Abstract
The conifer Picea abies (Norway spruce) defends itself against herbivores and pathogens with a terpenoid-based oleoresin composed chiefly of monoterpenes (C10) and diterpenes (C20). An important group of enzymes in oleoresin biosynthesis are the short-chain isoprenyl diphosphate synthases that produce geranyl diphosphate (C10), farnesyl diphosphate (C15), and geranylgeranyl diphosphate (C20) as precursors of different terpenoid classes. We isolated a gene from P. abies via a homology-based polymerase chain reaction approach that encodes a short-chain isoprenyl diphosphate synthase making an unusual mixture of two products, geranyl diphosphate (C10) and geranylgeranyl diphosphate (C20). This bifunctionality was confirmed by expression in both prokaryotic (Escherichia coli) and eukaryotic (P. abies embryogenic tissue) hosts. Thus, this isoprenyl diphosphate synthase, designated PaIDS1, could contribute to the biosynthesis of both major terpene types in P. abies oleoresin. In saplings, PaIDS1 transcript was restricted to wood and bark, and transcript level increased dramatically after methyl jasmonate treatment, which induces the formation of new (traumatic) resin ducts. Polyclonal antibodies localized the PaIDS1 protein to the epithelial cells surrounding the traumatic resin ducts. PaIDS1 has a close phylogenetic relationship to single-product conifer geranyl diphosphate and geranylgeranyl diphosphate synthases. Its catalytic properties and reaction mechanism resemble those of conifer geranylgeranyl diphosphate synthases, except that significant quantities of the intermediate geranyl diphosphate are released. Using site-directed mutagenesis and chimeras of PaIDS1 with single-product geranyl diphosphate and geranylgeranyl diphosphate synthases, specific amino acid residues were identified that alter the relative composition of geranyl to geranylgeranyl diphosphate.
Conifers are frequently subject to attack by herbivorous insects and fungal pathogens (Phillips and Croteau, 1999; Trapp and Croteau, 2001; Franceschi et al., 2005; Keeling and Bohlmann, 2006a). However, the long life span and evolutionary persistence of these trees suggest that they possess effective defense strategies. The best known example of conifer chemical defense is oleoresin, a viscous mixture of terpenoids found in specialized ducts. Oleoresin may be both a constitutive and an inducible defense. For example, in Picea abies (Norway spruce), resin ducts are found constitutively in bark and foliage. However, this species also forms new (traumatic) resin ducts in the wood in response to attack by stem-boring insects and their associated fungi or after trees are sprayed with methyl jasmonate (MJ). Traumatic ducts are believed to help resist attack by augmenting the constitutive resin flow to provide a stronger physical and chemical barrier against herbivores and pathogens (Nagy et al., 2000; Martin et al., 2002; Hudgins et al., 2004; Franceschi et al., 2005; Byun-McKay et al., 2006; Keeling and Bohlmann, 2006a).
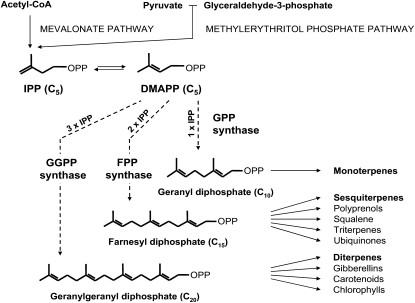
Terpenoids are the largest class of plant secondary metabolites, with more than 30,000 structural variants. Oleoresin consists mainly of monoterpenes (C10) and diterpene resin acids (C20) as well as smaller amounts of sesquiterpenes (C15; Langenheim, 2003). The biosynthesis of oleoresin, like all other terpenoids, begins with the synthesis of isopentenyl diphosphate (IPP) via the mevalonic acid pathway or the methylerythritol phosphate pathway (Gershenzon and Kreis, 1999; Fig. 1). IPP and its isomer, dimethylallyl diphosphate (DMAPP), are the five-carbon building blocks of terpenoids that undergo successive condensation reactions to form the larger intermediates geranyl diphosphate (GPP; C10), farnesyl diphosphate (FPP; C15), and geranylgeranyl diphosphate (GGPP; C20). These terpene diphosphate intermediates are in turn the precursors of monoterpenes, sesquiterpenes, and diterpenes, respectively, as well as many larger products (Fig. 1).
Figure 1.
Outline of terpenoid biosynthesis leading to the major conifer oleoresin components, monoterpenes and diterpenes, as well as to other classes of terpenes or compounds with terpene components. In the first phase of terpenoid biosynthesis, IPP and DMAPP are formed via the plastidial methylerythritol phosphate pathway and the cytosolic mevalonate pathway. The next phase consists of the reactions catalyzed by short-chain IDSs, GPP synthase, FPP synthase, and GGPP synthase. GPP synthase condenses one molecule of DMAPP and one molecule of IPP. FPP synthase condenses one molecule of DMAPP with two molecules of IPP in succession. GGPP synthase condenses one molecule of DMAPP with three molecules of IPP in succession. During these repeated condensations, the intermediate prenyl diphosphates are normally bound and not released by the enzymes. The PaIDS1 protein is believed to act like a GGPP synthase, but it releases a significant portion of the GPP formed as an intermediate. The remainder of the GPP is converted directly to GGPP without release of FPP. OPP indicates a diphosphate group.
The enzymes catalyzing the condensations of IPP and DMAPP to GPP, FPP, and GGPP are referred to collectively as short-chain isoprenyl diphosphate synthases (IDSs), members of a large enzyme class known as prenyltransferases (Kellogg and Poulter, 1997; Ogura and Koyama, 1998; Liang et al., 2002; Liang, 2009). IDSs have been frequently studied because they direct flux into different branches of terpenoid biosynthesis and so control product distribution. GPP, FPP, and GGPP are each formed by a specific, short-chain IDS: GPP synthase (EC 2.5.1.1) condenses DMAPP with one molecule of IPP; FPP synthase (EC 2.5.1.10) condenses DMAPP successively with two IPP molecules; and GGPP synthase (EC 2.5.1.30) condenses DMAPP successively with three IPP molecules (Gershenzon and Kreis, 1999; Fig. 1). Plant short-chain IDSs have been the subject of much research in recent years, but comparatively little attention has been paid to the enzymes in conifers (Hefner et al., 1998; Tholl et al., 2001; Burke and Croteau, 2002b; Martin et al., 2002; Schmidt et al., 2005; Schmidt and Gershenzon, 2007, 2008).
In addition to the short-chain IDSs, there are other types. Medium- and long-chain IDSs catalyze the formation of products with more than 20 carbon atoms, such as intermediates in ubiquinone, plastoquinone, dolichol, and rubber biosynthesis (Liang et al., 2002; Kharel and Koyama, 2003; Liang, 2009). IDS proteins can also be classified by the configuration of the double bond formed. The major short-chain prenyl diphosphate intermediates, such as GPP, FPP, and GGPP, all have (E)-double bonds, and most short-chain IDSs reported make products with (E)-double bonds. However, two enzymes making (Z)-products have recently been described (Sallaud et al., 2009; Schilmiller et al., 2009). IDSs producing (Z)-products show little sequence similarity to those making (E)-products (Wang and Ohnuma, 2000; Liang et al., 2002; Liang, 2009). The vast majority of (E)-product, short-chain IDSs have a homodimeric architecture, but the existence of heterodimeric GPP synthases has been well documented (Burke et al., 1999; Tholl et al., 2004). A recent study suggests that these enzymes may be widespread in the plant kingdom (Wang and Dixon, 2009).
Most of the short-chain, homodimeric IDSs described make only a single main product, usually GPP, FPP, or GGPP. However, products with one more or one fewer C5 unit than the main product are occasionally reported at low levels in in vitro assays. For example, FPP synthase from maize (Zea mays) produces up to 10% GGPP (Cervantes-Cervantes et al., 2006), and a GPP synthase from the orchid Phalaenopsis bellini produces nearly 20% FPP (Hsiao et al., 2008). However, it is not clear if these enzymes have such a broad product spectrum in vivo. And no example is yet known of such a short-chain IDS making two products that differ from each other in size by more than one C5 unit.
The amino acid sequences of short-chain plant IDSs making products with (E)-double bonds show significant amino acid similarity and contain two highly conserved regions with numerous Asp residues, designated the first and second Asp-rich motifs, respectively (Ashby et al., 1990; Fig. 2). Site-directed mutagenesis studies found that most of the Asp residues in these two highly conserved motifs are critical for substrate binding and catalysis (Koyama et al., 1995, 1996). The amino acid sequence features that determine the chain length of the product have been studied by random chemical mutagenesis and experiments on an avian FPP synthase, from which an x-ray structure has been obtained (Tarshis et al., 1994). Here, it was found that the fifth amino acid residue before the first Asp-rich motif is the key residue in determining product chain length. Replacement of the Phe at this position with smaller amino acids resulted in the product specificity shifting to GGPP or longer products. Conversely, replacement of a small amino acid with a larger one shortened the chain length of the product to GPP (Fernandez et al., 2000).
Figure 2.
Alignment of deduced amino acid sequences of PaIDS1 (accession no. GQ369788) and selected additional conifer IDSs, including PaIDS2 (EU432047), A. grandis GPP synthase1 (AgGPPS1; AAN01133), AgGPPS2 (AAN01134), AgGPPS3 (AAN01135), PaIDS5 (EU432050), PaIDS6 (EU432051), and AgGGPPS (AAL17614). Identical amino acids are boxed in black. The artificial translation initiation site for PaIDS1 is indicated by a triangle. The Asp-rich motifs conserved among IDSs are indicated by the black lines. The amino acid sequence used for the generation of peptide antibodies is marked by the dotted line. Dashes indicate sequence gaps introduced to optimize the alignment.
Here, we report the characterization of a new IDS from P. abies that possesses the unique ability to produce both GPP (C10) and GGPP (C20) but not FPP (C15). The product spectrum of this enzyme, its cellular location, and the expression pattern of the corresponding gene all suggest the involvement of this enzyme in oleoresin biosynthesis.
RESULTS
Isolation of a P. abies IDS cDNA Clone and Sequence Comparison
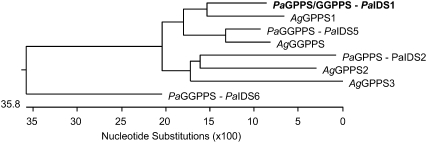
The isolation of IDS genes from P. abies was undertaken by PCR using primers designed from conserved regions of known plant GGPP synthases with P. abies RNA as the template. To find IDSs with a role in oleoresin formation, the fragments obtained were then employed to screen cDNA libraries constructed with mRNA isolated from saplings that had been treated with MJ to induce oleoresin accumulation. A full-length clone was obtained (PaIDS1) that was represented by four different sequences differing from each other only in the 3′ untranslated region, which consisted of 66 to 132 nucleotides between the stop codon and the poly(A)+ tail. The protein encoded by PaIDS1 consisted of 383 amino acids and had a calculated mass of 41.8 kD. The amino acid sequence had the highest similarity to other conifer GPP and GGPP synthase sequences (Table I), reaching 86% identity to the GPP synthase 1 of Abies grandis and 84% and 82% identity to GGPP synthases from P. abies (PaIDS5) and A. grandis, respectively (Burke and Croteau, 2002b; Schmidt and Gershenzon, 2007). Other conifer GPP synthase sequences, like PaIDS2 and PaIDS6 from P. abies and the GPP synthases 2 and 3 from A. grandis, showed 55% to 70% identity to PaIDS1 (Burke and Croteau, 2002b; Schmidt and Gershenzon, 2008). The two Asp-rich motifs, DDxxxxDxDDxRRG and DDxxD, found in other plant IDS proteins forming products with (E)-configurations are both conserved in the gene (Fig. 2). The TargetP 1.1 software (http://www.cbs.dtu.dk/services/TargetP; Emanuelsson et al., 2000) predicted a chloroplast signal peptide for PaIDS1 at the 5′ end of the cDNA (Fig. 3). When the PaIDS1 sequence was subjected to a phylogenetic analysis with the known conifer IDS genes showing GPP or GGPP synthase activity, it clustered closely together with the GPP synthase1 gene of A. grandis and associated at a greater distance with the GGPP synthase genes PaIDS5 and AgGGPP synthase (Fig. 3). The other GPP and GGPP synthase gene sequences isolated from P. abies and A. grandis clustered more distantly.
Table I.
Sequence relatedness of spruce and fir IDSs
| Sequence Relatedness | ||||||||
|---|---|---|---|---|---|---|---|---|
| PaIDS1 | 100 | |||||||
| PaIDS2 | 76.3 | 100 | ||||||
| AgGPPS1 | 86.1 | 74.1 | 100 | |||||
| AgGPPS2 | 74.6 | 76.3 | 74.1 | 100 | ||||
| AgGPPS3 | 71.6 | 72.2 | 71.4 | 75.0 | 100 | |||
| PaIDS5 | 83.6 | 75.4 | 83.2 | 75.8 | 74.4 | 100 | ||
| PaIDS6 | 69.3 | 63.2 | 66.5 | 63.6 | 63.1 | 67.0 | 100 | |
| AgGGPPS | 82.4 | 74.9 | 81.7 | 75.3 | 75.0 | 91.3 | 65.7 | 100 |
| PaIDS1 | PaIDS2 | AgGPPS1 | AgGPPS2 | AgGPPS3 | PaIDS5 | PaIDS6 | AgGGPPS | |
Figure 3.
Phylogenetic tree of the deduced amino acid sequences of PaIDS1 and other gymnosperm GPP synthase (GPPS) and GGPP synthase (GGPPS) sequences isolated from P. abies and A. grandis. The program DNA Lasergene (MegAlign) was used to create a neighbor-joining tree. The accession numbers of the sequences used are listed in Figure 2. Not all known conifer GPP synthase and GGPP synthase sequences were included.
Heterologous Expression in Escherichia coli and Characterization of PaIDS1
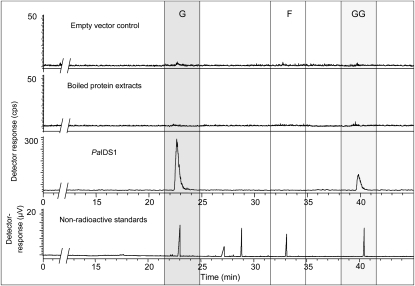
To study the enzymatic activity of the isolated clone, the protein encoded by PaIDS1 was expressed in E. coli after truncation of the signal sequence at the 5′ end of the coding region for directing chloroplast import. Analysis of crude bacterial extracts of the expressing cultures with SDS-PAGE revealed a recombinant protein band with an apparent molecular mass of approximately 40 kD. These extracts were subsequently purified on a nickel-nitrilotriacetic acid agarose column (Supplemental Fig. S1). When incubated with [1-14C]IPP and DMAPP as substrates, the purified recombinant PaIDS1 protein exhibited both GPP and GGPP synthase enzyme activity, whereas FPP was not detected (Fig. 4). The ratio of GPP to GGPP produced averaged approximately 9:1, taking into account the fact that 1 mol of GGPP (which is formed from three IPP units and one of DMAPP) should incorporate three times as much radioactivity from [1-14C]IPP as 1 mol of GPP (formed from one IPP and one DMAPP). Neither the empty vector controls nor heat-treated assays showed any significant enzyme activity.
Figure 4.
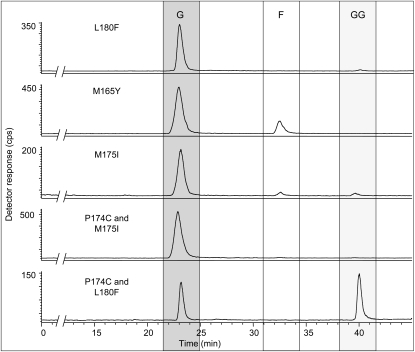
Catalytic activities of recombinant PaIDS1 protein heterologously expressed in E. coli and assayed with [1-14C]IPP and DMAPP. Reaction products were hydrolyzed enzymatically, and the resulting alcohols were analyzed by radio-gas chromatography. The main hydrolysis products detected were geraniol (G) and geranylgeraniol (GG), indicating that GPP and GGPP were the principal enzyme products. No release of FPP (detected as farnesol [F]) was measured (third panel from top). The ratio of GPP to GGPP was calculated as approximately 9:1, taking into account that 1 mol of GGPP incorporates three times as much radioactivity from IPP (three units) as GPP does (one IPP unit). The substrate IPP was hydrolyzed to some extent, but the resulting isopentenol eluted close to the solvent front and is not shown. Purified protein extracts of bacteria expressing the empty vector or boiled protein extracts of bacteria expressing PaIDS1 did not show any measurable activity (two top panels). Compounds were identified by coinjection of standards, as depicted in the thermal conductivity detector trace (bottom panel). Standards included geraniol (23 min), nerol (27 min), linalool (29 min), (E,E)-farnesol (33 min), and (E,E,E)-geranylgeraniol (40.5 min). At least five different replicates of each sample were analyzed, and the variance was below 5%.
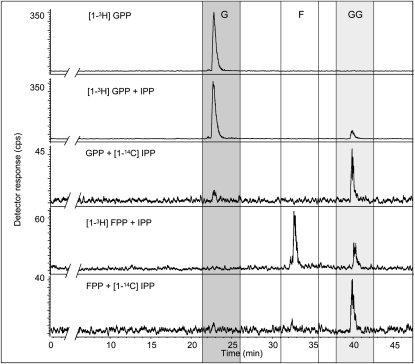
To explore the mechanism of GGPP (C20) formation, the shorter allylic diphosphates, GPP (C10) and FPP (C15), were offered as substrates. Like DMAPP (C5), both GPP and FPP were incorporated into GGPP in the presence of IPP (Fig. 5), suggesting that reaction proceeds by a sequential addition of IPP units as for other IDS proteins. The kinetic constants of the recombinant PaIDS1 (measured under linear conditions with respect to time and protein concentration) showed an apparent Km value of 390 μm for DMAPP, with the IPP concentration constant at 250 μm, and 170 μm for IPP, with the DMAPP concentration constant at 800 μm (for Lineweaver-Burk plots, see Supplemental Fig. S2). The Vmax was 4.2 nkat mg−1 and the kcat was 1.4 s−1. Activity required MgCl2 and reached its highest value at 10 mm MgCl2. At 5 or 15 mm MgCl2, activity was only 65% of wild-type PaIDS1, and no change of the product profile was observed (data not shown).
Figure 5.
Catalytic activities of recombinant PaIDS1 protein heterologously expressed in E. coli and assayed with combinations of radioactive and nonradioactive IPP, GPP, and FPP. Substrates and reaction products were hydrolyzed enzymatically, and the resulting alcohols were analyzed by radio-gas chromatography (for details of compound identification, analysis, and abbreviations, see Fig. 4). No formation of GGPP was found after using only [1-3H]GPP as a substrate (top panel). However, GPP or FPP together (radioactive or nonradioactive) with IPP were accepted as substrates for PaIDS1 and led to the formation of radioactively labeled GGPP (bottom panels), indicating that catalysis proceeds by sequential addition of IPP units. At least three biological replicates of each sample were analyzed, and the variance was below 5%.
PaIDS1 Expression in Transgenic P. abies Embryogenic Tissue
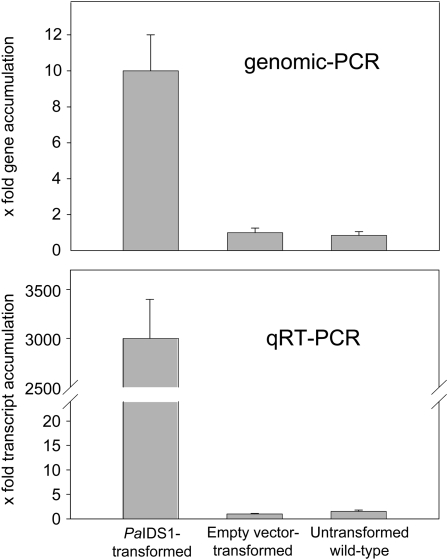
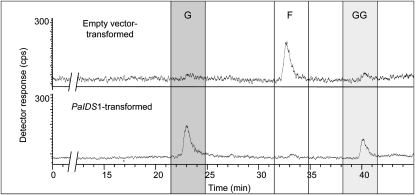
To confirm the surprising bifunctional GPP and GGPP synthase activity of PaIDS1 in an appropriate eukaryotic expression system, the full-length PaIDS1 gene was introduced into embryogenic tissue of P. abies under the control of the ubi1 promoter via Agrobacterium tumefaciens transformation. Four transgenic lines were obtained that tested positive for the presence of both the nptII and uidA genes (data not shown). The relative abundance of the PaIDS1 gene in pooled samples from these four lines was about 10-fold higher than in either the vector control or untransformed plants, and PaIDS1 mRNA transcript levels in embryogenic tissue were around 3,000-fold higher than in either the vector control or untransformed controls, indicating the success of the transformation (Fig. 6). Enzyme assays performed with crude protein extracts of PaIDS1-transformed embryogenic tissue exhibited both GPP and GGPP synthase enzyme activity when incubated with IPP and DMAPP as substrates, whereas only traces of FPP were observed, just as for the assay of PaIDS1 protein expressed in E. coli (Fig. 7). Crude protein extracts of empty vector control lines and untransformed embryogenic cultures of P. abies exhibited mainly FPP synthase enzyme activity, with only traces of GPP and GGPP.
Figure 6.
Relative abundance of PaIDS1 gene copies and mRNA transcripts in embryogenic tissue transformed with PaIDS1. Gene abundance was measured by quantitative genomic PCR (top), and transcript abundance of the IDS1 gene was measured by qRT-PCR (bottom), both of which employed SYBR Green for detection and the ubiquitin gene for normalization (for details, see text). Samples consisted of embryogenic tissue transformed with PaIDS1, tissue transformed with the empty vector, and wild-type tissue. Abundance of the vector control was set to 1.0. Each value is the average of four biological replicates, each of which is represented by at least four technical replicates.
Figure 7.
IDS activity of crude protein extract of embryogenic tissue transformed with a full-length construct of PaIDS1 and assayed in vitro with [1-14C]IPP and DMAPP. Reaction products were enzymatically hydrolyzed, and the resulting alcohols were analyzed by radio-gas chromatography. For details and abbreviations, see Figure 4. The main products of the PaIDS1 reaction were GPP (G) and GGPP (GG; bottom panel). Only traces of FPP (F) were detected. An extract of embryogenic tissue transformed with the empty vector control showed only FPP synthase activity (top panel). Compounds were identified by coinjection of standards, as depicted in the thermal conductivity detector trace (for details, see Fig. 4). At least four biological replicates of each sample were analyzed, and the variance was below 5%.
Comparison of PaIDS1 Transcript Levels among Organs and after MJ Treatment
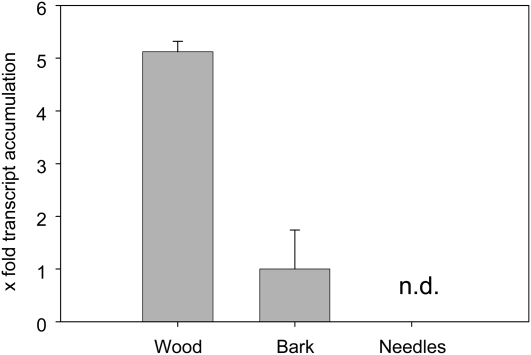
Terpene oleoresin is synthesized in ducts found in wood, bark, and needles. To determine whether PaIDS1 is localized in these organs, transcript levels were measured in P. abies saplings by quantitative real-time PCR (qRT-PCR). The highest transcript level was found in wood (Fig. 8). PaIDS1 transcripts in bark were only present at about 20% of the level of the wood, whereas no transcripts were detectable in needles.
Figure 8.
Relative abundance of PaIDS1 mRNA transcripts in different plant organs. Transcript abundance was measured by qRT-PCR using SYBR Green for detection and the ubiquitin gene for normalization (for details, see text). Each value is the average of three biological replicates, each of which is represented by three technical replicates. Values for wood and needles are expressed relative to bark, which was set to 1. n.d., Not detectable.
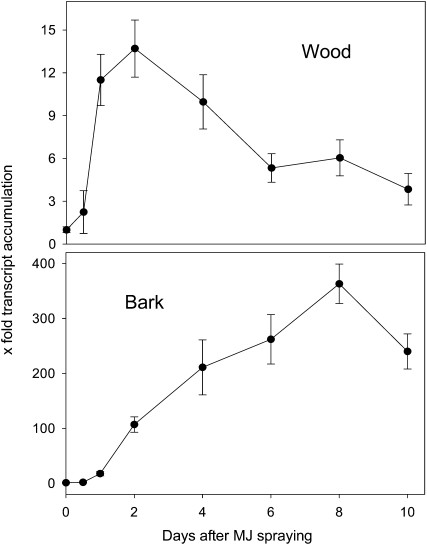
To determine whether the PaIDS1 enzyme had a role in induced oleoresin formation in P. abies stems, transcript levels were measured over a 10-d time course after saplings had been sprayed with MJ. In bark (and attached cambium), transcripts of the IDS1 gene increased continuously and reached a maximum at day 8, representing a 350-fold increase from the first time point (Fig. 9). In wood, the PaIDS1 genes also showed a transcript accumulation upon MJ spraying; however, the magnitude was less than in bark. Here, maximum induction, with a relative abundance of almost 14-fold relative to the first time point, occurred 2 d after MJ treatment (Fig. 9).
Figure 9.
Relative abundance of PaIDS1 mRNA transcripts in MJ-treated P. abies saplings. Transcript abundance of the IDS1 gene was measured by qRT-PCR using SYBR Green for detection and the ubiquitin gene for normalization (for details, see text). The time points measured were 0, 0.5, 1, 2, 4, 6, 8, and 10 d after the onset of treatment. Bark samples included the cambium. The time-zero measurement is the untreated control; its abundance was set to 1.0. Each value is the average of four biological replicates, each of which is represented by at least three technical replicates.
Localization of PaIDS1 Protein in Resin Duct Epithelial Cells
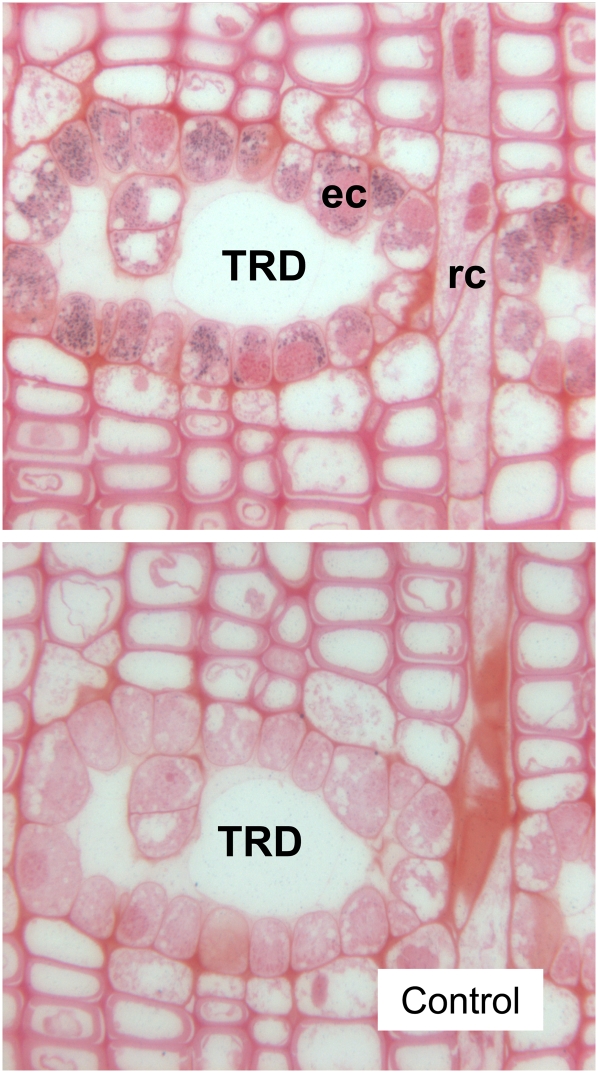
Antiserum raised against PaIDS1 gave a single strong band on western blots with extracts of E. coli heterologously expressing the recombinant protein, but it did not yield signals with extracts expressing PaIDS2 or PaIDS5, the most similar isolated IDS enzymes from P. abies (Schmidt and Gershenzon, 2007, 2008). The molecular mass of the detected band was approximately 42 kD on SDS-PAGE (data not shown), consistent with the predicted molecular mass of the PaIDS1 protein. In sections of P. abies stems treated with the fungus Ceratocystis polonica to induce oleoresin formation, immunogold labeling of the PaIDS1 antiserum appeared extensively in epithelial cells that line the inner surface of traumatic resin ducts (Fig. 10). These cells are thought to synthesize the oleoresin components and secrete them into the ducts. There was no reproducible immunogold labeling above the background signal in any other cell type or in sections treated with preimmune serum only. For comparison, antiserum raised against a P. abies FPP synthase (PaIDS4) that is not involved in induced oleoresin defense reactions (Schmidt and Gershenzon, 2007) was also tested. This did not show any signals, either in stem cells in C. polonica-treated tissue or in the untreated controls (data not shown).
Figure 10.
Immunogold labeling using a PaIDS1 polyclonal antibody applied to stem sections of P. abies that had been treated with the fungus C. polonica. Gold particles were observed only in epithelial cells (ec) lining the inner surface of traumatic resin ducts (TRD), which contain induced oleoresin, and not in the remaining xylem cells, including the ray channel (rc), a site of primary oleoresin secretion (top). The antibody serum did not generate any labeling above the background signal in tissue treated with preimmune serum only (bottom picture).
Alteration of PaIDS1 Product Specificity by Site-Directed Mutagenesis and the Creation of Chimeric IDS Proteins
To explore the contribution of individual amino acid residues to the unusual product spectrum of PaIDS1, site-directed mutagenesis was performed on selected positions in or around the regions previously shown to be important in the product length determination of IDS proteins (Ohnuma et al., 1996a, 1996b, 1997; Tarshis et al., 1996; Fernandez et al., 2000). In several cases, residues were changed to those found in previously characterized single-product GPP or GGPP synthases from conifers to see if this would make PaIDS1 a single-product enzyme.
The majority of the mutated proteins showed no difference in product composition from that of the wild-type enzyme (Table II). However, changes in residues at some positions within the first Asp-rich motif, known to be involved in substrate binding, did cause significant changes in the products of PaIDS1 catalysis. When a Met residue at position 175 (Fig. 11) was changed to Ile, as found in the sequence of the P. abies GPP synthase PaIDS2, the product distribution shifted toward more GPP. While the wild-type enzyme produced 90% GPP and 10% GGPP, the M175I mutant formed 95% GPP plus small amounts of both GGPP and FPP (Fig. 12). The immediately adjacent residue 174 is a Pro in the wild-type PaIDS1 and a Cys in PaIDS2. Mutation of Pro to Cys (P174C) caused no change in product composition, but when combined with the previously mentioned mutation (P174C and M175I), the product composition shifted completely to 100% GPP (Fig. 12).
Table II.
Effects of site-directed mutagenesis on the reaction rate and product composition of PaIDS1
| Mutation | Effect on Reaction Rate | Effect on Product Compositiona |
|---|---|---|
| M159I | Minor | None |
| I161M | Minor | None |
| M165C | Minor | None |
| M165Y | Minor | 90% GPP, 10% FPP |
| P173 and P174 deletion | Lower activityb | None |
| P174S | Minor | None |
| P174C | Minor | None |
| M175I | Minor | 95% GPP, 3% FPP, 2% GGPP |
| M175I and P174C | Minor | 100% GPP, 0% GGPP |
| M175I and P174S | Minor | None |
| L180F | Minor | 98% GPP, 2% GGPP |
| L180F and P174C | Minor | 65% GPP, 35% GGPP |
| V227M | Lower activityb | None |
| I235T | Lower activityb | 100% GPP, 0% GGPP |
| V240L | Lower activityb | None |
| G257D and P174C | Minor | None |
| L273F | Lower activityb | 100% GPP, 0% GGPP |
Wild-type PaIDS1 produces approximately 90% GPP and 10% GGPP upon heterologous expression.
About 20% of the activity of wild-type PaIDS1.
Figure 11.
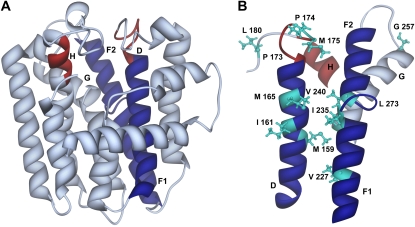
Three-dimensional ribbon model of the complete structure of wild-type PaIDS1 (A) and the substrate-binding region (B) constructed with reference to the avian FPP synthase crystal structure (Tarshis et al., 1994). Depicted are the α-helices D (amino acids 154–181), F1 + F2 + G (amino acids 222–277), and H (amino acids 309–313). Helices that are part of the predicted substrate-binding pocket are colored in blue (helices D + F), and the DDxxD motifs (amino acids 170–176 and 309–313) are indicated in red. Residues used for mutagenesis that are visible in this diagram are shown in turquoise. The model was displayed with the program UCSF Chimera (National Centre for Research Resources).
Figure 12.
Catalytic activities of mutated recombinant PaIDS1 proteins heterologously expressed in E. coli and assayed with [1-14C]IPP and DMAPP. Reaction products were enzymatically hydrolyzed, and the resulting alcohols were analyzed by radio-gas chromatography. For details of compound identification, analysis, and abbreviations, see Figure 4. At least three replicate assays of each mutant were analyzed, and the variance was below 5%.
Previous studies on IDS enzymes had shown that the fifth amino acid prior to the first Asp-rich motif (position 165 in PaIDS1) is particularly critical in determining product size (Fig. 11). Larger amino acids block extension of the chain, leading to a decrease in product length. Consistent with this trend, mutation of residue 165 from Met to the bulkier Tyr eliminated all GGPP formation, triggering the formation of 10% FPP with 90% GPP (Fig. 12). When the Met at 165 was mutated to the smaller Cys instead, there was no effect on product distribution. Two other sequence changes based on differences between PaIDS1 and other P. abies or A. grandis IDS, I235T and L273F, completely eliminated the formation of the longer GGPP, leaving only GPP as the product.
Mutations to residues found in single-product GGPP synthases of the conifers P. abies and A. grandis gave mixed results. When Leu at position 180 was changed to Phe, there was a significant increase in GPP and a decrease in GGPP. However, the double mutant, L180F and P174C, gave a significant increase in GGPP from 10% to 35% at the expense of GPP (Fig. 12).
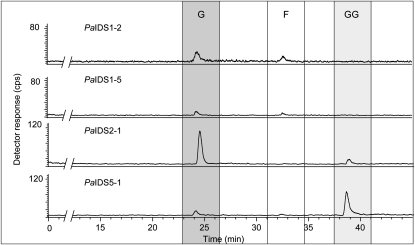
As another approach to investigating the structure-function relationships of PaIDS1, we created chimeric proteins assembled from PaIDS1 and two other P. abies IDS enzymes, the GPP synthase, PaIDS2, and the GGPP synthase, PaIDS5. Dividing the proteins between the two Asp-rich motifs at residues 213 to 216 (Fig. 2), we produced chimeras in which either the N-terminal or C-terminal portion of PaIDS1 was exchanged for the corresponding portion of PaIDS2 or PaIDS5. The proteins in which the PaIDS1 N-terminal sequence had been exchanged, PaIDS2-1 and PaIDS5-1, had product spectra according to the origin of their N-terminal sequences. PaIDS2-1, which contained the N-terminal sequence of PaIDS2 (a pure GPP synthase) and the C-terminal sequence of PaIDS1, produced 95% GPP and 5% GGPP (Fig. 13). Meanwhile, PaIDS5-1, which contained the N-terminal sequence of PaIDS5 (a pure GGPP synthase) and the C-terminal sequence of PaIDS1, produced 40% GPP and 60% GGPP. However, the PaIDS1-2 and PaIDS1-5 proteins, which contained the N-terminal sequence of PaIDS1 and the C-terminal sequence of either PaIDS2 or PaIDS1-5, did not produce any GGPP, but only 80% GPP and 20% FPP (Fig. 13). All of the chimeric proteins had only 10% to 15% of the activity of wild-type PaIDS1.
Figure 13.
Catalytic activities of chimeric recombinant PaIDS1 proteins heterologously expressed in E. coli. Either the N-terminal or C-terminal half of PaIDS1 was exchanged for the corresponding portion of the known GPP synthase, PaIDS2, or the known GGPP synthase, PaIDS5, and the resulting proteins were assayed with [1-14C]IPP and DMAPP. Reaction products were enzymatically hydrolyzed, and the resulting alcohols were analyzed by radio-gas chromatography. For details of compound identification, analysis, and abbreviations, see Figure 4. At least three replicate assays of each mutant were analyzed, and the variance was below 5%.
DISCUSSION
PaIDS1 Is an Unusual Short-Chain IDS That Produces Both GPP and GGPP
The short-chain IDSs combine the basic C5 units of the terpenoid pathway into C10 (GPP), C15 (FPP), and C20 (GGPP) intermediates and so control the levels of precursors for different classes of terpenes (Fig. 1). We have been investigating the genes encoding the short-chain IDSs of P. abies to learn more about the control of terpene oleoresin formation in conifers, which is a mixture of monoterpenes (C10) and diterpenes (C20). In previous investigations, we have isolated IDSs with GPP synthase, FPP synthase, and GGPP synthase activities (Schmidt and Gershenzon, 2007, 2008). Here, we report an unusual IDS (PaIDS1) that produces both GPP and GGPP in substantial amounts, but no FPP. This novel activity was first demonstrated by heterologous expression in E. coli (Fig. 4) and then confirmed by expression in embryonic tissue of P. abies that was transformed via A. tumefaciens with the PaIDS1 gene under the control of a constitutive promoter (Figs. 6 and 7). To our knowledge, this is the first report of A. tumefaciens-mediated gene transfer in conifers involving the overexpression of a characterized endogenous gene. Previously, the overexpression of an endogenous cinnamoyl alcohol dehydrogenase gene was achieved in Pinus radiata by particle bombardment, but this transformation caused cosuppression, resulting in lower activity of the encoded enzyme (Möller et al., 2003). In another study, even though overexpression of an endogenous peroxidase-like gene in P. abies showed a higher level of total peroxidase activity, no correlation to lignin polymer composition was found (Elfstrand et al., 2001). The ratio of GPP to GGPP formed by PaIDS1 was approximately 9:1 (Fig. 4).
PaIDS1 is, to our knowledge, the first short-chain, homodimeric IDS described that makes two major products with such a large difference in size. Reports on other bifunctional IDSs indicate that enzymes making predominantly a single product in in vitro assays can also catalyze the formation of lower amounts of products with one additional or one fewer C5 unit. For example, there are GPP synthases that also make low amounts of FPP (Burke and Croteau, 2002b; Hsiao et al., 2008), FPP synthases that also make GGPP (Cervantes-Cervantes et al., 2006), and GGPP synthases that produce minor amounts of FPP (Takaya et al., 2003). Interestingly, bifunctionality is not limited to plants. For example, Vandermoten et al. (2008) described a GPP synthase from Myzus persicae producing significant amounts of FPP. Other dual FPP/GGPP synthases have been reported for a protozoan and a hyperthermophilic archaeon (Chen and Poulter, 1994; Ling et al., 2007). However, there are no other reports of a homodimeric, short-chain IDS that makes multiple products differing by 10 or more carbon atoms.
Bifunctional activities observed for short-chain IDSs in general may be examples of “catalytic promiscuity,” an expression that was coined to describe the ability of some enzymes to display an adventitious secondary activity at the active site responsible for the primary activity (Copley, 2003). If this adventitious secondary activity becomes useful to the organism at some point, the enzyme may be recruited to provide more of the secondary product (Vandermoten et al., 2009). The existence of a bifunctional protein, such as PaIDS1, that produces precursors for both monoterpene and diterpene formation is not surprising, as monoterpenes and diterpenes occur together as principal constituents of oleoresin.
PaIDS1 Has the Properties of a GGPP Synthase That Releases a Substantial Portion of the Intermediate GPP
The enzymological properties of PaIDS1 are generally similar to those of single-product GPP and GGPP synthases characterized from P. abies and other conifers. All of these enzymes share a requirement for the same divalent metal ion, Mg2+. While the Km values of PaIDS1 for the substrates IPP and DMAPP are somewhat greater than those determined for other short-chain conifer IDSs, the ratio between the Km for DMAPP and that for IPP is approximately the same. The substrate specificity of PaIDS1, its ability to use other allylic diphosphates (Fig. 5), also parallels that of GGPP synthases from conifers and other species (Hefner et al., 1998; Ogura and Koyama, 1998; Takaya et al., 2003), suggesting that reaction also involves successive condensation of IPP units (Fig. 1). These successive condensations usually proceed without significant release of the intermediates GPP and FPP. PaIDS1, therefore, may be simply a GGPP synthase in which a large portion of the intermediate GPP is released, while the remainder continues along the reaction path toward GGPP. Unlike GPP, the intermediate FPP is not released.
To assess what structural features of PaIDS1 might be responsible for the simultaneous formation of GPP and GGPP, a program of site-directed mutagenesis was undertaken. This effort relied on modeling of PaIDS1 with reference to an avian FPP synthase (Tarshis et al., 1994), the only short-chain IDS for which a crystal structure is available. The sequence of this avian FPP synthase shares 30% amino acid identity to PaIDS1. Its structure is composed of 13 α-helices, of which 10 form a large central cavity (Fig. 11). The active site is located in the central cavity with two conserved Asp-rich motifs facing each other on opposite walls. These motifs have been demonstrated to be involved in substrate binding and catalysis via coordination with Mg2+, hence the requirement of this avian FPP synthase and other IDSs for this cofactor (Marrero et al., 1992; Joly and Edwards, 1993; Song and Poulter, 1994; Koyama et al., 1994, 1995, 1996). In PaIDS1, the first Asp-rich region spans amino acid residues 170 to 183 and is located in helix D and an adjacent loop, while the second region spans residues 309 to 313 and is located in helix H. These two helices, as well as helix F1, form a significant portion of the active site (depicted in Fig. 11).
In site-directed mutagenesis, selected residues of PaIDS1 were changed to those present in other conifer IDSs that form either GPP or GGPP as a single product. A double mutant at P174C and M175I that changed these residues to those present in single-product GPP synthases (Burke and Croteau, 2002b; Schmidt and Gershenzon, 2008) could catalyze only the formation of GPP and no GGPP. The residues at positions 174 and 175 are thought to have been inserted in the IDS sequence during evolution from bacteria to plants (Wang and Ohnuma, 1999) and are clearly important in determining product specificity. Mutation of the fifth amino acid prior to the first Asp-rich motif (M165Y) also caused the production of shorter products. This residue, which projects into the active site cavity (Fig. 11), has been shown to influence product length in other short-chain IDSs, with large, bulky R groups appearing to block additional condensation with IPP and thus to give shorter chain products (Ohnuma et al., 1996b; Fernandez et al., 2000; Soderberg et al., 2001). Here, mutagenesis from Met to Tyr gave an increased proportion of shorter chain products, while mutagenesis of Met to Cys had no effect on product distribution. Interestingly, there were two other residues where mutation (I235T and L273F) increased the proportion of the short-chain product GPP to 100%. These project into the central cavity from helix F, immediately opposite Met-165, and so may also prevent additional condensations by steric effects.
Mutagenesis of PaIDS1 sequence sites to those present in sequences of similar, single-product GGPP synthases was largely unsuccessful at altering product composition. The one exception was the double mutant, P174C and L180F, which gave a 65% GPP, 35% GGPP distribution compared with 90% GPP, 10% GGPP in the wild-type enzyme. Residues 174 and 180 are located in the first Asp-rich region in the loop attached to helix D and so are likely to be part of the active site. However, it is not obvious how modification at these positions can result in a greater percentage of additional IPP condensations to give more GGPP and less GPP.
The residues in and around the first Asp-rich region (residues 170–183) of PaIDS1 are clearly important in chain length determination. This conclusion is supported by experiments involving chimeras formed by exchanging the N- or C-terminal portion of PaIDS1 with corresponding portions of similar single-product GPP or GGPP synthases. When the N terminus (including the first Asp-rich region) was replaced by the N terminus of a single-product P. abies GPP synthase or a single-product GGPP synthase, the product distribution shifted accordingly to either GPP or GGPP (Fig. 13). Replacement of the C terminus in the same way did not have such an effect, but the fact that mutagenesis of residues 235 and 273 also affected the length of the product suggests that control of chain length resides at many places in the protein, not all of which are clearly known. Other open questions about PaIDS1 concern why a significant proportion of GPP, the C10 intermediate formed by the enzyme, is released but the next larger intermediate, FPP, remains bound. It is conceivable that as the product elongates from C10 to C15 in the active site, it twists in the cavity in a way that prevents release until it reaches C20 and cannot elongate further. The residues responsible for preventing release might be localized in the C-terminal domain of PaIDS1, because site-directed mutagenesis at positions 235 and 273 and chimeras with replacements of the C-terminal portion of PaIDS1 (residues 213–383) produced FPP as their final product instead of GGPP, albeit with a much lower activity. More work is needed to understand how PaIDS1 creates its unique bifunctional product spectrum.
PaIDS1 Transcript and Protein Are Associated with Oleoresin Biosynthesis
The ability of PaIDS1 to produce both GPP and GGPP suggests that this enzyme could have a biological role in both monoterpene and diterpene production. Since the major components of conifer oleoresin are monoterpenes and diterpenes, we looked for a connection between the occurrence of PaIDS1 in P. abies and oleoresin biosynthesis. The formation of oleoresin in this species can be induced by treatment with MJ, which mimics attack by bark beetles or pathogenic fungi (Franceschi et al., 2002; Martin et al., 2002; Hudgins et al., 2003; Byun-McKay et al., 2006; Erbilgin et al., 2006; Zeneli et al., 2006). MJ treatment increases the accumulation of resin monoterpenes and diterpenes and stimulates the formation of traumatic resin ducts in stems in the newly formed wood (xylem) just inside the vascular cambium. In this work, MJ treatment also dramatically increased the transcript abundance of PaIDS1 in wood and bark, compared with untreated controls, indicating that the encoded protein is involved in oleoresin formation. In untreated tissue, PaIDS1 transcript is found in wood and bark, but not needles, consistent with a role in resin formation only in traumatic ducts. Although traumatic resin ducts are located in wood, their development is initiated in cells that are still part of the vascular cambium (Martin et al., 2002; Franceschi et al., 2005). Since the cambium layer is usually included with the bark during sampling, it is not surprising to observe expression of a gene involved in traumatic resin duct formation in both the bark and wood.
In previous studies on P. abies short-chain IDSs, we have demonstrated that a single-product GPP synthase (PaIDS2) and a single-product GGPP synthase (PaIDS5) are also associated with oleoresin biosynthesis in stems (Schmidt and Gershenzon, 2007, 2008). While these single-product enzymes could suffice for the formation of the major resin terpenes, it is interesting that PaIDS1 transcript is induced much more significantly than those of PaIDS2 or PaIDS5 after MJ treatment and thus may have an especially important function in induced resin formation. Additional evidence for the role of PaIDS1 in induced oleoresin formation comes from our studies that localized this protein to epithelial cells lining the traumatic resin ducts in stems treated with the fungus C. polonica. Epithelial cells are thought to synthesize the resin and secrete it into the lumen of the ducts (Bannan, 1936). Antibodies to PaIDS1 did not react with antigens in any other type of cell in the stem.
At the subcellular level, PaIDS1 appears to be localized in the plastid by virtue of its predicted chloroplast signal peptide. A plastidial location is also consistent with current concepts on the intracellular compartmentalization of GPP- and GGPP-producing or -utilizing enzymes in plant cells (Parmryd et al., 1999; Welsch et al., 2000; Bouvier et al., 2002, 2005; Bick and Lange, 2003; Martin et al., 2004; Tholl et al., 2004; Keeling and Bohlmann, 2006b; Ro and Bohlmann, 2006).
PaIDS1 Has Strong Sequence Identity to Other Conifer GPP and GGPP Synthases and Is Likely to Have Evolved from a Single-Product Enzyme
A phylogenetic analysis of gene sequences for characterized plant short-chain IDSs shows that PaIDS1 clusters closely with most previously reported, single-product GPP and GGPP synthase genes from conifers. However, PaIDS1 shares no significant sequence similarity to a gene encoding one conifer GPP synthase that clusters most closely with several angiosperm GPP synthases thought to function in gibberellin biosynthesis (Bouvier et al., 2002; Van Schie et al., 2007; Schmidt and Gershenzon, 2008). It also possesses no significant similarity to the family of heterodimeric GPP synthases known from angiosperms (Burke and Croteau, 2002a; Tholl et al., 2004; Wang and Dixon, 2009).
Given its demonstrated ability to produce both GPP and GGPP, it is not surprising that PaIDS1 has highest sequence similarity to single-product GPP and GGPP synthases from conifers. Its kinetic properties and reaction mechanism also do not distinguish it from other members of this group. PaIDS1 may be considered a GGPP synthase that has the ability to release a substantial portion of the intermediate GPP, giving it a novel product spectrum. Our site-directed mutagenesis experiments showing facile shifts of GPP and GGPP after modification of one or two residues suggest that the evolutionary conversion from single-product GPP and GGPP synthases to an enzyme like PaIDS1 may require only a few steps. Having a single short-chain IDS that makes both the GPP and GGPP precursors for conifer resin in a fixed proportion may have been selected to allow better control of the ratio between monoterpenes and diterpenes and to facilitate increased synthesis of both products simultaneously. Other plant species that produce mixtures of terpene natural products may have similar dual-function short-chain IDSs.
MATERIALS AND METHODS
Chemicals
All chemicals and solvents were analytical grade and were obtained from Merck, Serva, or Sigma. [1-14C]IPP (55 Ci mol−1) was purchased from Biotrend, [1-3H]GPP (15 Ci mmol−1), [1-3H]FPP (20 Ci mmol−1), and [α-32P]dATP were obtained from Hartmann Analytic, and unlabeled DMAPP and IPP were obtained from Echelon Research.
Plant Material
The clonal 4-year-old Picea abies (Norway spruce) saplings used (Samenklenge und Pflanzgarten Laufen) are members of clone 3369-Schongau. Saplings were grown in standard soil in a climate chamber (Vötsch) under a 21°C-day/16°C-night temperature cycle, controlled light conditions (16 h per day at 150–250 μmol, obtained from a mixture of cool-white fluorescent and incandescent lamps), and a relative humidity of 70% in a climate chamber (Vötsch). For induction experiments, saplings were sprayed with 100 μm MJ in 0.5% (v/v) Tween 20 as detergent. When stem tissue was harvested, the uppermost internode (representing the previous year's growth) was used. Bark and wood from this zone were separated and frozen in liquid nitrogen in preparation for RNA isolation. The bark included the cambium layer that peeled off from the wood during separation.
The embryogenic culture of P. abies (line 186/3c VIII; kindly provided by Harald Kvaalen, Norwegian Forest and Landscape Institute) was initiated from mature zygotic embryos, dissected from seeds, and cultured on modified Litvay's (MLV) medium (Klimaszewska et al., 2001). Casein amino acids (Becton Dickinson) were added at 1 g L−1, l-Gln (Duchefa) at 0.5 g L−1, Suc at 10 g L−1, and Gelrite (Duchefa) at 4 g L−1 when a semisolid medium was required. The medium was supplemented with 10 μm 2,4-dichlorophenoxyacetic acid and 5.0 μm 6-benzyladenine. The pH of the medium was adjusted with KOH to 5.7 prior to sterilization in the autoclave. The pH of the l-Gln solution was adjusted to 5.7, and the solution was filter sterilized and added to the medium after sterilization. Unless stated otherwise, all cultures were kept in the dark at 24°C.
Amplification and Cloning of the PaIDS1 Sequence
Degenerate primers were designed for conserved regions of GGPP synthase sequences from Taxus canadensis, Helianthus annuus, Croton sublyratus, and Lupinus albus for different conserved regions: 5′-CIATG(A/C)G(G/T)TA(C/T)TCICTICTIGCIGG-3′ and 5′-CTTATCIG(C/T)(A/C)AICAAATC(C/T)TT(C/T)CC-3′. RT was carried out with pooled total RNA from MJ-treated P. abies bark harvested at various time points after treatment (6, 12, and 24 h and 2, 4, 6, 10, and 20 d) and oligo-(dT) primers using SuperScript II (Invitrogen), according to the manufacturer's instructions. Subsequently, PCR was performed with 35 cycles of 94°C for 1 min, annealing for 1 min, and then 72°C for 1 min in a thermocycler (Stratagene). To identify the optimal annealing temperature, a temperature gradient was used with 2°C intervals ranging from 42°C to 64°C. A 430-bp fragment was generated at 44°C and subcloned into pCR 4-TOPO (Invitrogen) according to the manufacturer's instructions.
Isolation of the PaIDS1 Clone
Total RNA was isolated according to a method developed by Wang et al. (2000) from P. abies bark harvested at various time points (as above) after being sprayed with 100 μm MJ. From total pooled RNA, poly(A)+ RNA was isolated using Dynabeads (Invitrogen). cDNA was synthesized using the SMART cDNA Library Construction Kit (Clontech). In vitro packaging with Gigapack III Gold (Stratagene) yielded a library of 8 × 107 plaque-forming units, which was subsequently screened with the [α-32P]dATP-labeled 430-bp fragment previously amplified by PCR. Single plaques were isolated from positive colonies, and λDNA was converted into pDNA by subcloning into pTriplEx2 vector and transformation in Escherichia coli BM25.8. Sequence analysis was carried out using an ABI 3100 automatic sequencer (Applied Biosystems). One sequence was obtained that was named PaIDS1.
Sequence and Phylogenetic Analyses
The DNAStar Lasergene program version 7.0 (MegAlign) was used to align and to calculate the deduced amino acid sequences of each full-length cDNA or of known sequences from other gymnosperms and angiosperms. The amino acid alignment was conducted by use of ClustalW (gonnet 250 matrix, gap penalty 10.00, gap length penalty 0.20, delay divergent sequences 30%, gap length 0.10, DNA transition weight 0.5). The same software was used to visualize the phylogenetic tree.
Functional Expression of PaIDS1
The entire coding sequence of clone PaIDS1 lacking the predicted signal peptide for chloroplast transport (based on analysis at http://www.cbs.dtu.dk/services) was amplified with primers that included the start and stop codons for translation. The primer combination 5′-ATGTGCTCAAACACAAATGCCCAG-3′ and 5′-GTTCTGTCTTTGTGCAATGTAATG-3′ was used to express PaIDS1. The amplification was carried out with the Expand High Fidelity PCR System (Roche Applied Science). PCR was performed for 2 min at 94°C initial denaturation, 10 cycles of 15 s at 94°C, 30 s at 55°C, and 55 s at 72°C, 20 cycles of 15 s at 94°C, 30 s at 55°C, and 55 s plus 5 s of elongation for each cycle at 72°C, and a final extension for 7 min at 72°C in a Tpersonal thermocycler (Biometra). The resulting cDNA fragments were cloned into the expression vector pCR-T7 CT TOPO (Invitrogen), which adds a tag containing six His residues to the C terminus. Positive clones were first transferred into E. coli strain TOP10F′ (Invitrogen) and then into strain BL21 (DE3)pLysS (Invitrogen). Bacterial cultures of 100 mL were grown to an optical density (OD600nm) of 0.6, and transformants were induced with 2 mm isopropylthio-β-galactoside as described by the manufacturer's instructions, except that they were grown exclusively at 18°C. Bacterial pellets were resuspended in 3 mL of buffer containing 20 mm 3-(N-morpholino)-2-hydroxypropanesulfonic acid (MOPSO), pH 7.0, 10% (v/v) glycerol, and 10 mm MgCl2 and sonicated using a Sonopuls HD 2070 (Bandelin) for 4 min, cycle 2, power 60%. The PaIDS1 protein was purified over nickel-nitrilotriacetic acid agarose columns (Qiagen) according to the manufacturer's instructions. Protein was eluted with 20 mL of 250 mm imidazole in the assay buffer, and 1.5-mL fractions were collected. After adding 2 mm dithiothreitol, each fraction was checked for purity by SDS-PAGE. Fraction 2, which contained the highest amount of recombinant protein, was used for the assay. The added 250 mm imidazole was not removed before the assay, as it did not affect the activity.
Assay of Recombinant PaIDS1 and Product Identification
Small-scale PaIDS1 assays were carried out in a final volume of 50 μL containing 20 mm MOPSO (pH 7.0), 10 mm MgCl2 (for PaIDS5), 10% (v/v) glycerol, 2 mm dithiothreitol, and 800 μm IPP or 250 μm DMAPP at saturated concentrations. For radioactive detection of products, 10 μm [1-14C]IPP (55 Ci mol−1) was added to each assay. The reaction was initiated by the addition of recombinant protein, and the assay mixture was overlaid with 1 mL of hexane and incubated for 20 min at 30°C. Assays were stopped by the addition of 10 μL of 3 n HCl and incubated for an additional 20 min at 30°C to solvolyze the acid-labile allylic diphosphates formed during the assay to their corresponding alcohols. Solvolysis products were extracted into the hexane phase by vigorous mixing for 15 s. After centrifugation, 300 μL of the hexane phase was used for total radioactivity determination by liquid scintillation counting. Protein concentration was measured according to Bradford (1976) using the Bio-Rad reagent with bovine serum albumin (BSA) as standard. The reaction was linear for up to 30 min at protein concentrations of 40 μg mL−1. Kinetic parameters were calculated with Michaelis-Menten kinetics and Lineweaver-Burk plotting using the enzyme kinetics program EkI 3 (Wiley-VCH). All assays were carried out in triplicate.
For the identification of reaction products of PaIDS1, larger scale assays were carried out in a final volume of 500 μL. The assay mixture was as described above, but concentrations of 40 μm [1-14C]IPP, 40 μm DMAPP, and a 1-mL layer of pentane were used. Assays were incubated overnight at 30°C. To stop the assay and hydrolyze all diphosphate esters, a 1-mL solution of 2 units of calf intestine alkaline phosphatase (Sigma) and 2 units of potato apyrase (Sigma) in 0.2 m Tris-HCl, pH 9.5, was added to each assay and followed by overnight incubation at 30°C. After enzymatic hydrolysis, the resulting isoprenyl alcohols were extracted into 2 mL of diethyl ether. Then, following the addition of a standard terpene mixture, the organic extracts were evaporated under N2 and used for radio-gas chromatography (GC) measurements. Radio-GC analysis was performed on a Hewlett-Packard HP 6890 gas chromatograph (injector at 220°C, thermal conductivity detector at 250°C) in combination with a Raga radioactivity detector (Raytest) using a DB 5-MS capillary column (J&W Scientific; 30 m × 0.25 mm with a 0.25-μm phase coating). Separation of the injected concentrated organic phase (1 μL) was achieved under an H2 flow rate of 2 mL min−1 with a temperature program of 3 min at 70°C, followed by a gradient from 70°C to 240°C at 6°C min−1 with a 3-min hold at 240°C. Products measured by radio-GC were identified by comparison of retention times with those of coinjected, nonradioactive, authentic terpene standards monitored via a thermal conductivity detector.
To study the reaction mechanism, substrates were used as follows in the assays depicted in Figure 5: 600 μm [1-3H]GPP (15 Ci mmol−1); 400 μm [1-3H]GPP (15 Ci mmol−1) and 400 μm IPP; 400 μm GPP, 400 μm IPP, and 40 μm [1-14C]IPP (55 Ci mol−1); 200 μm [1-3H]FPP (20 Ci mmol−1) and 600 μm IPP; and 200 μm FPP, 600 μm IPP, and 40 μm [1-14C]IPP (55 Ci mol−1).
Agrobacterium tumefaciens Strain and Culture Preparation
The disarmed A. tumefaciens strain C58/pMP90 (Koncz and Schell, 1986) containing either the binary vector pCAMGW∷IDS1 or pCAMBIA2301 (as a control) was used in the transformation experiment. For construction of the transformation vector, attR recombination sites (Gateway Technology, Invitrogen) were cloned into the vector pMJM (McKenzie et al., 1994), creating pHJMGW. A 4-kb-long cassette from pMJMGW containing the maize (Zea mays) ubiquinone promoter (ubi1) and the attR recombination sites was cloned into pCAMBIA2301. The resulting plasmid pCAMGW carries the neomycin phosphotransferase gene (nptII), which confers kanamycin resistance under the control of the double 35S cauliflower mosaic virus promoter and terminator and the GUS reporter gene (uidA), coding for GUS, under the control of the 35S cauliflower mosaic virus promoter and the nopaline synthase terminator. The complete open reading frame of IDS1 was cloned into pCAMGW upstream of the ubi1 promoter using Gateway Technology.
The bacterial culture was grown in yeast extract peptone liquid medium containing 50 μg mL−1 rifampicin, 50 μg mL−1 kanamycin sulfate, and 20 μg mL−1 gentamicin sulfate (all Duchefa) on a shaker at 250 rpm at 28°C for 16 h. Subsequently, the bacterial cells were pelleted by centrifugation and resuspended in liquid MLV medium to an OD600nm of 0.6.
Transformation Procedure for Embryogenic Tissue
Prior to the transformation experiment, embryogenic tissue was prepared and cultured on filter papers as described by Klimaszewska and Smith (1997). For the transformation experiments, the embryogenic tissue was harvested from the filter papers 7 d after subculture and resuspended in liquid MLV medium. Subsequently, an equal volume of A. tumefaciens culture (OD600nm = 0.6) in MLV was added to the cell suspension, resulting in 100 mg (fresh weight) of embryogenic tissue suspended in 1 mL of bacterial culture at OD600nm of 0.3 in MLV medium in a 100-mL Erlenmeyer flask. Control embryogenic tissue was treated the same way except that A. tumefaciens was omitted.
From the culture, 1 mL (100 mg of fresh material) was poured over a 5.5-cm sterile filter paper (Whatman no. 2) in a Büchner funnel. A short vacuum pulse (5 s, 45.7 cm of mercury) was applied to drain the liquid, and the filter was placed on a semisolid MLV medium in a 100-mm × 15-mm petri dish. A total of 10 petri dishes were used for each transformation experiment and the controls.
After 2 d of coculture on the semisolid medium supplemented with 50 μm acetosyringone, filter papers with cells from five petri dishes were placed in an Erlenmeyer flask (250 mL) with 50 mL of liquid MLV medium. The cells were then dislodged by manual shaking and collected on the new filter papers in a Büchner funnel. The filters papers with cells were then placed on fresh MLV medium with 250 mg L−1 cefotaxime (Duchefa). At the first signs of embryogenic tissue growth (approximately 2 d), the filters with cells were transferred to selective medium. Selective medium contained cefotaxime and 35 mg L−1 kanamycin. For maintenance, the transgenic embryogenic cultures of P. abies were subcultured every 14 d and cultivated as described above.
Quantitative Genomic PCR
For quantitative genomic PCR of embryogenic tissue, genomic DNA was isolated using the Plant DNA Isolation Kit (Qiagen) according to the manufacturer's instructions. Genomic PCR was performed with Brillant SYBR Green QPCR Master Mix (Stratagene) as described in the section on qRT-PCR below, but with the following changes. The denaturation step at the beginning was elongated to 13 min at 95°C. Gene abundance was normalized to the abundance of the ubiquitin gene, amplified with the forward primer (5′-CCCTCGAGGTAGAGTCATCG-3′) and the reverse primer (5′-CCAGAGTTCTCCCATCCTCC-3′). This quantity was the average of three independent biological replicates, each represented by four measurements on three technical replicates. Relative gene abundance was obtained by comparison with the gene abundance of the pCAMGW vector control.
Assay of PaIDS1 in Transgenic Embryogenic Tissue
Embryogenic tissue culture was dried on a filter paper, resuspended in assay buffer (see above), and treated with an Ultraturrax with maximal speed for 20 s at 4°C. After centrifugation at 20,000g for 10 min at 4°C, supernatant containing crude protein extract of embryogenic tissue was assayed. Protein concentration was measured according to Bradford (1976) using the Bio-Rad reagent with BSA as standard. Identical amounts of protein extracts were used in the larger scale assays as described for the assay of recombinant PaIDS1 and product identification.
qRT-PCR
Total RNA from plants at different time points after MJ treatment and from different organs was isolated according to the method of Wang et al. (2000), except that an additional DNA digestion step was included (RNase Free DNase Set; Qiagen). Using identical amounts of total RNA, template cDNA for subsequent PCR was generated using SuperScript III (Invitrogen) according to the manufacturer's instructions. qRT-PCR was performed with Brillant SYBR Green QPCR Master Mix (Stratagene). Specific qRT-PCR primers, forward (5′-GACATCTGGTATCATCACTC-3′) and reverse (5′-GTGACCTTCCCCTTTACTTG-3′), were designed using the following criteria: predicted melting temperature of at least 58°C, primer length of 22 to 24 nucleotides, guanosine-cytosine content of at least 48%, and an amplicon length of 120 to 150 bp. Primer specificity was confirmed by melting curve analysis, by an efficiency of product amplification of 1.0 ± 0.1, and by sequence verification of at least eight cloned PCR amplicons for each PCR procedure. Reactions with water instead of cDNA template were run with each primer pair as controls. A standard thermal profile of 95°C for 10 min, then 45 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s, was used. The fluorescence signal was captured at the end of each cycle, and a melting curve analysis was performed from 53°C to 95°C with data capture every 0.2°C during a 1-s hold. qRT-PCR was performed as described in the operator's manual using a Stratagene MX3000P. Transcript abundance was measured as the average of three biological replicates; each is represented by three determinations of three technical replicates. All amplification plots were analyzed with the MX3000P software to obtain threshold cycle values. Transcript abundance was normalized with the transcript abundance of the ubiquitin gene (GenBank accession no. EF681766), amplified with the forward primer (5′-GTTGATTTTTGCTGGCAAGC-3′) and reverse primer (5′-CACCTCTCAGACGAAGTAC-3′). Relative transcript abundance was obtained by calibration against the transcript abundance at the onset of treatment.
For qRT-PCR of embryogenic tissue, RNA was isolated using the Plant RNA Isolation Kit (Invitek) according to the manufacturer's instructions, including an additional DNA digestion step (RNase Free DNase Set; Qiagen). Further steps were carried out as above. Relative transcript abundance was calibrated against the transcript abundance of pCAMGW vector control plants.
Microscopy and Immunogold Labeling of PaIDS1
Stems of P. abies trees (5 years old) were inoculated with Ceratocystis polonica as described by Christiansen et al. (1999). Bark samples (2 × 3 mm, including phloem, vascular cambium, and xylem with traumatic resin ducts) were collected 36 d after inoculation. All samples were isolated from areas 5 cm above the inoculation site, well above the edge of necrotic lesions (Nagy et al., 2000). Fixation, dehydration, and embedding were performed as described previously (Nagy et al., 2000). Semithin sections (1 μm) were cut using a LKB 2128 Ultratome (Leica Microsystems) and dried onto Superfrost Plus slides (Menzel-Gläzer) for light microscopy.
Immunogold labeling was performed as described previously (Nagy et al., 2000). Polyclonal peptide antibodies for PaIDS1 were synthesized against amino acids 285 to 300 with the sequence 5′-GASDDEIERVRKYARC-3′ (Fig. 2) and affinity purified (Eurogentec). Polyclonal peptide antibodies for PaIDS4 were synthesized against the amino acids 172 to 187 with the sequence 5′-HEGATDLSKYKMPT-3′ (Schmidt and Gershenzon, 2007). The primary antibodies were diluted 1:4,000 in TBST/BSA buffer (10 mm Tris, pH 7.2, 0.15 m NaCl, 0.1% [w/v] Tween 20, and 2% [w/v] BSA), and the secondary antibody goat anti-rabbit IgG conjugated to 10-nm gold (British Biocell International) was diluted 1:4,000 in TBST/BSA buffer. The sections were counterstained with 0.5% Safranin O, air dried, mounted with Biomount (British Biocell International) for evaluation by a Leitz Aristoplan light microscope, and documented with a Leica DC300 digital camera. To ensure the specificity of the antibody labeling, controls included (1) incubation with the rabbit preimmune serum instead of the primary antibody and (2) incubation with the secondary antibody omitting the primary antibody step.
Three-Dimensional Structural Modeling of PaIDS1
The ribbon model of PaIDS1 was created with the program Swiss-Model (version 36.0003). The crystallographic structure of avian FPP synthase was used as a template to build the PaIDS1 model structure. The PaIDS1 sequence was entered under the Web address http://swissmodel.expasy.org and created as an image with DeepView SwissPdbViewer 3.7 (Sp5). Pairwise alignments were made between the PaIDS1 model structure and avian crystal structure to establish positions and nearby contacts of analogous catalytic residues located near the substrate-binding site. The image depicted (Fig. 11) was created with the program UCSF Chimera (National Centre for Research Resources).
Construction of Mutated PaIDS1
Site-directed mutagenesis was performed with the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer's instructions. The cloned PaIDS1 in the expression vector pCR-T7 CT TOPO served as a template for PCR amplification. Sequences of the primers used are listed in Supplemental Table S2. PCR was performed for 60 s at 95°C initial denaturation, 18 cycles of 60 s at 95°C, 60 s at 55°C, and 4 min at 68°C, and a final annealing for 7 min at 68°C in a Tpersonal thermocycler (Biometra). After amplification, the parental (methylated) DNA was digested with DpnI (10 units μL−1) for 1 h at 37°C. The DpnI-treated DNA was transformed in XL10-Gold Ultracompetent Cells (Stratagene). All mutations were confirmed by DNA sequencing, and the plasmid was transformed into E. coli BL21 (DE3)pLysS (Invitrogen).
Generation of Chimeric IDS
Chimeric versions of PaIDS1 assembled with the N-terminal or C-terminal portion of the coding sequence exchanged for the corresponding portion of the coding sequence of PaIDS2 or PaIDS5 were generated by separate PCR amplification followed by ligation. Amplification of the partial PaIDS2 and PaIDS5 sequences was performed using primers that included the BssSI restriction site at nucleotide position 652 for PaIDS2, the BanII site at position 717 for PaIDS5, and primers with the added appropriate restriction site for amplification of the PaIDS1 partial cDNA (Supplemental Table S2). The PCR conditions are as described for the functional expression of PaIDS1. Amplicons were restricted with the appropriate restriction enzyme, gel purified using of Wizard SV Gel and PCR Clean-Up System (Promega) following the manufacturer's instructions, and ligated using of T4 DNA Ligase (Invitrogen). The chimeric constructs were cloned into the vector pCR-T7 CT TOPO (Invitrogen) and transformed into One Shot Top 10 (Invitrogen) chemically competent cells. After sequencing, the constructs were transformed into E. coli BL21 (DE3)pLysS (Invitrogen).
The PaIDS1 sequence was deposited in GenBank with accession number GQ369788.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SDS-PAGE analysis with Coomassie Brilliant Blue staining of PaIDS1 recombinant protein expressed in E. coli and partially purified.
Supplemental Figure S2. Lineweaver-Burk plots of partially purified recombinant PaIDS1 protein expressed in E. coli for determination of Km and Vmax. for DMAPP and IPP.
Supplemental Table S1. Sequences of primers used for site-directed mutagenesis, including the melting temperatures and the positions of the changed amino acids.
Supplemental Table S2. Sequences of primers used for the construction of chimeric IDS, including their melting temperatures.
Supplementary Material
Acknowledgments
We thank Marion Stäger, Andrea Bergner, and Beate Rothe for excellent technical assistance, Mike Phillips for providing the pCAMGW plasmid, Harald Kvaalen for kindly providing the embryogenic culture of P. abies, Elin Ørmen and Nina Nagy for technical assistance with the immunocytochemistry, and Almuth Hammerbacher and Kimberly Falk for critical reading of the manuscript.
This work was supported by the Max Planck Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jonathan Gershenzon (gershenzon@ice.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ashby MN, Spear DH, Edwards PA (1990) Prenyltransferases from yeast to man. In AD Attic, ed, Molecular Biology of Atherosclerosis. Elsevier Science Publishers, Amsterdam, pp 27–34
- Bannan MW (1936) Vertical resin ducts in the secondary wood of the Abietineae. New Phytol 35 11–46 [Google Scholar]
- Bick JA, Lange BM (2003) Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch Biochem Biophys 415 146–154 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Rahier A, Camara B (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44 357–429 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Suire C, d'Harlingue A, Backhaus RA, Camara B (2002) Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J 24 241–252 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Burke C, Croteau R (2002. a) Interaction with the small subunit of geranyl diphosphate synthase modifies the chain length specificity of geranylgeranyl diphosphate synthase to produce geranyl diphosphate. J Biol Chem 277 3141–3149 [DOI] [PubMed] [Google Scholar]
- Burke C, Croteau R (2002. b) Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Arch Biochem Biophys 405 130–136 [DOI] [PubMed] [Google Scholar]
- Burke CC, Wildung MR, Croteau R (1999) Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc Natl Acad Sci USA 96 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun-McKay A, Godard A, Toudefallah M, Martin M, Alfaro R, King J, Bohlmann J (2006) Wound-induced terpene synthase gene expression in Sitka spruce that exhibit resistance or susceptibility to attack by the white pine weevil. Plant Physiol 140 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Cervantes M, Gallagher CE, Zhu C, Wurtzel ET (2006) Maize cDNAs expressed in endosperm encode functional farnesyl diphosphate synthase with geranylgeranyl diphosphate synthase activity. Plant Physiol 141 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Poulter CD (1994) Isolation and characterization of idsA: the gene for the short chain isoprenyl diphosphate synthase from Methanobacterium thermoautotrophicum. Arch Biochem Biophys 314 399–404 [DOI] [PubMed] [Google Scholar]
- Christiansen E, Krokene P, Berryman AA, Franceschi VR, Krekling T, Lieutier F, Lönneborg A, Solheim H (1999) Mechanical injury and fungal infection induce acquired resistance in Norway spruce. Tree Physiol 19 399–403 [DOI] [PubMed] [Google Scholar]
- Copley SD (2003) Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr Opin Chem Biol 7 265–272 [DOI] [PubMed] [Google Scholar]
- Elfstrand M, Fossdal CG, Sitbon F, Olsson O, Lönneborg A, von Arnold S (2001) Overexpression of the endogenous peroxidase-like spi 2 in transgenic Norway spruce plants results in increased total peroxidase activity and reduced growth. Plant Cell Rep 20 596–603 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak H, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Erbilgin N, Krokene P, Christiansen E, Zeneli G, Gershenzon JG (2006) Exogenous application of methyl jasmonate elicits defenses in Norway spruce (Picea abies) and reduces host colonization by the bark beetle Ips typographus. Oecologia 148 426–436 [DOI] [PubMed] [Google Scholar]
- Fernandez SM, Kellogg BA, Poulter CD (2000) Farnesyl diphosphate synthase: altering the catalytic site to select for geranyl diphosphate activity. Biochemistry 39 15316–15321 [DOI] [PubMed] [Google Scholar]
- Franceschi VR, Krekling T, Christiansen E (2002) Application of methyl jasmonate on Picea abies (Pinaceae) stems induces defense-related responses in phloem and xylem. Am J Bot 89 578–586 [DOI] [PubMed] [Google Scholar]
- Franceschi VR, Krokene P, Christiansen E, Krekling T (2005) Anatomical and chemical defenses of conifer bark against bark beetles and other pests. New Phytol 167 353–375 [DOI] [PubMed] [Google Scholar]
- Gershenzon J, Kreis JW (1999) Biochemistry of terpenoids: monoterpenes, sesquiterpenes, diterpenes, sterols, cardiac glycosides, and steroid saponins. In M Wink, ed Biochemistry of Plant Secondary Metabolism: Annual Plant Reviews 21. Sheffield Academic Press, Sheffield, UK, pp 222–299
- Hefner J, Ketchum E, Croteau R (1998) Cloning and functional expression of a cDNA encoding geranylgeranyl diphosphate synthase from Taxus canadensis and assessment of the role of this prenyltransferase in cells induced for taxol production. Arch Biochem Biophys 360 62–74 [DOI] [PubMed] [Google Scholar]
- Hsiao Y-Y, Jeng M-F, Tsai W-C, Chuang Y-C, Li C-Y, Wu T-S, Kuoh C-S, Chen W-H, Chen H-H (2008) A novel homodimeric geranyl diphosphate synthase from the orchid Phalaenopsis bellina lacking a DD(X)2-4 motif. Plant J 55 719–733 [DOI] [PubMed] [Google Scholar]
- Hudgins JW, Christiansen E, Franceschi VR (2003) Methyl jasmonate induces changes mimicking anatomical defenses in diverse members of the Pinaceae. Tree Physiol 23 361–371 [DOI] [PubMed] [Google Scholar]
- Hudgins JW, Christiansen E, Franceschi VR (2004) Induction of anatomically based defense responses in stems of diverse conifers by methyl jasmonate: a phylogenetic perspective. Tree Physiol 24 251–264 [DOI] [PubMed] [Google Scholar]
- Joly A, Edwards PA (1993) Effect of site-directed mutagenesis of conserved aspartate and arginine residues upon farnesyl diphosphate synthase activity. J Biol Chem 268 26983–26989 [PubMed] [Google Scholar]
- Keeling I, Bohlmann J (2006. a) Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol 170 657–675 [DOI] [PubMed] [Google Scholar]
- Keeling I, Bohlmann J (2006. b) Diterpene resin acids in conifers. Phytochemistry 67 2415–2423 [DOI] [PubMed] [Google Scholar]
- Kellogg BA, Poulter CD (1997) Chain elongation in the isoprenoid biosynthetic pathway. Curr Opin Chem Biol 1 570–578 [DOI] [PubMed] [Google Scholar]
- Kharel Y, Koyama T (2003) Molecular analysis of cis-prenyl chain elongating enzymes. Nat Prod Rep 20 111–118 [DOI] [PubMed] [Google Scholar]
- Klimaszewska K, Lachance D, Pelletier G, Lelu AM, Séguin A (2001) Regeneration of transgenic Picea glauca, P. mariana, and P. abies after cocultivation of embryogenic tissue with Agrobacterium tumefaciens. In Vitro Cell Dev Biol Plant 37 748–755 [Google Scholar]
- Klimaszewska K, Smith DR (1997) Maturation of somatic embryos of Pinus strobus is promoted by a high concentration of gellan gum. Physiol Plant 100 949–957 [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Koyama T, Obata S, Saito K, Takeshita-Koike A, Ogura K (1994) Structural and functional roles of the cysteine residues of Bacillus stearothermophilus farnesyl pyrophosphate synthase. Biochemistry 33 12644–12648 [DOI] [PubMed] [Google Scholar]
- Koyama T, Tajima M, Nishino T, Ogura K (1995) Significance of Phe-220 and Gln-221 in the catalytic mechanism of farnesyl diphosphate synthase of Bacillus stearothermophilus. Biochem Biophys Res Commun 212 681–686 [DOI] [PubMed] [Google Scholar]
- Koyama T, Tajima M, Sano H, Doi T, Koike-Takeshita A, Obata S, Nishino T, Ogura K (1996) Identification of significant residues in the substrate binding site of Bacillus stearothermophilus farnesyl diphosphate synthase. Biochemistry 35 9533–9538 [DOI] [PubMed] [Google Scholar]
- Langenheim JH (2003) Plant Resins: Chemistry, Evolution, Ecology, and Ethnobotany. Timber Press, Portland, OR
- Liang PH (2009) Reaction kinetics, catalytic mechanisms, conformational changes, and inhibitor design for prenyltransferases. Biochemistry 48 6562–6570 [DOI] [PubMed] [Google Scholar]
- Liang PH, Ko TP, Wang AH (2002) Structure, mechanism and function of prenyltransferases. Eur J Biochem 269 3339–3354 [DOI] [PubMed] [Google Scholar]
- Ling Y, Li ZH, Miranda K, Oldfield E, Moreno SN (2007) The farnesyl-diphosphate/geranylgeranyl-diphosphate synthase of Toxoplasma gondii is a bifunctional enzyme and a molecular target of bisphosphonates. J Biol Chem 282 30804–30816 [DOI] [PubMed] [Google Scholar]
- Marrero PF, Poulter CD, Edwards PA (1992) Effects of site-directed mutagenesis of the highly conserved aspartate residues in domain II of farnesyl diphosphate synthase activity. J Biol Chem 267 21873–21878 [PubMed] [Google Scholar]
- Martin D, Fäldt J, Bohlmann J (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135 1908–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Tholl D, Gershenzon J, Bohlmann J (2002) Methyl jasmonate induces traumatic resin ducts, terpenoid resin biosynthesis, and terpenoid accumulation in developing xylem of Norway spruce stems. Plant Physiol 129 1003–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie MJ, Jameson PE, Poulter RTM (1994) Cloning an ipt gene from Agrobacterium tumefaciens: characterization of cytokinins in derivative transgenic plant tissue. Plant Growth Regul 14 217–228 [Google Scholar]
- Möller R, McDonald AG, Walter C, Harris PJ (2003) Cell differentiation, secondary cell-wall formation and transformation of callus tissue of Pinus radiata D. Don. Planta 217 736–747 [DOI] [PubMed] [Google Scholar]
- Nagy NE, Franceschi R, Solheim H, Krekling T, Christiansen E (2000) Wound-induced traumatic resin duct development in stems of Norway spruce (Pinaceae): anatomy and cytochemical traits. Am J Bot 87 303–313 [PubMed] [Google Scholar]
- Ogura K, Koyama T (1998) Enzymatic aspects of isoprenoid chain elongation. Chem Rev 98 1263–1276 [DOI] [PubMed] [Google Scholar]
- Ohnuma S, Hirooka K, Hemmi H, Ishida C, Ohto C, Nishino T (1996. a) Conversion of product specificity of archaebacterial geranylgeranyl-diphosphate synthase. J Biol Chem 271 18831–18837 [DOI] [PubMed] [Google Scholar]
- Ohnuma S, Hirooka K, Ohto C, Nishino T (1997) Conversion from archaeal geranylgeranyl diphosphate synthase to farnesyl diphosphate synthase. J Biol Chem 272 5192–5198 [DOI] [PubMed] [Google Scholar]
- Ohnuma S, Narita K, Nakazawa T, Ishida C, Takeuchi Y, Ohto C, Nishino T (1996. b) A role of the amino acid residue located on the fifth position before the first aspartate-rich motif of farnesyl diphosphate synthase on determination of the final product. J Biol Chem 271 30748–30754 [DOI] [PubMed] [Google Scholar]
- Parmryd I, Andersson B, Dallner G (1999) Protein prenylation in spinach chloroplasts. Proc Natl Acad Sci USA 96 10074–10079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M, Croteau R (1999) Resin-based defenses in conifers. Trends Plant Sci 4 184–190 [DOI] [PubMed] [Google Scholar]
- Ro K, Bohlmann J (2006) Diterpene resin acid biosynthesis in loblolly pine (Pinus taeda): functional characterization of abietadiene/levopimaradiene synthase (PtTPS-LAS) cDNA and subcellular targeting of PtTPS-LAS and abietadienol/abietadienal oxidase (PtAO, CYP720B1). Phytochemistry 67 1572–1578 [DOI] [PubMed] [Google Scholar]
- Sallaud C, Rontein D, Onillon S, Jabès F, Duffé P, Giacalone C, Thoraval S, Escoffier C, Herbette G, Leonhardt N, et al (2009) A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21 301–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA 106 10865–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Gershenzon J (2007) Cloning and characterization of isoprenyl diphosphate synthases with farnesyl diphosphate and geranylgeranyl diphosphate synthase activity from Norway spruce (Picea abies) and their relation to induced oleoresin formation. Phytochemistry 68 2649–2659 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Gershenzon J (2008) Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry 69 49–57 [DOI] [PubMed] [Google Scholar]
- Schmidt A, Zeneli G, Hietala AM, Fossdal CG, Krokene P, Christiansen E, Gershenzon J (2005) Induced chemical defenses in conifers: biochemical and molecular approaches to studying their function. In JT Romeo, ed, Chemical Ecology and Phytochemistry of Forest Ecosystems: Recent Advances in Phytochemistry, Vol 39. Elsevier, Oxford, pp 1–28
- Soderberg T, Chen A, Poulter CD (2001) Geranylgeranylglyceryl phosphate synthase: characterization of the recombinant enzyme from Methanobacterium thermoautotrophicum. Biochemistry 40 14847–14854 [DOI] [PubMed] [Google Scholar]
- Song L, Poulter CD (1994) Yeast farnesyl-diphosphate synthase: site-directed mutagenesis of residues in highly conserved prenyltransferase domains I and II. Proc Natl Acad Sci USA 91 3044–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya A, Zhanga W, Asawatreratanakul K, Wititsuwannakul D, Wititsuwannakul R, Takahashi S, Koyama T (2003) Cloning, expression and characterization of a functional cDNA clone encoding geranylgeranyl diphosphate synthase of Hevea brasiliensis. Biochim Biophys Acta 1625 214–220 [DOI] [PubMed] [Google Scholar]
- Tarshis LC, Proteau PJ, Kellogg BA, Sacchettini JC, Poulter CD (1996) Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci USA 93 15018–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarshis LC, Yan M, Poulter CD, Sacchettini JC (1994) Crystal structure of recombinant farnesyl diphosphate synthase at 2.6-A resolution. Biochemistry 33 10871–10877 [DOI] [PubMed] [Google Scholar]
- Tholl D, Croteau R, Gershenzon J (2001) Partial purification and characterization of the short-chain prenyltransferases, geranyl diphospate synthase and farnesyl diphosphate synthase, from Abies grandis (grand fir). Arch Biochem Biophys 386 233–242 [DOI] [PubMed] [Google Scholar]
- Tholl D, Kish CM, Orlova I, Sherman D, Gershenzon J, Pichersky E, Dudareva N (2004) Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 16 977–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp C, Croteau B (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermoten S, Charloteaux B, Santini S, Sen SE, Béliveau C, Vandenbol M, Francis F, Brasseur R, Cusson M, Haubruge E (2008) Characterization of a novel aphid prenyltransferase displaying dual geranyl/farnesyl diphosphate synthase activity. FEBS Lett 582 1928–1934 [DOI] [PubMed] [Google Scholar]
- Vandermoten S Haubruge E, Cusson M (2009) New insights into short-chain prenyltransferases: structural features, evolutionary history and potential for selective inhibition. Cell Mol Life Sci 66 3685–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schie C, Lange T, Schmidt A, Haring M, Schuurink R (2007) Geranyl diphosphate synthase is required for the biosynthesis of gibberellins. Plant J 52 752–762 [DOI] [PubMed] [Google Scholar]
- Wang G, Dixon RA (2009) Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc Natl Acad Sci USA 106 9914–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Ohnuma S (1999) Chain-length determination mechanism of isoprenyl diphosphate synthases and implications for molecular evolution. Trends Biochem Sci 24 445–451 [DOI] [PubMed] [Google Scholar]
- Wang K, Ohnuma S (2000) Isoprenyl diphosphate synthases. Biochim Biophys Acta 1529 33–48 [DOI] [PubMed] [Google Scholar]
- Wang X, Hunter W, Plant A (2000) Isolation and purification of functional total RNA from woody branches and needles of Sitka and white spruce. Biotechniques 28 292–296 [DOI] [PubMed] [Google Scholar]
- Welsch R, Beyer P, Hugueney P, Kleinig H, von Lintig J (2000) Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 211 846–854 [DOI] [PubMed] [Google Scholar]
- Zeneli G, Krokene P, Christiansen E, Krekling T, Gershenzon J (2006) Methyl jasmonate treatment of mature Norway spruce (Picea abies) trees increases the accumulation of terpenoid resin components and protects against infection by Ceratocystis polonica, a bark beetle-associated fungus. Tree Physiol 26 977–988 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.