Abstract
The budding yeast Gcn5p is a prototypic histone acetyltransferase controlling transcription of diverse genes. Here we show that Gcn5p is itself regulated by Snf1p and Spt3p. Snf1p likely controls Gcn5p via direct interaction. Mutating four residues in the Gcn5p catalytic domain, T203, S204, T211, and Y212 (TSTY), phenocopies snf1 null cells, including Gcn5p hypophosphorylation, hypoacetylation at the HIS3 promoter, and transcriptional defects of the HIS3 gene. However, overexpressing Snf1p suppresses the above phenotypes associated with the phosphodeficient TSTY mutant, suggesting that it is the interaction with Snf1p important for Gcn5p to activate HIS3. A likely mechanism by which Snf1p potentiates Gcn5p function is to antagonize Spt3p, because the HIS3 expression defects caused by snf1 knockout, or by the TSTY gcn5 mutations, can be suppressed by deleting SPT3. In vitro, Spt3p binds Gcn5p, but the interaction is drastically enhanced by the TSTY mutations, indicating that a stabilized Spt3p–Gcn5p interaction may be an underlying cause for the aforementioned HIS3 transcriptional defects. These results suggest that Gcn5p is a target regulated by the competing actions of Snf1p and Spt3p.
THE Saccharomyces cerevisiae Gcn5p is one of the best-studied histone acetyltransferases. The structure, catalytic mechanism, and functions of Gcn5p are highly conserved through evolution (Roth et al. 2001). Gcn5p-mediated promoter histone H3 hyperacetylation is critical for transcriptional activation of multiple stress-related genes, including amino acid biosynthesis (Kuo and Allis 1998; Kuo et al. 2000), phosphate metabolism (Gregory et al. 1998; Vogelauer et al. 2000), sporulation (Burgess et al. 1999), and others (Chiang et al. 1996; Krebs et al. 1999; Verdone et al. 2002). In addition, Gcn5p contributes to global acetylation of histones H3 and H4 extending beyond the promoter region (Kuo et al. 2000), although the significance and mechanism of the global acetylation remains poorly understood. Under certain conditions, the H3 acetylation activity of Gcn5p spreads well into the open reading frame of several genes, and that acetylation is causally linked to H3 eviction on the path of the transcribing RNA polymerase II (Govind et al. 2007).
The structures of the catalytic domain of ciliate, yeast, and human Gcn5p have been solved by both NMR and X-ray crystallography (Clements et al. 1999; Lin et al. 1999; Rojas et al. 1999; Trievel et al. 1999). Kinetic and mutational studies identified residues essential for catalysis, acetyl coenzyme A binding, and histone substrate association (Kuo et al. 1998; Wang et al. 1998; Tanner et al. 1999; Langer et al. 2001; Poux and Marmorstein 2003). The yeast Gcn5p is the catalytic subunit of several chromatographically distinct complexes including SAGA, ADA (Grant et al. 1997), SALSA, and SLIK (Pray-Grant et al. 2002, 2005; Sterner et al. 2002a). All Gcn5p complexes except the smaller ADA complex share TBP-associated factors (TAFs) with TFIID (Grant et al. 1998). Indeed, SAGA and TFIID make overlapping contributions to the expression of yeast genes (Holstege et al. 1998; Lee et al. 2000; Huisinga and Pugh 2004). SAGA is recruited to the promoter by transcriptional activators (Kuo et al. 2000), and performs both HAT-dependent and HAT-independent functions (Roberts and Winston 1997; Sterner et al. 1999). The latter includes recruitment of TBP or the Swi/Snf complex (Dudley et al. 1999; Sterner et al. 1999; Belotserkovskaya et al. 2000; Lee et al. 2000; Larschan and Winston 2001, 2005; Bhaumik and Green 2002; Yu et al. 2003; Biswas et al. 2004; Qiu et al. 2004; Topalidou et al. 2004), regulation of histone H2B ubiquitinylation (Henry et al. 2003; Daniel et al. 2004; Wyce et al. 2004; Ingvarsdottir et al. 2005; Lee et al. 2005), stimulation of histone methylation (Pray-Grant et al. 2005; Govind et al. 2007), and H3/H4 eviction (Govind et al. 2007). Moreover, it appears that the histone acetyltransferase Gcn5p also possesses non-HAT functions. For example, microarray data showed that gcn5Δ cells displayed transcriptional defects in more genes than strains expressing a catalytically inactive mutant Gcn5p (Holstege et al. 1998; Huisinga and Pugh 2004). Tethering a catalytically inactive mutant of Gcn5p to subtelomeric regions is sufficient to counteract silencing (Jacobson and Pillus 2004). Consistent with these findings, a catalytically inactive mutant allele of Gcn5p, E173H, can be rescued by certain suppressors without changing the H3 hypoacetylation phenotype (Liu et al. 2005, and see below).
To date, very little is known about how Gcn5p itself may be regulated. The transcriptional coactivator function of Gcn5p depends critically on its stable association with Ada2p and Ada3p (Candau et al. 1997). These three proteins form a core for different Gcn5p-containing complexes. Upon associating with Ada2p and Ada3p, Gcn5p adopts the nucleosomal acetylation activity (Balasubramanian et al. 2002; Sterner et al. 2002b). Interestingly, Ada2p/Ada3p not only stimulate the enzymatic activity of Gcn5p, but also expand its in vitro substrate specificity from nearly exclusively K14 of free histone H3, to K18>K14>K9>K23 of H3 within oligonucleosomes (Sermwittayawong and Tan 2006). K36 of nucleosomal histone H3 is recently shown to be a conserved, novel target for the SAGA complex (Morris et al. 2007). The ability of Ada2/Ada3 to help Gcn5p expand its substrate specificity is reminiscent of, and possibly related to the observations that Mg2+ alone can convert an otherwise refractory nucleosomal array into a good acetylation substrate for recombinant Gcn5p (Tse et al. 1998). It is thus likely that transacting factors such as Ada2p/Ada3p and Mg2+ modulate the nucleosomal histone conformation in such a way that Gcn5p is less restrictive of selecting its substrate. Indeed, Ada2p possesses the chromatin-binding SANT domain (Aasland et al. 1996; Sterner et al. 2002b; Boyer et al. 2004). On the other hand, it remains possible that the molecular and enzymatic behaviors of Gcn5p may be modulated by its association with Ada2p, Ada3p, and even other proteins.
For transcriptional regulation, Gcn5p performs both synergistic and antagonistic functions with many factors, including Spt3p, Spt8p (Belotserkovskaya et al. 2000; Larschan and Winston 2001; Bhaumik and Green 2002; Yu et al. 2003; Helmlinger et al. 2008), TBP (Dudley et al. 1999; Barbaric et al. 2003), Swi/Snf complex components (Pollard and Peterson 1997; Syntichaki et al. 2000; Hassan et al. 2001, 2002), the histone H2A variant Htz1p (Santisteban et al. 2000), global chromatin regulators Nhp6p and Sin1p (Yu et al. 2003), histone deacetylases (Perez-Martin and Johnson 1998), and Snf1p (Lo et al. 2000, 2001). While some of these proteins may influence chromatin dynamics, which consequently affects Gcn5p functions, Snf1p and Spt3p appear to play more direct roles in regulating Gcn5p. For example, Gcn5p physically interacts with and is phosphorylated by Snf1p in vitro (Liu et al. 2005); overproduction of Snf1p rescues the E173H allele of Gcn5p selectively (Liu et al. 2005). Deleting SPT3, which encodes an integral component of the SAGA complex, partially rescues the transcriptional defects of HO in gcn5Δ (Yu et al. 2003). One explanation is that Spt3p inhibits TBP–TATA association (Belotserkovskaya et al. 2000; Yu et al. 2003) and that the Gcn5p action somehow stimulates TBP recruitment (Bhaumik and Green 2002). Conversely, transcriptional activation of GAL genes is enhanced by Spt3p (Eisenmann et al. 1992; Bhaumik and Green 2002). Spt8p and Spt3p can be crosslinked to TBP (Warfield et al. 2004; Sermwittayawong and Tan 2006; Mohibullah and Hahn 2008), consistent with the genetic evidence for a physical role of Spt3p for TBP recruitment in vivo (Eisenmann et al. 1992; Dudley et al. 1999; Larschan and Winston 2001; Yu et al. 2003; Laprade et al. 2007). It is thus likely that Gcn5p interacts with TBP in an indirect manner.
In the present work, the functional and physical relationships between Gcn5p, Snf1p, and Spt3p were characterized. Four previously uncharacterized residues within the catalytic domain of Gcn5p (T203, S204, T211, and Y212) are critical determinants for Snf1p-dependent Gcn5p phosphorylation both in vitro and in vivo. These residues are also important for the histone acetylation and transcriptional activation functions of Gcn5p. Interestingly, transcriptional defects resulting from alanine substitution of these residues or from deleting SNF1 can be suppressed by knocking out SPT3. Physically, Gcn5p interacts with Spt3p in vitro. These findings suggest a new regulatory mechanism for transcriptional activation by Gcn5p.
MATERIALS AND METHODS
Yeast strains and plasmids:
Yeast strains, plasmid constructs, and oligonucleotide primers used in this work are listed in Tables 1–3. All genetic methods were performed according to standard procedures (Sherman 1991). Yeast transformation was done using the lithium acetate method (Gietz et al. 1992).
TABLE 1.
Yeast strain list
| Strains | Relevant genotype | Source |
|---|---|---|
| yMK839 | MATatrp1 leu2-3,112 ura3-52 | Kuo et al. (1996) |
| yMK842 | MATatrp1 leu2-3,112 ura3-52 gcn5Δ∷hisG | Kuo et al. (1996) |
| yMK986 | MATatrp1 leu2-3,112 ura3-52 gcn5E173H | Liu et al. (2005) |
| yYL232 | MATatrp1 leu2-3,112 ura3-52 snf1Δ∷LEU2 | Liu et al. (2005) |
| yYL515 | MATatrp1 leu2-3,112 ura3-52 spt3Δ∷KanMX6 | This study |
| yYL516 | MATatrp1 leu2-3,112 ura3-52 gcn5E173H spt3Δ∷KanMX6 | This study |
| yYL590 | MATatrp1 leu2-3,112 ura3-52 GCN5-8xmyc∷TRP1 | This study |
| yYL591 | MATatrp1 leu2-3,112 ura3-52 SNF1-8xmyc∷TRP1 | This study |
| yMK1422 | MATatrp1 leu2-3,112 ura3-52 gcn5Δ∷hisG SNF1-8xmyc∷TRP1 | This study |
| yYL622 | MATatrp1 leu2-3,112 ura3-52 SPT7-13xmyc∷TRP1 | This study |
| yYL682 | MATatrp1 leu2-3,112 ura3-52 gcn5Δ∷hisG spt3Δ∷KanMX6 | This study |
| yYL683 | MATatrp1 leu2-3,112 ura3-52 snf1Δ∷LEU2 spt3Δ∷KanMX6 | This study |
| yYL782 | MATatrp1 leu2-3,112 ura3-52 gcn5TSTY/4A-8xmyc∷TRP1 | This study |
| yYL783 | MATatrp1 leu2-3,112 ura3-52 gcn5TSTY/4A-8xmyc∷TRP1 spt3Δ∷KanMX6 | This study |
| yYL786 | MATatrp1 leu2-3,112 ura3-52 spt8Δ∷URA3 snf1Δ∷LEU2 | This study |
| yYL787 | MATatrp1 leu2-3,112 ura3-52 spt8Δ∷URA3 GCN5-8xmyc∷TRP1 | This study |
| yYL788 |
MATatrp1 leu2-3,112 ura3-52 spt8Δ∷URA3 gcn5 TSTY/4A-8xmyc∷TRP1 |
This study |
TABLE 2.
Plasmid list
| Plasmid | Description | Source |
|---|---|---|
| pMK100 | pRSET–Gcn5–6xHis | Kuo et al. (1996) |
| pFW32 | YEp–SNF1 | Winston and Minehart (1986) |
| pYL41 | YEplac112–SNF1 | Liu et al. (2005) |
| pYL42 | pYEX-4T–GST–Snf1 | Hahn and Thiele (2004) |
| pYL43 | pYEX-4T–GST–Snf1K84R | Hahn and Thiele (2004) |
| pYL44 | pYEX-4T–GST | Liu et al. (2005) |
| pYL54 | pYEX-4T–Gcn5–TAP | Liu et al. (2005) |
| pYL55 | pYEX-4T–Gcn5E173H–TAP | This study |
| pYL67 | 8xmyc∷TRP1 for tagging proteins with 8 myc repeats | Liu et al. (2005) |
| pYL72 | pMK547Gcn5, 3xHA–Gcn5 | Liu et al. (2005) |
| pYL89 | pMK547Spt3, 3xHA–Spt3 | This study |
| pYL90 | pET21a–3xHA–Spt3 | This study |
| pYL93 | pYEX-4T–Gcn5TSTY/4A–TAP | This study |
| pYL98 | pRSET–Gcn5TSTY/4A–6xHis | This study |
| pMK284 | GCN5 integration construct | Liu et al. (2005) |
| pMK515 | pET21–6xHis–Gcn5 protein | Liu et al. (2005) |
| pMK625 |
pGEX5–HA–SPT3 and derivatives |
This study |
TABLE 3.
List of oligos
| Name | Sequence | Description |
|---|---|---|
| GCN5AAs | GACTATGTTAGAAATGCCGCGAACATAAAATATTTTTTG | GCN5 T203A/S204A, sense |
| GCN5AAas | CAAAAAATATTTTATGTTCGCGGCATTTCTAACATAGTC | GCN5 T203A/S204A, antisense |
| GCN5T203As | CTTAAAAGACTATGTTCGAAATGCCTCGAACATAAAATATT | GCN5 T203A, sense |
| GCN5T203Aas | AATATTTTATGTTCGAGGCATTTCGAACATAGTCTTTTAAG | GCN5 T203A, antisense |
| GCN5S204As | CTTAAAAGACTATGTTCGAAATACCGCGAACATAAAATATTT | GCN5 S203A, sense |
| GCN5S204Aas | AAATATTTTATGTTCGCGGTATTTCGAACATAGTCTTTTAAG | GCN5 S203A, antisense |
| T211As | CATAAAATATTTTTTGGCGTACGCAGATAATTACGCT | GCN5 T211A, sense |
| T211Aas | AGCGTAATTATCTGCGTACGCCAAAAAATATTTTATG | GCN5 T211A, antisense |
| T211/Y212As | CATAAAATATTTTTTGGCCGCGGCAGATAATTACGCT | GCN5 T211A/Y212A, sense |
| T211/Y212Aas | AGCGTAATTATCTGCCGCGGCCAAAAAATATTT TAT G | GCN5 T211A/Y212A, antisense |
| Y212As | CATAAAATATTTTTTGACTGCAGCAGATAATTACGCT | GCN5 Y212A, sense |
| Y212Aas | AGCGTAATTATCTGCTGCAGTCAAAAAATATTTTATG | GCN5 Y212A, antisense |
| spt3KX | GATCGGCGGAAACGAAAAGTAAAAAGTAAGGTTGAAGACAC | Replacing SPT3 with KanMX |
| TCTGGCTTCGTACGCTGCAGGTCG | ||
| spt3KXas | CATACCAGAAGGAAACCCATGCACCTCCATGATGAAATTATA CCATAGGCCACTAGTGGATCTG | Replacing SPT3 with KanMX |
| MHK232 | GCGATTTCGAAATCGTTC | Genomic PCR to verify gcn5 mutant integration |
| MHK234 | CTGAGAGAATAGGAGG | Genomic PCR to verify gcn5 mutant integration |
| MK80 | CTGTGGGAAAAACTTATC | HIS3 oligo for ChIP, −1 nucleosome |
| MK80as | AAAGGACTGTGTTATGAC | HIS3 oligo for ChIP, −1 nucleosome |
| MK81 | TGAGCAGGCAAGATAAAC | HIS3 oligo for ChIP, +1 nucleosome |
| MK81as | CCACCCTTTAAAGAGATC | HIS3 oligo for ChIP, +1 nucleosome |
| MK100-1 | CGGTAACATCGTTATGTCCG | ACT1 oligo for ChIP |
| MK100as | ACGATAGATGGACCACTTTCG | ACT1 oligo fpr ChIP |
| YLK92 | CTGCTCAGTGCGGCCGCTCTAGCTCTAGAATGATGGACAAG CATAAG | 3xHA–Spt3 construction |
| YLK93 | TGCAGGTCGACGGTATCGGGGGATCCACTATTACATGATAAT TGGTTTAG | 3xHA–Spt3 construction |
| YLK94 | TCCGAATTCGGCGGCCGCATCTTTTACCCATAC | HA–Spt3 PCR from pYL89 |
| YLK95 | CTACTCGAGCTACATGATAATTGGTTTAGAACTGAG | HA–Spt3 PCR from pYL89 |
| YLK107 | GTACAATCAATCAATCAATCATCACATAAAATGTTCAGCGAATTG ACCATGGCAATTCCC | Replacing Gcn5–TAP with HA- |
| Spt3–TAP in pYL54 | ||
| YLK108 | GACGGCTATGAAATTCTTTTTCCATCTTCTCTTTTCCATGGATGG TTTAGAACTGAGTC | Replacing Gcn5–TAP with HA- |
| Spt3–TAP in pYL54 | ||
| HIS3 5′RT | AGCTTTGCAGAGGCTAGCAG | HIS3 RT–PCR 5′ primer |
| HIS3 3′RT | GCGAGGTGGCTTCTCTTATG | HIS3 RT–PCR 3′ primer |
| PGK1 5′RT | TCATTGGTGGTGGTGACACT | PGK1 RT–PCR 5′ primer |
| PGK1 3′RT | GCAACACCTGGCAATTC | PGA1 RT-PCR 3′ primer |
| o415 | GCGAGTGGCACTGGAAATCCTTAAGGGTAGAGGTGGTGAAGATG ATTTGAAAAAAGC | SPT3(1–107) sense |
| o416 | GCTTTTTTCAAATCATCTTCACCACCTCTACCCTTAAGGATTTCCA GTGCCACTCGC | SPT3(1–107) anti-sense |
| o419 | GGCCGCTCTAGCTCTAGAATGATGGACAAGCTTCCTGGGGCAGG TGGTGAAGATGATTTG | SPT3(107–337) sense |
| o420 | CAAATCATCTTCACCACCTGCCCCAGGAAGCTTGTCCATCATTCTA GAGCTAGAGCGGCC | SPT3(107–337) anti-sense |
| o421 | GCCGCTCTAGCTCTAGAATGATGGACAAGCTTAATAATGACGACAAT GATGATATGGATG | SPT3(156–337) sense |
| o422 |
CATCCATATCATCATTGTCGTCATTATTAAGCTTGTCCATCATTCTA GAGCTAGAGCGGC |
SPT3(156–337) anti-sense |
The spt3Δ strains were created by introducing a PCR fragment containing the KanMX6 cassette flanked by SPT3 sequences outside the open reading frame (Wach et al. 1994). G418-resistant transformants were examined by genomic PCR to confirm the genotype.
The yeast construct pYL89 that expresses HA-tagged Spt3p was created by cotransforming XbaI-linearized pMK547 (Liu et al. 2005) (an ARS CEN TRP1 plasmid with ADH1 promoter and terminator flanking a multicloning sequence and an trimeric HA epitope tag), and a PCR-amplified SPT3 open reading frame fragment, resulting in N′–HA-tagged Spt3p. To create pMK625 that expresses GST–HA–Spt3p in bacteria, the EcoRI–XhoI HA–SPT3 fragment of pYL89 was gel purified and inserted into the EcoRI and NotI sites of pGEX-5X-2 (GE Life Sciences) to create pMK625. The GST fusion significantly enhances the solubility of the fusion protein in Escherichia coli and is essential for obtaining sufficient quantity for in vitro assays. Following the QuikChange method (Roche), pMK625 was further modified using primers o415–o422 (Table 3) to create selective fragments of Spt3p.
2μ HA–GCN5 wild type, TSTY/4A, and HA–SNF1 constructs (pMK681, pMK681 TSTY/4A, and pMK682, respectively) were generated by cotransforming to yeast the GCN5 and SNF1 open reading frame PCR fragments flanked with sequences homologous to the NotI-linearized pMK595. A trimeric HA tag was consequently fused to the NH3 terminus of Gcn5p and Snf1p.
To introduce GCN5 mutants to the native locus, a URA3 integrative construct, pMK284 (Liu et al. 2005) was used for site-directed mutagenesis. pMK284 bearing selective mutations of GCN5 was linearized by NgoMIV, and transformed into yMK986 possessing the E173H allele of GCN5. The EcoRI site spanning codons 169 and 170 had been eliminated upon creating the E173H mutation. Replacing the E173H allele with other mutations thus restored the EcoRI site, and the presence of which served as a screening criterion. After genomic PCR verification of the integration, yeast cells were selected by 5-FOA for URA3 pop-out. Colonies formed on 5-FOA plates were grown for genomic PCR amplification using primers MHK234 and MHK232. PCR fragments bearing an EcoRI site were further sequenced for verification.
Pro-Q Diamond staining:
For Pro-Q Diamond staining, 3 × 109 yeast cells expressing a TAP-tagged Gcn5p (expressed from pYL54 or pYL93) were collected from early log phase cultures. Whole cell lysates were prepared as previously described (Liu et al. 2005) except that 800 μl of PiPT buffer (50 mm potassium phosphate, pH 7.5; 140 mm potassium chloride; 0.1% Triton X-100; 1 mm DTT; 1 mm EDTA; 1 mm sodium orthovanadate; 10 mm sodium fluoride; complete protease inhibitor cocktail (Roche; 1 tablet/20 ml buffer); 1 mm PMSF) was used. Affinity purification was done by incubating 500 μl of whole cell lysates with 20 μl of IgG sepharose 6G beads (GE Life Sciences) at 4° for 1 hr. IgG beads were collected with gentle centrifugation and washed twice with PiPT buffer without protease inhibitors, followed by two more washes with PiPT buffer containing 500 mm KCl. If necessary, TEV digestion was conducted according to Rigaut et al. (1999). In most cases, the IgG-bound materials were directly boiled in 1× SDS–PAGE loading dye and resolved by electrophoresis. Alternatively, some of the beads were treated with λ phosphatase (New England Biolab) prior to boiling and electrophoresis.
Following SDS–PAGE, the gels were fixed with 100 ml of 50% methanol and 10% acetic acid (v/v) for 1 hr to overnight. The residual methanol and acetic acid was removed by washing with 50 ml deionized water for 10 min with gentle agitation. After repeating the wash three times, the gel was incubated with 50 ml of Pro-Q Diamond phosphoprotein gel staining solution (Molecular Probes) for 1 hr, with gentle agitation in dark. To reduce the background and nonspecific staining, the gel was treated with destaining solution [50 mm NaOAc, pH 4.0; 20% acetonitrile (v/v)] for 30 min with three repeats, and twice deionized water wash at room temperature for 5 min each. The staining was detected by Molecular Imager FX-PRO Plus (BioRad), and stained by Coomassie Blue R250.
Recombinant protein expression and purification:
Protocols for purification of His-tagged Gcn5p (amino acid residues 19–348), GST–Snf1p, and for in vitro phosphorylation of Gcn5p by Snf1p were as previously described (Liu et al. 2005). To express GST–HAx3–Spt3p (pMK625) and its truncated fragments, BL21 Codon-Plus strain was transformed with pMK625 and derivatives. Two hundred ml LB-Amp cultures grown to 0.6 OD600 were induced with 0.5 mm IPTG at 37° for 3 hr. Cells were then pelleted and suspended in 6 ml lysis buffer (50 mm NaPi, pH 7.5, 150 mm NaCl, 1 mm PMSF), and frozen at −80°. After two freeze-and-thaw (4°) cycles, cell slurry was sonicated on ice for 20 sec, seven times, with ice-water chilling in between each homogenization. Lysates were clarified by centrifugation at 10,000 × g for 15 min at 4°. One hundred fifty microliters of 1:1 glutathione beads slurry (Sigma) were added to the lysates and incubated under gentle agitation for 2 hr at 4°. Beads were pulse-spin collected and washed twice with 10 ml lysis buffer, and once with 1.5 ml lysis buffer. Bound proteins were eluted with 50 mm reduced glutathione (Sigma) in 150 μl lysis buffer at 4° for 30 min. A second elution was conducted exactly as the first one, and the two eluates were combined and stored at −80°.
Gcn5p–Spt3p interactions:
The interaction between Gcn5p and Spt3p was tested by the Farwestern approach. Pulldown assays using immobilized Gcn5p or Spt3p suffered from high background binding of both proteins to the matrix nonspecifically (data not shown). For Farwestern assays, His-tagged Gcn5p or Hmt1p, 0.5 μg each, were resolved by SDS–PAGE and blotted to PVDF membrane by standard blotting methods. The membrane was first blocked by 10% nonfat milk in TTBS (50 mm Tris–HCl, pH 7.5, 150 mm NaCl, and 0.1% Tween-20) for 0.5–1 hr at room temperature, followed by three washes of 100 ml of TTBS. All subsequent steps, except the final development, were carried out at 4°. For Spt3p–Gcn5p interaction, the membrane was incubated with 4 ml of TTBS containing protease inhibitors, 0.1% gelatin, and about 5 μm of GST or GST–HA–Spt3p recombinant proteins. Alternatively, crude bacterial lysates containing the Spt3 derivatives or GST were used for the binding. No clear difference was seen between the use of a highly purified Spt3 or crude bacterial lysates. The binding reaction was gently rocked at 4° overnight, followed by three 10-min TTBS washes. To detect the presence of HA-tagged Spt3p on the membrane, the 12CA5 monoclonal anti-HA antibodies (Roche), 1:1000 dilution, was incubated with the membrane (in TTBS supplemented with 0.1% gelatin) for 2 hr, followed by three TTBS washes and secondary antibody incubation (HRP-conjugated goat anti-mouse Ab, BioRad, 1:7500 dilution) (2 hr), and final washes (three TTBS washes, 10 min each). The Lumi-Light Western Blotting kit (Roche) was used for the development.
RNA preparation and RT–PCR:
Yeast cells were grown in appropriate selective media until OD600 reached 0.5. Cells were then collected by centrifugation (5000 × g, 5 min, 4°) and transferred to either YPD or synthetic minimal medium supplemented with required nutrients and 40 mm 3-AT for HIS3 induction. Cell cultures were further incubated at 37° for 2–3 hr before harvesting for RNA preparation. Procedures for RNA preparation were described previously (Liu et al. 2005). Ten micrograms of total RNA was treated with 10 units of DNaseI (Roche) in 100 μl (50 mm Tris-HCl, pH 7.5, 5 mm MgCl2), and incubated at 37° for 1 hr. cDNA was synthesized following the instruction of ImpronII reverse transcriptase kit (GE Life Sciences) using 30 ng of poly(dT) primer. Semiquantitative PCR reactions were conducted in 50 mm KCl, 10 mm Tris-HCl (pH 9.0, 25°), 1% Triton X-100, 2 mm MgCl2, 0.1 mm each dNTP, 0.5 μM each primer, and 1.25 units Taq DNA polymerase (Promega), and appropriately diluted DNA templates. PCR parameters were (94°, 4 min; 50°, 4 min; 72°, 30 sec) for 2 cycles; (94°, 45 sec; 50°, 45 sec; 72°, 30 sec) for 24 cycles; and 72°, 3 min. PCR products were resolved in polyacrylamide gels followed by ethidium bromide staining. Chromatin IP was conducted according to Kuo and Allis (1999).
RESULTS
Gcn5p is a phosphoprotein:
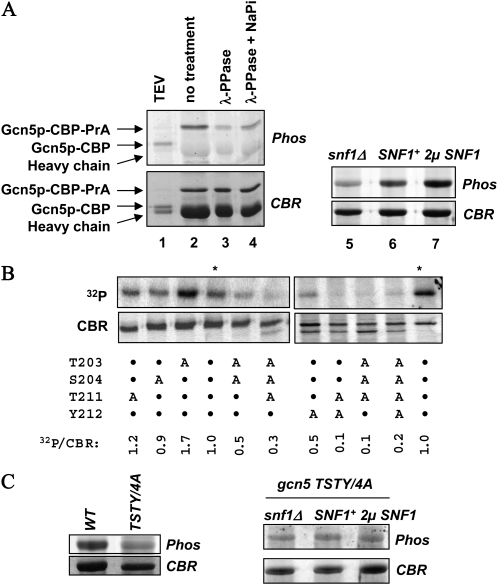
We previously reported that Snf1p phosphorylated Gcn5p in vitro, and that these two proteins were copurified from yeast whole cell lysates (Liu et al. 2005). These results suggested that Gcn5p was regulated by Snf1p via direct interaction and/or phosphorylation. To test this hypothesis, we first examined whether Snf1p controlled Gcn5p phosphorylation in vivo. We used the phosphate-specific Pro-Q Diamond fluorescence dye to assess the in vivo phosphorylation status of Gcn5p. Gcn5p was fused to the tandem affinity purification tag (TAP) consisting of (from the carboxyl end) the protein A fragment (PrA), endopeptidase TEV cleavage site, and the calmodulin binding protein (CBP) (Rigaut et al. 1999). Constitutively expressed Gcn5p–TAP was partially purified from yeast whole cell lysate by IgG beads, followed by SDS–PAGE and Pro-Q Diamond staining. Gcn5p was positively stained (Figure 1A, lane 2), and this staining was diminished in λ phosphatase-treated samples (compare lanes 2 and 3 in Figure 1A). The identity of Gcn5p was further verified by treating the IgG matrix-bound materials with the TEV protease (lane 1) that liberated Gcn5p from the IgG beads and concomitantly caused a faster mobility on SDS–PAGE. We thus conclude that Gcn5p is phosphorylated in vivo.
Figure 1.—
Gcn5p is a phosphorylated protein. (A) SNF1 dosage-dependent Gcn5p phosphorylation in vivo. TAP-tagged Gcn5p was purified from yeast and stained with the phosphate-specific fluorescence dye, Pro-Q Diamond. After capturing the fluorescent images, protein gels were stained with Coomassie Blue R250 (CBR). Phos, Pro-Q Diamond staining. (B) Mapping amino acid residues important for Gcn5p phosphorylation in vitro. Recombinant Gcn5p bearing the indicated alanine mutations was treated with yeast Snf1p in the presence of [γ-32P]ATP, followed by SDS–PAGE, CBR staining, and autoradiography. The radiolabeling efficiency was assessed by PhosphoImager. Asterisks indicate wild-type Gcn5p reaction products. The relative phosphorylation status was obtained by calculating the 32P-to-CBR staining intensities, with the latter acquired by National Institutes of Health (NIH) Image analysis. Two sets of samples were analyzed separately, each with the wild-type Gcn5p as the normalization standard. A, Ala substitution; •, residues unaltered. (C) The TSTY/4A quadruple mutant is hypophosphorylated (left; both from SNF1+ background) and insensitive to changes in Snf1p dosage in vivo (right). TAP-tagged Gcn5p bearing the TSTY/4A mutations was purified from strains with the indicated SNF1 dosage and stained by Pro-Q Diamond and Coomassie Blue.
To assess the role of Snf1p in Gcn5p phosphorylation in vivo, we examined the effects of varying the Snf1p dosage on Gcn5p phosphorylation (Figure 1A, lanes 5–7). Compared with the wild-type strain, snf1Δ cells produced a much lower phosphostaining intensity of Gcn5p (lane 5). In contrast, introducing a multicopy SNF1 plasmid to yeast (lane 7) caused significantly stronger Gcn5p phosphorylation. Thus, the in vivo phosphorylation of Gcn5p correlates positively to the dosage of Snf1p.
To explore further the relationship between Snf1p and Gcn5p, we attempted to map the amino acid residues of Gcn5p important for its phosphorylation. By comparing the Gcn5p catalytic domain to the consensus sequence shared by the yeast Snf1p and other AMP-activated protein kinases (AMPK), we found that residues T203, S204, and T211 resembled the AMPK consensus (Φ-x-K/R-x-x-S/T-x-x-x-Φ, Φ = hydrophobic residues) (Kuchin et al. 2000) (200VRNT*S*NIKYFLT*YADNYA). Of these three residues, T211 is conserved among the GNAT family members (Neuwald and Landsman 1997; Dyda et al. 2000), while T203 and S204 are present primarily in fungal homologs. The catalytic function of any of these residues has not been tested. In addition, T211 and the adjacent hydroxyl amino acid, Y212, are spatially equivalent to the suspected active center of Hat1p, E255, and D256 (Sternglanz and Schindelin 1999), raising an interesting possibility that phosphorylation at or around T211 may influence the HAT activity of Gcn5p. We thus created and purified a series of mutant Gcn5p in E. coli for [γ-32P]ATP labeling by Snf1p in vitro. The wild-type Gcn5p was phosphorylated by Snf1p strongly (Figure 1B, asterisk-marked lanes). Single alanine substitutions of T203, S204, or T211 had only modest effects. We repetitively observed stronger phosphorylation of the T203A mutant. The biochemical basis is unclear. Significantly, the Y212A single mutation reduced phosphorylation to ∼50% of the wild-type level, suggesting that Y212 was a critical residue for Snf1p to recognize Gcn5p for its phosphorylation. More severe defects were seen in two double mutants (T203A/S204A and T211A/Y212A), the T203A/S204A/T211A triple mutant (referred to as TST/3A hereafter), and the quadruple TSTY/4A mutant. We thus conclude that these four residues are collectively critical determinants for Gcn5p phosphorylation in vitro by Snf1p, and that Y212 is probably the most important among the four.
To examine whether T203, S204, T211, and Y212 were also important for Gcn5p phosphorylation in vivo, we introduced the TSTY/4A quadruple mutations to the TAP-tagged GCN5 construct and purified Gcn5p for Pro-Q Diamond staining. Contrary to the wild-type Gcn5p in which its phosphorylation correlated well with the dosage of Snf1p (Figure 1A), the TSTY/4A mutant showed weak Pro-Q Diamond staining intensity (Figure 1C) even in the presence of a multicopy SNF1 plasmid. Deleting SNF1 failed to cause further reduction of Gcn5p phosphorylation. We thus surmise that Snf1p is likely the major kinase that targets or depends on the TSTY region for Gcn5p phosphorylation in vivo. However, as shown below, we believe that Gcn5p phosphorylation per se plays a less important role in transcriptional activation than does the interaction with Snf1p in vivo.
TSTY mutants affect Gcn5p functions in vivo:
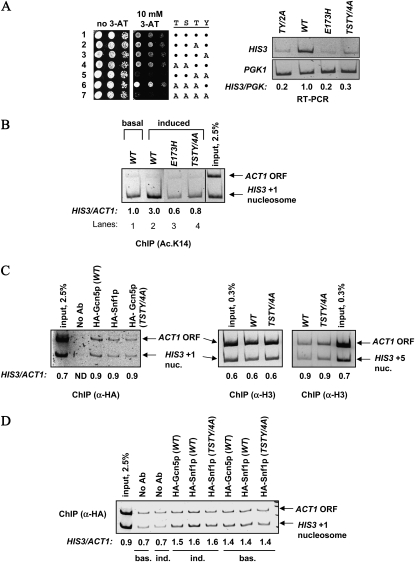
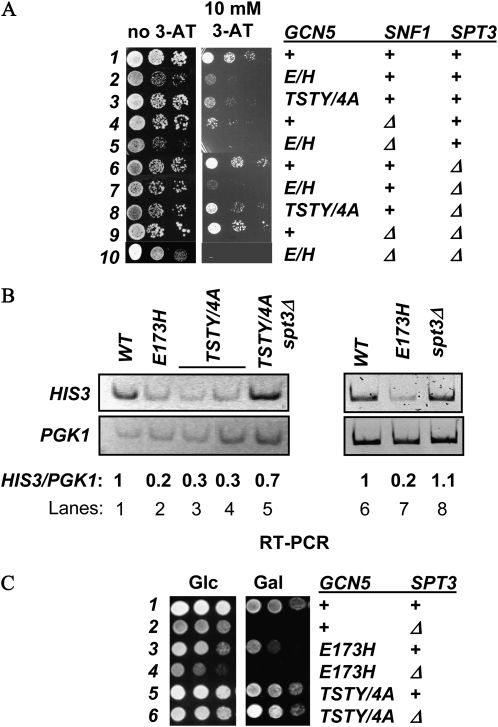
To test whether the TSTY residues were also important for the transcriptional activation function of Gcn5p, we created several mutants and integrated them into the GCN5 locus. The expression status of HIS3 was assessed by comparing the cellular resistance to 3-amino-triazole (3-AT), and by reverse-transcription PCR (RT–PCR) for quantifying the HIS3 mRNA levels. When expressed from the native GCN5 promoter, single alanine substitution of the conserved T211 and Y212 did not cause a discernible HIS3 expression defect; neither did T203A/S204A nor T203A/S204A/T211A mutants (Figure 2A left, rows 1–4 and 6). In contrast, the T211A/Y212A (dubbed TY/2A hereafter) and TSTY/4A mutants caused clear 3-AT hypersensitivity (rows 5 and 7) that also correlated with the diminishment of HIS3 transcription, as shown by RT–PCR (Figure 2A, right panel). The severity of the HIS3 expression defects was similar to the E173H mutant that targeted the catalytic center of Gcn5p (Liu et al. 2005).
Figure 2.—
Residues critical for Gcn5p phosphorylation are also important for transcriptional activation and promoter acetylation. (A) HIS3 expression defects are caused by TY/2A and TSTY/4A mutants. Left panel, cellular sensitivity to 3-AT was assessed by spotting serially diluted yeast strains to 10 mM 3-AT plates. Right panel, semiquantitative RT–PCR comparing HIS3 and PGK1 expression. The intensity of each band was quantified by NIH Image, and normalized to the wild-type samples. RT–PCR and ChIP experiments in this and subsequent figures were from at least two independent experiments. Although absolute values varied somewhat among different experiments, trends were highly reproducible (data from representative experiments are shown). All mRNA analyses and ChIP studies, unless otherwise noted, were conducted under amino acid starvation conditions that activated HIS3 expression. (B–D) gcn5 TSTY/4A mutant causes H3 hypoacetylation but does not affect the recruitment of Gcn5p, Snf1p, or histone H3 occupancy at the HIS3 promoter. Chromatin immunoprecipitation with the indicated antibodies was conducted using yeast cells harvested from minimal medium that induced HIS3 transcription or from the YPD medium that allowed basal expression. Semiquantitative multiplex PCR was used to compare the amount of HIS3 promoter and the ACT1 open reading frame associated with the indicated antigens. The relative immunoprecipitation efficiency, expressed as HIS3-to-ACT1 ratio, was quantified by NIH Image. Note that panel C only shows ChIP results from induced cultures. ND, not determined.
We further used chromatin immunoprecipitation to see whether the HIS3 transcriptional defects were coupled to histone H3 hypoacetylation. The TSTY/4A mutations reduced H3 K14 acetylation at the HIS3 promoter (compare lanes 2 and 4, Figure 2B). Hypoacetylation at another lysine of H3, K18, also was prominent in the TSTY/4A strain (data not shown). Neither the recruitment of Gcn5p nor the histone H3 occupancy was appreciably affected at the HIS3 promoter (Figure 2C). Similarly, Snf1p remained associated with the HIS3 promoter in both wild-type and gcn5 TSTY/4A background (Figure 2, C and D). Together, these data revealed new residues in the catalytic domain of Gcn5p that are important for its HAT and transcriptional activation activities. Of the four residues, T211 and Y212 appear to be more important than T203 and S204. On the other hand, double alanine substitutions of T203 and T204 caused ∼50% reduction of the in vitro phosphorylation of Gcn5p (Figure 1B), suggesting that these two residues may perform a hitherto unidentified function in vivo. With respect to HIS3 regulation, however, the TSTY/4A and TY/2A mutants displayed comparable phenotypes (e.g., Figure 2A and see below).
Snf1p is important for Gcn5p functions in vivo:
The above data reveal that the TSTY region of Gcn5p is important for the well-established histone acetylation and transcriptional activation activities of Gcn5p and for the Snf1p-dependent phosphorylation of Gcn5p in vivo. Snf1p has been shown to phosphorylate multiple chromatin proteins, including histone H3 (Lo et al. 2000, 2005) and possibly the Srb/mediator complex components (Kuchin et al. 2000) (although our previous work ruled out the involvement of histone H3 phosphorylation in HIS3 transcriptional activation; see Liu et al. 2005). It is tempting to speculate that Gcn5p phosphorylation is critical for the transcriptional activation of HIS3. On the other hand, if Snf1p is indeed the responsible kinase, these two proteins have to be engaged in a physical interaction, transiently or stably, on chromatin or in the nucleoplasm. It is therefore also possible that the physical interaction between Gcn5p and Snf1p plays a more direct role in activating HIS3 transcription. For example, Gcn5p may provide a docking site for Snf1p to get access to another protein(s) that is important for transcriptional activation. In this case, Gcn5p phosphorylation is a functionally redundant byproduct of this contact with Snf1p.
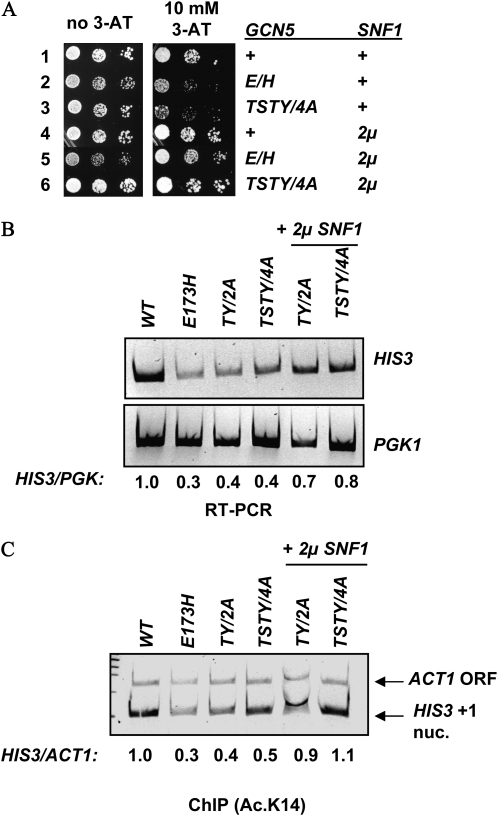
To see whether phosphorylation of Gcn5p or its association with Snf1p was more important for HIS3 activation, we examined the effects of overexpressing Snf1p in the TSTY/4A mutant. The Pro-Q Diamond staining (Figures 1C) showed that the TSTY/4A quadruple mutations quantitatively eliminated Snf1p-dependent Gcn5p phosphorylation. If phosphorylation of Gcn5p by Snf1p is critical for HIS3 activation, then the phosphodeficient gcn5 TSTY/4A mutant should remain hypersensitive to 3-AT even in the presence of a 2μ SNF1. In contrast, if the Gcn5p–Snf1p association is more important, increasing the concentration of Snf1p may augment its interaction with the TSTY/4A mutant of Gcn5p, thus upregulating HIS3 expression without causing Gcn5p hyperphosphorylation. Results in Figure 3 conform to the latter hypothesis. When transformed with a 2μ plasmid bearing the wild-type SNF1 gene, TSTY/4A mutant cells exhibited enhanced resistance to 3-AT (Figure 3A), consistent with about a twofold increase in HIS3 transcription (Figure 3B). Intriguingly, the 2μ SNF1 plasmid also rescued the E173H alleles of GCN5 (Figure 3A). This suppression is in contrast with our previous findings that overexpressing Snf1p did not rescue the HIS3 expression defects caused by the complete knockout or another catalytically inactive allele of Gcn5p, F221A (Liu et al. 2005). The allele specificity of the SNF1 high-copy suppressor is in agreement with the notion that Gcn5p and Snf1p interact directly. As the TSTY/4A phosphodeficient mutant was rescued by 2μ SNF1 without exhibiting discernible increase in its phosphorylation (Figure 1C), it is likely that Gcn5p phosphorylation either has a minor effect in HIS3 expression, or that the normal function of Gcn5p phosphorylation at or near the TSTY region is dispensable if there is sufficient amount of Snf1p available.
Figure 3.—
gcn5 TSTY/4A is suppressed by overexpressing Snf1p. (A) Cellular sensitivity to 3-AT. +, wild type without extra copies of SNF1; 2μ, multicopy plasmid introduced. (B) RT–PCR analysis of HIS3 expression. (C) 2μ SNF1 rescues the H3 hypoacetylation phenotype associated with gcn5 TSTY/4A. Shown are ChIP results. TY/2A: T211A Y212A; TSTY/4A: T203A, S204A, T211A, and Y212A.
In addition to restoring HIS3 transcription, 2μ SNF1 also suppressed the histone H3 hypoacetylation phenotype (Figure 3C). Chromatin IP using antibodies against histone H3 acetylated at K14 demonstrated that both the TY/2A and TSTY/4A mutant alleles resumed their ability to acetylate H3 at the HIS3 promoter in the presence of the multicopy SNF1 gene. These results suggest that one of the likely molecular mechanisms underlying the TSTY/4A and TY/2A phenotypes is a weakened Gcn5p–Snf1p interaction, resulting in the crippled HAT activity of Gcn5p (see below and Figure 5B) and consequently the compromised activation of HIS3 gene.
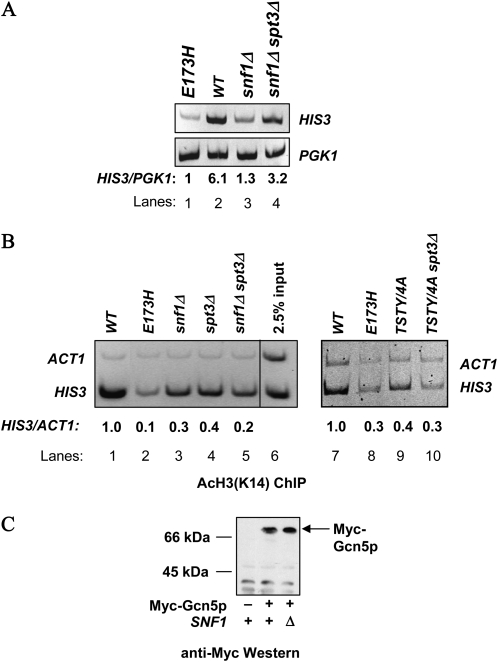
Figure 5.—
Deleting SPT3 suppresses HIS3 transcriptional defects caused by snf1Δ. (A) RT–PCR shows suppression of HIS3 transcriptional defects in snf1Δ cells, but (B) histone H3 remains to be hypoacetylated at the HIS3 promoter, as shown by ChIP analyses. (C) The steady state level of Gcn5p is not affected by deleting SNF1. Myc-tagged Gcn5p was expressed from its native chromosomal locus in SNF1+ or snf1Δ background. Yeast whole cell extracts were prepared and probed with anti-Myc antibodies in Western blotting assays.
GCN5, SNF1, and SPT3 interact genetically to control HIS3 expression:
GCN5 genetically interacts with many regulators for appropriate control of transcription (see the Introduction for details). Overexpressing Snf1p may reinforce the relationship of Gcn5p with a positive partner, or neutralizes a negative regulator that opposes Gcn5p. We focused our studies on Spt3p, because Gcn5p and Spt3p perform antagonistic functions at certain well-characterized genes. For example, deleting SPT3 rescues the transcriptional defects of HO caused by either a HAT-deficient, or a complete knockout allele of GCN5 (Yu et al. 2003). The Gcn5p-dependent basal expression of HIS3 is increased in spt3Δ cells (Sterner et al. 1999). We suspected that the negative effect by Spt3p may be one of the targets for Snf1p in facilitating Gcn5p functions.
To delineate the genetic relationship between GCN5 and SPT3, we deleted the latter in different gcn5− background and examined the expression status of HIS3. Figure 4A shows that Spt3p indeed was an allele-specific regulator of Gcn5p. Deleting SPT3, while imposing no discernible effect on 3-AT resistance (row 6), suppressed the TSTY/4A mutant (compare rows 3 and 8). However, the E173H (row 7) and complete knockout (data not shown) mutants were not rescued by deleting SPT3. Semiquantitative RT–PCR assays confirmed the suppression of HIS3 transcriptional defects (compare lanes 3, 4, and 5, Figure 4B). To understand further the GCN5–SPT3 genetic interactions, we examined cellular growth on galactose, as spt3Δ cells were reported by several groups to be unable to use galactose as the sole carbon source (Dudley et al. 1999; Larschan and Winston 2001; Bhaumik and Green 2002). spt3Δ cells in our hands also were Gal− (row 2, Figure 4C), whereas gcn5 TSTY/4A cells grew well on the galactose medium (compare rows 1, 3, and 5). Importantly, the gcn5 TSTY/4A spt3Δ double mutant cells displayed clear Gal+ growth (row 6). Thus, gcn5 TSTY/4A was the suppressor for the galactose auxotroph phenotype of spt3Δ cells. These results demonstrate that the antagonistic relationship between Gcn5p and Spt3 was maintained in cellular responses to both amino acid starvation (e.g., HIS3 activation) and galactose utilization. Similar to the observation that spt3Δ allele was unable to suppress the HIS3 expression defects caused by the E173H allele (row 7, Figure 4A), this gcn5 mutant cannot rescue the Gal− phenotypes of spt3Δ cells (row 4, Figure 4C), further supporting the notion that the genetic interactions between GCN5 and SPT3 were dependent on the target genes as well as the alleles of these two regulators.
Figure 4.—
SPT3 is a negative regulator of GCN5. Deleting SPT3 rescues the 3-AT hypersensitivity (A) and HIS3 transcriptional defects (B, shown are RT–PCR results). (C) gcn5 TSTY/4A suppresses the gal− phenotype caused by SPT3 deletion. Yeast cells were serially diluted and spotted to glucose (Glc) or galactose (Gal) medium.
That 2μ SNF1 and spt3Δ were common suppressors of the TSTY/4A allele of GCN5 prompted us to look more deeply into the interrelationship of these three. To this end, we combined different gcn5 alleles with snf1Δ or spt3Δ and analyzed HIS3 expression and acetylation. Yeast cells lacking Snf1p were defective in HIS3 activation (Figure 5A, lane 3; Liu et al. 2005). Consistently, the HIS3 promoter became hypoacetylated in the GCN5+ snf1Δ strain (Figure 5B, lane 3). Since the level of Gcn5p remained unchanged in the absence of Snf1p (Figure 5C), the HAT action of Gcn5p at the HIS3 promoter clearly depended on a functional Snf1p, a notion consistent with the genetic and physical interactions between Gcn5p and Snf1p (see Figures 1–3), as well as the discovery that Snf1p was present at the HIS3 promoter (Figure 2C, left panel). Furthermore, the GCN5–SNF1 genetic interaction appeared to involve SPT3 because the HIS3 transcriptional defect of snf1Δ cells was effectively reverted by deleting SPT3 (Figure 5A, compare lanes 3 and 4), suggesting that a key function of Snf1p was to antagonize a negative activity of Spt3p. This antagonism required a certain function(s) of Gcn5p, for snf1Δ spt3Δ cells became sensitive to 3-AT if GCN5 was replaced with the E173H allele (rows 9 and 10, Figure 4A). Intriguingly, deleting SPT3, though rescuing the HIS3 transcriptional defect of snf1Δ cells, did not suppress the hypoacetylation phenotype (lane 5, Figure 5B). Similarly, GCN5+ spt3Δ cells were hypoacetylated at the HIS3 promoter but exhibited near normal expression of HIS3 (Figure 4B, lane 8). Together, these results suggest that a histone H3 acetylation-independent function of Gcn5p is responsible for HIS3 activation in the snf1Δ spt3Δ background, and that this activity of Gcn5p may be suppressed by Spt3p under normal conditions.
Gcn5p interacts with Spt3p:
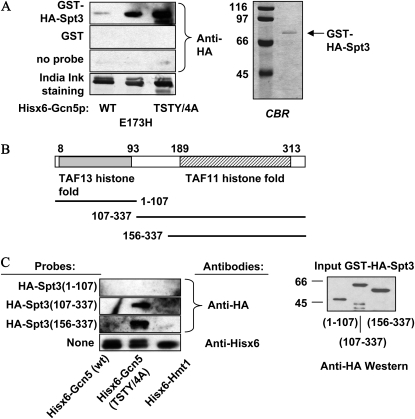
We next sought to test the possible molecular basis underlying the Gcn5p–Spt3p functional connection. Given the extensive genetic interactions between GCN5 and SPT3 in the regulation of multiple genes (see the Introduction), we suspected that direct association might exist between these two proteins. Since both Gcn5p and Spt3p are components of the SAGA and the SLIK/SALSA complexes, it would be difficult to detect the in vivo physical interaction between Gcn5p and Spt3p. Instead, we expressed Gcn5p and Spt3p in E. coli and tested whether these two proteins could interact directly in vitro. Wild type and two different mutant Gcn5p, E173H and TSTY/4A, were expressed as Hisx6-tagged proteins, resolved, and blotted to PVDF membrane. HA–Spt3p (Figure 6A, right panel) was then used as the probe to bind the immobilized Gcn5p. The anti-HA antibodies were used under a standard Western blotting condition to examine the relative amounts of HA–Spt3p bound by different Gcn5p.
Figure 6.—
Gcn5p and Spt3p interact directly in vitro. (A) Full-length Spt3p was double tagged by GST and HA and was purified from E. coli for Gcn5p binding in the Farwestern. Bacterially expressed, Hisx6-tagged wild-type and mutant Gcn5 proteins were resolved and immobilized on PVDF membrane and incubated with soluble GST–HA–Spt3p. Anti-HA antibodies were then used to quantify the relative amount of Spt3p trapped by different Gcn5p (top three panels, left). A fourth membrane strip was stained by India ink for Gcn5p loading control. Right panel: CBR staining of the purified Spt3p used in the Farwestern assays. (B) Schematics of Spt3p histone fold domains and the three fragments used for Farwestern assays. (C) The carboxyl histone fold domain of Spt3p is sufficient for Gcn5p interaction. Left: Farwestern results. Three different Spt3p fragments were expressed as GST- and HA-double tagged probes. The fragments used in each assay are listed on the left of each Western strip. Anti-His tag Western results (bottom strip, left) revealed comparable amounts of Hisx6–Gcn5p and Hisx6–Hmt1p. Right: Anti-HA Western blotting showing the relative amounts of the three GST–HA–Spt3p derivatives used in the Farwestern tests.
Figure 6A shows that the interaction between Gcn5p and Spt3p could be readily detected by the Farwestern approach. Intriguingly, the TSTY/4A and TY/2A (data not shown) Gcn5p trapped HA–Spt3p even more strongly than did the wild-type counterpart. The E173H allele (center lane, Figure 6A, left), also displayed enhanced affinity for Spt3p, although the enhancement seen in the E173H allele was substantially weaker than the TSTY/4A mutant. These biochemical data demonstrated the intrinsic affinity between Gcn5p and Spt3p and that certain mutations of Gcn5p may either increase the affinity or stabilize the association with Spt3p.
Homologs of Spt3p are found in fungi, insects, worm, and mammals (our BLAST search results; data not shown). Spt3p shares significant homology with the histone fold domains of two TBP-associated factors, TAF11 (a.k.a. TAFII28) and TAF13 (a.k.a. TAFII18) (Birck et al. 1998) (Figure 6B). We were interested in knowing which of the two histone fold domains of Spt3p, if separable, was critical for Gcn5p association. To this end, we prepared three GST–HA fusion fragments of Spt3p for E. coli production and in vitro binding assays (Figure 6B).
The Spt3 (1–107) and (156–337) fragments contained respectively the N′ and C′ histone fold domains; Spt3 (107–337) also included the bridging sequence in between. Comparable amounts of these three fragments were purified from E. coli and used as the probe in binding assays (Figure 6C). While Spt3 (1–107) showed very weak, if any, interaction with either allele of Gcn5p, both (107–337) and (156–337) fragments interacted positively with the TSTY/4A mutant. No significant interaction was seen with either the BSA internal control (data not shown), or another unrelated Hisx6-tagged protein, Hmt1p. We repetitively observed that the (156–337) fragment had the strongest affinity, suggesting that the N′ histone fold domain and the linker sequence may interfere with the Gcn5p–Spt3p association. We conclude that the C′ TAF11 histone fold domain contains the major interface for Gcn5p interaction, and that the TSTY/4A mutations of Gcn5p significantly enhance the association with the TAF11 histone fold domain of Spt3p.
DISCUSSION
Snf1p and Spt3p are respectively positive and negative regulators of Gcn5p:
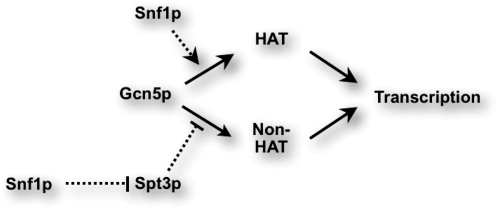
We report here a Gcn5p–Snf1p–Stp3p regulatory network that is critical for the transcriptional activation of HIS3 (Figure 7). Snf1p is an activator of Gcn5p for both the promoter acetylation and transcriptional activation of HIS3. Conversely, Spt3p is an inhibitor that becomes more potent in gcn5 TSTY or snf1Δ mutant cells, leading to transcriptional defects in these mutants. Since the spt3Δ suppressor only rescues the HIS3 transcriptional defect but not the promoter hypoacetylation phenotype (lane 5, Figure 5A), we suspect that Gcn5p exerts an H3 acetylation-independent function inhibited by Spt3p. This inhibition likely results from a direct interaction between Gcn5p and Spt3p. As to Snf1p, in addition to potentiating the HAT function of Gcn5p, it may dampen the negative effect of Spt3p. A probable scenario regarding the TSTY/4A quadruple mutant is that these mutations augment or stabilize the interaction with Spt3p, hence enhancing the repressive strength of Spt3p. Overproducing Snf1p may effectively compete against Spt3p for the same or overlapping binding site on Gcn5p. The observed interdigitating relationships among these three proteins demonstrate a delicate system that balances the action of the conserved histone acetyltransferase Gcn5p. However, given the multitude of genetic interactions between Gcn5p and other transcriptional and chromatin regulators, it is possible that Snf1p also negotiates with additional factors, such as histone deacetylases, in the control of Gcn5p. Further genetic tests and screens may yield clues for this hypothesis.
Figure 7.—
Model for Gcn5p, Snf1p, and Spt3p relationship for HIS3 transcriptional control. Dotted lines represent activating (with arrow) or inhibiting (with end bar) functions of Snf1p and Spt3p revealed in this work. See discussion for details.
Intriguingly, a low-resolution electron microscopy study showed that Gcn5p and Spt3p are spatially segregated by ∼14 nm within the purified SAGA complex (Wu et al. 2004). We do not believe that the electron microscopy image of a static SAGA complex and our model are mutually exclusive. We hypothesize that the interactions between Gcn5p, Snf1p, and Spt3p are transient but are critical for regulating the biochemical activities of the SAGA complex, i.e., chromatin modification and transcription activation. Given that SAGA is responsible for mostly stress-related transcription (Lee et al. 2000; Huisinga and Pugh 2004), it seems possible that SAGA exists as an inactive form prior to its engagement in transcriptional activation. In its inactive state, the SAGA complex positions Spt3p in a way that Gcn5p is inhibited. Stresses, such as nutrient deprivation and heat shock by which many genes are activated in a SAGA-dependent fashion (Huisinga and Pugh 2004), segregate Gcn5p and Spt3p and consequently instigate the chromatin modification and transcriptional activation activities of SAGA. This scenario is consistent with the observations that deleting SPT3 enhances the basal expression of HIS3 in rich medium in a GCN5-dependent manner (Sterner et al. 1999; Belotserkovskaya et al. 2000). A recent report by Winston and colleagues (Helmlinger et al. 2008) that Gcn5 and Spt8 proteins in the Schizosaccharomyces pombe SAGA complex play opposing roles in the control of proliferation-to-sexual differentiation switch also points to a dynamic interrelationship between SAGA subunits. It is worth noting that in our hands, spt3Δ and spt8Δ cells exhibited equivalent responses to genetic manipulations of GCN5 or SNF1 (data not shown). As to the Gcn5p activator Snf1p, it has been shown that the kinase activity of Snf1p is repressed when cells are grown in glucose-rich medium (Hardie et al. 1998) and becomes activated after brief centrifugation or wash of yeast cells (Smith et al. 1999). We suspect that Snf1p was inadvertently activated during the purification of SAGA and related Gcn5p complexes, hence awakening the nucleosomal HAT activity of Gcn5p. Indeed, Berger and colleagues (Belotserkovskaya et al. 2000) first reported that the composition and chromatographic behaviors of the SAGA complex were changed upon amino acid starvation.
Gcn5p phosphorylation by Snf1p:
It is interesting that the TSTY/4A phosphorylation-deficient mutant can be rescued by Snf1p overexpression (Figure 3). This suppression can be due to one of two reasons. While the Pro-Q Diamond phosphostaining did not detect an obvious phosphorylation change of the TSTY/4A mutant when Snf1p was overexpressed, we cannot rule out that a key, but quantitatively minor phosphorylated Gcn5p species can be augmented when the dosage of Snf1p increases. Alternatively, Snf1p may use Gcn5p as the docking site to get access to other target proteins, such as the Srb/mediator complex (Kuchin et al. 2000). In this case, Gcn5p phosphorylation is functionally redundant. Overproducing Snf1p may compensate for the crippled affinity for the TSTY/4A mutant Gcn5p, thus restoring HIS3 activation. Consistently, the complete knockout allele of GCN5 is insensitive to Snf1p overproduction for HIS3 activation (Liu et al. 2005).
Of the four residues tested, T211 and Y212 clearly play critical roles in both Gcn5p phosphorylation and in HIS3 activation. T203 and S204 appear to have an auxiliary function for phosphorylation (data not shown). When we mutated any of the four residues to aspartic acid, a commonly used phosphomimetic amino acid to assess the effect of constitutive phosphorylation, yeast cells became hypersensitive to 3-AT, suggesting a severe loss of Gcn5p function (data not shown). Intriguingly, all these phosphomimetic mutants were totally insoluble when expressed in bacteria and could not acetylate histones in the in-gel activity tests (data not shown). It is therefore possible that constitutive phosphorylation of Gcn5p is detrimental and causes a structural catastrophe. Phosphorylation of Gcn5p is thus likely a strictly regulated event. Protein phosphatases, such as Glc7p known to interact genetically with Snf1p (McCartney and Schmidt 2001; Hedbacker and Carlson 2008), may be part of the Gcn5p regulatory circuit as well.
It is also interesting that both Gcn5p and Snf1p appear to be associated with the HIS3 locus relatively constitutively (Figure 2D). This observation is consistent with our Ada2p ChIP results (M.-H. Kuo, unpublished data) and the notion that the enzymatic activities of Gcn5p and Snf1p are subjected to regulation (see above). On the other hand, the stable association of Gcn5p with HIS3 seems to contradict the observations that Gcn5p-dependent H3 hyperacetylation of the HIS3 promoter is triggered by amino acid starvation (Kuo et al. 1998) and that the SAGA complex is recruited to several loci including GAL1 and ARG1 (Henry et al. 2003; Govind et al. 2007) under certain induced conditions. One explanation is that the HAT activity of Gcn5p, instead of its recruitment, is upregulated in response to amino acid starvation at the HIS3 locus. Alternatively, two different Gcn5p-containing complexes may each be responsible for the basal and induced H3 acetylation.
Spt3p and Gcn5p regulation:
It remains to be seen how Spt3p inhibits Gcn5p at the biochemical and molecular level. One possibility is that Spt3p and Snf1p compete for an overlapping motif of Gcn5p for transcriptional regulation. In vitro, we did not observe a clear effect on the HAT activity of Gcn5p in the presence or absence of Spt3p (data not shown), suggesting that Spt3p does not directly influence the HAT action in a highly refined biochemical system. Similarly, we did not observe strong evidence for Spt3p acetylation by Gcn5p in vitro. To our surprise, the H3 hyperacetylation of the HIS3 promoter is lost in the spt3Δ strain (Figure 5A), even though HIS3 activation appears to be normal. Equally intriguing is that the snf1Δ spt3Δ strain exhibits a similar H3 hypoacetylation trait (Figure 5B). These results clearly demonstrate that the HAT activity of Gcn5p is not indispensable under certain conditions. Evidence presented in this work (e.g., Figure 5B) led us to speculate that Spt3p represses a non-H3 acetylation activity of Gcn5p. In the absence of Spt3p, this activity of Gcn5p is upregulated to an extent that the canonical H3 hyperacetylation is masked or no longer needed. Indeed, the H3 K14Q mutation, which mimics a constitutively acetylated state, triggers upregulation of a Gcn5p-driven reporter gene, but deleting GCN5 perturbs such enhancement (Zhang et al. 1998). The TSTY/4A mutant of Gcn5p may preserve this mystic, acetylation-independent function that is rendered active upon the removal of Spt3p.
Another function of Gcn5p is to evict histones H3 and H4 within the open reading frame during transcription (Govind et al. 2007). H3/H4 eviction depends on the HAT activity of Gcn5p (Govind et al. 2007). We have been using HIS3 as the model to understand how Gcn5p activates transcription. HIS3 is a small gene, with only five positioned nucleosomes covering the entire open reading frame. Probably because of the highly compact nature of this gene, we did not observe clear H3 eviction during HIS3 activation (Figure 2C). Consistently, the quadruple TSTY/4A mutant, though causing H3 hypoacetylation, does not affect histone H3 occupancy at the HIS3 locus. It will be interesting to examine whether H3 eviction is impaired by the TSTY/4A mutant at other longer Gcn5p target genes, and, if so, whether manipulating SNF1 and SPT3 can modulate this function of Gcn5p.
Finally, phenotypic comparison between two alleles of gcn5, E173H and TSTY/4A, further suggests molecular distinction of these mutants, even though both alleles cause H3 hypoacetylation and HIS3 transcriptional defects. For example, while TSTY/4A is rescued by overexpressing Snf1p and by deleting SPT3, the E173H allele does not respond to spt3Δ. Furthermore, the hypoacetylation phenotype of the TSTY/4A mutant can be suppressed by 2μ SNF1, but the E173H mutant remains hypoacetylated at HIS3. We attribute these differences to the facts that E173 is the active center for the HAT action, and that the TSTY residues are outside the active center (Tanner et al. 1999; Trievel et al. 1999) and may be important for maintaining a certain conformational isoforms of Gcn5p. Allele-specific suppression displayed by these two mutants underscores the value of genetic dissection, and provides clues for further examination that will likely lead to a better understanding of how Gcn5p performs its chromatin modification and transcriptional regulation functions.
Acknowledgments
We thank Marian Carlson, Dennis Thiele, and Fred Winston for generous supply of plasmid constructs, and Liping Gu and R. William Henry for their advice on Pro-Q Diamond staining. We are grateful for Steve Triezenberg, David Almy, and Jianjun Luo for valuable discussion and David Arnosti for critical reading of the manuscript. Y.L. was a recipient of the College of Natural Sciences thesis completion scholarship, Michigan State University. This work was supported by National Institutes of Health GM R01-62282.
References
- Aasland, R., A. F. Stewart and T. Gibson, 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21 87–88. [PubMed] [Google Scholar]
- Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant and S. Tan, 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277 7989–7995. [DOI] [PubMed] [Google Scholar]
- Barbaric, S., H. Reinke and W. Horz, 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23 3468–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya, R., D. E. Sterner, M. Deng, M. H. Sayre, P. M. Lieberman et al., 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20 634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik, S. R., and M. R. Green, 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22 7365–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birck, C., O. Poch, C. Romier, M. Ruff, G. Mengus et al., 1998. Human TAF (II)28 and TAF (II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell 94 239–249. [DOI] [PubMed] [Google Scholar]
- Biswas, D., A. N. Imbalzano, P. Eriksson, Y. Yu and D. J. Stillman, 2004. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex. Mol. Cell. Biol. 24 8312–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, L. A., R. R. Latek and C. L. Peterson, 2004. The SANT domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell. Biol. 5 158–163. [DOI] [PubMed] [Google Scholar]
- Burgess, S. M., M. Ajimura and N. Kleckner, 1999. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc. Natl. Acad. Sci. USA 96 6835–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau, R., J. X. Zhou, C. D. Allis and S. L. Berger, 1997. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, Y. C., P. Komarnitsky, D. Chase and C. L. Denis, 1996. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J. Biol. Chem. 271 32359–32365. [DOI] [PubMed] [Google Scholar]
- Clements, A., J. R. Rojas, R. C. Trievel, L. Wang, S. L. Berger et al., 1999. Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J. 18 3521–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, J. A., M. S. Torok, Z. W. Sun, D. Schieltz, C. D. Allis et al., 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279 1867–1871. [DOI] [PubMed] [Google Scholar]
- Dudley, A. M., C. Rougeulle and F. Winston, 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda, F., D. C. Klein and A. B. Hickman, 2000. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29 81–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney and F. Winston, 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6 1319–1331. [DOI] [PubMed] [Google Scholar]
- Gietz, D., A. St Jean, R. A. Woods and R. H. Schiestl, 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind, C. K., F. Zhang, H. Qiu, K. Hofmeyer and A. G. Hinnebusch, 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25 31–42. [DOI] [PubMed] [Google Scholar]
- Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell et al., 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11 1640–1650. [DOI] [PubMed] [Google Scholar]
- Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese et al., 1998. A subset of TAF (II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94 45–53. [DOI] [PubMed] [Google Scholar]
- Gregory, P. D., A. Schmid, M. Zavari, L. Lui, S. L. Berger et al., 1998. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell 1 495–505. [DOI] [PubMed] [Google Scholar]
- Hahn, J. S., and D. J. Thiele, 2004. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J. Biol. Chem. 279 5169–5176. [DOI] [PubMed] [Google Scholar]
- Hardie, D. G., D. Carling and M. Carlson, 1998. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67 821–855. [DOI] [PubMed] [Google Scholar]
- Hassan, A. H., K. E. Neely and J. L. Workman, 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104 817–827. [DOI] [PubMed] [Google Scholar]
- Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy et al., 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111 369–379. [DOI] [PubMed] [Google Scholar]
- Hedbacker, K., and M. Carlson, 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger, D., S. Marguerat, J. Villen, S. P. Gygi, J. Bahler et al., 2008. The S. pombe SAGA complex controls the switch from proliferation to sexual differentiation through the opposing roles of its subunits Gcn5 and Spt8. Genes Dev. 22 3184–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre et al., 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner et al., 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95 717–728. [DOI] [PubMed] [Google Scholar]
- Huisinga, K. L., and B. F. Pugh, 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13 573–585. [DOI] [PubMed] [Google Scholar]
- Ingvarsdottir, K., N. J. Krogan, N. C. Emre, A. Wyce, N. J. Thompson et al., 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, S., and L. Pillus, 2004. Molecular requirements for gene expression mediated by targeted histone acetyltransferases. Mol. Cell. Biol. 24 6029–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs, J. E., M. H. Kuo, C. D. Allis and C. L. Peterson, 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin, S., I. Treich and M. Carlson, 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97 7916–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, M. H., and C. D. Allis, 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20 615–626. [DOI] [PubMed] [Google Scholar]
- Kuo, M. H., and C. D. Allis, 1999. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods 19 425–433. [DOI] [PubMed] [Google Scholar]
- Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook et al., 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383 269–272. [DOI] [PubMed] [Google Scholar]
- Kuo, M. H., J. Zhou, P. Jambeck, M. E. Churchill and C. D. Allis, 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, M. H., E. vom Baur, K. Struhl and C. D. Allis, 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6 1309–1320. [DOI] [PubMed] [Google Scholar]
- Langer, M. R., K. G. Tanner and J. M. Denu, 2001. Mutational analysis of conserved residues in the GCN5 family of histone acetyltransferases. J. Biol. Chem. 276 31321–31331. [DOI] [PubMed] [Google Scholar]
- Laprade, L., D. Rose and F. Winston, 2007. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics 177 2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan, E., and F. Winston, 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan, E., and F. Winston, 2005. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 25 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. K., L. Florens, S. K. Swanson, M. P. Washburn and J. L. Workman, 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 25 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett et al., 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405 701–704. [DOI] [PubMed] [Google Scholar]
- Lin, Y., C. M. Fletcher, J. Zhou, C. D. Allis and G. Wagner, 1999. Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A. Nature 400 86–89. [DOI] [PubMed] [Google Scholar]
- Liu, Y., X. Xu, S. Singh-Rodriguez, Y. Zhao and M. H. Kuo, 2005. Histone H3 Ser10 phosphorylation-independent function of Snf1 and Reg1 proteins rescues a gcn5- mutant in HIS3 expression. Mol. Cell. Biol. 25 10566–10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, W. S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu et al., 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5 917–926. [DOI] [PubMed] [Google Scholar]
- Lo, W. S., L. Duggan, N. C. Emre, R. Belotserkovskya, R. Shiekhattar and S. L. Berger, 2001. Snf1–a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293 1142–1146. [DOI] [PubMed] [Google Scholar]
- Lo, W. S., E. R. Gamache, K. W. Henry, D. Yang, L. Pillus et al., 2005. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 24 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney, R. R., and M. C. Schmidt, 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276 36460–36466. [DOI] [PubMed] [Google Scholar]
- Mohibullah, N., and S. Hahn, 2008. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 22 2994–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, S. A., B. Rao, B. A. Garcia, S. B. Hake, R. L. Diaz et al., 2007. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 282 7632–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald, A. F., and D. Landsman, 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22 154–155. [DOI] [PubMed] [Google Scholar]
- Perez-Martin, J., and A. D. Johnson, 1998. Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, K. J., and C. L. Peterson, 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17 6212–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux, A. N., and R. Marmorstein, 2003. Molecular basis for Gcn5/PCAF histone acetyltransferase selectivity for histone and nonhistone substrates. Biochemistry 42 14366–14374. [DOI] [PubMed] [Google Scholar]
- Pray-Grant, M. G., D. Schieltz, S. J. McMahon, J. M. Wood, E. L. Kennedy et al., 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22 8774–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates, 3rd and P. A. Grant, 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433 434–438. [DOI] [PubMed] [Google Scholar]
- Qiu, H., C. Hu, S. Yoon, K. Natarajan, M. J. Swanson et al., 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 24 4104–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann et al., 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17 1030–1032. [DOI] [PubMed] [Google Scholar]
- Roberts, S. M., and F. Winston, 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, J. R., R. C. Trievel, J. Zhou, Y. Mo, X. Li et al., 1999. Structure of tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401 93–98. [DOI] [PubMed] [Google Scholar]
- Roth, S. Y., J. M. Denu and C. D. Allis, 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70 81–120. [DOI] [PubMed] [Google Scholar]
- Santisteban, M. S., T. Kalashnikova and M. M. Smith, 2000. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103 411–422. [DOI] [PubMed] [Google Scholar]
- Sermwittayawong, D., and S. Tan, 2006. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J. 25 3791–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., 1991. Getting started with yeast. Methods Enzymol. 194 3–21. [DOI] [PubMed] [Google Scholar]
- Smith, F. C., S. P. Davies, W. A. Wilson, D. Carling and D. G. Hardie, 1999. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 453 219–223. [DOI] [PubMed] [Google Scholar]
- Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya et al., 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D. E., R. Belotserkovskaya and S. L. Berger, 2002. a SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. USA 99 11622–11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon and S. L. Berger, 2002. b The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277 8178–8186. [DOI] [PubMed] [Google Scholar]
- Sternglanz, R., and H. Schindelin, 1999. Structure and mechanism of action of the histone acetyltransferase Gcn5 and similarity to other N-acetyltransferases. Proc. Natl. Acad. Sci. USA 96 8807–8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki, P., I. Topalidou and G. Thireos, 2000. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404 414–417. [DOI] [PubMed] [Google Scholar]
- Tanner, K. G., R. C. Trievel, M. H. Kuo, R. M. Howard, S. L. Berger et al., 1999. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 274 18157–18160. [DOI] [PubMed] [Google Scholar]
- Topalidou, I., M. Papamichos-Chronakis, G. Thireos and D. Tzamarias, 2004. Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. EMBO J. 23 1943–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel, R. C., J. R. Rojas, D. E. Sterner, R. N. Venkataramani, L. Wang et al., 1999. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 96 8931–8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse, C., E. I. Georgieva, A. B. Ruiz-Garcia, R. Sendra and J. C. Hansen, 1998. Gcn5p, a transcription-related histone acetyltransferase, acetylates nucleosomes and folded nucleosomal arrays in the absence of other protein subunits. J. Biol. Chem. 273 32388–32392. [DOI] [PubMed] [Google Scholar]
- Verdone, L., J. Wu, K. van Riper, N. Kacherovsky, M. Vogelauer et al., 2002. Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J. 21 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer, M., J. Wu, N. Suka and M. Grunstein, 2000. Global histone acetylation and deacetylation in yeast. Nature 408 495–498. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wang, L., L. Liu and S. L. Berger, 1998. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield, L., J. A. Ranish and S. Hahn, 2004. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes Dev. 18 1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, F., and P. L. Minehart, 1986. Analysis of the yeast SPT3 gene and identification of its product, a positive regulator of Ty transcription. Nucleic Acids Res. 14 6885–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P. Y., C. Ruhlmann, F. Winston and P. Schultz, 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15 199–208. [DOI] [PubMed] [Google Scholar]
- Wyce, A., K. W. Henry and S. L. Berger, 2004. H2B ubiquitylation and de-ubiquitylation in gene activation. Novartis Found. Symp. 259 63–73; discussion 73–67, 163–169. [PubMed] [Google Scholar]
- Yu, Y., P. Eriksson, L. T. Bhoite and D. J. Stillman, 2003. Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein. Mol. Cell. Biol. 23 1910–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., J. R. Bone, D. G. Edmondson, B. M. Turner and S. Y. Roth, 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17 3155–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]