Abstract
Stimulated by its physiological ligand, hepatocyte growth factor, the transmembrane receptor tyrosine kinase Met activates a signaling machinery that leads to mitogenic, motogenic, and morphogenic responses. Remarkably, the food-borne human pathogen Listeria monocytogenes also promotes autophosphorylation of Met through its virulence factor internalin B (InlB) and subsequently exploits Met signaling to induce phagocytosis into a broad range of host cells. Although the interaction between InlB and Met has been studied in detail, the signaling specificity of components involved in InlB-triggered cellular responses remains poorly characterized. The analysis of regulated phosphorylation events on protein kinases is therefore of particular relevance, although this could not as yet be characterized systematically by proteomics. Here, we implemented a new pyridopyrimidine-based strategy that enabled the efficient capture of a considerable subset of the human kinome in a robust one-step affinity chromatographic procedure. Additionally, and to gain functional insights into the InlB/Met-induced bacterial invasion process, a quantitative survey of the phosphorylation pattern of these protein kinases was accomplished. In total, the experimental design of this study comprises affinity chromatographic procedures for the systematic enrichment of kinases, as well as phosphopeptides; the quantification of all peptides based on the iTRAQTM reporter system; and a rational statistical strategy to evaluate the quality of phosphosite regulations. With this improved chemical proteomics strategy, we determined and relatively quantified 143 phosphorylation sites detected on 94 human protein kinases. Interestingly, InlB-mediated signaling shows striking similarities compared with the natural ligand hepatocyte growth factor that was intensively studied in the past. In addition, this systematic approach suggests a new subset of protein kinases including Nek9, which are differentially phosphorylated after short time (4-min) treatment of cells with the Met-activating InlB321. Thus, this quantitative phosphokinome study suggests a general, hypothesis-free concept for the detection of dynamically regulated protein kinases as novel signaling components involved in host-pathogen interactions.
The human food-borne pathogen Listeria monocytogenes has evolved mechanisms to cross the intestinal, placental, and blood-brain barriers with severe consequences for pregnant women, newborns, and immunocompromised individuals. As a facultative intracellular pathogen, L. monocytogenes invades host cells within minutes, thus escaping the humoral arm of adaptive immunity. In this protective host niche, the organism replicates and spreads from cell to cell through the formation of so-called membrane protrusions. L. monocytogenes utilizes two different molecular routes to invade non-professional phagocytotic cells. (i) Internalin A binds to the cell adhesion molecule E-cadherin, resulting in the initial penetration of intestinal tissue (1, 2). (ii) In contrast, internalin B (InlB)1 contributes to the systemic infection of the host, promoting the invasion of a broader range of cell types including hepatocytes (3) and endothelial cells (4). A basic GW motif at the C terminus mediates the attachment of InlB to the bacterial cell wall, but the non-covalent nature of this interaction also allows the partial release of InlB into the environment (5, 6). GW domains of soluble InlB interact with glycosaminoglycans (7) and the complement receptor qC1q-R (8) on the host cell surface, although these interactions seem to be dispensable for the process of listerial invasion. In contrast, the N-terminal region of InlB comprising the cap, leucine-rich repeat, and inter-repeat domains (termed InlB321) constitutes structural features that stimulate the bacterial ingestion into the host cell cytosol. The horseshoe-like shape of InlB321 allows binding to and activation of the transmembrane tyrosine kinase Met, which is also the receptor for the host growth factor, hepatocyte growth factor (HGF). Although InlB binds to a different region of Met compared with HGF, it exploits the Met signaling capabilities, ultimately leading to actin cytoskeleton rearrangements, membrane engulfment, and uptake of the pathogen. InlB induces a rapid autophosphorylation in the kinase domain of Met (9) followed by recruitment of specific adapter molecules initiating signal transduction via prominent downstream components such as PI3K and the Raf-Erk pathway (10). Moreover, immobilized InlB321 is sufficient to induce the efficient uptake of latex beads into the host cell (11, 12). Recently, the structure of the InlB321-Met complex was solved at the atomic level, unambiguously demonstrating that InlB321 is mandatory but also sufficient to activate Met signaling (13).
Numerous molecular studies of signaling components have been reported, and a complex protein network downstream of Met has been compiled (14). However, the molecular interactions defined so far are still insufficient to derive the InlB-induced signal transduction pathway resulting in uptake of Listeria. As a basic signaling principle, protein kinase-catalyzed phosphorylation regulates virtually every function of substrate proteins, i.e. protein-protein interactions, localization, activity, and stability. With more than 500 members, the superfamily of protein kinases is among the largest protein families encoded by the human genome (15). The functional mechanisms regulated by kinase-mediated phosphorylations on substrate proteins are also involved in the activity control of the kinases themselves. Studying these modifications directly at the kinase level enables classification of their activated states, and their systematic investigation by proteomics has already been used to detect and correlate kinases with potential functions in cell cycle control and cancer biology (16, 17). A detailed knowledge of InlB/Met-affected phosphorylation sites of proteins from the kinase superfamily would contribute to a better understanding of the listerial invasion strategy in addition to complementing our knowledge of the Met signaling pathway.
Phosphorylation sites can be detected during the process of automatic peptide sequencing in well established bottom-up proteome approaches. However, the substoichiometric nature and poor ionization properties of phosphopeptides usually require purification strategies such as IMAC to optimize analysis by mass spectrometry (18). Furthermore, the complexity of the total phosphoproteome requires the pre-enrichment of protein kinases as a prerequisite for characterization of the low abundance family members. We and others have demonstrated that the highly conserved ATP-binding region of protein kinases offers possibilities for their systematic purification based on immobilized ATP-competitive small molecule inhibitors with broad kinase selectivity. In combination with phosphopeptide enrichment, this strategy has proven to be highly appropriate for a comprehensive LC-MS/MS-based phosphorylation site analysis of these key signaling components (17, 19, 20). To characterize the role of protein kinases as key regulatory elements in signaling pathways, the acquisition of quantitative peptide data of both the phosphorylated and unmodified proteins is required. Powerful isotopic labeling approaches such as SILAC (21) and iTRAQTM (22) have been devised and successfully applied to dissect cell and signaling states mainly at the substrate protein level (23, 24), but they are also beginning to support the in-depth characterization of the human kinome (17, 20, 25). Because the detection of individually regulated phosphopeptides has to cope with the so-called “one-hit wonder” problem in proteomics, the interpretation of single peptide regulation requires that particular attention must be paid to the process of statistical raw data evaluation. We have recently established a validated statistical strategy for the quality control of quantitative MS methods used in this study (26). In total, this bioinformatics work flow normalizes unequal sample amounts, corrects isotopic impurities of iTRAQ labeling reagents, and importantly can calculate the reliability of regulatory data based on the actual signal-to-noise properties of the mass spectrometer used.
The synthesis of an optimized affinity resin as a base for a robust single step capture of protein kinases was the starting point in this study and allowed the systematic analyses of this enzyme class in human epithelial cells. In the following, we explain the biochemical strategy established for the quantitative characterization of phosphorylation events at these kinases in the context of infection. The dissection of one representative data set shows the potential of the selected strategy but also underscores the necessity of our statistical approach for evaluating the regulatory information based on iTRAQ reporter ions. Finally, we apply the total approach to analyze protein kinases systematically in the Met receptor kinase pathway exploited by the invasin InlB from L. monocytogenes. The majority of unambiguously regulated phosphorylation events are in accordance with our existing knowledge about the HGF/Met pathway. Furthermore, this study suggests novel candidates such as Nek9 involved in signal networks exploited in the process of listerial invasion.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Phospho-Met (Tyr1234/1235), phospho-Mek1/2 (Ser217/221), phospho-GSK3A/B (Ser21/9), phospho-Akt (Ser473), and phospho-p44/42 MAPK (Thr201/Tyr203; Erk1/2) were purchased from Cell Signaling Technology Inc., and anti-actin-(20–33) antibody was purchased from Sigma-Aldrich. To prevent phosphatase activity, the following substances were used: calyculin A (LC Laboratories), phosphatase inhibitor mixtures 1 and 2 (Sigma-Aldrich), sodium orthovanadate (Sigma-Aldrich), and sodium fluoride (Merck). The protease inhibitor mixture CompleteTM was purchased from Roche Applied Science. Triton X-100 and ethanolamine ReagentPlus were obtained from Sigma-Aldrich, and N,N-dimethylformamide (DMF), triethylammonium bicarbonate, and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC-HCl) were from Fluka. The organic solvents ethanol, methanol, and acetonitrile of pro analysis purity were purchased from J. T. Baker Inc.
Protein Purification
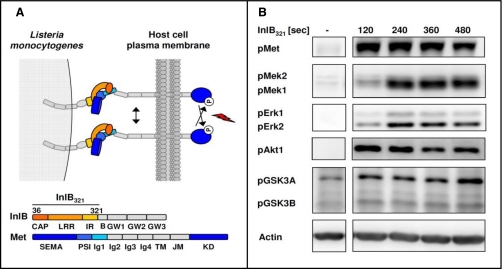
InlB321 was recombinantly expressed and purified as described elsewhere (27). In brief, DNA coding for InlB-(36–321) was cloned into the pGEX-6P-1 vector (GE Healthcare) and expressed as a GST fusion protein in Escherichia coli BL21 (Stratagene). Cells were disrupted using a French press. Purification of the proteins was performed using Glutathione-Sepharose followed by proteolytic cleavage of the GST tag in solution. Final purification was achieved through Mono Q (GE Healthcare) anion exchange chromatography. Two further InlB321 constructs were expressed and purified as StreptagII fusion proteins (28). In brief, DNA coding for InlB-(36–321) and InlB-(36–321) loss of function (11) were cloned into the pASK-IBA3-Plus vector (IBA GmbH) comprising a tet promoter for controlling InlB expression. Expression was performed in strain E. coli BL21. Recombinantly expressed InlB was purified from centrifuged French press lysates by StrepTactin-Sepharose affinity chromatography and competing elution with desthiobiotin. Final purification was again achieved through Mono Q (GE Healthcare) anion exchange chromatography. The recombinant proteins were dialyzed against PBS, concentrated to a final concentration of ∼2 mg/ml, filtered through a 0.2-μm membrane, and stored at −20 °C until use. Stability and purity of the recombinant proteins were investigated by SDS-PAGE. The bioactivity of the recombinantly expressed InlB321 constructs was confirmed in Western blot analyses using phosphorylation site-specific antibodies, indicating the activity state of Met as exemplified in Fig. 1B.
Fig. 1.
N-terminal variant InlB321 is a potent activator of Met signal transduction. A, schematic representation of the Met741-InlB321 complex according to Niemann et al. (13). Amino acids 36–321 from InlB are necessary for Met binding and activation. B, Western blot analyses of the activation of Met, Akt1, Mek1/2, and Erk1/2 and the inhibition of GSK3A/B by phosphorylation site-specific antibodies reveal a fast InlB321-dependent induction of the Met signaling machinery. Within the first minutes after addition, InlB321 activates the mitogen-activated protein kinase pathway and the PI3K/Akt1 module (pMet, pAkt, and pErk1/2, n = 3; pMek1/2 and pGSK3A/B, n = 2). IR, inter-repeat; LRR, leucine-rich repeat; TM, transmembrane; JM, juxtamembrane; KD, kinase domain; SEMA, semaphorin. B, B-repeat; PSI, plexin-semaphorin-integrin domain.
Western Blot Analysis
HeLa S3 cells (ATCC CCL 2.2) were grown in DMEM (Invitrogen) supplemented with 2 mm l-glutamine, 10% FBS Gold (PAA Laboratories), 50 units/ml penicillin (Invitrogen), and 50 μg/ml streptomycin (Invitrogen) in a 7.5% CO2 humidified 37 °C atmosphere. 4 × 105 cells were seeded onto tissue culture treated dishes (35 × 10 mm, BD Biosciences) and starved for 8 h in DMEM prior to stimulation with InlB321 at the indicated times. Supernatants were discarded, and cells were scraped off the dishes after addition of 50 μl of SDS sample buffer (0.24 m Tris, pH 6.8, 40% glycerin, 12% 2-mercaptoethanol, 0.27 m SDS, 0.004% bromphenol blue) containing 1 μl of Benzonase to each dish. After 5-min incubation at room temperature, the resulting cell lysates were heated to 80 °C for 10 min. 5 μl from each sample were applied to 10% SDS-PAGE. Protein transfer to polyvinylidene fluoride membranes (Immun-BlotTM PVDF membrane, 0.2 μm, Bio-Rad) was conducted in a cooled (8 °C) tank blotting device for 2.5 h with 200 V (blotting buffer: 40 mm glycine, 20 mm Tris, 2 mm SDS, 20% (v/v) MeOH). Membranes were blocked with 5% bovine serum albumin in TBS (20 mm Tris, pH7.5, 137 mm NaCl) supplemented with 0.1% (v/v) Tween 20 and probed with the indicated primary antibodies according to the manufacturer's recommendation at 4 °C overnight. After membrane washing (10 min each with TBS supplemented with (i) 0.1% (v/v) Tween 20, (ii) 0.5 m NaCl, (iii) 0.5% (v/v) Triton X-100, and (iv) 0.1% (v/v) Tween 20), the respective secondary antibody conjugated to HRP (goat anti-mouse HRP or goat anti-rabbit HRP, Dianova) was probed at room temperature for 1 h. Finally, membranes were washed with TBS supplemented with 0.1% (v/v) Tween 20, and the light reaction was started by addition of Lumi-Light Western blotting substrate (Roche Diagnostics). Luminescence signals were detected with a cooled charge-coupled device camera (Fujifilm Las3000, Raytest). Prior to incubation with further antibodies, the membranes were stripped for 10 min each in buffer A (0.2 m glycine, 0.5 m NaCl, pH 2) and buffer B (0.5 m Tris, pH 11) and reblocked with bovine serum albumin as indicated above. Exposures were saved in a 16-bit TIFF format, and AIDA software (version 4.06, Raytest) was used to ensure that the dynamic range of the charge-coupled device chip was not exceeded.
Synthesis of VI16743
Preparation of 2-{2-[4-(8-Cyclopentyl-7-oxo-7,8-dihydro-pyrido[2,3-d]pyrimidin-2-ylamino)-phenoxy]-ethyl}-isoindole-1,3-dione
A solution of 0.32 g (1.00 mmol) of 8-cyclopentyl-2-methanesulfonyl-8H-pyrido[2,3-d]pyrimidin-7-one (29), 0.28 g (1.00 mmol) of 2-[2-(4-amino-phenoxy)-ethyl]-isoindole-1,3-dione (30), and 0.13 g (0.17 ml, 1.00 mmol) of N-ethyldiisopropylamine in 20 ml of propan-2-ol was refluxed for 24 h under an argon atmosphere. The solvent was evaporated under reduced pressure, and the residue was partitioned between 30 ml of water and 30 ml of ethyl acetate. The separated water phase was extracted twice with 30 ml of ethyl acetate, the collected organic phase was washed with 30 ml of brine and dried with magnesium sulfate, and the solvent was evaporated under reduced pressure. The residue was crystallized from 10 ml of acetonitrile to give pure product.
Preparation of 2-[4-(2-Amino-ethoxy)-phenylamino]-8-cyclopentyl-8H-pyrido[2,3-d]pyrimidin-7-one (VI16743)
A solution of 0.52 g (1.00 mmol) of 2-{2-[4-(8-cyclopentyl-7-oxo-7,8-dihydro-pyrido[2,3-d]pyrimidin-2-ylamino)-phenoxy]-ethyl}-isoindole-1,3-dione and 0.52 g (0.48 ml, 10.00 mmol) of hydrazine hydrate in 25 ml of ethanol was refluxed for 3 h. The solvent was evaporated under reduced pressure, and the residue was partitioned between 30 ml of 3 n HCl in water and 30 ml of ethyl acetate. The separated water phase was extracted four times with 30 ml of ethyl acetate, the pH of the water phase was adjusted to 10 using solid sodium carbonate, and the water phase was extracted three times with 2:1 ethyl acetate:tetrahydrofuran. The organic phase was washed with 30 ml of brine and dried with magnesium sulfate, and the solvent was evaporated under reduced pressure. The residue was crystallized from 5 ml of acetonitrile to give a pure product.
Generation of Kinase Affinity Resin
VI16743 (MW 365.44) was dissolved in 50% DMF, EtOH at a concentration of 3 mm. For immobilization, this inhibitor solution was added (1 ml/tube) to washed and drained ECH-Sepharose 4B (0.5 ml/tube). The reaction was started by dropwise addition of 150 μl of 1 m EDC-HCl in 50% DMF, EtOH followed by overnight incubation at room temperature in the dark with gentle agitation. After two washes in 50% DMF, EtOH, 1 ml of DMF/EtOH/ethanolamine (1:1:1) was added to block the remaining reactive groups. The reaction was repeated by dropwise addition of 150 μl of 1 m EDC-HCl in 50% DMF, EtOH. Incubation was again conducted at room temperature in the dark overnight with gentle agitation. Sepharose was washed twice with 50% DMF, EtOH; twice with 50 mm HEPES-NaOH, pH 7.5, 0.5 M NaCl; and twice with double distilled H2O. For kinase affinity chromatography four Tricorn 5/50 columns (GE Healthcare) were each packed with 1 ml of VI16743 affinity resin.
Cell Culture, Treatment, and Lysis
HeLa S3 cells (ATCC CCL 2.2) were grown in DMEM (Invitrogen) supplemented with 2 mm l-glutamine, 10% FBS Gold (PAA Laboratories), 50 units/ml penicillin (Invitrogen), and 50 μg/ml streptomycin (Invitrogen) in a 7.5% CO2 humidified 37 °C atmosphere. For each quantitative phosphokinome experiment, 1.5 × 108 adherent cells were grown for 24 h on 50 dishes to 50% cell confluence (15-cm diameter, Corning) and then starved for 24 h in DMEM prior to stimulation. In total, four stimulation experiments were performed by treatment of one-half of the cells with 10 nm InlB321 for 4 min. The other half served as control and was either left untreated or incubated with 10 nm InlB321 loss of function, which is unable to bind and activate the receptor tyrosine kinase Met (11). Medium was rapidly discarded after stimulation, 1.5 ml of ice-cold high salt lysis buffer (50 mm HEPES-NaOH, pH 7.5, 1 m NaCl, 1 mm EGTA, Complete, 10 mm NaF, 2.5 mm Na3VO4, 50 ng/ml calyculin A, 1% phosphatase inhibitor mixture 1, 1% phosphatase inhibitor mixture 2, 1% Triton X-100) was added to each dish, and cells were immediately frozen by plunging the bottom of the dishes into liquid nitrogen before storing on dry ice. Samples from all dishes were rethawed in a 4 °C chamber, completely transferred to 50-ml plastic tubes, and solubilized by sonification (5 × 5 s, Bandelin Sonopuls HD200 MS73 lance, Bandelin Electronic). The resulting cell lysates were filtered using a 0.45-μm SterivexTM-HV syringe filter (Millipore) and centrifuged at 70,000 × g for 30 min (4 °C) to remove non-solubilized cell material. The protein concentration of each supernatant was determined by Bradford protein assay (Bio-Rad), and equal protein amounts from InlB-stimulated lysate and from control lysate were used for small molecule kinase affinity chromatography.
Kinase Affinity Chromatography
Two Tricorn 5/50 columns filled with VI16743-coupled Sepharose were connected in series for each lysate. Chromatography with both lysates from one experiment was performed in parallel with two GE Healthcare pump P500 systems connected to GE Healthcare valve V7. The columns were equilibrated with buffer A (50 mm HEPES-NaOH, pH 7.5, 1 m NaCl, 1 mm EGTA, 1 mm EDTA, 10 mm NaF, 0.1 mm Na3VO4, 0.1% Triton X-100). Lysates were applied to the columns with a flow rate of 3 ml/h, washed with 60 column volumes of buffer A (6 ml/h), and equilibrated with buffer B (50 mm HEPES-NaOH, pH 7.5, 1 mm EGTA, 1 mm EDTA, 10 mm NaF, 0.1 mm Na3VO4, 0.1% Triton X-100). The columns were disconnected, and each column was eluted separately with 0.5% SDS at room temperature (flow rate, 6 ml/h). Twelve 0.5-ml fractions per column were collected. Each fraction was immediately reduced by adding 10% (v/v) 200 mm DTT before incubation for 30 min at 56 °C. Carbamidomethylation was carried out by adding 10% (v/v) 500 mm iodoacetamide and incubating for 1 h in the dark. 10% of each fraction was resolved by SDS-PAGE, and protein detection was by Coomassie-fast silver staining (31).
Sample Preparation for LC-MS/MS Analysis
Protein-containing fractions of the eluates from each sample were pooled and concentrated under vacuum. Proteins were purified by chloroform/methanol precipitation (32). The two resulting protein pellets corresponding to InlB321-treated and control cells were resolved in 50 mm triethylammonium bicarbonate, 10% ACN. Before proteolytic digestion and further LC-MS sample preparations, an aliquot of each protein sample was analyzed on Coomassie-fast silver-stained SDS-polyacrylamide gels. Using a densitometric quantification procedure based on the LMW-SDS Marker kit (GE Healthcare) and AIDA (version 4.06), 120 μg of total protein from each sample were digested with sequencing grade modified trypsin (Promega) as recommended in a ratio of 1:50 at 37 °C overnight. Subsequently, digestion was completed by adding a further 1 μg of trypsin for 2 h to each sample. The peptide solutions were vacuum-dried, and peptides were resolved in 0.2% TFA in water; desalted on self-packed LiChroprep RP-18 (Merck) SPE columns; eluted with 0.2% TFA, 60% ACN in water; and again vacuum-dried.
iTRAQ Modification of Peptides
iTRAQ labeling was performed according to the manufacturer's protocol (Applied Biosystems). Briefly, dried peptides derived from 120-μg protein fractions of InlB-treated and control cells were resolved in 80 μl of iTRAQ dissolution buffer. To ensure complete iTRAQ labeling two portions of two different labeling reagents, sufficient to label 200 μg of total protein, were dissolved in 70 μl/portion ethanol and used for the labeling reaction that was always conducted for 80 min at room temperature in the dark. Following labeling, both samples from one experiment were combined, vacuum-dried, and desalted on self-packed LiChroprep RP-18 (Merck) SPE columns. The iTRAQ labels used are shown in Table I.
Table I. iTRAQ labels used in this study.
| Experiment | Control sample | InlB321-treated sample |
|---|---|---|
| 1 | 114.1 | 117.1 |
| 2 | 115.1 | 117.1 |
| 3 | 114.1 | 117.1 |
| 4 | 115.1 | 117.1 |
Phosphopeptide Enrichment
A Ga3+-based phosphopeptide enrichment kit (Pierce) was used to separate phosphopeptides from non-phosphorylated peptides. All the following steps were carried out in so-called inner surface polymer optimized sample tubes (Roth) to guarantee a high recovery of phosphorylated peptides in solution. Combined iTRAQ modified peptides derived from stimulated and control cells were solubilized in 100 μl of binding buffer (1:1:1 methanol:acetonitrile:H2O containing 2% acetic acid, pH 2.8), and three Ga3+-chelated gel disks were added. After 1 h of incubation at room temperature with gentle agitation, unbound peptides were removed by washing 10 times using binding buffer supplemented with 100 mm NaCl followed by washing 10 times with pure binding buffer. Phosphopeptides were specifically eluted with 10 repetitive washes each with 100 μl of 100 mm ammonium phosphate buffer (pH 4.5). The flow-through and all washings were combined and constituted the phosphopeptide-depleted fraction. The IMAC eluate comprising the phosphopeptide-enriched fraction and the phosphopeptide-depleted fraction were vacuum-dried. Both fractions were desalted on self-packed LiChroprep RP-18 (Merck) SPE columns.
Strong Cation Exchange Chromatography, LC-MS/MS Analyses, and Database Searching
The complex phosphopeptide-depleted fraction was further subfractionated by SCX chromatography. Dried peptides were resolved in SCX buffer A (0.065% formic acid, 25% acetonitrile) and separated on a Mono S PC1.6/5 column (GE Healthcare) via an Ettan micro-LC system (GE Healthcare) with a linear gradient from 0 to 35% SCX buffer B (0.065% formic acid, 25% acetonitrile, 0.5 m potassium chloride) in 30 min. The flow rate was 150 μl/min. 1-minute fractions were collected with a microfraction collector (SunCollect), and peptide elution was monitored at 214 nm. Peptide-containing fractions were vacuum-dried and desalted by RP C18 chromatography (μZipTip pipette tips, Millipore).
LC-MS/MS analyses of all desalted SCX fractions and all phosphopeptide-enriched fractions were performed on an Acquity ultraperformance LC system (Waters Corp.) connected to a Q-TOF microTM mass spectrometer (Waters Corp.). Peptides were flushed onto a C18 precolumn (5-μm Symmetry C18, 180 μm × 20 mm, Waters Corp.) with a flow rate of 15 μl/min and washed at constant flow for 3 min. Peptides were then separated on an analytical column (1.7-μm BEH130, 75 μm × 150 mm, Waters Corp.) with UPLC buffer A (0.1% formic acid in water) and UPLC buffer B (0.1% formic acid in acetonitrile) via linear 90- or 120-min gradients at a flow rate of 300 nl/min controlled with AcquityUPLC software V1.22. Eluting peptides were ionized using PicoTip emitter needles (New Objective Inc.) and voltages of ∼1800 kV. Data-dependent acquisition of MS and MS/MS data was under control of MassLynx software V4.1 (Waters Corp.). Doubly and triply charged peptide ions were automatically selected and fragmented with m/z-dependent collision energy settings optimized for iTRAQ labeled peptides with a maximum of 18 s per peptide. MS data were automatically processed by MassLynx V4.1, generating peak lists for protein identification by database searches. Database searches were carried out with Mascot Daemon 2.1.6 in the UniProtKB/Swiss-Prot database (release 55.0 of February 26, 2008 with 356,194 entries; taxonomy, Homo sapiens with 18,610 entries). Mascot Daemon was used to merge all peak lists from SCX fractions and IMAC eluate fractions corresponding to one experiment. Proteins were only accepted as identified when at least one unique peptide showed an individual score above 20, which indicated identity or extensive homology (p < 0.05) based on the search parameter settings used (enzyme, trypsin; maximum missed cleavages, 1; fixed modification: carbamidomethyl (Cys), iTRAQ (N terminus), iTRAQ (Lys), variable modifications, phosphorylation (Ser, Thr, Tyr) and oxidation (Met); peptide tolerance, 65 ppm; MS/MS tolerance, 0.15 Da). Phosphopeptides and phosphorylation sites were usually manually confirmed or rejected if the spectrum was of poor quality. Decoy searches were performed in a reversed UniProtKB/Swiss-Prot database with search parameters identical to those described above. False discovery rates (FDR) were calculated on the basis of FDR = FP/(FP + TP). False positive (FP) and true positive (TP) peptide matches were assigned by counting strictly unique peptide matches (rank 1 and red bold) with confident identification scores (p < 0.05) of target or decoy search results. We determined on average 12% TP, 0.1% FP, and false discovery rates of about 1%. Raw data information of all experiments can be provided on request as a Scaffold data file.
Peptide Quantification
To determine by-product impurities of the iTRAQ reagents a small aliquot from each label was analyzed on a Q-TOF2TM instrument (Micromass, Waters Corp.). Fragmentation was performed by a stepwise increase of collision energy from 10 to 45 eV. The relation of ion intensities of each reporter signal to the detected by-products was calculated from processed spectra. The percentage of impurities was used to determine the sample-specific reporter ion intensities. Peptide quantification was then performed by an in-house bioinformatics tool, named iTRAQassist, as described recently (26). In brief, the Mascot .dat file from a merged peak list from one experiment and the respective by-product calibration file were uploaded. The iTRAQ reporter and regulation base (control) used, as well as the isolation width for the iTRAQ reporter ion detection (0.05 Da) and peptide cutoff (20), were chosen to restrict the quantitative approach to unambiguously identified peptide sequences. iTRAQassist then performed a normalization of reporter ion intensities and provided statistical information for the regulatory data at the individual peptide and protein levels. A documentation of iTRAQassist functions, the program, and a test data set will be provided on request. Peptides were accepted as regulated if their corresponding likelihood curves showed a distinct separation from the majority of the remaining peptides belonging to the same protein. Finally, all detected regulated peptides were checked manually regarding peptide sequence quality and an unambiguous phosphorylation site assignment. Additionally, regulated candidates were further restricted to peptides that exhibited reporter ion intensities of at least 15 ion counts.
RESULTS
InlB321 Is an Effective Activator of Met-mediated Signaling
We used the structurally verified variant of InlB comprising amino acids 36–321 (InlB321) to ensure that only Met signaling was observed without cross-activation of co-receptors. InlB321 is sufficient to stimulate uptake of coated latex beads, and the recent structure of the Met ectodomain complexed with InlB321 revealed how the receptor is locked in a signaling-competent conformation (Fig. 1A). InlB321 expression was performed according to Schubert et al. (27), and its capacity to stimulate the Met receptor and its known downstream components was investigated by Western blot analyses. Cells were stimulated for 2, 4, 6, or 8 min by adding extracellular InlB321, and subsequently total cell lysates were probed with phosphorylation site-specific antibodies that indicated the activation of Met, Mek1/2, Erk1/2, and Akt and the inhibition of GSK3A/B (pMet, pMek1/2, pErk1/2, pAkt, and pGSK3A/B). Soluble InlB321 induced a pronounced autophosphorylation of tyrosines in the kinase domain of Met already within the first 2 min. This activation level of Met did not decrease significantly in the following 6 min. As expected, components of the PI3K/Akt pathway (pAkt and pGSK3A/B) exhibited a rapid phosphorylation site-specific response, confirming their intimate regulatory connection to Met. In contrast, the activation of the Raf-Erk cascade (pMek and pErk) downstream of Met was slightly delayed compared with the PI3K/Akt pathway, and phosphorylation signals were maximally induced 4 min after stimulation with InlB321 (Fig. 1B).
Thus, time-resolved analyses established recombinantly expressed InlB321 as a potent activator of proximal Met signaling. The InlB variant consisting of the leucine-rich repeat structure together with the cap and inter-repeat domains is sufficient to induce Met kinase activity and major Met-dependent downstream cascades. This demonstrates that InlB321 is a suitable effector for studying Met-mediated signaling systematically by proteomics. Because all investigated kinases were concomitantly induced after 4 min, we selected this time point for quantitative proteome analysis of the Met pathway.
Humane Kinome Affinity Chromatography Based on Immobilized VI16743
Based on the multitarget selectivity of small molecule kinase inhibitors directed against the conserved ATP-binding region, several affinity matrices have been generated that allow the systematic enrichment of protein kinases (20, 33, 34). The immobilized pyridopyrimidine class inhibitor PP58 was demonstrated as a promising compound for a comprehensive kinase prefractionation strategy from different cell lines (19, 35). However, structural investigations of PP58 in complex with the Src kinase revealed a preference of this inhibitor for protein kinases with small and hydrophilic “gatekeeper” (GK) amino acids at the bottom of the ATP binding pocket (36). Because about 75% of all protein kinases harbor larger side chains at the critical GK position, we modified PP58 to improve its properties as a broad spectrum kinase inhibitor (supplemental Data S1). The resulting molecule, VI16743, was immobilized on Sepharose beads using the free amino group. As previously shown for PP58, this coupling strategy fully retains the kinase binding properties (35).
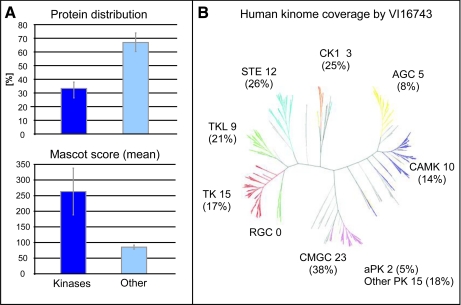
At first, we determined the kinase spectrum of our novel compound for the systematic analysis of InlB321/Met-dependent protein kinases. Whole cell lysates from the human epithelial cancer cell line HeLa S3 were applied to small molecule affinity chromatography (SMAC) based on immobilized VI16743. Binding, stringent washing, and elution steps were adapted from similar previous kinome analyses, and enriched proteins were identified by peptide sequencing (LC-MS/MS). In total, we could unambiguously identify 409 proteins in the eluted fractions of the VI16743 matrix. Among these, 102 proteins contained a putative kinase domain and were classified as kinases according to the human kinome annotation by Manning et al. (37) and the annotations of the Swiss-Prot database (supplemental Data S2). Although eight of these kinases interact with low molecular weight phosphate acceptors, the majority (94 kinases) transfer phosphate groups to proteins. Importantly, about half of all the unique peptides identified upon affinity purification were derived from protein kinases, indicating high kinase selectivity of immobilized VI16743. Moreover, the combined MS/MS data for the peptides derived from kinases further underscore the specificity of the SMAC strategy: kinases were identified on average by 10 peptides and a median Mascot score of greater than 250, whereas non-kinases were detected on average by only four peptides and a median Mascot score of 86 (Fig. 2A). This demonstrates the specific affinity features of VI16743 for proteins from the superfamily of kinases, which are furthermore substantiated by the high reproducibility of protein kinase identifications: 69% of all kinases were consistently identified in three independent replicate experiments. In contrast, only 37% of non-kinase proteins could be reproducibly identified over the same set of experiments. An alignment of identified protein kinases with the human kinome dendrogram (37) showed that members from virtually all subfamilies of the human kinome were purified by VI16743 affinity chromatography (Fig. 2B) with a slight preference for members of the CMGC (CDK/MAPK/GSK3/CLK family) group. A structural feature shared among many CMGC group members is the presence of a space-filling phenylalanine at the conserved GK position. Thus, compared with PP58, VI16743 exhibits considerably increased coverage for kinases possessing this structural element. Overall, analyses of GK residues in protein kinases purified by PP58 from three different cell lines indicated a clear preference for the small and hydrophilic threonine (19, 35), whereas VI16743 selected for kinases with the larger and hydrophobic leucine and phenylalanine (supplemental Data S1).
Fig. 2.
VI16743 affinity matrix permits comprehensive one-step purification of human protein kinases. A, evaluation of protein distribution and Mascot identification score in VI16743-purified fractions demonstrate the high selectivity of the immobilized inhibitor for protein kinases. Error bars indicate the standard deviation within four independent experiments. B, VI16743 enables purification of protein kinases from nearly all groups of the human kinome. aPK, atypical protein kinases; PK, protein kinases; CAMK, calcium/calmodulin-dependent kinases; TK, tyrosine kinases; TKL, tyrosine kinase-like kinases. STE, sterile homologue kinases; AGC, PKA/PKG/PKC-family kinases; RCG, receptor guanylate cyclases; CMGC, CDK/MAPK/GSK3/CLK family kinases.
According to estimates that around 300 distinct kinases are expressed in a mammalian cell (38), VI16743 allowed the purification of more than 30% of the human kinome (94 protein kinases) and is presently one of the most effective ATP-competitive ligands for SMAC of protein kinases starting with total cell lysates. Furthermore, VI16743 enabled direct access to Met itself and known downstream kinases such as GSK3A/B, Mek1/2, and Erk1/2 in the context of a systematic screen, suggesting that this resin is an excellent affinity matrix for the direct biochemical investigation of kinase-mediated signal transduction induced by InlB from Listeria.
Combination of SMAC, IMAC, iTRAQ, and LC-MS/MS Enables Quantitative Phosphoproteome Analyses of Protein Kinases
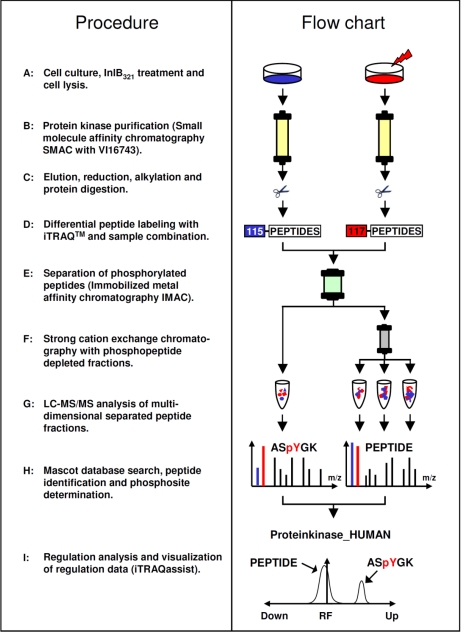
Quantitative phosphorylation site analyses of protein kinases can be highly informative in providing details of their activity states within signaling networks. This study addresses the systematic identification of InlB321/Met-dependent phosphorylation events occurring on members of the protein kinase superfamily. All the methods and technologies required are already available in cell biology and proteomics and are now combined to establish a quantitative work flow for phosphokinome analysis (Fig. 3). To reduce potential effects of endogenous HGF, HeLa S3 cells were starved in minimal medium for 24 h. Under these conditions the cells afforded excellent and reproducible responses to external InlB321 (Fig. 1B). Subsequently, the cells were stimulated with InlB321 for 4 min or left untreated for the same time as a control. iTRAQ labeled phosphorylated and non-phosphorylated peptides from the enriched kinase fractions of stimulated and control cells were finally combined and analyzed by SCX/LC-MS/MS and Mascot. The evaluation of iTRAQ reporter ion information was performed by iTRAQassist (26), an in-house developed software for the calculation and graphical presentation of regulatory events observed at the purified kinases.
Fig. 3.
Experimental strategy for MS/MS-based identification and quantification of ligand-induced phosphorylation events on protein kinases in cellular signaling. InlB321-treated and untreated HeLa S3 cells (1.5 × 108 per experiment) were lysed, and the resulting protein extracts were applied in parallel for protein kinase affinity chromatography. Bound proteins were eluted, alkylated, and digested. Peptides of each sample were modified by specific iTRAQ reagents prior to sample combination. Before LC-MS/MS analysis, IMAC was used to separate phosphorylated from non-phosphorylated peptides. The acquired fragmentation spectra were interpreted with the Mascot search algorithm. Quantification analysis was performed with the in-house developed software iTRAQassist (26).
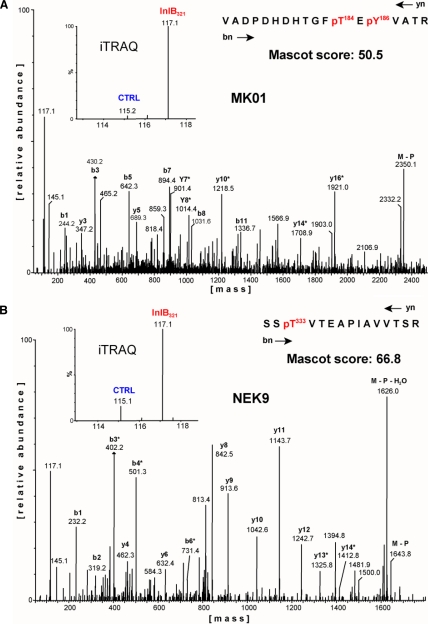
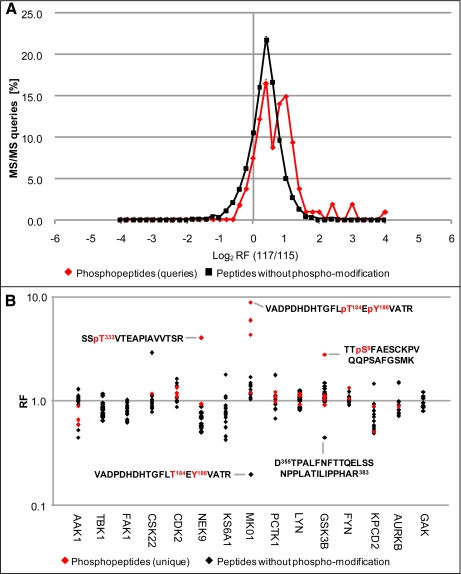
Particular attention has to be paid to the systematic and detailed inspection of all regulatory results, starting with the original information from the MS/MS raw data format as exemplified by fragmentation data belonging to the kinases MK01 and Nek9 (Fig. 4). Low energy collisions with argon atoms result in b- and y-ion-dominated fragmentation spectra that facilitate both the peptide sequencing and unambiguous determination of phosphorylation site positions. As recommended by Ross et al. (22), we moderately increased the fragmentation energy profiles such that the resulting iTRAQ reporter ions often exhibited intensities similar to those of the y- or b-ion series. Several protein kinases exhibited spectra indicating the induction of phosphorylations following the InlB321-mediated activation of the Met pathway. Fig. 4 displays two such cases where peptides derived from InlB321-treated cells were labeled with iTRAQ-117, whereas the control was labeled with iTRAQ-115. The ratios of 115 and 117 reporter ion signals strongly suggest an InlB321-dependent up-regulation of the depicted phosphorylated peptides. To obtain a general overview of all regulatory events we comparatively analyzed all non-modified peptides and phosphorylated peptides belonging to the same representative experiment (Fig. 5A). The majority of all MS/MS scans in this experiment suggested regulation factors (RFs) slightly above 1, underscoring the necessity of data normalization. Importantly, the non-modified peptides showed a normal distribution, whereas the phosphorylated peptide fraction revealed a tendency toward larger regulatory events and in addition exhibited a non-normal distribution of regulatory data with some distinct accumulations in the up-regulated region of the total data set. These results indicate that the principal mechanisms of signal transduction, i.e. the dynamic modification of pre-existing signaling components, are also of major relevance in the first steps of the InlB-induced Met pathway. However, the raw data presented in Fig. 5A are still compromised by iTRAQ by-product intensities and did not allow a cumulative view on regulatory peptide data belonging to individual protein kinases. Furthermore, raw data obtained by iTRAQ can vary significantly in quality, depending on the noise characteristics of the MS detector used. Therefore, it is important to account for these factors in a bioinformatics approach for data correction and the quality control of regulatory events. We have recently established a four-step strategy that takes into account all these aspects and finally provides rationalized presentations for evaluation of the reliability of quantitative results (26). First, a separate measurement of by-product impurities of the iTRAQ reagents is used to correct the actual ion intensity of each reporter. Second, iTRAQ reporter ion intensities of all MS/MS scans from compared samples are normalized by calculating their trimmed mean. Third, the algorithm computes a weighted cumulated regulation factor for each unique peptide even when it was identified several times.
Fig. 4.
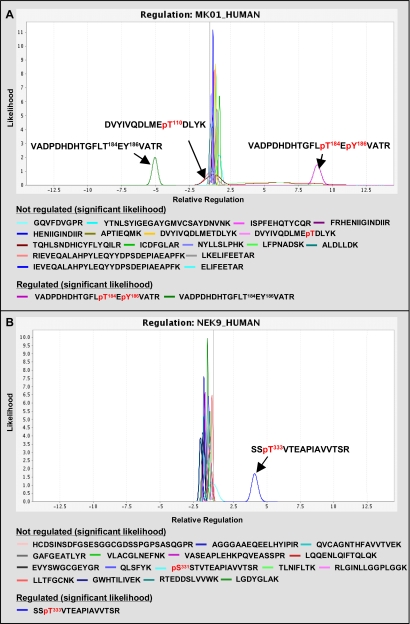
Qualitative and quantitative phosphosite analysis at protein kinases. Two MS/MS spectra represent the identification of already known (A) and novel (B) InlB321-induced phosphorylation sites. In A, the fragmentation spectrum is derived from a doubly phosphorylated peptide from the protein kinase MK01 (Erk2), indicating MK01 kinase activity. The magnification inset shows the low molecular mass range. Under MS/MS conditions, peptides from the InlB321-treated sample release the iTRAQ reporter ion with a mass of 117 Da, whereas the iTRAQ reporter ion with a mass of 115 Da is generated from peptides from the control (CTRL) approach. MK01 phosphorylation is strongly induced in the stimulated cell state. In B, a spectrum is shown from a tryptic peptide from Nek9 carrying a phosphate group at Thr333. This modification is significantly increased after treatment of cells with InlB321.
Fig. 5.
Cluster analysis of single peptide regulation factors revealed InlB321-induced alterations in protein kinases. A, distribution of peptide regulation factors calculated from raw signal intensities (117 Da, InlB321; 115 Da, control) from all MS/MS scans from one Met activation experiment is plotted. The majority of the analyzed 1833 MS/MS scans are not regulated. Phosphopeptide regulations show a non-normal and shifted distribution, indicating their pronounced up-regulation in comparison with non-modified peptides. B, exemplified view of relative peptide regulations of the 15 most robustly identified protein kinases from one typical experiment. All MS/MS scans from A were normalized and iTRAQ by-product-corrected, and RFs from scans corresponding to a unique peptide were accumulated by iTRAQassist. Resulting RFs indicate Met pathway-dependent regulations of specific peptides in this small set of protein kinases after 4 min of InlB321 treatment. The two marked peptides from MK01 (Erk2) correspond to the same amino acid sequence in its unmodified and phosphorylated form with oppositely directed RFs, whereas the two highlighted peptides from GSK3B have different sequences. The marked phosphopeptide from Nek9 comprising Thr333 and the doubly phosphorylated peptide from MK01 correspond to the raw data shown in Fig. 4.
At this analytical stage the process of iTRAQassist evaluation is still incomplete but allows a first representative view on the investigated protein kinases. Fig. 5B exemplifies the resulting peptide regulation data for the 15 protein kinases that were detected with the highest Mascot identification scores in a typical experiment. This depiction of data allows a cluster analysis of RF values of peptides belonging to the same protein. Most of these RF values from different kinases, such as FAK1, LYN, and GAK, produced noticeable RF clusters with only moderate deviations from an RF value of 1. In addition to these, several peptides exhibited RF values significantly different from the peptides of the main cluster of each protein kinase. Remarkably, and in addition to differentially regulated phosphorylated peptides, several of these peptides were identified as unmodified peptides as detected in CSK22 (CK2α), KPCD2 (PKD2), and AURKB (Aurora B). Because these peptide sequences were identified correctly and represent unique database hits, the data suggest an InlB321/Met pathway-dependent posttranslational modification in the corresponding protein regions. As the present study focused on phosphorylation events, we searched for reciprocally regulated phosphopeptides and their unmodified counterparts to support this general assumption. Indeed, we found some pairs representing both the phosphorylated and unmodified peptide species that exhibited reciprocal RF values as expected (supplemental Data S3A). Such cases were also detected for MK01 and GSK3B, confirming that regulated unmodified peptides can coincide with the stimulus-induced regulation of the corresponding phosphorylated amino acid sequence. The MK01 peptide represents a clear InlB321-stimulated induction of two phosphorylated residues at Thr184 and Tyr186 in a typical activation site motif, TEY, for mitogen-activate protein kinases. The detected up-regulated phosphosite in the kinase GSK3B presented in Fig. 5B corresponds to Ser9 that mediates kinase inhibition. In conclusion, iTRAQ-detected modifications caused by InlB321 in the PI3K/Akt pathway and the Raf-Erk cascade compare well with literature data and strongly support the reliability of this proteomics strategy for studying InlB321/Met-dependent phosphorylation events in protein kinases.
However, care must be taken in evaluating RF values. The negative RF value of one unmodified peptide also detected at GSK3B (Fig. 5B) suggests an additional regulation at this protein region, but inspection of the raw data revealed weak and noisy iTRAQ reporter ion signals. Thus, a simple RF value does not allow the evaluation of the robustness of the underlying MS data. Therefore, in a final fourth step, iTRAQassist performs a rational statistical evaluation of the quality of RF values that has been applied to all data sets of this study. iTRAQassist uses a signal-to-noise model algorithm that was established and evaluated previously for the LC-MS work flow used. This model allows prediction of possible variations for any iTRAQ reporter ion intensity and in turn can calculate the robustness of observed RFs at the individual peptide level. The result can be graphically presented as so-called likelihood curves. The top of the curves presents the most likely regulatory factor of individual peptides on the x axis, whereas the intensity on the y axis and the shape of these likelihood curves summarize statistically the underlying quality of the data. Robust regulations will always coincide with good signal-to-noise properties of iTRAQ ions with only a minor probability of variant regulations. Thus, high quality data are presented by small and intense likelihood curves. Contrarily, a flattened curve spanning a larger region of possible RF values refers to uncertainties concerning the actual regulation. In this way, the comparative inspection of likelihood curves belonging to the same protein offers an intuitive way to detect PTM-regulated protein regions as exemplified for MK01 and Nek9 (Fig. 6). This level of iTRAQ data evaluation facilitates the identification of clusters referring to the general expression state of whole proteins. Complementarily, likelihood curves from regulated peptides/phosphorylations will not overlap significantly with these “main clusters” usually observed near RF = 1 in signal transduction experiments.
Fig. 6.
Statistical evaluation of iTRAQ-based peptide regulations. Analyses were done by iTRAQassist as described previously (26). The most likely and possible regulations were calculated based on a work flow-specific noise model and were depicted as likelihood curves for every peptide. Significantly regulated phosphopeptides can be detected after 4-min InlB321 treatment of HeLa cells at MK01 (A) and Nek9 (B).
In the case of MK01 (Fig. 6A), the likelihood view confirms the statistical significance of the regulatory data from the phosphorylated peptides and the corresponding counter-regulated non-modified peptide. The likelihood curve of the detected phosphopeptide DVYIVQDLMEpT110DLYK (where pT is phosphothreonine) is depicted as part of the main cluster and, like unmodified peptides, exhibits no specific regulation. In contrast, the doubly phosphorylated peptide VADPDHDHTGFLpT184EpY186VATR (where pY is phosphotyrosine) is up-regulated (RF = 8.9) with a high robustness. Furthermore, alternatively singly and doubly phosphorylated peptides of the same sequence were also detected, but their regulations were of low significance as shown by the flatness of the curves (RF ranging from about 2.5 to 10). The reciprocally regulated, unmodified peptide VADPDHDHTGFLTEYVATR (RF = −5.1) also exhibits high robustness, corresponds to all phosphorylated forms with this amino acid sequence, and indicates that a relatively high fraction of MK01 was activated in response to InlB321. Applying iTRAQassist and likelihood curve presentations consequently in this study revealed pairs of such reciprocally regulated phosphorylated and non-modified peptides that can be termed significant only at GSK3A, MK01, GSK3B, and CDKL5 (supplemental Data S3B). However, not all regulated phosphorylated peptides might have been identified successfully by the selected IMAC/LC-MS approach, and other types of modifications were not considered in the data search strategy used in this study. Therefore, we also systematically looked for “unpaired” regulated non-modified peptides that may indicate dynamically regulated modifications occurring in these regions of the investigated protein kinases (Fig. 5B and supplemental Data S3C). Actually, a few non-modified regulated peptides with RF values between −2.4 and +8 could be observed in protein kinases, although their likelihood curves overlap partially with curves in the main cluster in most cases (e.g. KPCD2, GSK3B, and PCTK1). Noteworthy are the single peptides from protein kinases CSK22 and M3K3 that are clearly separated from the main cluster, highlighting a significant InlB321-induced modification in the corresponding protein region, although the specific type of modification could not be determined in this study. In conclusion, the sequential combination of SMAC, IMAC, iTRAQ, LC-MS/MS, and appropriate statistical data analysis strategies yielded proof of concept for the feasibility of quantitative phosphokinome analysis in an implementation that does not require previous metabolic labeling as in SILAC-based strategies (17).
InlB321/Met-dependent Signaling Components Identified by Quantitative Proteomics
During the InlB321/Met activation experiments of this study, we investigated a total of 94 protein kinases with a focus on phosphorylation site determination and quantification. Approximately half of all the identified protein kinases were identified with at least one posttranslational modification (supplemental Data S2). Hence, this study provides regulatory information for 143 unique phosphorylation sites derived from these protein kinases (supplemental Data S4). As expected, the frequency of phosphorylation sites on protein kinases is significantly higher compared with the general estimations that 30% of all proteins contain phosphorylated residues, although this work flow certainly has not detected every phosphorylation site present. The identified sites were detected at serine, threonine, and tyrosine in a ratio of 64:28:8. About one-third of the identified kinase phosphosites are currently not found in phosphosite databases such as Phosida (39), Phosphosite from Cell Signaling Technology Inc., or Phospho.ELM (40) and are described for the first time in the present study. Both phosphopeptide regulation and protein expression were analyzed together in a comprehensive statistical approach. iTRAQassist generated 1718 different likelihood plots derived from all identified proteins that were manually inspected to detect differentially regulated proteins and phosphorylation events in the InlB321/Met pathway.
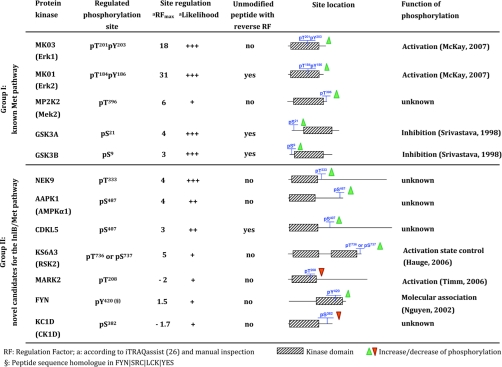
Table II summarizes two groups of protein kinases exhibiting regulation at individual phosphorylation sites following the InlB321-dependent Met activation. Differentially regulated phosphorylation sites of six kinases were already functionally characterized in previous studies (41–45). Group I comprises functionally well characterized kinases that were already described for the Met pathway and can also serve as proof of concept for this proteome study. In contrast, group II presents a candidate list of regulated kinases as novel Met signaling components that should be evaluated functionally during the process of InlB-mediated bacterial invasion. Because only one time point was investigated by this study, we also accepted kinases as candidates that might have been detected in the earliest or latest states of dynamic phosphosite regulations. For example, KC1D and Fyn showed moderate but statistically significant (iTRAQassist) regulations of phosphopeptides and might become fully regulated at other time points (see likelihood plots in supplemental Data S5). In addition to the InlB321-induced activation site investigated in our Western blot analysis (Fig. 1B), we detected a regulated phosphopeptide mapping to the outer C-terminal region of MP2K2 (Mek2), which belongs to the MAPK pathway. KS6A3, a so-called MAPK-activated protein kinase downstream of the MAPK cascade, was also found to harbor a differentially phosphorylated peptide. Manual inspection of the MS/MS data revealed that the modification might be located either at Thr736 or at Ser737. Thus, the systematic characterization of protein kinases by the presented proteomics approach shows that InlB321 is also a potent activator of the Met-dependent MAPK and the PI3K pathways similar to HGF. Furthermore, our data suggest a negative phosphorylation site regulation in response to InlB321 stimulation, namely Thr208 (corresponding to MARK2) in the activation loop of microtubule-associated protein/microtubule affinity-regulating kinase (MARK) kinase family members. We detected unique peptides from MARK1, MARK2, and MARK3 but not from MARK4, which is therefore most likely not expressed in HeLa S3 cells. Further regulated phosphorylation sites were detected on CDKL5 and AMPK. Interestingly, AMPK is involved in actin cytoskeleton dynamics, a process that is certainly contributing to the uptake of L. monocytogenes into the cell. However, the most robust and pronounced response was observed at Nek9 among all novel candidates. Its InlB321-induced regulation at position 333 (Thr333) could be identified unambiguously and provides a biochemical basis for functional investigations of this protein kinase. The modified residue is located close to a nuclear localization signal of Nek9, favoring a direct influence on the cellular localization of this kinase in response to Met activation.
Table II. InlB321-mediated regulation of phosphorylation sites on human protein kinases identified in the present study.
DISCUSSION
Protein kinases are key components involved in the control of virtually every signaling cascade. Kinase-mediated phosphorylation tightly regulates the activity, localization, and stability as well as the molecular interactions of substrate proteins. Thus, the systematic characterization of dynamic phosphorylation events is currently one primary goal in infection research as pathogens exploit and manipulate host signaling by effectors to realize individual steps of their infection cycle. InlB from L. monocytogenes mediates invasion by binding and activation of the host receptor kinase Met, but both the “mode of action” and the details of possible effects of the released form of InlB on neighboring cells and tissues remain unknown. Interestingly, neither the well characterized Met pathway nor the structural investigation of the InlB321-Met complex provides intuitive concepts to explain the induced uptake of Listeria into the cells. The InlB-Met interaction might therefore constitute novel signaling mechanisms. Study of the InlB-induced proximal signaling will most likely complement our understanding, and this is probably most easily accessed using chemical proteomics that already has strategies for the systematic analyses of human protein kinases. Efficient enrichment of kinases can be achieved by immobilized kinase inhibitors, allowing their affinity purification from highly complex protein samples and greatly facilitating the comprehensive analysis of posttranslationally modified forms by LC-MS/MS approaches. VI16743 used in this study is to our knowledge one of the most nonspecific ATP-competitive protein kinase inhibitors and by far exceeds the capture efficiency of the previously described PP58 based on the same chemical scaffold. In earlier studies, PP58-based affinity purifications permitted the identification of 84 distinct protein kinases from three different cell lines of which each was analyzed using 5 × 109 cells as starting material (19, 35). In contrast, VI16743 chromatography in combination with similar LC-MS instrumentation identified a total of 94 protein kinases derived from virtually every branch of the human kinome, although only one cell line and 10 times less total cell extract served as starting point in this study.
Whereas the identification of phosphorylation sites on kinases can already provide important clues for the understanding of disease-related processes, the detection of regulated phosphorylation events is particularly beneficial to reveal components participating in signal transduction or certain cellular processes (17, 20). In the present work, we combined an LC-MS/MS-based strategy for systematic phosphorylation site determination in the human kinome with a survey of quantitative data using iTRAQ peptide labeling. Several statistical methods for iTRAQ data evaluation have been presented previously (46–48), but these were mainly focused on the quantification of protein expression and not on PTM regulation. Therefore, and driven by the challenge to establish a proteome-based work flow for characterization of pathogen-induced host signaling pathways, we have recently described a rational statistical approach to unambiguously detect significantly regulated modifications occurring at individual peptides from one protein (26). In total, we investigated 143 unique phosphorylation sites at serine, threonine, and tyrosine with a ratio of 64:28:8. Thus, threonine and in particular tyrosine phosphorylation are highly over-represented regulatory modifications for kinases compared with their contribution to total cellular phosphorylation where a ratio of 90:10:0.5 was found (49).
Quantitative peptide sequencing in combination with iTRAQassist revealed 12 InlB321-induced regulated phosphorylation sites in different protein kinases (Table II). Furthermore, we observed a few unmodified peptides that show InlB321-dependent regulation factors, whereas the majority of unmodified peptides from the same protein kinase were not regulated (see MK01 in Fig. 6A). Daub et al. (17) recently showed that such non-modified peptide regulations can represent “counter-regulations” that coincide with the parallel increase or decrease of phosphorylations occurring in the same protein region. However, the present study could only reveal four pairs of phosphorylated and non-modified peptides of the same sequence that are significantly counter-regulated (supplemental Data S3, A and B). Hence, similar approaches will most likely facilitate the identification of protein regions regulated by PTMs other than phosphorylation. However, their success will depend on protein coverages achieved by bottom-up proteomics and the ratio of proteins actually participating in the process of modification/signal transduction. Actually, several non-modified peptide regulations seem to behave differently from the rest of the protein, but unfortunately these regulations often could not be termed significant in comparison with other peptides of the main cluster nor could this study identify the corresponding modified peptides (supplemental Data S3C).
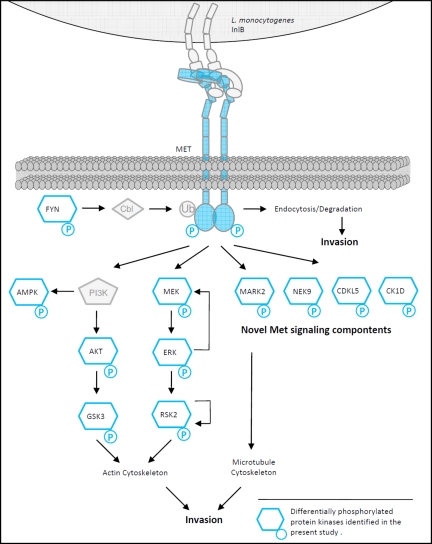
Focusing on phosphosite regulation, our approach identified known Met signaling events, thus substantiating the relevance of quantitative proteome data for signal transduction studies. More importantly, this study also revealed a new subset of Met-dependent protein kinases potentially exploited by InlB from the pathogen L. monocytogenes. Interestingly, several of these kinase functions can either be related to known HGF/Met-controlled cellular phenotypes or even be linked to the process of bacterial invasion (summarized in Fig. 7). One such promising candidate is the family of MARK kinases that controls the stability of microtubule (MT) filaments. Microtubule-destabilizing agents such as nocodazole strongly reduce invasion efficiency of L. monocytogenes into different cell lines (50–52). The maintaining of MT filaments therefore might be a feasible strategy of the pathogen to support the first steps of the infection cycle. A possible connection to cytoskeleton dynamics was also recently identified for the protein kinase AMPK that we found to be regulated. Besides its known function as an energy sensor (53), AMPK appears to be involved in actin cytoskeleton dynamics, cell polarity, and cell cycle-related processes (54, 55). We identified an up-regulation of Ser487 in the outer C-terminal region of the catalytic subunit of the heterotrimeric AMPK complex. As yet, the role of AMPK in the InlB-activated Met pathway has not been characterized. Its possible influence on the actin cytoskeleton might also contribute to listerial invasion efficiency. This is supported by the fact that Akt1, as a component of the PI3K/Akt1 pathway, contributes to the InlB/Met-dependent process of invasion (56, 57) and was identified as being responsible for the phosphorylation of the catalytic AMPK subunit at Ser487 (58, 59). Other kinases such as CDKL5 have poorly characterized functions, and no information is available about the identified regulated phosphorylation sites. iTRAQassist also indicated robust regulations at well characterized kinases such as Fyn and casein kinase 1. It was demonstrated that HGF induces the increased association of Fyn with Cbl, which mediates ubiquitination and consequently uptake and degradation of the Met receptor (60), also an indispensable process for Listeria invasion (14). Casein kinase 1 can also be linked to natural Met-dependent responses because recent studies substantiate its regulatory role in cell cycle progression and mitosis (61). However, time-resolved phosphorylation studies at both kinases must first verify the hypothesis that the observed weak regulations actually indicate a more intensive regulation at other time points.
Fig. 7.
Met signal transduction exploited by InlB from L. monocytogenes. InlB321-dependent differentially phosphorylated protein kinases identified in the present study are highlighted in blue. L. monocytogenes activates the PI3K and MAPK pathways, both essential for actin cytoskeleton remodeling as a prerequisite for invasion. The identified induction of phosphorylation events on RSK2 and MEK support a negative feedback theory in this signaling module. Dephosphorylation of MARK2 in the kinase domain as one novel finding is suggested to block MARK2 activation, which is probably essential for Listeria invasion. Active MARK2 leads to destabilization of MTs by phosphorylation of microtubule-associated proteins. Earlier studies already demonstrated that destabilization of MTs by nocodazole impaired efficient Listeria uptake by the host cell. The functional contribution of novel candidates such as Nek9 or CK1D to the InlB/Met invasion strategy will be the subject of further studies. Ub, ubiquitin.
Besides the already known Met signaling components, the most pronounced and robustly regulated phosphorylation site was detected at the cell cycle-related kinase Nek9. Phosphorylation at Thr333 was strongly up-regulated after 4-min treatment with InlB321. This modification site is located adjacent to a functional nuclear localization signal downstream of the kinase domain (62). Nek9 is described as a contributor to cell cycle progression (63) as well as DNA transcription (64), and a nuclear-cytoplasmic distribution was observed (63). The modification of Thr333 in the Met pathway might disrupt a supposed intramolecular loop (62), thus exposing the nuclear localization signal and resulting in nuclear shuttling of Nek9. This modification might prime Nek9 for dimerization, which is essential for autophosphorylation at Thr210 and correlates with kinase activation. Whereas an evaluation of all presented candidates by immunological methods should confirm the presented proteomics data, the functional investigation of Nek9 is already now obligatory based on the existing data quality. Prospective functional approaches (e.g. by RNA interference) will help to understand the individual contributions of protein kinases with respect to listerial invasion and physiological cell responses to Met activation. In addition, site-directed mutations of regulated phosphorylation sites should allow an evaluation of the influence of the identified phosphorylation sites in the InlB-Met system, thus gaining new insights into the function of these partially uncharacterized protein kinases.
Infection research has focused in the past on the detection and functional characterization of virulence factors from different human pathogens. Among these, numerous effectors have been described that manipulate the host and realize individual steps in the infection cycle (65). Interestingly, successful screenings for the host-interacting proteins and even structural studies do not necessarily explain effector-induced processes mechanistically. Internalin B from L. monocytogenes obviously exploits the Met signaling pathway to invade different cell types, but the signaling network involved that coordinates such a process has not been characterized in detail to date. This study presents a quantitative proteomics strategy that focuses on protein kinases that form an essential part of the host signaling network. The results of this study demonstrate modifications at known and potentially novel InlB/Met pathway components, providing a robust resource for Listeria research. Furthermore, the InlB-affected proximal host signaling raises questions whether these kinases might be involved in the natural physiological function of the Met pathway. It will be obligatory now to attempt the direct comparison of InlB versus HGF signaling, and time-resolved experiments will further facilitate the compilation of hierarchical protein kinase cascades. This information may define possible intervention points for preventing listerial invasions in immunocompromised patients and certainly will extend our knowledge of this fundamentally important host signaling pathway that is frequently found deregulated in several types of cancer.
Supplementary Material
Acknowledgments
We thank Kirsten Minkhart and Reiner Munder for technical assistance, Dr. Uwe Kärst for fruitful discussions, and Dr. Victor Wray for proofreading the manuscript. We thank Axel Ullrich for support of this study with funding from the Department of Molecular Biology, Max Planck Institute of Biochemistry.
Footnotes
* This work was supported by European Research Area ERAnet Project 0313939B (SPATELIS, Spatio-temporal analysis of Listeria-host protein interactions) and by funding from the Department of Molecular Biology, Max Planck Institute of Biochemistry.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Data S1–S5.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Data S1–S5.
1 The abbreviations used are:
- InlB
- internalin B
- HGF
- hepatocyte growth factor
- PI3K
- phosphatidylinositol-4,5-bisphosphate 3-kinase
- Erk
- extracellular signal-regulated kinase (Erk1 = MK03, Erk2 = MK01)
- GSK3
- glycogen synthase kinase 3
- Mek
- dual specificity mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (Mek1 = MP2K1, Mek2 = MP2K2)
- PTM
- posttranslational modification
- iTRAQ
- isobaric tag for relative and absolute quantification
- SILAC
- stable isotope labeling by amino acids in cell culture
- RF
- regulation factor
- SMAC
- small molecule affinity chromatography
- GK
- gatekeeper
- RP
- reversed phase
- MAPK
- mitogen-activated protein kinase
- DMF
- N,N-dimethylformamide
- EDC-HCl
- N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride
- DMEM
- Dulbecco's modified Eagle's medium
- HRP
- horseradish peroxidase
- SPE
- solid phase extraction
- SCX
- strong cation exchange
- UPLC
- ultraperformance LC
- FP
- false positive
- TP
- true positive
- p
- phospho-
- MARK
- microtubule-associated protein/microtubule affinity-regulating kinase
- AMPK
- AMP-activated protein kinase
- MT
- microtubule.
REFERENCES
- 1.Lecuit M., Vandormael-Pournin S., Lefort J., Huerre M., Gounon P., Dupuy C., Babinet C., Cossart P. ( 2001) A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292, 1722– 1725 [DOI] [PubMed] [Google Scholar]
- 2.Mengaud J., Ohayon H., Gounon P., Mege R.-M., Cossart P. ( 1996) E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84, 923– 932 [DOI] [PubMed] [Google Scholar]
- 3.Dramsi S., Biswas I., Maguin E., Braun L., Mastroeni P., Cossart P. ( 1995) Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol 16, 251– 261 [DOI] [PubMed] [Google Scholar]
- 4.Parida S. K., Domann E., Rohde M., Müller S., Darji A., Hain T., Wehland J., Chakraborty T. ( 1998) Internalin B is essential for adhesion and mediates the invasion of Listeria monocytogenes into human endothelial cells. Mol. Microbiol 28, 81– 93 [DOI] [PubMed] [Google Scholar]
- 5.Trost M., Wehmhöner D., Kärst U., Dieterich G., Wehland J., Jänsch L. ( 2005) Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics 5, 1544– 1557 [DOI] [PubMed] [Google Scholar]
- 6.Jonquières R., Bierne H., Fiedler F., Gounon P., Cossart P. ( 1999) Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol 34, 902– 914 [DOI] [PubMed] [Google Scholar]
- 7.Jonquières R., Pizarro-Cerdá J., Cossart P. ( 2001) Synergy between the N- and C-terminal domains of InlB for efficient invasion of non-phagocytic cells by Listeria monocytogenes. Mol. Microbiol 42, 955– 965 [DOI] [PubMed] [Google Scholar]
- 8.Marino M., Banerjee M., Jonquières R., Cossart P., Ghosh P. ( 2002) GW domains of the Listeria monocytogenes invasion protein InlB are SH3-like and mediate binding to host ligands. EMBO J 21, 5623– 5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Y., Naujokas M., Park M., Ireton K. ( 2000) InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103, 501– 510 [DOI] [PubMed] [Google Scholar]
- 10.Copp J., Marino M., Banerjee M., Ghosh P., van der Geer P. ( 2003) Multiple regions of internalin B contribute to its ability to turn on the Ras-mitogen-activated protein kinase pathway. J. Biol. Chem 278, 7783– 7789 [DOI] [PubMed] [Google Scholar]
- 11.Machner M. P., Frese S., Schubert W. D., Orian-Rousseau V., Gherardi E., Wehland J., Niemann H. H., Heinz D. W. ( 2003) Aromatic amino acids at the surface of InlB are essential for host cell invasion by Listeria monocytogenes. Mol. Microbiol 48, 1525– 1536 [DOI] [PubMed] [Google Scholar]
- 12.Braun L., Nato F., Payrastre B., Mazié J. C., Cossart P. ( 1999) The 213-amino-acid leucine-rich repeat region of the listeria monocytogenes InlB protein is sufficient for entry into mammalian cells, stimulation of PI 3-kinase and membrane ruffling. Mol. Microbiol 34, 10– 23 [DOI] [PubMed] [Google Scholar]
- 13.Niemann H. H., Jäger V., Butler P. J., van den Heuvel J., Schmidt S., Ferraris D., Gherardi E., Heinz D. W. ( 2007) Structure of the human receptor tyrosine kinase met in complex with the Listeria invasion protein InlB. Cell 130, 235– 246 [DOI] [PubMed] [Google Scholar]
- 14.Hamon M., Bierne H., Cossart P. ( 2006) Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol 4, 423– 434 [DOI] [PubMed] [Google Scholar]
- 15.Manning G. ( 2005) Genomic overview of protein kinases. WormBook 1– 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. ( 2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190– 1203 [DOI] [PubMed] [Google Scholar]
- 17.Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. ( 2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438– 448 [DOI] [PubMed] [Google Scholar]
- 18.Corthals G. L., Aebersold R., Goodlett D. R. ( 2005) Identification of phosphorylation sites using microimmobilized metal affinity chromatography. Methods Enzymol 405, 66– 81 [DOI] [PubMed] [Google Scholar]
- 19.Wissing J., Jänsch L., Nimtz M., Dieterich G., Hornberger R., Kéri G., Wehland J., Daub H. ( 2007) Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol. Cell. Proteomics 6, 537– 547 [DOI] [PubMed] [Google Scholar]
- 20.Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., Drewes G. ( 2007) Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol 25, 1035– 1044 [DOI] [PubMed] [Google Scholar]
- 21.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376– 386 [DOI] [PubMed] [Google Scholar]
- 22.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. ( 2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154– 1169 [DOI] [PubMed] [Google Scholar]
- 23.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. ( 2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635– 648 [DOI] [PubMed] [Google Scholar]
- 24.Chen Y., Choong L. Y., Lin Q., Philp R., Wong C. H., Ang B. K., Tan Y. L., Loh M. C., Hew C. L., Shah N., Druker B. J., Chong P. K., Lim Y. P. ( 2007) Differential expression of novel tyrosine kinase substrates during breast cancer development. Mol. Cell. Proteomics 6, 2072– 2087 [DOI] [PubMed] [Google Scholar]
- 25.Bantscheff M., Boesche M., Eberhard D., Matthieson T., Sweetman G., Kuster B. ( 2008) Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer. Mol. Cell. Proteomics 7, 1702– 1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hundertmark C., Fischer R., Reinl T., May S., Klawonn F., Jänsch L. ( 2009) MS-specific noise model reveals the potential of iTRAQTM in quantitative proteomics. Bioinformatics 25, 1004– 1011 [DOI] [PubMed] [Google Scholar]
- 27.Schubert W. D., Göbel G., Diepholz M., Darji A., Kloer D., Hain T., Chakraborty T., Wehland J., Domann E., Heinz D. W. ( 2001) Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain. J. Mol. Biol 312, 783– 794 [DOI] [PubMed] [Google Scholar]
- 28.Schmidt T. G., Koepke J., Frank R., Skerra A. ( 1996) Molecular interaction between the Strep-tag affinity peptide and its cognate target, streptavidin. J. Mol. Biol 255, 753– 766 [DOI] [PubMed] [Google Scholar]
- 29.Barvian M., Boschelli D. H., Cossrow J., Dobrusin E., Fattaey A., Fritsch A., Fry D., Harvey P., Keller P., Garrett M., La F., Leopold W., McNamara D., Quin M., Trumpp-Kallmeyer S., Toogood P., Wu Z., Zhang E. ( 2000) Pyrido[2,3-d]pyrimidin-7-one inhibitors of cyclin-dependent kinases. J. Med. Chem 43, 4606– 4616 [DOI] [PubMed] [Google Scholar]
- 30.Moloney G. P., Robertson A. D., Martin G. R., MacLennan S., Mathews N., Dodsworth S., Sang P. Y., Knight C., Glen R. ( 1997) A novel series of 2,5-substituted tryptamine derivatives as vascular 5HT1B/1D receptor antagonists. J. Med. Chem 40, 2347– 2362 [DOI] [PubMed] [Google Scholar]
- 31.Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., Righetti P. G. ( 2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327– 1333 [DOI] [PubMed] [Google Scholar]
- 32.Wessel D., Flügge U. I. ( 1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem 138, 141– 143 [DOI] [PubMed] [Google Scholar]
- 33.Godl K., Wissing J., Kurtenbach A., Habenberger P., Blencke S., Gutbrod H., Salassidis K., Stein-Gerlach M., Missio A., Cotten M., Daub H. ( 2003) An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc. Natl. Acad. Sci. U.S.A 100, 15434– 15439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brehmer D., Greff Z., Godl K., Blencke S., Kurtenbach A., Weber M., Müller S., Klebl B., Cotten M., Kéri G., Wissing J., Daub H. ( 2005) Cellular targets of gefitinib. Cancer Res 65, 379– 382 [PubMed] [Google Scholar]
- 35.Wissing J., Godl K., Brehmer D., Blencke S., Weber M., Habenberger P., Stein-Gerlach M., Missio A., Cotten M., Müller S., Daub H. ( 2004) Chemical proteomic analysis reveals alternative modes of action for pyrido[2,3-d]pyrimidine kinase inhibitors. Mol. Cell. Proteomics 3, 1181– 1193 [DOI] [PubMed] [Google Scholar]
- 36.Blencke S., Zech B., Engkvist O., Greff Z., Orfi L., Horváth Z., Kéri G., Ullrich A., Daub H. ( 2004) Characterization of a conserved structural determinant controlling protein kinase sensitivity to selective inhibitors. Chem. Biol 11, 691– 701 [DOI] [PubMed] [Google Scholar]
- 37.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. ( 2002) The protein kinase complement of the human genome. Science 298, 1912– 1934 [DOI] [PubMed] [Google Scholar]
- 38.Su A. I., Cooke M. P., Ching K. A., Hakak Y., Walker J. R., Wiltshire T., Orth A. P., Vega R. G., Sapinoso L. M., Moqrich A., Patapoutian A., Hampton G. M., Schultz P. G., Hogenesch J. B. ( 2002) Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U.S.A 99, 4465– 4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gnad F., Ren S., Cox J., Olsen J. V., Macek B., Oroshi M., Mann M. ( 2007) PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol 8, R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diella F., Cameron S., Gemünd C., Linding R., Via A., Kuster B., Sicheritz-Pontén T., Blom N., Gibson T. J. ( 2004) Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics 5, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKay M. M., Morrison D. K. ( 2007) Integrating signals from RTKs to ERK/MAPK. Oncogene 26, 3113– 3121 [DOI] [PubMed] [Google Scholar]
- 42.Srivastava A. K., Pandey S. K. ( 1998) Potential mechanism(s) involved in the regulation of glycogen synthesis by insulin. Mol. Cell. Biochem 182, 135– 141 [PubMed] [Google Scholar]
- 43.Hauge C., Frödin M. ( 2006) RSK and MSK in MAP kinase signalling. J. Cell Sci 119, 3021– 3023 [DOI] [PubMed] [Google Scholar]
- 44.Timm T., Matenia D., Li X. Y., Griesshaber B., Mandelkow E. M. ( 2006) Signaling from MARK to tau: regulation, cytoskeletal crosstalk, and pathological phosphorylation. Neurodegener. Dis 3, 207– 217 [DOI] [PubMed] [Google Scholar]
- 45.Nguyen T. H., Liu J., Lombroso P. J. ( 2002) Striatal enriched phosphatase 61 dephosphorylates Fyn at phosphotyrosine 420. J. Biol. Chem 277, 24274– 24279 [DOI] [PubMed] [Google Scholar]
- 46.Boehm A. M., Pütz S., Altenhöfer D., Sickmann A., Falk M. ( 2007) Precise protein quantification based on peptide quantification using iTRAQ. BMC Bioinformatics 8, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu J., Qian J., Borisov O., Pan S., Li Y., Liu T., Deng L., Wannemacher K., Kurnellas M., Patterson C., Elkabes S., Li H. ( 2006) Optimized proteomic analysis of a mouse model of cerebellar dysfunction using amine-specific isobaric tags. Proteomics 6, 4321– 4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin W. T., Hung W. N., Yian Y. H., Wu K. P., Han C. L., Chen Y. R., Chen Y. J., Sung T. Y., Hsu W. L. ( 2006) Multi-Q: a fully automated tool for multiplexed protein quantitation. J. Proteome Res 5, 2328– 2338 [DOI] [PubMed] [Google Scholar]
- 49.Mann M., Ong S. E., Grønborg M., Steen H., Jensen O. N., Pandey A. ( 2002) Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol 20, 261– 268 [DOI] [PubMed] [Google Scholar]
- 50.Kuhn M. ( 1998) The microtubule depolymerizing drugs nocodazole and colchicine inhibit the uptake of Listeria monocytogenes by P388D1 macrophages. FEMS Microbiol. Lett 160, 87– 90 [DOI] [PubMed] [Google Scholar]
- 51.Guzman C. A., Rohde M., Chakraborty T., Domann E., Hudel M., Wehland J., Timmis K. N. ( 1995) Interaction of Listeria monocytogenes with mouse dendritic cells. Infect. Immun 63, 3665– 3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greiffenberg L., Goebel W., Kim K. S., Weiglein I., Bubert A., Engelbrecht F., Stins M., Kuhn M. ( 1998) Interaction of Listeria monocytogenes with human brain microvascular endothelial cells: InlB-dependent invasion, long-term intracellular growth, and spread from macrophages to endothelial cells. Infect. Immun 66, 5260– 5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hardie D. G., Scott J. W., Pan D. A., Hudson E. R. ( 2003) Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett 546, 113– 120 [DOI] [PubMed] [Google Scholar]
- 54.Koh H., Chung J. ( 2007) AMPK links energy status to cell structure and mitosis. Biochem. Biophys. Res. Commun 362, 789– 792 [DOI] [PubMed] [Google Scholar]
- 55.Lee J. H., Koh H., Kim M., Kim Y., Lee S. Y., Karess R. E., Lee S. H., Shong M., Kim J. M., Kim J., Chung J. ( 2007) Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature 447, 1017– 1020 [DOI] [PubMed] [Google Scholar]
- 56.Ireton K., Payrastre B., Cossart P. ( 1999) The Listeria monocytogenes protein InlB is an agonist of mammalian phosphoinositide 3-kinase. J. Biol. Chem 274, 17025– 17032 [DOI] [PubMed] [Google Scholar]
- 57.Seveau S., Tham T. N., Payrastre B., Hoppe A. D., Swanson J. A., Cossart P. ( 2007) A FRET analysis to unravel the role of cholesterol in Rac1 and PI 3-kinase activation in the InlB/Met signalling pathway. Cell. Microbiol 9, 790– 803 [DOI] [PubMed] [Google Scholar]
- 58.Horman S., Vertommen D., Heath R., Neumann D., Mouton V., Woods A., Schlattner U., Wallimann T., Carling D., Hue L., Rider M. H. ( 2006) Insulin antagonizes ischemia-induced Thr172 phosphorylation of AMP-activated protein kinase alpha-subunits in heart via hierarchical phosphorylation of Ser485/491. J. Biol. Chem 281, 5335– 5340 [DOI] [PubMed] [Google Scholar]
- 59.Woods A., Vertommen D., Neumann D., Turk R., Bayliss J., Schlattner U., Wallimann T., Carling D., Rider M. H. ( 2003) Identification of phosphorylation sites in AMP-activated protein kinase (AMPK) for upstream AMPK kinases and study of their roles by site-directed mutagenesis. J. Biol. Chem 278, 28434– 28442 [DOI] [PubMed] [Google Scholar]
- 60.Taher T. E., Tjin E. P., Beuling E. A., Borst J., Spaargaren M., Pals S. T. ( 2002) c-Cbl is involved in Met signaling in B cells and mediates hepatocyte growth factor-induced receptor ubiquitination. J. Immunol 169, 3793– 3800 [DOI] [PubMed] [Google Scholar]
- 61.Pal G., Paraz M. T., Kellogg D. R. ( 2008) Regulation of Mih1/Cdc25 by protein phosphatase 2A and casein kinase 1. J. Cell Biol 180, 931– 945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roig J., Mikhailov A., Belham C., Avruch J. ( 2002) Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev 16, 1640– 1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roig J., Groen A., Caldwell J., Avruch J. ( 2005) Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol. Biol. Cell 16, 4827– 4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan B. C., Lee S. C. ( 2004) Nek9, a novel FACT-associated protein, modulates interphase progression. J. Biol. Chem 279, 9321– 9330 [DOI] [PubMed] [Google Scholar]
- 65.Bhavsar A. P., Guttman J. A., Finlay B. B. ( 2007) Manipulation of host-cell pathways by bacterial pathogens. Nature 449, 827– 834 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.