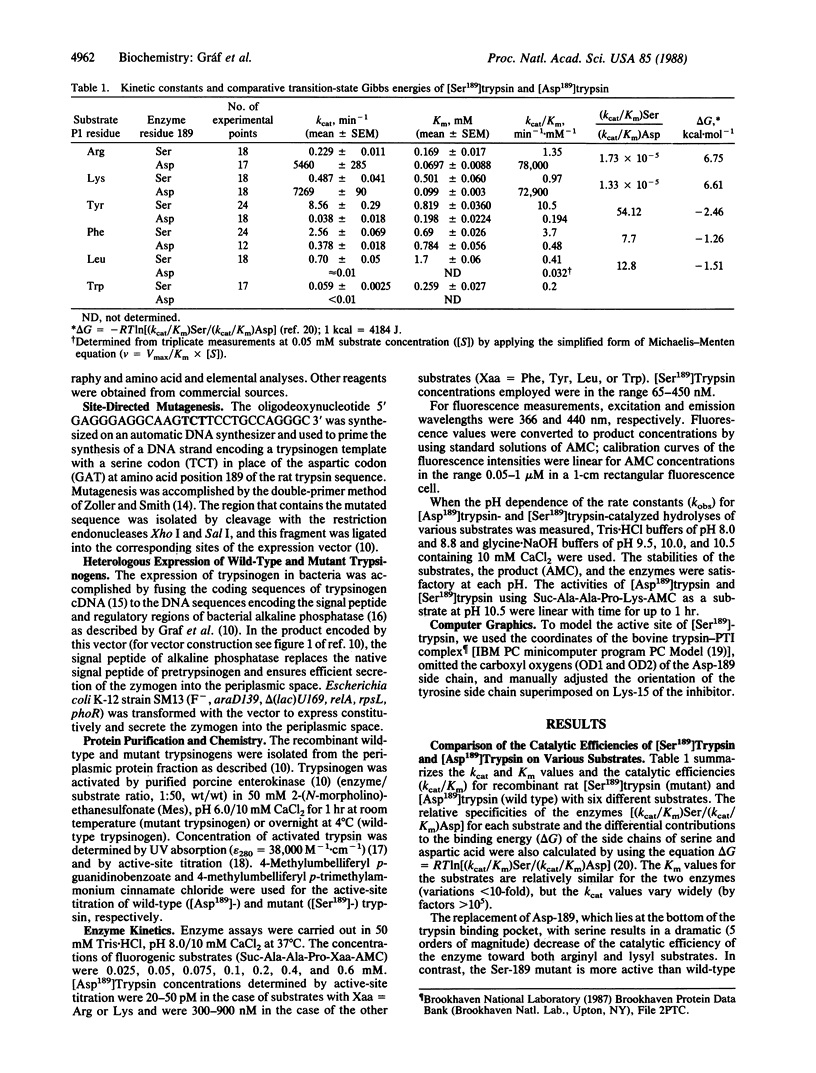
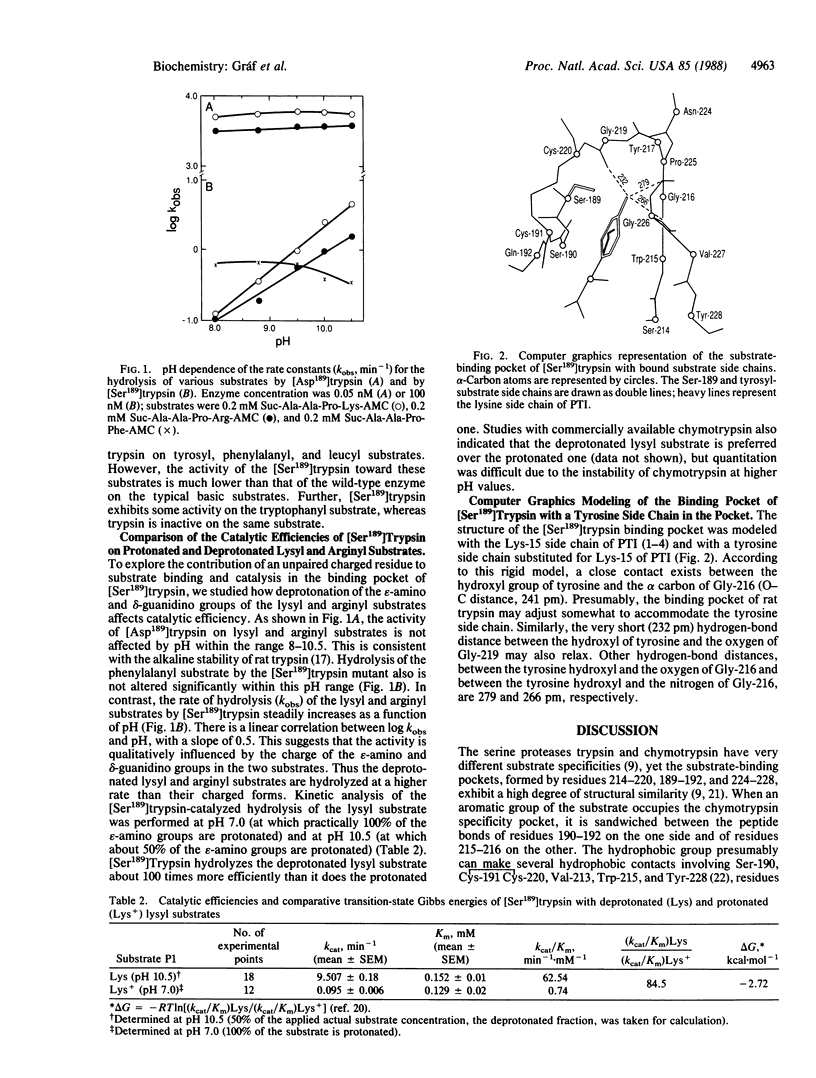
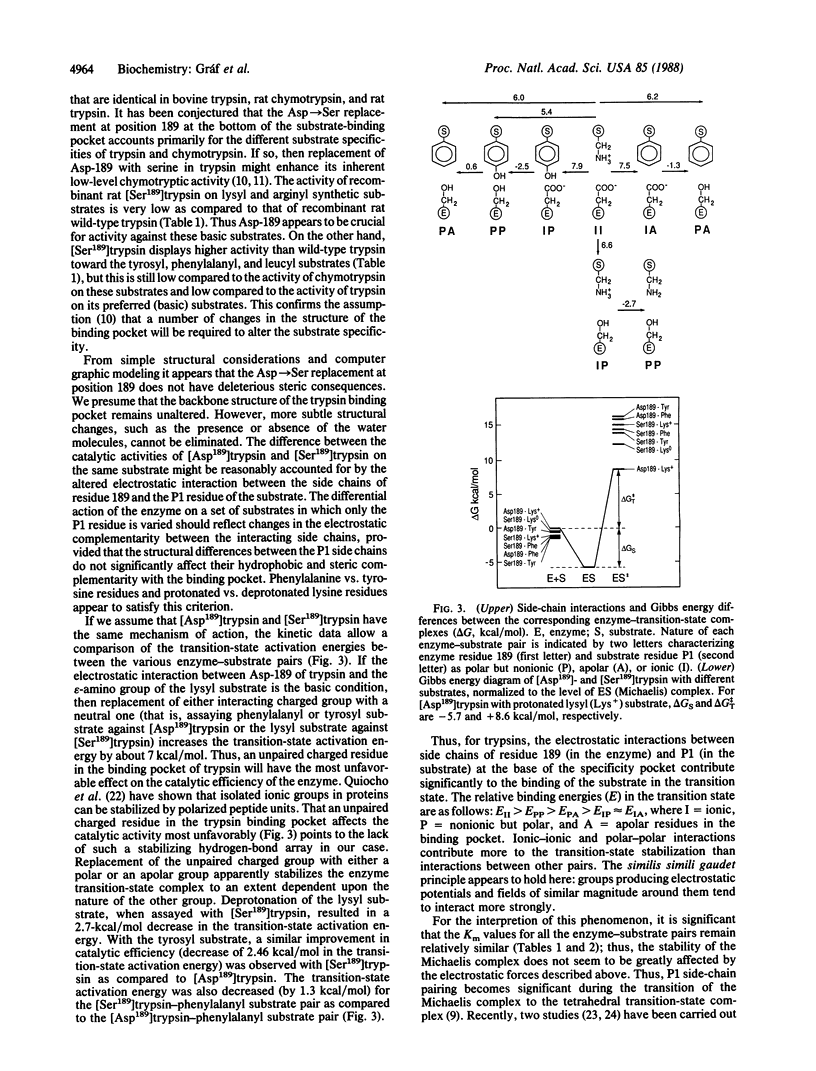
Abstract
The aspartic residue (Asp-189) at the base of the substrate-binding pocket of trypsin was replaced by serine (present in a similar position in chymotrypsin) through site-directed mutagenesis. The wild-type (with Asp-189 in the mature trypsin sequence) and mutant (Ser-189) trypsinogens were expressed in Escherichia coli, purified to homogeneity, activated by enterokinase, and tested with a series of fluorogenic tetrapeptide substrates with the general formula succinyl-Ala-Ala-Pro-Xaa-AMC, where AMC is 7-amino-4-methyl-coumarin and Xaa is Lys, Arg, Tyr, Phe, Leu, or Trp. As compared to [Asp189]trypsin, the activity of [Ser189]trypsin on lysyl and arginyl substrates decreased by about 5 orders of magnitude while its Km values increased only 2- to 6-fold. In contrast, [Ser189]trypsin was 10-50 times more active on the less preferred, chymotrypsin-type substrates (tyrosyl, phenylalanyl, leucyl, and tryptophanyl). The activity of [Ser189]trypsin on lysyl substrate was about 100-fold greater at pH 10.5 than at pH 7.0, indicating that the unprotonated lysine is preferred. Assuming the reaction mechanisms of the wild-type and mutant enzymes to be the same, we calculated the changes in the transition-state energies for various enzyme-substrate pairs to reflect electrostatic and hydrogen-bond interactions. The relative binding energies (E) in the transition state are as follows: EII greater than EPP greater than EPA greater than EIP approximately equal to EIA, where I = ionic, P = nonionic but polar, and A = apolar residues in the binding pocket. These side-chain interactions become prominent during the transition of the Michaelis complex to the tetrahedral transition-state complex.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bode W., Schwager P. The refined crystal structure of bovine beta-trypsin at 1.8 A resolution. II. Crystallographic refinement, calcium binding site, benzamidine binding site and active site at pH 7.0. J Mol Biol. 1975 Nov 15;98(4):693–717. doi: 10.1016/s0022-2836(75)80005-2. [DOI] [PubMed] [Google Scholar]
- Craik C. S., Choo Q. L., Swift G. H., Quinto C., MacDonald R. J., Rutter W. J. Structure of two related rat pancreatic trypsin genes. J Biol Chem. 1984 Nov 25;259(22):14255–14264. [PubMed] [Google Scholar]
- Craik C. S., Roczniak S., Largman C., Rutter W. J. The catalytic role of the active site aspartic acid in serine proteases. Science. 1987 Aug 21;237(4817):909–913. doi: 10.1126/science.3303334. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. Conformational equilibria in -and -chymotrypsin. The energetics and importance of the salt bridge. J Mol Biol. 1972 Mar 14;64(2):497–509. doi: 10.1016/0022-2836(72)90513-x. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Shi J. P., Knill-Jones J., Lowe D. M., Wilkinson A. J., Blow D. M., Brick P., Carter P., Waye M. M., Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985 Mar 21;314(6008):235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- Graf L., Craik C. S., Patthy A., Roczniak S., Fletterick R. J., Rutter W. J. Selective alteration of substrate specificity by replacement of aspartic acid-189 with lysine in the binding pocket of trypsin. Biochemistry. 1987 May 5;26(9):2616–2623. doi: 10.1021/bi00383a031. [DOI] [PubMed] [Google Scholar]
- Huber R., Bode W., Kukla D., Kohl U., Ryan C. A. The structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor III. Structure of the anhydro-trypsin-inhibitor complex. Biophys Struct Mech. 1975 May 30;1(3):189–201. doi: 10.1007/BF00535756. [DOI] [PubMed] [Google Scholar]
- Huber R., Kukla D., Bode W., Schwager P., Bartels K., Deisenhofer J., Steigemann W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. II. Crystallographic refinement at 1.9 A resolution. J Mol Biol. 1974 Oct 15;89(1):73–101. doi: 10.1016/0022-2836(74)90163-6. [DOI] [PubMed] [Google Scholar]
- INAGAMI T., STURTEVANT J. M. Nonspecific catalyses by alpha-chymotrypsin and trypsin. J Biol Chem. 1960 Apr;235:1019–1023. [PubMed] [Google Scholar]
- Inouye H., Michaelis S., Wright A., Beckwith J. Cloning and restriction mapping of the alkaline phosphatase structural gene (phoA) of Escherichia coli and generation of deletion mutants in vitro. J Bacteriol. 1981 May;146(2):668–675. doi: 10.1128/jb.146.2.668-675.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson G. W., Roberts D. V., Adams R. W., Kyle W. S., Elmore D. T. Determination of the operational molarity of solutions of bovine alpha-chymotrypsin, trypsin, thrombin and factor Xa by spectrofluorimetric titration. Biochem J. 1973 Jan;131(1):107–117. doi: 10.1042/bj1310107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331–358. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- Mares-Guia M. Hydrophobic interactions in the trypsin active center. The sensitivity of the hydrophobic binding site to side chain modifications in competitive inhibitors of the amidinium type. Arch Biochem Biophys. 1968 Sep 20;127(1):317–322. doi: 10.1016/0003-9861(68)90232-4. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A., Sack J. S., Vyas N. K. Stabilization of charges on isolated ionic groups sequestered in proteins by polarized peptide units. Nature. 1987 Oct 8;329(6139):561–564. doi: 10.1038/329561a0. [DOI] [PubMed] [Google Scholar]
- Russell A. J., Thomas P. G., Fersht A. R. Electrostatic effects on modification of charged groups in the active site cleft of subtilisin by protein engineering. J Mol Biol. 1987 Feb 20;193(4):803–813. doi: 10.1016/0022-2836(87)90360-3. [DOI] [PubMed] [Google Scholar]
- Rühlmann A., Kukla D., Schwager P., Bartels K., Huber R. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. Crystal structure determination and stereochemistry of the contact region. J Mol Biol. 1973 Jul 5;77(3):417–436. doi: 10.1016/0022-2836(73)90448-8. [DOI] [PubMed] [Google Scholar]
- Sanborn B. M., Hein G. E. The interaction of trypsin with neutral substrates and modifiers. Biochemistry. 1968 Oct;7(10):3616–3624. doi: 10.1021/bi00850a039. [DOI] [PubMed] [Google Scholar]
- Vajda T., Szabó T. Specificity of trypsin and alpha-chymotrypsin towards neutral substrates. Acta Biochim Biophys Acad Sci Hung. 1976;11(4):287–294. [PubMed] [Google Scholar]
- Wells J. A., Powers D. B., Bott R. R., Graycar T. P., Estell D. A. Designing substrate specificity by protein engineering of electrostatic interactions. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1219–1223. doi: 10.1073/pnas.84.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]


