Abstract
Agrobacterium tumefaciens T-DNA normally integrates into random sites in the plant genome. We have investigated targeting of T-DNA by nonhomologous end joining process to a specific double-stranded break created in the plant genome by I-CeuI endonuclease. Sequencing of genomic DNA/T-DNA junctions in targeted events revealed that genomic DNA at the cleavage sites was usually intact or nearly so, whereas donor T-DNA ends were often resected, sometimes extensively, as is found in random T-DNA inserts. Short filler DNAs were also present in several junctions. When an I-CeuI site was placed in the donor T-DNA, it was often cleaved by I-CeuI endonuclease, leading to precisely truncated targeted T-DNA inserts. Their structure requires that T-DNA cutting occurred before or during integration, indicating that T-DNA is at least partially double stranded before integration is complete. This method of targeting full-length T-DNA with considerable fidelity to a chosen break point in the plant genome may have experimental and practical applications. Our findings suggest that insertion at break points by nonhomologous end joining is one normal mode of entry for T-DNA into the plant genome.
Transformation is an important tool for the introduction of useful traits into crops. However, transgene expression is variable among transformants. To minimize variation caused by position effect and complex transgene structure, it is desirable to have a transformation method that targets a single copy of the desirable gene to a specific genomic location suitable for stable gene expression. Site-specific recombination systems such as Cre/lox, Flp/FRT, and ϕC31 or R4 integrase have been used successfully to introduce double-stranded (DS) DNA into predetermined sites in the mammalian genome (Baer and Bode, 2001; Thyagarajan et al., 2001). The Cre/lox site-specific recombination system has been used successfully in tobacco (Nicotiana tabacum) protoplasts to introduce genes into previously placed lox sites in the genome (Albert et al., 1995; Day et al., 2000), but regeneration of fertile plants from protoplasts is not feasible for most crops. Agrobacterium tumefaciens delivery proved a less tractable approach for Cre/lox targeting in Arabidopsis (Vergunst and Hooykaas, 1998), presumably because T-DNA is delivered in single-stranded form, whereas site-specific recombinases require DS DNA as substrate.
Homologous recombination (HR) between T-DNA and plant DNA, where each has an overlapping truncated part of a selectable marker, has been reported to occur at very low frequency in higher plants (Offringa et al., 1990). Only a few examples of targeted disruption of an endogenous gene by HR have been reported in higher plants (Miao and Lam, 1995; Kempin et al., 1997; Hanin et al., 2001; Terada et al., 2002). DS DNA breaks in the genome produced by endonucleases or transposons can increase the frequency of HR and gene targeting (Puchta et al., 1993; Chiurazzi et al., 1996; Puchta et al., 1996; Puchta, 1998; Xiao and Peterson, 2000). DS DNA breaks in the genome can be repaired by either HR or nonhomologous end joining (NHEJ). In plants, DS DNA breaks are mainly repaired by NHEJ (Gorbunova and Levy, 1999). Salomon and Puchta (1998) studied repair of an I-SceI-induced break in a negatively selectable marker after A. tumefaciens delivery of the I-SceI gene. Among the various deletions and insertions inactivating the marker, they found four instances of insertion of nearly full-length donor T-DNA. To date, there have been no further experiments aimed at testing directly whether a DS DNA break can be used to target the integration of T-DNA to a desired position in the plant genome. Here, we report an assessment of the number and quality of targeted insertions of a repair construct into a truncated NPT gene at a DS DNA break introduced into the tobacco genome by transiently expressing the rare cutting endonuclease I-CeuI. Kanamycin-resistant targeted events occur at a low but usable efficiency of one to several per experiment in cocultivations of A. tumefaciens with tobacco leaf segments (150–200 per experiment), with about one-half of the products exhibiting essentially the predicted structure (donor DNA with normal T-DNA borders or an I-CeuI truncation and deletion of 10 bp or less of target DNA). Possible implications of details of their structure for the mechanism of the process are discussed below.
RESULTS
Generation of Transgenic Target Lines
The target construct for this study, delivered as T-DNA from pNOV2729 (Fig. 1), contains an NPT (nonfunctional 5′-truncated neomycin phosphotransferease) gene flanked by a cut site for meganuclease I-CeuI. Target DNA was introduced into tobacco SR1 using A. tumefaciens-mediated transformation as described in “Materials and Methods.” Four single copy lines (T2729.2, T2729.10, T2729.26, and T2729.36) were identified by Southern-blot and segregation analyses. Seeds from self-pollinated T0 plants were selected on phosphinothrycin (PPT; active ingredient of herbicide Basta)-containing medium. This population of plants with hemizygous and homozygous target loci was the source of leaf pieces transformed in the following experiments.
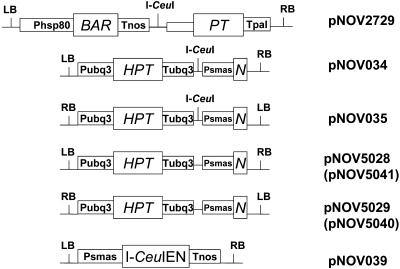
Figure 1.
Schematic drawing of vector T-DNA regions. LB, T-DNA left border repeat; RB, T-DNA right border repeat; BAR, Basta resistance gene; N, the 5′-region of NPT (neomycin phosphotransferase) gene conferring kanamycin resistance; PT, the 3′-end of NPT gene. The first intron of Arabidopsis CHC1 gene (http://www.chromdb.org/) was used to separate the 5′ and 3′ region of the NPT gene. HPT, Hygromycin phosphotransferase; I-CeuI, I-CeuI endonuclease cleavage sequence; Phsp80, Brassica oleracea HSP80 promoter; Pubq3, Arabidopsis ubiquitin 3 promoter; Psmas, super-MAS promoter; Tnos, nopaline synthase 3′-untranslated region (UTR); Tpal, Arabidopsis phenylalanine aminolyase-1 3′-UTR; Tubq3, Arabidopsis ubiquitin-3 3′-UTR; I-CeuIEN, synthetic I-CeuI endonuclease gene. The following vectors have VS1 origin of replication: pNOV034, pNOV039, pNOV2729, pNOV5028, and pNOV5040. These vectors have RK2 origin of replication: pNOV035, pNOV5025, and pNOV5041.
Targeted Integration of T-DNA
A. tumefaciens-mediated transformation was used to cotransform leaf explant tissues with donor vector carrying the NPT repair construct and an I-CeuI expression vector. The two constructs were placed on separate compatible T-DNA binary vectors such that they could be delivered by a single strain. Vectors for expression of synthetic I-CeuI were pNOV039 (Fig. 1) with a VS1 origin (Itoh and Haas, 1985) and pNOV040 with an RK2 origin (Rothstein et al., 1987). Donor T-DNAs (Fig. 1) were also put into either VS1 or RK2 vectors. Targeting donor and I-CeuI endonuclease vectors were cotransformed into A. tumefaciens LBA4404. We selected for targeted transformants by placing only the promoter and first exon of NPT on the donor DNA, followed by the 5′ end of an intron, situated to read outward through an adjacent right or left T-DNA border (Fig. 1). Such donor DNAs should produce kanamycin-resistant plant cells only by a targeting process, inserting in correct orientation at or near the target site positioned in the genome of the recipient plant.
Cocultivations were performed as described in “Materials and Methods,” using 150 to 200 leaf explants per experiment. Kanamycin-resistant shoots were recovered from selection/regeneration medium and rooted on selection rooting medium. To differentiate targeted shoots from escapes, a PCR assay was performed on genomic DNA using primers 1 and 2 (Fig. 2). Targeted transformants yielded a PCR product of about 3.5 kb. The number of events obtained (Table I) varied among experiments. We also tried to transform target lines with NHEJ donor vector (pNOV034, pNOV035, pNOV5028, or pNOV5029) alone, i.e. without codelivery of the I-CeuI endonuclease expression vector, but no targeted event was obtained (Table I). When I-CeuI expression vectors (pNOV039 and pNOV040) were used alone, no event was obtained either.
Figure 2.
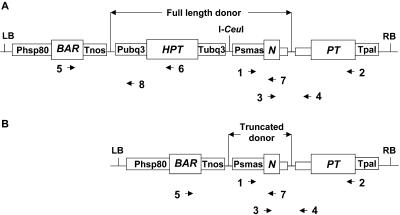
Schematic representation of targeted T-DNA insertion and primers used for PCR analysis. A, Whole-donor T-DNA is integrated into the cleaved I-CeuI site in the target locus if the I-CeuI site in the donor T-DNA is not cleaved. B, I-CeuI site in the donor T-DNA is cleaved, and the truncated T-DNA is integrated at the cleaved I-CeuI site of the target locus. PCR primers used for amplification: 1, PSMASFW2 (5′-CCG GTG AGT AAT ATT GTA CGG CTA AGA-3′); 2, NPTR6 (5′-AGA TCC TCA GAA GAA CTC GTC AAG AAG-3′); 3, NPTFA (5′-GAT CTC TAG AAT GAT TGA ACA AGA TGG ATT–3′); 4, INTBAFRV (5′-GCC GCG CTG CCT CGT CCT GAA AAA TTC AGA AA-3′) or NPTR2 (5′-GAA TAG TAC TAA TAC CTG GCA CTT CGC CCA ATA G-3′); 5, BAR78FW (5′-CAC TAC ATC GAG ACA AGC ACG GTC AAC T-3′); 6, HYG707RV (5′-CGG CCT CCA GAA GAA GAT GT–3′); 7, KAN97RV (5′-CAG AGC AGC CGA TTG TCT GT-3′); and 8, UBQPR182rv (5′-CTC CTC ACT CTT CTG CTA CAG ACT CGG AAC TC-3′).
Table I.
No. of PCR-positive targeted events
| Target Lines
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Transformation Vector(s)a | T2729.2
|
T2729.10
|
T2729.26
|
||||||||
| A | B | A | B | C | A | B | C | D | E | F | |
| pNOV034 | -b | - | - | - | 0 | - | 0 | - | - | - | - |
| pNOV040 | 0 | 0 | 0 | 0 | - | - | - | - | - | - | - |
| pNOV034 + pNOV040 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | - | - | - | - |
| pNOV035 | - | - | - | - | 0 | - | 0 | - | - | - | - |
| pNOV039 | 0 | 0 | 0 | 0 | - | 0 | - | - | - | - | - |
| pNOV035 + pNOV039 | 0 | 3 | 2 | 4 | 0 | 2 | 1 | - | - | - | - |
| pNOV5028 | - | - | - | - | - | - | - | 0 | - | - | - |
| pNOV5028 + pNOV040 | - | - | - | - | - | - | - | 0 | 0 | 0 | 0 |
| pNOV5029 | - | - | - | - | - | - | - | 0 | - | - | - |
| pNOV5029 + pNOV039 | - | - | - | - | - | - | - | 1 | 0 | - | 0 |
| pNOV5040 + pNOV040 | - | - | - | - | - | - | - | 0 | 0 | 1 | 0 |
| pNOV5041 + pNOV039 | - | - | - | - | - | - | - | 0 | 0 | - | 1 |
a If two plasmids were used, they were put into the same A. tumefaciens cell.
b Not tested. A to F, Individual cocultivation experiments.
In our initial experiments, we found that there were many more targeted events when we employed a donor strain with pNOV035 than one with pNOV034 (Table I, experiments A and B with plant lines T2729.2, T2729.10, and T2729.26). There are two salient features in which pNOV035 differs from pNOV034: orientation of donor DNA with respect to T-DNA borders (Fig. 1) and type of vector backbone. The more successful pNOV035 construct is in an RK2-type vector, and its repair 5′-NPT construct reads through the left border (LB) of donor T-DNA. We constructed four additional donor plasmids in an effort to determine what features foster targeting. In the first pair of new vectors, we simplified the donors by eliminating the internal I-CeuI site common to pNOV034 and pNOV035, forming pNOV5028 (from pNOV034) and pNOV5029 (from pNOV035). In the second set, derived from these simplified donors, we reversed the orientation of each donor DNA with respect to the T-DNA borders. pNOV5028, like its parent pNOV034, has the repair 5′-NPT construct reading through the T-DNA right border (RB) of a VS1-based binary vector. In pNOV5040, it is reversed, reading through LB. Conversely, pNOV5029, like its parent pNOV035, has 5′-NPT reading through the LB of an RK2-based binary. In pNOV5041, it is reversed, reading through RB. Targeting experiments with these new vectors yielded three new events. We note that two of the three new events were obtained from vectors with 5′-NPT that reads through LB, consistent with the earlier trend. Further, two of the three new events were obtained from RK2-based targeting donor vectors, again consistent with the earlier trend.
Sequence Analysis of Junctions of Targeted Events
In our experimental design, the coding region of NPT in both donor and target is separated from the putative junction area by intron sequence; thus, there is no direct selection for fidelity of the targeting process, except that the splicing junctions of the intron must be preserved and false splice sites avoided. To investigate the precision of integration of donor DNA at the cleaved I-CeuI recognition sequence in the chromosomal target, we cloned and sequenced the junctions of each putative targeted event. We designate the NPT side of the insertion, where donor DNA and target plant DNA are joined, as the “right junction” irrespective of whether the T-DNA border involved is an RB or LB (Fig. 2). Similarly, “left junction” designates the opposite side of the insert, in some cases truncated, where donor DNA joins to DNA of the target plant. Primers 3 and 4 (Fig. 2) were chosen to amplify the right junction region covering NPT exon 1 and intron 1. For a simple targeted event, these primers are expected to amplify an approximately 750-bp fragment containing the right (NPT) junction of donor T-DNA and the target DNA I-CeuI cleavage site (Fig. 2, A and B). Most events yielded PCR products of about this size, with the single exception of a 2.5-kb product from event 15CD.1 (Table II). The sequences of right junctions for pNOV035-derived events are depicted in a more detailed manner in Figure 3. Because the repair construct in pNOV035 reads through its left T-DNA border, the predicted junction would be a fusion of the LB cleavage site of donor to the I-CeuI cut in the target locus as shown in the model at the top of Figure 3. This model includes the 4-bp “sticky end” of the I-CeuI site, which survived in the junctions surprisingly often (see below). Initially, we did not anticipate that these four bases would be preserved because they are a 3′ extension and, thus, would not be a substrate for DNA polymerase filling-in. In Table II, we have scored junctions maintaining only the short arm of the cut site as zero, deletions from that point as negative numbers, and junctions maintaining the long arm of the I-CeuI cut site as +4, +3, etc., reflecting the number of bases preserved. The sequence of the cloned right junction PCR products revealed that resection from the I-CeuI cleavage site in the chromosomal target site is usually minimal. As summarized in the right half of Table II, in 10 of 18 events the right one-half of the I-CeuI cut site and 2 to 4 bp of the overhanging end are preserved. In six of the remaining eight cases, only 0 to 7 bp beyond the overhang was missing at the junction (Table II). There were only two instances of more extensive deletion (18 and 38 bp).
Table II.
Sequence alterations at the T-DNA integration junctions
| Event
|
Target Line
|
Targeting Vectors
|
Tnos-Donor Junctiona
|
NPT-Donor Junctiona
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Target | Filler | Donor T-DNA | Donor T-DNA | Filler | Target | |||||
| 7BH.1 | T2729.10 | PNOV035, pNOV039 | NDb | ND | ND | ND | LB | -22 | 15 | +4 |
| 7BH.2 | T2729.10 | pNOV035, pNOV039 | -105 | 180 | ICeuIc | -194 | LB | -9 | 0 | -5 |
| 9AH.1 | T2729.2 | pNOV035, pNOV039 | 0 | 46 | ICeuI | +3 | LB | -6 | 1 | +4 |
| 9AH.2 | T2729.2 | pNOV035, pNOV039 | ND | ND | ND | ND | LB | -6 | 0 | -7 |
| 9AH.4 | T2729.2 | pNOV035, pNOV039 | -2 | 20 | RB | -6 | LB | -39 | 0 | +4 |
| 9BG.1 | T2729.10 | pNOV034, pNOV040 | -1 | 348 | ICeuI | +4 | RB | -4 | 271 | +4 |
| 9BH.1 | T2729.10 | pNOV035, pNOV039 | +3 | 69 | ICeuI | -239 | LB | -26 | 7 | +4 |
| 9BH.2 | T2729.10 | pNOV035, pNOV039 | -2 | 0 | RB | -4 | LB | -9 | 0 | -5 |
| 9BH.3 | T2729.10 | pNOV035, pNOV039 | -3 | 6 | RB | -9 | LB | -123 | 8 | -4 |
| 9BH.4 | T2729.10 | pNOV035, pNOV039 | 0 | 0 | ICeuI | -4 | LB | -168 | 0 | +4 |
| 9CH.1 | T2729.26 | pNOV035, pNOV039 | ND | ND | ND | ND | LB | 0 | 0 | -18 |
| 9CH.2 | T2729.26 | pNOV035, pNOV039 | -2 | 0 | RB | -3 | LB | -129 | 10 | +3 |
| 12CE.1 | T2729.26 | pNOV034, pNOV040 | ND | ND | ND | ND | RB | -2 | 0 | +4 |
| 12CE.2 | T2729.26 | pNOV034, pNOV040 | +4 | 57 | ICeuI | +4 | RB | -20 | 0 | 0 |
| 12CG.1 | T2729.26 | pNOV035, pNOV039 | ND | ND | ND | ND | LB | -1 | 80 | +4 |
| 13CG.1 | T2729.26 | pNOV5029, pNOV039 | -4 | 0 | RB | -3 | LB | -6 | 0 | -6 |
| 15CD.1 | T2729.26 | pNOV5040, pNOV040 | ND | ND | ND | ND | LB | +1818 | 0 | -38 |
| 16CF.2 | T2729.26 | pNOV5041, pNOV039 | +2 | 4 | LB | -41 | RB | 0 | 1 | +2 |
a The model for the NPT junction is a fusion between the T-DNA LB or RB cut site and the short arm of the target locus I-CeuI endonuclease cleavage site. LB or RB indicates that this end of the donor T-DNA is proximal to the NPT junction. For events derived from T-DNA with internal I-CeuI site cleaved (see text), the junction at the Tnos side is a fusion between the target I-CeuI cleavage site and the T-DNA I-CeuI cleavage site. The length of donor DNA is scored from the short arm of the cut site; thus, a + 4 means retention of long arm sequence, etc. Where full-length T-DNA was integrated, the junction is between donor T-DNA cut site and short arm of target I-CeuI site.
b ND, Not determined due to difficulty in PCR or sequencing of PCR products.
c Donor T-DNA is cut at the I-CeuI site as determined by sequencing junctions.
Figure 3.
DNA sequence at right junctions of targeted insertions with pNOV035 T-DNA. Donor T-strand and host plant I-CeuI cleavage site are illustrated at the top of the figure, together with a theoretically “perfect” junction of T-strand LB to the long arm of the I-CeuI site. In the donor T-DNA, the LB repeat sequence is underlined, and its VirD2-nick site is indicated by a slash. In the target DNA, the I-CeuI recognition sequence is highlighted in red, and the 4-bp overhang sequence is highlighted in blue. Short filler sequences of unknown origin found in these events are shown in lowercase letters and highlighted in green. In several cases where 1 or 2 bp could be attributed either to the insert or to the host DNA (termed microhomology regions by other authors), we have assigned them to the host. This was noted in a minority of events and may be mechanistically significant but clearly is not a requirement for integration. Event 12CG.1 has an 80-bp of filler DNA inserted at “**” that is a direct repeat of part of the intron just beyond the I-CeuI target site.
On the donor T-DNA side, for events involving LB donor constructs, 22 bp of T-DNA border sequence should precede donor DNA in a perfect junction. Of our 18 events, 14 came from LB-type donors and only one (9CH.1) had all 22 bp of LB. The remaining events exhibit resections of a few base pairs for one-half and more extensive deletions of 22 to 129 bp for others. This range is typical for LBs of random T-DNA insertion events (for review, see Zambryski, 1992). With the caveat that the number of cases is small, the four events derived from RB-type donors appeared to preserve donor DNA better, exhibiting deletions of 0, 2, 4, and 20 bp from their border cut site (in events 16CF.2, 12CE.1, 9BG.1, and 12CE.2, respectively). Right T-DNA borders are normally better preserved than LBs in random T-DNA insertion events also (for review, see Zambryski, 1992).
The donor DNA of event 15CD.1 merits separate consideration. This event yielded a very large right junction PCR product because it resulted from “LB skipping” during T-DNA transfer: The donor T-strand was not cut at the LB and continued up to a presumably accidental breakage point, incorporating 1,818 contiguous bp of binary vector DNA. It seems remarkable that the NPT gene is functional despite this vector DNA, but, apparently, the splicing of the intron can eliminate this excess baggage and produce a correct mRNA sequence.
Short filler DNA was found in six of the right junctions, with lengths of 1, 1, 7, 8, 10, and 15 bp. Two events had larger filler DNAs: 12CG.1 had an 80-bp filler that was a direct repeat of DNA near the target site, and 9BG.1 had a 271-bp filler that exhibited no match to any sequence in GenBank.
The left junctions between donor DNA and target DNA likewise were amplified using several primer pairs, cloned, and sequenced. For events that incorporated full-length donor DNA, primers 5 and 8 (Fig. 2) amplified a fragment of approximately 1,250 bp. For events truncated to the I-CeuI site in some donor DNAs, primers 5 and 7 amplified a fragment of approximately 2,200 bp. PCR products were gel purified, cloned, and sequenced. Sequence information from these 12 clones is summarized in Table II. As noted at the RB, we found good preservation of target DNA in most of our events. For a perfect event, target DNA is expected to start at the staggered I-CeuI cut site. As at the right junctions, we scored borders preserving the long arm (3′ extension) of the site as +4, the short arm as 0, and deletions beyond that point as negative numbers of base pairs. In contrast to results at the RB, only one event, 12CE.2, scored a +4 on the plant side of the left junction. Most events whose left junctions could be amplified and sequenced preserved target DNA to a position ranging from +2 to –4. Event 7BH.2 was exceptional, with its target DNA resected to –105. Preservation of donor DNA at the left junction was also quite good in most cases, with nine of the 12 events exhibiting donor DNAs ending between +4 and –9. Notable donor deletions were suffered by events 7BH2 (–194), 9BH.1 (–239), and 16CF.2 (–41). Filler DNAs were commonly found in left junction fragments and tended to be longer, with only three small fillers (4, 6, and 20 bp) but five extended fillers ranging from 46 to 348 bp (Table II). Intriguingly, the latter group occurs exclusively in left junctions where the I-CeuI site in donor DNA was cleaved, leaving a free 3′-overhanging end. The presence of I-CeuI endonuclease selects against simple joining of donor and target cohesive ends; thus, filler DNA is expected in such junctions. Filler may be “recruited” by both 3′ extensions before sealing this left end break (see “Discussion”).
Five of the 18 events yielded no left junction PCR fragment. For event 12CE.1, the left junction was readily amplified and cloned, but the DNA could not be sequenced, presumably due to unusual structure. Southern analysis (data not shown) indicates that event 15CD.1 has a large insertion at the left junction that makes the PCR amplification unlikely with the condition used. The remaining events whose left junctions could not be amplified may have similar large filler DNA. Alternatively, they many have a deletion that removes a primer binding site, rendering PCR amplification unlikely. Southern-blot analyses were performed on DNA isolated from three events (13CG.1, 15CD.1, and 16CF.2) and showed that there is no gross DNA rearrangement around the target locus (data not shown). Taken together, these results confirm that donor DNA is attached to the single copy of target DNA.
Analysis of Inheritance of Targeted Events
Targeted plants were self-pollinated or pollinated with non-transgenic SR1, and their seeds were plated on kanamycin (200 mg L–1) or hygromycin (20 mg L–1). Table III shows the antibiotic resistance phenotype of the progeny populations. The results demonstrate that the kanamycin resistance phenotype of targeted events is heritable. Because all of the primary targeted events should be hemizygous for kanamycin resistance, the backcross population should show 1:1 and selfed population 3:1 segregation of resistant to sensitive phenotype. Most progeny populations are consistent with this prediction. For some events such as 16CF.2 that have one additional copy of donor T-DNA inserted (data not shown), the resistant to sensitive ratio for hygromycin is predicted to be higher than for kanamycin. Events with donor DNA truncated at the I-CeuI site (7BH.2 and 9BH.1) do not have the HPT gene integrated at the target locus. Nevertheless, both 7BH.2 and 9BH.1 are hygromycin resistant, suggesting that these events have at least one additional copy of donor T-DNA inserted in the genome and/or that the truncated T-DNA fragment with RB and HPT gene expression cassette has integrated elsewhere in the genome (Table III).
Table III.
Antibiotics resistance phenotype of the progeny from targeted events
| Lines and crosses
|
Kanamycin
|
Hygromycin
|
||
|---|---|---|---|---|
| Resistant seedlings | Sensitive seedlings | Resistant seedlings | Sensitive seedlings | |
| Target line T2729.26 | ||||
| 9CH.1 × SR1 | 59 | 63 | 61 | 45 |
| 9CH.1 selfed | 111 | 35 | 89 | 32 |
| 9CH.2 × SR1 | 40 | 30 | 27 | 28 |
| 12CE.1 selfed | 64 | 26 | 89 | 28 |
| 13CG.1 selfed | 80 | 20 | 277 | 64 |
| 16CF.2 selfed | 110 | 42 | 124 | 20 |
| Target line T2729.10 | ||||
| 7BH.1 × SR1 | 63 | 51 | 40 | 10 |
| 7BH.2 × SR1 | 91 | 85 | 72 | 50 |
| 9BH.1 selfed | 87 | 41 | 206 | 63 |
| 9BG.1 × SR1 | 26 | 28 | 29 | 25 |
DISCUSSION
Requirements for Targeted Integration
The results presented here demonstrate the feasibility of targeting T-DNA to a DS break in the plant genome. Targeted insertion in our experiments is DS break dependent: Without the I-CeuI endonuclease expression vector, no targeted event was ever recovered. We obtained targeted transformants with an efficiency of 1% to 2% with respect to the number of leaf fragments used in the cocultivation. Although this efficiency is usable, improvement may be possible by optimizing tissue culture conditions; for example, the ratio of donor DNA to I-CeuI expression cassette and concentration of A. tumefaciens, etc.
The various target plant lines used here had a single-copy insert containing, in order, a recognition sequence for I-CeuI endonuclease, the 3′ end of the aforementioned intron, and the remainder of the NPT gene plus a terminator sequence. Although selection is very powerful for targeted events, there is little constraint on the detailed structure at the integration site. The left side of the insert is under no selection at all, and the right side, the NPT junction, is situated within an intron. Fortuitous creation of a very efficient but incorrect splice site would probably render a targeted event unable to grow on kanamycin; apart from this, small insertions or deletions should not impair activity of the recombinant NPT gene. Based on this argument, any common structural features of the targeted integration events we examined presumably reflect the mechanism by which T-DNA insertion has occurred.
Common Features of Targeted Events
First, host plant DNA is well preserved. At the NPT junctions (Table II), target DNA is very often preserved perfectly, even to the extent of preserving the 4-bp 3′ extension (“long arm”) of the I-CeuI cut site. Of the 18 NPT junctions analyzed here, eight preserve all of the long arm, and four more have lost only 1, 2, or 4 bp of the 3′ extension. At the left junctions of our events, where there is no selective constraint, the target DNA is again generally well preserved. Of the 12 events for which we could get left junction sequence, three retain part of the long arm of the I-CeuI cut site (2, 3, and 4 bp, respectively), two retain the short arm of the site, six have very small resections (1–4 bp with an average of 2.3), and only one has suffered a substantial deletion (105 bp).
Second, T-DNAs that contained an internal I-CeuI site are frequently truncated. Considering only events derived from donors with an I-CeuI site, of 10 events characterized by sequencing, four have full-length T-DNA and six are truncated (four exactly and two roughly) to the I-CeuI site. Because I-CeuI is active only on DS DNA (data not shown), the donor DNA in these events must have become DS before or during the process of integration. The alternative idea that a deletion occurred after integration of full-length donor T-DNA does not account for the precise joining of the left edge of the target site in plant DNA to truncated donor in five of the six truncated events. The mechanism for the sixth truncated event, 07BH.2, is unclear because both sides of the junction are resected.
Third, T-DNAs are better preserved at the RB and more variable on the LB ends, just as for normal T-DNA inserts. The right T-DNA borders of our events exhibit an average of 5.7 bp resection from the VirD2 nicking site in the RB repeat. This value is within the range reported for T-DNA inserts in earlier studies: 5.7 bp in the study of Mayerhofer et al. (1991), 3.9 bp in the work of Gheysen et al. (1991), and 3.7 bp in transgenic aspen (Populus tremula; Kumar and Fladung, 2002). Our events exhibited an average of a 41.5-bp deletion at the LB, again within the range observed by others: 33.8 bp by Gheysen et al. (1991), 24.6 bp by Mayerhofer et al. (1991), and 6.7 bp in the aspen study (Kumar and Fladung, 2002).
Fourth, pNOV035 donor with 5′-NPT reading over the left T-DNA border produced more targeted insertions than pNOV034 with 5′-NPT reading over the RB. Table I shows that pNOV035 produced 12 events, whereas pNOV034 produced only three in parallel experiments. A possible explanation for this observation is provided in the next section.
Evidence for Single-Stranded and DS T-DNA Integration
The simplest way to account for the remarkable preservation of the 3′ extension of the target I-CeuI cut site in so many of our targeted events is to propose that these ends initiate the repair of the DS break by synthesis-dependent strand annealing (SDSA) mechanism of NHEJ, a model proposed for T-DNA integration by earlier investigators (Salomon and Puchta, 1998; Kumar and Fladung, 2002). According to this model, one of the 3′ ends of the genomic cut site (possibly further exposed by 5′ exonuclease activity on the sister strand) serves as primer for repair synthesis along the T-strand. Because of the polarity of T-DNA, this must initiate near the LB and copy toward the RB. The process is completed by action of the remaining 3′ end of the target site, which invades the T-strand duplex, synthesizing a patch. Repair synthesis and ligation can complete the integration. This model allows both 3′ ends of the target DNA to act as primers, preserving their sequence in the eventual product. Although we presume that either 3′ end of the plant target site could serve as initial primer for duplexing the T-strand, genetically it may appear that only one is used because only one will result in a T-DNA integrated in the correct orientation for NPT expression.
We found filler DNA at 53% of the junctions sequenced here. To account for these in the SDSA model, one can visualize an optional first step in which either 3′-end of genomic DNA may invade any nearby DS DNA and prime a short stretch of DNA, falling from the template shortly thereafter (perhaps evicted by reclosure of the invaded duplex). In this way, a short filler DNA may be introduced at either side of the I-CeuI cut site. Puchta and collaborators in earlier studies found no filler DNA in products of DS break repair in Arabidopsis versus 38% for tobacco (Kirik et al., 2000), a strong indication that filler DNA may be a host plant-dependent feature.
The above SDSA model accounts for the preservation of host plant DNA by allowing the 3′ end of the cut genomic DNA (i.e. the 3′ extension of the I-CeuI cut site) to serve as primer for synthesis of either filler DNA or T-DNA at both edges of the I-CeuI cleavage site. Likewise, the model can account for the loss of a few bases from both ends of the T-strand: These could be lost in the quest for microhomology with primers. The SDSA model predicts our finding that pNOV035, with 5′-NPT reading over the LB, gave more kanamycin-resistant events than did pNOV034, whose 5′-NPT reads over the RB. Because I-CeuI cleavage can occur after partial duplexing of the T-DNA, only LB-proximal genetic information survives in the truncated insertion event, and only pNOV035 has 5′-NPT situated to survive truncation. Only full-length pNOV034 T-DNA would be expected to survive selection. However, the pNOV034 products that we found were all truncated, not full length, as predicted by the SDSA model when only single-stranded T-strand is used directly as template for annealing. To account for these products, the donor T-strand must be truncated before the start of integration. The T-strand must be converted partially or totally into DS form to allow such truncation. We propose that the three pNOV034-derived targeted events have incorporated truncated T-DNA that is already partially or completely DS before SDSA process of NHEJ is initiated. We suggest that plant cells can use either single-stranded T-strand or duplexed T-DNA in the same mechanism of integration. It is plausible that the initial and most prevalent single-stranded T-strand is used more often than duplexed T-DNA, consistent with the observation that donor vector pNOV035 gave us more targeted events than pNOV034.
Is Integration at DS Breaks the Normal Mode of Random T-DNA Insertion?
As noted above, a salient characteristic of our events is the preservation of target DNA. It is plausible that the mechanism by which DS breaks are repaired should avoid further damage by deletion of flanking DNA. Does our collection of events differ from typical A. tumefaciens T-DNA insertions? If insertion of T-DNA into DS breaks in the plant genome is the normal mode of integration, we would expect similarity in the extent of target site deletion in the two cases. For the 12 events described here that could be sequenced at both junctions, the average target site deletion is 13 bp, with a range of 0 to 114 bp. Mayerhofer et al. (1991) found an average of 47.3 bp (with a range of 29–73 bp), whereas Gheysen et al. (1991) reported 18.6 bp (with a range of 13–28 bp) for Arabidopsis transformants. More recently, Kumar and Fladung (2002) analyzed 10 aspen transformants and found target deletions averaging 88.3 bp (with a range of 0–570 bp). Thus, our events appear to fall within the range of random insertion events in the extent of target site deletion, as well as the loss of donor DNA at left and RBs, as summarized above.
Evidence is accumulating that T-DNA may exploit its host cell's mechanism for repair of breaks. Some NHEJ proteins are required for A. tumefaciens-mediated random T-DNA integration in yeast (Saccharomyces cerevisiae; Van Attikum et al., 2001). A mutation in the Arabidopsis histone H2A gene blocks the process of T-DNA integration completely in Arabidopsis roots (Mysore et al., 2000). If there is more than one pathway for T-DNA integration, the existence of such mutants argues that there must be some shared steps. At least one pathway, probably not the only one, is insertion of T-DNA into DS breaks, as demonstrated earlier (Salomon and Puchta, 1998) and is analyzed more extensively in this study. The characteristics of products of this pathway are loss of several base pairs from the RB of T-DNA and deletion of little or no host plant DNA at the insertion site. Filler DNA may occur at the ends of T-DNA, apparently depending upon the plant host (Kirik et al., 2000). Whatever the mechanism of the normal T-DNA insertion process, it is clear from this work that T-DNA can opportunistically exploit DS breaks, as suggested by Salomon and Puchta (1998), or from another perspective, that DS breaks can trap T-DNA. The quality of the T-DNA insert is normal, and the damage of the process to target site DNA is minimal. From these practical perspectives, the approach described here has potential utility for genetic engineering of crop plants.
MATERIALS AND METHODS
Construction of I-CeuI Endonuclease Expression Vectors
Homing endonuclease I-CeuI protein sequence (Gauthier et al., 1991) was back translated into DNA sequence using the BackTranslate program of the GCG Wisconsin Package (Accelrys Inc., Burlington, MA) using maize (Zea mays) highly expressed gene codons except that AGC, CCC, and CTG were used for Ser, Pro, and Leu, respectively. Because expression of I-CeuI endonuclease is toxic to Escherichia coli, a 189-bp potato (Solanum tuberosum) ST-LS1 modified intron sequence (Narasimhulu et al., 1996) was inserted into the I-CeuI endonuclease coding sequence to prevent its expression in bacteria. Twelve oligonucleotides of 75 to 85 bp were used to construct the full-length synthetic gene in pBluescript KS+ (Stratagene, La Jolla, CA) forming pBSSynI-CeuI-Int. Details of construction can be obtained from the authors upon request. To form a plant expression cassette, pBH37 plasmid, a vector that contains a modified SMAS (super MAS) promoter (Ni et al., 1995) and Nos terminator (Bevan et al., 1983) with multiple cloning sites between, was digested with BglII, and the site was converted to MfeI by introduction of the following site conversion oligonucleotide: 5′-GAT CGG CAA TTG CC-3′. The resulting plasmid, pBH37M, was digested with MfeI in the presence of alkaline phosphatase. SynI-CeuI-Int was excised from pBSSynI-CeuI-Int as an EcoRI fragment and ligated into MfeI-cleaved pBH37M to form pSmasI-CeuI-Int. pSmasI-CeuI-Int was excised as an HindIII/EcoRI fragment and ligated into HindIII/EcoRI-digested pHINK078 or pNOV100 to form pNOV039 or pNOV040, respectively. pHINK078 is a binary vector with VS1 origin of replication and spectinomycin resistance gene in the backbone. pNOV100 is a binary vector with RK2 origin of replication and kanamycin resistance gene in the backbone (Rothstein et al., 1987).
Construction of Target Vector
The BAR coding region was amplified from pGSFR1 (D'Halluin et al., 1992) using two primers, BARCLA (5′-TCA TAT CGA TGA GCC CAG AAC GAC GCC-3′) and BARBGL (5′-TTT GAG ATC TTC ATA TCT CGG TGA CGG GCA GG-3′). The gel-purified PCR product was digested with BglII and inserted into SmaI/BamHI-digested pHSPnos to form pNOV2703. pHSPnos is a pSPORT1-derived vector (GIBCO BRL, Rockville, MD) containing the Brassica oleracea HSP80 promoter (Wilson and Brunke, 1997) followed by the nopaline synthase (Nos) terminator (Bevan et al., 1983). pNOV2703 was digested with NotI, filled in by Klenow fragment, digested with XhoI, and the 2.4-kb NotI(blunted)/XhoI fragment containing the B. napus HSP80 promoter-BAR-Tnos expression cassette was isolated. Binary vector pHINK078 was digested with ApaI, blunted with Klenow polymerase, and then cut with XhoI. The above 2.4-kb NotI(blunted)/XhoI HSP80 promoter-BAR-NOS 3′-UTR fragment was inserted into this vector to form pNOV2797. pNOV2797 was digested with BglII, filled in with Klenow fragment, and religated to form pNOV2706. The SacI-NcoI polylinker region (88 bp) from pQD1D1 was inserted into SacI/NcoI-digested pNOV2706 to form pNOV2722. pQD1D1 is a derivative of pLITMUS 284 (New England Biolabs, Beverly, MA) containing the SMAS promoter (Ni et al., 1995). pNOV2722 was cut with BglII and ligated with a BglII/BamHI-digested DNA fragment containing recognition sequence for I-CeuI endonuclease to form pNOV2724. The DNA fragment for the I-CeuI endonuclease recognition site was formed by annealing two oligonucleotides, ICEUBGL2 (5′-TCG AAG ATC TCT ATA ACG GTC GTA AGG TAG-3′) and ICEUBAM1 (5′-ACT TGG ATC CTC GCT ACC TTA GGA CCG TTA-3′), and filled in by Klenow polymerase. pNOV2720 is a pBluescript KS+ vector containing SMAS promoter, NPT with four Arabidopsis introns, and Arabidopsis PAL-1 3′-UTR (Tpal). pNOV2720 was digested with BglII and SacI to isolate a 3,054-bp BglII/SacI fragment containing a 5′-truncated NPT-Tpal. This fragment was inserted into BglII/SacI-digested pNOV2724 to form pNOV2729 (with I-CeuI site; Fig. 1).
Construction of Repair Donor Vectors
An EcoRV-XbaI (40 bp) fragment excised from pLITMUS28 (New England Biolabs) was cloned into pBluescript II KS+ at the corresponding sites to form pNOV2704. pNOV2705 is a pBluescript KS+ vector containing a hygromycin resistance cassette with the Arabidopsis Ubq3 promoter, hygromycin phosphotransferase gene (HPT), and Ubq3 3′-UTR (Tubq3). pNOV2704 was digested with NotI, blunted with Klenow polymerase, then cut with XbaI and ligated with the 3.1-kb KpnI(blunted)/XbaI fragment of pNOV2705 containing the Arabidopsis Ubq3 promoter-HPT-Tubq3 to create pNOV2726. pLITMUS28 was digested with BglII and ligated with a fragment-containing FRT site derived from annealed oligonucleotides FRTBGL2 (5′-GAT CTG AAG TTC CTA TTC TCT AGA AAG TAT AGG AAC TTC G-3′) and FRTBAM1 (5′-GAT CCG AAG TTC CTA TAC TTT CTA GAG AAT AGG AAC TTC A-3′) to create plasmid pNOV2727. pNOV2727 was digested with XhoI, blunted by filling in with Klenow, then cut with SacI to isolate the 2.8-kb XhoI(blunted)/SacI fragment. pNOV2720 was cut with ClaI, filled in with Klenow, then digested with SacI to isolate the 4.8-kb SacI/ClaI(blunted) fragment. The 2.8-kb XhoI (blunt)/SacI fragment of pNOV2727 was ligated with the 4.8-kb ClaI(blunted)/SacI fragment of pNOV2720 to create pNOV2732. pNOV2729 was digested with EcoRV and XhoI partially to isolate the 10-kb EcoRV/XhoI fragment. The fragment was ligated with the 3.2-kb SacI(blunted)/XhoI fragment of pNOV2726 to create pNOV2734. pNOV2734 was digested with Ecl136II and BglII partially to isolate the 10.4-kb Ecl136II/BglII fragment. pNOV2732 was digested with EcoRI, filled in with Klenow polymerase, and cut again with BglII partially to isolate the 1.4-kb EcoRI/BglII fragment. The above pNOV2734 Ecl136II/BglII (10.4 kb) and pNOV2732 EcoRI/BglII (1.4 kb) fragments were ligated to form pNOV2757. To eliminate all homology between donor DNA and the target locus, the BAR expression cassette was removed from pNOV2757 by digestion with XmaI and MluI. The plasmid was reclosed with an oligonucleotide pair (5′-CGC GGA GAT CTG GG-3′ and 5′-CCG GCC CAG ATC TC-3′) that introduced a BglII site while destroying XmaI and MluI sites to form NHEJ targeting vector pNOV034. To introduce the T-DNA of pNOV034 into an RK2 vector, pNOV034 was digested with MfeI, PacI, and XhoI. The two insert fragments were purified and ligated into a modified pNOV100 vector whose XbaI site had been converted to PvuI with the site conversion oligonucleotide 5′-CTAGGCGATCGC-3′. This modified vector was digested with PvuII and EcoRI in the presence of alkaline phosphatase and purified. Three-way ligation with the two T-DNA subfragments described above produced pNOV035.
Both pNOV034 and pNOV035 were cut with Bsu36I, filled in with Klenow polymerase, and religated to destroy the I-CeuI site to form pNOV5028 and pNOV5029, respectively. pNOV5028 was digested with BstXI, blunted with Klenow polymerase, and then cut with AscI to isolate the 4.7-kb fragment. Binary vector pHINK078 with VS1 origin of replication was cut with EcoRI, filled in with Klenow polymerase, and then recut with MluI to isolate the 4.6-kb vector. This 4.6-kb MluI/EcoRI(blunt) fragment of pHINK078 was ligated with the 4.7-kb BstXI(blunt)/AscI fragment of pNOV5028 to form pNOV5040. Binary vector pNOV100 with RK2 origin of replication was digested with BamHI, filled in with Klenow treatment, and then recut with MluI partially to isolate the 7.6-kb fragment, which was ligated with the 4.7-kb BstXI(blunt)/AscI fragment of pNOV5028 to form pNOV5041.
Plant Transformation
To establish target lines, actively growing tobacco (Nicotiana tabacum) seedlings (3–5 weeks old) were used for cocultivation with Agrobacterium tumefaciens strain LBA4404 containing pNOV2729. A. tumefaciens cells were grown in culture medium (5 g L–1 yeast [Saccharomyces cerevisiae] extract, 10 g L–1 peptone, and 5 g L–1 NaCl) with 50 mg L–1 spectinomycin for 24 to 36 h at 28°C. The A. tumefaciens culture was centrifuged, and the pellet was washed twice with 10 mL of Murashige Skoog medium with 30 g L–1 sucrose (MS3S) and resuspended in 15 mL of liquid MS3S, adjusted to a final A660 of 1.0, and employed for transformation. Leaves were excised from seedlings and cut into 1- to 2-mm strips while immersed in the A. tumefaciens cell suspension. Leaf strips were blotted dry on sterile filter paper and then placed on cocultivation medium (MS3S with naphthaleneacetic acid [0.1 mg L–1], 6-benzylaminopurine [1 mg L–1], and Gelrite agar [2.4 g L–1]) for 2 d. Leaf explants were then transferred to selection shooting medium (cocultivation medium with 5 mg L–1 PPT and 200 mg L–1 carbenicillin). Regenerated shoots were excised and rooted in selection rooting medium (MS3S with 5 mg L–1 PPT, 200 mg L–1 carbenicillin, 1 mg L–1 naphthaleneacetic acid, and 2.4 g L–1 Phyta-agar). Shoot tips were excised, and plantlets were rerooted in selection and shooting medium. PPT-resistant plantlets were assigned line numbers, transplanted into soil, and grown in a greenhouse. Plants were self-pollinated or backcrossed to non-transgenic SR1.
Selection of Targeted Transformants
For targeting experiments, seeds from single-copy T-DNA transgenic tobacco target lines were surface sterilized with 15% (v/v) clorox bleach for 15 min, washed with sterile distilled water three times, and dried in a laminar flow hood overnight. Dried seeds were plated on MS3S agar with 5 mg L–1 PPT. PPT-resistant seedlings were transferred to MS3S agar at 14 d after plating and grown under fluorescent light (12 h of light and 12 h of dark). Actively growing seedlings (3–5 weeks old) were used for cocultivation for 2 to 4 d with A. tumefaciens strain LBA4404 containing donor vectors. The procedure was as described above for generation of target lines except that 200 mg L–1 kanamycin instead of PPT was used as selection agent.
Molecular Analysis of Transgenic Plants
DNA was isolated from leaf tissues using a hexadecyl-trimethyl ammonium bromide protocol (Que et al., 1997). PCR was performed using a Perkin-Elmer 9600 thermocycler (Perkin-Elmer Cetus, Emeryville, CA). For some PCR experiments, Ready-To-Go PCR Beads (Amersham Pharmacia, Piscataway, NJ) were employed. Taq polymerase (Qiagen Inc, Valencia, CA) was used for other PCR amplifications. The Expand Long Template PCR amplification system (Roche Molecular Biochemicals, Indianapolis) was used for amplification of longer (>3 kb) products.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the materials. Obtaining any permission will be the responsibility of the requestor. Federal or international regulation may restrict shipment of viable transgenic seeds.
Acknowledgments
We thank Monique Turmel and Claude Lemieux for providing plasmids containing the native I-CeuI endonuclease gene. We are grateful to Dawn McNamara and Kelly Clark for taking care transgenic plants in the greenhouse. We are also thankful for helpful discussions with our colleagues, Genevieve Hansen, Leo Melchers, Jan Suttie, and Allan Wenck.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.026104.
References
- Albert H, Dale EC, Lee E, Ow DW (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7: 649–659 [DOI] [PubMed] [Google Scholar]
- Baer A, Bode J (2001) Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr Opin Biotechnol 12: 473–480 [DOI] [PubMed] [Google Scholar]
- Bevan M, Barnes WM, Chilton M-D (1983) Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res 11: 369–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurazzi M, Ray A, Viret J-F, Perera R, Wang X-H, Lloyd A, Signer ER (1996) Enhancement of somatic intrachromosomal homologous recombination in Arabidopsis by the HO endonuclease. Plant Cell 8: 2057–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CD, Lee E, Kobayashi J, Holappa LD, Albert H, Ow DW (2000) Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev 14: 2869–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Halluin K, De Block M, Denecke J, Janssens J, Leemans J, Reynaerts A, Botterman J (1992) The bar gene as selectable and screenable marker in plant engineering. Methods Enzymol 216: 415–426 [DOI] [PubMed] [Google Scholar]
- Gauthier A, Turmel M, Lemieux C (1991) A group I intron in the chloroplast large subunit rRNA gene of Chlamydomonas eugametos encodes a double-strand endonuclease that cleaves the homing site of this intron. Curr Genet 19: 43–47 [DOI] [PubMed] [Google Scholar]
- Gheysen G, Villarroe R, Van Montagu M (1991) Illegitimate recombination in plants: a model for T-DNA integration. Genes Dev 5: 287–297 [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Levy AA (1999) How plants make ends meet: DNA double-strand break repair. Trends Plant Sci 4: 263–269 [DOI] [PubMed] [Google Scholar]
- Hanin M, Volrath S, Bogucki A, Briker M, Ward E, Paszkowski J (2001) Gene targeting in Arabidopsis. Plant J 28: 671–677 [DOI] [PubMed] [Google Scholar]
- Itoh Y, Haas D (1985) Cloning vectors derived from the Pseudomonas plasmid pVS1. Gene 36: 27–36 [DOI] [PubMed] [Google Scholar]
- Kempin SA, Liljegren SJ, Block LM, Rounsley SD, Yanofsky MF, Lam E (1997) Targeted disruption in Arabidopsis. Nature 389: 802–803 [DOI] [PubMed] [Google Scholar]
- Kirik A, Salomon S, Puchta H (2000) Species-specific double-strand break repair and genome evolution in plants. EMBO J 19: 5562–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Fladung M (2002) Transgene integration in aspen: structures of integration sites and mechanism of T-DNA integration. Plant J 31: 543–551 [DOI] [PubMed] [Google Scholar]
- Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C (1991) T-DNA integration: a mode of illegitimate recombination in plants. EMBO J 10: 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z-H, Lam E (1995) Targeted disruption of the TGA3 locus in Arabidopsis thaliana. Plant J 7: 359–365 [DOI] [PubMed] [Google Scholar]
- Mysore KS, Nam J, Gelvin S (2000) An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc Natl Acad Sci USA 97: 948–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhulu SB, Deng X-B, Sarria R, Gelvin SB (1996) Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8: 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Cui D, Einstein J, Narasimhulu S, Vergara CE, Gelvin SB (1995) Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J 7: 661–676 [Google Scholar]
- Offringa R, de Groot MJA, Haagsman HJ, Does MP, van der Elzen PJM, Hooykaas PJJ (1990) Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO J 9: 3077–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H (1998) Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J 13: 331–339 [Google Scholar]
- Puchta H, Dujon B, Hohn B (1993) Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res 21: 5034–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B (1996) Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA 93: 5055–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein SJ, Lahners KN, Lotstein RJ, Carozzi NB, Jayne SM, Rice DA (1987) Promoter cassettes, antibiotic-resistance genes, and vectors for plant transformation. Gene 53: 153–161 [DOI] [PubMed] [Google Scholar]
- Que Q, Wang HY, English JJ, Jorgensen RA (1997) The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9: 1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S, Puchta H (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J 17: 6086–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S (2002) Efficient gene targeting by homologous recombination in rice. Nat Biotechnol 20: 1030–1034 [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP (2001) Site-specific genomic integration in mammalian cells mediated by phage ϕC31 integrase. Mol Cell Biol 21: 3926–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Attikum H, Bundock P, Hooykaas PJJ (2001) Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J 20: 6550–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst AC, Hooykaas PJJ (1998) Cre/lox-mediated site-specific integration of Agrobacterium T-DNA in Arabidopsis thaliana by transient expression of cre. Plant Mol Biol 38: 393–406 [DOI] [PubMed] [Google Scholar]
- Wilson SL, Brunke KJ March 18, 1997. Plant promoter. U.S. Patent No. 5,612,472
- Xiao Y-L, Peterson T (2000) Intrachromosomal homologous recombination in Arabidopsis induced by a maize transposon. Mol Gen Genet 263: 22–29 [DOI] [PubMed] [Google Scholar]
- Zambryski PC (1992) Chronicles from the Agrobacterium plant cell DNA transfer story. Annu Rev Plant Physiol Plant Mol Biol 43: 465–490 [Google Scholar]