Abstract
Many phytopathogenic bacteria use a type III secretion system to deliver type III effector proteins into the host plant cell. The Pseudomonas syringae type III effector AvrRpt2 is cleaved at a specific site when translocated into the host cell. In this study, we first demonstrate that the factor(s) required for AvrRpt2 cleavage is present in extracts from animal and yeast cells, as well as plant cells. The cleavage factor in animal and plant cell extracts was heat labile but relatively insensitive to protease inhibitors. Second, mutational analysis of AvrRpt2 was applied to identify features important for its cleavage. In addition to two of the amino acid residues in the immediate vicinity of the cleavage site, a large part of the region C-terminal to the cleavage site was required when AvrRpt2 was cleaved in animal cell extract. Most of these features were also important when AvrRpt2 was cleaved in plant cells. Third, we investigated the effect of cleavage in interactions of AvrRpt2 with plant cells. Cleavage of AvrRpt2 appeared to be important for proper interactions with Arabidopsis cells that lack the resistance gene product corresponding to AvrRpt2, RPS2. In addition, removal of the region N-terminal to the cleavage site was important for the correct localization of the C-terminal effector region of the protein in the host cell. We speculate that the virulence function of AvrRpt2 requires removal of the N-terminal region to redirect the effector protein to a specific subcellular location in the host cell after translocation of the protein.
The majority of phytopathogenic gram-negative bacteria require the type III protein secretion system (TTSS) for pathogenicity (Galán and Collmer, 1999). In most cases, the TTSS is also required for the elicitation of strong defense responses, such as the hypersensitive response (HR), in resistant plants. Therefore, genes encoding components or regulators of the TTSS were initially identified as HR and pathogenicity (hrp) mutations (Lindgren et al., 1986). Recently, the Xanthomonas campestris AvrBs2 protein was shown to be directly translocated into the host cell via the TTSS (Casper-Lindley et al., 2002). In addition, based on mounting indirect evidence and analogies with the role of the TTSS in some bacterial pathogens of animals, it is generally believed that phytopathogenic bacteria use the TTSS to translocate certain proteins into the host cell (Staskawicz et al., 2001). The bacterial proteins that are secreted or translocated via the TTSS are called type III effectors. It is believed that many type III effectors function as virulence factors in susceptible hosts (Galán and Collmer, 1999).
When a plant exhibits strong resistance to a pathogen, the resistance response is often conditioned by a single avirulence (avr) gene in the pathogen and the corresponding resistance (R) gene in the plant (Dangl and Jones, 2001). Hence, this type of resistance is called gene-for-gene resistance. When plants carry appropriate R genes, some of the translocated bacterial proteins are recognized as Avr proteins. In fact, many type III effectors of phytopathogens were initially identified as avr gene products. It is believed that many Avr proteins actually function as virulence factors in susceptible hosts. In fact, the virulence function of AvrRpt2 in rps2 Arabidopsis plants has been demonstrated (Chen et al., 2000; Guttman and Greenberg, 2001).
In the gene-for-gene relationship, the Pseudomonas syringae avr gene avrRpt2 corresponds to the Arabidopsis R gene RPS2 (Kunkel et al., 1993; Yu et al., 1993). AvrRpt2 is a type III effector (Mudgett and Staskawicz, 1999). AvrRpt2 must be translocated into the host cell for RPS2-mediated recognition to occur (Leister et al., 1996; Wu et al., 2003). The AvrRpt2 protein appears to be specifically cleaved between Gly71 and Gly72 by a presumed plant cytoplasmic protease (Mudgett and Staskawicz, 1999). These lines of evidence strongly suggest that AvrRpt2 is translocated via the TTSS into the plant cell and subsequently modified (cleaved). The region C-terminal to residue 81 of AvrRpt2 is sufficient to elicit the RPS2-dependent HR (Mudgett et al., 2000).
Some type III effectors are activated by host cells in a manner dependent on factors prevalent among eukaryotes but not specific to particular host species. Several P. syringae Avr proteins are myristoylated in the plant cell, and this modification is important for recognition by their corresponding R genes (Nimchuk et al., 2000; Shan et al., 2000; Tampakaki et al., 2002). The Yersinia pestis ser/thr protein kinase YpkA requires actin for its activation (Juris et al., 2000). Both the myristoylation machinery and actin are prevalent among eukaryotes.
Here, we report that the AvrRpt2 protein is cleaved accurately in animal and yeast cell extracts and that therefore the factor(s) required for AvrRpt2 cleavage is not plant specific but present in diverse eukaryotes. We also demonstrate that a large part of AvrRpt2 that is C-terminal to the cleavage site contributes to cleavage. These observations suggest that AvrRpt2 may make a major contribution to its own cleavage and that the eukaryotic factor(s) may play a relatively minor role. The cleavability of AvrRpt2 is important for a proper interaction with host plant cells. We show that cleavage is crucial in subcellular localization of AvrRpt2 in Arabidopsis cells. Cleavage of a type III effector in the host cell might be a common strategy for the redirection of proteins to a specific subcellular site after translocation.
RESULTS
AvrRpt2 Protein Was Cleaved Properly in Rabbit Reticulocyte Lysate
Initially, we intended to characterize AvrRpt2 cleavage to identify the recognition sequence for the presumed plant-specific protease so that we could use the sequence as a biotechnology tool in plant cells. We chose a rabbit reticulocyte lysate-based in vitro translation system coupled with T7 RNA polymerase-based in vitro transcription for the purpose of rapid production of AvrRpt2 derivative proteins. In this way, PCR products encoding avrRpt2 gene derivatives with the T7 promoter sequence at the 5′ ends can be used directly for protein production.
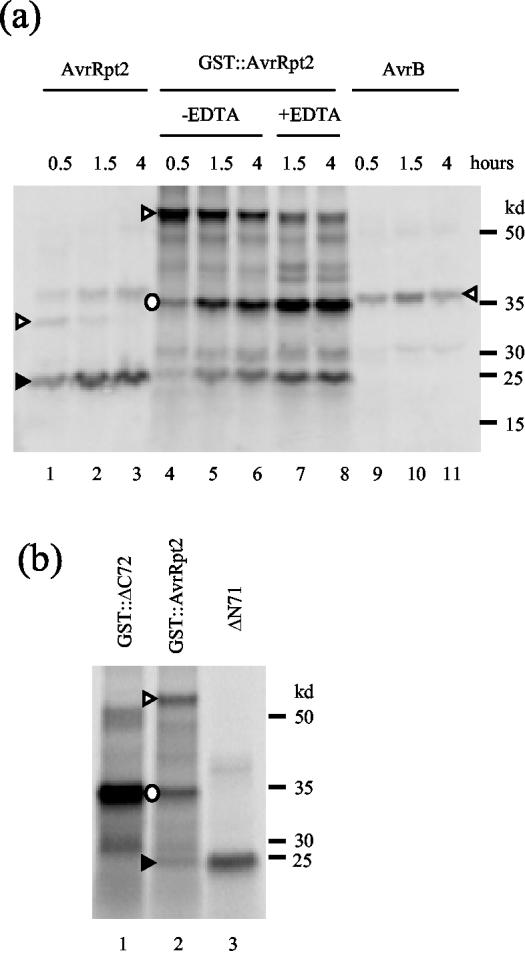
As shown in Figure 1a, when AvrRpt2 was produced and labeled with [35S]Met using the rabbit reticulocyte lysate in vitro translation system, the major product was approximately 25 kD (lanes 1–3, marked by a black triangle). The intensity of the band corresponding to the full-length size (apparently 32 kD, white triangle) decreased with prolonged incubation, whereas the intensity of the 25-kD band increased. In contrast, when the P. syringae effector protein AvrB was expressed, the only product observed corresponded to the expected full-length size of the protein (lanes 9–11, white triangle). When Avr-Rpt2 is cleaved in plant cells or by plant cell extracts, the approximate size of the C-terminal region of the protein is 25 kD (Mudgett and Staskawicz, 1999). Therefore, we speculated that a similar cleavage event occurs in the rabbit reticulocyte lysate. The small size of the N-terminal region of the protein that would result from such a cleavage event would have precluded its detection. Therefore, we fused glutathione S-transferase (GST) to the N terminus of AvrRpt2 (GST::AvrRpt2) to make the N-terminal region longer. When GST::AvrRpt2 was expressed in the in vitro translation system, major bands of 60, 35, and 25 kD, corresponding to the expected sizes of the full-length, N-terminal, and C-terminal regions, respectively, were observed (lanes 4–6, white triangle, white circle, and black triangle, respectively). We do not know the nature of the other minor bands in these lanes. With a longer incubation, the intensity of the 60-kD band decreased, whereas that of the 35- and 25-kD bands increased. The ratio of the intensities between the 35- and 25-kD bands is approximately 2:1, which is expected based on the relative Met contents of the predicted N- and C-terminal regions (10:5). Figure 1b shows that the 35- and 25-kD bands comigrate with the bands corresponding to the expected post-cleavage N- and C-terminal products.
Figure 1.
AvrRpt2 can be cleaved in rabbit reticulocyte lysate. a, Smaller products observed are cleaved products of AvrRpt2. AvrRpt2 (lanes 1–3), GST::AvrRpt2 (lanes 4–8), and AvrB (lanes 9–11) were translated and 35S-labeled using rabbit reticulocyte lysate. The durations of incubations are indicated. In lanes 7 and 8, EDTA was added to a final concentration of 20 mm after a 0.5-h in vitro translation reaction to stop the reaction. The hours indicated are the total of the reaction time and the incubation time following the EDTA addition. The labeled proteins were resolved by denaturing PAGE and were detected by phosphor imaging of the gel. b, AvrRpt2 is cleaved at the correct site in rabbit reticulocyte lysate. The cleaved products of the GST::AvrRpt2 reaction (lane 2) were compared with GST::ΔC72 (lane 1), which corresponds to the N-terminal region expected if the cleavage occurs between G71 and G72, and with ΔN71 (lane 3), which corresponds to the expected C-terminal region. White triangle, Full-length; black triangle, C-terminal region after cleavage; white circle, N-terminal region after cleavage. The N-terminal region was not observed with AvrRpt2 (a, lanes 1–3) due to its small size. The positions of molecular markers are indicated on the right.
To exclude the possibility that the 35- and 25-kD bands were produced by alternative translation initiation or premature translation termination, EDTA was added to the reaction after 30 min to stop protein synthesis, and the accumulation of the 35- and 25-kD bands was observed (Fig. 1a, lanes 7 and 8). A reduction of the 60-kD band intensity and an increase in the 35- and 25-kD band intensities was evident 1 h after addition of EDTA (lane 7), so the 35- and 25-kD bands must result from cleavage rather than translation. We conclude that AvrRpt2 is cleaved in rabbit reticulocyte lysate and that the cleavage site is very close to, if not exactly the same as, the cleavage site observed in Arabidopsis extracts.
Characterization of the Host Cleavage Factor(s)
Next, we tested whether rabbit reticulocyte lysate can cleave Escherichia coli-produced AvrRpt2 protein. C-terminal (his)6-tagged AvrRpt2 (AvrRpt2::his) was produced in E. coli and purified using nickel-chelated resin. The protein was incubated with rabbit reticulocyte lysate or Arabidopsis protein extracts and subjected to immunoblot analysis using an anti-his antibody. As shown in Figure 2a, both reticulocyte lysate (lanes 2 and 3) and Arabidopsis extracts (lanes 4 and 5) cleaved the E. coli-produced AvrRpt2::his protein to generate a C-terminal product of the expected size. We also reproducibly observed a slightly larger band using Arabidopsis extracts, but we do not know the nature of this band. This larger band was not observed in a previous study (Mudgett and Staskawicz, 1999). The AvrRpt2::his protein was also cleaved when incubated with yeast extracts, although the efficiency of cleavage was not very high (lane 8). Therefore, the factor(s) required for AvrRpt2 cleavage is present in plant, animal, and fungal cells and must be prevalent among eukaryotes.
Figure 2.
The cleavage factor(s) is prevalent among eukaryotes. a, E. coli-produced AvrRpt2::his is properly cleaved with rabbit reticulocyte lysate, Arabidopsis extracts, and yeast extracts. AvrRpt2::his was produced in E. coli and purified by affinity chromatography. The purified protein was incubated with buffer control (lanes 1 and 7), reticulocyte lysate (lanes 2 and 3), Arabidopsis extracts (lanes 4 and 5), or yeast extracts (lane 8) for the indicated time (2.5 h for lanes 7–9). Then the reactions were resolved by denaturing PAGE, and the gel was subjected to immunoblot analysis using anti-(his)3 antibody. ΔN71::his (lanes 6 and 9) was included as a molecular marker for the post-cleavage C terminus. The left and right panels represent separate experiments. b, The cleavage factors in rabbit reticulocyte lysate and in Arabidopsis extracts are heat labile and partially sensitive to protease inhibitors. Reticulocyte lysate (lanes 1–4) and Arabidopsis extracts (lanes 5–8) were treated with either heat (5 min in boiling water, lanes 1,2, 5, and 6) or a cocktail of protease inhibitors (lanes 3, 4, 7, and 8). E. coli-produced AvrRpt2::his was incubated with the treated extracts for the indicated time. The reactions were analyzed by immuno-blot as described in a. c, A bovine actin fraction contains a weak cleavage activity. AvrRpt2::his was incubated with an actin fraction for 1 h (lane 2) and was analyzed by immunoblot as in a. The full-length protein (lane 1) and the expected C-terminal region after cleavage (lane 3) were included as positional markers. The efficiency of cleavage can be compared by the ratio between the full-length and cleaved forms in each lane. White triangle, Full-length; black triangle, C-terminal region.
Figure 2b shows that the cleaving activities in reticulocyte lysate and Arabidopsis extracts were heat labile (lanes 1, 2, 5, and 6), which suggests that the cleavage was mediated by a protein factor. The activities were only partially blocked by a cocktail containing protease inhibitors of Ser proteases, Cys proteases, metalloproteases, and calpains (lanes 3, 4, 7, and 8).
Because the cleavage factor appears to be prevalent among eukaryotes, we speculated that AvrRpt2 may have a self-cleavage activity that can be activated by a prevalent eukaryotic factor. The Yersinia type III effector protein kinase YpkA is activated by a eukaryote-specific protein, actin (Juris et al., 2000). Although we did not find a putative actin-binding site in AvrRpt2, we tested a commercially available bovine muscle actin fraction for AvrRpt2::his cleavage (lane 2). The results in Figure 2c show that the actin fraction exhibited a weak cleavage activity. Considering that a large amount of actin fraction was added to the reaction (6 μg in a 20-μL reaction; it is unlikely that 2 μL of reticulocyte lysate, which was used in a 20-μL reaction and gave a better cleavage activity, contains a comparable amount of actin) and that it only gave a weak cleavage activity, it is likely that the required eukaryotic factor is not actin itself but is a contaminant in the fraction. The preparation procedure for the actin fraction includes acetone precipitation and extensive dialysis. Therefore, the required factor is unlikely to be a lipid or a small molecule.
Only Two Amino Acid Residues Near the Cleavage Site Are Required for Cleavage
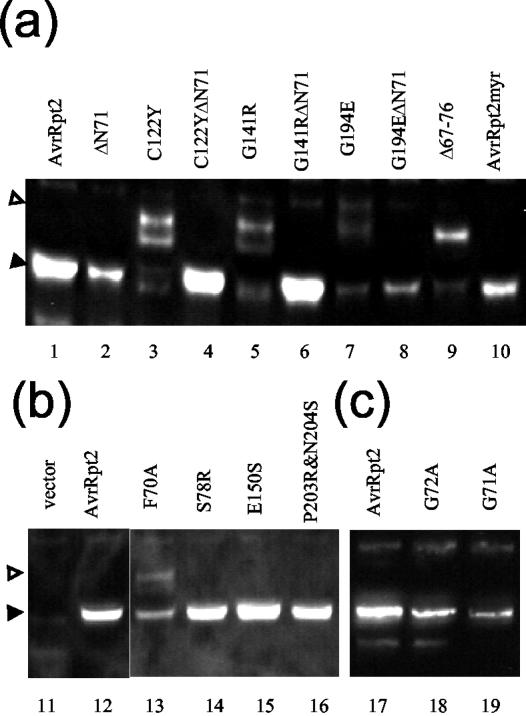
Taking advantage of the quick cleavage assay using rabbit reticulocyte lysate, we tested the cleavage activity of various mutant versions of AvrRpt2. We first tested a series of mutations in the vicinity of the cleavage site. All of the mutants were generated in the GST::AvrRpt2 context. Table I lists the mutants created and the results of the cleavage assay. The results indicate that the only mutants with a detectable cleavage deficiency are GST::F70A and GST::G71A. Therefore, F70 and G71 are the only two amino acid residues near the cleavage site that are required for cleavage.
Table I.
Only two amino acid residues near the cleavage site are important for cleavage in rabbit reticulocyte lysate
All of the mutants were generated in the GST::AvrRpt2 context (amino acid residue nos. refer to the position in AvrRpt2, which is 255 amino acid residues long, and the GST part is ignored for numbering purposes). Only amino acid residues near the cleavage site, which correspond to positions 61 to 82 in the wild type, are shown. The mutated residues are in bold. The arrowhead in the wild-type sequence indicates the cleavage site.
| Name | Amino Acid Residues 61 to 82 | Protein Accumulation | Cleavage |
|---|---|---|---|
| Wild type | GRHKIEVPAFG▾GWFKKKSSKHE | +++ | + |
| S78R | GRHKIEVPAFGGWFKKKRSKHE | +++ | + |
| K75 mol&K76 mol | GRHKIEVPAFGGWFLLKSSKHE | ++ | + |
| W73E&F74M&K75Q | GRHKIEVPAFGGEMQKKSSKHE | ++ | + |
| W73I&F74Y | GRHKIEVPAFGGIYKKKSSKHE | + | + |
| AvrRpt2myr | GRHKIEVPAFGGCV—SSKHE | ++ | + |
| G72A | GRHKIEVPAFGAWFKKKSSKHE | + | + |
| G71A | GRHKIEVPAFAGWFKKKSSKHE | +/- | - |
| F70A | GRHKIEVPAAGGWFKKKSSKHE | + | - |
| A69S | GRHKIEVPSFGGWFKKKSSKHE | ++ | + |
| V67A&P68A | GRHKIEAAAFGGWFKKKSSKHE | ++ | + |
| I65A&E66A | GRHKAAVPAFGGWFKKKSSKHE | ++ | + |
| H63A&K64A | GRAAIEVPAFGGWFKKKSSKHE | ++ | + |
| G61A&R62A | AAHKIEVPAFGGWFKKKSSKHE | ++ | + |
A Large Part of the C-Terminal Region of AvrRpt2 Is Required for Cleavage
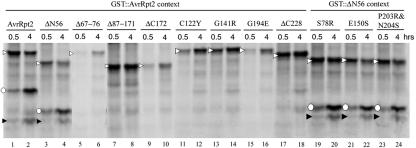
Because the “FG” sequence appears in many protein sequences and because the cleavage is highly specific, these two residues should not be the sole determinants of the cleavage specificity. Therefore, we studied other parts of the protein, beginning with analysis of four deletion mutants, ΔN56, Δ67–76, Δ87–171, and ΔC172, in the GST fusion context. Figure 3a shows that cleavage does not require the N-terminal 56 amino acid residues (GST::ΔN56; lanes 3 and 4). However, not only are the 10 amino acid residues surrounding the cleavage site (GST::Δ67–76; Fig. 3a, lanes 5 and 6) required, but deletions in the C-terminal region of the protein (GST::Δ87–171; Fig. 3a, lanes 7 and 8; GST::ΔC172; Fig. 3a, lanes 9 and 10) also lost cleavage activity.
Figure 3.
A large part of the C-terminal region of AvrRpt2 is required for cleavage. The AvrRpt2 derivatives indicated were translated and 35S-labeled in rabbit reticulocyte lysate. The reactions were incubated for the number of hours indicated. The reactions were analyzed as in Figure 1. The derivatives were made in either the GST::AvrRpt2 context (lanes 1–18) or the GST::ΔN56 context (lanes 19–24). White triangle, Full-length; black triangle, C-terminal region; white circle, N-terminal region.
Because a large part of the C-terminal region of the protein was required for cleavage, we decided to test other mutants within this region. We generated corresponding mutations based on mutants that had been previously identified (Axtell et al., 2001). These mutants were not recognized by RPS2 when directly expressed in plant cells. Only point mutations (GST::C122Y, GST::G141R, and GST::G194E) and the smallest C-terminal deletion mutation (GST::ΔC228) among them were chosen. One mutant gene identified by Axtell et al. (2001) contained two point mutations (C122Y and G131D). We were unable to create G131D by PCR for an unknown reason, and it was omitted from the assay. Three more mutants were included in the assay (GST::(ΔN56) S78R, GST::(ΔN56) E150S, and GST::(ΔN56) P203R&N204S). These mutants were created previously for unrelated purposes and produced in the Δ1–56 context to facilitate PCR. Figure 3b shows that all the mutants created based on the report by Axtell et al. (2001) were inactive in cleavage, whereas the three mutants that we added were active.
Important Molecular Features of AvrRpt2 for Cleavage Are Similar in the Reticulocyte Lysate and Plant Cells
To test whether our findings using reticulocyte lysate correlate with events in the plant cell, we expressed AvrRpt2 mutants in Arabidopsis protoplasts. All of the AvrRpt2 derivatives tested were fused at their C termini to green fluorescent protein (GFP) for stability of the proteins in protoplasts (Leister and Katagiri, 2000) and to a FLAG tag for detection by immunoblotting (AvrRpt2::GFPf). As shown in Figure 4a, the only detectable band for the wild-type AvrRpt2::GFPf comigrated with that of the artificially truncated derivative representing the cleaved size (Fig. 4a, lanes 1 and 2). This indicates that, in agreement with our prior observations, wild-type AvrRpt2::GFPf was efficiently cleaved in protoplasts (Leister and Katagiri, 2000). In contrast, C122Y::GFPf, G141R::GFPf, G194E::GFPf, and Δ67–76::GFPf were not cleaved efficiently (Fig. 4a, lanes 3, 5, 7, and 9). Bands that were larger than the expected cleaved size were observed in these lanes (compare with the artificially truncated derivatives; Fig. 4a, lanes 4, 6, and 8). C122Y::GFPf and G141R::GFPf produced multiple bands, including bands similar in size to the cleaved AvrRpt2::GFPf (Fig. 4a, black triangle), in addition to a band of the full-length size (Fig. 4a, white triangle). However, the sizes of the bands similar to the cleaved AvrRpt2::GFPf do not seem to be exactly the same as the size of the cleaved form. In contrast, G194E::GFPf and Δ67–76::GFPf produced bands that are apparently exactly the same size as that of the cleaved AvrRpt2::GFPf, in addition to bands of the full-length size. As shown in Figure 4, b and c, the S78R::GFPf, E150S::GFPf, P203R&N204S::GFPf, G72A::GFPf, G71A::GFPf, and F70A::GFPf mutants were tested similarly. S78R::GFPf, E150S::GFPf, and P203R&N204S::GFPf accumulated well, and only the cleaved size products were observed, consistent with the results using reticulocyte lysate. The G72A::GFPf, G71A::GFPf, and F70A::GFPf mutants did not accumulate well. G72A::GFPf and G71A::GFPf were cleaved efficiently, whereas F70A::GFPf was partially cleaved. We did not detect ΔC228::GFPf (data not shown). With the exception of G71A::GFPf, the effects of the mutations on cleavage were similar in Arabidopsis protoplasts and reticulocyte lysate, although cleavage in planta was generally more efficient.
Figure 4.
Cleavage of AvrRpt2 derivatives in the plant cell. The indicated AvrRpt2::GFPf derivatives were expressed in Arabidopsis rps2 mutant protoplasts. The extracts from the protoplasts were analyzed by immunoblot as in Figure 2 except that the anti-FLAG antibody was used. AvrRpt2::GFPf was included in every separate experiment as a positive control. a through c represent results from three different experiments. White triangle, Full-length; black triangle, C-terminal region.
Cleavage of AvrRpt2 Is Important for Its Interaction with the Host Plant Cell
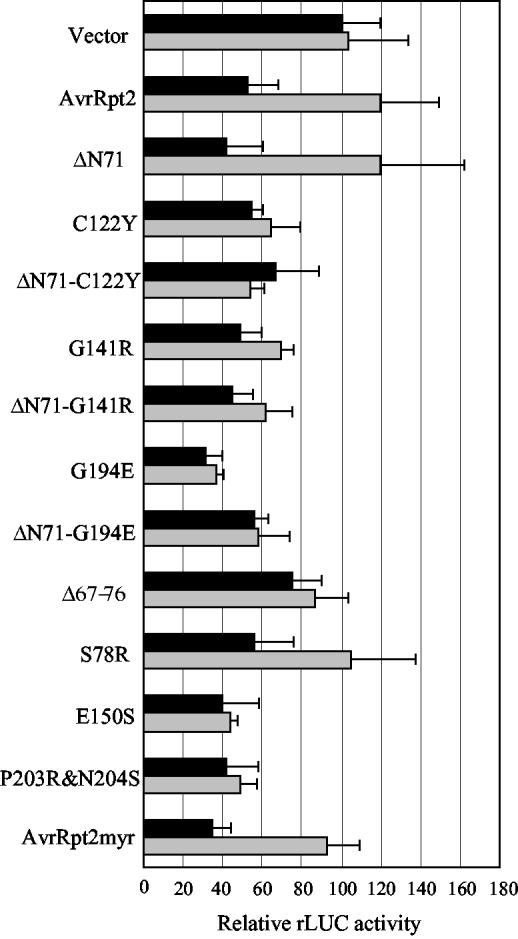
We tested whether cleavage is required for recognition by RPS2 using a transient expression assay (Leister et al., 1996). We also tested 71 amino acid residue deletions from the N terminus (ΔN71) in AvrRpt2 derivatives that are deficient in cleavage to examine whether the N-terminal region affects recognition by RPS2. The derivatives used in this experiment were all FLAG-tagged (AvrRpt2::f). Figure 5 shows the results of the transient expression assays with selected mutants. In this assay, gene-for-gene interactions were detected as decrease of a reporter gene activity dependent on both avr and R genes (see the result with AvrRpt2::f). For an unknown reason, many of the AvrRpt2 derivatives tested caused a decrease of the reporter gene activity in an RPS2-independent manner. We did not observe further reduction of the reporter activity in an RPS2-dependent manner. This could be correlated with a loss of recognition by RPS2 because G141R::f and G194E::f, which showed the RPS2-independent reporter activity decrease, were previously shown to be inactive when expressed in plant cells (Axtell et al., 2001). In any case, it is clear that these mutants showing RPS2-independent reporter activity decrease differ from wild-type AvrRpt2 in interactions with rps2 mutant cells. All mutations made in the C-terminal region, except S78R::f, led to the malfunction, and the artificial deletion of the N-terminal 71 amino acid residues did not restore the proper function. None of the mutations near the cleavage sites (G72A::f, G71A::f, and F70A::f led to a loss of recognition (data not shown). G72A and G71A were completely cleaved, and F70A was partially cleaved in planta (Fig. 4, b and c, lanes 13, 18, and19). Furthermore, whereas the portion of the protein C-terminal to amino acid residue 82 is sufficient for recognition by RPS2 (Mudgett et al., 2000), the cleavage deficient mutant Δ67–76::f did not function properly. These results suggest that the cleavage is necessary, but not sufficient, for a proper interaction with plant cells lacking RPS2.
Figure 5.
RPS2 recognition of AvrRpt2 derivatives. The indicated AvrRpt2::f derivatives were tested for recognition by RPS2 by a transient expression assay either with (black bars) or without (gray bars) RPS2. When a derivative is active, the relative rLUC activity with RPS2 is significantly lower than it is without RPS2. The mean rLUC activity value for vector with RPS2 was set to 100 to normalize the entire results. Error bar = sd. The number of replicates, n = 4, except for vector with RPS2 (n = 40), AvrRpt2::f with RPS2 (n = 40), vector without RPS2 (n = 36), and AvrRpt2::f without RPS2 (n = 36).
Targeting of AvrRpt2 to the Plasma Membrane Does Not Affect Recognition by RPS2
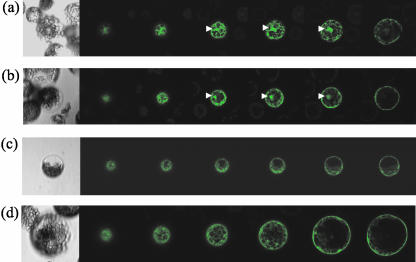
Several P. syringae type III effectors, including AvrRpm1 and AvrB, are myristoylated at their N termini in the plant cell, whereas AvrRpt2 is not. Myristoylation of AvrRpm1 and AvrB is required for their plasma membrane localization and quantitatively important for recognition by the corresponding R gene product, RPM1 (Nimchuk et al., 2000). As shown below, an AvrRpt2::GFP fusion does not appear to be targeted to the plasma membrane. We were interested in testing whether an AvrRpt2 derivative with a canonical myristoylation site can be localized to the plasma membrane and whether such a protein would be recognized more efficiently by RPS2. The observation that F70 and G71 are the only residues in the immediate vicinity of the cleavage site that are essential for cleavage in the reticulocyte lysate (Table I) allowed us to engineer a new post-cleavage N terminus. The amino acid sequence immediately after the cleavage site was changed to the AvrB myristoylation site (AvrRpt2myr: from G(72) WFKKKSSKH to G(72) CVSSKH). AvrRpt2myr::GFPf was cleaved efficiently in the plant cell (Fig. 4, lane 10). The localization of AvrRpt2myr::GFPf was tested by transient expression of the construct in Arabidopsis protoplasts followed by observation under a laser confocal microscope. Figure 6 shows that AvrRpt2myr::GFPf, like AvrB::GFP, appears to be largely localized in the plasma membrane (Fig. 6, c and d). The membrane-localization of AvrB::GFP was reported previously (Nimchuk et al., 2000). The localization of AvrRpt2myr::GFPf differs from that of AvrRpt2::GFPf and GFP::f, which are both distributed in the cytosol with a tendency to accumulate in the nucleus (Fig. 6, a and b). These results suggest that AvrRpt2myr::GFPf is myristoylated and that the myristoylation targets the protein to the plasma membrane. When RPS2-mediated recognition of AvrRpt2myr::f was tested using the transient expression assay, no significant difference compared with AvrRpt2::f was observed (Fig. 5). It appears that forced targeting of AvrRpt2 to the plasma membrane does not improve recognition by RPS2.
Figure 6.
AvrRpt2myr::GFPf is largely localized to the plasma membrane in the plant cell. GFP::f (a), AvrRpt2::GFPf (b), AvrRpt2myr::GFPf (c), and AvrB::GFP (d) were expressed in Arabidopsis protoplasts, and the GFP fluorescence was observed under a laser confocal microscope. In each panel, a light image is shown on the left, followed by six confocal optical section images from the bottom to the middle of the protoplast. Protoplasts exhibiting representative localization of the GFP-tagged proteins were chosen. Note that not all protoplasts express the derivatives. Nuclear staining is indicated by white arrowheads.
Cleavage Is Important for AvrRpt2 Localization in the Host Cell
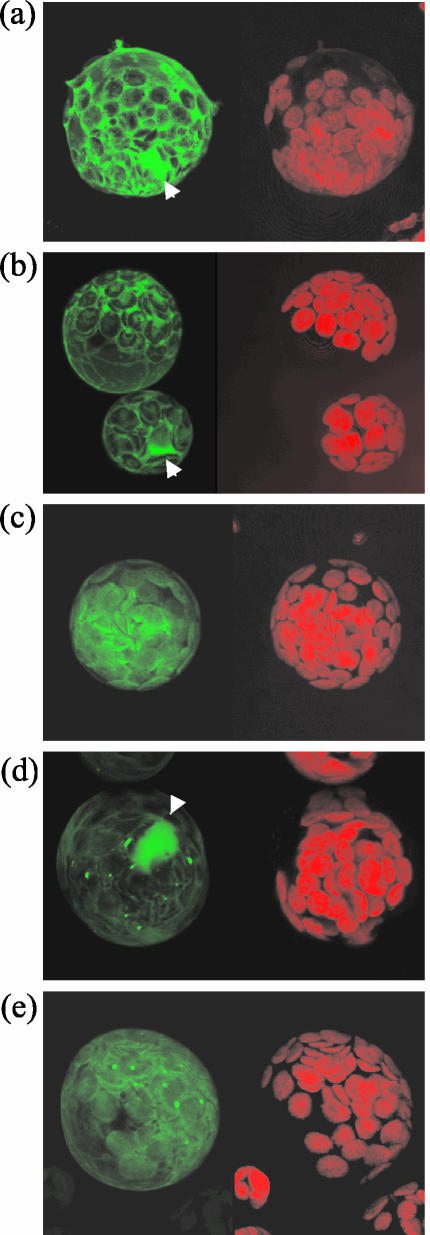
Does cleavage target AvrRpt2 to a specific site in the host cell? This question can be addressed by observing where AvrRpt2 would be localized in the plant cell if the N-terminal region were not removed. We tested localization of several cleavage-deficient mutants as C-terminal GFPf fusions (Fig. 7). In contrast to AvrRpt2::GFPf, which was localized in the cytosol and nucleus and excluded from the chloroplast (Fig. 7b), C122Y::GFPf was mainly localized in the chloroplast (Fig. 7c), whereas Δ67–76::GFPf was strongly localized in the nucleus (Fig. 7d). G141R::GFPf, G194E::GFPf, and ΔC228::GFPf did not accumulate to levels sufficient for observation (data not shown). The localization of S78R::GFPf, E150S::GFPf, P203R&N204S::GFPf, and G72A::GFPf was indistinguishable from that of AvrRpt2::GFPf (data not shown). F70A::GFPf was often detected in the chloroplast in addition to the cytosol and the nucleus (Fig. 7e). The localization of G71A::GFPf was similar to that of AvrRpt2::GFPf, but it was occasionally detected in chloroplasts (data not shown). Considering the low efficiency for detection of GFP in the chloroplast (Reed et al., 2001), this mutant protein may be partitioned in the chloroplast at a significant level. In summary, there is a good correlation between cleavability and subcellular localization similar to that of the wild-type AvrRpt2::GFPf protein.
Figure 7.
Cleavage of the N-terminal region of AvrRpt2 is important for subcellular localization in the host cell. GFP::f (a), AvrRpt2::GFPf (b), C122Y::GFPf (c), Δ67–76::GFPf (d), and F70A::GFPf (e) were expressed in Arabidopsis protoplasts, and the GFP fluorescence (green, on the left in each panel) and chlorophyll fluorescence (red, on the right in each panel) were observed under a laser confocal microscope. Chlorophyll fluorescence images indicate the positions of chloroplasts. The composite images of all the sections are shown. A representative fluorescence pattern for each derivative is shown. Nuclear staining is indicated by white arrowheads.
DISCUSSION
The AvrRpt2 protein is properly cleaved in plant, animal, and yeast cell extracts. Two amino acid residues immediately preceding the cleavage site and a large part of the C-terminal region of AvrRpt2 were required for cleavage. Most of these features of Avr-Rpt2 were also important when AvrRpt2 was cleaved inside plant cells. Cleavage deficiency altered the way AvrRpt2 interacts with rps2 plant cells. Cleavage also appears to be required for the proper localization of the C-terminal region of the protein in the host cell.
What Is the AvrRpt2 Cleavage Mechanism?
The factor(s) required for AvrRpt2 cleavage is prevalent among eukaryotes. It is unlikely that the specificity of highly selective proteases is conserved among plants, animals, and fungi. In addition, incubation of the eukaryotic cell extracts with a cocktail of protease inhibitors did not exhibit a strong effect on cleavage of AvrRpt2. Therefore, it is likely that the eukaryotic factor is not a specific protease. A large part of the C-terminal region of AvrRpt2 contributes to cleavage. Cleavage specificity could be determined by a protein-protein interaction between relatively large peptides. Alternatively, a large peptide could have self-cleaving activity. It has been shown that AvrPphB is homologous to cysteine proteases and that amino acid residues conserved among cysteine proteases are required for self-cleavage of AvrPphB (Shao et al., 2002). Although we failed to detect any appreciable protease homology in AvrRpt2, one attractive model is that AvrRpt2 has self-cleaving activity that is activated by a eukaryotic factor.
If AvrRpt2 cleaves itself, the reaction appears to occur in an intramolecular manner rather than an intermolecular manner. We used reticulocyte lysate to coexpress each of the cleavage-deficient mutants with mutations in the C-terminal region in the GST fusion context together with the cleavage site mutant GST::Δ67–76. We did not detect “trans”-cleavage of any of the cleavage-deficient mutants (data not shown).
AvrRpt2 may still be able to cleave other proteins, and cleavage of other plant proteins may be central in how AvrRpt2 affects plant defense. Recently, it was reported that RIN4, a protein involved in RPS2- and RPM1-mediated recognition, rapidly disappears in the presence of AvrRpt2 (Axtell and Staskawicz, 2003; Mackey et al., 2003). The AvrRpt2 mutants that we demonstrated to be cleavage deficient, C122Y, G141R, and G194E, did not cause the disappearance of RIN4 (Axtell and Staskawicz, 2003). It will be interesting to see whether AvrRpt2 in the presence of an appropriate eukaryotic factor can directly digest RIN4.
Identification of the required eukaryotic factor will provide insight into how the C-terminal region of AvrRpt2 functions in cleavage. Our characterization of the factor indicates that it is heat labile, acetone precipitable, and not removed by extensive dialysis. These observations strongly suggest that the factor is a protein.
Cleavage Is Important for Proper Interaction with Host Cells
We characterized three different classes of AvrRpt2 loss-of-function mutants. The first class contains E150S and P203R&N204S, which are cleaved well in the plant cell but exhibit an altered interaction with rps2 plant cells in the transient assay. The second class is composed of Δ67–76, which is deficient in both cleavage and interaction with rps2 plant cells, but appears to recover proper interactions with both RPS2 and rps2 plant cells when the N-terminal region is artificially removed (the portion C-terminal to amino acid residue 82 retains the ability to be recognized by RPS2 [Mudgett et al., 2000]). The third class contains C122Y, G141R, and G194E, which are deficient in both cleavage and interaction with rps2 plant cells, and in which artificial removal of the N-terminal region does not restore proper interactions with plant cells. Therefore, these residues are required for both cleavage and proper interactions with plant cells. All of these classes of mutants are consistent with the notion that the cleavage is necessary but not sufficient for proper AvrRpt2 function. This notion suggests that the N-terminal region of the protein has an adverse effect on the AvrRpt2 function because the C-terminal region of AvrRpt2 is the effector part of the protein. As discussed below, the adverse effect of the N-terminal region may be related to improper localization of the C-terminal region. Alternatively, cleavage may lead to a conformational change of the C-terminal region of the protein.
Where Is the Site of Action for AvrRpt2 in the Host Cell?
In general, defense responses induced by the AvrRpt2-RPS2 interaction are weaker/slower than those induced by the AvrRpm1/AvrB-RPM1 interaction, although the RPS2- and RPM1-mediated pathogen recognition mechanisms appear to share components (Ritter and Dangl, 1996; Leister and Katagiri, 2000; Axtell and Staskawicz, 2003; Mackey et al., 2003). The myristoylation and consequential plasma membrane-localization of AvrRpm1 and AvrB quantitatively contributes to the ability of those proteins to be recognized by RPM1 (Nimchuk et al., 2000). Therefore, the RPM1-mediated recognition mechanism is thought to operate at the plasma membrane (Boyes et al., 1998). Does the RPS2-mediated recognition mechanism also operate at the plasma membrane? It was of interest to see whether myristoylation and membrane localization of AvrRpt2myr would enhance RPS2 recognition. We did not observe any appreciable increase in the ability of AvrRpt2myr to be recognized by RPS2 (Fig. 5). Therefore, our observations do not appear to support the idea that RPS2 recognition occurs at the plasma membrane.
Axtell and Staskawicz (2003) recently demonstrated that when AvrRpt2 is expressed at a low level in Arabidopsis, it is associated with the membrane fraction. This and our observations appear to contradict each other. However, they are easy to reconcile if we assume that the membrane association of Avr-Rpt2 is mediated by a limited number of specific binding sites in the membrane. In this way, when AvrRpt2 is overexpressed, as in our experimental setup, the AvrRpt2-binding sites in the membrane would be quickly saturated, and the majority of Avr-Rpt2 protein would remain in the cytosol. Forming a complex with the binding site in the membrane could be important for AvrRpt2 recognition mediated by RPS2. If this is the case, forced targeting of excessive AvrRpt2 to the membrane by AvrRpt2myr would not significantly affect the level of the interaction between AvrRpt2 and RPS2.
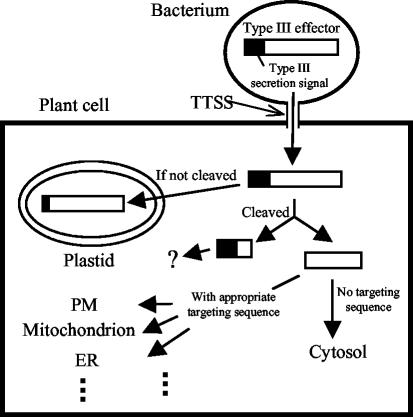
Is Cleavage a Mechanism to Redirect an Avr Protein after Translocation?
Various N-terminal signal peptides are known to target proteins in the eukaryotic cell, such as targeting to the secretion pathway or to organelles (Emanuelsson and von Heijne, 2001). It will be interesting to test whether AvrRpt2 derivatives that are engineered with various intracellular-targeting signals at the cleavage site will be localized to the intended sites. We demonstrated the feasibility of this by engineering a myristoylation site at the cleavage site (Fig. 6c). This also raises the question of whether many pathogen effector proteins are localized by posttranslocational host-cellular targeting (Fig. 8). It would be advantageous for a pathogen to have a means independent of the type III-protein translocation signal (usually located at the original N terminus of the protein) to target virulence proteins to specific sites in the host cell. The region of AvrPphB N-terminal to the cleavage site is important for type III-dependent translocation of the protein (Tampakaki et al., 2002). Once the N-terminal region is cleaved off, AvrPphB is myristoylated and localized to the plasma membrane of the plant cell (Nimchuk et al., 2000). In the case of AvrRpt2, AvrRpt2::GFPf localization in the plant cell is indistinguishable from GFP::f alone (Fig. 7). The wild-type AvrRpt2 may not have such a posttranslocational targeting function. However, Guttman et al. (2002) suggested that the C-terminal region of AvrRpt2 tends to keep proteins from organelle targeting. They also pointed out that many type III signal peptides are similar to plastid-targeting signal peptides. At least one cleavage-deficient mutant (C122Y::GFPf) was mainly localized in the chloroplast. Two mutants with limited cleavability (G71A::GFPf and F70A::GFPf) appeared partially localized to the chloroplast. Another cleavage mutant (Δ67–76::GFPf) was localized in the nucleus. It is not clear why this mutant is localized in the nucleus. We did not inadvertently create an obvious nuclear localization signal when we made the mutation. In any case, the inappropriate localization of the cleavage-deficient mutant proteins suggests that cleavage is crucial for proper localization of the C-terminal region of AvrRpt2.
Figure 8.
Type III effector redirection model. This model illustrates that cleaving off the N-terminal region, which contains the type III secretion signal, could redirect the type III effector to a particular subcellular location in the host plant cell. Many type III secretion signal sequences are similar to plastid-targeting signals, so an uncleaved type III effector could be transported into the plastid (If not cleaved). However, cleavage removes the cryptic plastid-targeting signal and exposes a new N terminus (Cleaved). If post-cleavage N terminus does not have a particular targeting signal, the C-terminal region stays in the cytosol, like AvrRpt2. If it has a targeting signal, the C-terminal region can be transported to an appropriate subcellular location. For example, AvrPphB is targeted to the plasma membrane via myristoylation of the new N terminus. PM, Plasma membrane; ER, endoplasmic reticulum.
Many type III secretion signal peptides of P. syringae are similar to plastid-targeting signal peptides, and it has been suggested that the site of action for many type III effectors may be in the plastid (Guttman et al., 2002). Cleavage of the N-terminal region of AvrRpt2 appears to remove the putative signal for plastid targeting. Some other Avr proteins are myristoylated at their N termini, leading to membrane localization (Nimchuk et al., 2000). It is possible that these proteins would otherwise be targeted to the plastid, which is not their presumed site of action. AvrPphB is both cleaved and myristoylated (Nimchuk et al., 2000; Shao et al., 2002). These observations raise questions such as how common it is for cleavage of N-terminal signal sequences to target type III effectors for redirection in host cells. Because signal peptides for various target sites in eukaryotic cells are commonly located at the N terminus of a protein (Emanuelsson and von Heijne, 2001), the process of exposing a new targeting signal peptide by cleaving off the N-terminal region of a protein could enable the targeting of proteins to a wide variety of intracellular sites in the host cell. This type of mechanism may be more broadly used by type III effectors than we currently realize.
MATERIALS AND METHODS
Plant Materials
Three types of Arabidopsis plants were used, all are the Columbia ecotype: RPM1 RPS2 (Col-0, wild type), RPM1 rps2 (rps2-101Cl Mindrinos et al., 1994), and rpm1 RPS2 (rps3-1; Innes et al., 1993). Plants were grown in Metro-Mix 200 soil (The Scotts Company, Marysville, OH) at 22°C in approximately 80% relative humidity with a 12-h-light/12-h-dark cycle in environment-controlled growth chambers.
Plasmids
The pKEx4tr and pExavrRpt2 constructs have been described previously (Mindrinos et al., 1994; Leister et al., 1996; Tao et al., 2000). The PCR products for GST::AvrRpt2 and its derivatives were amplified from a GST::AvrRpt2::MalE construct made in the pGEX-5X-3 vector (Pharmacia, Uppsala). Note that the MalE part was not included in the PCR products and is irrelevant to this study. The junction sequence between GST and AvrRpt2 in the construct is GGATCCCCATG (GGATCC is the BamHI site in the vector, and ATG is the start codon for AvrRpt2). The pET-avrRpt2-his construct has been described (Wu et al., 2003). AvrRpt2 derivatives were amplified by PCR from the templates used in the in vitro transcription/translation system with primers that incorporated a BamHI site at the 5′ end of the gene and a KpnI site at the 3′ end. The products were inserted into the vector pKEx-CFL (for in-frame C-terminal fusion to the FLAG epitope tag) or pKEx-GFPFL (for in-frame C-terminal fusion to GFP and the FLAG epitope tag). Both of these vectors are derived from pKEx4tr, with the FLAG and GFPFL inserts cloned between KpnI and NotI. In pKEx-CFL, two amino acids GT were added between the last amino acid of AvrRpt2 and the FLAG tag. In pKEx-GFPFL, two amino acids GT were added between the last amino acid of AvrRpt2 and GFP. Details of the cloning procedures are available upon request.
In Vitro Translation with Rabbit Reticulocyte Lysate
The templates for in vitro transcription were generated by PCR SOEing (Horton et al., 1990) in which the final 5′ primer contains the T7 promoter sequence. AVRT3 was used as the final 3′ primer. The primer pairs used in SOEing to create mutations are supplied in supplemental materials. Three mutant versions were cloned into a plasmid vector first for the purpose of unrelated experiments and then amplified from the plasmids.
For in vitro translation coupled with in vitro transcription, the TNT T7 Quick for PCR DNA kit (Promega, Madison, WI) was used according to the supplier's instructions. The proteins were labeled with [35S]Met and resolved by denaturing PAGE. The gels were dried and subjected to phosphor imaging (Molecular Dynamics, Sunnyvale, CA).
Cleavage of Escherichia coli Produced Protein
Production and purification of AvrRpt2 from E. coli was performed as described (Wu et al., 2003). For preparation of Arabidopsis extracts, one well-expanded leaf from a 4-week-old Col-0 wild-type plant was ground in 100 μL of extraction buffer (20 mm Tris-HCl, pH 7.5, 50 mm NaCl, and 0.01% [v/v] Triton X-100), and the extracts were collected after centrifugation at 16,000g for 2 min at 4°C. Yeast cell extracts were prepared from Brewer's yeast (Saccharomyces cerevisiae strain NAV203α) as follows. Yeast cells were grown in 5 mL of yeast peptone dextrose medium overnight at 30°C. Cells were harvested and disrupted by vortexing in breaking buffer (0.1 m Tris-HCl, pH 8, 20% [w/v] glycerol, and 1 mm dithiothreitol) with glass beads (PN. G8772, Sigma-Aldrich, St. Louis). The cleared lysate was used as the yeast cell extract. The E. coli-produced AvrRpt2 protein (2 μg) was incubated with either 2 μL of T7 TNT lysate (rabbit reticulocyte lysate), 3 μL of Arabidopsis extracts, 6 μL of yeast cell extracts, or 3 μL of 2 mg mL–1 bovine muscle actin (PN. A3653, Sigma-Aldrich) in total volume of 20 μL in 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, and 5 mm MgCl2 at 30°C for the indicated duration of time. Then proteins in 3 μL of the mixture were resolved by denaturing PAGE and blotted onto a nitrocellulose membrane by semidry blotting (Bio-Rad Laboratories, Hercules, CA). (His)6-tagged proteins were detected by immunoblot analysis using the anti-(his)3 antibody (Qiagen USA, Valencia, CA) as the primary antibody and the SuperSignal West Femto Maximum Sensitivity Substrate (Pierce, Rockford, IL) for detection, according to the manufacturers' instructions. The chemiluminescent image was captured by a ChemiImager (Alpha Innotech, San Leandro, CA).
AvrRpt2 Expression in Protoplasts
GFP and FLAG-tagged AvrRpt2 derivatives were expressed in Arabidopsis protoplasts by transient expression as previously described (Leister and Katagiri, 2000). Approximately 1.2 × 105 protoplasts were lysed in 30 μL of lysis solution (1× Tris-buffered saline, 0.5% [w/v] sodium deoxycholate, and 1% [v/v] IGEPAL CA-630) after an 18-h incubation. The lysate was subjected to immunoblot analysis as described above, except that the anti-FLAG M2 antibody (Sigma-Aldrich) was used as the primary antibody.
Biolistic Transient Assay for RPS2 Recognition of AvrRpt2
Biolistic transient assay for RPS2 recognition of AvrRpt2 was performed as previously described (Leister et al., 1996).
Microscopy
Confocal microscopy was performed using a Fluoview 5 confocal microscope (FV5-LSM, Olympus, Melville, NY).
Acknowledgments
We thank Jane Glazebrook for critical reading of the manuscript and Kim Campbell for helping to care for the plants.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.025999.
References
- Axtell MJ, McNellis TW, Mudgett MB, Hsu CS, Staskawicz BJ (2001) Mutational analysis of the Arabidopsis RPS2 disease resistance gene and the corresponding Pseudomonas syringae avrRpt2 avirulence gene. Mol Plant-Microbe Interact 14: 181–188 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95: 15849–15854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper-Lindley C, Dahlbeck D, Clark ET, Staskawicz BJ (2002) Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc Natl Acad Sci USA 99: 8336–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Boch J, Katagiri F, Kunkel BN (2000) The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol Plant-Microbe Interact 13: 1312–1321 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, von Heijne G (2001) Prediction of organellar targeting signals. Biochim Biophys Acta 1541: 114–119 [DOI] [PubMed] [Google Scholar]
- Galán JE, Collmer A (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Guttman DS, Greenberg JT (2001) Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol Plant-Microbe Interact 14: 145–155 [DOI] [PubMed] [Google Scholar]
- Guttman DS, Vinatzer BA, Sarkar SF, Ranall MV, Kettler G, Greenberg JT (2002) A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295: 1722–1726 [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai Z, Ho SN, Pease LR (1990) Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8: 528–535 [PubMed] [Google Scholar]
- Innes RW, Bisgrove SR, Smith NM, Bent AF, Staskawicz BJ, Liu YC (1993) Identification of a disease resistance locus in Arabidopsis that is functionally homologous to the RPG1 locus of soybean. Plant J 4: 813–820 [DOI] [PubMed] [Google Scholar]
- Juris SJ, Rudolph AE, Huddler D, Orth K, Dixon JE (2000) A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc Natl Acad Sci USA 97: 9431–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ (1993) RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT, Ausubel FM, Katagiri F (1996) Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc Natl Acad Sci USA 93: 15497–15502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT, Katagiri F (2000) A resistance gene product of the nucleotide binding site-leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J 22: 345–354 [DOI] [PubMed] [Google Scholar]
- Lindgren PB, Peet RC, Panopoulos NJ (1986) Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol 168: 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mudgett MB, Chesnokova O, Dahlbeck D, Clark ET, Rossier O, Bonas U, Staskawicz BJ (2000) Molecular signals required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc Natl Acad Sci USA 97: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB, Staskawicz BJ (1999) Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol Microbiol 32: 927–941 [DOI] [PubMed] [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101: 353–363 [DOI] [PubMed] [Google Scholar]
- Reed ML, Wilson SK, Sutton CA, Hanson MR (2001) High-level expression of a synthetic red-shifted GFP coding region incorporated into transgenic chloroplasts. Plant J 27: 257–265 [DOI] [PubMed] [Google Scholar]
- Ritter C, Dangl JL (1996) Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8: 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Thara VK, Martin GB, Zhou JM, Tang X (2000) The Pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell 12: 2323–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE (2002) A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109: 575–588 [DOI] [PubMed] [Google Scholar]
- Staskawicz BJ, Mudgett MB, Dangl JL, Galán JE (2001) Common and contrasting themes of plant and animal diseases. Science 292: 2285–2289 [DOI] [PubMed] [Google Scholar]
- Tampakaki AP, Bastaki M, Mansfield JW, Panopoulos NJ (2002) Molecular determinants required for the avirulence function of AvrPphB in bean and other plants. Mol Plant-Microbe Interact 15: 292–300 [DOI] [PubMed] [Google Scholar]
- Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F (2000) Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene. RPS2. Plant Cell 12: 2541–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wood MD, Tao Y, Katagiri F (2003) Direct delivery of bacterial avirulence proteins into resistant Arabidopsis protoplasts leads to hypersensitive cell death. Plant J 33: 131–137 [DOI] [PubMed] [Google Scholar]
- Yu GL, Katagiri F, Ausubel FM (1993) Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol Plant-Microbe Interact 6: 434–443 [DOI] [PubMed] [Google Scholar]