Abstract
Active disease surveillance during epidemics is of utmost importance in detecting and eliminating new cases quickly, and targeting such surveillance to high-risk individuals is considered more efficient than applying a random strategy. Contact tracing has been used as a form of at-risk targeting, and a variety of mathematical models have indicated that it is likely to be highly efficient. However, for fast-moving epidemics, resource constraints limit the ability of the authorities to perform, and follow up, contact tracing effectively. As an alternative, we present a novel real-time Bayesian statistical methodology to determine currently undetected (occult) infections. For the UK foot-and-mouth disease (FMD) epidemic of 2007, we use real-time epidemic data synthesized with previous knowledge of FMD outbreaks in the UK to predict which premises might have been infected, but remained undetected, at any point during the outbreak. This provides both a framework for targeting surveillance in the face of limited resources and an indicator of the current severity and spatial extent of the epidemic. We anticipate that this methodology will be of substantial benefit in future outbreaks, providing a compromise between targeted manual surveillance and random or spatially targeted strategies.
Keywords: epidemiology, statistics, Bayesian, contact tracing, foot-and-mouth disease, epidemic
1. Introduction
Epidemic control essentially focuses on reducing the risk of transmission between susceptible and infected individuals. Traditional methods such as vaccination or pre-emptive culling serve to decrease the size of the susceptible population and may be used with varying success (Anderson & May 1991; Keeling et al. 2001; Woolhouse & Donaldson 2001). An alternative strategy is to focus on infected individuals, either quarantining or culling to prevent onward transmission. In such cases, early detection is vital to reduce the time for which the individual is actively shedding infection (Howard & Donnelly 2000; Ferguson et al. 2001b; Woolhouse & Donaldson 2001). When infection occurs significantly before the onset of symptoms, mechanisms of contact tracing become vital, with the aim of identifying at-risk individuals by their interaction with known infectious cases (Riley & Ferguson 2006).
Contact tracing has traditionally and successfully been used for a variety of human diseases, including the 2003 SARS outbreak, when high-risk individuals were quarantined, and a variety of sexually transmitted infections, where at-risk contacts are tested and treated (Cowan et al. 1996; Donnelly et al. 2003). For foot-and-mouth disease (FMD), contact tracing from known infected premises (IPs) is mandatory, generating dangerous contacts (DCs) whose livestock must be culled. During the 2001 FMD outbreak in the UK, contact tracing only was deemed insufficient to control the epidemic—the high number of cases and the limited veterinary resources meant that contacts could not be identified and removed quickly enough (Ferguson et al. 2001a,b; Keeling et al. 2001). Indeed, subsequent studies have demonstrated that for epidemics where the number of cases and/or contacts is high, contact tracing loses its efficacy owing to resource constraints (Eames & Keeling 2003; Kao 2003; Kiss et al. 2005). Given these resource constraints, the contiguous premises cull policy (where animals on premises neighbouring IPs were culled) was implemented as a crude but rapid method of identifying and removing potentially at-risk premises (The Royal Society 2002; Haydon et al. 2004). During the 2007 FMD outbreak in Surrey, however, the number of cases was small and the policy was better developed in the wake of 2001, so that effective contact tracing was implemented. This led to a large number of DCs being identified and subsequently culled; to what extent this would have been kept up if the epidemic had been larger is unclear (Ryan et al. 2008).
Here, we perform a rigorous statistical analysis of the 2007 FMD outbreak, and, as the epidemic unfolds, we focus on predicting which premises may be infected but currently undetected—we term such premises occult infections. Such inference is, in general, complex owing to the necessity of dealing with the fact that the exact moment of infection is never directly observed. Here, in contrast to others (e.g. Chis-Ster & Ferguson 2007), we postulate that the time between infection and detection is a stochastic quantity and should, therefore, be modelled as such—this is particularly true when treating the farm as an individual since a wide variety of factors, such as stocking density, management system and stockmanship, can all affect how quickly a disease is detected. Furthermore, if an epidemic is to be analysed during its course, the presence of occult infections should be taken into account. If this is not done, then our analysis will be biased since disease transmission rates from only known IPs will appear artificially high. To include this complexity in our inference, we use a Bayesian framework that incorporates the unobserved data as parameters to be inferred. Using prior information determined from the 2001 FMD outbreak in the UK (Kypraios 2007), we introduce a completely different approach that makes use of new highly specialized Bayesian Markov chain Monte Carlo (MCMC) methodology designed for making quantitative risk predictions in real time as epidemics progress (O'Neill & Roberts 1999; Neal & Roberts 2004; Jewell et al. in press). This methodology therefore addresses two major epidemiological concerns: it predicts the uncertain scale of the current epidemic, and it rapidly identifies high-risk premises that can be targeted for further epidemiological investigation or control.
2. Data and methods
2.1. Data
The data required for this analysis fall into three categories, which are as follows.
— Covariate data describe the population in terms of an individual's attributes. For this analysis, we use location (as OSGB reference), number of cattle and number of sheep present on the farm as derived from the 2003 census. These data remain constant throughout the epidemic.
— Epidemiological data describe the disease states of individuals. These data are acquired from the field as the epidemic progresses and, for our purposes, comprise a daily list of detection times and cull times linked to affected farms. In practice, this requires Defra to notify us of any detections or culls that have occurred on a daily basis.
— Unobserved data which, if it were available, would allow straightforward inference. This refers to the infection times during the epidemic, which, owing to the lag between infection and the onset of clinical disease, are never directly observed. In addition, this lag means that at any point during an epidemic, there may be farms that are infected (and infectious) that have not yet been detected and which require accounting for in the analysis. We take particular trouble, therefore, to impute such data, allowing the statistical analysis to reflect the uncertainty surrounding this inability in observation.
2.2. Model construction
The model treats each premises as an individual that can be susceptible, infected, notified (i.e. detected and reported) or removed (i.e. culled), with each infected individual passing through all four states in sequence. We therefore assume that each infection will eventually be detected (even if the detection occurs in the future). The fundamental aim for statistical inference is then to estimate the rate of transition from susceptible to infected, and from infected to notified. Since we are interested in analysing an epidemic during its course, these states interact with the time of analysis (T obs) giving rise to three possible types of infected individuals:
— infections that have been notified and removed before T obs;
— infections that have been notified but not yet removed; and
— individuals of unknown disease status that are either currently presumed susceptible or have been prematurely culled as DCs. These individuals, if infected, are termed occults and will be imputed in the inference mechanism (see the electronic supplementary material, sections A.4 and B).
The infection rate is parametrized in terms of infected–susceptible and notified–susceptible pairs. With a knowledge of individual-level covariates (as described above), we fit parameters to describe the effects that each covariate has on the transmission rate between farms. We adopt a model similar to Keeling et al. (2001), which assumes heterogeneity in infectivity and susceptibility according to the species present on each premises, as well as using a spatial function to allow the probability of infection to vary over distance. Let 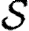 ,
, 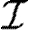 , and
, and 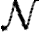 be the sets of susceptible, infected, and notified individuals respectively at the time of the analysis, with
be the sets of susceptible, infected, and notified individuals respectively at the time of the analysis, with 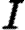 ,
, 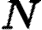 , and
, and 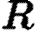 the respective vectors of infection, notification, and removal times. We thus parametrize the disease transmission rate between infected i and susceptible j as
the respective vectors of infection, notification, and removal times. We thus parametrize the disease transmission rate between infected i and susceptible j as
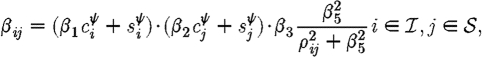 |
2.1 |
and the transmission rate between notified i and susceptible j as
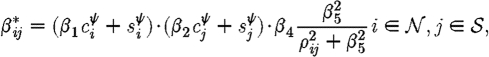 |
2.2 |
where c and s represent the number of cattle and sheep, respectively, on premises i and j, and ρ ij is the Euclidean distance between them. β 1 is then interpreted as the relative infectivity of cattle versus sheep, β 2 as the relative susceptibility of cattle versus sheep, β 3 and β 4 as the rate of spatial transmission for infected and notified premises, respectively, with β 5 governing its rate of decay with increasing distance. The difference between β 3 and β 4 therefore allows the measurement of the effect of control measures applied directly to a notified premises, such as restrictions on vehicles entering or leaving, and the general upregulation of biosecurity required.
We allow for a latent period in which an individual is infected but not yet infectious by multiplying β ij by a time-dependent infectivity function h(.), describing how an individual's infectivity increases from the moment of infection. The shape of this function is application dependent, and could range from a more biologically plausible smooth function to a more simplistic step function. Here, we choose the latter, with h(t)=0 for t<4 days and h(t)=1 otherwise, giving an ‘exposed’ (infected but not infectious) period of 4 days. The choice of 4 days reflects the underlying pathobiology of the disease as described by Defra Veterinary Surveillance (2006): it is assumed that animals are infectious once disease-associated lesions on the buccal mucosa and the coronary band have formed and ruptured, this occurring 2–3 days post-infection. An upper bound of 4 days is then used to allow some time for the within-herd/flock epidemic to establish, in accordance with the stated within-herd/flock incubation time of 2–6 days. This gives our model equivalence to the SEIR model specified in Keeling et al. (2001). The total infectious pressure on any susceptible j, immediately before its infection, is therefore
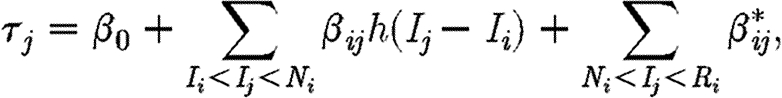 |
2.3 |
where I k, N k and R k denote the infection, notification and removal times of individual k, respectively. Note the incorporation of β 0 that accounts for a background infection rate owing to factors other than those explicitly modelled (e.g. another primary incursion).
To model the rate of notification conditional on an infection, we use the following distribution:
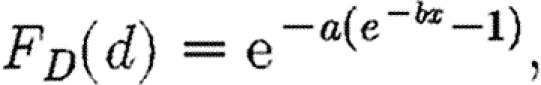 |
2.4 |
with parameters a=0.005 and b=0.6 fixed to give a mean infection to notification time of 7.5 days (Kypraios 2007).
2.3. Bayesian analysis
The methodology used in this analysis is based on recently developed Bayesian techniques (Jewell et al. in press) applied to more established models of disease dynamics. We use a Bayesian approach since it allows the incorporation of prior knowledge of the epidemic, and provides a very natural framework to treat each unobserved infection time. We then use a MCMC algorithm to iteratively sample from the conditional posterior distributions of the transmission parameters θ={β 0,…,β 5, ψ} and infection times I (detailed information available in the electronic supplementary material).
First, the Bayesian paradigm requires that both a likelihood function (i.e. the likelihood of the data given the parameters) and prior information (represented as probability distributions) are specified. These are now examined in turn.
The likelihood function is constructed by assuming that the infection times constitute a time-inhomogeneous Poisson process with the rate at any particular time point equal to the sum of infectious pressures on susceptibles at that point (equation (2.3)). This pressure changes throughout the epidemic as new infections occur, and current ones are notified and removed. Notification times are then modelled, conditionally on their respective infection times, by the distribution in equation (2.4). This function then provides a link between the observed notification times and unobserved information. Considering m known infections and [I]-m occults (including DCs), the form of the likelihood function is the following, with details of its derivation available in section A.5 in the electronic supplementary material:
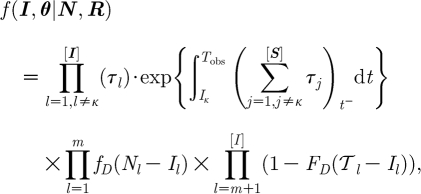 |
2.5 |
with κ the index case, such that the likelihood is conditional on the first detected case. f
D(.) is specified in equation (2.4), with F
D(.) the corresponding cumulative density function giving the probability that N
j occurs after 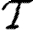 l, where
l, where
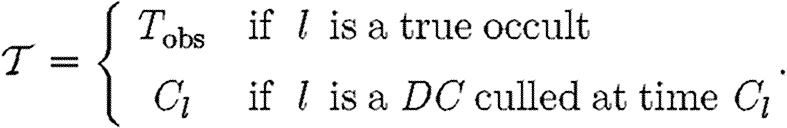 |
Prior distributions are now required for each of the parameters in the model. For this, we turn to previous studies as our source of prior information. Kypraios (2007) provided a Bayesian analysis of the UK foot-and-mouth outbreak in 2001 using a similar model from which we take priors for β 1, β 2 and ψ. A combination of the posterior distance kernel provided in Kypraios (2007) and the empirical kernel provided by Savill et al. (2006), which are remarkably consistent, inform our choice of β 3, β 4 and β 5. For each parameter, we use a gamma distribution with mean and variance chosen to match our prior information.
The joint posterior distribution of the unobserved information (parameters and infection times) is then proportional to the product of the likelihood and prior probability distribution functions. If it were possible to integrate this product over the whole parameter (including infection time) space in order to find the constant of proportionality, then a closed-form posterior would be available. However, since the likelihood is intractable to integration, an adaptive reversible-jump MCMC algorithm is used to simulate samples from the joint posterior (Jewell et al. in press).
The MCMC algorithm is an iterative process in which alternately samples from the conditional posterior of the parameters (given the data and current (simulated) infection times) and the conditional posterior of the infection times (given the data and current parameter values). The important feature of the MCMC for this study is that it is possible for the algorithm to explore the possibility of susceptible individuals, in fact, being infected—the occult infections. Briefly, occult infections can be regarded, from the point of view of infectious pressure, as individuals that have been infected, but have their notification and removal times in the future. Thus, the only infectious pressure they contribute to the system is, therefore, from their infected state up until the analysis time. The algorithm chooses a susceptible at random and proposes an infection time based on the distribution in equation (2.4). In concept, the likelihood is then queried to see, first, whether the amount of infectious pressure in the system at the proposed infection time supports the possibility of the infection having occurred, and, second, whether the subsequently increased infectious pressure supports the observed data. If the occult is consistent with the likelihood, then the proposal is accepted; if not, it is rejected. Importantly, the MCMC also proposes the reverse move—in other words, previously added occults are also removed, again, in a probabilistic way. Since infection times are treated as parameters in the Bayesian sense, adding or removing occults amounts to moving between models of different dimensions, and hence the amount of time that the model spends in any one dimension is equal to the probability of those occults existing. The necessity of capturing these occult infections is encapsulated in the likelihood where it is easy to see that for a given level of infectious pressure consistent with the observed data, the presence of occults requires a smaller per-pair infectious pressure than if they were ignored. This is then, of course, reflected in the estimated values of the parameters. Full details of how this algorithm works are presented in section B in the electronic supplementary material.
3. Results
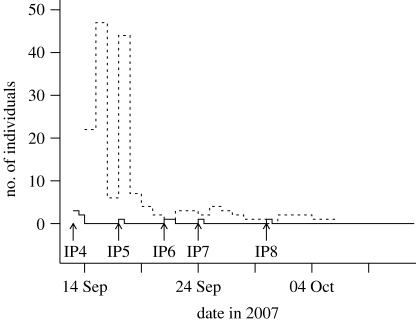
Our analysis of the 2007 FMD outbreak began once it became apparent that the disease was concentrating around the Surrey town of Egham. At a given time point, the analysis provides a current prediction for the number of occults, described as a probability distribution, given the data available up to that time. Beginning just after the detection of IP4, daily analyses were carried out. Figure 1 shows the number of notified individuals present on each day, as well as the median predicted number of occults provided by the analysis. For selected time points, figure 2 then gives the probability that apparently uninfected farms are, in fact, infected. This gives an estimate of the current extent of the epidemic given the uncertainties in parameter estimates and the routes of transmission. Figure 2 shows that at each stage of the epidemic, the greatest occult probabilities were associated with premises close to the known IPs, and were those with large numbers of cattle or sheep—in agreement with qualitative understanding. Taken together with figure 1, figure 2 also demonstrates how the algorithm learns about the epidemic as data are gathered from the field. The first map (13 September) is drawn based on only four data points. The dataset here contains very little information on spatial transmission since only two farms are closer than 10 km, and results in a very diffuse prediction which is heavily influenced by the priors (cf. the fast-moving 2001 FMD outbreak). As the epidemic progresses and the number of data points increases, so does the quantity of statistical information, leading to a more precise estimate (see Posteriors in the electronic supplementary material). This is shown most strikingly by the difference between 13 and 17 September where just the addition of one data point drastically reduces the median number of predicted undetected infections, and correspondingly reduces the extent of the distribution of high-probability premises. Moreover, knowing in hindsight the time frame of the epidemic, we see how the spatial distribution of high-risk farms localizes towards the end, reflecting the fact that the epidemic was being controlled effectively.
Figure 1.
The epidemic time line. The solid line shows the current number of notified farms. The dashed line gives the predicted median number of undetected infections, given the available information up to that time.
Figure 2.
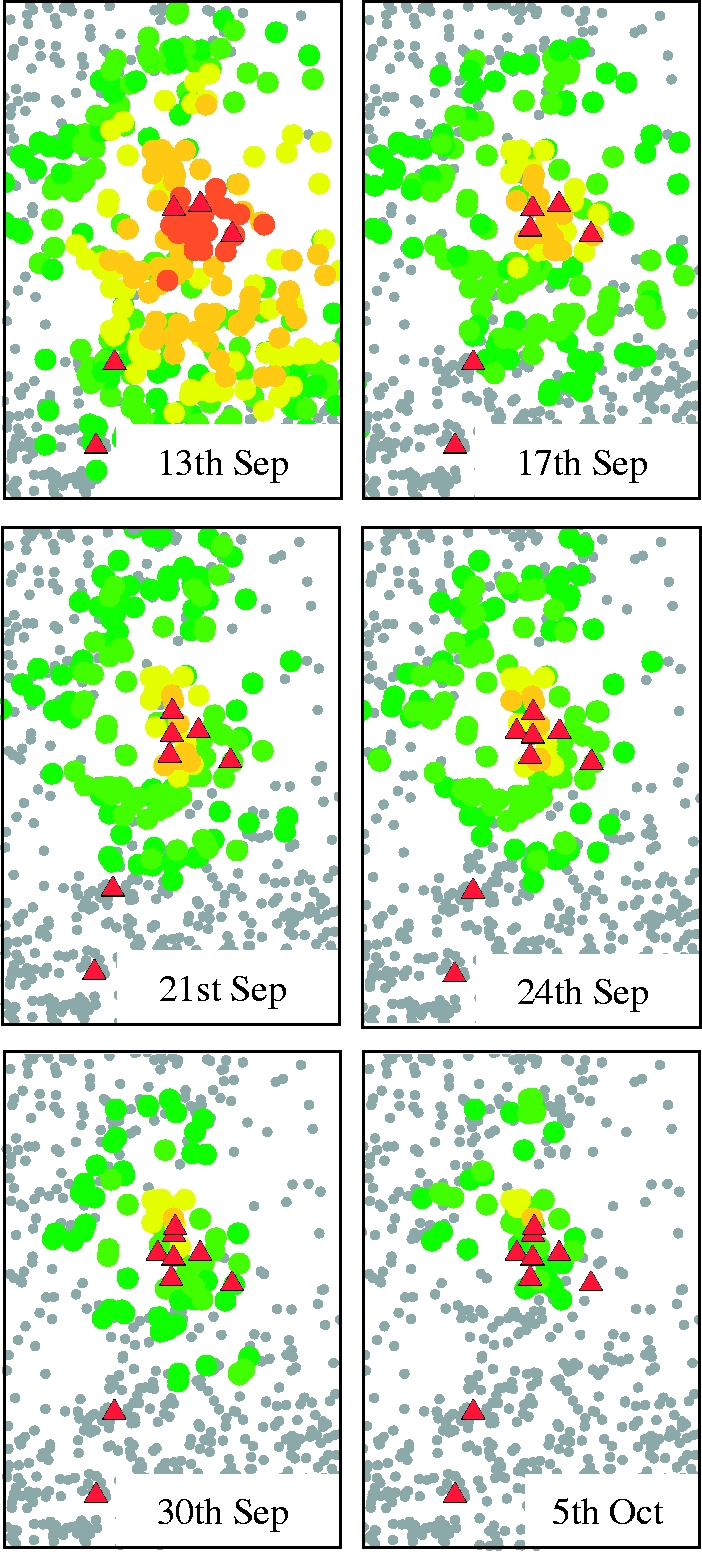
Probability of occult infections (dark green circles, 0.005–0.010; light green circles, 0.011–0.050; yellow circles, 0.051–0.100; light orange circles, 0.101–0.350; dark orange circles, 0.351–1.000; red triangles, known IP; grey circles, susceptibles) during the 2007 foot-and-mouth outbreak. For brevity, we show only a subset (after IPs 4–8, and a week after IP8) of our analyses.
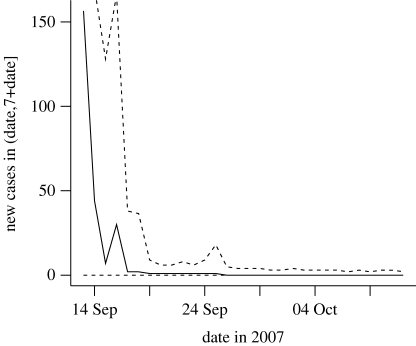
In terms of disease control, it is necessary to have an idea on the extent of the resources needed to cope with the outbreak. To answer this question, figure 3 shows the predicted number of new infections in a period of 7 days from the observation time. Consistent with the priors and large amount of uncertainty, this prediction is high early in the epidemic with wide 95% credibility intervals ranging from 0 to 1196 new cases. However, this is soon refined as the epidemic progresses to give the small value consistent with the final outcome of the epidemic. We postulate that, taken together, these results give an overview of the extent of the epidemic, the short-term risk posed by the presence of undetected infections and the resource required for control measures.
Figure 3.
Predicted numbers of new cases within a 7-day time period after the observation time, given the available information up to that time. Solid line, median; dashed line, 95% credibility interval.
From this ability to predict occult infections, we can consider whether the Bayesian occult probabilities provide a better guide for targeted control than other measures. Table 1 gives the occult ranking of the last four infected premises for dates in September before their detection. For the Bayesian analysis, this ranking simply sorts the undetected farms in the order of highest probability, and this is compared with ranking farms by their proximity to the closest known infected premises. Table 1 reflects the same patterns that were observed in figure 2. In addition, table 1 indicates the dynamic nature of risk to individual farms as the epidemic progresses, with an individual's rank increasing as time progresses (according to its likely infection date). For IP5, IP6 and IP8, the Bayesian algorithm clearly outperforms the spatial measure, and is roughly comparable for IP7. We also note that since this method incorporates individual farm characteristics, as well as spatial relationships, it should be capable of discriminating between two close farms with different characteristics. In considering this, we looked at two farms within 0.5 km of each other, of which IP7 was one. According to the database, IP7 holds just less than 20 cattle, whereas the other farm holds over 300 sheep1. IP7 is, therefore, consistently at higher risk than the other farm owing to the higher susceptibility of cattle compared with sheep (e.g. 25% (rank 15) occult probability for IP7 versus 11% (rank 53) on the 17 September; 21% (rank 13) versus 3% (rank 35) on 21 September).
Table 1.
Occult risk ranking. (Rankings are calculated from Bayesian analysis (ba) occult probabilities (in parentheses) and by spatial proximity to currently infected individuals (sp).) (Note that for an individual IP, the early (off-diagonal) ranks take into account the fact that there is a higher probability of it not having yet been infected. See also §3).
| IP | method | 13 Sep | 17 Sep | 21 Sep | 24 Sep |
|---|---|---|---|---|---|
| IP5 | ba | 6 (0.56) | |||
| sp | 11 | ||||
| IP6 | ba | 36 (0.18) | 20 (0.22) | ||
| sp | 14 | 30 | |||
| IP7 | ba | 38 (0.17) | 15 (0.25) | 13 (0.21) | |
| sp | 6 | 14 | 10 | ||
| IP8 | ba | 31 (0.20) | 28 (0.16) | 2 (0.36) | 1 (0.33) |
| sp | 24 | 42 | 2 | 10 |
This ranking system is an oversimplified summary of a complex Bayesian analysis and is only used for comparison with the ad hoc spatial ranking system. In reality, the use of these results requires consideration of the Bayesian occult probabilities themselves (rather than their ranks), with further consideration being given to other factors that influence effective surveillance, such as the centrality of occults in known contact networks (Jewell 2008). Such model-based assessment will, of course, never be a replacement for the detailed contact tracing performed by veterinarians when identifying DCs, but may help to direct additional resources.
4. Discussion
The ‘epidemiological triangle’ is an important concept in studying the spread of infectious diseases and states that the characteristics of an outbreak depend on the host, pathogen and environment in which they coexist (Morens et al. 2004). Changes over time in one or more of the three vertices are therefore likely to alter the dynamics of outbreaks of the same disease recurring in the population. This is demonstrated in the differences between FMD outbreaks in 1967 and 2001 where large differences in farming practices accounted for very different patterns of transmission (Defra FMD information http://www.defra.gov.uk/footandmouth), and also in the recent 2007 outbreak that occurred in a sparsely populated area of the country. This shows that by basing predictions of epidemic spread simply on past information, it is difficult to quantify how relevant they are to the current outbreak. On the other hand, early in the epidemic, there is inherently very little data from the current epidemic on which to base a prediction. A Bayesian analysis provides a framework for learning about a current disease outbreak. At the beginning of an epidemic, the prediction process begins with prior information based on our past experience and expert opinion, and then updates this by incorporating data obtained from the field during the epidemic under study. Using this methodology therefore provides a very natural way to make as accurate a prediction as possible, while quantifying the amount of uncertainty we have about the quantities on which the predictions are based.
The statistical methodology presented in this paper aims to identify the presence of occult infections, although it has at its core the aim of estimating disease transmission parameters for a specified epidemic model. When interpreting the results, it is important to realize that the occults are identified probabilistically, meaning that it is impossible to say from the data whether an individual is definitely infected or not, only that it might be infected with a given probability. Therefore, we do not claim this methodology to be more effective than manual contact tracing given the resource available with which to carry it out. However, as noted by Eames & Keeling (2003), if the resource requirement is not sufficient to match the speed of the epidemic, then the advantage of identifying DCs over culling on a distance basis is lost (though we note that this was not the case with FMD2007). It is in these scenarios, for example a large FMD 2001-style outbreak, that our statistical method of identifying likely infections is intended to improve upon a blanket-culling policy, allowing prioritization of disease surveillance so as to use suboptimal resources in the most efficient way. Moreover, if a certain amount of contact-tracing information were to be made available to us, it could readily be used in our framework to provide, as our preliminary results show, a marked increase in prediction accuracy.
In this study, we have used static covariate data taken from the 2006 agricultural census. However, it is known that this dataset is highly dynamic, particularly in the number of animals present on the farm (Robinson & Christley 2006). Inaccuracies in the covariate data will certainly have an effect on our predictions, which will be noticeable in the case that the associated parameter values are large. There are a number of possible ways to overcome this limitation. First, the most preferable solution would be to have the most up-to-date version of Defra's livestock database possible, requiring more efficient data handling than is currently implemented (Anderson 2008). Second, part of our current work is looking at Bayesian methods to incorporate a measure of uncertainty into the dataset itself. This will allow the algorithm to integrate over the uncertainty that exists in the dataset, and to reflect this in the joint posterior distribution (parameters, infection times and occult probabilities). Any predictions that we make will, of course, be more uncertain owing to the increased variance contributed by data uncertainty, but will remain a true reflection of what is currently known about the system. To improve on this uncertainty, dynamic data, available as a result of field investigations, should be included into the dataset. Our early results suggest that incorporating contact-tracing data into the analysis provides a large reduction in posterior uncertainty, and therefore provides a very important source of information. In practice, however, the usefulness of this as yet unpublished methodology would depend on efficient data acquisition from the field teams.
An important aspect of providing a ‘real-time’ analysis in any situation is algorithm run-time. For our methodology, run-time is approximately proportional to [S][I], with [I] including both known and occult infections. In order to deal with the potentially large dimension of the statistical calculations, the MCMC is parallelized by sharing the calculation of the likelihood among multiple processors. A relationship exists between the dimension of I and the number of processors, with better parallel scalability being achieved with larger datasets owing to Amdahl's Law (Kontoghiorghes 2006). However, the possibility of scaling on massively parallel cluster machines commonly available in research establishments means that even large-scale epidemics can be analysed using this algorithm. In this study, for 1 000 000 iterations of the MCMC on a twin dual-core 2.4 GHz AMD OpteronTM machine (Sun Fire X4100 server), our algorithm took 51 min for the dataset on 13 September, and 30 min on 5 October, showing the effect of the occult infections on algorithm run-time. Taking the former as an upper bound on run-time, a linear scaling indicates a run-time of approximately 18 days for a large outbreak of 2000 IPs such as FMD2001 on the same hardware. However, this can easily be shortened to below our required run-time by expanding the number of processors—even using 150 processors is well within the capability of typical university high-performance computers.
We have shown, therefore, that a fully Bayesian approach to real-time inference on disease epidemics is feasible in the agricultural context. To perform this, we have developed tailor-made MCMC methods, which, although computationally intensive, can be readily implemented on time scales short enough to inform policy on a day-to-day basis. This provides a framework for monitoring disease-control efforts, and provides information to adapt policy in the event that the outbreak does not behave as expected based on past experience. Although the example presented here is of a small outbreak that was contained quickly, the methodology is equally applicable to larger epidemics in more densely populated areas. In particular, since more epidemiological data are available in these situations, a likelihood-based approach such as ours will perform significantly better, reducing posterior uncertainty quickly, therefore giving an even greater prediction accuracy. Since this work demonstrates its capability in probabilistically identifying occult cases, it is anticipated that such techniques will form the basis of statistical contact tracing to provide a targeted surveillance strategy even in the face of limited contact-tracing resource.
Acknowledgements
We would like to thank the following individuals for their assistance with this work: Dr Mike Tildesley and Dr Theodore Kypraios. We thank Defra for the provision of data and funding for this research. We also thank the Wellcome Trust for funding part of this research (M.J.K.).
Footnotes
Data confidentiality prevents us from displaying precise farm-level covariates. Therefore, here, we provide only one example of many.
References
- Anderson I. London, UK: The Stationary Office; 2008. Foot and mouth disease 2007: a review and lessons learned. See http://www.cabinetoffice.gov.uk/fmdreview.aspx. [Google Scholar]
- Anderson R. M., May R. M.1991Infectious diseases of humans: dynamics and control New York, NY:Oxford University Press [Google Scholar]
- Chis-Ster I., Ferguson N.2007Transmission parameters of the 2001 foot and mouth epidemic in Great Britain. PLoS One 2, e502.10.1371/journal.pone.0000502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan F. M., French R., Johnson A. M.1996The role and effectiveness of partner notification in STD control: a review. Genitourin. Med 72, 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defra Veterinary Surveillance 2006. Full profile for foot and mouth disease. Electronic. See http://www.defra.gov.uk/animal/diseases/vetsurveillance/profiles/fmd-fullprofile.pdf
- Donnelly C. A., et al. 2003Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. The Lancet 361, 1761–1766doi:10.1016/S0140-6736(03)13410-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames K. T. D., Keeling M. J.2003Contact tracing and disease control. Proc. R. Soc. B 270, 2565–2571doi:10.1098/rspb.2003.2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson N. M., Donnelly C., Anderson R.2001aTransmission intensity and impact of control policies on the foot and mouth epidemic in Great Britain. Nature 413, 542–548doi:10.1038/35097116 [DOI] [PubMed] [Google Scholar]
- Ferguson N. M., Donnelly C. A., Anderson R. M.2001bThe foot-and-mouth epidemic in Great Britain: pattern of spread and impact of interventions. Science 292, 1155–1161doi:10.1126/science.1061020 [DOI] [PubMed] [Google Scholar]
- Haydon D. T., Kao R. R., Kitching R. P.2004The UK foot-and-mouth disease outbreak—the aftermath. Nat. Rev. Microbiol 2, 675–681doi:10.1038/nrmicro960 [DOI] [PubMed] [Google Scholar]
- Howard S. C., Donnelly C. A.2000The importance of immediate destruction in epidemics of foot and mouth disease. Res. Vet. Sci 69, 189–196doi:10.1053/rvsc.2000.0415 [DOI] [PubMed] [Google Scholar]
- Jewell C. P. Real-time inference and risk-predition for notifiable diseases of animals. 2008. PhD thesis, University of Warwick.
- Jewell C. P., Kypraios T., Roberts G. O., Christley R. A novel approach to real-time risk-prediction for emerging infectious diseases: a case study in Avian Influenza H5N1. Prev. Vet. Med. In press. [DOI] [PubMed]
- Kao R. R.2003The impact of local heterogeneity on alternative control strategies for foot-and-mouth disease. Proc. R. Soc. B 270, 2557–2564doi:10.1098/rspb.2003.2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M. J., et al. 2001Dynamics of the 2001 UK foot and mouth epidemic: stochastic dispersal in a heterogeneous landscape. Science 294, 813–818doi:10.1126/science.1065973 [DOI] [PubMed] [Google Scholar]
- Kiss I. Z., Green D. M., Kao R. R.2005Disease contact tracing in random and clustered networks. Proc. R. Soc. B 272, 1407.doi:10.1098/rspb.2005.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoghiorghes E. J.2006Handbook of parallel computing and statistics Boca Raton, FL:Chapman and Hall [Google Scholar]
- Kypraios T. Efficient bayesian inference for partially observed stochastic epidemics and a new class of semi-parametric time series models. 2007. PhD. thesis, Department of Mathematics and Statistics, Lancaster University, Lancaster.
- Morens D. M., Folkers G. K., Fauci A. S.2004The challenge of emerging and re-emerging infectious diseases. Nature 430, 242.doi:10.1038/nature02759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal P. J., Roberts G. O.2004Statistical inference and model selection for the 1861 Hagelloch measles epidemic. Biostatistics 5, 249–261doi:10.1093/biostatistics/5.2.249 [DOI] [PubMed] [Google Scholar]
- O'Neill P. D., Roberts G. O.1999Bayesian inference for partially observed stochastic epidemics. J. R. Stat. Soc. Ser. A 162, 121–129doi:10.1111/1467-985X.00125 [Google Scholar]
- Riley S., Ferguson N. M.2006From the cover: smallpox transmission and control: spatial dynamics in Great Britain. Proc. Natl Acad. Sci. USA 103, 12 637–12 642doi:10.1073/pnas.0510873103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S. E., Christley R. M.2006Identifying temporal variation in reported births, deaths and movements of cattle in britain. BMC Vet. Res 2, 11.doi:10.1186/1746-6148-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan E., et al. 2008Clinical and laboratory investigations of the outbreaks of foot-and-mouth disease in southern England in 2007. Vet. Rec 163, 139–147 [DOI] [PubMed] [Google Scholar]
- Savill N. J., Shaw D. J., Deardon R., Tildesley M. J., Keeling M. J., Woolhouse M. E., Brooks S. P., Grenfell B. T.2006Topographic determinants of foot and mouth disease transmission in the UK 2001 epidemic. BMC Vet. Res 2, 3.doi:10.1186/1746-6148-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Royal Society Inquiry into infectious diseases in livestock, Tech. rep. 2002London, UK:The Stationary Office [Google Scholar]
- Woolhouse M., Donaldson A.2001Managing foot-and-mouth. Nature 410, 515–516doi:10.1038/35069250 [DOI] [PubMed] [Google Scholar]