2R for the i2 type) as described previously by Rhead et al. (43).
The distribution of vacA polymorphisms is shown in Table 2. Of the 254 strains with complete epidemiological data, 225 strains were successfully genotyped for the vacA allele (Table 2 and see Table S1 in the supplemental material). The strains that were not successfully genotyped for vacA failed to yield PCR products or gave incorrectly sized bands and thus were not further analyzed. The genotyped strains were obtained from patients with a mean age of 50 years and an age range of 14 to 86 years. These patients included 111 females (49.3%), with a mean age of 52 years and an age range of 21 to 86 years, and 114 males (50.7%), with a mean age of 49 years and an age range of 14 to 82 years. Of these 225 strains, 206 strains (91.6%) carried an s1/i1/m1 vacA allele (mean patient age of 50 years and age range of 14 to 86 years). Of the strains carrying the s1/i1/m1 allele, 96 (46.6%) were from female patients (mean age of 51 years and age range of 21 to 86 years), and 110 (53.4%) were from male patients (mean age of 49 years and age range of 14 to 82 years). Eleven strains (4.9%) carried an s1/i1/m2 vacA genotype and were from patients with a mean age of 54 years and an age range of 38 to 82 years. Of these strains, 9 (81.8%) were obtained from female patients (mean age of 56 years and age range of 38 to 82 years), and 2 (18.2%) were obtained from male patients (mean age of 43 years and age range of 41 to 44 years). Eight strains (3.9%) carried the s1/i2/m2 vacA allele and were obtained from patients with a mean age of 57 years and an age range of 38 to 72 years. Six of these strains (75.0%) were obtained from female patients (mean age of 58 years with an age range of 38 to 72 years), and 2 (25.0%) were obtained from male patients (mean age of 52 years and age range of 43 to 61 years). Of note, neither s2 nor s1/i2/m1 vacA alleles were found.
Distribution of the vacA allele and gender.
Statistical analysis of allele distributions showed a significant association between the vacA allele and gender (P = 0.0233). Patients that carried non-s1/i1/m1 strains were 4.3 times more likely to be female than male. This difference is likely driven by the m region, since the distribution of any combination that contained the m region compared to gender was statistically significant (P = 0.005 for s/m and P = 0.0233 for i/m), whereas the combination lacking m (P = 0.1672 for s/i) was not significant. Moreover, when the distribution of polymorphisms within each region was analyzed alone versus gender, only the distribution of the m polymorphisms was significant (P = 0.008). In fact, if a patient carried an m2 allele, they were 3.75 times more likely to be female than male. This finding combined with the finding that the m1 allele appears to affect toxicity toward a larger variety of cells (23) may contribute to the finding that males are 1.5 to 2.5 times more likely to develop gastric cancer than females (reviewed in reference 44).
Associations among vacA, cagA, and disease.
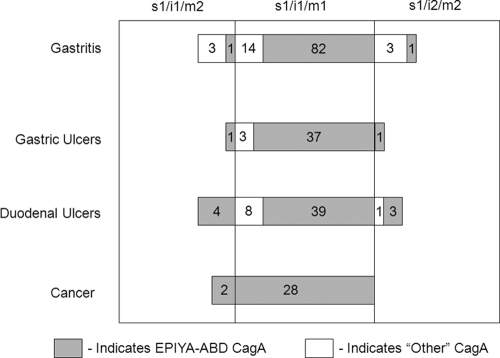
Given the diversity of the identified roles of the VacA toxin, we assessed whether the distribution of the vacA alleles had a direct impact on disease state. First, the individual regions were assessed for their impact on disease development. The distribution of polymorphisms in the m region and i region among disease states was not statistically significant (P = 0.5397 and P = 0.7399, respectively), and the distribution of polymorphisms in the s region in relationship to disease state could not be determined because only the s1 allele was found within this population. Statistical analysis for two-way associations using SASS software showed no statistical association between the distribution of the vacA alleles and disease state (P = 0.7499). However, log linear modeling taking into account age and gender did reveal a two-way association between the vacA allele and disease only in the East Asian (EPIYA-ABD) strains (P = 0.030). The majority of East Asian EPIYA-ABD CagA strains carrying non-s1/i1/m1 vacA alleles were associated with duodenal ulcers. Conversely, strains carrying non-s1/i1/m1 vacA alleles and any other genotype of CagA were associated with gastritis. A complete breakdown of the vacA allele and disease state is provided in Table 2.
VacA was previously suggested to interact synergistically with the H. pylori virulence factor CagA (2, 37). Thus, we next analyzed whether there was any association between the distribution of vacA alleles, the distribution of cagA alleles, and disease state. Of the 225 strains that were genotyped for the vacA allele, 224 of these strains had previously been genotyped for the cagA allele. Of these strains, 199 isolates (88.8%) can be classified as East Asian (carrying an EPIYA-D motif), and 25 isolates (11.2%) were determined to be classified as Western strains (carrying at least one EPIYA-C motif) (see Table S1 in the supplemental material). Eight East Asian strains carried an EPIYA motif other than a defined -ABD motif, either incomplete or containing the addition of one or more motifs, including -AABD, -BD, -BBD, and -ABAB*D, as well as -AB*D, where a mutation within the EPIYA-B motif is designated by the asterisk. Based on these distributions, the strains were subdivided based on the presence of a complete CagA EPIYA-ABD motif versus all other EPIYA motifs, yielding 33 isolates that were determined to have “other genotypes.”
When the distribution of the vacA alleles (s1/i1/m1, s1/i1/m2, and si/i2/m2) was assessed among the distribution of cagA alleles (EPIYA-ABD versus all other genotypes), a strong two-way association was identified (P = 0.0007) (Fig. 2 and Table 3). People infected with strains carrying non-EPIYA-ABD cagA alleles were associated with a two-times-higher probability of carrying the s1/i1/m2 vacA allele and a 10-times-higher probability of carrying the s1/i2/m2 vacA allele than people infected with the EPIYA-ABD cagA allele (Table 3). When combinations of the regions were compared, the distribution of polymorphisms among the cagA alleles was statistically significant for every combination, s/m (P = 0.0006), i/m (P = 0.0007), and s/i (0.0019), and the distributions of individual regions of vacA, m (P = 0.0019) and i (P = 0.0019), versus the cagA allele were also statistically significant. Once again, the distribution of polymorphisms in the s region among cagA alleles could not be determined because only the s1 allele was found within this population. This indicates that each of these regions is important for the distribution of the vacA allele with the cagA allele.
FIG. 2.
Schematic depiction of the distribution of the vacA genotypes stratified by disease state and cagA allele within this South Korean population. Shown is the distribution of vacA genotypes within the four different disease states: gastritis, gastric ulcers, duodenal ulcers, and cancer. The shaded portions within the disease state and vacA genotype subgroupings correspond to the isolates that carry a cagA EPIYA-ABD motif.
TABLE 3.
vacA genotype and cagA alleleb
| Parameter | Value |
|||
|---|---|---|---|---|
| vacA-genotyped isolates | vacA- and cagA-genotyped isolates |
cagA allele |
||
| EPIYA-ABD | Other genotypesa | |||
| Overall total | ||||
| No. of patients | 225 | 224 | 191 | 33 |
| Age range (yr) | 14-86 | 14-86 | 14-86 | 28-82 |
| Mean age (yr) | 50 | 50 | 50 | 49 |
| No. of females | 111 | 111 | 90 | 21 |
| Age range (yr) | 21-86 | 21-86 | 21-86 | 28-82 |
| Mean age (yr) | 52 | 52 | 52 | 50 |
| No. of males | 114 | 113 | 101 | 12 |
| Age range (yr) | 14-82 | 14-82 | 14-82 | 33-81 |
| Mean age (yr) | 49 | 49 | 49 | 49 |
| Strains carrying s1/i1/m1 | ||||
| No. of patients | 206 | 205 | 180 | 25 |
| Age range (yr) | 14-86 | 14-86 | 14-86 | 28-81 |
| Mean age (yr) | 50 | 50 | 50 | 47 |
| No. of females | 96 | 96 | 82 | 14 |
| Age range (yr) | 21-86 | 21-86 | 21-86 | 28-61 |
| Mean age (yr) | 51 | 51 | 52 | 46 |
| No. of males | 110 | 109 | 98 | 11 |
| Age range (yr) | 14-82 | 14-82 | 14-82 | 33-81 |
| Mean age (yr) | 49 | 49 | 49 | 49 |
| Strains carrying s1/i1/m2 | ||||
| No. of patients | 11 | 11 | 8 | 3 |
| Age range (yr) | 38-82 | 38-82 | 41-68 | 38-82 |
| Mean age (yr) | 54 | 54 | 52 | 58 |
| No. of females | 9 | 9 | 6 | 3 |
| Age range (yr) | 38-82 | 38-82 | 41-68 | 38-82 |
| Mean age (yr) | 56 | 56 | 56 | 58 |
| No. of males | 2 | 2 | 2 | 0 |
| Age range (yr) | 41-44 | 41-44 | 41-44 | NAc |
| Mean age (yr) | 43 | 43 | 43 | NA |
| Strains carrying s1/i2/m3 | ||||
| No. of patients | 8 | 8 | 3 | 5 |
| Age range (yr) | 38-72 | 38-72 | 54-72 | 38-68 |
| Mean age (yr) | 57 | 57 | 62 | 53 |
| No. of females | 6 | 6 | 2 | 4 |
| Age range (yr) | 38-72 | 38-72 | 54-72 | 38-68 |
| Mean age (yr) | 58 | 58 | 63 | 56 |
| No. of males | 2 | 2 | 1 | 1 |
| Age range (yr) | 43-61 | 43-61 | 61 | 43 |
| Mean age (yr) | 52 | 52 | NA | NA |
Indicates any other genotype besides EPIYA-ABD, including Western strains and EPIYA-AABD, -BD, -BBD, and -ABAB*D, as well as -AB*D, where a mutation within the EPIYA-B motif is designated by *.
There is a statistical two-way association between the distribution of cagA alleles and the distribution of vacA alleles (P = 0.0007).
NA, not applicable.
Given the strong correlation between the cagA allele and disease state that we previously observed (24), we next wondered if the vacA allele affected this distribution. Indeed, log linear modeling revealed a significant three-way association among vacA allele, cagA allele, and disease state (P = 0.027). As with the case of gender, the distribution of any combination that contained the m region when assessed via the distribution of the cagA allele and disease state was statistically significant (P = 0.004 for s/m and P = 0.025 for i/m), whereas the non-m combination (P = 0.586 for s/i) was not significant. Moreover, when the distribution of polymorphisms within each region was analyzed individually, only the distribution of the m polymorphism (P = 0.011) was significant. A complete breakdown of the strains based on disease state, vacA allele, and cagA allele is provided in Table 4.
TABLE 4.
vacA allele, cagA allele, and disease stateb
| Parameter | Value |
||||
|---|---|---|---|---|---|
| vacA- and cagA-genotyped isolates | Disease state of vacA-genotyped isolates |
||||
| Gastritis | Gastric ulcer | Duodenal ulcer | Gastric cancer | ||
| Overall total | |||||
| No. of patients | 224 | 103 | 42 | 49 | 30 |
| No. of EPIYA-ABD isolates | 192 | 83 | 38 | 40 | 30 |
| No. of isolates of other genotypesa | 32 | 20 | 4 | 9 | 0 |
| No. of females | 111 | 68 | 10 | 19 | 14 |
| No. of females with strains carrying EPIYA-ABD | 90 | 57 | 8 | 11 | 14 |
| No. of females with strains carrying other genotypesa | 21 | 11 | 2 | 8 | 0 |
| No. of males | 113 | 35 | 32 | 30 | 16 |
| No. of males with strains carrying EPIYA-ABD | 101 | 26 | 30 | 29 | 16 |
| No. of males with strains carrying other genotypesa | 12 | 9 | 2 | 1 | 0 |
| Strains carrying s1/i1/m1 | |||||
| No. of patients | 205 | 95 | 40 | 42 | 28 |
| No. of patients with strains carrying EPIYA-ABD | 180 | 81 | 37 | 34 | 28 |
| No. of patients with strains carrying other genotypesa | 25 | 14 | 3 | 8 | 0 |
| No. of females | 96 | 61 | 8 | 14 | 13 |
| No. of females with strains carrying EPIYA-ABD | 82 | 55 | 7 | 7 | 13 |
| No. of females with strains carrying other genotypesa | 14 | 6 | 1 | 7 | 0 |
| No. of males | 109 | 34 | 32 | 28 | 15 |
| No. of males with strains carrying EPIYA-ABD | 98 | 26 | 30 | 27 | 15 |
| No. of males with strains carrying other genotypesa | 11 | 8 | 2 | 1 | 0 |
| Strains carrying s1/i1/m2 | |||||
| No. of patients | 11 | 4 | 1 | 4 | 2 |
| No. of patients with strains carrying EPIYA-ABD | 8 | 1 | 1 | 4 | 2 |
| No. of patients with strains carrying other genotypesa | 3 | 3 | 0 | 0 | 0 |
| No. of females | 9 | 4 | 1 | 3 | 1 |
| No. of females with strains carrying EPIYA-ABD | 6 | 1 | 1 | 3 | 1 |
| No. of females with strains carrying other genotypesa | 3 | 3 | 0 | 0 | 0 |
| No. of males | 2 | 0 | 0 | 1 | 1 |
| No. of males with strains carrying EPIYA-ABD | 2 | 0 | 0 | 1 | 1 |
| No. of males with strains carrying other genotypesa | 0 | 0 | 0 | 0 | 0 |
| Strains carrying s1/i2/m2 | |||||
| No. of patients | 8 | 4 | 1 | 3 | 0 |
| No. of patients with strains carrying EPIYA-ABD | 3 | 1 | 0 | 2 | 0 |
| No. of patients with strains carrying other genotypesa | 5 | 3 | 1 | 1 | 0 |
| No. of females | 6 | 3 | 1 | 2 | 0 |
| No. of females with strains carrying EPIYA-ABD | 2 | 1 | 0 | 1 | 0 |
| No. of females with strains carrying other genotypesa | 4 | 2 | 1 | 1 | 0 |
| No. of males | 2 | 1 | 0 | 1 | 0 |
| No. of males with strains carrying EPIYA-ABD | 1 | 0 | 0 | 1 | 0 |
| No. of males with strains carrying other genotypesa | 1 | 1 | 0 | 0 | 0 |
Indicates any other genotype besides EPIYA-ABD, including Western strains and EPIYA-AABD, -BD, -BBD, -ABAB*D, as well as -AB*D strains, where a mutation within the EPIYA-B motif is designated by *.
There is a statistical three-way association between the distribution of cagA alleles, the distribution of vacA alleles, and disease state (P = 0.027).
Sequence analysis of the intermediate type 1 region.
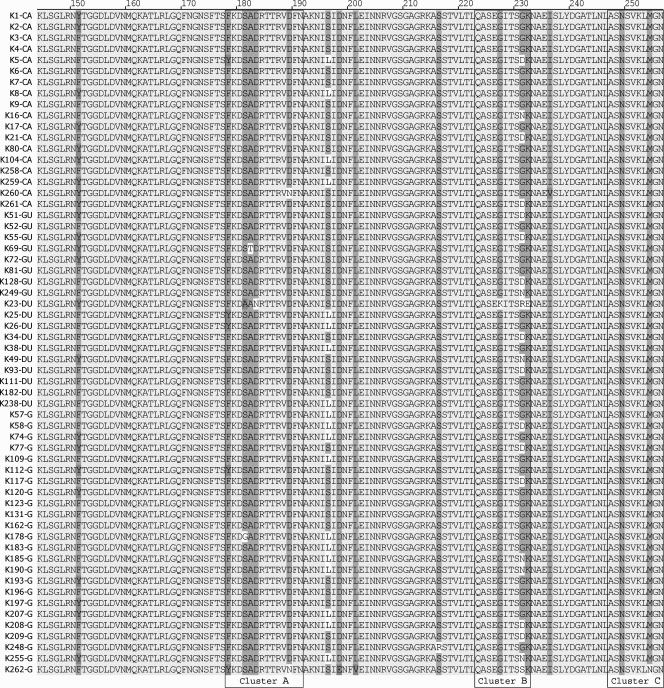
The i region was previously suggested to be the best indicator of pathology caused by VacA, and the i1 type is more virulent than i2 (2, 43). Two specific amino acid sequences, a phenylalanine at position 178 in cluster A and a methionine at position 254 in cluster C (Fig. 3), have been identified as markers within the Taiwanese population, and a particular amino acid substitution at position 231 in cluster B was previously suggested to affect disease severity (46). Given these reasons, we determined the i1 type amino acid sequence for 60 strains (18 from cancer patients, 8 from gastric ulcer patients, 10 from duodenal ulcer patients, and 24 from gastritis patients) (Fig. 3).
FIG. 3.
Amino acid alignment of the i1 type of VacA. This amino acid alignment is from 60 South Korean strains of various disease states: 24 from gastritis (G) patients, 10 from duodenal ulcer (DU) patients, 8 from gastric ulcer (GU) patients, and 18 from gastric cancer (CA) patients. The abbreviations listed after the strain correspond to the disease state of the strain. The three defined clusters of the i1 region, clusters A, B, and C, are indicated.
Sequence analysis of the i1 type in our strains revealed that a phenylalanine at position 178 in cluster A was present in 91.7% of the strains in the South Korean population. Additionally, the substitution of a methionine at position 254 in cluster C was well conserved in the South Korean population: 98.3% of strains carried this substitution.
The substitution of a glycine for a serine at position 231 in cluster B was previously suggested to be statistically linked to disease development within the Taiwanese population (46). However, at this site within strains among the South Korean population, neither the distribution of the amino acids with regard to disease (P = 0.8082) nor the distribution of glycine compared to any other amino acid with regard to disease (P = 0.5214) was statistically significant. Additionally, there was no statistical significance when the distribution of glycine was determined with regard to individual disease states: gastritis (P = 0.7871), gastric ulcer (P = 0.4329), duodenal ulcer (P = 0.7287), or cancer (P = 0.2414). However, there was a statistical association between the distribution of the glycine amino acid and the distribution of cagA alleles (P = 0.0318).
Sequence analysis of the i1 type revealed two additional amino acid polymorphisms across strains among the South Korean population. At position 151, the majority of the strains carried a phenylalanine (37 strains) or a tyrosine (23 strains). This distribution of amino acids at this position was not statistically linked to disease (P = 0.3886) or the distribution of the cagA allele (P = 0.6983). Additionally, polymorphism at position 196 leads to either a serine or a leucine at this position. The distribution of amino acids at this position had no statistical association with the distribution of cagA alleles (P = 1.0000) or disease state (P = 0.0669).
DISCUSSION
The majority (92%) of the South Korean isolates analyzed in this study carried the s1/i1/m1 vacA allele, which was previously suggested to be the most virulent form of the toxin (3, 25, 31-33, 43). The finding that the majority of H. pylori strains in this population carry the most toxic form of both VacA and CagA may explain the high rate of severe gastric disease among the South Korean population. When age and gender were taken into account, a two-way association between the distribution of vacA alleles and disease state was found for the strains carrying EPIYA-ABD CagA. Non-s1/i1/m1 vacA alleles were associated with duodenal ulcers within the population carrying East Asian EPIYA-ABD CagA and with gastritis within the population carrying any other genotype of CagA.
The distribution of the m alleles varied significantly across gender and the cagA allele and had a significant impact on the three-way association between the cagA allele and disease state. This suggests that polymorphism within the m region is the major contributor to the association of the vacA allele, the cagA allele, and disease state within this population. This is in concordance with a previously reported meta-analysis that found that the m1 region increased the risk for gastric cancer in Latin American (odds ratio [OR] = 3.59) and African (OR = 10.18) populations (48). The increased tropism of the m1 allele (38) combined with the finding that patients infected with H. pylori strains carrying the m2 allele are more likely to be female may explain why males are overall more likely to develop gastric cancer (reviewed in reference 44). To our knowledge, this is the first time that the m allele distribution has been linked to gender. To determine the role that the m allele has in the association between gender and disease state, populations where the m2 allele is more prevalent, such as in regions of China (40) and Poland (26), should be analyzed. However, it should be noted that this region alone is not a good predictor of disease for the South Korean population, since two of the strains that carried the m2 allele were obtained from cancer patients (Table 2).
Previous work with Western strains suggested that the i region of vacA is the major determinant of vacuolating activity and is the most important region for disease development (2, 13, 43). However, we found that within this population of predominantly East Asian strains, the i region was not a major determinant of disease state. This may indicate that the i region is more important within the context of strains that express the Western cagA allele or that there are other factors that mask the importance of this region in East Asian isolates.
Three clusters within the i1 region where sequence differences occur have been reported: clusters A, B, and C (43). Amino acid substitutions (tyrosine to phenylalanine in cluster A and asparagine to methionine in cluster C) are conserved and predicted to serve as a marker for Taiwanese VacA (46). Also, the substitution of a glycine for a serine at the ninth amino acid in cluster B was statistically linked to disease development in the Taiwanese population (46). The amino acid substitutions within clusters A and C were also conserved in the South Korean isolates, indicating that these substitutions are likely a marker of East Asian VacA. No correlation between the ninth amino acid substitutions in cluster B and disease severity was identified for our South Korean population, which suggests that this amino acid does not play a role in disease progression or is important in combination with another virulence factor.
Two additional positions within the i1 region that showed polymorphism were identified: positions 151 and 196. Neither the phenylalanine nor the tyrosine found at position 151 was linked to disease state or the distribution of the cagA allele. While the distribution of the amino acids at position 196 had no statistical association with the cagA allele or disease, there was a trend toward significance: ∼78% of strains from cancer patients and 100% of strains from gastric ulcer patients carry a serine at this position. This suggests that additional populations should be assessed to determine if a serine at position 196 has an impact on disease development and severity.
Previously reported studies have identified an association between vacA and cagA that appears to affect H. pylori toxicity and disease severity (55, 57). Basso et al. found that increasing numbers of CagA EPIYA-C motifs impacted cancer risk and that i region polymorphisms of VacA were a major indicator for the development of peptic ulcers (5). Additionally, infection with strains carrying CagA and s1/m1 VacA results in highly active corpus gastritis (33), which is linked to the development of gastric cancer (31-33). In our study, log linear modeling, taking into consideration age and gender, identified a two-way association between the vacA allele and disease state for East Asian (CagA EPIYA-ABD) strains. Not surprisingly, since the majority of these strains carried both CagA EPIYA-ABD and vacA s1/i1/m1, the majority of cancer strains (28 out of 30) carry this combination. This suggests that the role of the vacA allele could differentially affect disease progression based on other virulence factors. This could be due to the finding that VacA acts as an immune modulator (16, 52) and perhaps changes the immune response to the immunogenic CagA. Like CagA, VacA was found to disorganize the cytoskeleton of gastric epithelial cells, leading to increased cell spreading and growth (39). Thus, this phenotype may help compensate for the presence of a less virulent cagA allele or synergistically contribute to severe gastric maladies in conjunction with East Asian CagA. Evidence suggests that the combination of CagA and VacA may dampen the effect of each protein alone, possibly leading to an increased survival of infected host cells (2). This perhaps occurs through CagA preventing VacA-induced apoptosis (37, 56) or by inhibiting the autophagy pathway induced by VacA (50).
CagA and VacA are the two best-studied virulence factors of H. pylori. Interestingly, both toxins exhibit a high degree of polymorphism, and it is becoming increasingly evident that these polymorphisms, alone and in concert, affect H. pylori-induced disease. Indeed, the finding that the majority of South Korean H. pylori strains carry the most toxic forms of CagA and VacA may explain the reason for the high prevalence of gastric disease and mortality of patients with gastric cancer in South Korea. However, the reason why only a portion of the population develops gastric cancer still remains unclear. Other bacterial virulence factors as well as multiple host, dietary, and environmental factors have been indicated as being participants in H. pylori-induced disease (reviewed in references 8 and 53). Further study is required to determine which factors are involved and what role they have in the development of H. pylori-induced gastric cancer.
Supplementary Material
Acknowledgments
We thank Jang Gi Cho for help with graphics and Jeannette Whitmire and Beth Carpenter for critical reading of the manuscript.
This work was supported by a National Research Foundation of Korea grant funded by the South Korean Government (grant 2009-0073957) (to J.-H.C.) and by grant AI065529 from the NIAID (to D.S.M.).
Contents of the manuscript are the sole responsibility of the authors and do not necessarily represent the official views of the NIH or the DOD. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print on 2 December 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Amieva, M. R., and E. M. El-Omar. 2008. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology 134:306-323. [DOI] [PubMed] [Google Scholar]
- 2.Argent, R. H., R. J. Thomas, D. P. Letley, M. G. Rittig, K. R. Hardie, and J. C. Atherton. 2008. Functional association between the Helicobacter pylori virulence factors VacA and CagA. J. Med. Microbiol. 57:145-150. [DOI] [PubMed] [Google Scholar]
- 3.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 4.Atherton, J. C., T. L. Cover, R. J. Twells, M. R. Morales, C. J. Hawkey, and M. J. Blaser. 1999. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J. Clin. Microbiol. 37:2979-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basso, D., C. F. Zambon, D. P. Letley, A. Stranges, A. Marchet, J. L. Rhead, S. Schiavon, G. Guariso, M. Ceroti, D. Nitti, M. Rugge, M. Plebani, and J. C. Atherton. 2008. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 135:91-99. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1998. Helicobacter pylori and gastric diseases. BMJ 316:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 8.Brenner, H., D. Rothenbacher, and V. Arndt. 2009. Epidemiology of stomach cancer. Methods Mol. Biol. 472:467-477. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, B. M., T. K. McDaniel, J. M. Whitmire, H. Gancz, S. Guidotti, S. Censini, and D. S. Merrell. 2007. Expanding the Helicobacter pylori genetic toolbox: modification of an endogenous plasmid for use as a transcriptional reporter and complementation vector. Appl. Environ. Microbiol. 73:7506-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 11.Cover, T. L., and S. R. Blanke. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3:320-332. [DOI] [PubMed] [Google Scholar]
- 12.Cover, T. L., S. A. Halter, and M. J. Blaser. 1992. Characterization of HeLa cell vacuoles induced by Helicobacter pylori broth culture supernatant. Hum. Pathol. 23:1004-1010. [DOI] [PubMed] [Google Scholar]
- 13.Douraghi, M., Y. Talebkhan, H. Zeraati, F. Ebrahimzadeh, A. Nahvijoo, A. Morakabati, M. Ghafarpour, M. Esmaili, M. Bababeik, A. Oghalaie, N. Rakhshani, M. E. Hosseini, M. A. Mohagheghi, and M. Mohammadi. 2009. Multiple gene status in Helicobacter pylori strains and risk of gastric cancer development. Digestion 80:200-207. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.EUROGAST Study Group. 1993. Epidemiology of, and risk factors for, Helicobacter pylori infection among 3194 asymptomatic subjects in 17 populations. Gut 34:1672-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebert, B., W. Fischer, E. Weiss, R. Hoffmann, and R. Haas. 2003. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science 301:1099-1102. [DOI] [PubMed] [Google Scholar]
- 17.Gwack, J., A. Shin, C. S. Kim, K. P. Ko, Y. Kim, J. K. Jun, J. Bae, S. K. Park, Y. C. Hong, D. Kang, S. H. Chang, H. R. Shin, and K. Y. Yoo. 2006. CagA-producing Helicobacter pylori and increased risk of gastric cancer: a nested case-control study in Korea. Br. J. Cancer 95:639-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatakeyama, M. 2008. Linking epithelial polarity and carcinogenesis by multitasking Helicobacter pylori virulence factor CagA. Oncogene 27:7047-7054. [DOI] [PubMed] [Google Scholar]
- 19.Hatakeyama, M. 2004. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer 4:688-694. [DOI] [PubMed] [Google Scholar]
- 20.Higashi, H., A. Nakaya, R. Tsutsumi, K. Yokoyama, Y. Fujii, S. Ishikawa, M. Higuchi, A. Takahashi, Y. Kurashima, Y. Teishikata, S. Tanaka, T. Azuma, and M. Hatakeyama. 2004. Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J. Biol. Chem. 279:17205-17216. [DOI] [PubMed] [Google Scholar]
- 21.Higashi, H., R. Tsutsumi, A. Fujita, S. Yamazaki, M. Asaka, T. Azuma, and M. Hatakeyama. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. U. S. A. 99:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashi, H., R. Tsutsumi, S. Muto, T. Sugiyama, T. Azuma, M. Asaka, and M. Hatakeyama. 2002. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295:683-686. [DOI] [PubMed] [Google Scholar]
- 23.Ji, X., T. Fernandez, D. Burroni, C. Pagliaccia, J. C. Atherton, J. M. Reyrat, R. Rappuoli, and J. L. Telford. 2000. Cell specificity of Helicobacter pylori cytotoxin is determined by a short region in the polymorphic midregion. Infect. Immun. 68:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, K. R., Y. M. Joo, S. Jang, Y. J. Yoo, H. S. Lee, I. S. Chung, C. H. Olsen, J. M. Whitmire, D. S. Merrell, and J. H. Cha. 2009. Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. J. Clin. Microbiol. 47:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letley, D. P., and J. C. Atherton. 2000. Natural diversity in the N terminus of the mature vacuolating cytotoxin of Helicobacter pylori determines cytotoxin activity. J. Bacteriol. 182:3278-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maciorkowska, E., I. Roszko, O. Kowalczuk, M. Kaczmarski, L. Chyczewski, and A. Kemona. 2007. The evaluation of vacA gene alleles frequency in Helicobacter pylori strains in children and adults in Podlaskie region. Folia Histochem. Cytobiol. 45:215-219. [PubMed] [Google Scholar]
- 27.Manente, L., A. Perna, E. Buommino, L. Altucci, A. Lucariello, G. Citro, A. Baldi, G. Iaquinto, M. A. Tufano, and A. De Luca. 2008. The Helicobacter pylori's protein VacA has direct effects on the regulation of cell cycle and apoptosis in gastric epithelial cells. J. Cell. Physiol. 214:582-587. [DOI] [PubMed] [Google Scholar]
- 28.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed] [Google Scholar]
- 29.Matysiak-Budnik, T., and F. Megraud. 1997. Epidemiology of Helicobacter pylori infection with special reference to professional risk. J. Physiol. Pharmacol. 48(Suppl. 4):3-17. [PubMed] [Google Scholar]
- 30.McClain, M. S., P. Cao, H. Iwamoto, A. D. Vinion-Dubiel, G. Szabo, Z. Shao, and T. L. Cover. 2001. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J. Bacteriol. 183:6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meining, A., M. Stolte, R. Hatz, N. Lehn, S. Miehlke, A. Morgner, and E. Bayerdorffer. 1997. Differing degree and distribution of gastritis in Helicobacter pylori-associated diseases. Virchows Arch. 431:11-15. [DOI] [PubMed] [Google Scholar]
- 32.Miehlke, S., A. Hackelsberger, A. Meining, R. Hatz, N. Lehn, P. Malfertheiner, M. Stolte, and E. Bayerdorffer. 1998. Severe expression of corpus gastritis is characteristic in gastric cancer patients infected with Helicobacter pylori. Br. J. Cancer 78:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miehlke, S., C. Kirsch, K. Agha-Amiri, T. Gunther, N. Lehn, P. Malfertheiner, M. Stolte, G. Ehninger, and E. Bayerdorffer. 2000. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int. J. Cancer 87:322-327. [PubMed] [Google Scholar]
- 34.Montecucco, C., and R. Rappuoli. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat. Rev. Mol. Cell Biol. 2:457-466. [DOI] [PubMed] [Google Scholar]
- 35.Neel, B. G., H. Gu, and L. Pao. 2003. The ′Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 28:284-293. [DOI] [PubMed] [Google Scholar]
- 36.Neugut, A. I., M. Hayek, and G. Howe. 1996. Epidemiology of gastric cancer. Semin. Oncol. 23:281-291. [PubMed] [Google Scholar]
- 37.Oldani, A., M. Cormont, V. Hofman, V. Chiozzi, O. Oregioni, A. Canonici, A. Sciullo, P. Sommi, A. Fabbri, V. Ricci, and P. Boquet. 2009. Helicobacter pylori counteracts the apoptotic action of its VacA toxin by injecting the CagA protein into gastric epithelial cells. PLoS Pathog. 5:e1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pagliaccia, C., M. de Bernard, P. Lupetti, X. Ji, D. Burroni, T. L. Cover, E. Papini, R. Rappuoli, J. L. Telford, and J. M. Reyrat. 1998. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc. Natl. Acad. Sci. U. S. A. 95:10212-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pai, R., T. L. Cover, and A. S. Tarnawski. 1999. Helicobacter pylori vacuolating cytotoxin (VacA) disorganizes the cytoskeletal architecture of gastric epithelial cells. Biochem. Biophys. Res. Commun. 262:245-250. [DOI] [PubMed] [Google Scholar]
- 40.Pan, Z. J., D. E. Berg, R. W. van der Hulst, W. W. Su, A. Raudonikiene, S. D. Xiao, J. Dankert, G. N. Tytgat, and A. van der Ende. 1998. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J. Infect. Dis. 178:220-226. [DOI] [PubMed] [Google Scholar]
- 41.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127-1131. [DOI] [PubMed] [Google Scholar]
- 42.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhead, J. L., D. P. Letley, M. Mohammadi, N. Hussein, M. A. Mohagheghi, M. E. Hosseini, and J. C. Atherton. 2007. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 133:926-936. [DOI] [PubMed] [Google Scholar]
- 44.Roder, D. M. 2002. The epidemiology of gastric cancer. Gastric Cancer 5(Suppl. 1):5-11. [DOI] [PubMed] [Google Scholar]
- 45.Roovers, K., and R. K. Assoian. 2000. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays 22:818-826. [DOI] [PubMed] [Google Scholar]
- 46.Sheu, S. M., K. H. Hung, B. S. Sheu, H. B. Yang, and J. J. Wu. 2009. Association of nonsynonymous substitutions in the intermediate region of the vacA gene of Helicobacter pylori with gastric diseases in Taiwan. J. Clin. Microbiol. 47:249-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin, A., H. R. Shin, D. Kang, S. K. Park, C. S. Kim, and K. Y. Yoo. 2005. A nested case-control study of the association of Helicobacter pylori infection with gastric adenocarcinoma in Korea. Br. J. Cancer 92:1273-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugimoto, M., and Y. Yamaoka. 2009. The association of vacA genotype and Helicobacter pylori-related disease in Latin American and African populations. Clin. Microbiol. Infect. 15:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szabo, I., S. Brutsche, F. Tombola, M. Moschioni, B. Satin, J. L. Telford, R. Rappuoli, C. Montecucco, E. Papini, and M. Zoratti. 1999. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 18:5517-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terebiznik, M. R., D. Raju, C. L. Vazquez, K. Torbricki, R. Kulkarni, S. R. Blanke, T. Yoshimori, M. I. Colombo, and N. L. Jones. 2009. Effect of Helicobacter pylori's vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy 5:370-379. [DOI] [PubMed] [Google Scholar]
- 51.Tokudome, S., R. Ando, R. Ghadimi, T. Tanaka, N. Hattori, Z. Yang, M. Marumoto, H. Agawa, K. Arakawa, Y. Osaka, H. Tanaka, A. Hosono, and M. A. Moore. 2007. Are there any real Helicobacter pylori infection-negative gastric cancers in Asia? Asian Pac. J. Cancer Prev. 8:462-463. [PubMed] [Google Scholar]
- 52.Torres, V. J., S. E. VanCompernolle, M. S. Sundrud, D. Unutmaz, and T. L. Cover. 2007. Helicobacter pylori vacuolating cytotoxin inhibits activation-induced proliferation of human T and B lymphocyte subsets. J. Immunol. 179:5433-5440. [DOI] [PubMed] [Google Scholar]
- 53.Tsugane, S., and S. Sasazuki. 2007. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer 10:75-83. [DOI] [PubMed] [Google Scholar]
- 54.Tsutsumi, R., A. Takahashi, T. Azuma, H. Higashi, and M. Hatakeyama. 2006. Focal adhesion kinase is a substrate and downstream effector of SHP-2 complexed with Helicobacter pylori CagA. Mol. Cell. Biol. 26:261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Doorn, L. J., C. Figueiredo, F. Megraud, S. Pena, P. Midolo, D. M. Queiroz, F. Carneiro, B. Vanderborght, M. D. Pegado, R. Sanna, W. De Boer, P. M. Schneeberger, P. Correa, E. K. Ng, J. Atherton, M. J. Blaser, and W. G. Quint. 1999. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116:823-830. [DOI] [PubMed] [Google Scholar]
- 56.Willhite, D. C., and S. R. Blanke. 2004. Helicobacter pylori vacuolating cytotoxin enters cells, localizes to the mitochondria, and induces mitochondrial membrane permeability changes correlated to toxin channel activity. Cell. Microbiol. 6:143-154. [DOI] [PubMed] [Google Scholar]
- 57.Yamazaki, S., A. Yamakawa, T. Okuda, M. Ohtani, H. Suto, Y. Ito, Y. Yamazaki, Y. Keida, H. Higashi, M. Hatakeyama, and T. Azuma. 2005. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J. Clin. Microbiol. 43:3906-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.