Abstract
Targeting of proteins to their final destination is a prerequisite for living cells to maintain their homeostasis. Clathrin functions as a coat that forms transport carriers called clathrin-coated vesicles (CCVs) at the plasma membrane and post-Golgi compartments. In this study, we established an experimental system using Schneider S2 cells derived from the fruit fly, Drosophila melanogaster, as a model system to study the physiological roles of clathrin adaptors, and to dissect the processes of CCV formation. We found that a clathrin adaptor Drosophila GGA (dGGA), a homolog of mammalian GGA proteins, localizes to the trans-Golgi network (TGN) and is capable of recruiting clathrin from the cytosol onto TGN membranes. dGGA itself is recruited from the cytosol to the TGN in an ARF1 small GTPase (dARF79F)-dependent manner. dGGA recognizes the cytoplasmic acidic-cluster-dileucine (ACLL) sorting signal of Lerp (lysosomal enzyme receptor protein), a homolog of mammalian mannose 6-phosphate receptors. Moreover, both dGGA and another type of TGN-localized clathrin adaptor, AP-1 (adaptor protein-1 complex), are shown to be involved in the trafficking of Lerp from the TGN to endosomes and/or lysosomes. Taken together, our findings indicate that the protein-sorting machinery in fly cells is well conserved relative to that in mammals, enabling the use of fly cells to dissect CCV biogenesis and clathrin-dependent protein trafficking at the TGN of higher eukaryotes.
Keywords: AP-1, CIMPR, GGA, Lerp, Lysosome, Clathrin
Introduction
Most secretory proteins, integral membrane proteins and soluble proteins that function in the luminal space of organelles are synthesized in the endoplasmic reticulum (ER) and transported toward their final destinations through a succession of membrane-enclosed organelles. Movement of cargo proteins between these organelles is mediated by membranous structures termed ‘transport carriers’. Physical formation of transport carriers involves three major events: (i) concentration of the cargo proteins at exit sites of donor compartments, (ii) recruitment of ‘coat’ proteins from the cytoplasm to the exit sites, and (iii) budding and detachment of coated transport carriers containing the cargo proteins (Bonifacino and Lippincott-Schwartz, 2003; Hirst and Robinson, 1998).
The coat protein, clathrin, serves as a scaffold for formation of transport carriers at post-Golgi compartments such as the trans-Golgi network (TGN), plasma membrane and endosomes. Clathrin-associated proteins such as the heterotetrameric adaptor protein (AP) complexes and the monomeric Golgi-localized, γ-adaptin ear containing, ARF binding proteins (GGAs) function as ‘adaptors’, promoting both concentration of cargo and recruitment of clathrin at sites of transport-carrier formation (Robinson, 2004). AP-1 and GGAs, in particular, localize to the TGN and endosomes, and participate in protein trafficking between TGN and endosomes (Bonifacino and Lippincott-Schwartz, 2003; Robinson, 2004). However, their mechanisms of action and potentially cooperative functions remain to be fully elucidated.
The functions of the GGAs in protein sorting have been previously studied mainly using mammalian and yeast cells (Bonifacino, 2004). All the GGA proteins comprise: (i) a VHS domain that, in the mammalian GGAs, binds DXXLL-type, acidic-cluster-dileucine (ACLL) sorting signals (where D is aspartate, X is any amino acid, and L is leucine) (Puertollano et al., 2001a; Takatsu et al., 2001; Doray et al., 2002a); (ii) a GAT domain that interacts with class I ARF GTPases as well as Rabaptin-5, ubiquitin, TSG101 and PI4P (Puertollano et al., 2001b; Mattera et al., 2003; Mattera et al., 2004; Shiba et al., 2003; Puertollano et al., 2003; Wang et al., 2007); (iii) a largely unstructured hinge region that binds clathrin and, in some cases, other adaptors such as AP-1 (Puertollano et al., 2001a; Doray et al., 2002b); and (iv) a GAE domain that binds a cohort of accessory proteins having ΨG(P/D/E)(Ψ/L/M) motifs (where Ψ is an aromatic residue, G is glycine, P is proline, E is glutamate and M is methionine) (Hirst et al., 2001; Hirst et al., 2003; Lui et al., 2003; Mattera et al., 2003; Bonifacino, 2004; Ghosh and Kornfeld, 2004; Mardones et al., 2007).
The best-characterized cargos for mammalian GGAs are the cation-independent and cation-dependent mannose 6-phosphate receptors (CI- and CD-MPR). These type-I integral membrane proteins function as receptors for the sorting from the TGN to endosomes of lysosomal enzymes modified with mannose 6-phosphate groups (Ghosh et al., 2003a). Definitive demonstration of a physiological role of the GGAs in MPR sorting, however, has been hampered by the existence of three, potentially redundant, GGAs (GGA1-GGA3) in mammals. Indeed, RNAi of each individual GGA has been shown to cause mild impairment of lysosomal enzyme sorting (Mardones et al., 2007; Hida et al., 2007; Ghosh et al., 2003b). Combined RNAi of two or three GGAs has not resulted in more complete missorting, possibly due to the reduced efficiency of depletion of each GGA in the combination treatment.
In yeast, accumulating information on the physiological functions of yeast Gga proteins indicates that most of the fundamental physical properties such as cargo-binding, Arf and accessory protein interaction and clathrin binding, are conserved from yeast to mammals except that the ACLL Gga-binding motif in the cargo molecules is not conserved (Hirst et al., 2000; Dell'Angelica et al., 2000; Costaguta et al., 2001; Misra, 2002; Takatsu, 2002). Deletion of the two yeast Gga proteins causes defects in TGN-vacuole protein trafficking. In the Gga-deficient cells, missorting of vacuolar proteinases such as carboxypeptidase Y (CPY) to the extracellular space occurs due to a defect in the transport of the sorting receptor Vps10p that shuttles between the TGN and endosomes (Hirst et al., 2000; Dell'Angelica et al., 2000). The yeast Ggas are also involved in the Golgi-to-vacuole trafficking of ubiquitylated cargo molecules, such as the Gap1 and Fur4 permeases and of the ferrichrome transporter Arn1p, at the transport step of these membrane proteins into the internal vesicles of multivesicular bodies (MVB) (Scott et al., 2004; Deng et al., 2009; Lauwers et al., 2009).
In this study, we employed the Schneider S2 haemocytic cell line (hereafter referred to as S2) that is derived from the fruit fly Drosophila melanogaster in order to dissect the mechanisms of lysosomal protein trafficking. These fly cells offer several advantages over mammalian cells, the most important of which is the existence in their genome of single genes encoding GGA, each of the subunits of AP-1 complex, and Lerp (lysosomal enzyme receptor protein), an ortholog of mammalian CI-MPR (Boehm and Bonifacino, 2001; Dennes et al., 2005). Moreover, most mammalian proteins that are involved in cargo sorting and transport-carrier formation are also conserved in the fly genome, most often as single proteins (Boehm and Bonifacino, 2001), indicating that this organism possesses similar mechanisms of intracellular protein trafficking. Despite these advantages, molecular tools for the S2 cell line to study its TGN-endosome transport are not readily available. Therefore, we cloned Drosophila GGA (dGGA), the μ1 subunit of AP-1, Lerp and clathrin heavy chain (dCHC), and analyzed their function in S2 cells. The results allowed us to conclude that the Drosophila version of GGA functions in the recruitment of clathrin coats, and is involved in sorting of Lerp at the TGN. Moreover, these results establish S2 cells as a useful system for dissecting the precise function of each protein and for identifying novel factors that are involved in clathrin-dependent sorting at the TGN.
Results
Molecular characterization of Drosophila GGA
To examine the physiological roles of GGAs in vivo, we turned to Drosophila S2 cells, which express a single GGA (dGGA). Fig. 1 shows the sequence alignment between dGGA and the three human GGAs (hGGAs) that was generated with the ClustalW program (http://align.genome.jp/). Although the overall amino acid sequence identity between hGGAs and dGGA is relatively low (~26%), dGGA is also organized into VHS, GAT and GAE domains, as predicted by the BLASTP program and Conserved Domain Database (CDD) search service at NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).
Fig. 1.
Alignment of amino acid sequences of dGGA and three human GGAs. The deduced amino acid sequences of dGGA and hGGAs 1-3 were aligned using the ClustalW program. The regions with purple, yellow and blue underbars indicate VHS, GAT and GAE domains, respectively. Open boxes colored in purple, orange, red and blue indicate amino acid residues involved in the interaction of hGGA1 with ACLL motif, ARF1, ubiquitin and accessory molecules, respectively. The letters in red are putative clathrin-binding motifs and those in blue indicate putative internal dileucine motifs. The ‘*’, ‘:’ and ‘.’ characters indicate positions with fully, strongly and weakly conserved residues, respectively (more detailed information is available at http://align.genome.jp/clustalw/).
VHS domain
The VHS domain is the most conserved from dGGA to hGGAs. The VHS domain of dGGA (amino acids 8-145) showed approximately 60% similarity and 40% identity to that of hGGAs. Moreover, 11 out of the 12 amino acid residues in hGGA1 that are involved in the interaction with the ACLL motif (Shiba et al., 2002), are conserved in dGGA (Fig. 1, purple boxes).
GAT domain
The GAT domain of dGGA (amino acids 149-290) shows approximately 54% similarity and 27% identity with GAT of hGGAs. In the N-terminal region of the GAT domain, nine residues (Fig. 1, yellow boxes), which are required for the interaction of hGGAs with ARF1 (Shiba et al., 2003), are conserved in GAT of dGGA. As shown in Fig. 1, two out of the three amino acid residues in the C-terminal region of GAT (Fig. 1, red boxes) that are responsible for ubiquitin binding (Shiba et al., 2004) are also conserved in dGGA.
Hinge region
No significant similarity was observed in the hinge regions of dGGA and hGGAs. However, we found a sequence containing the ‘DLL’-type clathrin-binding box and its related ‘ELL’ sequence (Q323LLNELLGDLLIDGS337), and an internal ACLL motif-like sequence (V491DSIDDVPLLSD502) in this region of dGGA.
GAE (γ-ear) domain
The GAE domain of dGGA (amino acids 523-643) also shows significant conservation with approximately 54% similarity and 28% identity to the corresponding regions of hGGAs. However, the amino acid stretch that contains the basic amino acid cluster (blue box), which is required for the interaction with accessory molecules (Nogi et al., 2002; Miller et al., 2003; Kametaka et al., 2007), is not seen in the C-terminal region of the GAE domain of dGGA. These results show that each globular domain of dGGA and hGGAs is significantly conserved, and that some functional sites in the unstructured hinge region of hGGAs are also seen in dGGA, suggesting that dGGA is a structural counterpart of hGGAs in Drosophila.
Expression of HA-dGGA and EGFP-dGGA in S2 cells
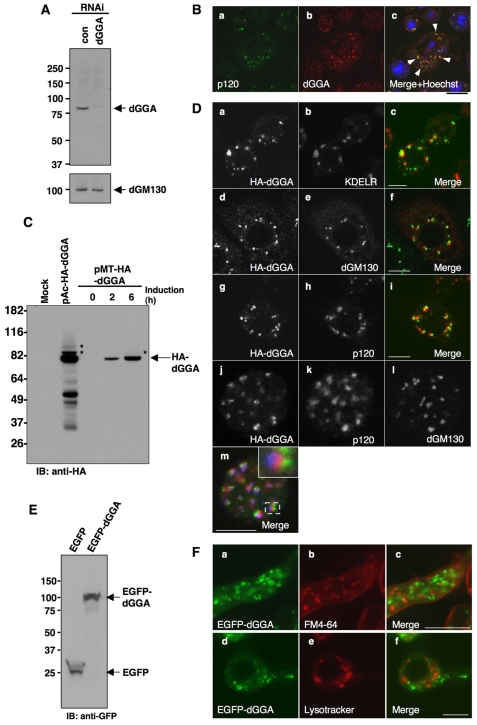
For detection of endogenous dGGA, we generated a specific antibody to dGGA. A protein of ~80 kDa was detected in S2 cell lysate with the antibody, and RNAi for dGGA caused loss of the signal, indicating that it specifically recognized the endogenous protein (Fig. 2A). To assess the cellular localization of dGGA in S2 cells, indirect immunofluorescence microscopy was performed. dGGA was mainly localized to large perinuclear puncta and fine dots scattered throughout the cytoplasm. The larger structures were adjacent to, but distinct from, the medial-Golgi compartments labeled by antibodies to p120 (Fig. 2B). For more precise analysis of the distribution of dGGA, an N-terminally HA-tagged dGGA was transiently expressed in S2 cells using the expression vector pAc5.1-HA-dGGA or the copper-inducible expression vector pMT-HA-dGGA. The tagged protein migrated as an ~80 kDa species with several degradation products on SDS-PAGE, as detected by immunoblotting (Fig. 2C). One or two higher molecular weight signals, which could be modified forms of dGGA, were also detected (Fig. 2C, asterisks). HA-dGGA was also localized to large perinuclear puncta and fine dots (Fig. 2B,D). The larger structures were adjacent to, but distinct from, the cis- and medial-Golgi compartments labeled by antibodies to the KDEL receptor (Fig. 2Da-c, cis), dGM130 (Fig. 2Dd-f, cis) and p120 (Fig. 2Dg-i, medial), respectively (Yano et al., 2005). Triple labeling of HA-GGA, dGM130 and p120 clearly revealed that HA-dGGA was always positioned at more proximal to p120 than to dGM130, indicating that HA-dGGA localizes to the trans side of the Golgi complex in S2 cells (Fig. 2Dj-m). To further characterize the dGGA-positive compartments, EGFP-tagged dGGA was expressed in S2 cells. The expression was confirmed by immunoblotting (Fig. 2E) and, as observed for HA-dGGA and endogenous dGGA, EGFP-dGGA appeared as large puncta and fine dots in the cytoplasm (Fig. 2F, Fig. 3D). The larger puncta were associated with the medial-Golgi marker, p120, by immunofluorescence microscopy (Fig. 3D). Time-lapse imaging of EGFP-dGGA showed that fine dots emerged from the Golgi-associated large EGFP-dGGA-positive structures, and that some of them showed long directional movement through the cytoplasm (see supplementary material Fig. S1 and Movie 1). To examine the relationship between these structures and endosomal and/or lysosomal structures, EGFP-dGGA was expressed in S2 cells and the membrane of endocytic compartments was labeled with the styryl dye FM4-64 or Lysotracker. The fluorescence of FM4-64 initially appeared at the plasma membrane (data not shown) and subsequently labeled intracellular endocytic organelles (Fig. 2Fa-c). The fine EGFP-dGGA-positive dots hardly colocalized with the FM4-64-positive endosomes or Lysotracker-positive lysosomes (Fig. 2F). These results suggest that the fine dots containing EGFP-dGGA are not endocytic compartments but might be transport intermediates derived from the trans-Golgi.
Fig. 2.
Localization of endogenous dGGA, HA-dGGA and EGFP-dGGA in S2 cells. (A) Detection of endogenous dGGA. S2 cell lysate prepared from untreated (con) and treated with siRNA for dGGA (dGGA) was subjected to immunoblotting with anti-dGGA antibody. dGM130 was used as a loading control. (B) S2 cells were co-stained with the anti-dGGA (Bb, red) and anti-p120 (Ba, green) antibodies. Arrows (Bc) indicate the dGGA foci associated with p120-positive Golgi compartment. Nuclei were counter-stained with Hoechst 33342 in Bc (blue). (C) HA-dGGA expressed in S2 cells. S2 cells were transiently transfected with pAc-HA-dGGA or pMT-HA-dGGA. HA-dGGA expression from the pMT-HA-dGGA was induced with 0.5 mM CuSO4 for the indicated time (hours). Total cell lysates prepared from these cells were subjected to immunoblotting with anti-HA. (D) Localization of HA-dGGA (Da,c,d,f,g,i,j,m: green in merged images) was compared with KDEL receptor (KDELR: ER and cis-Golgi marker) (Db and red in Dc); dGM130 (cis-Golgi marker) (De, red in Df,l and blue in Dm); and p120 (medial-Golgi marker) (Dh, red in Di,k and red in Dm). The typical set of triple-labeled stack of Golgi is shown in the inset of Dm. (E) EGFP-dGGA expressed in S2 cells. Lysate prepared from S2 cells transiently expressing EGFP or EGFP-dGGA was subjected to immunoblotting with anti-GFP antibody. (F) Colocalization analysis of EGFP-dGGA with endosomal and/or lysosomal markers. S2 cells stably expressing EGFP-dGGA (green) were stained with FM4-64 (Fa-c) for 30 minutes or Lysotracker (Fd-f) for 5 minutes at 25°C. Endosomal compartments labeled with FM4-64 (Fb,c), and Lysotracker-positive lysosomes (Fe,f) are shown in red. For A, C and E, numbers on the left indicate the positions of molecular mass markers (in kDa). Scale bars: 10 μm.
Fig. 3.
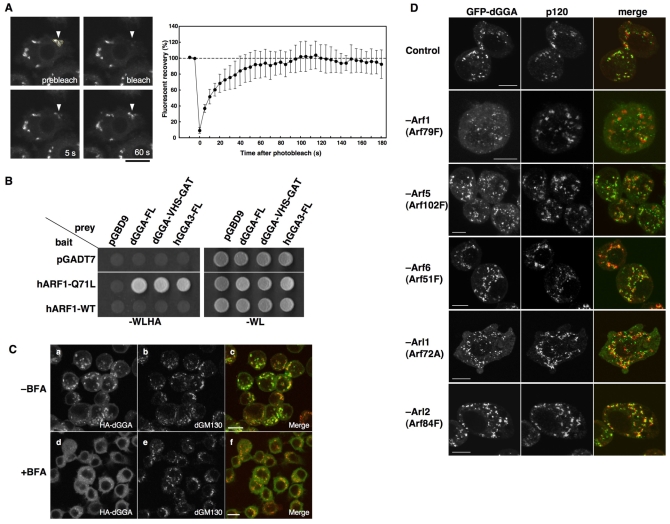
ARF-dependent Golgi localization of dGGA. (A) The kinetics of association of EGFP-dGGA with the Golgi were studied in stably transfected S2 cells by fluorescence recovery after photobleaching (FRAP, left panels). Quantification of the FRAP data from a set of three experiments is shown in the right panel. Values are the mean ± s.d. (B) Interaction between dGGA and human ARF1 was examined by a yeast two-hybrid experiment. (C) S2 cells stably expressing HA-dGGA were treated with 2 μg/ml BFA for 1 minute and co-stained with anti-HA (Ba,d, green in Bc,f) and anti-dGM130 (Bb,e, red in Bc,f) antibodies. Scale bars: 10 μm. (D) Effect of ARF protein depletion on EGFP-dGGA localization. S2 cells stably expressing EGFP-dGGA (left column) were mock-treated or treated with dsRNA for ARF79F, ARF102F, ARF51F, ARF72A and ARF84F. They were stained with anti-p120 antibody (middle column). Merged images are shown in the right column (green, EGFP-dGGA; red, p120). Scale bars: 5 μm.
ARF-dependent recruitment of dGGA onto the Golgi complex
In mammals, class I ARF small GTPases (ARF1 and ARF3 in human) cycle between Golgi membranes and cytosol in a process that is controlled by ARF-GAP and ARF-GEF activities (Nie et al., 2003). Because the association of GGAs with the Golgi complex mainly depends on the interaction with the membrane-bound form of class I ARFs, GGAs also cycle rapidly between the cytosol and Golgi membranes (Puertollano et al., 2001a; Shiba et al., 2003; Kametaka et al., 2005). To examine the dynamics of dGGA association with Golgi membranes, EGFP-dGGA expressed in S2 cells was analyzed by live-cell imaging combined with fluorescence recovery after photobleaching (FRAP). The Golgi-associated EGFP-dGGA showed quick recovery after photobleaching (t1/2=12 seconds, Fig. 3A and supplementary material Movie 2). Whereas the GTP-bound ‘active’ forms of ARFs are associated to the Golgi complex, GDP-bound ‘inactive’ forms are found in the cytosol. To examine whether dGGA is able to bind to active forms of ARFs, we performed yeast two-hybrid experiments. As shown in Fig. 3B, dGGA specifically interacts with the GTP-bound Q71L mutant, but not with wild-type or GDP-bound T31N mutant (data not shown) of human ARF1. Drosophila has a single class I ARF encoded by the Arf79F gene, and the amino acid sequence of ARF79F is highly homologous to that of human ARF1 [173 out of 181 (96%) identity; 178 out of 181 (98%) similarity in amino acid sequence], making it likely that dGGA also interacts with the membrane-bound, GTP form of ARF79F. Moreover, treatment with Brefeldin A (BFA), a potent inhibitor of the GTP-exchange factors (GEFs) for class I ARFs, caused quick dissociation of dGGA from the Golgi complex into the cytoplasm without affecting the dGM130-positive Golgi architecture (Fig. 3C), supporting a role of ARF79F in the association of dGGA with the Golgi complex. In the Drosophila genome, three ARF genes (Arf79F, class I; Arf102F, class II; and Arf51F, class III) and two ARF-related (ARL) genes (Arf72A, also known as Arl1; and Arf84F, also known as Arl2) are encoded. Therefore, we further assessed all the ARF and ARF-related proteins for their contribution to the dGGA recruitment onto the Golgi membrane using RNAi. Knocking down of each ARF or ARL genes was carried out in the EGFP-dGGA-expressing S2 cell line, and the localization of EGFP-dGGA was examined. Because dsRNA treatment for 4 days caused drastic redistribution of p120 or complete loss of p120 staining due to cell death (data not shown), we analyzed cells 40 hours after the beginning of the dsRNA treatment, a time when the p120 staining pattern is unaffected. As shown in Fig. 3D, depletion of Arf79F showed significant dissociation of EGFP-dGGA from the Golgi. Although depletion of Arf102F or Arf72A also slightly increase diffuse cytosolic signal of EGFP-dGGA, most of the punctate structures were still consistently associated with p120. Taken together, these results indicated that dGGA cycles between the Golgi and cytosol in an ARF79F-ARF1-regulated manner.
Clathrin interacts with the DLL sequence in the hinge region of dGGA
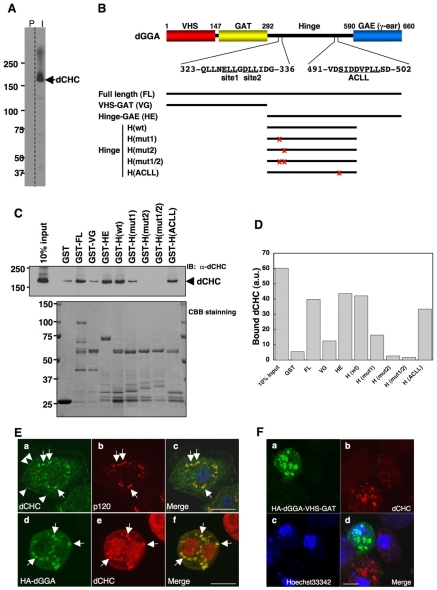
The trans-Golgi localization of dGGA suggested that this protein plays a role in the formation of clathrin-coated transport intermediates that transport integral membrane cargo proteins from the TGN to endosomes. To assess the involvement of clathrin in this process, an antibody to dCHC was raised (Fig. 4A). To examine the contribution of dGGA to the recruitment of dCHC onto the trans-Golgi, we performed in vitro pull-down assays to see whether dGGA interacts with dCHC. As shown in Fig. 4, a series of dGGA recombinant proteins was prepared as GST-fusion proteins. The full-length dGGA (GST-FL), Hinge and GAE region (GST-HE), and Hinge region (GST-H: wild) efficiently pulled-down dCHC from a cytosolic extract from S2 cells, whereas VHS-GAT (GST-VG) pulled-down dCHC at levels close to the control (GST; Fig. 4C,D). Thus, the binding site maps to the hinge region (amino acid position 292-590 in dGGA). We next introduced mutations in two putative clathrin binding sequences, D331LL to DAA (site 2 in Fig. 4B) and/or E327LL to EAA (site 1 in Fig. 4B), and in a putative ACLL motif, D496VPLL to DVPAA (ACLL in Fig. 4B). We observed that mutations in the two putative clathrin-binding boxes strongly affected the interaction with dCHC, whereas the putative ACLL motif was dispensable for this interaction. These results indicate that the D331LL and E327LL sequences in the hinge region of dGGA have crucial roles in interaction with dCHC.
Fig. 4.
Clathrin binding of recombinant dGGA and effect of dGGA-VHS-GAT overexpression in vivo. (A) Generation of anti-dCHC antibody. Total lysate prepared from the S2 cells was subjected to immunoblotting with pre-immune (P) and anti-dCHC (I) sera. (B) Schematic representation of the domains of dGGA. Putative clathrin-binding motifs (site1 and site2) and internal ACLL motif are shown. Domains used for the GST-pulldown experiment are depicted as black bars. Mutation sites are indicated by red x. (C) Pulldown of dCHC with recombinant dGGA proteins. Total lysate prepared from the S2 cells was incubated with 25 μg of GST or GST-fusion proteins (lower panel, CBB stained) and binding proteins were pulled-down with the glutathione Sepharose. Bound proteins were subjected to immunoblotting with anti-dCHC antibody (upper panel). 10% of input was loaded as a positive control. (D) Quantification of bound dCHC to each GST-fusion protein. (E) Localization of dCHC at the trans-Golgi. S2 cells (Ea-c) were stained with anti-dCHC (Ea,c, green) and anti-p120 (Eb,c, red) antibodies. Also, S2 cells transiently expressing HA-dGGA (Ed-f) were stained with anti-HA (Ed,f, green) and anti-dCHC (Ee,f, red) antibodies. (F) S2 cells transiently expressing HA-dGGA-VHS-GAT construct were stained with anti-HA (Fa,d, green) and anti-dCHC (Fb,d, red) antibodies. Nuclear DNA was stained with 1 μg/ml Hoechst 33342 (Fc,d, blue). For A and C, numbers on the left indicate the positions of molecular mass markers (in kDa). Scale bars: 10 μm.
Golgi-association of clathrin heavy chain
We also examined the intracellular localization of dCHC in S2 cells (Fig. 4E). The antibody preferentially labeled large perinuclear puncta (Fig. 4E upper panels, arrows) and smaller dots beneath the cell surface (Fig. 4E upper panels, arrowheads). The dCHC-positive, perinuclear large structures were juxtaposed to p120-positive structures (Fig. 4E upper panels) and colocalized with the signal for HA-dGGA (Fig. 4E lower panels, arrows), indicating that they correspond to the trans-Golgi compartments. By contrast, the small peripheral dots colocalized with AP50, a μ2 subunit of AP-2 (data not shown), indicating that they represent the plasma-membrane-associated clathrin-coated pits. These results indicated that a significant amount of cellular clathrin localizes to the trans-Golgi, together with dGGA.
In mammals, expression of the VHS-GAT domains of mammalian GGAs has been previously shown to exert a dominant-effect on clathrin localization and MPR sorting at the TGN (Puertollano et al., 2001a). Thus, to further assess the role of dGGA vis-à-vis clathrin in vivo, we examined the effect of overexpression of dGGA-VHS-GAT on clathrin localization in S2 cells. As shown in Fig. 4F, overexpression of the truncated dGGA reduced dCHC association with the Golgi compartment. Together with the above GST-pull down experiments, these results strongly suggest that dGGA has a role in clathrin recruitment at the TGN.
In vitro reconstitution of ARF-dependent recruitment of clathrin onto cellular membranes
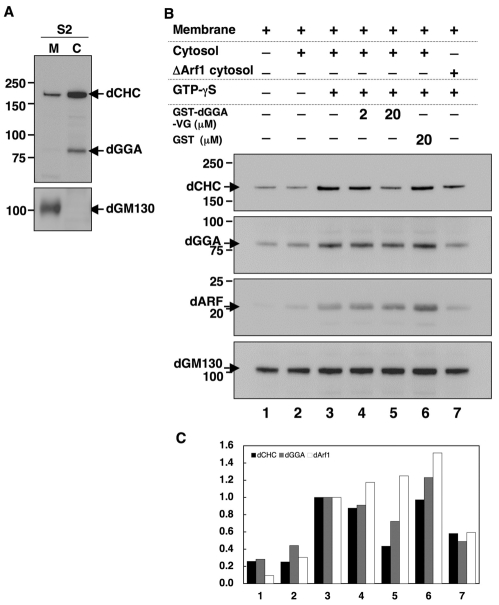
To further corroborate the role of dGGA as a clathrin adaptor, an in vitro membrane recruitment assay was performed. When cytosol and membrane fractions were prepared from S2 cells, most of the intracellular dCHC and dGGA proteins were present in the cytosolic fraction, probably due to the transient nature of their interaction with membranes (Fig. 5A) (Hirst et al., 2000; Kametaka et al., 2005) whereas dGM130, which is strongly associated with the Golgi, was fractionated with the membrane pellet under the same experimental conditions (Fig. 5A). The cytosolic dCHC was eluted in high molecular weight fractions in gel filtration analysis (supplementary material Fig. S2D), suggesting that the cytosolic dCHC form a large protein complex, most probably triskelions. The total membrane fraction isolated from S2 cells was incubated with S2 cytosol together with an ATP-regeneration system, in the presence or absence of GTP-γS, a non-hydrolysable analog of GTP. After incubation for 30 minutes at 25°C, the membrane was sedimented by centrifugation and the membrane-bound protein was analyzed by immunoblotting. In this experiment, anti-human ARF1 antibody was used to probe Drosophila ARFs in the immunoblot analysis. This antibody recognized Drosophila ARF79F, a fly ortholog of mammalian class I ARFs, as well as human ARF1 (supplementary material Fig. S3). Its specificity was also confirmed by the dramatic decrease upon RNAi treatment for Drosophila ARF79F in S2 cells (Fig. 7). As shown in Fig. 5B, larger amounts of dCHC and dGGA, as well as ARF79F, were recruited from the cytosol to membranes in the presence of GTP-γS than in its absence, suggesting that intracellular GTPases are involved in the recruitment of these soluble factors onto the membrane. When cytosol was prepared from ARF79F-depleted cells, the recruitment of dGGA and dCHC was decreased by approximately 50% (Fig. 5B, lanes 3 and 7), suggesting that the class I ARF is involved in that reaction in vitro as well as in vivo (Fig. 3C). Next, to determine whether dGGA is responsible for the recruitment of dCHC onto the membrane, GST-dGGA-VHS-GAT (GST-dGGA-VG) was added in the reaction mixture. Twenty μM of GST-dGGA-VG significantly decreased the recruitment of dGGA and dCHC (Fig. 5B, lanes 3 and 5), whereas GST itself had no inhibitory effect (Fig. 5B, lane 6). These results indicate that membrane-anchored, active ARF is required for dGGA recruitment onto the membrane, which is then a prerequisite for further recruitment of clathrin.
Fig. 5.
In vitro reconstitution of clathrin recruitment onto isolated membrane. (A) Subcellular fractionation of dGGA, dCHC and dGM130. The PNS fraction prepared from S2 was separated to membrane (M) and cytosol (C) fractions by ultracentrifugation. Each fraction was subjected to immunoblotting for dCHC and dGGA (upper panel), or dGM130 (lower panel). (B) Isolated membrane was incubated with the cytosol fractions prepared from S2 (lanes 2-6) or from S2 cells depleted of ARF79F (lane 7), plus ATP-regeneration system in the presence or absence of 30 μM of GTP-γS, or with GST or GST-dGGA-VHS-GAT (GST-dGGA-VG) at the indicated concentration. After incubation, membranes in the reaction mixture were precipitated with centrifugation at 20,000 g for 20 minutes and the membrane fractions subjected to immunoblotting with anti-dCHC, anti-dGGA, anti-hARF and anti-dGM130 antibodies. (C) Quantification of the membrane-bound proteins. For each protein, the ratio of the amount in each experimental condition to that in lane 3 was plotted.
Fig. 7.
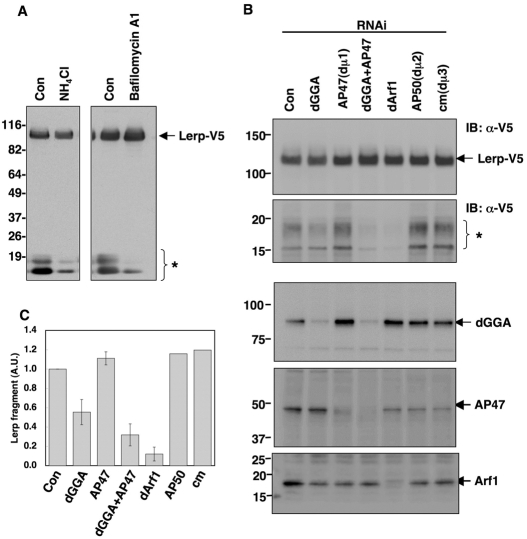
RNAi for dGGA causes delay in Lerp processing in vivo. (A) S2 cells transiently expressing Lerp-V5 were treated with 25 mM NH4Cl or 1 μM Bafilomycin A1 for 12 hours, and Lerp-V5 and its small fragments (asterisks) were detected with anti-V5 antibody. (B) S2 cells transiently expressing pMT-Lerp-V5 were treated with dsRNA for knocking down of each genes indicated for 4 days. After the dsRNA treatment, expression of Lerp-V5 was induced with 0.5 mM CuSO4 for 8 hours, and cellular proteins were subjected to immunoblotting with anti-V5 antibody. Top two panels show the upper and lower area of the same membrane blot, respectively. Numbers on the left indicate the positions of molecular mass markers (in kDa). Asterisks in A and B indicates the degradative fragments of Lerp-V5. (C) Quantification of the Lerp processing. Relative intensity of the Lerp fragments in the knocked-down cells to that of the negative control cells was plotted. The value indicate mean ± s.d. The data presented are representative of three independent experiments.
Lerp localizes to the trans-Golgi together with dGGA
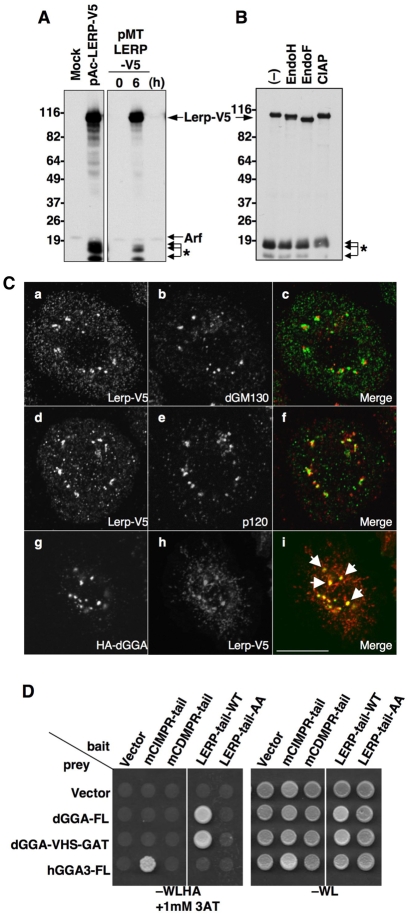
Recently, Lerp was identified as a Drosophila ortholog of the mammalian CI-MPR and was shown to function as a sorting receptor for lysosomal cathepsins when ectopically expressed in MPR-deficient mouse fibroblasts (Dennes et al., 2005). However, its properties and physiological function in Drosophila have not been elucidated. To determine the intracellular localization of Lerp in Drosophila, Lerp was expressed in S2 cells as a C-terminally V5-tagged construct (Lerp-V5, Fig. 6). Lerp-V5 migrated as an ~110 kDa protein on SDS-PAGE (Fig. 6A). Treatment with endoglycosidase H (EndoH) or PNGase (EndoF) increased the electrophoretic mobility of Lerp, indicating that this protein is glycosylated in vivo (Fig. 6B). In addition, C-terminal degradation products of Lerp-V5 ranging from 15 to 20 kDa were detected (Fig. 6A,B, asterisks). Immunofluorescence microscopy showed that, like dGGA, Lerp-V5 localized to large perinuclear puncta and fine cytoplasmic dots in S2 cells (Fig. 6C). Because the large puncta were adjacent to the signal for p120 or dGM130, and were well colocalized with HA-dGGA (Fig. 6Ci, arrows), we concluded that Lerp mainly localizes to the trans-Golgi.
Fig. 6.
Lerp is a Golgi-localized, glycosylated, transmembrane protein that interacts with dGGA. (A) C-terminal V5-tagged Lerp was expressed in S2 cells from a pAc-Lerp-V5 construct (left panel) or from a pMT-Lerp-V5 construct (right panel) induced with 0.5 mM CuSO4 for the indicated time (hour). The total cell lysates were subjected to immunoblotting with anti-V5 antibody. Asterisks indicate the degradation products of the Lerp-V5, as assessed in Fig. 7. (B) The S2 lysate containing Lerp-V5 was treated with buffer (−), endoglycosidase H (EndoH), PNGase (EndoF) or calf intestine alkaline phosphatase (CIAP), and the mobility shift on SDS-PAGE examined with immunoblotting. (C) Intracellular localization of Lerp-V5 was examined with anti-V5 antibody (green in merged images) together with dGM130 (Ca-c), p120 (Cd-f) and HA-dGGA (Cg-i) as cis-, medial- and trans-Golgi markers (red in merged images), respectively. (D) Molecular interaction between dGGA and the cytoplasmic region of Lerp was assessed with the yeast two-hybrid assay. Numbers on the left indicate the positions of molecular mass markers (in kDa). Scale bars: 10 μm.
dGGA interacts with Lerp through a C-terminal acidic-cluster-dileucine signal of Lerp
One of the salient features of mammalian GGAs is their ability to recognize the ACLL-type sorting signals present in cargo membrane proteins (Bonifacino, 2004). It was reported that the VHS domain of dGGA binds the Lerp cytoplasmic tail through the C-terminal ACLL signal in a pull-down experiment (Dennes et al., 2005). We confirmed the occurrence of this interaction using the yeast two-hybrid system (Fig. 6D). In addition, we showed that the interaction is eliminated by introducing mutations in the C-terminal hydrophobic residues to alanine (L885,886A, LERP-tail-AA in Fig. 6D). Furthermore, we found that the VHS-GAT region of dGGA was sufficient for the interaction. By contrast, we did not detect any interaction between dGGA and tails of two MPRs, or between hGGA3 and Lerp (Fig. 6D), suggesting that the binding mechanisms are slightly different between the species. These results indicate that dGGA specifically recognizes the ACLL signal at the Lerp C-terminus.
Involvement of dGGA in Lerp traffic in vivo
As mentioned above, C-terminal fragments ranging from 10 to 20 kDa appeared when Lerp-V5 was expressed in S2 cells (Fig. 6A and Fig. 7A). These fragments were most probably degradation products generated in endosomal and/or lysosomal compartments because treatment of the cells with ammonium chloride or bafilomycin A1 (both inhibitors of acidification) decreased the levels of these fragments (Fig. 7A). Because Lerp is a type-I integral membrane protein with a relatively short cytoplasmic tail of 40 amino acids, these fragments, which still possess the V5 epitope at the C-terminus, are most probably generated by processing of Lerp at its lumenal side. We took advantage of these fragments as indicators of post-Golgi trafficking of Lerp. To dissect the route by which Lerp is transported to the degradative compartments, S2 cells transiently transfected with the copper-inducible Lerp-V5 expression construct (Fig. 6A) were split and subjected to RNAi for dGGA, AP47 (μ1 subunit of AP-1 complex; see supplementary material Fig. S3), ARF79F (dARF1), AP50 (μ2 subunit of AP-2 complex), or carmine (μ3 subunit of AP-3 complex), under non-induction conditions. After dsRNA treatment, expression of Lerp-V5 was induced with 0.5 mM CuSO4 for 8 hours. We observed that depletion of dGGA reduced the levels of Lerp processing by approximate 50% (Fig. 7B,C). Moreover, to our surprise, knockdown of AP47 accentuated this inhibitory effect on Lerp processing caused by the depletion of dGGA, whereas depletion of AP47 itself had little effect on the processing (Fig. 7B,C). dsRNAs for AP50 and carmine, which had been successfully used in knockdown experiments (Chaudhuri et al., 2007), had no effect on the Lerp processing (Fig. 7B). In addition, depletion of ARF79F completely blocked Lerp processing (Fig. 7B), consistent with its role in the recruitment of dGGA and AP-1 complex to the TGN (Fig. 5B). This effect of ARF79F depletion, however, might not be completely specific due to its general function in maintenance of Golgi architecture, as described above.
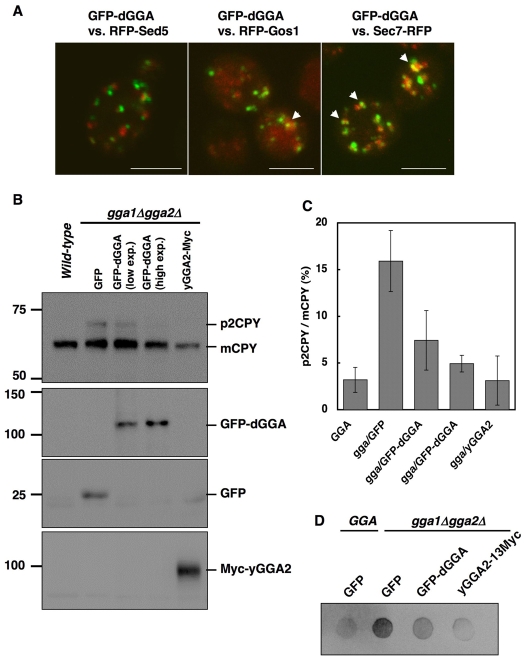
dGGA can suppress the defect in the vacuolar protein sorting of yeast gga1Δgga2Δ double disruptant
In yeast, Gga proteins have been well characterized and are known to play important roles in sorting of membrane proteins in the TGN- to-endosomes routes (Hirst et al., 2000; Dell'Angelica et al., 2000; Black and Pelham, 2000; Costaguta et al., 2001; Scott et al., 2004; Deng et al., 2009). The yeast mutant lacking both GGA genes is defective in both sorting of vacuolar soluble proteins such as CPY and processing of mating pheromones (Hirst et al., 2000; Dell'Angelica et al., 2000; Black and Pelham, 2000; Costaguta et al., 2001). To test whether dGGA is a functional homolog of yeast Gga proteins, we expressed EGFP-dGGA under control of a copper-inducible CUP1 promoter. First, we investigated the intracellular localization of dGGA in the wild-type cells. EGFP-dGGA exhibited a punctate Golgi pattern that was partially colocalized with RFP-Gos1 and Sec7-RFP, medial- and trans-Golgi markers, respectively, but not with RFP-Sed5, a cis-Golgi marker (Matsuura-Tokita et al., 2006; Costaguta et al., 2006) (Fig. 8A), suggesting that dGGA, like yeast Gga proteins (Fernandez and Payne, 2006), is localized on the medial- to trans-Golgi. Next, to see whether dGGA can suppress the CPY sorting defect in the gga1Δgga2Δ mutant, EGFP-dGGA was introduced into the gga deficient strain GPY2385 (Costaguta et al., 2001). GPY2385 transformed with p416Cu-GFP (GFP in Fig. 8B) vector showed p2CPY (Golgi form precursor of 69 kDa), whereas that transformed with pRS315-yGGA2-13Myc (yGGA2-Myc in Fig. 8B) as well as wild-type cells showed only mCPY (vacuolar mature form of 61 kDa). When EGFP-dGGA was expressed in GPY2385 from p416Cu-GFP-dGGA plasmid under non-inducing conditions (low expression), only a slight decrease in the ratio of p2CPY to mCPY was observed (Fig. 8B,C). Also, induction of EGFP-dGGA with copper sulfate further decreased the p2CPY:mCPY ratio (Fig. 8B,C). Consistent with this result, immunoblot analysis of secreted CPY showed the suppression of the CPY missorting phenotype by overexpression of EGFP-dGGA in GPY2385 (Fig. 8D). These results indicate that dGGA is a functional counterpart of yeast Gga proteins.
Fig. 8.
Functional complementation of the vps phenotype of gga-deficient yeast by overexpression of dGGA. (A) Distribution of EGFP-dGGA in the yeast. Wild-type (SEY6210) cells were co-transformed with EGFP-dGGA and mRFP-coupled Sed5 (cis-Golgi, left panel), Gos1 (medial-Golgi, center panel) or Sec7 (trans-Golgi, right panel) expression vectors. Confocal images from four slices with 1-μm interval in depth were projected for each panel. Colocalized foci are indicated with arrows. Scale bars: 5 μm. (B) Wild-type or gga1Δgga2Δ double disruptant (GPY2385) cells were transformed with plasmids for expression of EGFP, EGFP-dGGA or yeast GGA2-13Myc. EGFP-dGGA expression was also induced with 0.5 mM CuSO4 for 8 hours (high exp.). The transformants were harvested and subjected to immunoblotting with anti-CPY antibody. (C) Quantification of the CPY processing. The ratio of the intensity of p2CPY to that of mCPY was plotted. The value indicate mean ± s.d. (D) CPY secretion assay. Cells growing in mid-log phase were spotted onto a nitrocellulose membrane placed on a growth plate. After incubation overnight, cells were washed out and the membrane was subjected to immunoblotting for CPY.
Discussion
Drosophila as a novel system for dissecting the mechanisms of post-Golgi protein trafficking
Genome sequencing projects have revealed the conservation of clathrin adaptor genes throughout the eukaryotic kingdom, including well-established model organisms such as Drosophila melanogaster and Caenorhabditis elegans (Boehm and Bonifacino, 2001). Drosophila is a particularly appealing organism for analysis of GGA function because of the existence of a single dGGA, the ease of RNAi approaches and the conservation of Golgi function (Rabouille et al., 1999; Kondylis et al., 2001; Kondylis and Rabouille, 2003; Kondylis et al., 2005). Although the morphology of the Drosophila Golgi is different from that of mammals, the basic features of the organelle (e.g. polarity, number of cisternae per stack and function in the secretory pathway) are quite similar (Kondylis and Rabouille, 2003; Kondylis et al., 2005). The molecular machineries responsible for protein sorting from the Golgi complex to the endosomal system, however, have not been characterized in Drosophila. Dennes and coworkers showed that Lerp, a Drosophila homolog of the mammalian CI-MPR, could rescue the defects in the lysosomal cathepsin sorting in MPR-deficient fibroblasts (Dennes et al., 2005). However, the properties and functions of Lerp and its putative adaptor dGGA in Drosophila cells have remained uncharacterized to date. In this study we used Drosophila S2 cells to assess the role of dGGA and its regulators in the trafficking of Lerp.
dGGA appears to be a functional counterpart of mammalian GGAs
The results presented here indicate that dGGA meets the requirements to be a clathrin adaptor for membrane cargo molecules such as Lerp. Its ability to interact with GTP-ARF1, the ACLL motif of Lerp and the dCHC all imply that dGGA functions in CCV formation for trafficking of Lerp between the TGN and endosomal compartments. In addition to the molecular interaction between dGGA and Lerp in vitro, we also observed that mCherry-tagged Lerp and EGFP-dGGA depart together from the Golgi in vesicular structures in living cells (supplementary material Fig. S1 and Movie 1). Moreover, a dominant-negative form of dGGA caused redistribution of clathrin from the Golgi complex and also inhibited recruitment of clathrin in vitro. Furthermore, dGGA knockdown caused a decrease in the level of Lerp processing at endosomal and/or lysosomal compartments in vivo (Fig. 7). Thus, dGGA is likely to function at the exit step of Lerp from the TGN. Our assays, however, do not allow precise determination of dGGA function at a molecular level. More extensive biochemical and in vivo functional analyses will be required.
Both dGGA and dAP-1 contribute to Lerp trafficking
Despite the overlapping localization and common biochemical features of GGAs and AP-1, they are thought to function differently in mammalian cells. It has been shown that AP-1 and cargo molecules, but not GGAs, are concentrated in purified CCV fractions (Hirst et al., 2000). Moreover, knockdown of GGAs or AP-1 in mammalian cells only causes slight missorting of pro-cathepsin D to the extracellular space without detectable perturbation in the localization of its sorting receptors, MPRs. On the basis of these findings, it has been presumed that GGAs and AP-1 function cooperatively, but not at exactly the same step in cargo trafficking. Indeed, Kornfeld's group proposed a ‘hand-off’ model in which GGAs first prime the cargo molecule and AP-1 is subsequently recruited through direct interaction with GGAs (Bai et al., 2004). GGAs are thus replaced by AP-1 to execute clathrin-coated carrier formation (Ghosh et al., 2003b). In this model, GGA is displaced by AP-1 through a phosphorylation state-dependent structural change. Internal ACLL motifs in the hinge region of human GGA1 and GGA3 are thought to mediate this structural change (Doray et al., 2002c). Through the characterization of dGGA and Drosophila AP-1 (dAP-1) in the current study (Fig. S2), we found that many structural and functional features of these adaptors were strikingly conserved from mammals to fly. Like mammalian AP-1, dAP-1 was shown to localize to the TGN in S2 cells (Fig. S2) and the Golgi localization was sensitive to BFA. Moreover, knockdown of AP47, which encodes a μ1 subunit of dAP-1, accentuated the defect in processing of Lerp seen in dGGA-depleted cells (Fig. 7), even though knockdown of AP47 itself had little effect. These observations support the notion that dGGA and dAP-1 function closely together in Lerp trafficking in S2 cells.
We also noticed that dGGA has one putative internal ACLL sequence in the hinge region (Fig. 1 and Fig. 4, S493IDDVPLL500). Interestingly, this ACLL motif is preceded by a Ser493 residue that could be a target of casein kinase II (CKII). CKII is known to be involved in the phospho-regulation of CI-MPR trafficking in mammals through phosphorylation of the cytoplasmic tail of CI-MPR (Kato et al., 2003), human GGA1 and GGA3 (Ghosh and Kornfeld, 2003), and PACS-1 (Scott et al., 2004). Although this idea needs further assessment in the future, we often observe the dGGA signal as a doublet on immunoblotting (Fig. 2A, asterisks), so dGGA might be under phospho-regulation control like the mammalian GGAs. Taken together, these findings strengthen the idea that Drosophila possesses similar mechanisms of CCV formation at the TGN and of regulation of clathrin adaptors.
Concluding remarks
This is the first report that the putative lysosomal enzyme receptor Lerp and its sorting proteins dGGA and dCHC localize to trans-Golgi compartments in Drosophila cells, where Lerp is believed to be sorted and packaged into CCV destined for endosomal compartments. We also found that the entire molecular system responsible for post-Golgi protein trafficking in Drosophila is highly conserved relative to that in mammals. This conservation should enable genome-wide screens for novel factors involved in the complex processes of CCV formation and regulation of protein sorting at the TGN. Recently, Robinson's group (Cambridge Institute for Medical Research, Cambridge, UK) also showed with isolated CCVs that double knockdown of dGGA and dAP-1 caused significant reduction of Lerp incorporation into the CCVs in Dmel2, one of another Drosophila cell lines (Hirst et al., 2009). Although we need to assess the molecular relationship of these clathrin adaptors more carefully, their results support our current results showing the physiological consequence of dGGA in cargo sorting at the trans-Golgi. Thus, this approach using the insect systems will lead us to a better understanding of how those clathrin adaptors are important in the development of multicellular organisms and in the molecular basis for lysosomal diseases in higher organisms.
Materials and Methods
Cell cultures and transfection
The Schneider S2 cell line (Schneider, 1972) was obtained from Invitrogen (Carlsbad, CA) and maintained in Schneider's medium supplemented with 10% fetal bovine serum, 50 U/ml penicillin and 50 μg/ml streptomycin. Transfection of S2 cells was performed with FuGene6 HD (Roche Molecular Biochemicals, Indianapolis, IN) or cellfectin (Invitrogen). For isolation of stable transfectants, S2 cells were cotransfected with 2 μg of expression constructs and 0.2 μg of pCoBlast (Invitrogen). At 24 hours after transfection, cells were transferred to fresh culture medium containing 10 μg/ml blasticidin (Invitrogen) and maintained for an additional 10-14 days to obtain stable transfectants. For gene silencing, 5 μg of dsRNA was added to 2.5×105 cells in 0.5 ml complete medium and cultured for 3-5 days. For expression of proteins from the pMT/V5His (Invitrogen) backbone vectors, which harbor metallothionein promoter, 0.5 mM CuSO4 was added to the medium. Growth media and transformation methods for yeast cells were as described elsewhere (Kametaka et al., 1997). The S. cerevisiae strains SEY6210 (MAT a ura3-52 leu2-3, 112 his3-Δ200 trp1-Δ901 lys2-801 suc2- Δ 9) and GPY2385 (SEY6210 gga1Δ::TRP1 gga2Δ::HIS3, Costaguta et al., 2001) were kindly provided by Gregory Payne (UCLA, La Jolla, CA).
Antibodies
Antibodies to the V5 and HA epitope tags were purchased from Invitrogen and Covance (Princeton, NJ), respectively. Anti-human ARF antibody was from Abcam (Cambridge, MA). Rabbit polyclonal antibodies to dGGA, AP47 or dCHC were raised against the synthetic peptides CILPTQDRMPSFKRDPEK, CTTDSKILQEYITQEGHK or DDSTEHKNIIQMEPQLMC, respectively (MBL, Nagoya, Japan), and further purified by affinity chromatography (Fig. 4A, supplementary material Fig. S3, Fig. S2A). Anti-p120 rat monoclonal antibody and anti-dGM130 rabbit polyclonal antibody were gifts from Satoshi Goto and Masato Abe (Mitsubishi Life Science Institute, Tokyo, Japan) (Yano et al., 2005). Anti-CPY rabbit polyclonal antibody was kindly provided by Takahiro Shintani (Tohoku University, Sendai, Japan).
Reagents
Protease inhibitor cocktails, brefeldin A, bafilomycin A1, were purchased from Sigma-Aldrich (St Louis, MO). Drosophila cDNA clones were obtained from Drosophila Genome Resource Center (Bloomington, IN).
Cloning of Drosophila genes and construction of plasmids
cDNA clones, LD41311 for dGGA, LD05154 for LERP, and LD43101 for dCHC, were used as templates for PCR amplification of the ORFs. Construction of the HA-dGGA expression vector, pAc- or pMT-HA-dGGA-FL was performed by subcloning of the amplified HA-tagged dGGA full-length ORF into the EcoRI-XhoI sites of pAc5.1-A or pMT/V5His-A vector (Invitrogen), respectively. From pAc-HA-dGGA-FL, the EcoRI-SalI fragment that contains the dGGA-VHS-GAT (VG) region was subcloned into pAc5.1-A vector to generate pAc-HA-dGGA-VG. These EcoRI-XhoI HA-dGGA-FL and the EcoRI-SalI HA-dGGA-VG fragments were also inserted into pGEX6P-1 vector (GE Healthcare) to generate pGEX-HA-dGGA-FL and pGEX-HA-dGGA-VG, respectively. The EcoRI-XhoI HA-dGGA-FL and the EcoRI-SalI HA-dGGA-VG fragments were subcloned into pGAD10 vector (Clontech) to make pGAD-dGGA-FL and pGAD-dGGA-VG, respectively. The region containing the full-length Lerp coding sequence was subcloned into the KpnI-NotI sites of pAc5.1-A or pMT-A vectors to produce pAc- or pMT-Lerp-V5His, respectively. The DNA encoding the C-terminal cytoplasmic tail of Lerp was subcloned into pGBD9 to generate pGBD9-Lerp-tail, which was further used for substitution of the C-terminal dileucine with alanines. pGBKT7-human ARF1 plasmids (wt and Q71L) were described previously (Kametaka et al., 2005). The yeast vectors for expression of RFP-fused Sed5, Gos1or Sec7 were gifts from Akihiko Nakano (RIKEN, Japan) (Matsuura-Tokita et al., 2006). Copper-inducible EGFP fusion vector p416Cu-GFP and pRS315-GGA2-13Myc were provided by Takahiro Shintani (Tohoku University, Sendai, Japan) and Gregory Payne (UCLA, La Jolla, CA), respectively. The EcoRI-XhoI fragment of dGGA ORF was transferred to p416Cu-GFP to generate p416Cu-GFP-dGGA.
In vitro transcription of dsRNA
To prepare dsRNAs for silencing of Drosophila genes, fragments of 300-700 nucleotides were amplified by PCR with gene-specific primer pairs tagged with T7 promoter sequence at the 5′-ends. The resultant T7-bearing cDNA fragments were used as templates in the in vitro transcription reaction performed with Megascript in vitro transcription kit (Ambion, Austin, TX) following the manufacturer's instruction. The nucleotide sequences of the primers for preparation of the dsRNA are listed in supplementary material Table S1.
Immunofluorescence
Indirect-immunofluorescence microscopy was essentially performed as described previously (Kametaka et al., 2005). Briefly, S2 cells were cultured on cover slips precoated with poly-L-lysine, and fixed with 4% paraformaldehyde in 0.12 M phosphate buffer (pH 7.2) at room temperature for 20 minutes. After permeabilization with 0.1% Triton X-100 for 5 minutes, blocking was carried out with PBS containing 1 mg/ml bovine serum albumin at room temperature for 20 minutes. Then, the cells were incubated with appropriate primary antibody diluted in PBS for 1 hour, followed by treatment with secondary antibody conjugated with fluorescent dye (Invitrogen). When required, nuclei were additionally labeled with 1 μg/ml Hoechst 33342 (Invitrogen). Fluorescent images were captured with a Zeiss 510 confocal microscope (Carl Zeiss, Thornwood, NY) or FV1000 confocal microscope (Olympus, Japan). For quantitative analysis of fluorescence intensities, nonsaturated images were captured with an UPlanFL N 40× 1.30 NA objective (Olympus) and a fully open pinhole. Fluorescence quantification was performed as described previously (Kametaka et al., 2005).
Pull-down assay
Bacterially expressed GST-fusion proteins were purified and used for pull-down experiments as previously described (Kametaka et al., 2007). Briefly, 25 μg of purified GST or GST-dGGA fusion proteins were bound to glutathione Sepharose CL-4B (GE Healthcare, Piscataway, NJ) to prepare the ‘GST-beads’. S2 cells were lysed with PBS containing 0.5% Triton X-100 and 1× protease inhibitor cocktail (Roche) on ice for 30 minutes, and centrifuged at 20,000 g for 15 minutes at 4°C to prepare the total cell lysate. Then, the lysate was incubated with the GST-beads for 18 hours at 4°C with gentle rotation. After the incubation, beads were washed with PBS containing 0.25% Triton X-100 three times and the bound protein was eluted with 1× Laemmli's sample buffer containing 5% (v/v) β-mercaptoethanol and subjected to immunoblotting.
Subcellular fractionation and in vitro recruitment assay
S2 cells were suspended in ice-cold homogenization buffer (10 mM HEPES-KOH pH 7.4, 150 mM KCl, 0.5 mM MgSO4, 1 mM DTT, 1× Roche protease inhibitor cocktail) and homogenization was carried out by passing the cells through a 27-gauge needle 20 times. The homogenate was centrifuged at 800 g for 5 minutes to generate the post-nuclear supernatant (PNS) fraction. Then the PNS was further ultracentrifuged at 100,000 g for 1 hour in a Beckman TLA45 rotor to generate the total membrane fraction and the cytosolic fraction.
Membrane recruitment experiments were performed essentially as described before (Ooi et al., 1998) with slight modifications. Briefly, the total membrane isolated as above was incubated with or without the S2 cytosol in the presence of an ATP regeneration system (1 mM MgATP, 1 mM creatine phosphate, 150 U creatine kinase per reaction) at 25°C for 30 minutes, and then the membrane was re-sedimented by centrifugation at 20,000 g for 15 minutes. After transfer of the cytosol, the membrane pellet was rinsed with the homogenization buffer once and solubilized with 1× Laemmli's buffer.
Yeast two-hybrid and yeast rescue experiments
Yeast two-hybrid assays were performed essentially as described previously (Kametaka et al., 2005). For the rescue experiment, the expression vectors p416Cu-GFP, p416Cu-GFP-dGGA and pRS315-yGGA2-13Myc were introduced into GPY2385 or its parental wild-type strain SEY6210 cells with a TE/LiOAc/PEG method as described previously (Schiestl and Gietz, 1989). Expression of GFP-dGGA in these transformants was induced with 0.5 μM of CuSO4 for 6 hours. CPY maturation in these cells was examined with immunoblotting using anti-CPY antiserum.
CPY-secretion assay was performed as previously described (Roberts et al., 1991) by spotting freshly grown cells onto nitrocellulose membrane on selection medium for overnight growth. The membrane was subjected to immunoblotting for CPY.
Supplementary Material
Acknowledgments
We thank Satoshi Goto and Masato Abe for rabbit anti-dGM130 and rat anti-p120 antibodies and many helpful discussions. We thank Gregory Payne, Takahiro Shintani and Akihiko Nakano for yeast strains, antibodies and plasmids. We also thank Ai Kametaka for live-cell analysis, Atsuko Yabashi, Katsuyuki Kanno and Miyuki Mashiko for technical assistance, and all the members of the Waguri laboratory for helpful discussions. We also thank Margaret S. Robinson and Jennifer Hirst (Cambridge Institute for Medical Research, Cambridge, UK) for sharing unpublished results during the preparation of this manuscript. This work is supported by Grant-in-Aid for Young Scientists (B) (S.K., 20770159) and for Scientific Research (B) (S.W., 21390055) from Japan Society for the Promotion of Science, and Grant-in-Aid for Scientific Research on Priority Areas (S.K., 20034046) from Ministry of Education, Culture, Sports, Science and Technology of Japan. J.S.B. is supported by NICHD, NIH. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/3/460/DC1
References
- Bai H., Doray B., Kornfeld S. (2004). GGA1 interacts with the adaptor protein AP-1 through a WNSF sequence in its hinge region. J. Biol. Chem. 279, 17411-17417 [DOI] [PubMed] [Google Scholar]
- Black M. W., Pelham H. R. (2000). A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 151, 587-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M., Bonifacino J. S. (2001). Adaptins: the final recount. Mol. Biol. Cell 12, 2907-2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S. (2004). The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell. Biol. 5, 23-32 [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Lippincott-Schwartz J. (2003). Coat proteins: shaping membrane transport. Nat. Rev. Mol. Cell. Biol. 4, 409-414 [DOI] [PubMed] [Google Scholar]
- Chaudhuri R., Lindwasser O. W., Smith W. J., Hurley J. H., Bonifacino J. S. (2007). Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J. Virol. 81, 3877-3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costaguta G., Stefan C. J., Bensen E. S., Emr S. D., Payne G. S. (2001). Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell 12, 1885-1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Puertollano R., Mullins C., Aguilar R. C., Vargas J. D., Hartnell L. M., Bonifacino J. S. (2000). GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149, 81-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Guo Y., Watson H., Au W. C., Shakoury-Elizeh M., Basrai M. A., Bonifacino J. S., Philpott C. C. (2009). Gga2 mediates sequential ubiquitin-independent and ubiquitin-dependent steps in the trafficking of ARN1 from the trans-Golgi network to the vacuole. J. Biol. Chem. 284, 23830-23841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennes A., Cromme C., Suresh K., Kumar N. S., Eble J. A., Hahnenkamp A., Pohlmann R. (2005). The novel Drosophila lysosomal enzyme receptor protein mediates lysosomal sorting in mammalian cells and binds mammalian and Drosophila GGA adaptors. J. Biol. Chem. 280, 12849-12857 [DOI] [PubMed] [Google Scholar]
- Doray B., Bruns K., Ghosh P., Kornfeld S. (2002a). Interaction of the cation-dependent mannose 6-phosphate receptor with GGA proteins. J. Biol. Chem. 277, 18477-18482 [DOI] [PubMed] [Google Scholar]
- Doray B., Ghosh P., Griffith J., Geuze H. J., Kornfeld S. (2002b). Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science 297, 1700-1703 [DOI] [PubMed] [Google Scholar]
- Doray B., Bruns K., Ghosh P., Kornfeld S. A. (2002c). Autoinhibition of the ligand-binding site of GGA1/3 VHS domains by an internal acidic cluster-dileucine motif. Proc. Natl. Acad. Sci. USA 99, 8072-8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez G. E., Payne G. S. (2006). Laa1p, a conserved AP-1 accessory protein important for AP-1 localization in yeast. Mol. Biol. Cell 17, 3304-3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Kornfeld S. (2004). The cytoplasmic tail of the cation-independent mannose 6-phosphate receptor contains four binding sites for AP-1. Arch. Biochem. Biophys. 426, 225-230 [DOI] [PubMed] [Google Scholar]
- Ghosh P., Dahms N. M., Kornfeld S. (2003a). Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell. Biol. 4, 202-212 [DOI] [PubMed] [Google Scholar]
- Ghosh P., Griffith J., Geuze H. J., Kornfeld S. (2003b). Mammalian GGAs act together to sort mannose 6-phosphate receptors. J. Cell Biol. 163, 755-766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida T., Ikeda H., Kametaka S., Akazawa C., Kohsaka S., Ebisu S., Uchiyama Y., Waguri S. (2007). Specific depletion of GGA2 causes cathepsin D missorting in HeLa cells. Arch. Histol. Cytol. 70, 303-312 [DOI] [PubMed] [Google Scholar]
- Hirst J., Robinson M. S. (1998). Clathrin and adaptors. Biochim. Biophys. Acta 14, 173-193 [DOI] [PubMed] [Google Scholar]
- Hirst J., Lui W. W., Bright N. A., Totty N., Seaman M. N., Robinson M. S. (2000). A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 149, 67-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Lindsay M. R., Robinson M. S. (2001). GGAs: roles of the different domains and comparison with AP-1 and clathrin. Mol. Biol. Cell 12, 3573-3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Motley A., Harasaki K., Peak Chew S. Y., Robinson M. S. (2003). EpsinR: an ENTH domain-containing protein that interacts with AP-1. Mol. Biol. Cell 14, 625-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Motley A., Harasaki K., Peak Chew S. Y., Robinson M. S. (2009). Spatial and functional relationship of GGAs and AP-1 in Drosophila and HeLa cells. Traffic 10, 1696-1710 [DOI] [PubMed] [Google Scholar]
- Kametaka S., Mattera R., Bonifacino J. S. (2005). Epidermal growth factor-dependent phosphorylation of the GGA3 adaptor protein regulates its recruitment to membranes. Mol. Cell. Biol. 25, 7988-8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S., Moriyama K., Burgos P. V., Eisenberg E., Greene L. E., Mattera R., Bonifacino J. S. (2007). Canonical interaction of cyclin G associated kinase with adaptor protein 1 regulates lysosomal enzyme sorting. Mol. Biol. Cell 18, 2991-3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Misra S., Puertollano R., Hurley J. H., Bonifacino J. S. (2002). Phosphoregulation of sorting signal-VHS domain interactions by a direct electrostatic mechanism. Nat. Struct. Biol. 9, 532-536 [DOI] [PubMed] [Google Scholar]
- Kondylis V., Rabouille C. (2003). A novel role for dp115 in the organization of tER sites in Drosophila. J. Cell Biol. 162, 185-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., Goulding S. E., Dunne J. C., Rabouille C. (2001). Biogenesis of Golgi stacks in imaginal discs of Drosophila melanogaster. Mol. Biol. Cell 12, 2308-2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., Spoorendonk K. M., Rabouille C. (2005). dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol. Biol. Cell 16, 4061-4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Jacob C., André B. (2009). K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185, 493-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui W. W., Collins B. M., Hirst J., Motley A., Millar C., Schu P., Owen D. J., Robinson M. S. (2003). Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin. Mol. Biol. Cell 14, 2385-2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardones G. A., Burgos P. V., Brooks D. A., Parkinson-Lawrence E., Mattera R., Bonifacino J. S. (2007). The trans-Golgi network accessory protein p56 promotes long-range movement of GGA/clathrin-containing transport carriers and lysosomal enzyme sorting. Mol. Biol. Cell 18, 3486-3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Tokita K., Takeuchi M., Ichihara A., Mikuriya K., Nakano A. (2006). Live imaging of yeast Golgi cisternal maturation. Nature 441, 1007-1010 [DOI] [PubMed] [Google Scholar]
- Mattera R., Arighi C. N., Lodge R., Zerial M., Bonifacino J. S. (2003). Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J. 22, 78-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R., Puertollano R., Smith W. J., Bonifacino J. S. (2004). The trihelical bundle subdomain of the GGA proteins interacts with multiple partners through overlapping but distinct sites. J. Biol. Chem. 279, 31409-31418 [DOI] [PubMed] [Google Scholar]
- Miller G. J., Mattera R., Bonifacino J. S., Hurley J. H. (2003). Recognition of accessory protein motifs by the gamma-adaptin ear domain of GGA3. Nat. Struct. Biol. 10, 599-606 [DOI] [PubMed] [Google Scholar]
- Misra S., Puertollano R., Kato Y., Bonifacino J. S., Hurley J. H. (2002). Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415, 933-937 [DOI] [PubMed] [Google Scholar]
- Nie Z., Hirsch D. S., Randazzo P. A. (2003). Arf and its many interactors. Curr. Opin. Cell Biol. 15, 396-404 [DOI] [PubMed] [Google Scholar]
- Nogi T., Shiba Y., Kawasaki M., Shiba T., Matsugaki N., Igarashi N., Suzuki M., Kato R., Takatsu H., Nakayama K., et al. (2002). Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain. Nat. Struct. Biol. 9, 27-31 [DOI] [PubMed] [Google Scholar]
- Ooi C. E., Dell'Angelica E. C., Bonifacino J. S. (1998). ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J. Cell Biol. 142, 391-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Aguilar R. C., Gorshkova I., Crouch R. J., Bonifacino J. S. (2001a). Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292, 1712-1716 [DOI] [PubMed] [Google Scholar]
- Puertollano R., Randazzo P. A., Presley J. F., Hartnell L. M., Bonifacino J. S. (2001b). The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell 105, 93-102 [DOI] [PubMed] [Google Scholar]
- Puertollano R., van der Wel N. N., Greene L. E., Eisenberg E., Peters P. J., Bonifacino J. S. (2003). Morphology and dynamics of clathrin/GGA1-coated carriers budding from the trans-Golgi network. Mol. Biol. Cell 14, 1545-1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., Kuntz D. A., Lockyer A., Watson R., Signorelli T., Rose D. R., van den Heuvel M., Roberts D. B. (1999). The Drosophila GMII gene encodes a Golgi alpha-mannosidase II. J. Cell Sci. 112, 3319-3330 [DOI] [PubMed] [Google Scholar]
- Roberts C. J., Raymond C. K., Yamashiro C. T., Stevens T. H. (1991). Methods for studying the yeast vacuole. Meth. Enzymol. 194, 644-661 [DOI] [PubMed] [Google Scholar]
- Robinson M. S. (2004). Adaptable adaptors for coated vesicles. Trends. Cell Biol. 14, 167-174 [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. (1989). High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16, 339-346 [DOI] [PubMed] [Google Scholar]
- Schneider I. (1972). Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27, 353-365 [PubMed] [Google Scholar]
- Scott P. M., Bilodeau P. S., Zhdankina O., Winistorfer S. C., Hauglund M. J., Allaman M. M., Kearney W. R., Robertson A. D., Boman A. L., Piper R. C. (2004). GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 6, 252-259 [DOI] [PubMed] [Google Scholar]
- Shiba T., Kawasaki M., Takatsu H., Nogi T., Matsugaki N., Igarashi N., Suzuki M., Kato R., Nakayama K., Wakatsuki S. (2003). Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat. Struct. Biol. 10, 386-393 [DOI] [PubMed] [Google Scholar]
- Takatsu H., Katoh Y., Shiba Y., Nakayama K. (2001). Golgi-localizing, gamma-adaptin ear homology domain, ADP-ribosylation factor-binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J. Biol. Chem. 276, 28541-28545 [DOI] [PubMed] [Google Scholar]
- Wang J., Sun H. Q., Macia E., Kirchhausen T., Watson H., Bonifacino J. S., Yin H. L. (2007). PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol. Biol. Cell 18, 2646-2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H., Yamamoto-Hino M., Abe M., Kuwahara R., Haraguchi S., Kusaka I., Awano W., Kinoshita-Toyoda A., Toyoda H., Goto S. (2005). Distinct functional units of the Golgi complex in Drosophila cells. Proc. Natl. Acad. Sci. USA 102, 13467-13472 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.