Abstract
Gene expression in living cells is highly dynamic, but temporal patterns of gene expression in intact tissues are largely unknown. The mammalian pituitary gland comprises several intermingled cell types, organised as interdigitated networks that interact functionally to generate co-ordinated hormone secretion. Live-cell imaging was used to quantify patterns of reporter gene expression in dispersed lactotrophic cells or intact pituitary tissue from bacterial artificial chromosome (BAC) transgenic rats in which a large prolactin genomic fragment directed expression of luciferase or destabilised enhanced green fluorescent protein (d2EGFP). Prolactin promoter activity in transgenic pituitaries varied with time across different regions of the gland. Although amplitude of transcriptional responses differed, all regions of the gland displayed similar overall patterns of reporter gene expression over a 50-hour period, implying overall co-ordination of cellular behaviour. By contrast, enzymatically dispersed pituitary cell cultures showed unsynchronised fluctuations of promoter activity amongst different cells, suggesting that transcriptional patterns were constrained by tissue architecture. Short-term, high resolution, single cell analyses in prolactin-d2EGFP transgenic pituitary slice preparations showed varying transcriptional patterns with little correlation between adjacent cells. Together, these data suggest that pituitary tissue comprises a series of cell ensembles, which individually display a variety of patterns of short-term stochastic behaviour, but together yield long-range and long-term coordinated behaviour.
Keywords: Live-cell, Microscopy, Pituitary, Prolactin, Transcription
Introduction
Gene expression in living cells has become amenable to quantitative study in recent years with several technical advances, notably the use of unstable reporter genes such as firefly luciferase and fluorescent proteins. These approaches have confirmed earlier observations that gene expression is frequently pulsatile, and might be subject to stochastic regulation (Raj and van Oudenaarden, 2008). Using the mammalian anterior pituitary as a model system, large-amplitude fluctuations in prolactin gene expression have been observed in cell lines and primary cultures of dispersed pituitary cells (Semprini et al., 2009; Shorte et al., 2002; Takasuka et al., 1998), but little is known about the real-time transcriptional behaviour of cells in intact tissues.
The pituitary gland is a key endocrine tissue, which contains a series of differentiated hormone-secreting cells, each secreting specific hormones including prolactin (PRL), growth hormone (GH), adrenocorticotrophin, thyrotrophin and gonadotrophins. In addition, folliculostellate cells exist throughout the pituitary: they do not secrete classical hormones, but are thought to exert paracrine effects through production of growth factors (Winters and Moore, 2007). Immunohistochemical analyses have suggested that these cell types are differentially distributed throughout the anterior pituitary tissue (Nakane, 1970), but these studies did not address how these cells are functionally organised. More recent evidence that tissue architecture and/or organisation is important for pituitary cell function has included observations of functionally linked networks of folliculostellate cells (Fauquier et al., 2001), and of a co-ordinated somatotroph cell network, in which adherens junctions allow propagation of calcium signalling (Bonnefont et al., 2005). Lactotroph cells form a large proportion of the pituitary gland (Nolan and Levy, 2009), and prolactin production is critical for reproductive function in mammals. Adenomas arising from lactotroph cells are a common pituitary pathology in humans, and are a frequent cause of infertility. The spatiotemporal organisation and function of lactotroph cells is still not well understood and it remains to be established whether they are functionally interlinked.
Activity of both the prolactin and GH gene promoters, as measured by low light luminescence imaging of luciferase reporter gene expression, has been shown to exhibit dramatic dynamic heterogeneity in both cell lines (Friedrichsen et al., 2006; McFerran et al., 2001; Norris et al., 2003; Stirland et al., 2003; Takasuka et al., 1998) and primary pituitary cells (Shorte et al., 2002). In each case studied so far, large-amplitude, non-co-ordinated and non-circadian pulses have been observed in promoter activity, resulting in marked heterogeneity of gene expression at any single time-point, with the pulsing lasting for many hours in vitro. This work has shown that gene transcription is highly pulsatile in pituitary cells grown in culture, but the question arises as to whether such dynamic heterogeneity is a feature of cells in the intact tissue, where architectural constraints and paracrine factors might be expected to modify such behaviour in vivo.
In order to study the spatiotemporal organisation of gene expression in intact tissue and primary pituitary cells, transgenic Fischer-344 rats were generated, expressing either firefly luciferase or destabilised EGFP (d2EGFP) reporter genes under the control of extensive (>100 kb) regulatory regions of the human prolactin locus, using a bacterial artificial chromosome (BAC) recombination approach [hPRL-luciferase and hPRL-d2EGFP BAC-transgenic rats (Semprini et al., 2009)]. Quantitative time-course measurements of reporter gene expression were obtained from primary cultures of dispersed pituitary cells and 400 μm thick pituitary tissue slices in which cell-cell contacts were maintained. These studies suggested that a long-range, long-term (50 hours) synchronisation of transcriptional timing existed between different regions of intact pituitary tissue, which was lost when cells were dispersed in a primary culture system. Analysis of transcriptional profiles in individual cells over 15 hours suggested that regions of the tissue constitute a series of ensembles of cells with different temporal patterns in adjacent cells. This implied a complex pattern of transcriptional behaviour within pituitary tissue, in which diverse local responses sum together across longer distances to generate a more co-ordinated tissue response.
Results
Lactotroph organisation within the anterior pituitary
To determine the location of lactotroph cells within pituitary tissue, pituitary glands from male luciferase- or d2EGFP-expressing transgenic Fischer-344 rats were removed, sliced to a thickness of 400 μm in the coronal orientation and imaged by luminescence or fluorescence microscopy. Pituitary slices from PRL-luciferase transgenic rats revealed that lactotroph cells were restricted to the anterior pituitary, with no significant signal detected in the posterior pituitary (Fig. 1A). Signal intensity was greater around the periphery of the pituitary with maximum intensity detected in the lateral parts of the gland (Fig. 1A). Pituitary slices from PRL-d2EGFP transgenic rats imaged using confocal microscopy provided improved resolution of signal, and a similar pattern was again detected, with EGFP expression restricted to the anterior pituitary with increased signal in the lateral aspects of the gland (Fig. 1B). Immunohistochemical analysis of endogenous rat PRL protein expression in Fischer rat pituitary slices confirmed that the spatial organisation of lactotroph cells was similar to that detected by luminescence and fluorescence imaging, with a higher intensity around the periphery of the anterior pituitary (Fig. 1C).
Fig. 1.
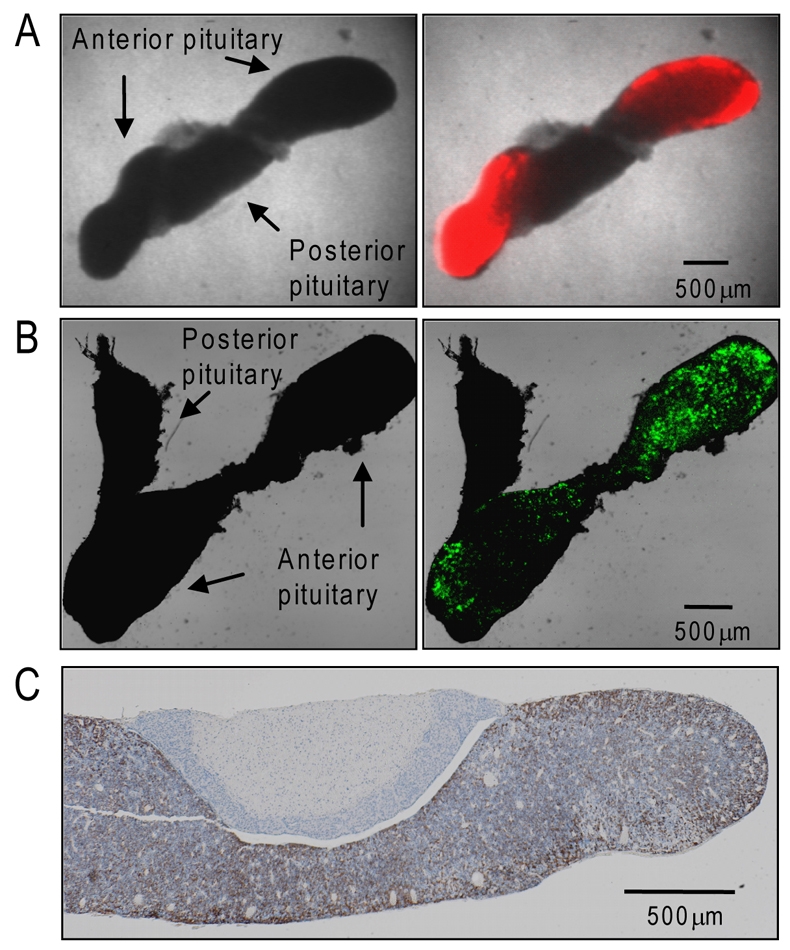
Lactotroph organisation within the anterior pituitary gland. Localisation of luminescent signal (A) and fluorescent signal (B) from 400 μm thick pituitary slices, taken from transgenic rats expressing PRL-luciferase or PRL-d2EGFP, respectively. Left panels show transmitted-light image. Right panels show transmitted-light image merged with localisation of reporter gene signal. (C) Immunohistochemistry of PRL protein expression (brown) in a wild type rat pituitary. Scale bars: 500 μm.
In order to confirm that the apparent peripheral localisation of the luciferase-expressing pituitary cells was not a result of restricted access of luciferin substrate or oxygen to the cells within the slice, rats were injected intraperitoneally with 1 mM luciferin 30 minute before death, to facilitate direct access of luciferin to the pituitary tissue from the bloodstream. Following pituitary removal and slicing, the luciferase signal again appeared more intense around the periphery, with signal decaying rapidly in the absence of luciferin in the surrounding medium (within 1.5 hours; supplementary material Fig. S1A,B). Supplementation of luciferin into the medium resulted in a rapid restoration of the luminescent signal (supplementary material Fig. S1A,B). Although the luminescent signal was again greatest at the periphery, there was detectable signal within the centre of the tissue, indicating adequate diffusion of luciferin substrate. In an adjacent 400 μm slice taken from the same pituitary, the lateral regions of the anterior lobes were dissected away to assess whether exposure of cells at the cut edge to the luciferin-containing medium resulted in an increase in luminescence (supplementary material Fig. S1C). Luminescence signal did not increase in these regions within a 10 hour period, indicating that limitation of luciferin or oxygen access was not responsible for the peripheral luminescence. These results therefore reflect genuine regional variations in activation of the PRL promoter within the anterior pituitary.
Ex vivo visualisation of temporal dynamics of prolactin expression in pituitary tissue using luciferase imaging
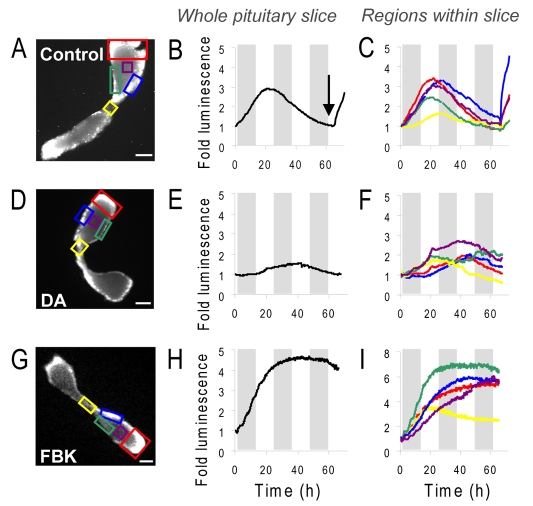
In order to assess temporal dynamics of promoter activity in living pituitary tissue, PRL-luciferase transgenic rat pituitary slices were subjected to long-term, real-time luminescence imaging for up to 72 hours. Pituitary slices were maintained in culture medium in a temperature-, CO2- and humidity-controlled imaging chamber for continuous microscopic luminescence imaging. In the absence of hormonal stimulation, there was a transient rise in luciferase expression in the pituitary tissue slices and then a decrease over 65 hours (Fig. 2A,B; supplementary material Fig. S2A). Different regions of the gland were compared to assess whether there were quantitative or qualitative differences in response (Fig. 2A,C). The absolute signal intensity and the amplitude of the change differed between central, lateral, dorsal and ventral regions of the gland, with the lowest signal and smallest amplitude responses seen in the central (midline) segments (yellow boxed area). The overall temporal pattern of reporter gene expression was closely similar within all the regions of the anterior pituitary lobe, with a rise in luminescence over 20-25 hours followed by a slow reduction in signal over the remaining 40-hour period. A slower rate of rise was detected in the central (midline) region (Fig. 2C). The slow decrease in luciferase activity occurring after 20-25 hours was not due to loss of luciferin substrate (supplementary material Fig. S1D). The viability of the tissue at the end of the time series was tested by addition of forskolin (FSK; an activator of cAMP signalling) at 65 hours, and a rise in luminescence signal was detected (Fig. 2B, arrow). As photon production by luciferase depends on the availability of intracellular ATP and oxygen, and an increase in luciferase expression requires active transcription and translation, this indicated maintenance of cellular viability and integrity.
Fig. 2.
Real-time luminescence imaging of PRL promoter-directed transcription in living transgenic pituitary tissue slices. (A,D,G) Luminescent images of 400 μm thick pituitary tissue slices, (B,E,H) graphs of luminescent photon flux from whole pituitary slices and (C,F,I) selected regions within the slices over approximately 3 days, with no treatment (control; A-C), 1 μM dopamine (D-F), or 5 μM forskolin and 0.5 μM BayK8644 (FBK; G,H,I). Arrow in B shows time of stimulation with 5 μM forskolin. Grey bars indicate 12-hour periods. Scale bars: 500 μm.
Dopamine acts in vivo to suppress PRL gene transcription (Elsholtz et al., 1991) and lactotroph proliferation through dopamine D2 receptors (Ben-Jonathan and Hnasko, 2001) and inhibits the secretion of PRL (Gonzalez-Iglesias et al., 2008) in all lactotroph subtypes in both male and female rats (Christian et al., 2007). We hypothesised that the transient increase in luciferase expression detected over the initial 25 hours of imaging in culture conditions resulted from the removal of the tissue from the in vivo tonic inhibition by dopamine. To test whether treatment with dopamine would inhibit the observed rise in luciferase activity, slices were treated with 1 μM dopamine within the culture medium. Under these conditions the transient rise in luminescence signal within the whole pituitary slice decreased (Fig. 2D,E; supplementary material Fig. S2A). Detailed analysis of regions within the pituitary slice showed that the basal luminescence signal remained suppressed for approximately 10-15 hours in the presence of dopamine before a rise was detected within some regions of the slice, potentially due to degradation of the dopamine in the medium (Fig. 2F). The pattern of signal varied across different regions, with the inhibitory effect being less pronounced within the central region (yellow boxed area; Fig. 2D,F), possibly reflecting differential effects of dopamine in different regions of the gland.
Treatment of pituitary tissue slices with the combined stimulus of forskolin and the calcium channel agonist BayK8644 [FBK; previously shown to induce a synergistic activation in pituitary gene expression (Szabo et al., 1990)] resulted in a prolonged rise in PRL promoter activity. The temporal pattern of response was uniform across the pituitary, but greater and more prolonged increases were observed in lateral, dorsal and ventral portions of the gland (Fig. 2G-I; supplementary material Fig. S2A). Indeed, in all three conditions tested (unstimulated, dopamine-treated and FBK-treated) the central region of the pituitary exhibited lower amplitude response patterns (Fig. 2, yellow traces). Similar patterns were seen in PRL transcription in all three conditions when pituitary slices from a female transgenic rat were imaged (supplementary material Fig. S2).
Ex vivo visualisation of prolactin promoter activity in dispersed pituitary cells in culture using luciferase imaging
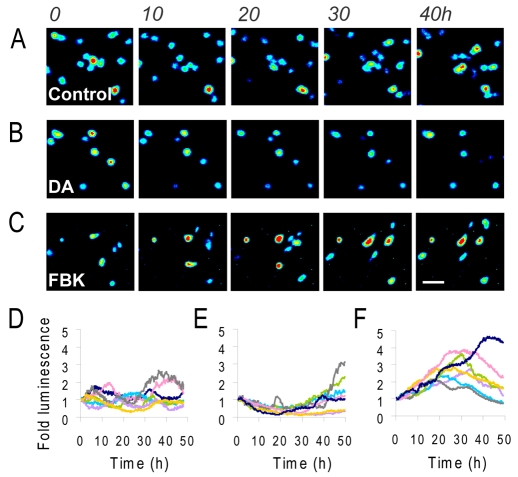
As regions within pituitary tissue slices showed relatively correlated patterns of PRL promoter activity in response to various stimuli, it was important to assess whether these transcriptional response patterns would be maintained when communication between neighbouring cells was disrupted. PRL-luciferase transgenic pituitary glands were subjected to enzymatic disaggregation to enable assessment of PRL transcription dynamics in dispersed lactotroph cells. In unstimulated primary cell cultures, dramatic fluctuations in promoter activity were observed in individual cells, with marked heterogeneity in timing between different cells and no apparent synchronisation (Fig. 3A,D). These data are in support of previous observations of heterogeneous PRL promoter activity in primary pituitary cells (Semprini et al., 2009; Shorte et al., 2002). In some cells, clear cycles in luciferase expression could be discerned, but there was no evidence of co-ordination amongst adjacent cells. These data suggested that the co-ordinated transcriptional response in intact pituitary slices was lost when the cells were disaggregated. Treatment with dopamine inhibited PRL promoter activity for around 20 hours, and then a rise in transcription was observed in some cells, although no clear pulses were observed over 50 hours of imaging (Fig. 3B,E). Stimulation with FBK induced a characteristic transient rise in PRL expression in most cells (Fig. 3C,F).
Fig. 3.
Real-time luminescence imaging of PRL transcription in living luminescent transgenic pituitary cells. (A-C) Representative luminescence images of individual cells dispersed from whole pituitary glands taken from luminescent transgenic rats with no treatment (control; A), 1 μM dopamine (B) or 5 μM forskolin and 0.5 μM BayK8644 (C). (D-F) Graphs of luminescent photon flux from individual cells with no treatment (control; D), 1 μM dopamine (E) or 5 μM forskolin and 0.5 μM BayK8644 (F). Each graph shows seven representative cell traces where luminescence signal per cell is normalised to the initial value per cell. Numbers above A show time in hours. Scale bar: 50 μm.
Patterns of prolactin transcriptional response in different cell areas, and in high- and low-copy transgenic lines
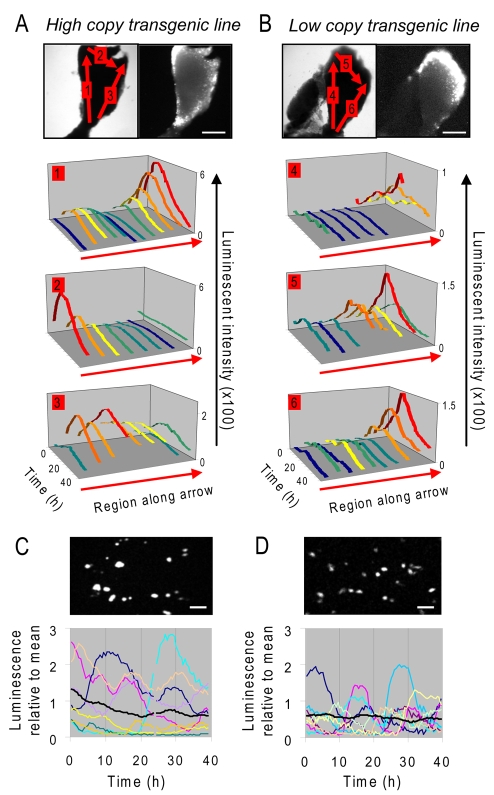
Analysis of the transcription dynamics from single lactotroph cells within living luciferase-transgenic pituitary tissue slices was difficult because of luminescent light scatter from adjacent cells detected by wide-field microscopy. To determine, instead, whether closely adjacent regions within the tissue showed co-ordinated PRL promoter activity, we compared two PRL-luciferase transgenic rat lines, one with a high copy number of the PRL-luciferase BAC transgene and one with a low transgene copy number [line 49 and line 37A respectively (Semprini et al., 2009)]. Small regions were analysed to assess possible co-ordination of transcriptional response on a sub-millimetre scale. The luminescence signal from ‘cell areas’ of approximately 50 μm (likely to contain groups of cells rather than individual cells) was acquired over periods of 50 hours. Cell areas within both high and low expressing transgenic lines showed similar patterns of PRL expression over time (Fig. 4A,B). It was noteworthy that the profiles from the cell areas within the low copy line were more ‘noisy’, suggesting that there was a smoothing effect within the high copy line when the expression from more transcription units was summed.
Fig. 4.
Regional expression of the time-course of PRL promoter-directed luciferase expression in unstimulated pituitary slices and dispersed cells from transgenic rats with differing expression levels. (A,B) Analysis of PRL expression in ‘cell areas’ (approximately 50 μm; represented by coloured lines on graphs) over a period of 50 hours. Comparison of responses in high copy (A) and low copy (B) 400 μm thick transgenic pituitary slices. The arrows on the images mark the orientation of the analysis. (C,D) Corresponding expression profiles from individual dispersed pituitary cells were analysed from the same high (C) and low (D) copy transgenic lines in unstimulated conditions. Each line represents an individual cell with the black lines representing the average response. Scale bars: 500 μm (A,B), 75 μm (C,D).
In both transgenic lines the lateral regions of the pituitary produced higher luciferase expression, although the intensity of signal was significantly lower in the low-copy line compared to the high-copy line (approximately one third of high-copy values; supplementary material Fig. S3). The temporal pattern of response was closely similar across the gland, with the largest amplitude changes seen in cells within the dorsal, ventral and lateral regions. Uncoordinated cyclical fluctuations in luciferase signal were observed in dispersed pituitary cells from both lines as before (Fig. 4C,D); although signal intensity was lower in the low-copy line (approximately 20% of high-copy values; supplementary material Fig. S3).
Transcriptional patterns in single living cells within intact tissue from EGFP-transgenic rats
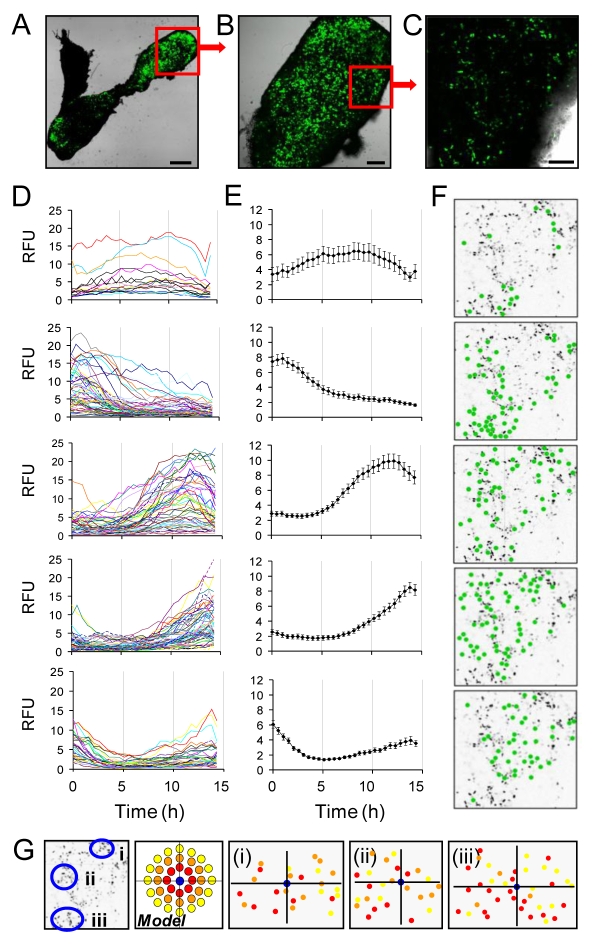
To achieve improved spatial resolution of the transcriptional responses of single cells within the tissue, pituitary tissue slices were taken from transgenic Fischer-344 rats expressing d2EGFP under the control of the hPRL locus [line 455; (Semprini et al., 2009)] and were visualised using confocal microscopy. Images of intact pituitary slices using a low magnification (2.5×) objective showed clear anterior-pituitary-specific transgene expression, with higher signal in the lateral aspects of the gland, in agreement with the pattern seen with the luciferase reporter (Fig. 1A and Fig. 5A). Higher magnification objectives enabled detection of single cells expressing the PRL-d2EGFP transgene within the pituitary tissue (10×, 20×; Fig. 5B,C). These cells were present throughout the tissue, although as with luciferase-transgenic rats, there was increased signal around the lateral aspects of the gland, which might reflect both a greater cell density and signal per cell.
Fig. 5.
Visualisation of the dynamics of PRL promoter-directed d2EGFP expression in single lactotroph cells within living transgenic pituitary tissue by confocal microscopy. (A) Whole fluorescent transgenic pituitary slice (2.5× objective; scale bar: 500 μm). Fluorescent (green) signal indicates PRL promoter expression. (B) Higher magnification of boxed region in A reveals single cell resolution (10× objective; scale bar: 150 μm). (C) A small region within the pituitary slice (boxed region in B) was imaged sequentially over 15 hours (20× objective; scale bar: 75 μm). (D) Single cell transcriptional responses from cells within the section of pituitary in C clustered into five groups based on transcriptional profile (data taken from 239 cells with the groups containing 18, 61, 57, 64 and 39 cells, respectively). (E) Average transcriptional response for each of the five groups. (F) Location of the cells showing each profile within the pituitary slice (cells shown by green dots). (G) Three cells were selected within the slice and the correlation was compared between the transcriptional profile of the chosen cells and the cells surrounding it (red denotes a correlation coefficient of 0.75-1.0; orange, 0.5-0.75; yellow <0.5).
Confocal fluorescence and transmitted light images from small regions of pituitary tissue were acquired every 15 minutes for 15 hours in order to assess the transcriptional behaviour of closely adjacent cells over time. It was possible to group cells into five distinct patterns of transcriptional response over the 15 hours of observation, as illustrated in Fig. 5D,E. This raised the question as to whether local, intercellular network or paracrine interactions might be enabling short-range co-ordination of temporal patterns of transcriptional activity between cells. However, analysis of these specific patterns of cell transcriptional response showed that they could not be attributed to adjacent groups of cells (Fig. 5F), but rather that these statistically definable ensembles of cells were dispersed across regions of 100-200 μm. To test whether co-ordinated responses were detected between adjacent cells within small cell clusters (as suggested in the model in Fig. 5G, where correlation between cells would be lost as the distance increased), correlation analysis was used to compare patterns seen in index cells with those of their neighbours. This indicated that there was no clear relationship between the responses of immediately adjacent cells (Fig. 5G).
Discussion
Gene expression is highly dynamic in living cells and stochastic regulation of expression is likely to be an important determinant of tissue phenotype (Raj and van Oudenaarden, 2008). Until now, most work on the dynamics of gene expression has relied on model systems using isolated cells in culture, and it has been unclear to what extent the architecture of living intact tissues could modify the spatial and temporal patterning of gene transcription. In this work we have used novel approaches that give insight into the spatiotemporal organisation of transcriptional behaviour in living cells in the context of tissue structure. Using reporter genes with a short half-life, such as luciferase and destabilised EGFP, quantitative measurements of transcription rate can be made in real time over prolonged periods by luminescence or fluorescence microscopy, but data so far have been limited to work on cell lines and dispersed cell cultures. In this report we describe, for the first time, patterns of transcription in living cells in intact tissue from transgenic rats. Our data show that individual cells within intact pituitary tissue display a variety of patterns of transcriptional behaviour over time, with long-range similarity across distant regions of the gland, but a variety of short-term patterns among neighbouring cells. Detailed analysis of these temporal patterns indicates that pituitary tissue can be viewed as a series of cell ensembles, in which individual cells display diverse short-term patterns of stochastic behaviour, but which together yield long-range and long-term coordinated behaviour.
The pituitary gland contains a series of different endocrine cell types, differentiated to produce specific hormones, such as PRL, GH and gonadotrophins. These cells are not homogeneously distributed across the gland, and different cell types show different regional distribution (Bonnefont et al., 2005; Fauquier et al., 2001; Nakane, 1970). An increasing body of evidence suggests that structural features of the pituitary exert an important influence on gland function, and for example the effects of paracrine and juxtacrine signalling have been well documented between hormonal and folliculostellate cells (Bonnefont et al., 2005; Denef, 2008; Renner et al., 1997). The relative proportion of lactotroph cells in our dispersed cell preparations was estimated prior to culture by flow cytometry using immunocytochemical analysis (26% of cells; mean of three animals) and within tissue slices by immunohistochemistry (31%; mean of three animals; data not shown). This was similar to previously published cell numbers (Phelps, 1986), and indicates that there is unlikely to be a major difference in relative proportions of cell populations in our preparations. Previous work has indicated, however, that even for specific cell types within the pituitary gland, cells might be functionally sub-specialised in different regions of the tissue. An example is GH, where there is evidence of cell networks linked with focal adherens junctions in transgenic mouse pituitaries (Bonnefont et al., 2005). Prolactin-secreting cells display different secretory responses to both dopamine inhibition and TRH stimulation, according to whether they reside in the central or peripheral zones of the gland (Boockfor and Frawley, 1987). Among lactotroph cells, electron microscopic analysis has shown that different subclasses of cells might be distinguished by size and appearance of secretory granules (Nogami and Yoshimura, 1982), and type I and type II lactotroph cells have been shown to respond differently to the acute effects of oestrogen (Christian and Morris, 2002). The mechanism behind this sub-specialisation is not known, nor whether cells located in different regions of the tissue are fixed in position and function, and whether the sub-differentiation is permanent or reversible. Our data show that lactotroph cells in different regions display different patterns and amplitudes of transcriptional response, but that there is some co-ordination of overall response across the whole tissue. The co-ordination within the tissue was reduced when treated with dopamine, consistent with the results of previous studies where lactotrophs were found to be differentially responsive to dopamine (Boockfor and Frawley, 1987; Kineman et al., 1994).
Both the reporter genes we have used to assess transcriptional regulation have specific advantages. Luciferase is highly sensitive, and low level expression can be detected using luminescence imaging equipment. The short half-life of luciferase enzyme activity enables faithful reporting of rapid dynamic changes in transcription rate. The d2EGFP reporter is destabilised to enhance its ability to report dynamic changes but it still has a longer half-life, estimated at 120 minutes. d2EGFP offers better spatial resolution for analysis of single cells within tissue slices and is amenable to both confocal and two-photon microscopy; high levels of luciferase expression can result in some light scatter amongst adjacent cells, which can preclude single cell analysis if the cells are closely packed, as in the intact adult pituitary gland. Our data reveal interesting new information about the nature of co-ordination of transcriptional patterns across the intact tissue. Luciferase expression was studied over long periods (up to 70 hours), comparing a series of 50 μm regions, each of which is likely to contain ten or more cells. Comparison of these groups of cells revealed similar response patterning across distant regions of the whole gland, albeit to varying intensities. Increased noise was detected when analysing the low copy transgenic line, because of a reduced signal (and therefore reduced light scatter) or a reduction in the number of active promoters within the area of interest. The quantification of transcriptional responses within single cells in living tissue was enabled using the d2EGFP reporter gene, where individual cells could be identified and distinguished from their neighbours, although imaging studies could only be conducted over shorter periods of 12-15 hours. The EGFP data showed that neighbouring cells could display different transcriptional patterns, with evidence for non-co-ordinated cycles. Thus small regions of cells would contain ensembles of cells that each behaved independently but whose summed behaviour, over longer time periods, was similar to that of far-distant cell ensembles. Thus we propose that in the pituitary gland, individual cells behave in a stochastic manner, not only when studied in dispersed primary cell cultures, but even when immediate inter-cellular architectural relationships are maintained. However, these stochastic patterns sum together to generate overall long-range response patterns that are similar in distant regions across distances of several millimetres, equivalent to many thousands of cell diameters apart.
In summary, we have used transgenic rat models to analyse the spatiotemporal patterns of gene expression in relation to tissue structure. The findings suggest that individual cells might display widely varying patterns of transcriptional response over short periods, and that different regions of the intact pituitary respond with different amplitudes to environmental stimuli. Whereas dispersion of cells in a primary culture allows cells to exhibit independent cycles of promoter activity, the context of tissue structure appears, at least partially, to constrain this behaviour. The transgenic reporter approach gives novel insights into the timing of gene expression in vivo, and also gives the opportunity to understand the effects of structural alterations in the pituitary. It will be important to understand how these patterns are established in the foetal pituitary, as the lactotroph cell network becomes established, and it will also become possible to study the nature of transcriptional patterning as the pituitary undergoes remodelling, for example during oestrogen-induced hyperplasia and adenoma formation.
Materials and Methods
Animals
For this study male transgenic rats expressing hPRL-luciferase and hPRL-d2EGFP were used; the characteristics of these were described previously [lines 49, 37A and 455 (Semprini et al., 2009)]. Animal studies were undertaken under UK Home Office License following review by the local ethics committee. All rats were given free access to water and Beekay rat chow (Special Diet Service, Witham, UK), and maintained under controlled conditions of temperature (21±1°C) and humidity (50±10%), under a 12-hour light:dark cycle.
Generation of pituitary tissue slice preparations and primary cultures
Pituitaries were resected from transgenic rats that were injected with luciferin intraperitoneally (150 mg/ml in physiological saline, 50 mg/kg) 30 minutes prior to death. Pituitaries were washed in PBS, suspended in low-melting point agarose and sliced to 400 μm thickness in the coronal orientation using a vibrating microtome (Campden Instruments). Pituitary slices were transferred to culture medium (DMEM + 4.5 g/l glucose, 10% foetal bovine serum, sodium pyruvate, penicillin-streptomycin and ultraglutamine) supplemented with 1 mM luciferin prior to imaging for luminescent tissue or unsupplemented for fluorescent tissue slices. For primary cultures, pituitaries were washed in PBS and then disaggregated in collagenase (2.5 mg/ml; w/v), DNase (0.4 mg/ml; w/v) and BSA (0.75%; w/v) for 30 minutes at 37°C. Cells were passed through a 70 μm cell strainer (BD Biosciences) and washed in PBS with centrifugation at 200 g for 5 minutes. Cells were resuspended in culture medium (DMEM + 4.5 g/l glucose, sodium pyruvate, penicillin/streptomycin and ultraglutamine) and left to recover for 72 hours before imaging.
Real-time imaging of luminescent pituitary tissue slices
Luminescent pituitary slices (400 μm thick) were immediately transferred to 35-mm glass-coverslip-based dishes (Iwaki), containing medium supplemented with 10% foetal calf serum and 1 mM luciferin (BioSynth). A sterilised culture plate insert (Millicell) was placed above the slice to gently reduce movement during imaging. The dish was transferred to the stage of a Carl Zeiss Axiovert 100M microscope in a dark room and maintained at 37°C in a Zeiss incubator with 5% CO2, 95% air. Bright-field images were taken before and after luminescence imaging. Luminescence images were obtained using either a Fluar 2.5× 0.12 NA objective and captured using a photon-counting charge-coupled device camera (Orca II; Hamamatsu Photonics) or a Fluar 5× 0.25 NA objective using a photon-counting Hamamatsu 2-stage VIM intensified camera. Where described, forskolin (5 μM) and BayK 8644 (0.5 μM) or dopamine (1 μM) were added directly to the dish. Sequential images (each integrated over 15-30 minutes, dependent upon the light intensity produced by the slice), were captured and analysed using Kinetic Imaging software AQM6 (Andor, Belfast, UK). Photon counts within the whole slice or specific regions of interest were obtained by measuring the mean intensity per pixel after subtracting the average instrument noise.
Real-time imaging of luminescent primary pituitary cultures
Collagenase-type-I-dispersed pituitaries (5×106 cells) were seeded onto 35 mm glass-coverslip-based dishes (Iwaki) and cultured in culture medium (see above) for 3 days. After 2 days the medium was replaced with fresh medium containing 1 mM luciferin. The cells were transferred to the incubator on the stage of a Carl Zeiss Axiovert 100M microscope, as described above. The cells were imaged using a Fluar 10× 0.5 NA objective. Where described, forskolin (5 μM) and BayK 8644 (0.5 μM) or dopamine (1 μM) were added directly to the dish. Sequential images were captured using 4×4 binning on an Orca II camera (Hamamatsu Photonics) with an exposure time of 30 minutes.
Real-time confocal imaging of fluorescent pituitary tissue slices
Fluorescent pituitary slices, 400 μm thick, were transferred to 35 mm glass-coverslip-based dishes (Iwaki), containing culture medium and imaged using a Carl Zeiss LSM510 Meta microscope equipped with an XL incubator (maintained at 37°C, 5% CO2, in humid conditions). Fluorescent images were taken using either a Fluar 2.5× 0.12 NA or a 10× 0.5 NA air objective (Carl Zeiss). Time-course confocal images were taken using a 20× 0.5 NA objective with sequential images captured every 15 minutes using timeseries software [AutoTimeserie v.1.19 LSM32 (Rabut and Ellenberg, 2004)] with autofocus.
Analysis of real-time imaging data
Analysis of luminescent tissue slices and dispersed cells was carried out using Kinetic Imaging software AQM6. Regions of interest of the tissue slice or single dispersed cells were drawn around and mean intensity data was collected. Average instrument dark count (corrected for the number of pixels being used) was subtracted from the luminescence signal. Timecourse data from fluorescent pituitary slices was analysed using Cell Tracker v0.6 software (Shen et al., 2006).
Immunofluorescence
Pituitaries were washed in PBS and fixed in 4% paraformaldehyde for 2 hours. They were washed in PBS and then transferred to 70% ethanol before being processed and embedded in wax. Pituitary sections (5 μm) were cut and mounted onto slides with incubation at 37°C for 48 hours. Sections were dewaxed and rehydrated prior to antigen retrieval in 0.01 M sodium citrate, pH 6.5 for 20 minutes. Slides were blocked for 1 hour at room temperature in 20% normal donkey serum in PBS containing 5% BSA. Slides were incubated with rabbit anti-ovine prolactin (PRL; MCNR51) used at 1:4000 (Invitrogen/Zymed) in blocking buffer for 18 hours at 4°C in a humidity chamber. PRL was detected using anti-rabbit Alexa Fluor 546 (1:200). Coverslips were mounted on the slides with Permafluor fluorescent mounting medium.
Supplementary Material
Acknowledgments
We thank N. Halliwell and A. Platt-Higgins for assistance with the work. A. Adamson, A. McNamara, R. Awais and J. Boyd advised on the manuscript. This work was funded by a Wellcome Trust Programme Grant (#67252). C.V.H. was funded by The Professor John Glover Memorial Postdoctoral Fellowship. S.S. was funded by a Wellcome Trust Intermediate Fellowship and a Research Excellence Award. A.S.M. and J.R.M. are supported by MRC Human Reproductive Sciences Unit funding G002.00007.01. J.J.M. is a Wellcome Trust Principal Research Fellow. We also acknowledge support from the Wellcome Trust Functional Genomics Initiative. The Centre for Cell Imaging has been supported through BBSRC REI grant BBE0129651. Hamamatsu Photonics and Carl Zeiss Limited provided technical support. Deposited in PMC for release after 6 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/3/424/DC1
References
- Ben-Jonathan N., Hnasko R. (2001). Dopamine as a prolactin (PRL) inhibitor. Endocr. Rev. 22, 724-763 [DOI] [PubMed] [Google Scholar]
- Bonnefont X., Lacampagne A., Sanchez-Hormigo A., Fino E., Creff A., Mathieu M. N., Smallwood S., Carmignac D., Fontanaud P., Travo P., et al. (2005). Revealing the large-scale network organization of growth hormone-secreting cells. Proc. Natl. Acad. Sci. USA 102, 16880-16885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boockfor F. R., Frawley L. S. (1987). Functional variations among prolactin cells from different pituitary regions. Endocrinology 120, 874-879 [DOI] [PubMed] [Google Scholar]
- Christian H. C., Morris J. F. (2002). Rapid actions of 17beta-oestradiol on a subset of lactotrophs in the rat pituitary. J. Physiol. 539, 557-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian H. C., Chapman L. P., Morris J. F. (2007). Thyrotrophin-releasing hormone, vasoactive intestinal peptide, prolactin-releasing peptide and dopamine regulation of prolactin secretion by different lactotroph morphological subtypes in the rat. J. Neuroendocrinol. 19, 605-613 [DOI] [PubMed] [Google Scholar]
- Denef C. (2008). Paracrinicity: the story of 30 years of cellular pituitary crosstalk. J. Neuroendocrinol. 20, 1-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsholtz H. P., Lew A. M., Albert P. R., Sundmark V. C. (1991). Inhibitory control of prolactin and Pit-1 gene promoters by dopamine. Dual signaling pathways required for D2 receptor-regulated expression of the prolactin gene. J. Biol. Chem. 266, 22919-22925 [PubMed] [Google Scholar]
- Fauquier T., Guerineau N. C., McKinney R. A., Bauer K., Mollard P. (2001). Folliculostellate cell network: a route for long-distance communication in the anterior pituitary. Proc. Natl. Acad. Sci. USA 98, 8891-8896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen S., Harper C. V., Semprini S., Wilding M., Adamson A. D., Spiller D. G., Nelson G., Mullins J. J., White M. R., Davis J. R. (2006). Tumor necrosis factor-alpha activates the human prolactin gene promoter via nuclear factor-kappaB signaling. Endocrinology 147, 773-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Iglesias A. E., Murano T., Li S., Tomic M., Stojilkovic S. S. (2008). Dopamine inhibits basal prolactin release in pituitary lactotrophs through pertussis toxin-sensitive and -insensitive signaling pathways. Endocrinology 149, 1470-1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kineman R. D., Gettys T. W., Frawley L. S. (1994). Paradoxical effects of dopamine (DA): Gi alpha 3 mediates DA inhibition of PRL release while masking its PRL-releasing activity. Endocrinology 135, 790-793 [DOI] [PubMed] [Google Scholar]
- McFerran D. W., Stirland J. A., Norris A. J., Khan R. A., Takasuka N., Seymour Z. C., Gill M. S., Robertson W. R., Loudon A. S., Davis J. R., et al. (2001). Persistent synchronized oscillations in prolactin gene promoter activity in living pituitary cells. Endocrinology 142, 3255-3260 [DOI] [PubMed] [Google Scholar]
- Nakane P. K. (1970). Classifications of anterior pituitary cell types with immunoenzyme histochemistry. J. Histochem. Cytochem. 18, 9-20 [DOI] [PubMed] [Google Scholar]
- Nogami H., Yoshimura F. (1982). Fine structural criteria of prolactin cells identified immunohistochemically in the male rat. Anat. Rec. 202, 261-274 [DOI] [PubMed] [Google Scholar]
- Nolan L. A., Levy A. (2009). The trophic effects of oestrogen on male rat anterior pituitary lactotrophs. J. Neuroendocrinol. 21, 457-464 [DOI] [PubMed] [Google Scholar]
- Norris A. J., Stirland J. A., McFerran D. W., Seymour Z. C., Spiller D. G., Loudon A. S., White M. R., Davis J. R. (2003). Dynamic patterns of growth hormone gene transcription in individual living pituitary cells. Mol. Endocrinol. 17, 193-202 [DOI] [PubMed] [Google Scholar]
- Phelps C. J. (1986). Immunocytochemical analysis of prolactin cells in the adult rat adenohypophysis: distribution and quantitation relative to sex and strain. Am. J. Anat. 176, 233-242 [DOI] [PubMed] [Google Scholar]
- Rabut G., Ellenberg J. (2004). Automatic real-time three-dimensional cell tracking by fluorescence microscopy. J. Microsc. 216, 131-137 [DOI] [PubMed] [Google Scholar]
- Raj A., van Oudenaarden A. (2008). Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner U., Gloddek J., Arzt E., Inoue K., Stalla G. K. (1997). Interleukin-6 is an autocrine growth factor for folliculostellate-like TtT/GF mouse pituitary tumor cells. Exp. Clin. Endocrinol. Diabetes 105, 345-352 [DOI] [PubMed] [Google Scholar]
- Semprini S., Friedrichsen S., Harper C. V., McNeilly J. R., Adamson A. D., Spiller D. G., Kotelevtseva N., Brooker G., Brownstein D. G., McNeilly A. S., et al. (2009). Real-time visualization of human prolactin alternate promoter usage in vivo using a double-transgenic rat model. Mol. Endocrinol. 23, 529-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Nelson G., Nelson D. E., Kennedy S., Spiller D. G., Griffiths T., Paton N., Oliver S. G., White M. R., Kell D. B. (2006). Automated tracking of gene expression in individual cells and cell compartments. J. R. Soc. Interface. 3, 787-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorte S. L., Leclerc G. M., Vazquez-Martinez R., Leaumont D. C., Faught W. J., Frawley L. S., Boockfor F. R. (2002). PRL gene expression in individual living mammotropes displays distinct functional pulses that oscillate in a noncircadian temporal pattern. Endocrinology 143, 1126-1133 [DOI] [PubMed] [Google Scholar]
- Stirland J. A., Seymour Z. C., Windeatt S., Norris A. J., Stanley P., Castro M. G., Loudon A. S., White M. R., Davis J. R. (2003). Real-time imaging of gene promoter activity using an adenoviral reporter construct demonstrates transcriptional dynamics in normal anterior pituitary cells. J. Endocrinol. 178, 61-69 [DOI] [PubMed] [Google Scholar]
- Szabo M., Staib N. E., Collins B. J., Cuttler L. (1990). Biphasic action of forskolin on growth hormone and prolactin secretion by rat anterior pituitary cells in vitro. Endocrinology 127, 1811-1877 [DOI] [PubMed] [Google Scholar]
- Takasuka N., White M. R., Wood C. D., Robertson W. R., Davis J. R. (1998). Dynamic changes in prolactin promoter activation in individual living lactotrophic cells. Endocrinology 139, 1361-1368 [DOI] [PubMed] [Google Scholar]
- Winters S. J., Moore J. P. (2007). Paracrine control of gonadotrophs. Semin. Reprod. Med. 25, 379-387 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.