Abstract
The housefly, Musca domestica, is an excellent model system to study the diversification of the pathway that specifies the sexual fate. A number of different mechanisms have been described in the housefly, which reflects in part the broad diversity of sex-determining strategies used in insects. In this study we present the molecular identification and characterization of F, which acts as the master switch in the housefly pathway. We provide evidence that F corresponds to the transformer ortholog in Musca (Mdtra), which, as a result of alternative processing, expresses functional products only in individuals committed to the female fate. We demonstrate that, once activated, a self-sustaining feedback loop will maintain the female-promoting functions of Mdtra. Absence of Mdtra transcripts in eggs of Arrhenogenic (Ag) mutant females suggests that maternally deployed Mdtra activity initiates this self-sustaining loop in the zygote. When an M factor is paternally transmitted to the zygote, the establishment of the loop is prevented at an early stage before cellularization and splicing of Mdtra shifts irreversibly to the male nonproductive mode. On the basis of the analysis of two mutant alleles we can explain the different sex-determining systems in the housefly largely as deviations at the level of Mdtra regulation. This plasticity in the housefly pathway may provide a suitable framework to understand the evolution of sex-determining mechanisms in other insect species. For instance, while sex determination in a close relative, the tsetse fly Glossina morsitans, differs at the level of the instructive signal, we find that its tra ortholog, Gmtra, is regulated in a mode similar to that of Mdtra.
PROPER sexual development is based on a binary decision between two alternative developmental programs. In insects, the genetic system underlying this decision has been most extensively studied in Drosophila melanogaster, providing profound insights in the molecular mechanisms that determine the sexual fate (Cline and Meyer 1996; Schutt and Nothiger 2000). Nevertheless, it appears that in many other insect species the primary instructive signal that specifies the sexual fate has diversified extensively. Alone in the housefly, Musca domestica, several types have been described in natural populations, ranging from dominant male determiners to female determiners and even the use of maternal signals (Dubendorfer et al. 2002). Since this spectrum, to a certain extent, reflects the variety of sex-determining signals found in insects, the Musca system appears particularly suited for studying evolutionary diversification of this key developmental process.
The “standard” type of sex determination in the housefly employs a dominant male-determining factor, M, which is located on the Y chromosome (Hiroyoshi 1964). In addition, naturally occurring strains exist where the M factor can be located on any of the five autosomes or even on the X chromosome (Denholm et al. 1983; Inoue et al. 1983). In some populations, all individuals are homozygous for the M factor, and the female fate is determined by the presence of a dominant female determiner FD (McDonald et al. 1978). Even more remarkably, the sex of the housefly can be determined by the maternal genotype. Such strains consist of arrhenogenic females (Ag/+) that produce only sons and of thelygenic females (+/+) that give rise to daughters only (Inoue and Hiroyoshi 1981). Our genetic analysis revealed that all these different systems have a common genetic basis and led us to propose the following model: In M. domestica the gene F acts as the key switch in sex determination. An active F is conceived as a female signal, whereas male development follows when F is inactive. Zygotic activation of F requires its own maternal activity, suggesting that this gene relies on a self-sustaining feedback loop to maintain its female-promoting function (Dubendorfer et al. 2002). The presence of a dominant male-determining factor M in the zygote prevents the activation of F, thereby promoting male development. The F factor is genetically defined by two alleles. The naturally occurring dominant female-determining allele FD is thought to act as a gain-of-function mutation, which is resistant to repression by M and no longer relies on the autoregulatory function (Dubendorfer and Hediger 1998). The recessive allele Fman, which spontaneously arose in a laboratory strain, acts as a strong hypomorphic mutation resulting in male development of homozygous Fman animals in the absence of the male-promoting M factor (Schmidt et al. 1997a).
The pivotal position of F at the top of the sex-determining hierarchy in Musca, and its function in selecting and maintaining the female fate through a positive feedback loop, resembles that of Sex-lethal (Sxl) in Drosophila. However, we previously showed that the Musca homolog of Sxl, MdSxl, is not sex-specifically expressed and thus an unlikely candidate for F (Meise et al. 1998). Studies in the fruit fly Ceratitis capitata identified the transformer ortholog, Cctra, as the main switch in the pathway that determines the sexual fate (Pane et al. 2002). Selection and maintenance of the female fate is based on a positive autoregulatory function of Cctra. The same mechanism might also be operational in other members of the Tephritidae (Lagos et al. 2007; Ruiz et al. 2007) and in Lucilia cuprina, a member of the Calliphoridae family (Concha and Scott 2009). Since Musca is phylogenetically more closely related to Tephritidae and Calliphoridae, transformer seems to be a more likely candidate for F.
In a previous study we showed that the Musca homolog of doublesex, Mddsx, acts as a main effector in the pathway downstream of F (Hediger et al. 2004). It produces a set of sex-specific protein isoforms that functionally correspond to the dsx variants in Drosophila. Sex-specific regulation of Mddsx is achieved at the level of splicing by a mechanism similar to that of dsx in Drosophila. The Musca homolog of Drosophila transformer2, Mdtra2, was proposed not only to participate as an essential cofactor in the regulation of Mddsx, but also to act as an upstream regulator of F on the basis of the finding that the dominant FD allele is largely resistant to Mdtra2 silencing (Burghardt et al. 2005). In F+/F+ females, on the other hand, transient silencing of Mdtra2 causes a complete and irreversible shift to male development, indicating that Mdtra2 is required for upholding the self-sustaining feedback loop of F (Burghardt et al. 2005). This situation is reminiscent of that in Ceratitis, where Salvemini and co-workers (Salvemini et al. 2009) recently reported that Cctra2 is required to maintain the productive female-specific splicing mode of Cctra. The presence of multiple clusters of putative TRA/TRA2 binding sites, intronic splice silencers (ISS), and RBP1 binding sites in the sex-specifically processed region suggested that CcTRA and CcTRA2 form a complex that directly associates with Cctra pre-mRNA to impose the female splice (Salvemini et al. 2009). Clustering of such putative binding sites was also previously observed in tra orthologs of other tephritids (Ruiz et al. 2007).
Here we present the molecular identification of the Musca transformer ortholog, Mdtra, and present evidence that this gene indeed corresponds to the key switch F. In addition, we isolated the tra ortholog in the close relative, the tsetse fly Glossina morsitans (Gmtra). In G. morsitans the instructive signal appears to be different from that in M. domestica. Although standard sex determination in the tsetse fly is also based on XX–XY male heterogamety, the Y chromosome does not have a sex-determining function. From the study of aneuploid sets of sex chromosomes it has been inferred that the sexual phenotype is based on the ratio of X to autosomes, as in Drosophila (Maudlin 1979). Nevertheless, sex-specific processing of Gmtra shares several features with Mdtra that are not found in Drosophila tra. Transgenic expression of the female variant of Mdtra is sufficient to activate and maintain female expression of endogenous Mdtra, invoking an autoregulatory function that serves to maintain the female-promoting functions of this gene. We show that the establishment of this feedback loop in the early zygote requires maternally deployed activity of Mdtra and that presence of M causes this loop to collapse shortly after fertilization. On the basis of small lesions found in the two known alleles of F we propose a model of how splicing regulation of Mdtra is achieved at the molecular level.
MATERIALS AND METHODS
Isolation of Mdtra:
Genomic DNA was isolated from two to three adult flies according to standard procedures. A “touchdown” PCR was performed using the degenerate primer pair Mddsx-70 (5′-NNN NTC ATC AAT CAA CA-3′) and Mddsx-69 (5′-NNN NTG TTG ATT GTT GT-3′). These primers were designed according to putative TRA/TRA2 binding sites found in Mddsx. The following concentrations and conditions were used for the PCR: 500 ng genomic DNA, 300 μm primer each, 10 mm dNTP each, and 25 mm Mg2+ in a total volume of 50 μl; denaturation at 94° for 2 min, followed by 16 cycles of 94° denaturation for 50 sec, annealing for 90 sec starting from 57° and then decreasing 1° every cycle to a touchdown of 41°, and extension at 72° for 2.5 min; and the subsequent 10 cycles were denaturation at 94° for 50 sec, annealing at 41° for 50 sec, and extension at 72° for 2.5 min; and finally extension at 72° for 5 min. Subcloning and sequencing of the candidate fragments were carried out by standard procedures.
To retrieve full-length transcripts 5′ and 3′ RACEs were performed using the BD SMART RACE cDNA amplification kit of Clontech or the 5′/3′ RACE kit from Boehringer Mannheim (Mannheim, Germany). The Expand long template system from Roche was used to isolate long genomic fragments according to the manufacturer's protocols. The genomic sequence has been submitted to GenBank (accession no. GU070694).
Isolation of Gmtra:
Searching the G. morsitans database (http://www.sanger.ac.uk/Projects/G_morsitans/) with the TRA-CAM domain identified a fragment (GMsg-2558) that showed sequence similarity in the 3′ part of the TRA-CAM domain and contained one putative TRA/TRA2 binding site. PCR on genomic DNA and 5′/3′ RACEs on RNA isolated from adult flies led to the identification of additional sequences spanning the entire Gmtra region. The Gmtra splice pattern was defined using standard PCR conditions and the gene-specific primers Gmtra-5 (5′-ACAGGTACATTGCAGTAGCTG-3′), Gmtra-13 (5′-CTTTACACAACAACGTGCCC-3′), Gmtra-3B (5′-TTTGCGCCAACGCATTCTG-3′), and Gmtra-18B (5′-TTAGCTTATAATTAGGTTTGGGG-3′). The genomic sequence has been submitted to GenBank (accession no. GU070695).
Isolation of Md-l(3)73Ah:
The degenerate primer pair F2 and R2, designed according to the D. melanogaster sequence of l(3)73Ah, were used to isolate part of the Musca homolog Md-l(3)73Ah. The sense primer F2 (5′-GAR TGS STS CAY ACS TTY TG-3′) is located at the end of exon 2, whereas the antisense primer R2 (5′-TTS ARS GTR TGS TCY TTS CC-3′) is located near the end of exon 5, which enabled us to isolate a 500-bp cDNA fragment of the 3′ part of the Musca-l(3)73Ah using standard PCR conditions.
RNA expression analysis:
Total RNA was either extracted from single adult flies (Musca or Glossina) according to the RNeasy Mini protocol of QIAGEN (Valencia, CA) or from 200 mg of adult flies, larvae, or embryos using the TRI REAGENT (Sigma, St. Louis) protocol. RT–PCR analysis was performed using the Enhanced Avian HS RT–PCR kit of Sigma and standard protocols for PCR reactions. The Mdtra splice pattern was defined using standard PCR conditions and the gene-specific primers Mdtra-16s, Mdtra-34as, and Mdtra-35as. Analyses for male-specific transcripts of the Mdtra gene were made using the primer pair Mdtra-16s and Mdtra-36as or Mdtra-12Bs and Mdtra-20as. Analyses for female-specific transcripts of the Mdtra gene were made using the primer pair Mdtra-16s and Mdtra-33as or Mdtra-9s and Mdtra-24as. For more information about all primers used to analyze the Mdtra splice pattern see Table S1.
Transcripts of the Mddsx gene were amplified using primers in the common exon (Mddsx-6s, 5′-CTAAAAGATGCCGGTGTTGAC-3′) and in the female-specific (Mddsx-11as, 5′-TGCAAGCACATTCATGTTTTG-3′) or the male-specific (Mddsx-46as, 5′-CCGCTGCACTTGCCGAC-3′) exon, respectively. Control transcripts of the Mdtra2 gene were amplified using the primer pair Mdtra2-16 (5′-TTGCTTGAGTTGCCTGCTGCATA-3′) and Mdtra2-9 (5′-CGTCCCCTGTAAACACCTGGG-3′). Control transcripts of the CYP6D3 gene were amplified using the primer pair CYP6D3-1 (5′-GTTCGGTAATATTTGGCTTGG-3′) and CYP6D3-2 (5′-CCCGTATTCCGTAGTTGAATT).
Musca strains and crosses:
Strains were reared as described previously (Schmidt et al. 1997a; Hediger et al. 2004). The strains were as follows: (1) wild-type strain, females XX; +/+, males XY; +/+; (2) autosomal M strain A, females XX; pw bwb/pw bwb, males XX; M pw+ bwb+/pw bwb; (3) autosomal M strain B, females XX; pw bwb w/pw bwb w, males XX; M pw+ bwb+ w/pw bwb w; (4) FD/Fman strain, females XX; FD Ba/Fman Ba+, males XX; Fman Ba+/Fman Ba+; (5) Fman strain, females XX; Fman Ba+/F+ Ba, males XX; Fman Ba+/Fman Ba+; (6) multimarked strain, females XX; ac/ac; ar/ar; bwb/bwb; ye/ye; snp/snp, males XY; ac/ac; ar/ar; bwb/bwb; ye/ye; snp/snp; and (7) Ag strain, females and males XX; Ag/+ or XX; +/+.
To obtain a pure female progeny, wild-type females (strain 1) were crossed to no-M males of strain 5. Pure male progeny was obtained by crossing wild-type females (strain 1) to males homozygous for the autosomal M factor.
To map Mdtra, females of strain 5 were crossed to males from the multimarked stock (strain 6). Subsequently, F1 males were backcrossed to females of strain 6. F2 flies heterozygous for only one of each marker were collected for further analysis.
To determine the genotype of females of strain 7 (Ag/+ vs. +/+), single females were crossed to no-M males of the same strain. Those that produced only sons were Ag/+ females, whereas those that produced only daughters were +/+ females.
For sequence analysis of the MdtraD allele, we used FD females collected from natural populations in various parts of the world: Japan (Inoue and Hiroyoshi 1982), Turkey (S. Cakir, unpublished data), Spain, France, Tanzania, South Africa (kindly provided by Leo Beukeboom, University of Groningen), North America, and Australia (kindly provided by Rhonda Hamm, Itaka University). To test for presence of FD, females were crossed to males homozygous for MIII (strain 2). We analyzed Mdtra sequences of only those females that produced male and female offspring.
Cryosections and in situ hybridizations:
Ovaries were embedded in tissue-freezing medium (Jung) and frozen in liquid nitrogen. Sections (30 μm) were made with a cryostat at −14° and collected on gelatinized microscope slides. Antisense digoxigenin riboprobes for the Mdtra gene were generated using standard methods. The female-specific probe included exons E2a, E4, E5, and E6, while the male-specific probe contained exons E2b and E3. RNA in situ hybridization visualized by alkaline phosphatase was performed as described by Vosshall et al. (1999), with the modification that hybridization was carried out at 55°.
Injection of dsRNA:
dsRNA was generated and injected into early blastoderm embryos as described earlier (Hediger et al. 2001, 2004). The fragment used for Mdtra silencing has a length of 621 bp and extends from exon 1 to exon E5, lacking the male-specific exons (E2b and E3) and the arginine/serine-rich domain in exon 5.
Transgenic constructs and germ-line transformation:
The pBac (3xP3-eGFP; hsp70-MdtraF1) transgene was cloned by introducing a 1.2-kb NotI fragment spanning the complete female-specific Mdtra-ORF (MdtraF1; PCR sequence from primer Mdtra-19 to Mdtra-33) into the NotI-digested pBacHsp70 vector (kindly provided by E. Wimmer). This vector contains the eGFP marker under the control of the 3xP3 eye-specific promoter and a “Dmhsp70-polylinker-Dmhsp70poly(A) trailer” cassette. Coprecipitation with the helper plasmid and injection into housefly embryos of strain 3 were performed as described previously (Hediger et al. 2004). Transformed flies were identified by the GFP expression in their pigmentless, white (w) mutant eyes. Expression of the MdtraF1 cDNA was induced by repeated heat-shock pulses given throughout development. Each cycle consisted of 1 hr at 45° followed by 4 hr at 25°.
RESULTS
Isolation of tra orthologs in Musca and Glossina:
Since TRA belongs to a family of rapidly evolving proteins with low complexity, a direct attempt to isolate a Musca ortholog on the basis of homology at the coding sequence level was expected to be severely hampered (Kulathinal et al. 2003; O'Neil and Belote 1992). Therefore we took a different strategy on the basis of the observation that the Mediterranean fruit fly ortholog Cctra is a target of Cctra2 and contains several clusters of well conserved TRA/TRA2 binding sites (Pane et al. 2002; Salvemini et al. 2009). Given that the gain-of-function allele FD cannot be silenced by Mdtra2 RNAi, we presumed that Mdtra, the prime candidate for F, may also be a direct target of Mdtra2 (Burghardt et al. 2005). We conducted a genomewide screen for sequences containing TRA/TRA2 binding sites using degenerate primers, which were designed from the three putative sites previously found in the Mddsx gene (Hediger et al. 2004). With primer pair MdDSX-69/MdDSX-70 we amplified a 360-bp genomic fragment that contains a cluster of nine sites (see materials and methods). Using the same set of degenerate primers on cDNAs prepared from male embryos, additional sequences flanking this genomic region were isolated. Within this fragment that encompasses exons E2b and E3, a total of 29 putative binding sites were identified (Figure 1 and supporting information, Figure S1).
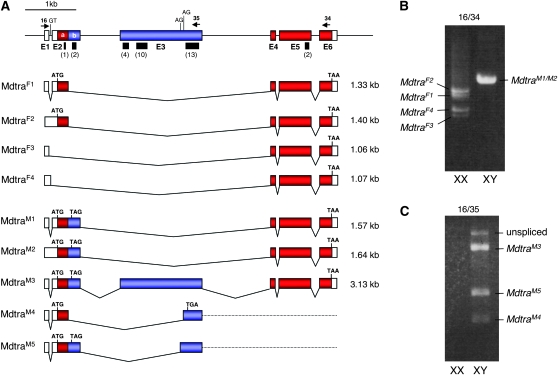
Figure 1.—
Gene organization and splice variants of Mdtra. (A) Exons in red (E2a, E4, E5, and E6) contain the long ORF that is intact in the female-specific transcripts MdtraF1 and MdtraF2. Exons in blue (E2b and E3) are male specific and contain several in-frame translation termination signals. Black bars indicate the location of clusters of putative TRA/TRA2 binding sites with the numbers of sites given in parentheses below (see Figure S1 for sequences). Arrows represent the position of primers used in RT–PCR experiments. (B) RT–PCR analysis of total RNA from male and female adults with primers Mdtra-16 and Mdtra-34. (C) Minor splice variants MdtraM3–M5 in males are detected by amplification with primers Mdtra-16 and Mdtra-35, which is located in the male-specific exon E3.
To retrieve full-length transcript sequences, 5′ and 3′ RACE were performed on cDNAs prepared from male and female adult houseflies. Alignment of the extended cDNA sequences to the corresponding genomic sequences revealed the presence of at least six exons (Figure 1). A genomic region of ∼6 kb in length harbors a diverse pattern of alternatively processed transcripts. Of particular interest was the presence of sex-specific splice variants at all developmental stages tested (Figure 1). The only transcripts that contain an intact long ORF, MdtraF1 and MdtraF2 (Figure 1A), were exclusively found in female XX individuals (Figure 1B). We identified two additional transcripts MdtraF3 and MdtraF4 that are present only in females. In contrast to MdtraF1 and MdtraF2, which both encode a full-length protein of 367 aa, these transcripts lack exon 2 sequences and the first translational start signal. The next available start signal is located in exon 6 and, as a result, these transcripts are expected to give rise to a truncated protein of 64 aa. Transcripts that are predominantly detected in males, on the other hand, contain additional sequences that introduce in-frame stop signals causing premature termination of translation (Figure 1A, blue boxes). In RNA preparations from males we generally find a more variable pattern of differently sized transcripts, suggesting that the male splice mode is less robust than that of the female. We identified at least five different male-specific transcripts, MdtraM1–M5, by sequence analysis, but additional low abundance variants may yet exist (Figure 1, B and C). Importantly, none of these transcripts contain a long ORF. Instead, all male-specific ORFs appear prematurely truncated and give rise to small and presumably nonfunctional peptides.
The exon–intron organization of this gene largely coincides with that of tra orthologs found in Tephritidae and Calliphoridae (Pane et al. 2002; Lagos et al. 2007; Ruiz et al. 2007; Concha and Scott 2009). Similarly to these tra orthologs, the sex-specific splicing regulation of Mdtra is mainly based on exon-skipping mechanisms and 5′ alternative splicing, rather than 3′ alternative splicing as observed for the Drosophila transformer gene. In addition, the housefly gene is flanked at the 3′ end by a divergently transcribed homolog of the l(3)73Ah gene (data not shown). Close linkage between tra and l(3)73Ah is also observed in D. melanogaster, C. capitata, B. oleae, and A. obliqua (Irminger-Finger and Nothiger 1995; Pane et al. 2002; Lagos et al. 2007; Ruiz et al. 2007).
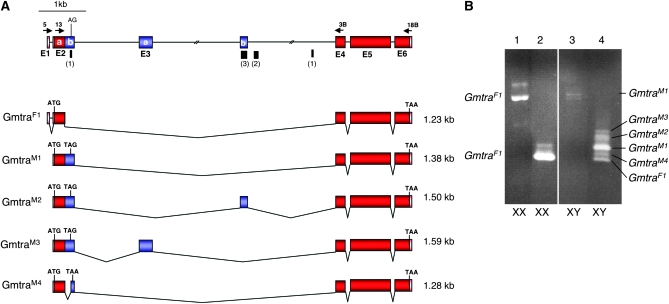
We extended the analysis of tra homologs in the Calyptratae group to the tsetse fly G. morsitans. A BLAST search with Mdtra sequences led to the identification of a partial cDNA in the Glossina EST database (http://www.sanger.ac.uk/Projects/G_morsitans/). With 3′ and 5′ RACEs this cDNA fragment was extended on both sides and revealed a transcriptional unit that has a genomic organization very similar to that of Mdtra (Figure 2A). Two observations make this Glossina gene a likely candidate for representing the tra ortholog in tsetse flies. First, this gene generates several splice variants, all of which are sex-specifically enriched (Figure 2B). Of these, only the GmtraF1 variant that is predominantly detected in female XX individuals has a long uninterrupted ORF. The four male-specific variants GmtraM1–M4 include additional sequences, which introduce translational stop signals shortly after the first AUG and thus encode small peptides. Second, four small clusters of putative TRA/TRA2 binding sites were found in the intron that is differentially spliced in the two sexes (Figure 2A). Given that this Glossina gene shares these critical features with the tra orthologs in tephritids and in Musca it is likely that it represents the tra ortholog in tsetse flies.
Figure 2.—
Gene organization and splice variants of Gmtra. (A) Exons in red (E2a, E4, E5, and E6) contain the long ORF that is intact in the female-specific transcripts GmtraF1. Exons in blue (E2b, E3a, and E3b) are male specific and contain several in-frame translation termination signals. Black bars indicate the location of small clusters of putative TRA/TRA2 binding sites with the numbers of sites given in parentheses (see Figure S1 for sequences). Arrows above the top line represent the position of primers used for expression analysis. (B) RT–PCR analysis of total RNA extracted from adult XX and XY flies. Results from female (XX) samples are shown amplified with primers 5 and 18B (lane 1) and with primers Gmtra-13 and Gmtra-3B (lane 2). RT–PCR results from male (XY) samples are shown with primers Gmtra-5 and Gmtra-18B (lane 3) and with primers Gmtra-13 and Gmtra-3B (lane 4).
Mdtra and Gmtra encode SR proteins with homology to TRA and FEM:
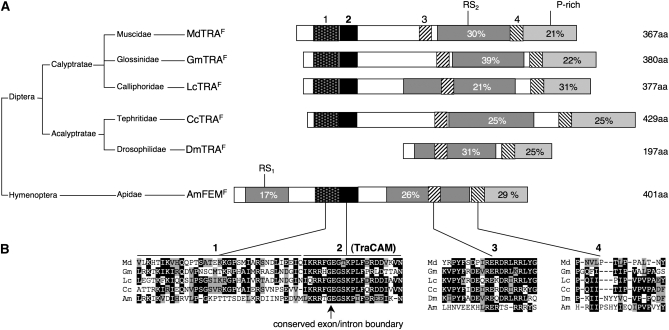
Alignment of the long ORF of female-specific MdTRAF1 and GmTRAF1 with TRA polypeptides of Drosophila, Ceratitis, Glossina, Lucilia, and the tra-like FEM protein in the honeybee, Apis mellifera, shows a low (but some) degree of similarity at the amino acid level (Figure S2). TRA belongs to a class of rapidly diverging splicing regulatory (SR) proteins, which share domains enriched in arginine/serine dipeptides (RS domain) and a proline-rich domain at the C-terminal end (Kulathinal et al. 2003; Hasselmann et al. 2008). Both of these structural features are also preserved in MdTRAF1 and GmTRAF1 (Figure 3). In addition, using the ClustalW alignment program we identified a short highly conserved domain of 21 aa in the amino-terminal regions of all these TRA isoforms. This domain was termed TRA-CAM (C, Ceratitis; A, Apis; and M, Musca). Ruiz and co-workers (Ruiz et al. 2007) showed that the N-terminal domain displays the highest level of similarity among the TRA homolog in the tephritids. Another strikingly preserved feature is the position of the sex-specifically spliced intron at the same position within this TRA-CAM domain. Apart from the TRA-CAM domain the ClustalW algorithm identified three additional blocks of sequence similarity (domains 1, 3, and 4; Figure 3). Domain 1 is present just upstream of the TRA-CAM domain and both domains are absent in DmTRAF. In addition, the TRA protein in dipterans reveals a short conserved motif, domain 3, which is located within or flanking the RS domain at the amino-terminal end. Likewise, domain 4, which is juxtaposed to the proline-rich domain in the carboxy-terminal region, occurs preferentially in the dipteran orthologs of tra.
Figure 3.—
Small conserved protein motifs in TRA homologs. (A) Schematic alignment of proteins encoded by the tra ortholog of Musca domestica (MdTRAF), Glossina morsitans (GmTRAF), Lucilia cuprina (LcTRAF), Ceratitis capitata (CcTRAF), Drosophila melanogaster (DmTRAF), and Apis mellifera (AmFEMF). The percentage of arginine/serine residues in the RS domains (dark shading) and the percentage of proline residues in the P-rich domains (light shading) are indicated within the boxes. The relative position and size of the four conserved motifs are indicated by differently shaded boxes (B) Sequence alignment of these small motifs. The conserved exon/intron boundary in motif 2 (TRA-CAM) is indicated by an arrow. In Musca, the glutamine (Q) directly upstream of the conserved TRA-CAM domain is replaced by an arginine (R) in the dominant allele MdtraD; see also Figure 7A and text.
Mdtra is necessary for female development:
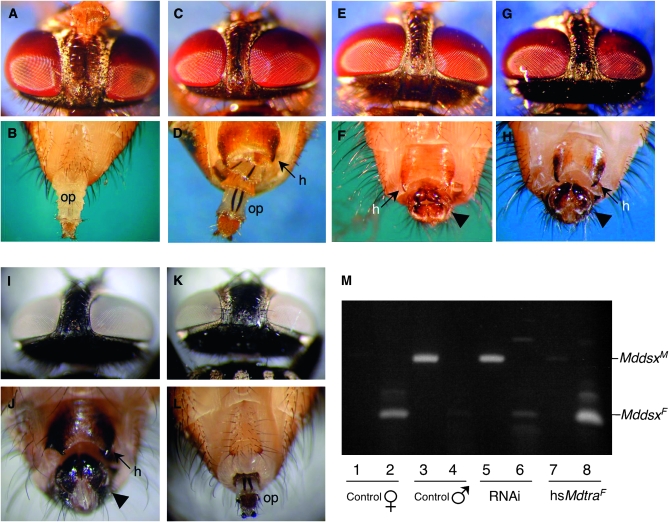
To test for potential function of Mdtra in sex determination, we injected dsRNA fragments corresponding to exons E2a and E4 and upstream sequences of exon E5 into early preblastoderm embryos of an autosomal M strain (see materials and methods), in which the male-determining factor M is linked to the wild-type alleles of bwb (brown body) and pw (pointed wings). This RNAi treatment causes a strong sex reversal phenotype only in flies with a female genotype (pw bwb/pw bwb) (Table 1 and Figure 4, A–H). A description of the sexual dimorphic structures is presented in the Figure 4 legend. The vast majority of these individuals (96%) developed into mosaic intersexuals displaying weak to strong degrees of masculinization (Figure 4, C and D). A small fraction of these no-M males (4%) were completely transformed into males (Figure 4, E and F). These no-M males were fertile and produced exclusively daughters when crossed to wild-type females, confirming the absence of the male determiner M. No detectable phenotypes were observed in genotypically male individuals (M pw+ bwb+/pw bwb) to which the same treatment was employed and in buffer-injected control flies of both sexes. The masculinization of RNAi-treated no-M animals correlates with a shift in the splice pattern of Mddsx from the female to the male mode (Figure 4M), placing Mddsx downstream of Mdtra.
TABLE 1.
Masculinization of flies with a female genotype caused by Mdtra RNAi
| Injected substance | No. of injected embryos | Total no. of hatched adults | Female genotype |
Male genotype |
|||
|---|---|---|---|---|---|---|---|
| Strain | No. | External sexual phenotype | No. | External sexual phenotype | |||
| F+a | Ringer | 2733 | 246 | 107 | Female | 139 | Male |
| Mdtra dsRNA | 2070 | 234 | 0 | Female | 0 | Female | |
| 105 | Intersexualc | 0 | Intersexual | ||||
| 4 | Male | 134 | Male | ||||
| FDb | Ringer | 950 | 44 | 24 | Female | 20 | Male |
| Mdtra dsRNA | 3227 | 189 | 89 | Femaled | 0 | Female | |
| 2 | Intersexualc | 0 | Intersexual | ||||
| 1 |
Male |
97 |
Male |
||||
Autosomal M strain: Female genotype is XX; bwb pw/bwb pw; F+/F+ (phenotype bwb pw), and male genotype is XX; M pw+ bwb+/pw bwb; F+/F+ (phenotype bwb+ pw+).
Strain FD/Fman: Female genotype is XX; FD Ba/Fman Ba+ (phenotype Ba), and male genotype is XX; Fman Ba+/Fman Ba+ (phenotype Ba+).
Flies with mixed external sexually dimorphic structures, e.g., with male genitalia but female eye distance, or flies with mixed external genitalia and male or female eye distances.
All with ovotestes.
Figure 4.—
Silencing or misexpression of Mdtra causes complete sex transformations in the housefly. Experiments were conducted with the MIII bwb+/ bwb strain to allow rapid and easy phenotypic distinction between M-bearing individuals (bwb+) and no-M individuals (bwb) (see materials and methods). Phenotypically, males can be recognized by a significantly narrower interocular distance. Also, males display a darkly pigmented copulatory apparatus (arrowhead) and exhibit characteristic horn-like structures (h) at the tip of sternite 5. The external female genitalia are characterized by the presence of an ovipositor (op). (A–H) Female–male sex reversion caused by injections of Mdtra dsRNA into syncytial embryos. (A) Head and (B) genital region of a control female (bwb) and (G and H) of a control male (bwb+), respectively, are shown. (C and D) Intersexual no-M individual (bwb) treated with Mdtra RNAi displaying a male-like interocular distance and sexually mixed genitalia composed of an almost complete ovipositor and a male-like sternite 5 with hornlike structures. (E and F) Completely sex-reverted no-M individual treated with Mdtra RNAi displaying normal male morphology in the head and genital region. (I–L) Male–female sex reversion caused by transgenic expression of MdtraF1. (I and J) Head and genital region of a nontransgenic control male (bwb+). (K and L) Complete feminization of an M-bearing individual (bwb+) after heat-induced induction of MdtraF1 expression. This fertile individual displays normal female morphology in the head and genital region. (M) Mddsx splicing patterns in sex-reverted individuals. Transcripts of Mddsx are analyzed by RT–PCR with primer pairs specific for the male splice variants (lanes 1, 3, 5, and 7) and the female splice variants (lanes 2, 4, 6, and 8). Total RNA was extracted from single flies: control female (lanes 1 and 2), control male (lanes 3 and 4), no-M individual (bwb) treated with Mdtra RNAi (lanes 5 and 6; see also E and F), and M-bearing individual (bwb+) expressing the MdtraF1 transgene (lanes 7 and 8; see also K and L).
To test whether Mdtra is needed for female development in animals carrying the dominant female-determining allele of F (FD), we injected the same dsRNA fragments into embryos collected from FD mothers. This time, the vast majority of FD individuals (97%) developed into normal-looking females and only few intersexes or males were recovered (Table 1). However, all of the RNAi-treated FD animals contained gonads that were clearly male-like in size and morphology. Microscopic sections through these testis-like gonads revealed the presence of different spermatogenic stages (Figure S3). The finding that in FD animals only the gonads are masculinized is likely due to a transient silencing effect of Mdtra in early development affecting primarily gonad differentiation.
Activation of Mdtra is sufficient to instruct female development:
The female-to-male transformation caused by loss of Mdtra activity reflects the situation in individuals that are homozygous for the loss-of-function allele of F, the key switch in sex determination of Musca. If Mdtra indeed corresponds to F, it is expected that forced expression of female Mdtra activity in genotypically male flies will impose female development, overriding the repression by M and, thus, mimicking the gain-of-function allele FD. To this end, a construct expressing the full-length protein MdTRAF1 under the control of the Drosophila hsp70 promoter, hs70∷MdtraF1, was generated and again introduced into a strain that allows simple phenotypic distinction of individuals with and without MIII. In two independent lines, we observed a strong male-to-female transformation in MIII-bearing individuals after exposure to repeated heat pulses throughout development. These individuals display typical female traits such as a wider distance between the eyes and several female structures in the genital region such as the ovipositor (Figure 4, K and L). In line 14.2 the feminizing effect of hs70∷MdtraF1 was the strongest, causing a complete sex reversion of MIII-bearing flies to fertile females. In this line, the transgene inserted onto chromosome 3 to a site close to the genetically mapped location of the MIII factor. We ruled out the possibility that feminization was a result of insertional inactivation of MIII by the transgene, by crossing these females to males homozygous for MIII. The offspring carrying the transgene developed into fertile females, indicating that it is the activity of the MdtraF1 transgene that overrules the male-promoting activity of MIII. In accordance with the expected position of Mdtra upstream of Mddsx, the splicing pattern of Mddsx is shifted from the male to the female mode in sex-reverted animals carrying the MdtraF1 transgene (Figure 4M). We can thus conclude that the activity of Mdtra is not only required but also sufficient to direct female splicing of Mddsx in a genotypically male background.
Flies homozygous for the partial loss-of-function allele of F, Fman, develop as males (Schmidt et al. 1997a). To test whether expression of MdTRAF1 can substitute for loss of F activity in these animals we crossed line 14.2 into an Fman homozygous background. All 165 animals homozygous for Fman, carrying the MdtraF1 transgene, developed into morphologically normal looking and fertile females. Hence, MdtraF1 expression not only overrules repression by M but also substitutes for lack of F function, suggesting that it acts downstream or at the level of F. Taken together our data provide evidence that Mdtra is not only essential for female development but also sufficient to instruct female differentiation and thus acts as a bona fide genetic on/off switch in the sex-determining pathway of the housefly.
Female-specific splicing of Mdtra requires Mdtra and Mdtra2:
In Drosophila sex-specific splicing of tra is controlled by Sxl, whereas in tephritids splicing regulation of the corresponding tra ortholog requires the activity of tra itself (Pane et al. 2002; Lagos et al. 2007). In addition, Salvemini and co-workers (Salvemini et al. 2009) recently reported that the tra2 ortholog in Ceratitis is also required for female-specific splicing of Cctra. The presence of multiple clusters of putative TRA/TRA2 sites, ISSs, and RBP1 binding sites in Cctra and other tephritid tra orthologs gives further support to the notion that tra in these species directly controls its activity by directing the female-specific splicing mode.
Likewise, we find multiple clusters of ISSs and RBP1 binding sites located within the transcribed sequences of Mdtra and Gmtra and, in particular, several clusters of TRA/TRA2 binding sites in the alternatively spliced region (Figure 5, A and B). Hence, splice regulation of tra in these Calyptratae species may also involve a direct autoregulatory activity. Consistent with this notion, a shift from the female to the male splice mode of endogenous Mdtra RNA was observed in genotypically female flies in which Mdtra was silenced by injecting dsRNA fragments into blastoderm embryos (Figure 5C). In the reciprocal experiment splicing of endogenous Mdtra nascent transcripts shifts from the male to the female mode in M-bearing animals in which MdTRAF1 is ectopically expressed (Figure 5C). In accordance with the presumed function of Mdtra2 as an essential cofactor in Mdtra-dependent splice regulation, silencing of Mdtra2 in genotypically female individuals by injecting dsRNA fragments of Mdtra2 also causes a shift in the splice pattern of Mdtra from the female to the male mode (Figure 5C). We conclude that the female splicing pattern of Mdtra requires functional products of Mdtra and Mdtra2. This is consistent with the findings in C. capitata where it has been shown that tra, once activated, is needed together with Cctra2 to maintain a continuous production of active products by a positive feedback mechanism (Salvemini et al. 2009). Likewise we propose that the activities of Mdtra and Mdtra2 are continuously required to maintain the active female splicing mode.
Figure 5.—
Mdtra splicing regulation. Sites of putative cis-regulatory regions in the genomic regions of Mdtra (A) and Gmtra (B) are shown. Boxes represent exons, lines represent introns. The locations of TRA/TRA2 (boxes), RBP1 types A and B (triangles), and TRA2-ISS (ovals) binding sites are shown (for sequences see Figure S1). If not otherwise indicated (numbers within the boxes), each mark represents one binding site. (C) Splicing patterns of Mdtra in no-M individuals (bwb) treated with either Mdtra or Mdtra2 RNAi and in M-bearing individuals expressing the hsMdtraF1. Total RNA was extracted from single flies: control bwb female (line 1), control bwb+ male (line 2), masculinized no-M individual treated either with Mdtra2 (line 3) or Mdtra (line 4) RNAi, and feminized M-bearing individual containing the hsMdtraF1 transgene (line 5). Endogenous Mdtra transcripts were specifically amplified with primer Mdtra-24, which is located in 3′-UTR Mdtra sequences not present in the transgene. Amplification of Mdtra2 and MdCYP6D3 transcripts serves as an internal standard and quality control of extracted RNA.
Maternal requirement for female-specific splicing of Mdtra:
Regarding Mdtra's autoregulatory function some important questions arise about when and how the productive female splicing mode of Mdtra is initiated in individuals with a female genotype. How does the positive feedback loop become established in the female zygote to maintain the selected female fate? One likely scenario is that the components necessary for activating the self-sustaining loop are maternally provided. In a previous report we described that Mdtra2 transcripts are abundantly present in unfertilized eggs (Burghardt et al. 2005). To test whether Mdtra also provides maternal products, we performed in situ hybridizations with DIG-labeled Mdtra RNA probes encompassing exons E2a, E4, E5, and E6. Sections through adult ovaries unveiled the presence of large amounts of Mdtra transcripts in nurse cells and oocytes (Figure 6A). The predominantly localized Mdtra transcripts in the cytoplasm are female processed variants, since no cytoplasmic labeling is observed with probes containing only male-specific sequences (Figure 6B). Instead, these MdtraM specific probes gave a distinct punctated staining pattern that is confined to sites within the large polyploid nuclei of the nurse cells (Figure 6C). These labeled foci most likely represent intranuclear deposits of unprocessed Mdtra RNA. Moreover, when examining transcripts in unfertilized eggs, exclusively female-processed MdtraF1 transcripts are detected (Figure 6D).
Figure 6.—
Maternal and early zygotic expression of Mdtra in wild-type and Ag mutant females. In situ hybridization on frozen sections of adult ovaries is shown with (A) a Mdtra-specific riboprobe containing female-specific sequences (exons E2a, E4, E5, and E6) and (B) a riboprobe containing exclusively male-specific sequences (exons E2b and E3). Hybridization signals were visualized with an alkaline phosphatase-based detection system. (A) Mdtra transcripts predominantly accumulate in the cytoplasm of nurse cells (NC) and in the developing oocyte (OC). (B) Male-specific Mdtra sequences are detected only within the polyploid nuclei of nurse cells (arrows). The punctated staining pattern depicts nuclear sites where unprocessed Mdtra RNA accumulates. (C) Enlarged view of the punctated staining pattern in nurse cell nuclei. (D) RT–PCR analysis of Mdtra transcripts in unfertilized eggs collected from wild-type females (wt, lane 6), from Ag/+ mutant females (Ag, lanes 3–5) that produced an all male progeny, and from +/+ females of the same strain (No-Ag, lanes 1 and 2) that produced only females. Total RNA was isolated from unfertilized eggs laid by a single female over a period of 24 hr. Primers Mdtra-16 and Mdtra-33 were used to amplify MdtraF1 transcripts, and Mdtra-16 and Mdtra-36 were used to amplify MdtraM1 transcripts. Amplification of MdCYP6D3 transcripts served as an internal control for amount and quality of RNA samples. (E) Profile of Mdtra transcripts during embryogenesis. Total RNA was isolated from staged wild-type egg collections and amplified with primers Mdtra-16 and Mdtra-33 for detection of MdtraF1 transcripts and with primers Mdtra-12B and Mdtra-20 to detect MdtraM1 transcripts. Amplification of Mdtra2 transcripts served as an internal control for amount and quality of RNA samples. Eggs, unfertilized eggs; h, hours after egg deposition.
In a mixed population of staged male and female embryos, male-specifically spliced transcripts of Mdtra are first detected within 2–3 hr after egg laying before cellularization of the blastoderm (Figure 6E). It seems likely that these male transcripts are generated and processed de novo, suggesting that Mdtra must already be transcribed at the early blastoderm stage. It is thus conceivable that maternal supplies of Mdtra and Mdtra2 activities present during the initial period of zygotic Mdtra transcription can impose the female processing mode in embryos with a female genotype. Likewise, it follows that, in embryos with a male genotype, M acts very early to prevent the establishment of the feedback loop by maternal Mdtra.
Further support for the conception that activation of zygotic Mdtra depends on maternally supplied Mdtra activity comes from studies of arrhenogenic mutant females (Ag/+). These females produce only male offspring due to a failure in activating zygotic F as shown in previous genetic studies (Schmidt et al. 1997b). We examined whether the dominant mutation Ag has any bearings on the maternal inheritance of MdtraF products. None of the eggs collected from Ag/+ females contained detectable levels of MdtraF (Figure 6D, lanes 3–5) whereas those collected from wild-type sibling females had normal levels of female transcripts (Figure 6D, lanes 1 and 2). Instead, low levels of male-processed MdtraM1 transcripts were found in eggs from mutant Ag females. We infer from these data that the Ag mutation prevents the production of a sufficient supply of maternal MdtraF needed to activate zygotic Mdtra. Altogether, these results are consistent with the notion that maternal supplies of female Mdtra transcripts and Mdtra2 transcripts are inherited to the zygote to serve as an initial source for directing female splicing of the zygotically active Mdtra gene.
Molecular lesions in Mdtra sequences derived from Fman and FD:
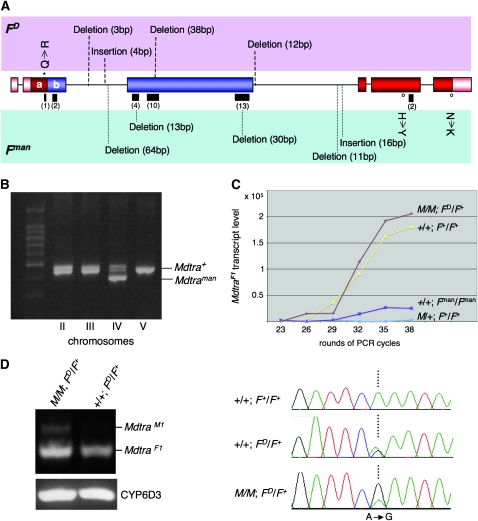
Besides the functional correspondence to F described above, there are also several structural features of Mdtra that make it a strong candidate for being F. The F gene was previously identified by two mutations: the recessive loss-of-function allele Fman (Schmidt et al. 1997a) and the dominant gain-of-function allele FD (Rubini et al. 1972). We isolated Mdtra sequences from these mutant backgrounds and aligned them with a reference sequence, which was assembled from Mdtra sequences from seven strains of different origins. While practically no polymorphisms were found in the reference sequences, the Mdtra sequences from Fman and FD animals displayed an unusually high number of small deletions and insertions in the region that is differentially processed (Figure 7A, Figure S4). Consistent with the different origins of the two mutant F strains, these indels do not overlap and thus must have arisen independently. We used the lesions in the Mdtra sequence derived from Fman as a molecular signature to assign the location of this gene to a specific chromosome in a multimarked background. The Mdtra variant of the Fman allele specifically segregates with markers on chromosome IV (Figure 7B), consistent with the genetic location of F (McDonald et al. 1978; Schmidt et al. 1997a).
Figure 7.—
Analysis and expression of Mdtra sequences in FD and Fman mutant backgrounds. (A) Molecular lesions in the Mdtra genomic region of FD and Fman animals. The positions of insertions and deletions are indicated by either dashed lines in the FD allele (top row) or dotted lines in the Fman allele (bottom row). Lengths of insertions and deletions are given in parentheses. Amino acid substitutions are marked with an open dot for the Fman allele and with a star for the FD allele. (B) The region encompassing a 64-bp deletion in the Fman allele was amplified from flies heterozygous for one of the four marked chromosomes II, III, IV, and V (see materials and methods). Only flies heterozygous for the marker on chromosome IV also contain both the Mdtra+ and the Mdtraman allele. (C) Semiquantitative analysis of MdtraF1 transcript levels in flies of different genotypes: control males (M/+; F+/F+), No-M males (+/+; Fman/Fman), control females (+/+; F+/F+), and FD females homozygous for M (M/M; FD/F+). Total RNA was isolated from five flies per genotype and RT–PCR was performed with primer pairs Mdtra-9 and Mdtra-20. After each of the indicated cycles, 5 μl of the amplification reaction was analyzed. Intensity of the MdtraF1 bands was normalized using levels of amplified Mdtra2 products as an internal control. (D) RT–PCR of RNA from M/M; FD/F+ females detects male MdtraM1 (primers Mdtra-18 and Mdtra-3) and female MdtraF1 transcripts (primers Mdtra-18 and Mdtra-20), while RT–PCR of RNA from +/+; FD/F+ females produces only female MdtraF1 transcripts (MdCYP6D internal standard). Analysis of the sequence chromatogram of the region encompassing the MdtraD-specific nucleotide substitution A to G in exon E2a allowed us to compare the relative amounts of F+ (A) and FD (G) -derived Mdtra transcripts in the absence or the presence of M and reveals that in M/M; FD/F+ females MdtraF1 transcripts are almost exclusively derived from the FD allele. In contrast, in +/+; FD/F+ females, both F alleles produce equal amounts of MdtraF1 transcripts.
We noticed that some of the small deletions in the Mdtra sequence of homozygous Fman individuals remove a substantial number of TRA/TRA2 binding sites (Figure S4). The loss of these putative binding sites correlates with a marked reduction of levels of female-specific Mdtra transcripts in homozygous Fman animals (Figure 7C). The strong hypomorphic character of Fman can thus be explained by impaired regulation of Mdtra to uphold the female-specific splicing mode due to the loss of binding sites. In addition, we detected several single-base-pair substitutions within the Mdtra sequence of the Fman allele. Two of the five single-base-pair substitutions that reside within the coding region lead to an amino acid exchange in exon 5 (H → Y) and exon 6 (N → K), respectively.
Regarding their effect on Mdtra functions the lesions found in the Mdtra sequence of the gain-of-function FD allele are less well explained. Among the deviations from the reference sequence we also find a small deletion that removes several TRA/TRA2 binding sites and a putative RBP1 type A binding site in one of the clusters (Figure S4). Nevertheless, normal levels of female Mdtra transcripts are detected in M/M; FD/F+ females (Figure 7C). A specific nucleotide substitution (A to G) in exon E2a, resulting in a Q to R aa change, allowed us to test whether female Mdtra transcripts are derived from the FD (G) and/or from the F+ (A) allele. In FD/F+ females with no M factor present, both alleles produce comparable levels of female transcripts (Figure 7D). However, in FD/F+ females, which are homozygous for the M factor on chromosome III, female transcripts are almost exclusively derived from the Mdtra allele on the FD chromosome. In this background some low levels of male transcripts are detected that presumably are generated by the Mdtra allele on the F+ chromosome. From these observations we conclude that M is not capable of preventing the female splicing of the Mdtra allele on the FD chromosome. On the other hand, this allele seems not to be able to transactivate the female splicing of the Mdtra allele on the F+ chromosome when an M factor is present. This is also consistent with the notion that the FD-derived Mdtra allele does not depend on autoregulation to exert its female-promoting activity. For instance, FD, in contrast to F+, can restore female differentiation at late stages after recovery from dsRNA-based silencing during embryonic development. In this regard the FD-derived Mdtra allele behaves exactly like FD in genetic tests where it has been demonstrated that zygotic FD does not require maternal F to be active (Dubendorfer and Hediger 1998).
DISCUSSION
Mdtra corresponds to the female determiner F:
Since Mdtra displays all the relevant features that have been specifically assigned to the female determiner F in genetic studies, we propose that these genes are identical. Changes in Mdtra activity precisely mimic the reciprocal phenotypic effects of the loss- and gain-of-function alleles of F, Fman, and FD. More compelling, female development can be restored in homozygous mutant Fman animals by transgenic expression of the female variant, MdtraF. This expression can impose female development even in the presence of the repressor M, reflecting the feminizing and M-resistant properties of the gain-of-function allele FD. Changes in the activity of F or Mdtra cause corresponding shifts in the sex-specific splicing pattern of Mdsx, placing them upstream of Mddsx in the pathway. On the other hand, F and Mdtra are both susceptible to the silencing effects of Mdtra2 RNAi, positioning them downstream and/or parallel to Mdtra2 (see below). Besides occupying the same relative position in the pathway, they behave similarly in several other aspects. For instance, it has been previously shown in clonal studies that the activity of F is irreversibly set around the blastoderm stage and that its female-promoting activity is continuously required from there on (Hilfiker-Kleiner et al. 1994). This is in line with our finding that transcription of Mdtra already starts in the early embryo before cellularization when the presence of an M factor irreversibly locks it into the male nonproductive mode of splicing. In partially transformed individuals we often observed a graded response to Mdtra RNAi with strongest effects close to the injection site and lesser effects farther away from the injection site. It thus seems that the RNAi response in the syncytial embryo is not even but depends on the local concentration of dsRNA, which at the single-nucleus level may or may not be sufficient to cause an irreversible collapse of the feedback loop. This early commitment to the female or male fate based on the stable setting of a key switch is reminiscent of the situation in Drosophila, where activity of Sxl is irreversibly set during the early cleavage stages prior to cellularization (Keyes et al. 1992). Another important feature that F and Mdtra have in common is their dependence on their own maternal contribution to activate the female-promoting function in the zygote (Dubendorfer and Hediger 1998). Our findings based on studies of the maternal effect of the Ag mutation suggest a requirement of maternal Mdtra activity for female processing of Mdtra in the zygote. Finally, we identified a specific set of lesions in Mdtra sequences derived from the two F mutant backgrounds, FD and Fman, which are likely to impinge on proper regulation of Mdtra as is discussed below.
Regulation of Mdtra:
In Drosophila regulation of tra is achieved at the level of splicing to generate functional products only in individuals committed to the female fate. Likewise, alternative splicing of tra seems to be a common regulatory strategy in other dipterans to restrict expression of functional proteins to females. However, unlike in Drosophila, this splicing regulation appears not to be controlled by Sxl (Meise et al. 1998; Saccone et al. 1998). It was first demonstrated in the Medfly, C. capitata, that regulation of the tra ortholog, Cctra, is fundamentally different from that in Drosophila in that it involves an autoregulatory activity to maintain its female mode of splicing (Pane et al. 2002). The participation of the auxiliary factor Cctra2 in Cctra splicing and the presence of TRA/TRA2 binding sites in Cctra pre-mRNA suggested that this regulation is direct (Salvemini et al. 2009). Here we report that Mdtra and Mdtra2 are both required to uphold female splicing of Mdtra. The presence of several clusters of putative TRA/TRA2 binding sites in the Mdtra pre-mRNA suggests that Mdtra and Mdtra2 are also directly involved in splicing regulation. In our initial survey for TRA/TRA2 binding sites in the genome we recovered only three fragments, of which one contained the Mdtra sequences. Clustering of these repeats in the Musca genome thus seems to be scarce and otherwise found only in Mddsx and the recently discovered fruitless ortholog, both of which are targets of Mdtra (this article and N. Meier and D. Bopp, unpublished results). Likewise, the clustering of putative ISS and RBP1 binding sites in Mdtra and Gmtra is an unusual structural feature that is shared with tra orthologs of tephritids (Ruiz et al. 2007). Whether these sequences are involved in splicing regulation, however, remains to be investigated. Support for the relevance of putative TRA/TRA2 binding sites in Mdtra splicing comes from sequence analysis of the hypomorphic allele Mdtraman. The only lesions found in this allele are several small deletions that remove a substantial number of putative TRA/TRA2 binding sites. It seems reasonable to assume that these deletions will lower the overall binding affinity of MdTRA/MdTRA2 complexes to Mdtra pre-mRNA and, thereby, decrease the efficacy of female splicing. The MdtraD-specific lesions, on the other hand, cause Mdtra to be locked into the female mode of splicing irrespective of whether M is present or not. In addition, we showed that female-specific processing of MdtraD is reestablished after transient silencing of Mdtra or Mdtra2, indicating that activation of this allele does not depend on an autoregulatory function. This is in accordance with genetic results, which demonstrated that the MdtraD allele is active in zygotes that lack maternal activity of Mdtra (Dubendorfer and Hediger 1998).
To reconcile the phenotypic effects with the lesions found in MdtraD and Mdtraman we propose a model in which splicing of Mdtra consists of three superimposed levels of control (Figure S5). An important inference of this model is that the most subordinate level, the male mode of splicing, does not simply represent a deregulated state but requires the interaction of Mdtra pre-mRNA with a specific set of yet unidentified male-splicing promoting factors (MPFs). This interaction is required to activate the male-specific splice sites. The association of these MPFs with Mdtra pre-mRNA is antagonized by maternally supplied products of Mdtra and Mdtra2. Binding of these maternal factors to the Mdtra pre-mRNA prevents MPFs from binding and instead the female-specific splice sites are preferentially utilized. This second level of control initiates the production of functional MdTRA, which will continually prevent MPFs from activating the male splice sites. However, when an M factor is introduced into the system, it will antagonize female processing of Mdtra, either by hindering MdTRA/MdTRA2 to repress the function of the MPFs or by stabilizing the association of MPFs with Mdtra pre-mRNA. According to this model, we postulate that the MdtraD lesions primarily affect cis-regulatory regions required for interaction with MPFs. As a result, the inclusion of the male-specific exons is no longer supported and instead female splice sites are preferentially used by default.
Putative TRA/TRA2 binding sites have also been found in the alternatively spliced regions of tra orthologs in other tephritids (Lagos et al. 2007; Ruiz et al. 2007), in L. cuprina (Concha and Scott 2009), and in G. morsitans (this article). From this it can be inferred that the dependence of female splicing of tra on its own activity is a common feature of tra regulation not only in members of the tephritids but also in the Calyptratae. In addition, this study provides evidence that expression of MdtraF alone has the capacity to shift the splicing mode from male to female. Hence, Mdtra acts as a bona fide switch in the pathway in that it is not only required but also sufficient to activate and maintain the female mode of development. This feedback mechanism may serve as a cellular memory that ensures the proper implementation of the female program throughout the life cycle. A similar strategy has been described in Drosophila, where it is the gene Sxl that uses a feedback mechanism to sustain its female-promoting activity throughout development (Cline and Meyer 1996).
Evolutionary diversification of the sex determination pathway in the housefly:
Although the process of sex determination is fundamental to all sexually reproducing organisms, the genetic logic underlying this process appears to be highly variable. Already in the insect world, an astounding variety of sex-determining strategies seem to exist. Nevertheless, it has been postulated that these differences may simply reflect variations on a common theme (Nöthiger and Steinmann-Zwicky 1985). These authors proposed that, in insects, a common principle underlies sex determination consisting of three basic components, a primary signal, a binary switch gene, which responds to the signal in an on/off manner, and a downstream bifunctional switch, which, depending on the setting of the binary switch, directs either overt male or female differentiation. These elements have been proposed to form the variables for rapid evolutionary diversification as observed for instance in the housefly (Dubendorfer et al. 2002). The molecular identification and characterization of the master switch F in Musca in this study gives us now a suitable framework to explain the mechanistic basis of this diversification. The findings presented here led us to propose the following model for sex determination in the housefly.
Mdtra acts as the central on/off switch in the pathway. Its continuous activity is required to properly implement female development. This perpetuation of Mdtra activity is normally ensured by a positive feedback loop that is established already in the very early zygote during the cleavage stages when zygotic Mdtra becomes first transcribed. Maternal deposits of functional Mdtra and Mdtra2 products will initiate and activate the loop in the zygote (Figure 8A). In principle, the loop is continually active in the female lineage, as the female-promoting Mdtra activity is incessantly passed from mothers to daughters. Hence, presence of these maternal Mdtra activators predisposes the eggs for female development and would lead to a continuous production of females from one generation to the other. To disrupt this “female” continuity the activation of the loop in the early zygote needs to be prevented, a compulsory measure for male development, which is usually achieved by a specific repressor such as the paternally transmitted M factor. Our model proposes that the sole function of M is to prevent the establishment of the female-promoting self-regulatory loop of Mdtra. Once the loop collapses, cells will not be able to resume a female identity and instead follow the alternative male fate (Figure 8B). The straightforwardness by which a male determiner can interfere with female development may explain the manifold occurrence of M factors in natural Musca populations (Hiroyoshi 1964; Rubini and Palenzona 1967; Wagoner 1969). Interference at any regulatory level of Mdtra, e.g., transcriptional, post-transcriptional, or post-translational, would be sufficient to prevent the establishment of the loop. However, the observed resistance of MdtraD to M repression suggests that M does not compromise overall functions of Mdtra, at least not those involved in Mddsx regulation. Rather it seems to specifically impair the functions that are involved in Mdtra splicing. M factors have been found at different genomic sites in natural housefly populations. It has not yet been resolved whether they are transposed versions of the same gene or whether these are different genes that have adopted the function of a dominant loop breaker. It is conceivable that M factors evolved from different genes by acquiring a dominant antimorphic or neomorphic mutation that specifically impinges on the autoregulatory loop of Mdtra.
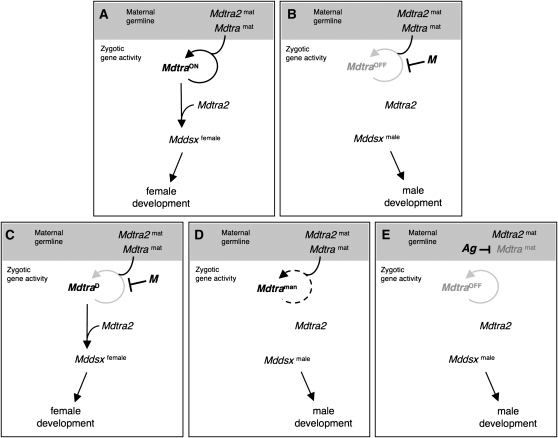
Figure 8.—
Model to account for the different sex-determining mechanisms in the housefly. (A) In a “standard” XX zygote, maternal Mdtra2mat and Mdtramat products activate zygotic Mdtra, which, once activated, maintains its productive (ON) mode of expression throughout development by a positive feedback loop. Together with Mdtra2, Mdtra sets its direct downstream target Mddsx into the female mode of expression, which leads to overt female differentiation. (B) In a standard M-containing zygote, activation or maintenance of the zygotic loop of Mdtra is prevented by the paternally transmitted M. As a result, Mdtra is locked into the nonproductive (OFF) mode of expression and Mddsx is set by default into the male mode of expression and male development ensues. (C) Dominant female determiner I: Presence of an MdtraD allele in the zygote uncouples the productive mode of Mdtra expression from the need of maternal factors for activation and from repression by M. This allele is by default locked into the productive mode of expression and sets Mddsx into the female mode of expression. (D) Dominant female determiner II: In Mdtraman mutant zygotes, female Mdtra expression is severely reduced to levels below the threshold required to set Mddsx into the female mode of expression. In this background, presence of a wild-type Mdtra allele will behave as a dominant female determiner by providing sufficiently high levels of zygotic Mdtra activity required to impose female development. (E) Maternal sex determination: Eggs laid by an Ag/+ mutant mother are devoid of maternal Mdtra products due to a dominant Mdtra repressing activity of Ag in the germ line. As a result, zygotic Mdtra cannot be activated and male development follows. On the other hand, +/+ females in this strain produce eggs with normal levels of maternal Mdtra products. When fertilized by male Ag/+ siblings, all zygotes will develop into females since Ag does not prevent somatic activation of Mdtra.
Musca strains that differ from the standard type described above can be explained on the basis of discrete mutational changes that primarily affect regulation of Mdtra. For instance, we propose that lesions in the dominant gain-of-function allele MdtraD uncoupled it from the requirement for Mdtra and Mdtra2 to initiate and maintain the female mode of expression. Consequently, its disengagement from the feedback loop made this allele resistant to repression by M and converted it into a dominant female determiner (Figure 8C). On the other hand, lesions in the Mdtraman allele have the opposite effect such that female splicing of this allele is severely reduced even when maternal Mdtra and Mdtra2 activities are supplied. Homozygosity for this allele leads to male development, since levels of female Mdtra products remain below a level that is required to promote female development (Figure 8D). In heterozygous animals, on the other hand, the presence of a wild-type Mdtra allele provides sufficient activity to implement female development. In this strain, hence, the wild-type allele of Mdtra behaves as a dominant female determiner. Finally, the maternal type of sex determination found in Musca can be derived from the dependence of zygotic Mdtra on maternally provided factors. We propose that the dominant Ag mutation in this maternal strain specifically represses expression of Mdtra in the female germ line without affecting the somatic functions of Mdtra. As a result, eggs of these arrhenogenic mothers are devoid of a sufficiently large supply of maternal Mdtra products to activate the self-sustaining loop in the zygote (Figure 8E). This is also in line with a previous postulate based on genetic data that Ag acts as an M derivative that specifically represses F in the female germ line without affecting F function in the soma (Schmidt et al. 1997b).
Taken together, these examples in Musca convincingly illustrate how subtle changes at the level of Mdtra regulation can lead to profound differences regarding the genetic logic of how the sexual fate is specified. This plasticity in the Musca system gives support to the proposition of Nothiger and Steinmann-Zwicky (1985) that seemingly different strategies can arise from small variations in an otherwise well conserved pathway. It seems from a number of studies in Acalyptratae and Calyptratae that a system that is primarily based on tra autoregulation represents the most common and possibly the ancestral mode of sex determination in higher Diptera. The most notable exceptions are found in the Drosophila lineage. In D. melanogaster, for instance, the sex determination cascade has been extended by incorporating Sxl as an upstream regulator of tra. The Drosophila system seems to be a derivative of the ancestral tra-based system in which the key functions, selection and maintenance of the sexual fate, are both delegated to Sxl. It has been proposed that Sxl was initially recruited as an additional coregulator of tra pre-mRNA splicing, thereby establishing a functionally redundant circuit to tra autoregulation (Siera and Cline 2008). In this context, Sxl may have gradually relieved tra from upholding the female mode of its splicing to become eventually the exclusive regulator of tra. As a result, the mechanism based on autoregulation of tra and its repression by M became obsolete and eventually completely disappeared in Drosophila.
Acknowledgments
We are indebted to Andreas Dübendorfer and Rolf Nöthiger for many stimulating and insightful discussions and Andreas Dübendorfer and Markus Niessen for helpful comments on the manuscript. Leo Beukeboom, Rhonda Hamm, Jeff Scott, and Sukran Cakir are gratefully acknowledged for providing samples of houseflies collected from natural populations in various parts of the world. We thank Claudia Brunner for technical assistance and Raymond Grunder and Hanna Nägeli for rearing of the housefly cultures. This work was supported by a grant from the Swiss National Science Foundation (31003A-109690) to D.B.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.109249/DC1.
References
- Burghardt, G., M. Hediger, C. Siegenthaler, M. Moser, A. Dubendorfer et al., 2005. The transformer2 gene in Musca domestica is required for selecting and maintaining the female pathway of development. Dev. Genes Evol. 215 165–176. [DOI] [PubMed] [Google Scholar]
- Cline, T. W., and B. J. Meyer, 1996. Vive la difference: males vs females in flies vs worms. Annu. Rev. Genet. 30 637–702. [DOI] [PubMed] [Google Scholar]
- Concha, C., and M. J. Scott, 2009. Sexual development in Lucilia cuprina (Diptera, Calliphoridae) is controlled by the transformer gene. Genetics 182 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholm, I., M. G. Franco, P. G. Rubini and M. Vecchi, 1983. Identification of a male determinant on the X chromosome of housefly (Musca domestica) populations in South-East England. Genet. Res. 42 311–322. [Google Scholar]
- Dubendorfer, A., and M. Hediger, 1998. The female-determining gene F of the housefly, Musca domestica, acts maternally to regulate its own zygotic activity. Genetics 150 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubendorfer, A., M. Hediger, G. Burghardt and D. Bopp, 2002. Musca domestica, a window on the evolution of sex-determining mechanisms in insects. Int. J. Dev. Biol. 46 75–79. [PubMed] [Google Scholar]
- Hasselmann, M., T. Gempe, M. Schiott, C. G. Nunes-Silva, M. Otte et al., 2008. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454 519–522. [DOI] [PubMed] [Google Scholar]
- Hediger, M., M. Niessen, E. A. Wimmer, A. Dubendorfer and D. Bopp, 2001. Genetic transformation of the housefly Musca domestica with the lepidopteran derived transposon piggyBac. Insect Mol. Biol. 10 113–119. [DOI] [PubMed] [Google Scholar]
- Hediger, M., G. Burghardt, C. Siegenthaler, N. Buser, D. Hilfiker-Kleiner et al., 2004. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev. Genes Evol. 214 29–42. [DOI] [PubMed] [Google Scholar]
- Hilfiker-Kleiner, D., A. Dubendorfer, A. Hilfiker and R. Nothiger, 1994. Genetic control of sex determination in the germ line and soma of the housefly, Musca domestica. Development 120 2531–2538. [DOI] [PubMed] [Google Scholar]
- Hiroyoshi, T., 1964. Sex-limited inheritance and abnormal sex ration in strains of the housefly. Genetics 50 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, H., and T. Hiroyoshi, 1981. A maternal effect mutant of the housefly that transforms sex. Jpn. J. Genet. 56 604–605. [Google Scholar]
- Inoue, H., and T. Hiroyoshi, 1982. A male-determining factor of autosomes 1 and occurrence of male-recombination in the housefly, Musca domestica L. Jpn. J. Genet. 57 221–229. [Google Scholar]
- Inoue, H., Y. Fukumori and T. Hiroyoshi, 1983. Mapping of autosomal male-determing factors of the housefly, Musca domestica L., by means of sex-reversal. Jpn. J. Genet. 58 451–461. [Google Scholar]
- Irminger-Finger, I., and R. Nothiger, 1995. The Drosophila melanogaster gene lethal(3)73Ah encodes a ring finger protein homologous to the oncoproteins MEL-18 and BMI-1. Gene 163 203–208. [DOI] [PubMed] [Google Scholar]
- Keyes, L. N., T. W. Cline and P. Schedl, 1992. The primary sex determination signal of Drosophila acts at the level of transcription. Cell 68 933–943. [DOI] [PubMed] [Google Scholar]
- Kulathinal, R. J., L. Skwarek, R. A. Morton and R. S. Singh, 2003. Rapid evolution of the sex-determining gene, transformer: structural diversity and rate heterogeneity among sibling species of Drosophila. Mol. Biol. Evol. 20 441–452. [DOI] [PubMed] [Google Scholar]
- Lagos, D., M. Koukidou, C. Savakis and K. Komitopoulou, 2007. The transformer gene in Bactrocera oleae: the genetic switch that determines its sex fate. Insect Mol. Biol. 16 221–230. [DOI] [PubMed] [Google Scholar]
- Maudlin, I., 1979. Chromosome polymorphism and sex determination in a wild population of tsetse. Nature 277 300–301. [DOI] [PubMed] [Google Scholar]
- McDonald, I. C., P. Evenson, C. A. Nickel and O. A. Johnson, 1978. Housefly genetics: isolation of a female determining factor on chromosome 4. Ann. Entomol. Soc. Am. 71 692–694. [Google Scholar]
- Meise, M., D. Hilfiker-Kleiner, A. Dubendorfer, C. Brunner, R. Nothiger et al., 1998. Sex-lethal, the master sex-determining gene in Drosophila, is not sex-specifically regulated in Musca domestica. Development 125 1487–1494. [DOI] [PubMed] [Google Scholar]
- Nothiger, R., and M. Steinmann-Zwicky, 1985. A single principle for sex determination in insects. Cold Spring Harbor Symp. Quant. Biol. 50 615–621. [DOI] [PubMed] [Google Scholar]
- O'Neil, M. T., and J. M. Belote, 1992. Interspecific comparison of the transformer gene of Drosophila reveals an unusually high degree of evolutionary divergence. Genetics 131 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane, A., M. Salvemini, P. Delli Bovi, C. Polito and G. Saccone, 2002. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129 3715–3725. [DOI] [PubMed] [Google Scholar]
- Rubini, P. G., and D. Palenzona, 1967. Response to selection for high number of heterochromosomes in Musca domestica L. Genet. Agrar. 21 101–110. [Google Scholar]
- Rubini, P. G., M. G. Franco and S. Vanossi Este, 1972. Polymorphisms for heterochromosomes and autosomal sex-determinants in Musca domestica L. Atti del IX Congresso Nazionale Italiano di Entomologia, Siena, Italy, pp. 341–352.
- Ruiz, M. F., A. Milano, M. Salvemini, J. M. Eirin-Lopez, A. L. Perondini et al., 2007. The gene transformer of anastrepha fruit flies (Diptera, tephritidae) and its evolution in insects. PLoS ONE 2 e1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone, G., I. Peluso, D. Artiaco, E. Giordano, D. Bopp et al., 1998. The Ceratitis capitata homologue of the Drosophila sex-determining gene sex-lethal is structurally conserved, but not sex-specifically regulated. Development 125 1495–1500. [DOI] [PubMed] [Google Scholar]
- Salvemini, M., M. Robertson, B. Aronson, P. Atkinson, L. C. Polito et al., 2009. Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int. J. Dev. Biol. 53 109–120. [DOI] [PubMed] [Google Scholar]
- Schmidt, R., M. Hediger, R. Nothiger and A. Dubendorfer, 1997. a The mutation masculinizer (man) defines a sex-determining gene with maternal and zygotic functions in Musca domestica L. Genetics 145 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R., M. Hediger, S. Roth, R. Nothiger and A. Dubendorfer, 1997. b The Y-chromosomal and autosomal male-determining M factors of Musca domestica are equivalent. Genetics 147 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt, C., and R. Nothiger, 2000. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127 667–677. [DOI] [PubMed] [Google Scholar]
- Siera, S. G., and T. W. Cline, 2008. Sexual back talk with evolutionary implications: stimulation of the Drosophila sex-determination gene sex-lethal by its target transformer. Genetics 180 1963–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall, L. B., H. Amrein, P. S. Morozov, A. Rzhetsky and R. Axel, 1999. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96 725–736. [DOI] [PubMed] [Google Scholar]
- Wagoner, D. E., 1969. Presence of male determining factors found on three autosomes in the house fly, Musca domestica. Nature 223 187–188. [DOI] [PubMed] [Google Scholar]