Abstract
In this study, genome-wide expression profiling based on Affymetrix ATH1 arrays was used to identify discriminating responses of Arabidopsis thaliana to five herbicides, which contain active ingredients targeting two different branches of amino acid biosynthesis. One herbicide contained glyphosate, which targets 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), while the other four herbicides contain different acetolactate synthase (ALS) inhibiting compounds. In contrast to the herbicide containing glyphosate, which affected only a few transcripts, many effects of the ALS inhibiting herbicides were revealed based on transcriptional changes related to ribosome biogenesis and translation, secondary metabolism, cell wall modification and growth. The expression pattern of a set of 101 genes provided a specific, composite signature that was distinct from other major stress responses and differentiated among herbicides targeting the same enzyme (ALS) or containing the same chemical class of active ingredient (sulfonylurea). A set of homologous genes could be identified in Brassica napus that exhibited a similar expression pattern and correctly distinguished exposure to the five herbicides. Our results show the ability of a limited number of genes to classify and differentiate responses to closely related herbicides in A. thaliana and B. napus and the transferability of a complex transcriptional signature across species.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-009-9590-y) contains supplementary material, which is available to authorized users.
Keywords: Differentiation of herbicides, Gene expression signature, Transfer from model to crop plant, Acetolactate synthase, 5-enolpyruvylshikimate-3-phosphate synthase
Introduction
Herbicides contain structurally diverse chemicals designed to inhibit targets in susceptible weeds, and, thus, help reduce crop yield losses or assist the selective growth of genetically modified plants. In general, important plant-specific processes, such as photosynthesis and the biosynthesis of cell walls, lipids and amino acids are the primary targets of these chemicals (Anderson 1996). Inhibitors of acetolactate synthase (ALS) have become one of the most important herbicide groups because of their large spectrum weed control activity, high crop selectivity, low application rates and low mammalian toxicity (Zhou et al. 2007). ALS is the first enzyme in the biosynthesis of the three branched chain amino acids- leucine, isoleucine, and valine. Several structurally divergent herbicide chemical families including sulfonylurea, imidazolinone, triazolopyrimidine, pyrimidinyl-thiobenzoate, and sulfonylamino-carbonyltriazolinone compounds possess ALS as their common primary target. Glyphosate is another amino acid biosynthesis inhibiting herbicide widely used on both grass and broadleaf weeds. It targets 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) involved in the biosynthesis of the aromatic amino acids phenylalanine, tyrosine, and tryptophan (Duke and Powles 2008). Glyphosate is widely used as a herbicide also in conjunction with genetically modified Roundup-Ready™ crops (http://monsanto.com).
A number of studies involving transcriptional analyses have been undertaken to understand the primary mode of action, detoxification mechanisms or diagnostic detection of different herbicides. Reverse transcription-PCR analysis in Arabidopsis thaliana identified three glutathione-S-transferases (GST) (AtGSTF2, AtGSTU1, AtGSTU24) and two isoforms of 12-oxophytodienoate reductase (OPR1, OPR2), which are putatively involved in the metabolism of chloroacetanilide herbicides and explosive compounds (Mezzari et al. 2005). Kelley et al. (2004, 2006) analyzed GH3, an auxin-regulated gene, as a potential candidate to assess and diagnose differential soybean injury caused by various plant growth regulator herbicides. Plant responses to various chemicals used as herbicides, herbicide-additives, or inhibitors have also been studied in Arabidopsis thaliana at the transcriptome level by using array hybridization technologies (affymetrix.arabidopsis.info/narrays/experimentbrowse.pl; Glombitza et al. 2004; Madhou et al. 2006; Manfield et al. 2004; Raghavan et al. 2005, 2006; Manabe et al. 2007). Glombitza et al. (2004) used a focused DNA array comprised of only 267 target sequences related to secondary metabolism to determine the differential effects of bromoxynil, primisulfuron and prosulfuron herbicides and other biotic and abiotic stresses on A. thaliana. Similarly, a microarray containing 600 barley cDNAs was used to study the impacts of 2,4-dichlorophenoxyacetic acid (2,4-D), cinidon-ethyl and tribenuron-methyl on wheat transcriptomes under controlled environment and field settings (Pasquer et al. 2006).
Affymetrix ATH1 expression analysis of Arabidopsis suspension cultured cells habituated to cellulose synthesis inhibitor isoxaben, implicated novel genes in cell wall assembly (Manfield et al. 2004). Treatment with 1 mM 2,4-D for 1 h regulated Arabidopsis genes involved in auxin response (IAA1, IAA5, IAA13, IAA19), ethylene signaling (ERS, CTR1, AtERF4, AtERF8), as well as ABA biosynthesis (NCED3), signaling and response (Raghavan et al. 2005). A follow-up study using a range of 2,4-D concentrations from auxinic (1 μM) to herbicidal (1 mM) activity revealed that in contrast to the response to high 2,4-D concentrations, both ABA biosynthesis and signaling was repressed at auxinic concentrations (Raghavan et al. 2006). By studying the global transcriptome expression of imidazolinone-sensitive (wild-type) and imidazolinone-resistant (csr1-2 D) mutant Arabidopsis plants, it has been observed that imazapyr did not significantly alter gene expression in csr1-2 D mutants and hence inferred that imidazolinone herbicides act solely by targeting CSR1, a catalytic subunit of ALS (Manabe et al. 2007). In contrast, their time course analysis in wild-type plants showed a dynamic change in gene expression pattern with induction of mostly detoxification genes at early time points, while genes related to amino acid biosynthesis, secondary metabolites and tRNA were induced at a later stage.
In contrast to these analyses, which either allowed single compound—single plant species interactions and/or considered only the active ingredients as the test herbicides, we compared Arabidopsis transcriptional responses to five related herbicides and attempted to transfer results obtained in the model plant to a related crop species. To allow better herbicidal uptake by the plants (cells) comparable to realistic field situations, field applicable commercial formulations were used that contains inert and non-toxic coformulants in addition to the active compounds. It is required by e.g., US EPA regulations that tests for non-target plant phytotoxicity are to be conducted with the typical end-use product and not with the technical grade of the active ingredient only (Electronic Code of Federal regulations, ECFR 2009). Four ALS-inhibiting herbicides and a glyphosate containing herbicide were chosen, targeting two different branches of plant amino acid biosynthesis (Table 1). Among the selected ALS-inhibiting herbicides two had active ingredients from the same chemical family, which contained a sulfonylurea backbone (sulfometuron and primisulfuron). The other two ALS-inhibiting herbicides contained active ingredients from other chemical families with completely unrelated structures: the imidazolinones and triazolopyrimidines. The major objectives of our study were: (a) to study the impact of these agronomically used herbicides on Arabidopsis transcriptome, (b) to identify a robust transcriptional signature in Arabidopsis that allows differentiating and classifying the response to closely related herbicides targeting the same enzyme (ALS), yet having different active compounds, or having even the same chemical backbones (sulfonylurea) and (c) to test if such a complex transcriptional signature identified in Arabidopsis could be transferred to a related, major agronomic species, Brassica napus. The latter objective might lay the groundwork for future research to apply gene expression signature in environmental risk characterization, such as non-target movement of herbicides. Moreover, it might also provide a general case of translational research from the model towards a related crop plant.
Table 1.
Characteristics of the five herbicides used in Arabidopsis thaliana (Col-0) and Brassica napus (Westar) gene expression studies
| Trade name | Manufacturer | Mode of action | Chemical family | Active ingredient (a.i.) |
|---|---|---|---|---|
| Arsenal™a | BASF Corporation, Research Triangle Park, NC, USA | ALS inhibitor | Imidazolinone | Imazapyr |
| Beacon™ | Syngenta Crop. Protection Inc., Greensboro, NC, USA | ALS inhibitor | Sulfonylurea | Primisulfuron-methyl |
| FirstRate™ | Dow AgroSciences LLC, Indianapolis, IN, USA | ALS inhibitor | Triazolopyrimidine | Cloransulam-methyl |
| Oust™ | E.I. du Pont de Nemours and Company, Wilmington, DE, USA | ALS inhibitor | Sulfonylurea | Sulfometuron methyl |
| Roundup original™ | Monsanto Company, St. Louis, MO, USA | EPSPS inhibitor | – | Glyphosate |
| Trade name | % a.i. | A.th. treatment concentration (g ha−1 a.i.)b | B.n. treatment concentration (g ha−1 a.i.)b | |
|---|---|---|---|---|
| Arsenal™a | 53.1 | 0.395 | 1.429 | |
| Beacon™ | 75.0 | 0.586 | 9.058 | |
| FirstRate™ | 84.0 | 0.163 | 0.549 | |
| Oust™ | 75.0 | 0.131 | 0.886 | |
| Roundup original™ | 41.0 | 10.652 | 79.745 |
Abbreviations: ALS acetolactate synthase, EPSPS 5-enolpyruvylshikimate-3-phosphate synthase, A.th. Arabidopsis thaliana, B.n. Brassica napus. Treatment concentrations were determined to result in comparable reductions in growth (“Materials and methods”). In addition to active ingredients, the herbicides contain inert components indicated in Material Safety Data Sheets
aApplicators concentrate
bThe herbicide rates [% of the field application rate active ingredient (f.a.r. a.i.)] estimated to produce a 50% reduction in shoot dry weight based on nonlinear regression analysis of data from a range-finding dose response study. A probit model was used to identify the EC50 values for A.th., while the Weibull model, which was developed later for use in these herbicide studies, was used with B.n.. For B.n. and Roundup Original™ the herbicide concentration used was enough to cause approximately 60% reduction in shoot dry weight based on the Weibull model
Results and discussion
Gene expression analyses in response to herbicides containing ALS- and EPSPS-inhibitors in A. thaliana
To identify effects of closely related herbicides at the gene expression level, Arabidopsis plants were treated with one of four ALS-inhibiting herbicides or a herbicide containing the EPSPS-targeting active ingredient glyphosate (Roundup Original™) at non-lethal concentrations producing an approximately 50% reduction in shoot dry weight (EC50, Table 1). Two of the ALS-targeting herbicides Oust™ and Beacon™, have active ingredients sulfometuron and primisulfuron respectively, with similar chemical structures derived from a sulfonylurea backbone. The other ALS-targeting herbicides contain completely unrelated active ingredients; Arsenal™ with the imidazolinone chemical imazapyr, and FirstRate™ with the triazolopyrimidine chemical cloransulam. Altered transcript levels in response to the herbicides were analyzed using Affymetrix ATH1 GeneChips. The array expression analyses were independently verified by quantitative real-time reverse transcription polymerase chain reaction. Although the magnitude of changes varied in a few cases between the two techniques, the majority (54/62) of cases either confirmed the directionality of change or verified the lack of a significant alteration (Table S1).
Common and distinct gene expression patterns induced by ALS- and EPSPS-inhibiting herbicides in A. thaliana
Only sixteen genes were significantly affected by Roundup Original™ application (Table S2), while 478 genes were significantly and coordinately regulated by all four ALS-inhibiting herbicides at the EC50 concentration (Table S3). In our study eight Roundup Original™ induced genes were linked to secondary metabolism and defense of which seven were also induced by ALS-inhibitors. All of them were potentially related to herbicide metabolism and detoxification, e.g., CYP71A13, UGT74E2, ABC transporter gene ATPDR12 and two MATE transporter genes (Table S2).
Among the 478 genes responsive to ALS-inhibiting herbicides, 82 were down-regulated and the remaining 396 were up-regulated. According to TAIR classifications, genes globally classified as cellular biosynthetic processes were enriched among differentially expressed genes (Table 2). The significant enrichment of included subcategories was more instructive. An effect on protein biosynthesis was noticed such as genes encoding ribosome associated proteins or protein synthesis initiation (eIF4A, eIF4E and eIF5), which were mostly up-regulated. Amino acid metabolic process was also found enriched, but at a lower stringency level (P-value 0.015). Only one gene (threonine ammonia lyase, AT3G10050) related to the BCCA group was induced along with six other genes related to amino acid biosynthesis. Other enriched categories included indole derivative metabolic process, camalexin metabolic process, and response to temperature, e.g., members of cytochrome P450 monooxygenases (CYP71A13, CYP71B15, CYP79B2) involved in camalexin biosynthesis (Mikkelsen et al. 2000; Nafisi et al. 2007) were up-regulated. Thus, specific branches of plant defense or general stress responses may be activated by ALS-inhibiting herbicides.
Table 2.
Functional classification according to TAIR ontology
| Functional category | Observed out of 477* | Observed frequency (%) | Expected out of 21,947 | Expected frequency (%) | P value |
|---|---|---|---|---|---|
| Cellular biosynthetic process | 57 | 11.90 | 1,304 | 5.90 | 8.87E-05 |
| Organelle organization and biogenesis | 34 | 7.10 | 652 | 3.00 | 5.80E-04 |
| Translation | 29 | 6.10 | 556 | 2.50 | 3.18E-03 |
| Ribonucleoprotein complex biogenesis and assembly | 23 | 4.80 | 201 | 0.90 | 2.04E-08 |
| Ribosome biogenesis and assembly | 20 | 4.20 | 122 | 0.60 | 4.48E-10 |
| RNA processing | 16 | 3.40 | 205 | 0.90 | 2.52E-03 |
| rRNA processing/metabolic process | 9 | 1.90 | 25 | 0.10 | 3.31E-07 |
| Indole and derivative metabolic process | 7 | 1.50 | 30 | 0.10 | 6.3E-04 |
| Camalexin metabolic process | 3 | 0.60 | 4 | 0.00 | 8.83E-03 |
| Response to temperature stimulus | 19 | 4.00 | 291 | 1.30 | 5.02E-03 |
| Syncytium formation | 4 | 0.80 | 10 | 0.00 | 9.17E-03 |
478 genes regulated similarly by four ALS-inhibiting herbicides were tested for over-representation against Arabidopsis thaliana ATH1 (23,334 genes) background population by using BioMaps tool at VirtualPlant (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb2/). Categories enriched at P < 0.01 are listed here. The method used to calculate P value of over-representation was hypergeometric distribution
* One gene was not assigned and hence total number of observed genes was reduced to 477
In addition, a further category also not directly related to the primary herbicide target was over-represented. Transcripts encoding proteins involved in cell wall modifications were classified under the enriched syncytium formation sub-category (Table 2). All of them were expansin family genes EXPA1, EXPA3, EXPA6, EXPA8 and were down regulated (Table S3). In addition to the over-representation of functional categories, several individual, but related transcripts were also changed. An α-fucosidase (AT1G67830) involved in controlling growth by cell wall loosening (Torre et al. 2002) was repressed. This was concomitant with the induction of genes related to ethylene biosynthesis and perception, such as ACC synthase (AT4G11280), ACC oxidase (AT1G05010, AT5G43450) and the transcriptional co-activator AtMBF1A (AT2G42680). Thus, these particular genes could be involved in the herbicide induced growth retardation and accelerated senescence.
An important factor controlling herbicide selectivity is the differential rate of herbicide detoxification in crop and weed species. The enzymatic systems involved in herbicides metabolism include cytochrome P450 monooxygenases (CYP), GST and UDP-glycosyltransferases (UGT), which are also involved in plant secondary metabolism (Wagner et al. 2002; Messner et al. 2003; Brazier-Hicks et al. 2007). Some of the most highly induced genes were members of these families with unknown substrates such as CYP89A2, CYP81D8, CYP91A1, GSTU9, GSTU24, GSTZ1, UGT74E2, UGT76B1, UGT87A2 (Table S3). They could be related to the metabolism of ALS-inhibiting herbicides irrespective of their chemical structure.
Changes in the expression of genes involved in central energy pathways were also observed. A number of genes encoding components of mitochondrial transporters indicated enhanced organellar membrane exchange and metabolic activity. The up-regulation of two mitochondrial alternative oxidases ATAOX1a (AT3G22370) and ATAOX1b (AT3G22360), along with two mitochondrial NADH dehydrogenases, NDB2 (AT4G05020) and NDB4 (AT2G20800) point to a specific, concerted action to prevent oxidative damage (Table S3). A similar co-expression pattern of ATAOX1a and NDB2 had previously been observed in response to imidazolinone (Manabe et al. 2007). The treatment with sulfonylurea and imidazolinone herbicides triggered induction of AOX enzyme activity in sycamore cells, while no such change was observed with glyphosate treatment (Aubert et al. 1997).
Molecular signature for differentiating herbicide responses
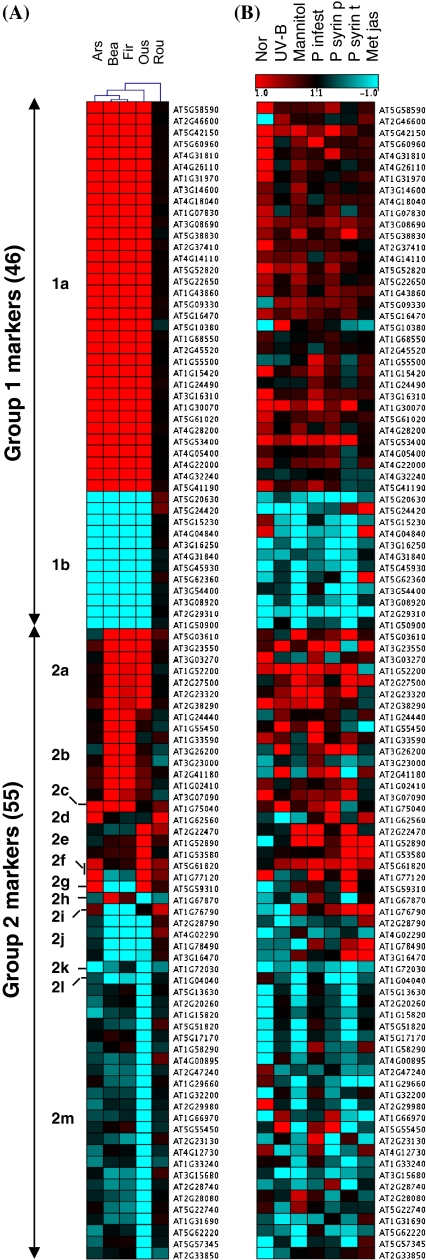
Besides providing insight regarding their molecular effects, herbicide-specific changes in gene expression provided an opportunity to analyze whether the combined expression signature of any genes could establish a collective molecular marker for differentiating responses among closely related herbicides. Metabolite profiling has been useful to diagnose herbicide treatments on plants (Aranibar et al. 2001; Sauter et al. 1988). However, metabolites may differ depending on the actual enzymatic activities in different plants or species and measuring mRNAs is much more standarized than quantifying metabolite levels. Therefore, identifying mRNA markers to develop a signature that is sensitive enough to classify even related herbicides that have either similar chemical backbone or a common primary target would be very useful to diagnose herbicide treatments on plants. To identify such a signature, we used stringent statistical thresholds to obtain a set of gene transcripts that could characterize each herbicide response (Fig. 1a, Table S4, “Materials and methods”). First, a change larger than twofold in at least one experiment with opposite or no response in at least one other treatment was required. Besides differentiating the herbicide responses, this measure also antagonized the selection of general stress responsive genes. Second, genes had to be well expressed in control experiments in order to select reliable alterations in transcription, although some potent markers may be eliminated thereby. A total of 101 candidate marker genes were selected and classified into two major groups. Group 1 contained 46 candidate genes that differentiated the ALS-inhibiting group from the EPSPS group, of which 34 were up-regulated and 12 were down-regulated. Group 2 contained 55 candidates that differentiated the responses among the four ALS-inhibiting herbicides, as well as towards glyphosate. Thirteen different combinations of gene expression patterns were observed for group 2 markers (Fig. 1a, Table S4).
Fig. 1.
Composite signature of transcriptional responses of 101 marker genes to herbicide treatments and selected stresses. a Clustering of the responses to five herbicide treatments (Table S4). Group 1 marker differentiates all ALS-inhibitors from EPSPS-inhibitor and group 2 marker differentiates among four ALS inhibitors and towards EPSPS-inhibitor. The TIGR/TAIR 6 genome annotation code was used as the gene identifier. b The two top correlated stress response patterns with each herbicide (Table S8) were assorted to the gene matrix of panel A showing a signature clearly distinct from all herbicides in all seven cases. Red color indicates up-regulation of transcripts in treatment group compared to control; blue indicates down-regulation and black indicates no change. Abbreviations: ARS [Arsenal™, imazapyr active ingredient (a.i.)], BEA (Beacon™, primisulfuron a.i.), FIR (FirstRate™, cloransulam a.i.), OUS (Oust™, sulfometuron a.i.), ROU (Roundup Original™, glyphosate a.i.), Nor norflurazon, UV-B ultraviolet B, P infest: Phytophthora infestans; P syrin p: Pseudomonas syringae pv. phaseolicola; P syrin t: Pseudomonas syringae pv. tomato avr Rpm1; Met jas: methyl jasmonate
Cluster analysis revealed a close similarity between Beacon™ (a.i. primisulfuron) and FirstRate™ (a.i. cloransulam) mediated transcriptional alteration. Arsenal™ (a.i. imazapyr) also was associated with this pair of herbicides, while Oust™ (a.i. sulfometuron) was not. Thus, surprisingly, the effects of two SU herbicides (Beacon™ and Oust™) on gene transcription could be differentiated. This analysis suggested that the composite signature from these 101 genes could differentiate the plant’s responses towards herbicides that even possess structurally related active compounds.
Testing the validity of the identified molecular signature
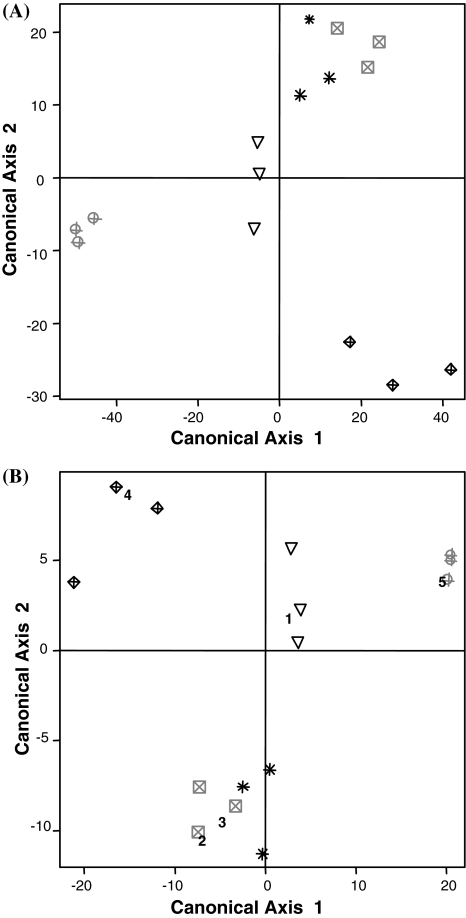
To test the potential of these diagnostic genes as a marker set of herbicide exposure in Arabidopsis, we first calculated correlations of individual herbicide experiments. Correlations between individual samples of different treatment groups were, except for FirstRate™ versus Beacon™, considerably lower than within group correlations (Table S5). Roundup Original™ did not show a correlation to any of the other treatment groups. To test the reproducibility of independently conducted experiments as well as a prediction of how well any single experiment would identify the correct herbicide signature we employed a classification strategy. Diagonal discriminant analysis has been tested useful in gene expression data (Dudoit et al. 2002). Therefore, this method was applied to our data set. The performance of the classification to the herbicide treatments was assessed by a leave-one-out cross-validation. The cross-validation rate was 100%, i.e., each 101-gene-signature of the biologically independent replicates was correctly classified to the corresponding herbicide group (Fig. 2a). Hence, the global similarity of FirstRate™ and Beacon™ as measured by their high correlations (Table S5) was resolved by using the supervised learning algorithm. These results confirmed the high reproducibility of the independent replicates as well as the distinctive properties and robustness of the signature provided by the 101 marker set.
Fig. 2.
Classification of independent Arabidopsis experiments based on leave-one-out cross-validation and association of B. napus signatures with reduced Arabidopsis data set. a Canonical coefficients of a diagonal discriminant analysis based on leave-one-out cross-validation with the transcriptional responses of 101 Arabidopsis marker genes. All 15 experiments were perfectly classified to the correct herbicide signature (“Materials and methods”). b Canonical coefficients of a diagonal discriminant analysis with the reduced data set of 23 Arabidopsis genes and orthologous B. napus data. Even with this reduced data set the Arabidopsis signatures are correctly classified with the respective herbicide treatments (symbols). Furthermore, the signatures of the orthologous B. napus genes (numbers) are correctly associated to the Arabidopsis herbicide treatments (symbols). With only two components displayed, both the B. napus BEA and FIR expression patterns seem to be related to the A. thaliana BEA data; however, with all four components they are correctly differentiated (not shown). Abbreviations: ARS [triangle 1, Arsenal™, imazapyr active ingredient (a.i.)], BEA (square, 2, Beacon™, primisulfuron a.i.); FIR (star, 3, FirstRate™, cloransulam a.i.); OUS (rhombus, 4: Oust™, sulfometuron a.i.); ROU (sphere, 5, Roundup Original™, glyphosate a.i.)
Distinguishing herbicide responses from other chemical and stress treatments
To compare the expression pattern of the identified molecular markers for herbicides with other major stress responses, expression data were collected from quality-controlled Affymetrix ATH1 microarray experiments for a wide range of abiotic, biotic and plant growth regulators stresses. In particular, the herbicide marker expression pattern was compared to a broad range of chemical compounds that inhibit diverse physiological processes, such as photosystem I (methyl viologen), photosystem II (PNO8), carotenoid biosynthesis (norflurazon), gibberellin biosynthesis (prohexadione, uniconazole, paclobutrazol, propiconazole), auxin transport (TIBA, NPA) and brassinosteroid biosynthesis (Brz220) (Table S6, “Materials and methods”). All selected experiments had been performed using leaves or seedlings of the Col-0 genetic background similar to this study. In addition, 72 out of 101 marker genes displayed a constitutive expression across four developmental stages (seedling until rosette) that cover all these experiments (Table S7).
Determination of Pearson correlation coefficients by comparing the expression patterns altered by herbicides and diverse stress treatments clearly demonstrated that expression data from none of the other stress experiments were similar to the studied herbicide treatments. Herbicide-to-herbicide correlations were significantly better in all cases (Tables S5, S8). Even an outlier herbicide replication for Roundup Original™ (Rou2) exhibited a greater tendency towards glyphosate correlation than to any other stress treatment. The transcriptional signatures of the stress treatments showing the two top correlations (Table S8) with each of the five herbicides were selected and displayed along with the herbicide signatures to visualize this differentiation. Clear distinctions of the expression patterns for herbicides were found (Fig. 1a vs. b). More importantly, the majority of the included chemical compounds inhibits diverse biochemical pathways (Table S6) and did not have overlapping expression signatures with the compounds used in this study.
Identification of Brassica genes showing similar expression patterns
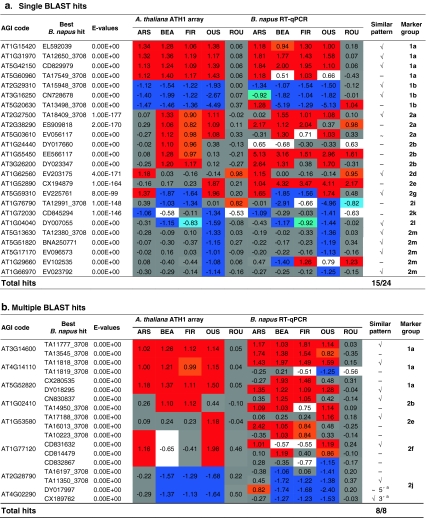
To independently validate the Arabidopsis transcript-based herbicide signature and to explore its translation into a related Brassica crop species, genes homologous to the 101 markers were searched within the TIGR EST transcript assemblies of Brassica napus (Childs et al. 2007) using the reciprocal best BLAST hit method based on nucleotide sequences. Out of the 101 query genes, 14 failed to report any EST sequences (Table S9). Single or multiple bi-directional best matches were found for 32 Arabidopsis markers covering 13 different expression patterns (Table 3). In total, marker group 1a had the highest number (seven) of B. napus homologs, while each of the groups 2d, 2f, 2g, 2i, 2k and 2l were represented by one candidate. For marker groups 2c and 2h, no B. napus homologs could be detected in the available sequence database (Table 3).
Table 3.
Comparative expression analyses of A. thaliana marker genes and B. napus best BLASTN hit ESTs
Changes in gene expression of herbicide treatment versus control are displayed as log2-values. Orange (>0.8) and red (>1) indicate induction, light blue (<−0.8) and blue (<−1) highlight suppression; non-significant changes (−0.5 < ratio < 0.5) are labeled grey
aNon-overlapping independent ESTs may represent 5′- and 3′-part of the orthologous gene. Abbreviations ARS [Arsenal™, imazapyr active ingredient (a.i.)], BEA (Beacon™, primisulfuron a.i.), FIR (FirstRate™, cloransulam a.i.), OUS (Oust™, sulfometuron, a.i.), ROU (Roundup Original™, glyphosate a.i.)
Due to the genome triplication in the Brassica lineage, B. napus homologs may not preserve all features of the corresponding Arabidopsis genes (Town et al. 2006; Lysak et al. 2007). To account for these complications, candidate B. napus genes had to be tested whether they exhibited similar expression patterns to the herbicides as observed for their respective counterparts in A. thaliana. In order to apply functionally comparable herbicide amounts, B. napus plants were also treated with EC50 concentrations of each herbicide (“Materials and methods”, Table 1) and the corresponding expression changes for each candidate were assessed by RT-qPCR analyses. Fifteen among the 24 single hit B. napus candidate genes (62.5%) showed similar response patterns as their A. thaliana counterparts (Table 3). It is possible that for the remaining 9 marker genes, a more conserved or functionally related copy has yet to be discovered in B. napus.
For eight Arabidopsis sequences, multiple hits with same e-values (0) were reported (Table 3). In all cases, however, a similar expression pattern could be pinpointed to a single representative. Thus, the expression pattern could be useful to distinguishing between the orthologous isoform that maintained the sub-functions for herbicide responsiveness and those that likely have lost this sub-function while retaining high sequence conservation. Moreover, in one case two non-overlapping hits were obtained, which were homologous to either the 5′- (DY017997) or 3′- (CX189762) part of AT4G02290. Interestingly, both homologs were expressed in a similar way like the A. thaliana gene suggesting that the transcript assemblies map to one B. napus locus.
Correct classification of B. napus herbicide exposure even based on a reduced set of genes
In total, 23 B. napus genes were identified that exhibited a similar response pattern to the herbicides like that of their homologous Arabidopsis genes. Since only three groups (groups 2c, 2h, 2i) out of fifteen different expression patterns in the original 101 marker set were not represented, it was examined whether such a reduced set could still allow a correct classification of the herbicide responses. Indeed, a diagonal discriminant analysis applying only 23 Arabidopsis genes again separated the five herbicides and a 100% cross-validation rate was found by a leave-one-out cross-validation assessment (Fig. 2b). Next, the B. napus genes herbicide-dependent expression patterns were classified with these Arabidopsis data and each Brassica pattern was correctly associated with the corresponding herbicide pattern. Thus, a robust gene expression signature for the differentiation of herbicide responses could be developed in A. thaliana and B. napus.
Conclusions
In this study changes in gene expression were identified, that provided enhanced molecular insight regarding the effects of five herbicides targeting two different branches of amino acid biosynthesis. A set of 101 gene transcripts could be extracted from these analyses that provided a composite diagnostic signature to differentiate among Arabidopsis responses to related herbicides, which was clearly distinct from those provoked by other stress treatments and chemical compounds. The possible specificity of the diagnostic signature was corroborated by the finding that none of the herbicide-to-stress correlations reached the quality of herbicide-to-herbicide correlations. Interestingly, even herbicides with active ingredients having the same enzymatic target (ALS) or similar chemical structures (sulfonylurea) could be distinguished. However, using the commercial herbicide formulations precludes a unambiguous assignment of these differences due to the active chemicals only, since we cannot rule out that non-active ingredients used in the herbicide formulation might have an effect on plant gene expression. On the other hand, in contrast to studies using pure active ingredients this work provided the opportunity to establish gene expression changes and marker signatures, which are associated with the formulated herbicides as they are used in agriculture and as they facilitate better access to the treated plant (cells). Furthermore, the successful transfer of a diagnostic composite transcript signature from the model plant into the crop species Brassica napus could be demonstrated—despite the triplicate nature of the Brassica genome and the lack of a complete genome or transcriptome sequence. In the particular case studied here a robust gene expression pattern has been established that allows the specific differentiation and classification of closely related herbicides. Its ability to classify the correct herbicidal exposures highlights future implications of such transcript-based signatures in environmental studies such as non-target movement of herbicides, or in a broader perspective in analogous analyses of the impact of chemicals on organisms.
Materials and methods
Herbicide treatment and isolation of RNA
Prior to carrying out gene expression studies, range-finding experiments were conducted to identify concentrations of the five herbicides that would produce similar growth responses in 14-day-old seedlings of Arabidopsis thaliana (accession Col-0) and Brassica napus cv. Westar (Table 1). Four of the herbicides were ALS-inhibitors. Two of them Oust™ and Beacon™, have active ingredients (a.i.), sulfometuron and primisulfuron, respectively, with similar chemical structures derived from a sulfonylurea backbone. The other ALS-targeting herbicides contain completely unrelated active ingredients; Arsenal™ with imazapyr, and FirstRate™ with cloransulam. The fifth herbicide Roundup Original™ contained an EPSPS-targeting active ingredient (glyphosate). A surfactant (Preference™, Agriliance, LLC, St. Paul, MN, USA) was added to each of the herbicide solutions as recommended on the label to enhance herbicide efficacy. An additional set of plants was sprayed only with the surfactant and used as a control. Plants were maintained at 22–23°C with a 16 h photo-period (175–210 μmol m−2 s−1).
For expression analyses plants were grown and treated with the concentration of each herbicide producing a 50% reduction in shoot dry weight (Table 1) or the surfactant solution as a control. Hence, the application of a comparable functional dose (EC50) would allow direct comparison of herbicide regulated gene expression patterns in these two species. Three independent biological replicates (individual pots) were used per herbicide or control treatment. Leaves without visible symptoms were collected at 24 h from at least ten plants post treatment. Total RNA was isolated from approximately 200 mg of pooled leaf tissue using TRIzol reagent (Invitrogen, USA), DNase treated and further purified with RNAeasy mini spin columns (Qiagen, USA).
RNA labeling, hybridization and scanning
RNA quality analyses (Agilent Bioanalyzer 2100, Agilent Technologies, USA) and array hydridizations were conducted at the Center for Genome Research and Biocomputing Core Laboratories of Oregon State University, USA. A. thaliana whole genome oligonucleotide microarray (ATH1, Affymetrix) were used for hybridization of each replicon and scanned with an Affymetrix GeneChip™ Scanner 3000.
Microarray data analysis
Array images were first visually screened following each scan to correct for signal artifacts, scratches or debris. Then the arrays were processed with MAS 5.0 algorithm to check for data quality based on the four important parameters: scale factor (<3), average background level (<100), percentage of genes present and ratios derived from 3′- and 5′-feature of housekeeping genes. CEL files were imported to ChipInspector (Genomatix, Germany) to check if dynamic range of any array deviates from the average. Data was normalized on the single probe level (Cohen et al. 2008). Each raw value of an array was adjusted to make the mean intensity the same for each individual array in an analysis. Probe-sets were defined following Haberer et al. (2006) based on TIGR/TAIR 6 genome annotation release. Statistical algorithm for permutated t-test with false discovery rate (FDR) calculation was similar to SAM (Tusher et al. 2001).
Multi-class analysis was performed in ChipInspector (Genomatix, Germany) based on exhaustive matching combination within an experimental class. Statistical thresholds applied were: FDR-0, median number of falsely called features—0, chosen delta value for regulation—0.14 and minimum probe coverage—3. Transcripts having ≥twofold (1.0 in log2) changes in abundance with P-values ≤0.05 (Tables S2, S3) were functionally categorized by MapMan 2.0.0 (Usadel et al. 2005) and TAIR (www.arabidopsis.org) gene classifications. The BioMaps tool at www.virtualplant.org was used to identify statistically over-represented groups based on TAIR classifications. Significance is based on P-values corrected for multiple testing by hypergeometric distribution (Table 2).
Stringent statistical cut-off criteria were applied to check whether transcript profiling could be used to identify markers for differentiating responses of A. thaliana to the applied herbicides. A change of ≥twofold (1.0 in log2) in at least one treatment group and opposite or no deregulation in at least one other treatment group at P ≤ 0.05 was set as an initial cut-off level (Table S4). In addition, an average expression threshold value of 7.0 (MAS 5.0, normalized, log scale, target value 200) was introduced to exclude transcripts of low abundance and low reliability. Hierarchical cluster analysis (HCL) of the markers was conducted by average linkage agglomerative clustering using the Euclidean distance (Fig. 1a) according to Rainer et al. (2006).
Candidate markers genes were analyzed for other stresses responses across the ATH1 Affymetrix based microarray experiments available from the NASCArrays database as described previously (Aceituno et al. 2008). Expression data from 17 abiotic, 14 biotic, 8 plant growth regulators and 14 chemical compound treatments at the 2-leaved rosette to early flowering stage were selected (Table S6). The basal expression patterns of the markers were checked at www.genevestigator.ethz.ch (Table S7).
The similarity of the transcriptional response to herbicides and selected stresses was analyzed by Pearson correlation coefficients (Tables S5, S8). For the statistical anaylsis and classification of samples into treatment groups based on microarray data several methods have been suggested (e.g., diagonal discriminant analysis (DDA), support vector machines (SVM), and k-nearest neighbour). Misclassifation rates for many discriminant methods have been compared using real or simulated data. Dudoit et al. (2002) found that DDA has smallest error rates. For typical data matrices as encountered with a small number of microarrays with numerous gene expression values the crucial methodological aspect is a stable estimation of the covariance matrix. In this situation DDA is a robust method, because only the diagonal of the covariance matrix (variances) is estimated.
The method diagonal discriminant analysis was trained with the gene marker sets given in Table S10 or in Table 3. Leave-one-out cross-validation (LOOCV) was performed to check the efficiency of the diagnostic markers in predicting the particular herbicide to which Arabidopsis plants had been exposed. It is a common choice for the small sample sizes usually encountered in microarray studies. Each sample is left out, one at a time. For each training set constructed in that way, i.e., 14 experiments in this project, a DDA was calculated and the discriminant function was used to predict the class of the left-out sample. This prediction was then counted as being correct or incorrect. By repeating this procedure for each sample the total number of classification errors was determined (Simon et al. 2003).
Quantitative real time RT-PCR (RT-qPCR) analyses
Fourteen Arabidopsis genes were selected for validation of microarray data by RT-qPCR using the same RNA samples as for the array experiments (Table S11). A QuantiTect SYBR Green RT-PCR kit (Qiagen, USA) was used for one-step real time RT-PCR amplification in an ABI prism 7700 (SDS 1.9.1, Applied Biosystems, USA). The herbicide-regulated expression of candidate B. napus homologs was also analyzed by RT-qPCR assay. Primers were designed based on EST sequences (Table S12). The real-time expression data were subjected to analysis by geNORM (Vandesompele et al. 2002), which identified ACTIN2 (AT3G18780) and UBQ10 (AT4G05320) as the two most stably expressed genes in A. thaliana, while ACTIN7 (AF111812) and Actin (AY570244) for B. napus. Hence, ACTIN2 and ACTIN7 were used as a single reference gene for data normalization for Arabidopsis and Brassica, respectively. As a reciprocal control, we checked for the relative ACTIN2 expression also among the ATH1 analyses; indeed the gene proved to be highly stable with relative expression levels of 0.952–0.974 in case of all five herbicide treatments. Relative fold change in gene expression was estimated following the ΔΔCt method with the carrier-sprayed sample as calibrator (Livak and Schmittgen 2001). Each value represents the mean of three biological replicates with two technical replicates each (Tables 3, S1).
Identification of homologous genes in Brassica napus
Candidate homologs between the Arabidopsis coding sequences (TAIR6 annotation; Poole 2007) and the TIGR transcript assemblies of Brassica napus comprising 35,460 singletons and tentative consensus sequences were identified (Childs et al. 2007). In the absence of synteny- or map-based genome data, candidate homologs between Arabidopsis and Brassica were defined as reciprocal best BLASTN hit (RBH; Hirsh and Fraser 2001; Haberer et al. 2006). BLASTN provides a higher selectivity by e.g., recognizing also synonymous changes in case of species from one family. In case of two or more reciprocal top scoring matches showing equal scores (e-values) between Brassica ESTs and an Arabidopsis gene model, we considered all top scoring matches for further analysis and designated such relations as ‘multiple’ (bi-directional best BLAST) hits.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was performed while MD held a National Research Council Associate Award at US Environmental Protection Agency’s National Health and Environmental Effect Research Laboratory, Corvallis, Oregon. This study was funded in part by the U.S. Environmental Protection Agency under contract number 68-D-01-005 to Dynamac Inc and also by FONDECYT 1060457 and NSF DBI 0445666 to R.A.G. It has been subject to the agency’s peer plus administrative review and has been approved for publication as an EPA document. Mention of trade names or commercial products does not constitute endorsement or recommendation for use. We sincerely thank Anne-Marie Girard, Caprice Rosato CGRB, OSU for providing technical support with the microarray experiments, Martin Irmler, Helmholtz Zentrum for help with microarray data evaluation, Dieter Ernst, Helmholtz Zentrum for his comments on the MS, E Henry Lee, EPA for statistical advises, John Fowler, OSU, Olivier Da Ines, Helmholtz Zentrum and Jim Westwood, Virginia Tech for fruitful discussions and Henry Miller, SEE employee, EPA for harvesting plants.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Author contributions
MD, DO, TF, JR, LW, and AS conceptualized the research; MD performed expression analyses in Arabidopsis and B. napus; JR assisted in expression analyses; DO and TP were responsible for herbicide treatments and related analyses; MD, TS, RG, FA, GH, MM, GW performed statistical and comparative data analyses, MD wrote the manuscript with contributions from AS, RG, and DO.
References
- Aceituno FF, Moseyko N, Rhee SY, Gutiérrez RA. The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genomics. 2008;9:438. doi: 10.1186/1471-2164-9-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson WP. Weed science: principles. 3. St. Paul: West Publishing Co.; 1996. p. 388. [Google Scholar]
- Aranibar N, Singh BK, Stockton GW, Ott KH. Automated mode-of-action detection by metabolic profiling. Biochem Biophys Res Commun. 2001;286:150–155. doi: 10.1006/bbrc.2001.5350. [DOI] [PubMed] [Google Scholar]
- Aubert S, Bligny R, Day DA, Whelan J, Douce R. Induction of alternative oxidase synthesis by herbicides inhibiting branched chain amino acid synthesis. Plant J. 1997;11:649–657. doi: 10.1046/j.1365-313X.1997.11040649.x. [DOI] [Google Scholar]
- Brazier-Hicks M, Offen WA, Gershater MC, Revett TJ, Lim EK, Bowles DJ, Davies GJ, Edwards R. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci USA. 2007;104:20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell R, Chan AP. The TIGR plant transcript assemblies database. Nucleic Acids Res. 2007;35:D846–D851. doi: 10.1093/nar/gkl785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CD, Lindenmeyer MT, Eichinger F, Hahn A, Seifert M, Moll AG, Schmid H, Kiss E, Gröne E, Gröne HJ, Kretzler M, Werner T, Nelson PJ. Improved elucidation of biological processes linked to diabetic nephropathy by single probe-based microarray data analysis. PLoS ONE. 2008;3(8):e2937. doi: 10.1371/journal.pone.0002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit S, Fridlyand J, Speed TP. Comparison of discriminant methods for the classification of tumors using gene expression data. J Am Stat Assoc. 2002;97:77–87. doi: 10.1198/016214502753479248. [DOI] [Google Scholar]
- Duke SO, Powles SB. Glyphosate: a once-in-a-century herbicide. Pest Manag Sci. 2008;64:319–325. doi: 10.1002/ps.1518. [DOI] [PubMed] [Google Scholar]
- ECFR (2009) Data requirements for pesticides. http://ecfr.gpoaccess.gov/cgi/t/text/text-idx?c=ecfr&sid=013b05537f6069487ae3f2252ae1d5a0&rgn=div5&view=text&node=40:23.0.1.1.9&idno=40#40:23.0.1.1.9.7.1.2
- Glombitza S, Dubuis PH, Thulke O, Welzl G, Bovet L, Götz M, Affenzeller M, Geist B, Hehn A, Asnaghi C, Ernst D, Seidlitz HK, Gundlach H, Mayer KF, Martinoia E, Werck-Reichhart D, Mauch F, Schäffner AR. Crosstalk and differential response to abiotic and biotic stressors reflected at the transcriptional level of effector genes from secondary metabolism. Plant Mol Biol. 2004;54:817–835. doi: 10.1007/s11103-004-0274-3. [DOI] [PubMed] [Google Scholar]
- Haberer G, Mader MT, Kosarev P, Spannagl M, Yang L, Mayer KFX. Large scale cis-element detection by analysis of correlated expression and sequence conservation between Arabidopsis and Brassica oleracea. Plant Physiol. 2006;142:1589–1602. doi: 10.1104/pp.106.085639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh AE, Fraser HB. Protein dispensability and rate of evolution. Nature. 2001;411:1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- Kelley KB, Lambert KN, Hager AG, Riechers DE. Quantitative expression analysis of GH3, a gene induced by plant growth regulator herbicides in soybean. J Agric Food Chem. 2004;52:474–478. doi: 10.1021/jf035134l. [DOI] [PubMed] [Google Scholar]
- Kelley KB, Zhang Q, Lambert KN, Riechers DE. Evaluation of auxin responsive genes in soybean for detection of off-target plant growth regulator herbicides. Weed Sci. 2006;54:220–229. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Kwok C, Michaela K, Petr B. Ancestral chromosomal blocks are triplicated in Brassicaceae species with varying chromosome number and genome size. Plant Physiol. 2007;145:402–410. doi: 10.1104/pp.107.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhou P, Raghavan C, Wells A, Stevenson TW. Genome-wide microarray analysis of the effect of a surfactant application in Arabidopsis. Weed Res. 2006;46:275–283. doi: 10.1111/j.1365-3180.2006.00511.x. [DOI] [Google Scholar]
- Manabe Y, Tinker N, Colville A, Miki B. CSR1, the sole target of imidazolinone herbicide in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:1340–1358. doi: 10.1093/pcp/pcm105. [DOI] [PubMed] [Google Scholar]
- Manfield IW, Orfila C, McCartney L, Harholt J, Bernal AJ, Scheller HV, Gilmartin PM, Mikkelsen JD, Knox JP, Willats WGT. Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J. 2004;40:260–275. doi: 10.1111/j.1365-313X.2004.02208.x. [DOI] [PubMed] [Google Scholar]
- Messner B, Thulke O, Schäffner AR. Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta. 2003;212:138–146. doi: 10.1007/s00425-002-0969-0. [DOI] [PubMed] [Google Scholar]
- Mezzari MP, Walters K, Jelinkova M, Shih MC, Just CL, Schnoor JL. Gene expression and microscopic analysis of Arabidopsis exposed to chloroacetanilide herbicides and explosive compounds. A phytoremediation approach. Plant Physiol. 2005;138:858–869. doi: 10.1104/pp.104.056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem. 2000;275:33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell. 2007;19:2039–2052. doi: 10.1105/tpc.107.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquer F, Ochsner U, Zarn J, Keller B. Common and distinct gene expression patterns induced by the herbicides 2, 4-dichlorophenoxyacetic acid, cinidon-ethyl and tribenuron-methyl in wheat. Pest Manag Sci. 2006;62:1155–1167. doi: 10.1002/ps.1291. [DOI] [PubMed] [Google Scholar]
- Poole RL. The TAIR database. Methods Mol Biol. 2007;406:179–212. doi: 10.1007/978-1-59745-535-0_8. [DOI] [PubMed] [Google Scholar]
- Raghavan C, Ong EK, Dalling MJ, Stevenson TW. Effect of herbicidal application of 2, 4-dichlorophenoxyacetic acid in Arabidopsis. Funct Integr Genomics. 2005;5:4–17. doi: 10.1007/s10142-004-0119-9. [DOI] [PubMed] [Google Scholar]
- Raghavan C, Ong EK, Dalling MJ, Stevenson TW. Regulation of genes associated with auxin, ethylene and ABA pathways by 2, 4-dichlorophenoxyacetic acid in Arabidopsis. Funct Integr Genomics. 2006;6:60–70. doi: 10.1007/s10142-005-0012-1. [DOI] [PubMed] [Google Scholar]
- Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and bioconductor- based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter H, Lauer M, Fritsch H. Metabolite profiling of plants—a new diagnostic technique. Abstr Pap Am Chem Soc. 1988;195:129. [Google Scholar]
- Simon RM, Korn EL, McShane LM, Radmacher MD, Wright GW, Zhao Y. Design and analysis of DNA microarray investigations. Series: statistics in biology and health. New York: Springer; 2003. [Google Scholar]
- Torre FDL, Sampedro J, Zarra I, Revilla G. AtFXG1, an Arabidopsis gene encoding α-l-fucosidase active against fucosylated xyloglucan oligosaccharides. Plant Physiol. 2002;128:247–255. doi: 10.1104/pp.010508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, Vigouroux M, Trick M, Bancroft I. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, Scheible WR, Gibon Y, Morcuende R, Weicht D, Meyer S, Stitt M. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005;138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, Preter KD, Pattyn F, Poppe B, Roy ND, Paepe AD, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:34.1–34.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner U, Edwards R, Dixon DP, Mauch F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol. 2002;49:515–532. doi: 10.1023/A:1015557300450. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Liu W, Zhang Y, Liu KK. Action mechanisms of acetolactate synthase inhibiting herbicides. Pest Biochem Physiol. 2007;89:89–96. doi: 10.1016/j.pestbp.2007.04.004. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.