Abstract
Changes in the distribution of methylcytosine residues along a transgene locus of tobacco (Nicotiana tabacum) in relation to the type of gene silencing were studied in parental plant leaves, calli, and regenerated plants derived thereof. Parental-silenced HeLo1 (hemizygous for locus 1) plants show posttranscriptional silencing of the residing nptII (neomycin phosphotransferase II) transgene and cytosine methylation restricted to the 3′ end and center part of the transcribed region. Here, we report that with an increasing number of cell cycles, DNA methylation changes gradually, and methylation is introduced into the promoter during cell culture and more slowly in vegetatively propagated plants. After 24 months of callus in vitro cultivation, an epigenetic variant, designated locus 1E, was obtained in which cytosine methylation of symmetrical (CG and CNG) sites was almost complete within the 5′ end of the nptII-transcribed region and the 35S promoter. Further, methylation of nonsymmetrical sites appeared de novo in the promoter, whereas this type of methylation was significantly reduced in the 3′ end of the transcribed region when compared with locus 1. The newly established epigenetic patterns were stably transmitted from calli into regenerated plants and their progeny. The protein and steady-state RNA levels remained low in locus 1E, whereas with nuclear run-on assays, no detectable amounts of primary transcripts were found along the nptII gene, indicating that the methylated promoter became inactivated. The results suggest that a switch between posttranscriptional and transcriptional gene silencing could be a mechanism leading to irrevocable shut down of gene expression within a finite number of generations.
In plants, genes can be silenced by transcriptional and posttranscriptional mechanisms. The posttranscriptional gene silencing (PTGS) process is characterized by several features, including normal transcriptional activity of the promoter, transcript instability, and ability to degrade homologous RNA (Vaucheret et al., 2001), whereas in transcriptional gene silencing (TGS), the promoter is inactivated and no or little transcripts are produced (Vaucheret and Fagard, 2001). Methylation of cytosine residues is associated with both TGS and PTGS; however, the location of methylcytosines within the silenced genes differs for each type of silencing. In TGS, transgenes are frequently methylated in the promoter region, whereas PTGS is correlated with methylation of the transcribed region, particularly at its 3′ end (Depicker and Van Montagu, 1997). The function of DNA methylation has been addressed by genetic and biochemical experiments in which induced hypomethylation leads to partial release of both TGS and PTGS in some, but not all, transgenic loci (Kilby et al., 1992; Kovarik et al., 2000; Morel et al., 2000; Bartee et al., 2001; Kloti et al., 2002).
The mechanisms of de novo methylation and of methylation spreading along the DNA are not fully understood. Most studies on plants show that relatively stable inheritance of methylation patterns is restricted to a particular region of the transgene without spreading into neighboring regions. For example, the promoter of a reporter gene was not influenced by methylation of a linked repeated element in a transgenic locus of Arabidopsis (Muller et al., 2002). Also, a tobacco (Nicotiana tabacum) retroelement was highly methylated de novo in transgenic lines of Nicotiana sylvestris without significant methylation of flanking sequences (Kunz et al., 2003). However, there is evidence for methylation spreading from integrated transposons in both plants and animals. In rapeseed (Brassica napus), genomic DNA flanking the integration site of a short interspersed element retroposon was methylated, but not its empty allele (Arnaud et al., 2000). Also, RNA-directed methylation of viroid cDNA resulted in low, but significant, levels of cytosine methylation of immediate flanking sequences (Pélissier et al., 1999). In virus-induced gene silencing and transgene-induced PTGS, spreading of RNA targeting from the initiator region to 5′ and 3′ regions and to the 5′ region only, respectively, was accompanied by cytosine methylation of the corresponding transgenic DNA (Vaistij et al., 2002; Van Houdt et al., 2003).
Inverted repeats (IRs) possess exceptional silencing capacities for their own and homologous genes (Selker, 1999). There is evidence that IRs can trigger methylation of themselves and also unlinked loci by a yet unknown mechanism. Methylation seems to be more dense at the IR center than at distal regions and may be associated with TGS or PTGS depending on the orientation and distance from the IR center (Stam et al., 1998). By hypomethylation of IR loci, either induced by a drug (Kovarik et al., 2000) or genetically (Bartee and Bender, 2001), methylcytosine residues are demethylated only partially, whereas other genomic regions are completely demethylated by the same treatment, suggesting that these structures may contain multiple methylation signals for de novo methylation in cis. Methylation-inducing dsRNA resulting from convergent transcription of sequences residing in IRs might be one of the signals (Mette et al., 2000). Also, loss of one gene copy from an IR locus that is convergently transcribed results in loss of silencing and methylation, suggesting that a palindromic arrangement is sufficient to trigger methylation (De Buck and Depicker, 2001).
DNA methylation was extensively studied in plant cell cultures and regenerated plants (for review, see Kaeppler et al., 2000). Both global and gene-specific methylation were shown to vary during culture processes, and, in some cases, these changes were transmitted to regenerated plants. Decrease in methylation of single-copy genes was observed more frequently than an increase (Olhoft and Phillips, 1999). However, analysis of the global methylation levels revealed their significant increase under stress conditions (Kovarik et al., 1997; Schmitt et al., 1997). These studies have provided evidence that DNA methylation is less stable in cell culture than in plants growing from seeds.
Here, we addressed the question of whether the established PTGS and methylation are stable in a tobacco transgene locus that contains a T-DNA IR in the course of long-term propagation in cell culture and subsequent plant regeneration. Methylation was studied in both promoter and transcribed region using restriction enzymes sensitive to CG, CNG, and nonsymmetrical methylation. The reporter gene expression was evaluated by determining the NPTII protein content and steady-state RNA levels and the type of gene silencing by performing nuclear run-on assays. We show that tissue culture leads reproducibly to a switch from PTGS to TGS, correlated with hypermethylation of the promoter.
RESULTS
Hypermethylation Is Induced in the Promoter Region of Posttranscriptionally Silenced Transgenes during Cell Culture
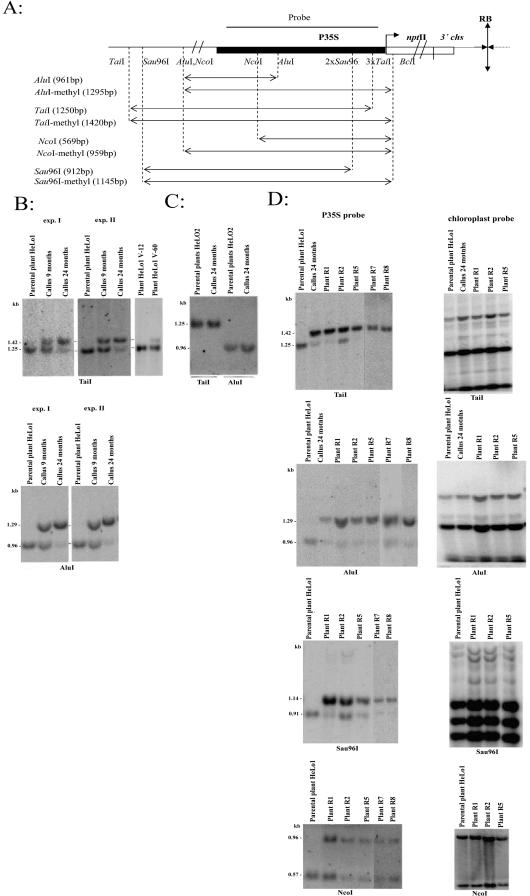
The posttranscriptionally silenced locus 1 in the line hemizygous for locus 1 (HeLo1) contains a T-DNA IR, and expression of the residing nptII genes was more than 100-fold reduced at the protein level compared with a non-silenced line hemizygous for locus 2 (HeLo2) bearing a single-copy insertion of the T-DNA (Van Houdt et al., 2000). From both a silenced and a non-silenced transgenic plant, two independent callus cultures were established, referred to as experiments I and II. Methylation in different regions of the nptII transgenes was monitored by methylation-sensitive restriction enzymes in parental HeLo1 plants, in 9- and 24-month-old callus cultures, in plants regenerated from 24-month-old calli, and in parental plants multiplied by stem cuts. The promoter region was analyzed by Southern-blot hybridization using restriction enzymes AluI (recognition site at –286 relative to the +1 transcription start site [TSS] of the nptII gene) and TaiI (recognition sites at –79, –119, and –130, Table I; Fig. 1A). Genomic DNA samples were predigested with methylation-insensitive BclI to avoid interference with methylation of sites in the nptII transcribed region. The 1.25-kb TaiI and 0.96-kb AluI bands were expected in case the promoter was not methylated, whereas a 1.29-kb AluI band was expected for methylated molecules (Fig. 1A). Because the three TaiI sites in the 35S promoter are separated by less than 50 nucleotides located close to the +1 TSS (Fig. 1A), a 1.42-kb band can be generated only under complete methylation of all three TaiI sites. In the parental HeLo1 plant, a very pronounced 1.25-kb band could be visualized after TaiI digestion (Fig. 1B). Also, AluI digestion yielded a strong 0.96-kb band, indicating no or very little methylation in the 35S promoter of locus 1. In 9-month-old callus DNA, upper bands became clearly visible in both digests, and in 24-month-old calli, the 1.42-kb TaiI and 1.29-kb AluI were dominant bands in the hybridization profiles, suggesting that de novo methylation of these sites had been completed. The near absence of the 1.25-kb TaiI band in 24-month-old callus DNA indicates heavy methylation of both copies of the 35S promoter (three fully methylated TaiI sites per copy). The more distally located sites (with respect to the IR center) tested were partially methylated (HpaII at –923) or non-methylated (CfoI at –1,146 and SmaI at –1,636) in both callus and plants (Fig. 2). The same results were obtained in an independently established and propagated callus culture from the same HeLo1 plant (experiment II, Fig. 1B). In vegetatively propagated HeLo1 plants, there was no change in methylation patterns after 12 months of cultivation on agar (line HeLo1-V12 in Fig. 1B). However, DNA extracted from a plant grown for 60 months showed a faint 1.42-kb TaiI band (line HeLo1-V60). Its intensity was about 16% of the total hybridization signal (Fig. 2). In a non-silenced locus 2 callus line (HeLo2), the promoter remained non-methylated during the whole time span of subcultivation (Fig. 1C).
Table I.
Relative position of the analyzed restriction sites and methylation sensitivities of the enzymes (Nelson et al., 1993)
| Enzyme | Position Relative to TSSa | Distance from IR Center | Site Cutting | Methylation Sensitivity | Sequence Context |
|---|---|---|---|---|---|
| kb | |||||
| Promoter region approximately 300/+1b | |||||
| TaiI-site a | -79 | ∼1.5 | ACGT | AmCGT | CG |
| TaiI-siteb | -119 | ∼1.6 | ACGT | AmCGT | CG |
| TaiI-sitec | -130 | ∼1.6 | ACGT | AmCGT | CG |
| Sau96-site a | -163 | ∼1.6 | gGGTCCad | GGWmCC, GGWCmC | Nonsymmetrical |
| Sau96I-siteb | -175 | ∼1.6 | gGGACCa | GGWmCC, GGWCmC | Nonsymmetrical |
| AluI | -286 | ∼1.7 | tAGCTg | AGmCT | CWG |
| NcoI | -522 | ∼2.0 | CCATGG | CmCATGG | Nonsymmetrical |
| HpaII | -923 | ∼2.4 | CCGG | CmCGG, mCCGG | CG, CCG |
| MspI | -923 | ∼2.4 | CCGG | mCCGG | CCG |
| CfoI | -1146 | ∼2.6 | GCGC | GmCGC | CG |
| SmaI | -1636 | ∼3.1 | CCCGGG | CCmCGGG | CG |
| Transcribed region +1/+1,035c | |||||
| ClaI | +37 | ∼1.4 | ATCGAT | ATmCGAT | CG |
| BamI-II | +881 | ∼0.6 | gGGATCCt | GGATmCC | Nonsymmetrical |
| SmaI | +884 | ∼0.6 | CCCGGG | CCmCGGG | CG |
| Eco47III | +1,211 | ∼ 0.2 | AGCGCT | AGmCGCT | CG |
a TSS is at nucleotide +1, corresponding to 1,447 bp from the right border.
b The 35S cauliflower promoter securing efficient transcription is approximately 300 bp (Meyer et al., 1994).
c The polyadenylation site from the chalcon synthase gene (accession no. X03710) is at + 1,041, 408 bp from the right border.
d The small letters indicate nucleotides flanking the recognition site that are relevant to the methylation context.
Figure 1.
Methylation analysis of the 35S promoter region. A, Physical map of the restriction sites in locus 1 and strategy for methylation analysis of the promoter. Evidence for the IR character of the T-DNA insertions has been given elsewhere (Van Houdt et al., 2000); SmaI and BamHI digestions in Figure 2B are in agreement with these previous observations. One of two symmetrically positioned transcription units is shown. The sizes of restriction fragments (in parentheses) were calculated from the T-DNA sequence. P35S, Promoter of the cauliflower mosaic virus; nptII, neomycin phosphotransferase II; RB, T-DNA right border; polyA, transcription termination sequence from 3′-untranslated region of a chalconsynthase gene (3′ chs) from Antirrhinium majus. B, Southern-blot hybridization analysis of DNA from the HeLo1 line carrying the T-DNA IR (locus 1). Analysis of two independent calli (experiment I, II) is presented. Double digestions of genomic DNAs were performed with BclI/TaiI and BclI/AluI restriction enzymes, and the blots were hybridized against the P35S promoter probe. HeLo1-V12 and HeLo1-V60, HeLo1 plant multiplied vegetatively for 12 and 60 months, respectively. C, Southern-blot hybridization analysis of DNA from the non-silenced HeLo2 line carrying a single-copy insertion of the T-DNA. Double digestions of genomic DNAs were performed with BclI/TaiI and BclI/AluI restriction enzymes, and the blot was hybridized against the P35S promoter probe. D, DNA from R1, R2, R5, R7, and R8 plants regenerated from a single HeLo1 callus was digested with BclI/TaiI, BclI/AluI, BclI/Sau96I, and BclI/NcoI enzymes and hybridized against the P35S promoter probe. Hybridization against the pTB29 chloroplast probe (right) confirmed completeness of DNA digestion and equivalent loading of samples.
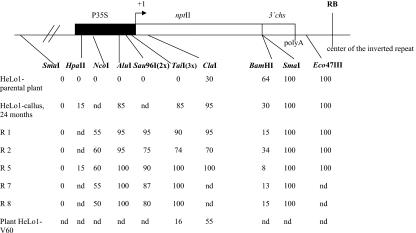
Figure 2.
Summary of methylation analysis of locus 1 in the parental plant, derived callus tissue, and epigenetic variants (R1, R2, R5, R7, and R8) of plants regenerated from callus. Relative methylation levels (percentage) in restriction sites indicated were calculated from hybridization profiles in Figures 1, B to D, and 3B. Using ImageQuant software, intensities of radioactive signals in individual bands were determined. Relative methylation was then expressed as band signal corresponding to methylated DNA relative to the total signal in each lane. Bands with partially methylated DNA in BamHI digestions were excluded from calculations. nd, Not determined.
All five tobacco plants (R1, R2, R5, R7, and R8) regenerated from 24-month-old HeLo1 calli (experiment I) maintained methylation in the analyzed sites in the promoter, although the extent was slightly heterogeneous among the different regenerated plants (Fig. 1D). For example, in R1, R5, R7, and R8 regenerants, the TaiI and AluI sites were completely methylated, whereas in R2, approximately 20% of the molecules could still be digested with TaiI, indicating the absence of methylation in at least one of these sites (Fig. 2). Plants are known to contain, in addition to symmetrical methylation (at CG and CNG), some nonsymmetrical methylation. To reveal nonsymmetrical methylation, DNA was digested with Sau96I (GGWCC cutting at positions –163 and –175, Table I) and hybridized on the blots to the 35S promoter probe (Fig. 1D). The 1.14-kb Sau96I band is generated only when both target sites in the promoter proximal region are methylated; the 0.91-kb band represents partially methylated or non-methylated molecules (Fig. 1A). It is evident that although in the parental leaf there is no signal in the 1.1-kb region, in regenerated plants, the probe hybridized strongly to the 1.14-kb fragment. Similarly, analysis of a more distal nonsymmetrical motif at the NcoI site (at –522, Table I) showed methylation in regenerated plants, although in this case, methylation was not so heavy (Fig. 1D). The right panel of Figure 1D shows rehybridized blots with a chloroplast probe to monitor completeness of the digests. It has been reported that callus culture may be mutagenic (Kaeppler et al., 2000). To exclude possible mutation of restriction endonuclease cleavage sites during the long-term cultivation in cell culture, a section of the 35S promoter (from –1,331 to –1,662) was PCR amplified and sequenced. The sequences obtained from the parental plant and 24-month-old callus were identical, suggesting that the observed resistance of the 35S promoter sequences toward methylation-sensitive restriction endonucleases was not caused by mutation in the target sites.
The described epigenetic variant was designated locus 1E in the calli—and derived R1, R2, R5, R7, and R8 sublines. We conclude that methylation is induced in the promoter region of the posttranscriptionally silenced nptII genes upon propagation in cell culture.
Methylation Distribution Changes in the Transcribed Sequences of the nptII Genes in Locus 1
In addition to the analysis of de novo methylation patterns in promoter sequences of the nptII genes in locus 1 induced by cell propagation in callus, we also investigated whether cell culture cultivation would affect methylation of sites in the transcribed region of the nptII transgenes and more downstream sequences (Eco47III site). Methylation was studied at ClaI (located at +37), BamHI (+881), SmaI (+884), and Eco47III (downstream region) in the parental HeLo1 plants, calli (24 months), and regenerated R1, R2, R5, R7, and R8 plants (Table I; Fig. 3A).
Figure 3.
Methylation analysis of the nptII-transcribed region. A, Physical map of the restriction sites in locus 1 and strategy for methylation analysis of the nptII-transcribed region. Details as in Figure 1A. B, DNAs from HeLo1 plant, Helo1 callus, and regenerated plants were analyzed by digestion with ClaI/TruI (5′ of the nptII-coding sequence), BamHI, and SmaI (3′ of the nptII-coding sequence). Blots were probed with the nptII-coding sequence. Two independently established HeLo1 calli were analyzed by BamHI digestion (experiments I and II). The result of SmaI-digested HeLo2 DNA is presented to show the position of hybridizing fragment obtained after digestion of DNA without methylation of the SmaI restriction site. HeLo1-V12 and HeLo1-V60, HeLo1 plant multiplied vegetatively for 12 and 60 months, respectively. Right, rehybridization of selected samples with the chloroplast probe.
Methylation in the 5′ half of the nptII-transcribed region was assayed by double digestion with TruI (methylation insensitive) and ClaI (sensitive to CG methylation). In the parental plant leaves, the 2.0-kb TruI band was digested with ClaI into two 1.0-kb bands, of which one hybridized to the nptII gene probe (Fig. 3B). Hypomethylation of the ClaI site at+37 was consistent with our previous findings of low levels of cytosine methylation at the 5′ end of the nptII transgene (Van Houdt et al., 2000). However, the DNA from 24-month-old callus and from regenerated plants was completely (callus and sublines R1 and R5) or partially (subline R2) resistant to ClaI digestion, indicating that similar to the promoter, the site in the 5′-transcribed region was subjected to de novo methylation upon callus propagation, which is maintained in the regenerated plants. The 2.0-kb TruI/ClaI band became more intense relative to the 1.0-kb band in the vegetatively multiplied HeLo1 plant (line HeLo1-V60 in Fig. 3B) as compared with the original plant, indicating increased methylation of the CG dinucleotide after prolonged cultivation.
Methylation of the BamHI site just at the 3′ end of the nptII-coding region has been correlated with PTGS (Van Houdt et al., 1997, 2000; Kovarik et al., 2000). Partial methylation of the site yielded three bands of 2.5, 3.6, and 6.2 kb after hybridization with the nptII probe. The 2.5-kb band corresponded to non-methylated, the 3.6-kb band to a fragment with one methylated BamHI site, and the 6.2-kb band to a fragment with two coordinately methylated BamHI sites within the IR center (Fig. 3A). In parental leaf DNA, approximately 60% to 70% of the signal was found in the 6.2-kb band, whereas in callus and regenerated plants, the intensity of the 6.2-kb band was significantly reduced, and the 2.5-kb band was stronger. In R1, R5, R7, and R8, the 6.2-kb hybridization fraction was hardly visible but still clearly detectable in the R2 subline, suggesting differences among the sublines (Fig. 3B). Thus, the level of nonsymmetrical methylation of the BamHI sites downstream from the coding sequence (but in the transcribed region) appeared to be inversely correlated with that of ClaI in the 5′-untranslated region and promoter sites in all DNAs analyzed. However, sites in the 3′-untranslated region and more downstream-containing symmetrical CG motifs showed a different picture: The hybridization patterns of DNAs digested with SmaI (located next to BamHI in the 3′-untranslated region) and Eco47III (downstream from the polyadenylation signal) were indistinguishable in the parental plant, calli, and regenerated plants and showed complete CG methylation (Fig. 3B; data not shown).
The methylation changes in locus 1 induced upon cell culture propagation and long-term cultivation as revealed by the relative methylation levels of particular sites or groups of sites are summarized in Figure 2.
Expression of the nptII Reporter Gene in Epigenetic Variants of Locus 1
Next, we investigated the effect of promoter methylation on the expression of the two linked nptII genes in locus 1E. The levels of the NPTII protein in cell extracts were determined by NPTII ELISA. Figure 4A shows results obtained for different periods of cell culture. Expression of the nptII gene in the HeLo1 line remained low throughout cultivation of callus and in the regenerated R5 plant. In the non-silenced line HeLo2, the nptII gene expression was high in both plant and callus. The steady-state nptII RNA levels measured by northern-blot hybridization were in good agreement with those of the protein assay because only for the HeLo2 plant was an intense signal detected (Fig. 4B).
Figure 4.
Expression analysis of the nptII reporter gene at the protein (A), steady-state (B), and nascent (C) RNA levels in the original and epimutated locus 1. A, NPTII protein accumulation levels were determined by ELISA. The cell extracts from the non-silenced HeLo2 line had >100-fold higher content of NPTII protein compared with samples from the silenced lines. Ordinate, Concentration of NPTII per milligram of protein in log; SR-1, wild-type tobacco. B, Northernblot analysis of RNA samples prepared from the parental silenced HeLo1 plant, the R5 regenerant, the non-silenced HeLo2 (positive control), and the non-transgenic SR-1 (negative control). Five micrograms of total RNA was loaded per lane and hybridized to the nptII DNA probe. C, Representative slot-blot hybridization with nascent RNA synthesized by run-on transcriptions with nuclei from the parental HeLo1, its epimutant form R5, and the non-silenced HeLo2 plant. The 5Sr DNA and rbcS (small subunit of Rubisco gene) hybridizations served as positive controls; plasmid vector (pBluescript; Promega, Madison, WI) hybridization was a negative control.
The experiments indicate that the nptII gene expression in loci 1 and 1E was silenced but not at what level silencing was installed. To determine the dynamic status of transcription, we carried out nuclear run-on assays using nuclei from the silenced HeLo1, R5, and non-silenced HeLo2 lines. The 32P-UTP-labeled nascent RNAs were hybridized to filters containing plasmids carrying the nptII gene, the control genes coding for ribulose-1,5-bisphosphate carboxylase (rbcS) and 5S rRNA, and the empty pBluescript vector (Fig. 4C). With RNAs from HeLo1 and Helo2 nuclei, the nptII gene hydridized strongly, confirming a high transcriptional activity of the 35S promoter in both genotypes. In contrast, RNA isolated from R5 nuclei did not hybridize to the nptII gene, whereas it did with the positive controls rbcS and 5S rDNA, suggesting that transcription of the nptII genes was blocked in the locus 1E epimutant. We conclude that the nptII genes in locus 1E of the R5 plant are silenced by TGS, whereas those in the original locus 1 are silenced by PTGS.
DISCUSSION
In the present study, we have followed the epigenetic changes of the IR transgene locus 1 (Kovarik et al., 2000; Van Houdt et al., 2000) that are induced during cell culture cultivation and long-term vegetative propagation of plants. Initially, in plants and freshly induced callus (up to 3 months), the methylation was limited to a few hundred base pairs from the IR center in the 3′ part of the transcribed region of the nptII gene (Kovarik et al., 2000; Van Houdt et al., 2000). However, after longer cultivation either in cell culture or in a vegetatively propagated plant, methylation appeared in distal regions. In callus and regenerated plants, the newly established epigenetic state involved complete methylation of the analyzed sites in the 35S promoter and 5′-transcribed region, whereas sites located distally (up to 2.3 kb) were only partially or not methylated (Fig. 2). This observation indicates that the detected methylation changes are most likely not the result of methylation spreading from the chromosomal sequences flanking the T-DNA integration site. The cell culture-induced methylation was maintained in regenerated plants (with slight variability between individual plants) and S1 progeny (not shown), suggesting that epigenetic changes are mitotically and meiotically inherited and are not grossly influenced by cell differentiation. The non-silenced locus 2 remained non-methylated irrespective of the callus culture cultivation and plant regeneration processes, implying that the induced methylation changes are a locus 1-specific phenomenon that cannot be attributed to global epigenetic changes induced, for instance, by stress (Kovarik et al., 1997; Schmitt et al., 1997), although limited stability of DNA modifications has already been described in plant cell cultures (Kaeppler et al., 2000). Further, de novo methylation cannot be explained by DNA pairing with the homologous locus because the HeLo1 line used for these experiments is hemizygous for the transgene locus. Based on the known doubling mass of tobacco cells in culture (10–12 d), approximately 70 to 80 cell cycles are estimated to occur during the 24 months of callus culture; during that time, complete methylation of sites, located between 1.4 and 1.7 kb from the IR center, is installed. In a parallel experiment, in which plants were multiplied vegetatively from stem cuts for up to 24 months, no methylation change was observed, whereas 60-month cultivation resulted in weak methylation of sites in the promoter and increased methylation of the 5′-transcribed region. It may be that induction of methylation changes in cis is a regular process activated upon the application of environmental stresses such as in vitro cultivation and that this is strongly enhanced in dedifferentiated cells. In particular for an IR configuration, methylation spreading from the IR center is a highly favorable explanation for the observed methylation change. More detailed analyses of single-cell cloned lines over the prolonged culture period are warranted to be conclusive.
De Novo Methylation of the Promoter Is Correlated with Its Inactivity
Our data indicate that in regenerated plants, methylation of the 35S promoter is heavy and includes methylation of both symmetrical and nonsymmetrical sites, a pattern similar to that found in a transcriptionally silenced 35S promoter in other transgenic lines (Meyer et al., 1994; Park et al., 1996). In contrast to the parental locus 1, no primary transcripts were found in locus 1E epimutant by nuclear run-on assays (Fig. 4C). Thus, we may conclude that silencing occurring at locus 1 has evolved from PTGS to TGS. The TGS phenotype of locus 1E could either have developed de novo in cell culture or was preexisting in the parental plant representing an epigenetic chimera with variegated phenotypes, the PTGS being most abundant. In the literature, there is evidence for preexisting epigenetic variability in primary explants (Matzke and Matzke, 1996). Therefore, it cannot be excluded that HeLo1 plant cells with a preexisting state of promoter methylation are present and selectively amplified during callus growth, resulting in calli and regenerated plants with a transcriptionally silent locus 1. However, no selective growth advantage has been observed in non-PTGS calli over PTGS calli (data not shown). It is our estimate that Southern-blot hybridization cannot detect less than 5% of differentially methylated molecules in a total of 10 μg of genomic DNA. As a consequence, a fraction of molecules with preexisting promoter methylation and TGS phenotype cannot be a priori excluded. If so, then there would be a balance of competing forces; those keeping promoter free of methylation and those supporting its de novo methylation and inactivation (Turker, 2002). Perhaps the blocking forces are weakened in cell culture, thereby allowing the installation of methylation in the promoter. Using bisulfite genomic sequencing, Meyer et al. (1994) observed about 7% of symmetrical CG sites methylated in the active 35S promoter, whereas in an inactive one, this level reached 97%. Mosaic methylation was also observed in a mutant 35S promoter deficient for symmetrical methylation sites, and the levels were significantly increased in a silenced state (Dieguez et al., 1998). Thus, it is likely that low level of preexisting methylation (if present) probably would not affect transcription activity of strong 35S promoter. However, preexisting methylation could be important for attracting factors that increase and spread methylation over the region (Pradhan and Esteve, 2003; see last section).
Dependence of promoter activity on methylation density has already been demonstrated in mammalian systems (Boyes and Bird, 1991). Strong promoters required higher level of methylation for inactivation than the weaker ones. The TaiI site at –79 that is completely methylated in the epimutated locus 1E is part of a binding site for a transcription factor from the ASF-1 family (Lam et al., 1989). Perhaps a mechanism that inactivates the 35S promoter via methylation might involve failure of factor binding to critical site(s) and/or altered protein-binding activity (Kloti et al., 2002). It would be interesting to determine whether methylation changes and loss of gene activity in locus 1E are correlated with a modified chromatin configuration and histone modifications.
PTGS of several transgene loci has recently been shown to be released in callus cultures of tobacco cells (Mitsuhara et al., 2002). Perhaps the switch of PTGS to TGS might explain why locus 1 remained silenced throughout callus cultivation.
Inverse Correlation between Methylation of Several Sites in the Promoter and a Nonsymmetrical Site in the Transcribed Region
The increase of cytosine methylation in the promoter (Fig. 1, B and D) and in the 5′ part of the transcribed region was accompanied by a decrease in methylation of the BamHI site in the 3′-transcribed region (Fig. 3B). This correlation was fairly regular and was reflected by a subtle variability in methylation patterns between individually regenerated plants. The decrease of methylation at the 3′ end in the epimutated locus 1E seems to be restricted to a subset of cytosines because the SmaI and Eco47III sites retained full methylation in both locus 1 and locus 1E (Fig. 2). Digestion with BamHI is inhibited by methylation of cytosines that occur in nonsymmetrical contexts on both strands (mCCT and mCCC; Kovarik et al., 2000). In contrast, both SmaI and Eco47III are sensitive to methylation of symmetrical CG dinucleotides. We propose that this is due to the fact that maintenance of methylation in nonsymmetrical sites at the 3′ end of the transcribed region might be mediated by different mechanisms than that of cytosines in symmetrical contexts. It should be noted that the met1/ddm2 mutation had a stronger hypomethylation effect in IR than in singlet PAI loci, suggesting that different enzyme activities could have different methylation targets in the genome (Bartee and Bender, 2001). In addition, removing of the 35S promoter by action of Cre recombinase resulted in preferential loss of nonsymmetrical methylation at the invertedly repeated transgenes in Arabidopsis (Aufsatz et al., 2002). Thus, maintenance of nonsymmetrical methylation may depend on the continuous presence of a methylation-provoking activity, whereas maintenance of symmetrical methylation could be ensured by a maintenance DNA methyltransferase activity. RNA molecules have been proposed to function in de novo methylation of corresponding DNA sequences (Depicker and Van Montagu, 1997; Pélissier et al., 1999), and nptII-specific 21- to 25-nucleotide-long RNA molecules have already been detected in the parental locus 1 (locus X; Van Houdt et al., 2003). The observed promoter inactivity of locus 1E would probably result in decreased production of the silencing-inducing RNA molecules and, consequently, in reduced methylation of nonsymmetrical motifs. Interestingly, nonsymmetrical methylation appeared in the promoter of epimutated locus 1E (Sau96I and NcoI sites), suggesting redistribution of nonsymmetrical methylation during the epigenetic change. It is not clear whether this methylation is also RNA dependent or whether it is a consequence of an altered chromatin configuration.
Methylation Dynamics in the IR Locus. Possible Mechanisms
Although heavy methylation across IR loci has been well documented in the literature, the mechanism by which IR sequences become methylated is not known. Further, it is unclear what directs methylation of promoter sequences in an IR induced upon cell culture propagation as described here.
First, the question arises as to whether the observed methylation of the 35S promoter in locus 1E can be ascribed to an RNA-directed mechanism (Pélissier et al., 1999; Mette et al., 2000; Wassenegger, 2000). Transcription of the 35S promoter in locus 1 would require read-through double-stranded transcripts of at least 3 kb in length. We have no evidence for promoter transcription or longer read-through transcripts as analyzed by RT-PCR or run-on transcription (data not shown). Moreover, the RNA-induced methylation spreading described by Vaistij et al. (2002) and Van Houdt et al. (2003) was installed immediately after introduction of the silencer trigger, whereas in the described system, the changes were gradual, and numerous cell divisions were required to install promoter methylation.
Second, several DNA-based models could explain the observed methylation changes. In vitro studies with purified DNA methyltransferase enzymes have been shown to direct methylation of adjacent cytosines in synthetic single-stranded DNA templates (Christman et al., 1995). Arnaud et al. (2000) studying integrated transposon of the long interspersed nuclear element (LINE) in rapeseed proposed that heavily methylated single-stranded DNA structures formed at replication forks might attract methyltransferase(s) to originally non-methylated cytosine residues located in cis. Although the dense methylation of locus 1 at the 3′ end of the nptII-coding region makes this possibility attractive, differences exist in methylation patterns between locus 1 and sequences flanking the LINE insertion. Although the LINE-induced spreading mainly involves CG methylation that is limited to several tens of base pairs from the LINE insertion site, in our study, CG (TaiI, ClaI), CNG (AluI), and nonsymmetrical (Sau96I, NcoI) motifs were highly methylated, and the region affected by de novo methylation spanned across several hundreds of base pairs.
Third, spreading of methylation may possibly be directed by chromatin modification originally imposed on an IR center by double-stranded RNA. Heterochromatin is preferentially assembled on IRs in fruitfly (Drosophila melanogaster; Dorer and Henikoff, 1994). In this organism, heterochromatin may also spread up to distances of several kilobase pairs. Perhaps spreading of heterochromatin in locus 1 precedes DNA methylation. Possibly, the IR center of locus 1 attracts methyltransferases via interaction with proteins associated with heterochromatin.
MATERIALS AND METHODS
Plant Material and Cell Culture Conditions
The transgenic tobacco (Nicotiana tabacum) plants hemizygous for locus 1 (designated HeLo1) were obtained by crossing a plant homozygous for locus 1 (designated HoLo1) with an untransformed SR1 tobacco plant as described previously (Van Houdt et al., 2000). The structure of the silenced locus 1 in HeLo1 together with relevant restriction sites are depicted in Figures 1A and 3A. Plants were grown either in greenhouse or on 0.7% (w/v) agar medium containing B5 salts. Plants growing under sterile conditions were vegetatively multiplied from stem cuts every 2 months.
Calli were established from leaf explants by hormonal treatment and grown in 0.7% (w/v) agar containing B5 salts supplemented with Suc (30 g L–1), α-napthaleneacetic acid (2.0 mg L–1), and 6-benzylaminopurine (0.2 mg L–1). Callus was transferred onto fresh agar medium every 3 to 4 weeks, when its weight had increased 4-fold in comparison with that of the inoculum.
To obtain regenerated plants, calli were transferred onto shoot-inducing medium containing α-napthaleneacetic acid (0.1 mg L–1) and 6-benzylaminopurine (2.0 mg L–1). About eight to 10 plantlets were obtained after two subcultures (one month each). After the rooting phase on growth medium without hormones, the plantlets were transferred into greenhouse conditions. Regenerated plants were crossed to non-transgenic tobacco to obtain stable epimutated HeLo1 lines.
DNA Probes
The following probes and plasmids were used in the hybridization experiments: The nptII coding sequence probe was an approximately 830-bp insert of the pGEMnptII plasmid (Van Houdt et al., 1997), the 35S promoter probe was an approximately 980-bp insert of the PGSJ290 plasmid (Van Houdt et al., 1997), the pTB29 plasmid carrying a 3.4-kb chloroplast genome insert (Sugiura et al., 1986), the pGEMrbcS plasmid carrying a 2.2-kb insert of the short subunit of rbcS (Rubisco gene; Ingelbrecht et al., 1994), and the p5SrDNA-NT plasmid carrying an approximately 120-bp fragment of the 5S rRNA gene from tobacco (Fulnecek et al., 1998).
DNA Isolation and Southern-Blot Hybridization
Total genomic DNA was isolated from approximately 10 g of leaves or lyophilized calli by a cetyltrimethylammonium bromide method as described previously (Kovarik et al., 2000). DNA methylation was analyzed with methylation-sensitive restriction endonucleases. Genomic DNA (10 μg) was digested with an excess of the enzymes (5 units μg–1 DNA) and subjected to electrophoresis on 0.8% (w/v) agarose gel. After electrophoresis, the gels were alkali blotted onto Hybond-XL membranes (Amersham Biosciences, Little Chalfont, UK) and hybridized with 32P-labeled DNA probes (DekaLabel kit, MBI, Fermentas, Vilnius, Lithuania). After washing under high-stringency conditions, the hybridization bands were visualized with a PhosphorImager (Storm, Molecular Dynamics, Sunnyvale, CA), and the data were processed by ImageQuant software (Molecular Dynamics). Completeness of digestion was monitored by rehybridization of the blots with a chloroplast probe. Relative methylation levels of particular sites or groups of sites were determined by PhosphorImager quantification and comparison of the hybridization signals of bands corresponding to methylated and non-methylated DNA molecules.
Protein Extraction and NPTII ELISA
To extract soluble proteins, the lyophilized callus tissue (20–40 mg dry weight) was ground with 200 to 400 μL extraction buffer (0.25 m Tris-HCl [pH 8.0] and 1 mm phenylmethylsulfonylfluoride) and centrifuged at 20,000g to remove cell debris. The protein concentration of the extracts was determined according to the Bio-Rad protein assay (Bio-Rad, Hercules, CA). The NPTII ELISA was done according to the manufacturer's instructions (5 Prime→3 Prime, Boulder, CO). The microtiter plates were read at 405 nm on the ELISA reader (Amersham Biosciences) using a kinetic program (5-min intervals for 2 h).
RNA Isolation and Northern-Blot Hybridization
Total RNA was isolated from young leaves using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The RNA quality was checked by electrophoresis on a 1% (w/v) agarose gel. RNA was subjected to electrophoresis in 1.2% (w/v) formaldehyde-agarose gels according to a Qiagen-recommended protocol. After electrophoresis, the gel was washed for 10 min in sterile water to remove formaldehyde, and RNA was denatured in 0.05 m NaOH and blotted onto Hybond-XL membrane (Amersham Biosciences) in 20× standard saline citrate (150 mm NaCl and 15 mm Na3-citrate [pH 7.0]). Hybridization with a 32P-labeled probe was executed in ULTRAhyb buffer (Ambion, Austin, TX) for 24 h at 40°C. After washing under low-stringency conditions (2× for 15 min in 2× standard saline citrate + 0.1% [w/v] SDS at 50°C), the hybridization bands were visualized using a PhosphorImager (Storm, Molecular Dynamics).
Run-On Transcription
Run-on transcription in isolated nuclei was performed as described by van Blokland et al. (1994). Young plant leaves (10–12 g) were ground in liquid nitrogen and resuspended in extraction buffer (10 mm NaCl, 10 mm MES [pH 6.0], 5 mm EDTA, 0.15 m spermidin, 20 mm β-mercaptoethanol, 0.6% [w/v] Triton X-100, and 0.25 m Suc). After purification on a Percoll gradient, nuclei were resuspended in 1.2 mL of storage buffer [10 mm Tris-HCl, 100 mm (NH4)2SO4, 10 mm MgCl2, and 5 mm β-mercaptoethanol] and stored with an equal amount of 99% (w/v) glycerol at –80°C.
For the run-on, 800 μL of nuclei was thawed and washed in storage buffer to remove glycerol. The reaction was started by adding nucleotide mix (final concentration 0.5 mm for each C, G, and ATP) and 100 μCi of [α-32P]UTP (6,000 Ci mmol–1; ICN Biomedicals, Irvine, CA) and stopped by SDS (final concentration 3% [w/v]) after incubation at 27°C for 30 min. Labeled RNA was purified by DNaseI treatment and phenol extractions and finally recovered by ethanol precipitation. The pellet was resuspended in 100 μL of sterile water and used as a probe for hybridization (approximately 2 × 106 cpm).
Plasmids carrying relevant inserts were linearized, denatured, and about 1 to 2 μg was bound to the nylon membrane (Hybond XL, Amersham Biosciences). Hybridization was performed in ULTRAhyb buffer (Ambion) for 48 h at 42°C. Membranes were washed in standard saline phosphate/EDTA + 0.1% (w/v) SDS at room temperature for 15 min and in the same solution at 60°C for 30 min. After overnight exposure to the screen, the radioactive signals were visualized using a PhosphorImager (Storm, Molecular Dynamics).
Distribution of Materials
Upon request, all novel materials described in this publication will be available in a timely manner for noncommercial purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank L. Jedlickova (Institute of Biophysics, Brno, Czech Republic) for technical assistance and Dr. Hans Peter Schoeb (Friedrich-Miescher Institute, Basel) for advice on the run-on assay protocol.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.023796.
This work was supported by the Grant Agency of the Czech Republic (grant nos. 521/01/0037 and Z5004920 to A.K. and 521/01/P042 to M.F.), by the Fund for Scientific Research (Flanders; visiting fellowship to A.K.), and by the Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (postdoctoral fellowship to H.V.H.).
References
- Arnaud P, Goubely C, Pélissier T, Deragon JM (2000) SINE retroposons can be used in vivo as nucleation centers for de novo methylation. Mol Cell Biol 20: 3434–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, van der Winden J, Matzke AJ, Matzke M (2002) RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA Suppl 99: 16499–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L, Bender J (2001) Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family. Nucleic Acids Res 29: 2127–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J, Bird A (1991) DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64: 1123–1134 [DOI] [PubMed] [Google Scholar]
- Christman JK, Sheikhnejad G, Marasco CJ, Sufrin JR (1995) 5-Methyl-2′-deoxycytidine in single-stranded DNA can act in cis to signal de novo DNA methylation. Proc Natl Acad Sci USA 92: 7347–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck S, Depicker A (2001) Disruption of their palindromic arrangement leads to selective loss of DNA methylation in inversely repeated gus transgenes in Arabidopsis. Mol Genet Genomics 265: 1060–1068 [DOI] [PubMed] [Google Scholar]
- Depicker A, Van Montagu M (1997) Posttranscriptional gene silencing in plants. Curr Opin Cell Biol 9: 373–382 [DOI] [PubMed] [Google Scholar]
- Dieguez MJ, Vaucheret H, Paszkowski J, Mittelsten Scheid O (1998) Cytosine methylation at CG and CNG sites is not a prerequisite for the initiation of transcriptional gene silencing in plants, but it is required for its maintenance. Mol Gen Genet 259: 207–215 [DOI] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S (1994) Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77: 993–1002 [DOI] [PubMed] [Google Scholar]
- Fulnecek J, Matyasek R, Kovarik A, Bezdek M (1998) Mapping of 5-methylcytosine residues in Nicotiana tabacum 5S rRNA genes by genomic sequencing. Mol Gen Genet 259: 133–141 [DOI] [PubMed] [Google Scholar]
- Ingelbrecht I, Van Houdt H, Van Montagu M, Depicker A (1994) Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc Natl Acad Sci USA 91: 10502–10506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43: 179–188 [DOI] [PubMed] [Google Scholar]
- Kilby NJ, Leyser HM, Furner IJ (1992) Promoter methylation and progressive transgene inactivation in Arabidopsis. Plant Mol Biol 20: 103–112 [DOI] [PubMed] [Google Scholar]
- Kloti A, He X, Potrykus I, Hohn T, Futterer J (2002) Tissue-specific silencing of a transgene in rice. Proc Natl Acad Sci USA 99: 10881–10886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A, Koukalova B, Bezdek M, Opatrny Z (1997) Hypermethylation of tobacco heterochromaic loci in response to osmotic stress. Theor Appl Genet 95: 301–306 [Google Scholar]
- Kovarik A, Van Houdt H, Holy A, Depicker A (2000) Drug-induced hypomethylation of a posttranscriptionally silenced transgene locus of tobacco leads to partial release of silencing. FEBS Lett 467: 47–51 [DOI] [PubMed] [Google Scholar]
- Kunz C, Narangajavana J, Jakowitsch J, Park YD, Rene Delon T, Kovarik A, Koukalova B, van der Winden J, Aufsatz W, Mette F et al. (2003) Studies on the effects of a flanking repetitive sequence on the expression of single copy transgenes in Nicotiana sylvestris and in N. sylvestris-N. tomentosiformis hybrids. Plant Mol Biol 52: 203–215 [DOI] [PubMed] [Google Scholar]
- Lam E, Benfey PN, Gilmartin PM, Fang RX, Chua NH (1989) Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci USA 86: 7890–7894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19: 5194–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Niedenhof I, ten Lohuis M (1994) Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrida. EMBO J 13: 2084–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhara I, Shirasawa-Seo N, Iwai T, Nakamura S, Honkura R, Ohashi Y (2002) Release from posttranscriptional gene silencing by cell proliferation in transgenic tobacco plants: possible mechanism for noninheritance of the silencing. Genetics 160: 343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Mourrain P, Beclin C, Vaucheret H (2000) DNA methylation and chromatin structure affect transcriptional and posttranscriptional transgene silencing in Arabidopsis. Curr Biol 10: 1591–1594 [DOI] [PubMed] [Google Scholar]
- Muller A, Marins M, Kamisugi Y, Meyer P (2002) Analysis of hypermethylation in the RPS element suggests a signal function for short inverted repeats in de novo methylation. Plant Mol Biol 48: 383–399 [DOI] [PubMed] [Google Scholar]
- Nelson M, Raschke E, McClelland M (1993) Effect of site-specific methylation on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res 21: 3139–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olhoft PM, Phillips RL (1999) Genetic and epigenetic instability in tissue culture and regenerated progenies. In HR Lerner, ed, Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization. Marcel Decker, New York, pp 111–148
- Park YD, Papp I, Moscone EA, Iglesias VA, Vaucheret H, Matzke AJ, Matzke MA (1996) Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J 9: 183–194 [DOI] [PubMed] [Google Scholar]
- Pélissier T, Thalmeir S, Kempe D, Sanger HL, Wassenegger M (1999) Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res 27: 1625–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Esteve PO (2003) Allosteric activator domain of maintenance human DNA (Cytosine-5) methyltransferase and its role in methylation spreading. Biochemistry 42: 5321–5332 [DOI] [PubMed] [Google Scholar]
- Selker EU (1999) Gene silencing: repeats that count. Cell 97: 157–160 [DOI] [PubMed] [Google Scholar]
- Schmitt F, Oakeley EJ, Jost JP (1997) Antibiotics induce genome-wide hypermethylation in cultured Nicotiana tabacum plants. J Biol Chem 272: 1534–1540 [DOI] [PubMed] [Google Scholar]
- Stam M, Viterbo A, Mol JN, Kooter JM (1998) Position-dependent methylation and transcriptional silencing of transgenes in inverted T-DNA repeats: implications for posttranscriptional silencing of homologous host genes in plants. Mol Cell Biol 18: 6165–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M, Shinozaki K, Zaita N, Kusuda M, Kumano M (1986) Clone bank of the tobacco (Nicotiana tabacum) chloroplast genome as a set of overlapping restriction endonuclease fragments: mapping of eleven ribosomal protein genes. Plant Sci 44: 211–216 [Google Scholar]
- Turker MS (2002) Gene silencing in mammalian cell and the spread of DNA methylation. Oncogene 21: 5388–5393 [DOI] [PubMed] [Google Scholar]
- Vaistij FE, Jones L, Baulcombe DC (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blokland R, van der Geest N, Mol JN, Kooter JM (1994) Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J 6: 861–877 [Google Scholar]
- Van Houdt H, Bleys A, Depicker A (2003) RNA target sequences promote spreading of RNA silencing. Plant Physiol 131: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt H, Ingelbrecht I, Van Montagu M, Depicker A (1997) Posttranscriptional silencing of a neomycin phosphotransferase II transgene correlates with the accumulation of unproductive RNAs and with increased cytosine methylation of 3′ flanking regions. Plant J 12: 379–392 [Google Scholar]
- Van Houdt H, Kovarik A, Van Montagu M, Depicker A (2000) Cross-talk between posttranscriptionally silenced neomycin phosphotransferase II transgenes. FEBS Lett 467: 41–46 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Beclin C, Fagard M (2001) Posttranscriptional gene silencing in plants. J Cell Sci 114: 3083–3091 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Fagard M (2001) Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet 17: 29–35 [DOI] [PubMed] [Google Scholar]
- Wassenegger M (2000) RNA-directed DNA methylation. Plant Mol Biol 43: 203–220 [DOI] [PubMed] [Google Scholar]