Abstract
Transcriptional cofactors are essential to the regulation of transforming growth factor β (TGFβ) superfamily signaling and play critical and widespread roles during embryonic development, including craniofacial development. We describe the cleft secondary palate 1 (csp1) N-ethyl-N-nitrosourea-induced mouse model of non-syndromic cleft palate (NSCP) that is caused by an intronic Prdm16 splicing mutation. Prdm16 encodes a transcriptional cofactor that regulates TGFβ signaling, and its expression pattern is consistent with a role in palate and craniofacial development. The cleft palate (CP) appears to be the result of micrognathia and failed palate shelf elevation due to physical obstruction by the tongue, resembling human Pierre Robin sequence (PRS)-like cleft secondary palate. PRDM16 should be considered a candidate for mutation in human clefting disorders, especially NSCP and PRS-like CP.
INTRODUCTION
Non-syndromic orofacial clefts are among the most common human birth defects, occurring with a frequency of 1–2 per 1000 newborns, largely dependent on geographical origin, racial and ethnic background and socioeconomic status (1). Isolated cleft palate (CP) and cleft lip (CL) with or without CP (CL/P) are believed to be genetically and developmentally distinct defects that exhibit complex inheritance. CP occurs with a birth prevalence of about 1 in 1500 births (1). In addition to intrinsic palate defects, CP may occur as a consequence of abnormal developmental events that do not primarily involve the palate, such as micrognathia and respiratory distress due to glossoptosis observed in the Pierre Robin sequence (PRS) (OMIM 261800) (2). Isolated CP has a complex etiology, and to date only three loci with significant association have been identified, Chr. 2p13 (TGFa) (3), Chr. 2q32 (SATB2) (4) and TBX22 (5,6). TBX22 mutations cause X-linked CP with ankyloglossia (CPX) and are one of the most common causes of CP (5,7,8). Mutations in human and mouse SATB2 and SOX9 cause PRS-like clefting (4,9–12). Suggestive linkages to 1p34, 2p24–25 and 12q21 were identified in a Finnish population, which has a much higher CP incidence (1/1000 live births) (13). Identifying the genetic basis for the remainder of non-syndromic orofacial clefting cases remains a challenge. Candidate loci for human clefting have been proposed on the basis of known syndromic clefting loci and mouse models of clefting that are the result of spontaneous, targeted, insertional or chemical mutagenesis. At least 175 mouse loci have been reported to include CP as a component of their mutant phenotype (Mouse Genome Informatics, http://www.informatics.jax.org), as have deletions and duplications of every human chromosome, suggesting that clefting is a common endpoint for many developmentally disruptive events.
The mandible is derived at embryonic day (E) 11.5 from early cranial neural crest cells that comprise the ectomesenchyme of the mandibular component of the first branchial arch. The caudal (aboral) part gives rise to Meckel's cartilage (reviewed in 14). Meckel's cartilage first appears at E12.5 and later serves as a template cartilage upon which the mandible bone develops. Bone begins to form at around E13.5 through intramembranous ossification, which proceeds in a proximo-distal direction to lengthen the mandible. Proximally, the secondary cartilages, coronoid, condylar and angular, undergo endochondral ossification and help to form the proximal part of the mandible. The forward extension of Meckel's cartilage and subsequent outgrowth of the mandible is thought to be critical to the coordinated downward movement of the tongue within the oral cavity to allow the elevation and fusion of the palate shelves. Therefore, developmental defects affecting any of these processes, including mandibular defects such as micrognathia observed in PRS, may physically interfere with normal palate shelf movement and result in CP. Several mouse gene mutations have been identified that result in mandible defects, including many affecting transforming growth factor β (TGFβ) superfamily signaling (15–22).
The influence of TGFβ family members during orofacial development has been clearly demonstrated in mouse and human (20,23–29). This broad family of growth factor ligands includes various forms of TGFβs, activins and BMPs that bind heterodimeric transmembrane serine/threonine kinase receptors. Propagation of signals occurs via receptor-activated SMADs (R-SMADs), which associate with Co-SMADs and translocate to the nucleus where they regulate transcription of downstream target genes. The intrinsic DNA-binding ability of Smads is weak, and interaction with transcriptional co-activators and co-repressors is essential for high affinity binding to promoter DNA; this likely mediates the cell- and tissue-specific effects of TGFβ signaling (reviewed in 30).
In humans, PRDM16 is the paralog of myelodysplastic syndrome 1/ecotropic viral integration site 1 (MDS1/EVI1) and has homologous zinc finger DNA-binding domains (7 and 3 C2H2-type zinc fingers, respectively) and positive regulatory (PR), repressor and acidic domains (31). Translocations resulting in increased expression of isoforms of PRDM16 or EVI1 that lack the PR domain are common causes of myelodysplastic syndrome or acute myeloid leukemia (31,32). Prdm16 is expressed in murine embryonic orofacial tissue and, like Evi1, has the ability to bind SMADs in vitro (33); therefore, Prdm16 is likely to play a similar downstream regulatory role in mediating TGFβ signaling. Prdm16 has been recently shown to direct brown fat differentiation and also plays a critical role in regulating the switch between smooth muscle and brown adipose tissue in common Myf-5-expressing progenitor cells (34,35). In this report, we describe the cleft secondary palate 1 (csp1) mouse model of human PRS-like cleft secondary palate that is caused by an intronic mutation that affects normal splicing of Prdm16. A gene trap allele of Prdm16 fails to complement the csp1 mutant phenotype. Our analysis suggests that the mechanism of clefting in csp1 mutants is impaired palate shelf elevation, likely due to palate-extrinsic factors. Prdm16 expression is consistent with a presumptive role in TGFβ superfamily signaling in craniofacial structures, including the primary and secondary palates, Meckel's cartilage and teeth, in addition to other non-craniofacial structures, and we show that expression of TGFβ pathway molecules is perturbed in vivo in the absence of Prdm16.
RESULTS
Characterization of cleft secondary palate in csp1 mutant mice
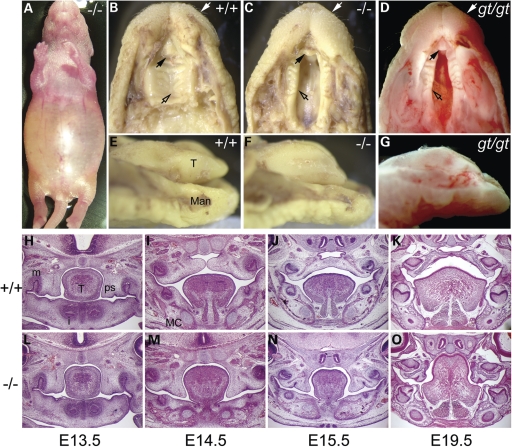
The csp1 mutation was identified in a screen of N-ethyl-N-nitrosourea (ENU)-mutagenized mice for late embryonic recessive mutant phenotypes likely to model human congenital abnormalities (36). Newborn homozygous csp1 mutant pups were identified as having a wide cleft of the secondary palate and abnormal positioning and morphology of the tongue with no obvious additional gross phenotypic abnormalities (Fig. 1B, C, E and F). Primary palate formation appears normal. Mutant pups are born alive but die within 24 h, exhibiting gasping, signs of respiratory distress and extremely distended abdomen in which the stomach and intestines become filled with air, as is commonly seen in newborn mice with cleft secondary palate (Fig. 1A). The CP phenotype occurs with virtually complete penetrance in mutant csp1 pups, and ∼6% of heterozygous animals exhibit CP. The CP penetrance in homozygous csp1 mice drops to 9% (4/45 pups) after four generations of backcross onto the C57BL/6J strain. Interestingly, 93% of these mutants still die shortly after birth due to respiratory failure, suggesting an additional role for Prdm16 in the respiratory and/or circulatory systems. Some incidence of posterior submucous CP is evident in these pups.
Figure 1.
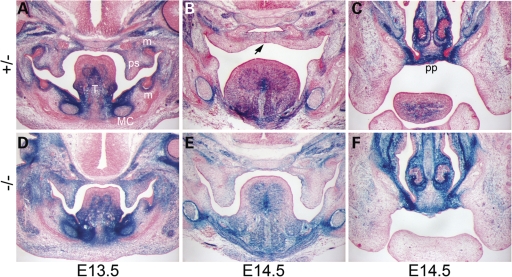
The csp1 and Prdm16Gt683Lex mutants exhibit CP. (A–G) Newborn csp1 and Prdm16Gt683Lex mutant pups die shortly after birth with a distended abdomen (A). Gross examination of mutant embryos identified the presence of a wide CP (open arrows) and pointed snout (white arrows) (B–D) accompanied by abnormal tongue position and morphology (E–G). Black arrows show the unaffected primary palate. (H–O) Histological analysis shows normal palate shelves at E13.5 that fail to elevate, remaining at the sides of the tongue at late E14.5 (I and M). Palate shelves remain in this position after normal palate fusion occurs by E15.5 (J and N) and do not undergo delayed elevation as evidenced by the failure of elevation at E19.5 (K and O) and postnatally (data not shown). T, tongue; Man, mandible; m, molar; i, incisor; MC, Meckel's cartilage.
To investigate the nature of the CP in csp1 mutants, we performed a histological analysis at multiple stages of palate development. In mice, lip fusion is complete by E11.5. Secondary palate development begins with palate shelf outgrowth from the maxillary prominences at E12.5. This is followed by downward growth along either side of the tongue and then concurrent rapid shelf elevation and flattening of the tongue at approximately E14. Fusion occurs between the medial edge epithelium (MEE) of the two palate shelves through a combination of epithelial–mesenchymal transformation, cell migration and apoptosis (37–39). Whereas apposition and fusion of the palatal shelves at the midline occurs by E14.5 in most mouse strains, palate development appears delayed by a day in wild-type FVB/NJ mice. Primary palate formation and secondary palate shelf formation and outgrowth appear normal in csp1 mutants relative to their wild-type littermates at E13.5 (Fig. 1H and L). By E14.5–E15, a specific defect in palate shelf elevation is evident and persists to birth, and the tongue remains elevated within the oral cavity (Fig. 1I–K and M–O).
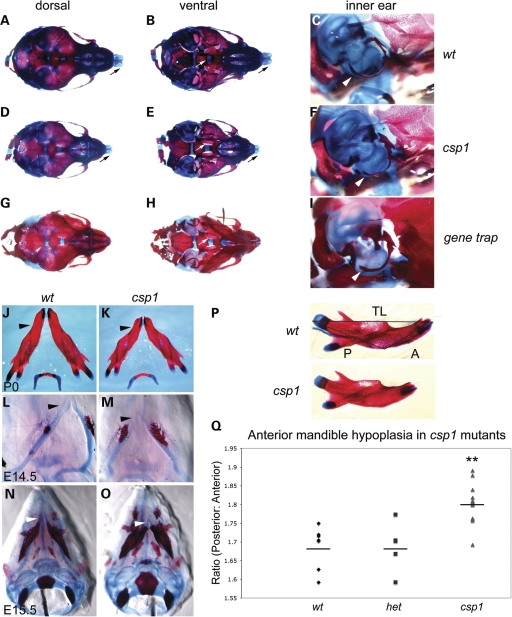
We examined skeletal preparations of newborn wild-type and mutant csp1 pups to address whether the mechanism by which palate shelf elevation is disrupted is due to a primary defect intrinsic to the palate shelves or due to secondary influences, such as physical obstruction by the tongue or by crowding caused by abnormal craniofacial bone development, as seen in humans with the PRS (OMIM 261800). Mutants show unfused palatine bones (Fig. 2B and E) and shortened and narrowed snout, including the nasal and maxillary bones and nasal cartilage (Fig. 2A and D). Mutants also showed variable mandibular hypoplasia, accompanied by abnormal curvature (Fig. 2J and K), with a disproportionate reduction in the size of the anterior body of the mandible (Fig. 2P and Q). All other craniofacial structures appear morphologically normal despite the variable reduction in overall size. We performed in vitro suspension palate culture to determine whether csp1 mutant palate shelves are capable of elevation and fusion in the absence of the tongue and mandible (40). csp1 mutant palate shelves were elevated in this assay, although fusion was variable (Supplementary Material, Fig. S1). These data suggest that the palatal shelf elevation defect observed in csp1 mutants is secondary to physical obstruction by the tongue and/or reduced size of craniofacial bones. We looked at early craniofacial skeletal development in csp1 mutants, and the anterior shortening of Meckel's cartilage and mandible is evident at E14.5 (Fig. 2L and M) and E15.5 (Fig. 2N and O). Premature ossification of the early mandible is apparent in mutants as well.
Figure 2.
csp1 and Prdm16Gt683Lex mutants exhibit craniofacial skeleton defects with anterior-specific mandibular hypoplasia. (A–I) Abnormalities in the craniofacial skeleton in newborn csp1 mutants are evident in dorsal (A, D and G) and ventral (B, E and H) views of Alcian blue/Alazarin-stained heads. In addition, hypoplasia of the mutant tympanic rings is evident (C, F and I). Newborn mutants show failure of palatine bone fusion (white arrows in B, E and H), variably shortened frontonasal region (D and G) and abnormal nasal cartilage formation (black arrows in A, B, D and E) and shortening and abnormal curvature of the anterior mandible (black arrowheads in J and K). Anterior shortening of Meckel's cartilage is evident early during craniofacial bone formation at E14.5 (L and M) and E15.5 (N and O). In addition, csp1 mutant mandibles appear smaller than wild-type counterparts, and ossification appears to be more robust. Morphometric analysis using measurements of the posterior (P in P) and anterior (A in P) aspects of newborn csp1 (n = 14), heterozygous (n = 5) and wild-type (n = 7) mandible bones, followed by calculation of the posterior:anterior (P/A) ratios, detects an anterior-specific mandibular hypoplasia (Q). P/A ratios were significantly greater in csp1 mutants (mean: 1.802, standard deviation: 0.050) compared with wild-type (mean: 1.687, standard deviation: 0.056) and heterozygous pups (mean: 1.688, standard deviation: 0.066) using ANOVA (P < 0.0001). In the chart (Q), the double asterisk designates statistical significance, and horizontal lines denote the means for each genotype class.
A mutation in Prdm16 causes the csp1 mutant phenotype
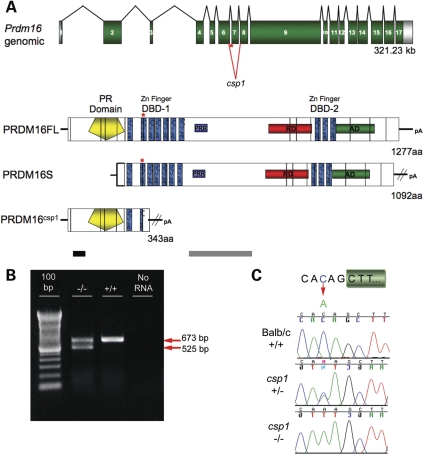
The csp1 mutation was originally localized to distal mouse chromosome 4 (36). High-resolution analysis of affected recombinant mutants refined the csp1 mutation to an approximately 550 kb interval containing five candidate genes. Coding sequences for each candidate gene were screened for mutations by comparison of reverse transcriptase (RT)–PCR products amplified from total RNA isolated from wild-type and csp1 newborn heads. One primer pair from the Prdm16 gene amplified an alternate small RT–PCR product in addition to the wild-type product in RNA samples obtained from multiple csp1 mutant mice (Fig. 3B). Direct sequencing of these RT–PCR products revealed the small product to be derived from a mutant transcript in which exon 7 is absent. The consequence of this aberrant splicing event is a frame-shift leading to premature termination of PRDM16 within exon 8 and inclusion of 46 out-of-frame amino acids. If stable, the resultant truncated protein would include the PR domain and the first zinc finger of DBD-1; however, no shortened protein is detectable by western analysis, suggesting this mutant peptide is unstable (Fig. 3A).
Figure 3.
A splicing mutation in Prdm16 causes the csp1 mutant phenotype. (A) Prdm16 genomic structure and the domain structures of wild-type PR-containing (FL), PR-minus (S) and csp1 mutant PRDM16 protein structure. Green numbered boxes depict exons, and black splicing lines show the intervening introns roughly to scale. Positive regulatory (PR), zinc finger DNA-binding domains 1 and 2 (DBD-1 and DBD-2) and repressor (RD), acidic (AD) and proline-rich (PRR) domains of PRDM16 with homology to EVI1 are shown. The intronic exon 7 splice acceptor mutation (red asterisk and splicing lines), identified in csp1 mutants, causes variable skipping of exon 7. The putative truncated PRDM16csp1 mutant protein is depicted. Exons (verticle black lines) are superimposed upon the PRDM16 protein. The PRDM16 peptides used to generate N-terminal and middle anti-PRDM16 polyclonal antibodies are depicted by black and gray bars, respectively. (B) RT–PCR amplification of Prdm16 from mutant and wild-type postnatal day 0 (P0) heads. An aberrant short splice product (red arrow) produced by exon 7 skipping is observed in csp1 mutants, but not in wild-type pups, in addition to the wild-type splice product (red arrow). (C) A recessive C-to-A mutation in the base pair preceding the AG-dinucleotide of the exon 7 SA is permissive for normal splicing with variably reduced efficiency in csp1 mutants.
Prdm16 consists of 17 exons spanning approximately 325 kb of genomic DNA. The Prdm16 transcript contains 4394 nucleotides and an open-reading frame that encodes a 1277 amino acid protein (Fig. 3A) (31). No mutations were detected in Prdm16 coding sequences; however, analysis of the sequence flanking the affected exons revealed a cytosine (C) to adenosine (A) mutation within intron 6 at the base-pair position immediately preceding the invariant AG-dinucleotide splice acceptor site (Fig. 3C). The decreased, but not abrogated, splicing efficiency at exon 7 is explained by the replacement of a frequently occurring C at this position in mammalian splice acceptor sites to a rarely occurring A (41). The influence that this mutation has on efficiency is likely variable and may contribute to phenotypic variability that we see in csp1 homozygous and heterozygous mice.
To validate an etiologic role for Prdm16 in the ENU-induced csp1 mutant line, we analyzed the Prdm16Gt683Lex strain, which carries an SA-bgeo-PGK-Btk 1st exon-STOP-SD gene trap insertion within the first intron of Prdm16. Newborn homozygous Prdm16Gt683Lex mutant pups exhibit a gross phenotype virtually identical to homozygous csp1 mutants (Fig. 1D and G). In a complementation analysis, all csp1+/−, Prdm16Gt683Lex/+ doubly heterozygote newborn pups (n = 5) had CP and additional anomalies, identical to those seen in the csp1 and Prdm16Gt683Lex homozygous mutant phenotype. These observations confirm causality for the Prdm16 exon 7 splicing mutation in csp1 mutants.
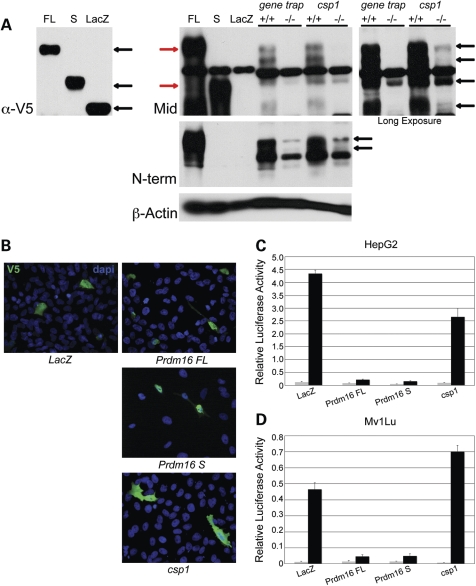
We generated polyclonal antibodies against peptides corresponding to the N-terminal and middle regions of PRDM16 (Fig. 3A) and performed western blot analysis to measure PRDM16 expression in csp1 and Prdm16Gt683Lex mutants (Fig. 6A). Western blot data using the middle-specific anti-PRDM16 antibody are shown. In nuclear lysates from wild-type and mutant Prdm16Gt683Lex E14.5 heads, there is complete loss of protein products in the mutant that are consistent in size with those detected in lysates from HepG2 cells transfected with epitope-tagged PRDM16 expression constructs. With longer exposure, detectable levels of these protein products become apparent in csp1 mutants (Fig. 6A). This is consistent with the Prdm16Gt683Lex allele being a null mutation and the csp1 allele hypomorphic. The two antibodies show specificity for the full-length, PR-containing (FL) and short, PR-minus (S) isoforms in vitro, but it is unclear whether additional protein products showing reduced levels in mutant lysates are the result of endogenous alternative Prdm16 transcripts, post-translational modifications or degradation products.
Prdm16 expression during mouse embryonic development
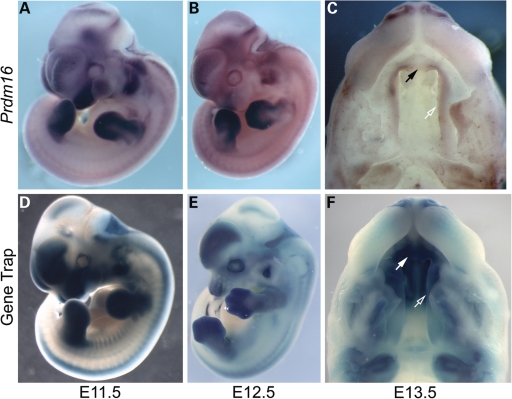
We utilized in situ hybridization to characterize the spatial and temporal expression pattern of Prdm16 during mouse embryonic development. We also analyzed β-galactosidase activity via 5-bromo-4-chloro-3-inodyl-β-d-galactopyranoside (X-gal) staining of embryos from the Prdm16Gt683Lex strain. Prdm16 expression in the otic vesicle, lens of the eye, limb buds, craniofacial primordia and neural tube is evident by E9.5 (data not shown). By E11.5, strong expression is observed in the forebrain, hindbrain, choroid plexi within the brain ventricle, lens and retina of the eye, mesenchyme of the medial and lateral nasal prominences, maxillary and mandibular prominences, otic vesicle, limb bud mesenchyme, somites, cranial and peripheral nerves and the left ventricle of the heart (Fig. 4A and D). As development continues, expression is seen in developing cartilage, epithelium-lined structures of the nervous, respiratory, circulatory and digestive systems and developing muscle (data not shown). In the developing limbs, we observe a gradual restriction of Prdm16 expression from the mesenchyme of the limb bud (E9.5–E11.5) to the presumptive cartilage condensation of the limb digits (E12.5) to the perichondria surrounding the bones and joints of the digits (E13.5–E15.5). Expression is also evident within the knee joint (Supplementary Material, Fig. S2), and it is also seen in craniofacial cartilage, including Meckel's cartilage, bones of the inner ear, nasal cartilage and calvarium. Expression persists in the forebrain, choroid plexus epithelium, as well as in Rathke's pouch and the pituitary gland. Cranial and spinal ganglia and associated neurons, including the facial acoustic and trigeminal ganglia, show Prdm16 expression as well (Fig. 4A, B, D and E). Expression within the developing palate shelves is strongest in mesenchymal cells and appears strongest in the anterior secondary palate, primary palate and upper incisors (Fig. 4C and F). While Prdm16-βgeo reporter expression appears more intense, it is consistent with the endogenous Prdm16 expression pattern visualized via in situ hybridization.
Figure 4.
Comparison of whole-mount in situ hybridization analysis of endogenous Prdm16 expression with reporter expression visualized by whole-mount X-gal staining in Prdm16Gt683Lex heterozygous gene trap embryos. (A, B, D and E) Overlapping expression of endogenous Prdm16 and reporter expression in E11.5 (A and D) and E12.5 (B and E) is observed in developing orofacial structures including the nasal, maxillary and mandibular prominences, forebrain, choroid plexi, cranial nerves, dorsal root ganglia, dermomyotome, forelimb and hindlimb mesenchymes, otic vesicle, the eye and ventricles of the heart. Prdm16 expression in Prdm16Gt683Lex embryos appears more intense in all structures. Craniofacial expression becomes more restricted to frontonasal region by E12.5. (C and F) Palate shelf expression in whole-mount E13.5 embryonic heads via in situ hybridization in csp1-FVB/NJ strain background (C) and X-gal staining of Prdm16Gt683Lex-mixed 129/SvEvBrd × C57BL6/J strain background (F) Prdm16 expression is visible in the secondary palate (open arrows), primary palate (solid arrows), incisor teeth mesenchyme and nasal cartilage.
Prdm16 is expressed in the palate, tongue, teeth and Meckel's cartilage during mouse embryonic orofacial development
During mouse palate development, Prdm16 reporter expression is evident primarily in the mesenchyme of early secondary palate shelves through palate shelf outgrowth, elevation and fusion at E13.5–E14.5 (Fig. 5). Prdm16 is expressed in a posterior-to-anterior gradient of increased expression within the palate shelves (Supplementary Material, Fig. S3). Prdm16 is also expressed in chondrocytes of Meckel's cartilage and surrounding perichondrium, tongue musculature, salivary ducts and glands and mesenchyme of the molar and incisor teeth, consistent with abnormal phenotypes described previously. Prdm16 is detected at higher levels on the oral side of the palate shelves and in the hinge region (Fig. 5 A, B and D, E; Supplementary Material, Fig. S2A–C, E–G and I–K, M–O) but is expressed throughout the palate shelf mesenchyme and, to a lesser extent, within the oral epithelium anteriorly (Supplementary Material, Fig. S1C, G and K, O). Upon apposition and fusion of the palate shelves, Prdm16 is expressed in the cartilage primordia of the primary and secondary palate in a pattern that is consistent with a role in chondrogenesis and bone formation (Fig. 5B and C; Supplementary Material, Fig. S3I–L). Although csp1 mutants do not have CL, it is interesting that very strong Prdm16 expression is evident bilaterally at the points of fusion between the anterior-most portion of the secondary palate and the primary palate (Fig. 5C and F; Supplementary Material, Fig. S3D, H, L and P). Expression remains strong within the muscles of the tongue, Meckel's cartilage and tooth mesenchyme (Fig. 5; Supplementary Material, Fig. S3).
Figure 5.
The Prdm16Gt683Lex mutation causes recessive CP, and reporter expression in craniofacial structures is consistent with this phenotype. (A–F) Histological analysis of coronal sections through the medial aspect of the secondary palate and the primary palate in heterozygous control and homozygous Prdm16Gt683Lex E13.5 and E14.5 embryos. Reporter expression is dose-sensitive. (A and D) At E13.5, before palate shelf elevation and fusion, Prdm16 reporter expression is strongest in the mesenchyme at the oral side of the palate shelves (ps), although it is visible throughout the palate shelf and hinge region. Expression within the tongue (T) musculature, molar tooth mesenchyme (m) and within Meckel's cartilage (MC) and surrounding perichondrium is also detected. Expression in palate epithelia is difficult to detect. (B and E) This expression pattern is maintained at E14.5, after palate shelf elevation and fusion, although it appears weaker. Black arrow, shown in B, depicts epithelial seam at the point of fusion between the palate shelves. Prdm16 expression levels are highest in the primary palate (C and F).
Prdm16 negatively regulates TGFβ signaling in vitro
To examine the role of PRDM16 in TGFβ signaling, we utilized an in vitro assay to measure TGFβ signaling activity. EVI-1, the PRDM16 paralog, has been shown to be a negative regulator of TGFβ signaling using the 3TP-lux TGFβ-responsive luciferase reporter (42). We used this reporter in HepG2 and Mv1Lu cells to determine whether PRDM16 plays a similar inhibitory role on TGFβ signaling. We tested three isoforms of PRDM16: the PRDM16 FL, PRDM16 S, whose N-terminus begins at ATG597, and CSP1, the truncated protein product predicted to result from the csp1 mutant allele (Fig. 3A). To date, we have not conclusively detected the PRDM16 S isoform in the mouse, but since its activated overexpression in human results in AML, we included it in our analyses (31,32). Immunocytochemical analysis of transfected Mv1Lu cells using an anti-V5-tag antibody verified the normal nuclear localization of transfected PRDM16 isoforms, with the exception of CSP1, which was localized to all cellular compartments, thereby defining a nuclear localization signal downstream of the csp1 mutation in the second Zn finger of DBD1 (Fig. 6B).
Figure 6.
The csp1 and Prdm16Gt683Lex mutations are loss-of-function alleles that affect normal PRDM16 function in the negative regulation of TGFβ signaling in a TGFβ-responsive luciferase assay. (A) Western blot analysis using a novel polyclonal anti-PRDM16 antibody (Mid) (Fig. 3A, gray). Comparison of protein products detected in nuclear lysates isolated from HepG2 cells transfected with N-terminal V5-tagged PRDM16 FL and S isoform- and LacZ control-expressing plasmids using an anti-V5 antibody (black arrows on left) with the protein products detected from the same lysates using the anti-PRDM16 Mid antibody (red arrows, right) shows that the anti-PRDM16 antibody binds PRDM16 specifically. Western blot analysis of PRDM16 expression in Prdm16 loss-of-function mutants shows loss of endogenous PRDM16 in Prdm16GT683Lex and csp1 homozygous mutants in nuclear lysates isolated from wild-type and mutant embryonic head tissue. The doublet (black arrows on right) present in wild-type samples that disappears in the mutant samples is most consistent with the PRDM16 FL isoform and may represent post-translational modification of PRDM16 in vivo. Strong bands in the middle of this blot are cross-reacting background bands consistently observed when using this antibody. (B) V5-tagged LacZ, Prdm16FL, Prdm16S and Prdm16csp1 expression plasmids transfected into Mv1Lu cells show localization of PRDM16 isoforms to the nucleus, but this subcellular localization is abolished in the truncated CSP1 mutant protein. HepG2 cells (C) and Mv1 Lu cells (D) were co-transfected with pRL-TK, p3TP-Lux and plasmids expressing LacZ (control), Prdm16FL, Prdm16S or the Prdm16csp1 mouse mutant. Relative luciferase activity (RLA) was measured (Firefly luciferase: Renilla luciferase ratio) in the presence (black) or absence (gray) of TGFβ1. In both cell lines, both wild-type PRDM16 isoforms strongly inhibited TGFβ signaling, whereas CSP1 fails to abrogate TGFβ signaling. RLA values and error bars represent the means and standard deviations, respectively, for three separate experiments.
Expression of PRDM16 FL or S isoforms markedly inhibits TGFβ activity in HepG2 and Mv1Lu cells (Fig. 6C and D). These data are similar to those observed in the analysis of MDS1/EVI1 and EVI-1, in which both forms have a repressive effect on TGFβ signaling (43,44). Overexpression of the truncated Prdm16csp1 protein product failed to inhibit TGFβ signaling efficiently (Fig. 6C and D). These results are somewhat counterintuitive, as it is known that the PR-containing and PR-minus forms of EVI1 can exert opposing effects on the transcriptional regulation of downstream target genes (44,45). It is likely that the availability of co-activators and co-repressors within a specific cell type is critical to the regulation of TGFβ signaling by PRDM16 and EVI-1, given that in vitro studies measuring TGFβ responsiveness show only inhibitory effects. Despite this, these results indicate that PRDM16 isoforms can antagonize TGFβ signaling, and this regulation is abrogated by the csp1 loss-of-function mutation.
Loss of Prdm16 results in reduced TGFβ signaling during craniofacial development
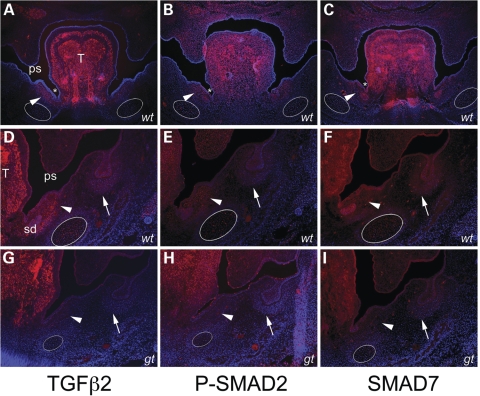
To follow up our investigation of the regulatory role for PRDM16 during TGFβ signaling, we examined the consequence of loss of Prdm16 on key molecules in the TGFβ pathway by the immunohistochemistry of embryonic craniofacial structures (Fig. 7; Supplementary Material, Fig. S4). We used antibodies against TGFβ ligands (TGFβ2 and TGFβ3), against the activated regulatory SMADs (Phospho-SMAD2 and Phospho-SMAD1, 5 and 8) and against the TGFβ-specific inhibitory SMAD (SMAD7). Protein expression for each of these molecules is evident in regions affected by loss of Prdm16 in csp1 and gene trap null mutant embryos, including the palate shelves, tongue, Meckel's cartilage and its surrounding perichondrium, salivary glands and ducts and teeth (Fig. 7A–F; Supplementary Material, Fig. S4A–F, J and M). Specifically, Prdm16Gt683Lex gene trap reporter expression overlaps with these TGFβ molecules in Meckel's cartilage and surrounding perichondrium and in a defined region at the base of the tongue in close proximity to the invaginating oral sulci, the salivary ducts and undifferentiated mesenchyme superior and medial to Meckel's cartilage perichondrium (Fig. 5A and D). Of note, given the Meckel's cartilage/mandible and tongue defects observed in Prdm16 mutants, is that expression of TGFβ2, TGFβ3, Phospho-SMAD2, Phospho-SMAD1, 5, 8 and SMAD7 is dramatically decreased in the gene trap null embryos in these structures (Fig. 7G–I; Supplementary Material, Fig. S4G–I, N, O). We also observe a subtle decrease in fluorescent intensity in the palate shelves of gene trap mutants (Fig. 7D–I; Supplementary Material, Fig. S4D–I, L–O).
Figure 7.
Loss of Prdm16 expression perturbs in vivo TGFβ signaling in the mandible. Immunofluorescent detection of TGFβ2, Phospho-SMAD2 and SMAD7 protein expression in wild-type (wt) (A–F) versus Prdm16Gt683Lex gene trap null mutant (gt) (G–I) E13.5 embryos. Expression of each protein in wild-type embryos is evident in secondary palate shelves (ps), tongue (T), Meckel's cartilage (white ovals in A–I), molar teeth (white arrows) and the undifferentiated mesenchyme between the oral sulci (white asterisks) at the sides of the tongue and the perichodrium surrounding Meckel's cartilage which contains the salivary ducts (sd) (A–F). A dramatic reduction in protein expression levels for each protein is observed in Prdm16Gt683Lex mutants in and around Meckel's cartilage and the region near the base of the tongue, consistent with the mandibular and salivary gland hypoplasia and gross tongue abnormalities observed in these mutants.
Prdm16 mutations cause additional non-craniofacial phenotypic anomalies consistent with Prdm16 expression
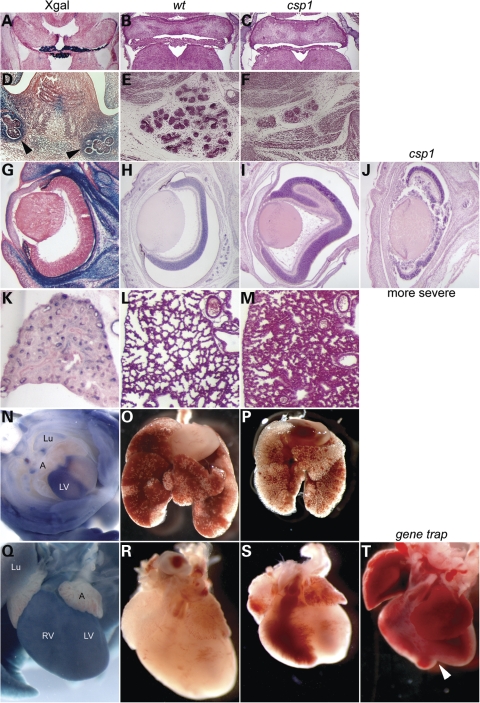
In addition to the palate, Meckel's cartilage, mandible and tongue defects described, detailed examination of csp1 mutants uncovered additional abnormalities in the nervous system, heart, lungs and eye that are consistent with the expression pattern of Prdm16 that is revealed by the reporter in heterozygous gene trap mutant embryos (Fig. 8). Severe choroid plexus hypoplasia is evident in the brain ventricles, which corresponds to our observation of intense reporter expression in choroid plexi epithelia (Fig. 8A–C). Severe hypoplasia of the submandibular and sublingual salivary glands and ducts is evident and consistent with Prdm16 expression (Fig. 8D–F; Supplementary Material, Fig. S3A and E). Abnormal retinal folds of variable severity are observed in the eye postnatally, consistent with Prdm16 expression in the retinal pigment epithelium and retina (Fig. 8G–J). The lungs appear reduced in size compared with wild-type, and histologic defects are apparent at just before birth (E19.5), consistent with Prdm16 expression in the lung epithelium (Fig. 8K–N). Finally, ventricles of the heart appear grossly abnormal and reduced in size, often exhibiting a cleft at their apex, corresponding to the ventricle-specific Prdm16 expression that is restricted to the left ventricle starting at about E10.5 and broadening to the entirety of each ventricle by P0 (Fig. 8N, Q–T). Preliminary histologic examination failed to identify obvious structural defects in the cardiac septa, valves or major blood vessels.
Figure 8.
Additional phenotypic abnormalities in csp1 and Prdm16Gt683Lex mutants correlate with Prdm16 expression. Prdm16 is expressed in choroid plexi epithelium (A), and csp1 mutants exhibit dramatic choroid plexi hypoplasia (B and C). Prdm16 is expressed in the developing salivary glands (black arrowheads in D), and csp1 mutants have severe salivary gland hypoplasia (E and F). csp1 mutants exhibit abnormal retinal folds of variable severity (H–J). Although in situ hybridization shows that endogenous Prdm16 is expressed in the retina in increasingly high levels as embryos progress to birth (data not shown), the Prdm16 gene trap reporter expression is only very weak in the retina, and strong in the pigmented layer of the retina (rpe) (G). We cannot explain this inconsistency. We observe histologic abnormalities in lungs of late embryonic (E19.5) mutants embryos (L and M) and general lung hypoplasia (O and P) when compared with wild-type embryos accompanied by Prdm16 expression in the lung and airway epithelium (K and N). Prdm16 expression in the heart is primarily restricted to the left ventricle as early as E10.5 (N, E13.5 ventral view) and gradually becomes expressed throughout the ventricles, evident postnatally (Q, P3 shown). This expression pattern correlates with gross cardiac ventricular hypoplasia (S), which is more severe in Prdm16Gt683Lex mutants that can exhibit a severe cleft between the ventricles as shown (white arrowhead in T). Lu (lung), LV (left ventricle), RV (right ventricle).
DISCUSSION
In this study, we have identified an ENU-induced single base-pair mutation in the zinc finger transcription factor Prdm16 that causes recessive CP in csp1 mice. An aberrant transcript, resulting from the perfect skipping of exon 7, creates a frame-shift and premature termination of PRDM16. The splicing defect is not complete, in that wild-type transcript can be amplified from homozygous csp1 mutant RNA. This incomplete splicing defect reflects a reduction in splicing efficiency that is likely explained by the nature of the C-to-A change at the base pair immediately adjacent to the invariable AG-dinucleotide splice acceptor site of exon 7 (reviewed in 41). Our data suggest that this is a strong loss-of-function mutation.
The most obvious phenotype in homozygous csp1 mutants is CP due to failed palate shelf elevation. The tongue remains elevated, and it is difficult to determine whether this is the result of an intrinsic tongue defect, crowding due to abnormal development of the orofacial region or merely due to increased freedom of movement subsequent to the cleft. CP stemming from failed palate shelf elevation can result from both palate-intrinsic and -extrinsic causes. Jag2, Irf6 and Fgf10 null palate shelves fail to elevate due to aberrant fusion of MEE to tongue and oral epithelia (24,28,46). The Foxf2 null mouse mutation and human TBX22 mutations (CPX) cause CP, in large part due to tongue anomalies that impair normal coordinated movement of the tongue, preventing elevation, apposition and fusion (5,7,8,47). In addition, Gli3 null mutants were recently reported to have highly penetrant CP on a C57BL/6 strain background due to improper positioning of the tongue in the absence of Meckel's cartilage and mandibular hypoplasia (48). The human condition most similar to the csp1 mutant phenotype is PRS, characterized by CP secondary to micrognathia and glossoptosis (OMIM 261800). A primarily non-palate-specific mechanism of clefting in csp1 mutants is consistent with our observation that mutant palate shelves are able to elevate and fuse in in vitro suspension palate culture experiments. The mandible and frontonasal region of csp1 mutants are often narrow and shortened, and the tongue remains elevated in the oral cavity above the palate shelves. Abnormal morphology and abundant Prdm16 expression within Meckel's cartilage, craniofacial cartilage and tongue, in addition to the palate shelves, support a palate-extrinsic role for Prdm16 during palate elevation. Prdm16 is also expressed in craniofacial musculature and cranial and spinal ganglia, and it is notable that a mouse mutant deficient in skeletal muscle and loss of spinal and cranial motor neurons exhibits a small mandible and CP (49).
It is clear from mouse models that many TGFβ superfamily signaling pathway molecules play critical roles in most aspects of craniofacial and secondary palate development (20,25–27,29,50). In addition to mouse mutations in Hoxa2, Egfr and the dominant Dmm allele of Col2a1 (51–53), TGFβ superfamily signaling is critical to normal Meckel's cartilage and mandible development. Many reported mutants exhibit generalized hypoplasia of Meckel's cartilage and mandible or severe proximal- or distal-specific insufficiencies. Tgfβ2 null mutants show mandibular hypoplasia and obvious proximal defects, conditional loss of Bmp4 in mandibular ectoderm causes dramatic mandible defects with anterior aspects most severely affected, whereas increased BMP activity resulting from the loss of BMP antagonists, Chordin (Chd) and Noggin (Nog) causes variable defects ranging from the absence of the anterior mandible to complete agnathia (17,20,21). Complete or conditional inactivation of Ctgf, a downstream modifier of TGFβ signaling, and the TGFβ/activin receptors, Alk5, Tgfbr2 and Acvr2a, results in mandibular hypoplasia with primarily proximal-specific Meckel's cartilage and mandible defects that lead to CP (16,18,19,22).
Less severe anterior-specific Meckel's cartilage and mandible defects similar to the mandible phenotype observed in csp1 mutants result from the conditional ablation of the type I BMP receptor Alk2, but there is likely crosstalk between different gene types and major signaling pathways (15). For instance, the Aristaless-like homeobox genes, Alx3, Alx4, Prx1 and Prx2, are expressed in the distal-medial mandibular arch, and decreasing dosage of these genes in Alx3/Alx4 and Prx1/Prx2 double-mutants exhibit anterior-specific truncation of Meckel's cartilage and the mandible, among other craniofacial skeletal anomalies (54,55). Similarly, single and double mutants for Hand1/Hand2 transcription factors and Snail1/Snail2 exhibit anterior-specific mandibular hypoplasia reminiscent of csp1 mutant mandibles (56,57). Finally, haploinsufficiency of Satb2 and Sox9 demonstrates a requirement for these genes in anterior Meckel's cartilage and mandible development and in the etiology of PRS-like CP in mice and humans (4,9–12,58).
The function of PRDM16 can potentially be inferred from studies of its paralog EVI1. EVI1 is a zinc finger transcription factor that binds (GATA)n sequences and acts as a bi-functional regulator of transcription in the TGFβ signaling pathway (59,60). An intergenic splicing event that involves the MDS1 and EVI1 genes produces the MDS1/EVI1 gene product; this differs from EVI1 by the addition of a PR domain to its N-terminus (61). The PR domain is proposed to be involved in the regulation of protein–protein interactions and chromatin-mediated gene expression, similar to the SET domain in Drosophila (62,63). These two protein isoforms can have opposing effects, and EVI1 can repress transcriptional activation mediated by MDS1/EVI1, although these opposing effects on transcription are not universally observed, possibly due to the use of different cell lines (43,45,64). EVI1 was first shown to repress TGFβ signaling via physical interaction with SMAD3 through its repressor domain (65,66), and further studies have demonstrated the ability to interact with all SMADs (64). The Evi1 null phenotype and its expression pattern are consistent with a regulatory role in TGFβ signaling and in the repression of TGFβ-mediated inhibition of cell proliferation (67,68). We predict an overlapping but unique role for Prdm16. Recent reports describing a similar broad SMAD-binding ability for PRDM16 and the inclusion of consensus target DNA-binding sequences for PRDM16 DBD-1 and DBD-2 within known consensus sequences for EVI1 support this prediction (33,60,69,70).
Our results suggest that Prdm16 can affect embryonic craniofacial development through the modulation of TGFβ signaling. We have demonstrated that Prdm16 negatively regulates TGFβ signaling in vitro using a TGFβ-responsive reporter assay. These assays do not necessarily reflect the effect on TGFβ signaling during embryonic development, which may be inhibitory or excitatory depending upon the presence of specific Smads and transcriptional cofactors. Thus, although Prdm16 has a negative effect on TGFβ signaling in vitro, and a Prdm16 loss-of-function mutation restores TGFβ activity, the in vivo developmental consequences seen in csp1 and Prdm16Gt683Lex mutants are likely due to an overall decrease in TGFβ pathway function, as their palate and craniofacial phenotypes are consistent with those observed in many TGFβ superfamily gene loss-of-function mouse mutants. Moreover, activated or repressed levels of TGFβ activity in vivo may disrupt the delicate balance between cell proliferation and differentiation in a temporally and spatially dependent manner with similar developmental consequences. One prime candidate for an in vivo downstream target of Prdm16 is Smad7. We hypothesized that Prdm16 loss-of-function mutations could relieve Smad7 repression and perpetuate Smad7 activity and negative feedback on TGFβ signaling. Our analysis of TGFβ2 and TGFβ3 ligand expression and TGFβ/activin and BMP pathway activity, as measured by Phospho-SMAD2 and Phospho-SMAD1, 5, 8 expression, respectively, identified a specific downregulation of these TGFβ signaling molecules in tissues within the mandible that are most dramatically affected by the absence of Prdm16. We found that SMAD7 expression was also reduced in these structures, contrary to our hypothesis that SMAD7 expression levels would increase in the absence of PRDM16-mediated transcriptional repression. Since SMAD7 expression is induced in the presence of TGFβ to initiate a negative feedback on TGFβ levels, and we observed dramatically decreased levels of TGFβ pathway molecules, it is possible that SMAD7 expression is not induced in these cells.
Interestingly, we have identified rare PRDM16 sequence variants in three NSCL/P patients, one of which disrupts a putative MAPK phosphorylation site and abrogates normal repression of TGFβ signaling by PRDM16 in vitro (B.C.B., Vieira et al., manuscript in preparation). It will be important to undertake similar genetic analyses of PRS-like CP cases to uncover a potential role for PRDM16 in the etiology of PRS-like CP.
MATERIALS AND METHODS
Derivation and genotyping of mutant mice
csp1 mice were originally generated by ENU mutagenesis of BALB/c mice, and the mutant locus mapped to distal chromosome 4 (line 27) (36). Genetic fine-mapping of recombination events in affected newborn pups was performed using a combination of characterized and novel microsatellite markers. The affected recombinant interval was narrowed to a 550 kb region between novel markers oBB96/97 and oBB120/121. We screened the coding sequences of the five genes within this interval, Trp73, Wdr8, Megf6, Arhgef16 and Prdm16, for causative mutations by direct sequencing of RT–PCR products derived from wild-type or csp1 mutant total RNA derived from P0 heads. The csp1 mutation described in Results was the only mutation identified, was confirmed in multiple mutants and is not present in the parental BALB/c strain. csp1 mice were outcrossed to FVB/NJ mice for at least eight generations and are now maintained by intercross matings. csp1 mice were outcrossed to C57BL6/J mice for four generations to explore strain-specific phenotypic variability. Genotyping was performed by PCR using primers that flank the intron 6 csp1 mutation, followed by HindIII restriction enzyme digestion and electrophoresis through 2% agarose gel to visualize the RFLP created by the csp1 single base substitution. The Prdm16Gt683Lex mutant mouse strain was generated by Lexicon Genetics, Inc. and obtained via the Mouse Mutant Resource Repository Center (MMRRC) at the University of California-Davis. They were maintained on a mixed 129SvEvBrd × C57BL/6J mixed strain background by intercross matings. Genotyping was performed as described (http://www.mmrrc.org/strains/11760/011760.html). Complementation tests were performed by examination of F1 progeny generated from heterozygous csp1 × heterozygous Prdm16Gt683Lex matings for the presence of CP at birth. Embryos were genotyped using genomic DNA isolated from the yolk sac tissue, and mutant embryos older than E14.5 could be distinguished by the presence of CP. All animals were housed in accordance with the Harvard Medical School ARCM regulations. Timed matings were monitored by examination of females for the presence of a vaginal plug each morning: noon on the day the plug was identified was considered E0.5.
In vitro suspension palate culture
To examine palate shelf elevation, E13.5 embryos from timed csp1 heteterozygous intercross matings were harvested. Heads were separated from the bodies, and the mandibles, tongues and brains were removed before placing groups of five to six heads in 50 ml glass bottles (Wheaton) filled with 8 ml BGJb medium (Gibco) supplemented with 1% penicillin–streptomycin and sealed with aluminum caps with rubber seals following the protocol reported by Shiota et al. (71). In brief, bottles were flushed with 5% CO2/95% O2 medical gas mixture for 2 min every 24 h and media was changed only upon observing an obvious color change. Bottles were incubated at 37°C while rolling at approximately 20–25 r.p.m. on a bottle roller (Wheaton) for about 3 days. Palate shelf elevation and fusion were evaluated after 3 days of culture, and the tissue was removed from each head for genomic DNA extraction and genotyping.
Skeletal preparations and mandible measurements
Newborn wild-type and csp1 mutant embryos were collected, skinned and eviscerated before placement into 95% ethanol for at least 1 day. All steps were performed with slow rocking at room temperature. Ethanol was replaced with Alcian blue staining solution (0.03% Alcian blue, 80% ethanol, 20% acetic acid) for 2–3 days, followed by a 6–12 h wash in 95% ethanol. Ethanol was replaced with 2% KOH solution for 24 h or until the tissue is appropriately cleared, followed by staining in Alizarin Red solution (0.03% Alazarin Red, 1% KOH, water) for 24 h. Skeletons were cleared in 1% KOH/20% glycerol solution and transferred to a 1:1 glycerol:95% ethanol solution. For the quantification of the anterior mandible shortening, mandibles were removed from the skull and separated into left and right mandible bones. Mandibles were visualized using a Leica MZ12.5 dissecting scope and digital photographs were taken using a Leica DC500 camera. Mandible measurements were taken in arbitrary units as shown in Figure 2P for each P0 newborn pup embryo using ImageJ software (NIH). The XLSTAT (Addinsoft) software package for Microsoft Excel was utilized to perform statistical analysis by ANOVA.
Histological analysis, whole-mount in situ hybridization and X-gal staining
Embryos to be used for histological analysis were fixed with Bouin's fixative for at least 48 h, rinsed thoroughly in multiple changes of 70% ethanol and processed for paraffin embedding using a Leica TP1020 automated tissue processor. Sections were cut at a thickness of 7 μM and with hematoxylin and eosin using standard protocols. Whole-mount in situ hybridization was performed on the basis of previously described protocols (72) using a BioLane HTI for post-hybridization washes and antibody incubations, followed by visualization with BM Purple (Roche, Indianapolis, IN, USA). Prdm16 coding sequence (Prdm16× 4-8F, 5′-CTGGCTCAAGTACATCCGTGT-3′; Prdm16× 4-8R, 5′-CGTGCTGTGGATATGCTTGT-3′) and 3′-UTR-specific (Prdm16-3′-UTR-F, 5′-CACCCTCAACACCTCCACTT-3′; Prdm16-3′-UTR-R, 5′-AGGTGTGGGTTTGCCAATAA-3′) probes were used in this analysis and they show identical patterns of expression. In brief, RT–PCR products amplified from total RNA isolated from wild-type P0 heads were cloned into pPCR-script Amp SK+ (Stratagene, Cedar Creek, TX, USA), and digested plasmid DNA was in vitro-transcribed to produce antisense (BamHI, T3) and sense (NotI, T7) digoxigen-labeled probes. Whole-mount and section X-gal staining was performed as described (73). E13.5–E15.5 embryos were fixed in 2% formaldehyde, 0.2% glutaraldehyde for 1.5–2 h on ice, infused with 30% sucrose for at least 24 h, cryoembedded in OCT, cut into 15 μm sections and stained.
Antibodies, western blotting and immunofluorescence
N-terminal and middle Prdm16-specific cDNA was amplified from a murine full-length Prdm16 expression plasmid (N-terminal: exons 1–2, amino acids 4–62; middle: exon 9, amino acids 451–678) using PfuUltra II Fusion HS DNA Polymerase (Stratagene) using primers with EcoRI and HindIII restriction sites at the 5′ ends (Prdm16-Nterm-F, 5′-GAATTCAGGCGAGGGCGAGGAAG-3′; Prdm16-Nterm-R, 5′-AAGCTTGGAGTGAAGTCCTCGCTGGTG-3′; Prdm16-mid-F, 5′-GAATTCATTACACGCCTGGCAGCATC-3′; Prdm16-mid-R, 5′-AAGCTTGGTGGCGGGAAGAAGGAAT-3′). Amplified cDNAs were digested with EcoRI and HindIII, ligated into the pET-30c expression vector and sequenced to verify high fidelity amplification. Peptide expression was induced by IPTG in BL21 cells, affinity-purified and submitted for polyclonal antibody production (ProSci Incorporated, Poway, CA, USA). PRDM16-specific antiserum was affinity-purified before use. Western blots were performed using established protocols. Nuclear fractions were isolated from embryonic heads as directed using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce, Rockford, IL, USA). Nuclear fractions (100 µg) were separated on a 6% polyacrylamide gel, transferred to PVDF membrane for 2 h at 600 mA and incubated in the presence of PRDM16 N-terminal (1:7500), middle (1:7500) or β-actin (1:2000, Sigma-Aldrich) antisera, followed by antibody detection using the SuperSignal West Femto Maximum Sensitivity Substrate (Pierce). For immunfluorescent antibody detection of Prdm16 expression, constructs transfected into Mv1Lu cells (2000 cells/chamber) were seeded in eight-chamber CultureSlides (BD Biosciences, Bedford, MA, USA) 1 day prior to transfection. After 24 h, cells were fixed in 3.7% paraformaldehyde for 10 min followed by permeabilization in PBS/0.1% Triton X-100. Cells were blocked in 3% evaporated milk in PBS/0.1% Triton for 20 min and incubated with an anti-V5 tag antibody (1:200, Invitrogen) for 1 h at room temperature. After PBS washes, cells were incubated with an FITC-conjugated goat anti-mouse secondary antibody (1:500, Sigma) for 30 min at room temperature, washed again in PBS, coverslipped with Vectashield (Vector Labs) containing DAPI for nuclear detection and visualized using a Zeiss Axiophot microscope. Immunofluorescent antibody detection in mouse embryos was performed as follows. Cryosections (10 µm) were rinsed in PBS and blocked for 1 h with 10% goat serum in PBS containing 1% BSA, 0.01% Thimerosal and 0.3% Triton. Sections were incubated with anti-TGFβ2 (Santa Cruz Biotechnology, 1/200), anti-TGFβ3 (Santa Cruz Biotechnology, 1/200), anti-Phosho-Smad1/Smad5/Smad8 (Cell Signaling Technology, 1/100), anti-Phospho-Smad2 (Zymed Laboratories, 1/200) and anti-SMAD7 (Santa Cruz Biotechnology, 1/100) overnight at 4°C. Secondary antibody was AlexaFluor 594 goat anti-rabbit (Molecular Probes), diluted 1/500 and incubated with sections for 1 h at room temperature. All antibodies were diluted in PBS containing 1% BSA, 0.01% Thimerosal and 0.3% Triton. Slides were mounted with Vectashield mounting medium containing DAPI (Vector Laboratories) and visualized using either a Zeiss Axiophot or Axio Imager Z1 microscope.
Expression constructs
All wild-type Prdm16 expression constructs were generated by PCR amplification from a full-length mouse EST clone (GenBank Accession No. CB248179.1) using PfuUltra II HS Fusion DNA polymerase (Stratagene), followed by TOPO-PCR cloning into the pENTR-SD/D-TOPO Gateway entry plasmid (Invitrogen) and subsequent LR Clonase recombination into the pcDNA-DEST40 mammalian destination expression plasmid (Invitrogen) to create C-terminally 6XHis-, V5-tagged PRDM16 fusion proteins. Specifically, Prdm16FL was amplified using primers specific to amino acids 1–1276, including the alternatively spliced 16th exon (Prdm16FL-5′F, 5′-CACCATGCGATCCAAGGCGAGGGCGA-3′; Prdm16FL-3′R, 5′-GAGGTGGTTGATGGGGTTAAAGGCT-3′); Prdm16S, amino acids 186–1276, corresponding to human ATG597 (70) (Prdm16S-5′F, 5′-CACCATGTGTCAGATCAACGAACAGAT-3′; Prdm16FL-3′R). The Prdm16csp1 expression plasmid was generated by RT–PCR amplification from csp1 mutant total RNA isolated from P0 head tissue (Prdm16FL-5′F; Prdm16csp1-3′R, 5′-CCGTGCTGTGGATATGCTTGTGCTGTTTG-3′), gel purification of the csp1-specific product and cloning as described earlier.
TGFβ-responsive luciferase assays
Transfections were performed using FuGene HD and FuGene6 transfection reagents (Roche) for HepG2 and Mv1Lu cells, respectively. Luciferase assays were performed as described previously (44). Cells were seeded in 24-well plates at a density of 1.5 × 105 cells/well (HepG2) and 0.5 × 05 cells/well (Mv1Lu) and were transfected 24 h later. We transiently co-transfected HepG2 cells with 100 ng of the reporter plasmid, p3TP-Lux (Firefly luciferase), 100 ng of the transfection control plasmid, pRL-TK (Promega) and 800 ng of the indicated effector expression constructs. For Mv1Lu cells, the plasmid DNA quantities were halved. For dose–response experiments, effector plasmids were co-transfected in the indicated ratio, with total quantity remaining constant as described earlier. Twenty-four hours post-transfection, medium containing either 5 ng human recombinant TGFβ1 (PeproTech, Rocky Hill, NJ, USA) or no TGFβ1 was added to each well to induce TGFβ activity. Luciferase activity was measured after 18 h using the Dual Luciferase Assay System (Promega) and the Veritas Microplate Luminometer (Turner Biosystems, Sunnyvale, CA, USA). Firefly luciferase levels were normalized for transfection efficiency by calculating the ratio of Firefly to Renilla luciferase.
SUPPLEMENTARY MATERIAL
FUNDING
This research was supported by National Institutes of Health (F32 HD045066 and K12 DE014528 to B.C.B. and R01 DE015246 and R01 HD036404 to D.R.B.).
AUTHOR CONTRIBUTIONS
B.C.B. carried out most of the experiments. A.T.-D. performed the immunofluorescence analysis of TGFβ signaling. M.P. performed mouse colony management, genotyping and embryo dissections and performed skeletal preparations. B.J.H. generated the csp1 mutant and mapped the csp1 locus to Chr. 4. B.C.B. and D.R.B. designed the experiments and wrote this manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Joan Massague for the 3TP-Lux reporter plasmid. We thank H. Qiu and K. Parker for technical assistance, and William Pu for examination of Prdm16 mutants for histologic cardiac defects. We thank Patrick Seale and Heather Conroe from Bruce Spiegelman's Lab for providing Prdm16Gt683Lex embryos for some of our immunofluorescence experiments. We thank J. Murray, A. Vieira, K. Christensen, R. Stottman, P. Tran, D. Manning and E. Cozzi for critical review of this manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Mossey P.A., Little J. Epidemiology of oral clefts: an international perspective. In: Wyszynski D.F., editor. Cleft Lip and Palate. From Origin to Treatment. New York: Oxford University Press; 2002. pp. 127–158. [Google Scholar]
- 2.Wyszynski D.F. Cleft Lip and Palate: From Origin to Treatment. 1st edn. Oxford: Oxford University Press; 2002. [Google Scholar]
- 3.Shiang R., Lidral A.C., Ardinger H.H., Buetow K.H., Romitti P.A., Munger R.G., Murray J.C. Association of transforming growth-factor alpha gene polymorphisms with nonsyndromic cleft palate only (CPO) Am. J. Hum. Genet. 1993;53:836–843. [PMC free article] [PubMed] [Google Scholar]
- 4.FitzPatrick D.R., Carr I.M., McLaren L., Leek J.P., Wightman P., Williamson K., Gautier P., McGill N., Hayward C., Firth H., et al. Identification of SATB2 as the cleft palate gene on 2q32–q33. Hum. Mol. Genet. 2003;12:2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- 5.Marcano A.C., Doudney K., Braybrook C., Squires R., Patton M.A., Lees M.M., Richieri-Costa A., Lidral A.C., Murray J.C., Moore G.E., et al. TBX22 mutations are a frequent cause of cleft palate. J. Med. Genet. 2004;41:68–74. doi: 10.1136/jmg.2003.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suphapeetiporn K., Tongkobpetch S., Siriwan P., Shotelersuk V. TBX22 mutations are a frequent cause of non-syndromic cleft palate in the Thai population. Clin. Genet. 2007;72:478–483. doi: 10.1111/j.1399-0004.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- 7.Braybrook C. The T-box transcription factor gene TBX22 is mutated in X-linked cleft palate and ankyloglossia. Nat. Genet. 2001;29:179–183. doi: 10.1038/ng730. [DOI] [PubMed] [Google Scholar]
- 8.Braybrook C., Lisgo S., Doudney K., Henderson D., Marcano A.C., Strachan T., Patton M.A., Villard L., Moore G.E., Stanier P., et al. Craniofacial expression of human and murine TBX22 correlates with the cleft palate and ankyloglossia phenotype observed in CPX patients. Hum. Mol. Genet. 2002;11:2793–2804. doi: 10.1093/hmg/11.22.2793. [DOI] [PubMed] [Google Scholar]
- 9.Benko S., Fantes J.A., Amiel J., Kleinjan D.J., Thomas S., Ramsay J., Jamshidi N., Essafi A., Heaney S., Gordon C.T., et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 10.Britanova O., Depew M.J., Schwark M., Thomas B.L., Miletich I., Sharpe P., Tarabykin V. Satb2 haploinsufficiency phenocopies 2q32–q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am. J. Hum. Genet. 2006;79:668–678. doi: 10.1086/508214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakobsen L.P., Ullmann R., Christensen S.B., Jensen K.E., Molsted K., Henriksen K.F., Hansen C., Knudsen M.A., Larsen L.A., Tommerup N., et al. Pierre Robin sequence may be caused by dysregulation of SOX9 and KCNJ2. J. Med. Genet. 2007;44:381–386. doi: 10.1136/jmg.2006.046177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velagaleti G.V., Bien-Willner G.A., Northup J.K., Lockhart L.H., Hawkins J.C., Jalal S.M., Withers M., Lupski J.R., Stankiewicz P. Position effects due to chromosome breakpoints that map approximately 900 kb upstream and approximately 1.3 Mb downstream of SOX9 in two patients with campomelic dysplasia. Am. J. Hum. Genet. 2005;76:652–662. doi: 10.1086/429252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koillinen H., Lahermo P., Rautio J., Hukki J., Peyrard-Janvid M., Kere J. A genome-wide scan of non-syndromic cleft palate only (CPO) in Finnish multiplex families. J. Med. Genet. 2005;42:177–184. doi: 10.1136/jmg.2004.019646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miletich I., Sharpe P.T. Neural crest contribution to mammalian tooth formation. Birth Defects Res. C. Embryo Today. 2004;72:200–212. doi: 10.1002/bdrc.20012. [DOI] [PubMed] [Google Scholar]
- 15.Dudas M., Sridurongrit S., Nagy A., Okazaki K., Kaartinen V. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech. Dev. 2004;121:173–182. doi: 10.1016/j.mod.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Ivkovic S., Yoon B.S., Popoff S.N., Safadi F.F., Libuda D.E., Stephenson R.C., Daluiski A., Lyons K.M. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development. 2003;130:2779–2791. doi: 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W., Selever J., Murali D., Sun X., Brugger S.M., Ma L., Schwartz R.J., Maxson R., Furuta Y., Martin J.F. Threshold-specific requirements for Bmp4 in mandibular development. Dev. Biol. 2005;283:282–293. doi: 10.1016/j.ydbio.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 18.Matzuk M.M., Kumar T.R., Bradley A. Different phenotypes for mice deficient in either activins or activin receptor type II. Nature. 1995;374:356–360. doi: 10.1038/374356a0. [DOI] [PubMed] [Google Scholar]
- 19.Oka K., Oka S., Sasaki T., Ito Y., Bringas P., Jr, Nonaka K., Chai Y. The role of TGF-beta signaling in regulating chondrogenesis and osteogenesis during mandibular development. Dev. Biol. 2007;303:391–404. doi: 10.1016/j.ydbio.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanford L.P., Ormsby I., Gittenberger-de Groot A.C., Sariola H., Friedman R., Boivin G.P., Cardell E.L., Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stottmann R.W., Anderson R.M., Klingensmith J. The BMP antagonists Chordin and Noggin have essential but redundant roles in mouse mandibular outgrowth. Dev. Biol. 2001;240:457–473. doi: 10.1006/dbio.2001.0479. [DOI] [PubMed] [Google Scholar]
- 22.Zhao H., Oka K., Bringas P., Kaartinen V., Chai Y. TGF-beta type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner. Dev. Biol. 2008;320:19–29. doi: 10.1016/j.ydbio.2008.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vieira A.R. Unraveling human cleft lip and palate research. J. Dent. Res. 2008;87:119–125. doi: 10.1177/154405910808700202. [DOI] [PubMed] [Google Scholar]
- 24.Ingraham C.R., Kinoshita A., Kondo S., Yang B., Sajan S., Trout K.J., Malik M.I., Dunnwald M., Goudy S.L., Lovett M., et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat. Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito Y., Yeo J.Y., Chytil A., Han J., Bringas P., Jr, Nakajima A., Shuler C.F., Moses H.L., Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- 26.Kaartinen V., Voncken J.W., Shuler C., Warburton D., Bu D., Heisterkamp N., Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial–mesenchymal interaction. Nat. Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- 27.Proetzel G., Pawlowski S.A., Wiles M.V., Yin M., Boivin G.P., Howles P.N., Ding J., Ferguson M.W., Doetschman T. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat. Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson R.J., Dixon J., Malhotra S., Hardman M.J., Knowles L., Boot-Handford R.P., Shore P., Whitmarsh A., Dixon M.J. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat. Genet. 2006;38:1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- 29.Xu X., Han J., Ito Y., Bringas P., Jr, Urata M.M., Chai Y. Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev. Biol. 2006;297:238–248. doi: 10.1016/j.ydbio.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Massague J., Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mochizuki N., Shimizu S., Nagasawa T., Tanaka H., Taniwaki M., Yokota J., Morishita K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–3214. [PubMed] [Google Scholar]
- 32.Testoni N., Borsaru G., Martinelli G., Carboni C., Ruggeri D., Ottaviani E., Pelliconi S., Ricci P., Pastano R., Visani G., et al. 3q21 and 3q26 cytogenetic abnormalities in acute myeloblastic leukemia: biological and clinical features. Haematologica. 1999;84:690–694. [PubMed] [Google Scholar]
- 33.Warner D.R., Horn K.H., Mudd L., Webb C.L., Greene R.M., Pisano M.M. PRDM16/MEL1: a novel Smad binding protein expressed in murine embryonic orofacial tissue. Biochim. Biophys. Acta. 2007;1773:814–820. doi: 10.1016/j.bbamcr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 34.Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scime A., Devarakonda S., Conroe H.M., Erdjument-Bromage H., et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seale P., Kajimura S., Yang W., Chin S., Rohas L.M., Uldry M., Tavernier G., Langin D., Spiegelman B.M. Transcriptional control of brown fat determination by PRDM16. Cell. Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herron B.J., Lu W., Rao C., Liu S., Peters H., Bronson R.T., Justice M.J., McDonald J.D., Beier D.R. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat. Genet. 2002;30:185–189. doi: 10.1038/ng812. [DOI] [PubMed] [Google Scholar]
- 37.Ferguson M.W. Palate development. Development. 1988;103:41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- 38.Moore K.L., Persaud T.V.N. Before We Are Born: Essentials of Embryology and Birth Defects. 4 edn. Philadelphia: W.B. Saunders Company; 1993. [Google Scholar]
- 39.Chai Y., Maxson R.E., Jr Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- 40.Kosazuma T., Hashimoto S., Ohno H., Chou M.J., Shiota K. Organ culture of the fetal mouse palate for screening the developmental toxicity of chemicals: a validation study. Congenit. Anom. (Kyoto) 2004;44:60–71. doi: 10.1111/j.1741-4520.2004.00010.x. [DOI] [PubMed] [Google Scholar]
- 41.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 42.Carcamo J., Zentella A., Massague J. Disruption of transforming growth factor beta signaling by a mutation that prevents transphosphorylation within the receptor complex. Mol. Cell. Biol. 1995;15:1573–1581. doi: 10.1128/mcb.15.3.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nitta E., Izutsu K., Yamaguchi Y., Imai Y., Ogawa S., Chiba S., Kurokawa M., Hirai H. Oligomerization of Evi-1 regulated by the PR domain contributes to recruitment of corepressor CtBP. Oncogene. 2005;24:6165–6173. doi: 10.1038/sj.onc.1208754. [DOI] [PubMed] [Google Scholar]
- 44.Sood R., Talwar-Trikha A., Chakrabarti S.R., Nucifora G. MDS1/EVI1 enhances TGF-beta1 signaling and strengthens its growth-inhibitory effect but the leukemia-associated fusion protein AML1/MDS1/EVI1, product of the t(3;21), abrogates growth-inhibition in response to TGF-beta1. Leukemia. 1999;13:348–357. doi: 10.1038/sj.leu.2401360. [DOI] [PubMed] [Google Scholar]
- 45.Soderholm J., Kobayashi H., Mathieu C., Rowley J.D., Nucifora G. The leukemia-associated gene MDS1/EVI1 is a new type of GATA-binding transactivator. Leukemia. 1997;11:352–358. doi: 10.1038/sj.leu.2400584. [DOI] [PubMed] [Google Scholar]
- 46.Jiang R., Lan Y., Chapman H.D., Shawber C., Norton C.R., Serreze D.V., Weinmaster G., Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang T., Tamakoshi T., Uezato T., Shu F., Kanzaki-Kato N., Fu Y., Koseki H., Yoshida N., Sugiyama T., Miura N. Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev. Biol. 2003;259:83–94. doi: 10.1016/s0012-1606(03)00176-3. [DOI] [PubMed] [Google Scholar]
- 48.Huang X., Goudy S.L., Ketova T., Litingtung Y., Chiang C. Gli3-deficient mice exhibit cleft palate associated with abnormal tongue development. Dev. Dyn. 2008;237:3079–3087. doi: 10.1002/dvdy.21714. [DOI] [PubMed] [Google Scholar]
- 49.Grieshammer U., Lewandoski M., Prevette D., Oppenheim R.W., Martin G.R. Muscle-specific cell ablation conditional upon Cre-mediated DNA recombination in transgenic mice leads to massive spinal and cranial motoneuron loss. Dev. Biol. 1998;197:234–247. doi: 10.1006/dbio.1997.8859. [DOI] [PubMed] [Google Scholar]
- 50.Knight A.S., Schutte B.C., Jiang R., Dixon M.J. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev. Dyn. 2006;235:1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- 51.Gendron-Maguire M., Mallo M., Zhang M., Gridley T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- 52.Miettinen P.J., Chin J.R., Shum L., Slavkin H.C., Shuler C.F., Derynck R., Werb Z. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat. Genet. 1999;22:69–73. doi: 10.1038/8773. [DOI] [PubMed] [Google Scholar]
- 53.Ricks J.E., Ryder V.M., Bridgewater L.C., Schaalje B., Seegmiller R.E. Altered mandibular development precedes the time of palate closure in mice homozygous for disproportionate micromelia: an oral clefting model supporting the Pierre-Robin sequence. Teratology. 2002;65:116–120. doi: 10.1002/tera.10022. [DOI] [PubMed] [Google Scholar]
- 54.Beverdam A., Brouwer A., Reijnen M., Korving J., Meijlink F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development. 2001;128:3975–3986. doi: 10.1242/dev.128.20.3975. [DOI] [PubMed] [Google Scholar]
- 55.ten Berge D., Brouwer A., Korving J., Martin J.F., Meijlink F. Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- 56.Barbosa A.C., Funato N., Chapman S., McKee M.D., Richardson J.A., Olson E.N., Yanagisawa H. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev. Biol. 2007;310:154–168. doi: 10.1016/j.ydbio.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray S.A., Oram K.F., Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–1797. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- 58.Bi W., Huang W., Whitworth D.J., Deng J.M., Zhang Z., Behringer R.R., de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc. Natl Acad. Sci. USA. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kreider B.L., Orkin S.H., Ihle J.N. Loss of erythropoietin responsiveness in erythroid progenitors due to expression of the Evi-1 myeloid-transforming gene. Proc. Natl Acad. Sci. USA. 1993;90:6454–6458. doi: 10.1073/pnas.90.14.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morishita K., Suzukawa K., Taki T., Ihle J.N., Yokota J. EVI-1 zinc finger protein works as a transcriptional activator via binding to a consensus sequence of GACAAGATAAGATAAN1-28 CTCATCTTC. Oncogene. 1995;10:1961–1967. [PubMed] [Google Scholar]
- 61.Fears S., Mathieu C., Zeleznik-Le N., Huang S., Rowley J.D., Nucifora G. Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc. Natl Acad. Sci. USA. 1996;93:1642–1647. doi: 10.1073/pnas.93.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang S., Shao G., Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J. Biol. Chem. 1998;273:15933–15939. doi: 10.1074/jbc.273.26.15933. [DOI] [PubMed] [Google Scholar]
- 63.Xie M., Shao G., Buyse I.M., Huang S. Transcriptional repression mediated by the PR domain zinc finger gene RIZ. J. Biol. Chem. 1997;272:26360–26366. doi: 10.1074/jbc.272.42.26360. [DOI] [PubMed] [Google Scholar]
- 64.Alliston T., Ko T.C., Cao Y., Liang Y.Y., Feng X.H., Chang C., Derynck R. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J. Biol. Chem. 2005;280:24227–24237. doi: 10.1074/jbc.M414305200. [DOI] [PubMed] [Google Scholar]
- 65.Kurokawa M., Mitani K., Imai Y., Ogawa S., Yazaki Y., Hirai H. The t(3;21) fusion product, AML1/Evi-1, interacts with Smad3 and blocks transforming growth factor-beta-mediated growth inhibition of myeloid cells. Blood. 1998;92:4003–4012. [PubMed] [Google Scholar]
- 66.Kurokawa M., Mitani K., Irie K., Matsuyama T., Takahashi T., Chiba S., Yazaki Y., Matsumoto K., Hirai H. The oncoprotein Evi-1 represses TGF-beta signalling by inhibiting Smad3. Nature. 1998;394:92–96. doi: 10.1038/27945. [DOI] [PubMed] [Google Scholar]
- 67.Hoyt P.R., Bartholomew C., Davis A.J., Yutzey K., Gamer L.W., Potter S.S., Ihle J.N., Mucenski M.L. The Evi1 proto-oncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mech. Dev. 1997;65:55–70. doi: 10.1016/s0925-4773(97)00057-9. [DOI] [PubMed] [Google Scholar]
- 68.Perkins A.S., Fishel R., Jenkins N.A., Copeland N.G. Evi-1, a murine zinc finger proto-oncogene, encodes a sequence-specific DNA-binding protein. Mol. Cell. Biol. 1991;11:2665–2674. doi: 10.1128/mcb.11.5.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Funabiki T., Kreider B.L., Ihle J.N. The carboxyl domain of zinc fingers of the Evi-1 myeloid transforming gene binds a consensus sequence of GAAGATGAG. Oncogene. 1994;9:1575–1581. [PubMed] [Google Scholar]
- 70.Nishikata I., Sasaki H., Iga M., Tateno Y., Imayoshi S., Asou N., Nakamura T., Morishita K. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood. 2003;102:3323–3332. doi: 10.1182/blood-2002-12-3944. [DOI] [PubMed] [Google Scholar]
- 71.Shiota K., Kosazuma T., Klug S., Neubert D. Development of the fetal mouse palate in suspension organ culture. Acta Anat. (Basel) 1990;137:59–64. doi: 10.1159/000146859. [DOI] [PubMed] [Google Scholar]
- 72.Wilkinson D.G. In Situ Hybridization: A Practical Approach. 2nd edn. Oxford University Press; 1999. [Google Scholar]
- 73.Nagy A., Gertsenstein M., Vintersten K., Behringer R. Manipulating the Mouse Embryo. 3rd edn. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.