Abstract
Plant root architecture is highly responsive to changes in nutrient availability. However, the molecular mechanisms governing the adaptability of root systems to changing environmental conditions is poorly understood. A screen for abnormal root architecture responses to high nitrate in the growth medium was carried out for a population of ethyl methanesulfonate-mutagenized Arabidopsis (Arabidopsis thaliana). The growth and root architecture of the arm (for anion altered root morphology) mutant described here was similar to wild-type plants when grown on low to moderate nitrate concentrations, but on high nitrate, arm exhibited reduced primary root elongation, radial swelling, increased numbers of lateral roots, and increased root hair density when compared to the wild-type control. High concentrations of chloride and sucrose induced the same phenotype. In contrast, hypocotyl elongation in the dark was decreased independently of nitrate availability. Positional cloning identified a point mutation in the AtCTL1 gene that encodes a chitinase-related protein, although molecular and biochemical analysis showed that this protein does not possess chitinase enzymatic activity. CTL1 appears to play two roles in plant growth and development based on the constitutive effect of the arm mutation on primary root growth and its conditional impact on root architecture. We hypothesize that CTL1 plays a role in determining cell wall rigidity and that the activity is differentially regulated by pathways that are triggered by environmental conditions. Moreover, we show that mutants of some subunits of the cellulose synthase complex phenocopy the conditional effect on root architecture under nonpermissive conditions, suggesting they are also differentially regulated in response to a changing environment.
Root systems exhibit a high degree of architectural plasticity in response to water and nutrient availability. Root architecture is a genetically defined and environmentally regulated process. In particular, the growth and development of lateral roots (LRs) is greatly influenced by environmental factors such as mineral nutrient abundance (Casimiro et al., 2003; López-Bucio et al., 2003; Nibau et al., 2008; Iyer-Pascuzzi et al., 2009; Péret et al., 2009). Nitrate availability is one of the major determinants of root morphology (Zhang and Forde, 2000; Hermans et al., 2006; Gojon et al., 2009). Low nitrate levels in the soil stimulate LR development, which substantially increases the root surface area available for nutrient acquisition. Conversely, high levels of nitrate inhibit LR elongation by preventing LR meristematic activation at postemergence (Zhang et al., 1999) but generally have no impact on the primary root (PR) growth. Interestingly, when roots of nitrogen-deficient plants contact nitrate, LR outgrowth is enhanced within the nitrate-rich patch (Zhang and Forde, 1998).
Several sensing and signaling pathways are thought to be involved in root nitrate responses in Arabidopsis (Arabidopsis thaliana). LR responses to external nitrate abundance have been associated with the MADS-box transcription factor ARABIDOPSIS NITRATE-REGULATED1 (ANR1; Zhang and Forde, 1998, 2000; Zhang et al., 1999) and with a systemic signal, possibly Gln, through a basic Leu zipper (bZIP) and a LIM transcription factor (Tranbarger et al., 2003). Nitrate transporters may also be components of signaling pathways, as observed for NRT1.1 and NRT2.1, which have been shown to function as nitrate sensors. Perception of nitrate availability could occur in part through NRT2.1, independent of its nitrate uptake function in the root (Malamy and Ryan, 2001; Little et al., 2005; Miller et al., 2007). NRT2.1 seems to be directly involved in the regulation of LR initiation when nitrate is limiting and Suc is abundant. It has also been reported that the dual-affinity nitrate transporter NRT1.1 acts upstream of ANR1 in mediating the stimulatory effect of a localized nitrate supply on LR proliferation (Liu et al., 1999; Remans et al., 2006).
MicroRNAs have also been shown to modulate LR emergence in response to nitrogen (Gifford et al., 2008). The microRNA 167a/b, specifically expressed in pericycle cells and LR cap, was shown to be repressed by increased nitrate supply. Accordingly, this resulted in the induction of one miR167a/b target, AUXIN RESPONSE FACTOR8, which in turn reduces emergence of initiated LRs (Gifford et al., 2008; Gojon et al., 2009). In addition to external ion availability, systemic inhibition of LR development has been observed when tissue nitrate concentrations are high, and this response has been linked to abscisic acid (Signora et al., 2001). Long-distance signals mediating the shoot response to nitrate perception in roots may also involve cytokinins. It is possible that the reduction in cytokinins observed during N deficiency (Takei et al., 2004) relieves a general inhibition of root growth by this hormone and that an increase in auxin stimulates cell division and LR development. In the latter case, auxin reprograms cells overlaying LR primordia to facilitate organ emergence, and genes encoding several Arabidopsis cell wall remodeling enzymes have been reported to be expressed in these cells (Péret et al., 2009, and refs. therein). Finally, the impact of the cell wall on cell shape and size is a critical determinate in plant growth and development (Cosgrove, 1999).
In the experiments reported here, we screened for root architecture mutants that produce LRs in the presence of high nitrate. These conditions repress LR growth in wild-type seedlings. We describe the physiological and genetic characteristics of the arm (for anion altered root morphology) mutant, whose phenotype is not only conditional on high, external nitrate, but also on other environmental cues.
RESULTS
Isolation of the arm Mutant with Altered Response to High Nitrate
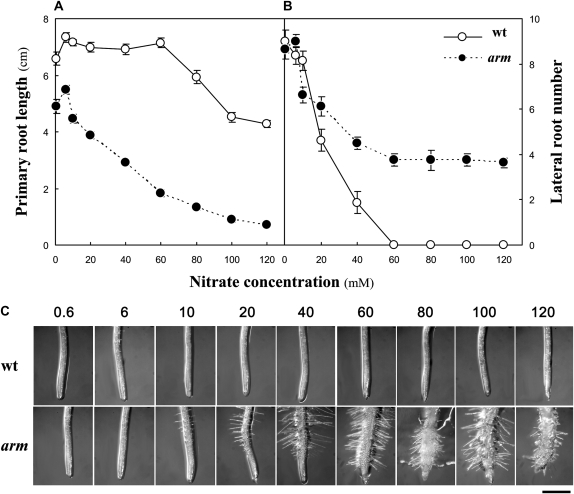
This screen was designed to identify mutants impaired in morphological responses of seedling roots to nitrate abundance. The standard Murashige and Skoog growth medium (Murashige and Skoog, 1962), which is commonly used and adapted in nutritional screens (Hauser et al., 1995; Schneider et al., 1997; Malamy and Ryan, 2001), was modified by eliminating NH4NO3 and limiting KNO3 to a range of concentrations that bracketed the presence or absence of LRs in wild-type plants. Thirteen days after germination, wild-type Columbia-0 (Col-0) seedlings grown on vertical plates with high nitrate (60 mm KNO3) had a single PR, and no or very few LRs (Figs. 1, A and B, and 2). Growth at 120 mm KNO3 decreased PR length and completely repressed the lateral branching (Fig. 1, A and B). In contrast, plants grown on low (0.6 mm) or moderate (6 mm) KNO3 had developed many LRs (Figs. 1, A and B, and 2). We screened ethyl methanesulfonate-mutagenized Arabidopsis seedlings for the presence of LRs when grown on normally restrictive levels (60 mm) of KNO3. One mutant showing conspicuous features that were conditional on high nitrate is described here. The phenotype of this mutant grown on 60 mm KNO3 included reduced PR length (Fig. 1A), high numbers of LRs (Fig. 1B), radial swelling, and increased root hair length and density (Fig. 1C). At 0.6 and 6 mm nitrate, PR length of mutant seedlings was decreased by one-fourth compared to the wild type, but no difference in the number of emerged LRs was observed. High concentrations of KNO3 inhibited PR elongation by more than one-half and induced the emergence of LRs, root hair, and radial swelling. Similar observations were apparent after prolonged growth (27 d after germination) in these restrictive conditions (Fig. 2). Because of these characteristics (and chloride sensitivity described below), the mutant was named arm. It is noteworthy that the total length of LRs elaborated by the arm mutant at 60 mm KNO3 does not compensate for the decrease in PR length compared to the wild type at high nitrate supply (Supplemental Fig. S1, A and B). Yet, because of root cell swelling, the root-to-shoot dry biomass ratio is not affected by the arm mutation (Supplemental Fig. S1C).
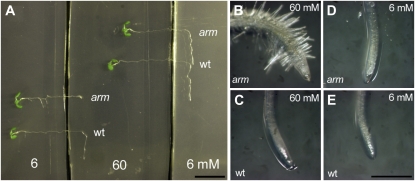
Figure 1.
Effect of nitrate availability in the growth medium on root system architecture of wild-type (wt) and arm plants. Seedlings were grown vertically on Murashige and Skoog medium with increasing nitrate (KNO3) supply for 13 d. A, PR length. B, LR (>1 mm) number; n = 8 ± se. Black symbols, wild type; white symbols, arm mutant. C, Picture of root tip. Bar = 0.5 mm.
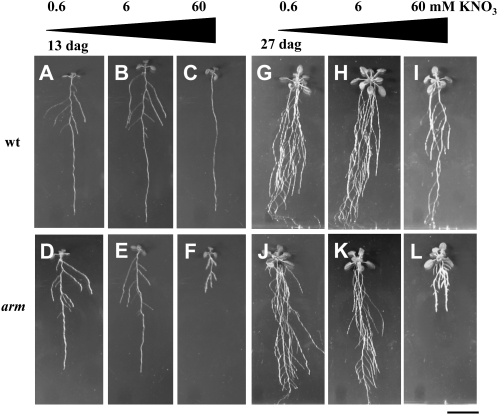
Figure 2.
Conditional phenotypes of wild-type (wt) and arm plants to nitrate supply. Pictures of seedlings grown vertically on Murashige and Skoog medium with three nitrate concentrations: 0.6 mm (A, D, G, and J), 6 mm (B, E, H, and K), and 60 mm (C, F, I, and L) KNO3 for 13 d (A–E) and 27 d (G–L). A to C and G to I, Wild type; D, E, and J to L, arm mutant. Bar = 2 cm. dag, Days after germination.
Specificity of the arm Phenotype to Sugars and Minerals
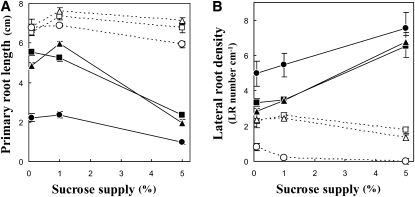
Several studies have shown that Suc levels can influence morphological responses and in particular to nitrate availability (Zhang et al., 1999; Malamy and Ryan, 2001; MacGregor et al., 2008). Therefore, we examined root responses to Suc supplies in the growth medium. Nine combinations of nitrate (0.6, 6, and 60 mm KNO3) and Suc (0.1%, 1%, and 5% w/w) were tested. High Suc supply decreased PR length and increased LR density in the arm mutant across all nitrate concentrations (Fig. 3; Supplemental Fig. S2). The combination of high (5%) Suc and high (60 mm) nitrate had the strongest negative effect, resulting in an 85% reduction of PR length in arm plants compared to the wild type. Overall, high Suc induces changes in root morphology that are similar to patterns observed on high nitrate.
Figure 3.
Effect of the carbon/nitrogen ratio on root system architecture of wild-type and arm plants. Seedlings were grown at nine different combinations of nitrate (squares, 0.6 mm; triangles, 6 mm; circles, 60 mm KNO3) and Suc (0.1%, 1%, and 5% w/w) concentrations. PR length (A) and LR density (B; no. of LRs divided by the PR length between hypocotyl and last observed branching) were measured at day 13. White symbols, wild type; black symbols, arm mutant. n = 8 to 14 ± se.
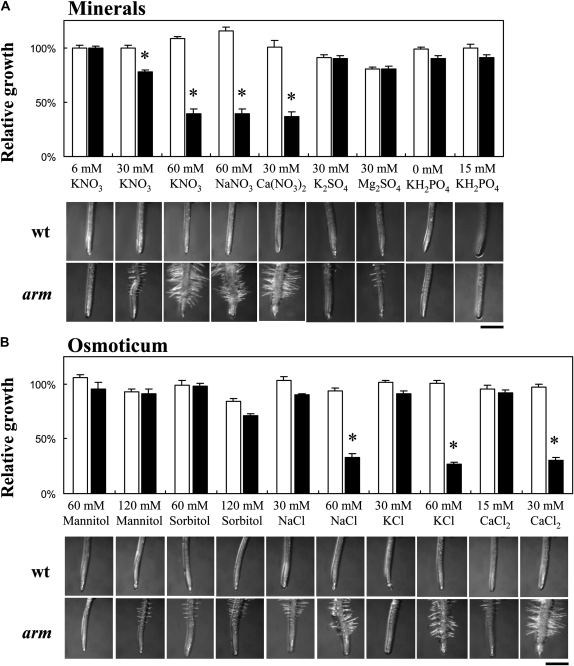
Since the mutants identified in this screen exhibited abnormal root architecture in response to high KNO3 in the growth medium, it was necessary to determine the role of cations, anions, and osmotic potential on the mutant phenotype. Thus, 6-d-old seedlings grown on 6 mm KNO3 (considered as the reference substrate) were transferred to fresh media enriched with or depleted of a variety of ions and molecules for an additional 6 d of growth (Fig. 4). Sixty millimolar NaNO3 and 30 mm Ca(NO3)2 impacted root morphology to the same extent as 60 mm KNO3. In contrast, 30 mm K2SO4 or MgSO4 had little effect (Fig. 4A). This shows that the arm mutant is responsive to nitrate rather than the potassium ions of KNO3. Unfortunately, high phosphate was limited to 15 mm because of precipitation at higher concentrations. Nevertheless, a 10-fold increase of the phosphate concentration had no impact on root architecture. Sixty millimolar mannitol or sorbitol had no effect, but a slight decrease of PR length was observed at 120 mm sorbitol (Fig. 4B), suggesting this is not an osmotic effect on root architecture. Chloride salts (30 mm CaCl2, 60 mm KCl, and 60 mm NaCl) induced exactly the same root responses as nitrate salts (Fig. 4B), implicating monovalent anions as the active molecular species since high sulfate and phosphate had little impact and, thus, the rationale behind the mutant's name (arm, anion altered root morphology). We also tested if the anion-responsive root phenotype is caused by pH changes in the agar medium, as a consequence of the high anion influx. Media buffered with 2.5 mm MES resulted in the same mutant phenotype as described before.
Figure 4.
Effect of minerals and sugars on PR elongation of wild-type (wt) and arm plants. Six-day-old seedlings grown on moderate (6 mm KNO3) medium were transferred to diverse substrates enriched in minerals (A), sugars (B), and chloride salts (B). The relative root growth was calculated by dividing the PR elongation 6 d after transfer on a particular substrate by the average elongation observed on moderate nitrate medium (considered as the reference growth condition). White bars, wild type; black bars, arm mutant. n = 8 to 14 ± se. Asterisks indicate significant differences between genotypes at level <0.05. Bars = 0.5 mm.
Local Mineral Sensing and Mineral Content
Given the dramatic impact of high nitrate and chloride on the root system, we wanted to determine if this is a localized or systemic response. Seedlings grown on uniformly moderate (6 mm KNO3) nitrate supply were transferred onto plates divided into regions of high and moderate KNO3 concentrations (6 and 60 mm; Fig. 5). When plants were placed such that the root tip zone alone contacted high nitrate, PR growth was completely inhibited and radial swelling of the root tip was apparent in the arm mutant after 2 d of transfer. When the root tip zone was on medium containing moderate nitrate, while a substantial portion of the mature root zone was on high nitrate, no inhibition of PR elongation was observed (Fig. 5A). These observations demonstrate a local response to nitrate ions in the induction of the arm root phenotype.
Figure 5.
Local sensing of nitrate availability in root tip of wild-type (wt) and arm plants. A, Six-day-old seedlings germinated on moderate (6 mm KNO3) nitrate substrate were transferred onto plates divided by a high (60 mm KNO3) nitrate strip band. Seedlings were transferred so that the older root zone or the root tip alone was in contact with 60 mm KNO3. B to E, Close-ups of root tip: arm (B) and wild type (C) contacting high nitrate; arm (D) and wild type (E) contacting moderate nitrate. Pictures were taken 2 d after transfer. Bars = 1 cm in A and 1 mm B to E. The experiment was repeated with a minimum of 10 seedlings of each genotype. [See online article for color version of this figure.]
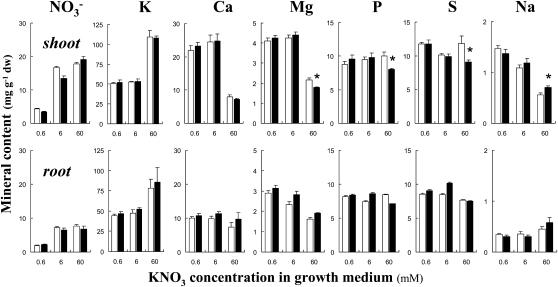
Since the arm mutant is responsive to changes in mineral ion availability, we examined the effect of the arm mutation on the ionome (Lahner et al., 2003). The mineral content was analyzed in root and shoot of seedlings grown in vitro with low, moderate, and high KNO3 conditions. No conspicuous differences in tissue nitrate content that would indicate a severe dysfunction in ion homeostasis was observed between the wild type and arm (Fig. 6). Likewise, little difference was observed for tissue concentrations of other minerals that included potassium, calcium, magnesium, phosphorus, sulfur, and sodium. It is noteworthy that the addition of 60 mm KNO3 in the growth medium resulted in higher potassium and lower calcium, magnesium, and sodium tissue concentrations in the shoots of both genotypes (Fig. 6).
Figure 6.
Mineral profile in tissues of wild-type and arm plants. Seedlings were grown at three nitrate supplies (0.6, 6, and 60 mm KNO3). Mineral content in shoot and root was measured 13 d after germination. P and S refer to total phosphorus and sulfur content. White bars, wild type; black bars, arm mutant. n = 5 to 7 independent biological replicates (20–25 pooled plants) ± se. Asterisks indicate significant differences between genotypes at level <0.05. dw, Dry weight.
Positional Cloning of the arm Mutation
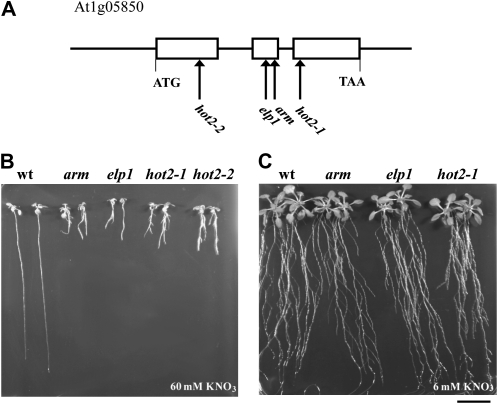
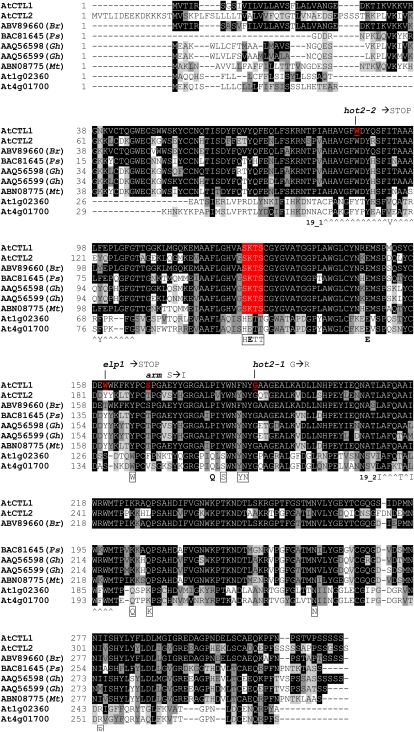
Prior to physiological and genetic characterization, homozygous M4 seeds were rescreened for inhibited PR length and increased LR density on high nitrate and then back-crossed five times to wild-type Col-0 to remove unlinked mutations caused by the chemical mutagenesis. The mutant phenotype in the F2 progeny for each back-cross segregated in a 3:1 (wild type:mutant) ratio (337:94, χ2 = 1.75 observed for the first cross), demonstrating the recessive monogenic nature of the mutation. A map-based cloning approach was used to identify the mutation responsible for the arm phenotype. An F2 population was generated from a cross between the arm mutant in the Col-0 background and wild-type Landsberg erecta (Ler-1). The DNA samples of plants showing the mutant phenotype on high nitrate were analyzed. An initial mapping procedure with 22 individuals indicated that the mutation was positioned at the top of chromosome 1 around the single sequence length polymorphism (SSLP) marker F21M12 (Supplemental Fig. S3). Higher-resolution mapping with about 500 plants narrowed the region to a 140-kb chromosomal section within BAC clones F3F20 and T20M3. That region contains At1g05850 (Fig. 7A), which encodes chitinase-like protein1 (CTL1). The first mutant allele pom1 isolated by Hauser et al. (1995) and other alleles, including elp1 (Zhong et al., 2002) and hot2-1 and hot2-2 (Kwon et al., 2007), phenocopy the arm mutant phenotype observed at high nitrate (Table I, Fig. 7B). A single nucleotide substitution G→T was found in the second exon of At1g05850 in the arm mutant (Fig. 7A), resulting in a change of Ser-168 into Ile (Fig. 8). Significantly, we could rescue the root phenotype of elp1 and hot2 mutant lines on moderate (6 mm) nitrate and 1% Suc medium (Fig. 7C). If AtCTL1 is responsible for the arm phenotype, it should be allelic to elp1 and hot2; indeed, the F1 progeny of arm crossed to either mutant exhibited the arm phenotype on high nitrate substrate, indicating that arm is allelic to elp1 and hot2 (data not shown).
Figure 7.
Positional cloning of arm mutation and root morphology of mutant alleles of AtCTL1 as a function of nitrate supply. A, A single nucleotide mutation in the second exon of AtCTL1 gene (At1g05850) was found in the arm mutant. B and C, Seedlings of the wild type (wt), arm, and previously described ctl1 alleles that include elp1 and hot2 were grown on high nitrate (60 mm KNO3) for 13 d (B) and moderate nitrate (6 mm KNO3) over 25 d (C). Bar = 2 cm.
Table I.
Description of allelic mutants of AtCTL1 and the nitrate concentrations in the growth medium used in these reports
| Mutant Allele | Phenotype | NO3− Concentration | Reference(s) |
|---|---|---|---|
| pom1-1 | Principal axis of root expansion is radial rather than longitudinal. Radial dimensions of epidermis and cortex cells increased. | 39 mm | Hauser et al. (1995) |
| erh2/pom1-12 | ectopic root hair2 mutant was isolated in a screen for identifying genes in the specification of epidermal cell fate in PR. More than one-third of its root hair in ectopic deposition. | 39 mm | Schneider et al. (1997) |
| Inability to respond to low doses of propyzamide (microtubule-binding drug). | 19.5 mm | Yuen et al. (2005) | |
| pom1-14, pom1-15 | Detected as revertants of the phytochrome phyB-1 mutation by restoring short length hypocotyls. | 39 mm | Reed et al. (1998) |
| pom1-2, pom1-21 | Shorter hypocotyls, severe reduction in cellulose synthesis. Partial revertant of the1-3 mutation (mutant allele of THESEUS1, a plasma-membrane-bound receptor-like kinase probably acting as a cell wall integrity censor). | 9 mm | Mouille et al. (2003); Hématy et al. (2007) |
| hot2-1, hot2-2 | sensitive to hot temperature2 mutants were isolated based on a lesser thermotolerance in a hypocotyl elongation assay. | 9 mm | Hong and Vierling (2000); Hong et al. (2003) |
| Hypersensitivity to salt. | 39 mm | Kwon et al. (2007) | |
| elp1 | ectopic deposition of lignin in pith1 mutant was identified in a screen for ectopic deposition of lignin in inflorescence stems. | 39 mm | Zhong et al. (2000, 2002) |
| 3G-12/pom1-22 | Isolated from a screen of callus for enhanced shoot development at suboptimal concentrations of cytokinin. | 12.5 mm | Cary et al. (2001) |
| pom1-26 | Isolated from a screen for ectopically lignification. | 39 mm | Rogers et al. (2005) |
Figure 8.
Multiple alignments of class II chitinases from Arabidopsis and other species. Comparison of AtCTL1-At1g05850 (chosen as consensus sequence), AtCTL2-At3g16920, ABV89660 (Brassica rapa), BAC81645 (Pisum sativum), GhCTL1-AAQ56598, GhCTL2-AAQ56599 (Gossypium hirsutum), ABN08775 (Medicago truncatula), and two other Arabidopsis class II chitinases, At1g02360 and At4g01700, using the alignment program VectorNTI. Gaps (-) are introduced to optimize alignment, and the degree of shading of boxes represents the level of similarity: identical residues (black), conservative (dark gray), weakly similar (light gray), and different (white). The “^” symbols indicate chitinase 19_1 signature C-x(4,5)-F-Y-[ST]-x(3)-[FY]-[LIVMF]-x-A-x(3)-[YF]-x(2)-F-[GSA] and 19_2 signature [LIVM]-[GSA]-F-x-[STAG](2)-[LIVMFY]-W-[FY]-W-[LIVM] (Passarinho and de Vries, 2002). Residues in bold are essential for catalytic activity and boxed residues putatively bind the substrate in class I chitinases (Hahn et al., 2000; Passarinho and de Vries, 2002). The putative secretion signal in AtCTL1 is underlined. The residue substitutions in arm and previously described (hot2-1, hot2-2, and elp1) mutants are indicated. [See online article for color version of this figure.]
Expression Profile of the CTL1 Gene and Chitinase Activity Test of Recombinant CTL1 Protein
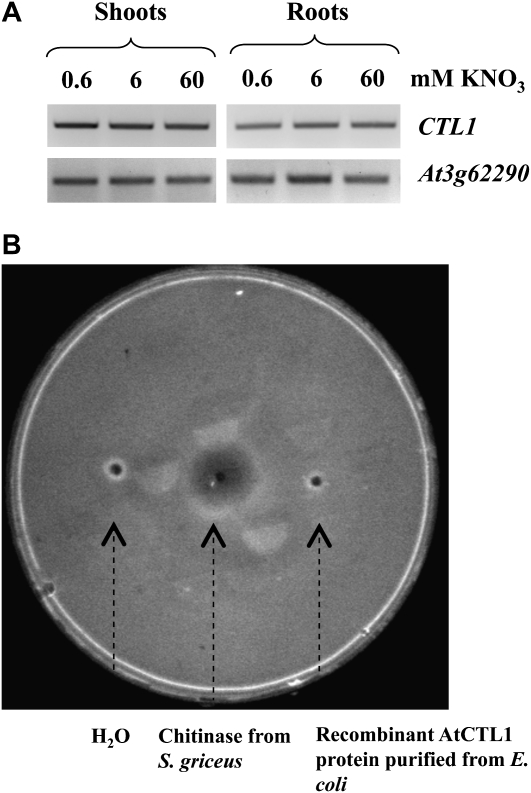
As a first step to dissecting the role of AtCTL1 in plant development, we analyzed the expression of this gene in response to nitrate availability. Reverse transcription-PCR was carried with cDNA isolated from root and shoot of wild-type plants grown on 0.6, 6, and 60 mm KNO3, and no significant difference in expression was documented (Fig. 9A). Likewise, no change in message abundance was observed for CTL1 in the arm mutant for all nitrate treatments (data not shown).
Figure 9.
Expression of the CTL1 gene and chitinase activity of CTL1 protein. A, Effect of nitrate supply on the expression of CTL1 gene in wild-type Arabidopsis: reverse transcription-PCR analysis of AtCTL1 transcript abundance in root and shoot tissues. Seedlings were grown at three different nitrate concentrations (0.6, 6, and 60 mm KNO3) for 13 d prior to harvest. Equal loading of cDNA sample in each lane was verified with the constitutive At3g62290 (ADP-RIBOSYLATION FACTOR A1E) gene. The experiment was duplicated, and similar results were obtained. B, Detection of chitinase activity in plate. Sterile distilled water, 10 μg chitinase from Streptimyces griceus, and 10 μg purified of recombinant CTL1 purified from E. coli were loaded into an agarose gel containing glycol chitin as substrate. After incubation and staining with calcofluor white M2R, the zone of clearance around the well left by the degradation of chitin by chitinase activity was detected under UV light.
AtCTL1 is designated as chitinase-like protein because it shows significant amino sequence similarity to a group of chitinases (GH19 glycoside hydrolases; Graham and Sticklen, 1994). However, among class II members of the chitinases family, AtCTL1 (together with AtCTL2) does not possess conserved amino acid residues that are essential for chitinase activity, in particular the H-E-T-T motif, which is replaced by S-K-T-S (Passarinho and de Vries, 2002; Fig. 8). The absence of these essential residues suggests CTL1 is not a chitinase. We tested recombinant CTL1 protein purified from Escherichia coli for chitinase activity in a simple plate assay but were unable to demonstrate any activity (Fig. 9B). This result is in agreement with comments from Zhong et al. (2002) who referred to unpublished data yielding a similar outcome.
Cell Elongation Defects in ctl1 Mutant Hypocotyls Are Not Conditional on Nitrate Abundance
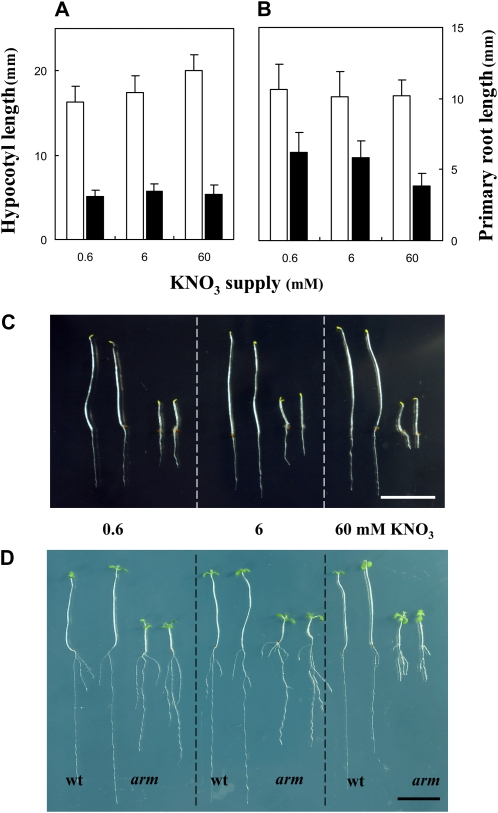
Short hypocotyls and cell elongation defects were previously reported in ctl1 mutant alleles (Reed et al., 1998; Mouille et al., 2003; Hématy et al., 2007; Table I). Given the impact of high nitrate on root cell swelling in the arm mutant, we wanted to know if the hypocotyl elongation defect is also conditional on the nitrate supply in the growth medium. To this end, we measured the hypocotyl length of seedlings germinated at three nitrate concentrations (0.6, 6, and 60 mm KNO3) in the dark. Five days after germination in the dark, wild-type seedlings had long hypocotyls (Fig. 10). Under the same conditions, the arm mutant had significantly shorter hypocotyls than the wild type, and the defect was observed across all nitrate concentrations (Fig. 10, A and C), whereas inhibition of PR growth was only observed on high nitrate (Fig. 10B). The phenomenon was still observed when dark-grown seedlings were returned to light (Fig. 10D).
Figure 10.
Effect of nitrate availability on hypocotyl and root length in dark-grown seedlings. Seedlings were grown vertically at three nitrate (KNO3) supplies for 5 d in darkness. A, Hypocotyl length. B, PR length n > 55 ± sd. White bars, wild type; black bars, arm mutant. All differences between genotypes are statistically significant at level P < 0.01. C, Picture of seedlings after 5 d growth in darkness. D, Picture of seedlings grown for 5 d in darkness and returned to light for 7 additional days. Bars = 1 cm. wt, Wild type. [See online article for color version of this figure.]
The Phenotype of Cellulose Biosynthesis Mutants Is Conditional on Nitrate Abundance
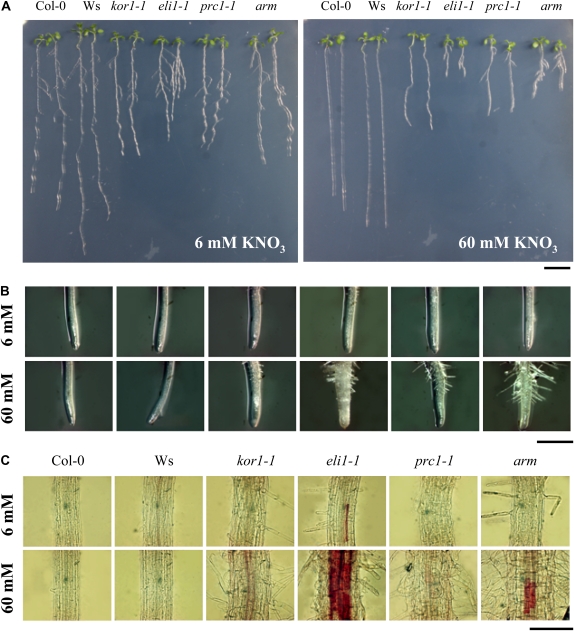
A direct role of CTL1 in cell wall synthesis seems likely because (1) a 75% reduction of cellulose synthesis was observed in pom1-21 mutant seedlings compared to the wild type (Mouille et al., 2003), (2) incomplete cell walls were observed in elp1 inflorescence stems (Zhong et al., 2002), and (3) AtCTL1 expression is highly coordinated with cellulose synthesis in primary walls, based on the expression patterns of cellulose synthases CESA1, CESA3, and CESA6 (Persson et al., 2005). Interestingly, mutations in CESA3 and CESA6 but also in KORRIGAN1 (KOR1)/RADIALLY SWOLLEN2, encoding a membrane-bound endo-1,4-β-d-glucanase, lead to similar reductions of PR growth and, to some extent, radial swelling as described in the arm mutant (Hauser et al., 1995; Fagard et al., 2000; Desprez et al., 2002; Cano-Delgado et al., 2003; Refregier et al., 2004; Hématy et al., 2007). Therefore, we tested the effect of nitrate availability in the growth medium on the CESA3 (eli1-1), CESA6 (prc1-1), and KOR1 (kor1-1) mutant phenotypes (Fig. 11, A and B). At moderate nitrate (6 mm KNO3) concentration, the mutants displayed slightly shorter PRs. The same conditional effects on root architecture in the presence of high nitrate (60 mm KNO3) as described for the arm mutant was observed in eli1-1 and prc1-1 mutants (Fig. 11; Supplemental Fig. S4). We note that despite a reduction in PR length, kor1-1 mutants did not elaborate LRs on high nitrate.
Figure 11.
Effect of nitrate supply on the root morphology and lignin deposition in cellulose synthesis mutants. Pictures of whole seedlings (A) and root tip (B) of wild-type Col-0, Wassilewskija (Ws), cellulose biosynthesis mutants (kor1-1 [KOR1]; eli1-1 [CESA3]; prc1-1 [CESA6]), and arm grown at two different nitrate concentrations (6 and 60 mm KNO3) for 13 d. All mutants are in the Col-0 background, except kor1-1, which is in Wassilewskija. C, Phloroglucinol staining of root tissues (1 cm above root tip) of seedlings grown in the same conditions. Bars = 1 cm in A, 0.5 mm in B, and 500 μm in C.
Another phenotype of ctl1 mutants was the ectopic deposition of lignin in the pith cells of stems and the root endodermis that would not normally accumulate this polymer (Zhong et al., 2000, 2002; Table I). Interestingly, reduction of cellulose synthesis by inhibitors or in the eli1-1 (cesa3) mutant background resulted in the accumulation of ectopic lignin deposition (Cano-Delgado et al., 2003; Rogers et al., 2005). Thus, we examined the conditional nature of the lignin deposition in roots on the nitrate availability. Lignin accumulation in roots of 13-d-old wild-type, arm, and cellulose synthesis mutant seedlings was visualized with phloroglucinol, which stains lignin a magenta color (Fig. 11C). Substantial evidence of lignin deposition was visible in endodermal cells of arm roots grown with high nitrate. However, no staining was observed in wild-type and arm roots grown at moderate levels of KNO3 (Fig. 11C), indicating that lignin deposition is also conditional on the nitrate supply. In the cellulose biosynthesis mutant eli1-1, nitrate-induced lignin deposition was more widespread than observed in the arm mutant (Fig. 11C).
DISCUSSION
Earlier Descriptions of Allelic Mutants of CTL1 and the Discovery of Permissive Growth Conditions
Chitin, a polymer of N-acetyl-d-glucosamine, is an elicitor of plant defense during fungal infection. In response to fungal attack, the plant activates chitinases that catalyze the hydrolysis of 1,4-β-linkages inside chitin in order to degrade the fungal cell wall and to limit invasion (Eckardt, 2008). At first, the finding in the present screen of the CTL1 gene involved in root development and nutritional signaling seems to be puzzling. However, numerous reports point to a broader spectrum of roles for GH19 glycoside hydrolase (chitinase) family than just in defense against pathogens. These roles are related to abiotic stress responses, as various as frost, heat, mineral deficiencies, heavy metals, or UV exposure (Lima et al., 2002; Kasprzewska, 2003; Bekesiova et al., 2008) but also to developmental processes (Passarinho and de Vries, 2002).
Key features of the root morphology of the arm mutant grown under restrictive conditions as described in this study include (1) inhibition of PR growth, (2) increased lateral branching, (3) radial swelling of the root tip and elongation zone, and (4) increased root hair density. Phenotypically, allelic mutants of AtCTL1 have reduced rosette growth and display abnormal patterns of root growth (Hauser et al., 1995; Schneider et al., 1997; Zhong et al., 2002; Yuen et al., 2005; Kwon et al., 2007; summarized in Table I). The insight of the results reported here is the finding of permissive growth conditions that allow normal root development of ctl1 mutants (Figs. 2 and 7C). It is noteworthy that relatively high nitrate and ammonium concentrations were used in the majority of the previous descriptions of ctl1 mutants (Table I), and those studies using a relatively low nitrate concentration focused primarily on the hypocotyl and not the root phenotype (Mouille et al., 2003; Hématy et al., 2007). In certain cases, the ctl1 mutant phenotype was conditional on high Suc in the growth medium (Hauser et al., 1995; Supplemental Fig. S2). Low Suc blocked the abnormal radial expansion of the PR but not completely the inhibition of root length, something that Zhong et al. (2002) reported to some extent with inhibitors of ethylene perception. Here, we showed that several features of ctl1 mutants, such as the promotion of LR proliferation, radial swelling, high root hair density (Figs. 1 and 2) and lignin deposition (Fig. 11), are conditional on the environmental conditions (such as high NO3−, Cl−, and Suc). Therefore, some earlier conclusions regarding CLT1's role in cell wall formation and lignin biosynthesis at least in roots (Table I) may need to be revised.
AtCTL1 Plays a Role in Plant Growth and Development
We propose that AtCTL1 plays a role in plant growth and development wherein (1) it has a constitutive effect on cell wall synthesis and cell elongation and (2) its function is modulated by environmental stimuli that alter organ morphology.
(1) Slight inhibition of PR length under permissive conditions implicates CTL1 in primary cell wall synthesis and cell elongation. Hauser et al. (1995) showed that the principal axis of expansion is radial rather than longitudinal in pom1 and that the radial dimension is augmented in epidermis and cortex cells but decreased in endodermis cells (see also Fig. 11). While not as dramatic as ctl1-dependent changes in the presence of high nitrate or chloride, there is a measurable decrease in PR elongation under permissive conditions (Figs. 1 and 2). Cell wall remodeling during elongation requires cell wall-modifying enzymes, including those involved in the depolymerization of 1,4-β-linkages in the preexisting matrix (Cosgrove, 2005; Somerville, 2006). AtCTL1 could play a role in this process if it possesses such a catalytic activity (see later in “Discussion”). Furthermore, CTL1 orthologs in various plant species have been shown to be essential for cell wall structure and cellulose synthesis in both primary and secondary cell walls (Mouille et al., 2003; Aspeborg et al., 2005; Persson et al., 2005; Geisler-Lee et al., 2006). Nevertheless, the exact role of CTL1 in defining the physical properties of the wall remains to be determined.
(2) The ctl1 mutants described here and in previous screens (Table I) have a conditional impact on root architecture and defects in cell elongation in hypocotyls triggered by darkness. If these were constitutive phenotypes expressed under all conditions and in every organ, it would be easy to ascribe this effect to a direct role in the enzymology of cell wall synthesis or remodeling. However, we have demonstrated here that the dramatic effect on root morphology is inducible by a variety of environmental factors, including high levels of anions and Suc. One might suggest osmotic potential is the unifying variable, yet significant changes in root morphology are not induced when roots are transferred onto media containing high concentrations of sorbitol or mannitol, where comparable levels of nitrate and chloride still have a striking impact (Figs. 4 and 5). It is noteworthy that 30 mm K2SO4 has no effect on root morphology in contrast to decreased root growth in the presence of equivalent concentrations of osmotically active ions in Ca(NO3)2. This observation, in conjunction with the other ion pairs tested in Figure 4, suggests ion concentrations and electrostatic interactions in the cell wall are not responsible for the conditional phenotype. Multiple environmental variables have been shown to induce the ctl1 phenotype, including high nitrate and chloride (Figs. 1, 2, and 4; Table I), high salt (Kwon et al., 2007), and high temperature (Hong et al., 2003; hot2, perhaps also linked to high chloride). In addition, high Suc can induce the ctl1 phenotype (Fig. 3; Supplemental Fig. S2; Hauser et al., 1995). Taken together, these data show that CTL1 is part of multiple response pathways that alter root morphology.
The morphology of wild-type roots grown under a variety of environmental conditions that induce the arm phenotype described here includes a single long PR with the inhibition of LR primordia development at postemergence (Fig. 2C; Malamy and Ryan, 2001; Signora et al., 2001; Liang and Harris, 2005; Qi et al., 2007). Since the inhibition of LR development under nonpermissive conditions does not occur in ctl1 mutants, even in loss-of-function alleles (hot2-2 and elp1), we interpret that as a removal of a repressor. In the context of a biochemical activity, nonpermissive conditions (such as high nitrate or chloride) in wild-type roots might rely on CTL1-dependent stiffening of the cell wall to prevent LR emergence. Likewise, in the wild-type PR, CTL1-dependent stiffening of specific regions of the cell wall could lead to cell elongation versus radial swelling (as seen in the ctl1 mutants). Thus, the conditional role of CTL1 in changing root architecture is also linked to cell wall synthesis/remodeling and cell elongation. Mutations in some cellulose synthase subunits (CESA3 and CESA6) phenocopy the arm mutant under nonpermissive conditions, suggesting their activity is also altered in response to environmental conditions. This is not too surprising assuming that regulation of the cell wall structure plays a pivotal role in controlling cell shape and, ultimately, root morphology.
Enzymatic Function of CTL1
A key question regarding CTL1's role in altering root growth and development in the context of the diversity of mutant phenotypes, e.g. hypersensitivity to salt, decreased thermotolerance, reduced synthesis of cellulose, and ectopic deposition of lignin in stems and roots (Table I), is determining the enzymatic function of this protein. On first inspection, AtCTL1 belongs to the class II family of chitinases that lack a Cys-rich lectin domain in the N-terminal region of the protein that has a role in chitin binding (Meins et al., 1994; Passarinho and de Vries, 2002). AtCTL1 also has a putative secretion sequence at the N terminus, suggesting that the protein may be secreted into the cell wall compartment (Zhong et al., 2002; Kwon et al., 2007). According to Hahn et al. (2000), two Glu residues are required for catalysis in the GH19 family. The first one, within the H-E-T-T active site (Glu-127 replaced by Lys-127 in AtCTL1), acts as a general acid, and the second one (Glu-149 in AtCTL1) functions as a base to activate a water molecule for the single-step displacement reaction. The acid moiety of the catalytic site is highly conserved among chitinases between species (Bishop et al., 2000). That residue is replaced by Lys-127 in AtCTL1 as well as in all of the CTL1 ortholog proteins (Aspeborg et al., 2005; Fig. 8). Taken together, these data strongly suggest that CTL1 is not a functional chitinase and that conclusion is supported by the absence of measurable chitinase activity in recombinant CTL1 (Fig. 9B). To date, no endogenous substrates have been established for CTL1.
CONCLUSION
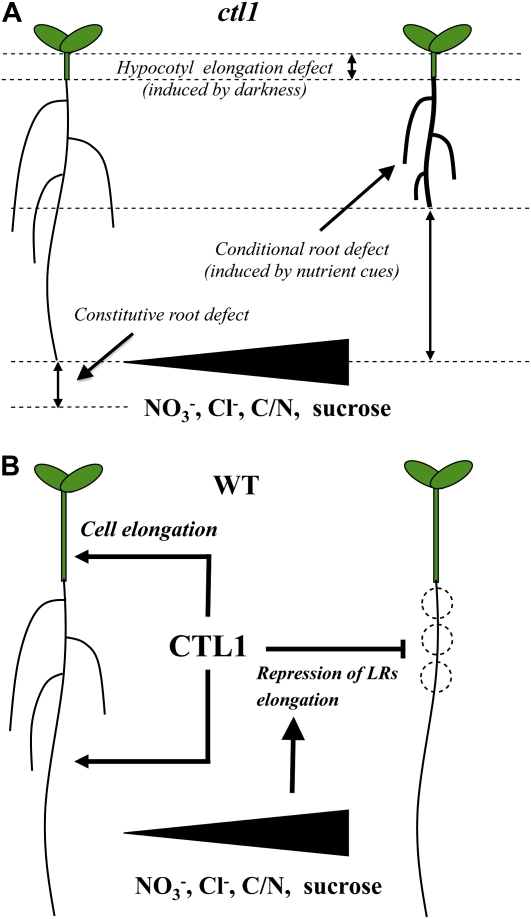
In the results reported here, we show that CTL1 plays a role in plant growth and development (Fig. 12). It is a constitutive player in cell elongation, as demonstrated by a decrease in PR growth under permissive conditions. Additionally, CTL1 functions as a component of multiple response pathways that control developmental plasticity in root morphology in response to environmental stimuli. The cell elongation defects in hypocotyls (e.g. triggered by darkness) and the conditional impact on root architecture (e.g. in the presence of high concentrations of monovalent anions and Suc) appear to be driven by CTL1-mediated changes in cell wall structure. Since CTL1 is constitutively expressed across a range of permissive and restrictive conditions, we hypothesize that CTL1-mediated regulation is not dependent on changes in CTL1 protein abundance and is probably a function of differentially regulated enzymatic activity.
Figure 12.
Proposed model of CTL1 role in plant growth and development. A, Summary scheme of the ctl1/arm phenotype. Mutations in the chitinase-like CTL1 gene in Arabidopsis result in cell elongation defects in the hypocotyls (induced by darkness) and in roots (upon permissive and restrictive conditions). While the elongation defect in the PR is constitutive, dramatic changes in root architecture are conditional on anion (nitrate and chloride) supply, high carbon/nitrogen (C/N) ratio, and Suc. The conditional root phenotype includes LR elongation, radial swelling, and increased root hair density. B, Suggested role of CTL1 in wild-type (WT) seedlings: (1) it plays a role in cell wall synthesis/structure and cell elongation in various organs, and (2) it is a component of multiple environmental response pathways (probably involving cellulose synthase subunits CESA3 and CESA6) that influence root morphology by altering cell wall rigidity. [See online article for color version of this figure.]
MATERIALS AND METHODS
Mutant Isolation and Plant Growth Conditions
M2 ethyl methanesulfonate-mutagenized seeds (Col-0) were purchased from Lehle Seeds. Seeds were surface sterilized with ethanol 70% (v/v) during 10 min and in a 20% (v/v) HClO during 5 min. They were plated on 1× Murashige and Skoog strength with the nitrogen concentration modified to 60 mm KNO3, 1% Suc, and 0.8% agar. Seeds were stratified at 4°C for 2 d in the dark and then incubated vertically in a culture chamber at a temperature of 22°C and a daylight regime of 16 h (75 μmol photons m−2 s−1)/8 h darkness. Thirteen-day-old seedlings grown vertically were then screened for the presence of LRs at high nitrate concentration.
Genetic Mapping
A total of 480 F2 from a cross between arm in the Col-0 background and wild-type Ler-1 and showing the mutant phenotype at high nitrate were used to map the mutation. DNA was isolated from each plant and analyzed for recombination events with SSLP markers and cleaved-amplified polymorphic sequence markers. Primer pairs for newly found SSLP markers between Col-0 and Ler-1 were designed during this mapping process: CSU01, forward, 5′-TTCCAAATACGATTTCAGGTGAG-3′; reverse, 5′-CAAGTGACGGGATCCAAAAT-3′; CSU02, 5′-CTGCTCTCATGCCTCTCTTG-3′; reverse, 5′-AATTAACTTTTAGTGATGTGACAGGAA-3; CSU03, forward, 5′-TGTTCGCATAGCTTTTGCTG-3′; reverse, 5′-ACAATCCACAGGACGGTGAG-3′; and CSU04, forward, 5′-ATTGTAAAGCAAGCGACTGGA-3′; reverse, 5′-GAAAATCTTAACACCACTTTCAGATA-3′. The description of other registered markers is available from The Arabidopsis Information Resource (www.arabidopsis.org).
Mineral Analysis
Seedlings vertically grown in petri dishes for 13 d were harvested and dried at 80°C prior to mineral analysis. Nitrate content was measured with a Flow Solution 3000 (OI Analytical) using cadmium reduction. For other mineral analyses, samples were digested with nitric acid and assayed by inductively coupled plasma mass spectrometry at the Purdue Ionomics Information Management Systems, Purdue University.
Cloning of CTL1 Gene and Expression in Escherichia coli
The CTL1 gene (At1g05850) was amplified using the primers 5′-TAGTTGTAAGCCATGGTGACAATCAGGAGT-3′ (forward) and 5′-GATATATCATAAGGATCCCGAAGAGGAAGA-3′ (reverse), which contained NcoI and BamHI restriction sites (underlined). Easy A High Fidelity Enzyme (Stratagene) was used according to the manufacturer's protocol. The 1,100-bp fragment was gel purified, digested with NcoI and BamHI, and ligated into pET32a(+) vector. In this vector, CTL1 is expressed as a thioredoxin fusion protein. The resulting plasmid was transformed into E. coli Rosetta2 competent cells (Novagen) to construct recombinant plasmid. For protein expression, one colony was grown overnight in Luria-Bertani medium containing ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL) at 37°C with vigorous shaking. The overnight culture was diluted (1:100) into fresh Luria-Bertani medium containing ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL) and grown at 37°C with vigorous shaking until OD550 was approximately 0.6 to 0.8. The recombinant protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside (0.5 mm) for 4 h, with vigorous shaking, at room temperature. The cells were harvested by centrifugation at 3,000g for 10 min at 4°C and frozen at −80°C.
Purification of Recombinant CTL1 Protein
The purification of the recombinant protein was as described by Kirubakaran and Sakthivel (2007), except that the dialysis was performed progressively at 6, 4, 2, and 1 m of urea. After renaturation of the protein from inclusion bodies, a batch purification of 6xHis-tagged proteins under native conditions was performed using Ni-NTA agarose (Qiagen) following the manufacturer's protocol (Qiagen). Protein analysis was performed on 12% SDS-PAGE (Bio-Rad). Protein quantification was done following the Bradford method (Bradford, 1976).
Chitinase Activity Assay
Glycol chitin was obtained by acetylation of glycol chitosan (Sigma-Aldrich) after the method of Trudel and Asselin (1989). Glycol chitin (0.01%) was mixed with melted 0.8% agarose and poured into a petri plate. After solidification, 20 μL of sterile distilled water, 10 μg of chitinase from Streptomyces griceus (Sigma-Aldrich), and 10 μg of recombinant CTL1 were pipetted into separate wells in the plate. Plates were incubated at 37°C for 2 h, stained with 0.01% of Calcofluor White M2R (Sigma-Aldrich), washed with distilled water, and visualized under UV light to detected the zone of clearance around the well left by the degradation of chitin by chitinase activity (Anil et al., 2007).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At1g05850.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Physiological characterization of wild-type and arm plants.
Supplemental Figure S2. Effect of different combinations of nitrate and Suc on root system architecture of the wild type and arm mutant.
Supplemental Figure S3. Positional cloning of arm mutation.
Supplemental Figure S4. Root architecture parameters of the wild type, cellulose synthesis mutants, and arm plants.
Supplementary Material
Acknowledgments
We thank S.-W. Hong for sharing hot2 seeds, Z.-H. Ye for elp1, and H. Höfte for kor, eli1-1, and e112.
This work is supported by a grant from the U.S. National Science Foundation to D.R.B. and from the Belgian Science Policy Office (BelSPo project IAPVI/33) to N.V. C.H. is currently a postdoctoral fellow of the Fonds National de la Recherche Scientifique and previously of the Federal Belgian Science Policy (BelSPo, Return grant).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Daniel R. Bush (dbush@colostate.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Anil K, Seshagirirao K, Podile AR (2007) A simple, rapid and yet less expensive method to detect chitinase in agarose plates. J Biochem Biophys Methods 70 683–684 [DOI] [PubMed] [Google Scholar]
- Aspeborg H, Schrader J, Coutinho PM, Stam M, Kallas A, Djerbi S, Nilsson P, Denman S, Amini B, Sterky F, et al (2005) Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol 137 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekesiova B, Hraska S, Libantova J, Moravcikova J, Matusikova I (2008) Heavy-metal stress induced accumulation of chitinase isoforms in plants. Mol Biol Rep 35 579–588 [DOI] [PubMed] [Google Scholar]
- Bishop JG, Dean AM, Mitchell-Olds T (2000) Rapid evolution in plant chitinases: molecular targets of selection in plant pathogen coevolution. Proc Natl Acad Sci USA 97 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34 351–362 [DOI] [PubMed] [Google Scholar]
- Cary A, Uttamchandani SJ, Smets R, Van Onckelen HA, Howell SH (2001) Arabidopsis mutants with increased organ regeneration in tissue culture are more competent to respond to hormonal signals. Planta 213 700–707 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8 165–171 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1999) Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol 50 391–417 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6 850–861 [DOI] [PubMed] [Google Scholar]
- Desprez T, Vemhettes S, Fagard M, Refregier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt NA (2008) Chitin signaling in plants: insights into the perception of fungal pathogens and rhizobacterial symbionts. Plant Cell 20 241–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunneras S, et al (2006) Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol 140 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutiérrez GA, Coruzzi G, Birnbaum K (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A, Nacry P, Davidian JC (2009) Root uptake regulation: a central process for NPS homeostasis in plants. Curr Opin Plant Biol 12 328–338 [DOI] [PubMed] [Google Scholar]
- Graham LS, Sticklen MB (1994) Plant chitinases. Can J Bot 72 1057–1083 [Google Scholar]
- Hahn M, Hennig M, Schlesier B, Höhne W (2000) Structure of jack bean chitinase. Acta Crystallogr D Biol Crystallogr 56 1096–1099 [DOI] [PubMed] [Google Scholar]
- Hauser MT, Morikami A, Benfey P (1995) Conditional root expansion mutants of Arabidopsis. Development 121 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, Pelletier S, Renou JP, Höfte H (2007) A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol 17 922–931 [DOI] [PubMed] [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11 610–617 [DOI] [PubMed] [Google Scholar]
- Hong SW, Lee U, Vierling E (2003) Arabidopsis hot mutants define multiple functions required for acclimation to high temperature. Plant Physiol 132 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A, Simpson J, Herrera-Estrella L, Benfey PN (2009) Functional genomics of root growth and development in Arabidopsis. Curr Opin Plant Biol 12 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzewska A (2003) Plant chitinases-regulation and function. Cell Mol Biol Lett 8 809–824 [PubMed] [Google Scholar]
- Kirubakaran SI, Sakthivel N (2007) Cloning and overexpression of antifungal barley chitinase gene in Escherichia coli. Protein Expr Purif 52 159–166 [DOI] [PubMed] [Google Scholar]
- Kwon YR, Kim SH, Jung MS, Kim MS, Oh JE, Ju HW, Kim K, Vierling E, Lee H, Hong SW (2007) Arabidopsis hot2 encodes an endochitinase-like protein that is essential for tolerance to heat salt and drought stresses. Plant J 49 184–193 [DOI] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, Guerinot ML, Harper JF, Ward JM, McIntyre L, et al (2003) Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol 21 1215–1221 [DOI] [PubMed] [Google Scholar]
- Liang Y, Harris JM (2005) Response of root branching to abscisic acid is correlated with nodule formation both in legumes and nonlegumes. Am J Bot 92 1675–1683 [DOI] [PubMed] [Google Scholar]
- Lima VM, Magioli C, B de A Gerhardt L, Tarré E, Menezes RMG, Sachetto-Martins G, Margis-Pinheiro M (2002) Bean class IV chitinase promoter is modulated during plant development and under abiotic stress. Physiol Plant 116 512–521 [Google Scholar]
- Little DY, Rao H, Olivia S, Daniel-Vedele F, Krapp A, Malamy JE (2005) The putative high nitrate transporter NRT21 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA 102 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6 280–287 [DOI] [PubMed] [Google Scholar]
- MacGregor DR, Deak KI, Ingram PA, Malamy JE (2008) Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 20 2643–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Ryan S (2001) Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol 127 899–909 [PMC free article] [PubMed] [Google Scholar]
- Meins F, Fritig B, Linthorst HJM, Mikkelsen JD, Neuhaus JM, Ryals J (1994) Plant chitinase genes. Plant Mol Biol Rep 12 22–28 [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM (2007) Nitrate transport and signaling. J Exp Bot 58 2297–2306 [DOI] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. Plant J 35 393–404 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC (2008) Branching out in new directions: the control of root architecture by lateral root formation. New Phytol 179 595–614 [DOI] [PubMed] [Google Scholar]
- Passarinho PA, de Vries SC (2002) Arabidopsis chitinases: a genomic survey. In The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/101199/tab0023, http://www.aspb.org/publications/arabidopsis/
- Péret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, Beeckman T, Bennett M (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 114 399–408 [DOI] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Sommerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarrays data sets. Proc Natl Acad Sci USA 102 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Wu Z, Li J, Mo X, Wu S, Chu J, Wu P (2007) AtCYT1-INV1, a neutral invertase, is involved in osmotic stress-induced inhibition of lateral root growth in Arabidopsis. Plant Mol Biol 64 575–587 [DOI] [PubMed] [Google Scholar]
- Reed JW, Elumalai RP, Chory J (1998) Suppressors of an Arabidopsis thaliana phyB mutation identity genes that control light signaling and hypocotyls elongation. Genetics 148 1295–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refregier G, Pelletier S, Jaillard D, Höfte H (2004) Interaction between wall deposition and cell elongation in dark-grown hypocotyls cells in Arabidopsis. Plant Physiol 135 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A (2006) A central role for the nitrate transporter NRT21 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LA, Dubos C, Surman C, Willment J, Cullis IF, Mansfield SD, Campbell MM (2005) Comparison of lignin deposition in three ectopic lignification mutants. New Phytol 168 123–140 [DOI] [PubMed] [Google Scholar]
- Schneider K, Wells B, Dolan L, Roberts K (1997) Structural and genetic analysis of epidermal cell differentiation in Arabidopsis primary roots. Development 124 1789–1798 [DOI] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28 655–662 [DOI] [PubMed] [Google Scholar]
- Somerville CR (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol 22 53–78 [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H (2004) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45 1053–1062 [DOI] [PubMed] [Google Scholar]
- Tranbarger TJ, Al-Ghazi Y, Muller B, Teyssendier de la Serve B, Doumas P, Touraine B (2003) Transcription factor genes with expression correlated to nitrate-related root plasticity of Arabidopsis thaliana. Plant Cell Environ 26 459–469 [Google Scholar]
- Trudel J, Asselin A (1989) Detection of chitinase activity after gel electrophoresis. Anal Biochem 178 362–366 [DOI] [PubMed] [Google Scholar]
- Yuen CYL, Sedbrook JC, Perrin RM, Carroll KL, Masson PH (2005) Loss-of-function mutations of ROOT HAIR DEFECTIVE3 suppress root waving skewing and epidermal cell file rotation in Arabidopsis. Plant Physiol 138 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51 51–59 [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG (1999) Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA 96 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Kays SJ, Schroeder BP, Ye ZH (2002) Mutation of a chitinase-like gene causes ectopic deposition of lignin aberrant cell shapes and overproduction of ethylene. Plant Cell 14 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ripperger A, Ye ZH (2000) Ectopic deposition of lignin in the pith of stems of two Arabidopsis mutants. Plant Physiol 123 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.