Full text
PDF








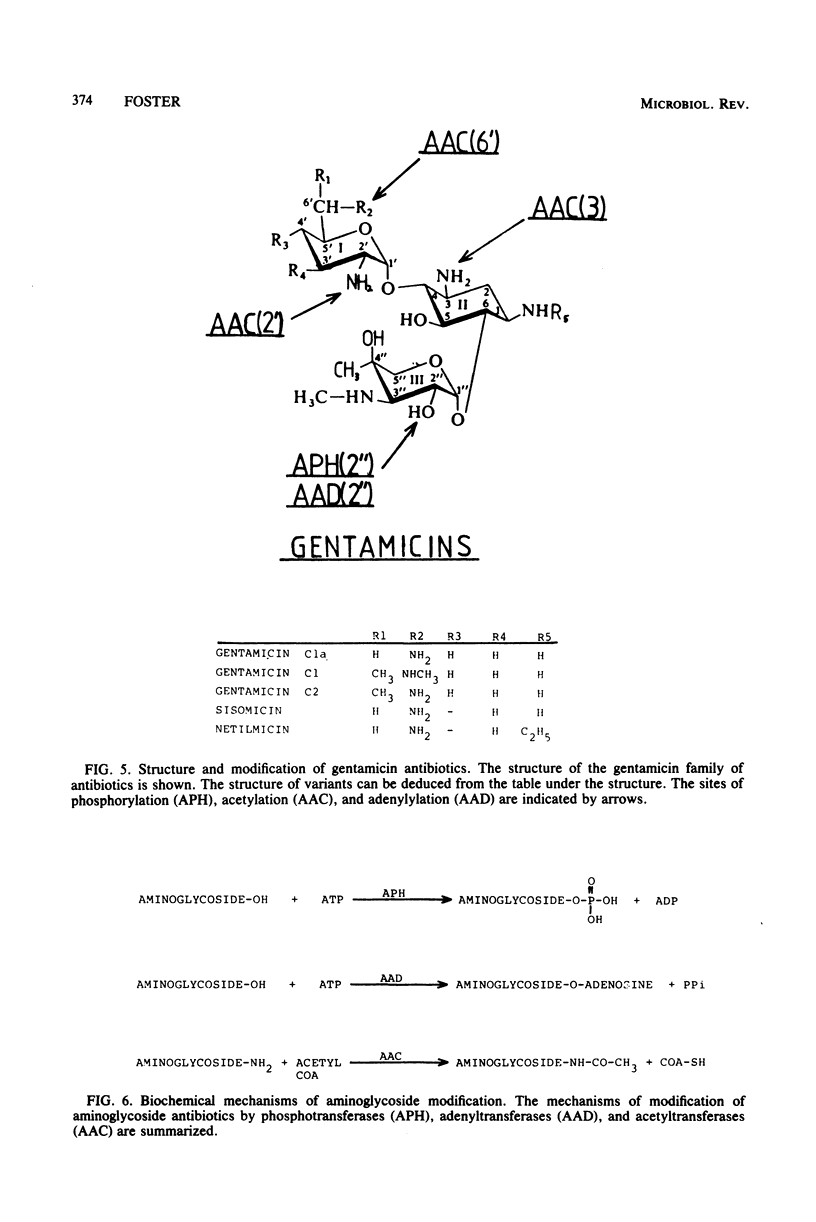
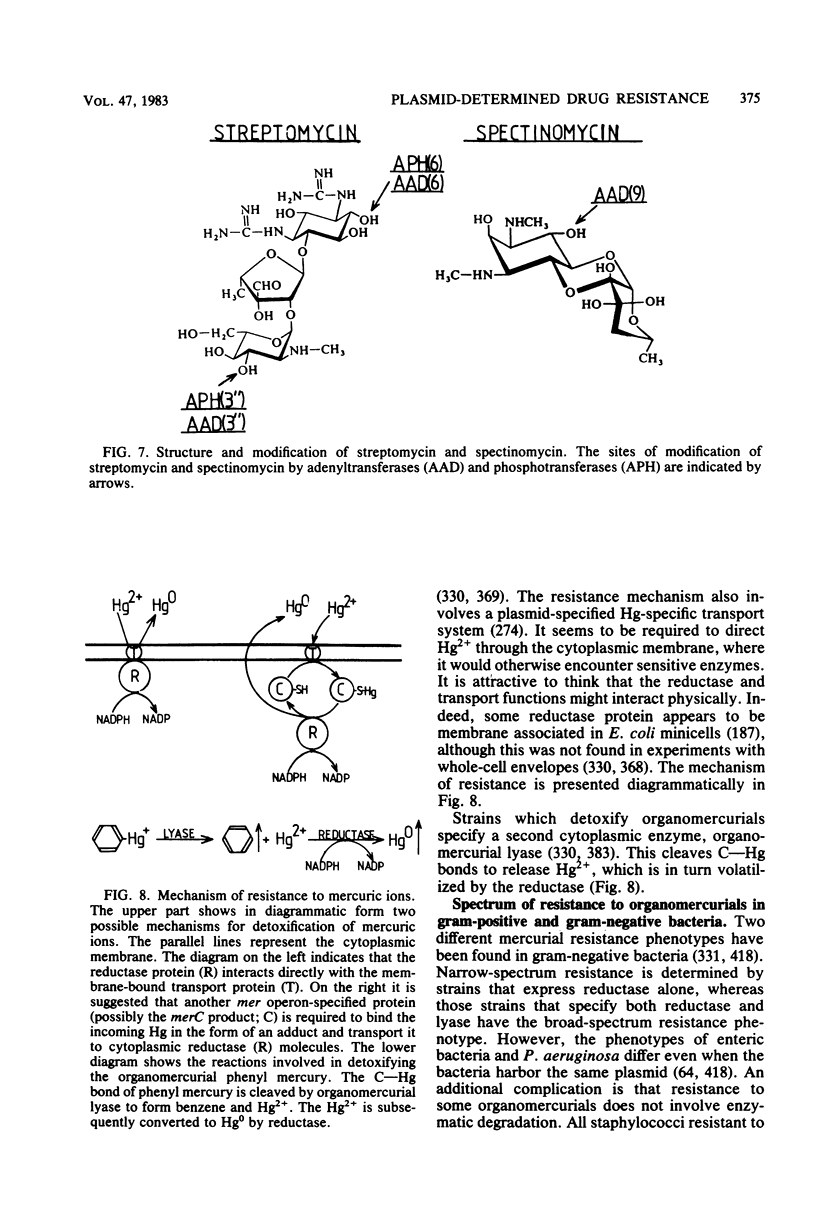
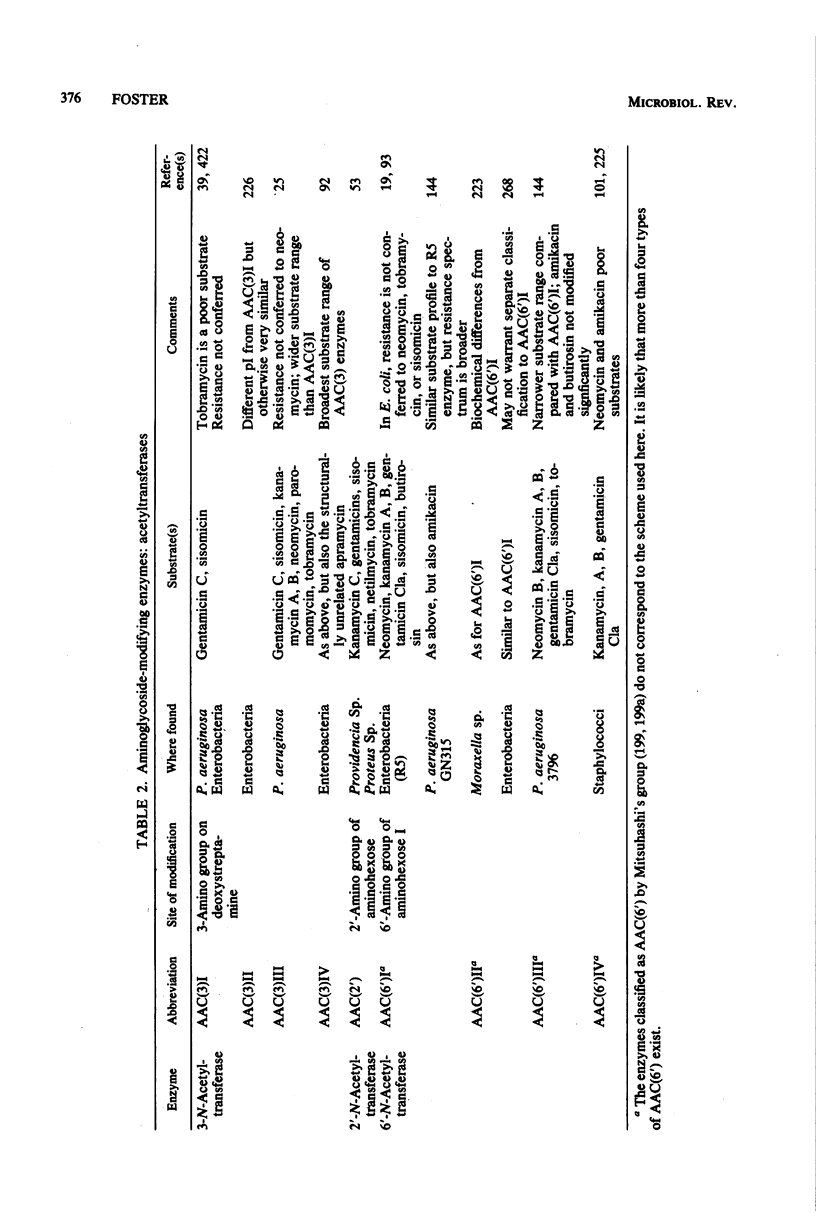
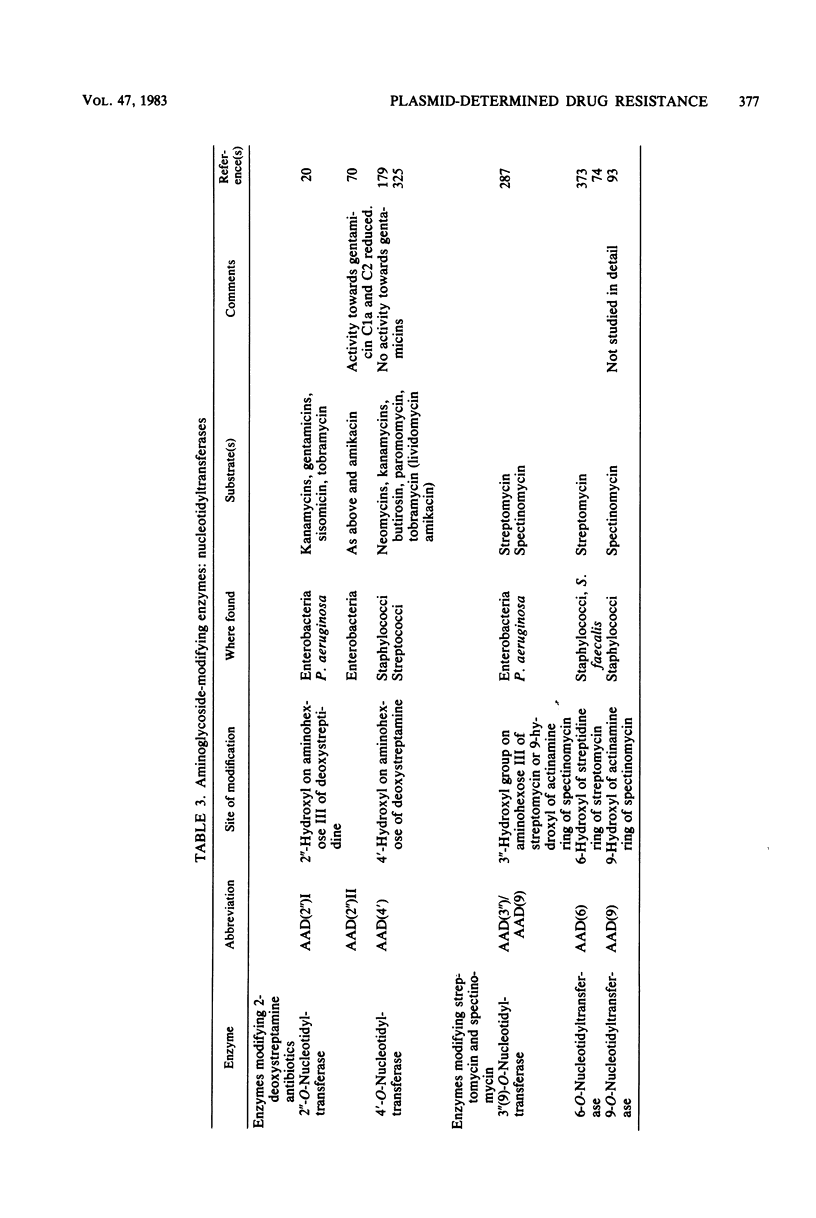









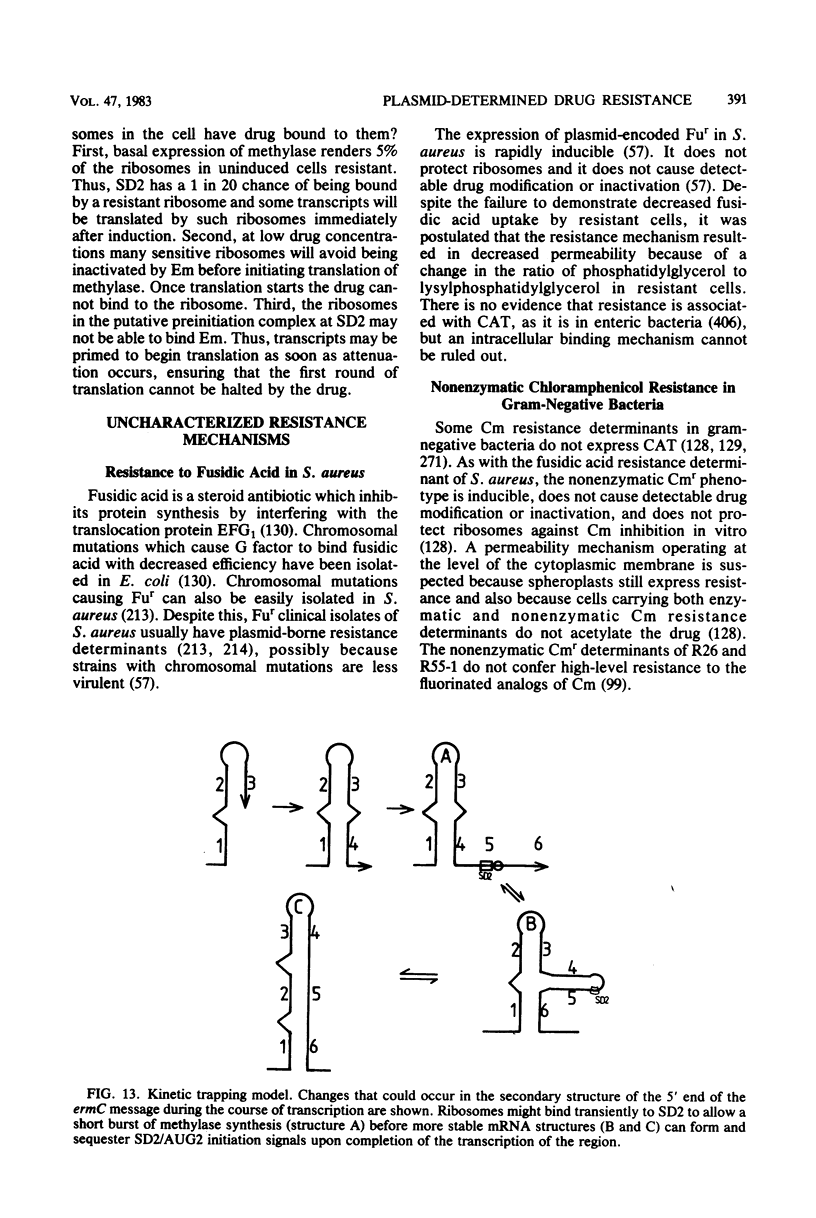

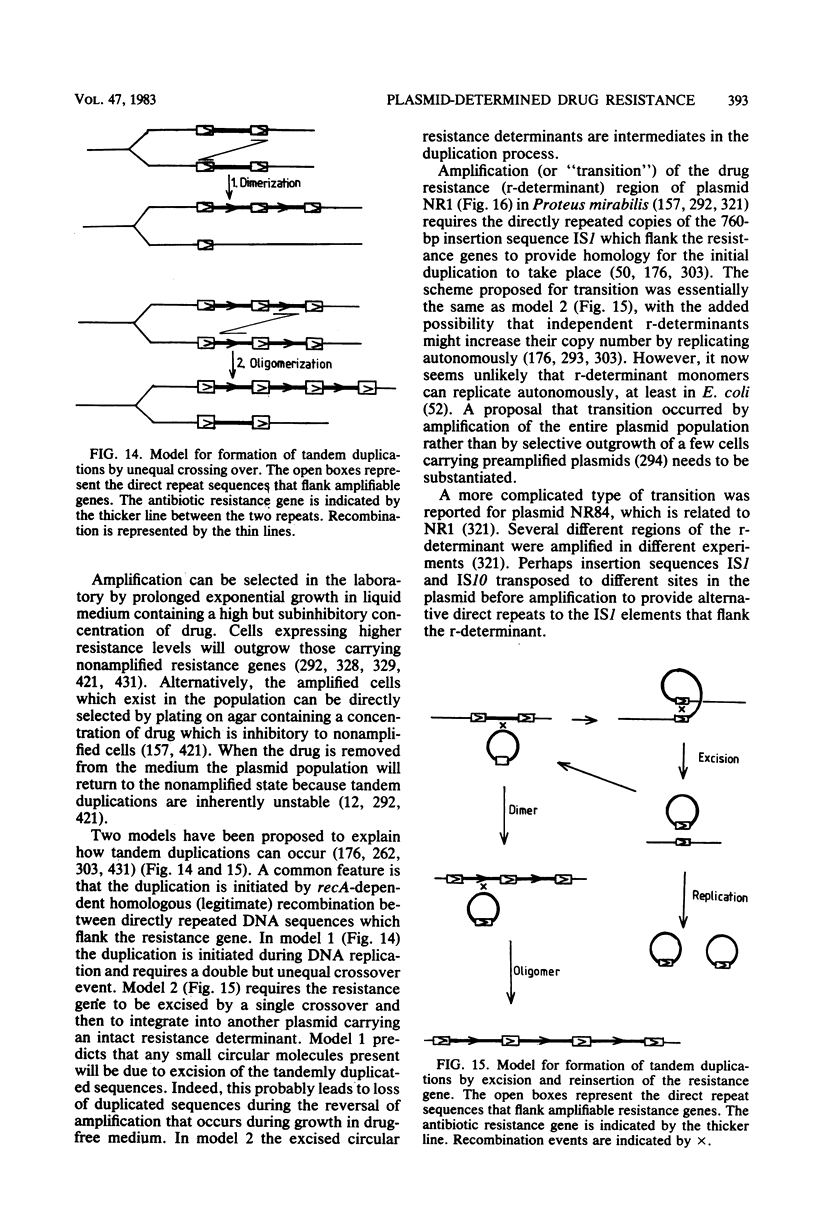
















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad M. H., Rechenmacher A., Böck A. Interaction between aminoglycoside uptake and ribosomal resistance mutations. Antimicrob Agents Chemother. 1980 Nov;18(5):798–806. doi: 10.1128/aac.18.5.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen N. E., Alborn W. E., Jr, Hobbs J. N., Jr, Kirst H. A. 7-Hydroxytropolone: an inhibitor of aminoglycoside-2"-O-adenylyltransferase. Antimicrob Agents Chemother. 1982 Nov;22(5):824–831. doi: 10.1128/aac.22.5.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen N. E. Macrolide resistance in Staphylococcus aureus: inducers of macrolide resistance. Antimicrob Agents Chemother. 1977 Apr;11(4):669–674. doi: 10.1128/aac.11.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbuchner J., Schmid K., Schmitt R. Tn1721-encoded tetracycline resistance: mapping of structural and regulatory genes mediating resistance. J Bacteriol. 1983 Jan;153(1):116–123. doi: 10.1128/jb.153.1.116-123.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Ambler R. P., Scott G. K. Partial amino acid sequence of penicillinase coded by Escherichia coli plasmid R6K. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3732–3736. doi: 10.1073/pnas.75.8.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P. The amino acid sequence of Staphylococcus aureus penicillinase. Biochem J. 1975 Nov;151(2):197–218. doi: 10.1042/bj1510197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Amyes S. G., Smith J. T. R-factor trimethoprim resistance mechanism: an insusceptible target site. Biochem Biophys Res Commun. 1974 May 20;58(2):412–418. doi: 10.1016/0006-291x(74)90380-5. [DOI] [PubMed] [Google Scholar]
- Amyes S. G., Smith J. T. The purification and properties of the trimethoprim-resistant dihydrofolate reductase mediated by the R-factor, R388. Eur J Biochem. 1976 Jan 15;61(2):597–603. doi: 10.1111/j.1432-1033.1976.tb10055.x. [DOI] [PubMed] [Google Scholar]
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Annear D. I., Mee B. J., Bailey M. Instability and linkage of silver resistance, lactose fermentation, and colony structure in Enterobacter cloacae from burn wounds. J Clin Pathol. 1976 May;29(5):441–443. doi: 10.1136/jcp.29.5.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asheshov E. H. The genetics of tetracycline resistance in Staphylococcus aureus. J Gen Microbiol. 1975 May;88(1):132–140. doi: 10.1099/00221287-88-1-132. [DOI] [PubMed] [Google Scholar]
- BROWN G. M. The biosynthesis of folic acid. II. Inhibition by sulfonamides. J Biol Chem. 1962 Feb;237:536–540. [PubMed] [Google Scholar]
- Ball P. R., Chopra I., Eccles S. J. Accumulation of tetracyclines by Escherichia coli K-12. Biochem Biophys Res Commun. 1977 Aug 22;77(4):1500–1507. doi: 10.1016/s0006-291x(77)80148-4. [DOI] [PubMed] [Google Scholar]
- Ball P. R., Shales S. W., Chopra I. Plasmid-mediated tetracycline resistance in Escherichia coli involves increased efflux of the antibiotic. Biochem Biophys Res Commun. 1980 Mar 13;93(1):74–81. doi: 10.1016/s0006-291x(80)80247-6. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Datta N., Hedges R. W., Grinter N. J. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J Bacteriol. 1976 Mar;125(3):800–810. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C. F., Mutzel R., Barbé J., Müller W. A multifunctional gene (tetR) controls Tn10-encoded tetracycline resistance. J Bacteriol. 1982 May;150(2):633–642. doi: 10.1128/jb.150.2.633-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste R., Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2276–2280. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste R., Davies J. Enzymatic acetylation of aminoglycoside antibiotics by Escherichia coli carrying an R factor. Biochemistry. 1971 May 11;10(10):1787–1796. doi: 10.1021/bi00786a009. [DOI] [PubMed] [Google Scholar]
- Benveniste R., Davies J. R-factor mediated gentamicin resistance: A new enzyme which modifies aminoglycoside antibiotics. FEBS Lett. 1971 May 20;14(5):293–296. doi: 10.1016/0014-5793(71)80282-x. [DOI] [PubMed] [Google Scholar]
- Beppu T., Arima K. Induction by mercuric ion of extensive degradation of cellular ribonucleic acid in Escherichia coli. J Bacteriol. 1969 Jun;98(3):888–897. doi: 10.1128/jb.98.3.888-897.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström S., Olsson O., Normark S. Common evolutionary origin of chromosomal beta-lactamase genes in enterobacteria. J Bacteriol. 1982 May;150(2):528–534. doi: 10.1128/jb.150.2.528-534.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddlecome S., Haas M., Davies J., Miller G. H., Rane D. F., Daniels P. J. Enzymatic modification of aminoglycoside antibiotics: a new 3-N-acetylating enzyme from a Pseudomonas aeruginosa isolate. Antimicrob Agents Chemother. 1976 Jun;9(6):951–955. doi: 10.1128/aac.9.6.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham A. H., Bruton C. J., Atkinson T. Isolation and partial characterization of four plasmids from antibiotic-resistant thermophilic bacilli. J Gen Microbiol. 1979 Oct;114(2):401–408. doi: 10.1099/00221287-114-2-401. [DOI] [PubMed] [Google Scholar]
- Bobrowski M. M., Matthew M., Barth P. T., Datta N., Grinter N. J., Jacob A. E., Kontomichalou P., Dale J. W., Smith J. T. Plasmid-determined beta-lactamase indistinguishable from the chromosomal beta-lactamase of Escherichia coli. J Bacteriol. 1976 Jan;125(1):149–157. doi: 10.1128/jb.125.1.149-157.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongaerts G. P., Kaptijn G. M. Aminoglycoside phosphotransferase-II-mediated amikacin resistance in Escherichia coli. Antimicrob Agents Chemother. 1981 Sep;20(3):344–350. doi: 10.1128/aac.20.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg P. D., Rainnie D. J. The effect of silver ions on the respiratory chain of Escherichia coli. Can J Microbiol. 1974 Jun;20(6):883–889. doi: 10.1139/m74-135. [DOI] [PubMed] [Google Scholar]
- Britz M. L., Wilkinson R. G. Chloramphenicol acetyltransferase of Bacteroides fragilis. Antimicrob Agents Chemother. 1978 Jul;14(1):105–111. doi: 10.1128/aac.14.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan L. E., Haraphongse R., Van den Elzen H. M. Gentamicin resistance in clinical-isolates of Pseudomonas aeruginosa associated with diminished gentamicin accumulation and no detectable enzymatic modification. J Antibiot (Tokyo) 1976 Jul;29(7):743–753. doi: 10.7164/antibiotics.29.743. [DOI] [PubMed] [Google Scholar]
- Bryan L. E., Kwan S. Mechanisms of aminoglycoside resistance of anaerobic bacteria and facultative bacteria grown anaerobically. J Antimicrob Chemother. 1981 Dec;8 (Suppl 500):1–8. doi: 10.1093/jac/8.suppl_d.1. [DOI] [PubMed] [Google Scholar]
- Bryan L. E., Nicas T., Holloway B. W., Crowther C. Aminoglycoside-resistant mutation of Pseudomonas aeruginosa defective in cytochrome c552 and nitrate reductase. Antimicrob Agents Chemother. 1980 Jan;17(1):71–79. doi: 10.1128/aac.17.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan L. E., Van Den Elzen H. M. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob Agents Chemother. 1977 Aug;12(2):163–177. doi: 10.1128/aac.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan L. E., Van den Elzen H. M. Streptomycin accumulation in susceptible and resistant strains of Escherichia coli and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1976 Jun;9(6):928–938. doi: 10.1128/aac.9.6.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinska M., Benveniste R., Davies J., Daniels P. J., Weinstein J. Gentamicin resistance in strains of Pseudomonas aeruginosa mediated by enzymatic N-acetylation of the deoxystreptamine moiety. Biochemistry. 1972 Feb 29;11(5):761–765. doi: 10.1021/bi00755a013. [DOI] [PubMed] [Google Scholar]
- Buckel P., Buchberger A., Böck A., Wittmann H. G. Alteration of ribosomal protein L6 in mutants of Escherichia coli resistant to gentamicin. Mol Gen Genet. 1977 Dec 14;158(1):47–54. doi: 10.1007/BF00455118. [DOI] [PubMed] [Google Scholar]
- Burchall J. J., Hitchings G. H. Inhibitor binding analysis of dihydrofolate reductases from various species. Mol Pharmacol. 1965 Sep;1(2):126–136. [PubMed] [Google Scholar]
- Burdett V., Inamine J., Rajagopalan S. Heterogeneity of tetracycline resistance determinants in Streptococcus. J Bacteriol. 1982 Mar;149(3):995–1004. doi: 10.1128/jb.149.3.995-1004.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHABBERT Y. Antagonisme in vitro entre l'érythromycine et la spiramycine. Ann Inst Pasteur (Paris) 1956 Jun;90(6):787–790. [PubMed] [Google Scholar]
- Cabello F., Timmis K., Cohen S. N. Replication control in a composite plasmid constructed by in vitro linkage of two distinct replicons. Nature. 1976 Jan 29;259(5541):285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- Campbell B. D., Kadner R. J. Relation of aerobiosis and ionic strength to the uptake of dihydrostreptomycin in Escherichia coli. Biochim Biophys Acta. 1980 Nov 5;593(1):1–10. doi: 10.1016/0005-2728(80)90002-x. [DOI] [PubMed] [Google Scholar]
- Cella R., Vining L. C. Action of streptomycin on the growth of Streptomyces griseus. Can J Microbiol. 1974 Nov;20(11):1591–1597. doi: 10.1139/m74-246. [DOI] [PubMed] [Google Scholar]
- Cella R., Vining L. C. Rsistance to streptomycin in a producing strain of Streptomyces griseus. Can J Microbiol. 1975 Apr;21(4):463–472. doi: 10.1139/m75-065. [DOI] [PubMed] [Google Scholar]
- Cerná J., Rychlík I., Pulkrábek P. The effect of antibiotics on the coded binding of peptidyl-tRNA to the ribosome and on the transfer of the peptidyl residue to puromycin. Eur J Biochem. 1969 May 1;9(1):27–35. doi: 10.1111/j.1432-1033.1969.tb00571.x. [DOI] [PubMed] [Google Scholar]
- Chabbert Y. A., Scavizzi M. R. Chelocardin-inducible resistance in Escherichia coli bearing R plasmids. Antimicrob Agents Chemother. 1976 Jan;9(1):36–41. doi: 10.1128/aac.9.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Allet B., Gallay E., Boy de La Tour E., Caro L. Involvement of IS1 in the dissociation of the r-determinant and RTF components of the plasmid R100.1. Mol Gen Genet. 1977 Jun 24;153(3):289–295. doi: 10.1007/BF00431594. [DOI] [PubMed] [Google Scholar]
- Chandler M., Silver L., Frey J., Caro L. Suppression of an Escherichia coli dnaA mutation by the integrated R factor R100.1: generation of small plasmids after integration. J Bacteriol. 1977 Apr;130(1):303–311. doi: 10.1128/jb.130.1.303-311.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., de la Tour E. B., Willems D., Caro L. Some properties of the chloramphenicol resistance transposon Tn9. Mol Gen Genet. 1979 Oct 3;176(2):221–231. doi: 10.1007/BF00273216. [DOI] [PubMed] [Google Scholar]
- Chevereau M., Daniels P. J., Davies J., LeGoffic F. Aminoglycoside resistance in bacteria medicated by gentamicin acetyltransferase II, an enzyme modifying the 2'-amino group of aminoglycoside antibiotics. Biochemistry. 1974 Jan 29;13(3):598–603. doi: 10.1021/bi00700a030. [DOI] [PubMed] [Google Scholar]
- Chiang S. J., Clowes R. C. Intramolecular transposition and inversion in plasmid R6K. J Bacteriol. 1980 May;142(2):668–682. doi: 10.1128/jb.142.2.668-682.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Ball P. Transport of antibiotics into bacteria. Adv Microb Physiol. 1982;23:183–240. doi: 10.1016/s0065-2911(08)60338-0. [DOI] [PubMed] [Google Scholar]
- Chopra I. Decreased uptake of cadmium by a resistant strain of Staphylococcus aureus. J Gen Microbiol. 1970 Oct;63(2):265–267. doi: 10.1099/00221287-63-2-265. [DOI] [PubMed] [Google Scholar]
- Chopra I., Eccles S. J. Diffusion of tetracycline across the outer membrane of Escherichia coli K-12: involvement of protein Ia. Biochem Biophys Res Commun. 1978 Jul 28;83(2):550–557. doi: 10.1016/0006-291x(78)91025-2. [DOI] [PubMed] [Google Scholar]
- Chopra I., Howe T. G. Bacterial resistance to the tetracyclines. Microbiol Rev. 1978 Dec;42(4):707–724. doi: 10.1128/mr.42.4.707-724.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Lacey R. W., Connolly J. Biochemical and genetic basis of tetracycline resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1974 Oct;6(4):397–404. doi: 10.1128/aac.6.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I. Mechanism of plasmic-mediated resistance to cadmium in Staphylococcus aureus. Antimicrob Agents Chemother. 1975 Jan;7(1):8–14. doi: 10.1128/aac.7.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I. Mechanisms of resistance to fusidic acid in Staphylococcus aureus. J Gen Microbiol. 1976 Oct;96(2):229–238. doi: 10.1099/00221287-96-2-229. [DOI] [PubMed] [Google Scholar]
- Chopra I., Shales S., Ball P. Tetracycline resistance determinants from groups A to D vary in their ability to confer decreased accumulation of tetracycline derivatives by Escherichia coli. J Gen Microbiol. 1982 Apr;128(4):689–692. doi: 10.1099/00221287-128-4-689. [DOI] [PubMed] [Google Scholar]
- Clark D. L., Weiss A. A., Silver S. Mercury and organomercurial resistances determined by plasmids in Pseudomonas. J Bacteriol. 1977 Oct;132(1):186–196. doi: 10.1128/jb.132.1.186-196.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B. Plasmids, drug resistance, and gene transfer in the genus Streptococcus. Microbiol Rev. 1981 Sep;45(3):409–436. doi: 10.1128/mr.45.3.409-436.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Bauer B. Plasmid-determined tetracycline resistance in Streptococcus faecalis: evidence for gene amplification during growth in presence of tetracycline. Proc Natl Acad Sci U S A. 1975 May;72(5):1720–1724. doi: 10.1073/pnas.72.5.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. A., Pratt R. F. Inactivation of Bacillus cereus beta-lactamase I by 6 beta-bromopencillanic acid: mechanism. Biochemistry. 1980 Aug 19;19(17):3996–4003. doi: 10.1021/bi00558a017. [DOI] [PubMed] [Google Scholar]
- Coleman D. C., Chopra I., Shales S. W., Howe T. G., Foster T. J. Analysis of tetracycline resistance encoded by transposon Tn10: deletion mapping of tetracycline-sensitive point mutations and identification of two structural genes. J Bacteriol. 1983 Feb;153(2):921–929. doi: 10.1128/jb.153.2.921-929.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman D. C., Foster T. J. Analysis of the reduction in expression of tetracycline resistance determined by transposon Tn10 in the multicopy state. Mol Gen Genet. 1981;182(1):171–177. doi: 10.1007/BF00422786. [DOI] [PubMed] [Google Scholar]
- Coombe R. G., George A. M. New plasmid-mediated aminoglycoside adenylyltransferase of broad substrate range that adenylylates amikacin. Antimicrob Agents Chemother. 1981 Jul;20(1):75–80. doi: 10.1128/aac.20.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Cheng K. J. The role of the bacterial cell envelope in antibiotic resistance. J Antimicrob Chemother. 1975 Dec;1(4):363–377. doi: 10.1093/jac/1.4.363. [DOI] [PubMed] [Google Scholar]
- Courvalin P. M., Shaw W. V., Jacob A. E. Plasmid-mediated mechanisms of resistance to aminoglycoside-aminocyclitol antibiotics and to chloramphenicol in group D streptococci. Antimicrob Agents Chemother. 1978 May;13(5):716–725. doi: 10.1128/aac.13.5.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P., Davies J. Plasmid-medicated aminoglycoside phosphotransferase of broad substrate range that phosphorylates amikacin. Antimicrob Agents Chemother. 1977 Apr;11(4):619–624. doi: 10.1128/aac.11.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P., Weisblum B., Davies J. Aminoglycoside-modifying enzyme of an antibiotic-producing bacterium acts as a determinant of antibiotic resistance in Escherichia coli. Proc Natl Acad Sci U S A. 1977 Mar;74(3):999–1003. doi: 10.1073/pnas.74.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowlesmith I., Howe T. G. Characterization of beta-lactamase-deficient (bla) mutants of the R plasmid R1 in Escherichia coli K-12 and comparison with similar mutants of RP1. Antimicrob Agents Chemother. 1980 Nov;18(5):667–674. doi: 10.1128/aac.18.5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowlesmith I., Howe T. G. Quantitative correlation between penicillin resistance and beta-lactamase activity specified by the R plasmids R1, R1 bla-45, and RP1 in Escherichia coli K-12. Antimicrob Agents Chemother. 1980 Nov;18(5):675–679. doi: 10.1128/aac.18.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundliffe E., Thompson J. The mode of action of nosiheptide (multhiomycin) and the mechanism of resistance in the producing organism. J Gen Microbiol. 1981 Sep;126(1):185–192. doi: 10.1099/00221287-126-1-185. [DOI] [PubMed] [Google Scholar]
- Curiale M. S., Levy S. B. Two complementation groups mediate tetracycline resistance determined by Tn10. J Bacteriol. 1982 Jul;151(1):209–215. doi: 10.1128/jb.151.1.209-215.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis N. A., Brown C., Boxall M., Boulton M. G. Modified peptidoglycan transpeptidase activity in a carbenicillin-resistant mutant of Pseudomonas aeruginosa 18s. Antimicrob Agents Chemother. 1978 Aug;14(2):246–251. doi: 10.1128/aac.14.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis N. A., Orr D., Boulton M. G., Ross G. W. Penicillin-binding proteins of Pseudomonas aeruginosa. Comparison of two strains differing in their resistance to beta-lactam antibiotics. J Antimicrob Chemother. 1981 Feb;7(2):127–136. doi: 10.1093/jac/7.2.127. [DOI] [PubMed] [Google Scholar]
- Curtis N. A., Richmond M. H. Effect of R-factor-mediated genes on some surface properties of Escherichia coli. Antimicrob Agents Chemother. 1974 Dec;6(6):666–671. doi: 10.1128/aac.6.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J. W., Smith J. T. R-factor-mediated beta-lactamases that hydrolyze oxacillin: evidence for two distinct groups. J Bacteriol. 1974 Aug;119(2):351–356. doi: 10.1128/jb.119.2.351-356.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. M., Ward D. C. Mercurated polynucleotides: new probes for hybridization and selective polymer fractionation. Biochemistry. 1975 Jun 3;14(11):2458–2469. doi: 10.1021/bi00682a028. [DOI] [PubMed] [Google Scholar]
- Damper P. D., Epstein W. Role of the membrane potential in bacterial resistance to aminoglycoside antibiotics. Antimicrob Agents Chemother. 1981 Dec;20(6):803–808. doi: 10.1128/aac.20.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang-Van A., Tiraby G., Acar J. F., Shaw W. V., Bouanchaud D. H. Chloramphenicol resistance in Streptococcus pneumoniae: enzymatic acetylation and possible plasmid linkage. Antimicrob Agents Chemother. 1978 Apr;13(4):577–583. doi: 10.1128/aac.13.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Dacey S., Hughes V., Knight S., Richards H., Williams G., Casewell M., Shannon K. P. Distribution of genes for trimethoprim and gentamicin resistance in bacteria and their plasmids in a general hospital. J Gen Microbiol. 1980 Jun;118(2):495–508. doi: 10.1099/00221287-118-2-495. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Becker D., Davies J. Plasmid-determined fusidic acid resistance in the Enterobacteriaceae. J Gen Microbiol. 1974 Jul;83(0):191–196. doi: 10.1099/00221287-83-1-191. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Datta N., Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature. 1965 Oct 16;208(5007):239–241. doi: 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- Davies J. E., Benveniste R. E. Enzymes that inactivate antibiotics in transit to their targets. Ann N Y Acad Sci. 1974 May 10;235(0):130–136. doi: 10.1111/j.1749-6632.1974.tb43262.x. [DOI] [PubMed] [Google Scholar]
- Davies J., O'Connor S. Enzymatic modification of aminoglycoside antibiotics: 3-N-acetyltransferase with broad specificity that determines resistance to the novel aminoglycoside apramycin. Antimicrob Agents Chemother. 1978 Jul;14(1):69–72. doi: 10.1128/aac.14.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Smith D. I. Plasmid-determined resistance to antimicrobial agents. Annu Rev Microbiol. 1978;32:469–518. doi: 10.1146/annurev.mi.32.100178.002345. [DOI] [PubMed] [Google Scholar]
- De Crombrugghe B., Pastan I., Shaw W. V., Rosner J. L. Stimulation by cyclic AMP and ppGpp of chloramphenicol acetyl transferase synthesis. Nat New Biol. 1973 Feb 21;241(112):237–239. doi: 10.1038/newbio241237a0. [DOI] [PubMed] [Google Scholar]
- Demain A. L. How do antibiotic-producing microorganisms avoid suicide? Ann N Y Acad Sci. 1974 May 10;235(0):601–612. doi: 10.1111/j.1749-6632.1974.tb43294.x. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B., Willetts N. S. Plasmid co-integrates of prophage lambda and R factor R100. J Bacteriol. 1976 Apr;126(1):166–176. doi: 10.1128/jb.126.1.166-176.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie P., Bryan L. E., Pickard M. A. Effect of enzymatic adenylylation on dihydrostreptomycin accumulation in Escherichia coli carrying an R-factor: model explaining aminoglycoside resistance by inactivating mechanisms. Antimicrob Agents Chemother. 1978 Oct;14(4):569–580. doi: 10.1128/aac.14.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty A., Grandi G., Grandi R., Gryczan T. J., Shivakumar A. G., Dubnau D. Naturally occurring macrolide-lincosamide-streptogramin B resistance in Bacillus licheniformis. J Bacteriol. 1981 Jan;145(1):129–137. doi: 10.1128/jb.145.1.129-137.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C. J., Foster T. J. Nonenzymatic chloramphenicol resistance determinants specified by plasmids R26 and R55-1 in Escherichia coli K-12 do not confer high-level resistance to fluorinated analogs. Antimicrob Agents Chemother. 1982 Nov;22(5):912–914. doi: 10.1128/aac.22.5.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty T. J., Koller A. E., Tomasz A. Penicillin-binding proteins of penicillin-susceptible and intrinsically resistant Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1980 Nov;18(5):730–737. doi: 10.1128/aac.18.5.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowding J. E. Mechanisms of gentamicin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1977 Jan;11(1):47–50. doi: 10.1128/aac.11.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyke K. G., Parker M. T., Richmond M. H. Penicillinase production and metal-ion resistance in Staphylococcus aureus cultures isolated from hospital patients. J Med Microbiol. 1970 Feb;3(1):125–136. doi: 10.1099/00222615-3-1-125. [DOI] [PubMed] [Google Scholar]
- Edlund T., Grundström T., Normark S. Isolation and characterization of DNA repetitions carrying the chromosomal beta-lactamase gene of Escherichia coli K-12. Mol Gen Genet. 1979 Jun 7;173(2):115–125. doi: 10.1007/BF00330301. [DOI] [PubMed] [Google Scholar]
- El Solh N., Ehrlich S. D. A small cadmium resistance plasmid isolated from Staphylococcus aureus. Plasmid. 1982 Jan;7(1):77–84. doi: 10.1016/0147-619x(82)90029-4. [DOI] [PubMed] [Google Scholar]
- Engberg B., Nordström K. Replication of R-factor R1 in Scherichia coli K-12 at different growth rates. J Bacteriol. 1975 Jul;123(1):179–186. doi: 10.1128/jb.123.1.179-186.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayolle F., Privitera G., Sebald M. Tetracycline transport in Bacteroides fragilis. Antimicrob Agents Chemother. 1980 Oct;18(4):502–505. doi: 10.1128/aac.18.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrazza D., Levy S. B. Biochemical and immunological characterization of an R plasmid-encoded protein with properties resembling those of major cellular outer membrane proteins. J Bacteriol. 1980 Oct;144(1):149–158. doi: 10.1128/jb.144.1.149-158.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J., Belasco J. G., Khosla S., Knowles J. R. beta-Lactamase proceeds via an acyl-enzyme intermediate. Interaction of the Escherichia coli RTEM enzyme with cefoxitin. Biochemistry. 1980 Jun 24;19(13):2895–2901. doi: 10.1021/bi00554a012. [DOI] [PubMed] [Google Scholar]
- Fisher J., Charnas R. L., Bradley S. M., Knowles J. R. Inactivation of the RTEM beta-lactamase from Escherichia coli. Interaction of penam sulfones with enzyme. Biochemistry. 1981 May 12;20(10):2726–2731. doi: 10.1021/bi00513a004. [DOI] [PubMed] [Google Scholar]
- Fitton J. E., Shaw W. V. Comparison of chloramphenicol acetyltransferase variants in staphylococci. Purification, inhibitor studies and N-terminal sequences. Biochem J. 1979 Feb 1;177(2):575–582. doi: 10.1042/bj1770575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fling M. E., Elwell L. P. Protein expression in Escherichia coli minicells containing recombinant plasmids specifying trimethoprim-resistant dihydrofolate reductases. J Bacteriol. 1980 Feb;141(2):779–785. doi: 10.1128/jb.141.2.779-785.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fling M. E., Walton L., Elwell L. P. Monitoring of plasmid-encoded, trimethoprim-resistant dihydrofolate reductase genes: detection of a new resistant enzyme. Antimicrob Agents Chemother. 1982 Nov;22(5):882–888. doi: 10.1128/aac.22.5.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S., Welch J. W. Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5342–5346. doi: 10.1073/pnas.79.17.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Nakahara H., Weiss A. A., Silver S. Transposon A-generated mutations in the mercuric resistance genes of plasmid R100-1. J Bacteriol. 1979 Oct;140(1):167–181. doi: 10.1128/jb.140.1.167-181.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Walsh A. Phenotypic characterization of R-factor tetracycline resistance determinants. Genet Res. 1974 Dec;24(3):333–343. doi: 10.1017/s0016672300015330. [DOI] [PubMed] [Google Scholar]
- Fox B., Walsh C. T. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem. 1982 Mar 10;257(5):2498–2503. [PubMed] [Google Scholar]
- Fox C. L., Jr, Modak S. M. Mechanism of silver sulfadiazine action on burn wound infections. Antimicrob Agents Chemother. 1974 Jun;5(6):582–588. doi: 10.1128/aac.5.6.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol. 1981 Jan;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa Y., Weisblum B. A family of r-determinants in Streptomyces spp. that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J Bacteriol. 1981 May;146(2):621–631. doi: 10.1128/jb.146.2.621-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth A. J. Purification and properties of a constitutive beta-lactamase from Pseudomonas aeruginosa strain Dalgleish. Biochim Biophys Acta. 1975 Feb 19;377(2):431–443. doi: 10.1016/0005-2744(75)90323-x. [DOI] [PubMed] [Google Scholar]
- GARROD L. P. The erythromycin group of antibiotics. Br Med J. 1957 Jul 13;2(5036):57–63. doi: 10.1136/bmj.2.5036.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney D. F., Cundliffe E., Foster T. J. Chloramphenicol resistance that does not involve chloramphenicol acetyltransferase encoded by plasmids from gram-negative bacteria. J Gen Microbiol. 1981 Jul;125(1):113–121. doi: 10.1099/00221287-125-1-113. [DOI] [PubMed] [Google Scholar]
- García-Lobo J. M., Ortiz J. M. Tn292l, a transposon encoding fosfomycin resistance. J Bacteriol. 1982 Jul;151(1):477–479. doi: 10.1128/jb.151.1.477-479.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayda R. C., Tanabe J. H., Knigge K. M., Markovitz A. Identification by deletion analysis of an inducible protein required for plasmid pSC101-mediated tetracycline resistance. Plasmid. 1979 Jul;2(3):417–425. doi: 10.1016/0147-619x(79)90025-8. [DOI] [PubMed] [Google Scholar]
- Georgopapadakou N. H., Smith S. A., Bonner D. P. Penicillin-binding proteins in a Staphylococcus aureus strain resistant to specific beta-lactam antibiotics. Antimicrob Agents Chemother. 1982 Jul;22(1):172–175. doi: 10.1128/aac.22.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey A. J., Bryan L. E. Mutation of Pseudomonas aeruginosa specifying reduced affinity for penicillin G. Antimicrob Agents Chemother. 1982 Feb;21(2):216–223. doi: 10.1128/aac.21.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey A. J., Bryan L. E., Rabin H. R. beta-Lactam-resistant Pseudomonas aeruginosa with modified penicillin-binding proteins emerging during cystic fibrosis treatment. Antimicrob Agents Chemother. 1981 May;19(5):705–711. doi: 10.1128/aac.19.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffic F. L., Moreau N., Siegrist S., Goldstein F. W., Acar J. C. La resistance plasmidique de Haemophilus sp. aux antibiotiques aminoglycosidiques : isolement et etude d'une nouvelle phosphotransférase. Ann Microbiol (Paris) 1977 May-Jun;128A(4):383–391. [PubMed] [Google Scholar]
- Goldman P. R., Northrop D. B. Preparation of stable gentamicin adenylyl transferase of high specific activity. Biochem Biophys Res Commun. 1975 Oct 27;66(4):1408–1413. doi: 10.1016/0006-291x(75)90516-1. [DOI] [PubMed] [Google Scholar]
- Goldman P. R., Northrop D. B. Purification and spectrophotometric assay of neomycin phosphotransferase II1;. Biochem Biophys Res Commun. 1976 Mar 8;69(1):230–236. doi: 10.1016/s0006-291x(76)80297-5. [DOI] [PubMed] [Google Scholar]
- Graham M. Y., Weisblum B. 23S ribosomal ribonucleic acid of macrolide-producing streptomycetes contains methylated adenine. J Bacteriol. 1979 Mar;137(3):1464–1467. doi: 10.1128/jb.137.3.1464-1467.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., van Embden J., Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974 Feb;117(2):619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Davis C. E. Incompatibility and host range of pGD10 from Capnocytophaga ochraceus, formerly Bacteroides ochraceus. Plasmid. 1982 Mar;7(2):196–198. doi: 10.1016/0147-619x(82)90078-6. [DOI] [PubMed] [Google Scholar]
- Haag R., Süssmuth R., Lingens F. The chloramphenicol resistance of a chloramphenicol-degrading soil bacterium. FEBS Lett. 1976 Mar 15;63(1):62–64. doi: 10.1016/0014-5793(76)80195-0. [DOI] [PubMed] [Google Scholar]
- Haas M., Biddlecome S., Davies J., Luce C. E., Daniels P. J. Enzymatic modification of aminoglycoside antibiotics: a new 6'-N-acetylating enzyme from a Pseudomonas aeruginosa isolate. Antimicrob Agents Chemother. 1976 Jun;9(6):945–950. doi: 10.1128/aac.9.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Grandi G., Gryczan T. J., Dubnau D. Translational attenuation of ermC: a deletion analysis. Mol Gen Genet. 1982;186(2):204–216. doi: 10.1007/BF00331851. [DOI] [PubMed] [Google Scholar]
- Hakenbeck R., Tarpay M., Tomasz A. Multiple changes of penicillin-binding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980 Mar;17(3):364–371. doi: 10.1128/aac.17.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Knowles J. R. Directed selective pressure on a beta-lactamase to analyse molecular changes involved in development of enzyme function. Nature. 1976 Dec 23;264(5588):803–804. doi: 10.1038/264803a0. [DOI] [PubMed] [Google Scholar]
- Hall B. M. Mercury resistance of Staphylococcus aureus. J Hyg (Lond) 1970 Mar;68(1):121–129. doi: 10.1017/s0022172400028576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton-Miller J. M. A novel method for evaluating K/K and its application to the competitive inhibition of staphylococcal penicillinase by cephalosporins. Biochem J. 1966 Dec;101(3):40C–42C. doi: 10.1042/bj1010040c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton-Miller J. M. beta-Lactamases and their clinical significance. J Antimicrob Chemother. 1982 Feb;9 (Suppl B):11–19. doi: 10.1093/jac/9.suppl_b.11. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. Aminoglycoside uptake and mode of action--with special reference to streptomycin and gentamicin. I. Antagonists and mutants. J Antimicrob Chemother. 1981 Oct;8(4):249–276. doi: 10.1093/jac/8.4.249. [DOI] [PubMed] [Google Scholar]
- Hancock R. E. Aminoglycoside uptake and mode of action-with special reference to streptomycin and gentamicin. II. Effects of aminoglycosides on cells. J Antimicrob Chemother. 1981 Dec;8(6):429–445. doi: 10.1093/jac/8.6.429. [DOI] [PubMed] [Google Scholar]
- Harder K. J., Nikaido H., Matsuhashi M. Mutants of Escherichia coli that are resistant to certain beta-lactam compounds lack the ompF porin. Antimicrob Agents Chemother. 1981 Oct;20(4):549–552. doi: 10.1128/aac.20.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood J., Smith D. H. Catabolite repression of chloramphenicol acetyl transferase synthesis in E. coli K12. Biochem Biophys Res Commun. 1971 Jan 8;42(1):57–62. doi: 10.1016/0006-291x(71)90361-5. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Baumberg S. Resistance to arsenic compounds conferred by a plasmid transmissible between strains of Escherichia coli. J Bacteriol. 1973 Jul;115(1):459–460. doi: 10.1128/jb.115.1.459-460.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Hedstrom R. C., Crider B. P., Eagon R. G. Comparison of kinetics of active tetracycline uptake and active tetracycline efflux in sensitive and plasmid RP4-containing Pseudomonas putida. J Bacteriol. 1982 Oct;152(1):255–259. doi: 10.1128/jb.152.1.255-259.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz C., Braun C. B., Rosano C. L. Role of ribosome recycling in uptake of dihydrostreptomycin by sensitive and resistant Escherichia coli. Biochim Biophys Acta. 1981 Jan 29;652(1):168–176. doi: 10.1016/0005-2787(81)90220-3. [DOI] [PubMed] [Google Scholar]
- Höltje J. V. Induction of streptomycin uptake in resistant strains of Escherichia coli. Antimicrob Agents Chemother. 1979 Feb;15(2):177–181. doi: 10.1128/aac.15.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje J. V. Streptomycin uptake via an inducible polyamine transport system in Escherichia coli. Eur J Biochem. 1978 May 16;86(2):345–351. doi: 10.1111/j.1432-1033.1978.tb12316.x. [DOI] [PubMed] [Google Scholar]
- Iida S., Arber W. Plaque forming specialized transducing phage P1: isolation of P1CmSmSu, a precursor of P1Cm. Mol Gen Genet. 1977 Jun 24;153(3):259–269. doi: 10.1007/BF00431591. [DOI] [PubMed] [Google Scholar]
- Iida S., Meyer J., Linder P., Goto N., Nakaya R., Reif H. J., Arber W. The kanamycin resistance transposon Tn2680 derived from the R plasmid Rts1 and carried by phage P1Km has flanking 0.8-kb-long direct repeats. Plasmid. 1982 Sep;8(2):187–198. doi: 10.1016/0147-619x(82)90056-7. [DOI] [PubMed] [Google Scholar]
- Imsande J. Genetic regulation of penicillinase synthesis in Gram-positive bacteria. Microbiol Rev. 1978 Mar;42(1):67–83. doi: 10.1128/mr.42.1.67-83.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Matthew M. The distribution of beta-lactamase genes on plasmids found in Pseudomonas. Plasmid. 1979 Jan;2(1):41–47. doi: 10.1016/0147-619x(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Jahn G., Laufs R., Kaulfers P. M., Kolenda H. Molecular nature of two Haemophilus influenzae R factors containing resistances and the multiple integration of drug resistance transposons. J Bacteriol. 1979 May;138(2):584–597. doi: 10.1128/jb.138.2.584-597.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., Dyke K. G. Ethidium bromide resistance, a new marker on the staphylococcal penicillinase plasmid. J Bacteriol. 1969 Dec;100(3):1413–1414. doi: 10.1128/jb.100.3.1413-1414.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahan F. M., Kahan J. S., Cassidy P. J., Kropp H. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci. 1974 May 10;235(0):364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J., Brown P. C., Schimke R. T. Amplified dihydrofolate reductase genes in unstably methotrexate-resistant cells are associated with double minute chromosomes. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5669–5673. doi: 10.1073/pnas.76.11.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe H., Kobayashi F., Yamaguchi M., Utahara R., Mitsuhashi S. 3"-phosphoryldihydrostreptomycin produced by the inactivating enzyme of Pseudomonas aeruginosa. J Antibiot (Tokyo) 1971 Sep;24(9):651–652. doi: 10.7164/antibiotics.24.651. [DOI] [PubMed] [Google Scholar]
- Kawabe H., Kondo S., Umezawa H., Mitsuhashi S. R factor-mediated aminoglycoside antibiotic resistance in Pseudomonas aeruginosa: a new aminoglycoside 6'-N-acetyltransferase. Antimicrob Agents Chemother. 1975 May;7(5):494–499. doi: 10.1128/aac.7.5.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe H., Naito T., Mitsuhashi S. Acetylation of amikacin, a new semisynthetic antibiotic, by Pseudomonas aeruginosa carrying an R factor. Antimicrob Agents Chemother. 1975 Jan;7(1):50–54. doi: 10.1128/aac.7.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keggins K. M., Lovett P. S., Duvall E. J. Molecular cloning of genetically active fragments of Bacillus DNA in Bacillus subtilis and properties of the vector plasmid pUB110. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1423–1427. doi: 10.1073/pnas.75.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitts P., Symington L., Burke M., Reed R., Sherratt D. Transposon-specified site-specific recombination. Proc Natl Acad Sci U S A. 1982 Jan;79(1):46–50. doi: 10.1073/pnas.79.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Knott-Hunziker V., Petursson S., Waley S. G., Jaurin B., Grundström T. The acyl-enzyme mechanism of beta-lactamase action. The evidence for class C Beta-lactamases. Biochem J. 1982 Nov 1;207(2):315–322. doi: 10.1042/bj2070315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi F., Koshi T., Eda J., Yoshimura Y., Mitsuhashi S. Lividomycin resistance in staphylococci by enzymatic phosphorylation. Antimicrob Agents Chemother. 1973 Jul;4(1):1–5. doi: 10.1128/aac.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu Y., Murakami K., Nishikawa T. Penetration of moxalactam into its target proteins in Escherichia coli K-12: comparison of a highly moxalactam resistant mutant with its parent strain. Antimicrob Agents Chemother. 1981 Nov;20(5):613–619. doi: 10.1128/aac.20.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo I., Ishikawa T., Nakahara H. Mercury and cadmium resistances mediated by the penicillinase plasmid in Staphylococcus aureus. J Bacteriol. 1974 Jan;117(1):1–7. doi: 10.1128/jb.117.1.1-7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K., Okuyama A., Tanaka N. Differential effects of antibiotics on peptidyl transferase reactions. Biochem Biophys Res Commun. 1972 Jun 9;47(5):1196–1202. doi: 10.1016/0006-291x(72)90961-8. [DOI] [PubMed] [Google Scholar]
- Labia R., Guionie M., Barthélémy M. Properties of three carbenicillin-hydrolysing beta-lactamases (CARB) from Pseudomonas aeruginosa: identification of a new enzyme. J Antimicrob Chemother. 1981 Jan;7(1):49–56. doi: 10.1093/jac/7.1.49. [DOI] [PubMed] [Google Scholar]
- Lacey R. W. Antibiotic resistance plasmids of Staphylococcus aureus and their clinical importance. Bacteriol Rev. 1975 Mar;39(1):1–32. doi: 10.1128/br.39.1.1-32.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey R. W., Chopra I. Evidence for mutation to streptomycin resistance in clinical strains of Staphylococcus aureus. J Gen Microbiol. 1972 Nov;73(1):175–180. doi: 10.1099/00221287-73-1-175. [DOI] [PubMed] [Google Scholar]
- Lacey R. W., Grinsted J. Linkage of fusidic acid resistance to the penicillinase plasmid in Staphylococcus aureus. J Gen Microbiol. 1972 Dec;73(3):501–508. doi: 10.1099/00221287-73-3-501. [DOI] [PubMed] [Google Scholar]
- Lacey R. W., Rosdahl V. T. An unusual "penicillinase plasmid" in staphylococcus aureus; evidence for its transfer under natural conditions. J Med Microbiol. 1974 Feb;7(1):1–9. doi: 10.1099/00222615-7-1-1. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Dahlberg J. E., Weisblum B. Structure of an inducibly methylatable nucleotide sequence in 23S ribosomal ribonucleic acid from erythromycin-resistant Staphylococcus aureus. Biochemistry. 1973 Jan 30;12(3):457–460. doi: 10.1021/bi00727a015. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci U S A. 1971 Apr;68(4):856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. J., Weisblum B., Fahnestock S. R., Nomura M. Alteration of 23 S ribosomal RNA and erythromycin-induced resistance to lincomycin and spiramycin in Staphylococcus aureus. J Mol Biol. 1973 Feb 15;74(1):67–72. doi: 10.1016/0022-2836(73)90355-0. [DOI] [PubMed] [Google Scholar]
- Lane D., Chandler M. Mapping of the drug resistance genes carried by the r-determinant of the R100.1 plasmid. Mol Gen Genet. 1977 Nov 29;157(1):17–23. doi: 10.1007/BF00268682. [DOI] [PubMed] [Google Scholar]
- Le Goffic F., Martel A., Capmau M. L., Baca B., Goebel P., Chardon H., Soussy C. J., Duval J., Bouanchaud D. H. New plasmid-mediated nucleotidylation of aminoglycoside antibiotics in Staphlococcus aureus. Antimicrob Agents Chemother. 1976 Aug;10(2):258–264. doi: 10.1128/aac.10.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goffic F., Martel A. La résistance aux aminosides provoquée par une isoenzyme la kanamycine acétyltransférase. Biochimie. 1974;56(6-7):893–897. doi: 10.1016/s0300-9084(74)80512-2. [DOI] [PubMed] [Google Scholar]
- Le Goffic F., Martel A., Witchitz J. 3-N enzymatic acetylation of gentamicin, tobramycin, and kanamycin by Escherichia coli carrying an R factor. Antimicrob Agents Chemother. 1974 Dec;6(6):680–684. doi: 10.1128/aac.6.6.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grice S. F., Matzura H., Marcoli R., Iida S., Bickle T. A. The catabolite-sensitive promoter for the chloramphenicol acetyl transferase gene is preceded by two binding sites for the catabolite gene activator protein. J Bacteriol. 1982 Apr;150(1):312–318. doi: 10.1128/jb.150.1.312-318.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Lee L. N. Characterization of two tetracycline resistance determinants in Streptococcus faecalis JH1. J Bacteriol. 1982 May;150(2):835–843. doi: 10.1128/jb.150.2.835-843.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboul J., Davies J. Enzymatic modification of hygromycin B in Streptomyces hygroscopicus. J Antibiot (Tokyo) 1982 Apr;35(4):527–528. doi: 10.7164/antibiotics.35.527. [DOI] [PubMed] [Google Scholar]
- Lerch K. Copper metallothionein, a copper-binding protein from Neurospora crassa. Nature. 1980 Mar 27;284(5754):368–370. doi: 10.1038/284368a0. [DOI] [PubMed] [Google Scholar]
- Levesque R., Roy P. H., Letarte R., Pechère J. C. A plasmid-mediated cephalosporinase from Achromobacter species. J Infect Dis. 1982 May;145(5):753–761. doi: 10.1093/infdis/145.2.753. [DOI] [PubMed] [Google Scholar]
- Levesque R., Roy P. H. Mapping of the plasmid (pLQ3) from Achromobacter and cloning of its cephalosporinase gene in Escherichia coli. Gene. 1982 Apr;18(1):69–75. doi: 10.1016/0378-1119(82)90057-9. [DOI] [PubMed] [Google Scholar]
- Levy S. B., McMurry L. Plasmid-determined tetracycline resistance involves new transport systems for tetracycline. Nature. 1978 Nov 2;276(5683):90–92. doi: 10.1038/276090a0. [DOI] [PubMed] [Google Scholar]
- León J., García-Lobo J. M., Ortiz J. M. Fosfomycin resistance plasmids do not affect fosfomycin transport into Escherichia coli. Antimicrob Agents Chemother. 1982 Apr;21(4):608–612. doi: 10.1128/aac.21.4.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar M. K., Majumdar S. K. Relationship between alkaline phosphatase and neomycin formation in Streptomyces fradiae. Biochem J. 1971 May;122(4):397–404. doi: 10.1042/bj1220397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malke H., Reichardt W., Hartmann M., Walter F. Genetic study of plasmid-associated zonal resistance to lincomycin in Streptococcus pyogenes. Antimicrob Agents Chemother. 1981 Jan;19(1):91–100. doi: 10.1128/aac.19.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. C., Robishaw E. E. Effects of macrolides on peptide-bond formation and translocation. Biochemistry. 1971 May 25;10(11):2054–2061. doi: 10.1021/bi00787a014. [DOI] [PubMed] [Google Scholar]
- Mao J. C., Robishaw E. E. Erythromycin, a peptidyltransferase effector. Biochemistry. 1972 Dec 5;11(25):4864–4872. doi: 10.1021/bi00775a035. [DOI] [PubMed] [Google Scholar]
- Marcoli R., Iida S., Bickle T. A. The DNA sequence of an IS/-flanked transposon coding for resistance to chloramphenicol and fusidic acid. FEBS Lett. 1980 Jan 28;110(1):11–14. doi: 10.1016/0014-5793(80)80011-1. [DOI] [PubMed] [Google Scholar]
- Martel A., Moreau N., Capmau M. L., Soussy C. J., Duval J. 2"-O-phosphorylation of gentamicin components by a Staphylococcus aureus strain carrying a plasmid. Antimicrob Agents Chemother. 1977 Jul;12(1):26–30. doi: 10.1128/aac.12.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mates S. M., Eisenberg E. S., Mandel L. J., Patel L., Kaback H. R., Miller M. H. Membrane potential and gentamicin uptake in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6693–6697. doi: 10.1073/pnas.79.21.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew M., Hedges R. W., Smith J. T. Types of beta-lactamase determined by plasmids in gram-negative bacteria. J Bacteriol. 1979 Jun;138(3):657–662. doi: 10.1128/jb.138.3.657-662.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew M. Plasmid-mediated beta-lactamases of Gram-negative bacteria: properties and distribution. J Antimicrob Chemother. 1979 Jul;5(4):349–358. doi: 10.1093/jac/5.4.349. [DOI] [PubMed] [Google Scholar]
- Matthew M., Sykes R. B. Properties of the beta-lactamase specified by the Pseudomonas plasmid RPL11. J Bacteriol. 1977 Oct;132(1):341–345. doi: 10.1128/jb.132.1.341-345.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L. M., Cullinane J. C., Levy S. B. Transport of the lipophilic analog minocycline differs from that of tetracycline in susceptible and resistant Escherichia coli strains. Antimicrob Agents Chemother. 1982 Nov;22(5):791–799. doi: 10.1128/aac.22.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L., Levy S. B. Two transport systems for tetracycline in sensitive Escherichia coli: critical role for an initial rapid uptake system insensitive to energy inhibitors. Antimicrob Agents Chemother. 1978 Aug;14(2):201–209. doi: 10.1128/aac.14.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L., Petrucci R. E., Jr, Levy S. B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J., Iida S. Amplification of chloramphenicol resistance transposons carried by phage P1Cm in Escherichia coli. Mol Gen Genet. 1979 Oct 3;176(2):209–219. doi: 10.1007/BF00273215. [DOI] [PubMed] [Google Scholar]
- NOVICK R. P. Staphylococcal penicillinase and the new penicillins. Biochem J. 1962 May;83:229–235. doi: 10.1042/bj0830229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y., Mitsuhashi S. New type of R factors incapable of inactivating chloramphenicol. J Bacteriol. 1972 Jan;109(1):1–7. doi: 10.1128/jb.109.1.1-7.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae R., Nakae T. Diffusion of aminoglycoside antibiotics across the outer membrane of Escherichia coli. Antimicrob Agents Chemother. 1982 Oct;22(4):554–559. doi: 10.1128/aac.22.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Kinscherf T. G., Silver S., Miki T., Easton A. M., Rownd R. H. Gene copy number effects in the mer operon of plasmid NR1. J Bacteriol. 1979 Apr;138(1):284–287. doi: 10.1128/jb.138.1.284-287.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Silver S., Miki T., Rownd R. H. Hypersensitivity to Hg2+ and hyperbinding activity associated with cloned fragments of the mercurial resistance operon of plasmid NR1. J Bacteriol. 1979 Oct;140(1):161–166. doi: 10.1128/jb.140.1.161-166.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzan Y., Gozhansky S. Chloramphenicol binding site of an fi- R-factor-specified variant of chloramphenicol acetyltransferase. Arch Biochem Biophys. 1980 Apr 15;201(1):115–120. doi: 10.1016/0003-9861(80)90494-4. [DOI] [PubMed] [Google Scholar]
- Noel D., Nikaido K., Ames G. F. A single amino acid substitution in a histidine-transport protein drastically alters its mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry. 1979 Sep 18;18(19):4159–4165. doi: 10.1021/bi00586a017. [DOI] [PubMed] [Google Scholar]
- Nordström K., Ingram L. C., Lundbäck A. Mutations in R factors of Escherichia coli causing an increased number of R-factor copies per chromosome. J Bacteriol. 1972 May;110(2):562–569. doi: 10.1128/jb.110.2.562-569.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Edelman I., Schwesinger M. D., Gruss A. D., Swanson E. C., Pattee P. A. Genetic translocation in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):400–404. doi: 10.1073/pnas.76.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Murphy E., Gryczan T. J., Baron E., Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979 Jan;2(1):109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Roth C. Plasmid-linked resistance to inorganic salts in Staphylococcus aureus. J Bacteriol. 1968 Apr;95(4):1335–1342. doi: 10.1128/jb.95.4.1335-1342.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent M. E., Hedges R. W. The nature of the genetic determinant for the SHV-1 beta-lactamase. Mol Gen Genet. 1979 Oct 1;175(3):239–243. doi: 10.1007/BF00397222. [DOI] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H., Azuma A., Suzuki Y., Hashimoto Y. Factors involved in beta-lactam antibiotic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1977 Oct;12(4):537–539. doi: 10.1128/aac.12.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olafson R. W., Abel K., Sim R. G. Prokaryotic metallothionein: preliminary characterization of a blue-green alga heavy metal-binding protein. Biochem Biophys Res Commun. 1979 Jul 12;89(1):36–43. doi: 10.1016/0006-291x(79)90939-2. [DOI] [PubMed] [Google Scholar]
- Ozanne B., Benveniste R., Tipper D., Davies J. Aminoglycoside antibiotics: inactivation by phosphorylation in Escherichia coli carrying R factors. J Bacteriol. 1969 Nov;100(2):1144–1146. doi: 10.1128/jb.100.2.1144-1146.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo D., Rosset R. A new ribosomal mutation which affects the two ribosomal subunits in Escherichia coli. Mol Gen Genet. 1977 Jun 8;153(2):199–204. doi: 10.1007/BF00264736. [DOI] [PubMed] [Google Scholar]
- Pattishall K. H., Acar J., Burchall J. J., Goldstein F. W., Harvey R. J. Two distinct types of trimethoprim-resistant dihydrofolate reductase specified by R-plasmids of different compatibility groups. J Biol Chem. 1977 Apr 10;252(7):2319–2323. [PubMed] [Google Scholar]
- Perlin M. H., Lerner S. A. Decreased susceptibility to 4'-deoxy-6'-N-methylamikacin (BB-K311) conferred by a mutant plasmid in Escherichia coli. Antimicrob Agents Chemother. 1982 Jul;22(1):78–82. doi: 10.1128/aac.22.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin M. H., Lerner S. A. Localization of an amikacin 3'-phosphotransferase in Escherichia coli. J Bacteriol. 1981 Aug;147(2):320–325. doi: 10.1128/jb.147.2.320-325.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Rownd R. H. Transition of R factor NR1 in Proteus mirabilis: molecular structure and replication of NR1 deoxyribonucleic acid. J Bacteriol. 1975 Sep;123(3):1013–1034. doi: 10.1128/jb.123.3.1013-1034.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Rownd R. H. Two origins of replication in composite R plasmid DNA. Nature. 1976 Jan 29;259(5541):281–284. doi: 10.1038/259281a0. [DOI] [PubMed] [Google Scholar]
- Perlman D., Stickgold R. Selective amplification of genes on the R plasmid, NR1, in Proteus mirabilis: an example of the induction of selective gene amplification. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2518–2522. doi: 10.1073/pnas.74.6.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. D., Silver S. Cadmium and manganese transport in Staphylococcus aureus membrane vesicles. J Bacteriol. 1982 May;150(2):973–976. doi: 10.1128/jb.150.2.973-976.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. Antibiotics as probes of ribosome structure: binding of chloramphenicol and erythromycin to polyribosomes; effect of other antibiotics. Antimicrob Agents Chemother. 1974 Mar;5(3):255–267. doi: 10.1128/aac.5.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piendl W., Böck A. Ribosomal resistance in the gentamicin producer organism Micromonospora purpurea. Antimicrob Agents Chemother. 1982 Aug;22(2):231–236. doi: 10.1128/aac.22.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwowarski J. M., Shaw P. D. Streptomycin resistance in a streptomycin-producing microorganism. Antimicrob Agents Chemother. 1979 Aug;16(2):176–182. doi: 10.1128/aac.16.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak J., Novick R. P. Closely related plasmids from Staphylococcus aureus and soil bacilli. Plasmid. 1982 Mar;7(2):152–162. doi: 10.1016/0147-619x(82)90074-9. [DOI] [PubMed] [Google Scholar]
- Porter F. D., Silver S., Ong C., Nakahara H. Selection for mercurial resistance in hospital settings. Antimicrob Agents Chemother. 1982 Nov;22(5):852–858. doi: 10.1128/aac.22.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privitera G., Sebald M., Fayolle F. Common regulatory mechanism of expression and conjugative ability of a tetracycline resistance plasmid in Bacteroides fragilis. Nature. 1979 Apr 12;278(5705):657–659. doi: 10.1038/278657a0. [DOI] [PubMed] [Google Scholar]
- Proctor G. N., Rownd R. H. Rosanilins: indicator dyes for chloramphenicol-resistant enterobacteria containing chloramphenicol acetyltransferase. J Bacteriol. 1982 Jun;150(3):1375–1382. doi: 10.1128/jb.150.3.1375-1382.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICHMOND M. H. DOMINANCE OF THE INDUCIBLE STATE IN STRAINS OF STAPHYLOCOCCUS AUREUS CONTAINING TWO DISTINCT PENICILLINASE PLASMIDS. J Bacteriol. 1965 Aug;90:370–374. doi: 10.1128/jb.90.2.370-374.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashtchian A., Nouravarsani R., Miller G. R., Gerlach E. H. Increased production of beta-lactamase under anaerobic conditions in some strains of Escherichia coli. Antimicrob Agents Chemother. 1979 Dec;16(6):772–775. doi: 10.1128/aac.16.6.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve E. C. Evidence that there are two types of determinant for tetracycline resistance among R-factors. Genet Res. 1978 Feb;31(1):75–84. doi: 10.1017/s001667230001781x. [DOI] [PubMed] [Google Scholar]
- Richmond M. H., Clark D. C., Wotton S. Indirect method for assessing the penetration of beta-lactamase-nonsusceptible penicillins and cephalosporins in Escherichia coli strains. Antimicrob Agents Chemother. 1976 Aug;10(2):215–218. doi: 10.1128/aac.10.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond M. H., Curtis N. A. The interplay of beta-lactamases and intrinsic factors in the resistance of gram-negative bacteria to penicillins and cephalosporins. Ann N Y Acad Sci. 1974 May 10;235(0):553–568. doi: 10.1111/j.1749-6632.1974.tb43290.x. [DOI] [PubMed] [Google Scholar]
- Richmond M. H. Factors influencing the antibacterial action of beta-lactam antibiotics. J Antimicrob Chemother. 1978 Jul;4(B):1–14. doi: 10.1093/jac/4.suppl_b.1. [DOI] [PubMed] [Google Scholar]
- Richmond M. H., Sykes R. B. The beta-lactamases of gram-negative bacteria and their possible physiological role. Adv Microb Physiol. 1973;9:31–88. doi: 10.1016/s0065-2911(08)60376-8. [DOI] [PubMed] [Google Scholar]
- Roberts M., Corney A., Shaw W. V. Molecular characterization of three chloramphenicol acetyltransferases isolated from Haemophilus influenzae. J Bacteriol. 1982 Aug;151(2):737–741. doi: 10.1128/jb.151.2.737-741.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. M., Reeve E. C. Analysis of the resistance mediated by several R-factors to tetracycline and minocycline. Genet Res. 1972 Oct;20(2):239–252. doi: 10.1017/s0016672300013744. [DOI] [PubMed] [Google Scholar]
- Rodgers W. H., Springer W., Young F. E. Cloning and expression of a Streptomyces fradiae neomycin resistance gene in Escherichia coli. Gene. 1982 May;18(2):133–141. doi: 10.1016/0378-1119(82)90111-1. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Tebar A., Rojo F., Dámaso D., Vázquez D. Carbenicillin resistance of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1982 Aug;22(2):255–261. doi: 10.1128/aac.22.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland S., Ferone R., Harvey R. J., Styles V. L., Morrison R. W. The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J Biol Chem. 1979 Oct 25;254(20):10337–10345. [PubMed] [Google Scholar]
- Rosdahl V. T. Naturally occurring constitutive -lactamase of novel serotype in Staphylococcus aureus. J Gen Microbiol. 1973 Jul;77(1):229–231. doi: 10.1099/00221287-77-1-229. [DOI] [PubMed] [Google Scholar]
- Rosenkranz H. S., Carr H. S. Silver sulfadiazine: effect on the growth and metabolism of bacteria. Antimicrob Agents Chemother. 1972 Nov;2(5):367–372. doi: 10.1128/aac.2.5.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin L. G., Medeiros A. A., Yolken R. H., Moxon E. R. Ampicillin treatment failure of apparently beta-lactamase-negative Haemophilus influenzae type b meningitis due to novel beta-lactamase. Lancet. 1981 Nov 7;2(8254):1008–1010. doi: 10.1016/s0140-6736(81)91214-9. [DOI] [PubMed] [Google Scholar]
- STACEY K. A., SIMSON E. IMPROVED METHOD FOR THE ISOLATION OF THYMINE-REQUIRING MUTANTS OF ESCHERICHIA COLI. J Bacteriol. 1965 Aug;90:554–555. doi: 10.1128/jb.90.2.554-555.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands L. C., Shaw W. V. Mechanism of chloramphenicol resistance in staphylococci: characterization and hybridization of variants of chloramphenicol acetyltransferase. Antimicrob Agents Chemother. 1973 Feb;3(2):299–305. doi: 10.1128/aac.3.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santanam P., Kayser F. H. Purification and characterization of an aminoglycoside inactivating enzyme from Staphylococcus epidermidis FK109 that nucleotidylates the 4'- and 4''-hydroxyl groups of the aminoglycoside antibiotics. J Antibiot (Tokyo) 1978 Apr;31(4):343–351. doi: 10.7164/antibiotics.31.343. [DOI] [PubMed] [Google Scholar]
- Schmitt R., Altenbuchner J., Wiebauer K., Arnold W., Pühler A., Schöffl F. Basis of transposition and gene amplification by Tn1721 and related tetracycline-resistance transposons. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):59–65. doi: 10.1101/sqb.1981.045.01.011. [DOI] [PubMed] [Google Scholar]
- Schmitt R., Bernhard E., Mattes R. Characterisation of Tn1721, a new transposon containing tetracycline resistance genes capable of amplification. Mol Gen Genet. 1979 Apr 17;172(1):53–65. doi: 10.1007/BF00276215. [DOI] [PubMed] [Google Scholar]
- Schottel J. L. The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J Biol Chem. 1978 Jun 25;253(12):4341–4349. [PubMed] [Google Scholar]
- Schottel J., Mandal A., Clark D., Silver S., Hedges R. W. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature. 1974 Sep 27;251(5473):335–337. doi: 10.1038/251335a0. [DOI] [PubMed] [Google Scholar]
- Schöffl F., Pühler A. Intramolecular amplification of the tetracycline resistance determinant of transposon Tn1771 in Escherichia coli. Genet Res. 1979 Jun;33(3):253–260. doi: 10.1017/s0016672300018395. [DOI] [PubMed] [Google Scholar]
- Shales S. W., Chopra I., Ball P. R. Evidence for more than one mechanism of plasmid-determined tetracycline resistance in Escherichia coli. J Gen Microbiol. 1980 Nov;121(1):221–229. doi: 10.1099/00221287-121-1-221. [DOI] [PubMed] [Google Scholar]
- Shannon K., Phillips I. Mechanisms of resistance to aminoglycosides in clinical isolates. J Antimicrob Chemother. 1982 Feb;9(2):91–102. doi: 10.1093/jac/9.2.91. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., Sporn P. Tn402: a new transposable element determining trimethoprim resistance that inserts in bacteriophage lambda. J Bacteriol. 1977 Mar;129(3):1632–1635. doi: 10.1128/jb.129.3.1632-1635.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Bentley D. W., Sands L. Mechanism of Chloramphenicol Resistance in Staphylococcus epidermidis. J Bacteriol. 1970 Dec;104(3):1095–1105. doi: 10.1128/jb.104.3.1095-1105.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Bouanchaud D. H., Goldstein F. W. Mechanism of transferable resistance to chloramphenicol in Haemophilus parainfluenzae. Antimicrob Agents Chemother. 1978 Feb;13(2):326–330. doi: 10.1128/aac.13.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Brodsky R. F. Characterization of chloramphenicol acetyltransferase from chloramphenicol-resistant Staphylococcus aureus. J Bacteriol. 1968 Jan;95(1):28–36. doi: 10.1128/jb.95.1.28-36.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit Rev Biochem. 1983;14(1):1–46. doi: 10.3109/10409238309102789. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Hopwood D. A. Chloramphenicol acetylation in Streptomyces. J Gen Microbiol. 1976 May;94(1):159–166. doi: 10.1099/00221287-94-1-159. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Packman L. C., Burleigh B. D., Dell A., Morris H. R., Hartley B. S. Primary structure of a chloramphenicol acetyltransferase specified by R plasmids. Nature. 1979 Dec 20;282(5741):870–872. doi: 10.1038/282870a0. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. The enzymatic acetylation of chloramphenicol by extracts of R factor-resistant Escherichia coli. J Biol Chem. 1967 Feb 25;242(4):687–693. [PubMed] [Google Scholar]
- Shaw W. V. The problems of drug-resistant pathogenic bacteria. Comparative enzymology of chloramphenicol resistance. Ann N Y Acad Sci. 1971 Jun 11;182:234–242. doi: 10.1111/j.1749-6632.1971.tb30660.x. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Unowsky J. Mechanism of R factor-mediated chloramphenicol resistance. J Bacteriol. 1968 May;95(5):1976–1978. doi: 10.1128/jb.95.5.1976-1978.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Dubnau D. Characterization of a plasmid-specified ribosome methylase associated with macrolide resistance. Nucleic Acids Res. 1981 Jun 11;9(11):2549–2562. doi: 10.1093/nar/9.11.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Grandi G., Kozlov Y., Dubnau D. Posttranscriptional regulation of an erythromycin resistance protein specified by plasmic pE194. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3903–3907. doi: 10.1073/pnas.77.7.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Budd K., Leahy K. M., Shaw W. V., Hammond D., Novick R. P., Willsky G. R., Malamy M. H., Rosenberg H. Inducible plasmid-determined resistance to arsenate, arsenite, and antimony (III) in escherichia coli and Staphylococcus aureus. J Bacteriol. 1981 Jun;146(3):983–996. doi: 10.1128/jb.146.3.983-996.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Keach D. Energy-dependent arsenate efflux: the mechanism of plasmid-mediated resistance. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6114–6118. doi: 10.1073/pnas.79.20.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair M. I., Holloway B. W. A chromosomally located transposon in Pseudomonas aeruginosa. J Bacteriol. 1982 Aug;151(2):569–579. doi: 10.1128/jb.151.2.569-579.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner R. H., Cundliffe E. Resistance to the antibiotics viomycin and capreomycin in the Streptomyces species which produce them. J Gen Microbiol. 1980 Sep;120(1):95–104. doi: 10.1099/00221287-120-1-95. [DOI] [PubMed] [Google Scholar]
- Sköld O. R-factor-mediated resistance to sulfonamides by a plasmid-borne, drug-resistant dihydropteroate synthase. Antimicrob Agents Chemother. 1976 Jan;9(1):49–54. doi: 10.1128/aac.9.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sköld O., Widh A. A new dihydrofolate reductase with low trimethoprim sensitivity induced by an R factor mediating high resistance to trimethoprim. J Biol Chem. 1974 Jul 10;249(13):4324–4325. [PubMed] [Google Scholar]
- Smith C. J., Markowitz S. M., Macrina F. L. Transferable tetracycline resistance in Clostridium difficile. Antimicrob Agents Chemother. 1981 Jun;19(6):997–1003. doi: 10.1128/aac.19.6.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. W. Arsenic resistance in enterobacteria: its transmission by conjugation and by phage. J Gen Microbiol. 1978 Nov;109(1):49–56. doi: 10.1099/00221287-109-1-49. [DOI] [PubMed] [Google Scholar]
- Smith K., Novick R. P. Genetic studies on plasmid-linked cadmium resistance in Staphylococcus aureus. J Bacteriol. 1972 Nov;112(2):761–772. doi: 10.1128/jb.112.2.761-772.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. L., Stone D., Novak P., Baccanari D. P., Burchall J. J. R plasmid dihydrofolate reductase with subunit structure. J Biol Chem. 1979 Jul 25;254(14):6222–6225. [PubMed] [Google Scholar]
- Stone D., Smith S. L. The amino acid sequence of the trimethoprim-resistant dihydrofolate reductase specified in Escherichia coli by R-plasmid R67. J Biol Chem. 1979 Nov 10;254(21):10857–10861. [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M., Kobayashi H., Nimi O., Nomi R. Susceptibility of protein synthesis to streptomycin in streptomycin-producing Streptomyces griseus. FEBS Lett. 1980 Feb 11;110(2):250–252. doi: 10.1016/0014-5793(80)80084-6. [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Mochizuki H., Nimi O., Nomi R. Mechanism of protection of protein synthesis against streptomycin inhibition in a producing strain. J Antibiot (Tokyo) 1981 Sep;34(9):1183–1188. doi: 10.7164/antibiotics.34.1183. [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Nimi O., Nomi R. Susceptibility of protein synthesis to neomycin in neomycin-producing Streptomyces fradiae. J Gen Microbiol. 1980 Dec;121(2):477–478. doi: 10.1099/00221287-121-2-477. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Jacoby G. A. Plasmid-determined resistance to boron and chromium compounds in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1978 Apr;13(4):637–640. doi: 10.1128/aac.13.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Jacoby G. A. Plasmid-determined resistance to tellurium compounds. J Bacteriol. 1977 Jan;129(1):276–281. doi: 10.1128/jb.129.1.276-281.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Kight-Olliff L. Tn1 generated mutants in the mercuric ion reductase of the Inc P plasmid, R702. Mol Gen Genet. 1980;180(1):91–97. doi: 10.1007/BF00267356. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Knight-Olliff L., Slater C. Effect of catabolite repression on the mer operon. J Bacteriol. 1982 Jan;149(1):191–197. doi: 10.1128/jb.149.1.191-197.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Silver S. Mercury resistance in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1972 Dec;112(3):1228–1236. doi: 10.1128/jb.112.3.1228-1236.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Silver S. Microbial transformations of metals. Annu Rev Microbiol. 1978;32:637–672. doi: 10.1146/annurev.mi.32.100178.003225. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Sugarman L. I. Cell-free mercury(II)-reducing activity in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1974 Jul;119(1):242–249. doi: 10.1128/jb.119.1.242-249.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Okamoto S. The enzymatic acetylation of chloramphenicol by the multiple drug-resistant Escherichia coli carrying R factor. J Biol Chem. 1967 Oct 25;242(20):4722–4730. [PubMed] [Google Scholar]
- Swedberg G., Sköld O. Characterization of different plasmid-borne dihydropteroate synthases mediating bacterial resistance to sulfonamides. J Bacteriol. 1980 Apr;142(1):1–7. doi: 10.1128/jb.142.1.1-7.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes R. B., Matthew M. The beta-lactamases of gram-negative bacteria and their role in resistance to beta-lactam antibiotics. J Antimicrob Chemother. 1976 Jun;2(2):115–157. doi: 10.1093/jac/2.2.115. [DOI] [PubMed] [Google Scholar]
- Tai P. C., Wallace B. J., Davis B. D. Selective action of erythromycin on initiating ribosomes. Biochemistry. 1974 Oct 22;13(22):4653–4659. doi: 10.1021/bi00719a029. [DOI] [PubMed] [Google Scholar]
- Tait R. C., Boyer H. W. On the nature of tetracycline resistance controlled by the plasmid pSC101. Cell. 1978 Jan;13(1):73–81. doi: 10.1016/0092-8674(78)90139-3. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Systematic difference in the methylation of ribosomal ribonucleic acid from gram-positive and gram-negative bacteria. J Bacteriol. 1975 Aug;123(2):771–774. doi: 10.1128/jb.123.2.771-774.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. P., Greenberg J., Rownd R. H. Generation of miniplasmids from copy number mutants of the R plasmid NR1. J Bacteriol. 1977 Dec;132(3):986–995. doi: 10.1128/jb.132.3.986-995.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennhammar-Ekman B., Sköld O. Trimethoprim resistance plasmids of different origin encode different drug-resistant dihydrofolate reductases. Plasmid. 1979 Jul;2(3):334–346. doi: 10.1016/0147-619x(79)90017-9. [DOI] [PubMed] [Google Scholar]
- Tezuka T., Tonomura K. Purification and properties of a second enzyme catalyzing the splitting of carbon-mercury linkages from mercury-resistant Pseudomonas K-62. J Bacteriol. 1978 Jul;135(1):138–143. doi: 10.1128/jb.135.1.138-143.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Then R. L., Angehrn P. Trapping of nonhydrolyzable cephalosporins by cephalosporinases in Enterobacter cloacae and Pseudomonas aeruginosa as a possible resistance mechanism. Antimicrob Agents Chemother. 1982 May;21(5):711–717. doi: 10.1128/aac.21.5.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Skinner R. H., Thompson J., Ward J. M., Hopwood D. A., Cundliffe E. Biochemical characterization of resistance determinants cloned from antibiotic-producing streptomycetes. J Bacteriol. 1982 Aug;151(2):678–685. doi: 10.1128/jb.151.2.678-685.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Ward J. M., Hopwood D. A. DNA cloning in Streptomyces: resistance genes from antibiotic-producing species. Nature. 1980 Jul 31;286(5772):525–527. doi: 10.1038/286525a0. [DOI] [PubMed] [Google Scholar]
- Thompson J., Cundliffe E. Resistance to thiostrepton, siomycin, and sporangiomycin in actinomycetes that produce them. J Bacteriol. 1980 May;142(2):455–461. doi: 10.1128/jb.142.2.455-461.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorbjarnardóttir S. H., Magnúsdóttir R. A., Eggertsson G. Mutations determining generalized resistance to aminoglycoside antibiotics in Escherichia coli. Mol Gen Genet. 1978 Apr 25;161(1):89–98. doi: 10.1007/BF00266619. [DOI] [PubMed] [Google Scholar]
- Timmis K. N., Cabello F., Cohen S. N. Cloning and characterization of EcoRI and HindIII restriction endonuclease-generated fragments of antibiotic resistance plasmids R6-5 and R6. Mol Gen Genet. 1978 Jun 14;162(2):121–137. doi: 10.1007/BF00267869. [DOI] [PubMed] [Google Scholar]
- Tipper D. J., Strominger J. L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobian J. A., Macrina F. L. Helper plasmid cloning in Streptococcus sanguis: cloning of a tetracycline resistance determinant from the Streptococcus mutans chromosome. J Bacteriol. 1982 Oct;152(1):215–222. doi: 10.1128/jb.152.1.215-222.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich P. K., An F. Y., Clewell D. B. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1980 Mar;141(3):1366–1374. doi: 10.1128/jb.141.3.1366-1374.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner K. J. A clinical isolate of Escherichia coli owing its trimethoprim resistance to a chromosomally-located trimethoprim transposon. J Antimicrob Chemother. 1981 Feb;7(2):157–162. doi: 10.1093/jac/7.2.157. [DOI] [PubMed] [Google Scholar]
- Tynecka Z., Gos Z., Zajac J. Energy-dependent efflux of cadmium coded by a plasmid resistance determinant in Staphylococcus aureus. J Bacteriol. 1981 Aug;147(2):313–319. doi: 10.1128/jb.147.2.313-319.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynecka Z., Gos Z., Zajac J. Reduced cadmium transport determined by a resistance plasmid in Staphylococcus aureus. J Bacteriol. 1981 Aug;147(2):305–312. doi: 10.1128/jb.147.2.305-312.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid. 1977 Nov;1(1):1–7. doi: 10.1016/0147-619x(77)90003-8. [DOI] [PubMed] [Google Scholar]
- Umezawa Y., Yagisawa M., Sawa T., Takeuchi T., Umezawa H. Aminoglycoside 3'-phosphotransferase III, a new phosphotransferase. Resistance mechanism. J Antibiot (Tokyo) 1975 Nov;28(11):845–853. doi: 10.7164/antibiotics.28.845. [DOI] [PubMed] [Google Scholar]
- Vastola A. P., Altschaefl J., Harford S. 5-epi-Sisomicin and 5-epi-Gentamicin B: substrates for aminoglycoside-modifying enzymes that retain activity against aminoglycoside-resistant bacteria. Antimicrob Agents Chemother. 1980 May;17(5):798–802. doi: 10.1128/aac.17.5.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran P. S., Wu H. C. Isolation and characterization of a phosphonomycin-resistant mutant of Escherichia coli K-12. J Bacteriol. 1972 Jun;110(3):935–944. doi: 10.1128/jb.110.3.935-944.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völker T. A., Iida S., Bickle T. A. A single gene coding for resistance to both fusidic acid and chloramphenicol. J Mol Biol. 1982 Jan 25;154(3):417–425. doi: 10.1016/s0022-2836(82)80004-1. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Padgett R. A., Stark G. R. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979 Sep 10;254(17):8679–8689. [PubMed] [Google Scholar]
- Walker J. B., Skorvaga M. Phosphorylation of streptomycin and dihydrostreptomycin by Streptomyces. Enzymatic synthesis of different diphosphorylated derivatives. J Biol Chem. 1973 Apr 10;248(7):2435–2440. [PubMed] [Google Scholar]
- Walker M. S., Walker J. B. Streptomycin biosynthesis and metabolism. Enzymatic phosphorylation of dihydrostreptobiosamine moieties of dihydro-streptomycin-(streptidino) phosphate and dihydrostreptomycin by Streptomyces extracts. J Biol Chem. 1970 Dec 25;245(24):6683–6689. [PubMed] [Google Scholar]
- Watanakunakorn C. Antibiotic-tolerant Staphylococcus aureus. J Antimicrob Chemother. 1978 Nov;4(6):561–568. doi: 10.1093/jac/4.6.561. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Holder S. B., Halling S. M. Deoxyribonucleic acid sequence common to staphylococcal and streptococcal plasmids which specify erythromycin resistance. J Bacteriol. 1979 Jun;138(3):990–998. doi: 10.1128/jb.138.3.990-998.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A. A., Murphy S. D., Silver S. Mercury and organomercurial resistances determined by plasmids in Staphylococcus aureus. J Bacteriol. 1977 Oct;132(1):197–208. doi: 10.1128/jb.132.1.197-208.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A. A., Silver S., Kinscherf T. G. Cation transport alteration associated with plasmid-determined resistance to cadmium in Staphylococcus aureus. Antimicrob Agents Chemother. 1978 Dec;14(6):856–865. doi: 10.1128/aac.14.6.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. W., Northrop D. B. Purification and properties of gentamicin acetyltransferase I. Biochemistry. 1976 Jan 13;15(1):125–131. doi: 10.1021/bi00646a019. [DOI] [PubMed] [Google Scholar]
- Willsky G. R., Malamy M. H. Effect of arsenate on inorganic phosphate transport in Escherichia coli. J Bacteriol. 1980 Oct;144(1):366–374. doi: 10.1128/jb.144.1.366-374.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. R., Skinner S. E., Shaw W. V. Analysis of two chloramphenicol resistance plasmids from Staphylococcus aureus: insertional inactivation of Cm resistance, mapping of restriction sites, and construction of cloning vehicles. Plasmid. 1981 May;5(3):245–258. doi: 10.1016/0147-619x(81)90002-0. [DOI] [PubMed] [Google Scholar]
- Wittmann-Liebold B., Greuer B. The primary structure of protein S5 from the small subunit of the Escherichia coli ribosome. FEBS Lett. 1978 Nov 1;95(1):91–98. doi: 10.1016/0014-5793(78)80059-3. [DOI] [PubMed] [Google Scholar]
- Wray L. V., Jr, Jorgensen R. A., Reznikoff W. S. Identification of the tetracycline resistance promoter and repressor in transposon Tn10. J Bacteriol. 1981 Aug;147(2):297–304. doi: 10.1128/jb.147.2.297-304.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Clewell D. B. Plasmid-determined tetracycline resistance in Streptococcus faecalis: tandemly repeated resistance determinants in amplified forms of pAMalpha1 DNA. J Mol Biol. 1976 Apr 15;102(3):583–600. doi: 10.1016/0022-2836(76)90336-3. [DOI] [PubMed] [Google Scholar]
- Yaguchi M., Wittmann H. G., Cabezón T., DeWilde M., Villarroel R., Herzog A., Bollen A. Alteration of ribosomal protein S17 by mutation linked to neamine resistance in Escherichia coli. II. Localization of the amino acid replacement in protein S17 from a meaA mutant. J Mol Biol. 1976 Jul 5;104(3):617–620. doi: 10.1016/0022-2836(76)90124-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Nohara C., Fukunaga Y., Yamagishi S. Transposition of the oxacillin-hydrolyzing penicillinase gene. J Bacteriol. 1981 Feb;145(2):808–813. doi: 10.1128/jb.145.2.808-813.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Yamagata S., Horii K., Yamagishi S. Comparison of transcription of beta-lactamase genes specified by various ampicillin transposons. J Bacteriol. 1982 Apr;150(1):269–276. doi: 10.1128/jb.150.1.269-276.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Yokota T. Beta-lactamase-directed barrier for penicillins of Escherichia coli carrying R plasmids. Antimicrob Agents Chemother. 1977 Jun;11(6):936–940. doi: 10.1128/aac.11.6.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. L., Zubay G., Levy S. B. Synthesis of an R plasmid protein associated with tetracycline resistance is negatively regulated. Proc Natl Acad Sci U S A. 1976 May;73(5):1509–1512. doi: 10.1073/pnas.73.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidenzaig Y., Fitton J. E., Packman L. C., Shaw W. V. Characterization and comparison of chloramphenicol acetyltransferase variants. Eur J Biochem. 1979 Oct 15;100(2):609–618. doi: 10.1111/j.1432-1033.1979.tb04208.x. [DOI] [PubMed] [Google Scholar]
- Zaidenzaig Y., Shaw W. V. The reactivity of sulfhydryl groups at the active site of an F-factor--specified variant of chloramphenicol acetyltransferase. Eur J Biochem. 1978 Feb;83(2):553–562. doi: 10.1111/j.1432-1033.1978.tb12123.x. [DOI] [PubMed] [Google Scholar]
- Zighelboim S., Tomasz A. Penicillin-binding proteins of multiply antibiotic-resistant South African strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980 Mar;17(3):434–442. doi: 10.1128/aac.17.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R. A., Moellering R. C., Jr, Weinberg A. N. Mechanism of resistance to antibiotic synergism in enterococci. J Bacteriol. 1971 Mar;105(3):873–879. doi: 10.1128/jb.105.3.873-879.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolg J. W., Hänggi U. J. Characterization of a R plasmid-associated, trimethoprim-resistant dihydrofolate reductase and determination of the nucleotide sequence of the reductase gene. Nucleic Acids Res. 1981 Feb 11;9(3):697–710. doi: 10.1093/nar/9.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupancic T. J., King S. R., Pogue-Geile K. L., Jaskunas S. R. Identification of a second tetracycline-inducible polypeptide encoded by Tn10. J Bacteriol. 1980 Oct;144(1):346–355. doi: 10.1128/jb.144.1.346-355.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Treeck U., Schmidt F., Wiedemann B. Molecular nature of a streptomycin and sulfonamide resistance plasmid (pBP1) prevalent in clinical Escherichia coli strains and integration of an ampicillin resistance transposon (TnA). Antimicrob Agents Chemother. 1981 Mar;19(3):371–380. doi: 10.1128/aac.19.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]


