Abstract
Understanding the metal ion content of plant mitochondria and metal ion interactions with the proteome are vital for insights into both normal respiratory function and the process of protein damage during oxidative stress. We have analyzed the metal content of isolated Arabidopsis (Arabidopsis thaliana) mitochondria, revealing a 26:8:6:1 molar ratio for iron:zinc:copper:manganese and trace amounts of cobalt and molybdenum. We show that selective changes occur in mitochondrial copper and iron content following in vivo and in vitro oxidative stresses. Immobilized metal affinity chromatography charged with Cu2+, Zn2+, and Co2+ was used to identify over 100 mitochondrial proteins with metal-binding properties. There were strong correlations between the sets of immobilized metal affinity chromatography-interacting proteins, proteins predicted to contain metal-binding motifs, and protein sets known to be oxidized or degraded during abiotic stress. Mitochondrial respiratory chain pathways and matrix enzymes varied widely in their susceptibility to metal-induced loss of function, showing the selectivity of the process. A detailed study of oxidized residues and predicted metal interaction sites in the tricarboxylic acid cycle enzyme aconitase identified selective oxidation of residues in the active site and showed an approach for broader screening of functionally significant oxidation events in the mitochondrial proteome.
Transition metal ions are essential in myriad biochemical functions by being incorporated into or associating with proteins to elicit functions in living cells. In plant mitochondria, key functions of metal cofactors include metabolism, electron transport, ATP synthesis, and the detoxification of reactive oxygen species (ROS). For example, copper (Cu) and iron (Fe) ions facilitate the transfer of electrons in the electron transport chain (ETC; Bligny and Douce, 1977; Pascal and Douce, 1993), proteins of the tricarboxylic acid (TCA) cycle utilize metal ion cofactors to catalyze primary metabolic reactions (Miernyk and Randall, 1987; Jordanov et al., 1992), manganese (Mn) and Fe are required for antioxidant defense enzymes (Alscher et al., 2002), and zinc (Zn) is required for the protein import apparatus in both carrier protein transport to the inner membrane (Lister et al., 2002) and presequence degradation (Moberg et al., 2003). Cobalt (Co) is known to substitute for other metal ions in the activation of NAD-malic enzyme and succinyl-CoA ligase from plant mitochondrial extracts (Palmer and Wedding, 1966; Macrae, 1971), but it is not known whether there is an in vivo requirement for trace amounts of Co for plant respiratory metabolism.
Metal ions, however, can also be highly toxic to cells and cell organelle functions. The redox-inactive heavy metal cadmium exhibits strong affinity for oxygen, nitrogen, and sulfur atoms (Nieboer and Richardson, 1980) and can inhibit enzyme activity by direct blocking of protein function or displacement of natural metal centers. There are numerous reports of heavy metals depleting cellular glutathione pools, leading to diminished antioxidant protection in the cell and resulting in ROS accumulation (Schutzendubel and Polle, 2002). Cadmium has been reported to both directly and indirectly inhibit plant mitochondrial function (Kesseler and Brand, 1994; Smiri et al., 2009), as have Co complexes (Guzhova et al., 1979). Redox-active metal catalysts such as Cu and Fe cations can also be cytotoxic, as they react with ROS via the Haber-Weiss reaction or Fenton-type reactions to produce the hydroxyl anion (Stohs and Bagchi, 1995). Inhibitory effects of exogenously added Cu and Fe on plant respiratory function have been reported (Kampfenkel et al., 1995; Padua et al., 1996, 1999). Therefore, the presence of free metal cations, redox active or inactive, in mitochondria may significantly contribute to the initiation and perpetuation of oxidative stress.
One of the best described mechanisms for metal-linked damage is metal-catalyzed oxidation (MCO) of proteins, which involves the oxidation of susceptible amino acids such as Arg, Lys, Pro, and His, among a plethora of other poorly characterized consequences (Stadtman, 1990). It has been proposed that MCO of proteins can be a highly specific event where proteins are more susceptible to damage if they bind metal ions and when the site of protein oxidation can be defined on the protein surface that binds to the metal ions (Stadtman, 1990). One of the major consequences of MCO is the irreversible formation of reactive carbonyls on amino acid side chains (Stadtman, 1990). Such carbonyls are known to accumulate in the wheat (Triticum aestivum) mitochondrial proteome during environmental stress, even more so than in other ROS-producing subcellular organelles of plants (Bartoli et al., 2004). The selectivity of protein susceptibility to MCO was also demonstrated in rice (Oryza sativa), where distinct subpopulations of the mitochondrial matrix proteome were carbonylated following Cu2+ and hydrogen peroxide (H2O2) treatment (Kristensen et al., 2004). The targeted damage of select sets of plant mitochondrial proteins has also been observed in other studies, but without clear linkage to the role of metal ions. For example, altered protein abundance has been observed in Arabidopsis (Arabidopsis thaliana; Sweetlove et al., 2002) and pea (Pisum sativum; Taylor et al., 2005) mitochondria after the initiation of oxidative or environmental stress. Additionally, inhibition of respiratory metabolism by the lipid peroxidation by-product 4-hydroxy-2-nonenal has been shown to operate through modification of a specific subset of proteins (Taylor et al., 2002; Winger et al., 2005, 2007). However, the mechanisms of targeted oxidative modification, the role of metals, and the consequences for mitochondrial metabolic function are not very well understood. Furthermore, whether or not selectivity of protein damage in mitochondria is based on relative metal ion affinity and if the sites of damage can be predicted by the sites of metal ion binding are not known.
In this study, we investigated metal homeostasis in the Arabidopsis mitochondrion during oxidative stress. The interactions between metal ions and proteins were also investigated using immobilized metal affinity chromatography (IMAC). Functional assays were used to determine the targets and consequences of metal ion interaction in the mitochondrion and to explore the linkages to the redox nature of the metal and the loss of mitochondrial functions. Finally, a detailed study of the oxidized peptides of aconitase was undertaken to probe the linkage between metal-binding sites, the oxidation of amino acids, and the inactivation of this critical TCA cycle enzyme.
RESULTS
The Plant Mitochondrial Metallome and Its Changes during Oxidative Stress
The metal content of mitochondria isolated from Arabidopsis cell culture was analyzed by inductively coupled plasma mass spectrometry (ICP-MS). This showed Fe, Cu, Zn, and Mn to be the predominant species of transition metals in Arabidopsis mitochondria, with trace levels of Co and molybdenum (Mo) also detected (Table I). Fractionating mitochondria into soluble and membrane components and further isolation of an integral membrane portion revealed that the integral membrane proteome contained 3-fold more Cu and Fe than the soluble proteome on a protein basis (Table I). This is likely due to the abundance of Cu- and Fe-containing ETC components and their enrichment in the integral membrane fraction. Mn was evenly distributed between the soluble and integral membrane compartments (Table I).
Table I.
The Arabidopsis cell suspension culture mitochondrial metallome
The top shows ICP-MS quantification of transition metal content of isolated Arabidopsis cell culture mitochondria. Data are expressed as both ng and pmol of metal per mg of protein ± se (n = 3 biological replicates). Percentage transition metallome values are calculated on a molar basis. At bottom, the metal contents of whole mitochondria (total), soluble fraction (soluble), and the Na2CO3-treated insoluble fraction (integral membrane) are expressed as ng of metal per mg of protein ± se (n = 3 biological replicates).
| Element | ng Metal per mg Protein | pmol Metal per mg Protein | Percentage Transition Metallome |
|---|---|---|---|
| Co | 0.3 ± 0.05 | 5.5 ± 0.5 | 0.1 |
| Cu | 50 ± 4 | 750 ± 50 | 14.5 |
| Fe | 180 ± 10 | 3,200 ± 200 | 62.5 |
| Mn | 7.0 ± 0.5 | 125 ± 5 | 2.5 |
| Mo | 2.0 ± 1.5 | 20 ± 15 | 0.4 |
| Zn |
70 ± 30 |
1,000 ± 500 |
20 |
| Element |
Total |
Soluble |
Integral Membrane |
| Cu | 100 ± 30 | 75 ± 10 | 240 ± 40 |
| Fe | 200 ± 40 | 200 ± 40 | 600 ± 80 |
| Mn | 15 ± 3 | 25 ± 5 | 30 ± 1 |
The redox-cycling metals, Cu and Fe, accounted for approximately 75% of the mitochondrial metallome (Table I). As labile metals are a likely cause of oxidative damage to proteins, ICP-MS was then used to quantify changes in metal composition of mitochondria isolated from cells under oxidative stress. The chemical treatments of cells and timing of analyses were selected based on previous studies of plant mitochondrial oxidative damage, in which the products of lipid peroxidation were measured to peak at 8 h post treatment (Winger et al., 2005).
Antimycin A is an inhibitor of complex III that ultimately leads to superoxide production within mitochondria (Maxwell et al., 1999), while menadione treatment of cells is reported to induce a broad cellular superoxide production from membranes (Hollensworth et al., 2000). Treatment of cells with either chemical was compared with methanol as well as an untreated control to ensure that the carrier solvent was not responsible for the changes observed. Both chemicals induced a 1.5- to 2-fold accumulation of Fe in mitochondria on a protein basis (Table II). This accumulation of Fe in mitochondria isolated from stress-treated cells could not be traced to either the soluble protein fraction or the integral membrane protein fraction from these mitochondrial extracts (Table II), suggesting that it was probably in the peripheral membrane fraction that was stripped from the membranes by carbonate extraction prior to ICP-MS analysis. Menadione treatment also elicited a slight reduction in mitochondrial integral membrane Cu content (Table II), suggesting damage to membrane-embedded cuproproteins. The accumulation of Fe following both menadione and antimycin A treatments may suggest a common mechanism induced by superoxide, regardless of the cellular source.
Table II.
Changes in the metallome of mitochondria by oxidative stress
At top, the metal contents of mitochondria isolated from cell suspension cultures treated with chemical elicitors of oxidative stress for 8 h were determined by ICP-MS. Data are expressed as ng of metal per mg of protein ± se (n ≥ 4). Statistical significance is indicated as follows: ** P < 0.01, * P < 0.05, # P < 0.1. At bottom, isolated mitochondria were subjected to no treatment (control) or 100 μm H2O2 treatment for 5 min at room temperature. The mitochondria were then fractionated into total mitochondria, soluble fraction, and integral membrane fraction for ICP-MS analysis. Data represent average metal contents expressed as ng of metal per mg of protein ± se (n = 3). Statistical significance is indicated as follows: # P < 0.1.
| Treatment | Cu | Fe | Mn |
|---|---|---|---|
| Total mitochondria | |||
| Control | 100 ± 20 | 180 ± 30 | 15 ± 2 |
| H2O2 | 60 ± 10* | 100 ± 15 | 15 ± 1 |
| Methanol | 130 ± 10 | 165 ± 10 | 20 ± 2 |
| Antimycin A | 100 ± 10 | 400 ± 75# | 20 ± 3 |
| Menadione | 120 ± 10 | 330 ± 30** | 20 ± 2 |
| Soluble fraction | |||
| Control | 80 ± 5 | 200 ± 30 | 25 ± 3 |
| H2O2 | 50 ± 5* | 85 ± 10* | 15 ± 1* |
| Methanol | 80 ± 10 | 120 ± 5 | 18 ± 1 |
| Antimycin A | 100 ± 20 | 130 ± 15 | 17 ± 2 |
| Menadione | 95 ± 10 | 150 ± 25 | 17 ± 2 |
| Integral membrane fraction | |||
| Control | 230 ± 30 | 600 ± 75 | 28 ± 1 |
| H2O2 | 130 ± 30* | 400 ± 120 | 25 ± 7 |
| Methanol | 350 ± 75 | 300 ± 25 | 25 ± 2 |
| Antimycin A | 200 ± 20 | 600 ± 100 | 25 ± 4 |
| Menadione |
160 ± 20# |
500 ± 100 |
25 ± 4 |
| Fraction |
Cu |
Fe |
Mn |
| Total | |||
| Control | 60 ± 20 | 200 ± 40 | 20 ± 4 |
| H2O2 | 40 ± 5 | 120 ± 2# | 15 ± 1 |
| Soluble | |||
| Control | 35 ± 5 | 160 ± 30 | 30 ± 5 |
| H2O2 | 35 ± 10 | 60 ± 30# | 25 ± 10 |
| Integral membrane | |||
| Control | 140 ± 20 | 160 ± 70 | 10 ± 2 |
| H2O2 | 140 ± 40 | 260 ± 80 | 10 ± 4 |
Damage to metalloproteins from oxidative treatment of cells was most apparent following H2O2 treatment. In isolated mitochondria, H2O2 induced a 40% decrease in Cu content (Table II). This loss of Cu could be traced to both the soluble and integral membrane protein fractions (Table II), which implies damage to mitochondrial soluble cuproproteins and also to cuprocomponents of the membrane-bound ETC. A 40% to 50% reduction of Fe and Mn content was also observed in the soluble protein fraction of mitochondria from H2O2-treated cells (Table II). This suggested that matrix metalloproteins such as Fe-S containing aconitase and Mn-containing superoxide dismutase (SOD) are sensitive to damage by H2O2. The observed metal losses are consistent with the fact that H2O2 is known to be able to damage proteins by fragmentation, destroying metal coordination sites (Hunt et al., 1988), and with evidence that plant aconitase is easily inactivated by H2O2 (Verniquet et al., 1991).
To provide further evidence of direct oxidative damage to metalloproteins causing loss of metal ions, mitochondria isolated from untreated cells were directly treated with H2O2 in vitro, and the metal content of whole mitochondria and across different mitochondrial compartments was measured by ICP-MS (Table II). In total mitochondrial samples, a significant reduction in Fe content was observed, and this loss was localized to the soluble fraction and not the integral membrane protein fraction. This apparent release of Fe from the soluble compartment could not be detected in low molecular mass fractions obtained by size-exclusion chromatography of mitochondrial extracts, but this is probably due to the detection limitations of the ICP-MS (data not shown).
Inhibition of Respiratory Function by Labile Metal Cations
Transition metal ions generally do not exist as free cations in cells but are sequestered by organic acids or protein ligands to minimize damage resulting from redox cycling with ROS (Rauser, 1999). However, in this study, ICP-MS has provided evidence that metal homeostasis can be disrupted during oxidative stress. During this time of increased metal flux into and out of the mitochondria, it is likely that the metal ions exist in a labile state and hence bind to and could have toxic effects on other proteins. Historical studies have noted that exogenous addition of metal ions affects respiratory complexes and consequently mitochondrial energy production (Skulachev et al., 1967; Kesseler and Brand, 1994; Padua et al., 1996; Kuznetsova et al., 2005). Hence, the respiratory capacity of mitochondria was measured following the addition of Cu2+, a redox-active ion, or Zn2+, a redox-inert metal ion, to characterize the differential effects of free metal ions on mitochondrial respiratory function.
A dose-response procedure was undertaken using treatments for 5 min with metal ions at 10, 50, and 100 μm concentrations compared with untreated control samples. This gave metal ion-protein ratios of 83, 415, and 830 nmol metal ions mg−1 protein, respectively. At 10 μm metal ions, no detrimental effects on respiration could be observed. However, at 50 and 100 μm, metal ions inhibit mitochondrial respiratory rate (Table III). The ETC and TCA cycle substrates used were succinate, NADH, and Glu + malate. Each of these substrates initiates mitochondrial respiration via a different pathway: succinate-dependent respiration measures the activity of the succinate dehydrogenase complex and the subsequent ETC; NADH-dependent respiration measures external NADH dehydrogenase entry to the ETC, bypassing complex I; while the organic acids Glu + malate are substrates for the TCA cycle, which ultimately produces NADH in the matrix that enters the ETC at complex I and/or the internal alternative NADH dehydrogenases.
Table III.
Metal ion inhibition of respiratory rate of isolated mitochondria
Isolated mitochondria (120 μg) were treated with various concentrations of Cu2+ or Zn2+ for 2 min prior to assaying the respiratory rates. Respiratory substrates Glu/malate, NADH, and succinate/ATP were added to drive respiration. State 3 (total) respiration was achieved by the addition of ADP. Cytochrome oxidase-dependent respiration was measured by adding n-propyl gallate to ensure that the rate observed was cytochrome oxidase dependent. Alternative oxidase-dependent respiration was measured by adding KCN. Normalized data are expressed as percentage of control ± se (n ≥ 3; * P < 0.05, ** P < 0.01, comparing each sample with the total oxygen consumption in untreated mitochondria). Concentrations of 10, 50, and 100 μm represent metal ion-protein ratios of 83, 415, and 830 nmol per mg mitochondrial proteins, respectively.
| Treatment | [Cu2+] | [Zn2+] | ||||
|---|---|---|---|---|---|---|
| μm | μm | |||||
| Total respiration | ||||||
| Substrate | 10 | 50 | 100 | 10 | 50 | 100 |
| Glu/malate | 105 ± 20 | 60 ± 15 | 60 ± 15 | 70 ± 10 | 75 ± 15 | 60 ± 15 |
| NADH | 65 ± 20 | 30 ± 1** | 20 ± 5** | 80 ± 10 | 50 ± 2** | 60 ± 2** |
| Succinate + ATP | 110 ± 15 | 55 ± 5** | 30 ± 10** | 100 ± 10 | 55 ± 5** | 65 ± 5** |
| Cytochrome oxidase-dependent respiration | ||||||
| Substrate | 10 | 50 | 100 | 10 | 50 | 100 |
| Glu/malate | 100 ± 20 | 55 ± 20 | 60 ± 20 | 100 ± 15 | 55 ± 15* | 30 ± 15* |
| NADH | 50 ± 25 | 30 ± 15* | 35 ± 10* | 70 ± 20 | 55 ± 10* | 40 ± 5** |
| Succinate + ATP | 140 ± 15 | 70 ± 10* | 35 ± 15* | 60 ± 20 | 55 ± 5** | 65 ± 5** |
| Alternative oxidase-dependent respiration | ||||||
| Substrate | 10 | 50 | 100 | 10 | 50 | 100 |
| NADH | 85 ± 10 | 20 ± 5** | 10 ± 10** | 60 ± 10 | 30 ± 1** | 60 ± 15 |
| Succinate + ATP | 130 ± 30 | 15 ± 5** | 5 ± 5** | 80 ± 15 | 40 ± 15* | 45 ± 10** |
Succinate-dependent respiration was susceptible to Cu2+ and Zn2+ treatment, resulting in at least a 40% decline in respiration at both 50 and 100 μm of each metal ion (Table III). In contrast, external NADH-dependent respiration was diminished differentially by Cu2+ and Zn2+. The toxicity of Cu2+ toward NADH-dependent respiration was more pronounced, as almost 80% inhibition of respiration was observed at both 50 and 100 μm Cu2+ concentrations. On the other hand, Zn2+ could only induce 40% inhibition of respiration under the same metal ion concentrations examined. Cu2+ and Zn2+ did not significantly affect Glu/malate-dependent respiration.
To determine if the cytochrome or alternative pathway was differentially affected by metal ion treatment, inhibitors were used to analyze each one independently. Cytochrome pathway inhibition by metal ions could be observed at 50 and 100 μm concentrations but did not occur in a dose-dependent manner, as the extent of the inhibition appeared to plateau (Table III). A 40% to 60% inhibition of respiration via the cytochrome pathway with either succinate or NADH as a substrate was observed following the addition of either metal ion. Zn2+ but not Cu2+ was found to significantly inhibit the Glu/malate-driven cytochrome pathway respiration. The alternative oxidase pathway was inhibited sharply by Cu2+ at 50 μm, and this metal ion could almost completely abolish respiration at 100 μm during both succinate- and NADH-driven alternative oxidase-dependent respiration. Zn2+ was also able to inhibit alternative pathway respiration, but to a lesser extent (Table III).
Selective Interaction of Mitochondrial Proteins with Immobilized Metal Cations
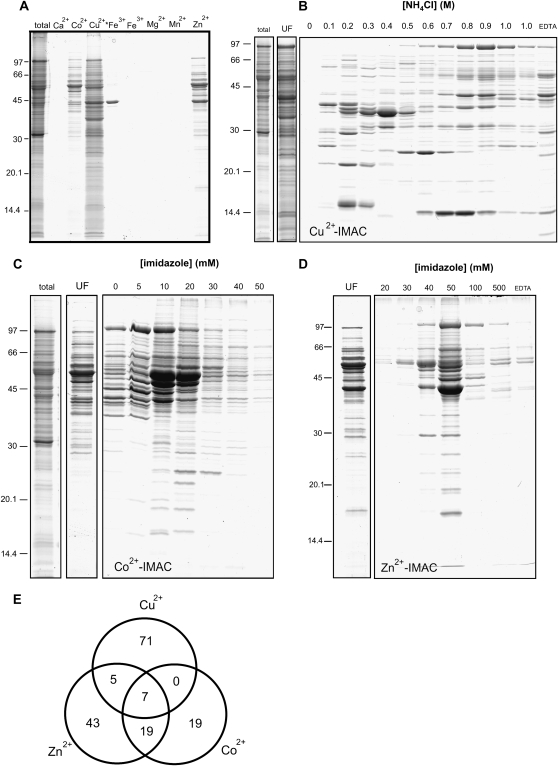
The interaction of metal ions with mitochondrial proteins that might be responsible for these toxic effects was investigated in vitro using IMAC to trap metal-binding proteins. Initial studies used Ca2+, Co2+, Cu2+, Fe3+, Mg2+, Mn2+, or Zn2+ to charge the IMAC resin (Fig. 1A). Arabidopsis mitochondrial proteins solubilized in 0.1% (v/v) Triton X-100 were introduced into IMAC columns charged with metal ions of interest. Proteins that were metal binding were retained on the IMAC resin, while unbound proteins were removed in wash steps. The bound proteins were eluted from the IMAC resin by stripping the metal ions with EDTA. The eluent was concentrated, desalted, and analyzed by SDS-PAGE and then compared with total mitochondrial protein extracts to determine the specificity of protein enrichment. This showed that only Co2+, Cu2+, Zn2+, and Fe3+ retained subsets of mitochondrial proteins (Fig. 1A). These metal ions were then used to explore the differential interactions of divalent metal ions with proteins and how these interactions potentially modulate protein function. To further eliminate apparent nonspecific binding of proteins to IMAC, fractionation of the proteins into weakly and strongly interacting sets by electrostatic or competitive displacement with NH4Cl and imidazole, respectively, was conducted. The enriched bands were excised for protein identification by MS. For MS protein identification of SDS-PAGE bands, multiple proteins are often identified, and only the major proteins identified from each band were reported (Supplemental Tables S1–S4).
Figure 1.
IMAC affinity purification of proteins from Arabidopsis mitochondria. A, Selective purification of mitochondria protein subsets using different metals to charge IMAC resin and EDTA elution. B, Different strengths of Cu2+ affinity to IMAC using NH4Cl step gradient elution. C and D, Different strengths of Co2+ (C) and Zn2+ (D) using imidazole gradient elution. E, Venn diagram of proteins identified from each metal-binding set using MS (for details, see Supplemental Tables S1–S4).
Cu2+ Interaction Proteins
Initial time-course studies of proteins binding to Cu2+-IMAC revealed that proteins bound rapidly and that 1-min incubations were sufficient to permit binding (data not shown). However, the protein profile of bound proteins was very complex and showed poor selectivity compared with whole mitochondrial samples (Fig. 1A). Various concentrations of imidazole were investigated but resulted in poor fractionation of proteins based on strength of binding (data not shown). Ammonium chloride step gradients allowed electrostatic displacement of the Cu2+-bound proteins and improved protein fractionation, providing distinct protein profiles based on the strength of protein affinity to Cu2+-IMAC (Fig. 1B). Thirty-five proteins were identified in bands from fractions between 0.1 and 0.6 m NH4Cl and were designated weakly associating proteins (Supplemental Fig. S1; Supplemental Table S1), while 48 strong Cu2+-IMAC-interacting proteins were identified from 0.8 to 1 m NH4Cl fractions and also from the EDTA-stripped IMAC resin (Supplemental Fig. S1; Supplemental Table S2). Some of the proteins identified were also found by Kung et al. (2006) in their study of Cu-binding proteins from whole Arabidopsis root extracts, notably glutathione-S-transferases (At2g30860 and At2g30870), ATP synthase β-subunit (At5g08670), and aconitase (At4g26870). Additionally, 25 proteins that have not previously been identified by MS from plant mitochondria were enriched by Cu2+-IMAC and identified. These previously uncharacterized proteins are indicated in Supplemental Tables S1 and S2. The experimentally derived Cu-interacting protein subset was very different from bioinformatic predictions, which have estimated that 0.5% (approximately 150 proteins) of the Arabidopsis proteome consists of Cu-binding proteins, with the largest number belonging to the Cu-dependent oxidoreductase class (Andreini et al., 2008). Instead, Cu-interacting proteins were often involved in redox reactions, such as cytochrome c (At1g22840 and At4g10040), electron transport flavoprotein (At1g50940), and a plethora of non-Cu-dependent dehydrogenases and reductases (Supplemental Tables S1 and S2). Interestingly, the list of Cu2+-binding proteins identified here is remarkably similar to the reported list of rice matrix proteins that are carbonylated after Cu2+-induced oxidation of rice mitochondrial extracts (Kristensen et al., 2004).
Co2+ Interaction Proteins
Initially, unfractionated proteins that were bound to Co2+-IMAC were analyzed by SDS-PAGE and revealed an enrichment of some protein bands when compared with the total mitochondrial protein sample (Fig. 1, A and C). The 10 bands that were enriched in the unfractionated sample were excised and identified (Supplemental Fig. S2; Supplemental Table S3). The separation of weak and strong Co2+-interacting proteins was attempted using both competitive and electrostatic displacement of proteins. Imidazole fractionation of Co2+-IMAC-bound proteins showed that most eluted at concentrations of 10 to 20 mm. Five extra protein bands were enriched in the 20 mm imidazole fraction and excised for protein identification (Fig. 1C; Supplemental Fig. S2; Supplemental Table S3). Fractionation via electrostatic displacement was also conducted. The majority of Co2+-binding proteins could be displaced from the resin using 0.1 m NH4Cl, showing that the binding of proteins to Co2+-IMAC is significantly weaker than that of Cu2+-IMAC-binding proteins (data not shown). Using Co2+-IMAC, 45 proteins involved in detoxification, DNA synthesis, protein fate, protein synthesis, signal transduction, and unknown functions were identified. However, energy production and metabolism proteins were by far the best represented functional category (Supplemental Table S3). Co2+-IMAC was able to purify proteins that were found using both Cu2+ and Zn2+, but the binding of proteins to Co2+ appears more similar to Zn, as the proteins NADH dehydrogenase subunit 9, cytochrome c oxidase subunit 5b, dihydrolipoamide dehydrogenase, malic enzyme, 2-oxoglutarate dehydrogenase, and nucleoside diphosphate kinase were found in common between the Co2+ and Zn2+ sets but were not in the Cu2+ set. Co2+-IMAC was also able to purify 19 proteins that could not be purified by the other divalent cations (Fig. 1E). Examples of proteins exclusively purified by Co2+-IMAC include DAG proteins, subunits of complex I, methylcrotonyl-CoA carboxylase α-subunit, arginase, and four other metabolic enzymes. Co2+-IMAC was also shown to be an effective tool in enriching low-abundance proteins, as nine proteins previously uncharacterized by MS to be in mitochondria were identified (Supplemental Table S3).
Zn2+ Interaction Proteins
Unfractionated proteins that were bound and eluted from Zn2+-IMAC were analyzed by SDS-PAGE, which revealed enrichment of specific protein bands when compared with the total mitochondrial protein sample (Fig. 1A). The 11 bands that were enriched in the unfractionated fraction were excised for identification (Supplemental Fig. S3; Supplemental Table S4). The separation of weak and strong Zn2+-interacting proteins was also attempted using both competitive and electrostatic displacement of proteins. As with Cu2+- and Co2+-IMAC, imidazole was not able to efficiently fractionate proteins bound to Zn2+-IMAC (Fig. 1D). Despite poor fractionation of all the IMAC protein sets with imidazole, relative protein-binding strength could be determined based on the concentration of imidazole needed to elute a large portion of the proteins. Proteins bound to Zn2+-IMAC more tightly than Co2+-IMAC, as the majority of proteins were displaced at 50 mm imidazole in Zn2+-IMAC compared with 10 to 20 mm imidazole in Co2+-IMAC (Fig. 1, C and D). Electrostatic displacement of Zn2+-interacting proteins using ammonium chloride also verified stronger protein binding to Zn2+ compared with Co2+, as proteins tended to elute at 0.7 m NH4Cl compared with 0.1 m NH4Cl in Co2+-IMAC (data not shown). Eleven additional protein bands observed to have eluted from Zn2+-IMAC in the 50 mm imidazole fraction were also excised and identified (Supplemental Fig. S3; Supplemental Table S4). Zn2+-IMAC appeared to have no success in the enrichment of the well-characterized Zn-dependent proteins such as translocases of the inner membrane (Lister et al., 2002) or the metalloprotease PreP (Moberg et al., 2003). Instead, the 74 proteins identified are likely to associate with Zn through a variety of mechanisms. Some examples of these include proteins that contain Zn fingers (cytochrome c oxidase 5b; Kubo et al., 2006), that use Zn as a cofactor (nucleoside diphosphate kinase, mitochondrial processing peptidase β; Parks and Agarwal, 1973; Luciano and Geli, 1996), that are inhibited by Zn (methylcrotonyl-CoA carboxylase, 2-oxoglutarate dehydrogenase; Diez et al., 1994; Brown et al., 2000), or that interact with divalent cations (malic enzyme, Glu dehydrogenase, succinate dehydrogenase Fe-S subunits; Massarini and Cazzulo, 1975; Kindt et al., 1980). Proteins with no known association with Zn ions, such as the ATP synthase subunits, were also found. Interestingly, Zn2+-IMAC was able to purify 11 subunits of the ATP synthase complex, which was greater than any of the other metal-IMAC resins used.
Fe3+ Interaction Proteins
Enrichment of proteins with Fe3+-IMAC was quite variable. The major protein enriched in Figure 1A was identified as malate dehydrogenase (At1g53240), but this pattern and specificity can be changed and greatly dependent on the pH and ionic strength of the medium used. Our data indicated that we were largely looking at a pseudocation exchange effect, which was in agreement with the findings by Zachariou and Hearn (1996; for more details, see Supplemental Fig. S4).
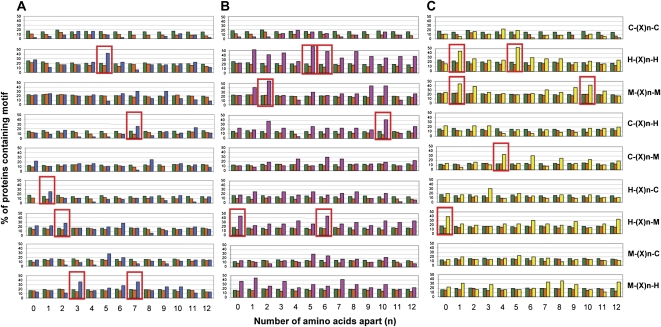
Metal-Binding Motifs in Mitochondrial IMAC-Interacting Protein Sets
Metals often bind proteins at specific coordination sites involving Cys, His, and Met residues (Harding, 2004). Hence, an analysis of the sequences of the proteins in our IMAC subsets was performed to determine if putative metal-binding motifs were more common that expected by random chance in these proteins. For simplicity, the metal-binding motif parameters used were the same as those used by Kung et al. (2006) to validate their metal-binding proteins. Kung et al. (2006) found that the top six statistically enriched motifs in their protein subsets could account for nearly 90% of the proteins identified. Kung et al. (2006) were also able to show Cu binding to synthetic peptides carrying a range of these putative Cu-binding motifs, but they did not characterize the biological significance of such motifs. In our data, 10, 45, and 21 Cys-His-Met motifs were found to be significantly greater in frequency in the Cu, Co, and Zn subsets, respectively, when compared with the entire Arabidopsis proteome or the known Arabidopsis mitochondrial proteome (Supplemental Table S5). Figure 2 shows the top six enriched motifs for each of the divalent metal cation-binding sets. In total, 72% of proteins from our Cu-binding subset contained one or more of the top six Cu-binding motifs, compared with 96% and 89% for Co and Zn, respectively (Supplemental Fig. S5), which is comparable to the findings of Kung et al. (2006) using this same method. One of the top six motifs, H-(X)5-H, was common to all three data sets and also to Kung et al. (2006), while HM was shared between our Co2+ and Zn2+ data sets. His residues were significantly more enriched in the Co-binding protein subset, suggesting that His motifs may complex Co more readily than Cu or Zn.
Figure 2.
Metal-binding motif analysis of proteins. Combinations of His (H), Cys (C), and Met (M) residues within proximity of 12 amino acid residues were considered putative metal-binding motifs. X represents any amino acid, and n denotes the number of amino acids apart. The frequency of motifs occurring in (A) Cu2+-IMAC isolated subset (n = 36 proteins; blue), (B) Co2+-IMAC isolated subset (n = 27 proteins; pink), and (C) Zn2+-IMAC isolated subset (n = 36 proteins; yellow) was compared with that of the Arabidopsis proteome (n = 26,702 proteins; green) and the Arabidopsis mitochondrial proteome (n = 716 proteins; orange). The top six statistically significant motifs are highlighted in the red boxes.
Relationship between Metal Ion Interaction and Modulation of Protein Function
A range of major TCA cycle enzymes, which were identified to interact in IMAC here and have been reported to be carbonylated in the literature, was selected for activity measurements. The aim was to determine if there was a clear relationship between observed metal ion interactions using IMAC, reports of protein oxidation, and modulation of enzyme activity.
Aconitase was found to interact with Cu2+ (Supplemental Table S2) and is reported to be oxidatively modified by Cu-catalyzed mechanisms (Kristensen et al., 2004). Aconitase was inhibited 25% by H2O2 treatment (Table IV), and this is in agreement with previous studies in plants and mammals (Verniquet et al., 1991; Brazzolotto et al., 1999). Cu2+ was able to diminish aconitase activity by 80%, and the primary mechanism of this inhibition appeared to be independent of reducing or oxidizing agents (Table IV). Fe3+ and Zn2+ did not affect aconitase activity directly or by MCO. Interestingly, the presence of Fe3+ or Zn2+ in the H2O2 treatment was in fact able to protect aconitase activity (Table IV). This may be due to the metal catalysts degrading H2O2 to products that are less toxic to aconitase. The addition of labile Fe3+ to the mitochondria may have been beneficial to aconitase activity through restoring of the Fe-S cluster, damaged by H2O2, from the inactive [3Fe4S]+ state to the active [4Fe4S]2+ state (Brazzolotto et al., 1999).
Table IV.
Modulation of mitochondrial enzyme activities by metal ions and metal-catalyzed oxidation
Data are expressed as the average nmol of product per min per mg of protein ± se (n ≥ 4; * P < 0.05, ** P < 0.01, comparing each treatment with the appropriate enzyme activity in untreated mitochondria). The measured product was aconitate for ACON, fumarate for FUM, and NADH for α-ketoglutarate dehydrogenase (KGDH), pyruvate dehydrogenase complex (PDC), malate dehydrogenase (MDH), and isocitrate dehydrogenase (ICDH). Metal ion interactions were as determined by IMAC experiments. Metal ions were added at 1.5 μmol per mg of mitochondrial protein. n/a, Not applicable.
| Treatment |
ACON |
FUM |
KGDH |
PDC |
MDH |
ICDH |
|---|---|---|---|---|---|---|
| Metal ion interaction: | Cu2+ | n/a | Co2+, Zn2+ | Co2+, Cu2+ | Cu2+, Fe3+ | Zn2+ |
| Control | 116 ± 12 | 83 ± 9 | 55 ± 4 | 80 ± 7 | 3,100 ± 400 | 32 ± 4 |
| Ascorbate | 132 ± 10 | 96 ± 10 | 57 ± 5 | 82 ± 8 | 3,900 ± 500 | 25 ± 4 |
| H2O2 | 75 ± 9* | 71 ± 17 | 60 ± 2 | 74 ± 5 | 3,900 ± 300 | 34 ± 4 |
| Cu2+ | 22 ± 7** | 82 ± 21 | 11 ± 4** | 25 ± 3** | 3,300 ± 400 | 37 ± 3 |
| Cu2+ + ascorbate | 29 ± 10** | 91 ± 14 | 14 ± 4** | 30 ± 10** | 2,800 ± 300 | 32 ± 3 |
| Cu2++ H2O2 | 28 ± 3** | 97 ± 23 | 8 ± 2** | 17 ± 8** | 3,100 ± 500 | 30 ± 6 |
| Fe3+ | 128 ± 6 | 48 ± 11* | 56 ± 6 | 73 ± 7 | 3,700 ± 500 | 25 ± 3 |
| Fe3+ + ascorbate | 108 ± 8 | 61 ± 5* | 55 ± 4 | 72 ± 14 | 3,000 ± 500 | 27 ± 5 |
| Fe3+ + H2O2 | 103 ± 7 | 43 ± 8** | 53 ± 3 | 79 ± 5 | 2,900 ± 400 | 26 ± 4 |
| Zn2+ | 99 ± 7 | 68 ± 14 | 22 ± 3** | 48 ± 2** | 3,900 ± 400 | 36 ± 4 |
| Zn2+ + ascorbate | 100 ± 6 | 70 ± 20 | 21 ± 2** | 42 ± 10* | 3,500 ± 500 | 25 ± 5 |
| Zn2+ + H2O2 | 90 ± 11 | 83 ± 17 | 25 ± 3** | 31 ± 12** | 4,100 ± 400 | 37 ± 7 |
The activities of pyruvate dehydrogenase complex and α-ketoglutarate dehydrogenase complex showed similar patterns of inhibition by metal ions. Both activities were significantly affected by Cu2+ and Zn2+ by direct mechanisms and were found to be resistant to MCO under the conditions examined (Table IV). The observed toxicity of Cu2+ toward pyruvate and α-ketoglutarate dehydrogenase complex activities agrees with in vivo and in vitro studies in mammalian mitochondria (Sheline and Choi, 2004). The toxicity of Zn2+ toward α-ketoglutarate dehydrogenase complex activity could at least in part explain the inhibition of Glu/malate-driven cytochrome oxidase-dependent respiration by Zn2+ (Table III). Subunits of pyruvate dehydrogenase complex were found by IMAC to interact with Co2+ and Cu2+, and subunits of α-ketoglutarate dehydrogenase complex were found to interact with Co2+ and Zn2+, indicating mechanisms for the direct effect of metal ions on these enzymes.
Fumarase was not found as a metal-interacting protein in our IMAC studies but has been reported to be carbonyl modified in rice mitochondria by Cu2+-catalyzed oxidation (Kristensen et al., 2004). In our hands, the activity of this enzyme was not affected by MCO (Table IV). However, it was the only enzyme investigated that showed sensitivity to ferric ions, which caused a 30% decrease in activity (Table IV).
The TCA cycle enzymes isocitrate dehydrogenase and malate dehydrogenase were also found to be putatively oxidized in rice mitochondria (Kristensen et al., 2004). As with fumarase, enzyme assays of Arabidopsis mitochondria showed both isocitrate dehydrogenase and malate dehydrogenase activities to be resistant to H2O2 and MCO under the conditions investigated (Table IV). None of the metal ions used was able to modulate either isocitrate dehydrogenase or malate dehydrogenase activity, despite experimental data from IMAC suggesting interactions with Zn2+ and Cu2+/Fe3+ for each enzyme.
Hence, while a number of TCA cycle enzymes that were selected for analysis showed a functional impact of incubation with metal ions, there was a complex relationship with the metal interaction data (Table IV). As MCO of proteins may not necessarily occur at the active site, protein carbonylation may not be noticeably detrimental in protein function assays. The specificity of carbonylation was further investigated to assess the susceptibility of putative metal-binding sites to oxidation using aconitase as a case study.
Linking Metal-Binding Sites and Oxidized Residues to Protein Structure of Aconitase
Aconitase interacts with Cu2+-IMAC (Fig. 1B; Supplemental Fig. S1; Supplemental Table S2), contains all but one of the top six putative Cu2+-binding motifs, has reduced activity on Cu2+/H2O2 exposure (Table IV), and is reported to be oxidatively modified by Cu-catalyzed mechanisms (Kristensen et al., 2004). The three-dimensional (3D) structures of the Arabidopsis mitochondrial aconitases (At2g05710 and At4g26970) can also be predicted using the crystal structure of the human cytosolic aconitase (Protein Data Bank accession no. 2b3y; Dupuy et al., 2006), which shares approximately 60% amino acid identity with the Arabidopsis mitochondrial aconitase proteins.
The locations of the putative Cu-binding motifs H-X5-H, M-X3-H, C-X7-H, H-X1-C, M-X7-H, and H-X2-M were mapped onto the aconitase structure using a BLAST alignment of the protein sequences (Supplemental Fig. S6). Based on the 2b3y crystal structure, the two coordinating His residues of the H-X5-H motif are 18.93 Å and are not likely to form a metal-coordinating pocket. The M-X3-H motif and the conserved C-X7-H and H-X1-C motifs are clustered together on the protein surface, indicating that this region is likely to attract Cu ions; however, the distances between the respective coordinating residues are 6.51, 11.58, and 5.25 Å for M-X3-H, C-X7-H, and H-X1-C, respectively, which is relatively large, as the criteria for metal coordination normally require the electron donor to be within approximately 0.75 Å of the metal ion (Harding, 2004). H-X2-M was the only motif found within the core of the protein, near the aconitase catalytic and substrate recognition sites. The predicted distance between the coordinating residues of this motif was 5.75 Å. Whether this site is responsible for Cu inactivation of aconitase activity remains to be investigated. However, it is unlikely that the motif is responsible for aconitase binding to the Cu2+-IMAC resin, as the site is buried within the protein core and is likely to have restricted access to the immobilized Cu2+.
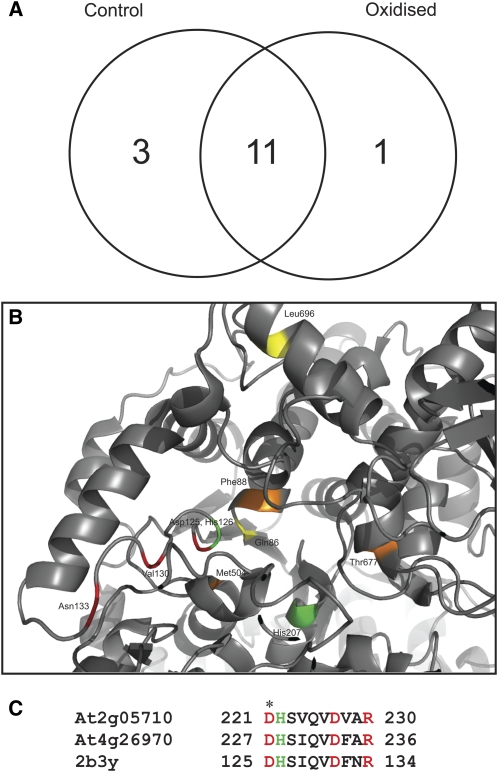
To test whether sites near putative metal-interacting motifs had an enhanced susceptibility to MCO, the soluble protein fraction from Arabidopsis mitochondria was treated with 100 μm Cu2+ and H2O2 to elicit MCO. The band corresponding to aconitase on SDS-PAGE was excised and trypsin digested, and peptides were analyzed by MS. The variable modifications of oxidation of the amino acids Cys, Asp, Phe, Lys, Met, Asn, Pro, Arg, and Tyr were used to determine if MCO elicited any detectable changes between mock and treated extracts. In triplicate experiments, there were 11 tryptic peptides from aconitase consistently found in both mock- and Cu2+-treated samples; in addition, three peptides from aconitase were consistently found only in the mock samples and one multiply oxidized peptide was found consistently and only in the Cu2+ treated samples (Fig. 3A). This latter peptide is in a highly conserved region of aconitase and includes an Asp and a His residue of the active site (Fig. 3, B and C). A range of other oxidized peptides of aconitase from the oxidized samples were found in single experiments but were not able to be repeatedly observed. These apparently random events typically mapped to the surface of the 3D structure, but they are generally not in close proximity to the metal-binding motifs (Supplemental Table S6; Supplemental Fig. S6).
Figure 3.
Peptides identified for plant mitochondrial aconitase (At2g05710). A, Number of peptides consistently identified by MS common to both mock- and H2O2-treated enzymes or unique to each treatment. n = 3 to 4 experiments. Peptides noted appeared in at least two experiments. B, Mapping identified peptides onto the 3D structure of the mammalian aconitase. C, BLAST alignment of the region containing the oxidized peptide in plant mitochondrial aconitases and human IRP1 (2b3y). Oxidized (red) and active site (green) residues are indicated in B and C. Substrate recognition site (yellow) and Fe-S cluster ligation site (orange) residues are shown in B. *, Asp-125 (in C) is a conserved active site residue and was found to be oxidized.
DISCUSSION
Metals are important cofactors in many biological reactions, but to date, there has been little systematic analysis of the metal composition of subcellular organelles in plants. Our screen of the metallome of Arabidopsis cell culture mitochondria is, to our knowledge, the first multielement profiling of a subcellular organelle in plants. Fractionation of mitochondrial samples revealed that the integral membrane fraction had a 6-fold greater Cu and Fe content than the soluble protein compartment on a protein basis, consistent with the redox transition metals involved in the ETC. The Arabidopsis mitochondrial Fe and Mn contents of 3.2 and 0.12 nmol mg−1 protein, respectively, are relatively similar to those of yeast mitochondria, which were found to be 5 to 10 and 0.16 to 0.36 nmol mg−1 protein, respectively (Luk and Culotta, 2001; Luk et al., 2003, 2005; Yang et al., 2006). The discovery of trace amounts of Co2+ and Mo2+ in Arabidopsis mitochondria was somewhat unexpected; however, reports of Co2+ substituting for other metals in metal-dependent enzyme reactions in plant mitochondria (Palmer and Wedding, 1966; Macrae and Moorhouse, 1970; Macrae, 1971) and of a mitochondrial Mo2+ carrier protein in Arabidopsis (Baxter et al., 2008) are consistent with these data.
Little is known regarding the subcellular perturbations in the metal content during oxidative stress, despite the general acceptance that MCO is a common source of oxidative modification in biological macromolecules (Stohs and Bagchi, 1995). Comparing the impact of oxidative stress on the metallome of the Arabidopsis mitochondria using the same chemical elicitors reported by Sweetlove et al. (2002) and Winger et al. (2005, 2007) allowed the changes in respiratory activity, lipid peroxidation, and protein degradation reported in these studies to be considered in light of the metal-catalyzed reactions investigated here. H2O2 treatment resulted in a detectable loss of Cu from total mitochondria (Table II). Upon fractionation of the mitochondria, a decrease in membrane-bound Cu levels was observed, suggesting damage to Cu-containing enzymes such as cytochrome c oxidase. There was also a loss of Cu, Fe, and Mn observed in the soluble compartment, suggesting damage to matrix metalloproteins following H2O2 treatment. Sweetlove et al. (2002) observed a decreased respiratory rate after H2O2 addition and breakdown of a series of proteins, including Fe-containing aconitase, complex I, and SOD, as well as the breakdown of a set of matrix enzymes that are shown here to bind to IMAC resins (Supplemental Tables S1–S4). Menadione treatment, like H2O2, was able to induce the loss of Cu in the membrane fraction (Table II), indicating possible damage to the cytochrome c oxidase complex. The proteome changes induced by both these chemical stresses were similar, according to Sweetlove et al. (2002).
Antimycin A and menadione both appeared to induce a 2-fold accumulation of Fe in mitochondria (Table II). Transcriptional changes in Fe metabolism during oxidative stress suggest that induction of ferroproteins such as cytochrome c and alternative oxidase may account at least in part for these Fe changes following antimycin A treatment (Yu et al., 2001). Winger et al. (2007) has reported similar patterns of HNE-modified proteins following antimycin A and menadione treatments, and lipid peroxidation leading to HNE is known to be an Fe2+-stimulated process. The disturbance of Fe and Cu homeostasis by menadione has also been supported in a study of hamster fibroblasts, in which menadione treatment increased the lability of both these redox-active transition metals (Calderaro et al., 1993).
This study is also in agreement with wider proteomic studies investigating the changes in protein abundance in Arabidopsis during abiotic stress. Aconitase is known to release ferrous ions when oxidatively damaged (Verniquet et al., 1991; Brazzolotto et al., 1999) and has been reported to decrease in abundance in cadmium-stressed (Sarry et al., 2006), salt-stressed (Jiang et al., 2007), and salt- and osmotic-stressed (Ndimba et al., 2005) plants. Another mitochondrial metalloprotein that has been shown to have altered abundance following oxidative stress is the Zn-containing mitochondrial processing peptidase (At3g02090.1), which increased in abundance in salt and osmotic stress (Ndimba et al., 2005). The increase in abundance of mitochondrial processing peptidase may contribute to the import of new proteins to replace degraded proteins. In contrast, while we have not seen any evidence of increases in Mn content in mitochondria, MnSOD has been observed to increase in abundance in salt-stressed (Jiang et al., 2007) and in salt- and osmotic-stressed (Ndimba et al., 2005) Arabidopsis plants.
While the release of metal ions from metalloproteins and the import of new metal cofactors into the mitochondrion to synthesize or repair metalloproteins are likely to contribute to the changes in the mitochondrial metallome, they also raise the need to study the way in which metals interact with the proteome as a whole. Examining this metal-protein interactome is extremely complicated and fraught with technical challenges. Here, we have used IMAC in an attempt to mimic protein binding to free metal ions in vivo. Under the native conditions explored, only exposed metal-binding surfaces are studied, and there is no assessment of metal-coordinating sites that are buried in the protein core (Ueda et al., 2003). In addition, steric hindrance caused by the use of immobilized metal ions on a fixed ligand will limit access to metal ion-binding sites in some proteins (Ueda et al., 2003). Additionally, the strength of metal coordination in a native metalloprotein may be too strong to allow for an exchange of metal ion cofactors with the IMAC resin. Like the IMAC study of Kung et al. (2006), we have found that IMAC selects subsets of proteins that contain significant numbers of putative metal interaction motifs on their surfaces. In the metal-protein interaction studies of Cu ions and liver disease (She et al., 2003; Smith et al., 2004), nickel hypersensitivity in human B cells (Heiss et al., 2005), and Cu ion homeostasis in Arabidopsis roots (Kung et al., 2006), no attempt was made to localize the metal-binding motifs on homologous proteins. However, the surface localization of the putative metal-binding motifs is supported by reports of the use of IMAC to assist in protein crystallization through surface His residues (Frey et al., 1996) and reports of protein contaminants in metal affinity purification of His-tagged recombinant proteins due to natural surface metal-binding motifs (Cai et al., 2004; Bolanos-Garcia and Davies, 2006). While the functional significance of these putative surface metal-interacting sites remains undefined, we noted that there was a good correlation between these proteins and experimentally observed sets of proteins that are carbonyl tagged (Kristensen et al., 2004), lipid peroxide tagged (Winger et al., 2007), and/or degraded (Sweetlove et al., 2002) during oxidative stresses. We thus hypothesized that they might have had an increased susceptibility to MCO due to their affinity for metal ions.
Detailed analysis of the activity of major matrix-located enzymes following exposure to oxidative conditions revealed very different responses (Table IV), indicating that the impact of oxidation on function is highly variable, dependent on different sets of conditions and different metal ions; thus, specific studies will be required to assess the impact of chemical stresses on each protein (Table IV). In support of this, studies in castor bean (Ricinus communis) peroxisomal proteins have also demonstrated a lack of correlation between the extent of protein oxidation and the inhibition of protein function following MCO (Nguyen and Donaldson, 2005). The prospect of having to perform functional analysis on all oxidized proteins to infer effects is daunting. However, our study of aconitase revealed that MS analysis of the peptides from damaged proteins and layering onto 3D structures (Fig. 3; Supplemental Table S6) can provide important information. For example, by revealing putative active site damage consistent with the loss of function of this enzyme during oxidative conditions as well as a range of surface-localized oxidation sites.
The general proposition that electron donor-dense regions on protein surfaces coordinate metal ions and, in the presence of ROS, the specific metal-coordinating residue(s) are oxidized (Stadtman, 1993) is supported by a series of specific studies. For Gln synthetase, it was observed that in a peptide containing a stretch of oxidation-prone amino acids, Met-268-His-269-Cys-270-His-271-Met-272, only the metal-coordinating His-269 was oxidized (Farber and Levine, 1986). The theory of “caged MCO” has been applied to the characterization of Cu-binding residues of the cuproproteins Cu/ZnSOD and azurin (Bridgewater et al., 2006a) and the known metal-interacting proteins angiotensin I and bacitracin (Bridgewater et al., 2006a, 2006b). The sites of oxidation that were found to be in the vicinity of the Cu-interacting site were determined by manual interpretation of the product ions derived from both MS/MS and MS/MS/MS analysis of peptide fragments (Bridgewater et al., 2006a, 2006b). Also, specific surface His residues of the cuproprotein ceruloplasmin have been shown to coordinate Cu and promote the oxidation of low-density lipoprotein (Mukhopadhyay et al., 1997). Our analyses to date have not provided evidence for a link between metal-binding sites and MCO in the case of the plant mitochondrial aconitase. However, studies of complex lysates have yet to be performed in any system to demonstrate MCO specificity on a large scale and thus determine if this concept is the exception or the rule in understanding metal-induced damage to proteins.
While it is generally considered that oxidative modification leads to protein dysfunction by affecting the structural integrity of the protein, promoting the formation of protein aggregates, and potentially damaging the active site (Starke-Reed and Oliver, 1989), protein oxidation may not be entirely detrimental to protein function. Hence, among the protein oxidation events in plant mitochondria may be important triggers for the stress response of the organelle. Protein oxidation can promote protein degradation and turnover (Rivett, 1985; Davies and Lin, 1988; Marcillat et al., 1988). It can also provide protective outcomes for the cell; for example, a bacterial transcription factor has been identified that senses and promotes an appropriate cellular response to an oxidative environment through MCO (Lee and Helmann, 2006). In addition, Met residues on protein surfaces have been shown to act as antioxidants to protect the active site of enzymes (Levine et al., 1996), and oxidation of Met residues can block phosphorylation-induced regulation of proteins (Hardin et al., 2009).
This study attempted to better define the protein interactions with metal ions and the associated modulation of protein functions in plant mitochondria. While the site specificity of MCO could not be confirmed, it was demonstrated that the metal content of mitochondria is dynamic and changes during oxidative stress, that different proteins have varying metal affinity and varying susceptibility to inactivation by H2O2, metal ions, or MCO, and that oxidative treatments do not equally and/or detrimentally affect a variety of mitochondrial enzyme activities. Defining the sites of oxidation and mapping them to the functional regions of protein sequences en masse in the future will be essential to uncover the broader oxidative modulation of enzyme activities in plant mitochondria.
MATERIALS AND METHODS
Induction of Oxidative Stress in Arabidopsis and Isolation of Mitochondria
Seven-day-old dark-grown heterotrophic Arabidopsis (Arabidopsis thaliana Landsberg erecta) cell suspension cultures (May and Leaver, 1993) were treated with 10 mm H2O2, 400 μm menadione prepared in methanol, or 25 μm antimycin A prepared in methanol for 8 h. The concentrations of antimycin A and menadione have previously been optimized (Winger et al., 2005). Equivalent volumes of either water or methanol were added as controls for the respective treatments. Mitochondria were isolated as described previously (Millar et al., 2001).
ICP-MS Analysis of Metal Content
To determine the sites of altered metal content, mitochondria were fractionated into soluble protein and integral membrane protein fractions. Intact isolated mitochondria were suspended in milliQ water before lysis by three freeze/thaw cycles. Soluble proteins were collected in the supernatant following centrifugation at 20,000g. Peripherally attached proteins were depleted from the total membrane fraction using 0.1 m Na2CO3 treatment to produce the integral membrane protein fraction (Fujiki et al., 1982). Redistilled concentrated HNO3 (kindly provided by Prof. John Watling, Centre for Forensic Science, University of Western Australia) was used to break down organic material by heating at 160°C for 1 h. The acid digest was diluted to less than 5% (v/v) HNO3 and passed through 0.22-μm filters (Millipore). Metal speciation and quantification were performed by the Centre for Forensic Science using the Perkin-Elmer Elan 5000 ICP-MS system calibrated against elements of interest.
Respiratory Assays
Oxygen consumption by cell suspension cultures and isolated mitochondria was measured by a Clark-type oxygen electrode (Hansatech Instruments). Respiratory data were collected and analyzed using OxyGraph Plus version 1.01 software (Hansatech Instruments). Liquid phase calibration was performed by adding excess sodium dithionite to 1 mL of autoclaved water to remove all oxygen at 25°C. Mitochondrial respiration assays were conducted at 25°C by adding 100 to 120 μg of mitochondrial protein to 1 mL of respiration medium (0.3 m Suc, 5 mm KH2PO4, 10 mm TES, 10 mm NaCl, 2 mm MgSO4, and 0.1% [w/v] bovine serum albumin, pH 7.2). Respiration via complex II was initiated by adding succinate and ATP. Respiration initiated through the external NADH dehydrogenase pathway was measured using NADH, CaCl2, and rotenone. Respiration initiated through matrix NADH dehydrogenases was measured using malate, Glu, CoA, TPP, and NAD+. In all respiratory assays, following the addition of substrates to initiate respiration, ADP was added to induce maximum state 3 respiratory rates. Respiration via the alternative and cytochrome oxidase pathways was determined by the addition of the inhibitors KCN and n-propyl gallate, respectively.
Enzyme Assays
Assays were performed at 25°C using a temperature-controlled spectrophotometer (U-2810 spectrophotometer; Hitachi), and data were collected by UV Solutions software version 2.1 (Hitachi). For inhibition assays, mitochondrial samples equivalent to 50 μg of protein were treated with 100 μm of metal ions for 5 min prior to the assay. The metal ions were added at an excess of 1.5 μmol mg−1 protein to induce maximal toxicity. Equimolar ascorbate or H2O2 was added to catalyze MCO. The protein samples were then directly added to the reaction medium for enzyme assays. All assays were conducted according to published methods (Lee et al., 2008).
Statistical Analyses
Unless stated otherwise, all data obtained from experiments were expressed as means ± se. Statistical significance was evaluated by the two-tailed unpaired Student's t test using Microsoft Office Excel 2003 or Kaleidagraph version 3.6 (Synergy Software) where appropriate.
IMAC
A HiTrap Chelating HP 1-mL column (Amersham Biosciences) consisting of prepacked iminodiacetic acid conjugated to agarose beads was used. All chromatography was conducted manually using syringes with the flow rate maintained at approximately 1 mL min−1. The column was charged with 1 column volume (CV) of 0.1 m metal ion solution, and excess metals were removed with 5 CV of milliQ water. The charged column was equilibrated with 10 CV of binding buffer (20 mm NaH2PO4, 0.5 m NaCl, and 0.1% [v/v] Triton X-100) unless stated otherwise. Prior to sample loading, 2 mg of pelleted mitochondrial proteins was suspended in 500 μL of milliQ water and subjected to three freeze/thaw cycles. The lysate was passed through a 0.22-μm filter (Millipore), and 500 μL of 2-fold concentrated binding buffer was added to the filtered lysate so that the proteins were suspended in 1 mL of lysate. The lysate was injected into the column, allowed to incubate at room temperature for 1 to 5 min, and then washed with at least 10 CV of binding buffer to remove unbound proteins. Proteins were fractionated with varying concentrations of NH4Cl or imidazole in the binding buffer. All remaining proteins were removed from the column by stripping the metal ions with an eluent containing 50 mm EDTA and 50 mm NaCl. Due to the high salt content of the eluent fractions, the proteins were concentrated using 5-kD cutoff centrifugal filter units (Millipore), and the excess salt was diluted following buffer exchange with milliQ water.
Metal-Binding Motif Analysis
Metal-binding motif analysis was based on analysis of similar work on Arabidopsis root Cu-binding proteins where His, Met, and Cys residues in any combination and up to 12 amino acids apart were considered putative metal-binding motifs (Kung et al., 2006). The algorithm for motif screening was written in MySQL, and 117 potential metal-binding motifs were analyzed. The gene accession codes of the Arabidopsis proteome, a total of 26,738 putative proteins, were extracted from The Arabidopsis Information Resource (TAIR). The list of accession codes was obtained from SUBA, where parameters for mitochondrial proteins were “annotated in SwissProt” or “found in mitochondria by MS” or “found in mitochondria by GFP,” culminating in 742 proteins (Heazlewood et al., 2005). The standard normal distribution (Z score) statistical analysis was conducted on the frequency of occurrence of the IMAC motifs comparing with the entire Arabidopsis and Arabidopsis mitochondrial proteomes. Removal of IMAC-interacting proteins from the Arabidopsis and Arabidopsis mitochondrial proteome lists was performed in order to conduct statistical analyses on two independent samples.
MCO of Proteins
The Cu ion/ascorbate/oxygen system is the preferred mechanism for MCO, as the major product of this MCO system is the formation of 2-oxohistidine (Uchida and Kawakishi, 1986). Dose-response assays determined that 1 nmol metal ion mg−1 protein was optimum to induce maximal metal-catalyzed protein oxidation with minimal protein degradation. Equimolar ascorbate was added to catalyze MCO. All MCO treatments were performed at room temperature with constant agitation for 5 min. The MCO reaction was stopped by the addition of 5 mm EDTA. The metal ions were then removed from the proteins by overnight dialysis at 4°C against 50 mm HEPES-NaOH, pH 7.2.
In-Gel Protein Digestion and Peptide Extraction
Gel plugs from protein bands of interest were excised and destained twice for 45 min with 50% (v/v) acetonitrile in 25 mm NH4HCO3. The plugs were dehydrated at 50°C on a dry block heater for 30 min, rehydrated with 12.5 μg mL−1 trypsin in 25 mm NH4HCO3, and incubated overnight at 37°C. Peptides were extracted by adding 15 μL of acetonitrile with vigorous shaking for 15 min, removing liquid, and adding 15 μL of 50% acetonitrile and 5% formic acid to the gel plugs followed by another 15 min of shaking. The second extraction step was repeated, and all the samples were pooled after each extraction step and lyophilized.
Peptide MS and Protein Identification
Samples extracted from in-gel trypsin digestion were loaded onto self-packed Microsorb (Varian) C18 (5 μm, 100  ) reverse-phase columns (0.5 × 50 mm) using an Agilent Technologies 1100 series capillary liquid chromatography system and eluted into a XCT Ultra IonTrap mass spectrometer with an electrospray ionization (ESI) source equipped with a low-flow nebulizer in positive mode and controlled by Chemstation (Rev B.01.03 [204]; Agilent Technologies) and MSD Trap Control version 6.0 (Build 38.15) software (Bruker Daltonik). Peptides were eluted from the C18 reverse-phase column at 10 μL min−1 using a 9-min acetonitrile gradient (5%–60%) in 0.1% formic acid at a regulated temperature of 50°C. The method used for initial ion detection utilized a mass range of 200 to 1,400 mass-to-charge ratio (m/z) with scan mode set to standard (8,100 m/z per s) and ion charge control conditions set at 250,000, with three averages taken per scan. Smart mode parameter settings were employed using a target of 800 m/z, a compound stability factor of 90%, a trap drive level of 80%, and optimize set to normal. Ions were selected for MS/MS after reaching an intensity of 80,000 cps, and two precursor ions were selected from the initial MS scan. MS/MS conditions employed SmartFrag for ion fragmentation, a scan range of 70 to 2,200 m/z using an average of three scans, the exclusion of singly charged ions option, and ion charge control conditions set to 200,000 in ultrascan mode (26,000 m/z per s). Resulting MS/MS spectra were exported from the DataAnalysis for LC/MSD Trap version 3.3 (Build 149) software package (Bruker Daltonik) using default parameters for AutoMS(n) and compound export. For identification of proteins excised from SDS-PAGE gels, spectra were queried against the Arabidopsis protein set (TAIR version 7.0) using the Mascot search engine (Matrix Science; Perkins et al., 1999) utilizing error tolerances of ±1.2 D for MS and ±0.6 D for MS/MS, “Max Missed Cleavages” set to 1, with variable modifications of oxidation (M) unless stated otherwise. Instrument was set to ESI-TRAP, and peptide charge was set at 2+ and 3+. Results were filtered using standard scoring, “Max Number of Hits” set to 20, significance threshold at P < 0.05, and ion score cutoff at 0. The top three protein hits were reported unless stated otherwise.
) reverse-phase columns (0.5 × 50 mm) using an Agilent Technologies 1100 series capillary liquid chromatography system and eluted into a XCT Ultra IonTrap mass spectrometer with an electrospray ionization (ESI) source equipped with a low-flow nebulizer in positive mode and controlled by Chemstation (Rev B.01.03 [204]; Agilent Technologies) and MSD Trap Control version 6.0 (Build 38.15) software (Bruker Daltonik). Peptides were eluted from the C18 reverse-phase column at 10 μL min−1 using a 9-min acetonitrile gradient (5%–60%) in 0.1% formic acid at a regulated temperature of 50°C. The method used for initial ion detection utilized a mass range of 200 to 1,400 mass-to-charge ratio (m/z) with scan mode set to standard (8,100 m/z per s) and ion charge control conditions set at 250,000, with three averages taken per scan. Smart mode parameter settings were employed using a target of 800 m/z, a compound stability factor of 90%, a trap drive level of 80%, and optimize set to normal. Ions were selected for MS/MS after reaching an intensity of 80,000 cps, and two precursor ions were selected from the initial MS scan. MS/MS conditions employed SmartFrag for ion fragmentation, a scan range of 70 to 2,200 m/z using an average of three scans, the exclusion of singly charged ions option, and ion charge control conditions set to 200,000 in ultrascan mode (26,000 m/z per s). Resulting MS/MS spectra were exported from the DataAnalysis for LC/MSD Trap version 3.3 (Build 149) software package (Bruker Daltonik) using default parameters for AutoMS(n) and compound export. For identification of proteins excised from SDS-PAGE gels, spectra were queried against the Arabidopsis protein set (TAIR version 7.0) using the Mascot search engine (Matrix Science; Perkins et al., 1999) utilizing error tolerances of ±1.2 D for MS and ±0.6 D for MS/MS, “Max Missed Cleavages” set to 1, with variable modifications of oxidation (M) unless stated otherwise. Instrument was set to ESI-TRAP, and peptide charge was set at 2+ and 3+. Results were filtered using standard scoring, “Max Number of Hits” set to 20, significance threshold at P < 0.05, and ion score cutoff at 0. The top three protein hits were reported unless stated otherwise.
For the identification of posttranslational modifications of aconitase, peptide extracts were analyzed on an Agilent 6510 Q-TOF mass spectrometer with an HPLC Chip Cube source. The Chip consisted of a 40-nL enrichment column (Zorbax 300SB-C18 5 u) and a 150-mm separation column (Zorbax 300SB-C18 5 u) driven by the Agilent Technologies 1100 series nano/capillary liquid chromatography system. Both systems were controlled by MassHunter Workstation Data Acquisition for Q-TOF (version B.01.02, Build 65.4, Patches 1,2,3,4; Agilent Technologies). Results were queried against the Arabidopsis protein set (TAIR version 7.0) using the Mascot search engine (Matrix Science) utilizing error tolerances of ±1.2 for MS and ±0.6 for MS/MS, Max Missed Cleavages set to 1, with variable modifications of oxidation (M) unless stated otherwise. Instrument was set to ESI-Q-TOF, and peptide charge was set at 2+ and 3+. Results were filtered using standard scoring, Max Number of Hits set to AUTO, significance threshold at P < 0.05, and ion score cutoff at 0. MASCOT Daemon version 2.0 was used to automate searches using variable modifications already submitted to the Unimod database. The variable modifications of oxidation of the amino acids Cys, Asp, Phe, Lys, Met, Asn, Pro, Arg, and Tyr were used to determine if MCO elicited any detectable changes between control and oxidized protein samples.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Annotated ammonium chloride step gradient fractionation of Cu2+-IMAC-bound proteins.
Supplemental Figure S2. Annotated imidazole step gradient fractionation of Co2+-IMAC-bound proteins.
Supplemental Figure S3. Annotated imidazole step gradient fractionation of Zn2+-IMAC-bound proteins.
Supplemental Figure S4. Effects of binding buffer pH and ionic strength on protein binding to Fe3+-IMAC.
Supplemental Figure S5. Percentage coverage of IMAC protein sets by identified metal-binding motifs.
Supplemental Figure S6. Localization of metal-binding motifs and oxidized residues.
Supplemental Table S1. List of weak Cu2+-IMAC-binding proteins eluted at 0.1 m to 0.6 m NH4Cl.
Supplemental Table S2. List of strong Cu2+-IMAC-binding proteins eluted at 0.8 m to 1 m NH4Cl and by EDTA.
Supplemental Table S3. List of Co2+-IMAC-interacting proteins.
Supplemental Table S4. List of Zn2+-IMAC-interacting proteins.
Supplemental Table S5. Frequency of occurrence of metal-binding motifs.
Supplemental Table S6. Peptides identified from control and oxidized aconitase.
Supplemental Data S1. Animation showing oxidized peptides and putative Cu-binding sites on the rotating 3D structure of aconitase.
Supplementary Material
This work was supported by the Australian Research Council (ARC; grant no. CE0561495 to A.H.M.). Y.-F.T. was the recipient of an Australian Postgraduate Award, N.L.T. was supported as an ARC Australian Postdoctoral Fellow, and A.H.M. was supported as an ARC Australian Professorial Fellow.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: A. Harvey Millar (harvey.millar@uwa.edu.au).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53 1331–1341 [PubMed] [Google Scholar]
- Andreini C, Banci L, Bertini I, Rosato A (2008) Occurrence of copper proteins through the three domains of life: a bioinformatic approach. J Proteome Res 7 209–216 [DOI] [PubMed] [Google Scholar]
- Bartoli CG, Gomez F, Martinez DE, Guiamet JJ (2004) Mitochondria are the main target for oxidative damage in leaves of wheat (Triticum aestivum L.). J Exp Bot 55 1663–1669 [DOI] [PubMed] [Google Scholar]
- Baxter I, Muthukumar B, Park HC, Buchner P, Lahner B, Danku J, Zhao K, Lee J, Hawkesford MJ, Guerinot ML, et al (2008) Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genet 4 e1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligny R, Douce R (1977) Mitochondria of isolated plant cells (Acer pseudoplatanus L.). II. Copper deficiency effects on cytochrome c oxidase and oxygen uptake. Plant Physiol 60 675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia VM, Davies OR (2006) Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim Biophys Acta 1760 1304–1313 [DOI] [PubMed] [Google Scholar]
- Brazzolotto X, Gaillard J, Pantopoulos K, Hentze MW, Moulis JM (1999) Human cytoplasmic aconitase (iron regulatory protein 1) is converted into its [3Fe-4S] form by hydrogen peroxide in vitro but is not activated for iron-responsive element binding. J Biol Chem 274 21625–21630 [DOI] [PubMed] [Google Scholar]
- Bridgewater JD, Lim J, Vachet RW (2006. a) Using metal-catalyzed oxidation reactions and mass spectrometry to identify amino acid residues within 10 A of the metal in Cu-binding proteins. J Am Soc Mass Spectrom 17 1552–1559 [DOI] [PubMed] [Google Scholar]
- Bridgewater JD, Lim J, Vachet RW (2006. b) Transition metal-peptide binding studied by metal-catalyzed oxidation reactions and mass spectrometry. Anal Chem 78 2432–2438 [DOI] [PubMed] [Google Scholar]
- Brown AM, Kristal BS, Effron MS, Shestopalov AI, Ullucci PA, Sheu KF, Blass JP, Cooper AJ (2000) Zn2+ inhibits alpha-ketoglutarate-stimulated mitochondrial respiration and the isolated alpha-ketoglutarate dehydrogenase complex. J Biol Chem 275 13441–13447 [DOI] [PubMed] [Google Scholar]
- Cai Y, Moore M, Goforth R, Henry R, Beitle R (2004) Genomic data for alternate production strategies. I. Identification of major contaminating species for cobalt(+2) immobilized metal affinity chromatography. Biotechnol Bioeng 88 77–83 [DOI] [PubMed] [Google Scholar]
- Calderaro M, Martins EA, Meneghini R (1993) Oxidative stress by menadione affects cellular copper and iron homeostasis. Mol Cell Biochem 126 17–23 [DOI] [PubMed] [Google Scholar]
- Davies KJ, Lin SW (1988) Degradation of oxidatively denatured proteins in Escherichia coli. Free Radic Biol Med 5 215–223 [DOI] [PubMed] [Google Scholar]
- Diez TA, Wurtele ES, Nikolau BJ (1994) Purification and characterization of 3-methylcrotonyl-coenzyme-A carboxylase from leaves of Zea mays. Arch Biochem Biophys 310 64–75 [DOI] [PubMed] [Google Scholar]
- Dupuy J, Volbeda A, Carpentier P, Darnault C, Moulis JM, Fontecilla-Camps JC (2006) Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure 14 129–139 [DOI] [PubMed] [Google Scholar]
- Farber JM, Levine RL (1986) Sequence of a peptide susceptible to mixed-function oxidation: probable cation binding site in glutamine synthetase. J Biol Chem 261 4574–4578 [PubMed] [Google Scholar]
- Frey W, Schief WR Jr, Pack DW, Chen CT, Chilkoti A, Stayton P, Vogel V, Arnold FH (1996) Two-dimensional protein crystallization via metal-ion coordination by naturally occurring surface histidines. Proc Natl Acad Sci USA 93 4937–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol 93 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzhova NV, Novodarova GN, Kolosova EM, Vol'pin ME (1979) Effect of cobalt and copper complexes with o-phenanthroline on the respiratory activity of mitochondria. Biokhimiia 44 1369–1376 [PubMed] [Google Scholar]
- Hardin SC, Larue CT, Oh MH, Jain V, Huber SC (2009) Coupling oxidative signals to protein phosphorylation via methionine oxidation in Arabidopsis. Biochem J 422 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding MM (2004) The architecture of metal coordination groups in proteins. Acta Crystallogr D Biol Crystallogr 60 849–859 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini J, Verboom RE, Millar AH (2005) Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol 139 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss K, Junkes C, Guerreiro N, Swamy M, Camacho-Carvajal MM, Schamel WW, Haidl ID, Wild D, Weltzien HU, Thierse HJ (2005) Subproteomic analysis of metal-interacting proteins in human B cells. Proteomics 5 3614–3622 [DOI] [PubMed] [Google Scholar]
- Hollensworth SB, Shen C, Sim JE, Spitz DR, Wilson GL, LeDoux SP (2000) Glial cell type-specific responses to menadione-induced oxidative stress. Free Radic Biol Med 28 1161–1174 [DOI] [PubMed] [Google Scholar]
- Hunt JV, Simpson JA, Dean RT (1988) Hydroperoxide-mediated fragmentation of proteins. Biochem J 250 87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Harris NS, Deyholos MK (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58 3591–3607 [DOI] [PubMed] [Google Scholar]
- Jordanov J, Courtois-Verniquet F, Neuburger M, Douce R (1992) Structural investigations by extended x-ray absorption fine structure spectroscopy of the iron center of mitochondrial aconitase in higher plant cells. J Biol Chem 267 16775–16778 [PubMed] [Google Scholar]
- Kampfenkel K, Van Montagu M, Inzé D (1995) Effects of iron excess on Nicotiana plumbaginifolia plants (implications to oxidative stress). Plant Physiol 107 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesseler A, Brand MD (1994) Quantitative determination of the regulation of oxidative phosphorylation by cadmium in potato tuber mitochondria. Eur J Biochem 225 923–935 [DOI] [PubMed] [Google Scholar]
- Kindt R, Pahlich E, Rasched I (1980) Glutamate dehydrogenase from peas: isolation, quaternary structure, and influence of cations on activity. Eur J Biochem 112 533–540 [DOI] [PubMed] [Google Scholar]
- Kristensen BK, Askerlund P, Bykova NV, Egsgaard H, Moller IM (2004) Identification of oxidised proteins in the matrix of rice leaf mitochondria by immunoprecipitation and two-dimensional liquid chromatography-tandem mass spectrometry. Phytochemistry 65 1839–1851 [DOI] [PubMed] [Google Scholar]
- Kubo N, Arimura S, Tsutsumi N, Kadowaki K, Hirai M (2006) Isolation and characterization of the pea cytochrome c oxidase Vb gene. Genome 49 1481–1489 [DOI] [PubMed] [Google Scholar]
- Kung CC, Huang WN, Huang YC, Yeh KC (2006) Proteomic survey of copper-binding proteins in Arabidopsis roots by immobilized metal affinity chromatography and mass spectrometry. Proteomics 6 2746–2758 [DOI] [PubMed] [Google Scholar]
- Kuznetsova SS, Azarkina NV, Vygodina TV, Siletsky SA, Konstantinov AA (2005) Zinc ions as cytochrome c oxidase inhibitors: two sites of action. Biochemistry (Mosc) 70 128–136 [DOI] [PubMed] [Google Scholar]
- Lee CP, Eubel H, O'Toole N, Millar AH (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol Cell Proteomics 7 1297–1316 [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD (2006) The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440 363–367 [DOI] [PubMed] [Google Scholar]
- Levine RL, Mosoni L, Berlett BS, Stadtman ER (1996) Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA 93 15036–15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mowday B, Whelan J, Millar AH (2002) Zinc-dependent intermembrane space proteins stimulate import of carrier proteins into plant mitochondria. Plant J 30 555–566 [DOI] [PubMed] [Google Scholar]
- Luciano P, Geli V (1996) The mitochondrial processing peptidase: function and specificity. Experientia 52 1077–1082 [DOI] [PubMed] [Google Scholar]
- Luk E, Carroll M, Baker M, Culotta VC (2003) Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc Natl Acad Sci USA 100 10353–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk E, Yang M, Jensen LT, Bourbonnais Y, Culotta VC (2005) Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J Biol Chem 280 22715–22720 [DOI] [PubMed] [Google Scholar]
- Luk EE, Culotta VC (2001) Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p. J Biol Chem 276 47556–47562 [DOI] [PubMed] [Google Scholar]
- Macrae AR (1971) Isolation and properties of a ‘malic’ enzyme from cauliflower bud mitochondria. Biochem J 122 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae AR, Moorhouse R (1970) The oxidation of malate by mitochondria isolated from cauliflower buds. Eur J Biochem 16 96–102 [DOI] [PubMed] [Google Scholar]
- Marcillat O, Zhang Y, Lin SW, Davies KJ (1988) Mitochondria contain a proteolytic system which can recognize and degrade oxidatively-denatured proteins. Biochem J 254 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarini E, Cazzulo JJ (1975) On the role of divalent cations in the reaction mechanism of malic enzyme. Experientia 31 1126–1128 [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May MJ, Leaver CJ (1993) Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol 103 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA, Randall DD (1987) Some kinetic and regulatory properties of the pea mitochondrial pyruvate dehydrogenase complex. Plant Physiol 83 306–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Liddell A, Leaver CJ (2001) Isolation and subfractionation of mitochondria from plants. Methods Cell Biol 65 53–74 [DOI] [PubMed] [Google Scholar]
- Moberg P, Stahl A, Bhushan S, Wright SJ, Eriksson A, Bruce BD, Glaser E (2003) Characterization of a novel zinc metalloprotease involved in degrading targeting peptides in mitochondria and chloroplasts. Plant J 36 616–628 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay CK, Mazumder B, Lindley PF, Fox PL (1997) Identification of the prooxidant site of human ceruloplasmin: a model for oxidative damage by copper bound to protein surfaces. Proc Natl Acad Sci USA 94 11546–11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndimba BK, Chivasa S, Simon WJ, Slabas AR (2005) Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5 4185–4196 [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Donaldson RP (2005) Metal-catalyzed oxidation induces carbonylation of peroxisomal proteins and loss of enzymatic activities. Arch Biochem Biophys 439 25–31 [DOI] [PubMed] [Google Scholar]
- Nieboer E, Richardson DHS (1980) The replacement of the nondescript term ‘heavy metal’ by a biologically significant and chemically significant classification of metal ions. Environ Pollut B 1 3–26 [Google Scholar]
- Padua M, Aubert S, Casimiro A, Bligny R (1996) Arrest of mitochondrial biogenesis in copper-treated sycamore cells. FEBS Lett 398 248–252 [DOI] [PubMed] [Google Scholar]
- Padua M, Aubert S, Casimiro A, Bligny R, Millar AH, Day DA (1999) Induction of alternative oxidase by excess copper in sycamore cell suspensions. Plant Physiol Biochem 37 131–137 [Google Scholar]
- Palmer JM, Wedding RT (1966) Purification and properties of succinyl-CoA synthetase from Jerusalem artichoke mitochondria. Biochim Biophys Acta 113 167–174 [DOI] [PubMed] [Google Scholar]
- Parks JRE, Agarwal RP (1973) Nucleoside Diphosphokinases, Ed 3. Academic Press, New York
- Pascal N, Douce R (1993) Effect of iron deficiency on the respiration of sycamore (Acer pseudoplatanus L.) cells. Plant Physiol 103 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20 3551–3567 [DOI] [PubMed] [Google Scholar]
- Rauser WE (1999) Structure and function of metal chelators produced by plants: the case for organic acids, amino acids, phytin, and metallothioneins. Cell Biochem Biophys 31 19–48 [DOI] [PubMed] [Google Scholar]
- Rivett AJ (1985) Preferential degradation of the oxidatively modified form of glutamine synthetase by intracellular mammalian proteases. J Biol Chem 260 300–305 [PubMed] [Google Scholar]
- Sarry JE, Kuhn L, Ducruix C, Lafaye A, Junot C, Hugouvieux V, Jourdain A, Bastien O, Fievet JB, Vailhen D, et al (2006) The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 6 2180–2198 [DOI] [PubMed] [Google Scholar]
- Schutzendubel A, Polle A (2002) Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J Exp Bot 53 1351–1365 [PubMed] [Google Scholar]
- She Y, Narindrasorasak S, Yang S, Spitale N, Roberts EA, Sarkar B (2003) Identification of metal-binding proteins in human hepatoma lines by immobilized metal affinity chromatography and mass spectrometry. Mol Cell Proteomics 2 1306–1318 [DOI] [PubMed] [Google Scholar]
- Sheline CT, Choi DW (2004) Cu2+ toxicity inhibition of mitochondrial dehydrogenases in vitro and in vivo. Ann Neurol 55 645–653 [DOI] [PubMed] [Google Scholar]
- Skulachev VP, Chistyakov VV, Jasaitis AA, Smirnova EG (1967) Inhibition of the respiratory chain by zinc ions. Biochem Biophys Res Commun 26 1–6 [DOI] [PubMed] [Google Scholar]
- Smiri M, Chaoui A, El Ferjani E (2009) Respiratory metabolism in the embryonic axis of germinating pea seed exposed to cadmium. J Plant Physiol 166 259–269 [DOI] [PubMed] [Google Scholar]
- Smith SD, She Y, Roberts EA, Sarkar B (2004) Using immobilized metal affinity chromatography two-dimensional electrophoresis and mass spectrometry to identify hepatocellular proteins with copper-binding ability. J Proteome Res 3 834–840 [DOI] [PubMed] [Google Scholar]
- Stadtman ER (1990) Metal ion-catalyzed oxidation of proteins: biochemical mechanism and biological consequences. Free Radic Biol Med 9 315–325 [DOI] [PubMed] [Google Scholar]
- Stadtman ER (1993) Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu Rev Biochem 62 797–821 [DOI] [PubMed] [Google Scholar]
- Starke-Reed PE, Oliver CN (1989) Protein oxidation and proteolysis during aging and oxidative stress. Arch Biochem Biophys 275 559–567 [DOI] [PubMed] [Google Scholar]
- Stohs SJ, Bagchi D (1995) Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med 18 321–336 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32 891–904 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Day DA, Millar AH (2002) Environmental stress causes oxidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J Biol Chem 277 42663–42668 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH (2005) Differential impact of environmental stresses on the pea mitochondrial proteome. Mol Cell Proteomics 4 1122–1133 [DOI] [PubMed] [Google Scholar]
- Uchida K, Kawakishi S (1986) Selective oxidation of imidazole ring in histidine residues by the ascorbic acid-copper ion system. Biochem Biophys Res Commun 138 659–665 [DOI] [PubMed] [Google Scholar]
- Ueda EK, Gout PW, Morganti L (2003) Current and prospective applications of metal ion-protein binding. J Chromatogr A 988 1–23 [DOI] [PubMed] [Google Scholar]
- Verniquet F, Gaillard J, Neuburger M, Douce R (1991) Rapid inactivation of plant aconitase by hydrogen peroxide. Biochem J 276 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger AM, Millar AH, Day DA (2005) Sensitivity of plant mitochondrial terminal oxidases to the lipid peroxidation product 4-hydroxy-2-nonenal (HNE). Biochem J 387 865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger AM, Taylor NL, Heazlewood JL, Day DA, Millar AH (2007) The cytotoxic lipid peroxidation product 4-hydroxy-2-nonenal covalently modifies a selective range of proteins linked to respiratory function in plant mitochondria. J Biol Chem 282 37436–37447 [DOI] [PubMed] [Google Scholar]
- Yang M, Cobine PA, Molik S, Naranuntarat A, Lill R, Winge DR, Culotta VC (2006) The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J 25 1775–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Nickels R, McIntosh L (2001) A genome approach to mitochondrial-nuclear communication in Arabidopsis. Plant Physiol Biochem 39 345–353 [Google Scholar]
- Zachariou M, Hearn MT (1996) Application of immobilized metal ion chelate complexes as pseudocation exchange adsorbents for protein separation. Biochemistry 35 202–211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.